Fong et al. examined cotranscriptional pre-mRNA splicing using RNA polymerase II mutants that change average elongation rates genome-wide. Slow and fast elongation affected constitutive and alternative splicing and often both increased or both decreased inclusion of a particular exon or retained intron. These results suggest that an optimal rate of transcriptional elongation is required for normal cotranscriptional pre-mRNA splicing.
Keywords: alternative splicing, intron retention, kinetic coupling, transcription elongation rate, cotranscriptional splicing
Abstract
Alternative splicing modulates expression of most human genes. The kinetic model of cotranscriptional splicing suggests that slow elongation expands and that fast elongation compresses the “window of opportunity” for recognition of upstream splice sites, thereby increasing or decreasing inclusion of alternative exons. We tested the model using RNA polymerase II mutants that change average elongation rates genome-wide. Slow and fast elongation affected constitutive and alternative splicing, frequently altering exon inclusion and intron retention in ways not predicted by the model. Cassette exons included by slow and excluded by fast elongation (type I) have weaker splice sites, shorter flanking introns, and distinct sequence motifs relative to “slow-excluded” and “fast-included” exons (type II). Many rate-sensitive exons are misspliced in tumors. Unexpectedly, slow and fast elongation often both increased or both decreased inclusion of a particular exon or retained intron. These results suggest that an optimal rate of transcriptional elongation is required for normal cotranscriptional pre-mRNA splicing.
The cellular transcription and mRNA processing machineries cooperate in order to couple the reactions they perform in space and time. Coupling in space comprises assembly of processing complexes at the site of transcription through “recruitment” mechanisms that often involve binding to the RNA polymerase II (Pol II) C-terminal domain (CTD) (Bentley 2014). Coupling in time means that processing complexes assemble and often carry out their reactions before transcription is completed (Beyer and Osheim 1988; Wetterberg et al. 1996; Pandya-Jones and Black 2009; Han et al. 2011; Khodor et al. 2011; Tilgner et al. 2012). It is not known whether temporal coupling is an accident of similar kinetics for transcription and processing or instead is a result of evolutionary selection. Because transcription is polar, it enforces order on cotranscriptional events; for example, upstream introns tend to be spliced before downstream introns in a “first come, first served” (Aebi et al. 1986) manner. Transcription elongation rate could in theory affect cotranscriptional pre-mRNA processing by setting the delay between synthesis of elements in the nascent RNA that compete for protein factors or complementary RNA sequences. Slow elongation will lengthen the window of opportunity for an upstream event to occur on the nascent RNA before facing competition from a sequence element further downstream (Kornblihtt et al. 2004; Dujardin et al. 2013; Bentley 2014).
Alternative splicing occurs on >90% of human pre-mRNAs (Wang et al. 2008; Nilsen and Graveley 2010), and its disruption causes misexpression of many genes in cancer and other disease states (Venables et al. 2009; David and Manley 2010; Germann et al. 2012; Wang et al. 2012a). If alternative splice sites compete for assembly of committed complexes, then slow elongation will favor processing at upstream sites, resulting in inclusion of alternative cassette exons (de la Mata et al. 2003). Conversely, fast elongation is predicted to reduce the competitive advantage of upstream splice sites and favor exon skipping (Fig. 1A). It has also been suggested that the “window of opportunity” model can influence alternative splicing by biasing competition between splicing enhancers and silencers (Dujardin et al. 2014).
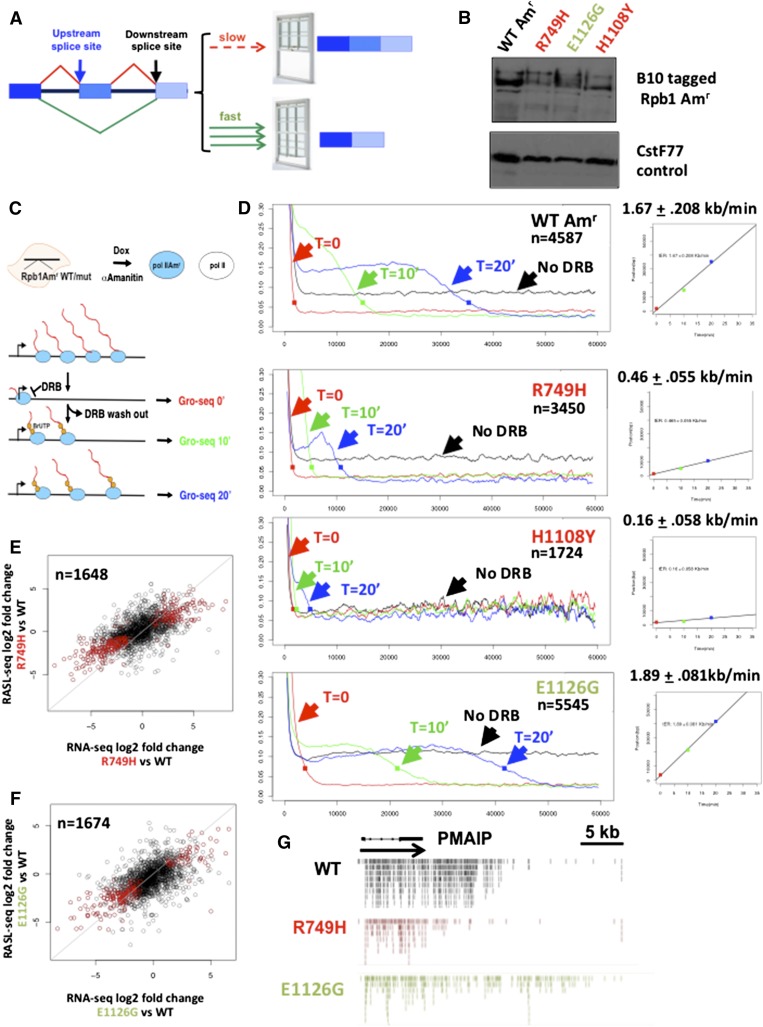
Figure 1.
Testing how elongation rate affects alternative splicing. (A) The “window of opportunity” model for how elongation rate affects alternative splicing. Slow elongation expands the window for recognition of the upstream 3′ splice site before it must compete with the downstream site, thereby favoring exon inclusion over skipping. (B) Expression of epitope-tagged human wild-type (WT) Amr and mutants with slow (red) and fast (green) elongation rates. Western blot of extracts from doxycycline-induced HEK293 Flp-in cell lines with anti B10 antibody and CstF77 loading control. (C) Scheme for measurement of elongation rates. Amanitin-resistant Rpb1 expression was induced with doxycycline, and cells were then treated with α-amanitin for 42 h to inhibit endogenous Pol II. Transcription was inhibited with DRB, which arrests Pol II at the TSS. Nuclei were harvested from DRB-inhibited cells (t = 0) and at 10 min (t = 10) and 20 min (t = 20) after washing out the inhibitor. Nuclei were processed for GRO-seq with BrUTP labeling of nascent transcripts. (D) Profiles of mean GRO-seq read counts for groups of the most highly transcribed genes with smoothing over 1-kb bins. Average elongation rates were calculated from the composite plots. (E,F) Correlation between fold change in cassette exon inclusion/exclusion in slow and fast Pol II mutants versus wild type as determined by RNA-seq and RASL-seq of α-amanitin-treated cells. Red dots correspond to exons with significant changes in inclusion (FDR < 0.05). (G) GRO-seq reads on the highly transcribed PMAIP gene that exhibits elongation-sensitive splicing (CI) (Supplemental Table 2) at the 10-min time point after DRB washout. Note that reads from the slow R749H mutant do not extend as far downstream from the TSS as wild type and that reads from the fast E1126G mutant extend further than wild type.
Other regulatory influences on alternative splicing operate independently of elongation rate, and the general importance of kinetic effects on alternative splicing is an open question (Lynch 2006; House and Lynch 2008; Nilsen and Graveley 2010). “Kinetic competition” between the splice sites was proposed on the basis of experiments using a slow Pol ΙΙ mutant (C4/R749H) that stimulates inclusion of alternative exons in Fibronectin (de la Mata et al. 2003) and several other genes (Ip et al. 2011). It is not known whether elongation rate can affect alternative intron retention (Bell et al. 2010; Bicknell et al. 2012; Wong et al. 2013), but this possibility is suggested by the fact that constitutive splicing in yeast and Drosophila is improved by a slow Pol II mutant and, in yeast, is inhibited by a fast mutant (Khodor et al. 2011; Braberg et al. 2013). A caveat of previous studies, however, is that it is not known whether the mutations that alter Pol II elongation rate on naked DNA templates have similar effects on natural genes in vivo.
How generally elongation rate alters alternative splicing outcomes and how well the “window of opportunity” model accounts for these effects are poorly understood. It is also unclear whether faster elongation has the opposite effect on splicing of slower elongation as predicted by the model. In vivo elongation rates of wild-type Pol II vary between ∼0.5 and >4 kb/min and may be regulated (Danko et al. 2013; Veloso et al. 2014), but it is not known whether cotranscriptional alternative splicing decisions are sensitive to changes in elongation rate within this range. We examined splicing in human cells expressing Rpb1 point mutants that either accelerate or decelerate average rates of elongation. Our results show that the “window of opportunity” model can account for only a fraction of the effects of elongation rate on splicing. Surprisingly, we found that fast and slow mutants often disrupt splicing in the same way, suggesting that an optimal rate of elongation is required to achieve a normal balance of spliced isoforms.
