Abstract
Background
Eucalyptus species are the most widely planted hardwood species in the world and are renowned for their rapid growth and adaptability. In Brazil, one of the most widely grown Eucalyptus cultivars is the fast-growing Eucalyptus urophylla x Eucalyptus grandis hybrid. In a previous study, we described a chemical characterization of these hybrids when subjected to flavonoid supplementation on 2 distinct timetables, and our results revealed marked differences between the wood composition of the treated and untreated trees.
Results
In this work, we report the transcriptional responses occurring in these trees that may be related to the observed chemical differences. Gene expression was analysed through mRNA-sequencing, and notably, compared to control trees, the treated trees display differential down-regulation of cell wall formation pathways such as phenylpropanoid metabolism as well as differential expression of genes involved in sucrose, starch and minor CHO metabolism and genes that play a role in several stress and environmental responses. We also performed enzymatic hydrolysis of wood samples from the different treatments, and the results indicated higher sugar contents and glucose yields in the flavonoid-treated plants.
Conclusions
Our results further illustrate the potential use of flavonoids as a nutritional complement for modifying Eucalyptus wood, since, supplementation with flavonoids alters its chemical composition, gene expression and increases saccharification probably as part of a stress response.
Electronic supplementary material
The online version of this article (doi:10.1186/s12870-014-0301-8) contains supplementary material, which is available to authorized users.
Keywords: Eucalyptus, Lignin, Phenylpropanoid metabolism, Syringyl/guaiacyl ratio, Gene expression, Hydrolysis, Stress
Background
Trees constitute the majority of the lignocellulosic biomass on Earth and are expected to play a significant role in the future as a renewable and environmentally cost-effective alternative feedstock for biofuel production, a source of fibers and solid wood products and a major sink for excess atmospheric CO2 [1-3]. In Brazil, the pulp and paper industries have been efficiently fed by Eucalyptus forests due to their rapid growth, adaptability and wood quality, but with the dramatic increase in industrial demands and the interest in second-generation biofuels and renewable chemicals, the quality and quantity of wood produced must also increase [4,5].
Wood is a highly variable material that differs among trees and is composed of the secondary xylem, a specialized type of conductive and structural support tissue produced through the lateral growth and differentiation of the meristematic vascular cambium [6]. Most of the genes expressed during the formation of the secondary xylem (xylogenesis) are involved in determining the physical and chemical properties of wood [2,7]. Despite the progress that has been made in defining the molecular and cellular events involved in xylogenesis, the mechanisms regulating the rate of this process and the variation in wood properties remain largely unknown [8-10].
The secondary xylem cell wall of Eucalyptus trees is mostly composed by cellulose (β-1,4-glucan), lignin (a phenolic polymer) and hemicelluloses (heterogeneous polysaccharides), in an approximate ratio of 2:1:1 [11]. During tree growth, cellulose microfibrils give the cell walls tensile strength, and the lignin encasing the cellulose microfibrils imparts rigidity to the cell walls. Despite its importance during growth, lignin becomes problematic during postharvest, cellulose-based wood processing because it must be extracted during industrial handling through a complicated process, resulting in an enormous expenditure of energy and chemicals and strain on the environment [11,12]. Thus, it is of major interest to investigate the molecular basis of lignification to further increase our overall comprehension of this metabolic process for better adaptation of industrial processes.
Lignin synthesis is a relatively well-understood process that begins with the assembly of radicals produced during the single-electron oxidation of monolignols [10,13,14]. The industrial exploitation of wood to obtain cellulose depends mostly on the composition of lignins because lignins determine the rigidity of the wood and the feasibility of cellulose extraction, which are of major concern in the paper and pulp industries. In angiosperms, lignin is composed of 2 major units: the guaiacyl (G) and syringyl (S) units, which are derived from corresponding monolignol precursors, the coniferyl and sinapyl alcohols, respectively [1,15]. The S/G ratio dictates the degree and nature of polymeric cross-linking; an increased G content leads to highly cross-linked lignin (more rigid wood), whereas S subunits are typically linked through more labile ether bonds at the 4-hydroxyl position [16-18]. Thus, S-rich lignins are much easier to dissociate from cellulosic content, resulting in a much cleaner and cheaper process [18]. The S/G ratio is variable among species and is commonly used to evaluate the quality of wood in commercial tree plantations [19,20].
The formation of lignin monomers begins with the catalytic step performed by the 4-coumaroyl:CoA-ligase (4CL) enzyme, which likely represents the most important branch point in the central phenylpropanoid biosynthesis pathway in plants [21,22]. Through 4CL activity, cells can produce the precursors for either flavonoids or the G and S lignin precursors [23]. The product of 4CL, p-coumaroyl-CoA, is the substrate of the enzyme chalcone synthase (CHS) [24], which carries out the committing step in flavonoid biosynthesis. This pathway is reviewed in detail elsewhere [10,24].
The flavonoids naringenin-chalcone and naringenin, which are synthesized by the enzymes chalcone synthase (CHS) and chalcone isomerase (CHI), respectively, are the primary C15 intermediates in flavonoid biosynthesis [25,26]. This metabolic pathway appears to be a promising target for improving wood quality in Eucalyptus trees, as shown by our previous work [27] demonstrating that flavonoid supplementation of the fast-growing Eucalyptus urophylla x Eucalyptus grandis hybrid, hereafter referred to as E. urograndis, changes its wood composition, reduces its extractive contents and alters its syringyl monomer composition.
In this context, the objective of the present work was to perform further studies on the effects of flavonoid supplementation on E. urograndis trees by analyzing gene expression in xylem tissue from treated and non-treated trees and by measuring the effect on sugar accessibility through enzymatic hydrolysis. We analyzed the obtained data with special emphasis on results that might be correlated with the previously observed changes in wood composition [27].
Results
RNA sequencing and differential gene expression
A total of over 335 million reads were generated from 8 samples: 3 samples from the control group (CT); 3 from the naringenin-supplemented groups (2 NAR and 1 NARSTOP); and 2 from the naringenin-chalcone supplemented groups (1 CH and 1 CHSTOP). The number of reads per sample ranged from 32 to 54 million (total) and 30 to 48 million (after filtering). The reads were mapped against the greater splice variants (44,974 sequences) of the E.grandis gene predictions from Phytozome 7.0 (54,935 transcripts) using the SOAP2 alignment software package [28] (Additional file 1).
Heat map clustering of all transcripts was performed using Expander software [29], resulting in 2 major groups: 1 formed by the 3 control sample replicates and the other by the flavonoid-supplemented samples (Figure 1).
Figure 1.
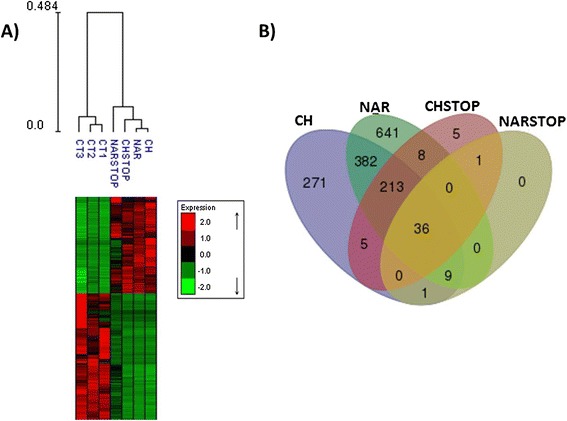
Heat map clustering and Venn diagram of differentially expressed genes. A) Heat map clustering of differentially expressed transcripts and comparison of the estimated log2 fold change correlations between each group subjected to differential expression analyses. B) Venn diagram of differentially expressed genes. CH- prolonged narigenin-chalcone supp; NAR – prolonged naringenin supp; CHSTOP- short-term naringenin-chalcone supp; NARSTOP – short-termnaringenin sup.
The read counts from each sample were used to test the differential expression of the genes between the control (CT) and supplemented (CH, NAR, CHSTOP and NARSTOP) treatments using the baySeq package [30]. A total of 1,573 genes were considered to be differentially expressed (FDR ≤0.01), which were distributed among the treatments (917 CH; 1,289 NAR; 268 CHSTOP; 47 NARSTOP) (Additional file 2).
The gene expression patterns observed for the supplemented and control groups were distinct, while similar profiles were observed within treatments, indicating similarities among the different types of flavonoid supplementation studied here. Most of the differences were observed in the long-term supplementation treatments, which comprised almost all of the genes that were differentially expressed in the short-term treatments as well. The NAR-supplemented plants displayed the greatest number of genes that were differentially expressed, while the NARSTOP-supplemented plants had fewer, which may indicate that naringenin supplementation has a stronger, but short-lasting impact on gene expression, whereas naringenin-chalcone has a smaller but more durable impact.
Functional analyses
To determine the biological functions of the genes responding to flavonoid supplementation, functional analyses were performed using the web-based tools Blast2GO and Mapman. The genes considered differentially expressed in each treatment were mapped to their corresponding metabolic pathways, and the treatments were tested for enrichment of particular metabolic responses.
Only 36 genes were differentially expressed in all four treatments, including genes encoding several heat-shock proteins, sequences with no hits and unknown proteins (Table 1).
Table 1.
