Abstract
Montelukast and Zafirlukast are known leukotriene receptor antagonists prescribed in asthma treatment. However, these fall short as mono therapy and are frequently used in combination with inhaled glucocorticosteroids with or without long acting beta 2 agonists. Therefore, it is of interest to apply ligand and structure based virtual screening strategies to identify compounds akin to lead compounds Montelukast and Zafirlukast. Hence, compounds with structures having 95% similarity to these compounds were retrieved from NCBI׳s PubChem database. Compounds similar to lead were grouped and docked at the antagonist binding site of cysteinyl leukotriene receptor 1. This exercise identified compounds UNII 70RV86E50Q (Pub Cid 71587778) and Sure CN 9587085 (Pub Cid 19793614) with higher predicted binding compared to Montelukast and Zafirlukast. It is shown that the compound Sure CN 9587085 showed appreciable ligand receptor interaction compared to UNII 70RV86E50Q. Thus, the compound Sure CN 9587085 is selected as a potent antagonist to cysteinyl leukotriene receptor 1 for further consideration in vitro and in vivo validation.
Keywords: Leukotriene antagonists, Virtual screening, Molecular docking, Pharmacophoric identification
Background
Leukotrienes are synthesized in response to many triggers, including antigen-antibody interaction; receptor activation, physical stimuli, and any stimulation that increases intercellular calcium [1]. These molecules are formed endogenously by the breakdown of arachidonic acid, via the 5- lipoxygenase enzyme pathway [2]. This pathway ultimately generates several species of leukotrienes with various physiological activities, including increased vascular permeability, influx of eosinophils into the airway tissues, enhanced mucous production – generalized inflammatory effects associated with asthma pathogenecity [3, 4]. The cysteinyl leukotreienes were also shown to have been recovered in excess concentrations during asthma exacerbations from bronchoalviolar lavage fluid and urine of patients with asthma [5, 6]. These observations suggest that leukotrienes could play a role in the pathogenesis of asthma; therefore, drugs that antagonize the activity of the cysteinyl leukotrienes at their receptor cysLT1 have been developed with the goal of attenuating the leukotrienes' effects in inflammation related to asthma [7].
Antileukotriene agents are non-steroidal anti-inflammatory bronchoconstriction preventers administered for treatment of asthma. Montelukast and Zafirlukast are most common antagonists which are widely prescribed in treatment of asthma [8]. These drugs competitively antagonize cysLt1 receptor mediated bronchoconstriction, increased vascular permeability and recruitment of eosinophils and bring about bronchodilation and suppression of bronchial inflammation. Though overall efficacy is lower than inhaled steroids, LTRAs may obviate need for the latter, and may be more acceptable in children. In severe asthma, they may permit reduction in steroid dose and need for rescue of beta 2 agonist inhalations. According to current guidelines, antileukotriene agents are recommended in the long-term treatment of asthma. In combination with inhaled glucocorticosteroids, they may allow to reduce the dose of steroids used and to control the disease in patients with moderate to severe asthma [9– 13]. Nonetheless, they are not preferred first choice therapy because use of antileukotrienes in adult asthma patients as a single asthma controller is usually less effective than inhaled glucocorticosteroids [14, 15]. Moreover, they are usually less effective than long acting beta 2 agonists in relieving bronchospasm [16– 19]. In the present study structure and ligand based approaches have been implemented to identify drug, which we anticipate to have better antagonizing potential than Montelukast and Zafirlukast
Methodology
Selection of multiclass antagonists:
Established leukotriene antagonists - Montelukast [20] and Zafirlukast [21] were selected as leads for virtual screening.