Results
In vivo validation of human Pol II elongation rate mutants
To investigate how transcription rate affects splicing in human cells, we constructed cell lines that express mutants (Fig. 1B) in conserved residues of the human Pol II large subunit (Rpb1) implicated in elongation control (Supplemental Fig. 1A). We previously made a human version of the slow Drosophila C4 mutant (Chen et al. 1996) by substituting R749H in the funnel domain (de la Mata et al. 2003). This mutation was introduced into an expression vector for α-amanitin-resistant (N792D) Rpb1 that we refer to as wild-type Amr. A similar strategy was used to make human versions of two well-characterized mutations in the trigger loop of yeast Rpb1 that accelerated or decelerated transcription in vitro (Malagon et al. 2006; Kaplan et al. 2008). H1085Y in the yeast Rpb1 mutates a contact with the β-phosphate of incoming NTPs and slows transcription. E1103G in the yeast protein stabilizes a triple α helix of the trigger loop and bridge helix and accelerates transcription with a loss of fidelity that is partially rescued by α-amanitin (Kaplan et al. 2008; Kireeva et al. 2008). We made isogenic doxycycline-inducible HEK293 cell lines of the homologous H1108Y and E1126G mutants as well as C4/R749H by Flp-mediated integration. The mutant proteins were expressed at levels similar to that of wild-type Amr (Fig. 1B).
It is important to verify that the effects of Pol II mutants on elongation rate in vitro also apply to chromosomal genes in vivo. To determine in vivo elongation rates, we used reversible inhibition of transcription by DRB (Singh and Padgett 2009) combined with genome-wide nuclear run-on sequencing (GRO-seq) (Fig. 1C; Core et al. 2008). Cells were induced with doxycycline and then treated with α-amanitin for 42 h so that the only Pol II engaged on genes was the resistant mutant (see the Materials and Methods). DRB was then added to arrest the Amr polymerases at the transcription start site (TSS) and clear them from gene bodies. When the DRB is washed out, a wave of transcription travels through genes, and we followed its progress by sequencing nascent RNA from nuclei harvested 10 and 20 min after the medium change. The average elongation rate of wild-type Amr Pol II under these conditions is ∼1.7 kb/min (Fig. 1D), in good agreement with previous estimates (Boireau et al. 2007; Jonkers et al. 2014; Veloso et al. 2014). Although we cannot exclude the possibility that the N792D mutation has some effect on elongation rate, we compared this “wild-type” Amr Pol II with the mutants under the same assay conditions. The R749H mutation reduced the average elongation rate to ∼0.5 kb/min, and H1108Y caused an even more severe elongation defect (Fig. 1D). It is possible that the latter mutation inhibits recovery from a DRB block. Conversely, the E1126G mutation caused a small but significant increase in the average elongation rate to ∼1.9 kb/min. Similar results were obtained using a smaller subset of common genes for meta-analysis (Supplemental Fig. 2A). The effects of the mutants on the average elongation rates are supported by the results for individual highly expressed genes (Fig. 1G; Supplemental Fig. 1B–F). In summary, these experiments establish that mutations in conserved residues of yeast and fly Rpb1 faithfully maintain their altered elongation properties when transposed into human Pol II. These results therefore validate our experimental approach by showing that the in vitro elongation phenotypes of the Pol II mutants apply widely in vivo on natural genes. In addition to their effects on elongation rate, the mutants could have additional unknown effects on polymerase function but are unlikely to affect protein:protein interactions with Pol II directly, as they are buried in the core of the enzyme.
Elongation rate mutants profoundly affect both exon inclusion and skipping
We tested the “window of opportunity” model (Fig. 1A) by RNA sequencing (RNA-seq) of polyA+ RNA from wild-type Amr and Rpb1 mutant cells treated with α-amanitin for 42 h. Sequence reads were analyzed using a negative binomial generalized linear model (GLM) implemented by a modified edgeR approach (see the Materials and Methods; Robinson et al. 2010) to identify changes in alternative exon inclusion. Exons whose splicing conforms to the simplest form of the “window of opportunity” model (Fig. 1A) would be included when elongation is slow (slow-included [SI]) and excluded when it is fast (fast-excluded [FE]), and we refer to these as type I. Exons that do not conform with the model would be slow-excluded (SE) and fast-included (FI), and we refer to them as type II. Consistent with the model, we identified 1007 examples (false discovery rate [FDR] < 0.05) of enhanced inclusion in the C4/R749H slow mutant (type I) relative to wild-type Amr out of 13,294 events sampled with more than nine total reads (Fig. 2A; Table 1). Note that under these conditions, changes in the abundance of long-lived splicing isoforms are likely to be underestimated. Unexpectedly, we found an even greater number of type II cases, 1148, in which exon skipping was enhanced by the slow mutant (Fig. 2A). A similar ratio of enhanced exon inclusion to skipping in C4/R749H was obtained by MATS analysis (Shen et al. 2012) of the same RNA-seq data sets. In two biological replicates, MATS identified 192 and 244 cases (P < 0.05) of enhanced inclusion (159 in common) and 192 and 265 cases (129 in common) of enhanced skipping in the slow mutant. In contrast, no significant differences in alternative exon inclusion were found between the replicates of the wild-type Amr RNA. MATS identified fewer examples in part because it filters out exons with <10% inclusion. We conclude that, contrary to the prediction of the simple “window of opportunity” model (Fig. 1A), slow Pol II elongation enhanced skipping of alternative exons as frequently as inclusion.
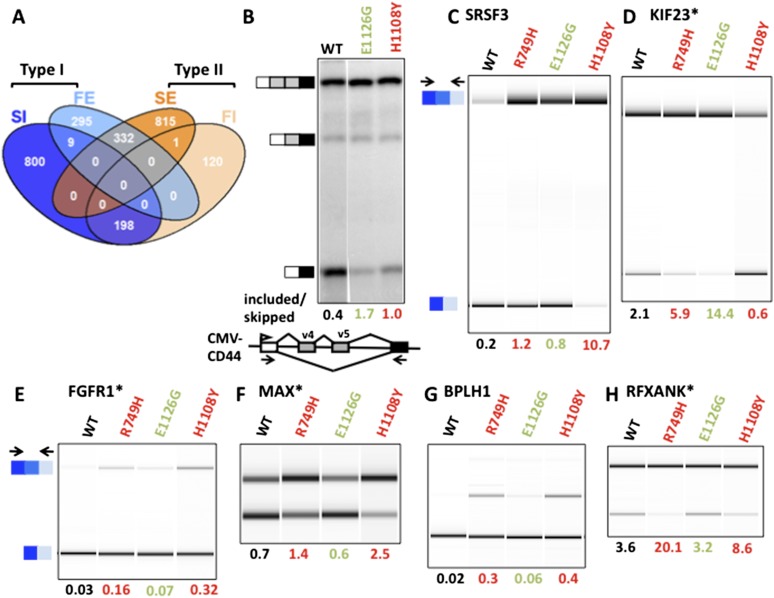
Figure 2.
Enhanced exon inclusion in fast and slow Pol II mutants. (A) Venn diagram of elongation rate-sensitive cassette exons detected by RNA-seq of polyA+ RNA from α-amanitin-treated cells expressing fast (E1126G) and slow (C4/R749H) mutants compared with wild-type (WT) Amr Pol II. Events that are significantly different (FDR < 0.05) in mutants relative to wild-type Amr were identified by the GLM likelihood ratio test (see the Materials and Methods). See also Table 1. Note extensive overlap between SI and FI (common included [CI]) and between SE and FE (common excluded [CE]) exons. (B) Fast and slow Pol II trigger loop mutants both increase inclusion of the alternatively spliced CD44 exons v4 v5. RT–PCR analysis of transcripts from α-amanitin-treated HEK293 cells transfected with the pcDNA3-CD44 minigene (Stickeler et al. 2001) and expression vectors for wild type and slow and fast Rpb1 mutants. 32P-labeled RT–PCR products were quantified by PhosphorImager and normalized for 32P- content. (C–H) Validation of selected examples of enhanced cassette exon inclusion in the slow mutant, C4/R749H. RT–PCR was performed on total RNA from α-amanitin-treated wild-type and Pol II mutant cells, and products were visualized and quantified by Bioanalyzer (Agilent). Primer coordinates and sequences are in Supplemental Table 1. Biological replicates are shown in Supplemental Figure 3. Note that in most cases, these exons are also more included in the H1108Y slow mutant and remarkably are often also more included in the fast E1126G mutant. Exons that are misspliced in cancer cells (Venables et al. 2009) are marked with an asterisk.
Table 1.
