Abstract
Candida albicans, a major human fungal pathogen, is the primary cause of invasive candidiasis in a wide array of immunocompromised patients. C. albicans virulence requires the ability to undergo a reversible morphological transition from yeast to filaments in response to a variety of host environmental cues. These cues are sensed by the pathogen and activate multiple signal transduction pathways to induce filamentation. Reversible phosphorylation events are critical for regulation of many of these pathways. While a variety of protein kinases are known to function as components of C. albicans filamentous growth signal transduction pathways, considerably little is known about the role of phosphatases. Here we demonstrate that PPG1, encoding a putative type 2A-related protein phosphatase, is important for C. albicans filament extension, invasion, and virulence in a mouse model of systemic candidiasis. PPG1 is also important for downregulation of NRG1, a key transcriptional repressor of C. albicans filamentous growth, and is shown to affect the expression of several filament-specific target genes. An epistasis analysis suggests that PPG1 controls C. albicans filamentation via the cyclic AMP-protein kinase A (cAMP-PKA) signaling pathway. We demonstrate that Ppg1 possesses phosphatase activity and that a ppg1 catalytic mutant shows nearly equivalent filamentation, invasion, and virulence defects compared to those of a ppg1Δ/Δ strain. Overall, our results suggest that phosphatases, such as Ppg1, play critical roles in controlling and fine-tuning C. albicans filament extension and virulence as well as signal transduction pathways, transcriptional regulators, and target genes associated with these processes.
INTRODUCTION
Candida albicans, the most commonly isolated human fungal pathogen, is the leading cause of candidiasis worldwide and the fourth leading cause of hospital-acquired bloodstream infections in the United States (1–3). As a commensal, C. albicans is part of the normal microbiota of the oral cavity and gastrointestinal and genitourinal tracts of healthy individuals (3). Immunocompromised individuals, such as AIDS and cancer patients, neonates, organ transplant recipients, and patients with indwelling catheters, can develop disseminated candidiasis, a systemic form of the disease with an approximately 40% mortality rate (2, 4, 5). Candida infections have become more difficult to treat due to a limited number of antifungal therapies and increased frequency of drug-resistant isolates (6, 7).
C. albicans possesses multiple virulence properties, including the ability to undergo a reversible transition from yeast (single ovoid, budding cells) to pseudohyphal and hyphal filaments (elongated cells attached end to end) (3, 8, 9). This morphological transition allows for efficient tissue invasion, immune evasion (including macrophage lysis), and biofilm formation (3, 10–14). C. albicans strains that are unable to undergo a reversible yeast-filament transition are highly attenuated for virulence (12, 15, 16). The C. albicans morphological transition is known to be induced by a variety of cues in the host environment, including serum, temperature of 37°C, neutral pH, high CO2, low O2, embedded/matrix conditions, and nitrogen and carbon starvation, as well as N-acetylglucosamine (3, 17–19). These environmental stimuli are sensed by the pathogen and cause activation of a variety of signal transduction pathways that target specific filamentous growth transcriptional regulators (20, 21). The regulators, in turn, direct the activation of a filamentous growth transcriptional program consisting of genes specifically important for filament development as well as other virulence-related processes (22, 23). A conserved mitogen-activated protein (MAP) kinase signaling pathway responds to a variety of environmental conditions, including nitrogen starvation, and results in activation of the Cph1 transcription factor (21, 24). A Ras-cyclic AMP-protein kinase A (Ras-cAMP-PKA) pathway is also known to respond to serum, 37°C, and high CO2 and directs activation of Efg1, a key transcriptional regulator of the filamentous growth program (21, 25, 26). Although the MAP kinase and Ras-cAMP-PKA pathways are the best characterized and appear to respond to a majority of host environmental cues, additional signaling pathways have also been identified, which direct C. albicans filamentation in response to neutral pH, GlcNAc, and embedded/matrix conditions (20, 21).
While a number of protein kinases in the MAP kinase and Ras-cAMP-PKA pathways function as critical filamentous growth signaling components, considerably less is known about the role of phosphatases in controlling C. albicans morphology. However, sequence and domain searches have indicated the presence of 28 putative phosphatases in C. albicans, based on comparison with Saccharomyces cerevisiae homologs (27). Phosphatases can regulate the activity of filamentous growth signaling pathways by dephosphorylating the kinases. As an example, a tyrosine phosphatase, Cpp1, is a repressor of filamentation and appears to function by inhibiting the MAP kinase pathway component Cek1 (28). PTC8, a member of the protein phosphatase M (PPM) family (Mg2+ dependent), is induced in response to growth in serum at 37°C and is important for filamentation (29) but has not been linked to any known filamentous growth signaling pathways. The Sit4 phosphatase has previously been shown to affect C. albicans filamentation and appears to function by controlling protein translation, cell wall biogenesis, and osmosensing, targeting the Hog1 MAP kinase pathway (30). While the sit4Δ/Δ mutant is highly attenuated for virulence in a mouse model of systemic candidiasis, it is unclear whether this defect can be attributed to reduced filamentation per se or a variety of other pleiotropic phenotypes. In addition, expression of the large majority of filament-specific genes was not affected in the sit4Δ/Δ mutant. Finally, another important phosphatase, calcineurin, plays a key role in controlling C. albicans virulence and antifungal susceptibility but does not appear to be required for the yeast-filament transition (31–33).
