Abstract
Background & objectives:
The human gut microbiota play a significant role in nutritional processes. The concept of probiotics has led to widespread consumption of food preparations containing probiotic microbes such as curd and yogurt. Curd prepared at home is consumed every day in most homes in southern India. In this study the home-made curd was evaluated for lactic acid bacteria (LAB) with probiotic potential.
Methods:
Fifteen LAB (12 lactobacilli, 1 Lactococcus, 2 Leuconostoc) and one yeast isolated from home-made curd were evaluated for resistance to acid, pepsin, pancreatin and bile salts; antimicrobial resistance; intrinsic antimicrobial activity; adherence to Caco-2 epithelial cells; ability to block pathogen adherence to Caco-2 cells; ability to inhibit interleukin (IL)-8 secretion from HT-29 epithelial cells in response to Vibrio cholerae; and ability to induce anti-inflammatory cytokine expression in THP-1 monocyte cells.
Results:
Lactobacillus abundance in fermenting curd peaked sharply at 12 h. Nine of the strains survived exposure to acid (pH 3.0) for at least one hour, and all strains survived in the presence of pancreatin or bile salts for 3 h. None showed haemolytic activity. All were resistant to most antimicrobials tested, but were sensitive to imipenem. Most strains inhibited the growth of Salmonella Typhimurium while five inhibited growth of V. cholerae O139. Seven strains showed adherence to Caco-2 cells ranging from 20-104 per cent of adherence of an adherent strain of Escherichia coli, but all inhibited V. cholerae adherence to Caco-2 cells by 20-100 per cent. They inhibited interleukin-8 secretion from HT-29 cells, in response to V. cholerae, by 50-80 per cent. Two strains induced IL-10 and IL-12 messenger ribonucleic acid (mRNA) expression in THP-1 cells.
Interpretation & conclusions:
LAB in curd had properties consistent with probiotic potential, but these were not consistent across species. LAB abundance in curd increased rapidly at 12 h of fermentation at room temperature and declined thereafter.
Keywords: Antimicrobial resistance, cytokines, fermented milk, gastrointestinal pathogen, lactobacilli, yogurt
The lumen of the gut contains a complex consortium of microbes. The gut microbiome, that these microbes collectively constitute, profoundly influences nutritional, physiologic and protective processes in the human host. Their activity includes the fermentation of unabsorbed dietary carbohydrate with production of short chain fatty acids, production of vitamins like biotin and vitamin K, mediation of immune responses and oral tolerance, and protection of the host against invasion by pathogenic microbes1. In healthy individuals, there is a balance between beneficial and pathogenic constituents of the microbiota. Alterations in this balance can lead to a state of dysbiosis that contributes to illness and morbidity2,3. Metchnikoff was the first to associate lactic acid bacteria (LAB) in fermented yogurt with health and longevity of certain Balkan communities4. The concept of probiotic microbes that he introduced has led to the widespread consumption of food preparations containing LAB and/or bifidobacteria, with the expectation that they will confer health benefits including reduction of cholesterol levels, improvement of immune function, resistance to infectious diseases, and prevention of colon cancer5. The Food and Agricultural Organization (FAO) defines probiotics as live microorganisms, which when administered in adequate amounts, confer a health benefit on the host and suggests that a number of in vitro characteristics may be used to predict whether a specific microbe will have probiotic properties, before subjecting them to more rigorous tests to prove that they provide a health benefit6. These are echoed in regional and societal guidelines7,8.
Fermented foods containing LAB are traditionally used every day as food in many cultures. Curd, prepared by fermentation of milk with an inoculum of previously made curd, is used in most households in southern India where it constitutes a significant part of the daily diet. The LAB that ferment the milk are likely to differ slightly in each household as there is no standardized starter culture used to prepare curd. Although curd is believed to have probiotic properties9,10, there is little documentation of this in the literature. This study was, therefore, undertaken to evaluate the LAB from home-made curd in southern India for probiotic properties.
Material & Methods
Bacterial strains and source: Clinical isolates of a pathogenic Escherichia coli, Vibrio cholerae O139, Salmonella Typhimurium strain and Shigella flexneri were kindly donated by the Department of Clinical Microbiology, Christian Medical College and Hospital (CMCH), Vellore, Tamil Nadu, India.
Growth medium: Vibrio was grown in alkaline peptone water while the remaining pathogenic strains were grown in brain heart infusion broth (Hi-media, Mumbai, India) at 37°C. Total viable counts of the LAB isolates were determined using de Man Rogosa Sharpe (MRS) agar at 37°C. Shigella and Salmonella were enumerated on Salmonella Shigella (SS) agar (Hi-media, Mumbai, India), E. coli on MacConkey agar (Hi-media, Mumbai, India) and V. cholerae on thiosulphate-citrate-bile salts-sucrose (TCBS) agar (Hi-media, Mumbai, India) after 18 h at 37°C.
Isolation of LAB and their identification by API 50CH: The study was carried out in the department of Gastrointestinal Sciences, CMCH, Vellore, Tamil Nadu, India between 2008 and 2010. Samples of freshly made curd were obtained from 16 households within a 50 km radius of Vellore and transported immediately to the laboratory. The households chosen belonged to laboratory researchers who made curd each day at home using a small portion of curd to inoculate milk which was then left covered overnight at room temperature. Curd samples were transported to the laboratory in the morning and were plated on MRS agar and incubated overnight at 37°C. Microbial colonies that grew out in culture were identified by Gram stain, catalase test and biochemical characterization11. API 50 CH was used in conjunction with API 50 CHL (bioMerieux, Chennai, India) medium for the identification of LAB. Strains identified by the API 50CH system were subjected to growth curve analysis to determine the time period during which exponential growth was seen. LAB were cultured on Lactobacillus MRS broth (Himedia, Mumbai, India) for 16 h at 37°C with 10 per cent CO2 under anaerobic conditions and used for testing.