Molecular Modeling of Cysteinyl Leukotriene Receptor1:
Since the three dimensional structure of human cysteinyl leukotriene receptor 1 was not available in Protein Data Bank, the structure was generated in silico [22]. The sequence of receptor was retrieved from NCBI (Accession Number: NP_006630.1) which was then blasted against Protein Data Bank database. The position specific iterated BLASTp program resulted in 36 % identity and query coverage of 55 % with template of Mu-Opioid receptor crystal structure (PDB ID: 4DKLA) [23]. Protein modeling was performed using Deep View/Swiss PDB Viewer and Swiss Model server [24]. The primary polypeptide chain of the receptor was aligned on the backbone of template obtained from PDB database, which was then followed by side chain optimization using the simultaneous global optimization of the energy for all nonidentical residues. Structural validation of the modeled receptor was assessed with structure validation tool Procheck [25] and Ramachandran plot [26]. Figure 1 shows the modeled structure of cysteinyl leukotriene receptor 1.
Figure 1.
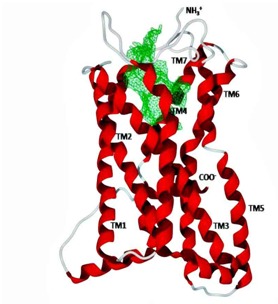
Structure of the 337 residues long cystenyl leukotrine receptor modeled using template structure of Mu-Opioid receptor crystal structure (PDB ID: 4DKLA). Binding site in the protein was detected using Gaussian filter enabled DoGSite server [27]. The N terminal of the protein is extracellular while C terminal lies in the cytoplasmic region of the cell. Lipid environment of membrane is majorly occupied by the polar transmembrane helices (TM1-TM7) and the interconnecting loops between the transmembrane helices are either cytoplasmic or extracellular.
Active site prediction:
Antagonist binding site was detected with DoGSite [27] active site prediction server. The server calculates potential active sites on the protein surface based on the 3D coordinates of a protein. DoGSite is a grid-based function prediction method which uses a difference of Gaussian filter to detect potential pockets on the protein surface and splits them into subpockets. Subsequently, global properties, describing the size, shape and chemical features are also calculated for the predicted pockets. As a convention, the cavity with relatively greater surface area and volume was considered as an active (antagonist binding) site (Figure 1). The calculated descriptors of the pocket are represented in Table 1 (see supplementary material).
Protein and lead preparation:
The selected leads (Montelukast and Zafirlukast) were uploaded in .sdf format in the PubChem structure search engine and similar compounds were retrieved with ≥95% structure. All the similar structures were optimized and cleaned with Marvin View v 5.6.0.2 (ChemAxon Ltd.) [28]. Threedimensional structure of modeled receptor was prepared by removing all bound water molecules and ligands. Explicit hydrogens, bond orders, hybridization and charges were assigned to protein structure if missing.
Virtual screening parameters:
Molecular docking program Molegro Virtual Docker (MVD) which incorporates highly efficient PLP (Piece wise Linear Potential) and MolDock [29, 30] scoring function provided a flexible docking platform. The leads (Montelukast and Zafirlukast) and similar chemical structures were docked in predicted antagonist binding site of cysteinyl leukotriene receptor 1. Docking parameters were set to 0.20 Å as grid resolution, maximum iteration of 1500 and maximum population size of 50. Energy minimization and hydrogen bonds were optimized after the docking. Simplex evolution was set at maximum steps of 300 with neighborhood distance factor of 1. Binding affinity and interactions of similar compounds with protein were evaluated on the basis of the internal ES (Internal electrostatic Interaction), internal hydrogen bond interactions and sp2-sp2 torsions. On the basis of MolDock rerank score [31] best interacting compound was selected from each class.
Softwares, Suites and Webservers used:
Pubchem database from NCBI was used to search and prepare library of similar chemical compounds for virtual screening. All the chemical structures were optimized in MarvinSketch 5.6.0.2, (1998-2011, Copyright © ChemAxon Ltd). Flexible Molecular docking of the compounds with target was completed using Molegro Virtual Docker 2010.4.0.0. Accelrys Discovery Studio® Visualizer 3.5.0.12158 (Copyright© 2005-12, Accelyrys Software Inc.) was used for visualization.
