Abstract
Objective:
To determine the proportion of children with herpes simplex encephalitis (HSE) displaying TLR3 deficiency, the extent of TLR3 allelic heterogeneity, and the specific clinical features of TLR3 deficiency.
Methods:
We determined the sequence of all exons of TLR3 in 110 of the 120 patients with HSE enrolled in our study who do not carry any of the previously described HSE-predisposing mutations of TLR3 pathway genes (TLR3, UNC93B1, TRIF, TRAF3, and TBK1). All the new mutant TLR3 alleles detected were characterized experimentally in-depth to establish the causal relationship between the genotype and phenotype.
Results:
In addition to the 3 previously reported TLR3-deficient patients from the same cohort, 6 other children or young adults with HSE carry 1 of 5 unique or extremely rare (minor allele frequency <0.001) missense TLR3 alleles. Two alleles (M374T, D592N) heterozygous in 3 patients are not deleterious in vitro. The other 3 are deleterious via different mechanisms: G743D+R811I and L360P heterozygous in 2 patients are loss-of-function due to low levels of expression and lack of cleavage, respectively, and R867Q homozygous in 1 patient is hypomorphic. The 3 patients' fibroblasts display impaired TLR3 responses and enhanced herpes simplex virus 1 susceptibility. Overall, TLR3 deficiency is therefore found in 6 (5%) of the 120 patients studied. There is high allelic heterogeneity, with 3 forms of autosomal dominant partial defect by negative dominance or haploinsufficiency, and 2 forms of autosomal recessive defect with complete or partial deficiency. Finally, 4 (66%) of the 6 TLR3-deficient patients had at least 1 late relapse of HSE, whereas relapse occurred in only 12 (10%) of the total cohort of 120 patients.
Conclusions:
Childhood-onset HSE is due to TLR3 deficiency in a traceable fraction of patients, in particular the ones with HSE recurrence. Mutations in TLR3 and TLR3 pathway genes should be searched and experimentally studied in children with HSE, and patients with proven TLR3 deficiency should be followed carefully.
TLR3 is one of the most highly conserved TLRs in humans that have evolved under the strongest purifying selection.1 TLR3 recognizes double-stranded RNA (dsRNA), a by-product produced during the viral replication of most viruses, including herpes simplex virus 1 (HSV-1).2 The most common known clinical consequence of human TLR3 deficiency is childhood herpes simplex encephalitis (HSE). Childhood HSE is a rare life-threatening complication of primary infection with HSV-1, a common neurotropic dsDNA virus that is innocuous in most children.3 HSE is the most common form of sporadic viral encephalitis in Western countries.4,5 The pathogenesis of HSE had long remained unclear. Our recent studies have demonstrated that HSE may result from single-gene inborn errors of TLR3-mediated immunity in some children,6 with homozygous or heterozygous mutations of a TLR3 pathway gene (TLR3, UNC93B1, TRIF, TRAF3, and TBK1).6 TLR3 is expressed on CNS-resident cells that are permissive for HSV-1 infection.7 High susceptibility to HSV-1 infection in patient-specific induced pluripotent stem cell (iPSC)–derived UNC-93B- and TLR3-deficient neurons and oligodendrocytes has been demonstrated recently.7 Impaired CNS–intrinsic TLR3–dependent interferon (IFN)-α/β and IFN-λ immunity to HSV-1 may therefore underlie HSE in children with TLR3 pathway deficiencies.7 However, only 10 of the 120 children or young adults with HSE studied by our group to date have been found to carry mutations affecting the TLR3 pathway. Two have autosomal dominant (AD) partial TLR3 deficiency8 and a third has autosomal recessive (AR) complete TLR3 deficiency.9 We investigated the morbid allelic diversity at the TLR3 locus and the proportion of patients with HSE carrying TLR3 mutations, by sequencing TLR3 in the remaining 110 patients. We describe new forms of TLR3 deficiency in human patients with early age-onset recurrent HSE.
METHODS
Patients.
Inclusion criteria were (1) age between 3 months and 15 years at the time of the first episode of HSE, or young adults developing HSE due to primary HSV-1 infection, and (2) clinically (signs of meningoencephalitis), radiologically (detectable lesions on cerebral CT scan or MRI), and virologically (CSF PCR positive for HSV-1, or detectable HSV-1-specific antibodies in serum and CSF) confirmed HSE. Clinical case reports on the 6 patients carrying novel mutations in TLR3 are provided on the Neurology® Web site at Neurology.org.
Standard protocol approvals, registrations, and patient consents.
Informed consent was obtained from patients, who were followed up in their home country, according to local regulation. All experiments were performed at Rockefeller University in the United States and Institut National de la Santé et de la Recherche Médicale in France under local regulations and the institutional review board approvals of each institution.
Molecular genetics.
Genomic DNA was extracted from leukocytes and primary fibroblasts. The exons of TLR3 were amplified by PCR and sequenced with the BigDye Terminator Cycle Sequencing Kit (Applied Biosystems, Foster City, CA). Whole exome sequencing was performed as previously described.7
Cell culture and stable transfections.
Human primary and SV40-immortalized fibroblasts were cultured as previously described.8 The TLR3-deficient P2.1 fibrosarcoma cell line was provided by Douglas W. Leaman.10 The vector encoding the C-terminally hemagglutinin (HA)–tagged pUNO-hTLR3 was purchased from InvivoGen (San Diego, CA). Mutants of the TLR3 gene were generated by site-directed mutagenesis. SV40-fibroblasts and P2.1 cells were transfected in the presence of X-tremeGENE9 Reagent (Roche, South San Francisco, CA). Blasticidin (Invitrogen, Carlsbad, CA, 5 μg/mL) was added on media for selection.
Immunoblots.