Growth kinetics of LAB: The profile of LAB growth during curd formation was explored through time course studies involving laboratory fermentation of milk using curd as starter culture. Inocula of fresh curd were used to seed three (two pasteurized, one unpasteurized) 150 ml aliquots of milk at 1 per cent (w/v) concentration. The milk was allowed to ferment for 48 h at room temperature and a 1.5 ml aliquot removed every 4 h for culture and for deoxyribonucleic acid (DNA) isolation. The samples were streaked on MRS agar, MacConkey agar and blood agar plates. MRS agar plates were incubated for 24 h at 37°C in anaerobic conditions with 10 per cent CO2, while MacConkey agar and blood agar plates were incubated for 48 h at 37°C. After incubation, colonies were enumerated and variations in colony morphology at different durations were noted. DNA was isolated from the curd samples12 and subjected to real time polymerase chain reaction (PCR) using primers targeted at 16S rRNA gene sequences specific to Lactobacillus genus13,14. Amplification of lactobacillus sequences was expressed relative to the amplification of sequences conserved for domain bacteria (universal).
Studies done with LAB: The following experiments were carried out when the strains were in early stationary phase. The isolates were screened for attributes consistent with probiotic properties. These included resistance to acid and pepsin, bile salt and pancreatin tolerance, antimicrobial resistance, intrinsic antimicrobial production, adhesion to (and inhibition of pathogen adhesion to) Caco-2 intestinal epithelial cells, modulation of IL-8 response in HT-29 colonic epithelial cells and modulation of cytokine expression in THP-1 monocyte cells.
Tolerance to acid, pepsin, bile and pancreatin and intrinsic haemolytic activity: The ability of the organisms to survive adverse conditions was assessed by incubating the organisms in MRS broth culture in presence of these adverse influences, and removing aliquots at specified times for plating on MRS agar plates to ascertain growth. The ability of the isolates to survive in the presence of hydrochloric acid (pH 1.0 and pH 3.0), pepsin (3 mg/ml, pH 2.0), pancreatin (1 mg/ml, pH 8.0) and bile salts (0.3% w/v Ox Gall) was measured. The reagents for these tests were obtained from Sigma Chemical Co., USA. Intrinsic haemolytic activity was determined by culture on Columbia blood agar plates for 48 h and observing haemolysis around the colonies.
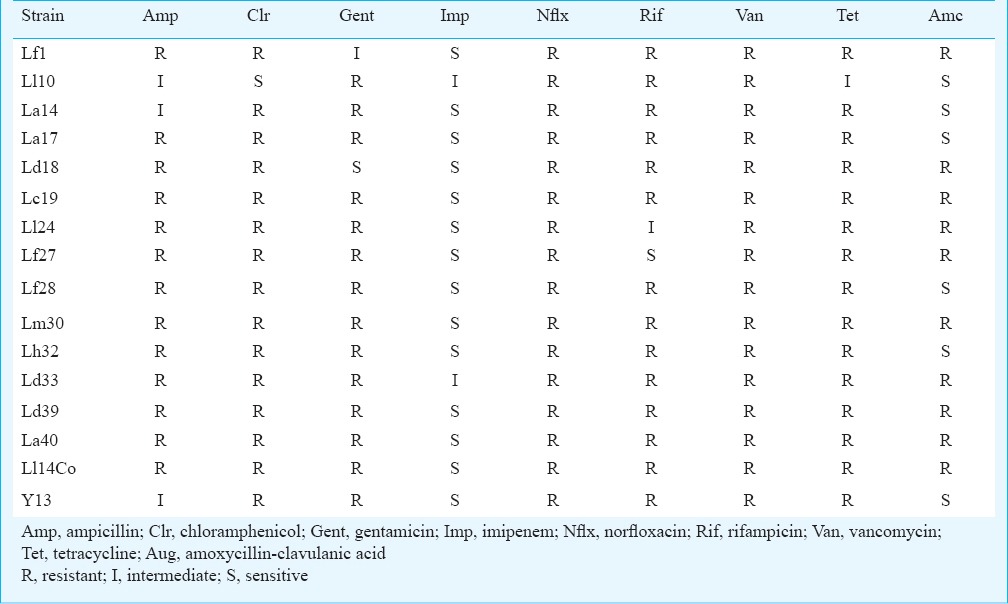
Antimicrobial susceptibility and activity: Antimicrobial sensitivity to ampicillin, co-amoxyclav, gentamicin, chloramphenicol, rifampicin, norfloxacin, vancomycin and imipenem were determined using the Kirby-Bauer disc diffusion method15 (Himedia, Mumbai, India) with standard definitions for antimicrobial resistance or sensitivity16. The discs contained the following amounts per disc (in µg): Ampicillin (10), co-amoxyclav (30), gentamicin (10). chloramphenicol (30), rifampicin (5), norfloxacin (10), vancomycin (30), imipenem (10), tetracycline (30).
Intrinsic antimicrobial activity against E. coli, S. Typhimurium, V. cholerae and S. flexneri was determined by well-diffusion in BHI soft agar plates and zones of inhibition were measured.
Adherence to Caco-2 cells: The ability of the bacteria to adhere to Caco-2 cells17, a colon cancer cell line with small intestinal differentiation was tested. Caco-2 cells were grown in Dulbecco's modified Eagle medium (DMEM) (Sigma Chemical Co., USA), supplemented (v/v) with 10 per cent foetal calf serum inactivated at 56°C for 30 min, 1 per cent non-essential amino acids, 1 per cent glutamine and 20 µg/ml of streptomycin and penicillin, at 37°C in 10 per cent CO2/90 per cent air. The cells were seeded into 24-well tissue culture plates (BD, Gurgaon, India) at a concentration of 1 million cells per ml, and incubated for seven days with change of medium every two days. Two ml volumes of early stationary growth phase broth cultures were pelleted, washed and suspended in non-supplemented DMEM to a concentration corresponding to McFarland standard 3. One ml of suspension containing 108 bacteria was overlaid onto the Caco-2 monolayers, and plates incubated at 37°C in 10 per cent CO2/90 per cent air for 90 min. The microbial suspension was aspirated, the monolayers washed twice, and 1 ml of 0.1 per cent Triton X-100 added for ten minutes to detach adherent microbial cells. The cells were plated on MRS agar at 1:100 and 1:10000 dilutions and incubated at 37°C in 10 per cent CO2/90 per cent air for 24 h, and colonies enumerated18. E. coli was used as a positive control and the adhesion of the test strains was expressed relative to the E. coli adhesion value.