Gene ID, FPKM values and annotation of the 36 genes that found to be differentially expressed in all tested conditions
| FPKM | ||||||
|---|---|---|---|---|---|---|
| Gene ID | Annotation | CT | CH | NAR | CHSTOP | NARSTOP |
| Eucgr.F04479.1 | HSP20 | 0.12 | 35.55 | 40.44 | 23.84 | 57.92 |
| Eucgr.K02389.1 | Unknown | 0.04 | 13.69 | 10.18 | 9.89 | 26.99 |
| Eucgr.K02399.1 | Unknown | 0.08 | 18.09 | 18.67 | 19.48 | 53.82 |
| Eucgr.G01188.2 | EGY3 | 2.78 | 45.78 | 41.56 | 33.40 | 76.35 |
| Eucgr.J01979.1 | HSP18.2 | 0.34 | 17.61 | 19.94 | 14.63 | 27.21 |
| Eucgr.K02410.1 | Unknown | 0.13 | 14.27 | 10.66 | 12.52 | 29.61 |
| Eucgr.J01980.1 | HSP18.2 | 0.02 | 12.20 | 11.59 | 9.15 | 26.39 |
| Eucgr.L02233.1 | no hit | 1.29 | 41.47 | 38.15 | 29.24 | 118.40 |
| Eucgr.F02898.1 | HSP20 | 2.76 | 525.99 | 343.17 | 280.68 | 342.59 |
| Eucgr.J01985.1 | HSP18.2 | 0.31 | 22.25 | 16.62 | 13.21 | 33.33 |
| Eucgr.K03553.1 | STS | 0.04 | 4.11 | 4.59 | 2.83 | 6.87 |
| Eucgr.L03261.1 | HSP18.2 | 1.44 | 47.41 | 27.81 | 19,98 | 75,67 |
| Eucgr.C03449.1 | HSFA2 | 0.29 | 14.55 | 12.98 | 14.36 | 18.58 |
| Eucgr.C00684.1 | HSP17.6II | 2.00 | 341.30 | 299.30 | 243.65 | 272.90 |
| Eucgr.K02384.1 | unknown | 0.07 | 14.56 | 10.59 | 10.60 | 27.23 |
| Eucgr.J01969.1 | HSP20 | 4.89 | 192.23 | 134.53 | 103.09 | 310.44 |
| Eucgr.K03472.1 | ARATH | 0.07 | 109.57 | 84.15 | 62.04 | 20.94 |
| Eucgr.H04513.1 | HSP70 | 0.23 | 15.72 | 18.79 | 11.56 | 21.62 |
| Eucgr.A00595.1 | PEBP | 0.10 | 81.17 | 76.66 | 57.33 | 98.83 |
| Eucgr.E02421.1 | Unknown | 0.19 | 260.04 | 220.12 | 119.63 | 51.05 |
| Eucgr.H04692.1 | HSP21 | 2.97 | 83.31 | 59.57 | 43.11 | 313.22 |
| Eucgr.G02440.1 | UGT73B2 | 0.00 | 5.46 | 5.80 | 3.33 | 4.06 |
| Eucgr.G02259.1 | UGT73B3 | 0.00 | 2.73 | 2.14 | 1.18 | 3.10 |
| Eucgr.J01959.1 | HSP18.2 | 3.19 | 142.90 | 89.76 | 58.22 | 148.21 |
| Eucgr.K00295.1 | HSP90.1 | 2.11 | 46.13 | 38.25 | 35.52 | 62.69 |
| Eucgr.A01833.1 | AAC3 | 0.13 | 32.00 | 24.43 | 15.08 | 10.47 |
| Eucgr.C03071.1 | HSP17.6II | 3.64 | 517.08 | 509.51 | 451.66 | 324.54 |
| Eucgr.B03843.1 | No hit | 1.45 | 93.17 | 63.39 | 67.61 | 20.77 |
| Eucgr.C03320.1 | DUF1677 | 0.38 | 24.39 | 18.22 | 14.01 | 9.78 |
| Eucgr.B00176.2 | PIMT2 | 3.86 | 153.25 | 109.01 | 82.97 | 57.39 |
| Eucgr.J02588.1 | No hit | 3.20 | 225.90 | 182.76 | 194.01 | 130.04 |
| Eucgr.C00690.1 | HSP17.6II | 2.48 | 563.68 | 498.40 | 514.67 | 458.94 |
| Eucgr.K00237.1 | PEBP | 0.04 | 115.83 | 64.41 | 61.47 | 11.49 |
| Eucgr.F03196.1 | GSTU25 | 1.43 | 292.73 | 240.63 | 168.71 | 38.29 |
| Eucgr.I02136.1 | HSP20 | 1.68 | 226.73 | 147.43 | 90.02 | 259.35 |
| Eucgr.H04427.1 | MEE32 | 49.92 | 0.52 | 0.85 | 1.32 | 13.93 |
A total of 36 genes were differentially expressed in all four conditions. FPKM -fragments per kilobase of exon per million fragments mapped. CT – control; CH – prolonged naringenin-chalcone supp; NAR – prolonged naringenin supp; CHSTOP- short-term naringenin-chalcone supp; NARSTOP – short-term naringenin supp.
Abbreviations: HSP20 HSP20-like chaperone superfamily protein, unknown unknown protein, EGY3 ethylene-dependent gravitropism-deficient and yellow-green-like 3, HSP18.2 heat shock protein 18.2, HSP20 HSP20-like chaperones superfamily protein, STS stachyose synthase, HSFA2 heat shock transcription factor A2, HSP17.6II 17.6 kDa class II heat shock protein, ARATH Adenine nucleotide alpha hydrolases-like superfamily protein, HSP70 BIP1heat shock protein 70 family protein, PEBP phosphatidylethanolamine-binding protein family protein, HSP21 heat shock protein 21, UGT73B2 UDP-glucosyltransferase 73B2, UGT73B3UDP glucosyl transferase 73B3, HSP90.1 heat shock protein 90.1, AAC3 ADP/ATP carrier 3, DUF1677 protein of unknown function, PIMT2 protein-l-isoaspartate methyltransferase 2, GSTU25 glutathione S-transferase TAU 25, MEE32 dehydroquinate dehydratase, putative/shikimate dehydrogenase.
Each supplemented group was analysed individually. Common categories between different treatments are shown in Figure 2, and all affected GO categories are listed in Additional file 3.
Figure 2.
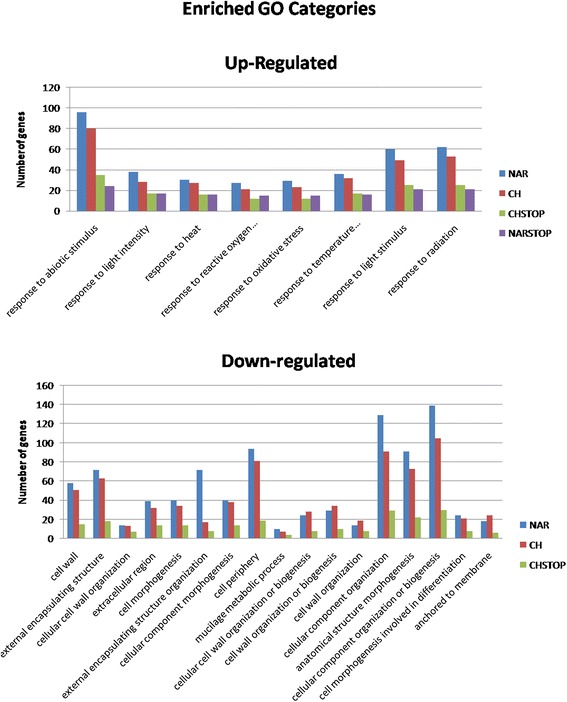
GO analysis. Common GO categories that were enriched (p-values ≤0.05) between treatments. CH – prolonged naringenin-chalcone supp; NAR – prolonged naringenin supp; CHSTOP- short term naringenin-chalcone supp; NARSTOP – short-term naringenin supplementation.
Many of the down-regulated categories that were common to all treatments are involved in cell wall formation and development. On the other hand, the common up-regulated categories are all related to stress and environmental responses. Interestingly, NARSTOP, which resulted in fewer differentially expressed genes, only led to enriched GO categories among up-regulated genes.
Mapman analyses of all of the differentially expressed genes also indicated down-regulation of cell wall-related genes and phenylpropanoid pathways, whereas flavonoid, minor CHO and starch and sucrose metabolism and stress response were associated with the most genes up-regulated (Figure 3).
Figure 3.
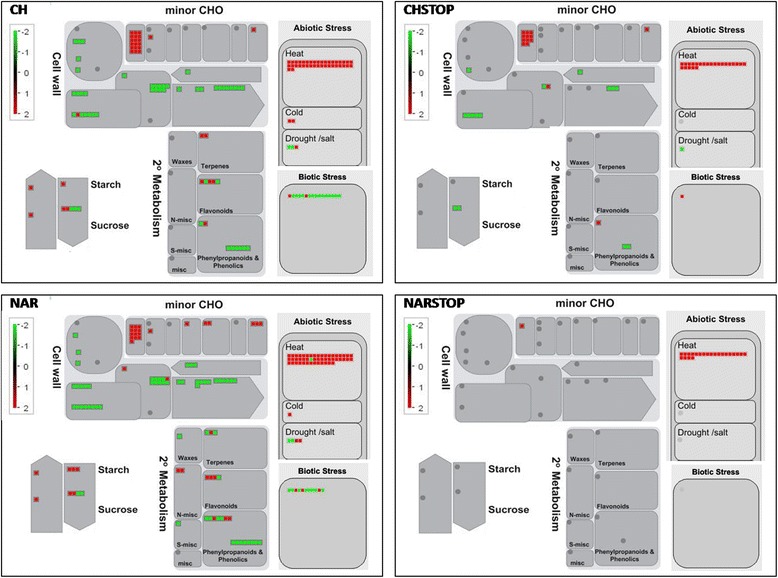
MapMan analysis. MapMan overview of the metabolism- and cellular response-related genes among the 1,573 genes that were differentially expressed under the four different flavonoid treatments. The presented values are the fold changes between the treatment and control groups. CH – prolonged naringenin-chalcone supp; NAR – prolonged naringenin supp; CHSTOP- short-term naringenin-chalcone supp; NARSTOP – short-term naringenin supp.
The phenylpropanoid genes
To further analyze the impact of flavonoid supplementation on lignification, a broader analysis was performed on the genes from the phenylpropanoid pathway, especially those related to lignin biosynthesis.
Several phenylpropanoid genes were differentially expressed between the treated samples and controls (Table 2), including the following genes that are directly related to lignin synthesis: 4CL, HCT, 2 OMT-methyltransferases, CCR and 2 CAD genes; 4CL, HCT and CCR were down-regulated, while the 2 methyltransferases and CAD genes were up-regulated. Additionally, several laccases were down-regulated among the treatments. These results are highly significant in terms of explaining the higher S/G ratio found in supplemented plants.
Table 2.
Differentially expressed phenylpropanoid-related genes
| FPKM | ||||||
|---|---|---|---|---|---|---|
| Gene ID | Annotation | CT | CH | NAR | CHSTOP | NARSTOP |
| Eucgr.C00859.1 | U91A1 | 0.00 | 1.60* | 0.83 | 0.28 | 0.00 |
| Eucgr.K00903.1 | AAT | 0.38 | 3.91 | 4.61* | 3.45 | 1.26 |
| Eucgr.K00901.1 | AAT | 0.83 | 0.03* | 0.98 | 1.18 | 1.51 |
| Eucgr.E01250.1 | PRR1 | 38.77 | 3.72* | 4.68* | 7.66 | 21.15 |
| Eucgr.B03781.1 | AA | 24.04 | 0.04* | 0.86 | 1.39 | 9.77 |
| Eucgr.D02454.1 | DFR | 0.05 | 3.10* | 2.73* | 1.32 | 0.48 |
| Eucgr.G02325.1 | DFR | 2.51 | 28.24* | 20.16* | 21.06 | 9.55 |
| Eucgr.F04163.1 | LAC14 | 6.22 | 0.15 | 0.32* | 0.57 | 1.82 |
| Eucgr.F02646.1 | LAC14 | 1.99 | 0.00* | 0.02* | 0.06* | 0.44 |
| Eucgr.F04162.1 | LAC14 | 1.95 | 0.09 | 0.04* | 0.06 | 0.72 |
| Eucgr.H04937.1 | LAC14 | 13.67 | 0.02* | 0.15* | 0.21 | 4.66 |
| Eucgr.F04160.1 | LAC14 | 17.17 | 0.04* | 0.25* | 0.26* | 3.08 |
| Eucgr.F02674.1 | LAC14 | 7.13 | 0.28 | 0.27* | 0.42 | 2.90 |
| Eucgr.H04936.1 | LAC14 | 8.51 | 0.04* | 0.03* | 0.08 | 2.50 |
| Eucgr.B02796.1 | LAC4 | 12.04 | 0.28* | 1.95 | 3.31 | 5.69 |
| Eucgr.K00957.1 | ATOMT1 | 1.36 | 17.54 | 17.19* | 10.55 | 15.16 |
| Eucgr.A01877.1 | OMT-like | 0.00 | 0.31 | 0.79 | 1.75* | 0.22 |
| Eucgr.J00363.1 | HCT | 88.68 | 3.76* | 16.43 | 29.57 | 53.35 |
| Eucgr.B00137.1 | 4CL | 12.31 | 1.83 | 2.50* | 4.21 | 6.24 |
| Eucgr.E00270.1 | CCR | 30.29 | 3.21 | 3.29* | 4.03 | 8.00 |
| Eucgr.G01350.2 | CAD5 | 23.73 | 146.29* | 126.30* | 144.55 | 62.96 |
| Eucgr.E01110.2 | CAD1 | 4.34 | 59.21 | 49.17* | 51.45 | 27.12 |
FPKM -fragments per kilobase of exon per million fragments mapped. CT – control; CH – prolonged naringenin-chalcone supp; NAR – prolonged naringenin supp; CHSTOP- short-term naringenin-chalcone supp; NARSTOP – short-term naringenin sup *Denotes differential expression.