Elongation rate-sensitive cassette exon and retained intron splicing events detected by RNA-seq and RASL-seq of polyA+ RNA from α-amanitin-treated cells expressing fast (E1126G) and slow (C4/R749H and H1108Y) mutants and wild-type Amr Pol II
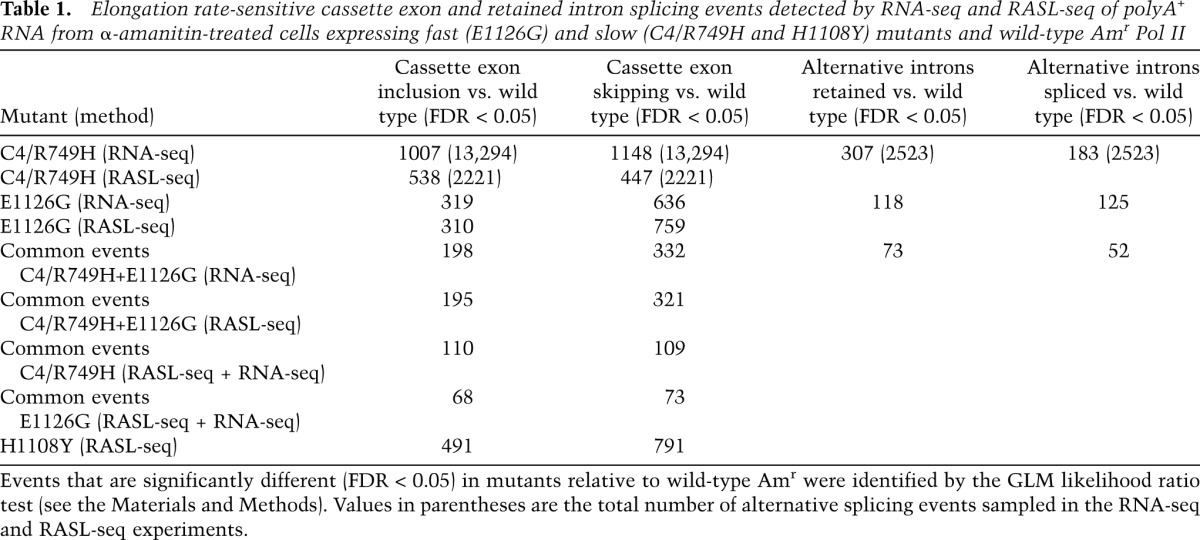
The fast E1126G mutant also caused numerous changes in alternative splicing of cassette exons. By edgeR analysis, we identified 636 exons where skipping was enhanced relative to wild type (type I), consistent with the “window of opportunity” model. On the other hand, E1126G also promoted inclusion of 319 exons (type II) (Fig. 2A; Table 1). MATS analysis also revealed an ∼2:1 ratio of enhanced skipping over inclusion in this fast mutant. In some cases, alternative splicing outcomes could be affected indirectly by altered expression of splicing or mRNA stability factors (e.g., SRSF3) (Fig. 2C) in cells expressing mutant Pol II. Most changes in the abundance of 173 splicing-related mRNAs (Supplemental Table 2) were less than twofold, however (Supplemental Fig. 2B,C).
To assess alternative splicing of cassette exons by an independent method, we conducted RASL-seq (RNA-mediated oligonucleotide annealing, selection, and ligation with next-generation sequencing) of polyA+ RNA from α-amanitin-treated cells expressing wild-type Amr and C4/R749H, H1108Y, and E1126G mutants. This method queries splicing of cassette exons by hybridization of oligonucleotides abutting alternative splicing junctions followed by ligation, PCR amplification, and high-throughput sequencing of the products (Li et al. 2012; Zhou et al. 2012). We analyzed 2221 cassette exons with more than nine sequencing reads for both the long and short forms in two biological replicates of the wild-type Amr and the mutants by the edgeR method. The results in Table 1 and Supplemental Table 2 confirmed that the C4/R749H slow mutant enhanced both inclusion (538 exons; FDR < 0.05) and skipping (447 exons), relative to wild type. The slow H1108Y mutant also frequently elevated exon inclusion (491 exons) and skipping (791 exons) relative to wild type (Table 1; Supplemental Table 2). In agreement with the RNA-seq results, RASL-seq of the fast E1126G mutant revealed a bias in favor of enhanced exon skipping (759 exons) over inclusion (310 exons). Overall, there was a good correlation between the fold changes in cassette exon inclusion detected by RNA-seq and RASL-seq for C4/R749H and E1126G relative to wild type (Fig. 1E,F).
Slow and fast elongation rates have overlapping effects on alternative exon inclusion
Unexpectedly, we observed many instances in which exon inclusion was perturbed in the same way by both the slow and fast Pol II mutants. Of the 319 exons detected by RNA-seq with enhanced inclusion in the fast E1126G mutant, 198 were also more highly included in the slow C4/R749H mutant (Table 1; Fig. 2A). This observation was reproduced in a transfected CD44 minigene driven by the CMV promoter (Stickeler et al. 2001), where inclusion of alternative exons v4 and v5 was favored by both the E1126G and H1108Y mutants (Fig. 2B). This result also shows that effects of these Pol II mutants on alternative splicing can operate independently of natural promoters. Similarly, of the 636 exons skipped in the fast mutant, 332 were also skipped in the slow mutant (Figs. 2A, 3; Table 1). RASL-seq confirmed the extensive overlap between cassette exons with enhanced inclusion (common included [CI]) or skipping (common excluded [CE]) relative to wild type in both the fast and slow Pol II mutants (Supplemental Tables 1, 3). Together, the RNA-seq and RASL-seq experiments detected 581 cases of enhanced skipping and 368 cases of enhanced inclusion that were common to the fast and slow mutants. In contrast, we found only 108 cases (only 10 by RNA-seq) (Fig. 2A) in which slow and fast elongation had opposite effects on inclusion of a cassette exon.
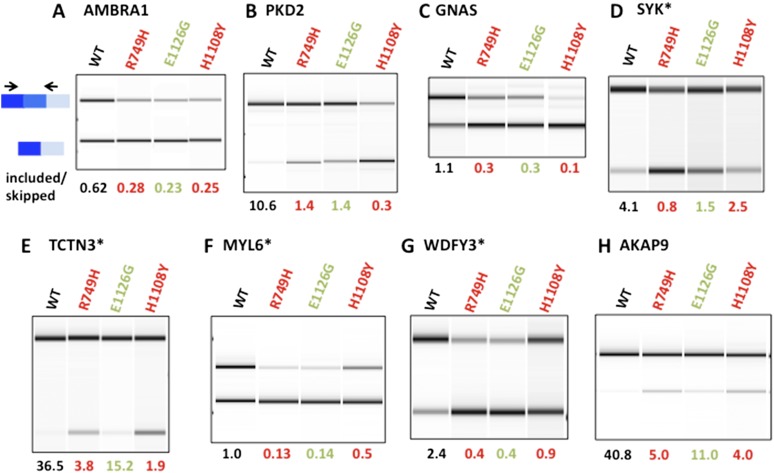
Figure 3.
Enhanced exon skipping in fast and slow Pol II mutants. Validation of selected examples of cassette exons with enhanced skipping in the slow mutant, C4/R749H. RT–PCR was as in Figure 2, and biological replicates are shown in Supplemental Figure 4. Note that in most cases, skipping of these exons is also promoted by the H1108Y slow mutant and the fast E1126G mutant. Exons that are misspliced in cancer cells (Venables et al. 2009) are marked with an asterisk.
We tested a subset of the affected cassette exons identified by RNA-seq and RASL-seq using RT–PCR with flanking primers. The sequencing results were confirmed in 60 out of 60 cases tested where RNA abundance permitted detection of both spliced isoforms (Supplemental Table 1). This high rate of validation is consistent with the stringent criteria used to identify affected cassette exons. The examples in Figures 2 and 3 illustrate enhanced exon inclusion and skipping, respectively, in the slow and fast mutants. These results were reproduced in biological replicate RNA samples (Supplemental Figs. 3, 4). The two slow mutants, C4/R749H and H1108Y, usually altered splicing of a given cassette exon in the same direction relative to wild type (49 out of 60 cases) but often differed in the magnitude of the effect (Figs. 2, 3; Supplemental Figs. 3, 4). On the other hand, the two slow mutants sometimes had opposite effects on exon inclusion (Fig. 2D; Supplemental Fig. 3C), underlining the specificity of splicing phenotypes associated with different Pol II mutants. In some cases, the effects of the slow Pol II mutants on cassette exon inclusion were mimicked by overexpression of a TFIIS dominant-negative mutant (R Sheridan, N Fong, DL Bentley, in prep.) that is predicted to impede elongation by a different mechanism through impaired backtracking (Sigurdsson et al. 2010). Several examples in which slow and fast elongation had opposite effects on inclusion of a cassette exon were also validated by RT–PCR (Fig. 4A–D; Supplemental Fig. 5A–D). Unlike our previous results with a minigene in Hep3B cells (de la Mata et al. 2003), inclusion of the fibronectin EDI exon was not altered significantly by slow or fast Pol II mutants in HEK293 lines (Supplemental Fig. 5E), reflecting possible cell type specificity or a difference between endogenous and transfected genes.
Figure 4.
Properties of different functional classes of elongation rate-sensitive exons. (A–D) A few cassette exons are included in slow mutants and skipped in fast mutants. RT–PCR products are shown as in Figure 2, and biological replicates are shown in Supplemental Figure 5, A–D. (E) Functional classes of elongation rate-sensitive cassette exons differ in exon and flanking intron length. Exons were classified according to the effects of the slow C4/R749H and fast E1126G mutants on their inclusion in RNA-seq experiments (Supplemental Table 2). Boxes show median and interquartile range. CI, n = 198; CE, n = 332; SI, n = 809; SE, n = 816; FI, n = 121; FE, n = 304; unaffected (U), n = 10,714. Note that the type I SI and FE exons (blue) have shorter flanking introns than unaffected (purple) or type II SE and FI exons (brown). The significance of differences in mean lengths of exons and flanking introns relative to unaffected exons were calculated by the Wilcoxon rank sum test. (*) P-value < 0.05; (**) P-value < 0.01; (***) P-value < 0.001. (F) Functional classes of elongation rate-sensitive cassette exons (as in E) differ in the strengths of their flanking splice sites. Splice site strength was calculated according to Yeo and Burge (2004). Note that the type I SI and FE exons have weaker 3′ splice sites than unaffected or type II SE and FI exons marked as in E. Box plots are as in E. Differences from unaffected exons are P-value < 0.05 (*) and P-value < 0.001 (***); Wilcoxon rank sum test.
