The visual system is a powerful model for probing the development, connectivity, and function of local and long-range neural circuits. Here, Wernet et al. review recent progress in the molecular genetic dissection of visual circuitry in Drosophila and mice, with a focus on the strengths and limitations of each model system.
Keywords: cell types, circuitry, function, vision
Abstract
The visual system is a powerful model for probing the development, connectivity, and function of neural circuits. Two genetically tractable species, mice and flies, are together providing a great deal of understanding of these processes. Current efforts focus on integrating knowledge gained from three cross-fostering fields of research: (1) understanding how the fates of different cell types are specified during development, (2) revealing the synaptic connections between identified cell types (“connectomics”) by high-resolution three-dimensional circuit anatomy, and (3) causal testing of how identified circuit elements contribute to visual perception and behavior. Here we discuss representative examples from fly and mouse models to illustrate the ongoing success of this tripartite strategy, focusing on the ways it is enhancing our understanding of visual processing and other sensory systems.
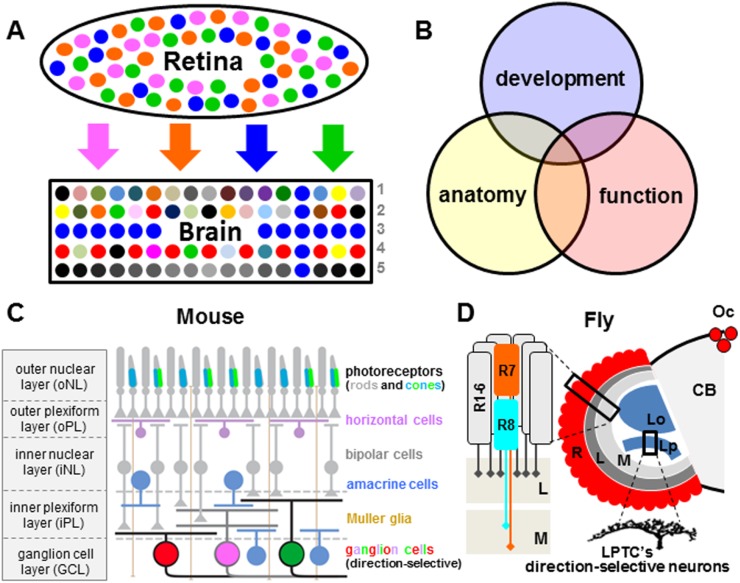
For many decades, the visual systems of both vertebrates and invertebrates have been a favorite arena for understanding how neural circuits are built and function. A considerable body of work has focused on the specification of cell types of the retina; for instance, the designation of different classes of photoreceptors with distinct spectral sensitivities in the Drosophila retina (Rister and Desplan 2011) and the establishment of the various cell types that comprise the vertebrate retina: photoreceptors, Muller glia, interneurons (horizontal, amacrine, and bipolar cells), and retinal ganglion cells (RGCs) (Livesey and Cepko 2001; Mu et al. 2004; Poché and Reese 2006). With that knowledge in hand, focus in recent years has expanded to understanding how the circuits formed by these retinal cells are linked to the staggering number of diverse visual neurons in the brain, which in turn enables sophisticated and diverse computational tasks. The ultimate goal is to understand how cellular identity, function, and connectivity relate to visually guided behaviors. (Fig. 1A).
Figure 1.
Visual circuitry: general problems, approach, and introduction to the visual system. (A) Schematic model depicting two central problems in the molecular genetic investigation of visual circuitry: stochastic inputs and extreme cell type diversity. The retina contains a limited number of photoreceptor neurons with varying spectral sensitivities or receptive fields (four classes are depicted). In many cases, these input channels are distributed randomly across the epithelium. Their output information is transmitted to the brain, where large numbers of diverse cell types can be organized either randomly or in repetitive structures like columns or strata (layers). Based on their functional properties, different cell types can represent very specific and highly divergent stimulus features (symbolized by different colors), which must have been extracted computationally through synaptic interactions within larger circuits. (B) The molecular genetic investigation of visual circuitry depends on three synergistic approaches. The anatomical reconstruction of neuronal cell types provides high-resolution information about their morphological classification as well as their synaptic connections. The developmental history of each cell type reveals the genes and signaling pathways that together form the molecular mechanism shaping cellular diversity. Characterization of circuit elements on a single-cell level (using electrophysiology or genetically encoded, activity-dependent calcium sensors) or in the behaving animal provides direct access to their functional role within the circuit. (C) Summary of the basic organization, laminar architecture (three cell layers and two synaptic layers) and seven major cell types of the mouse retina. Note that the diagram is representative of the major cell types but is not exhaustive of all retinal cell subtypes. (D) Introduction to the Drosophila visual system. In the retina (R), six outer photoreceptor (R1–6) axons of each unit eye (ommatidium) project to the lamina neuropile (L), while inner photoreceptors (R7 and R8) project to the medulla (M). The two remaining neuropiles are not innervated by photoreceptors: the lobula (Lob) and lobula plate (Lp). The first direction-selective neurons are found in the lobula plate. Outside the eyes, three Ocelli (Oc) on the vertex of the head also harbor photoreceptor cells.
Mice have been a prominent model for studies of visual circuit development and plasticity for well over two decades (e.g., Gordon and Stryker 1996). More recently, visual neuroscience has shifted to also understanding the function of the mature mouse visual system (for review, see Huberman and Niell 2011; Baker 2013). In theory, and as work in Drosophila has demonstrated, powerful genetic tools for the visualization and perturbation of neuronal activity can lead to a detailed functional description of the cells and circuits that enable visual perception. In addition, as computational tools for parsing large ultrastructure (electron microscopy [EM]) data sets have become available, a detailed understanding of the underlying microcircuits has become possible. For example, serial EM reconstruction has revealed precise maps of local circuits for sensing direction in the fly brain and mouse retina (Briggman et al. 2011; Helmstaedter et al. 2013; Takemura et al. 2013; Kim et al. 2014). Thus, the fields of mammalian and Drosophila visual neuroscience are converging in terms of techniques, and, potentially, common themes will emerge as to how circuits wire up and work at a cellular level (Fig. 1B).
Mouse and fly visual systems share only a very distant common ancestor, which had extremely limited visual functions. Nevertheless, despite important differences, both mice and flies evolved to solve the same general task: to extract crucial information from the visual world and funnel it into adaptive behavioral responses. In both species, large numbers of cell types have now been identified based on morphological, physiological, and functional criteria (Fischbach and Dittrich 1989; Chalupa and Werner 2004).
The mammalian eye includes various cell types that are organized into precise circuits to extensively process visual signals before sending them off to the brain. The retinal cell types are grouped into seven major classes: photoreceptors (rods and cones), horizontal cells, bipolar cells, amacrine cells, Muller glia, and RGCs (Fig. 1C). Each of those cell classes in turn is made up of anywhere from three subtypes (e.g., photoreceptors) to 40 subtypes (e.g., the amacrine cells) that each have unique morphologies, connections, and specific functions. The mouse retina contains rods, which are sensitive to dim achromatic light, and cones, which have different spectral tunings based on their differential expression of opsin photopigments. Amacrine and horizontal cells are the source of inhibitory drive in the retina, expressing either GABA or glycine and, in some cases, also acetylcholine (ACh) or other neuromodulators. Together, these cell types play a key role in shaping the intrinsic processing and output signals of the retina. RGCs are the output neurons of the eye: Their job is to transmit retinal processing of visual signals to the correct circuits in the brain. RGCs include ∼20 subtypes, each responding best to a specific feature in the visual scene, such as motion in specific directions, luminance, or color opponency (Dhande and Huberman 2014). RGCs transmit visual information to a large number of brain targets with diverse functions. At present, close to 50 unique retinorecipient nuclei have been identified in the mouse brain (Morin and Studholme 2014).
The striking regularity of the Drosophila eye, with ∼800 unit eyes or ommatidia, makes it a powerful model to understand the genetic control of cell fate decisions (Johnston 2013). Eight Drosophila photoreceptor neurons per ommatidium are specified from an undifferentiated larval epithelial primordium (Fig. 1D). They send direct axon projections to visual ganglions (neuropiles) in the brain that form the optic lobes. Six “outer photoreceptors” (R1–6) are largely identical between all ommatidia (Wolff and Ready 1993). Their axons terminate in the first optic ganglion, the lamina, whereas inner photoreceptors R7 and R8 project deeper, to the medulla. The two remaining neuropiles do not receive direct input from photoreceptors: The lobula and lobula plate are innervated medulla neurons processing visual information downstream from photoreceptors. The adult medulla neuropil alone comprises at least 70 different cell types (Fischbach and Dittrich 1989; Morante and Desplan 2008; Takemura et al. 2008, 2013), some of which can be reliably subdivided into further subtypes based on current three-dimensional (3D) imaging techniques (Meinertzhagen et al. 2009; Karuppudurai et al. 2014). What makes any of these cell types unique? This vast diversity of cell types makes a comprehensive study of visual circuitry both a daunting challenge and an exciting opportunity to parse a complex set of local circuits with a general-purpose function.
Here we review recent progress in the molecular genetic dissection of visual circuitry in Drosophila and mice. We focus on specific examples that emphasize the strengths and limitations of each model system. The combination of anatomy (which cells), function (what the cells do), and development (where the cells come from) serves as a powerful approach toward solving one of the central questions of neuroscience (How do we see?) and stands to reveal many important new principles common to both model organisms.
Of mosaics, layers, and columns: cell type diversity in the fly and mouse visual systems
The organization of the mammalian and fly retinae shows important similarities in the collective circuitry of the eyes and underlying neuropiles (for review, see Sanes and Zipursky 2010). Below, we discuss the conspicuous similarities in the organization and specification of cell types in Drosophila and mice.
The organization of the Drosophila visual system
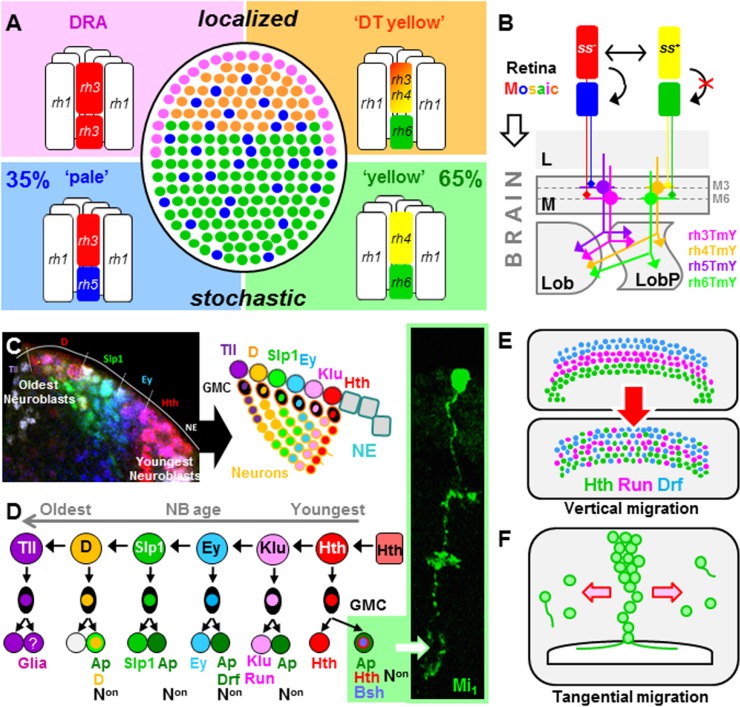
Despite the homogeneous appearance of Drosophila’s ∼800 unit eyes (or ommatidia), important functional differences exist between them (Hardie 1985). The expression of different Rhodopsin (Rh) genes in “inner photoreceptors” R7 and R8, the equivalent of vertebrate cone cells, defines at least four distinct ommatidial subtypes (Fig. 2A; Rister and Desplan 2011). One subtype forms a narrow band of morphologically distinct ommatidia in the “dorsal rim area” (DRA) of the retina (Tomlinson 2003; Wernet et al. 2003), where it measures the vector of polarized light emanating from the sky for navigation (Wolf et al. 1980; Weir and Dickinson 2012; Wernet et al. 2012). The remainder of the retina consists of two randomly distributed subtypes named “pale” (p; 35%) and “yellow” (y; 65%) (Franceschini et al. 1981), expressing distinct combinations of Rhodopsin genes in R7 and R8 (Chou et al. 1996, 1999; Papatsenko et al. 1997; Rister and Desplan 2011; Johnston 2013). yR7 cells in the dorsal third of the retina coexpress both UV Rhodopsins (Rh3 and Rh4), forming a fourth subtype (dorsal third yellow) of ommatidia. Its function remains unknown, yet a possible role of these ommatidia in detecting unpolarized UV-rich skylight has been proposed (Mazzoni et al. 2008; Thanawala et al. 2013). Interestingly, such coexpression of visual pigments is also observed in cone photoreceptors in the mouse retina (see below). The Drosophila retinal mosaic is reminiscent of the stochastic distribution of green and red cones in the human retina, although these two processes have evolved independently (for review, see Johnston and Desplan 2010; Rister and Desplan 2011).
Figure 2.
Cell types in the Drosophila visual system. (A) The retinal mosaic of Drosophila (shown schematically, in the center) contains at least four subtypes of unit eyes (ommatidia) expressing different combinations of rhodopsin genes in their inner photoreceptors, R7 (top) and R8 (bottom). Two subtypes are always found in the dorsal half of the eye: DRA ommatidia, as a narrow band in the periphery, serve as specialized detectors for linearly polarized light originating from the sky. “DT yellow” ommatidia are less -specialized UV detectors found adjacent to the DRA and whose function is currently unknown. The two remaining subtypes, named “pale” and “yellow,” are distributed stochastically at an uneven ratio (35/65), creating a mosaic of chromatic sensitivities. (B) Stochastically distributed ommatidial subtypes are set up in R7 cells by expression of the gene spineless (ss) in the “yellow” subtype (choice). Only “pale”-type R7 cells instruct underlying R8 cells to acquire the same subtype fate (instruction), while “yellow” R8 cells choose their fate by default. Little is known about postsynaptic elements. Specific synaptic partners to pR7, yR7, pR8, and yR8 photoreceptors have recently been described: morphologically similar TmY cells in the medulla (rh3TmY, rh4TmY, rh5TmY, and rh6TmY, respectively). It remains unclear how such neurons would be specified in response to a stochastic input. (C, left) Surface view of the developing medulla neuropil of Drosophila. Rows of NBs and their progeny sequentially express overlapping sets of transcription factors (Hth is red, Ey is blue, slp1 is green, and Dichaete [D] is pink). (Right) Schematic summarizing the cellular progeny arising from medulla NBs over time. Sequential recruitment of different transcription factors (same color code as above, except Klu is pink, Dichaete [D] is yellow, and Tll is purple) leads to cellular diversity in the NB progeny (modified from Li et al. 2013a). (D) Simplified flow chart summarizing how cellular diversity among NB progeny is achieved in the developing medulla neuropil. Asymmetric cell divisions give rise to glia as well as different classes of neurons expressing different combinations of transcription factors. In rare cases, these progenitor classes can be traced to the adult neuron. Shown is a GAL4/GFP flip-out clone of an Mi1 neuron originating from the Hth lineage (Hth+, Ap+, and Bsh+ Non). (Drf) Drifter/ventral veins lacking. (Adapted from Li et al. 2013a.) (E) In order for developing medulla neurons to adopt their adult morphology and location within the circuit, they move vertically, resulting in a mixing of the layers of cells labeled by transcription factor combinations (Hth, Run, and Drf). (Adapted from Hasegawa at al. 2011.) (F) Similarly, developing neurons migrate horizontally to their final position within the circuit. The schematic shows a mitotic clone of larval medulla NBs with cells detaching and migrating to the periphery. (Adapted from Hasegawa at al. 2011.)