In order to gain more insight into the poorly understood role of phosphatases in controlling C. albicans morphology and filamentous growth signaling pathways, we examined morphology data collected from a previous large-scale homozygous mutant analysis (34). One particular putative phosphatase mutant identified by this analysis, ppg1Δ/Δ, was not defective for proliferation but appeared to have a significant defect in morphogenesis and showed reduced abundance in mouse kidneys when combined with a large pool of bar-coded mutants. PPG1 encodes a putative type 2A serine/threonine phosphatase. Protein phosphatases in this subclass are known to regulate a variety of key cellular processes in many organisms, including cell wall integrity, actin cytoskeleton organization, auxin signaling, and polar movement in plants (35–37). In S. cerevisiae, the PPG1 ortholog is required for glycogen accumulation (38). Here, we demonstrate that C. albicans PPG1 plays an important role in controlling filament extension, invasion, and virulence. In addition, we examine the specific requirement of Ppg1 phosphatase activity for these processes as well as the relationship between PPG1 and several known C. albicans filamentous growth target genes and signal transduction pathways as well as a key transcriptional regulator.
MATERIALS AND METHODS
Strains and DNA constructions.
The genotypes for all strains used in this study are shown in Table S1 in the supplemental material. The wild-type (WT) control strain (DK318) has been described previously (39). A fusion PCR strategy (40) was used to generate the ppg1Δ/Δ strain. PPG1 5′ and 3′ flanking fragments were generated using primers MAO7 and MAO8 for the downstream flank as well as primers MAO5 and MAO6 for the upstream flank (all primers used in this study are described in Table S2 in the supplemental material). A second round of fusion PCR was then performed using the PPG1 5′ and 3′ flanks, as well as HIS1 and LEU2 markers, generated with primers RZO37/RZO38 and plasmids pSN52 and pSN40 (40), to yield ppg1Δ::HIS1 and ppg1Δ::LEU2 PCR products. SN152 (40) was transformed with ppg1Δ::HIS1 as well as wild-type alleles of C. albicans ARG4 (made using primers DKO400/DKO401) and LEU2 (made using primers DKO404/DKO405) in order to generate the ppg1Δ/+ strain (MAY7). SN152 was sequentially transformed with ppg1Δ::HIS1, ppg1Δ::LEU2, and C. albicans wild-type ARG4 to generate the ppg1Δ/Δ strain (MAY34). We used an SAT1 split-marker approach (41) to construct the ppg1Δ/Δ::PPG1 add-back strain (MAY50). Briefly, a PCR product containing PPG1 5′ upstream sequences, open reading frame (ORF), and terminator (T) was amplified from SC5314 genomic DNA using primers MAO155/MAO157. The 3′-flanking sequences downstream of the PPG1 terminator were also amplified with primers MAO158/MAO159. Partial, overlapping SAT1 cassettes were generated by PCR using plasmid pSFS2 (42) as a template and primers GSO128/GSO129 for the 5′ fragment and GSO127/GSO130 for the 3′ fragment. The 5′ PPG1 flank + ORF + T as well as the 5′ partial SAT1 cassette were digested with KpnI and ligated together. A similar ligation reaction was used to join the 3′ PPG1 terminator flank and the 3′ partial SAT1 cassette following digestion with SacII. Primers MAO56 and GSO129 were used to generate an ∼5-kb 5′ split-marker fragment (PPG1 5′-flanking region, ORF, and Terminator plus a 3.6-kb 5′ SAT1 partial cassette). Primers MAO160 and GSO130 were used to generate an ∼2-kb fragment (the PPG1 3′-flanking region downstream of Terminator plus a 1.4-kb 3′ SAT1 partial cassette). Each split-marker fragment was purified with the GeneElute PCR cleanup kit (Sigma) and concentrated by ethanol precipitation. DNA pellets (0.5 to 1 μg) were resuspended in 5 μl sterile ultrapure water for transformation into strain ppg1Δ/Δ (MAY34).