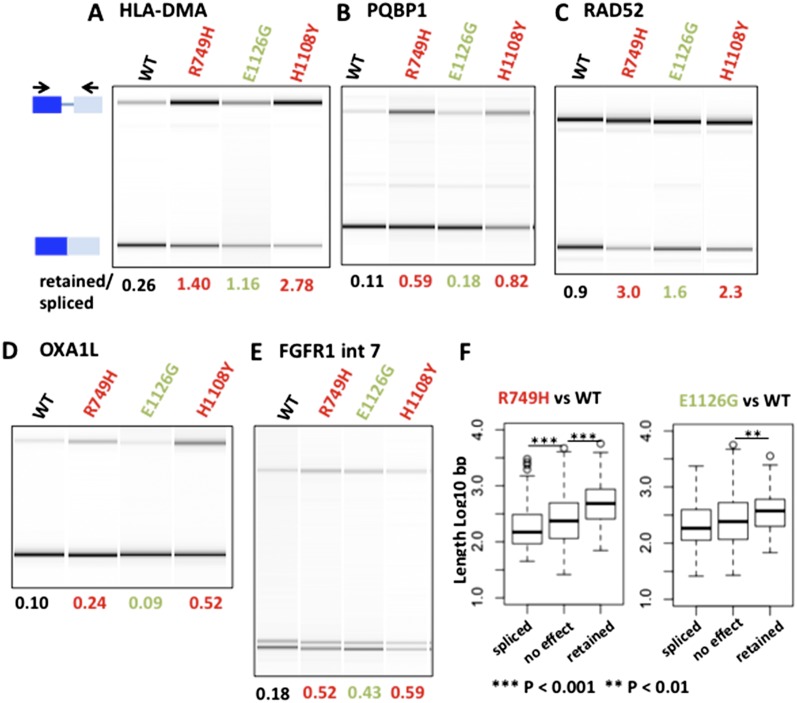
A number of elongation rate-sensitive exons were previously identified as abnormally spliced in cancer cells. Notably, the 405 previously identified exon-skipping events that are misregulated in breast and ovarian tumors (Venables et al. 2009) are significantly enriched for elongation rate-sensitive cassette exons over unaffected cassette exons in our RNA-seq analysis (P-value = 5.628 × 10−6, one-sided Fisher’s exact test). We identified 106 elongation rate-sensitive exons (Supplemental Table 2) in HEK293 cells that are abnormally included or skipped in these tumors relative to normal tissue. Ninety-three of these exons are sensitive to slow elongation (60 are excluded, and 33 are included), and 44 are sensitive to fast elongation (37 are excluded, and seven are included). The elongation rate effects on several cassette exons misregulated in tumors were validated by RT–PCR (Figs. 2, 3; Supplemental Figs. 3, 4), including the transcription factor MAX, the autophagy regulator WDFY3, the hedgehog signal transducer TCTN3, the tyrosine kinase tumor suppressor SYK, and fibroblast growth factor receptor FGFR1. These exons could represent a particular subset in which the splicing reaction is close to a “tipping point” that is easily perturbed by elongation rate or cellular transformation. An alternative possibility is that abnormal elongation could underlie some of the missplicing of cassette exons in tumor cells.
In summary, the sequencing and RT–PCR results (Table 1; Figs. 2, 3) show that a simple “window of opportunity” model (Fig. 1A) does not easily account for many of the changes in cassette exon splicing caused by Pol II elongation rate mutants. Contrary to the model, slow and fast elongation usually do not have opposite effects on the inclusion of a given alternative exon. Instead, many of the alternative cassette exon-splicing decisions that are sensitive to elongation rate are biased in the same direction when elongation rate deviates above or below that of wild-type Amr Pol II.
Elongation rate-dependent alternative splicing events are marked by distinct intron lengths and sequence motifs
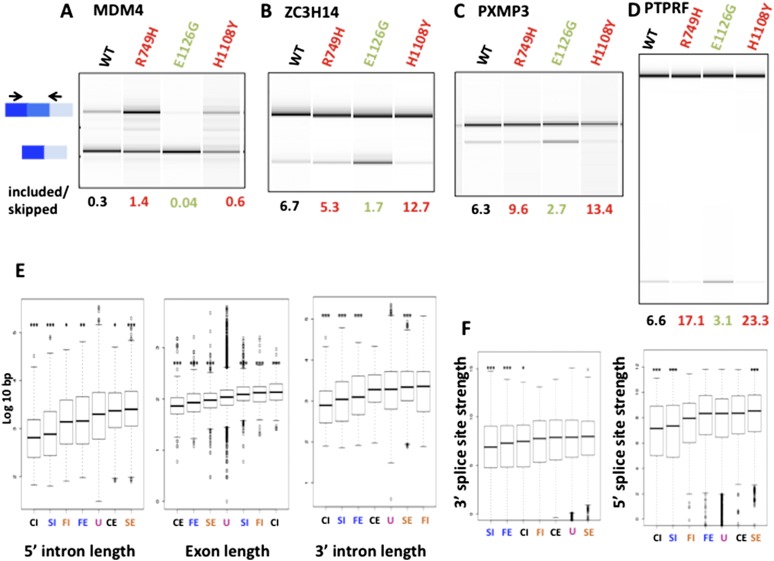
To identify features associated with elongation rate-dependent exon inclusion or skipping, we examined intron length because it is a potentially important determinant of the delay between the synthesis of competing splice sites. SI and CI exons, whose inclusion is enhanced by both slow and fast mutants, have shorter flanking introns than cassette exons that are unaffected by elongation rate (Fig. 4E). FE exons are also flanked by shorter introns. Conversely, SE and CE exons have longer upstream introns than unaffected exons (Fig. 4E). The results suggest that the competitive advantage of a 3′ splice site adjacent to a cassette exon is more likely to be enhanced by slow elongation or diminished by fast elongation when cross-intron distances and corresponding transcription times are short. For reasons that we do not understand, exons included in slow and fast mutants (SI, FI, and CI) are longer than those that are excluded (SE, FE, and CE) (Fig. 4E), suggesting that exon definition may be more robust for longer exons when elongation rate is perturbed.
We examined the strengths of splice sites flanking elongation rate-sensitive exons (http://genes.mit.edu/burgelab/maxent/download; Yeo and Burge 2004). Notably, 3′ and 5′ splice sites at SI and CI exons are weaker than those at unaffected or SE exons (Fig. 4F). The 3′splice sites of FE exons are also weak. These results are specific to splice sites flanking rate-sensitive cassette exons. The strengths of distal splice sites at the 5′ and 3′ ends of the introns upstream of and downstream from affected cassette exons do not differ significantly from unaffected alternative exons (data not shown). Weak splice sites therefore appear to predispose type I cassette exons for inclusion when elongation is slow and for skipping when elongation is fast. This observation suggests that slow elongation enhances and fast elongation inhibits the efficacy of a subset of weak splice sites that flank cassette exons. Conversely, type II SE exons have somewhat stronger splice sites than unaffected exons. At these sites, slow transcription appears to be suboptimal for efficient processing, possibly because nonproductive splicing complexes are formed.
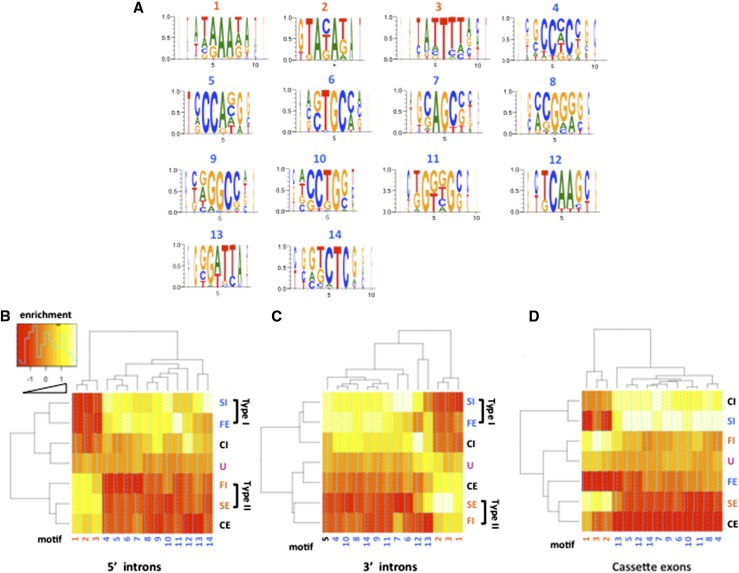
To elucidate additional features that distinguish exons that are included or skipped when elongation rate is altered, we looked for sequence elements enriched around these exons. We identified 14 consensus sequence motifs that are significantly enriched within 200 bases surrounding elongation rate-sensitive exons relative to unaffected exons (FDR < 0.01, Fisher’s exact test) (Fig. 5A). Among them, motifs 1, 3, 4, 5, 6, 10, 11, and 12 overlap conserved exonic splicing regulatory (ESR) sequences (Goren et al. 2006). Motifs 1 and 11 correspond to previously identified exonic and intronic splicing enhancers, respectively (Fairbrother et al. 2004; Wang et al. 2012b). The UG-rich motif 11 resembles binding sites for several regulators of alternative splicing, including ESRP1, ETR-3, and hnRNPF/H (Dujardin et al. 2010; Dittmar et al. 2012; Huelga et al. 2012). Motifs 1, 3, 8, 10, 11, and 13 overlap exonic silencers (Wang et al. 2004; Culler et al. 2010; Ke et al. 2011). Similarly, motifs 3, 4, and 7 resemble the intronic splicing silencers ISS-H, ISS-A, and ISS-B, respectively, that bind several hnRNPs (Wang et al. 2012c).
Figure 5.
