Abstract
Cryptococcus neoformans produces extracellular vesicles containing a variety of cargo, including virulence factors. To become extracellular, these vesicles not only must be released from the plasma membrane but also must pass through the dense matrix of the cell wall. The greatest unknown in the area of fungal vesicles is the mechanism by which these vesicles are released to the extracellular space given the presence of the fungal cell wall. Here we used electron microscopy techniques to image the interactions of vesicles with the cell wall. Our goal was to define the ultrastructural morphology of the process to gain insights into the mechanisms involved. We describe single and multiple vesicle-leaving events, which we hypothesized were due to plasma membrane and multivesicular body vesicle origins, respectively. We further utilized melanized cells to “trap” vesicles and visualize those passing through the cell wall. Vesicle size differed depending on whether vesicles left the cytoplasm in single versus multiple release events. Furthermore, we analyzed different vesicle populations for vesicle dimensions and protein composition. Proteomic analysis tripled the number of proteins known to be associated with vesicles. Despite separation of vesicles into batches differing in size, we did not identify major differences in protein composition. In summary, our results indicate that vesicles are generated by more than one mechanism, that vesicles exit the cell by traversing the cell wall, and that vesicle populations exist as a continuum with regard to size and protein composition.
INTRODUCTION
Extracellular vesicles are produced by all branches of microbial life (1). Most thoroughly studied in Gram-negative bacteria, the functions of these vesicles ranges from disposal of waste products and misfolded proteins to secretion of quorum-sensing signals to delivery of virulence-associated molecules (2). In the fungal kingdom, extracellular vesicles were first described in Cryptococcus neoformans and have subsequently been discovered in a variety of basidio- and ascomycetes (3–5). Recently, extracellular vesicles have also been described in Gram-positive bacteria (6).
The source of these eukaryotic extracellular vesicles is more complex than for bacterial extracellular vesicles, which are limited to the plasma membrane or outer membrane for source material. In addition to the plasma membrane, fungi and protozoan extracellular vesicles can additionally redirect intracellular organelle membranes for export. Current evidence supports multiple mechanisms of fungal extracellular vesicle formation. This includes work in the model yeast Saccharomyces cerevisiae, where single deletion of genes involved with Golgi-to-plasma membrane transport or multivesicular body (MVB) formation decreased but did not halt extracellular vesicle production (7). Electron tomography has also revealed C. neoformans extracellular vesicles forming from the plasma membrane, though from membrane invagination and subsequent scission rather than the outward budding associated with bacterial outer membrane vesicles (8). Higher eukaryotes, such as Caenorhabditis elegans and Drosophila melanogaster, utilize both plasma membrane and MVBs as membrane origin sources (9, 10), suggesting that eukaryotic microbes may also rely on redundant mechanisms for extracellular vesicle formation.
An important difference between microbial fungi and both other vesicle-producing eukaryotic microbes and metazoan systems is the presence of a cell wall bordering the plasma membrane. This adds an additional barrier through which vesicles must pass before being released into the extracellular environment. Electron microscopy (EM) has captured images of vesicles in the C. neoformans cell wall (3), but the mechanism by which these large hydrophobic objects are able to pass through remains unsolved.
Here, we utilized electron microscopy techniques to observe vesicles exiting C. neoformans cells through a variety of mechanisms, which involved both single and multiple vesicle-leaving events. We correlated size and event type, observing larger vesicles leaving in multiple leaving events. We used differential centrifugation to enrich larger vesicles to test the hypothesis that these sizes serve discrete cellular functions, and we observed that vesicle cargo is homogenously distributed, with no functional correlation between enriched subpopulations.
MATERIALS AND METHODS
Strains and media.
C. neoformans strains H99 and Cap67 were maintained in YPD broth (1% yeast extract, 2% peptone, 2% dextrose) (Difco). For all electron microscopy work and vesicle studies, strains were grown in a defined minimal medium (29.4 mM KH2PO4, 10 mM MgSO4, 13 mM glycine, 15 mM dextrose, and 3 μM thiamine-HCl) at 30°C for 3 to 5 days.
Vesicle purification.
Vesicle purification was performed as previously described (3). Cap67 was used for proteomic studies because of an increased vesicle yield in the absence of capsule (data not shown). Briefly, a 1-liter culture was inoculated with approximately 5 × 104 cells/ml of C. neoformans and grown for 5 days at 30°C. The cells were removed by centrifugation and two filtrations with a 0.45-μm pore filter. The cell-free supernatant was concentrated using an Amicon concentration unit with a 100-kDa-cutoff membrane. The concentrate was pelleted by ultracentrifugation at 100,000 × g for 1 h at 4°C. Pellets were combined and resuspended in a total of 3 ml phosphate-buffered saline (PBS), and 0.5 ml was set aside. The remainder was pelleted at 100,000 × g for 5 min at 4°C. The supernatant was moved to a fresh tube and pelleted at 100,000 × g for 10 min at 4°C. This was repeated with subsequent spins for 20 min and 60 min. The 0.5 ml set aside was spun at 100,000 × g for 1 h at 4°C to collect all possible vesicles. Pellets were resuspended in 0.2 ml PBS for use in further assays.
EM.
For transmission electron microscopy (TEM), the samples were first fixed with 2.5% glutaraldehyde and 2% paraformaldehyde in 0.1 M sodium cacodylate buffer, then postfixed with 1% osmium tetroxide followed by 2% uranyl acetate, and finally dehydrated through a graded series of ethanol solutions and embedded in LX112 resin (LADD Research Industries, Burlington, VT). Ultrathin sections were cut on a Reichert Ultracut UCT, stained with uranyl acetate followed by lead citrate, and viewed on a JEOL 1200EX transmission electron microscope at 80 kV. For scanning EM (SEM), the samples were first fixed in 2.5% glutaraldehyde–0.1 M sodium cacodylate–0.2 M sucrose–5 mM MgCl2 (pH 7.4) and then dehydrated through a graded series of ethanol solutions. Samples were critical-point dried using liquid carbon dioxide in a Tousimis Samdri 795 critical-point drier (Tousimis, Rockville, MD) and sputter coated with chromium in a Quorum EMS 150T ES (Quorum Technologies Ltd., United Kingdom). A Zeiss Supra field emission scanning electron microscope (Carl Zeiss Microscopy, LLC North America) was used to examine the samples, using an accelerating voltage of 1.5 kV. For cryo-scanning EM, the samples were fixed in 2.5% glutaraldehyde–0.1 M sodium cacodylate–0.2 M sucrose–5 mM MgCl2 (pH 7.4). After a water rinse, they were plunge-frozen in liquid ethane, transferred to liquid nitrogen, and finally transferred to Gatan Alto 2500 Cryotransfer (Warrendale, PA). Each sample was fractured at −120°C, sublimed at −100°C, and sputtered with chromium. A Zeiss Supra field emission scanning electron microscope (Carl Zeiss Microscopy, LLC North America) was used to examine the samples, using an accelerating voltage of 1.5 kV.
