In C. elegans, dosage compensation is achieved by twofold down-regulation of gene expression from both X chromosomes in hermaphrodites. Sharma et al. show that in males, the single X chromosome interacts with nuclear pore proteins, while in hermaphrodites, the dosage compensation complex impairs this interaction and alters X localization.
Keywords: dosage compensation, nuclear organization, C. elegans, nuclear pores
Abstract
The adjustment of X-linked gene expression to the X chromosome copy number (dosage compensation [DC]) has been widely studied as a model of chromosome-wide gene regulation. In Caenorhabditis elegans, DC is achieved by twofold down-regulation of gene expression from both Xs in hermaphrodites. We show that in males, the single X chromosome interacts with nuclear pore proteins, while in hermaphrodites, the DC complex (DCC) impairs this interaction and alters X localization. Our results put forward a structural model of DC in which X-specific sequences locate the X chromosome in transcriptionally active domains in males, while the DCC prevents this in hermaphrodites.
Throughout the animal kingdom, varied strategies have evolved to equalize expression of the X chromosome genes between sexes with different X to autosomes ratios, a process called dosage compensation (DC) (for review, see Ferrari et al. 2014). In Caenorhabditis elegans, genetic screens and RNA quantifications showed that DC occurs in hermaphrodites by twofold down-regulation of X-linked transcripts from both Xs (for review, see Strome et al. 2014). A number of mutants were isolated in which overexpression of X-linked genes led to hermaphrodite-specific defects (dpy genes) or sex determination and DC deficiency (sdc genes). Remarkably, all proteins of the sex-specific Dpy and Sdc classes interact and form a single complex, the DC complex (DCC). Structurally, the DCC is similar to condensin I and loaded on the X chromosome at rex (recruitment element on X) sites characterized by a 12-base-pair (bp) MEX (motif enriched on X) consensus sequence (Ercan et al. 2009; Jans et al. 2009). Thirty-eight rex sites have been experimentally demonstrated, and sequence analysis suggests the presence of 100–300 sites on the X chromosome (Jans et al. 2009). This estimation is due to the fact that the DCC moves and spreads along the X chromosome from its nucleation sites (Csankovszki et al. 2004). The DCC accumulates at promoters upstream of transcription start sites; however, the relationship between DCC accumulation and transcriptional regulation remains disputed (Ercan et al. 2009; Jans et al. 2009). Genome-wide run-on experiments have shown that the DCC reduces transcription from the X chromosome, although RNA polymerase chromatin immunoprecipitation (ChIP) does not show a significant reduction compared with autosomes (Kruesi et al. 2013). How DCC loading regulates RNA polymerase II (Pol II) function still remains unknown. Compared with autosomes, compensated X chromatin is depleted for the histone variant HTZ-1 and H4K16 acetylation, likely a consequence of lower transcription, and carries high H4K20 monomethylation (H4K20me1), spreading with the DCC (Whittle et al. 2008; Petty et al. 2011; Vielle et al. 2012; Wells et al. 2012). Inside the nuclear space, the compensated X displays a peculiar tridimensional conformation: While all autosomes have high interactions of both arms with the nuclear lamina, the X chromosome is only loosely interacting with the periphery at telomeres (Fig. 1D; Ikegami et al. 2010; Towbin et al. 2012). In males, no specific chromosome organization or X-specific chromatin marks have been described. Given the similarity between the DCC and condensins, the presence of a specific higher-order structure of the X chromosomes facilitated by the DCC has been suggested as a model for years but never tested directly (Ferrari et al. 2014).
Figure 1.
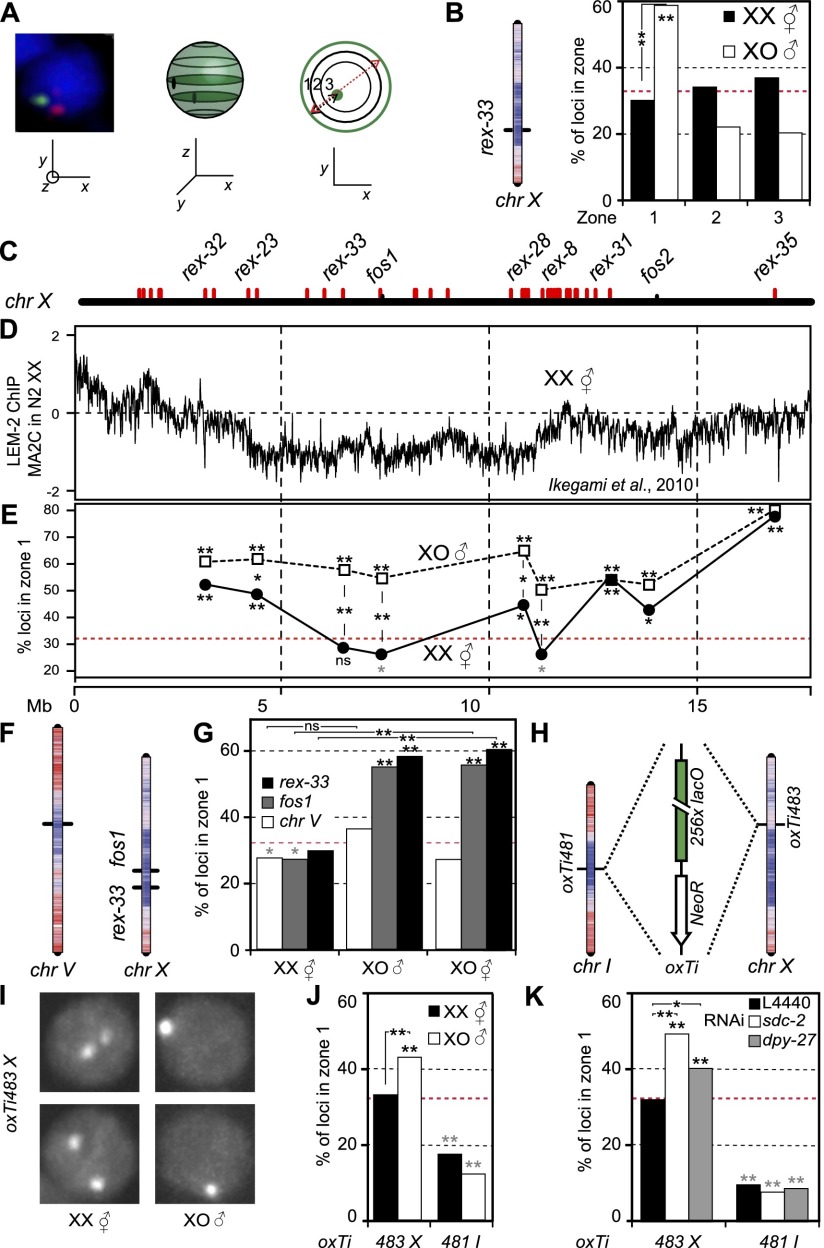
