To investigate the contexts in which the tumor suppressor versus oncogenic activity of miR-26 predominates in vivo, Zeitels et al. generated miR-26a transgenic mice. Despite measurable repression of Pten, elevated miR-26a levels were not associated with malignancy in transgenic animals. miR-26a expression potently suppressed intestinal adenoma formation in Apcmin/+ mice. This study reveals a tumor suppressor role for miR-26 in intestinal cancer that overrides putative oncogenic activity.
Keywords: microRNA, miR-26, intestine, colon cancer
Abstract
Down-regulation of miR-26 family members has been implicated in the pathogenesis of multiple malignancies. In some settings, including glioma, however, miR-26-mediated repression of PTEN promotes tumorigenesis. To investigate the contexts in which the tumor suppressor versus oncogenic activity of miR-26 predominates in vivo, we generated miR-26a transgenic mice. Despite measureable repression of Pten, elevated miR-26a levels were not associated with malignancy in transgenic animals. We documented reduced miR-26 expression in human colorectal cancer and, accordingly, showed that miR-26a expression potently suppressed intestinal adenoma formation in Apcmin/+ mice, a model known to be sensitive to Pten dosage. These studies reveal a tumor suppressor role for miR-26 in intestinal cancer that overrides putative oncogenic activity, highlighting the therapeutic potential of miR-26 delivery to this tumor type.
MicroRNAs (miRNAs) are a class of ∼18- to 23-nucleotide (nt) noncoding RNAs that negatively regulate mRNA stability and translation. Over the last 10 years, an important role for miRNAs in cancer pathogenesis has been uncovered (Di Leva et al. 2014). Studies have revealed nearly ubiquitous dysregulation of miRNA expression in human tumors, and miRNA expression profiles have proven useful for tumor classification. Clear examples of miRNAs that act as oncogenes or tumor suppressors through their ability to regulate key aspects of neoplastic transformation, including proliferation, apoptosis, and metastasis, have been documented.
A potential tumor suppressor role for the miR-26 family, comprising miR-26a and miR-26b, was first suggested by the discovery that expression of these miRNAs impairs MYC-mediated transformation (Chang et al. 2008). Subsequent studies demonstrated down-regulation of miR-26 across multiple tumor types, including lymphoma, hepatocellular carcinoma, breast cancer, and nasopharyngeal carcinoma (Ji et al. 2009; Kota et al. 2009; Lu et al. 2011; Zhang et al. 2011). Ectopic expression of miR-26 inhibits proliferation, induces apoptosis, and/or suppresses tumorigenicity in multiple cancer settings. The tumor suppressor activity of miR-26 is mediated through repression of direct targets that include Cyclin D2 (CCND2), Cyclin E2 (CCNE2), and Enhancer of zeste homolog 2 (EZH2) (Sander et al. 2008; Kota et al. 2009; Lu et al. 2011; Zhang et al. 2011). In other contexts, however, including glioma, lung cancer, and T-cell lymphoblastic leukemia, miR-26 exhibits oncogenic activity through its ability to repress PTEN (Huse et al. 2009; Mavrakis et al. 2011; Liu et al. 2012). Further investigation of the tumor suppressor versus oncogenic activity of miR-26 in vivo is therefore needed to dissect its role in cancer pathogenesis and explore the therapeutic potential and possible adverse consequences of miR-26 delivery.
One cancer setting in which the contribution of miR-26 has remained poorly defined is colorectal cancer (CRC). While previous studies have documented anti-tumorigenic effects of miR-26 expression in CRC cell lines (Ma et al. 2011; Zhang et al. 2013), the significance of these results has remained unclear, since miRNA profiling studies have inconsistently detected miR-26 down-regulation in human colorectal tumors (Schepeler et al. 2008; Schetter et al. 2008; Motoyama et al. 2009; Ma et al. 2011; Gaedcke et al. 2012). The cellular heterogeneity of normal and neoplastic intestinal tissue can confound the detection of abnormally expressed genes within tumor epithelial cells, the relevant transformed cell type. Further analysis of miR-26 expression specifically within CRC-derived epithelial cells is therefore needed to resolve whether miR-26 is frequently down-regulated in this tumor type.
In order to examine the pro- and anti-tumorigenic effects of miR-26 expression in vivo, we generated inducible miR-26a transgenic mice. Despite broad overexpression of miR-26a and a corresponding decrease in Pten expression, transgenic animals developed normally and did not exhibit elevated rates of malignancy. Moreover, we documented reduced miR-26 in human colorectal tumor epithelial cells and showed that enforced miR-26a expression strongly suppressed intestinal adenoma formation in Apcmin/+ mice, a model in which reduced Pten dosage is known to accelerate tumorigenesis (Shao et al. 2007). The tumor suppressor activity of miR-26a in intestinal epithelium is mediated through the repression of anti-proliferative targets that include Ccnd2, high-mobility group AT hook 1 (Hmga1), Myc-binding protein (Mycbp), SET domain containing 8 (Setd8), and Ezh2. These findings demonstrate that the tumor suppressor activity of miR-26 predominates within the intestinal epithelium, supporting the potential therapeutic efficacy of delivery of this miRNA to CRC.