Type I and type II classes of elongation rate-sensitive cassette exons are enriched in different sequence motifs. Type I exons are defined as those that behave as predicted by the “window of opportunity” model (i.e., slow inclusion and fast exclusion) (Fig. 1A), whereas type II exons do not conform with the model (i.e., slow exclusion and fast inclusion). (A) Fourteen consensus sequences assembled by hierarchical clustering of enriched hexamers (FDR < 0.005) within elongation rate-sensitive exons and 200 bases (excluding splice sites) of adjacent 5′ and 3′ introns relative to the same regions at unaffected (U) cassette exons (see the Materials and Methods). Y-axes represent relative frequency. (B–D) Enrichment of motifs 1–14 in the 5′ intron and 3′ intron and exon sequences of different functional classes of elongation rate-sensitive exons as in Figure 4E. Note that motifs 1–3 are generally enriched at type II SE and FI exons, whereas motifs 4–14 are more enriched at type I SI and FE exons. Heat maps were plotted using heatmap.2 in gplots of the R package. The distance of the solid green line from the dotted center line of each color cell is proportional to the measured enrichment.
Although the sequence elements enriched around type I and type II exons resemble splicing regulators, their functions in elongation rate-dependent splicing are still unclear. What is evident, however, is that sequence elements enriched at type I and type II exons differ from one another. Motifs 1–3 are overrepresented within type II SE and FI exons and their 5′ and 3′ flanking introns. A different set of motifs, 4–14, is enriched within type I SI and FE exons and their flanking introns (Fig. 5B–D). In summary, these observations show that exons whose inclusion is affected in different ways by slow and fast elongation are distinct classes distinguished by different sets of sequence motifs.
Elongation rate also affects intron retention
Unlike splicing of cassette exons, intron retention does not involve obvious competition between upstream and downstream splice sites. For this reason, one might expect intron inclusion to be insensitive to transcription elongation rate. We were therefore surprised to discover many examples of altered intron retention in Rpb1 mutants (Table 1). In the slow C4/R749H mutant, we identified 307 cases of enhanced intron retention (FDR < 0.05) out of 2523 events sampled and 183 cases of enhanced splicing (Table 1). In the fast E1126G mutant, 118 introns were preferentially retained, and 125 were excised relative to wild type (Table 1). By RT–PCR, we confirmed these effects of elongation rate on intron retention in 15 out of 15 cases in which RNA abundance permitted both PCR products to be detected by ethidium bromide staining (Fig. 6A–E; Supplemental Fig. 6; Supplemental Table 1). In some cases, the slow and fast mutants both promoted retention of the same intron (Fig. 6; Supplemental Fig. 6A,C,E). We cannot exclude the possibility that the stability of some transcripts with retained introns is altered due to changes in nonsense-mediated RNA decay (NMD) in Pol II mutant lines, but the results are not consistent with a generalized effect of this type, as the mutants enhanced intron retention or excision in a gene-specific way (Table 1). We also noted that both slow and fast Rpb1 mutants favored retention of longer alternatively spliced introns (Fig. 6F).
Figure 6.
Transcription elongation rate effects on alternative intron retention. (A–E) Validation of selected examples of retained introns with reduced splicing in the slow mutant, C4/R749H. RT–PCR was as in Figure 2 ,and biological replicates are shown in Supplemental Figure 6. Note that retention of these introns is also promoted by the H1108Y slow mutant and, in some cases, also by the fast E1126G mutant. (F) Intron retention in both fast and slow mutants correlates with intron length. Boxes show the median and interquartile range of intron length.
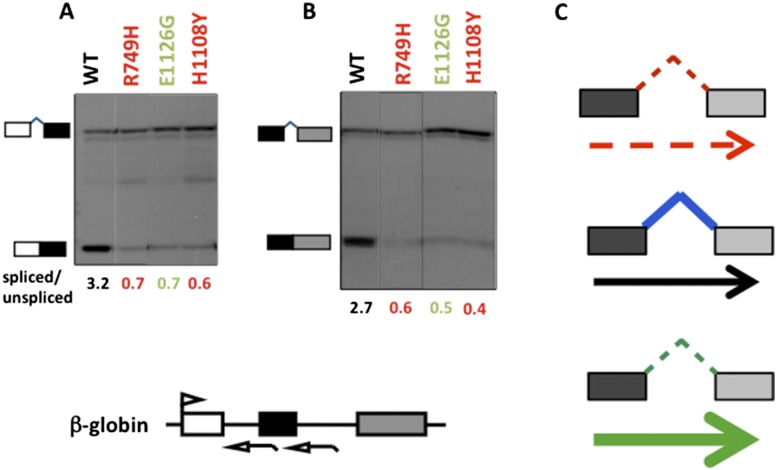
Since elongation rate can influence excision of retained introns, we investigated whether constitutive intron removal might also be affected. HEK293 cells were transiently transfected with expression vectors for the Amr Rpb1 mutants and a β-globin reporter. Splicing was monitored by RNase protection using antisense probes spanning intron 1 and 2 splice sites. Splicing of these constitutive introns was inhibited by both slow and fast mutants relative to wild-type Amr (Fig. 7A, B). This observation is reminiscent of reduced splicing at relatively strong splice sites flanking the SE class of cassette exons (Fig. 4F) and reduced splicing of long retained introns in slow and fast mutants (Fig. 6F). The inhibition of constitutive splicing by slow elongation appears to operate on chromosomal genes also, as the fraction of RNA-seq reads from within constitutive introns was elevated in libraries from the C4/R749H-expressing cells (4.1%) relative to wild-type Amr (2.9%). We did not observe enrichment of constitutive intron reads in the fast mutant but note that RNA-seq libraries from polyA+ RNA are probably underrepresented for intron-containing transcripts. These results in human cells are consistent with the finding that constitutive splicing in yeast is inhibited by fast elongation, but, in contrast to our results, slow elongation was reported to enhance constitutive splicing in yeast and Drosophila (Khodor et al. 2011; Braberg et al. 2013). Like alternative splicing, it therefore seems likely that fast and slow elongation can affect different constitutive splicing events differently.
Figure 7.
Elongation rate effects on constitutive splicing. (A,B) Splicing of intron 1 (A) and intron 2 (B) of human β-globin transcripts is inhibited by fast and slow elongation. RNase protection of transcripts from α-amanitin-treated HEK293 cells transfected with the pSVβ128 globin reporter and expression vectors for wild type (WT), slow R749H mutants, or fast E1126G Rpb1 mutants. Antisense RNA probes are indicated on the map. Ratios of spliced:unspliced transcripts normalized for 32P-U content are shown below each lane. (C) A “Goldilocks” model for kinetic coupling of transcription with splicing, reflecting our observations that at many splice sites, an optimal elongation rate is required to achieve a “normal” processing outcome.
Discussion
The compatible rates of splicing and transcription permit mRNA processing to occur simultaneously or cotranscriptionally. In principle, concurrent synthesis and splicing of the nascent transcript may or may not be mechanistically coupled to one another (Lazarev and Manley 2007). If it were coupled, however, the rate of elongation could potentially change the outcome of splicing reactions. In this study, we investigated how transcriptional elongation rate affects alternative splicing using mutants in conserved residues of the Pol II large subunit funnel (C4/R749H) and trigger loop (H1108Y and E1126G) domains. These mutants slow down (C4/R749H and H1108Y) or speed up (E1126G) in vitro elongation by Drosophila and yeast Pol II (Chen et al. 1996; Malagon et al. 2006; Kaplan et al. 2008). They faithfully displayed the same phenotypes in vivo when transposed into human Pol II, as shown by GRO-seq after release from a DRB block (Fig. 1C,D,G; Supplemental Fig. 1B–F). RNA-seq and RASL-seq analysis of Pol II mutant cell lines revealed significant changes in inclusion of ∼15%–40% of the cassette exons examined (Table 1; Figs. 2–4; Supplemental Figs. 3–5) in both slow and fast mutants relative to wild-type Pol II. These values probably underestimate the total number of elongation rate-sensitive splicing events, as our results are not saturating and are limited to HEK293 cells. Previously identified FN EDI and CFTR exon 9 that are affected by slow elongation in other cell types (de la Mata et al. 2003; Dujardin et al. 2014) were not identified as significant hits. In addition, several cases of elongation rate-sensitive cassette exons identified by RASL-seq and confirmed by RT–PCR were not identified as significant by RNA-seq with our cutoff of FDR < 0.05. Alternative intron retention was also affected by elongation rate in many cases (Table 1; Fig. 6; Supplemental Fig. 6). Abnormally slow and fast elongation rates are also both suboptimal for splicing of some constitutive introns (Fig. 7A, B). Previous work has documented widespread cotranscriptional splicing of nascent transcripts in several systems, including human cell lines (Khodor et al. 2011, 2012; Bhatt et al. 2012; Tilgner et al. 2012; for review, see Brugiolo et al. 2013). The pervasive effects of elongation rate on splicing reported here strongly suggest that mechanistic coupling between concurrent transcription and nascent pre-mRNA splicing is quite general.
We show that a relatively modest increase or decrease in elongation rate can have substantial effects on splicing of alternative exons and introns. This finding begs the question of whether physiological regulation of elongation rate exerts a significant influence on alternative splicing. In favor of this possibility, elongation rates vary between genes and within genes (Danko et al. 2013; Jonkers et al. 2014; Veloso et al. 2014) over a range (∼0.5–4 kb/min) that overlaps the average rates of the slow C4/R749H and fast E1126G mutants used here (0.5–1.9 kb/min) (Fig. 1D; Supplemental Fig. 2A). Furthermore, elongation rates have been reported to change in response to physiological stimuli (Danko et al. 2013) that could thereby regulate alternative splicing. Intriguingly, many elongation rate-sensitive cassette exons in our HEK293 cell lines are also abnormally included or skipped in breast and ovarian tumors (Figs. 2, 3; Supplemental Table 2; Venables et al. 2009). The importance of this observation remains to be tested; however, it is consistent with the possibility that abnormal elongation contributes to the widely documented pathology of alternative splicing in cancer and other diseases (David and Manley 2010; Germann et al. 2012). In this respect, it is noteworthy that several oncogene products are implicated in control of transcription elongation (Rahl et al. 2010; Smith et al. 2011).