One of the most fascinating questions about this retinal patterning is to understand at the molecular level how the initial stochastic decision is executed in R7 cells. While the genetic specification of stochastic cell fates might appear paradoxical, it has been studied extensively from bacteria to neurons (Losick and Desplan 2008; Johnston and Desplan 2010). The recent dissection of the Drosophila spineless locus, the gene that controls the stochastic expression of Rhodopsins, has provided important new insight. spineless encodes a PAS basic helix–loop–helix (bHLH) transcription factor (Wernet et al. 2006; Thanawala et al. 2013; Johnston and Desplan 2014) that directly activates the yR7 Rhodopsin 4 (Rh4) and indirectly represses the pR7 Rhodopsin 3 (Rh3). It appears that complex interplay between cis-regulatory sequences in the spineless locus and interchromosomal communication lead to robust stochastic cell fate decisions in R7 photoreceptors (Johnston and Desplan 2014). Initially, each spineless allele appears to make its own stochastic choice, which is the result of two silencer sequences interfering with an R7-specific activator. When two opposing decisions are made (one allele of spineless is “ON,” while the other is “OFF”), the expression state between alleles is coordinated over long distances (Johnston and Desplan 2014). Although the exact mechanism of this interchromosomal communication remains unknown, the regulatory sequences responsible have been identified, and the exact molecular logic behind stochastic specification events can now be elucidated (Johnston and Desplan 2014).
The specification of photoreceptor cell fates within stochastically distributed ommatidia has been studied in detail, culminating in a model of at least two genetically separable steps. First, R7 cells in “yellow” ommatidia (yR7) stochastically express spineless. In a second step, spineless also represses an unknown signal in R7 cells of the “yellow” subtype (Fig. 2B). In cells that do not express spineless (pR7), this signal instructs the underlying R8 cell to be of the “pale” type, leading to expression of the pR8 Rhodopsin (Rh5). By default, yR8 cells that have not been instructed by R7 choose the yR8 Rhodopsin Rh6. Finally, the “pale” (Rh5) versus “yellow” (Rh6) decision in R8 is further consolidated by a bistable regulatory loop (Mikeladze-Dvali et al. 2005) involving the Hippo/warts tumor suppressor pathway and the growth regulator Melted (Jukam and Desplan 2011; Jukam et al. 2013) as well as a network of transcription factors (Rister and Desplan 2011; Johnston 2013). It must be emphasized that at least two crucial Drosophila transcription factors involved in shaping the retinal mosaic are homologs of vertebrate photoreceptor determination factors: Orthodenticle/CRX and Traffic jam/NRL (Jukam et al. 2013). Therefore, in some instances, the same factors are recruited to define retinal cell fates in both flies and humans (Rister and Desplan 2011). In Drosophila, the identification of most crucial factors now allows for the complete re-engineering of retinal mosaics for functional studies (Wernet et al. 2007).
An important problem arises from such stochastically specified photoreceptors: How are their postsynaptic targets specified in the brain so as to provide an accurate “match” in connectivity? Based on their rhodopsin expression, “pale” and “yellow” R7 and R8 transmit information of different chromatic content to the optic lobes, where comparison between R7 and R8 activity from the same ommatidium as well as comparison between neighboring ommatidia lead to color discrimination (Gao et al. 2008; Morante and Desplan 2008; Yamaguchi et al. 2010; Schnaitmann et al. 2013; Melnattur et al. 2014). Do postsynaptic elements receive input from all ommatidia regardless of their “pale” and “yellow” fates, extracting information through a population code? Since the Drosophila visual system appears to be strictly hardwired (Scott et al. 2003; Hiesinger et al. 2006), it is difficult to imagine how this would be achieved. However, a recent study using transsynaptic tracing in combination with in vivo calcium imaging has demonstrated the existence of distinct neuron types postsynaptic to each of these stochastically specified photoreceptors (Jagadish et al. 2014). The four identified cell types all belong to the class of TmY neurons, a group of medulla neurons with bifurcated axons terminating in the lobula as well as the lobula plate (Fischbach and Dittrich 1989; Jagadish et al. 2014). How are the target neurons in the optic lobes that are contacted by photoreceptors specified? In the future, it will be critical to figure out how these target cells (named rh3TmY, rh4TmY, rh5TmY, and rh6TmY) are programmed to receive appropriate connections with the correct photoreceptor subtype (Fig. 2B).
Until recently, only the very early stages of optic lobe development in Drosophila were known (Campos-Ortega and Hofbauer 1990). However, this has now been addressed systematically in several new studies, leading to a description of how cellular diversity in the fly visual system is generated (Li et al. 2013a; Suzuki et al. 2013; Bertet et al. 2014; for review, see Li et al. 2013b). The mechanisms of neuronal specification in the medulla neuropil share surprising similarities with that of the fly embryonic ventral nerve cord, where each neural stem cell (called a neuroblast [NB]) expresses sequentially a series of transcription factors (Fig. 2C; for review, see Jacob et al. 2008). In the medulla, NBs are converted from a neuroepithelium (NE), where cells divide symmetrically, in a wave of neurogenesis (Egger et al. 2011). At least six transcription factors are expressed sequentially in medulla NBs as they age—from youngest to oldest: Homothorax (Hth), Klumpfuss (Klu), Eyeless (Ey), Sloppy paired 1 and 2 (Slp1/2), Dichaete (D), and Tailless (Tll) (Fig. 2C). These transcription factors regulate each other’s expression as well as downstream genes, thereby creating medulla cells with different transcriptional repertoires depending on the age of the NB that generated them. Asymmetric cell divisions ensure NB self-renewal and produce ganglion mother cells (GMCs) that divide once to give rise to two medulla neurons. As we discuss below, increasing evidence suggests that very similar mechanisms create the cellular diversity described in the mammalian retina (for review, see Cepko 2014). The cellular diversity is further increased by Notch-dependent asymmetric cell division of GMCs—one with Notch activity (NotchON) and one that is NotchOFF (Fig. 2D). An important functional role for this Notch function during asymmetric cell division was recently demonstrated in a different part of the developing optic lobes, the tips of the outer proliferation center (tOPC) (Bertet et al. 2014). In this region of the developing optic lobes, either NotchON or NotchOFF progeny are lost by apoptosis, depending on their time of birth. Astonishingly, different apoptotic genes are used, with reaper killing NotchON, while hid kills NotchOFF cells.
The final stage of medulla cell differentiation involves migration, during which cells from NB-derived concentric zones adopt their final positions within the topographically arranged columnar units (Hasegawa et al. 2011). During pupation, concentric layers of cells expressing Hth, Runt (Run), and Drifter (Drf) can be observed early but then intermingle later (Fig. 2E; Hasegawa et al. 2011). Similarly, when clones of neurons originating from a single NB are labeled as a vertical column reminiscent of the temporal series of their birth order, tangential migration later leads to their dispersion throughout the neuropil (Fig. 2F; Morante et al. 2011). The signals that guide these cells toward their correct location within the developing circuits are not known, but Ey, which is also part of the NB transcription factor sequence, is required for medulla cell migration (Morante et al. 2011). Taken together, the sequential expression of transcription factors that gives rise to different classes of developing medulla cells followed by their morphological differentiation creates the functional building blocks from which visual circuits with specific computational tasks will assemble. It must be pointed out that increasing evidence points toward this temporal mechanism being a general concept used to generate cellular diversity in both different Drosophila brain tissues (Isshiki et al. 2001; Bayraktar and Doe 2013; Li et al. 2013a,b; Bertet et al. 2014) and vertebrates (for review, see Cepko 2014).
To date, only a small subset of adult medulla neuron types can be linked to a specific combination of transcription factors inducing their specific developmental history. For instance, the oldest, Tll-positive NBs specifically give rise to medulla neuropil glia (Colonques et al. 2007; Soustelle and Giangrande 2007; Li et al. 2013a). The youngest, Hth-expressing NBs give rise to cells expressing the factors Bsh (brain-specific homeodomain) and Ap (Apterous) (Hasegawa et al. 2013). These cells will then mature into a medulla cell type called Mi1, which is involved in the perception of motion stimuli as shown through both serial EM reconstruction and electrophysiology, as discussed below (Takemura et al. 2013; Behnia et al. 2014). Future studies will assign every cell type in the medulla neuropil a unique NB origin and transcriptional signature.
Cell type specification in the mammalian visual system
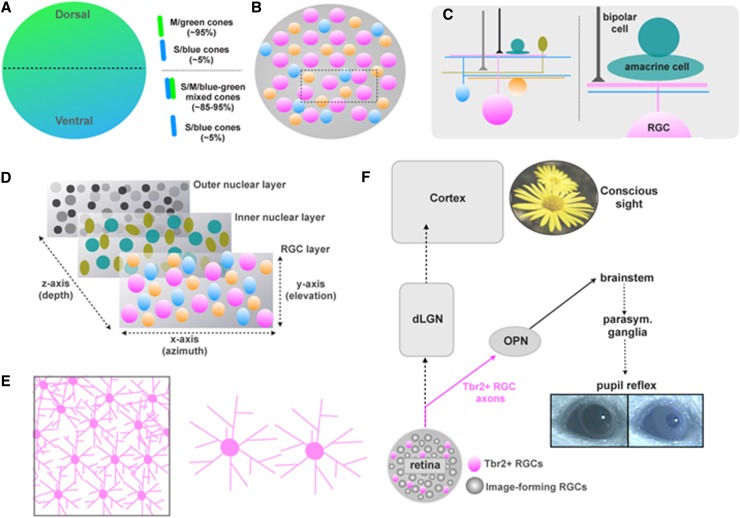
Mammalian retinal photoreceptors also represent an attractive focus for understanding the genetic programs that designate cell type specification and function. In mice, the photoreceptors are also regionally distributed: While rods are found throughout the mouse retina, mouse cone photoreceptor types occupy different spatial locations in a manner that is optimized for the average statistics of the visual fields. The dorsal retina (which views the lower visual field) contains >95% medium-wavelength-sensitive (M/green) cones but very few (∼5%) pure short-wavelength-sensitive (S/blue) cones. In the ventral retina (upper visual field), on the other hand, most cones exhibit mixed S/M opsin expression, and the remaining 5% are pure S/blue cones (Fig. 3A; Szél et al. 1992; Applebury et al. 2000; Haverkamp et al. 2005). Recent work suggests that the regional variation in these cone distributions is optimized for sensing not just colors but also different contrast levels that tend to predominate in the “sky” versus “ground” of the visual field (Baden et al. 2013). The coexpression of M and S opsins in the ventral mouse eye that faces the sky is reminiscent of the coexpression of the two UV opsins in the dorsal Drosophila eye that also faces the sky (Mazzoni et al. 2008). However, it is worth noting that such a distribution of cones is not observed in primates that instead have a fovea where most cones are located.
Figure 3.
Retinal cell types, regional variations, and select functions in mice. (A) Photoreceptor distributions vary by dorsal-ventral location in the mouse retina, with M/green cones dominating the dorsal retina, and mixed S/M/blue-green cones dominating the ventral retina. Pure S/blue cones are scattered throughout the retina, ranging from 5% (dorsal) to 15% (ventral). (B) Barring a few exceptions, such as cone photoreceptors (see Fig. 5B), most of the 100+ cell types in the retina (often called subtypes) are distributed in mosaics with relatively even spacing of neuronal cell bodies. Three mosaics are schematized (orange, blue, and pink), each representing a distinct retinal cell subtype. (C) Connectivity between the processes and dendrites of retinal neurons is highly laminar to allow interactions between specific subsets of neurons; e.g., a narrow-field amacrine (turquoise) and monostratified RGC (pink) and bipolar cell (gray). (D) Each cell layer of the retina (outer, inner, and RGC) contains mosaics of specific cell types (also see Fig. 1C) and are interconnected in a highly cell type-specific way: Processing of visual scenes arises from repeated connectivity between defined subsets of cells across the depth (Z-axis) of the retina and across the two planes of the retina that view azimuth (X-axis) and elevation (Y-axis) in the visual scene. (E) The processes of individual retinal cell subtypes (pink) cover or “tile” the spaces between their somas to ensure capture of input from specific presynaptic cells. (F) Tbr2-expressing (also called Eomes) RGCs (magenta) project to the OPN, a critical station in the pupillary light reflex pathway (OPN > brainstem Edinger Westfal nucleus > parasympathetic ciliary ganglion > iris constriction) that is anatomically separate from the image-forming pathway from other RGCs to the dLGN, which relays retinal signals to the cortex for conscious perception of visual scenes. Genetic deletion of Tbr2+ RGCs partially eliminates the pupil constriction reflex (Sweeney et al. 2014).
The genetic programs that specify rods versus cones and S versus M cones are becoming clear: S cones expressing the transcription factor Rorβ represent the photoreceptor “default state.” The thyroid hormone receptor TRβ2 directs otherwise S-cone-fated cells toward M-cone identity and differentiation (for review, see Swaroop et al. 2010; Rister et al. 2013). Given the preponderance of mixed S/M-cone distributions described above, the expectation is that S/M cones coexpress Rorβ and TRβ2 to varying degrees. Rods are generated from distinct progenitors that express the transcription factor Nrl and the nuclear receptor Nr2e3, which repress Rorβ. Amacrine and horizontal cells share a common developmental program that depends on the expression of the transcription factor Foxn4 (Li et al. 2004; Fujitani et al. 2006). In the absence of another transcription factor, Ptf1a, both cell types are dramatically reduced, and there is an overproduction of ganglion cells (Fujitani et al. 2006; Nakhai et al. 2007). Finally Prox1 promotes the horizontal cell fate at the expense of amacrine cells: In its absence, horizontal cells fail to form, while ectopic expression of Prox1 drives ectopic horizontal cell production (Dyer et al. 2003).