Results and Discussion
A doxycycline (dox)- and Cre-inducible miR-26a transgenic mouse
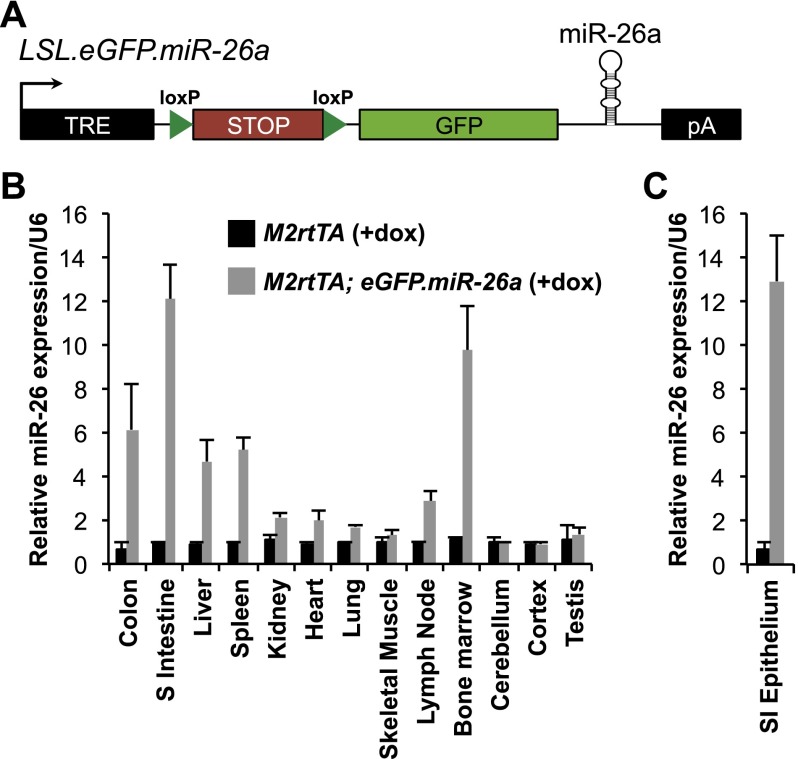
To assess the oncogenic versus tumor-suppressing activity of miR-26 in vivo, we generated a transgenic mouse with dox- and Cre-regulatable miR-26a expression (Fig. 1A). The transgene consists of a dox-regulatable promoter driving expression of a lox-stop-lox (LSL) cassette, which terminates transcription, followed by a green fluorescent protein (GFP) ORF and the human pre-miR-26a-2 hairpin with ∼100 base pairs (bp) of flanking sequence from the endogenous miR-26a-2 locus. The transgene, termed LSL.eGFP.miR-26a, was inserted downstream from the Col1A locus in embryonic stem cells (Beard et al. 2006). These embryonic stem cells also harbor a widely expressed reverse tetracycline transactivator transgene integrated at the Rosa26 locus (M2rtTA).
Figure 1.
A dox- and Cre-inducible miR-26a transgenic mouse. (A) Schematic of the LSL.eGFP.miR-26a transgene. (TRE) Tetracycline-responsive element; (pA) poly(A) signal. (B) Quantitative RT–PCR measurement of miR-26a expression in the indicated tissues of M2rtTA and M2rtTA; eGFP.miR-26a mice normalized to U6 after 2 wk of dox treatment. (C) miR-26a expression in purified small intestinal epithelium of dox-treated transgenic or control mice. For B and C, average values from three independent mice of each genotype are shown. Error bars for this and all subsequent figures represent standard deviations.
LSL.eGFP.miR-26a mice were crossed to CMV-Cre mice (Schwenk et al. 1995) to remove the LSL cassette and broadly activate miR-26a expression. Similar to other transgenes generated using this approach (Beard et al. 2006), the strongest expression was observed in the epithelial cells of the small and large intestine, bone marrow, spleen, thymus, and liver (Fig. 1B,C; Supplemental Fig. S1). Expression of the transgene was fully compatible with normal development and was well tolerated in adults. No overt histopathologic abnormalities or increased mortality were observed in a large cohort of transgenic animals after 1 yr of transgene expression (Supplemental Fig. S2A; data not shown).
Given the absence of increased rates of malignancy in M2rtTA; eGFP.miR-26a mice, we questioned whether expression of the dosage-sensitive Pten tumor suppressor was decreased by enforced miR-26a expression. Western blotting nevertheless confirmed that, in general, tissues with the highest miR-26a expression, such as intestinal epithelium and the liver, exhibited measureable PTEN down-regulation (Supplemental Fig. S3).
miR-26a suppresses intestinal tumorigenesis in Apcmin/+ mice
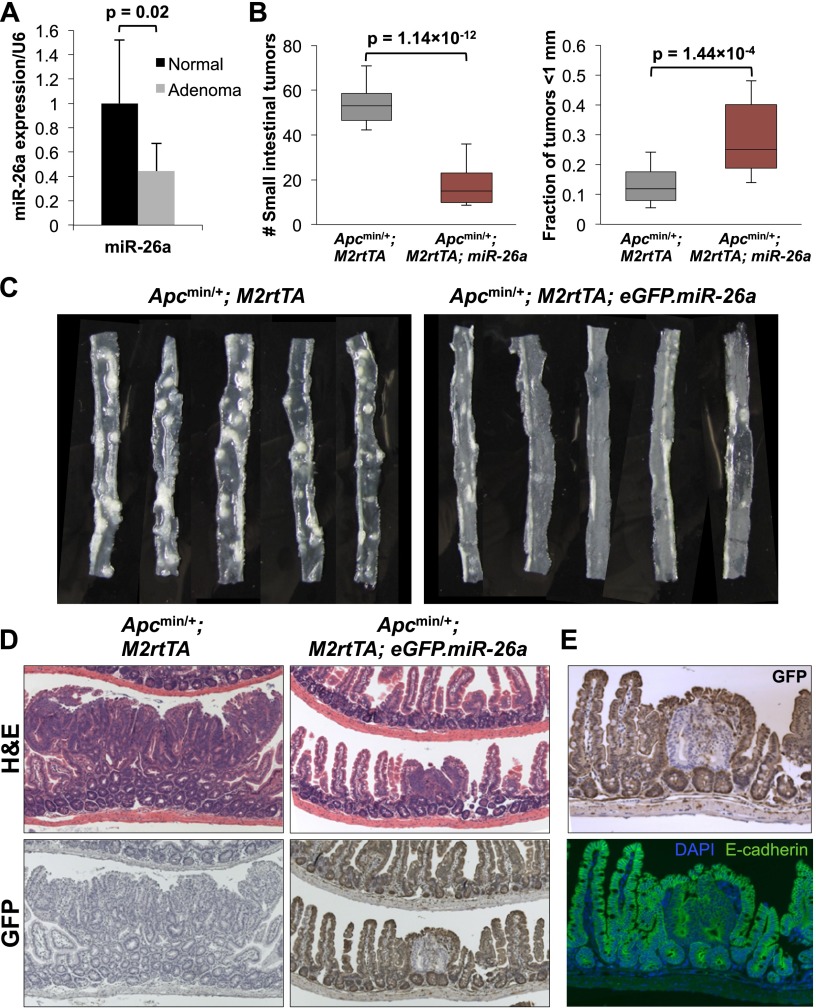
Robust transgene expression in the intestinal epithelium and a corresponding down-regulation of PTEN within this cell type provided an opportunity to directly assess the oncogenic versus tumor suppressor activity of miR-26a in this tissue. Normally, expression of miR-26 family members increases as intestinal stem cells differentiate, as revealed by analysis of miRNA levels in sorted LGR5+ stem cells (Supplemental Fig. S4), suggesting that miR-26 may regulate proliferation and/or differentiation of this cell type. Nevertheless, strongly enforced expression of miR-26a had no effect on baseline intestinal histology (Supplemental Fig. S2B). M2rtTA; eGFP.miR-26a mice were therefore bred to the Apcmin/+ model of intestinal tumorigenesis (Moser et al. 1990). Although Pten haploinsufficiency has previously been shown to strongly enhance tumor initiation and progression in this model (Shao et al. 2007), we observed reduced abundance of miR-26a in Apcmin/+ adenomas (Fig. 2A).