Inhibition of pathogen adhesion: The ability of the strains to inhibit pathogen adhesion to Caco-2 cells was also tested in vitro19. Briefly, Caco-2 monolayers in 24 well plates were overlaid with the test strains for 90 min, after which the bacteria were removed and the cell monolayer washed thrice with DMEM. One ml of DMEM containing 108 V. cholerae or S. Typhimurium cells was overlaid on the monolayers, incubated for 90 min, and adherent V. cholera or S. Typhimurium enumerated using the procedure described above.
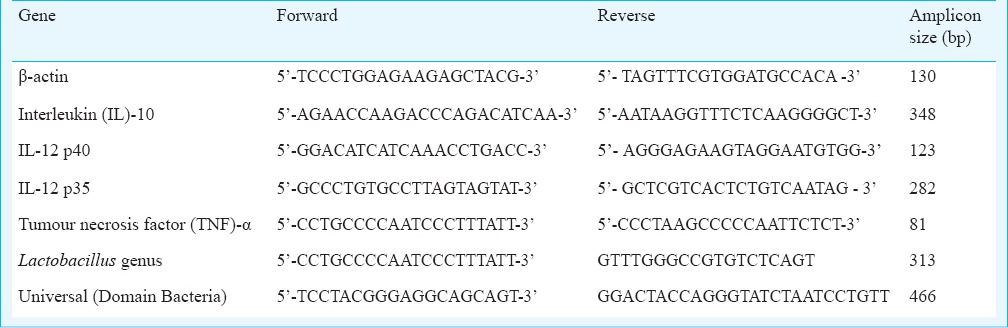
Relative expression of interleukin (IL)-8, IL-10, IL-12, and tumour necrosis factor (TNF)-α by real time PCR and ELISA: The bacteria were tested for modulation of cytokine expression or release from immune cells. Modulation of interleukin-8 release from HT-29 cells in response to V. cholerae was used as a test system to determine putative probiotic properties in the isolated lactic acid bacteria20. Pre-exposure to the test isolates (108/ml) for 90 min was followed by exposure of the cell monolayers to 108 V. cholerae cells for four h, after which supernatant and cells were separately saved for analysis. IL-8 concentrations in the supernatant were estimated by ELISA (Opt EIA Set, BD Biosciences, USA). Modulation of the cytokine response in THP-1 monocyte-macrophage cell lines was used to investigate the LAB for potential probiotic properties. THP-1 is a human mononuclear leukaemia cell line that can secrete the anti-inflammatory cytokine IL-10 and was grown in flasks, dissociated, washed and resuspended in medium prior to the experiments. THP-1 cell suspension (1×106 cells/ml/well) was placed in 24-well tissue culture plates (Falcon Milliwell, BD, USA), and incubated for one hour. Bacterial cells suspended in non-supplemented RPM1 (108 cells/ml) were added and incubated at 37°C in 10 per cent CO2/90 per cent air for two h. The microbial suspension was aspirated from the wells, one ml of fresh RPMI medium containing 20 µg gentamicin added to each well, and incubated at 37°C in 10 per cent CO2/90 per cent air for 4 h. The wells were aspirated and the suspensions were centrifuged at 7500 × g for 10 min at 4°C. To the pellets, 300 µl of TRI reagent was added and stored at -80°C for RNA isolation. RNA was isolated in batches using TRI reagent and converted to cDNA using a reverse transcriptase core kit (Eurogentec, Liege, Belgium). Expression of the housekeeping gene actin was monitored to confirm cDNA conversion. Real-time PCR using SYBR green20 was carried out to quantitate IL-10, IL-12 and TNF-α gene expression relative to expression of β-actin, using appropriate primers (Table I).
Table I.
Primers used in real time PCR
Results
Growth kinetics of LAB: Fig. 1 shows the proportion of Lactobacillus 16S rRNA genes as a proportion of the total 16S rRNA pool in curd and indicates that Lactobacillus formed the major microbial population at 12 h. The isolated strains were cultured and aliquots removed at different time points and suspended in phosphate buffer saline (PBS) and compared against McFarland standards. This indicated that microbial concentration in the cultures plateaued at around 16 h (Fig. 2). This was the point of time when the microbes were harvested for studies.
Fig. 1.
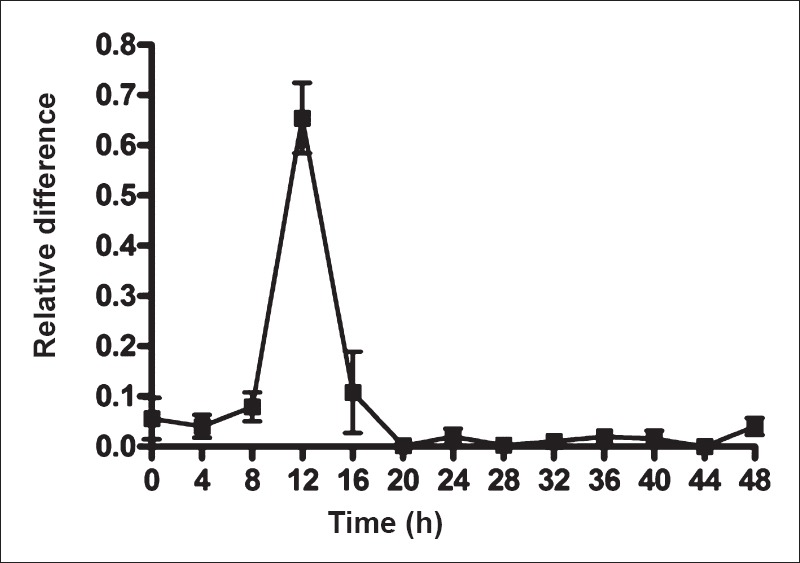
Growth of lactic acid bacteria (LAB) during milk fermentation to curd at room temperature. The values shown are mean ± SEMof three fermentations. Lactobacilli were quantitated relative to total bacteria by real time polymerase chain reaction targeting 16S rRNA gene sequences specific for lactobacilli.
Fig. 2.