In order to generate the ppg1Δ/Δ::ppg1H248A H173A D90L strain (MAY55), we first used site-directed mutagenesis to introduce mutations into the PPG1 ORF as follows: the D90L mutation was generated using upstream nonmutagenic primer MAO77 and internal mutagenic primer MAO133, the H173A mutation was generated using internal nonmutagenic primer MAO150 and internal mutagenic primer MAO151, and the H248A mutation was generated using internal nonmutagenic primer MAO152 and internal mutagenic primer MAO153. These three fragments were combined with an additional PCR fragment containing the 3′ end of the ORF (generated using the internal mutagenic primer MAO154 and downstream nonmutagenic primer MAO78) in an overlap PCR to make a final fragment containing the ppg1 ORF with all three mutations. A PCR fragment containing the PPG1 5′-flanking sequences, mutated ORF, and terminator was then used in the SAT1 split-marker approach described above to generate strain ppg1Δ/Δ::ppg1H248A H173A D90L (MAY55). The SAT1 split-marker approach was also used to construct strains with TPK1 deleted. The TPK1 5′ upstream and 3′ downstream flanking regions were PCR amplified using primers MAO138/MAO139 and MAO140/MAO141, respectively. The 5′ and 3′ TPK1 flanks were fused to the partial SAT1 cassettes as described above. Primers MAO148 and GSO129 were used in a PCR to generate an ∼4.2-kb 5′ split-marker fragment (TPK1 5′-flanking region + 3.6-kb 5′ SAT1 partial cassette). Primers MAO149 and GSO130 were used to generate an ∼2-kb 3′ split-marker fragment (TPK1 3′-flanking region + 1.4-kb 3′ SAT1 partial cassette). Following purification and concentration as described above, these fragments were used to transform both wild-type (SN152) and ppg1Δ/Δ (MAY34) strains. Homozygous tpk1Δ/Δ and ppg1Δ/Δ tpk1Δ/Δ deletion mutations were generated using the SAT flipper method (42) (DK318 served as the starting strain for the tpk1Δ/Δ mutant) followed by retransformation with the 5′ and 3′ TPK1-SAT1 split-marker fragments. The ppg1Δ/Δ efg1Δ/Δ mutant was generated using the SAT flipper method by transforming the ppg1Δ/Δ (MAY34) strain with a previously described efg1Δ::SAT1 deletion fragment (43).
To construct MAY73 (tetO-PPG1), primers MAO126/MAO128 were used to generate a PPG1 upstream fragment (positions −566 to −98 relative to the PPG1 start codon), which was digested with KpnI and cloned into plasmid pEL9 (44). Primers MBO127/MBO129 were used to generate a second PCR product from positions −33 to +466 relative to the start ATG of PPG1, which was cloned into the resulting plasmid digested with SacII to generate pEL9-PPG1. This plasmid was used as a template to amplify an ∼12-kb fragment using PhireTaq (Thermo Scientific), which included the tetR transactivator, SAT1 marker, tetO, and PPG1 flanks. This fragment was transformed into strain DK318 to generate tetO-PPG1 strain MAY73. Correct integration of all constructs as well as absence of the ORF in homozygous deletion mutants was verified by whole-cell PCR. For recombinant bacterial expression, WT PPG1 and ppg1H248A H173A D90L were cloned into the BamHI and NcoI restriction sites of expression vector pAG10H (kindly provided by P. John Hart), a modified pET19d vector with a tobacco etch virus-cleavable His-10 tag fused to the N terminus of the target protein (Ppg1) (45).
Media and growth conditions.
Yeast extract-peptone-dextrose (YEPD) medium (46) at 30°C was used as the standard non-filament-inducing growth conditions for all strains. Solid YEPD medium plus 10% fetal bovine serum (FBS), Spider medium, and Lee's pH 6.8 medium were prepared as previously described (24, 39, 47). Liquid serum and temperature induction experiments were performed by growing strains overnight in YEPD medium at 30°C to an optical density at 600 nm (OD600) of ∼4.0 and diluting 1:10 into 50 ml of prewarmed medium as described previously (39). Aliquots of cells were harvested at specific postinduction time points for RNA preparation and microscopy. For the liquid serum and temperature induction epistasis experiment, strains were grown overnight in YEPD medium at 30°C to saturated density and then diluted in prewarmed YEPD medium at 30°C or YEPD medium plus 10% serum at 37°C and harvested at the 3-h postinduction time point. The tetO-PPG1 strain was grown in both liquid and solid YEPD media at 30°C in the presence or absence of 20 μg/ml doxycycline (Dox; Sigma-Aldrich, St. Louis, MO). Liquid cultures were grown overnight to an OD600 of ∼1.0, and aliquots of cells were taken for microscopy and RNA extraction.