Results
Number of similar compounds screened with ≥90 similarity corresponding to each lead antagonist is listed in Table 2 (see supplementary material). Interestingly, similar compounds identified had better affinity than their respective leads. Evident from rerank score, compound UNII-70RV86E50Q (Pubcid: 71587778) (Figure 2a) showed 1.13 folds better interaction than its lead Montelukast, whereas, compound Sure CN 9587085 (Pubcid: 19793614) (Figure 2b) was 1.15 folds better interacting compound than its lead Zafirlukast (Table 2). In further investigation we observed that compound Sure CN 9587085 was marginally (1.01 folds) better interacting drug than UNII-70RV86E50Q (table 2). Sure CN9587085 not only showed better interaction against the cysteinyl leukotriene receptor 1 than its parent compound Zafirlukast but also showed superior binding affinity than montelukast and remaining virtually screened molecules (Table 2). The overall ligand receptor affinity score i.e. rerank score of Sure CN9587085 with the receptor (a function of stearic interactions, electrostatic interactions, H bond interactions etc) is shown in Table 3 (see supplementary material). Comprehensively shown in Figure 3a, the molecule demonstrated van der waals interactions with Glu 175, Phe 174 & 158, Thr 8,100 & 83 , Val 277, Tyr 83, 241 & 104, His 190, Phe 246 & 158, Leu 103,107,189, 107 & 281 , Ala 284 and shows electrostatic interaction with Lys 162, Arg 79, Ser 193, His 250. The ligand binding pattern of Sure CN9587085 in the active site is shown in Figure 3b. Hydrogen bonding of Sure CN9587085 with the receptor is shown if Figure 4a and the complete hydrogen bonding profile are shown in Table 4 (see supplementary material). Further, Figure 4b shows the hydrophobic intensities of the antagonist binding site upon binding of Sure CN9587085.
Figure 2.
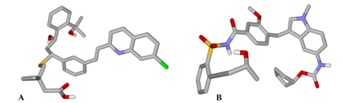
Structure of compound (A) UNII70RV86E50Q (Pubcid: CID: 71587778 ); (B) Sure CN9587085 (Pubcid: 19793614) selected after virtual screening.
Figure 3.
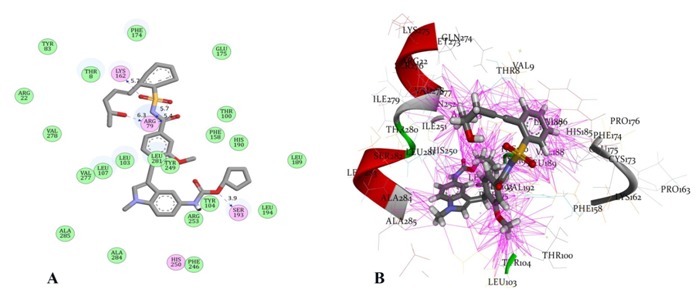
A) Interactions of Sure CN 9587085 with cysteinyl leukotrine receptor 1. Residues circled in green participate in van der waals interaction with the ligand while residues in pink forms electrostatic interactions. Hydrogen bonds with bond lengths are shown as blue arrows between ligand and residues Lys 162, Arg 79 and Ser 193; B) Binding pattern of Sure CN 9587085 with cysteinyl leukotrine receptor 1. The pink lines represent various interactions like electrostatic, van der Waals, stearic, hydrogen bonding and hydrophobic interactions that enable energetically favourable binding of the ligand in the cavity.
Figure 4.
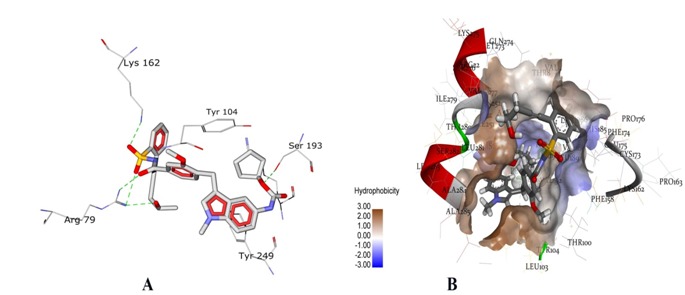
A) Hydrogen bonds (green dotted lines) formed between Sure CN 9587085 and cysteinyl leukotrine receptor 1; B) The active site of receptor is shown with hydrophobic intensities. The hydrophobic intensities of the binding site ranges from -3.00 (least hydrophobic area - blue shade) to 3.00 (highly hydrophobic area –brown shade).