The diversification of retinal cell subtypes is less well understood; however, some of the signals that promote the development of specific amacrine subtypes are now known. For example, the transcription factors NeuroD, Math3, and Barhl2 act together to drive cells toward a glycinergic amacrine fate, whereas Isl1 drives them toward a cholinergic fate. The cholinergic fate is particularly relevant to our understanding of retinal processing because cholinergic amacrine cells are an instrumental component of the retinal circuits that create direction-selective RGCs (DSGCs) (discussed in detail below). Bhlhb5 establishes several GABAergic amacrine subtypes and ON-type bipolar cells that are glutamatergic interneurons (Feng et al. 2006). Our current understanding of amacrine and bipolar diversity arises in large part from morphologic studies of their branching and stratification patterns, but as the number of transgenic markers for labeling unique retinal cell subtypes is expanding, so is our understanding of how those subtypes are specified. For example, Kay et al. (2011a) used transgenic labeling and gene profiling of amacrine cell subsets to discover that NeuroD6 defines a previously unrecognized population of non-GABAergic, nonglycinergic amacrine cells.
Although the programs that define the remaining 30+ amacrine and 12+ bipolar subtypes are still being defined (Reese 2011), the general principle that emerges from these studies is that progenitors turn on transcriptional programs that, over time, become more restricted toward the generation of certain cell types (Voinescu et al. 2009; Cepko 2014). That understanding, combined with the availability of a large number of transgenic lines for marking specific retinal cell types, raises the expectation that the transcriptional programs leading to the full diversity of retinal interneurons will be deciphered in the near future.
One hallmark feature of retinal cell types in the vertebrate retina is mosaicism: Each neuronal subtype is arranged in a regularly spaced manner in the plane of the retina, with the degree of regularity and spacing of their somas varying between cell types (Fig. 3B). Indeed, mosaic spacing is one of the fundamental units defining cell types in the retina (Field and Chichilnisky 2007; Field et al. 2010) and is often related to dendritic tiling or the extent to which the dendrites of retinal cell types avoid one another or overlap (Fig. 3E). Tiling of retinal cell types can vary from a coverage factor of 1 (equating to tip-to-tip dendritic coverage with no overlap) to ≥20 (significant overlap) (Peichle and Wassle 1979). Recent work indicates that the mosaic spacing of RGCs and some amacrine cells is mediated by DSCAM (Down syndrome cell adhesion molecule) and the closely related DSCAM1 (Fuerst et al. 2008, 2009), genes that play critical roles in neuronal self-avoidance in flies and laminar specificity in the chick retina (for review, see Hattori et al. 2008; Yamagata and Sanes 2008). Even more recently, Kay et al. (2012) discovered that MEGF10 and MEGF11 (genes homologous to Drosophila Draper that control cell engulfment in macrophages) are also critical for establishing cell–cell avoidance, leading to mosaic spacing of a subset of retinal interneurons, the cholinergic starburst amacrine cells (SACs). Molecular control specifically over the dendritic tiling process has also explained the generation of cell types with specific coverage factors. Lefebvre et al. (2012) discovered that within a given retinal neuron subtype, each individual cell expresses a unique and specific combination of protocadherin isoforms that causes the branches of its dendrites to avoid each other when those branches arise from the same cell (self-avoidance) but permits overlap of dendrites between neighboring neurons of the same subtype due to the expression of different combinations/splice variants of protocadherins. Much work is still needed to resolve, at the molecular level, the precise constraints on cell soma spacing and the dendritic overlap/coverage factor in the retina. Nonetheless, new concepts in this area are emerging quickly, including the demonstration of molecular control over outer retinal cell type tiling by guidance cue signaling. Kolodkin and coworkers (Matsuoka et al. 2012) recently showed that the repellant guidance cues Sema6a and PlexinA4 are important for self-avoidance of horizontal cell processes. The independence of such phenotypes between specific retinal neuron subtypes and their expression of distinct ligands, receptors, and isoform profiles reinforces the idea that while mosaic spacing and tiling are ubiquitous features in retinal neurons, each retinal neuron subtype may employ a different set of molecular signals to achieve these key features of retinal organization.
The other hallmark feature of the retina is vertical connectivity. Each individual retinal neuron is connected to its synaptic partners at a highly specific depth (or layer) (Fig. 3C,D), which provides numerous parallel channels for processing of visual information (Roska and Werblin 2001). The net result is a modular arrangement of repeating vertical circuits that together ensure adequate sampling of each visual feature across space (Fig. 3D; Masland 2012). For example, DSGCs achieve their namesake property due to asymmetric inhibition from OFF and/or ON SACs that stratify within specific sublaminae of the inner retina (see below; for review, see Demb 2007; Briggman et al. 2011; Wei and Feller 2011). Major progress has recently been made in understanding the receptor–ligand interactions that define laminar specificity of RGCs in mice, such as ON-type DSGCs , and ON SACs, and specific ON bipolar cells have recently been elucidated (Matsuoka et al. 2011a,b). This study from Kolodkin and coworkers (Matsuoka et al. 2011a,b; Sun et al. 2013) demonstrates that repellant interactions play an essential role in defining laminar connectivity by restricting cell–cell contacts and synaptic interactions to specific depths within the retinal neuropil. They have also shown that the expression of specific ligand–receptor menus controlling laminar specificity also defines new subgroups of amacrine cells that would otherwise be molecularly indistinguishable (Sun et al. 2013). The prominent role for repellants in establishing laminar-specific connectivity in the mouse retina is noteworthy given that, in the chick retina, adhesive interactions mediated by DSCAMs, sidekicks, and contactins appear to exert these roles (Yamagata et al. 2002; Yamagata and Sanes 2008, 2012). Very recently, evidence in mice has shown that type II cadherins mediate adhesive cell–cell interactions, leading to precise synaptic connectivity in the inner retina (Duan et al. 2014). Still, the molecular signals leading to precise cell–cell connectivity for most of the known circuits of the mouse retina remain a mystery, and while laminar specificity and synaptic specificity are undoubtedly linked processes at some level, they also display surprising independence. Even in mutants where laminar specificity of a given RGC subtype is severely altered by mutation to semaphorin/plexin signaling, the dendrites of that RGC still connect with the correct presynaptic partner (Matsuoka et al. 2011a). Thus, an additional level of genetic control over retinal wiring must exist to ensure cell–cell synaptic precision independently of laminar depth.
Several of the transcriptional programs that regulate RGC fate in retinal progenitors have been identified. Math5 is essential for the production of most RGCs, while Brn3a, Brn3b, and Brn3c are involved in broadly differentiating the RGC population into different RGC groups, albeit not into functionally or morphologically distinct subtypes (Badea et al. 2009). Although Math5 mutants lack most RGCs, the remaining cells include intrinsically photosensitive RGCs (ipRGCs) (Lin et al. 2004), which, as their name suggests, act as photoreceptors and express melanopsin (also called OPN4) (Berson et al. 2002; Hattar et al. 2002, 2003; Panda et al. 2002). These cells project to the suprachiasmatic nucleus (SCN) of the hypothalamus(the master circadian pacemaker of the brain) and to various thalamic and pretectal nuclei involved in modulation of the circadian clock as well as pupil reflexes and food-based entrainment of endogenous physiological rhythms (Hattar et al. 2002, 2003). As many as five subtypes of ipRGCs have been described, and their unique functions and developmental profiles are gradually becoming clear (e.g., Ecker et al. 2010; Chen et al. 2011; Estevez et al. 2012). Recently, Feldheim and coworkers (Sweeney et al. 2014) discovered that a subset of ipRGCs that project to the regions of the brain controlling pupil reflexes express the transcription factor Tbr2/Eomes (Fig. 3F) and that conditional loss of those Tbr2 RGCs leads to a selective removal of this cell type and connections (Sweeney et al. 2014). This is exciting because it stands as one of the first examples of transcriptional control over RGC subtype-specific development and as a proof of concept that, with the ever-growing number of transgenic mouse lines for marking specific RGCs, transcriptome profiling and Cre-based approaches for conditionally manipulating gene expression in those specific RGC types will soon lead to the discovery of the mechanisms by which specific RGC subtypes develop and function.
Finally, just as in flies, where specific photoreceptors must wire up with the correct targets and cell types in the brain, the various RGC subtypes must also wire up their axons to the correct central targets to enable specific retinal signals to drive the appropriate behaviors. Is the specification of visual brain targets in any way linked to the specification of specific targets or target neuron types in the brain (Fig. 1A)? At this point, nothing is definitively known about this important issue in mice. Still, it is noteworthy that Lumsden and coworkers (Delogu et al. 2012) recently discovered that the transcription factor Sox14 is critical for the development of several functionally distinct subcortical visual nuclei, all of which are targeted by ipRGCs and are rich with GABAergic target neurons. It is unknown whether ipRGCs also express Sox14, but it is tempting to speculate that, by coupling the timing and location of transcription factor expression in defined categories of RGC subtypes and in their target neurons in the brain, accurate ipRGC connectivity is ensured, a model very similar to what has been proposed in flies (see Fig. 2D). Meanwhile, there is also growing interest in defining the receptor–ligand interactions downstream from specific transcriptional programs that govern the specific wiring connectivity between RGC subtypes and their target choices in the brain. Osterhout et al. (2011) showed that a specific type II cadherin, Cadherin-6 (Cdh6), is expressed by the RGCs that target regions of the brain involved in pupil reflexes; in the absence of Cdh6, the axons of those RGCs mistarget to other retinorecipient areas.
The role of identified cell types in guiding specific visual behaviors in flies and mice
Over the last few years, much work has focused on understanding how the contributions of identified cellular units within the visual system of both flies and mice are related to the behavior of the animal. This work was crucial for understanding the functional role of cell types previously described in anatomical or developmental studies and linking them with functional studies using electrophysiology or in vivo calcium imaging.
Genetic dissection of the behavioral contributions by cell types in Drosophila
In the Drosophila visual system, the accepted model was that two general pathways process different kinds of visual information: (1) motion and form, involving the outer photoreceptors R1–6, and (2) color and polarization, for which inner photoreceptors R7 and R8 play the major role. This was supported by behavior experiments using inactivated R1–6 or R7 and R8 photoreceptors (Heisenberg and Buchner 1977; Yamaguchi et al. 2010). However, recent results have revealed rather unexpected retinal contributions to different visual behaviors: Color photoreceptor R8 has been implicated in the detection of motion (Wardill et al. 2012), while achromatic R1–6 appear to play a role in the detection of color (Schnaitmann et al. 2013) and linearly polarized reflections (Wernet et al. 2012). Hence, the complex interactions between those photoreceptor cells projecting to the lamina part of the optic lobes (R1–6) and those projecting to the medulla (R7 and R8) indicate that functional separation between these subtypes is far less absolute than previously assumed (Bausenwein et al. 1992) and that information about different visual qualities like color and motion begins to be integrated at a very early stage.
By developing a series of behavioral paradigms using tethered single flies in a virtual flight arena or walking flies on an air-suspended ball as well as population-based approaches, important functional properties could be assigned to a number of identified optic lobe neurons for which detailed morphological classifications exist (Fischbach and Dittrich 1989; Meinertzhagen and O’Neil 1991; Takemura et al. 2008). These studies took advantage of cell type-specific GAL4 driver lines to genetically perturb synaptic transmission in combination with behavioral assays. The most exciting progress has been made in the optic lobes, yet much attention has also focused on structures in the central complex in the central brain, where complex features of the visual world are represented (Hanesch et al. 1989; Liu et al. 2006; Ofstad et al. 2011). Using new cell type-specific labeling tools (Jenett et al. 2012), these cell types can now be specifically targeted for anatomical reconstruction, functional characterization, and the dissection of their contribution to behavior.
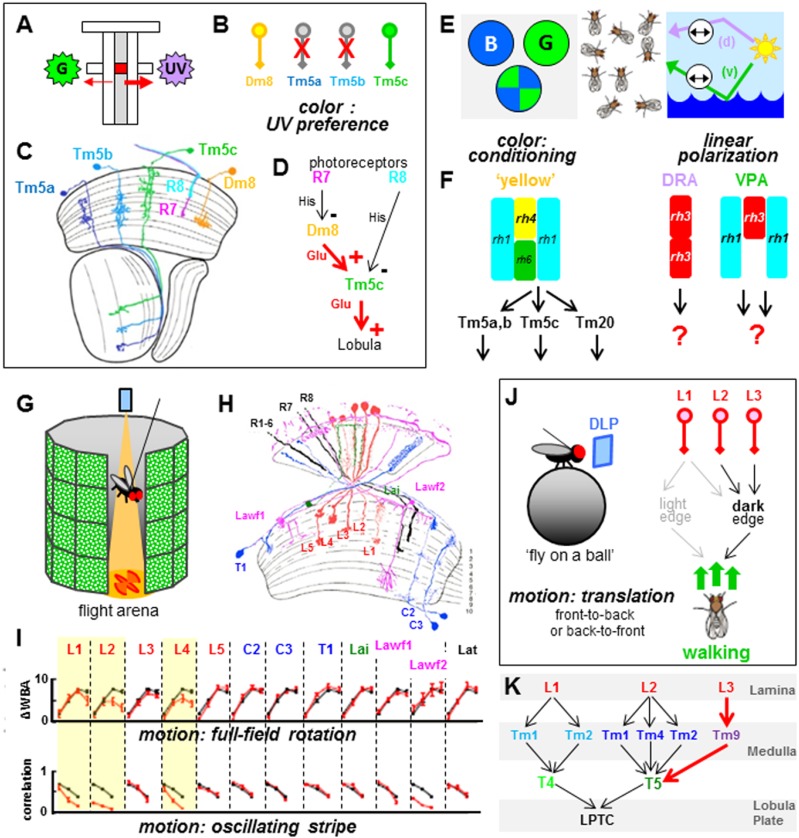
Considerable attention has been given to Drosophila’s ability to detect different qualities of light, like color. For instance, it was recently shown that one specific class of medulla wide-field amacrine cells called Dm8 (Fischbach and Dittrich 1989) as well as one type of small-field projection neurons named Tm5 are required for mediating spectral discrimination of green versus UV light (UV preference) (Fig. 4A,B; Gao et al. 2008; Karuppudurai et al. 2014; Ting et al. 2014). More recently, the behavioral contribution of Tm5 cells could be narrowed down to one morphological subtype: Tm5c (Fig. 4C; Karuppudurai et al. 2014). These studies culminated in the description of a hardwired, glutamatergic circuit downstream from inner photoreceptors R7 and R8 (involving Dm8 and Tm5c), relaying UV light information to the lobula (Fig. 4D). This model serves as the cellular basis for the innate escape behavior observed in many insects that fly toward the UV-rich sky to avoid perilous encounters. Finally, another recent study described new cell types required for the phototactic response to specific wavelengths of light (Otsuna et al. 2014). The placement of these cells into the above-mentioned circuits is hampered by the fact that some of them have not been characterized in previous morphological studies (Fischbach and Dittrich 1989). Learning assays were also used to investigate additional aspects of color vision, like the distinction of blue versus green quadrants (Fig. 4E). In these experiments, wavelength discrimination appears to crucially depend on not only inner photoreceptors sensitive to UV, green, or blue but also the broadband outer photoreceptors R1–6. This provides further evidence that color vision and motion detection use overlapping input channels (Schnaitmann et al. 2013). This study also revealed that only one class of “yellow” ommatidia was sufficient to mediate the wavelength discrimination task (more specifically, R1–6 + yR7) (Fig. 4F).