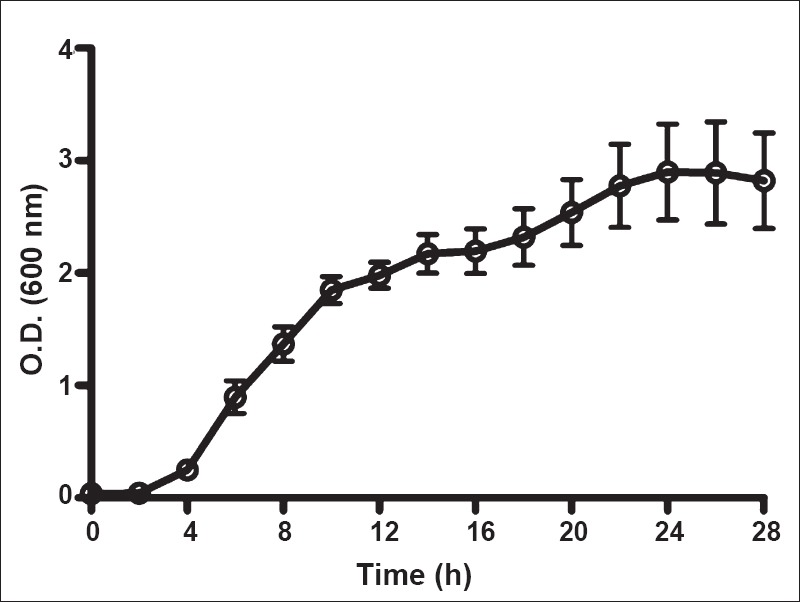
Growth curve of the LAB in culture. Optical density at 600 nm was used to assess density of growth. Values shown are ± SEMof 14 observations.
Isolation and identification of LAB: LAB were characterized for biochemical reactions using the Lactobacillus API 50CH (bioMerieux India, Chennai) test panel. Following 48 h of incubation, changes in medium colour in each of the 50 cupules were noted, and the strains were then identified using the apiweb™ software (bioMerieux SA, France). Growth curve analysis revealed that L. fermentum 1, Leuconostoc lactis, L. acidophilus 1, L. confusus and L. delbrueckii ssp. bulgaricus strains showed exponential growth between 4 and 10 h, and the others (L. acidophilus 3, L. delbrueckii ssp. delbrueckii, L. lindneri, Leuconostoc mesenteroides ssp. cremoris and L. helveticus) between 6 and 24 h. Thirty five Gram positive bacterial strains were isolated from the curd specimens, 15 of which were clearly identified as distinct species using the API 50CH. These 15 LAB (12 lactobacilli, 1 Lactococcus and 2 Leuconostoc) and one yeast, each from a separate sample of curd, were taken up for study (Table II).
Table II.
List of microbial (lactic acid bacteria) species taken up for characterization
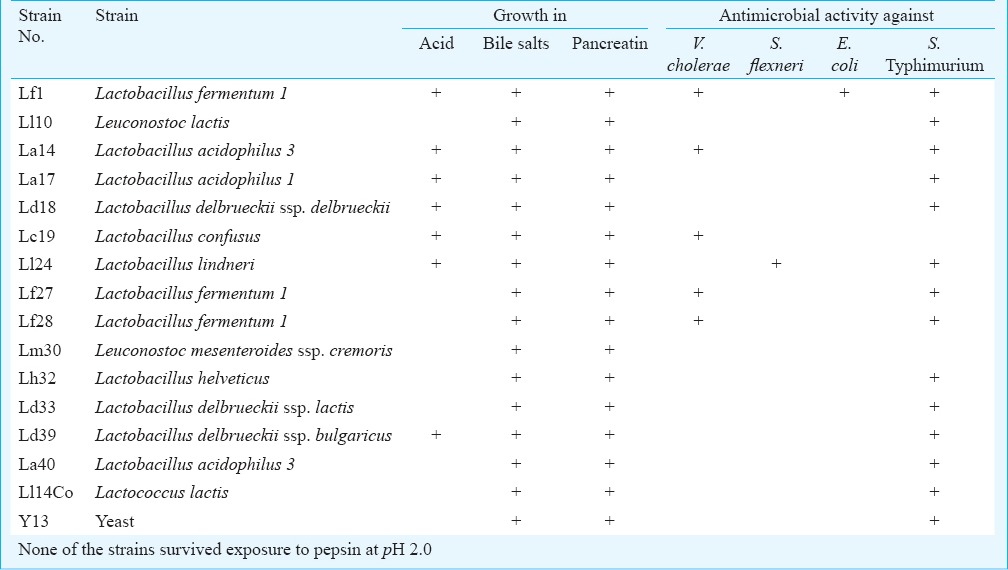
Ability to survive adverse culture conditions: On exposure to pH 3.0, seven of the survived for one hour and two for 3 h. The per cent of input bacteria that survived ranged from 0 to 6 per cent at the end of 3 h in pH 3.0, indicating that the microbiota isolated from curd were not capable of survival in low pH. On exposure to pH 1.0, none of the strains survived for an hour. None of the strains, except for one of Lactobacillus delbrueckii ssp. delbrueckii, survived exposure to pepsin at pH 2.0. All the strains survived four h of exposure to pancreatin at pH 8.0. Similarly, all strains survived exposure to bile salts for 3 h. In the case of pancreatin and bile salts, the survival after 3 h of the individual strains ranged from 77 to 120 per cent of the input bacteria for the different strains.
Haemolytic capacity and antimicrobial susceptibility in vitro: None of the strains tested exhibited any sign of haemolysis when grown on blood agar for 24 h under anaerobic conditions. The strains were also tested for evidence of resistance to commonly used antimicrobial classes using antimicrobial disk diffusion and growth for 24 h. All the tested strains were resistant to most of the antimicrobials tested including vancomycin, norfloxacin, ampicillin, chloramphenicol, gentamicin, rifampicin, tetracycline and amoxicillin-clavulanic acid (Table III). All strains were sensitive to imipenem.
Table III.
Antimicrobial susceptibility pattern of the LAB isolated from curd samples.
Intrinsic antimicrobial activity against pathogens: The intrinsic antimicrobial activity of the isolated strains was tested against four different pathogens using the well diffusion method. Fourteen of the 16 strains tested inhibited the growth of S. Typhimurium (Fig. 3). Five strains showed an inhibitory effect towards V. cholerae 0139. One strain was able to inhibit E. coli and another strain inhibited Shigella.
Fig. 3.