Discussion
Regardless of progress achieved in the understanding the inflammatory process in the pathogenesis of asthma the mortality and morbidity associated with this disease have not appreciably declined therefore, it becomes obvious to explore for new avenues to asthma therapy. Antileukotrienes have emerged as first new treatment strategy for chronic asthma in 20 years. To cite evidence, in a multitude of randomized studies of 12-24 weeks duration, patients treated with antileukotriene drugs showed clinically significant improvements in lung function and asthma symptoms compared with patients who received placebo. In another study, exacerbation rates decreased by 30% to 60% in patients with asthma treated with antileukotriene drugs [31– 33]. Thus it becomes imperative from these studies that there is quite commendable improvement in quality of life scores in patients treated with antileukotrine drugs. Although inhaled corticosteroids are the preferred therapy for chronic asthma because of the long term experience with this drug and their documented anti-inflammatory effect, they are not without safety concerns. In addition, in patients treated with low or moderate doses of inhaled corticostreroids are still symptomatic wherein antileukotrine drug offer an attractive option as add on therapy. Despite the advantages, antileukotrienes, is still not prescribed as a single therapy for symptomatic treatment of asthma, instead always used in combination with corticosteroids ± beta 2 agonists. This indicates that antileukotrienes though being efficacious, but not always a good choice in severe exacerbations unlike ICS and beta 2 agonist.
The molecular docking based screening of compounds that are alike Montelukast and Zafirlukast against cysteinyl leukotriene receptor 1 helped to identify Sure CN 9587085 from NCBI׳s PubChem database. The identified compound showed improved binding features with the receptor compared to the known antagonist lead compounds (Montelukast and Zafirlukast). The compound showed appreciable H bond interaction profile, electrostatic and hydrophobic interactions for further consideration.
Conclusion
There is a need for better Leukotrierne antagonists as Montelukast and Zafirlukast falls short as mono-therapy. Hence, molecular docking based screening of compounds against cysteinyl leukotriene receptor 1 was completed to identify Sure CN 9587085 from NCBI׳s PubChem database for further consideration using in vitro analysis.
Supplementary material
Acknowledgments
We extend our sincere thanks to Mr. Prem Singh, Khushboo Sharma and Amulya Narra, Project Fellows at Institute of Genetics, Hyderabad. We also thank the medical superintendent Dr. A. Sai Kumar, all physicians and supporting staffs at the Government general and Chest Hospital, Hyderabad.
Footnotes
Citation:Bandaru et al, Bioinformation 10(10): 652-657 (2014)
References
- 1.Scow DT, et al. Am Fam Physician. 2007;75:65. [PubMed] [Google Scholar]
- 2.Samuelsson B, et al. Science. 1987;237:1171. doi: 10.1126/science.2820055. [DOI] [PubMed] [Google Scholar]
- 3.Henderson WR, et al. Ann Intern Med. 1994;121:684. doi: 10.7326/0003-4819-121-9-199411010-00010. [DOI] [PubMed] [Google Scholar]
- 4.Lewis RA, et al. N Engl J Med. 1990;323:645. doi: 10.1056/NEJM199009063231006. [DOI] [PubMed] [Google Scholar]