Figure 2.
miR-26a suppresses tumorigenesis in Apcmin/+ mice. (A) miR-26a expression normalized to U6 in adenomas and paired normal tissue from Apcmin/+ mice. n = 10 samples per condition. P-values for A and B were calculated by two-tailed t-test. (B) Quantification of total tumor number (left) and the fraction of tumors that were <1 mm in size (right) in mice of the indicated genotypes. The ends of the boxes represent the 25th and 75th percentiles, the bars indicate the 10th and 90th percentiles, and horizontal lines within the boxes represent the median. n = 21 Apcmin/+; M2rtTA animals and 15 Apcmin/+; M2rtTA; eGFP.miR-26a animals. (C) Representative segments of small intestine from mice of the indicated genotypes. (D) H&E and anti-GFP staining of representative adenomas from mice of the indicated genotypes. (E) Magnified image of adenoma from the Apcmin/+; M2rtTA; eGFP.miR-26a mouse in D illustrating GFP-negative staining of E-cadherin-positive adenoma cells.
Apcmin/+; M2rtTA; eGFP.miR-26a mice and Apcmin/+; M2rtTA controls were given dox at 28 d of age and sacrificed at 150 d. Enforced miR-26a expression dramatically reduced tumor number and size (Fig. 2B,C). Interestingly, GFP immunohistochemistry revealed that the small number of adenomas that arose in Apcmin/+; M2rtTA; eGFP.miR-26a mice uniformly lacked expression of the transgene (Fig. 2D,E). Costaining of E-cadherin confirmed that GFP-negative cells were of epithelial origin in adenomas (Fig. 2E). Thus, adenoma formation in Apcmin/+; M2rtTA; eGFP.miR-26a mice confers strong selection for silencing or deletion of the transgene. These results indicate that despite resulting in measureable repression of PTEN in the intestinal epithelium, enforced miR-26a expression strongly suppresses tumorigenesis in this setting.
Reduced expression of miR-26 in human CRC
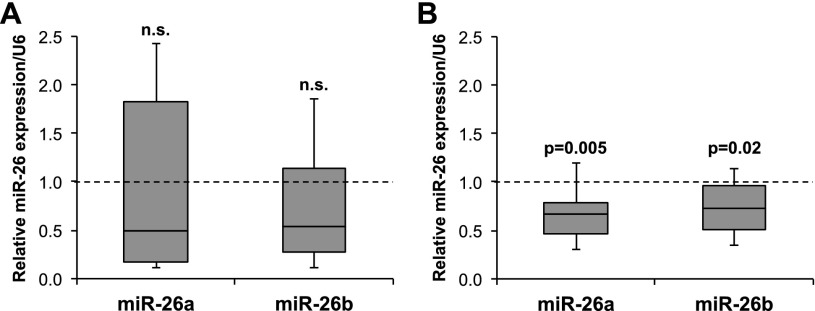
Given the reduced expression of miR-26a in Apcmin/+ intestinal adenomas and its dramatic anti-tumorigenic activity in this model, we sought to determine whether down-regulation of miR-26 family members is a common feature of human CRC. While previous studies of CRC have inconsistently detected miR-26 down-regulation (Schepeler et al. 2008; Schetter et al. 2008; Motoyama et al. 2009; Ma et al. 2011; Gaedcke et al. 2012), the heterogeneous cellular composition of colorectal tumors can confound these measurements. We therefore obtained biopsies of human CRCs and paired normal colons and measured miR-26 levels in the unfractionated tissue as well as in purified epithelium. While neither miR-26a nor miR-26b were detectably reduced in unfractionated tumor tissue (Fig. 3A), clearly lower levels of these miRNAs were observed when comparing purified tumor epithelial cells with paired normal colonic epithelial cells (Fig. 3B). Given our finding that miR-26 expression increases as intestinal epithelial cells differentiate (Supplemental Fig. S4), it is possible that reduced miR-26 levels in tumors is a secondary consequence of impaired differentiation of tumor epithelial cells. Nevertheless, taken together with our functional data from Apcmin/+ mice and previous studies that have demonstrated anti-tumorigenic effects of miR-26 expression in human CRC cell lines (Ma et al. 2011; Zhang et al. 2013), these data strongly suggest a tumor suppressor role for miR-26 in human colon cancer.
