Abstract
Background
Mozambique implemented artemisinin-based combinations therapy (ACT) using artemether-lumefantrine (AL) as the first-line treatment for uncomplicated malaria in 2009. AL remains highly efficacious, but widespread use may soon facilitate emergence of artemisinin tolerance/resistance. The prevalence of pfmdr1 different alleles in Maputo and Mozambique is not known, either after or before the introduction of ACT. Pfmdr1 molecular markers related to Plasmodium falciparum susceptibility were analysed before and after transition to ACT.
Methods
A first group of samples was collected between June 2003 and June 2005 and a second group in the period between March 2010 and March 2012. Three alleles were analysed by PCR-RFLP: N86Y, Y184F and D1246Y, in the pfmdr1 gene.
Results
Alleles N86, 184F and D1246 increased from 19.5, 19.6 and 74.4% in 2003–2005 to 73.2, 22.7 and 96.7% in 2010–2012, respectively. After implementation of ACT (2010–2012), pfmdr1 haplotypes, either two- and three-codon basis, were generally less diverse than before the implementation of ACT (2003–2005). The prevalence of haplotypes N86-184Y, N86-D1246 and 184Y-D1246 increased from 12,2, 27.3 and 71.7% in 2003–2005 to 59.4, 84.3 and 78.6% in 2010–2012. The three-codon basis haplotypes NFD and NYD also increased significantly during the same period.
Conclusion
The alleles N86 and 184 F and the triple haplotype N86-184 F-D1246 showed a significantly increased prevalence after introduction of ACT.
Keywords: Malaria, Mozambique, pfmdr1, ACT
Background
The Plasmodium falciparum multi-drug resistance gene 1 (pfmdr1) and particularly, single nucleotide polymorphisms (SNPs) resulting in an amino acid change in codons 86 (N86Y), 184 (Y184F), and 1246 (D1246Y) have been associated with changes in parasite susceptibility to various drugs, including artemisinin-based combination therapy (ACT) [1–4]. Initially, SNPs in pfmdr1 were associated with chloroquine (CQ) and amodiaquine (AQ) resistance [5]. For instance, the pfmdr1 86Y mutation has been associated with high CQ resistance [6, 7], and the combination of pfmdr1 86Y, Y184, and 1246Y is likely selected by AQ monotherapy and associated with increased risk of treatment failure [1, 2]. Although SNPs at positions 1034 and 1042 of pfmdr1 were described as being related to phenotype modulation by altering a drug pocket in PfMDR1 [8], they are very infrequent in Africa.
ACT drug resistance has recently been reported at the Thai-Cambodian border [9–11]. Historical evidence shows that the emergence of drug-resistant P. falciparum strains first originated in Southeast Asia and then, spread to Africa [12]. East Africa has been a major focus of drug resistance spread, and development in sub-Saharan Africa [12, 13], probably originated at Southeast Asia [13–15].
Surveillance of changes in prevalence of pfmdr1 SNPs may serve as an early warning tool of emerging P. falciparum tolerance/resistance to ACT [16]. Mozambique, an Eastern Africa country, with a population of 20.2 million, is amongst the ten most affected countries by malaria in the world. Here, malaria represents one of the major public heath challenges, being responsible for nearly 45% of registered disease episodes, 56% of cases admitted to paediatric health facilities and about 26% of deaths in hospitals [17]. During the last decade, African countries have changed first-line treatment of uncomplicated falciparum malaria to ACT due to the development of resistance to successively introduced anti-malarial drugs. In Mozambique, CQ was replaced by sulphadoxine-pyrimethamine (SP) in 2002 (in combination with AQ) and in 2004 AQ was replaced by artesunate (ATN) in that combination. Since 2009, this combination was replaced by artemether-lumefantrine (AL) [17, 18].
Apart from the work of Raman and colleges [19], the prevalence of pfmdr1 different alleles in Maputo and Mozambique is still scarce, either after or before the introduction of ACT. In line with this, molecular markers related to P. falciparum susceptibility were analysed in samples from Maputo for the period before and after transition to ACT.
Methods
Biological samples
DNA samples included in this study were collected from patient blood spots obtained at three health facilities in Maputo area. One study was conducted at Hospital Central de Maputo, Centro de Saúde de Bagamoio and Centro de Saúde de Boane between June 2003 and June 2005 and the other at Centro de Saúde de Boane and Centro de Saúde 1° de Maio between March 2010 and March 2012. Both studies were reviewed and approved by the Ethical Committees of the Ministry of Health of Mozambique and informed consent was obtained at the time.