Dynamic light scattering (DLS).
Vesicles in PBS were measured in a 90Plus/BI-MAS multiangle particle sizing analyzer (Brookhaven Instruments). The sample was illuminated with laser monochromatic light, which was scattered by the Brownian motion of the vesicles. These light fluctuations were detected at a 90° angle and analyzed by the autocorrelation function C(t): C(t) = Ae2Γt + B, where t is time delay, A and B are optical constants, and Γ is related to the relaxation of the fluctuations by Γ = Dq2. D is derived from D = (KBT)/3πη(t)d, which assumes each scattering particle to be a sphere, where KB is Boltzmann's constant (1.38054E−23 J degree−1), T is the temperature in K (303 K), η(t) is the viscosity of the liquid in which the particles are moving, and d is the particle diameter. The parameter q is derived from the scattering angle ϴ, the laser light wavelength λ0, and the solvent refractive index (n) from the equation q = (2πn/λ0)2sin(ϴ/2). Data are expressed as the average of 10 runs of 1 min per run.
Enzymatic assays.
Vesicles in 30 μl PBS solution were aliquoted to a 96-well plate. One hundred microliters of enzyme reaction buffer was added, and plates were stored in the dark at 37°C for 16 h before being read with an enzyme-linked immunosorbent assay (ELISA) plate reader. For urease, the reaction buffer consisted of 1% peptone, 0.1% dextrose, 0.5% NaCl, 0.2% KH2PO4, 2% urea, and 0.0012% phenol red. The urease reaction was read at 540 nm. For phosphatase, the reaction buffer consisted of 1 mg/ml p-nitrophenyl phosphate (PNPP) in 100 mM sodium acetate buffer, pH 5.5. The phosphatase reaction was read at 405 nm. For laccase, the reaction buffer consisted of 12.5 mM L-dopa in PBS. The laccase reaction was read at 450 nm. Specific activity was assessed as absorbance/nmol phospholipid. Phospholipid content was measured using the EnzyChrom phospholipid assay kit (BioAssay Systems, Hayward, CA).
Proteomics.
Protein cargo in vesicle suspensions was identified as previously described (11). Briefly, protein from vesicles in 100 μl PBS was precipitated by adding 6 volumes of ice-cold acetone and incubating overnight at −20°C. Proteins were then reduced with 10 mM dithiothreitol (DTT), and the cysteine residues were subsequently alkylated with 10 mM iodoacetamide. Protein digestion was carried out with mass spectrometry (MS)-grade trypsin (1:20, wt/wt) overnight at 37°C. The peptide mixture was desalted, concentrated on Zip-Tip (Millipore), and then analyzed using Nanoflow liquid chromatography-tandem mass spectrometry (LC-MS/MS). The peptides were loaded onto a 0.3- by 5-mm C18 precolumn and then eluted with a linear gradient of 5 to 90% acetonitrile in a 0.1% aqueous solution of formic acid. The gradient elution was performed over 120 min using a NanoLC 1D Plus (Eksigent) at a flow rate of 200 nl/min through a 75-μm by 15-cm fused silica capillary C18 high-performance liquid chromatography column (LC Packings) to a stainless steel nanobore emitter (Proxeon). The peptides were scanned and fragmented with an LTQ XL linear ion trap mass spectrometer (ThermoFinnigan) operated in data-dependent and MS/MS switching mode using the 3 most intense precursor ions detected in a survey scan from 400 to 1,600 atomic mass units (amu). A database containing the NCBI Cryptococcus neoformans sequences was searched using Mascot software (version 2.3; Matrix Science) for protein identification. Search criteria included trypsin specificity with one missed cleave allowed, methionine oxidation as a variable modification, a minimum precursor and fragment ion mass accuracy of 1.2 and 0.3 Da, respectively, and a requirement of at least one bold red peptide (i.e., highest-score peptide matching to protein with highest total score). Cutoff values for Mascot protein scores were set at 29 (P > 0.05) to be considered an accurate identification. Proteins identified with only one peptide were inspected manually.
Statistics.
Student's t test was used to compare data sets. All statistical analyses were done using GraphPad Prism.