Figure 3.
miR-26a is down-regulated in human CRC cells. (A,B) miR-26a and miR-26b expression in unfractionated colorectal tumors relative to paired normal tissue (A) or purified epithelial cells from tumors relative to paired normal colonic epithelial cells (B). n = 10 paired samples for each analysis. P-values were calculated by one-sample t-test. Box plots are defined as in Figure 2B.
miR-26a expression results in a cell-autonomous decrease in intestinal epithelial migration and proliferation
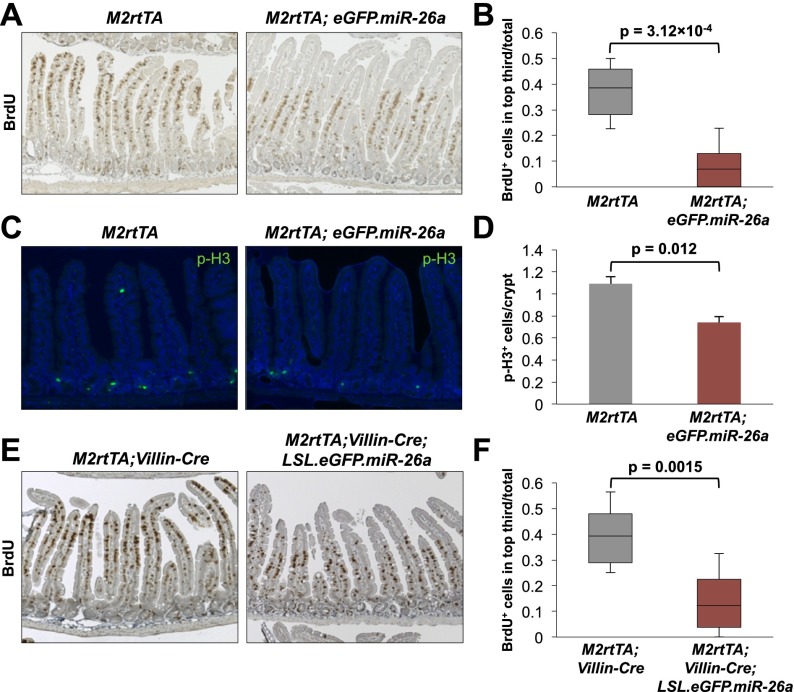
The uniform loss of transgene expression in adenomas of Apcmin/+; M2rtTA; eGFP.miR-26a mice precluded the use of tumor tissue from these animals to investigate the mechanism of tumor suppression. We therefore more closely examined the intestinal epithelium of transgenic animals to determine whether a relevant phenotype was detectable in preneoplastic tissue. Although expression of miR-26a does not result in any overt histologic abnormality in the intestine (Supplemental Fig. S2B), bromodeoxyuridine (BrdU) pulse-chase assays revealed a dramatic decrease in epithelial turnover in transgenic mice, indicative of an epithelial migration defect (Fig. 4A,B; Supplemental Fig. S5). This effect was accompanied by a decrease in phosphorylated histone 3 (p-H3)-positive mitotic cells within intestinal crypts, demonstrating decreased proliferation (Fig. 4C,D). Reduced migration and proliferation was not associated with precocious or impaired differentiation of epithelial cells (Supplemental Fig. S6; data not shown). Notably, there is precedent for a lack of global changes in intestinal morphology in the setting of altered crypt proliferation, as we observed (Haigis et al. 2006).
Figure 4.
miR-26a cell-autonomously inhibits intestinal epithelial proliferation. (A) Distribution of BrdU+ cells in small intestines of dox-treated mice of the indicated genotypes 72 h after BrdU administration. (B) Quantification of the fraction of BrdU+ cells in the upper third of villi. For all panels, 30 villi per mouse and three mice per genotype were quantified, and P-values were calculated using a nested ANOVA. Box plots are defined as in Figure 2B. (C,D) Representative p-H3 staining (C) and quantification (D). (E,F) BrdU staining (E) and quantification (F) as described in A and B.
To assess whether miR-26a inhibited proliferation and migration in a cell-autonomous manner, the Cre-regulatable transgene (LSL.eGFP.miR-26a) was activated specifically in intestinal epithelium by crossing to Villin-Cre mice (Supplemental Fig. S7; Madison et al. 2002). Similar to the effect of broad miR-26a expression, epithelial-restricted miR-26a expression was sufficient to significantly delay transit of BrdU+ cells (Fig. 4E,F). These data document potent cell-autonomous anti-proliferative and anti-migratory effects of enforced miR-26a expression within the intestinal epithelium.
miR-26a represses expression of proproliferative transcripts, including Ccnd2, Hmga1, Mycbp, Setd8, and Ezh2
In order to identify the targets of miR-26a that mediate its anti-proliferative effects, microarrays were used to assay gene expression in purified epithelial cells from dox-treated M2rtTA or M2rtTA; eGFP.miR-26a mice. One-hundred-sixty-six transcripts were down-regulated and 132 transcripts were up-regulated by ≥1.5-fold in miR-26a transgenic animals (Supplemental Table S1). Sylamer analysis (van Dongen et al. 2008) revealed dramatic enrichment of hexamers that are complementary to the miR-26 seed sequence within the 3′ untranslated regions (UTRs) of down-regulated genes (Supplemental Fig. S8). Furthermore, gene set enrichment analysis (GSEA) (Subramanian et al. 2005) demonstrated significant down-regulation of miR-26 targets predicted by TargetScan (Supplemental Fig. S9A; Grimson et al. 2007). These findings demonstrate strong engagement of canonical miR-26a targets in the intestinal epithelium of transgenic animals.