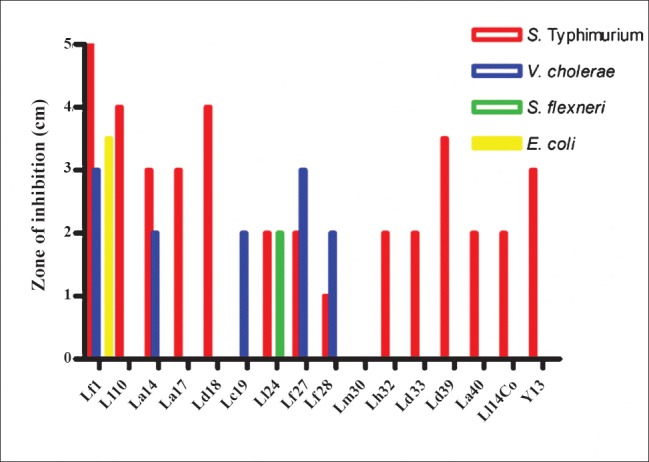
Intrinsic antimicrobial activity of the LAB strains against several gastrointestinal pathogens. Zones of inhibition (cm) are shown.
Microbial adherence to epithelial cells and inhibition of pathogen adherence: All the tested strains except Lactobacillus delbrueckii ssp. lactis were capable of adhering to Caco-2 cells (Fig. 4). Seven of the strains exhibited good adherence ranging from 20-104 per cent when compared to the control E. coli used to test epithelial cell adherence which showed adherence of approximately 105 bacteria per well. Adherence intensity of the remaining strains varied from 2-20 per cent compared to the control E. coli. Most of the strains were able to inhibit the adherence of V. cholerae O139 to Caco-2 cells (Fig. 5). Vibrio adherence was inhibited almost completely by two strains - L. acidophilus 1 and L. delbrueckii ssp. delbrueckii - and by 80 per cent by two other strains. The remaining strains showed varying degrees of inhibition of Vibrio adherence, ranging from 20 to 60 per cent. Most of the strains did not inhibit adherence of S. Typhimurium (Fig. 5).
Fig. 4.
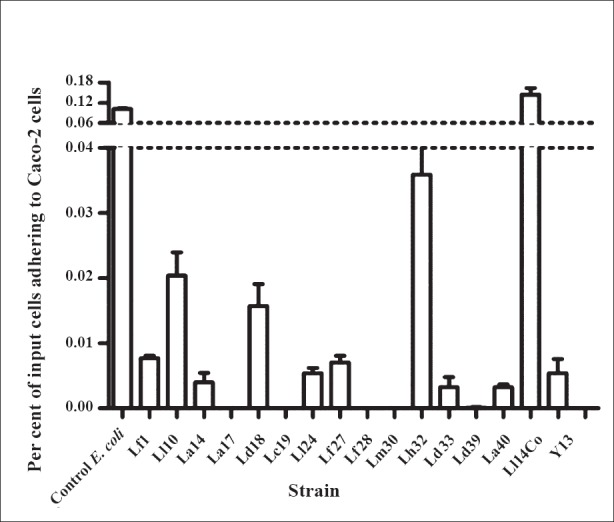
Adherence of LAB to Caco-2 cells. The Y-axis shows mean ± SE per cent of input bacteria adhering to the epithelial cell monolayer (n=3 experiments).
Fig. 5.
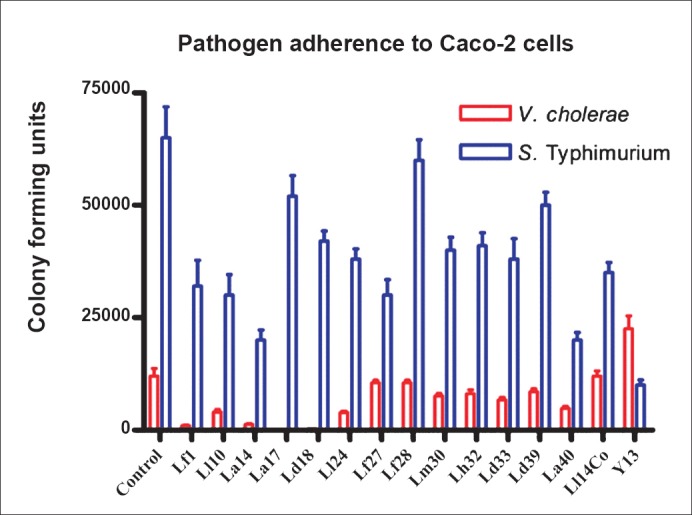
Inhibition of V. cholerae and S. Typhimurium adherence to Caco-2 cells by LABisolated in this study. Values shown are mean±SE colony forming units of pathogen adhering to Caco-2 cells after addition of test microbial strains which are named on the X-axis. Experiments were done in duplicate.
Effects on cytokine secretion by cell lines: HT-29 cells were exposed to the test strains prior to exposure to V. cholerae O139 that is known to induce a brisk IL-8 secretion from these cells. Most of the LAB strains could reduce the IL-8 response to V. cholerae by 50-80 per cent (Fig. 6). Two strains each induced increased expression of IL-10 and IL-12 mRNA in THP-1 cells, while the others had no perceptible effect. Most strains induced some expression of TNF-α mRNA in THP-1 cells (Fig. 7).
Fig. 6.
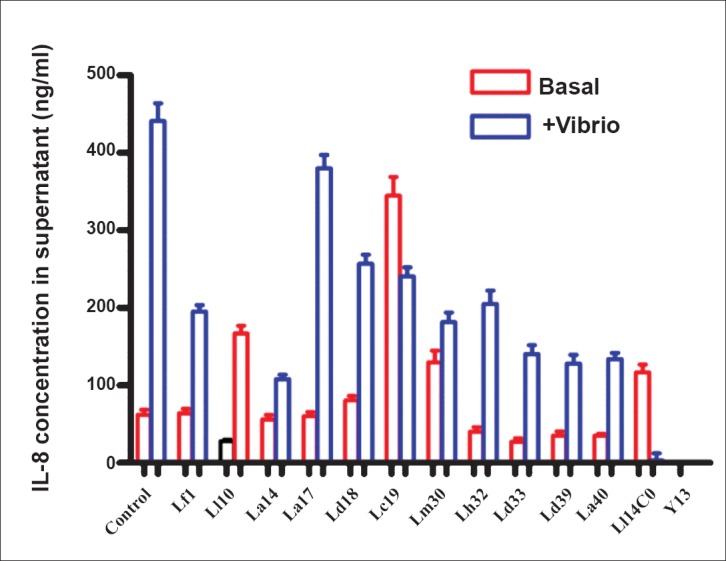
Interleukin (IL)-8 secretion in μg/ml (mean+SE) from HT-29 cells in response to exposure to V. cholerae and the effect of LAB.