RESULTS
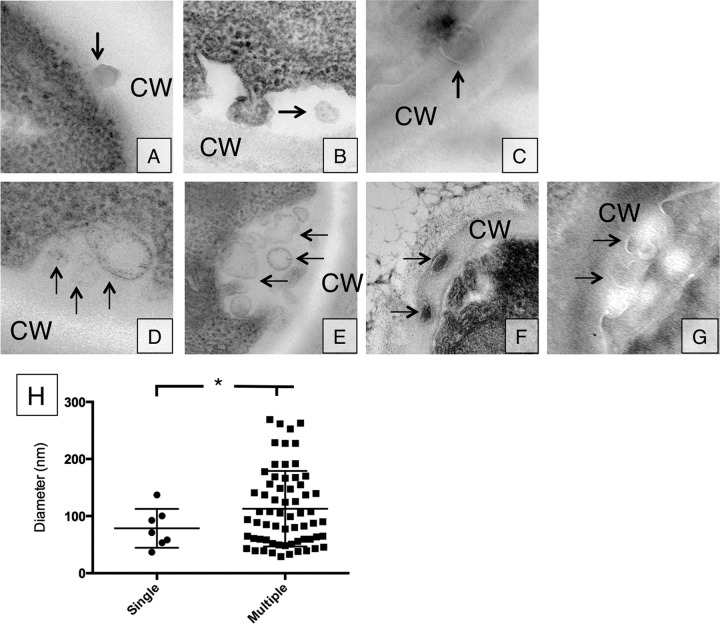
Our first goal was to investigate whether C. neoformans formed vesicles using multiple mechanisms, as described for S. cerevisiae. The approach was to visualize these structures with electron microscopy to ascertain their size and location in the cells. Using electron microscopy, we observed single vesicle-like structures in the cell wall or in the space between the plasma membrane and the cell wall (Fig. 1A to C). Vesicles were identified as circular structures with double membranes. We further identified what appeared to be MVB-to-plasma membrane fusion events in several cells, which resulted in the simultaneous release of several vesicles into the space between the plasma membrane and the cell wall (Fig. 1D to G). In some cases, we observed vesicles into the cell wall (Fig. 1F). These observations suggest that vesicles can be released either as single or as multiple vesicle events. We measured vesicle diameters, and found that larger vesicles (larger than 100 nm) were associated with multiple vesicle events, while smaller vesicles could be found leaving both as single or multiple vesicle events. All measurements were corrected by a factor of 1.273 to more closely reflect the three-dimensional (3D) nature of the 2D image (12).
FIG 1.
Vesicle release occurs in either single or multiple release events. (A to G) Single vesicle release events (A to C) as well as multiple vesicle release events (D to G) were observed in TEM section. Panels A, B, D, and E were obtained through conventional TEM, while panels C, F, and G were obtained with cryo-TEM. Arrows indicate vesicles outside the plasma membrane. (H) Vesicle density distribution for single and multiple vesicle release (single versus multiple, P < 0.05).
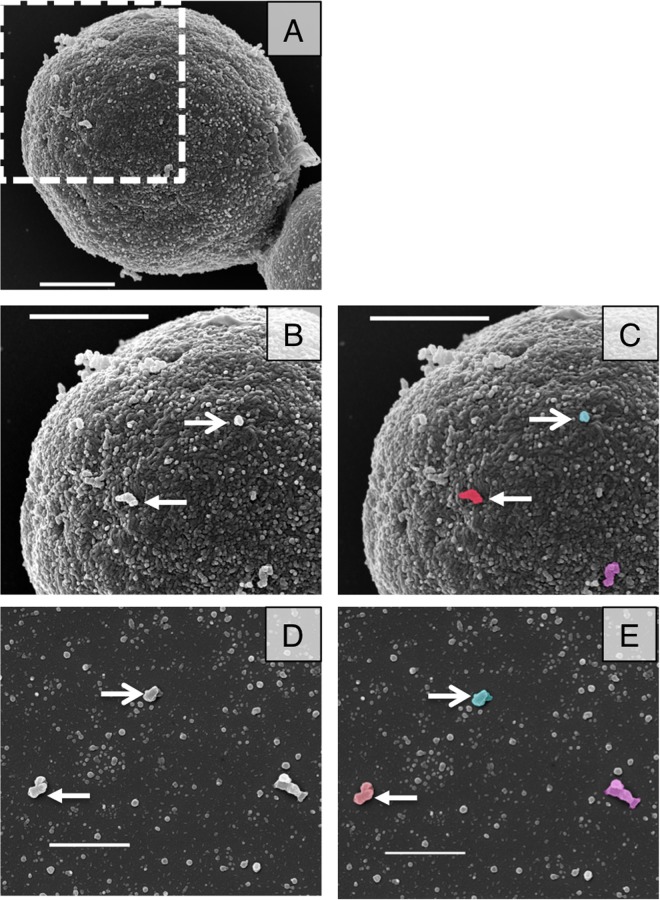
The observation that both single and multiple vesicle release events occurred led to the question of how these vesicles crossed the cell wall. To investigate this question, we used SEM imaging of acapsular Cap67 cells to view the cell wall surface. The surface contains many electron-dense areas in a speckled pattern that resemble small vesicles prepared from the same strain (Fig. 2). Because of the irregular nature of the cell wall, it is impossible to decisively identify these structures as vesicles, but we made a presumptive identification that these are vesicles from their rounded shape and dimensions. We noted several larger protrusions (Fig. 2B and C, arrows) that were similar in size and morphology to purified vesicle images (Fig. 2D and E, arrows). These cell wall surface vesicles may represent larger vesicles, leaving in a single vesicle release event, or several smaller vesicles aggregated together during the fixation process or brought together by exit though a single cell wall channel. These images show that we can identify vesicle-like structures on the outside of the cell wall.
FIG 2.
SEM images of acapsular cells shows vesicle-like structures at their surface. (A) SEM was used to investigate Cap67 cells. (B and C) Magnification of area marked in panel A, showing vesicle-like structures protruding from the surface of an acapsular cell. (D and E) Purified Cap67 vesicles. Arrows indicate structures on the cell wall surface similar to those in vesicle purifications. Panels C and E are false-colored images of panels B and D, respectively. Scale bars = 1 μm in all panels.
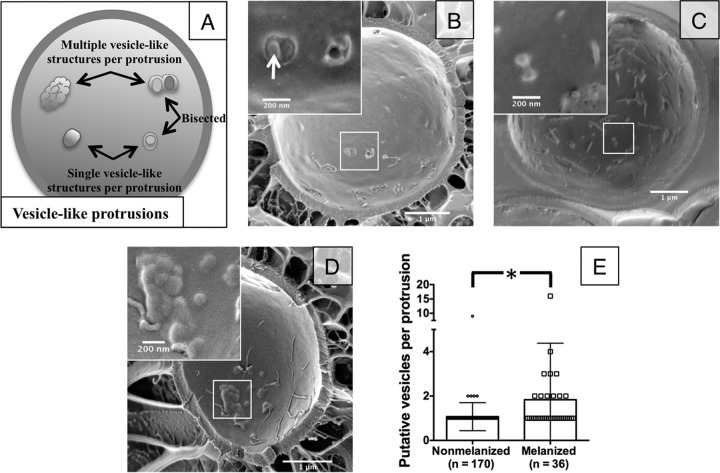
We attempted to further refine the process of vesicles crossing the cell wall using cryo-SEM to bypass potential loss of structural details during fixation. Our fractionated sample yielded many cells split between the plasma membrane bilayer, displaying typical thin aggregates or caverns along the P or E face of the membrane (Fig. 3). Some of the intramembrane protrusions were bisected, revealing vesicle-like structures within. In these bisected structures, we captured both multiple and single leaving events (Fig. 3B and C). This material, in the size and shape of vesicles, was found trapped between the plasma membrane and cell wall and was confirmed to contain additional vesicles by chance bisected material (Fig. 3B).