Genetic characterization of the parasites
Included samples came from PCR-confirmed P. falciparum infections [20]. DNA was extracted from filter paper blood spots using Chelex as described elsewhere [21]. Plasmodium falciparum mutations in pfmdr1 gene were typed by PCR-RFLP as described elsewhere [21, 22], primers sequences, amplification cycles and restrictions enzymes for 86Y and 1246Y are described in [21] and for Y184 in [23].
Statistical analysis
Allele and haplotype prevalence in the groups were compared by Chi-squared tests and, when appropriate, a residual analysis was performed in order to determine reasons for the rejection of independency between the variables under study. Statistical significance was set at p ≤ 0.05. All calculations were performed with IBM SPSS 20.0.
Results
In this study, the presence of alleles of the pfmdr1 gene (codons 86, 184 and 1246) in patients with uncomplicated falciparum malaria, before (2003–2005) and after the implementation of ACT in Maputo area (2010–2012) were analysed. A total of 133 samples were included in the group 2003–2005 (only 56 of the 133 samples were analysed for the codon 184) and 351 in 2010–2012.
The typing efficiency for each codon was as follows: 2003–2005 group, 100% of samples for the N86Y, 100% for Y184F (only 56 of the 133 samples were analysed) and 91% for D1246Y; 2003–2005 group, 96.9% of samples for the N86Y, 99.1% for Y184F and 96.0% for D1246Y.
The individual prevalence of SNPs at codons 86, 184 and 1246 in 2003–2005 and 2010–2012, including mixed codon infections, are shown in Figure 1 with Table 1 showing individual SNPs and haplotypes.
Figure 1.
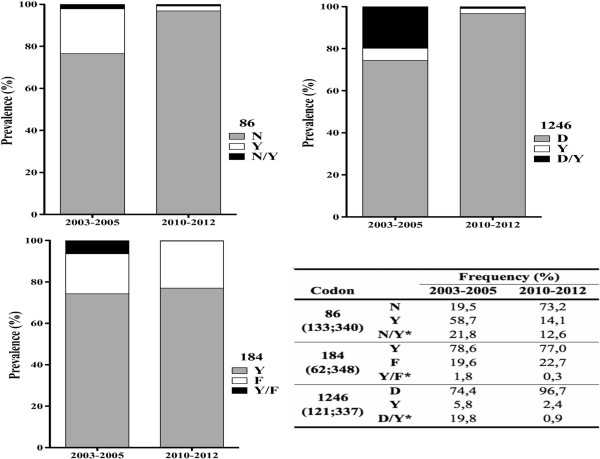
Temporal changes of prevalence at codons 86, 184 and 1246. Codons frequencies in each sample group are presented in the inserted table (numbers in brackets represent number of samples corresponding to 2003–2005, 2010–2012 and *representing mixed genotype infections).
Table 1.
Prevalence of Pfmdr1 haplotypes at codon N86Y, Y184F and D1246Y in Maputo, before and after the introduction of artemisinin-based combination therapy
| Codon | Haplotype | 2003-2005* | 2010-2012* |
|---|---|---|---|
| 86/184 (49;293) | NF | 10.2 | 24.3 |
| YF | 8.2 | 0.3 | |
| NY | 12.2 | 59.4 | |
| YY | 69.4 | 16.0 | |
| 86/1246 (77;286) | ND | 27.3 | 84.4 |
| NY | 1.3 | 0.3 | |
| YD | 64.9 | 13.6 | |
| YY | 6.5 | 1.7 | |
| 184/1246 (53;330) | FY | 0.0 | 0.0 |
| FD | 20.8 | 23.3 | |
| YY | 7.5 | 2.4 | |
| YD | 71.7 | 74.3 | |
| 86/184/1246 (45;282) | NFD | 11.1 | 24.8 |
| NFY | 0.0 | 0.0 | |
| NYY | 2.2 | 0.4 | |
| NYD | 11.1 | 59.2 | |
| YYY | 4.4 | 1.8 | |
| YFD | 8.9 | 0.4 | |
| YFY | 0.0 | 0.0 | |
| YYD | 62.2 | 13.5 |
*Sample collection period. Numbers in brackets represent number of samples collectedduring the correspondent periods 2003–2005;2010–2012.