Equal amounts of total cell protein extracts from each sample were subjected to immunoprecipitation with a goat antihuman TLR3 antibody directed against the human TLR3 ectodomain (R&D Systems, Minneapolis, MN). The immunoblot procedure was performed as previously described.9 Anti-TLR3, anti-HA (InvivoGen), and anti-GAPDH (Sigma-Aldrich, St. Louis, MO) were used.
TLR3 agonist stimulation and HSV-1 infection.
Polyinosine:polycytidylic acid [poly(I:C)], a TLR-3 agonist, was used at various concentrations as indicated. Cells were infected with HSV-1 (strain KOS-1) at an MOI of 1, or HSV-1-GFP (strain KOS),11 at various MOI. Cells and supernatants were harvested and cytokine production at mRNA or protein level were determined by qPCR or ELISA, respectively, as previously described.9 The green fluorescent protein (GFP) fluorescence of the HSV-1-GFP-infected samples was quantified at various time points. For assays of cell protection upon viral infection, cells were treated with IFN-α2b (Intron A, Schering-Plough, Kenilworth, NJ) at a concentration of 104 IU/mL for 18 hours before infection.
RESULTS
Five new TLR3 mutant alleles in 6 unrelated patients with HSE.
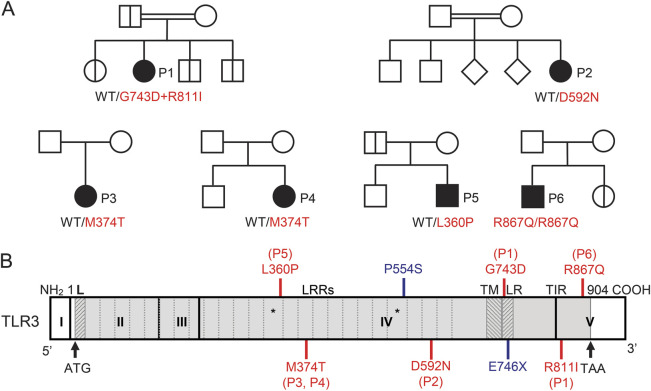
Using Sanger sequencing, we determined the sequence of all exons of TLR3 in 110 patients with HSE enrolled in our study who do not carry any of the previously described HSE-associated mutations of TLR3 pathway genes (TLR3, UNC93B1, TRIF, TRAF3, and TBK1). In 6 unrelated patients, we identified 5 rare missense mutations, and one of these patients had 2 missense mutations (figure 1A, table 1, figure e-1A, supplementary text e-1). Four of the 5 mutant alleles—G743D+R811I, D592N, M374T, and L360P—have not been reported in public databases, including the Single Nucleotide Polymorphism database (dbSNP), 1,000 genomes, which contains 1,128 human whole exomes, and the Exome Variant Server, which contains 6,503 human exomes. They were not found in our in-house whole-exome sequencing (WES) database, which contains data for 1,098 patients. Furthermore, they were not found in the Centre d'Etude du Polymorphisme Humain–Human Genome Diversity Project (CEPH-HGDP) panel, which contains DNA samples from 1,050 healthy individuals from 51 different populations from around the world.12 The fifth mutant allele, R867Q, was reported in dbSNP (rs199768900) with a minor allele frequency of ∼0.0009 and 3 DNA samples from the CEPH-HGDP DNA panel are heterozygous for the R867Q mutation, which was, however, homozygous in P6. We further performed WES in the 6 patients investigated here, and found no mutations in other essential TLR3 pathway genes, nor any homozygous HSE-relevant mutation in patient 1 or patient 2, who were born to consanguineous parents (supplementary text e-2, table e-1, table e-2). The TLR3 mutations in the 6 patients therefore appeared to be the most likely HSE-predisposing mutations present.
Figure 1. Five novel TLR3 mutant alleles in 6 unrelated patients with herpes simplex encephalitis.
(A) Family pedigrees with allele segregation in the 6 families. The patients are indicated in black. Healthy TLR3 wild-type relatives of patients 1 and 5 and heterozygous parents of patient 6 are shown in white. Heterozygous carriers of the patient 1 and 5 mutation and a homozygous sibling of patient 6 are indicated by bold vertical lines in each pedigree, respectively. The other family members of patients 2, 3, and 4 were not tested. (B) Schematic diagram of the human TLR3 gene, with the previously reported (blue) and new (red) mutations indicated at the corresponding location of each mutation. The coding exons are numbered with Roman numerals and delimited by a vertical bar. The regions corresponding to the leader sequence (L), leucine-rich repeats (LRR), transmembrane domain (TM), linker region (LR), and Toll/interleukin-1 receptor (TIR) domain are shaded in light gray and are delimited by dark gray lines. The 2 LRRs with an insertion are indicated by asterisks.
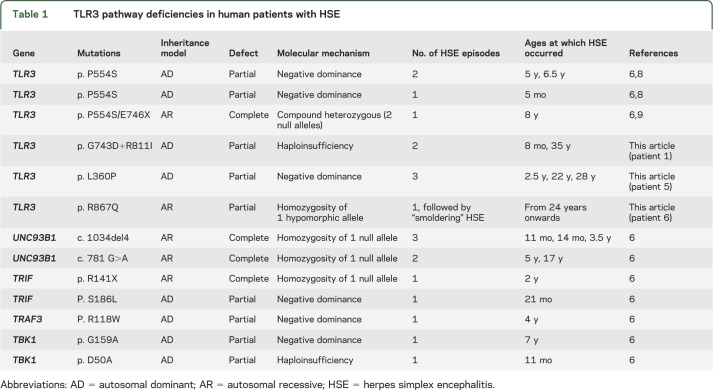
Table 1.
TLR3 pathway deficiencies in human patients with HSE
In silico study of the mutations.