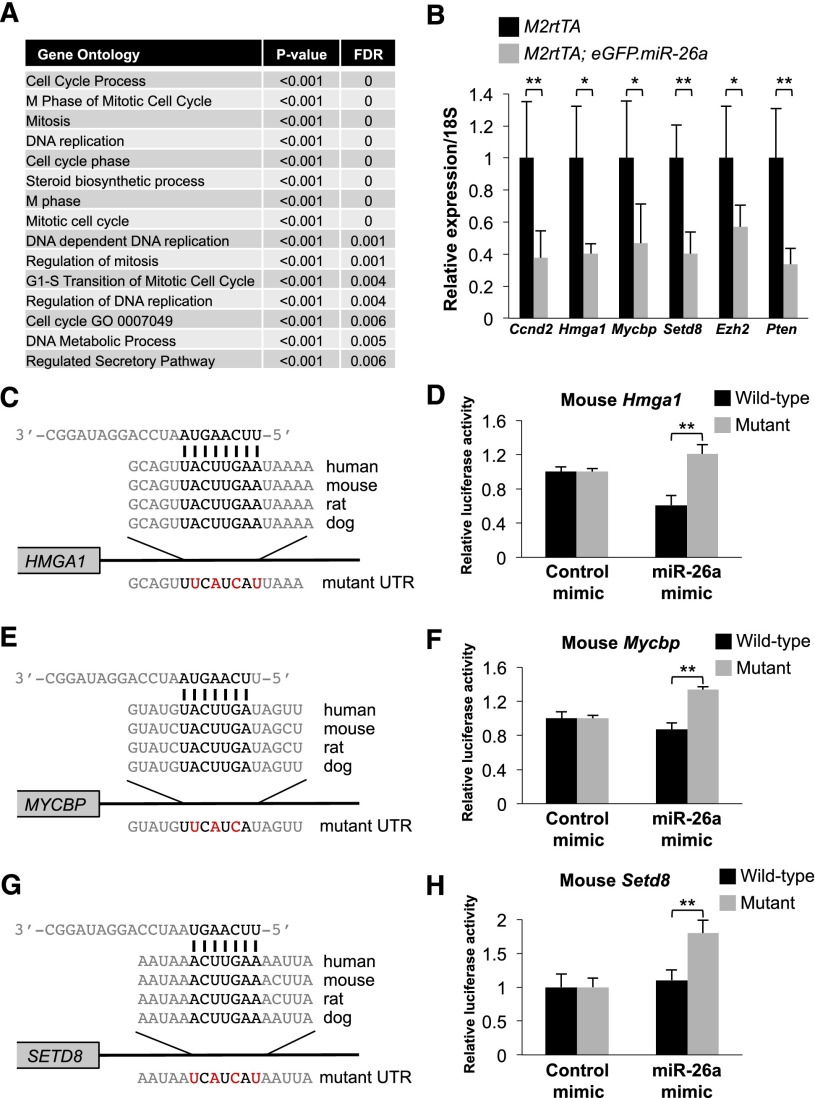
GSEA analysis also revealed broad repression of proproliferative transcripts in miR-26a-expressing epithelial cells. Thirteen of 15 of the top gene ontology biological process gene sets that were repressed in transgenic animals are related to cell cycle control, mitosis, or DNA replication (Fig. 5A). To identify direct miR-26a targets that contribute to the anti-proliferative activity of this miRNA, we compiled a list of genes that are predicted to be miR-26 targets by TargetScan and were repressed by ≥1.5-fold by miR-26a (Supplemental Table S1). On the basis of their previously documented roles in proliferation of colorectal or other types of cancer cells or involvement in Wnt signaling, a key driver of proliferation in this tissue, we selected Ccnd2, Hmga1, Mycbp, and Setd8 for additional study. Ccnd2, an established miR-26a target (Kota et al. 2009), is commonly overexpressed in colorectal tumors (Mermelshtein et al. 2005) and is essential for tumorigenesis in Apcmin/+ mice (Myant and Sansom 2011a). Hmga1 is an oncogenic chromatin remodeling factor that is broadly overexpressed in human malignancies, including CRC. Transgenic overexpression of Hmga1 drives intestinal epithelial proliferation and polyp formation, whereas knockdown in colon cancer cell lines reduces tumor-forming ability (Belton et al. 2012). Although not previously studied in the context of the intestinal epithelium, Mycbp is believed to act as a coactivator of MYC-mediated transcription, and its depletion impairs proliferation of breast cancer cells (Taira et al. 1998; Xiong et al. 2010). MYC is a critical Wnt/β-catenin target gene that is essential for intestinal homeostasis and tumorigenesis (Myant and Sansom 2011b). Moreover, MYCBP itself has been reported to be a Wnt/β-catenin target, further highlighting its potential importance in intestinal epithelial proliferation (Jung and Kim 2005). Setd8 is a critical cofactor for Wnt target gene activation (Li et al. 2011) and, accordingly, GSEA analysis revealed significant down-regulation of β-catenin targets in miR-26a transgenic epithelium (Supplemental Fig. S9B). Last, although microarray analysis showed a <1.5-fold decrease in expression, we included Ezh2 in validation studies because GSEA analysis showed significant repression of EZH2 target gene sets in transgenic mice (Supplemental Fig. S9C). EZH2 is a verified miR-26 target that is overexpressed in CRCs, and its depletion impairs proliferation of human CRC cells (Fluge et al. 2009; Fussbroich et al. 2011).
Figure 5.
miR-26a represses proproliferative targets in intestinal epithelium. (A) The top 15 gene ontology biological process terms enriched among miR-26a-repressed transcripts identified by GSEA. (B) Expression of the indicated transcripts, normalized to 18S rRNA, in purified intestinal epithelium from dox-treated mice of the indicated genotypes. Means and standard deviations from n = 3–5 animals per genotype shown. (C,E,G) Schematic representation of the miR-26-binding sites in the 3′ UTRs of HMGA1 (C), MYCBP (E), and SETD8 (G). Mutations introduced into reporter constructs are shown below each alignment and are highlighted in red. (D,F,H) Relative firefly luciferase activity of reporter constructs containing the indicated miR-26-binding site or its mutated version following transfection into HCT116 cells with control or miR-26a mimic. n = 3 replicates per condition. (*) P < 0.05; (**) P < 0.01 (two-tailed t-test).