To examine for temporal changes in prevalence of SNPs at codons 86, 184 and 1246, mixed infections (both alleles present) were analysed together with the polymorphism not associated with ACT tolerance/resistance, i.e., pfmdr1 86Y, Y184, 1246Y. The wild type allele N86 significantly recovered in prevalence from 19.5% in 2003–2005 to 73.2% 2010–2012 (p < 0.0001). Although allele 184F also increased in prevalence from 19.6% in 2003–2005 to 22.7% in 2010–2012, it was not statistically significant (p = 0.7300). The prevalence of allele D1246 also increased significantly from 74.4 to 96.7% in 2010–2012 (p < 0.0001). In 2010–2012, mixed infections were less frequent, a 58% decrease in multiple infections was observed for codon 86 and more than 80% for codons 184 and 1246 (Figure 1).
Haplotypes were compared on a two- and three-codon basis (Table 1) and analysed for temporal prevalence change. Minority haplotypes (<5%) and mixed infections were excluded from the analysis. After implementation of ACT (2010–2012), pfmdr1 haplotypes, either two- and three-codon basis, were generally less diverse than before the implementation of ACT (2003–2005) (Table 1). From 2003–2005 to 2010–2012, haplotypes N86-184Y and N86-F184 significantly increased from 12.2 to 59.4% and 10.2 to 24.3%, respectively (p < 0.0001). On the contrary, 86Y-Y184 decreased from 69.4% in 2003–2005 to 16.0% in 2010–2012 (p < 0.0001) (Table 1). Comparison of the codons combination 86–1246 showed a significant increase of the N86-D1246 haplotype from 27.3% in 2003–2005 to 84.4% in 2010–2012 (p < 0.0001). Haplotype 86Y-D1246 of the same combination, decreased from 64.9 to 13.6% (p < 0.0001). Although the prevalence of haplotypes F184-D1246 and 184Y-D1246 increased from 2003–2005 to 2010–2012 (Table 1) it did not reach statistical significance (p = 0.7455). The three-codon basis haplotype analysis revealed that, except for NFD and NYD, all the other codon combination decreased in prevalence from 2003–2005 to 2010–2012 (Table 1). This tendency was highly significant for YYD (from 62.2 to 13.5%; p < 0.0001). Haplotypes YFY and NFY were not detected either in 2003–2005 or 2010–2012 samples. In 2010–2012, more than half of the infections carried the NYD, showing a significant trend of increase from 11.1 to 59.2% (p < 0.0001). The haplotype NFD (associated with the ability to withstand higher lumefantrine concentrations) also presented a significant increase in prevalence from 11.1% in 2003–2005 to 24.8% in 2010–2012 (p < 0.0001).
Discussion
There are signs of decreasing malaria prevalence (5.168,684 cases in 2005 and 3.381,371 cases in 2010) in Mozambique, but the disease remains a major cause of morbidity and mortality [17, 18]. Continued surveillance of molecular markers of drug resistance and particularly, potential markers of ACT drug tolerance are important tools for the success of malaria control programmes.
In Mozambique, CQ was replaced by SP in combination with AQ in 2002, followed by SP + AS in 2004 and then SP + AS was replaced by AL as first line in 2009, because of the high levels of SP resistance. Nevertheless, from 2004 AL has been the second line of treatment of uncomplicated malaria in the country [17, 18]. In the present study, the prevalence and temporal changes of N86Y, Y184F and D1246Y pfmdr1 alleles were studied in Maputo area.
The present study found a high prevalence of mutant type 86Y and wild-type Y184 in samples from 2003–2005 (58.7 and 74.3%, respectively). This is in accordance with other studies performed in Africa where 86Y and Y184 frequencies were also high before the introduction of ACT [3–5, 16, 24–30]. Decreasing prevalence of the 86Y after introduction of ACT in Mozambique was also observed during two recent studies from Inhambane and Gaza, Mozambique [19, 31]. In line with this tendency, a high prevalence of wild type N86 and D1246 and the mutant 184Y was detected in the 2010–2012 group of samples (Figure 1), which is again in accordance withother studies in Africa [3–5, 16, 24–31]. Overall, these results suggest that the prevalence of 86Y and Y184 was high when SP and AQ were in use on a large-scale basis, but when these drugs were substituted by ACT their prevalence decreased significantly, which might indicate that this haplotype does not confer a fitness advantage upon ACT pressure [32].