The X chromosome is peripherally located in males, and DC impairs this localization. (A) Three-zone assay to quantify the radial position of a locus inside the nucleus. (B) Location of rex-33 on chr X with LEM-2 ChIP–chip profile overlay ([red] high LEM-2 interaction; [blue] low LEM-2 interaction) and three-zone scoring for the same locus in hermaphrodites and males. Statistical data for all scorings in Supplemental Table 1: (*) P < 0.05; (**) P < 0.01; (asterisks on the bars) P-value against random in χ2 test; (gray) significantly internal; (black) significantly peripheral; (asterisks between bars) P-value between samples in Fisher exact test. (C) Localization of analyzed rex and non-rex (fos) sites along chr X. (D) LEM-2 ChIP–chip profile along the hermaphrodite X chromosome (data from Ikegami et al. 2010). (E) Zone 1 scorings for sites in C in hermaphrodite and male. (Red dashed line) Random distribution (33%). (F) Location of scored loci on chr V and chr X, with overlay of the LEM-2 ChIP–chip profile. (G) Zone 1 scorings for the sites in F in XO males and XX and XO hermaphrodites. (H) Location of lacO repeats insertions on chr I and chr X, with overlay of the LEM-2 ChIP–chip profile. (I) Representative images of nuclei in male and hermaphrodite embryos for the chr X insertion. (J) Zone 1 scorings for the insertions in H in XX hermaphrodites and XO males. (K) Zone 1 scorings for the insertions in H in XX hermaphrodites grown on control (L4440), sdc-2, and dpy-27 RNAi.
Results and Discussion
We asked whether DC has an effect on X chromosome tridimensional organization by carrying out fluorescence in situ hybridization (FISH) for a rex site located in the center of the chr X (rex-33) (Fig. 1B). Radial distribution of this locus was scored in 40- to 150-cell stage male and hermaphrodite embryos (after DC establishment) (Chuang et al. 1994; Dawes et al. 1999) using the three-zone scoring assay (Fig. 1A; representative image of FISH data in Supplemental Fig. S1,; Askjaer et al. 2014). For each FISH signal, in the Z-plane in which the spot is the brightest, the nuclear section is divided into three zones of equal surface. The spot is then binned into one of these three zones. In hermaphrodite embryos, rex-33 is randomly localized, although the probe overlaps with a site previously shown as highly enriched for nuclear pore proteins (nucleoporins) (Fig. 1B; Ikegami and Lieb 2013). As the subnuclear localization of this site was not tested previously, our data suggest that pore–chromatin interaction occurs in the nuclear lumen or that this interaction is present in a minor fraction of cells. In males, rex-33 was preferentially located in zone 1 (58% of spots) (Fig. 1B). Scoring of eight additional loci distributed along the X chromosome (Fig. 1C,E) showed that in hermaphrodites, our measurements largely agree with LEM-2 ChIP data performed in hermaphrodites: Loci with high LEM-2 enrichment are preferentially located in zone 1 (Fig. 1E), while regions with low LEM-2 enrichment show random distribution inside the nucleus. In male embryos, all sites show highly significant enrichment in the peripheral-most zone (at least 50%) (Fig. 1E). The difference between males and hermaphrodites is particularly visible in the center of the chromosome, as telomeres appear to be anchored at the nuclear envelope similarly in both sexes (Ferreira et al. 2013). Moreover, peripheral positioning does not appear to be rex site-specific, as both rex and non-rex loci are peripherally located (Fig. 1E). It is, however, specific to the X chromosome, as an autosomal site located in the middle of chr V is similarly positioned in males and hermaphrodites (Fig. 1F,G). Our data therefore indicate a sex-specific X chromosome localization.
To ensure that the observed differences in X chromosome organization are due to DC and not an outcome of differential gene expression between sexes, we scored the X chromosome position in noncompensated XO hermaphrodite embryos (TY2205) (Csankovszki et al. 2009). These animals have one X chromosome, express a hermaphrodite transcriptional program, and do not initiate DC. Two sites located in the middle of chr X were located similarly in males, while the position of the center region of chr V was internal or random for all genotypes (Fig. 1F,G). Differences in X chromosome organization are therefore not a consequence of global gene expression differences between sexes but rather reflect DC acting on X. This implies that the “default” localization of the noncompensated X chromosome is at the nuclear periphery as in males and that this localization is impaired by the loading of the DCC on X chromosomes in hermaphrodites, leading to an internal location.