Figure 4.
Behavioral contributions of identified Drosophila neurons. (A–J) Summary of different behavioral assays linking identified cell types in the Drosophila visual system to specific visual tasks. (A) Summary of a population assay addressing one important aspect of color vision, which is the spontaneous attraction of flies to UV light (UV preference) when presented with a choice between UV and green light (adapted from Karuppudurai et al. 2014 with permission from Elsevier). (B,C). Crucial cellular components in the optic lobes were identified and could be narrowed down to a morphological subclass of a specific transmedullary neuron cell type (Dm8 and Tm5c), excluding a role for highly similar counterparts (Tm5a,b) (adapted from Karuppudurai et al. 2014 with permission from Elsevier). (D) Using this behavioral paradigm, a hardwired glutamatergic circuit downstream from inner photoreceptors R7 and R8, involving medulla cell types Dm8 and Tm5c, was identified to mediate UV preference. (E) Different population-based assays using freely walking flies to identify cellular contributions to different tasks, like conditioning to blue versus green colors (left), or the spontaneous orientation of the body axis in response to the incident e-vector (polarization vision) (right). (F) The distinction of blue versus green quadrants was dependent on broadband R1–6 photoreceptors and yR7 UV receptors in “yellow” ommatidia, while different retinal detectors were identified for celestial or reflected polarization vision stimuli. Interestingly, polarization vision stimuli perceived with the ventral eye depend on R1–6 + pR7, a retinal substrate that is complementary to what color vision employs. Dorsal polarization vision stimuli have their separate detectors in DRA ommatidia. While no information exists on their downstream targets, parallel medulla pathways (Tm5a,b, Tm5c, and Tm22) have been described for blue versus green choice using single flies in a flight simulator. (G–K) Behavioral assays addressing the perception of motion. (G–I) Summary of one study using single flies suspended in a virtual flight arena (G) (adapted from Tuthill et al. 2013 with permission from Elsevier) using LED panels to test the contribution of 12 lamina cell types (H) (modified from Tuthill et al. 2013 with permission from Elsevier) to different motion vision tasks, like full-field rotation and oscillating stripes. (I) Of the 12 cell types in the lamina neuropil that were tested, two output neurons (monopolar cells L2 and L4) were found to be crucial using this assay, while the remaining cells most likely serve to modulate and sculpt the behavioral response (modified from Tuthill et al. 2013 with permission from Elsevier). (J,K) Summary of a similar study using single flies walking on an air-suspended ball while being presented with visual stimuli projected on surrounding screens (shown in J, adapted from Clark et al. 2011 with permission from Elsevier). Special attention was given to translational motion stimuli (front-to-back or back-to-front motion). Focusing on lamina monopolar cells L1–L4, an important contribution of L3 was described for translational dark edges (adapted from Silies at al. 2013 with permission from Elsevier). However, a role for lamina monopolar cell L4 was not identified in this study. (K) Anatomical reconstruction confirms synaptic convergence of L2 targets (Tm1,2,4) and L3 targets (Tm8) onto the same lobula plate target, T5 (adapted from Shinomiya et al. 2014 with permission from Elsevier).
In the DRA of the fly retina, monochromatic R7 and R8 cells that both express the UV Rhodopsin Rh3 compare light using two orthogonal polarization detectors (R7 vs. R8). Their conserved function is to measure the e-vector of linearly polarized skylight, important for navigation (Wehner 2001). A population assay allowed the identification of a second group of retinal polarization detectors in the ventral half of the eye (Wernet et al. 2012). Specialized ommatidia in this ventral polarization area (VPA) likely detect polarized light reflected from water surfaces that are to be avoided (Fig. 4E; Wehner 2001). Interestingly, VPA ommatidia belong to the “pale” subtype and appear to involve comparison between R1–6 and Rh3-expressing pR7 (Fig. 4F). Therefore, “pale” and “yellow” ommatidia distributed stochastically over a larger area in the ventral fly retina mediate two very different behavioral tasks: color versus polarization vision. This is the first demonstration of stochastically distributed photoreceptor cell types guiding separate behavioral responses, adding an important new aspect to the functional role of retinal mosaics. Interestingly, while strong variations exist in the ratios between stochastically distributed cones in the human retina, these individual differences appear to have no behavioral consequence when faced with standard color vision tests. (Rister and Desplan 2011). In flies, the genetic dissection of downstream neural circuits mediating “pale”-specific versus “yellow”-specific functions is in progress, revealing several parallel pathways relaying information to deeper brain centers (Fig. 4F; Melnattur et al. 2014; Velez et al. 2014).
In recent years, great progress has been made toward understanding the contributions of individual cell types to the perception of moving edges (motion vision). A number of studies found that, on the input side of the motion vision circuit, lamina monopolar cell L1 feeds into a pathway for the detection of moving light edges (ON), while L2 detects moving dark edges (OFF) (Rister et al. 2007; Reiff et al. 2010; Joesch et al. 2010, 2013; Clark et al. 2011; Eichner et al. 2011). On the output side of the circuit, electrophysiological and anatomical data point toward lobula plate neurons T4 and T5 as being the outputs that feed onto the first cell type to be truly direction-selective, the so-called lobula plate tangential cells (LPTCs) (Douglass and Strausfeld 2000, 2003). In recent years, the identification of cell types located between the lamina and lobula plate were the subject of intense research, as we see below. Furthermore, a series of recent behavior studies has begun to readdress the role of lamina monopolar cells in mediating two distinct computations of ON (bright edge) and OFF (dark edge) signals for motion vision (Fig. 4G–K; Borst 2014).
One study has systematically tested the behavioral contribution of 12 cell types in the lamina (Tuthill et al. 2013) using sophisticated stimulation regimes on single flies in a virtual flight arena (Fig. 4G,H). Specific roles for monopolar cells L1, L2, and L4 in the detection of motion could be demonstrated (Fig. 4I). Manipulation of feedback neurons C2 and C3 also resulted in specific phenotypes, revealing important new aspects in motion vision architecture. The remaining eight lamina cell types appear to modulate and sculpt the behavioral responses (Tuthill et al. 2013, 2014). Another study using individual flies walking on an air-suspended ball (Fig. 3J) also assigned a crucial role for L3 in the detection of translational motion, specifically for dark edges (Silies et al. 2013). The convergence of L2 and L3 signals in this OFF pathway via different types of medulla neurons was recently confirmed using anatomical reconstruction at EM resolution (Fig. 4K; Shinomiya et al. 2014). Interestingly, this study found no role for L4, while contributions of L1 varied strongly between turning responses and forward walking. This illustrates how difficult it is to assign a specific function to a given neuron, since numerous parameters need to be accounted for, from fly genetics to stimulus design, experimental setup, and data analysis. Furthermore, the microcircuitry of lamina cell types is complex; it includes lateral inhibition (Freifeld et al. 2013; Takemura et al. 2013) and a wide distribution of electrical synapses (Joesch et al. 2010; Wardill et al. 2012), which warrants an even more careful interpretation of experimental data. The growing collections of cell type-specific GAL4 databases (Pfeiffer et al. 2008; Gohl et al. 2011; Jenett et al. 2012; Kvon et al. 2014) combined with optogenetic or circuit-breaking tools (de Vries and Clandinin 2012; Haikala et al. 2013) offer a promising basis for further improving the behavioral dissection of visual behavior.
Genetic analysis of cells types that control visual behavior in mice
Studies of visual circuits in the vertebrate retina and subcortical targets date back many decades with the classic description of “what the frog’s eye tells the frog’s brain” (Lettvin et al. 1968), which classified RGCs primarily according to their functional properties as spot, edge, or local motion detectors. In recent years, new genetic tools in mice have allowed labeling and recording from each of the 20 or so RGC types. This finally enabled researchers to understand how the receptive field properties, morphology, and stratification of an RGC in the retina relate to its patterns of central targeting, which has direct bearing on the specific function of an RGC for visual processing (for review, see Dhande and Huberman 2014).
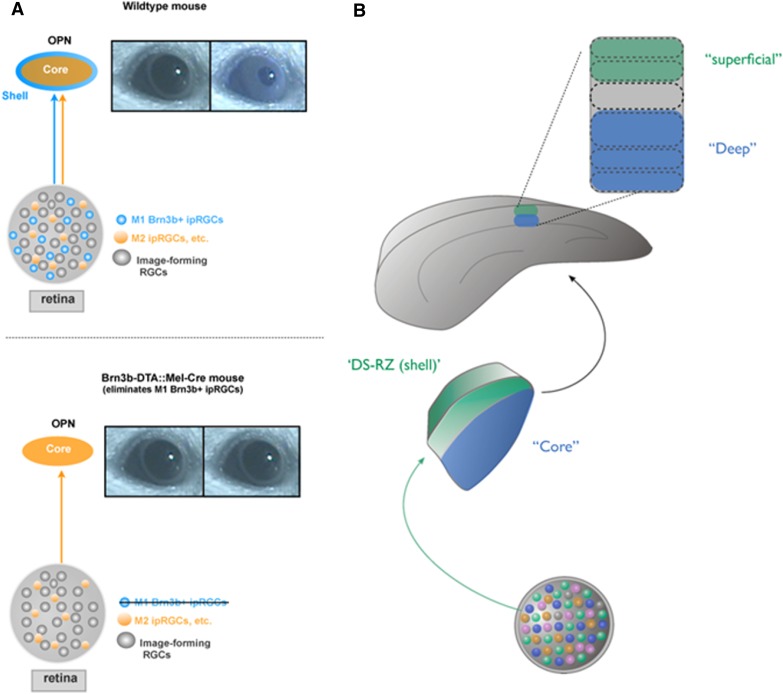
Ultimately, the goal is to causally link the features extracted by specific retinal circuits and delivered to the brain by specific RGC subtypes to defined categories of visual behavior. This will allow the determination of the precise contribution of each retinal output channel to visual perception and visually driven innate behaviors, much in the same way that genetic studies of Drosophila photoreceptor, lamina, and medulla neurons are leading to a rich understanding of their contributions to vision (see above). On the one hand, experimental demonstrations of the contribution of retinal output channels to visual perception are profoundly lacking for mice, which, perhaps, is not surprising, since most of the molecular markers for RGC subtypes were only discovered within the last 5 years. However, a handful of such experiments have been carried out and demonstrated that the approach of genetic identification of RGC subtypes can lead to successful causal linking of cell type and function and whole-animal behavior in not just flies but also mice: Hattar et al. (2002, 2003) have shown that type 1 ipRGC subtypes, the M1s, target the hypothalamic SCN and the “shell” of the olivary pretectal nucleus (OPN), suggesting that they control photic resetting of the circadian clock and pupil reflexes, respectively. M2s project to the OPN “core” and, because the OPN shell neurons are the crucial output cells for pupil reflexes, very likely contribute indirectly to pupil reflexes. Genetic knockout of the melanopsin photopigment or targeted ablation of ipRGCs by genetic approaches (Goz et al. 2008; Güler et al. 2008; Hatori et al. 2008) indeed linked M1 and M2 ipRGCs to clock entrainment or pupil reflexes. This represents an important milestone for the field of visual neuroscience, as it is the first instance in which the gene signature (OPN4) of a specific mammalian RGC type (ipRGCs) was used to link a receptive field property (intrinsic photosensitivity), a morphology (wide-field, densely overlapping dendrites), and central axon targeting (targeting the SCN and OPN) to a discrete set of measurable behaviors (circadian locomotor activity and pupil constriction). Hattar and coworkers (Chen et al. 2011) then went on to explore the expression of specific transcription factors in different classes of M1 RGCs and discovered that the M1 ipRGCs that target the OPN express Brn3b. That unique signature enabled them (using Brn3b-Cre mice) to kill only the M1s that target the OPN “shell” region and thereby abolish pupil constriction without tampering with circadian entrainment/SCN innervations (Fig. 5A,B; Chen et al. 2011). This stands as the only published example to date of causal link between a single defined RGC subtype, a specific central target, and a visually driven behavior or reflex.
Figure 5.
Genetic insights into cell type-specific circuits for controlling different aspects of visual circuit function in mice. (A) The OPN core and shell subregions are innervated by different sets of RGCs, including ipRGCs. Which RGCs and OPN subregions drive the pupil reflex was made clear from the discovery that Brn3b-expressing M1 ipRGCs project to the OPN shell but not the core. Expressing diphtheria toxin (DTA) to kill M1 ipRGCs cells expressing Brn3b leads to the loss of pupil constriction in the presence of light, even when the OPN core is spared (see Chen et al. 2011 for details). (B) Two parallel disynaptic circuits for carrying directional and nondirectional motion in the mouse visual system. Genetic labeling of ON–OFF DSGCs revealed that they project to the dLGN shell or direction-selective recipient zone (DS-RZ; green) (Huberman et al. 2009; Kay et al. 2011b; Krahe et al. 2011; Rivlin-Etzion et al. 2011); rabies virus transsynaptic mapping from the cortex to the retina shows that the dLGN neurons that receive ON–OFF DSGC input in turn project specifically to superficial layers of the primary visual cortex (V1) to deliver direction-selective (and orientation-selective) information to cells/processes that reside in superficial V1 layers. In contrast, non-direction-tuned RGCs (blue) project to the dLGN “core” (Huberman et al. 2008, 2009). Neurons in the core project to deeper layers of V1, thereby establishing a parallel circuit (see Cruz-Martín et al. 2014 for details).