Abbreviations: U91A1 UDP-Glycosyltransferase superfamily protein, AAT HXXXD-type acyl-transferase family protein, PRR1 pinoresinol reductase, AA Plant L-ascorbate oxidase, DFR Dihydroflavonol-4-reductase, LAC14 laccase 14, LAC4 laccase 4, ATOMT1 O-methyltransferase 1, OMT-like O-methyltransferase family protein, HCT hydroxycinnamoyl-CoA shikimate transferase, 4 CL 4 coumarate CoA ligase, CCR cinnamoyl-CoA reductase, CAD cinnamyl alcohol dehydrogenase.
Interestingly, no gene related to the phenylpropanoid pathway was differentially expressed as a result of NARSTOP treatment.
Secondary cell wall genes
In addition to genes from the phenylpropanoid pathway, many genes related to secondary cell wall formation were differentially expressed in response to flavonoid supplementation (Table 3). Among these genes, we observed sucrose synthases, cellulose synthases and many glucosylases and transferases, most of which were down-regulated following the prolonged supplementation treatments. However, we also observed the up-regulation of several genes related to secondary cell wall formation after both prolonged and short-term flavonoid supplementation, including galactinol synthase, stachyose synthase, raffinose synthase and starch synthase.
Table 3.
Differentially expressed secondary cell wall genes
| FPKM | ||||||
|---|---|---|---|---|---|---|
| Gene ID | Annotation | CT | CH | NAR | CHSTOP | NARSTOP |
| Eucgr.C03199.1 | SUS4 | 1,532.75 | 66.91* | 100.38* | 132.04 | 213.15 |
| Eucgr.C01715.1 | SPS1F | 3.32 | 63.28* | 54.72 | 35.80* | 28.00 |
| Eucgr.F00464.1 | SUT4 | 28.18 | 85.70 | 85.04 | 82.49* | 36.89 |
| Eucgr.D01765.2 | CSLG3 | 0.07 | 3.26 | 6.28* | 7.07* | 1.24 |
| Eucgr.F04010.1 | CSLC05 | 7.51 | 0.11* | 0.35 | 0.47* | 3.06 |
| Eucgr.J00420.1 | CSLA2 | 41.08 | 2.63* | 5.07* | 5.87* | 25.59 |
| Eucgr.E00226.1 | CSLD3 | 10.13 | 0.53 | 0.78 | 0.68 | 2.11 |
| Eucgr.E00821.1 | CSLG2 | 3.07 | 0.38* | 0.35 | 0.78* | 3.03 |
| Eucgr.J02497.1 | AMR1 | 1.00 | 5.25 | 6.14 | 6.11* | 2.68 |
| Eucgr.J02407.1 | MUR1 | 74.28 | 18.89 | 17.82 | 19.95* | 38.28 |
| Eucgr.B03204.1 | MUR2 | 13.62 | 55.51 | 54.92 | 47.91* | 32.70 |
| Eucgr.J01663.1 | XTH5 | 97.41 | 1.21* | 1.87* | 3.31* | 75.71 |
| Eucgr.B03348.1 | XTH33 | 26.89 | 0.21 | 0.15* | 0.07* | 9.47 |
| Eucgr.K00883.2 | XTH9 | 607.60 | 16.06* | 29.74 | 37.61* | 288.85 |
| Eucgr.C00184.1 | XTH23 | 45.26 | 0.45 | 0.72* | 0.22* | 37.20 |
| Eucgr.H02634.1 | XTH16 | 386.73 | 21.78* | 47.14 | 72.69 | 396.09 |
| Eucgr.D01294.1 | XTH8 | 10.82 | 1.01 | 1.54 | 1.66* | 7.36 |
| Eucgr.J00827.1 | GSL12 | 0.04 | 0.90* | 1.56* | 0.98* | 0.76* |
| Eucgr.A02002.1 | GSL7 | 0.20 | 1.24 | 2.13 | 1.38* | 0.79 |
| Eucgr.A02008.1 | GSL7 | 0.16 | 1.07 | 1.89 | 1.66* | 0.83 |
| Eucgr.K02988.2 | GH | 16.20 | 90.51* | 69.09* | 53.94 | 47.27* |
| Eucgr.H00494.1 | PWD | 6.13 | 20.24 | 26.24* | 27.22 | 7.21 |
| Eucgr.H03767.1 | BAM9 | 39.25 | 225.21 | 235.09* | 217.12 | 115.07 |
| Eucgr.E00460.1 | TPS | 0.08 | 5.75* | 6.21* | 4.26 | 0.34 |
| Eucgr.K00387.1 | SS | 9.88 | 66.76* | 54.46* | 39.19 | 19.93 |
| Eucgr.C04266.1 | RafS | 26.09 | 1,317.97* | 1007.17 | 544.49 | 193.24 |
| Eucgr.K03553.1 | STS | 0.04 | 4.11* | 4.59* | 2.83* | 6.87* |
| Eucgr.H00997.1 | STS | 0.81 | 37.45* | 29.83* | 17.09* | 7.06* |
| Eucgr.K03563.1 | GoSL1 | 0.23 | 4.48 | 11.12* | 12.00 | 8.48* |
| Eucgr.L00249.1 | GoSL2 | 0.34 | 280.16* | 135.36* | 52.37* | 1.25 |
| Eucgr.L00243.1 | GoSL2 | 0.02 | 29.83* | 19.77* | 9.75* | 1.37* |
| Eucgr.L00251.1 | GoSL2 | 0.21 | 325.56* | 149.21* | 61.46* | 3.38 |
| Eucgr.L03245.1 | GoSL2 | 0.07 | 190.81* | 124.71* | 44.86* | 2.79* |
| Eucgr.L00240.1 | GoSL2 | 0.02 | 34.50* | 22.77* | 10.27* | 0.66 |
| Eucgr.L00248.1 | GoSL2 | 0.17 | 162.07* | 86.93* | 32.28* | 0.69 |
| Eucgr.L03244.1 | GoSL2 | 0.12 | 279.83* | 137.40* | 63.66* | 2.99* |
| Eucgr.L00235.1 | GoSL2 | 0.04 | 73.19* | 38.88* | 15.60* | 0.03 |
| Eucgr.L00245.1 | GoSL2 | 1.83 | 287.87* | 164.66 | 80.81* | 5.28 |
| Eucgr.F01661.1 | Invertase | 0.15 | 2.82* | 2.16 | 2.39* | 1.33 |
| Eucgr.J00457.2 | Invertase | 5.69 | 47.20* | 42.83 | 33.08* | 18.78 |
| Eucgr.G01751.1 | Invertase | 4.83 | 0.23* | 0.42 | 1.17* | 2.23 |
| Eucgr.A02888.1 | Invertase | 7.36 | 0.04* | 0.08* | 0.08* | 1.55 |
FPKM -fragments per kilobase of exon per million fragments mapped. CT – control; CH – prolonged naringenin-chalcone supp; NAR – prolonged naringenin supp; CHSTOP- short-term naringenin-chalcone supp; NARSTOP – short-term naringenin sup *Denotes differential expression.
Abbreviations: Sus4 sucrose synthase 4, SPS1F sucrose phosphate synthase 1 F, SUT4 sucrose transporter 4, CSLG3 cellulose synthase like G3, CSLD3 cellulose synthase-like D3, CSLC05 Cellulose-synthase-like C5, CSLA2 cellulose synthase-like A02, CSLG2 cellulose synthase like G2, CSLG3 cellulose synthase like G3, CESA3 cellulose synthase family protein, AMR1 ascorbic acid mannose pathway regulator 1, MUR1 GDP-mannose 4,6 dehydratase 2, MUR2 fucosyltransferase 1, XTH5 xyloglucan endotransglucosylase/hydrolase 5, XTH33 xyloglucosyl transferase 33, XTH9 xyloglucan endotransglucosylase/hydrolase 9, XTH23 xyloglucan endotransglycosylase 6, XTH16 xyloglucan endotransglucosylase/hydrolase 16, XTH8 xyloglucan endotransglucosylase/hydrolase 8, GSL12 glucan synthase-like 12, GSL7 glucan synthase-like 7, GH glycoside hydrolase, PWD phosphoglucan water dikinases, BAM9 beta-amylase 9, TPS trehalose-6-phosphate synthase, SS starch synthase, Rafs raafinose synthase, STS stachyose synthase, GoSL1 galactinol synthase 1, GoSL2 galactinol synthase 2.
Stress-related genes
Some of the most differentially expressed genes belonged to stress-related gene categories, which were up-regulated in all of the supplemented groups. These genes included several encoding heat-shock proteins and UDP-glycosil transferases (Table 4).
Table 4.