In silico study of the 5 TLR3 mutants showed that the L360, G743, R811, and R867 residues were strictly conserved in all 32 species studied, whereas the M374 and D592 residues were not conserved (figure e-1B). As such, the L360P, G743D, R811I, and R867Q mutations were predicted to be highly damaging by both Polymorphism Phenotyping v2 (PolyPhen-2)13 and Sorting Intolerant From Tolerant (SIFT),14 whereas the M374T and D592N mutations were predicted to be benign. The TLR3 protein structure is represented in figure 1B. The ectodomain (ECD) of TLR3 is essential for ligand binding-triggered multimerization,15–22 and the L360P, M374T, and D592N mutations are located in the ECD of TLR3. The linker region bridges the transmembrane domain and the Toll/interleukin-1 receptor (TIR) domain, and the G743D mutation is located within. The TIR domain is essential for the recruitment of TRIF, the only known adaptor of TLR3, and thus for downstream signaling.23,24 The R811I and R867Q mutations are located in the TIR domain. Collectively, the TLR3 mutations identified, particularly L360P, G743D, R811I, and R867Q, are likely to result in a loss of TLR3 function by various molecular mechanisms, underlying AR or AD TLR3 deficiency.
Expression of the mutant TLR3 alleles in a TLR3-deficient cell line.
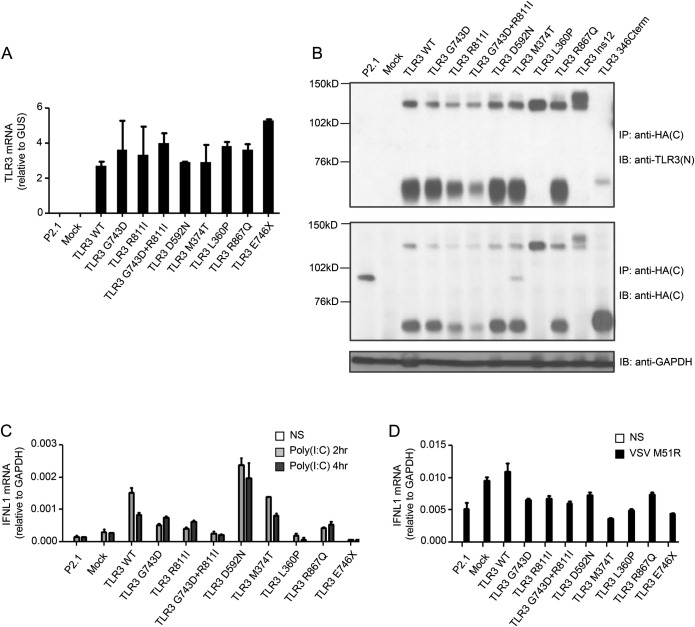
We investigated the functional impact of the 5 TLR3 mutants, by generating P2.1 cell lines stably transfected with constructs encoding C-terminally HA-tagged wild-type (WT) or mutated TLR3 proteins. P2.1 is a TLR3-deficient fibrosarcoma cell line that does not express functional TLR3 and does not respond to extracellular stimulation with poly(I:C).10 Similar levels of TLR3 mRNA were detected in P2.1 cells transfected with the WT and mutant TLR3 alleles, but not in cells with and without mock vector transfection (figure 2A). The WT TLR3 proteins were detected at 2 molecular weights (figure 2B)—about 130 kDa and about 70 kDa—corresponding to the uncleaved full-length protein and the cleaved C-terminal part of the TLR3 protein.25,26 The D592N, M374T, and R867Q TLR3 proteins were produced in similar amounts to the WT TLR3, and displayed the same cleavage pattern. The same expression pattern was also observed with the G743D, R811I, and G743D+R811I alleles, whereas the G743D and R811I alleles were expressed at very low levels, and the G743D+R811I allele was particularly weakly expressed (figure 2B). Interestingly, the L360P TLR3 appeared to be uncleaved, as we detected the 130 kDa protein but not the 70 kDa product (figure 2B). This suggests that the L360P mutation, which is located in the immediate vicinity of the TLR3 cleavage site (residues 323–356), may render the mutant protein resistant to cleavage by lysosomal cathepsins.25,26
Figure 2. Expression and function of the mutant TLR3 alleles.
(A) TLR3 mRNA levels were determined by quantitative reverse transcription PCR (RT-qPCR) in P2.1 TLR3-deficient fibrosarcoma cells with or without transfection with various TLR3 alleles (WT TLR3, G743D, R811I, G743D+R811I, D592N, M374T, L360P, R867Q, E746X mutant TLR3), or a mock vector. GUS was included for normalization. (B) TLR3 expression, as assessed by immunoblotting (IB) after immunoprecipitation (IP), in P2.1 TLR3-deficient fibrosarcoma cells not transfected (P2.1) or stably transfected with wild-type (WT) or mutant TLR3, or mock vector, with an anti-TLR3 N-terminal (N) antibody and an antihemagglutinin C-terminal tag antibody. Ins12 served as an uncleavable form lacking the entire LRR12 insertion and 346Cterm served as a C-terminal cleaved fragment, as previously established and characterized.26,27 The experiment shown is representative of 6 experiments performed. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used as an internal expression control for immunoblotting. (C) IFNL1 mRNA induction without stimulation (NS) or after 2 and 4 hours of stimulation with 25 μg/mL polyinosine:polycytidylic acid [poly(I:C)], as assessed by RT-qPCR, in P2.1 TLR3-deficient fibrosarcoma cells not transfected (P2.1) or transfected with WT TLR3, G743D, R811I, G743D+R811I, D592N, M374T, L360P, R867Q or E746X mutant TLR3, or mock vector. The E746X mutant served as a loss-of-function control. All transfections generated stable cell lines. GAPDH was included for normalization. (D) IFNL1 mRNA induction without stimulation (NS) or after 16 hours of stimulation with vesicular stomatitis virus (VSV) M51R at a multiplicity of infection (MOI) of 1. Mean values ± SD were calculated from 2 (A, D) or 3 (C) independent experiments.