What about the functions of the other 19 or so RGC subtypes? What do they contribute to visual perception and visually mediated behaviors? Aside from pupil reflexes, mice exhibit reflexive eye movements (e.g., see Dhande et al. 2013; http://www.hubermanlab.com/movies.html), innate freezing, or escape responses to looming stimuli (Yilmaz and Meister 2013) and can be trained to discriminate bars of different contrast levels or orientations (Busse et al. 2011; Glickfield et al. 2013). Which RGCs serve these behaviors? For some RGCs that project exclusively to a defined subset of central brain targets whose structure and function are clear, one can raise meaningful hypotheses about causal links between the afferent RGC types and behaviors. Still, definitive causal links remain to be demonstrated. The best such example is slow-velocity-tuned ON and ON–OFF direction-selective ganglion cells, which have now been tagged with molecular markers (Yonehara et al. 2009; Dhande et al. 2013). The slow-tuned DSGCs send axons specifically to the brainstem nuclei controlling horizontal and vertical image stabilization and not to other central targets (Simpson 1984; Dhande et al. 2013; Dhande and Huberman 2014), making it almost certain that slow-tuned DSGCs are the retinal neurons crucial for compensatory eye movements leading to image stabilization. However, establishing this will require selective killing or, ideally, reversible silencing/activation of these RGCs, an experiment that ought to be possible once the transcriptome of these cells is known and proper Cre-based drivers are generated. Selective immunotoxin-based ablation of SACs, which eliminates direction-tuned responses from all DSGCs (slow-tuned and fast-velocity-tuned) en masse, causes severe defects in image-stabilizing eye movements in response to full-field motion (Yoshida et al. 2001).
The functional relevance of the remaining RGC subtypes, including the heavily studied fast-tuned DSGCs and αRGCs (the latter comprising the presumed homologs to parasol cells in the primate retina), is unknown. Solid hypotheses about their function for vision will likely arrive from better understanding of their central targets and what they do. For example, transsynaptic viral circuit mapping of fast-tuned ON–OFF DSGCs indicate that these RGCs are disynaptically wired to superficial layers of the primary visual cortex (V1) through a specialized compartment of the dorsal lateral geniculate nucleus (dLGN) called the shell (Krahe et al. 2011), whereas non-direction-tuned RGCs are wired into deeper V1 through the dLGN core (Fig. 5B; Cruz-Martín et al. 2014). Still, the functional relevance of all of this to visual processing circuits in mouse V1 and perception remains rather opaque. Perceptual and visual behavioral assays that clearly require activation of V1 cells and long-range circuits are needed. Recently, high-throughput tests of pattern or contrast discrimination were created for mice (for orientation, see Glickfield et al. 2013; http://www.hubermanlab.com/movies.html; for contrast, see Busse et al. 2011; Glickfield et al. 2013), and, in one published instance, a causal role for the visual cortex was established for these behaviors (Glickfield et al. 2013). Thus, because they feed the geniculo–V1 pathway, fast-tuned DSGCs as well as other non-direction-tuned RGCs must contribute to perception of orientation and/or directional motion, but their influence on neurons within different laminar compartments of V1 and their role in vision remain to be tested.
What do the cells do? Functional characterization of cell types in flies and mice
In addition to behavioral dissection and anatomical reconstruction, the characterization of cellular contributions to neural circuit function requires the visualization of neuronal activity, and this can be accomplished by using electrophysiology or genetically encoded calcium sensors. In combination with data collected from anatomic reconstruction of the cells’ synaptic connections as well as their behavioral contribution, their computational role can be deduced.
Visualizing the functional properties of identified Drosophila cell types in vivo
In Drosophila, the most important progress once again came from studies focusing on motion vision. An influential computational model for the perception of visual motion was proposed as early as 1956 (Hassenstein and Reichardt 1956). This model of elementary movement detectors (EMDs) was deduced from experiments with the walking beetle Chlorophanus and resulted in strong mathematical predictions, including time delays and multiplication steps (Fig. 6A), that are currently being investigated using molecular genetic tools (Borst and Euler 2011). A comprehensive wiring diagram for the cellular elements of the EMDs between the input-level neurons L1/L2 and the output cells T4/T5 predicted a bifurcated pathway for both the L1/ON pathway (transmitted by medulla cells Mi1 and Tm3) and the L2/OFF pathway, predicted to take a different path across the medulla neuropil via Tm1 and Tm2 cells (Fig. 6B; Takemura et al. 2013). Three recent studies have now characterized these candidate cell types.
Figure 6.
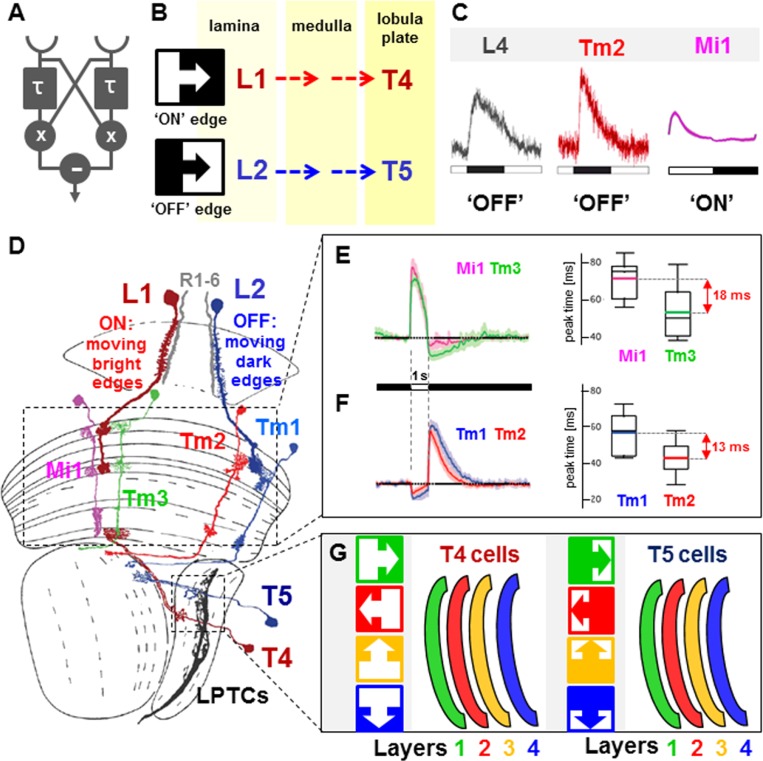
Functional characterization of Drosophila optic lobe neurons. (A) Summary of the computational model for EMDs receiving input from two points in space (semicircles) first proposed by Hassenstein and Reichardt (1956). (τ) Time constant of a delay channel; (×) multiplication; (−) subtraction. (Adapted from Borst and Euler 2011 with permission from Elsevier). (B) Based on previous work, moving light edges (ON) and dark edges (OFF) are detected by lamina monopolar cells L1 and L2, respectively. The information is then transmitted to the lobula plate, where T4 and T5 cells relay ON and OFF signals onto direction-selective output cells. (C) Characterization of two cell types in the lamina (L4) and medulla neuropil (Tm2) using genetically encoded calcium sensor GCaMP5 and two-photon microscopy. Both cell types are activated specifically by OFF edges, thereby placing them in the OFF pathway between L2 and T5 (adapted from Meier et al. 2014 with permission from Elsevier; adapted from Strother et al. 2014 with permission from Elsevier). (D) Based on large-scale anatomical reconstruction of optic lobe tissue using serial EM reconstruction, the cellular substrates for the ON/OFF pathways leading from the retina to the movement-sensitive LPTCs were proposed. One arm of the pathway is specialized for moving bright edges (R1–6 → L1 → Mi1/Tm3 → T4), and the other one is specialized for moving bright edges (R1–6 → L2 → Tm1/Tm2 → T5). (E) Electrophysiological characterization of the Mi1/Tm3 ON arm of the elementary motion detector. Both cells are hyperpolarized by light increments, and a significant time delay (18 msec) exists between the two cell types (adapted from Behnia et al. 2014). (F) In contrast, Tm1/Tm2 cells of the OFF arm are activated by light decrements and also manifest a significant delay (13 msec) (adapted from Behnia et al. 2014). (G) Functional characterization of direction-selective cells in the lobula plate using calcium imaging. Dendrites of T4 cells are excited maximally by moving bright edges, while T5 dendrites respond to moving dark edges. Dendrites responding to different cardinal directions locate to different layers. (Green) right; (red) left; (yellow) up; (blue) down. (Adapted by permission from Macmillan Publishers Ltd. from Maisak et al. 2013).
Using genetically encoded calcium sensors in combination with two-photon microscopy, one study showed that transmedullary neuron Tm2 and lamina monopolar cell L4 both increase their activity in response to dark edges, hence placing them into the L2 → T5 OFF pathway (Meier et al. 2014). Another study using a similar approach places Tm1 within the same OFF pathway, while medulla-intrinsic neuron Mi1 appears to fall into the L1 → T4 ON pathway (Fig. 6C; Strother et al. 2014). It should be pointed out that in the lamina (the periphery of the motion vision circuit), monopolar cells L2, L3, and L4 fall into the OFF pathway, whereas ON edges are transmitted via L1 alone. The reason for these differences in complexity are currently not known.
All four medulla cell types (Mi1/Tm3 and Tm1/Tm2) have now been characterized electrophysiologically (Behnia et al. 2014). Indeed, Mi1/Tm3 are depolarized by light increments (ON), placing them in the L1 pathway. In contrast, Tm1/Tm2 fall into the L2 OFF pathway, since they respond more specifically to light decrements (Fig. 6D–F), in good agreement with calcium imaging results done with Tm2 (Meier et al. 2014). Interestingly, a significant delay was measured between the responses of cells in both the ON and OFF pathways (18 msec between the delayed response of Mi1 and the direct response of Tm3; 14 msec for Tm1 vs. Tm2). In agreement with the model first formulated by Hassenstein and Reichardt (1956), modeling the response properties of these cells demonstrated that the measured delays could be sufficiently large to account for the optimal temporal frequency, which represents the speeds the EMD can detect (Fig. 6E,F; Behnia et al. 2014). However, the involvement of additional cell types cannot be excluded.
The second important prediction from the Drosophila connectome is that the two anatomical ON/OFF pathways connect to dendrites of LPTCs via T4 cells (L1 → Mi1/Tm3 → T4) and T5 cells (L2 → Tm1/Tm2 → T5), both of which fall into four different morphological classes (T4a,b,c,d and T5a,b,c,d, respectively) and have stratified projections into four distinct lobula plate layers. The functional relevance of these cells has been shown using in vivo calcium imaging (Maisak et al. 2013): Each of the four layers is sensitive to either brightness increments (T4) or brightness decrements (T5) moving in one of the four cardinal directions: front to back (layer 1), back to front (layer 2), up (layer 3), or down (layer 4) (Fig. 6G). Hence, information about moving light edges and dark edges is computed in separate pathways and converges into the same layer in the lobula plate, where the direction-selective LPTCs are located.
Taken together, these impressive studies combining anatomy and physiology revealed the implementation at the cellular level of a computational model describing motion detection. Although some functional gaps still need to be filled in this diagram and more pathways involved in more subtle aspects of motion detection will be discovered, the current results are in good agreement with previous morphological and physiological studies that had predicted some of these findings (Buchner et al. 1984; Bausenwein and Fischbach 1992; Douglass and Strausfeld 2003; Fischbach and Hiesinger 2008).
Defining the precise wiring diagram underlying specific aspects of retinal processing
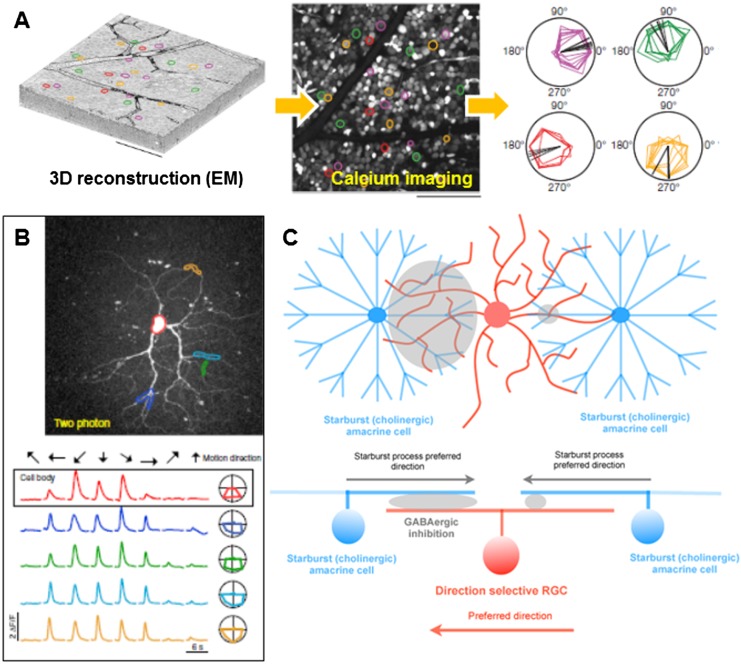
Just as in flies, it is essential to define the exact cell types and architectures of synaptic connections that give rise to circuits in the mouse retina that encode specific features in the visual scene. While it is fairly easy to explain how a cone or rod or an M1 ipRGC achieves its intrinsic photosensitivity (expression of specific photopigment), defining the cell–cell connections that drive the ability to detect more elaborate features detected by retinal cells is a more formidable challenge. In this context, DSGCs offer a powerful example of how electrophysiology and a growing number of circuit-mapping techniques, including EM, are combining to yield great depth of knowledge for how these cells achieve their unique tuning properties. In a heroic set of experiments, Briggman et al. (2011) combined calcium imaging of DSGC responses followed by complete serial EM reconstruction of the recorded and surrounding cells to confirm the asymmetric wiring model for direction tuning; they showed at the ultrastructural level that, while different starburst amacrine neurons may contact various sides of the dendritic arbors of DSGCs, only starburst cells located on the null side of the DSGC make bone fide synaptic connections with it (Fig. 7A). Other models for direction tuning of retinal neurons have been put forth on the basis of serial EM reconstruction, including, very recently, a “space–time” model that hinges on the observation that the sloping of starburst dendrites allows the cell to capture different sets of bipolar cell inputs along its arbor and thereby become centrifugally tuned (Kim et al. 2014). However, that model still requires experimental validation. What is now very clear from recent work using both electrophysiology and calcium imaging is that bipolar inputs to DSGCs are not tuned for direction (Fig. 7B). Dual-patch clamp recordings from DSGCs and SACs (Fried et al. 2002; Wei et al. 2011) revealed a strong asymmetric wiring onto one side of the dendritic arbor of the direction-selective ganglion cell. Combined with the centrifugal tuning of SACs (Euler et al. 2002), this asymmetric wiring should, in theory, be sufficient to confer direction selectivity on the RGC (Fig. 7C; Demb 2007; Wei and Feller 2011; Vaney et al. 2012). Rather, asymmetric wiring from SACs alone is sufficient to generate direction-selective responses in both ON DSGCs and ON–OFF direction-selective ganglion cells (Yonehara et al. 2013; Park et al. 2014). It should be noted, however, that other wiring schemes not involving SACs but that instead rely on RGC dendritic arbor shape can also create direction selectivity (Kim et al. 2008).