A consequence of the DC-positioning hypothesis above is that down-regulation of the DCC in hermaphrodites should lead to perinuclear localization of the X chromosome. As the localization in XO hermaphrodites suggests, mutation of components of the DCC (sdc-3) leads to relocation of the X chromosome to the periphery (Fig. 1G). To test this hypothesis more directly, we used lacO repeats inserted in the central region of chr X and a control insertion in the center of chr I. Expression in trans of GFP-lacI leads to the formation of a visible spot of which radial position inside the nucleus can be scored (Fig. 1H,I; Askjaer et al. 2014). Upon DCC knockdown, transcription in the vicinity of these lacO insertions is up-regulated for the X chromosome locus and stable around the chr I insertion site (Supplemental Fig. S2; Kruesi et al. 2013). As in our FISH analysis, the X-linked locus is randomly positioned in hermaphrodites and significantly enriched at the nuclear periphery in males. In contrast, the locus on chr I is internally positioned in both sexes (Fig. 1J). We depleted either SDC-2 or DPY-27, both subunits of the DCC, using RNAi. Phenotypically, sdc-2(RNAi) has a strong effect, with most animals showing a Dumpy phenotype, while dpy-27(RNAi) is less penetrant (data not shown). Upon down-regulation of sdc-2 or dpy-27, the chr X lacO insertion showed a 16% or 10% increase in the outermost zone 1 compared with control RNAi, respectively, while the same treatment did not change the position of the autosomal site (Fig. 1K). This implies that the DCC prevents the association of the chr X with the nuclear periphery.
Our results show that in rapidly cycling male embryonic blastomeres, the X chromosome is localized at the nuclear periphery. Such a conformation strongly suggests the existence of anchoring sequences on the X chromosome and anchor sites at the nuclear rim. A striking feature of the X chromosome is its enrichment for the MEX motif, a motif recognized by the DCC for loading (McDonel et al. 2006; Jans et al. 2009). We asked whether a single MEX motif would be able to direct an autosomal locus to the nuclear periphery in males specifically by creating strains with an ectopic autosomal insertion of a MEX motif (rex-1•33) (Fig. 2A; McDonel et al. 2006). This locus was preferentially positioned internally in hermaphrodites, while in males, the same site showed significant enrichment in zone 1 (Fig. 2B). Insertion of a similar construct with a mutated MEX motif led to a slightly internal localization similar in both sexes. We conclude that in males, the MEX motif is sufficient for perinuclear targeting of an otherwise internal locus. As many MEX motifs are present on the X chromosome (Jans et al. 2009), MEX periphery interaction could position the entire X chromosome close to the nuclear rim in males.
Figure 2.
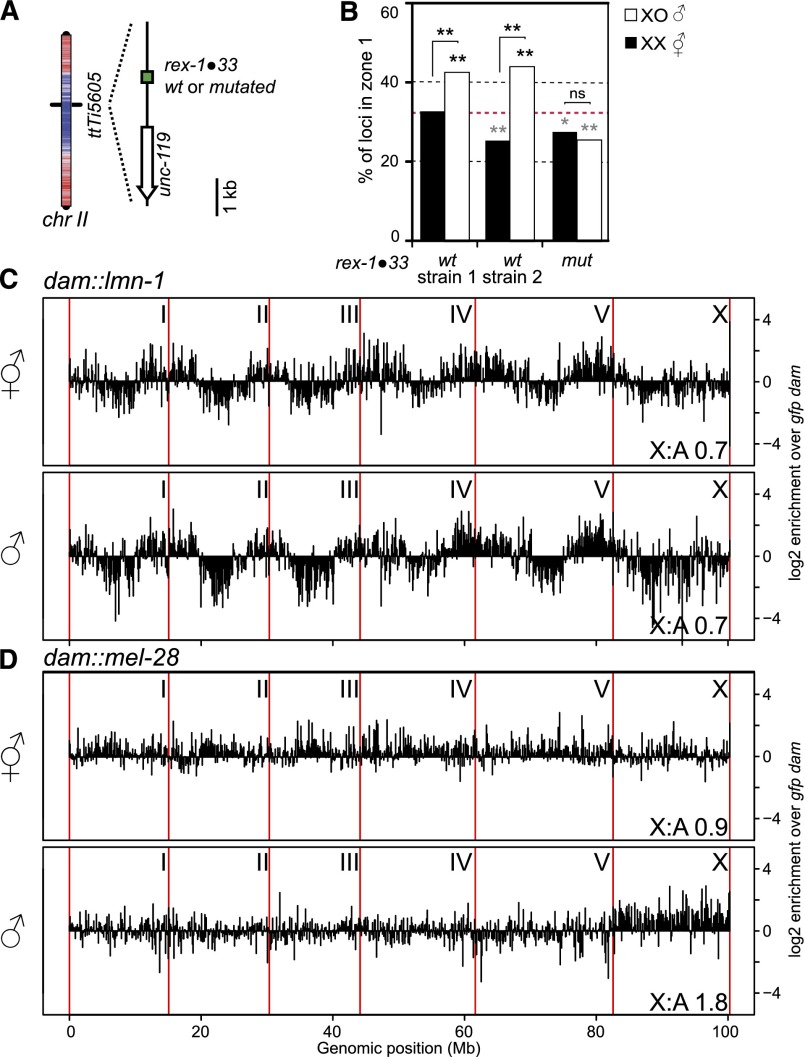
An ectopic MEX motif targets an autosomal locus to the nuclear periphery, and the male X chromosome shows widespread interaction with nuclear pores. (A) Location of the MEX motif insertion on chr II, with overlay of LEM-2 ChIP–chip profile (color as in Fig. 1B). (B) Zone 1 scorings for the insertion in A in two strains with a wild-type (WT) and one strain with a mutated rex site in male and hermaphrodite embryos. (C) DamID-seq profile of lamin interaction with the entire C. elegans genome in hermaphrodites and males. Chromosomes are separated by red vertical lines. The signal is averaged over 100-kb windows and normalized by free Dam-GFP control. (D) DamID profile as in C of nucleoporin MEL-28.