Function of the TLR3 mutants expressed in a TLR3-deficient cell line.
We then studied the poly(I:C) response in P2.1 cells transfected with the WT or a mutant TLR3. Unlike WT or the D592N, or M374T mutant TLR3 alleles, the G743D+R811I and L360P alleles could not confer poly(I:C) responsiveness in P2.1 cells as measured by IFNL1 mRNA production,8,9 and the G743D, R811I, and R867Q allele only marginally confer a poly(I:C) response (figure 2C). IFNL1 mRNA was induced to comparable levels (figure 2D) in all cells after infection with a mutant of vesicular stomatitis virus (VSV M51R), a potent inducer of IFN in many human cells.27 Thus, consistent with in silico predictions, the D592N and M374T TLR3 proteins functioned normally in our overexpression system, whereas the G743D+R811I (and both G743 and R811I individually), L360P, and R867Q TLR3 proteins were dysfunctional. The function of the L360P TLR3 was severely impaired, probably because the L360P mutant could not be cleaved and the primary signaling protein was not formed.25,26 The G743D+R811I TLR3 was produced in only small amounts and its function was impaired. The R867Q mutant was produced in normal amounts but was hypomorphic in terms of function. Thus, 3 of the 5 novel TLR3 mutants had severely impaired functions.
Severely impaired poly(I:C) responses in fibroblasts from patients 1, 5, and 6.
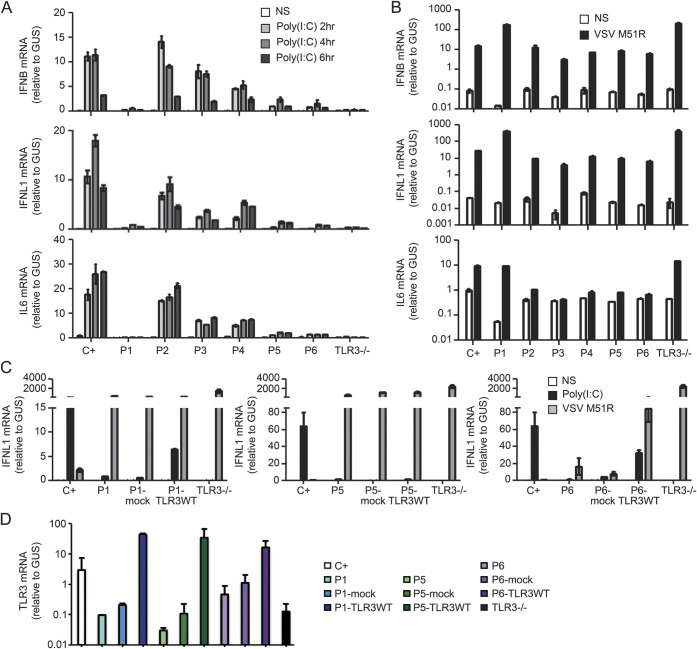
Human dermal fibroblasts display a TLR3-dependent response to extracellular stimulation with poly(I:C).8 The production of IFNB, IFNL1, and IL6 mRNA was almost abolished in heterozygous G743D+R811I fibroblasts (patient 1), and severely impaired in L360P heterozygous (patient 5) and in R867Q homozygous fibroblasts (patient 6) (figure 3A). The level of mRNA induction for IFNB, IFNL1, and IL6 in D592N and M374T heterozygous fibroblasts (patients 2, 3, and 4) was similar to that of the healthy control cells (figure 3A). As a control, mRNA for IFNB, IFNL1, and IL6 was induced to similar levels in the fibroblasts of all patients and controls, after infection with VSV M51R (figure 3B). These results were further confirmed by measuring the secretion of the IFN-β, IFN-λ, and IL-6 proteins (figure e-1C). The impaired response to poly(I:C) was further rescued in TLR3 G743D+R811I heterozygous fibroblasts from patient 1, and R867Q homozygous fibroblasts from patient 6, and partially rescued in heterozygous L360P fibroblasts from patient 5, when an exogenous WT TLR3 allele was stably expressed in these cells, as shown by the mRNA and protein production of IFN-λ (figures 3C and e-1D). Overexpression of WT TLR3 was confirmed by RT-qPCR in patient 1, 5, and 6 fibroblasts (figure 3D). Thus, these results validated our hypothesis that the heterozygous G743D+R811I and L360P alleles were responsible for AD partial TLR3 deficiency in patients 1 and 5, respectively, and that the homozygous R867Q allele was responsible for AR partial TLR3 deficiency in patient 6.
Figure 3. Impaired poly(I:C) responses in SV40 fibroblasts from patients 1, 5, and 6 and rescue of the poly(I:C) phenotype by wild-type TLR3 in SV40 fibroblasts from patients 1 and 6.
(A) IFNB, IFNL1, and IL6 mRNA levels in SV40 fibroblasts from a control (C+), patients 1–6, and a TLR3−/− patient, not stimulated (NS) or stimulated for 2, 4, and 6 hours with 25 μg/mL polyinosine:polycytidylic acid [poly(I:C)]. GUS was included for normalization. (B) IFNB, IFNL1, and IL6 mRNA induction without stimulation (NS) or after 16 hours of stimulation with vesicular stomatitis virus (VSV) M51R at a multiplicity of infection (MOI) of 1. (C) IFNL1 mRNA induction without stimulation (NS), after 4 hours of stimulation with 25 μg/mL poly(I:C), or after 16 hours of stimulation with VSV M51R. Fibroblasts from patients 1, 5, and 6 were left untransfected, mock-transfected (mock), or transfected with a vector encoding hemagglutinin–tagged wild-type (WT) TLR3. (D) TLR3 mRNA levels were assessed by quantitative reverse transcription PCR. Mean values ± SD were calculated from 3 (A, B) or 2 (C, D) independent experiments.
Three TLR3 mutants cause TLR3 deficiency by different mechanisms.