Differentially expressed stress-related genes
| FPKM | ||||||
|---|---|---|---|---|---|---|
| Gene ID | Annotation | CT | CH | NAR | CHSTOP | NARSTOP |
| Eucgr.H05081.4 | ALDH3I1 | 4.84 | 0.93* | 1.11* | 1.60 | 5.34 |
| Eucgr.C00112.1 | CIA2 | 0.70 | 12.29* | 14.05* | 17.42* | 3.57 |
| Eucgr.K01387.2 | COL9 | 3.33 | 26.78* | 22.63* | 18.02* | 2.56 |
| Eucgr.C03449.1 | HSFA2 | 0.29 | 14.55* | 12.98* | 14.36* | 18.58* |
| Eucgr.C03456.1 | HSFA2 | 0.09 | 2.67* | 3.64* | 2.25* | 1.45 |
| Eucgr.C00873.1 | HSFA2 | 1.92 | 24.38* | 18.82* | 14.75* | 7.94 |
| Eucgr.C03434.1 | HSFA2 | 0.31 | 6.75* | 6.22* | 6.19* | 2.03 |
| Eucgr.C01043.1 | HSFC1 | 2.92 | 122.26* | 116.87* | 96.98* | 13.96 |
| Eucgr.J01981.1 | HSP18.2 | 2.00 | 34.43 | 50.34 | 69.86* | 105.48* |
| Eucgr.J01980.1 | HSP18.2 | 0.02 | 12.20* | 11.59* | 9.15* | 26.39* |
| Eucgr.J01959.1 | HSP18.2 | 3.19 | 142.90* | 89.76* | 58.22* | 148.21* |
| Eucgr.J01958.1 | HSP18.2 | 2.68 | 115.32* | 90.34* | 65.56 | 100.03 |
| Eucgr.J01979.1 | HSP18.2 | 0.34 | 17.61* | 19.94* | 14.63* | 27.21* |
| Eucgr.J01958.1 | HSP18.2 | 2.68 | 115.32* | 90.34* | 65.56 | 100.03 |
| Eucgr.J01979.1 | HSP18.2 | 0.34 | 17.61* | 19.94* | 14.63* | 27.21* |
| Eucgr.J01977.1 | HSP18.2 | 0.37 | 9.43* | 8.59* | 7.08 | 19.26* |
| Eucgr.J01964.1 | HSP18.2 | 13.65 | 145.21* | 119.05* | 111.04 | 251.88* |
| Eucgr.J01985.1 | HSP18.2 | 0.31 | 22.25* | 16.62* | 13.21* | 33.33* |
| Eucgr.J01982.1 | HSP18.2 | 0.21 | 8.65* | 6.06* | 4.24 | 13.03* |
| Eucgr.F04479.1 | HSP20-like | 0.12 | 35.55* | 40.44* | 23.84* | 57.92* |
| Eucgr.I02136.1 | HSP20-like | 1.68 | 226.73* | 147.43* | 90.02* | 259.35* |
| Eucgr.J01969.1 | HSP20-like | 4.89 | 192.23* | 134.53* | 103.09* | 310.44* |
| Eucgr.G00061.1 | HSP20-like | 9.45 | 834.94* | 858.21* | 726.74* | 814.87 |
| Eucgr.E00433.1 | HSP20-like | 3.35 | 295.94 | 205.55* | 185.03 | 165.89 |
| Eucgr.F02898.1 | HSP20-like | 2.76 | 525.99* | 343.17* | 280.68* | 342.59* |
| Eucgr.A01416.1 | HSP21 | 0.08 | 12.40* | 7.60* | 4.34* | 0.29 |
| Eucgr.H04692.1 | HSP21 | 2.97 | 83.31* | 59.57* | 43.11* | 313.22* |
| Eucgr.J03127.1 | Hsp70b | 8.27 | 1576.75 | 1347.55* | 881.92 | 925.42 |
| Eucgr.H03518.1 | HSP70T-2 | 6.06 | 282.04* | 218.12* | 152.91 | 249.47 |
| Eucgr.K00295.1 | HSP90-1 | 2.11 | 46.13* | 38.25 | 35.52* | 62.69* |
| Eucgr.F03704.1 | MSL6 | 1.45 | 27.74 | 21.67* | 11.53* | 10.03 |
| Eucgr.H02896.1 | MYB305 | 0.07 | 6.57* | 8.20* | 6.34* | 1.14 |
| Eucgr.C00618.1 | Oleosin | 0.50 | 38.75* | 24.66* | 18.36* | 2.85 |
| Eucgr.F01003.1 | PAT1 | 2.30 | 43.54* | 47.40* | 43.89* | 6.02 |
| Eucgr.K00237.1 | PEBP | 0.04 | 115.83* | 64.41* | 61.47* | 11.49* |
| Eucgr.B00176.2 | PIMT2 | 3.86 | 153.25* | 109.01* | 82.97* | 57.39* |
| Eucgr.G01510.1 | RLK | 1.71 | 10.40* | 11.17* | 11.20* | 6.80 |
| Eucgr.F01854.1 | TRX1 | 4.16 | 598.03* | 295.93* | 191.61* | 25.01 |
| Eucgr.G02440.1 | UGT73B2 | 0.00 | 5.46* | 5.80* | 3.33* | 4.06* |
| Eucgr.L03261.1 | UGT73B3 | 1.44 | 47.41* | 27.81* | 19.98* | 75.67* |
| Eucgr.G02259.1 | UGT73B3 | 0.00 | 2.73* | 2.14* | 1.18* | 3.10* |
| Eucgr.I00409.1 | UGT73B3 | 0.06 | 3.27* | 4.18* | 2.91* | 0.60 |
| Eucgr.B02291.1 | UGT76E11 | 10.86 | 52.79* | 42.69* | 39.45 | 45.69 |
| Eucgr.K01389.2 | XERICO | 19.81 | 1,348.64* | 796.35* | 569.70* | 120.14 |
FPKM -fragments per kilobase of exon per million fragments mapped. CT – control; CH – prolonged naringenin-chalcone supp; NAR – prolonged naringenin supp; CHSTOP- short-term naringenin-chalcone supp; NARSTOP – short-term naringenin sup *Denotes differential expression.
Abbreviations: ALDH3I1 –aldehyde dehydrogenase 3I1, CIA2 chloroplast import apparatus 2, COL9 CONSTANS-like 9, HSFA2 heat shock transcription factor A2, HSFC1 heat shock transcription factor C1, HSP18.2 heat shock protein 18.2, HSP20 like chaperones superfamily protein, HSP21 heat shock protein 21, Hsp70b heat shock protein 70B, HSP70T-2 heat-shock protein 70 T-2, HSP90.1 heat shock protein 90.1, MSL6 mechanosensitive channel of small conductance-like 6, MYB305 myb domain protein 305, Oleosin family protein; PAT1 GRAS family transcription factor, PEBP –phosphatidylethanolamine-binding protein family protein, RLK receptor lectin kinase, TRX1 thioredoxin H-type 1, UGT73B2 UDP-glucosyltransferase 73B2, UGT73B3 UDP-glucosyl transferase 73B3, UGT76E11 UDP-glucosyl transferase 76E11, XERICO RING/U-box superfamily protein.
Enzymatic hydrolysis
To verify the effects of flavonoid supplementation on sugar yields and saccharification in Eucalyptus wood, enzymatic hydrolysis was performed. The hydrolysates were analyzed for total sugar contents (‘reduced sugars’), which included most of the pentoses and hexoses from the hemicellulose fraction, and glucose content (‘glucose’), allowing an estimate of the percent of saccharification to be obtained.
Flavonoid-supplemented plantlets showed increased sugar and glucose values compared to the control groups. The reduced sugar content was increased from 50% (CH) to 250% (NARSTOP), and the glucose content was increased from 43% (CH) to 253% (NARSTOP). With the exception of the naringenin-chalcone prolonged supplementation treatment (CH), all of the treatment values were considered statistically significant (Table 5).
Table 5.
Total sugar and glucose values
| n | Reduced sugars (mg/ml) | Reduced sugar yield % | Glucose (mg/ml) | |
|---|---|---|---|---|
| CT | 7 | 1.17 (0.67) | 5.69 (3.23) | 0.39 (0.23) |
| CH | 4 | 1.8 (0.33) | 9.06 (1.83) | 0.56 (0.07) |
| NAR | 3 | 2.54 (0.005)** | 12.72 (0.66)** | 0.87 (0.10)** |
| CHSTOP | 3 | 2.32 (0.24)* | 11.81 (1.08)* | 0.85 (0.29)* |
| NARSTOP | 3 | 3 (0.85)** | 14.59 (4.34)** | 0.99 (0.09)** |
Mean values and standard deviations (parentheses) for total sugar and glucose levels. n –number of biological replicates; CT – control; CH – prolonged naringenin-chalcone supp; NAR – prolonged naringenin supp; CHSTOP –short-term naringenin-chalcone supp; NARSTOP – short-term naringenin supp. *p-value <0.05; **p-value <0.01.
Discussion
The metabolism of phenylpropanoids follows 2 main pathways: the lignin branch and the flavonoid branch. The two pathways share common substrates and enzymes, and these shared components lead to a high level of interdependence between the pathways. Considering the economic interest in Eucalyptus trees for paper and pulp production, and given that flavonoids are known to have a direct influence on lignification and wood formation in several species [31,32], including Eucalyptus species, as previously demonstrated by our group [27], it is of high interest to verify the effects of flavonoid supplementation on gene expression, especially concerning genes related to wood formation. Additionally, there is a pressing interest in expanding the industrial uses of Eucalyptus because Eucalyptus forest cultures are well-established in Brazil and may affect other strategic sectors, such as second-generation biochemicals. In this case, Eucalyptus wood could be employed as lignocellulosic biomass for biological fermentation [33,34].
With this objective, we designed the present work to investigate the molecular basis of the differences in wood observed in flavonoid-supplemented E. urograndis trees. Additionally, in light of our previous findings, we paid special attention to the expression of genes involved with lignin and secondary cell wall formation and to the possible association between gene expression and the chemical composition of wood in Eucalyptus.
We analyzed the whole genome (44,974 genes) of Eucalyptus plants following supplementation with different flavonoids. A total of 1,573 (3,5%) differentially expressedgenes were identified, which were distributed among the supplementation groups: 963 genes were down-regulated and 610 genes were up-regulated. Most of the differentially expressed genes were associated with the prolonged supplementation groups (1,289 for NAR and 917 for CH), while the short-term supplementation groups displayed fewer differentially expressed genes (268 for CHSTOP and 47 for NARSTOP). Most of the differentially expressed genes in the CHSTOP and NARSTOP groups were also differentially expressed in the NAR and CH groups. Thus, naringenin supplementation appears to have had a stronger but less durable effect, while naringenin-chalcone supplementation has a longer-lasting effect on gene expression.
GO enrichment analyses demonstrated that there were several categories involved in cell wall formation that were down-regulated in all of the supplemented groups, including the phenylpropanoid pathway in the NAR-supplemented samples. The up-regulated gene categories included many responses to stress and the environment as well as genes related to sugar alcohols, through being involved in polyol, hexitol and alditol metabolism (minor CHOs), in the CH group. This pattern could also be observed in the mapping analysis of differentially expressed genes performed using MapMan software, in which several pathways, most notably those associated with the cell wall and phenylpropanoids, were down-regulated, while the metabolic pathways associated withminor CHOs, flavonoids, sucrose and starch displayed up-regulated genes. Furthermore, there was strong evidence that stress may play a major role, as several stress-related gene categories were found to be enriched via GO analysis, even in the groups subjected to short-term supplementation.