FIG 3.
Cryo-SEM reveals differences in putative vesicle release. (A) Schematic explaining how vesicle-like protrusions were counted. (B) Nonmelanized cells show aggregations between the plasma membrane and cell wall that reveal multiple vesicle cargo when bisected (arrow). (C and D) Nonmelanized cells show many individual protrusions (C), while melanized cells show an increased number of putative vesicles per protrusion (D). (E) Distribution of the number of putative vesicles per protrusion in melanized versus nonmelanized cells (P < 0.005).
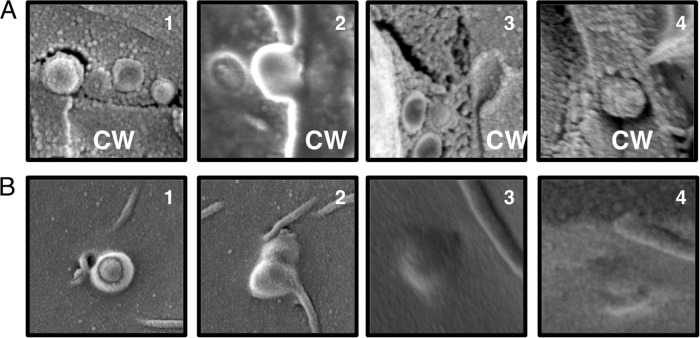
To aid our cryo-SEM studies, we examined melanized cells. Melanization changes the pore size, charge, and hydrophobicity of fungal cell walls, which we anticipated might interfere with vesicle transit through the cell wall and “trap” vesicles to facilitate visualization (13, 14). Indeed, we noticed an increase in the number of round, vesicle-shaped structures in aggregated protrusions between the plasma membrane and cell wall (Fig. 3D and E), although no increase in the overall number of trapped protrusions (data not shown), suggesting an increase in vesicle entrapment but not necessarily in vesicle production. Using melanized cells, we were able to identify single vesicles directly crossing the cell wall (Fig. 4A) and multiple vesicles crossing into the cell wall (Fig. 4B and C). This lends further support to multiple mechanisms of vesicle release from the cell body. Additionally, the vesicles observed in the cell wall did not appear to be aligned or surrounded by any obvious channel, suggesting another means of cell wall remodeling to facilitate vesicle transit.
FIG 4.
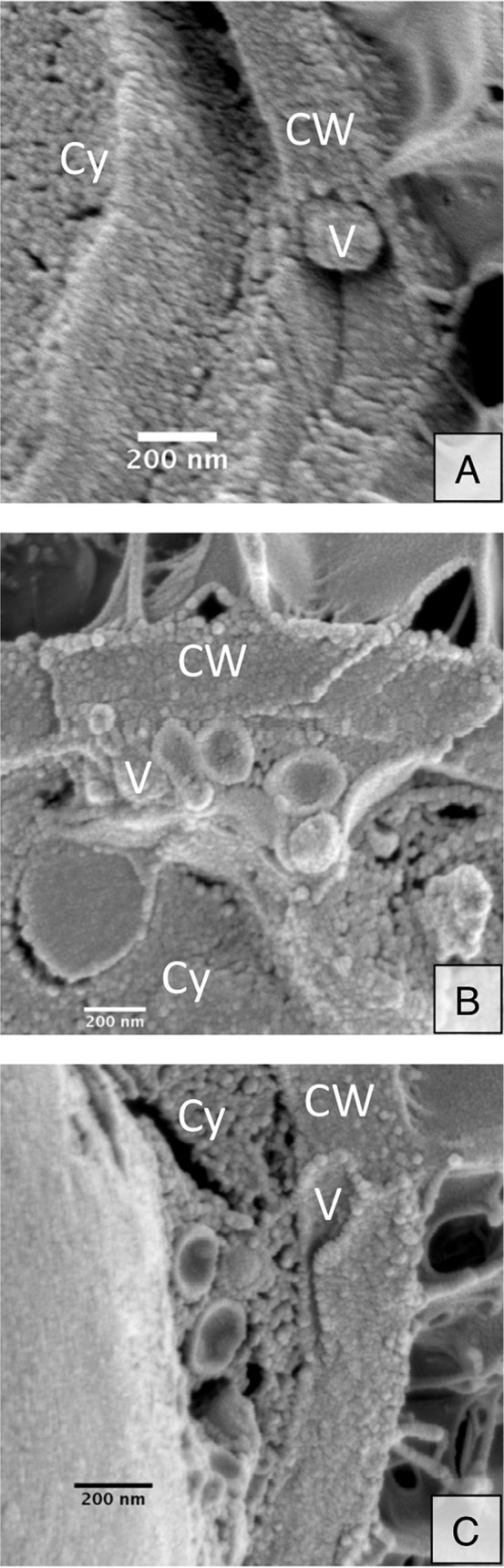
Cryo-SEM of melanized cells captures vesicle-like structures (V) in the cell wall (CW) and cytoplasm (Cy). Vesicles exit in both single leaving events (A) and multiple leaving events (B and C).
From these cryo-SEM images, we were able to assemble a panel of micrographs suggesting potential phases in cell wall transit. Vesicles near the perimeter fuse with the plasma membrane and transit through the cell wall, without the need of any obvious transit structures or channels (Fig. 5A and B).
FIG 5.