TLR3 multimerizes after binding dsRNA and several TLR3 mutations affecting the ECD, including the P554S mutation that we previously identified in 3 patients with HSE,8,9 are dominant negative (DN).17–19 The loss-of-function, uncleavable L360P TLR3 could also be DN. Control fibroblasts stably transfected with the L360P TLR3 allele lost their ability to respond to poly(I:C), in terms of IFNB and IFNL1 mRNA production, whereas transfection with the WT and G743D+R811I alleles had no such impact (figure 4A). These results were further confirmed by measuring the production of IFN-β and IFN-λ proteins in those cells (figure 4B). The expression of exogenous TLR3 was confirmed at the mRNA level (figure 4C). The loss-of-expression G743D+R811I TLR3 allele had no detectable DN effect in fibroblasts from healthy controls, suggesting that this allele underlies AD partial TLR3 deficiency (in patient 1) by haploinsufficiency. Thus, 3 novel TLR3 mutant alleles (including a double-mutated allele) define 3 novel forms of partial TLR3 deficiency in 3 patients: the uncleavable L360P mutant causes AD TLR3 deficiency due to a DN mechanism differing from that of P554S. The G743D+R811I mutant protein caused AD TLR3 deficiency due to haploinsufficiency. Finally, the hypomorphic R867Q TLR3 allele resulted in a partial form of AR TLR3 deficiency. Overall, along with the 2 previously published forms of TLR3 deficiency,8,9 there are now 5 different forms of human TLR3 deficiency (table 1).
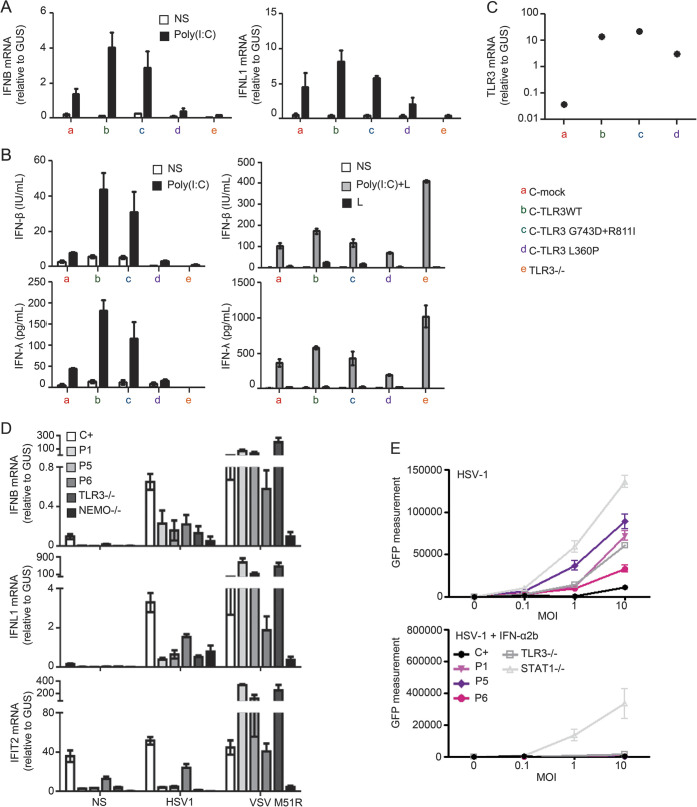
Figure 4. Dominant-negative effect of P5 allele and susceptibility of the patients' fibroblasts to herpes simplex virus 1.
(A) The induction of mRNA for IFNB and IFNL1 was assessed by quantitative reverse transcription PCR (RT-qPCR) in the absence of stimulation (NS) or after 4 hours of stimulation with 25 μg/mL polyinosine:polycytidylic acid [poly(I:C)] in SV40 fibroblasts from a healthy control transfected with an empty vector (C-mock) or with various TLR3 alleles, and in cells from a TLR3−/− patient. (B) Production of interferon (IFN)-β and IFN-λ in the absence of stimulation (NS), after 24 hours of stimulation with 25 μg/mL poly(I:C) in the presence of lipofectamine [poly(I:C)+L] or without lipofectamine [poly(I:C)], or lipofectamine alone (L), as assessed by ELISA. (C) The production of TLR3 mRNA was assessed by RT-qPCR in SV40 fibroblasts from healthy controls stably transfected with various TLR3 alleles (wild-type [WT] TLR3, G743D+R811I, L360P mutant TLR3), or a mock vector. GUS was included for normalization. (D) Induction of mRNA for IFNB, IFNL1, and IFIT2 in the absence of stimulation (NS) or after 24 hours of stimulation with herpes simplex virus 1 (HSV-1) at an multiplicity of infection (MOI) of 1, in SV40 fibroblasts from 3 healthy controls, the patients (1, 5, and 6), a TLR3−/− patient, and a NEMO−/− patient. (E) HSV-1 replication, quantified by green fluorescent protein (GFP) measurement, in SV-40 fibroblasts from 3 healthy controls, the patients, a TLR3−/− patient, and a STAT1−/− patient, 24 hours after HSV-1 GFP infection at MOI of 0.1, 1, and 10, with (lower panel) or without (upper panel) 16 hours of pretreatment with IFN-α2b. The data shown are representative of 3 (A, D, E) or 2 (B, C) independent experiments.
Enhanced susceptibility to HSV-1 in the patients' fibroblasts.