- 5.Wenzel SE, et al. Am Rev Respir Dis. 1990;142:112. doi: 10.1164/ajrccm/142.1.112. [DOI] [PubMed] [Google Scholar]
- 6.Drazen JM, et al. Am Rev Respir Dis. 1992;146:104. doi: 10.1164/ajrccm/146.1.104. [DOI] [PubMed] [Google Scholar]
- 7.Spector SL, et al. Ann Allergy Asthma Immunol. 1995;75:463. [PubMed] [Google Scholar]
- 8.Wenzel SE, et al. JAMA. 1998;280:2068. doi: 10.1001/jama.280.24.2068. [DOI] [PubMed] [Google Scholar]
- 9.Laviolette M, et al. Am J Respir Crit Care Med. 1999;160:1862. doi: 10.1164/ajrccm.160.6.9803042. [DOI] [PubMed] [Google Scholar]
- 10.Löfdahl CG, et al. BMJ. 1999;319:87. doi: 10.1136/bmj.319.7202.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Virchow C, et al. Am J Respir Crit Care Med. 2000;162:578. doi: 10.1164/ajrccm.162.2.9905041. [DOI] [PubMed] [Google Scholar]
- 12.Price DB, et al. Thorax. 2003;58:211. doi: 10.1136/thorax.58.3.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vaquerizo MJ, et al. Thorax. 2003;58:204. doi: 10.1136/thorax.58.3.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bleecker, et al. J Allergy Clin Immunol. 2000;105:1123. doi: 10.1067/mai.2000.106043. [DOI] [PubMed] [Google Scholar]
- 15.Laviolette M, et al. Am J Respir Crit Care Med. 1999;160:1862. doi: 10.1164/ajrccm.160.6.9803042. [DOI] [PubMed] [Google Scholar]
- 16.Bjermer L, et al. BMJ. 2003;327:891. doi: 10.1136/bmj.327.7420.891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nelson HS, et al. J Allergy Clin Immunol. 2000;106:1088. doi: 10.1067/mai.2000.110920. [DOI] [PubMed] [Google Scholar]
- 18.Fish JE, et al. Chest. 2001;120:423. doi: 10.1378/chest.120.2.423. [DOI] [PubMed] [Google Scholar]
- 19.Ringdal N, et al. Respir Med. 2003;97:234. doi: 10.1053/rmed.2003.1436. [DOI] [PubMed] [Google Scholar]
- 20.Leff JA, et al. N Engl J Med. 1998;339:147. doi: 10.1056/NEJM199807163390302. [DOI] [PubMed] [Google Scholar]
- 21.Mastalerz L, et al. Pol Arch Med Wewn. 2010;120:103. [PubMed] [Google Scholar]
- 22.Nayarisseri A, et al. Interdiscip Sci. 2013;5:4. doi: 10.1007/s12539-013-0183-8. [DOI] [PubMed] [Google Scholar]
- 23.Tabassum A, et al. Interdiscip Sci. 2014;6:1. doi: 10.1007/s12539-014-0190-4. [DOI] [PubMed] [Google Scholar]
- 24.Bandaru S, et al. Curr Top Med Chem. 2013;13:14. doi: 10.2174/15680266113139990115. [DOI] [PubMed] [Google Scholar]
- 25.Laskowski RA, et al. J Mol Biol. 1993;231:283. doi: 10.1006/jmbi.1993.1351. [DOI] [PubMed] [Google Scholar]
- 26.Ramachandran GN, et al. J Mol Biol. 1963;7:95. doi: 10.1016/s0022-2836(63)80023-6. [DOI] [PubMed] [Google Scholar]
- 27.Csizmadia F, et al. J Chem Inf Comput Sci. 2000;40:323. doi: 10.1021/ci9902696. [DOI] [PubMed] [Google Scholar]
- 28.Yadav M, et al. Interdiscip Sci. 2013;5:2. doi: 10.1007/s12539-013-0162-0. [DOI] [PubMed] [Google Scholar]
- 29.Thomsen, et al. J Med Chem. 2006;49:3315. doi: 10.1021/jm051197e. [DOI] [PubMed] [Google Scholar]
- 30.Yang JM, et al. Proteins. 2004;55:288. doi: 10.1002/prot.20035. [DOI] [PubMed] [Google Scholar]
- 31.Drazen, et al. N Engl J Med. 1999;340:197. doi: 10.1056/NEJM199901213400306. [DOI] [PubMed] [Google Scholar]
- 32.Bacharier, et al. Allergy. 2008;63:5. [Google Scholar]
- 33.Cruz AA, et al. Allergy. 2007;22:62. doi: 10.1111/j.1398-9995.2007.01241.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