Vesicle-like structures in the cell wall. The figure shows a series of micrographs suggesting potential phases in cell wall transit as viewed from a cross section of the cell wall (CW) (A) and from the interior of the cell (B). Panels 3 and 4 in panel A are close-up images of Fig. 4A and C, respectively.
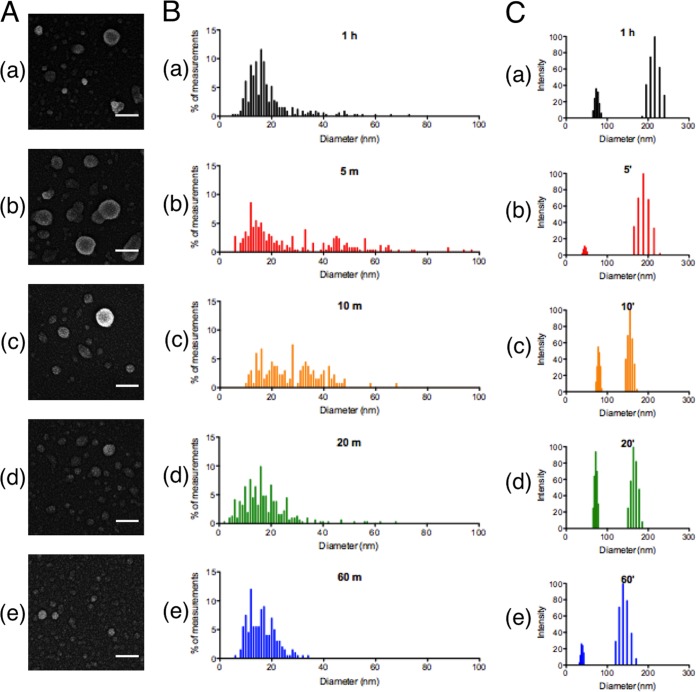
Our next goal was to investigate functional differences between vesicle subpopulations using differential sedimentation. Because larger vesicles were associated with multiple vesicle release events, we reasoned that we could enrich this larger population and test for functional differences from the smaller population. To date, cryptococcal vesicle preparations in our lab and others have been collected with a 1-h ultracentrifugation step spun at 100,000 × g, resulting in a vesicle population of sizes ∼20 nm to ∼200 nm (Fig. 6A). We took a vesicle suspension and progressively spun it at 100,000 × g in four subsequent spins: 5, 10, 20, and 60 min. We hypothesized that vesicle size and mass would be directly related and that the most massive vesicles would sediment after only a 5-min spin and that smaller vesicles would take longer centrifugation times to sediment.
FIG 6.
Vesicle fractions by differential sedimentation. (A, B, and C) SEM images, histograms of SEM diameter distributions, and dynamic light scattering measurements, respectively. (a to e) Vesicle preparations from a traditional 1-h sedimentation (a) and fractions of vesicles after subsequent sedimentations lasting 5 min (b), 10 min (c), 20 min (d), or 60 min (e). The results show that vesicle preparations can be enriched by progressive differential sedimentation.
Using this method, we purified populations that we named by their total centrifugation time (1-h historical control or 5 min, 10 min, 20 min, and 60 min; in this nomenclature the 1 h represents the vesicle preparation obtained by the regular protocol and the 5 min, 10 min, 20 min, and 60 min represent the vesicle preparations that result after the 1-h vesicle preparation is resuspended and the vesicular contents are fractionated by subsequent centrifugation lasting the designated times.) Examination by SEM revealed an enrichment of larger vesicles (>40 nm) as well as a number of smaller vesicles (<40 nm) in the 5-min and 10-min samples (Fig. 6A and B). Subsequent spins resulted in collection of vesicle populations consisting solely of smaller vesicles (<40 nm) (Fig. 6A and B). We further verified that spin time correlated with decreasing size using dynamic light scattering (DLS), which utilizes light scattered from Brownian motion fluctuations to estimate the size of small particles in suspension. By measuring the subpopulations resulting from a representative preparation, we confirmed that the vesicle sizes decreased with increasing sedimentation times (Fig. 6C). Thus, we were able to enrich vesicle populations of different sizes, but each enriched sample remained heterogeneous, consistent with a continuous population of vesicle sizes.
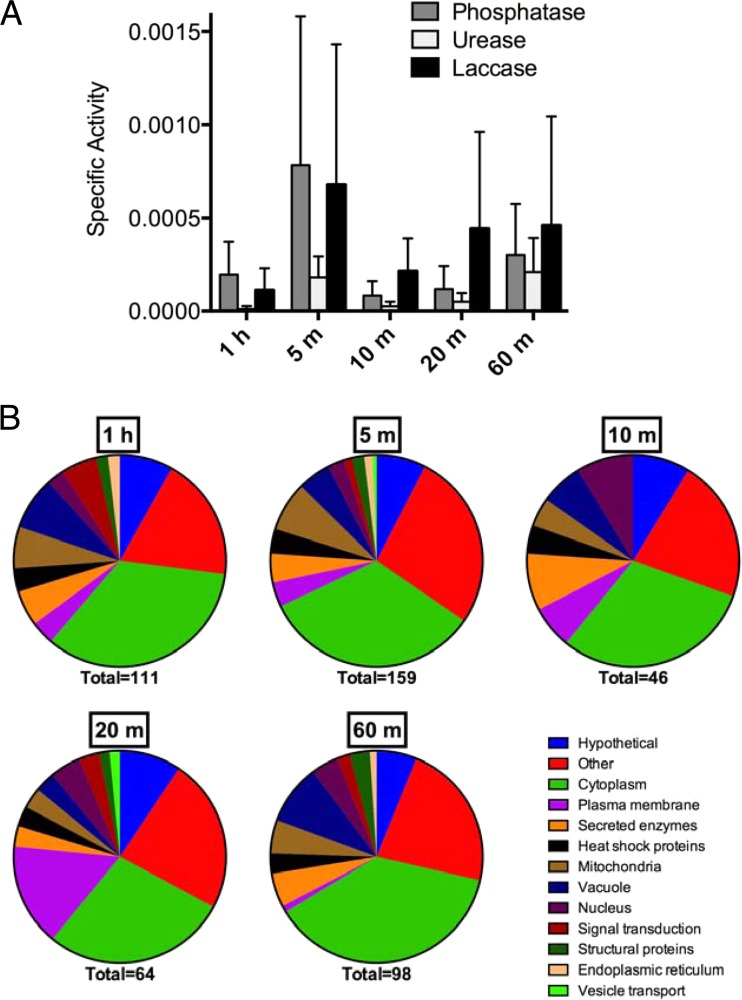
We hypothesized that vesicle size differences would correlate with functional differences, potentially reflecting their release origin differences. Secreted vesicles exhibit urease, phosphatase, and laccase activity (15, 16), so we tested the vesicle samples obtained from different centrifugation times for all three enzymatic activities. The highest specific activity for all three assays was in the 5-min and 60-min samples, but these differences were not significant (Fig. 7A). To ask whether protein cargo other than virulence enzymes may differ between vesicle subpopulations, we submitted vesicles for proteomic analysis. We identified 202 individual proteins among all five samples, with each sample containing between 46 and 159 proteins. Of the 202 identified C. neoformans proteins, 55 were previously reported as vesicle associated through proteomics (15), while 147 are newly identified in this report. Both the previously identified and newly identified proteins are extremely varied in function and cell localization (Fig. 7B; see Table S1 in the supplemental material). While the number of identified proteins differed between the size-enriched populations, the gene ontology distributions were nearly identical in all samples tested (Fig. 7B; see Table S1 in the supplemental material). We concluded that cargo loading does not discriminate between vesicle size or likely vesicle origin.