Before ACT implementation (2003–2005) the most common triple haplotype (pfmdr1 86,184 and 1246) was YYD (62.2%), indicating that the previous first-line treatment based in quinolines (i.e., CQ or AQ) mainly selected pfmdr1 86Y and 184Y. The second most common haplotypes were NYD and NFD (11.1%), indicating that N86 and D1246 were already being selected by the decreasing use of quinolines and the introduction of AS in combination with SP. This is in line with the observations that, AS per se potentially selects for pfmdr1 N86 and D1246, which have been associated with decreased susceptibility to artemisinins in vitro [33, 34]. Importantly however, no such selection has been shown after monotherapy with artemisinin derivatives in vivo. Another reason for the increased N86 and D1246 prevalence might be that SNPs associated with AQ resistance (86Y and 1246Y) cause a fitness cost to the parasite [26], which would affect the selection pattern under different drug pressures. That is, when quinolines pressure was released, wild type alleles (N86, D1246) immediately begin to rise in the parasite population. The combination of SNPs 86Y-1246Y was rare in 2010–2012 group of samples (Table 1), suggesting again that they may be associated with a significant fitness cost as observed previously [26].
Five to seven years after implementation of ACT in Mozambique, a significant selection of NFD and NYD was observed in the Maputo area. Other studies with African-derived samples support the idea that haplotype N86, 184F and D1246 is AL selected [3–5], while 86Y, 184Y and 1246Y is AQ or CQ selected [2, 3, 35–37]. Additionally, a study from Thailand suggested that selection of the haplotype N86-184F was likely caused by AS [38]. Generally, these results are consistent with a recent study performed in samples from people living in a touristic area in Inhambane, Mozambique [31] with those from numerous studies in Africa [3–5, 16, 29, 30, 39].
Despite the challenges of validating candidate markers of drug resistance, namely for ACT (a combination of chemically unrelated molecules) [10, 40–43], several works point to pfmdr1 SNPs as possible modulators for ACT response mainly based on the partner drug [2–5, 8, 23, 28–31, 35, 42]. In the absence of definite marker for artemisinin (ART) resistance, molecular monitoring of its partner drug markers may contribute to predict the effectiveness of ACT. Currently, the K13-propeller mutations (recently proposed by Ariey and colleagues [44] as a molecular marker for ACT resistance) may be a useful marker for large-scale surveillance efforts to contain and prevent global spread of ACT resistance.
Conclusion
A sustained success in malaria control is strongly dependent on continued effectiveness of first line treatment. The results of the present work are in line with data found recently in other African countries, where the wild type allele N86 showed increased prevalence after introduction of AL. Furthermore, it is shown a temporal increase of 184F mutant type after introduction of AL, and a significant increase of the triple haplotype N86-184F-D1246. Therefore, there is cause for concern/attention and close continued surveillance of pfmdr1 SNPs, which remain highly relevant as a marker of reduced susceptibility to AL.
Acknowledgements
The authors acknowledge the people from Maputo, Mozambique and the collaboration of the local laboratory workers from Hospital Central de Maputo, Centro de Saúde de Bagamoio, Centro de Saúde de Boane and Centro de Saúde 1° de Maio. This study was supported by Centro de Malaria e OutrasDoençasTropicais (CMDTLA), InstitutoPortuguês de ApoioaoDesenvolvimento (IPAD) and FundaçãoGulbenkian.
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
EL, SR, PF, LL, and SP participated in laboratory procedures. EL, NF and FN participated in overall study conception and design, data collection, analysis, interpretation and manuscript preparation. BdS participated in the statistical analysis, interpretation and manuscript preparation. All authors read and approved the final manuscript.
Contributor Information
Elsa Lobo, Email: loboelsa@yahoo.com.br.
Bruno de Sousa, Email: bruno.desousa@fpce.uc.pt.
Soraia Rosa, Email: soraiarocharosa@gmail.com.
Paula Figueiredo, Email: pauladfigueiredo@gmail.com.
Lis Lobo, Email: liscoelho5@hotmail.com.