Fig. 7.
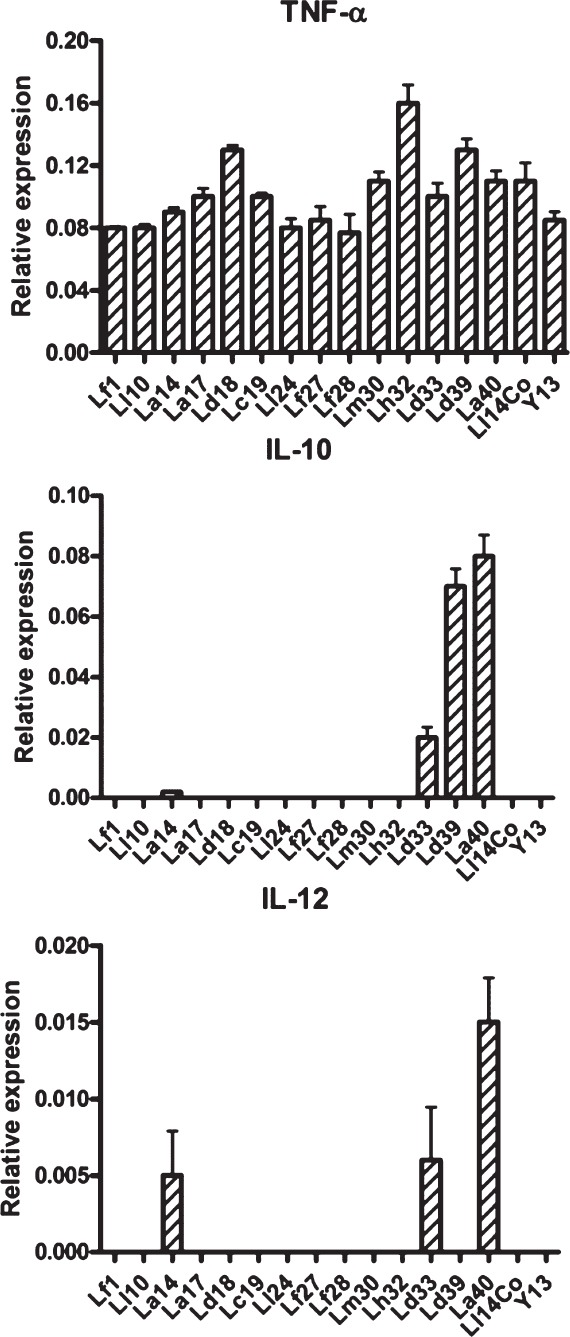
Messenger RNA expression of cytokines in THP-1 cells exposed to the various lactic acid bacilli (LAB). The values are shown as mean±SE (n=3) expression relative to that of beta-actin.
Discussion
Curd (fermented milk), freshly made by inoculation with starter curd, is a good source of bioavailable vitamin, minerals and folate21, and has less lactose and galactose than milk. Consumption of curd may enhance the immune response in the elderly. These health benefits have been linked to the presence of live bacteria22. It is not hitherto known whether LAB present in home-made curd have the potential to confer health benefits.
The Joint FAO/WHO Working Party Report in 20026 suggested that microbes should have a minimum set of characteristics that could predict probiotic potential. These included the ability to resist passage through the stomach in the presence of acid and pepsin, and the ability to grow in the proximal small intestine in the presence of pancreatin and bile salts. Highly potent probiotic strains would have additional properties including antibiotic resistance, adhesion to intestinal epithelia, antimicrobial activity, inhibition of pathogen adhesion, and immunomodulation potential. The FAO/WHO Working Party recommended testing for these properties using reliable in vitro screening methods, followed by clinical studies to confirm putative health benefits.
Only one of the strains studied survived exposure to pepsin although several survived exposure to acid for varying periods of time. However, it is unlikely that bacteria ingested in curd will be stressed to the extent as done in the present study, because the food with which curd is eaten is likely to buffer gastric acid and to limit pepsin effect. The pH in human stomach ranges from 1.0 during fasting to 4.5 after a meal, and most of the examined strains were resistant to pH 3.0 even after 3 h of exposure. These results are consistent with reports of ability of lactobacilli to retain their viability when exposed to pH values of 2.5-4.023,24. None of the lactobacilli survived in pepsin at pH 2.0, in consonance with published literature25. Probiotic bacteria in curd will have the advantage of the presence of curd proteins which have a protective effect on the starters and support bacterial survival in the acidic environment of the stomach23,25. Even strains not able to survive at pH 1.0 in vitro may exhibit substantial viability when consumed as starters or adjuncts in a matrix of fermented milk. In contrast to pepsin, all the strains examined in this study survived in the presence of pancreatin or bile salts simulating the near neutral small intestine environment.
All LAB tested fulfilled the safety requirement that a probiotic strain should not have β-haemolytic activity. The LAB strains exhibited resistance to a wide variety of antimicrobials. It is most likely that this reflects intrinsic resistance (rather than genetic resistance which can be transferred) to the specified antimicrobials, due to lack of targets in the microbes for these antimicrobials. This property also makes them useful in combination with antibiotics, to prevent antibiotic-induced diarrhoea.
Probiotic bacteria prevent the growth of pathogenic species by competing for nutrition and attachment sites on the colonic epithelium, and through production of organic acids and bacteriocins26. Several of the curd-derived LAB in this study inhibited growth of enteric pathogens. This is a useful property, especially to prevent the development of food-borne illness in tropical warm climates. Inhibition was noted specifically against S. Typhimurium and V. cholerae O139, but not against pathogenic E. coli or S. flexneri.