FIG 7.
Enzymatic activity and protein composition of vesicle fractions shown in Fig. 6. (A) Vesicles were assayed for phosphatase, urease, and laccase activities. (B) Protein composition as determined by proteomic analysis of vesicle subpopulations. The total number of proteins identified for each population is indicated below the graph. Differential centrifugation concentrates vesicle enzymatic activities but does not drastically change protein composition.
DISCUSSION
Our results show that extracellular vesicles in C. neoformans can be released outside the cell by different means, thus echoing similar findings with S. cerevisiae (7). We also observed that while smaller extracellular vesicles could be observed leaving both alone and in tandem with other vesicles, the largest vesicles observed exiting the cell were those associated with multiple vesicles, likely in an MVB-to-plasma-membrane fusion event. In an attempt to enrich for vesicle subpopulations by size using differential sedimentation, we observed similar protein compositions in all subpopulations, reflecting homogeneity in vesicle loading.
Vesicle transport by C. neoformans was first suggested decades ago after observation of yeast cells by freeze-etching (17, 18). Extracellular vesicular release was first reported in 2007 as a mechanism for the transport of large molecules to the extracellular space through the cell wall and for the delivery of concentrated preparations of components associated with virulence (3, 15). Despite numerous studies in recent years describing the composition of extracellular vesicles (15, 19) and their role in infection (20, 21), how these large membranous particles cross the cell wall remains a mystery. The three most obvious hypotheses are (i) movement though a channel, (ii) remodeling of the wall to facilitate transit, and (c) mechanical pressure to force vesicles through small cell wall pores. Our studies identified vesicles directly in the cell wall without any obvious trans-cell wall structures or changes in surrounding cell wall electron density (Fig. 1 and 4). This argues against the presence of a channel used to guide vesicles to the extracellular space. Figure 1F shows vesicles in the cell wall next to an area of damaged cell wall. This could indicate that vesicles are released where the cell wall has been compromised; however, vesicle release could also be stimulated under such circumstances to aid in cell wall repair. Overall, we found no support for a cell wall-spanning channel to aid in vesicle release.
Ultrastructural analysis using electron microscopy revealed that C. neoformans released extracellular vesicles by several mechanisms. Although EM provides only static images, we interpret the images to represent vesicles forming both by the pinching off of the plasma membrane and through fusion of multivesicular bodies with the plasma membrane (Fig. 1 and 4). These images complement the genetic studies done with S. cerevisiae, which have shown that mutations in both MVB formation pathways and Golgi-to-plasma membrane trafficking pathways result in stunted vesicle production (7). These data also suggest an explanation why a true vesicle-null strain may be very difficult to identify, as several pathways appear to converge to generate these extracellular vesicles.
Our fungal cell studies are complemented by recent metazoan extracellular vesicle work. Of note, the pinching off of the plasma membrane generates a vesicle with electron density similar to that of its cytoplasmic origin, while the putative MVB fusion vesicles are more electron lucid. While nomenclature is not uniform across all fields, those vesicles originating from the plasma membrane are referred to as microvesicles or ectosomes and retain similar cytoplasmic appearances; those vesicles originating from MVB fusion events are referred to as exosomes and are generally less dense (22). C. neoformans vesicles have previously been morphologically characterized (15). Our images suggest that the approximately 40% observed to be electron dense may stem directly from the plasma membrane, either by pinching off or by the recently described “inverted macropinocytosis” (8), while the rest may represent vesicles from other origins.
Our proteomic analyses identified 147 new vesicle-associated cryptococcal proteins in addition to those previously described by Rodrigues et al. (15), most likely due to an increased sensitivity of this method since the cited study. Like for those previously described in C. neoformans and other fungal species, we found a large functional distribution of identified hits (23–27). The distribution carried through all the size-enriched populations, suggesting that any loading mechanism is size independent. Although they were detected enzymatically, we did not identify laccase or urease by proteomic analysis, suggesting that this methodology has too low a sensitivity to measure all protein cargo. However, the presence of other potential virulence factors, such as amylase (28) and superoxide dismutase (29), confirms that these extracellular vesicles can act as “virulence bags” to deliver a high payload concentration under the right circumstances. Also of note is the high proportion of ribosomal and translation-related proteins, making up 20 to 30% of each sample. These, in conjunction with potential RNA cargo (30), are suggestive of a mechanism to influence host cell translation, particularly in the case of fungal cell internalization. Fungal small interfering RNA (siRNA) has recently been shown to interact with host cell RNA interference pathways (31), setting a precedent for involvement between molecular machinery of host and fungus cells.