Sara Pateira, Email: sc_pateira@hotmail.com.
Natercia Fernandes, Email: naterciaf@yahoo.com.br.
Fatima Nogueira, Email: fnogueira@ihmt.unl.pt.
References
- 1.Holmgren G, Hamrin J, Svärd J, Mårtensson A, Gil JP, Björkman A. Selection of pfmdr1 mutations after amodiaquine monotherapy and amodiaquine plus artemisinin combination therapy in East Africa. Infect Genet Evol. 2007;7:562–569. doi: 10.1016/j.meegid.2007.03.005. [DOI] [PubMed] [Google Scholar]
- 2.Humphreys GS, Merinopoulos I, Ahmed J, Whitty CJM, Mutabingwa TK, Sutherland CJ, Hallett RL. Amodiaquine and artemether-lumefantrine select distinct alleles of the Plasmodium falciparum mdr1 gene in Tanzanian children treated for uncomplicated malaria. Antimicrob Agents Chemother. 2007;51:991–997. doi: 10.1128/AAC.00875-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sisowath C, Ferreira PE, Bustamante LY, Dahlström S, Mårtensson A, Björkman A, Krishna S, Gil JP. The role of pfmdr1 in Plasmodium falciparum tolerance to artemether-lumefantrine in Africa. Trop Med Int Health. 2007;12:736–742. doi: 10.1111/j.1365-3156.2007.01843.x. [DOI] [PubMed] [Google Scholar]
- 4.Sisowath C, Strömberg J, Mårtensson A, Msellem M, Obondo C, Björkman A, Gil JP. In vivo selection of Plasmodium falciparum pfmdr 186 N coding alleles by artemether-lumefantrine (Coartem) J Infect Dis. 2005;191:1014–1017. doi: 10.1086/427997. [DOI] [PubMed] [Google Scholar]
- 5.Duraisingh MT, Drakeley CJ, Muller O, Bailey R, Snounou G, Targett GAT, Greenwood BM, Warhurst DC. Evidence for selection for the tyrosine-86 allele of the pfmdr 1 gene of Plasmodium falciparum by chloroquine and amodiaquine. Parasitology. 1997;114:205–211. doi: 10.1017/S0031182096008487. [DOI] [PubMed] [Google Scholar]
- 6.Khalil IF, Alifrangis M, Tarimo DS, Staalsø T, Satti GMH, Theander TG, Rønn AM, Bygbjerg IC. The roles of the pfcrt 76 T and pfmdr 186Y mutations, immunity and the initial level of parasitaemia, in predicting the outcome of chloroquine treatment in two areas with different transmission intensities. Ann Trop Med Parasitol. 2005;99:441–448. doi: 10.1179/136485905X46441. [DOI] [PubMed] [Google Scholar]
- 7.Duraisingh MT, Cowman AF. Contribution of thepfmdr1 gene to antimalarial drug-resistance. Acta Trop. 2005;94:181–190. doi: 10.1016/j.actatropica.2005.04.008. [DOI] [PubMed] [Google Scholar]
- 8.Ferreira PE, Holmgren G, Veiga MI, Uhlén P, Kaneko A, Gil JP. PfMDR1: mechanisms of transport modulation by functional polymorphisms. PLoS One. 2011;6:e23875. doi: 10.1371/journal.pone.0023875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dondorp AM, Nosten F, Yi P, Das D, Phyo AP, Tarning J, Lwin KM, Ariey F, Hanpithakpong W, Lee SJ, Ringwald P, Silamut K, Imwong M, Chotivanich K, Lim P, Herdman T, An SS, Yeung S, Singhasivanon P, Day NP, Lindegardh N, Socheat D, White NJ. Artemisinin resistance in Plasmodium falciparum malaria. N Engl J Med. 2009;361:455–467. doi: 10.1056/NEJMoa0808859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lim P, Alker A, Khim N, Shah N, Incardona S, Doung S, Yi P, Bouth D, Bouchier C, Puijalon O, Meshnick SR, Wongsrichanalai C, Fandeur T, Le Bras J, Ringwald P, Ariey F. Pfmdr1copy number and arteminisin derivatives combination therapy failure in falciparum malaria in Cambodia. Malar J. 2009;8:11. doi: 10.1186/1475-2875-8-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Alker AP, Lim P, Sem R, Shah NK, Yi P, Bouth DM, Tsuyuoka R, Maguire JD, Fandeur T, Ariey F, Wongsrichanalai C, Meshnick SR. Pfmdr1 and in vivo resistance to artesunate-mefloquine in falciparum malaria on the Cambodian–Thai border. Am J Trop Med Hyg. 2007;76:641–647. [PubMed] [Google Scholar]