A similar HSE-related cellular phenotype has been observed in all the 5 previously reported genetic etiologies of HSE, with mutations in TLR3, UNC93B1, TRIF, TRAF3, and TBK1 in a total of 10 patients (table 1). Virus-induced IFN-β and IFN-λ production was abnormally weak in fibroblasts from patients with TLR3 pathway deficiencies, following infection with HSV-1 and VSV, and the impairment of IFN production in turn leads to enhanced viral replication, and enhanced cell death in patients' fibroblasts.8,9 Like fibroblasts from the AR complete TLR3-deficient patient,9 fibroblasts from patients 1, 5, and 6 displayed impaired mRNA production of IFNB, IFNL1, and IFN-inducible genes, such as IFIT2, specifically after infection with HSV-1, while the same cells were able to produce high levels of IFNB, IFNL1, and IFIT2 mRNA after infection with VSV M51R (figure 4D). Like fibroblasts from a patient with AR complete TLR3 deficiency and a patient with AR complete STAT1 deficiency,9,28 fibroblasts from patients 1, 5, and 6 displayed higher levels of HSV-1 replication than cells from healthy controls (figure 4E). When cells were treated with IFN-α2b 16 hours before viral infection, rescue of the HSV-1 phenotype was observed in fibroblasts from patients 1, 5, and 6 and the AR TLR3-deficient patient, but not in AR STAT1-deficient cells, which have impaired responses to IFN-α, -β, and -λ29 (figure 4E). By inference, this fibroblast phenotype may account for the molecular pathogenesis of HSE in CNS-resident cells in the 3 patients with novel forms of inborn errors of TLR3 immunity, as recently shown for iPSC-derived neurons and oligodendrocytes from other UNC-93B- and TLR3-deficient patients.7
DISCUSSION
We report 3 novel forms of TLR3 deficiency in 3 patients with recurrent HSE. Together with the TLR3-deficient patients previously reported,8,9 this brings the total number of TLR3-deficient patients identified among the 120 HSE patients evaluated by our group to date to 6 (5%). Our studies provide compelling evidence that inborn errors of CNS-intrinsic TLR3 immunity may underlie the pathogenesis of HSE in the course of primary HSV-1 infection, at least in some children. There appears to be genetic heterogeneity at the population level (1 gene per patient, multiple morbid genes in the cohort), but physiopathologic homogeneity at the cellular level (the TLR3-IFN pathway being the core morbid axis, which is represented by so far 13 patients [11% of the HSE patients studied] with TLR3 pathway deficiencies). Not only is there locus heterogeneity, there is also allelic heterogeneity, because 5 types of TLR3 defects have been found in 6 patients: 2 forms of AR TLR3 deficiency, either complete or partial; 2 types of AD TLR3 deficiency by DN mechanisms, depending on whether TLR3 is excessively cleaved or not cleaved at all; and 1 type of AD TLR3 deficiency by haploinsufficiency (table 1). The clinical penetrance of TLR3 deficiencies as well as that of other human TLR3 pathway deficiencies (UNC-93B, TRIF, TBK1) is incomplete for HSE.6 Diverse factors may be causative of the incomplete penetrance of human TLR3 pathway deficiencies, including environmental factors, pathogen-related (viral infection load and virus strain) or pathogen-unrelated (another infection) factors, or host factors, including genetic (modifiers) or epigenetic (age at infection) factors. In any event, our finding that a small albeit sizeable fraction of children with HSE (5%) carry morbid mutations in TLR3 suggests that this is a core morbid gene, defining a core morbid pathway. WES and whole-genome sequencing in children with HSE will be instrumental for testing whether the TLR3 signaling pathway is mutated in most children with HSE.
Despite the incomplete clinical penetrance of the TLR3 pathway deficiencies in HSE, a high proportion of TLR3-deficient patients with HSE had recurrent HSE with late relapse. Overall, HSE recurrence is rare, being reported in only about 10% of affected children.4,30,31 It may occur early (before 18 months) or late (after 18 months), reflecting different mechanisms. Late relapses of HSE are particularly rare and their underlying mechanism remains unclear.30 The 3 patients with TLR3 deficiency reported in this study, like one of the 3 previously reported TLR3-deficient patients,6 had recurrent HSE, with intervals of about 2 to 26 years between HSE episodes (table 1). Late relapses of HSE have occurred in 4 of the 6 (67%) TLR3-deficient patients identified to date. Interestingly, among the other 8 patients with HSE with other deficiencies of the TLR3 pathway, both AR UNC-93B-deficient patients had also presented late relapses of HSE. Thus, in total, 6 patients with TLR3 pathway deficiencies, from a total of 13 such patients (46.15%), had recurrent HSE. HSE therefore seems to recur more frequently in patients with inborn errors of TLR3 immunity—an observation that requires confirmation in a larger number of patients with inborn errors of TLR3 immunity. We can speculate that TLR3 deficiency is associated with inefficient virus control in the brain, leading to incomplete viral latency in the CNS itself,32–34 in turn leading to a high rate of HSE recurrence due to virus reactivation. Additional clinical and CNS cellular studies are required to test this hypothesis. Meanwhile, our findings suggest that children with HSE due to TLR3 deficiency should be carefully followed up given the risk of relapse, and the threshold for antiviral treatment should be low in such patients. Moreover, IFN-α2b, in addition to acyclovir, might improve the prognosis of HSE if given early in the course of infection.
Supplementary Material
ACKNOWLEDGMENT
The authors thank the patients and their families for participating in this study and the past and present members of the Laboratory of Human Genetics of Infectious Diseases for discussions and technical, bioinformatics, or administrative assistance.