Size enrichment did not distinguish functional or proteomically enriched subclasses among vesicle subpopulations. Unlike recent studies in the eukaryotic microbe Trypanosoma cruzi, we observed no correlation between size and origin (exosomes versus microvesicles) (32). T. cruzi subpopulations were differentiated by a 2-h versus a 16-h ultracentrifugation step, suggesting that technical changes in protocols may account for our differences. Alternatively, this may be due to the requirement of the endosomal sorting complex required for transport (ESCRT) proteins for both MVB and ectosome formation (22, 33), as ostensibly some of the same loading or marking characteristics may exist between these mechanisms. If this is the case, it may have implications for loading cell wall glycans and capsular polysaccharide in a homogeneous way as well.
Our studies provide strong evidence that vesicles gain access to the outside of cells by traversing the cell wall. The mechanism by which vesicles travel across the cell wall remains to be determined. Our results provide insight into this process by demonstrating vesicle-like structures interacting with the cell wall and the presence of vesicles in the cell wall. The lack of obvious channels lends support toward cell wall remodeling, perhaps by degradative enzymes associated with vesicles or released from the MVB lumen along with vesicles. Our results also imply limits to how we may learn from standard ultrastructural studies and suggest the need for either improved methods or new methodologies to gain additional insight into the process of vesicular transport across the cell wall.
Supplementary Material
ACKNOWLEDGMENTS
J.W. and A.C. are supported through NIH grant 5R01AI33774-20. J.E.-M. and J.L.L.-G. are supported through grant CTQ2010-18644 (MINECO).
Special thanks go to Ben Clark, Leslie Cummings, and Geoff Perumal in the Einstein Analytical Imaging Facility for assistance with electron microscopy sample preparation and imaging.
We declare no financial, commercial, or other relationships that could be construed as a potential conflict of interest. There has been no payment for any aspects of the submitted work, nor are patents or copyrights pending regarding this work.
Footnotes
Published ahead of print 6 June 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/EC.00111-14.
REFERENCES
- 1.Deatherage BL, Cookson BT. 2012. Membrane vesicle release in bacteria, eukaryotes, and archaea: a conserved yet underappreciated aspect of microbial life. Infect. Immun. 80:1948–1957. 10.1128/IAI.06014-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.MacDonald IA, Kuehn MJ. 2012. Offense and defense: microbial membrane vesicles play both ways. Res. Microbiol. 163:607–618. 10.1016/j.resmic.2012.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rodrigues ML, Nimrichter L, Oliveira DL, Frases S, Miranda K, Zaragoza O, Alvarez M, Nakouzi A, Feldmesser M, Casadevall A. 2007. Vesicular polysaccharide export in Cryptococcus neoformans is a eukaryotic solution to the problem of fungal trans-cell wall transport. Eukaryot. Cell 6:48–59. 10.1128/EC.00318-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nosanchuk JD, Nimrichter L, Casadevall A, Rodrigues ML. 2008. A role for vesicular transport of macromolecules across cell walls in fungal pathogenesis. Commun. Integr. Biol. 1:37–39. 10.4161/cib.1.1.6639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vallejo MC, Matsuo AL, Ganiko L, Medeiros LC, Miranda K, Silva LS, Freymuller-Haapalainen E, Sinigaglia-Coimbra R, Almeida IC, Puccia R. 2011. The pathogenic fungus Paracoccidiodes brasiliensis exports extracellular vesicles containing highly immunogenic alpha-Galactosyl epitopes. Eukaryot. Cell 10:343–351. 10.1128/EC.00227-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rivera J, Cordero RJ, Nakouzi AS, Frases S, Nicola A, Casadevall A. 2010. Bacillus anthracis produces membrane-derived vesicles containing biologically active toxins. Proc. Natl. Acad. Sci. U. S. A. 107:19002–19007. 10.1073/pnas.1008843107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Oliveira DL, Nakayasu ES, Joffe LS, Guimaraes AJ, Sobreira TJ, Nosanchuk JD, Cordero RJ, Frases S, Casadevall A, Almeida IC, Nimrichter L, Rodrigues ML. 2010. Characterization of yeast extracellular vesicles: evidence for the participation of different pathways of cellular traffic in vesicle biogenesis. PLoS One 5:e11113. 10.1371/journal.pone.0011113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rodrigues ML, Franzen AJ, Nimrichter L, Miranda K. 2013. Vesicular mechanisms of traffic of fungal molecules to the extracellular space. Curr. Opin. Microbiol. 16:414–420. 10.1016/j.mib.2013.04.002. [DOI] [PubMed] [Google Scholar]
- 9.Colombo M, Moita C, van Niel G, Kowal J, Vigneron J, Benaroch P, Manel N, Moita LF, Thery C, Raposo G. 2013. Analysis of ESCRT functions in exosome biogenesis, composition and secretion highlights the heterogeneity of extracellular vesicles. J. Cell Sci. 126:5553–5565. 10.1242/jcs.128868. [DOI] [PubMed] [Google Scholar]
- 10.Tan SS, Yin Y, Lee T, Lai RC, Yeo RW, Zhang B, Choo A, Lim SK. 23 December 2013. Therapeutic MSC exosomes are derived from lipid raft microdomains in the plasma membrane. J. Extracell. Vesicles 10.3402/jev.v2i0.22614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Prados-Rosales R, Baena A, Martinez LR, Luque-Garcia J, Kalscheuer R, Veeraraghavan U, Camara C, Nosanchuk JD, Besra GS, Chen B, Jimenez J, Glatman-Freedman A, Jacobs WR, Jr, Porcelli SA, Casadevall A. 2011. Mycobacteria release active membrane vesicles that modulate immune responses in a TLR2-dependent manner in mice. J. Clin. Invest. 121:1471–1483. 10.1172/JCI44261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kong M, Bhattacharya RN, James C, Basau A. 2004. A statistical approach to estimate the 3D size distribution of spheres from 2D size distributions. Geol. Soc. Am. Bull. 117:244–249. 10.1130/B25000.1. [DOI] [Google Scholar]