- 12.Naidoo I, Roper C. Following the path of most resistance: dhps K540E dispersal in African Plasmodium falciparum. Trends Parasitol. 2010;26:447–456. doi: 10.1016/j.pt.2010.05.001. [DOI] [PubMed] [Google Scholar]
- 13.Clarke VD. Chloroquine-resistant malaria acquired in Kenya and Tanzania- Denmark, Georgia, New York. Cent Afr J Med. 1979;25:39–40. [PubMed] [Google Scholar]
- 14.Wongsrichanalai C, Pickard AL, Wernsdorfer WH, Meshnick SR. Epidemiology of drug-resistant malaria. Lancet Infect Dis. 2002;2:209–218. doi: 10.1016/S1473-3099(02)00239-6. [DOI] [PubMed] [Google Scholar]
- 15.Payne D. Spread of chloroquine resistance in Plasmodium falciparum. Parasitol Today. 1987;3:241–246. doi: 10.1016/0169-4758(87)90147-5. [DOI] [PubMed] [Google Scholar]
- 16.Lekana-Douki JB, Boutamba SDD, Zatra R, Edou SEZ, Ekomy H, Bisvigou U, Toure-Ndouo FS. Increased prevalence of the Plasmodium falciparum Pfmdr 186 N genotype among field isolates from Franceville, Gabon after replacement of chloroquine by artemether–lumefantrine and artesunate–mefloquine. Infect Genet Evol. 2011;11:512–517. doi: 10.1016/j.meegid.2011.01.003. [DOI] [PubMed] [Google Scholar]
- 17.ProgramaNacional de Controlo da Malária. InquéritoNacionalsobreIndicadores de MaláriaemMoçambiqu. 2007. http://www.malariasurveys.org/documents/IMM Inquerito Malaria 2007(Portuguese).pdf
- 18.ProgramaNacional de Controlo da Malária. Plano Estratégico da Malária 2012–2016. http://www.nationalplanningcycles.org/sites/default/files/country_docs/Mozambique/malaria_plano_estrategico_draftfinal_jan_2012.pdf
- 19.Raman J, Mauff K, Muianga P, Mussa A, Maharaj R, Barnes KI. Five years of antimalarial resistance marker surveillance in Gaza Province, Mozambique, following artemisinin-based combination therapy roll out. PLoS One. 2011;6:e25992. doi: 10.1371/journal.pone.0025992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Singh B, Bobogare A, Cox-Singh J, Snounou G, Abdullah MS, Rahman HA. A genus- and species-specific nested polymerase chain reaction malaria detection assay for epidemiologic studies. Am J Trop Med Hyg. 1999;60:687–692. doi: 10.4269/ajtmh.1999.60.687. [DOI] [PubMed] [Google Scholar]
- 21.Lopes D, Nogueira F, Gil JP, Ferreira C, do Rosario VE, Cravo P. pfcrt and pfmdr1 mutations and chloroquine resistance in Plasmodium falciparumfrom Sao Tome and Principe, West Africa. Ann Trop Med Parasitol. 2002;96:831–834. doi: 10.1179/000349802125002284. [DOI] [PubMed] [Google Scholar]
- 22.Figueiredo P, Benchimol C, Lopes D, Bernardino L, Do-Rosario V, Varandas L, Nogueira F. Prevalence of pfmdr1, pfcrt, pfdhfr and pfdhps mutations associated with drug resistance, in Luanda, Angola. Malar J. 2008;7:236. doi: 10.1186/1475-2875-7-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Duraisingh MT, Roper C, Walliker D, Warhurst DC. Increased sensitivity to the antimalarials mefloquine and artemisinin is conferred by mutations in the pfmdr1 gene of Plasmodium falciparum. Mol Microbiol. 2000;36:955–961. doi: 10.1046/j.1365-2958.2000.01914.x. [DOI] [PubMed] [Google Scholar]
- 24.Schneider AG, Premji Z, Felger I, Smith T, Abdulla S, Beck H-P, Mshinda H. A point mutation in codon 76 of pfcrt of P. falciparumis positively selected for by Chloroquine treatment in Tanzania. Infect Genet Evol. 2002;1:183–189. doi: 10.1016/S1567-1348(01)00021-1. [DOI] [PubMed] [Google Scholar]