GLOSSARY
- AD
autosomal dominant
- AR
autosomal recessive
- CEPH-HGDP
Centre d'Etude du Polymorphisme Humain–Human Genome Diversity Project
- dbSNP
Single Nucleotide Polymorphism database
- DN
dominant negative
- ds
double-stranded
- ECD
ectodomain
- GFP
green fluorescent protein
- HA
hemagglutinin
- HSE
herpes simplex encephalitis
- HSV-1
herpes simplex virus 1
- IFN
interferon
- iPSC
induced pluripotent stem cell
- poly(I:C)
polyinosine:polycytidylic acid
- TIR
Toll/interleukin-1 receptor
- VSV
vesicular stomatitis virus
- WES
whole-exome sequencing
- WT
wild-type
Footnotes
Editorial, page 1882
Supplemental data at Neurology.org
AUTHOR AFFILIATIONS
From the St. Giles Laboratory of Human Genetics of Infectious Diseases (H.K.L., M.J.C., Y.I., F.G.L., W.D., M.B., E.P., Y.R., E.J., L.A., J.-L.C., S.-Y.Z.), Rockefeller Branch, The Rockefeller University, New York, NY; the Laboratory of Human Genetics of Infectious Diseases (H.K.L., L.L., X.C., S.B., E.J., L.A., J.-L.C., S.-Y.Z.), Necker Branch, INSERM U1163, Paris; Paris Descartes University (H.K.L., X.C., S.B., E.J., L.A., J.-L.C., S.-Y.Z.), Sorbonne Paris Cite, Imagine Institute, France; the Immunodeficiency Unit, Division of Infectious Diseases, Department of Medicine (M.S.), and the Department of Neurology (R.R., P.J.T.), Helsinki University Central Hospital; the Department of Internal Medicine (T.H.), Oulu University Hospital; the Department of Pathology (A.P.), University of Helsinki and HUSLAB, Finland; Virology (P.L., F.R.), Cochin-Saint-Vincent de Paul Hospital, Paris Descartes University; Pediatric Neurology (M.T.), Bicêtre Hospital, Paris Sud University, France; the Departments of Pediatric Immunology-Allergy and Pediatric Hematology (A.Y.), School of Medicine, Ondokuz Mayis University, Samsun, Turkey; Brussels Free University and Infectious Diseases Unit (A.V.), Hôpital Universitaire des Enfants Reine Fabiola, Belgium; Meyer Children's Hospital (A.E.), Haifa, Israel; Molecular Neurology (R.R., P.J.T.), Research Programs Unit, Biomedicum, University of Helsinki, Finland; the Pediatric Immuno-Hematology Unit (J.-L.C.), Necker Hospital, Assistance Publique-Hôpitaux de Paris, Necker Hospital, France; and Howard Hughes Medical Institute (J.-L.C.), New York, NY.
AUTHOR CONTRIBUTIONS
H.K.L. performed the experiments, analyzed the data, and wrote the paper. M.S. performed the experiments, contributed patient samples, and collected clinical data. T.H. performed the experiments, contributed patient samples, and collected clinical data. M.J.C. performed the experiments. Y.I. analyzed the data. F.G.L. performed the experiments. W.D. performed the experiments. L.L. performed the experiments. MB. performed the experiments. E.P. performed the experiments. Y.R. performed the experiments. X.C. performed the experiments. S.B. performed the experiments. E.J. performed the experiments. A.P. performed the experiments, contributed patient samples, and collected clinical data. P.L. performed the experiments, contributed patient samples, and collected clinical data. F.R. performed the experiments, contributed patient samples, and collected clinical data. M.T. performed the experiments, contributed patient samples, and collected clinical data. L.A. analyzed the data. A.Y. performed the experiments, contributed patient samples, and collected clinical data. A.V. performed the experiments, contributed patient samples, and collected clinical data. R.R. performed the experiments, contributed patient samples, and collected clinical data. A.E. performed the experiments, contributed patient samples, and collected clinical data. P.J.T. performed the experiments, contributed patient samples, and collected clinical data. J.L.C. supervised the research and wrote the paper. S.Y.Z. supervised the research and wrote the paper.
STUDY FUNDING
Supported by the National Center for Advancing Translational Sciences (NCATS), NIH, Clinical and Translational Science Award (CTSA) program grant UL1TR000043, NIH grant 5R01AI088364, the Rockefeller University, INSERM, Paris Descartes University, the ANR (French National Agency for Research), the St Giles Foundation, the Thrasher Research Fund, the European Research Council (grant ERC-2010-AdG-268777), Helsinki University Central Hospital, and the Finnish Academy. Y.I. was supported by the AXA Research Fund, F.L. by the New York Stem Cell Foundation, and M.B. by the Charles H. Revson Foundation. The plasmids containing Ins12 or 346Cterm mutant TLR3 were a gift from Dr. Serge Lebecque.
DISCLOSURE
The authors report no disclosures relevant to the manuscript. Go to Neurology.org for full disclosures.