It was therefore clear that lignification and the phenylpropanoid pathway are affected by a great number of factors, and we believe that our work can help to clarify some of these factors. The interdependence of the phenylpropanoid, flavonoid and lignin branches has been explored in other studies. For example, it has been reported that 4CL activity is inhibited by some flavonoids, such as naringenin-chalcone and naringenin, which are the products of the chalcone synthase (CHS) and chalcone isomerase (CHI) enzymes, respectively [31,35]. The same study demonstrated that the administration of flavonoids suppressed the growth of 20 plant species, although the sensitivities of the plants to flavonoids were different.
In addition, the activation of the lignin precursor cinnamic acid (catalyzed by C4H) and p-coumaroyl-CoA (catalyzed by 4CL) is, to some extent, regulated by the activity of the CHS enzyme, which is involved in the first step of flavonoid biosynthesis [35]. It has also been reported that CHS is associated with growth suppression via the regulation of 4CL. This association has major importance in lignin biosynthesis in a great number of species [32,35].
As demonstrated by our results, several genes involved in the phenylpropanoid pathway were differentially expressed in plants subjected to supplementation with flavonoids (Table 2; Figure 3). Our most noteworthy findings revealed the differential expression of genes directly related to lignin synthesis. The NAR-supplemented group presented down-regulation of both the 4CL and CCR genes, whereas the ATOMT1 and 2 CAD genes were up-regulated. The CH-supplemented group exhibited HCT down-regulation and 1 CAD gene that was up-regulated. In the CHSTOP-supplemented group, 1 methyltransferase was up-regulated. No genes from the phenylpropanoid pathway were differentially expressed following supplementation with NARSTOP.
Surprisingly, the gene encoding F5H, which is one of the key enzymes involved in the synthesis of the monolignol sinapyl alcohol and, ultimately, the S lignin moiety, was not found to be differentially expressed on our analyses. This result is particularly interesting in light of our finding that the S/G ratios in all of the flavonoid-supplemented groups were higher than that of the control group. Thus, we expected a change in the expression of F5H following flavonoid treatment. Because phenylpropanoid metabolism is complex, it is likely that the differential regulation of other enzymatic steps, such as those encoded by the 4CL, HCT, CCR, ATOM1 and CAD genes, may underlie this response.
Some findings reported in the literature support this possibility. For example, 4CL plays a major role in phenylpropanoid metabolism, as its product, p-coumaroyl-CoA, is a substrate that is common to the flavonoid and lignin synthesis pathway. HCT silencing in Arabidopsis represses lignin synthesis and plant growth, and the metabolic flux is redirected toward flavonoids by chalcone synthase activity [24]. CCR catalyzes the reduction of hydroxycinnamoyl-CoA thioesters to the corresponding aldehydes; this reaction is considered to be a potential control point that regulates the overall carbon flux in favor of lignin [36]. Arabidopsis ATOMT1 knock-out mutants lack S units [37], and CAD catalyzes the reduction of cinnamaldehydes to cinnamyl alcohols, which is the last step in the biosynthesis of the monolignols, thus playing a pivotal role in determining the lignin monomer composition and increasing S contents [13].
There are also several laccases that have been demonstrated to be involved in lignification [38], and many laccases were found to be down-regulated in the NAR-, CH- and CHSTOP-supplemented samples.
Our results further corroborate those of [39], who suggested that Arabidopsis responds to the accumulation of 1 or more intermediates from the flavonoid pathway by down-regulating either the whole phenylpropanoid pathway or the specific branch leading to monocyclic phenolic compounds. According to our results, it is possible that the accumulation of naringenin-chalcone and naringenin, the products of CHS and CHI, respectively, due to exogenous supplementation, results in the down-regulation of genes from the phenylpropanoid pathway, with the exception of 2 genes involved in the final steps of sinapilic acid synthesis (ATOMT1 and CAD). This down-regulation may at least partially explain the higher S/G ratios observed in the supplemented samples and is in agreement with the findings of [40] that the reduction of total flux through the entire monolignol pathway affects G-lignin resulting in higher S/G ratio.
While the NARSTOP-supplemented plants did not show differential expression of any genes that are related to lignin synthesis according to our statistical analyses, they exhibited FPKM values that were similar to those of the NAR-, CH- and CHSTOP-supplemented groups, but closer to the control values than the other groups. This indicates that an early impact on gene expression may be sufficient to promote the phenotypic differences observed in this group.
Another possibility is that factors other than the genes from the lignification pathway per se influence the lignin monomer composition. Cook and collaborators [41] reported that the levels of cellulose, xylan and lignin are not completely dependent on the transcription of the genes involved in these metabolic pathways. Thus, the regulation of cell wall biosynthesis occurs at different levels, not only at the transcriptional level [41].
Additionally, other genes that have not yet been discovered may be causing the observed differences, as many no hits and unknown proteins were found among the most differentially expressed genes following flavonoid treatment. The stress and environmental response pathways were significantly enriched and associated with lignification; thus, these pathways may play major roles in the alterations of lignin composition after flavonoid supplementation.
Stress and lignification are closely related. Many of the products of the phenylpropanoid pathway are induced by biotic and abiotic stress [42]. Both flavonoids and sinapate esters, which are used for lignin synthesis, are important for UV protection [39]. Arabidopsis mutants with reduced levels of CHS and CHI activity show up to 60% higher levels of sinapate esters [39,42].
Moreover, a large number of phenylpropanoids are induced by stress, such as those derived from the C15 flavonoid skeleton that are synthesized via the chalcone synthase (CHS)-mediated condensation of p-coumaroyl-coenzyme A (CoA) and three molecules of malonyl-CoA [43]. In most plant families, the initial product of CHS is a tetrahydroxychalcone, which is further converted to other flavonoid classes, such as flavones, flavanones, flavanols, anthocyanins and 3-deoxyanthocyanidins, all of which are compounds that are important in the response to stress [44].
The observation that several genes related to stress responses are differentially expressed in flavonoid-supplemented trees confirms the importance of stress responses in defining Eucalyptus wood properties as has been previously shown by our group [45]. In this work, a comparison between three Eucalyptus species revealed differential expression of stress-related genes in E. urophylla, which could explain the higher plasticity and adaptability of this species compared to E. grandis and E. globulus, the two other studied species.
In the present study, the stress-related genes that were differentially expressed following flavonoid treatments included a noteworthy group composed of several UDP-glucosyltransferases (UGTs), which were up-regulated in all of our treatments. In plants, UGTs utilize UDP-glucose, UDP-galactose, and UDP-rhamnose as sugar donors and are involved in the modulation of plant architecture and the water stress response in Arabidopsis [46]. The glucosylation of coniferyl aldehyde and sinapyl aldehyde may regulate both lignin biosynthesis and the metabolism of other phenylpropanoids, such as ferulic acid, 5-hydroxyferulic acid, sinapic acid and their derivatives [47]. Thus, the presence of up-regulated UGTs in all of the groups is another interesting result that might help elucidate the chemical differences present in the flavonoid-supplemented trees.
Another notable finding regarding cell wall formation was the differential expression of genes involved in the metabolism of sucrose, starch, CHOs and minor sugars. Despite the down-regulation of sucrose synthase (Sus) and cellulose synthase (CesA), there were several other enzymes involved in this metabolic pathway that were up-regulated in the prolonged flavonoid treatments, even in the short-term treatments. Starting with galactinol synthase (GolS2), which was one of the most differentially expressed genes, all of the downstream genes in this pathway were differentially expressed (up-regulated), most of which were up-regulated after both the prolonged and short-term treatments.
The most notable of these genes was stachyose synthase (STS), which converts raafinose (a trisaccharide of galactose, fructose and glucose) into stachyose (a tetrasaccharide) by transferring a galactosyl moiety from galactinol, [48]. Raafinose synthase (RafS) was also differentially up-regulated. Additionally, sucrose phosphate synthase (SPS1F), which catalyses the conversion of UDP-glucose and D-fructose 6-phosphate into UDP and sucrose 6-phosphate [49], was also differentially up-regulated (Figure 4).
Figure 4.
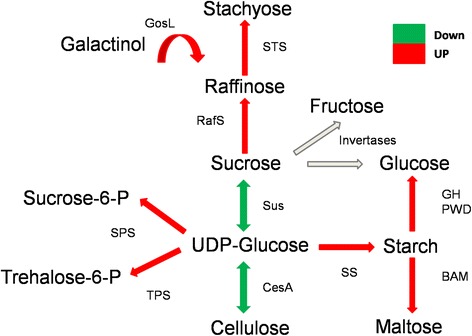
Effects of flanonoid supplementantion on secondary cell wall related genes. The effects of flavonoid supplementation on the expression of secondary cell wall-related genes. Sus –sucrose synthase; SPS –sucrose phosphate synthase; CesA –cellulose synthase;; GH –glycoside hydrolase; PWD –phosphoglucan water dikinases; BAM –beta-amylase; TPS –trehalose-6-phosphate synthase; SS –starch synthase; Rafs – raafinose synthase; STS –stachyose synthase; GoSL –galactinol synthase.
Starch synthase (SS) was up-regulated as well, as were enzymes involved in the degradation of starch into maltose (beta-amylase; BAM) and glucose (glycoside hydrolase; GH and phosphoglucan water dikinases; PWD). These results suggest a shift from sucrose and cellulose production to the synthesis of starch and minor sugars (galactinol, raffinose and stachyose).
Thus, to verify the possible effects of these transcriptional responses on cell wall formation and sugar accessibility, enzymatic hydrolysis was performed in wood samples from all of the experimental and control plants. The results revealed an increase in sugar contents (up to 250% in the NARSTOP group) and glucose yields of all of the flavonoid-supplemented Eucalyptus plants.
This may be the result of the plants producing more sugar or a result of the increased digestibility of lignin due to modifications of lignin structure and other cell wall components. The total values should increase exponentially with the use of pre-treatments and the extension of milling times [50,51].
Galactinol, raffinose and stachyose have been described as being involved in freezing and stress tolerance [52,53]. The levels of galactinol and raffinose are increased in the leaves of Arabidops is plants over-expressing HsfA2, a heat shock factor, suggesting a possible role for these compounds in protection from oxidative damage [54]. Moreover, this factor may also constitute another link between the stress response and cell wall formation in flavonoid-supplemented Eucalyptus plants because all of our treatment groups showed more than 1 up-regulated HsfA2 gene.
Our results are also in agreement with those of [55], who verified that in poplar trees, the over-expression of GolS and its product, galactinol, may serve as a molecular signal that initiates metabolic changes associated with combating stress, culminating in the formation of tension wood and increased glucose contents. Additionally, over-expression of raffinose synthase was found to result in increased biomass and total cellulose contents, while the total contents of lignin and xylose moieties were slightly reduced. Furthermore, the total amount of glucose was commensurately increased in the transgenic trees, by from ~1 to 4% [55]. Moreover, repression of the flavonoid pathway in Arabidopsis increases starch levels [56], and a chalcone isomerase-deficient Arabidopsis mutant exhibits increased levels of starch and soluble sugars in its leaves.