- 25.Thomsen TT, Ishengoma DS, Mmbando BP, Lusingu JP, Vestergaard LS, Theander TG, Lemnge MM, Bygbjerg IC, Alifrangis M. Prevalence of single nucleotide polymorphisms in the Plasmodium falciparum multidrug resistance gene (Pfmdr-1) in Korogwe District in Tanzania before and after introduction of artemisinin-based combination therapy. Am J Trop Med Hyg. 2011;85:979–983. doi: 10.4269/ajtmh.2011.11-0071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Froberg G, Jornhagen L, Morris U, Shakely D, Msellem M, Gil J, Bjorkman A, Martensson A. Decreased prevalence of Plasmodium falciparum resistance markers to amodiaquine despite its wide scale use as ACT partner drug in Zanzibar. Malar J. 2012;11:321. doi: 10.1186/1475-2875-11-321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gadalla N, Abdallah T, Atwal S, Sutherland C, Adam I. Selection of pfdhfr/pfdhps alleles and declining artesunate/sulphadoxine-pyrimethamine efficacy against Plasmodium falciparum eight years after deployment in eastern Sudan. Malar J. 2013;12:255. doi: 10.1186/1475-2875-12-255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Amor A, Toro C, Fernandez-Martinez A, Baquero M, Benito A, Berzosa P. Molecular markers in Plasmodium falciparumlinked to resistance to anti-malarial drugs in samples imported from Africa over an eight-year period (2002–2010): impact of the introduction of artemisinin combination therapy. Malar J. 2012;11:100. doi: 10.1186/1475-2875-11-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Duah N, Matrevi S, de Souza D, Binnah D, Tamakloe M, Opoku V, Onwona C, Narh C, Quashie N, Abuaku B, Duplessis C, Kronmann KC, Koram KA. Increased pfmdr1 gene copy number and the decline in pfcrt and pfmdr1 resistance alleles in Ghanaian Plasmodium falciparum isolates after the change of anti-malarial drug treatment policy. Malar J. 2013;12:377. doi: 10.1186/1475-2875-12-377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dokomajilar C, Nsobya SL, Greenhouse B, Rosenthal PJ, Dorsey G. Selection of Plasmodium falciparumpfmdr1 alleles following therapy with artemether-lumefantrine in an area of Uganda where malaria is highly endemic. Antimicrob Agents Chemother. 2006;50:1893–1895. doi: 10.1128/AAC.50.5.1893-1895.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thomsen TT, Madsen LB, Hansson HH, Tomás EVE, Charlwood D, Bygbjerg IC, Alifrangis M. Rapid selection of Plasmodium falciparum chloroquine resistance transporter gene and multidrug resistance gene-1 haplotypes associated with past chloroquine and present artemether-lumefantrine use in Inhambane District, Southern Mozambique. Am J Trop Med Hyg. 2013;88:536–541. doi: 10.4269/ajtmh.12-0525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fröberg G, Ferreira PE, Mårtensson A, Ali A, Björkman A, Gil JP. Assessing the cost-benefit effect of a Plasmodium falciparum drug resistance mutation on parasite growth in vitro. Antimicrob Agents Chemother. 2013;57:887–892. doi: 10.1128/AAC.00950-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reed MB, Saliba KJ, Caruana SR, Kirk K, Cowman AF. Pgh1 modulates sensitivity and resistance to multiple antimalarials in Plasmodium falciparum. Nature. 2000;403:906–909. doi: 10.1038/35002615. [DOI] [PubMed] [Google Scholar]