The nuclear periphery is a contrasted environment with silent chromatin anchored at the nuclear lamina (Ikegami et al. 2010; Towbin et al. 2012; Gonzalez-Aguilera et al. 2014) and active domains located close to nuclear pores by transcription-dependent mechanisms (Ikegami and Lieb 2013; Rohner et al. 2013). We therefore asked which domain the X chromosome is interacting with using DNA adenine methyltransferase identification (DamID) followed by sequencing (DamID-seq). Male or hermaphrodite L4 animals expressing Dam fusions to either lamin (LMN-1) (Towbin et al. 2012), the nucleoporin MEL-28/ELYS (Fernandez and Piano 2006; Galy et al. 2006), or GFP (diffusible control) were used to map genomic regions in close proximity to these nuclear landmarks. For lamin DamID in hermaphrodites, we obtained profiles very similar to previously published data, showing the characteristic high interaction pattern of the autosomal arms (Fig. 2C; Gonzalez-Aguilera et al. 2014). In males, the lamin DamID profile was remarkably similar to hermaphrodites, except for the X chromosome, which showed slightly more interactions with the nuclear lamina (Fig. 2C). For MEL-28/ELYS DamID in hermaphrodites, the signal was very similar between autosomes and the X chromosome (Fig. 2D). A previous study found discrete interaction sites of chromatin with nucleoporins (Ikegami and Lieb 2013); however, the resolution of our low animal number DamID impairs such fine-scale analysis. In males, however, at the chromosome scale, the single X chromosome shows broad interactions with MEL-28 (Fig. 2D). This is highly reminiscent of Drosophila males, in which the activating DCC (MSL/MOF) coating the X chromosome contains Nup153 and TPR/Mtor, two nucleoporins that show widespread interaction with the X chromosome (Mendjan et al. 2006; Vaquerizas et al. 2010). Upon loading, the MSL/MOF complex changes the chromatin conformation of X, although this conformation was unaffected upon knockdown of Nup153 or TPR/Mtor (Grimaud and Becker 2009). The interaction of nucleoporins with chromatin has been shown in a variety of systems to increase transcriptional output (Casolari et al. 2004; Capelson et al. 2010; Kalverda et al. 2010; Liang et al. 2013). While in these studies, a significant proportion of interactions between nucleoporins and chromatin took place in the nuclear interior, our FISH data suggest that the interactions with MEL-28 observed by DamID to a large extent represent events at nuclear pores.
Together, this allows us to draw a model in which the X chromosome in males is targeted to the nuclear rim by MEX motifs. Targeting to nuclear pores may be achieved through Pol III transcribed noncoding RNA (ncRNA) sequences (Ikegami and Lieb 2013), which are particularly enriched on the X chromosome (ratios of annotated ncRNA to coding sequences of 1.2 for X and 0.3 for all autosomes [WormBase data; http://www.wormbase.org]). Nuclear pore-mediated interaction has been shown in yeast to depend on incorporation of the histone variant HTZ1 (H2A.Z) (Light et al. 2010). Interestingly, the same variant HTZ-1 is underrepresented on hermaphrodite X chromosomes (Whittle et al. 2008; Petty et al. 2011), suggesting a possible mechanism for pore-mediated activation of X-linked genes in males by histone exchange and promotion of HTZ-1 incorporation.
In hermaphrodites, one hypothesis for the mechanism of transcriptional down-regulation of X chromosomes is that the DCC and H4K20me1 increase chromosome compaction by physically entangling chromatin, thereby impairing access of the RNA polymerases to X-linked genes (Chuang et al. 1994; Vielle et al. 2012). In this model, a given genomic length on the X chromosome should be more compact in dosage-compensated hermaphrodites than in males and more compact than a similarly sized autosomal region. To evaluate the difference in chromatin compaction levels, we analyzed the physical distance between pairs of sites separated by a genomic distance of 1 or 4 Mb (Fig. 3A; Supplemental Fig. S1A,B). Using projections along the Z-axis, we measured the projected distances between these loci in male and hermaphrodite embryos after DC establishment. Distances between loci were normalized to the nuclear diameter to correct for size differences between nuclei (Fig. 3B). Data in Figure 3 are presented as a cumulative frequency distribution (the proportion of measurements [in the Y-axis] below a given threshold [on the X-axis]) (alternative box plot representation in Supplemental Fig. S3). Irrespective of the sex of the animal and for both 1- and 4-Mb distances, regions located on the X chromosome were more compact than similar distances measured on an autosome (Fig. 3C,D). For autosomes, no difference in the distance distribution could be observed between males and hermaphrodites, although compaction slightly varied between the two autosomal regions probed. Comparing X chromosome measurements between hermaphrodites and males showed a slightly higher compaction in hermaphrodites (Fig. 3C,D), while X compaction is similar between XO hermaphrodites and XO males (Fig. 3E). This strongly suggests that the higher hermaphrodite X compaction is due to the DCC. Our measurements also imply a DCC-independent mechanism that increases compaction of the X chromosome relative to autosomes in males. This may be due to a non-sex-specific loading of non-DCC condensins, which have a clear preference for loading at ncRNA genes (Kranz et al. 2013), particularly abundant on the X chromosome. Alternatively, nuclear pore anchoring might also lead to increased compaction of the X chromosome (see below). Altogether, it appears unlikely that the small increase in compaction is solely responsible for the restriction of RNA Pol II access and transcription down-regulation of X-linked genes in hermaphrodites.
Figure 3.