A commonly accepted model for the relation between elongation rate and splicing proposes that slow transcription will expand the window of opportunity for recognition of the 3′ splice site upstream of an alternative exon so that it competes effectively with the next downstream 3′ splice site, resulting in inclusion of that exon (Fig. 1A; de la Mata et al. 2003). Transcriptome analysis of Pol II mutant cells permitted us to test how widely the “window of opportunity” model applies to splicing at endogenous genes. In accord with this mechanism, we found that slow elongation by the C4/R749H mutant promoted alternative exon inclusion in many cases, confirming previous studies that were limited to fewer genes (de la Mata et al. 2003; Ip et al. 2011). In particular, we found that exon inclusion was favored by slow elongation in cases in which the exons are relatively long, the flanking introns are short, and the proximal 3′ and 5′ splice sites are weak (Fig. 4E,F). In many but not all cases, a second slow mutant, H1108Y, behaved similarly to C4/R749H with respect to cassette exon inclusion (Table 1; Figs. 2–4; Supplemental Figs. 3–5).
Whether fast elongation also affects alternative splicing has not been examined previously, but it is predicted to constrain the window of opportunity for recognition of upstream 3′ splice sites, thereby favoring exon skipping (Fig. 1A). We found that fast elongation did indeed favor exon skipping about twice as frequently as inclusion (Table 1). The short flanking introns and weak splice sites characteristic of SI and FE type I exons (Fig. 4E,F) are features predicted to reduce the delay between synthesis of competing splice sites while increasing the time required for cassette exon recognition. These features of type I exons are predicted by the “window of opportunity” model to make alternative splicing sensitive to elongation rate. On the other hand, we also found numerous cases of type II exons in which slow elongation provoked skipping (SE), and fast elongation provoked inclusion (FI) (Table 1; Figs. 2, 3). The behavior of this class of exons is not explained by the “window of opportunity” model in a simple way. Type II exons have longer 3′ flanking introns and stronger 3′ splice sites than type I exons (Fig. 4E,F). Remarkably, type II exons share a common set of enriched sequence motifs that is distinct from those found at type I exons (Fig. 5). Together, the functional properties and sequence content of type I and type II exons suggest that they constitute two fundamentally distinct classes of alternative exon.
One rationale for the paradoxical behavior of type II exons is that splicing silencer or enhancer function associated with the motifs around these exons can be strengthened or weakened by altered elongation rate, resulting in more or less exon inclusion. For example, the results are consistent with the idea that silencing activity associated with elements enriched at type II exons (e.g., motifs 1–3) (Fig. 5) is strengthened by slow elongation, causing skipping, and weakened by fast elongation, causing inclusion. The behavior of type II exons could equally be explained if local enhancers were weakened by slow elongation or strengthened by fast elongation. It is possible that silencers/enhancers associated with sequence elements around type I exons (e.g., motifs 4–14 in Fig. 5) are also modulated by elongation rate. In the future, it will be of interest to test these ideas by asking whether association of RNA-binding proteins with sites near sensitive exons is affected by elongation rate. While this study was in preparation, evidence for an elongation rate effect was reported for a silencer near CFTR exon 9. In intron 8 of this gene, binding of the silencer protein ETR-3 was enhanced when elongation was inhibited by camptothecin, resulting in exon skipping (Dujardin et al. 2014).
A critical test of the “window of opportunity” model is whether slow and fast elongation have opposite effects on the splicing of individual cassette exons. A major finding of this study is the high frequency, ∼50% (Figs. 2A, 3; Table 1), of cases in which fast and slow mutants both perturbed alternative exon inclusion relative to wild-type Pol II but did so in the same direction rather than having opposing effects. Although we did find examples of opposing effects of slow and fast elongation (Fig. 4A–D; Supplemental Fig. 5A–D; Supplemental Table 2) on cassette exon splicing, they are a relatively small minority. We also found that fast and slow mutants influence constitutive splicing and alternative intron retention, events that do not involve competition between splice sites (Figs. 6, 7). Together, these diverse observations demonstrate that elongation rate frequently affects splicing in ways not predicted by the “window of opportunity” model.
If the “window of opportunity” model cannot account for the many cases in which slow and fast elongation bias alternative and constitutive splicing in the same direction, then what mechanisms might be responsible? We suggest that coupling of splicing with transcription is a “Goldilocks”-like phenomenon that requires an optimal elongation rate (Fig. 7C). This situation would arise if two or more parameters that influence a given splicing event vary as a function of elongation rate in different ways. Two such parameters, which are not completely independent, are nascent RNA folding and protein binding to splice sites, silencers, and enhancers. Cotranscriptional RNA folding is well known to be affected by the rate of growth of the RNA chain, with slow elongation favoring sequential folding, and fast elongation favoring nonsequential pathways with production of bigger loops (Pan and Sosnick 2006). RNA folding can, in turn, restrict access to processing or regulatory sites in the nascent RNA (Eperon et al. 1988; Libri et al. 1995; Buratti and Baralle 2004). It is also possible that the rate of elongation could influence protein binding to the nascent transcript independently of RNA folding. In the future, it will be of interest to determine how these important aspects of nascent RNA metabolism are influenced by elongation rate. We speculate that an optimal range of elongation rates for RNA Pol II evolved under selection pressure for effective kinetic coupling with cotranscriptional splicing, an idea that is consistent with the conservation of residues in the large subunit that limit the speed of transcription (Batada et al. 2004).
Materials and methods
Plasmids
The B10 epitope-tagged human Rpb1 wild-type Amr cDNA sequence from pAT7h1αAmr (Nguyen et al. 1996) was inserted into the HindIII–Not1 site of pcDNA5/FRT/TO (Invitrogen). The R749H, H1108Y, and E1126G mutations were introduced by mismatch overlap extension PCR. The pSVβ128 human β-globin reporter has been described (Fong and Bentley 2001).
Transient transfection and RNA analysis
HEK293 cells were transfected with 5 μg of reporter plasmid, 2.5 μg of Rpb1 expression vectors, and 0.5 μg of pSPVA Pol III transcribed loading control (data not shown) by CaPO4 precipitation. α-Amanitin (2.5μg/mL) was added after 12–16 h, and cells were harvested ∼30 h later. RNase protection was as described (Fong and Bentley 2001). Fixed, dried gels were quantified by PhosphorImager using Imagequant software. Ratios of spliced:unspliced and cleaved:uncleaved transcripts were corrected for 32P-U content of the protected fragments.
RT–PCR
RT reactions (2.0 μg total RNA in 20 μL) primed with random nonamers were incubated with Protoscript II reverse transcriptase (New England Biolabs) and dNTPs for 60 min at 42°C and then diluted to 60 μL. We amplified 1.0 μL of the cDNA in 50-μL PCR reactions for 30 cycles (15 sec at 94°C, 15 sec at 55°C, and 1 min at 72°C). Primers are described in Supplemental Table 1. In all cases, RT-minus controls were negative (data not shown).
Cell lines
pcDNA5/FRT/TO-Rpb1 Amr constructs were integrated into Flp-In-293 cells (Invitrogen) using Flp recombinase-mediated site-specific recombination. The cells were maintained in DMEM supplemented with 10% FBS, 200 μg/mL hygromycin B, 6.5 μg/mL blasticidin, and penicillin/streptomycin. All experiments were performed after induction with 2.0 μg/mL doxycycline for 12–16 h and treatment with 2.5 μg/mL α-amanitin for a further 42 h, at which time all cell lines were viable, and endogenous Pol II was inactive.
GRO-seq
Doxycycline-induced, α-amanitin-treated (42 h) cells were treated with 100 μM DRB for 3 h. Nuclei were harvested at this time (t = 0) and 10 and 20 min after washing out the DRB by washing three times in PBS and replacing with fresh the medium. GRO-seq with BrUTP labeling was as described (Core et al. 2008). Sequencing reads were trimmed at their 3′ ends to remove adapters, including polyA sequences, with the cutadapt program. Sequences >16 base pairs (bp) were mapped to the human genome (version hg18) by Bowtie with parameters “-l25 -n2 -e 200-best-strata.” Mapped reads with multiple locations were filtered out, and multiple reads with same genomic locations were kept, up to four. University of California at Santa Cruz refGene was used as gene annotation, and for genes with the same TSS, the longest one was kept. Reads were visualized on the Integrative Genomics Viewer browser (Fig. 1G; Supplemental Fig. 1B–F). To make the composite GRO-seq map, the genes were aligned at TSSs, and mapped reads were counted in 100-bp bins. The counts were normalized to 1 million total reads per sample. For each cell line, reads per kilobase of sequence per million mapped reads (RPKM) was calculated for the control sample without DRB treatment, and only the genes with RPKM >0.5 were kept for meta-analysis. The binned counts for all genes were averaged by position and normalized by the relative gene expression in the “no DRB” sample. The meta-profiles were smoothed with a 1-kb moving window. The front of the Pol II wave at each time point after DRB release was computed as the position at which the signal reached half of that in the “no DRB” control sample. These front positions relative to the TSS for the three time points (DRB-3h as 0, 10 min, 20 min) were linearly fitted to estimate the elongation rate.