Based on our results, flavonoid supplementation causes a stress response in E. urograndis, greatly affecting cell wall development, modifying lignification by affecting the expression of genes involved in the phenylpropanoid pathway and altering sugar metabolismin favor of starch and minor sugar synthesis, resulting in increased sugar accessibility and saccharification.
Conclusions
The effects of flavonoid supplementation on cell wall development in Eucalyptus plants are most likely due to a combination of transcriptional changes in several distinct pathways. The down-regulation of the phenylpropanoid pathway, combined with the up-regulation of ATOMT1 and CAD, results in a higher S/G ratio, which in turn, increases lignin solubility and facilitates access to cellulose and hemicellulose. Subsequently, as a result of the stress response, sugar metabolism is shifted towards starch and minor sugars, culminating in the increased sugar and saccharification levels identified due to hydrolysis.
Given the importance of Eucalyptus in several industrial sectors, there is great interest in expanding the use of these species as a resource for cellulose, paper and pulp production and as an alternative source of biomass for second-generation biochemicals. Our results contribute not only to our understanding of the molecular responses involved in wood formation but will also have a significant impact on the use of Eucalyptus as biomass. Finally, we expect our findings to guide future genetic manipulation and nutritional supplementation analyses of Eucalyptus trees aimed at achieving significant improvements in their productivity yields.
Methods
Plant materials and tissue harvesting
Plantlets of a 6-month-old commercial clone of Eucalyptus urograndis were provided by International Paper (Mogi-Guaçu, Brazil) and grown in a greenhouse. The plantlets were divided into 5 groups according to supplementation conditions (apart from the standard nutritional solution supplied to all groups), as follows: control group (CT); experimental group 1 (CH), supplemented with 0.1 mmol of naringenin-chalcone for 5 months; experimental group 2 (NAR), supplemented with 0.1 mmol of naringenin for 5 months; experimental group 3 (CHSTOP), supplemented with 0.1 mmol of naringenin-chalcone for only the first month and then given the standard nutrition solution for the next 5 months; and experimental group 4 (NARSTOP), supplemented with 0.1 mmol of naringenin for only the first month and then given the standard nutrition solution for the next 5 months. Approximately 100–150 mL of each solution was administered via root application daily. The treatments lasted 5 months. The composition of the standard nutritional solution has been described previously [57]. At the end of the experiment, all 5 groups of plantlets were cut, and their stems were debarked, immediately frozen in liquid nitrogen and kept at −80°C for analysis; no growth differences were observed between the control and treatment groups (Additional file 4). All samples were analyzed 5 months after the beginning of the experiment, regardless of the applied supplementation.
Naringenin (4′-,5-,7-trihydroxyflavanone, 95%) and naringenin-chalcone (1,3-diphenyl-2-propen-1-one, 97%) were purchased from Sigma-Aldrich Co. (Tokyo, Japan) and AcrosOrganics Co. (Tokyo, Japan), respectively.
Total RNA extraction
Total RNA was extracted according to the protocol described by [58], with the modifications proposed by [59]. The obtained RNA concentration and quality were verified using a Nanodrop 2000 spectrophotometer (Thermo Scientific).
mRNA sequencing
mRNA sequencing was performed at the High-Throughput Sequencing Facility of the Carolina Center for Genome Sciences (University of North Carolina, USA). From each xylem sample, 10 μg of total RNA was used to prepare an mRNAseq library according to the protocol provided by Illumina. The gel extraction step was modified by dissolving excised gel slices at room temperature to avoid underrepresentation of AT-rich sequences [60]. Quality control and quantification of the libraries were performed using a DNA 1000 series II Bioanalyzer Chip (Agilent). For each library, single-end sequences of 36 or 50 bp were generated in a single lane using an Illumina Genome Analyzer IIx. A total of 8 libraries were generated: 3 biological replicates of the control group (CT); 2 biological replicates of the 5-month naringenin-supplemented groups (NAR); and 1 library for each remaining group (subjected to 1 month of supplementation with naringenin (NARSTOP), 5 months of supplementation with naringenin-chalcone (CH) or 1 month of supplementation with naringenin-chalcone (CHSTOP). Each library was constructed from a sample pooled from 3 individual trees. The complete dataset of RNA-seq reads has been deposited in SRA under accession numbers SRS716289; SRS716288, SRS716285; SRS716286; SRS716284.
Read alignment
The obtained Illumina reads were filtered to exclude ribosomal sequences (using the SILVA database [61] and low quality reads (phred ≥20). The remaining reads were aligned against the greater splice variants of E. grandis transcripts from Phytozome 7.0 (44,974 sequences) available at (http://www.phytozome.net/) [62] using the SOAP2 alignment software package [28]. To prepare the data for Genebrowser analysis, the read were aligned to the E. grandis genome using the TopHat aligner [63] to allow for spliced alignments. Both programs were configured to allow up two mismatches (because SNPs can generate mismatches in the alignments, especially in cases such as the present analysis, where the sequences come from different species), discard sequences with ambiguities (Ns) and return only reads with unique alignments.
Gene annotation
The Autofact program [64] was used to perform an automatic annotation of all E. grandis transcripts. The main feature of Autofact is its ability to perform gene annotation based on sequence similarity searches of several databases. BLASTx [65] (e-value cutoff of 1e-5) was used to align the obtained contigs against the following public databases: the NCBI non-redundant (NR) database; the Uniref90 and Uniref100 databases, which contain clustered sets of proteins from Uniprot [66]; the KEGG database of metabolic pathways [67]; and TAIR (version 10), an Arabidopsis proteins database. Functional annotation (GO) was performed using BLAST2GO [68] and MaPMAN [69] with the default parameters.
Determination of gene expression levels
Gene expression was measured via the FPKM (fragments per kilobase of exon per million fragments mapped) method using only one read alignment for each transcript, as described by [70]. The FPKM values for all transcripts are available for searching in the EUCANEXT database (www.lge.ibi.unicamp.br/eucalyptusdb).
Enzymatic hydrolysis
Substrate preparation
Samples from each Eucalyptus treatment were frozen in liquid nitrogen and then dried in FreeZone6 (Labconco) at - 51°C and 25 Pa for 48 hours. Subsequently, the lignocellulosic material was reduced through 1 cycle of 5 minutes in a ball-milling reactor. The milled material was used as a substrate for fungal growth and hydrolysis assays.
Secretome production for enzymatic hydrolysis
The Neurospora crassa wide strain St.L. 74A (Missouri University, Kansas City, http://www.fgsc.net/) was used for secretome production. Conidia preparation was performed by inoculating the fungus in 100 mL of minimal medium plus Vogel’s salts supplemented with 143 μL of biotin (biotin 5 mg, ethanol 50% (v/v), 143 μL of a trace element solution (5 g monohydrate citric acid, 5 g of ZnSO4.7H2O, 1 g of Fe (NH4)2.6H2O, 0.25 g of CuSO4.5H2O, 0.05 g MnSO4.H2O, 0.05 g H3BO3, 0.05 g Na2MoO4.2H2O, qsp 1,000 mL), 1.5% agar and 2% sucrose. The prepared samples were grown for 3 days at 30°C in the dark and then for 7 days in the light at 25°C. A conidial suspension was then inoculated in 100 mL of the same medium described above without agar [71] containing as the only carbon source 2% of a substrate blend of 3 Eucalyptus species: E. grandis, E. urograndis and E. urophylla, in a ratio of 3:3:1, prepared as described above.
Eucalyptus hydrolysis
Hydrolysis was performed as described by [72] with the following modifications: enzymatic hydrolysis was performed in 2 mL tubes shaken at 1,000 rpm at 30°C in a Thermomixer (Eppendorf) for 48 hours. Approximately 10 mg of substrate from each substrate preparation was diluted in 400 μL of 50 mM sodium acetate buffer pH 5.5, and 100 μL of the N. crassa secretome was then added. The protein concentration of the secretome was 0.4 μg/μL, as determined in a Bradford Kit assay (BioRad) with BSA as a standard. This temperature and pH were optimal for the hydrolysis of carboxymethyl cellulose, xylan and ß-glucan, as determined by testing the temperatures of 25–40°C and pH levels of 4.0 − 9.0. All hydrolysis reactions were performed in triplicate. For determination of the reducing sugar concentration and glucose production in supernatants derived from Eucalyptus hydrolysis, 2 mL tubes were centrifuged at 20,000 × g for 10 minutes at 4°C. The supernatant was then recovered, and 100 μL of each reaction was used to determine the content of reducing sugars by adding 100 μL of the dinitrosalicylic acid (DNS) assay reagent [73] heated to 99°C. A 100 μl aliquot of the sample was next transferred to an ELISA plate, and its absorbance was measured at 540 nm using a Tecan Infinite M200 microplate reader, referring to calibration curves generated from glucose solutions. To calculate glucose contents, 20 μL of the supernatant was added to 100 μL of a working solution from a Glucose Oxidase Kit (Laborlab) in ELISA plates. The reaction was subsequently incubated at 37°C for 10 minutes, and its absorbance and measured at 505 nm (in a Tecan Infinite M200 microplate reader). Glucose concentrations were calculated with a factor referring to a standard solution of glucose at 1 mg/mL. A blank reaction containing only buffer and substrate was subtracted from the measurements obtained for each assay. To verify significant differences between the controls and the flavonoid-supplemented groups, a one way ANOVA test was performed between the control and each supplemented group. The results were considered significant if p < 0.05 and were classified as follows: *, p < 0.05; **, p < 0.01.
Supporting data
The data set(s) supporting the results of this article is (are) included within the article (and its additional file (s)). The complete dataset of RNA-seq reads has been deposited in SRA under accession numbers: SRS716289; SRS716288, SRS716285; SRS716286; SRS716284. The FPKM values for all transcripts are available for searching in the EUCANEXT database (www.lge.ibi.unicamp.br/eucalyptusdb).
Acknowledgements
This project was funded by the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES) and the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPQ) and Grant #2012/22652-7, São Paulo Research Foundation (FAPESP). We would like to acknowledge the contributions of the Center for Computational Engineering and Sciences at UNICAMP SP Brazil (FAPESP/CEPID project #2013/08293-7), and we would especially like to thank International Paper do Brasil for their assistance.
Additional files
All expressed genes on any condition.
All differential expressed genes on all conditions.
Complete gene onthology analysis.
Height and diameter of samples.
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
JLN designed and carried field experiments, analyzed RNA-seq data, performed sacharification experiments, and wrote the manuscript. LCN provided bioinformatic support, designed the database and automatic annotation, and reviewed the manuscript. MMS helped with field experiment, analyze RNA-seq data and reviewed the manuscript. ELOC designed and helped with field experiments, participated on the manuscript conception and reviewed the manuscript. JPFC designed and performed sacharification experiments and reviewed the manuscript. PJT prepared libraries and performed RNA sequencing. WLM helped with field experiments and sampling of material and RNA. FMS designed and coordinated enzymatic hydrolysis experiments. PM designed and coordinated RNA-seq experiments and data acquisition. ACD supervised all experiments and helped draft and review the manuscript with substantial contribution to data interpretation. GAGP conceived the study, and participated in its design and coordination and helped to draft the manuscript and participated in data analysis. All authors read and approved the final manuscript.