Adherence of lactobacilli to the intestinal epithelium is considered to be essential for the exertion of a beneficial (probiotic) effect in the large intestine27,28. Most of the LAB tested in this study displayed adherence to Caco-2 cells, in numbers similar to that previously reported by others28. These LAB also inhibited pathogen adherence to Caco-2 cells. Such inhibition of pathogen adherence to eukaryotic cell lines has been reported for strains such as L. johnsonii La1, Bifidobacterium CA1 and F9, and L. acidophilus LB29,30,31. In many cases, direct inhibitory activity has been linked to possible bacteriocin production by the probiotic strains and/or to their competitive adherence to epithelial cells, resulting in the inability of the pathogen to bind to its normal attachment site32. The adhesion of V. cholerae O139 to Caco-2 cells was reduced to more than 50 per cent. Strains with higher adhesive ability might well occupy common adhesion sites and thus prevent the further adhesion of the pathogenic bacterium through a process of exclusion26. On the other hand, even a weakly binding Lactobacillus strain on a Caco-2 cell could stimulate cellular mechanisms (immunomodulation), which may lead to increased colonization resistance or may interfere with pathogen adhesion to epithelium through mechanisms such as direct antagonism26. Further studies are required to elucidate the mechanisms underlying probiotic adherence and inhibition of pathogen adherence.
Epithelial cells are major constituents of the intestinal mucosal barrier and participate in innate immune and inflammatory responses in the gut. Modulation of interleukin-8 secretion by epithelial cell line HT-29 is consistent with probiotic potential of the LAB tested. Sub-epithelial cells, including monocytes, also participate in inflammatory responses in the bowel, secreting inflammatory products such as Th1 type cytokines, chemokines and several active oxides. Expression of anti-inflammatory cytokines in the monocyte cell line THP-1 was upregulated by two of the LAB strains indicating the possibility of an anti-inflammatory effect. This needs to be confirmed by further study. Lactobacilli are known to have anti-inflammatory effects in animal models of colitis33.
It is likely that the daily ingestion of curd in certain cultures and communities confers health benefits beyond nutrient intake in curd. A recent study using an animal model and parallel human studies showed that though long term consumption of fermented milk products did not change faecal microbiota composition, it significantly changed the expression of microbial enzymes involved in many metabolic pathways, particularly those related to carbohydrate fermentation34.
In conclusion, the home-made curds that were evaluated possessed a spectrum of LAB that had the potential to exert probiotic effects in individuals consuming the curd. These microbes had properties that predicted their ability to survive passage in the upper gut and to modulate gut mucosal innate immune reactions and, therefore, it is likely they have the potential to significantly influence health status. Further studies should focus on the contribution of daily curd intake to health in India.
Acknowledgment
Authors acknowledge the financial support received from the Department of Biotechnology, Government of India, New Delhi.
References
- 1.Kau AL, Ahern PP, Griffin NW, Goodman AL, Gordon JI. Human nutrition, the gut microbiome and the immune system. Nature. 2011;474:327–36. doi: 10.1038/nature10213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hawrelak JA, Myers SP. The causes of intestinal dysbiosis: a review. Altern Med Rev. 2004;9:180–97. [PubMed] [Google Scholar]
- 3.Henao-Mejia J, Elinav E, Jin C, Hao L, Mehal WZ, Strowig T, et al. Inflammasome-mediated dysbiosis regulates progression of NAFLD and obesity. Nature. 2012;482:179–85. doi: 10.1038/nature10809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Metchnikoff E. London: William Heinemann; 1907. The prolongation of life: optimistic studies. [Google Scholar]
- 5.Nagpal R, Kumar A, Kumar M, Behare PV, Jain S, Yadav H. Probiotics, their health benefits and applications for developing healthier foods: a review. FEMS Microbiol Lett. 2012;334:1–15. doi: 10.1111/j.1574-6968.2012.02593.x. [DOI] [PubMed] [Google Scholar]
- 6.Guidelines for the evaluation of probiotics in food. London, Ontario: Food and Agriculture Organization; 2002. Food and Agriculture Organization and World Health Organization. [Google Scholar]
- 7.Indian Council of Medical Research Task Force; Co-ordinating Unit ICMR; Co-ordinating Unit DBT. ICMR-DBT Guidelines for Evaluation of Probiotics in Food. Indian J Med Res. 2011;134:22–5. [PMC free article] [PubMed] [Google Scholar]
- 8.Guarner F, Khan AG, Garisch J, Eliakim R, Gangl A, Thomson A, et al. World Gastroenterology Organization. World Gastroenterology Organisation Global Guidelines: Probiotics and Prebiotics October 2011. J Clin Gastroenterol. 2012;46:468–81. doi: 10.1097/MCG.0b013e3182549092. [DOI] [PubMed] [Google Scholar]
- 9.Dewan P, Kaur I, Chattopadhya D, Faridi MMA, Agarwal KN. A pilot study on the effects of curd (dahi) and leaf protein concentrate in children with protein energy malnutrition (PEM) Indian J Med Res. 2007;126:199–203. [PubMed] [Google Scholar]
- 10.Saran S, Gopalan S, Krishna TP. Use of fermented foods to combat stunting and failure to thrive. Nutrition. 2002;18:393–6. doi: 10.1016/s0899-9007(01)00790-0. [DOI] [PubMed] [Google Scholar]
- 11.Holt JG. 9th ed. Baltimore: Williams & Wilkins; 1994. Bergey's manual of determinative bacteriology. [Google Scholar]