Quantitative PCR confirmed significant down-regulation of Ccnd2, Hmga1, Mycbp, Setd8, and Ezh2 in miR-26a-expressing intestinal epithelium (Fig. 5B). Consistent with its effects on PTEN protein abundance (Supplemental Fig. S3), miR-26a also strongly repressed Pten transcript levels in epithelial cells. Multiple studies have previously validated direct targeting of Ccnd2 and Ezh2 by miR-26 (Sander et al. 2008; Kota et al. 2009; Lu et al. 2011). Both Hmga1 and Mycbp have a single highly conserved predicted miR-26-binding site in their 3′ UTRs, whereas Setd8 has two conserved predicted sites (Fig. 5C,E,G; Supplemental Fig. S10A). Consistent with a recent report (Lin et al. 2013), we documented that the human and mouse HMGA1 miR-26-binding sites, but not mutated versions, conferred miR-26a-mediated repression when placed in the 3′ UTR of a luciferase reporter transcript (Fig. 5D; Supplemental Fig. S11A). Similar experiments validated the functionality of the MYCBP and one of the SETD8 miR-26-binding sites (Fig. 5F,H; Supplemental Figs. S10B,C, S11B,C).
The tumor suppressor activity of miR-26 predominates in intestinal cancer
The inducible miR-26a transgenic mouse described here provides a valuable model for the evaluation of miR-26 functions in various tissue contexts in vivo. Given the previous findings of both pro- and anti-tumorigenic activity of miR-26, it has remained unclear whether delivery of this miRNA would be a safe and efficacious therapeutic approach for specific cancers. We now show that broad overexpression of miR-26a for prolonged periods is well tolerated and does not result in any overt malignancy. Nevertheless, as reported (Huse et al. 2009), we confirmed that miR-26a represses the dosage-sensitive tumor suppressor Pten. The intestinal epithelium is a site of particularly high expression of miR-26a in M2rtTA; eGFP.miR-26a mice, exhibits a corresponding strong down-regulation of PTEN, and has been shown to be prone to spontaneous tumor development in the setting of Pten haploinsufficiency (Di Cristofano et al. 1998). The absence of any baseline intestinal pathology in transgenic animals and the strong suppression of tumorigenesis in Apcmin/+ mice demonstrate that the tumor suppressor activity of miR-26a overrides the potential oncogenic consequences of Pten repression in this tissue. Importantly, the balance between miR-26-mediated tumor suppression versus oncogenic activity may differ in distinct tissue or tumor contexts, likely explaining why expression of miR-26 accelerates tumorigenesis in settings such as PDGF-driven gliomagenesis in mice (Huse et al. 2009). Combining the LSL.eGFP.miR-26a mouse with various tissue-specific tTA or Cre driver lines and tumor models offers a flexible experimental approach for elucidating the effects of miR-26 expression in different cancer settings in vivo.
miR-26 represses a broad proproliferative gene expression program in intestinal epithelium, likely accounting for its potent anti-proliferative effects and tumor suppressor activity in this tissue. In particular, the miR-26 targets Ccnd2, Hmga1, and Ezh2 are all known to be important for intestinal tumorigenesis (Fussbroich et al. 2011; Myant and Sansom 2011a; Belton et al. 2012). Our finding that Mycbp, a Wnt/β-catenin target gene demonstrated to be important for cell proliferation in other cell types (Jung and Kim 2005; Xiong et al. 2010), is directly repressed by miR-26 suggests that it may also participate in intestinal neoplasia. Likewise, impairment of Wnt signaling through down-regulation of Setd8 (Li et al. 2011), another newly identified miR-26 target, would be expected to potently inhibit intestinal epithelial migration, proliferation, and neoplastic transformation. In aggregate, the predicted result of coordinated depletion of these targets could plausibly overcome the protumorigenic consequences of Pten down-regulation in this tissue.
In addition to establishing the tumor suppressor activity of miR-26 in the mouse intestine, our analysis of human CRC specimens has clarified previous work that has inconsistently detected reduced expression of this miRNA family in human colorectal tumors (Ma et al. 2011; Gaedcke et al. 2012; Zheng et al. 2013). Through the analysis of purified CRC epithelial cells, the relevant transformed cell type in this malignancy, we documented a clear reduction of miR-26a and miR-26b in these cancer cells. These findings are consistent with a tumor suppressor role for miR-26 in human CRC. Given that broad miR-26a expression was well tolerated in mice and potently suppressed intestinal tumorigenesis, these findings suggest that miR-26 delivery could be an efficacious and nontoxic therapeutic strategy for colon cancer.
Materials and methods
Mouse strains
CMV-Cre (Schwenk et al. 1995), Apcmin/+ (Moser et al. 1990), Villin-Cre (Madison et al. 2002), and Lgr5+/eGFP (Barker et al. 2007) mice were obtained from Jackson Laboratory. Strains used in this study were backcrossed to C57BLJ/6 mice for at least 10 generations. Dox (2 mg/mL, supplemented with 10 mg/mL sucrose) was administered in the drinking water to induce transgene expression. All experiments were approved by the Institutional Animal Care and Use Committees of the Johns Hopkins University School of Medicine and The University of Texas Southwestern Medical Center.
Apcmin/+ adenoma quantification
Apcmin/+; M2rtTA and Apcmin/+; M2rtTA; eGFP.miR-26a mice were given dox at 28 d of age. At 150 d of age, animals were sacrificed, and intestines were fixed overnight in 10% neutral buffered formalin. A dissecting microscope with a reticle was used to quantify adenoma number and size in a genotype-blind manner.
Isolation of human and mouse intestinal epithelial cells
Mouse and human intestinal epithelial isolation and sorting of LGR5+ cells from Lgr5+/eGFP mice were carried out as previously described (Chivukula et al. 2014). Tissue from colorectal tumors and paired normal colons was obtained through the University of Texas Southwestern Tissue Resource.