RNA-seq and RASL-seq
RNA-seq libraries from doxycycline-induced α-amanitin-treated cells expressing wild-type Amr, C4/R749H, and E1126G Rpb1 were made using the Illumina TruSEQ RNA library kit version 2 (catalog no. RS-122-2001) from polyA+ RNA with random priming. Paired-end (100-bp) RNA-seq reads from biological replicate samples (Supplemental Table 3) were mapped individually without merging by TopHat (version 1.4.1) with a default option. For DJ analysis, we pooled the replicate samples of R749H and E1126G and compared them with individual wild-type replicate samples. Coordinates of target exons and introns with their surrounding exons were calculated from the Human.Ensembl.GRCh37.65.gtf file. For alternate exon analysis, junction and nonjunction reads across these target exons and introns were counted using MATS (Shen et al. 2012), and junction reads only were analyzed by a new pipeline that we developed called DJ. For retained intron analysis, nonjunction counts were used in DJ.
RASL-seq was performed as described (Li et al. 2012) with the following modifications. A pool of oligonucleotides was designed to detect 5582 alternative splicing events. For each event, a set of three oligonucleotides was designed for detecting two alternative isoforms, including one oligonucleotide against common donor/acceptor of the splicing junction, while two other oligonucleotides targeted the acceptors/donors of two unique isoforms, respectively. The sequencing reads were first decoded by multiplex sample barcodes and were then mapped using Bowtie to a customized sequence index based on the expected donor–acceptor pair sequences. Up to two mismatches were allowed, with a maximum of one mismatch in either donor or acceptor sequences. The mapped number of reads for each junction pair was counted as the level of the corresponding isoform.
We developed DJ in order to handle variation among replicates and calculate the significance of mean fold changes. This approach has the advantage of using the GLM (McCullagh and Nelder 1989), which was applied to test relative exclusion and inclusion of alternatively spliced sequence elements. Given a target event i of a cassette exon or retained intron, the probability of observing read count Yig in experiment group g can be calculated by the negative binomial Yig ∼ NB(μig, ϕi), in which the mean value μig and a tag-wise dispersion ϕi are estimated from the pool of replicates. Using the edgeR package in R (Robinson et al. 2010), we tested the significance of relative inclusion and exclusion effects in Pol II mutants compared with the wild-type Amr control group. For this purpose, we designed a GLM model represented by μ = β0 + β1e + β2g + β3ge, where e indicates event indicator (1 indicates inclusion, and 0 indicates exclusion), and g indicates group indicator (1 indicates test group, and 0 indicates background group). The parameter β0 indicates global mean, and β1 and β2 indicate the main effect of an event (inclusion or exclusion) and group (mutation or control). The interaction term β3 indicates the effect of a slow/fast mutation on relative inclusion of a target exon or intron. Using the null model μ = β0 + β1e + β2g, we performed a likelihood ratio test. The multiple P-values were corrected with the Benjamini-Hochberg method using the p.adjust method in R.
Motif analysis
We identified enriched hexamer motifs in different classes of elongation rate-sensitive exons and 200-bp regions upstream and downstream, excluding the last 20 bp of the 5′ intron and the first 6 bp of the 3′ intron as described in Fairbrother et al. (2002). A total of 698 6mers were enriched (Fisher’s exact test, FDR < 0.005) in these regions relative to unaffected exons, and we organized them into 14 groups using hierarchical clustering with the average agglomeration method option (hclust and cutree with threshold = 3, in the R package) where each group has six or more members. The sequences within each group were aligned to one another by clustalw2 (Larkin et al. 2007), and position weight matrices (Stormo et al. 1982) were made from the alignments using a uniform background distribution. The 4-by-l position weight matrix (PWM) for a particular 6mer motif alignment contains an element PWM = logMij/bi, where l is a length of an alignment (more than six when gap is allowed at the edge), and Mij is a frequency of nucleotide j at site index i. The PWM score of a sequence was calculated as
 |
Heat maps were plotted using heatmap.2 in gplots of the R package.
Accession numbers
All sequencing data have been deposited at NCBI Gene Expression Omnibus under accession number GSE63375.
Supplementary Material
Acknowledgments
We thank J. Hooper and B. Gao for helpful suggestions and sequencing, J. Jackson and B. Erickson for technical help, S. Berget for pcDNA3-CD44, and M. Poirier, T. Blumenthal, R. Davis, J. Hesselberth, and members of our laboratory for helpful discussions and comments on the manuscript. This work was supported by National Institutes of Health grants GM58613 (to D.L.B.) and GM49369 (to X.-D.F.), and a seed grant from the University of Colorado at Denver Next-Generation sequencing facility to D.L.B.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.252106.114.
References
- Aebi M, Hornig H, Padgett RA, Reiser J, Weissmann C. 1986. Sequence requirements for splicing of higher eukaryotic nuclear pre-mRNA. Cell 47: 555–565. [DOI] [PubMed] [Google Scholar]
- Batada NN, Westover KD, Bushnell DA, Levitt M, Kornberg RD. 2004. Diffusion of nucleoside triphosphates and role of the entry site to the RNA polymerase II active center. Proc Natl Acad Sci 101: 17361–17364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell TJ, Miyashiro KY, Sul J-Y, Buckley PT, Lee MT, McCullough R, Jochems J, Kim J, Cantor CR, Parsons TD, et al. 2010. Intron retention facilitates splice variant diversity in calcium-activated big potassium channel populations. Proc Natl Acad Sci 107: 21152–21157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley DL. 2014. Coupling mRNA processing with transcription in time and space. Nat Rev Genet 15: 163–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beyer AL, Osheim YN. 1988. Splice site selection, rate of splicing, and alternative splicing on nascent transcripts. Genes Dev 2: 754–765. [DOI] [PubMed] [Google Scholar]
- Bhatt DM, Pandya-Jones A, Tong AJ, Barozzi I, Lissner MM, Natoli G, Black DL, Smale ST. 2012. Transcript dynamics of proinflammatory genes revealed by sequence analysis of subcellular RNA fractions. Cell 150: 279–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bicknell AA, Cenik C, Chua HN, Roth FP, Moore MJ. 2012. Introns in UTRs: why we should stop ignoring them. BioEssays 34: 1025–1034. [DOI] [PubMed] [Google Scholar]
- Boireau S, Maiuri P, Basyuk E, de la Mata M, Knezevich A, Pradet-Balade B, Backer V, Kornblihtt A, Marcello A, Bertrand E. 2007. The transcriptional cycle of HIV-1 in real-time and live cells. J Cell Biol 179: 291–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braberg H, Jin H, Moehle EA, Chan YA, Wang S, Shales M, Benschop JJ, Morris JH, Qiu C, Hu F, et al. 2013. From structure to systems: high-resolution, quantitative genetic analysis of RNA polymerase II. Cell 154: 775–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brugiolo M, Herzel L, Neugebauer KM. 2013. Counting on co-transcriptional splicing. F1000Prime Rep 5: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buratti E, Baralle FE. 2004. Influence of RNA secondary structure on the pre-mRNA splicing process. Mol Cell Biol 24: 10505–10514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Chafin D, Price DH, Greenleaf AL. 1996. Drosophila RNA polymerase II mutants that affect transcription elongation. J Biol Chem 271: 5993–5999. [PubMed] [Google Scholar]
- Core LJ, Waterfall JJ, Lis JT. 2008. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science 322: 1845–1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culler SJ, Hoff KG, Voelker RB, Berglund JA, Smolke CD. 2010. Functional selection and systematic analysis of intronic splicing elements identify active sequence motifs and associated splicing factors. Nucleic Acids Res 38: 5152–5165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danko CG, Hah N, Luo X, Martins AL, Core L, Lis JT, Siepel A, Kraus WL. 2013. Signaling pathways differentially affect RNA polymerase II initiation, pausing, and elongation rate in cells. Mol Cell 50: 212–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David CJ, Manley JL. 2010. Alternative pre-mRNA splicing regulation in cancer: pathways and programs unhinged. Genes Dev 24: 2343–2364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Mata M, Alonso CR, Kadener S, Fededa JP, Blaustein M, Pelisch F, Cramer P, Bentley D, Kornblihtt AR. 2003. A slow RNA polymerase II affects alternative splicing in vivo. Mol Cell 12: 525–532. [DOI] [PubMed] [Google Scholar]
- Dittmar KA, Jiang P, Park JW, Amirikian K, Wan J, Shen S, Xing Y, Carstens RP. 2012. Genome-wide determination of a broad ESRP-regulated posttranscriptional network by high-throughput sequencing. Mol Cell Biol 32: 1468–1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dujardin G, Buratti E, Charlet-Berguerand N, Martins de Araujo M, Mbopda A, Le Jossic-Corcos C, Pagani F, Ferec C, Corcos L. 2010. CELF proteins regulate CFTR pre-mRNA splicing: essential role of the divergent domain of ETR-3. Nucleic Acids Res 38: 7273–7285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dujardin G, Lafaille C, Petrillo E, Buggiano V, Gomez Acuna LI, Fiszbein A, Godoy Herz MA, Nieto Moreno N, Munoz MJ, Allo M, et al. 2013. Transcriptional elongation and alternative splicing. Biochim Biophys Acta 1829: 134–140. [DOI] [PubMed] [Google Scholar]
- Dujardin G, Lafaille C, de la Mata M, Marasco LE, Munoz MJ, Le Jossic-Corcos C, Corcos L, Kornblihtt AR. 2014. How slow RNA polymerase II elongation favors alternative exon skipping. Mol Cell 54: 1–8. [DOI] [PubMed] [Google Scholar]