Contributor Information
Jorge Lepikson-Neto, Email: lepikson@lge.ibi.unicamp.br.
Leandro C Nascimento, Email: leandro@lge.ibi.unicamp.br.
Marcela M Salazar, Email: msalazar@lge.ibi.unicamp.br.
Eduardo LO Camargo, Email: ecamargo@lge.ibi.unicamp.br.
João PF Cairo, Email: jpcairo@lge.ibi.unicamp.br.
Paulo J Teixeira, Email: Paulo@lge.ibi.unicamp.br.
Wesley L Marques, Email: wesleymarques@hotmail.com.
Fabio M Squina, Email: fabio.squina@me.com.
Piotr Mieczkowski, Email: piotr_Mieczkowski@med.unc.edu.
Ana C Deckmann, Email: ana.deckmann@gmail.com.
Gonçalo AG Pereira, Email: goncalo@unicamp.br.
References
- 1.Boudet AM, Kajita S, Grima-Pettenati J, Goffner D. Lignins and lignocellulosics: a better control of synthesis for new and improved uses. Trends Plant Sci. 2003;8:576–581. doi: 10.1016/j.tplants.2003.10.001. [DOI] [PubMed] [Google Scholar]
- 2.Plomion C, Leprovost G, Stokes A. Wood formation in trees. Plant Physiol. 2001;127:1513–1523. doi: 10.1104/pp.010816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rengel D, San Clemente H, Servant F, Ladouce N, Paux E, Wincker P, Couloux A, Sivadon P, Grima-Pettenati J. A new genomic resource dedicated to wood formation in Eucalyptus. BMC Plant Biol. 2009;9:36. doi: 10.1186/1471-2229-9-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Paiva JAP, Prat E, Vautrin S, Santos MD, San-Clemente H, Brommonschenkel S, Fonseca PGS, Grattapaglia D, Song X, Ammiraju JSS, Kudrna D, Wing RA, Freitas AT, Bergès H, Grima-Pettenati J. Advancing eucalyptus genomics: identification and sequencing of lignin biosynthesis genes from deep-coverage BAC libraries. BMC Genomics. 2011;12:137. doi: 10.1186/1471-2164-12-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grattapaglia D, Vaillancourt RE, Shepherd M, Thumma BR, Foley W, Külheim C, Potts BM, Myburg AA. Progress in myrtaceae genetics and genomics: eucalyptus as the pivotal genus. Tree Genet Genomes. 2012;8:463–508. doi: 10.1007/s11295-012-0491-x. [DOI] [Google Scholar]
- 6.Fukuda H. Xylogenesis: initiation, progression, and cell death. Annu Rev Plant Physiol Plant Mol Biol. 1996;47:299–325. doi: 10.1146/annurev.arplant.47.1.299. [DOI] [PubMed] [Google Scholar]
- 7.Paux E, Carocha V, Marques C, Mendes De Sousa A, Borralho N, Sivadon P, Grima-Pettenati J. Transcript profiling of Eucalyptus xylem genes during tension wood formation. New Phytol. 2005;167:89–100. doi: 10.1111/j.1469-8137.2005.01396.x. [DOI] [PubMed] [Google Scholar]
- 8.Kirst M, Myburg AA, De León JPG, Kirst ME, Scott J, Sederoff R. Coordinated genetic regulation of growth and lignin revealed by quantitative trait locus analysis of cDNA microarray data in an interspecific backcross of eucalyptus. Plant Physiol. 2004;135:2368–2378. doi: 10.1104/pp.103.037960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Foucart C, Jauneau A, Gion J-M, Amelot N, Martinez Y, Panegos P, Grima-Pettenati J, Sivadon P. Overexpression of EgROP1, a Eucalyptus vascular-expressed Rac-like small GTPase, affects secondary xylem formation in Arabidopsis thaliana. New Phytol. 2009;183:1014–1029. doi: 10.1111/j.1469-8137.2009.02910.x. [DOI] [PubMed] [Google Scholar]
- 10.Vanholme R, Demedts B, Morreel K, Ralph J, Boerjan W. Lignin biosynthesis and structure. Plant Physiol. 2010;153:895–905. doi: 10.1104/pp.110.155119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hu WJ, Harding SA, Lung J, Popko JL, Ralph J, Stokke DD, Tsai CJ, Chiang VL. Repression of lignin biosynthesis promotes cellulose accumulation and growth in transgenic trees. Nat Biotechnol. 1999;17:808–812. doi: 10.1038/11758. [DOI] [PubMed] [Google Scholar]
- 12.Fu D, Mazza G, Tamaki Y. Lignin extraction from straw by ionic liquids and enzymatic hydrolysis of the cellulosic residues. J Agric Food Chem. 2010;58:2915–2922. doi: 10.1021/jf903616y. [DOI] [PubMed] [Google Scholar]
- 13.Baucher M, Halpin C, Petit-conil M, Boerjan W. Lignin: genetic engineering and impact on pulping. Crit Rev Biochem Mol Biol. 2003;38:305–350. doi: 10.1080/10409230391036757. [DOI] [PubMed] [Google Scholar]
- 14.Li X, Ximenes E, Kim Y, Slininger M, Meilan R, Ladisch M, Chapple C. Lignin monomer composition affects Arabidopsis cell-wall degradability after liquid hot water pretreatment. Biotechnol Biofuels. 2010;3:27. doi: 10.1186/1754-6834-3-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boudet A-M. Lignins and lignification: selected issues. Plant Physiol Biochem. 2000;38:81–96. doi: 10.1016/S0981-9428(00)00166-2. [DOI] [Google Scholar]
- 16.Dixon RA, Lamb CJ, Masoud S, Sewalt VJH, Paiva NL. Metabolic engineering: prospects for crop improvement through the genetic manipulation of phenylpropanoid biosynthesis and defense responses — a review. Gene. 1996;179:61–71. doi: 10.1016/S0378-1119(96)00327-7. [DOI] [PubMed] [Google Scholar]
- 17.Guo D, Chen F, Inoue K, Blount JW, Dixon R a. Downregulation of caffeic acid 3-O-methyltransferase and caffeoyl CoA 3-O-methyltransferase in transgenic alfalfa. Impacts on lignin structure and implications for the biosynthesis of G and S lignin. Plant Cell. 2001;13:73–88. doi: 10.1105/tpc.13.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ferrer J-L, Austin MB, Stewart C, Noel JP. Structure and function of enzymes involved in the biosynthesis of phenylpropanoids. Plant Physiol Biochem. 2008;46:356–370. doi: 10.1016/j.plaphy.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rodrigues J, Meier D, Faix O, Pereira H. Determination of tree to tree variation in syringyl/guaiacyl ratio of Eucalyptus globulus wood lignin by analytical pyrolysis. J Anal Appl Pyrolysis. 1999;48:121–128. doi: 10.1016/S0165-2370(98)00134-X. [DOI] [Google Scholar]
- 20.Papermaking Properties of Eucalyptus Trees, Woods, and Pulp Fibers Celso Foelkel. In [http://www.eucalyptus.com.br/eucaliptos/ENG14.pdf]
- 21.Voo KS, Whetten RW, O’Malley DM, Sederoff RR. 4-coumarate:coenzyme a ligase from loblolly pine xylem. Isolation, characterization, and complementary DNA cloning. Plant Physiol. 1995;108:85–97. doi: 10.1104/pp.108.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Campbell MM, Sederoff RR. Variation in lignin content and composition (mechanisms of control and implications for the genetic improvement of plants) Plant Physiol. 1996;110:3–13. doi: 10.1104/pp.110.1.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vogt T. Phenylpropanoid biosynthesis. Mol Plant. 2010;3:2–20. doi: 10.1093/mp/ssp106. [DOI] [PubMed] [Google Scholar]
- 24.Besseau S, Hoffmann L, Geoffroy P, Lapierre C, Pollet B, Legrand M. Flavonoid accumulation in Arabidopsis repressed in lignin synthesis affects auxin transport and plant growth. Plant Cell. 2007;19:148–162. doi: 10.1105/tpc.106.044495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moustafa E. Purification and properties of chalcone-flavanone isomerase from soya bean seed. Phytochemistry. 1967;6:625–632. doi: 10.1016/S0031-9422(00)86001-X. [DOI] [Google Scholar]
- 26.Chen W-J, Yun M-S, Deng F, Yogo Y. Chalcone suppresses lignin biosynthesis in illuminated soybean cells. Weed Biol Manag. 2011;11:49–56. doi: 10.1111/j.1445-6664.2011.00404.x. [DOI] [Google Scholar]
- 27.Lepikson-Neto J, Alves A, Deckmann AC, Camargo LO, Salazar MM, Carolina Scatollin Rio M, Nascimento LC, Pereira GAG, Rodrigues JC. Flavonoid supplementation reduces extractives content and increases S/G ratio on Eucalyptus grandis x Eucalyptus urophylla hybrid trees. Bioresources. 2013;8:1–11. doi: 10.15376/biores.8.2.1747-1757. [DOI] [Google Scholar]
- 28.Li R, Yu C, Li Y, Lam T-W, Yiu S-M, Kristiansen K, Wang J. SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics. 2009;25:1966–1967. doi: 10.1093/bioinformatics/btp336. [DOI] [PubMed] [Google Scholar]
- 29.Shamir R, Maron-Katz A, Tanay A, Linhart C, Steinfeld I, Sharan R, Shiloh Y, Elkon R. EXPANDER–an integrative program suite for microarray data analysis. BMC Bioinformatics. 2005;6:232. doi: 10.1186/1471-2105-6-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hardcastle TJ, Kelly KA. BaySeq: empirical Bayesian methods for identifying differential expression in sequence count data. BMC Bioinformatics. 2010;11:422. doi: 10.1186/1471-2105-11-422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen W-J, Yun M-S, Deng F, Yogo Y, L S. Effects of root-applied naringenin and chalcone on the growth of annual plants. Weed Biol Manag. 2004;238(August):235–238. doi: 10.1111/j.1445-6664.2004.00143.x. [DOI] [Google Scholar]
- 32.Deng F, Aoki M, Yogo Y. Effect of naringenin on the growth and lignin biosynthesis of gramineous plants. Weed Biol Manag. 2004;4:49–55. doi: 10.1111/j.1445-6664.2003.00119.x. [DOI] [Google Scholar]
- 33.Mizrachi E, Mansfield SD, Myburg AA. Cellulose factories: advancing bioenergy production from forest trees. New Phytol. 2012;194:54–62. doi: 10.1111/j.1469-8137.2011.03971.x. [DOI] [PubMed] [Google Scholar]
- 34.Mansfield SD, Kang K-Y, Chapple C. Designed for deconstruction–poplar trees altered in cell wall lignification improve the efficacy of bioethanol production. New Phytol. 2012;194:91–101. doi: 10.1111/j.1469-8137.2011.04031.x. [DOI] [PubMed] [Google Scholar]
- 35.Chen W, Yun M-S, Deng F, Yogo Y. The rates of maize growth and lignin biosynthesis change after root-applied chalcone. Weed Biol Manag. 2005;5:118–122. doi: 10.1111/j.1445-6664.2005.00165.x. [DOI] [Google Scholar]