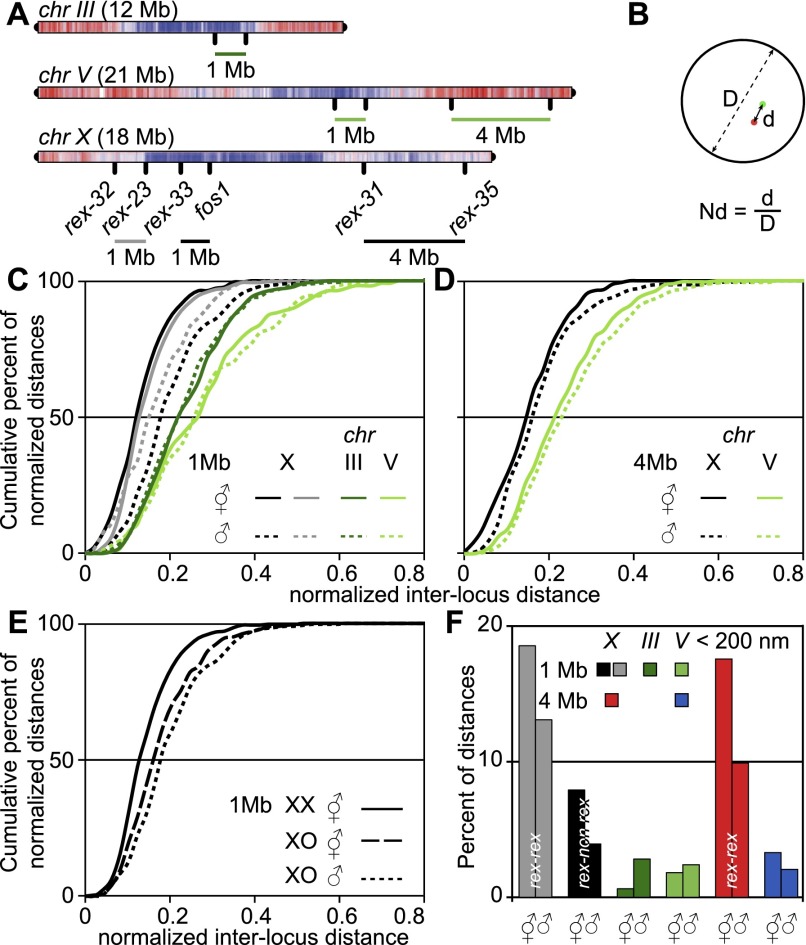
The X chromosome shows a higher degree of compaction compared with autosomes in males and hermaphrodites. (A) Probes used to assess chromatin compaction were rex-31/rex-35 (4 Mb, X), rex-32/rex-23 and rex33/fos1 (1 Mb, X), and controls on chr III and chr V (1 and 4 Mb). (B) Interlocus distance normalization to the nuclear diameter. (C,D) Cumulative frequency plot of normalized interlocus distances for 1- and 4-Mb probe combinations described in A. (E) Cumulative frequency plot of normalized interlocus distances for rex-33/fos1 in males, hermaphrodites, and non-dosage-compensated XO hermaphrodites. (F) Percentage of nonnormalized physical distances <200 nm measured in both sexes for all probe combinations. Statistical data are in Supplemental Table 1.
How is X compaction achieved? Random or rex site-specific interactions between chromosomal sites could be mediated by the DCC in hermaphrodites or the rex anchor sites at the nuclear rim in males, leading to the formation of a “rosette”-type structure in which clustered rex sites make the center of the rosette, while inter-rex sequences loop out (Jans et al. 2009; Chow and Heard 2010). When measuring absolute distances between two rex sites (rex-23/rex-32) and a rex and a non-rex site (rex-33/fos1, both 1 Mb distance) (Fig. 3A), the proportions of distances <200 nm was smaller between rex sites than between rex and non-rex sites for both males and hermaphrodites (Fig. 3F). Moreover, these proportions were higher in hermaphrodites than in males, reflecting the higher compaction observed in hermaphrodites. In line with the difference in compaction between autosomes and X, very few measurements <200 nm were observed for a 1- and a 4-Mb genomic distance on chr III or chr V (Fig. 3F). Given the low frequency of spatially close rex sites, DC is unlikely to function by generalized clustering of rex sites, but as rex sites are abundant on the X chromosome, a loose rosette organization might arise with a subset of randomly interacting rex sites (Bohn and Heermann 2010). Therefore, the increased colocalization of rex sites compared with rex–non-rex sites in hermaphrodites might be due to inter-rex interactions. Alternatively, the higher proportion of very close sites could be the result of the DCC-dependent global increase of X chromosome compaction. In males, the increased proportions of spatially close sites might reflect clustering to the same anchor site at the nuclear periphery.