Figure 7.
Mammalian retinal direction selectivity circuit. (A) Summary of high-resolution EM-based 3D reconstruction of synaptic connections in the mouse retina (adapted by permission from Macmillan Publishers Ltd. from Briggman et al. 2011). When combined with two-photon-based visualization of neuronal activity using genetically encoded calcium sensors, the direction-selective signals can be correlated with the morphological data. (B) Demonstration that dendritic segments of ON DSGCs are direction-selective (adapted from Yonehara et al. 2013 with permission from Elsevier). (Top) Two-photon image of the recorded neuron (targeted dendritic segments are shown in different colors). (Bottom) Calcium transients recorded from the above labeled cellular segments identify direction-selective signals in both the cell body and the dendrites. (C) SACs (blue) are tuned for centrifugal (soma > tip; “radial”) motion along their processes and provide GABAergic inhibitory synapses onto the dendrites of the RGCs that costratify at the same depth of the inner plexiform layer (e.g., the RGCs are shown in red). The inhibition from SACs is asymmetric onto the RGC dendrites; it is much greater on one side of the RGC’s dendritic arbor than the other side, which establishes a “null” response to visual stimuli moving in one direction (here the null direction of the RGC is rightward motion), but because inhibition is minimal from SACs on the other side of the RGC’s dendritic arbor, the excitatory response of the RGC is preserved when stimuli move in the direction opposite the null direction (corresponding here to leftward motion; red arrow), which renders the RGC direction selective. Adapted by permission from Macmillan Publishers Ltd. from Wei et al. (2011) and by permission from Macmillan Publishers Ltd. from Briggman et al. (2011). The excitatory drive to RGCs arises from bipolar cell inputs (not shown here) that are not direction-tuned (Yonehara et al. 2013). The same tuning circuit applies to both monostratified ON DSGCs (as shown here) and ON–OFF DSGCs, with the same pattern repeated in the OFF sublayer between OFF SACs (Park et al. 2014).
The organization of the motion detection pathways in the mouse retina is reminiscent of the Drosophila motion-sensing circuits. DSGCs resemble the LPTCs in the fly lobula plate that receive inputs from T4 and T5; the organization of the circuits with an ON pathway and an OFF pathway is also similar in both species, with fly L1-Mi1/Tm3-T4 and L2-Tm1/Tm2-T5 representing circuits analogous to the ON and OFF RGCs. Although it is very likely that these two visual systems do not share a common ancestry, the same computational problems are implemented by similar processes through convergent evolution. This makes the detailed study of Drosophila motion detection and the definition of the cellular implementation of the EMD highly relevant to the system in mammals
Concluding remarks
The progress reviewed here reveals how recent technological innovations in the fields of developmental genetics, anatomy, and functional studies have paved the way toward the dissection of neural circuitry at the cellular level. In both flies and mice, most progress has been made in the field of motion vision, where the cellular basis of the computational model by Hassenstein and Reichardt (1956) is taking shape. One immediate challenge is to extend this high-resolution circuit analysis toward different visual behaviors, like color vision.
Furthermore, important challenges remain. For instance, the complex problem of how correct synaptic connections between identified circuit elements are established, maintained, and regulated remains unsolved. Another major challenge lies in putting together the pieces of the puzzle by linking the developmental specification of cell types to the connectome as well as their functional role in the behaving animal. For instance, the Drosophila medulla cell type Mi1 discussed here serves as an exciting first demonstration that this strategy is successful: A complete picture of this neuron now exists, from the transcription factor code necessary for its initial specification to its adult morphology and the role it plays in the perception of motion. In mice, the Brn3b expression of OPN shell-projecting ipRGCs is the best link between gene, cell type, circuit, and behavior. As yet, however, whether Brn3b is crucial for the specification of these cells is unknown. The systematic extension of these studies to the entire repertoire of cell types in the visual system will ultimately provide a complete description of how visual circuitry informs visual perception on a cellular level.
One additional opportunity for future progress lies in the transcriptional profiling of identified circuit elements throughout the brain. Using cell type-specific molecular genetic tools, each cell type can be targeted, and its transcriptome can be solved (Hobert et al. 2010; Malone and Hobert 2011). In combination with existing knowledge about the development, function, and anatomy of these cell types, these transcriptomes will induce a new era of cell type-specific molecular manipulation. The molecular signature that defines neuronal identity will also provide a new testable hypothesis about gene networks and signaling pathways regulating the formation of neural circuits. Furthermore, in a synthesis with the progress reviewed above, transcriptomics will lead toward an even more complete description of visual circuitry, bridging the wide gap from the role that single molecules play in defining the shape of a neuron all the way to the functional role that this neuron and its neighbors play in the behaving animal.
Acknowledgments
We apologize for any work not cited due to constraints regarding manuscript length. We thank Xin Li for sharing unpublished material. Furthermore, we thank Onkar Dhande and Xin Li as well as three anonymous reviewers for helpful suggestions and comments on the text.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.248245.114.
References
- Applebury ML, Antoch MP, Baxter LC, Chun LL, Falk JD, Farhangfar F, Kage K, Krystolik MG, Lyass LA, Robbins JT. 2000. The murine cone photoreceptor: a single cone type expresses both S and M opsins with retinal spatial patterning. Neuron 27: 513–523. [DOI] [PubMed] [Google Scholar]
- Badea TC, Cahill H, Hattar S, Nathans J. 2009. Distinct roles of transcription factors brn3a and brn3b in controlling the development, morphology, and function of retinal ganglion cells. Neuron 61: 852–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baden T, Schubert T, Chang L, Wei T, Zaichuk M, Wissinger B, Euler T. 2013. A tale of two retinal domains: near-optimal sampling of achromatic contrasts in natural scenes through asymmetric photoreceptor distribution. Neuron 80: 1206–1217. [DOI] [PubMed] [Google Scholar]
- Baker M. 2013. Neuroscience: through the eyes of a mouse. Nature 502: 156–158. [DOI] [PubMed] [Google Scholar]
- Bausenwein B, Fischbach KF. 1992. Activity labeling patterns in the medulla of Drosophila melanogaster caused by motion stimuli. Cell Tissue Res 270: 25–35. [DOI] [PubMed] [Google Scholar]
- Bausenwein B, Dittrich AP, Fischbach KF. 1992. The optic lobe of Drosophila melanogaster. II. Sorting of retinotopic pathways in the medulla. Cell Tissue Res 267: 17–28. [DOI] [PubMed] [Google Scholar]
- Bayraktar OA, Doe CQ. 2013. Combinatorial temporal patterning in progenitors expands neural diversity. Nature 498: 449–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behnia R, Clark DA, Carter AG, Clandinin TR, Desplan C. 2014. Processing properties of ON and OFF pathways for Drosophila motion detection. Nature 512: 427–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berson DM, Dunn FA, Takao M. 2002. Phototransduction by retinal ganglion cells that set the circadian clock. Science 295: 1070–1073. [DOI] [PubMed] [Google Scholar]
- Bertet C, Li X, Erclik T, Cavey M, Wells B, Desplan C. 2014. Temporal patterning of neuroblasts controls notch-mediated cell survival through regulation of Hid or Reaper. Cell 158: 1173–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borst A. 2014. Neural circuits for elementary motion detection. J Neurogenet 10: 1–13. [DOI] [PubMed] [Google Scholar]
- Borst A, Euler T. 2011. Seeing things in motion: models, circuits, and mechanisms. Neuron 71: 974–994. [DOI] [PubMed] [Google Scholar]
- Briggman KL, Helmstaedter M, Denk W. 2011. Wiring specificity in the direction-selectivity circuit of the retina. Nature 471: 183–188. [DOI] [PubMed] [Google Scholar]
- Buchner E, Buchner S, Bülthoff I. 1984. Deoxyglucose mapping of nervous activity induced in Drosophila brain by visual movement. J Comp Physiol A Neuroethol Sens Neural Behav Physiol 155: 471–483. [Google Scholar]
- Busse L, Ayaz A, Dhruv NT, Katzner S, Saleem AB, Schölvinck ML, Zaharia AD, Carandini M. 2011. The detection of visual contrast in the behaving mouse. J Neurosci 31: 11351–11361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos-Ortega A, Hofbauer A. 1990. Proliferation pattern and early differentiation of the optic lobes in Drosophila melanogaster. Rouxs Arch Dev Biol 198: 264–274. [DOI] [PubMed] [Google Scholar]
- Cepko C. 2014. Intrinsically different retinal progenitor cells produce specific types of progeny. Nat Rev Neurosci 15: 615–627. [DOI] [PubMed] [Google Scholar]
- Chalupa LM, Werner JS. 2004The visual neurosciences, volumes 1 and 2. MIT Press, Cambridge. [Google Scholar]
- Chen SK, Badea TC, Hattar S. 2011. Photoentrainment and pupillary light reflex are mediated by distinct populations of ipRGCs. Nature 476: 92–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou WH, Hall KJ, Wilson DB, Wideman CL, Townson SM, Chadwell LV, Britt SG. 1996. Identification of a novel Drosophila opsin reveals specific patterning of the R7 and R8 photoreceptor cells. Neuron 17: 1101–1115. [DOI] [PubMed] [Google Scholar]
- Chou WH, Huber A, Bentrop J, Schulz S, Schwab K, Chadwell LV, Paulsen R, Britt SG. 1999. Patterning of the R7 and R8 photoreceptor cells of Drosophila: evidence for induced and default cell-fate specification. Development 126: 607–616. [DOI] [PubMed] [Google Scholar]
- Clark DA, Bursztyn L, Horowitz MA, Schnitzer MJ, Clandinin TR. 2011. Defining the computational structure of the motion detector in Drosophila. Neuron 70: 1165–1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colonques J, Ceron J, Tejedor FJ. 2007. Segregation of postembryonic neuronal and glial lineages inferred from a mosaic analysis of the Drosophila larval brain. Mech Dev 124: 327–340. [DOI] [PubMed] [Google Scholar]
- Cruz-Martín A, El-Danaf RN, Osakada F, Sriram B, Dhande OS, Nguyen PL, Callaway EM, Ghosh A, Huberman AD. 2014. A dedicated circuit links direction-selective retinal ganglion cells to the primary visual cortex. Nature 507: 358–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delogu A, Sellers K, Zagoraiou L, Bacianowska-Zbrog A, Mandal S, Guimera J, Rubenstein JL, Sugden D, Jessell TM, Lumsden A. 2012. Subcortical visual shell nuclei targeted by ipRGCs develop from a Sox14-GABAergic progenitor and require Sox14 to regulate daily activity rhythms. Neuron 75: 648–662. [DOI] [PubMed] [Google Scholar]
- Demb JB. 2007. Cellular mechanisms for direction selectivity in the retina. Neuron 55: 179–186. [DOI] [PubMed] [Google Scholar]
- de Vries SE, Clandinin TR. 2012. Loom-sensitive neurons link computation to action in the Drosophila visual system. Curr Biol 22: 353–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhande OS, Huberman AD. 2014. Retinal ganglion cell maps in the brain: implications for visual processing. Curr Opin Neurobiol 24: 133–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhande OS, Estevez ME, Quattrochi LE, El-Danaf RN, Nguyen PL, Berson DM, Huberman AD. 2013. Genetic dissection of retinal inputs to brainstem nuclei controlling image stabilization. J Neurosci 33: 17797–17813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglass JK, Strausfeld NJ. 2000. Optic flow representation in the optic lobes of Diptera: modeling the role of T5 directional tuning properties. J Comp Physiol A Neuroethol Sens Neural Behav Physiol 186: 783–797. [DOI] [PubMed] [Google Scholar]
- Douglass JK, Strausfeld NJ. 2003. Anatomical organization of retinotopic motion-sensitive pathways in the optic lobes of flies. Microsc Res Tech 62: 132–150. [DOI] [PubMed] [Google Scholar]
- Duan X, Krishnaswamy A, De la Huerta I, Sanes JR. 2014. Type II cadherins guide assembly of a direction-selective retinal circuit. Cell 158: 793–807. [DOI] [PubMed] [Google Scholar]
- Dyer MA, Livesey FJ, Cepko CL, Oliver G. 2003. Prox1 function controls progenitor cell proliferation and horizontal cell genesis in the mammalian retina. Nat Genet 34: 53–58. [DOI] [PubMed] [Google Scholar]
- Ecker JL, Dumitrescu ON, Wong KY, Alam NM, Chen SK, LeGates T, Renna JM, Prusky GT, Berson DM, Hattar S. 2010. Melanopsin-expressing retinal ganglion-cell photoreceptors: cellular diversity and role in pattern vision. Neuron 67: 49–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egger B, Gold KS, Brand AH. 2011. Regulating the balance between symmetric and asymmetric stem cell division in the developing brain. Fly (Austin) 5: 237–241. [DOI] [PubMed] [Google Scholar]
- Eichner H, Joesch M, Schnell B, Reiff DF, Borst A. 2011. Internal structure of the fly elementary motion detector. Neuron 70: 1155–1164. [DOI] [PubMed] [Google Scholar]
- Estevez ME, Fogerson PM, Ilardi MC, Borghuis BG, Chan E, Weng S, Auferkorte ON, Demb JB, Berson DM. 2012. Form and function of the M4 cell, an intrinsically photosensitive retinal ganglion cell type contributing to geniculocortical vision. J Neurosci 32: 13608–13620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Euler T, Detwiler PB, Denk W. 2002. Directionally selective calcium signals in dendrites of starburst amacrine cells. Nature 418: 845–852. [DOI] [PubMed] [Google Scholar]
- Feng L, Xie X, Joshi PS, Yang Z, Shibasaki K, Chow RL, Gan L. 2006. Requirement for Bhlhb5 in the specification of amacrine and cone bipolar subtypes in the mouse retina. Development 133: 4815–4825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field GD, Chichilnisky EJ. 2007. Information processing in the retina: circuitry and coding. Annu Rev Neurosci 30: 1–30. [DOI] [PubMed] [Google Scholar]