Microarray data have been deposited in the Gene Expression Omnibus repository under accession number GSE62831.
Supplementary Material
Acknowledgments
We thank Chip Hawkins in the Johns Hopkins Transgenic Core for assistance with generation of LSL.eGFP.miR-26a mice, John Shelton in the University of Texas Southwestern Molecular Pathology Core for assistance with histopathology, Cheryl Lewis in the University of Texas Southwestern Tissue Resource Core for assistance with human tissue specimens, Quan-Zhen Li in the University of Texas Southwestern Microarray Core for assistance with gene expression profiling, and Jeanetta Marshburn-Wynn for mouse colony maintenance. Kathryn O’Donnell, Tsung-Cheng Chang, and Kenneth Chen provided valuable technical assistance and advice. This work was supported by grants from Cancer Prevention Research Institute of Texas (CPRIT) (R1008 to J.T.M.) and the National Institutes of Health (R01CA120185 and P01CA134292 to J.T.M., 5P30CA142543 to the University of Texas Southwestern Simmons Cancer Center, and 5T32GM007309 to L.R.Z. and R.R.C.). J.T.M. is a CPRIT Scholar in Cancer Research.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.250951.114.
References
- Barker N, van Es JH, Kuipers J, Kujala P, van Born M, Cozijensen M, Haegebarth A, Korving J, Begthel H, Peters PJ, et al. . 2007. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature 449: 1003–1007. [DOI] [PubMed] [Google Scholar]
- Beard C, Hochedlinger K, Plath K, Wutz A, Jaenisch R. 2006. Efficient method to generate single-copy transgenic mice by site-specific integration in embryonic stem cells. Genesis 44: 23–28. [DOI] [PubMed] [Google Scholar]
- Belton A, Gabrovsky A, Bae YK, Reeves R, Iacobuzio-Donahue C, Huso DL, Resar LM. 2012. HMGA1 induces intestinal polyposis in transgenic mice and drives tumor progression and stem cell properties in colon cancer cells. PLoS ONE 7: e30034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang TC, Yu D, Lee YS, Wentzel EA, Arking DE, West KM, Dang CV, Thomas-Tikhonenko A, Mendell JT. 2008. Widespread microRNA repression by Myc contributes to tumorigenesis. Nat Genet 40: 43–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chivukula RR, Shi G, Acharya A, Mills EW, Zeitels LR, Anandam JL, Abdelnaby AA, Balch GC, Mansour JC, Yopp AC, et al. . 2014. An essential mesenchymal function for miR-143/145 in intestinal epithelial regeneration. Cell 157: 1104–1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Cristofano A, Pesce B, Cordon-Cardo C, Pandolfi PP. 1998. Pten is essential for embryonic development and tumour suppression. Nat Genet 19: 348–355. [DOI] [PubMed] [Google Scholar]
- Di Leva G, Garofalo M, Croce CM. 2014. MicroRNAs in cancer. Annu Rev Pathol 9: 287–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fluge O, Gravdal K, Carlsen E, Vonen B, Kjellevold K, Refsum S, Lilleng R, Eide TJ, Halvorsen TB, Tveit KM, et al. . 2009. Expression of EZH2 and Ki-67 in colorectal cancer and associations with treatment response and prognosis. Br J Cancer 101: 1282–1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fussbroich B, Wagener N, Macher-Goeppinger S, Benner A, Falth M, Sultmann H, Holzer A, Hoppe-Seyler K, Hoppe-Seyler F. 2011. EZH2 depletion blocks the proliferation of colon cancer cells. PLoS ONE 6: e21651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaedcke J, Grade M, Camps J, Sokilde R, Kaczkowski B, Schetter AJ, Difilippantonio MJ, Harris CC, Ghadimi BM, Moller S, et al. . 2012. The rectal cancer microRNAome—microRNA expression in rectal cancer and matched normal mucosa. Clin Cancer Res 18: 4919–4930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. 2007. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell 27: 91–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haigis K, Sage J, Glickman J, Shafer S, Jacks T. 2006. The related retinoblastoma (pRb) and p130 proteins cooperate to regulate homeostasis in the intestinal epithelium. J Biol Chem 281: 638–647. [DOI] [PubMed] [Google Scholar]
- Huse JT, Brennan C, Hambardzumyan D, Wee B, Pena J, Rouhanifard SH, Sohn-Lee C, le Sage C, Agami R, Tuschl T, et al. . 2009. The PTEN-regulating microRNA miR-26a is amplified in high-grade glioma and facilitates gliomagenesis in vivo. Genes Dev 23: 1327–1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung HC, Kim K. 2005. Identification of MYCBP as a β-catenin/LEF-1 target using DNA microarray analysis. Life Sci 77: 1249–1262. [DOI] [PubMed] [Google Scholar]
- Ji J, Shi J, Budhu A, Yu Z, Forgues M, Roessler S, Ambs S, Chen Y, Meltzer PS, Croce CM, et al. . 2009. MicroRNA expression, survival, and response to interferon in liver cancer. N Engl J Med 361: 1437–1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kota J, Chivukula RR, O’Donnell KA, Wentzel EA, Montgomery CL, Hwang HW, Chang TC, Vivekanandan P, Torbenson M, Clark KR, et al. . 2009. Therapeutic microRNA delivery suppresses tumorigenesis in a murine liver cancer model. Cell 137: 1005–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Nie F, Wang S, Li L. 2011. Histone H4 Lys 20 monomethylation by histone methylase SET8 mediates Wnt target gene activation. Proc Natl Acad Sci 108: 3116–3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Chen H, Hu Z, Mao Y, Xu X, Zhu Y, Xu X, Wu J, Li S, Mao Q, et al. . 2013. miR-26a inhibits proliferation and motility in bladder cancer by targeting HMGA1. FEBS Lett 587: 2467–2473. [DOI] [PubMed] [Google Scholar]