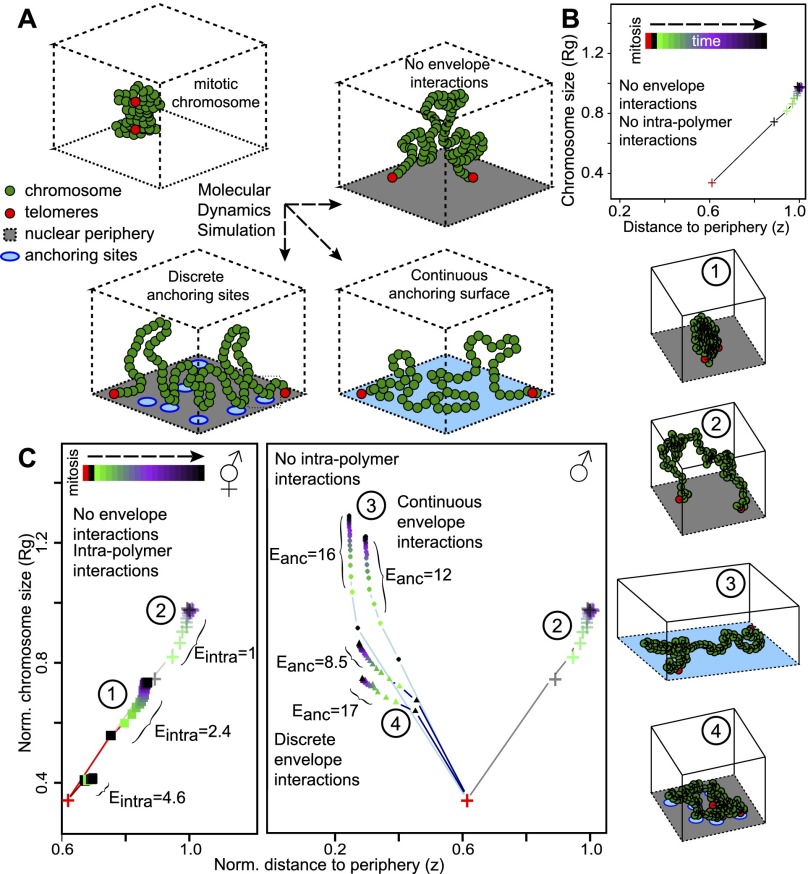
Our observation that the X chromosome is more compact than autosomes in both sexes is surprising. While in hermaphrodites, compaction of the X chromosome is likely achieved by the DCC, it remains unclear why this is the case in males. As the male X chromosome interacts with nucleoporins at the nuclear periphery, we asked whether anchoring could lead to compaction. We modeled the behavior of chromosomes as a bead spring polymer tethered at both telomeres to a surface representing the nuclear periphery (Fig. 4A; Supplemental Fig. S4; Kremer and Grest 1992). Using molecular dynamics simulations, we studied the time evolution of conformations (distance to the periphery [z] and size of the chromosome [Rg]) starting from a highly compact conformation: the mitotic chromosome (Fig. 4B). In the absence of interaction with the periphery (other than telomere tethering), higher self-affinity leads to more compact globular conformations (Fig. 4C, left, configurations 1 and 2; Doi and Edwards 1988). As condensins are known to cross-link chromatin fibers together, this could explain the observed compaction difference between autosomes and the hermaphrodite X chromosome. We then asked whether nuclear pore anchoring as in males would influence X compaction. In the absence of self-affinity, we introduced an affinity of the chromosome for anchor sites at the nuclear envelope. Unsurprisingly, this led to more peripheral conformations (Fig. 4C). When the chromosome was allowed to interact with any point on the surface, the interaction led to spreading (Fig. 4C, configuration 3; Metzger et al. 2003). In contrast, strong interaction with a limited number of anchor sites corresponding to the density of nuclear pores (about three pores per square micrometer) (Rohner et al. 2013) led to a limited spatial expansion of the chromosome compared with nonanchored chromosomes (Fig. 4C, right, configuration 4). The polymer is then stabilized in a compact conformation partly expanded from the dense mitotic state. As cells in embryos are rapidly dividing, steady state is not reached before the next cell division, and discrete anchoring of a chromosome to the nuclear pores entraps partially decondensed compact mitotic conformations. Modeling therefore indicates that strong chromosome anchoring to discrete sites is sufficient to explain the observed higher compaction of the X chromosome in males compared with autosomes.
Figure 4.
Theoretical modeling supports the competing roles of DCC loading and nuclear pore anchoring in X chromosome organization in males and hermaphrodites. (A) Bead spring polymeric model of chromosomes with intrachain attractive interactions and interactions with anchoring sites located at the nuclear envelope. Relaxation dynamics from an initial “mitotic-like” compact state were computed by molecular dynamics simulations. Chromosomal spatial organization is characterized by the typical size of the polymer, the radius of gyration (Rg), and the mean distance to the envelope (z). (B) Time evolution of Rg(t) and z(t) in the case of no anchoring and weak intrachain interactions. (C, left) Hermaphrodite situation: Rg(t) versus z(t) for different intrachain interactions: (+) E = 1; (▪) E = 2.4 and 4.6. As a result of DCC loading (Eintra > 1), the X chromosome folds into a more compact structure than autosomes (Eintra = 1). (Right) Male situation: Rg(t) versus z(t) for different anchoring conditions and weak intrachain interactions (Eintra = 1): (+) no interaction; (▲) interactions with discrete anchors (three sites per square micrometer); (●) continuous interactions with highly dense anchors. Example configurations for the different situations (1–4) are pictured at the right.
A structural model for DC
Many hypotheses were put forward to mechanistically explain how a condensin-like DCC hinders RNA Pol II recruitment to X-linked genes. Here we show that loading of the DCC onto the X chromosome impairs its perinuclear localization and interaction with nuclear pore proteins. Based on our experiments, we suggest a structural model for DC. In males, the X chromosome interacts with nuclear pore components, which increases the transcriptional output of X-linked genes compared with autosomes, observed in two genome-wide studies (Deng et al. 2011; Kruesi et al. 2013) and similar to the up-regulation observed in male mammalian cells and Drosophila (Deng et al. 2011). In hermaphrodites, loading of the DCC on the X chromosome (Csankovszki et al. 2004; Ercan et al. 2009; Vielle et al. 2012) impairs its perinuclear localization by masking pore interaction sites, thereby inhibiting pore-mediated gene activation and reducing transcription levels on a global chromosomal scale. Similar subnuclear localization-induced gene regulation has been observed in yeast, in which relocation of the HXK1 gene away from nuclear pores decreases transcriptional output of the gene (Taddei et al. 2006). The validity of this location-induced gene regulation for an entire chromosome remains to be tested. Our DC model offers a simple explanation for the observation that in hermaphrodites carrying X:autosome fusions, the DCC and associated H4K20 methylation spread from the X onto the autosome for megabases, but little difference was observed for the transcriptional levels of the autosomal genes (Ercan et al. 2009; Pferdehirt et al. 2011; Vielle et al. 2012). As the autosomal part of the chromosome fusion is devoid of rex sites, it does not undergo pore-mediated up-regulation; therefore, DCC binding is expected to have little effect on transcriptional levels. Altogether, this study leads to an updated model for DC in C. elegans, with a mechanism much more similar to Drosophila DC. As DC has evolved independently in Drosophila and nematodes, it is noticeable that in both species, DC leads to changes of the tridimensional structure of the chromosome, providing a paradigm to study the link between the creation of nuclear subdomains and the coordinated fine-tuning of the expression of a large number of genes.