- Field GD, Gauthier JL, Sher A, Greschner M, Machado TA, Jepson LH, Shlens J, Gunning DE, Matieson K, Dabrowski W, et al. 2010. Functional connectivity in the retina at the resolution of photoreceptors. Nature 467: 673–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischbach KF, Dittrich APM. 1989. The optic lobe of Drosophila melanogaster. I. A Golgi analysis of wild-type structure. Cell Tissue Res 258: 441–475. [Google Scholar]
- Fischbach KF, Hiesinger PR. 2008. Optic lobe development. Adv Exp Med Biol 628: 115–136. [DOI] [PubMed] [Google Scholar]
- Franceschini N, Kirschfeld K, Minke B. 1981. Fluorescence of photoreceptor cells observed in vivo. Science 213: 1264–1267. [DOI] [PubMed] [Google Scholar]
- Freifeld L, Clark DA, Schnitzer MJ, Horowitz MA, Clandinin TR. 2013. GABAergic lateral interactions tune the early stages of visual processing in Drosophila. Neuron 78: 1075–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fried SI, Münch TA, Werblin FS. 2002. Mechanisms and circuitry underlying directional selectivity in the retina. Nature 420: 411–414. [DOI] [PubMed] [Google Scholar]
- Fuerst PG, Koizumi A, Masland RH, Burgess RW. 2008. Neurite arborization and mosaic spacing in the mouse retina require DSCAM. Nature 451: 470–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuerst PG, Bruce F, Tian M, Wei W, Elstrott J, Feller MB, Erskine L, Singer JH, Burgess RW. 2009. DSCAM and DSCAML1 function in self-avoidance in multiple cell types in the developing mouse retina. Neuron 64: 484–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujitani Y, Fujitani S, Luo H, Burlison J, Long Q, Kawaguchi Y, Edlund H, MacDonal RJ, Furukawa T, Fujikado T, et al. 2006. Ptf1a determines horizontal and amacrine cell fates during mouse retinal development. Development 133: 4439–4450. [DOI] [PubMed] [Google Scholar]
- Gao S, Takemura SY, Ting CY, Huang S, Lu Z, Luan H, Rister J, Thum AS, Yang M, Hong ST, et al. 2008. The neural substrate of spectral preference in Drosophila. Neuron 60: 328–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glickfield LL, Histed MH, Maunsell JH. 2013. Mouse primary visual cortex is used to detect both orientation and contrast changes. J Neurosci 33: 19416–19422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon JA, Stryker MP. 1996. Experience-dependent plasticity of binocular responses in the primary visual cortex of the mouse. J Neurosci 16: 3274–3286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gohl DM, Silies MA, Gao XJ, Bhalerao S, Luongo FJ, Lin CC, Potter CJ, Clandinin TR. 2011. A versatile in vivo system for directed dissection of gene expression patterns. Nat Methods 8: 231–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goz D, Studholme K, Lappi DA, Rollag MD, Provencio I, Morin LP. 2008. Targeted destruction of photosensitive retinal ganglion cells with a saporin conjugate alters the effects of light on mouse circadian rhythms. PLoS ONE 3: e3153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Güler AD, Ecker JL, Lall GS, Haq S, Altimus CM, Liao HW, Barnard AR, Cahill H, Badea TC, Zhao H, et al. 2008. Melanopsin cells are the principal conduits for rod-cone input to non-image-forming vision. Nature 453: 102–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haikala V, Joesch M, Borst A, Mauss AS. 2013. Optogenetic control of fly optomotor responses. J Neurosci 33: 13927–13934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanesch U, Fischbach KF, Heisenberg M. 1989. Neuronal architecture of the central complex in Drosophila melanogaster. Cell Tissue Res 257: 343–366. [Google Scholar]
- Hardie RC. 1985. Functional organization of the fly retina. In Progress in sensory physiology, (ed. H. Autrum, et al.), pp 1–79. Springer, Berlin. [Google Scholar]
- Hasegawa E, Kitada Y, Kaido M, Takayama R, Awasaki T, Tabata T, Sato M. 2011. Concentric zones, cell migration and neuronal circuits in the Drosophila visual center. Development 138: 983–993. [DOI] [PubMed] [Google Scholar]
- Hasegawa E, Kaido M, Takayama R, Sato M. 2013. Brain-specific-homeobox is required for the specification of neuronal types in the Drosophila optic lobe. Dev Biol 377: 90–99. [DOI] [PubMed] [Google Scholar]
- Hassenstein B, Reichardt W. 1956. Systemtheoretische analyse der zeit, reihenfolgen und vorzeichenauswertung bei der bewegungsperzeption des rüsselkiifers Chlorophanus. Z Naturforsch IIb: 513–524. [Google Scholar]
- Hatori M, Le H, Vollmers C, Keding SR, Tanaka N, Buch T, Waisman A, Schmedt C, Jegla T, Panda S. 2008. Inducible ablation of melanopsin-expressing retinal ganglion cells reveals their central role in non-image forming visual responses. PLoS ONE 3: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattar S, Liao HW, Takao M, Berson DM, Yau KW. 2002. Melanopsin-containing retinal ganglion cells: architecture, projections, and intrinsic photosensitivity. Science 295: 1065–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattar S, Lucas RJ, Mrosovsky N, Thompson S, Douglas RH, Hankins MW, Lem J, Biel M, Hofmann F, Foster RG, et al. 2003. Melanopsin and rod-cone photoreceptive systems account for all major accessory visual functions in mice. Nature 424: 76–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattori D, Millard SS, Wojtowicz WM, Zipursky SL. 2008. Dscam-mediated cell recognition regulates neural circuit formation. Annu Rev Cell Dev Biol 24: 597–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haverkamp S, Wassle H, Duebel J, Kuner T, Augustine GJ, Feng G, Euler T. 2005. The primordial, blue-cone color system of the mouse retina. J Neurosci 25: 5438–5445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heisenberg M, Buchner E. 1977. The role of retinula cell types in visual behavior of Drosophila melanogaster. J Comp Physiol 117: 127–162. [Google Scholar]
- Helmstaedter M, Briggman KL, Turaga SC, Jain V, Seung HS, Denk W. 2013. Connectomic reconstruction of the inner plexiform layer in the mouse retina. Nature 500: 168–174. [DOI] [PubMed] [Google Scholar]
- Hiesinger PR, Zhai RG, Zhou Y, Koh TW, Mehta SQ, Schulze KL, Cao Y, Verstreken P, Clandinin TR, Fischbach KF, et al. 2006. Activity-independent prespecification of synaptic partners in the visual map of Drosophila. Curr Biol 16: 1835–1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobert O, Carrera I, Stefanakis N. 2010. The molecular and gene regulatory signature of a neuron. Trends Neurosci 33: 435–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huberman AD, Niell CM. 2011. What can mice tell us about how vision works? Trends Neurosci 34: 464–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huberman AD, Manu M, Koch SM, Susman MW, Lutz AB, Ullian EM, Baccus SA, Barres BA. 2008. Architecture and activity-mediated refinement of axonal projections from a mosaic of genetically identified retinal ganglion cells. Neuron 59: 425–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huberman AD, Wei W, Elstrott J, Stafford BK, Feller MB, Barres BA. 2009. Genetic identification of an on–off direction-selective retinal ganglion cell subtype reveals a layer-specific subcortical map of posterior motion. Neuron 62: 327–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isshiki T, Pearson B, Holbrook S, Doe CQ. 2001. Drosophila neuroblasts sequentially express transcription factors which specify the temporal identity of their neuronal progeny. Cell 106: 511–521. [DOI] [PubMed] [Google Scholar]
- Jacob J, Maurange C, Gould AP. 2008. Temporal control of neuronal diversity: common regulatory principles in insects and vertebrates? Development 135: 3481–3489. [DOI] [PubMed] [Google Scholar]
- Jagadish S, Barnea G, Clandinin TR, Axel R. 2014. Identifying functional connections of the inner photoreceptors in drosophila using Tango-Trace. Neuron 83: 630–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenett A, Rubin GM, Ngo TT, Shepherd D, Murphy C, Dionne H, Pfeiffer BD, Cavallaro A, Hall D, Jeter J, et al. 2012. A GAL4-driver line resource for Drosophila neurobiology. Cell Reports 2: 991–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joesch M, Schnell B, Raghu SV, Reiff DF, Borst A. 2010. ON and OFF pathways in Drosophila motion vision. Nature 468: 300–304. [DOI] [PubMed] [Google Scholar]
- Joesch M, Weber F, Eichner H, Borst A. 2013. Functional specialization of parallel motion detection circuits in the fly. J Neurosci 33: 902–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston RJ., Jr 2013. Lessons about terminal differentiation from the specification of color-detecting photoreceptors in the Drosophila retina. Ann N Y Acad Sci 1293: 33–44. [DOI] [PubMed] [Google Scholar]
- Johnston RJ Jr, Desplan C. 2010. Stochastic mechanisms of cell fate specification that yield random or robust outcomes. Annu Rev Cell Dev Biol 26: 689–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston RJ, Desplan C. 2014. Interchromosomal communication coordinates an intrinsically stochastic expression descision between alleles. Science 343: 661–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jukam D, Desplan C. 2011. Binary regulation of Hippo pathway by Merlin/NF2, Kibra, Lgl, and Melted specifies and maintains postmitotic neuronal fate. Dev Cell 21: 874–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jukam D, Xie B, Rister J, Terrell D, Charlton-Perkins M, Pistillo D, Gebelein B, Desplan C, Cook T. 2013. Opposite feedbacks in the Hippo pathway for growth control and neural fate. Science 342: 1238016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karuppudurai T, Lin TY, Ting CY, Pursley R, Melnattur KV, Diao F, White BH, Macpherson LJ, Gallio M, Pohida T, et al. 2014. A hard-wired glutamatergic circuit pools and relays UV signals to mediate spectral preference in Drosophila. Neuron 81: 603–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kay JN, Voinescu PE, Chu MW, Sanes JR. 2011a. Neurod6 expression defines new retinal amacrine cell subtypes and regulates their fate. Nat Neurosci 14: 965–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kay JN, De la Huerta I, Kim IJ, Zhang Y, Yamagata M, Chu MW, Meister M, Sanes JR. 2011b. Retinal ganglion cells with distinct directional preferences differ in molecular identity, structure, and central projections. J Neurosci 31: 7753–7762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kay JN, Chu MW, Sanes JR. 2012. MEGF10 and MEGF11 mediate homotypic interactions required for mosaic spacing of retinal neurons. Nature 483: 465–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim IJ, Zhang Y, Yamagata M, Meister M, Sanes JR. 2008. Molecular identification of a retinal cell type that responds to upward motion. Nature 452: 478–482. [DOI] [PubMed] [Google Scholar]
- Kim JS, Greene MJ, Zlateski A, Lee K, Richardson M, Turaga SC, Purcaro M, Balkam M, Robinson A, Behababi BF, et al. 2014. Space-time wiring specificity supports direction selectivity in the retina. Nature 509: 331–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krahe TE, El-Danaf RN, Dilger EK, Henderson SC, Guido W. 2011. Morphologically distinct classes of relay cells exhibit regional preferences in the dorsal lateral geniculate nucleus of the mouse. J Neurosci 31: 17437–17448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kvon EZ, Kazmar T, Stampfel G, Yáñez-Cuna JO, Pagani M, Schernhuber K, Dickson BJ, Stark A. 2014. Genome-scale functional characterization of Drosophila developmental enhancers in vivo. Nature 512: 91–95. [DOI] [PubMed] [Google Scholar]
- Lefebvre JL, Kostadinov D, Chen WV, Maniatis T, Sanes JR. 2012. Protocadherins mediate dendritic self-avoidance in the mammalian nervous system. Nature 488: 517–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lettvin JY, Maturana HR, McCullough WS, Pitts WH. 1968. What the frog’s eye tells the frog’s brain. In The mind: biological approaches to its functions (ed. Corning WM, Balaban M), pp 233–258. Intersciences Publishers, New York. [Google Scholar]
- Li S, Mo Z, Yang X, Price SM, Shen MM, Xiang M. 2004. Foxn4 controls the genesis of amacrine and horizontal cells by retinal progenitors. Neuron 43: 795–807. [DOI] [PubMed] [Google Scholar]
- Li X, Erclik T, Bertet C, Chen Z, Voutev R, Venkatesh S, Morante J, Celik A, Desplan C. 2013a. Temporal patterning of Drosophila medulla neuroblasts controls neural fates. Nature 498: 456–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Chen Z, Desplan C. 2013b. Temporal patterning of neural progenitors in Drosophila. Curr Top Dev Biol 105: 69–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin B, Wang SW, Masland RH. 2004. Retinal ganglion cell type, size, and spacing can be specified independent of homotypic dendritic contacts. Neuron 43: 475–485. [DOI] [PubMed] [Google Scholar]
- Liu G, Seiler H, Wen A, Zars T, Ito K, Wolf R, Heisenberg M, Liu L. 2006. Distinct memory traces for two visual features in the Drosophila brain. Nature 439: 551–556. [DOI] [PubMed] [Google Scholar]
- Livesey FJ, Cepko CL. 2001. Vertebrate neural cell-fate determination: lessons from the retina. Nat Rev Neurosci 2: 109–118. [DOI] [PubMed] [Google Scholar]
- Losick R, Desplan C. 2008. Stochasticity and cell fate. Science 320: 65–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maisak MS, Haag J, Ammer G, Serbe E, Meier M, Leonhardt A, Schilling T, Bahl A, Rubin GM, Nern A, et al. 2013. A directional tuning map of Drosophila elementary motion detectors. Nature 500: 212–216. [DOI] [PubMed] [Google Scholar]
- Malone JH, Hobert O. 2011. Microarrays, deep sequencing and the true measure of the transcriptome. BMC Biol 9: 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masland RH. 2012. The neuronal organization of the retina. Neuron 76: 266–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuoka RL, Nguyen-Ba-Charvet KT, Parray A, Badea TC, Chédotal A, Kolodkin AL. 2011a. Transmembrane semaphorin signalling controls laminar stratification in the mammalian retina. Nature 470: 259–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuoka RL, Chivatakarn O, Badea TC, Samuels IS, Cahill H, Katayama K, Kumar SR, Suto F, Chédotal A, Peachey NS, et al. 2011b. Class 5 transmembrane semaphorins control selective Mammalian retinal lamination and function. Neuron 71: 460–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuoka RL, Jiang Z, Samuels IS, Nguyen-Ba-Charvet KT, Sun LO, Peachy NS, Chedotal A, Yau KW, Kolodkin AL. 2012. Guidance-cue control of horizontal cell morphology, lamination and synapse formation in the mammalian outer retina. J Neurosci 32: 6859–6868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazzoni EO, Celik A, Wernet MF, Vasiliauskas D, Johnston RJ, Cook TA, Pichaud F, Desplan C. 2008. Iroquois complex genes induce co-expression of rhodopsins in Drosophila. PLoS Biol 6: e97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier M, Serbe E, Maisak MS, Haag J, Dickson BJ, Borst A. 2014. Neural circuit components of the Drosophila OFF motion vision pathway. Curr Biol 24: 385–392. [DOI] [PubMed] [Google Scholar]