- Liu B, Wu X, Liu B, Wang C, Liu Y, Zhou Q, Xu K. 2012. miR-26a enhances metastasis potential of lung cancer cells via AKT pathway by targeting PTEN. Biochim Biophys Acta 1822: 1692–1704. [DOI] [PubMed] [Google Scholar]
- Lu J, He ML, Wang L, Chen Y, Liu X, Dong Q, Chen YC, Peng Y, Yao KT, Kung HF, et al. . 2011. miR-26a inhibits cell growth and tumorigenesis of nasopharyngeal carcinoma through repression of EZH2. Cancer Res 71: 225–233. [DOI] [PubMed] [Google Scholar]
- Ma YL, Zhang P, Wang F, Moyer MP, Yang JJ, Liu ZH, Peng JY, Chen HQ, Zhou YK, Liu WJ, et al. . 2011. Human embryonic stem cells and metastatic colorectal cancer cells shared the common endogenous human microRNA-26b. J Cell Mol Med 15: 1941–1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madison BB, Dunbar L, Qiao XT, Braunstein K, Braunstein E, Gumucio DL. 2002. Cis elements of the villin gene control expression in restricted domains of the vertical (crypt) and horizontal (duodenum, cecum) axes of the intestine. J Biol Chem 277: 33275–33283. [DOI] [PubMed] [Google Scholar]
- Mavrakis KJ, Van Der Meulen J, Wolfe AL, Liu X, Mets E, Taghon T, Khan AA, Setty M, Rondou P, Vandenberghe P, et al. . 2011. A cooperative microRNA-tumor suppressor gene network in acute T-cell lymphoblastic leukemia (T-ALL). Nat Genet 43: 673–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mermelshtein A, Gerson A, Walfisch S, Delgado B, Shechter-Maor G, Delgado J, Fich A, Gheber L. 2005. Expression of D-type cyclins in colon cancer and in cell lines from colon carcinomas. Br J Cancer 93: 338–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moser AR, Pitot HC, Dove WF. 1990. A dominant mutation that predisposes to multiple intestinal neoplasia in the mouse. Science 247: 322–324. [DOI] [PubMed] [Google Scholar]
- Motoyama K, Inoue H, Takatsuno Y, Tanaka F, Mimori K, Uetake H, Sugihara K, Mori M. 2009. Over- and under-expressed microRNAs in human colorectal cancer. Int J Oncol 34: 1069–1075. [DOI] [PubMed] [Google Scholar]
- Myant K, Sansom O. 2011a. Efficient Wnt mediated intestinal hyperproliferation requires the cyclin D2–CDK4/6 complex. Cell Div 6: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myant K, Sansom OJ. 2011b. Wnt/Myc interactions in intestinal cancer: partners in crime. Exp Cell Res 317: 2725–2731. [DOI] [PubMed] [Google Scholar]
- Sander S, Bullinger L, Klapproth K, Fiedler K, Kestler HA, Barth TF, Moller P, Stilgenbauer S, Pollack JR, Wirth T. 2008. MYC stimulates EZH2 expression by repression of its negative regulator miR-26a. Blood 112: 4202–4212. [DOI] [PubMed] [Google Scholar]
- Schepeler T, Reinert JT, Ostenfeld MS, Christensen LL, Silahtaroglu AN, Dyrskjot L, Wiuf C, Sorensen FJ, Kruhoffer M, Laurberg S, et al. . 2008. Diagnostic and prognostic microRNAs in stage II colon cancer. Cancer Res 68: 6416–6424. [DOI] [PubMed] [Google Scholar]
- Schetter AJ, Leung SY, Sohn JJ, Zanetti KA, Bowman ED, Yanaihara N, Yuen ST, Chan TL, Kwong DL, Au GK, et al. . 2008. MicroRNA expression profiles associated with prognosis and therapeutic outcome in colon adenocarcinoma. JAMA 299: 425–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwenk F, Baron U, Rajewsky K. 1995. A cre-transgenic mouse strain for the ubiquitous deletion of loxP-flanked gene segments including deletion in germ cells. Nucleic Acids Res 23: 5080–5081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao J, Washington MK, Saxena R, Sheng H. 2007. Heterozygous disruption of the PTEN promotes intestinal neoplasia in APCmin/+ mouse: roles of osteopontin. Carcinogenesis 28: 2476–2483. [DOI] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. . 2005. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci 102: 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taira T, Maeda J, Onishi T, Kitaura H, Yoshida S, Kato H, Ikeda M, Tamai K, Iguchi-Ariga SM, Ariga H. 1998. AMY-1, a novel C-MYC binding protein that stimulates transcription activity of C-MYC. Genes Cells 3: 549–565. [DOI] [PubMed] [Google Scholar]
- van Dongen S, Abreu-Goodger C, Enright AJ. 2008. Detecting microRNA binding and siRNA off-target effects from expression data. Nat Methods 5: 1023–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong J, Du Q, Liang Z. 2010. Tumor-suppressive microRNA-22 inhibits the transcription of E-box-containing c-Myc target genes by silencing c-Myc binding protein. Oncogene 29: 4980–4988. [DOI] [PubMed] [Google Scholar]
- Zhang B, Liu XX, He JR, Zhou CX, Guo M, He M, Li MF, Chen GQ, Zhao Q. 2011. Pathologically decreased miR-26a antagonizes apoptosis and facilitates carcinogenesis by targeting MTDH and EZH2 in breast cancer. Carcinogenesis 32: 2–9. [DOI] [PubMed] [Google Scholar]
- Zhang C, Tong J, Huang G. 2013. Nicotinamide phosphoribosyl transferase (Nampt) is a target of microRNA-26b in colorectal cancer cells. PLoS ONE 8: e69963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng G, Wang H, Zhang X, Yang Y, Wang L, Du L, Li W, Li J, Qu A, Liu Y, et al. . 2013. Identification and validation of reference genes for qPCR detection of serum microRNAs in colorectal adenocarcinoma patients. PLoS ONE 8: e83025. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.