- 34.Mwai L, Kiara SM, Abdirahman A, Pole L, Rippert A, Diriye A, Bull P, Marsh K, Borrmann S, Nzila A. In vitro activities of piperaquine, lumefantrine, and dihydroartemisinin in Kenyan Plasmodium falciparum isolates and polymorphisms in pfcrt andpfmdr1. Antimicrob Agents Chemother. 2009;53:5069–5073. doi: 10.1128/AAC.00638-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Holmgren G, Gil JP, Ferreira PM, Veiga MI, Obonyo CO, Björkman A. Amodiaquine resistant Plasmodium falciparum malaria in vivo is associated with selection of pfcrt 76 T and pfmdr1 86Y. Infect Genet Evol. 2006;6:309–314. doi: 10.1016/j.meegid.2005.09.001. [DOI] [PubMed] [Google Scholar]
- 36.Kublin JG, Cortese JF, Njunju EM, Mukadam GRA, Wirima JJ, Kazembe PN, Djimdé AA, Kouriba B, Taylor TE, Plowe CV. Reemergence of chloroquine-aensitive Plasmodium falciparummalaria after cessation of chloroquine use in Malawi. J Infect Dis. 2003;187:1870–1875. doi: 10.1086/375419. [DOI] [PubMed] [Google Scholar]
- 37.Sutherland CJ, Alloueche A, Curtis J, Drakeley CJ, Ord R, Duraisingh M, Greenwood BM, Pinder M, Warhurst D, Targett GAT. Gambian children successfully treated with chloroquine can harbor and transmit Plasmodium falciparum gametocytes carrying resistance genes. Am J Trop Med Hyg. 2002;67:578–585. doi: 10.4269/ajtmh.2002.67.578. [DOI] [PubMed] [Google Scholar]
- 38.Mungthin M, Suwandittakul N, Chaijaroenkul W, Rungsrihirunrat K, Harnyuttanakorn P, Seugorn A, Na Bangchang K. The patterns of mutation and amplification of Plasmodium falciparumpfcrt and pfmdr1 genes in Thailand during the year 1988 to 2003. Parasitol Res. 2010;107:539–545. doi: 10.1007/s00436-010-1887-x. [DOI] [PubMed] [Google Scholar]
- 39.Price RN, Uhlemann A-C, Brockman A, McGready R, Ashley E, Phaipun L, Patel R, Laing K, Looareesuwan S, White NJ, Nosten F, Krishna S. Mefloquine resistance in Plasmodium falciparumand increased pfmdr1 gene copy number. Lancet. 2004;364:438–447. doi: 10.1016/S0140-6736(04)16767-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Simpson JA, Jamsen KM, Anderson TJ, Zaloumis S, Nair S, Woodrow C, White NJ, Nosten F, Price RN. Nonlinear mixed-effects modelling of in vitro drug susceptibility and molecular correlates of multidrug resistant Plasmodium falciparum. PLoS One. 2013;8:e69505. doi: 10.1371/journal.pone.0069505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pradines B, Bertaux L, Pomares C, Delaunay P, Marty P. Reduced in vitro susceptibility to artemisinin derivatives associated with multi-resistance in a traveller returning from South-East Asia. Malar J. 2011;10:268. doi: 10.1186/1475-2875-10-268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mungthin M, Khositnithikul R, Sitthichot N, Suwandittakul N, Wattanaveeradej V, Ward SA, Na-Bangchang K. Association between the pfmdr1 gene and in vitro artemether and lumefantrine sensitivity in Thai isolates of Plasmodium falciparum. Am J Trop Med Hyg. 2010;83:1005–1009. doi: 10.4269/ajtmh.2010.10-0339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Valecha N, Srivastava P, Mohanty SS, Mittra P, Sharma SK, Tyagi PK, Pradhan K, Dev V, Singh R, Dash AP, Sharma YD. Therapeutic efficacy of artemether-lumefantrine in uncomplicated falciparum malaria in India. Malar J. 2009;8:107. doi: 10.1186/1475-2875-8-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ariey F, Witkowski B, Amaratunga C, Beghain J, Langlois AC, Khim N, Kim S, Duru V, Bouchier C, Ma L, Lim P, Leang R, Duong S, Sreng S, Suon S, Chuor CM, Bout DM, Ménard S, Rogers WO, Genton B, Fandeur T, Miotto O, Ringwald P, Le Bras J, Berry A, Barale JC, Fairhurst RM, Benoit-Vical F, Mercereau-Puijalon O, Ménard D. A molecular marker of artemisinin-resistant Plasmodium falciparum malaria. Nature. 2014;505:50–55. doi: 10.1038/nature12876. [DOI] [PMC free article] [PubMed] [Google Scholar]