- 13.Pihet M, Vandeputte P, Tronchin G, Renier G, Saulnier P, Georgeault S, Mallet R, Chabasse D, Symoens F, Bouchara JP. 2009. Melanin is an essential component for the integrity of the cell wall of Aspergillus fumigatus conidia. BMC Microbiol. 9:177–188. 10.1186/1471-2180-9-177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jacobson ES, Ikeda R. 2005. Effect of melanization upon porosity of the cryptococcal cell wall. Med. Mycol. 43:327–333. 10.1080/13693780412331271081. [DOI] [PubMed] [Google Scholar]
- 15.Rodrigues ML, Nakayasu ES, Oliveira DL, Nimrichter L, Nosanchuk JD, Almeida IC, Casadevall A. 2008. Extracellular vesicles produced by Cryptococcus neoformans contain protein components associated with virulence. Eukaryot. Cell 7:58–67. 10.1128/EC.00370-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eisenman HC, Frases S, Nicola AM, Rodrigues ML, Casadevall A. 2009. Vesicle-associated melanization in Cryptococcus neoformans. Microbiology 155:3860–3867. 10.1099/mic.0.032854-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Takeo K, Uesaka I, Uehira K, Nishiura M. 1973. Fine structure of Cryptococcus neoformans grown in vivo as observed by freeze-etching. J. Bacteriol. 113:1449–1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Takeo K, Uesaka I, Uehira K, Nishiura M. 1973. Fine structure of Cryptococcus neoformans grown in vitro as observed by freeze-etching. J. Bacteriol. 113:1442–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Oliveira DL, Nimrichter L, Miranda K, Frases S, Faull KF, Casadevall A, Rodrigues ML. 2009. Cryptococcus neoformans cryoultramicrotomy and vesicle fractionation reveals an intimate association between membrane lipids and glucuronoxylomannan. Fungal Genet. Biol. 46:956–963. 10.1016/j.fgb.2009.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oliveira DL, Freire-de-Lima CG, Nosanchuk JD, Casadevall A, Rodrigues ML, Nimrichter L. 2010. Extracellular vesicles from Cryptococcus neoformans modulate macrophage functions. Infect. Immun. 78:1601–1609. 10.1128/IAI.01171-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang SH, Wu CH, Chang YC, Kwon-Chung KJ, Brown RJ, Jong A. 2012. Cryptococcus neoformans-derived microvesicles enhance the pathogenesis of fungal brain infection. PLoS One 7:e48570. 10.1371/journal.pone.0048570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wehman AM, Poggioli C, Schweinsberg P, Grant BD, Nance J. 2011. The P4-ATPase TAT-5 inhibits the budding of extracellular vesicles in C. elegans embryos. Curr. Biol. 21:1951–1959. 10.1016/j.cub.2011.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rodrigues ML, Nakayasu ES, Almeida IC, Nimrichter L. 2014. The impact of proteomics on the understanding of functions and biogenesis of fungal extracellular vesicles. J. Proteomics 97:177–186. 10.1016/j.jprot.2013.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang J, Wang F, Feng Y, Mi K, Chen Q, Shang J, Chen B. 2013. Comparative vesicle proteomics reveals selective regulation of protein expression in chestnut blight fungus by a hypovirus. J. Proteomics 78:221–230. 10.1016/j.jprot.2012.08.013. [DOI] [PubMed] [Google Scholar]
- 25.Silva BM, Prados-Rosales R, Espadas-Moreno J, Wolf JM, Luque-Garcia JL, Goncalves T, Casadevall A. 2014. Characterization of Alternaria infectoria extracellular vesicles. Med. Mycol. 52:202–210. 10.1093/mmy/myt003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vallejo MC, Nakayasu ES, Matsuo AL, Sobreira TJ, Longo LV, Ganiko L, Almeida IC, Puccia R. 2012. Vesicle and vesicle-free extracellular proteom of Paracoccidiodes brasiliensis: comparative analysis with other pathogenic fungi. J. Proteome Res. 11:1676–1685. 10.1021/pr200872s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Albuquerque PC, Nakayasu ES, Rodrigues ML, Frases S, Casadevall A, Zancope-Oliveira RM, Almeida IC, Nosanchuk JD. 2008. Vesicular transport in Histoplama capsulatum: an effective mechanism for trans-cell wall transfer of protiens and lipids in ascomycetes. Cell Microbiol. 10:1695–1720. 10.1111/j.1462-5822.2008.01160.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Camacho E, Sepulveda VE, Goldman WE, San-Blas G, Nino-Vega GA. 2012. Expression of Paracoccidiodes brasiliensis AMY1 in a Histoplasma capsulatum amy1 mutant, relates to an alpha-(1,4)-amylase to cell wall alpha-(1,3)-glucan synthesis. PLoS One 7:e50201. 10.1371/journal.pone.0050201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cox GM, Harrison TS, McDade HC, Taborda CP, Heinrich G, Casadevall A, Perfect JR. 2003. Superoxide dismutase influences the virulence of Cryptococcus neoformans by affecting growth within macrophages. Infect. Immun. 71:173–180. 10.1128/IAI.71.1.173-180.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nicola AM, Frases S, Casadevall A. 2009. Lipophilic dye staining of Cryptococcus neoformans extracellular vesicles and capsule. Eukaryot. Cell 8:1373–1380. 10.1128/EC.00044-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weiberg A, Wang M, Lin F, Zhao H, Zhang Z, Kaloshian I, Huang HD, Jin H. 2013. Fungal small RNAs suppress plant immunity by hijacking host RNA interference pathways. Science 342:118–123. 10.1126/science.1239705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bayer-Santos E, Aguilar-Bonavides C, Pessini Rodrigues S, Mauricio Cordero E, Ferreira Marques A, Varela-Ramirez A, Choi H, Yoshida N, Franco da Silveira J, Almeida IC. 2013. Proteomic analysis of Trypanosoma cruzi secretome: characterization of two populations of extracellular vesicles and soluble proteins. J. Proteome Res. 12:883–897. 10.1021/pr300947g. [DOI] [PubMed] [Google Scholar]
- 33.Henne WM, Buchkovich NJ, Emr SD. 2011. The ESCRT pathway. Dev. Cell 21:77–91. 10.1016/j.devcel.2011.05.015. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.