REFERENCES
- 1.Barreiro LB, Ben-Ali M, Quach H, et al. Evolutionary dynamics of human toll-like receptors and their different contributions to host defense. PLoS Genet 2009;5:e1000562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jacobs BL, Langland JO. When two strands are better than one: the mediators and modulators of the cellular responses to double-stranded RNA. Virology 1996;219:339–349. [DOI] [PubMed] [Google Scholar]
- 3.Whitley RJ, Kimberlin DW. Herpes simplex encephalitis: children and adolescents. Semin Pediatr Infect Dis 2005;16:17–23. [DOI] [PubMed] [Google Scholar]
- 4.Abel L, Plancoulaine S, Jouanguy E, et al. Age-dependent Mendelian predisposition to herpes simplex virus type 1 encephalitis in childhood. J Pediatr 2010;157:623–629. [DOI] [PubMed] [Google Scholar]
- 5.De Tiege X, Rozenberg F, Heron B. The spectrum of herpes simplex encephalitis in children. Eur J Paediatr Neurol 2008;12:72–81. [DOI] [PubMed] [Google Scholar]
- 6.Zhang SY, Herman M, Ciancanelli MJ, et al. TLR3 immunity to infection in mice and humans. Curr Opin Immunol 2013;25:19–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lafaille FG, Pessach IM, Zhang SY, et al. Impaired intrinsic immunity to HSV-1 in human iPSC-derived TLR3-deficient CNS cells. Nature 2012;491:769–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang SY, Jouanguy E, Ugolini S, et al. TLR3 deficiency in patients with herpes simplex encephalitis. Science 2007;317:1522–1527. [DOI] [PubMed] [Google Scholar]
- 9.Guo Y, Audry M, Ciancanelli M, et al. Herpes simplex virus encephalitis in a patient with complete TLR3 deficiency: TLR3 is otherwise redundant in protective immunity. J Exp Med 2011;208:2083–2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sun Y, Leaman DW. Ectopic expression of toll-like receptor-3 (TLR-3) overcomes the double-stranded RNA (dsRNA) signaling defects of P2.1 cells. J Interferon Cytokine Res 2004;24:350–361. [DOI] [PubMed] [Google Scholar]
- 11.Desai P, Person S. Incorporation of the green fluorescent protein into the herpes simplex virus type 1 capsid. J Virol 1998;72:7563–7568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cann HM, de Toma C, Cazes L, et al. A human genome diversity cell line panel. Science 2002;296:261–262. [DOI] [PubMed] [Google Scholar]
- 13.Adzhubei I, Schmidt S, Peshkin L, et al. A method and server for predicting damaging missense mutations. Nat Methods 2010;7:248–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc 2009;4:1073–1081. [DOI] [PubMed] [Google Scholar]
- 15.Choe J, Kelker MS, Wilson IA. Crystal structure of human toll-like receptor 3 (TLR3) ectodomain. Science 2005;309:581–585. [DOI] [PubMed] [Google Scholar]
- 16.Bell JK, Botos I, Hall PR, et al. The molecular structure of the Toll-like receptor 3 ligand-binding domain. Proc Natl Acad Sci U S A 2005;102:10976–10980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bell JK, Askins J, Hall PR, Davies DR, Segal DM. The dsRNA binding site of human Toll-like receptor 3. Proc Natl Acad Sci U S A 2006;103:8792–8797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de Bouteiller O, Merck E, Hasan UA, et al. Recognition of double-stranded RNA by human toll-like receptor 3 and downstream receptor signaling requires multimerization and an acidic pH. J Biol Chem 2005;280:38133–38145. [DOI] [PubMed] [Google Scholar]
- 19.Ranjith-Kumar CT, Miller W, Xiong J, et al. Biochemical and functional analyses of the human Toll-like receptor 3 ectodomain. J Biol Chem 2007;282:7668–7678. [DOI] [PubMed] [Google Scholar]
- 20.Takada E, Okahira S, Sasai M, Funami K, Seya T, Matsumoto M. C-terminal LRRs of human Toll-like receptor 3 control receptor dimerization and signal transmission. Mol Immunol 2007;44:3633–3640. [DOI] [PubMed] [Google Scholar]
- 21.Liu L, Botos I, Wang Y, et al. Structural basis of toll-like receptor 3 signaling with double-stranded RNA. Science 2008;320:379–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Botos I, Segal DM, Davies DR. The structural biology of Toll-like receptors. Structure 2011;19:447–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oshiumi H, Matsumoto M, Funami K, Akazawa T, Seya T. TICAM-1, an adaptor molecule that participates in Toll-like receptor 3-mediated interferon-beta induction. Nat Immunol 2003;4:161–167. [DOI] [PubMed] [Google Scholar]
- 24.Yamamoto M, Sato S, Hemmi H, et al. Role of adaptor TRIF in the MyD88-independent toll-like receptor signaling pathway. Science 2003;301:640–643. [DOI] [PubMed] [Google Scholar]
- 25.Toscano F, Estornes Y, Virard F, et al. Cleaved/associated TLR3 represents the primary form of the signaling receptor. J Immunol 2013;190:764–773. [DOI] [PubMed] [Google Scholar]
- 26.Garcia-Cattaneo A, Gobert FX, Muller M, et al. Cleavage of Toll-like receptor 3 by cathepsins B and H is essential for signaling. Proc Natl Acad Sci U S A 2012;109:9053–9058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stojdl DF, Lichty BD, tenOever BR, et al. VSV strains with defects in their ability to shutdown innate immunity are potent systemic anti-cancer agents. Cancer Cell 2003;4:263–275. [DOI] [PubMed] [Google Scholar]
- 28.Chapgier A, Wynn RF, Jouanguy E, et al. Human complete Stat-1 deficiency is associated with defective type I and II IFN responses in vitro but immunity to some low virulence viruses in vivo. J Immunol 2006;176:5078–5083. [DOI] [PubMed] [Google Scholar]
- 29.Chapgier A, Kong XF, Boisson-Dupuis S, et al. A partial form of recessive STAT1 deficiency in humans. J Clin Invest 2009;119:1502–1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Spiegel R, Miron D, Yodko H, Lumelsky D, Habib A, Horovitz Y. Late relapse of herpes simplex virus encephalitis in a child due to reactivation of latent virus: clinicopathological report and review. J Child Neurol 2008;23:344–348. [DOI] [PubMed] [Google Scholar]
- 31.Valencia I, Miles DK, Melvin J, et al. Relapse of herpes encephalitis after acyclovir therapy: report of two new cases and review of the literature. Neuropediatrics 2004;35:371–376. [DOI] [PubMed] [Google Scholar]
- 32.Asenbauer B, McEntagart M, King MD, Gallagher P, Burke M, Farrell MA. Chronic active destructive herpes simplex encephalitis with recovery of viral DNA 12 years after disease onset. Neuropediatrics 1998;29:120–123. [DOI] [PubMed] [Google Scholar]
- 33.Nicoll JA, Love S, Kinrade E. Distribution of herpes simplex virus DNA in the brains of human long-term survivors of encephalitis. Neurosci Lett 1993;157:215–218. [DOI] [PubMed] [Google Scholar]
- 34.Lellouch-Tubiana A, Fohlen M, Robain O, Rozenberg F. Immunocytochemical characterization of long-term persistent immune activation in human brain after herpes simplex encephalitis. Neuropathol Appl Neurobiol 2000;26:285–294. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.