- 36.Lacombe E, Hawkins S, Van Doorsselaere J, Piquemal J, Goffner D, Poeydomenge O, Boudet AM, Grima-Pettenati J. Cinnamoyl CoA reductase, the first committed enzyme of the lignin branch biosynthetic pathway: cloning, expression and phylogenetic relationships. Plant J. 1997;11:429–441. doi: 10.1046/j.1365-313X.1997.11030429.x. [DOI] [PubMed] [Google Scholar]
- 37.Goujon T, Sibout R, Pollet B, Maba B, Nussaume L, Bechtold N, Lu F, Ralph J, Mila I, Barrière Y, Lapierre C, Jouanin L. A new arabidopsis thaliana mutant deficient in the expression of O-methyltransferase impacts lignins and sinapoyl esters. Plant Mol Biol. 2003;51:973–989. doi: 10.1023/A:1023022825098. [DOI] [PubMed] [Google Scholar]
- 38.Berthet S, Demont-Caulet N, Pollet B, Bidzinski P, Cézard L, Le Bris P, Borrega N, Hervé J, Blondet E, Balzergue S, Lapierre C, Jouanin L. Disruption of LACCASE4 and 17 results in tissue-specific alterations to lignification of Arabidopsis thaliana stems. Plant Cell. 2011;23:1124–1137. doi: 10.1105/tpc.110.082792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li J, Ou-Lee TM, Raba R, Amundson RG, Last RL. Arabidopsis flavonoid mutants are hypersensitive to UV-B irradiation. Plant Cell. 1993;5:171–179. doi: 10.1105/tpc.5.2.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang JP, Naik PP, Chen H-C, Shi R, Lin C-Y, Liu J, Shuford CM, Li Q, Sun Y-H, Tunlaya-Anukit S, Williams CM, Muddiman DC, Ducoste JJ, Sederoff RR, Chiang VL. Complete proteomic-based enzyme reaction and inhibition kinetics reveal how monolignol biosynthetic enzyme families affect metabolic flux and lignin in populus trichocarpa. Plant Cell. 2014;26:894–914. doi: 10.1105/tpc.113.120881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cook CM, Daudi A, Millar DJ, Bindschedler LV, Khan S, Bolwell GP, Devoto A. Transcriptional changes related to secondary wall formation in xylem of transgenic lines of tobacco altered for lignin or xylan content which show improved saccharification. Phytochemistry. 2012;74(null):79–89. doi: 10.1016/j.phytochem.2011.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dixon RA, Paiva NL. Stress-induced phenylpropanoid metabolism. Plant Cell. 1995;7:1085–1097. doi: 10.1105/tpc.7.7.1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Harbone J. The Flavonoids: Advances in Research Since 1980. Chapman Hall: Springer; 1988. pp. 621–624. [Google Scholar]
- 44.Holton TA, Cornish EC. Genetics and biochemistry of anthocyanin biosynthesis. Plant Cell. 1995;7:1071–1083. doi: 10.1105/tpc.7.7.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Salazar MM, Nascimento LC, Camargo EL, Gonçalves DC, Lepikson-Neto J, Marques WL, Teixeira PJ, Mieczkowski P, Mondego JM, Carazzolle MF, Deckmann AC, Pereira GA. Xylem transcription profiles indicate potential metabolic responses for economically relevant characteristics of Eucalyptus species. BMC Genomics. 2013;14:201. doi: 10.1186/1471-2164-14-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tognetti VB, Van Aken O, Morreel K, Vandenbroucke K, van de Cotte B, De Clercq I, Chiwocha S, Fenske R, Prinsen E, Boerjan W, Genty B, Stubbs KA, Inzé D, Van Breusegem F. Perturbation of indole-3-butyric acid homeostasis by the UDP-glucosyltransferase UGT74E2 modulates Arabidopsis architecture and water stress tolerance. Plant Cell. 2010;22:2660–2679. doi: 10.1105/tpc.109.071316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lim E-K, Jackson RG, Bowles DJ. Identification and characterisation of Arabidopsis glycosyltransferases capable of glucosylating coniferyl aldehyde and sinapyl aldehyde. FEBS Lett. 2005;579:2802–2806. doi: 10.1016/j.febslet.2005.04.016. [DOI] [PubMed] [Google Scholar]
- 48.Peterbauer T, Mucha J, Mach L, Richter A. Chain Elongation of raffinose in pea seeds. Isolation, characterization, and molecular cloning of mutifunctional enzyme catalyzing the synthesis of stachyose and verbascose. J Biol Chem. 2002;277:194–200. doi: 10.1074/jbc.M109734200. [DOI] [PubMed] [Google Scholar]
- 49.Chua TK, Bujnicki JM, Tan T-C, Huynh F, Patel BK, Sivaraman J. The structure of sucrose phosphate synthase from Halothermothrix orenii reveals its mechanism of action and binding mode. Plant Cell. 2008;20:1059–1072. doi: 10.1105/tpc.107.051193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen F, Dixon RA. Lignin modification improves fermentable sugar yields for biofuel production. Nat Biotechnol. 2007;25:759–761. doi: 10.1038/nbt1316. [DOI] [PubMed] [Google Scholar]
- 51.Inoue H, Yano S, Endo T, Sakaki T, Sawayama S. Combining hot-compressed water and ball milling pretreatments to improve the efficiency of the enzymatic hydrolysis of eucalyptus. Biotechnol Biofuels. 2008;1:2. doi: 10.1186/1754-6834-1-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Saravitz DM, Pharr DM, Carter TE. Galactinol synthase activity and soluble sugars in developing seeds of four soybean genotypes. Plant Physiol. 1987;83:185–189. doi: 10.1104/pp.83.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Taji T, Ohsumi C, Iuchi S, Seki M, Kasuga M, Kobayashi M, Yamaguchi-Shinozaki K, Shinozaki K. Important roles of drought- and cold-inducible genes for galactinol synthase in stress tolerance in Arabidopsis thaliana. Plant J. 2002;29:417–426. doi: 10.1046/j.0960-7412.2001.01227.x. [DOI] [PubMed] [Google Scholar]
- 54.Nishizawa A, Yabuta Y, Shigeoka S. Galactinol and raffinose constitute a novel function to protect plants from oxidative damage. Plant Physiol. 2008;147:1251–1263. doi: 10.1104/pp.108.122465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Unda F, Canam T, Preston L, Mansfield SD. Isolation and characterization of galactinol synthases from hybrid poplar. J Exp Bot. 2012;63:2059–2069. doi: 10.1093/jxb/err411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rubin G, Tohge T, Matsuda F, Saito K, Scheible W-R. Members of the LBD family of transcription factors repress anthocyanin synthesis and affect additional nitrogen responses in Arabidopsis. Plant Cell. 2009;21:3567–3584. doi: 10.1105/tpc.109.067041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sarruje J. Soluções nutritivas. Summa Phytopathol. 1975;1:231–233. [Google Scholar]
- 58.Zeng Y, Yang T. RNA isolation from highly viscous samples rich in polyphenols and polysaccharides. Plant Mol Biol Rep. 2002;20:417–417. doi: 10.1007/BF02772130. [DOI] [Google Scholar]
- 59.Le Provost G, Paiva J, Pot D, Brach J, Plomion C. Seasonal variation in transcript accumulation in wood-forming tissues of maritime pine (Pinus pinaster Ait.) with emphasis on a cell wall glycine-rich protein. Planta. 2003;217:820–830. doi: 10.1007/s00425-003-1051-2. [DOI] [PubMed] [Google Scholar]
- 60.Quail MA, Kozarewa I, Smith F, Scally A, Stephens PJ, Durbin R, Swerdlow H, Turner DJ. A large genome center’s improvements to the Illumina sequencing system. Nat Methods. 2008;5:1005–1010. doi: 10.1038/nmeth.1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig W, Peplies J, Glöckner FO. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 2007;35:7188–7196. doi: 10.1093/nar/gkm864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Myburg AA, Grattapaglia D, Tuskan GA, Hellsten U, Hayes RD, Grimwood J, Jenkins J, Lindquist E, Tice H, Bauer D, Goodstein DM, Dubchak I, Poliakov A, Mizrachi E, Kullan ARK, Hussey SG, Pinard D, van der Merwe K, Singh P, van Jaarsveld I, Silva-Junior OB, Togawa RC, Pappas MR, Faria DA, Sansaloni CP, Petroli CD, Yang X, Ranjan P, Tschaplinski TJ, Ye C-Y, et al. The genome of Eucalyptus grandis. Nature. 2014;509:356–362. doi: 10.1038/nature13308. [DOI] [PubMed] [Google Scholar]
- 63.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Koski LB, Gray MW, Lang BF, Burger G. AutoFACT: an automatic functional annotation and classification tool. BMC Bioinformatics. 2005;6:151. doi: 10.1186/1471-2105-6-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Suzek BE, Huang H, McGarvey P, Mazumder R, Wu CH. UniRef: comprehensive and non-redundant UniProt reference clusters. Bioinformatics. 2007;23:1282–1288. doi: 10.1093/bioinformatics/btm098. [DOI] [PubMed] [Google Scholar]
- 67.Kanehisa M. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Conesa A, Götz S, García-Gómez JM, Terol J, Talón M, Robles M. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005;21:3674–3676. doi: 10.1093/bioinformatics/bti610. [DOI] [PubMed] [Google Scholar]
- 69.Thimm O, Bläsing O, Gibon Y, Nagel A, Meyer S, Krüger P, Selbig J, Müller LA, Rhee SY, Stitt M. MAPMAN: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J. 2004;37:914–939. doi: 10.1111/j.1365-313X.2004.02016.x. [DOI] [PubMed] [Google Scholar]
- 70.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 71.Phillips CM, Iavarone AT, Marletta MA. Quantitative proteomic approach for cellulose degradation by Neurospora crassa. J Proteome Res. 2011;10:4177–4185. doi: 10.1021/pr200329b. [DOI] [PubMed] [Google Scholar]
- 72.Bragatto J, Segato F, Cota J, Mello DB, Oliveira MM, Buckeridge MS, Squina FM, Driemeier C. Insights on how the activity of an endoglucanase is affected by physical properties of insoluble celluloses. J Phys Chem B. 2012;116:6128–6136. doi: 10.1021/jp3021744. [DOI] [PubMed] [Google Scholar]
- 73.Miller G. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Analytical chemistry. Anal Chem. 1959;31:426–428. doi: 10.1021/ac60147a030. [DOI] [Google Scholar]