- Meinertzhagen IA, O’Neil SD. 1991. Synaptic organization of columnar elements in the lamina of the wild type in Drosophila melanogaster. J Comp Neurol 305: 232–263. [DOI] [PubMed] [Google Scholar]
- Meinertzhagen IA, Takemura SY, Lu Z, Huang S, Gao S, Ting CY, Lee CH. 2009. From form to function: the ways to know a neuron. J Neurogenet 23: 68–77. [DOI] [PubMed] [Google Scholar]
- Melnattur KV, Pursley R, Lin TY, Ting CY, Smith PD, Pohida T, Lee CH. 2014. Multiple redundant medulla projection neurons mediate color vision in Drosophila. J Neurogenet 28: 374–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikeladze-Dvali T, Wernet MF, Pistillo D, Mazzoni EO, Teleman AA, Chen YW, Cohen S, Desplan C. 2005. The growth regulators warts/lats and melted interact in a bistable loop to specify opposite fates in Drosophila R8 photoreceptors. Cell 122: 775–787. [DOI] [PubMed] [Google Scholar]
- Morante J, Desplan C. 2008. The color-vision circuit in the medulla of Drosophila. Curr Biol 18: 553–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morante J, Erclik T, Desplan C. 2011. Cell migration in Drosophila optic lobe neurons is controlled by eyeless/Pax6. Development 138: 687–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin LP, Studholme KM. 2014. Retinofugal projections in the mouse. J Comp Neurol 522: 3733–3753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu X, Beremand PD, Zhao S, Pershad R, Sun H, Scarpa A, Liang S, Thomas TL, Klein WH. 2004. Discrete gene sets depend on POU domain transcription factor Brn3b/Brn-3.2/POU4f2 for their expression in the mouse embryonic retina. Development 131: 1197–1210. [DOI] [PubMed] [Google Scholar]
- Nakhai H, Sel S, Favor J, Mendoza-Torres L, Paulsen F, Duncker GI, Schmid RM. 2007. Ptf1a is essential for the differentiation of GABAergic and glycinergic amacrine cells and horizontal cells in the mouse retina. Development 134: 1151–1160. [DOI] [PubMed] [Google Scholar]
- Ofstad TA, Zuker CS, Reiser MB. 2011. Visual place learning in Drosophila melanogaster. Nature 474: 204–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterhout JA, Josten N, Yamada J, Pan F, Wu SW, Nguyen PL, Panagiotakos G, Inoue YU, Egusa SF, Volgyi B, et al. 2011. Cadherin-6 mediates axon-target matching in a non-image-forming visual circuit. Neuron 71: 632–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsuna H, Shinomiya K, Ito K. 2014. Parallel neural pathways in higher visual centers of the Drosophila brain that mediate wavelength-specific behavior. Front Neural Circuits 8: 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panda S, Sato TK, Castrucci AM, Rollag MD, DeGrip WJ, Hogenesch JB, Provencio I, Kay SA. 2002. Melanopsin (Opn4) requirement for normal light-induced circadian phase shifting. Science 298: 2213–2216. [DOI] [PubMed] [Google Scholar]
- Papatsenko D, Sheng G, Desplan C. 1997. A new rhodopsin in R8 photoreceptors of Drosophila: evidence for coordinate expression with Rh3 in R7 cells. Development 124: 1665–1673. [DOI] [PubMed] [Google Scholar]
- Park SJ, Kim IJ, Looger LL, Demb JB, Borghuis BG. 2014. Excitatory synaptic inputs to mouse on–off direction-selective retinal ganglion cells lack direction tuning. J Neurosci 34: 3976–3981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peichle L, Wassle H. 1979. Size, scatter and coverage of ganglion cell receptive field centres in the cat retina. J Physiol 291: 117–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer BD, Jenett A, Hammonds AS, Ngo TT, Misra S, Murphy C, Scully A, Carlson JW, Wan KH, Laverty TR, et al. 2008. Tools for neuroanatomy and neurogenetics in Drosophila. Proc Natl Acad Sci 105: 9715–9720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poché RA, Reese BE. 2006. Retinal horizontal cells: challenging paradigms of neural development and cancer biology. Development 136: 2141–2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reese BE. 2011. Development of the retina and optic pathway. Vision Res 51: 613–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiff DF, Plett J, Mank M, Griesbeck O, Borst A. 2010. Visualizing retinotopic half-wave rectified input to the motion detection circuitry of Drosophila. Nat Neurosci 13: 973–978. [DOI] [PubMed] [Google Scholar]
- Rister J, Desplan C. 2011. The retinal mosaics of opsin expression in invertebrates and vertebrates. Dev Neurobiol 71: 1212–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rister J, Pauls D, Schnell B, Ting CY, Lee CH, Sinakevitch I, Morante J, Strausfeld NJ, Ito K, Heisenberg M. 2007. Dissection of the peripheral motion channel in the visual system of Drosophila melanogaster. Neuron 56: 155–170. [DOI] [PubMed] [Google Scholar]
- Rister J, Desplan C, Vasiliauskas D. 2013. Establishing and maintaining gene expression patterns: insights from sensory receptor patterning. Development 140: 493–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivlin-Etzion M, Zhou K, Wei W, Elstrott J, Nguyen PL, Barres BA, Huberman AD, Feller MB. 2011. Transgenic mice reveal unexpected diversity of on–off direction-selective retinal ganglion cell subtypes and brain structures involved in motion processing. J Neurosci 31: 8760–8769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roska B, Werblin F. 2001. Vertical interactions across ten parallel, stacked representations in the mammalian retina. Nature 410: 583–587. [DOI] [PubMed] [Google Scholar]
- Sanes JR, Zipursky SL. 2010. Design principles of insect and vertebrate visual systems. Neuron 66: 15–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnaitmann C, Garbers C, Wachtler T, Tanimoto H. 2013. Color discrimination with broadband photoreceptors. Curr Biol 23: 2375–2382. [DOI] [PubMed] [Google Scholar]
- Scott EK, Reuter JE, Luo L. 2003. Dendritic development of Drosophila high order visual system neurons is independent of sensory experience. BMC Neurosci 4: 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinomiya K, Karuppudurai T, Lin TY, Lu Z, Lee CH, Meinertzhagen IA. 2014. Candidate neural substrates for off-edge motion detection in Drosophila. Curr Biol 24: 1062–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silies M, Gohl DM, Fisher YE, Freifeld L, Clark DA, Clandinin TR. 2013. Modular use of peripheral input channels tunes motion-detecting circuitry. Neuron 79: 111–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson JI. 1984. The accessory optic system. Annu Rev Neurosci 7: 13–41. [DOI] [PubMed] [Google Scholar]
- Soustelle L, Giangrande A. 2007. Novel gcm-dependent lineages in the postembryonic nervous system of Drosophila melanogaster. Dev Dyn 236: 2101–2108. [DOI] [PubMed] [Google Scholar]
- Strother JA, Nern A, Reiser MB. 2014. Direct observation of ON and OFF pathways in the Drosophila visual system. Curr Biol 24: 976–983. [DOI] [PubMed] [Google Scholar]
- Sun LO, Jiang Z, Rivlin-Etzion M, Hand R, Brady CM, Matsuoka RL, Yau KW, Feller MB, Kolodkin AL. 2013. On and off retinal circuit assembly by divergent molecular mechanisms. Science 342: 1241974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T, Kaido M, Takayama R, Sato M. 2013. A temporal mechanism that produces neuronal diversity in the Drosophila visual center. Dev Biol 380: 12–24. [DOI] [PubMed] [Google Scholar]
- Swaroop A, Kim D, Forrest D. 2010. Transcriptional regulation of photoreceptor development and homeostasis in the mammalian retina. Nat Rev Neurosci 11: 563–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweeney NT, Tierney H, Feldheim DA. 2014. Tbr2 is required to generate a neural mediating the pupillary light reflex. J Neurosci 34: 5447–5453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szél A, Röchlich P, Caffé AR, Juliusson B, Aguirre G, Van Veeen T. 1992. Unique topographic separation of two spectral classes of cones in the mouse retina. J Comp Neurol 325: 327–342. [DOI] [PubMed] [Google Scholar]
- Takemura SY, Lu Z, Meinertzhagen IA. 2008. Synaptic circuits of the Drosophila optic lobe: the input terminals to the medulla. J Comp Neurol 509: 493–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takemura SY, Bharioke A, Lu Z, Nern A, Vitaladevuni S, Rivlin PK, Katz WT, Olbris DJ, Plaza SM, Winston P, et al. 2013. A visual motion detection circuit suggested by Drosophila connectomics. Nature 500: 175–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thanawala SU, Rister J, Goldberg GW, Zuskov A, Olesnicky EC, Flowers JM, Jukam D, Purugganan MD, Gavis ER, Desplan C, et al. 2013. Regional modulation of a stochastically expressed factor determines photoreceptor subtypes in the Drosophila retina. Dev Cell 25: 93–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ting CY, McQueen PG, Pandya N, Lin TY, Yang M, Reddy OV, O’Connor MB, McAuliffe M, Lee CH. 2014. Photoreceptor-derived activin promotes dendritic termination and restricts the receptive fields of first-order interneurons in Drosophila. Neuron 81: 830–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomlinson A. 2003. Patterning the peripheral retina of the fly: decoding a gradient. Dev Cell 5: 799–809. [DOI] [PubMed] [Google Scholar]
- Tuthill JC, Nern A, Holtz SL, Rubin GM, Reiser MB. 2013. Contributions of the 12 neuron classes in the fly lamina to motion vision. Neuron 79: 128–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuthill JC, Nern A, Rubin GM, Reiser MB. 2014. Wide-field feedback neurons dynamically tune early visual processing. Neuron 82: 887–895. [DOI] [PubMed] [Google Scholar]
- Vaney DI, Sivyer B, Taylor WR. 2012. Direction selectivity in the rat retina: symmetry and asymmetry in structure and function. Nat Rev Neurosci 13: 194–208. [DOI] [PubMed] [Google Scholar]
- Velez MM, Gohl D, Clandinin TR, Wernet MF. 2014. Differences in neural circuitry guiding behavioral responses to polarized light presented to either the dorsal or ventral retina in Drosophila. J Neurogenet 8: 1–13. [DOI] [PubMed] [Google Scholar]
- Voinescu PE, Kay JN, Sanes JR. 2009. Birthdays of retinal amacrine cell subtypes are systematically related to their molecular identity and soma position. J Comp Neurol 517: 737–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wardill TJ, List O, Li X, Dongre S, McCulloch M, Ting CY, O’Kane CJ, Tang S, Lee CH, Hardie RC, et al. 2012. Multiple spectral inputs improve motion discrimination in the Drosophila visual system. Science 336: 925–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wehner R. 2001. Polarization vision—a uniform sensory capacity? J Exp Biol 204: 2589–2596. [DOI] [PubMed] [Google Scholar]
- Wei W, Feller MB. 2011. Organization and development of direction-selective circuits in the retina. Trends Neurosci 34: 638–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei W, Hamby AM, Zhou K, Feller MB. 2011. Development of asymmetric inhibition underlying direction selectivity in the retina. Nature 469: 402–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weir PT, Dickinson MH. 2012. Flying Drosophila orient to sky polarization. Curr Biol 22: 21–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wernet MF, Labhart T, Baumann F, Mazzoni EO, Pichaud F, Desplan C. 2003. Homothorax switches function of Drosophila photoreceptors from color to polarized light sensors. Cell 115: 267–279. [DOI] [PubMed] [Google Scholar]
- Wernet MF, Mazzoni EO, Celik A, Duncan DM, Duncan I, Desplan C. 2006. Stochastic spineless expression creates the retinal mosaic for colour vision. Nature 440: 174–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wernet MF, Celik A, Mikeladze-Dvali T, Desplan C. 2007. Generation of uniform fly retinas. Curr Biol 17: R1002–R1003. [DOI] [PubMed] [Google Scholar]
- Wernet MF, Velez MM, Clark DA, Baumann-Klausener F, Brown JR, Klovstad M, Labhart T, Clandinin TR. 2012. Genetic dissection reveals two separate retinal substrates for polarization vision in Drosophila. Curr Biol 22: 12–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf R, Gebhardt B, Gademann R, Heisenberg M. 1980. Polarization sensitivity of course control in Drosophila melanogaster. J Comp Physiol 139: 177–191. [Google Scholar]
- Wolff T, Ready DF. 1993. Pattern formation in the Drosophila retina. In The development of Drosophila melanogaster (ed. Bate MMA), pp. 1277–1325. Cold Spring Harbor Laboratory Press, Cold Spring Harbor. [Google Scholar]
- Yamagata M, Sanes JR. 2008. Dscam and Sidekick proteins direct lamina-specific synaptic connections in vertebrate retina. Nature 451: 465–469. [DOI] [PubMed] [Google Scholar]
- Yamagata M, Sanes JR. 2012. Expanding the Ig superfamily code for laminar specificity in retina: expression and role of contactins. J Neurosci 32: 14402–14414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagata M, Weiner JA, Sanes JR. 2002. Sidekicks: synaptic adhesion molecules that promote lamina-specific connectivity in the retina. Cell 110: 649–660. [DOI] [PubMed] [Google Scholar]
- Yamaguchi S, Desplan C, Heisenberg M. 2010. Contribution of photoreceptor subtypes to spectral wavelength preference in Drosophila. Proc Natl Acad Sci 107: 5634–5639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yilmaz M, Meister M. 2013. Rapid innate defensive responses of mice to looming visual stimuli. Curr Biol 23: 2011–2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonehara K, Ishikane H, Sakuta H, Shintani T, Nakamura-Yonehara K, Kamiji NL, Usui S, Noda M. 2009. Identification of retinal ganglion cells and their projections involved in central transmission of information about upward and downward image motion. PLoS ONE 4: e4320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonehara K, Farrow K, Ghanem A, Hillier D, Balint K, Teixeira M, Juttner J, Noda M, Neve RL, Conzelmann KK, et al. 2013. The first stage of cardinal direction selectivity is localized to the dendrites of retinal ganglion cells. Neuron 79: 1078–1085. [DOI] [PubMed] [Google Scholar]
- Yoshida K, Watanabe D, Ishikane H, Tachibana M, Pastan I, Nakanishi S. 2001. A key role of starburst amacrine cells in originating retinal directional selectivity and optokinetic eye movement. Neuron 30: 771–780. [DOI] [PubMed] [Google Scholar]