- Eperon LP, Graham IR, Griffiths AD, Eperon IC. 1988. Effects of RNA secondary structure on alternative splicing of pre-mRNA: is folding limited to a region behind the transcribing RNA polymerase? Cell 54: 393–401. [DOI] [PubMed] [Google Scholar]
- Fairbrother WG, Yeh RF, Sharp PA, Burge CB. 2002. Predictive identification of exonic splicing enhancers in human genes. Science 297: 1007–1013. [DOI] [PubMed] [Google Scholar]
- Fairbrother WG, Yeo GW, Yeh R, Goldstein P, Mawson M, Sharp PA, Burge CB. 2004. RESCUE-ESE identifies candidate exonic splicing enhancers in vertebrate exons. Nucleic Acids Res 32: W187–W190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong N, Bentley D. 2001. Capping, splicing and 3′ processing are independently stimulated by RNA polymerase II: different functions for different segments of the CTD. Genes Dev 15: 1783–1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Germann S, Gratadou L, Dutertre M, Auboeuf D. 2012. Splicing programs and cancer. J Nucleic Acids 2012: 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goren A, Ram O, Amit M, Keren H, Lev-Maor G, Vig I, Pupko T, Ast G. 2006. Comparative analysis identifies exonic splicing regulatory sequences—the complex definition of enhancers and silencers. Mol Cell 22: 769–781. [DOI] [PubMed] [Google Scholar]
- Han J, Xiong J, Wang D, Fu X-D. 2011. Pre-mRNA splicing: where and when in the nucleus. Trends Cell Biol 21: 336–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- House AE, Lynch KW. 2008. Regulation of alternative splicing: more than just the ABCs. J Biol Chem 283: 1217–1221. [DOI] [PubMed] [Google Scholar]
- Huelga SC, Vu AQ, Arnold JD, Liang TY, Liu PP, Yan BY, Donohue JP, Shiue L, Hoon S, Brenner S, et al. 2012. Integrative genome-wide analysis reveals cooperative regulation of alternative splicing by hnRNP proteins. Cell Reports 1: 167–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ip JY, Schmidt D, Pan Q, Ramani AK, Fraser AG, Odom DT, Blencowe B. 2011. Global impact of RNA polymerase II elongation inhibition on alternative splicing regulation. Genome Res 21: 390–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonkers I, Kwak H, Lis JT. 2014. Genome-wide dynamics of Pol II elongation and its interplay with promoter proximal pausing, chromatin, and exons. eLife 3: e02407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan CD, Larsson KM, Kornberg RD. 2008. The RNA polymerase II trigger loop functions in substrate selection and is directly targeted by α-amanitin. Mol Cell 30: 547–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke S, Shang S, Kalachikov SM, Morozova I, Yu L, Russo JJ, Ju J, Chasin LA. 2011. Quantitative evaluation of all hexamers as exonic splicing elements. Genome Res 21: 1360–1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khodor YL, Rodriguez J, Abruzzi KC, Tang CH, Marr MT 2nd, Rosbash M. 2011. Nascent-seq indicates widespread cotranscriptional pre-mRNA splicing in Drosophila. Genes Dev 25: 2502–2512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khodor YL, Menet JS, Tolan M, Rosbash M. 2012. Cotranscriptional splicing efficiency differs dramatically between Drosophila and mouse. RNA 18: 2174–2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kireeva ML, Nedialkov YA, Cremona GH, Purtov YA, Lubkowska L, Malagon F, Burton ZF, Strathern JN, Kashlev M. 2008. Transient reversal of RNA polymerase II active site closing controls fidelity of transcription elongation. Mol Cell 30: 557–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornblihtt AR, de la Mata M, Fededa JP, Munoz MJ, Nogues G. 2004. Multiple links between transcription and splicing. RNA 10: 1489–1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23: 2947–2948. [DOI] [PubMed] [Google Scholar]
- Lazarev D, Manley JL. 2007. Concurrent splicing and transcription are not sufficient to enhance splicing efficiency. RNA 13: 1546–1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Qiu J, Fu X-D. 2012. RASL-seq for massively parallel and quantitative analysis of gene expression. Curr Protoc Mol Biol. 98: 4.13.1–4.13.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libri D, Stutz F, McCarthy T, Rosbash M. 1995. RNA structural patterns and splicing: molecular basis for an RNA-based enhancer. RNA 1: 425–436. [PMC free article] [PubMed] [Google Scholar]
- Lynch KW. 2006. Cotranscriptional splicing regulation: it’s not just about speed. Nat Struct Mol Biol 13: 952–953. [DOI] [PubMed] [Google Scholar]
- Malagon F, Kireeva ML, Shafer BK, Lubkowska L, Kashlev M, Strathern JN. 2006. Mutations in the Saccharomyces cerevisiae RPB1 gene conferring hypersensitivity to 6-azauracil. Genetics 172: 2201–2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCullagh P, Nelder J. 1989. Generalized linear models. CRC Press, Boca Raton, FL. [Google Scholar]
- Nguyen VT, Giannoni F, Dubois MF, Seo SJ, Vigneron M, Kedinger C, Bensaude O. 1996. In vivo degradation of RNA polymerase II largest subunit triggered by α-amanitin. Nucleic Acids Res 24: 2924–2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsen TW, Graveley BR. 2010. Expansion of the eukaryotic proteome by alternative splicing. Nature 463: 457–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan T, Sosnick T. 2006. RNA folding during transcription. Annu Rev Biophys Biomol Struct 35: 161–175. [DOI] [PubMed] [Google Scholar]
- Pandya-Jones A, Black DL. 2009. Co-transcriptional splicing of constitutive and alternative exons. RNA 15: 1896–1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahl PB, Lin CY, Seila AC, Flynn RA, McCuine S, Burge CB, Sharp PA, Young RA. 2010. c-Myc regulates transcriptional pause release. Cell 141: 432–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen S, Park JW, Huang J, Dittmar KA, Lu ZX, Zhou Q, Carstens RP, Xing Y. 2012. MATS: a Bayesian framework for flexible detection of differential alternative splicing from RNA-Seq data. Nucleic Acids Res 40: e61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigurdsson S, Dirac-Svejstrup AB, Svejstrup JQ. 2010. Evidence that transcript cleavage is essential for RNA polymerase II transcription and cell viability. Mol Cell 38: 202–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh J, Padgett RA. 2009. Rates of in situ transcription and splicing in large human genes. Nat Struct Mol Biol 16: 1128–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith E, Lin C, Shilatifard A. 2011. The super elongation complex (SEC) and MLL in development and disease. Genes Dev 25: 661–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stickeler E, Fraser SD, Honig A, Chen AL, Berget SM, Cooper TA. 2001. The RNA binding protein YB-1 binds A/C-rich exon enhancers and stimulates splicing of the CD44 alternative exon v4. EMBO J 20: 3821–3830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stormo GD, Schneider TD, Gold L, Ehrenfeucht A. 1982. Use of the ‘Perceptron’ algorithm to distinguish translational initiation sites in E. coli. Nucleic Acids Res 10: 2997–3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilgner H, Knowles DG, Johnson R, Davis CA, Chakrabortty S, Djebali S, Curado J, Snyder M, Gingeras TR, Guigo R. 2012. Deep sequencing of subcellular RNA fractions shows splicing to be predominantly co-transcriptional in the human genome but inefficient for lncRNAs. Genome Res 22: 1616–1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veloso A, Kirkconnell KS, Magnuson B, Biewen B, Paulsen MT, Wilson TE, Ljungman M. 2014. Rate of elongation by RNA polymerase II is associated with specific gene features and epigenetic modifications. Genome Res 24: 896–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venables JP, Klinck R, Koh C, Gervais-Bird J, Bramard A, Inkel L, Durand M, Couture S, Froehlich U, Lapointe E, et al. 2009. Cancer-associated regulation of alternative splicing. Nat Struct Mol Biol 16: 670–676. [DOI] [PubMed] [Google Scholar]
- Wang Z, Rolish ME, Yeo G, Tung V, Mawson M, Burge CB. 2004. Systematic identification and analysis of exonic splicing silencers. Cell 119: 831–845. [DOI] [PubMed] [Google Scholar]
- Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB. 2008. Alternative isoform regulation in human tissue transcriptomes. Nature 456: 470–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Zhang J, Li K, Zhao W, Cui Q. 2012a. SpliceDisease database: linking RNA splicing and disease. Nucleic Acids Res 40: D1055–D1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Ma M, Xiao X, Wang Z. 2012b. Intronic splicing enhancers, cognate splicing factors and context-dependent regulation rules. Nat Struct Mol Biol 19: 1044–1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Xiao X, Zhang J, Choudhury R, Robertson A, Li K, Ma M, Burge CB, Wang Z. 2012c. A complex network of factors with overlapping affinities represses splicing through intronic elements. Nat Struct Mol Biol 20: 36–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wetterberg I, Bauren G, Wieslander L. 1996. The intranuclear site of excision of each intron in Balbiani ring 3 pre-mRNA is influenced by the time remaining to transcription termination and different excision efficiencies for the various introns. RNA 2: 641–651. [PMC free article] [PubMed] [Google Scholar]
- Wong JJ-L, Ritchie W, Ebner OA, Selbach M, Wong JWH, Huang Y, Gao D, Pinello N, Gonzalez M, Baidya K, et al. 2013. Orchestrated intron retention regulates normal granulocyte differentiation. Cell 154: 583–595. [DOI] [PubMed] [Google Scholar]
- Yeo G, Burge CB. 2004. Maximum entropy modeling of short sequence motifs with applications to RNA splicing signals. J Comput Biol 11: 377–394. [DOI] [PubMed] [Google Scholar]
- Zhou Z, Qiu J, Liu W, Zhou Y, Plocinik RM, Li H, Hu Q, Ghosh G, Adams JA, Rosenfeld MG, et al. 2012. The Akt-SRPK-SR axis constitutes a major pathway in transducing EGF signaling to regulate alternative splicing in the nucleus. Mol Cell 47: 422–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.