- 12.Apajalahti JH, Särkilahti LK, Mäki BR, Heikkinen JP, Nurminen PH, Holben WE. Effective recovery of bacterial DNA and per cent-guanine-plus-cytosine-based analysis of community structure in the gastrointestinal tract of broiler chickens. Appl Environ Microbiol. 1998;64:4084–8. doi: 10.1128/aem.64.10.4084-4088.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Balamurugan R, Janardhan HP, George S, Raghava MV, Muliyil J, Ramakrishna BS. Molecular studies of fecal anaerobic commensal bacteria in acute diarrhea in children. J Pediatr Gastroenterol Nutr. 2008;46:514–9. doi: 10.1097/MPG.0b013e31815ce599. [DOI] [PubMed] [Google Scholar]
- 14.Rekha R, Rizvi MA, Paul J. Designing and validation of genus-specific primers for human gut flora study. Electron J Biotechnol. 2006;9:505–11. [Google Scholar]
- 15.Bauer AW, Kirby WM, Sherris JC, Turck M. Antibiotic susceptibility testing by a standardized single disk method. Am J Clin Pathol. 1966;45:493–6. [PubMed] [Google Scholar]
- 16.Clinical and Laboratory Standards Institute (CLSI) 11th ed. Wayne PA, USA: CLSI; 2012. Performance standards for antimicrobial disk susceptibility tests; approved standard. [Google Scholar]
- 17.Jacobsen CN, Rosenfeldt Nielsen V, Hayford AE, Moller PL, Michaelsen KF, Paerregaard A, et al. Screening of probiotic activities of forty-seven strains of Lactobacillus spp. by in vitro techniques and evaluation of the colonization ability of five selected strains in humans. Appl Environ Microbiol. 1999;65:4949–56. doi: 10.1128/aem.65.11.4949-4956.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Burkholder KM, Bhunia AK. Salmonella enterica serovar Typhimurium adhesion and cytotoxicity during epithelial cell stress is reduced by Lactobacillus rhamnosus GG. Gut Pathog. 2009;1:14. doi: 10.1186/1757-4749-1-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Santos A, San Mauro M, Sanchez A, Torres JM, Marquina D. The antimicrobial properties of different strains of Lactobacillus spp. isolated from kefir. Syst Appl Microbiol. 2003;26:434–7. doi: 10.1078/072320203322497464. [DOI] [PubMed] [Google Scholar]
- 20.Nandakumar NS, Pugazhendhi S, Ramakrishna BS. Effects of enteropathogenic bacteria & amp; lactobacilli on chemokine secretion & amp; Toll like receptor gene expression in two human colonic epithelial cell lines. Indian J Med Res. 2009;130:170–8. [PubMed] [Google Scholar]
- 21.Cano PG, Aguero G, Perdigon G. Immunological effects of yogurt addition to a re-nutrition diet in a malnutrition experimental model. J Dairy Res. 2002;69:303–16. doi: 10.1017/s0022029902005411. [DOI] [PubMed] [Google Scholar]
- 22.Bourlioux P, Pochart P. Nutritional and health properties of yogurt. World Rev Nutr Diet. 1998;56:217–58. doi: 10.1159/000416229. [DOI] [PubMed] [Google Scholar]
- 23.Conway PL, Gorbach SL, Goldin BR. Survival of lactic acid bacteria in the human stomach and adhesion to intestinal cells. J Dairy Sci. 1987;70:1–12. doi: 10.3168/jds.S0022-0302(87)79974-3. [DOI] [PubMed] [Google Scholar]
- 24.Dunne C, O’Mahony L, Murphy L, Thornton G, Morrissey D, O’Halloran S, et al. In vitro selection criteria for probiotic bacteria of human origin: correlation with in vivo findings. Am J Clin Nutr. 2001;73(2 Suppl):386S–92S. doi: 10.1093/ajcn/73.2.386s. [DOI] [PubMed] [Google Scholar]
- 25.Charteris WP, Kelly PM, Morelli L, Collins JK. Development of an in vitro methodology to determine the transit tolerance of potentially probiotic Lactobacillus and Bifidobacterium species in the upper human gastrointestinal tract. J Appl Microbiol. 1998;84:759–68. doi: 10.1046/j.1365-2672.1998.00407.x. [DOI] [PubMed] [Google Scholar]
- 26.Tuomola EM, Salminen SJ. Adhesion of some probiotic and dairy Lactobacillus strains to Caco-2 cell cultures. Int J Food Microbiol. 1998;41:45–51. doi: 10.1016/s0168-1605(98)00033-6. [DOI] [PubMed] [Google Scholar]
- 27.Matijasic BB, Narat M, Zoric M. Adhesion of two Lactobacillus gasseri probiotic strains on Caco-2 cells. Food Technol Biotechnol. 2003;41:83–8. [Google Scholar]
- 28.Bernet-Camard MF, Liévin V, Brassart D, Neeser JR, Servin AL, Hudault S. The human Lactobacillus acidophilus strain LA1 secretes a nonbacteriocin antibacterial substance(s) active in vitro and in vivo. Appl Environ Microbiol. 1997;63:2747–53. doi: 10.1128/aem.63.7.2747-2753.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Coconnier MH, Liévin V, Lorrot M, Servin AL. Antagonistic activity of Lactobacillus acidophilus LB against intracellular Salmonella enterica serovar Typhimurium infecting human enterocyte-like Caco-2/TC-7 cells. Appl Environ Microbiol. 2000;66:1152–7. doi: 10.1128/aem.66.3.1152-1157.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liévin-Le Moal V, Amsellem R, Servin AL, Coconnier MH. Lactobacillus acidophilus (strain LB) from the resident adult human gastrointestinal microflora exerts activity against brush border damage promoted by a diarrhoeagenic Escherichia coli in human enterocyte-like cells. Gut. 2002;50:803–11. doi: 10.1136/gut.50.6.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ouwehand AC, Tuomola EM, Tölkkö S, Salminen S. Assessment of adhesion properties of novel probiotic strains to human intestinal mucus. Int J Food Microbiol. 2001;64:119–26. doi: 10.1016/s0168-1605(00)00440-2. [DOI] [PubMed] [Google Scholar]
- 32.Preidis GA, Hill C, Guerrant RL, Ramakrishna BS, Tannock GW, Versalovic J. Probiotics, enteric and diarrheal diseases, and global health. Gastroenterology. 2011;140:8–14. doi: 10.1053/j.gastro.2010.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nanda Kumar NS, Balamurugan R, Jayakanthan K, Pulimood A, Pugazhendhi S, Ramakrishna BS. Probiotic administration alters the gut flora and attenuates colitis in mice administered dextran sodium sulfate. J Gastroenterol Hepatol. 2008;23:1834–9. doi: 10.1111/j.1440-1746.2008.05723.x. [DOI] [PubMed] [Google Scholar]
- 34.McNulty NP, Yatsunenko T, Hsiao A, Faith JJ, Muegge BD, Goodman AL, et al. The impact of a consortium of fermented milk strains on the gut microbiome of gnotobiotic mice and monozygotic twins. Sci Transl Med. 2011;3:106ra106. doi: 10.1126/scitranslmed.3002701. [DOI] [PMC free article] [PubMed] [Google Scholar]