Materials and methods
Strains and culture conditions
Nematodes were grown on NG2 medium seeded with OP50 at 22.5°C unless otherwise stated.
Probe preparation, FISH, and microscopy
Probe preparation, FISH, and microscopy are detailed in the Supplemental Material.
DamID
L4 worms expressing fusions to lmn-1, mel-28, or gfp, grown at 25°C, were lysed and digested with DpnI. Fragments were ligated to double-stranded adapters before amplification and library preparation. Sequencing data can be accessed at Gene Expression Omnibus: GSE56270.
Polymer modeling
A chromosome was modeled by a self-avoiding bead and spring chain confined in a cubic box in which one face is the nuclear envelope. Two types of interactions were considered: intrachain nonspecific interactions between beads and specific interactions between beads and sites at the nuclear envelope.
Supplementary Material
Acknowledgments
We thank members of the Meister and Ochsenreiter laboratories for numerous discussions, Sonia Karaz and Julie Campos for expert technical help, Mylène Docquier and the iGE3 genomics platform of the University of Geneva for next-generation sequencing (NGS) and advice, and Iskra Katic and the Friedrich Miescher Institute (FMI) C. elegans laboratories for help and discussions. Some strains were provided by the Caenorhabditis Genetics Center (CGC), which is funded by National Institutes of Health Office of Research Infrastructure Programs (P40 OD010440). Our laboratories are supported by the Swiss National Science Foundation (SNF assistant professor grant PP00P3_133744), the Swiss Foundation for Muscle Diseases Research, and the University of Bern (to P.M.); the French Centre National de la Recherche Scientifique and the Ecole Normale Supérieure de Lyon (to C.V. and D.J.); the Spanish Ministry of Economy and Competitiveness (BFU2010-15478), the European Regional Development Fund (to P.A.); Nederlandse Organisatie voor Wetenschappelijk Onderzoek (NWO)-Aard en Levenswetenschappen (ALW) Veni (to J.K.); and ZonMW (Medische wetenschappen)-TOP (to B.v.S.). D.J. and C.V. acknowledge the Pôle Scientifique de Modélisation Numérique for computing resources
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.248864.114.
References
- Askjaer P, Ercan S, Meister P. 2014. Modern techniques for the analysis of chromatin and nuclear organization in C. elegans. In WormBook (ed. The C. elegans Research Community) WormBook, doi: 10.1895/wormbook.1.169.1. http://www.wormbook.org. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohn M, Heermann DW. 2010. Diffusion-driven looping provides a consistent framework for chromatin organization. PLoS ONE 5: e12218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capelson M, Liang Y, Schulte R, Mair W, Wagner U, Hetzer MW. 2010. Chromatin-bound nuclear pore components regulate gene expression in higher eukaryotes. Cell 140: 372–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casolari JM, Brown CR, Komili S, West J, Hieronymus H, Silver PA. 2004. Genome-wide localization of the nuclear transport machinery couples transcriptional status and nuclear organization. Cell 117: 427–439. [DOI] [PubMed] [Google Scholar]
- Chow JC, Heard E. 2010. Nuclear organization and dosage compensation. Cold Spring Harb Perspect Biol 2: a000604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang PT, Albertson DG, Meyer BJ. 1994. DPY-27: a chromosome condensation protein homolog that regulates C. elegans dosage compensation through association with the X chromosome. Cell 79: 459–474. [DOI] [PubMed] [Google Scholar]
- Csankovszki G, McDonel P, Meyer BJ. 2004. Recruitment and spreading of the C. elegans dosage compensation complex along X chromosomes. Science 303: 1182–1185. [DOI] [PubMed] [Google Scholar]
- Csankovszki G, Collette K, Spahl K, Carey J, Snyder M, Petty E, Patel U, Tabuchi T, Liu H, McLeod I, et al. 2009. Three distinct condensin complexes control C. elegans chromosome dynamics. Curr Biol 19: 9–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawes HE, Berlin DS, Lapidus DM, Nusbaum C, Davis TL, Meyer BJ. 1999. Dosage compensation proteins targeted to X chromosomes by a determinant of hermaphrodite fate. Science 284: 1800–1804. [DOI] [PubMed] [Google Scholar]
- Deng X, Hiatt JB, Nguyen DK, Ercan S, Sturgill D, Hillier LW, Schlesinger F, Davis CA, Reinke VJ, Gingeras TR, et al. 2011. Evidence for compensatory upregulation of expressed X-linked genes in mammals, Caenorhabditis elegans and Drosophila melanogaster. Nat Genet 43: 1179–1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doi M, Edwards SF, ed. 1988. The theory of polymer dynamics. Oxford University Press, Oxford. [Google Scholar]
- Ercan S, Dick LL, Lieb JD. 2009. The C. elegans dosage compensation complex propagates dynamically and independently of X chromosome sequence. Curr Biol 19: 1777–1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez AG, Piano F. 2006. MEL-28 is downstream of the Ran cycle and is required for nuclear-envelope function and chromatin maintenance. Curr Biol 16: 1757–1763. [DOI] [PubMed] [Google Scholar]
- Ferrari F, Alekseyenko AA, Park PJ, Kuroda MI. 2014. Transcriptional control of a whole chromosome: emerging models for dosage compensation. Nat Struct Mol Biol 21: 118–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira HC, Towbin BD, Jegou T, Gasser SM. 2013. The shelterin protein POT-1 anchors Caenorhabditis elegans telomeres through SUN-1 at the nuclear periphery. J Cell Biol 203: 727–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galy V, Askjaer P, Franz C, Lopez-Iglesias C, Mattaj IW. 2006. MEL-28, a novel nuclear-envelope and kinetochore protein essential for zygotic nuclear-envelope assembly in C. elegans. Curr Biol 16: 1748–1756. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Aguilera C, Ikegami K, Ayuso C, de Luis A, Iniguez M, Cabello J, Lieb JD, Askjaer P. 2014. Genome-wide analysis links emerin to neuromuscular junction activity in Caenorhabditis elegans. Genome Biol 15: R21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimaud C, Becker PB. 2009. The dosage compensation complex shapes the conformation of the X chromosome in Drosophila. Genes Dev 23: 2490–2495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikegami K, Lieb JD. 2013. Integral nuclear pore proteins bind to Pol III-transcribed genes and are required for Pol III transcript processing in C. elegans. Mol Cell 51: 840–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikegami K, Egelhofer TA, Strome S, Lieb JD. 2010. Caenorhabditis elegans chromosome arms are anchored to the nuclear membrane via discontinuous association with LEM-2. Genome Biol 11: R120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jans J, Gladden JM, Ralston EJ, Pickle CS, Michel AH, Pferdehirt RR, Eisen MB, Meyer BJ. 2009. A condensin-like dosage compensation complex acts at a distance to control expression throughout the genome. Genes Dev 23: 602–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalverda B, Pickersgill H, Shloma VV, Fornerod M. 2010. Nucleoporins directly stimulate expression of developmental and cell-cycle genes inside the nucleoplasm. Cell 140: 360–371. [DOI] [PubMed] [Google Scholar]
- Kranz AL, Jiao CY, Winterkorn LH, Albritton SE, Kramer M, Ercan S. 2013. Genome-wide analysis of condensin binding in Caenorhabditis elegans. Genome Biol 14: R112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremer K, Grest GS. 1992. Simulations for structural and dynamic properties of dense polymer systems. J Chem Soc, Faraday Trans 88: 1707–1717. [Google Scholar]
- Kruesi WS, Core LJ, Waters CT, Lis JT, Meyer BJ. 2013. Condensin controls recruitment of RNA polymerase II to achieve nematode X-chromosome dosage compensation. eLife 2: e00808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Y, Franks TM, Marchetto MC, Gage FH, Hetzer MW. 2013. Dynamic association of NUP98 with the human genome. PLoS Genet 9: e1003308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Light WH, Brickner DG, Brand VR, Brickner JH. 2010. Interaction of a DNA zip code with the nuclear pore complex promotes H2A.Z incorporation and INO1 transcriptional memory. Mol Cell 40: 112–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonel P, Jans J, Peterson BK, Meyer BJ. 2006. Clustered DNA motifs mark X chromosomes for repression by a dosage compensation complex. Nature 444: 614–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendjan S, Taipale M, Kind J, Holz H, Gebhardt P, Schelder M, Vermeulen M, Buscaino A, Duncan K, Mueller J, et al. 2006. Nuclear pore components are involved in the transcriptional regulation of dosage compensation in Drosophila. Mol Cell 21: 811–823. [DOI] [PubMed] [Google Scholar]
- Metzger S, Muller M, Binder K, Baschnagel J. 2003. Surface excess in dilute polymer solutions and the adsorption transition versus wetting phenomena. J Chem Phys 118: 8489–8499. [Google Scholar]
- Petty E, Laughlin E, Csankovszki G. 2011. Regulation of DCC localization by HTZ-1/H2A.Z and DPY-30 does not correlate with H3K4 methylation levels. PLoS ONE 6: e25973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pferdehirt RR, Kruesi WS, Meyer BJ. 2011. An MLL/COMPASS subunit functions in the C. elegans dosage compensation complex to target X chromosomes for transcriptional regulation of gene expression. Genes Dev 25: 499–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohner S, Kalck V, Wang X, Ikegami K, Lieb JD, Gasser SM, Meister P. 2013. Promoter- and RNA polymerase II-dependent hsp-16 gene association with nuclear pores in Caenorhabditis elegans. J Cell Biol 200: 589–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strome S, Kelly WG, Ercan S, Lieb JD. 2014. Regulation of the X chromosomes in Caenorhabditis elegans. Cold Spring Harb Perspect Biol 6: a018366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taddei A, Van Houwe G, Hediger F, Kalck V, Cubizolles F, Schober H, Gasser SM. 2006. Nuclear pore association confers optimal expression levels for an inducible yeast gene. Nature 441: 774–778. [DOI] [PubMed] [Google Scholar]
- Towbin BD, Gonzalez-Aguilera C, Sack R, Gaidatzis D, Kalck V, Meister P, Askjaer P, Gasser SM. 2012. Step-wise methylation of histone H3K9 positions heterochromatin at the nuclear periphery. Cell 150: 934–947. [DOI] [PubMed] [Google Scholar]
- Vaquerizas JM, Suyama R, Kind J, Miura K, Luscombe NM, Akhtar A. 2010. Nuclear pore proteins nup153 and megator define transcriptionally active regions in the Drosophila genome. PLoS Genet 6: e1000846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vielle A, Lang J, Dong Y, Ercan S, Kotwaliwale C, Rechtsteiner A, Appert A, Chen QB, Dose A, Egelhofer T, et al. 2012. H4K20me1 contributes to downregulation of X-linked genes for C. elegans dosage compensation. PLoS Genet 8: e1002933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells MB, Snyder MJ, Custer LM, Csankovszki G. 2012. Caenorhabditis elegans dosage compensation regulates histone H4 chromatin state on X chromosomes. Mol Cell Biol 32: 1710–1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittle CM, McClinic KN, Ercan S, Zhang X, Green RD, Kelly WG, Lieb JD. 2008. The genomic distribution and function of histone variant HTZ-1 during C. elegans embryogenesis. PLoS Genet 4: e1000187. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.