RNA preparation and Northern analysis.
RNA extractions were carried out using the hot acid phenol protocol (48). Probe preparation and Northern analysis (using 3 μg of total RNA from each sample) were carried out as described previously (39). Blots were scanned using a phosphorimager and visualized as described previously (39). The primers used to generate probes for Northern analysis are indicated in Table S2 in the supplemental material.
Invasion assays.
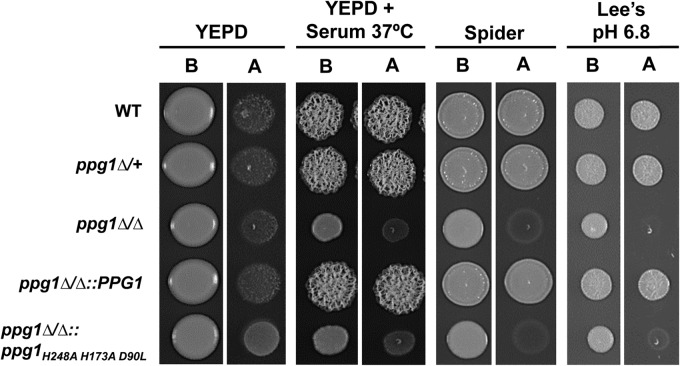
Invasion assays were performed as described previously (49) with minor modifications. Briefly, saturated overnight cultures were diluted to an OD600 of 1.0, and four additional 1:10 serial dilutions were performed. Three microliters from each dilution was spotted onto the specific solid agar medium (see Fig. 4), and cells were grown for 2 days. Spot images were taken before and after the plates were washed with double-distilled water (ddH2O).
FIG 4.
Ppg1 phosphatase activity is important for agar invasion under a variety of filament-inducing conditions. The indicated strains were grown in YEPD at 30°C overnight. A total of 6 × 103 cells of each strain were spotted onto the indicated solid media. Following 2 days of growth at 30°C (or 37°C for the YEPD + serum condition), images were taken before (B) and after (A) each plate was washed with ddH2O.
Expression of Ppg1 in E. coli.
Expression constructs for His-tagged Ppg1 and Ppg1H248A H173A D90L were transformed into an Escherichia coli Rosetta 2(DE3) strain (Stratagene). Expression strains were induced by the addition of 0.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG; Sigma-Aldrich), followed by a 16-h incubation at 19°C. Cells were harvested, and extracts were prepared using the nickel-nitrilotriacetic acid (Ni-NTA) Fast Start kit protocol (Qiagen). Recombinant proteins were purified from the supernatant of bacterial extracts using Ni-NTA agarose affinity chromatography. Eluted fractions were pooled and analyzed using SDS-PAGE (data not shown). Protein concentrations of the fractions were determined using a Bradford assay (50). Western analysis was performed as described previously (51) using 20 μg of recombinant purified protein, a 1:20,000 dilution of mouse monoclonal anti-His antibody (GenScript), and a 1:5,000 dilution of secondary goat anti-rabbit IgG antibody (Invitrogen).
Phosphatase activity assay.
Phosphatase assays were performed according to the manufacturer's instructions as follows: 20 μg of recombinant Ppg1 protein samples was incubated with 100 μl of p-nitrophenyl phosphate (pNPP) substrate solution (Sigma-Aldrich, St. Louis, MO). The colorimetric pNPP substrate turns yellow upon the release of a phosphate group by the phosphatase. The pNPP reaction was stopped after 15 min by the addition of 50 μl of 3 M NaOH solution. Activity was measured by spectrophotometer as absorbance at 405 nm. All assays were performed in biological duplicate and technical quadruplicate.
Virulence and histological assays.
Virulence in the mouse model of systemic candidiasis was assessed, and subsequent histological analysis of kidney sections was performed as described previously (39). All animal experimentation was conducted following the National Institutes of Health guidelines for housing and care of laboratory animals and performed in accordance with institutional regulations after pertinent review and approval by the Institutional Animal Care and Use Committee at The University of Texas at San Antonio. Briefly, overnight cultures of each strain were washed three times in sterile pyrogen-free saline, and 200 μl of cell suspension (containing the appropriate inoculum size) was used to inject individual 6- to 8-week-old female BALB/c mice by lateral tail vein. Survival was monitored after infection. Moribund mice were sacrificed, and their deaths were recorded on the next day. Survival data and differences between groups were analyzed using the Kaplan-Meier log rank test. Analyses were performed using Prism by GraphPad Prism ver. 6.04 (GraphPad Software Inc., San Diego, CA).
RESULTS
C. albicans Ppg1 possesses phosphatase activity that is abolished upon mutation of the putative catalytic site.
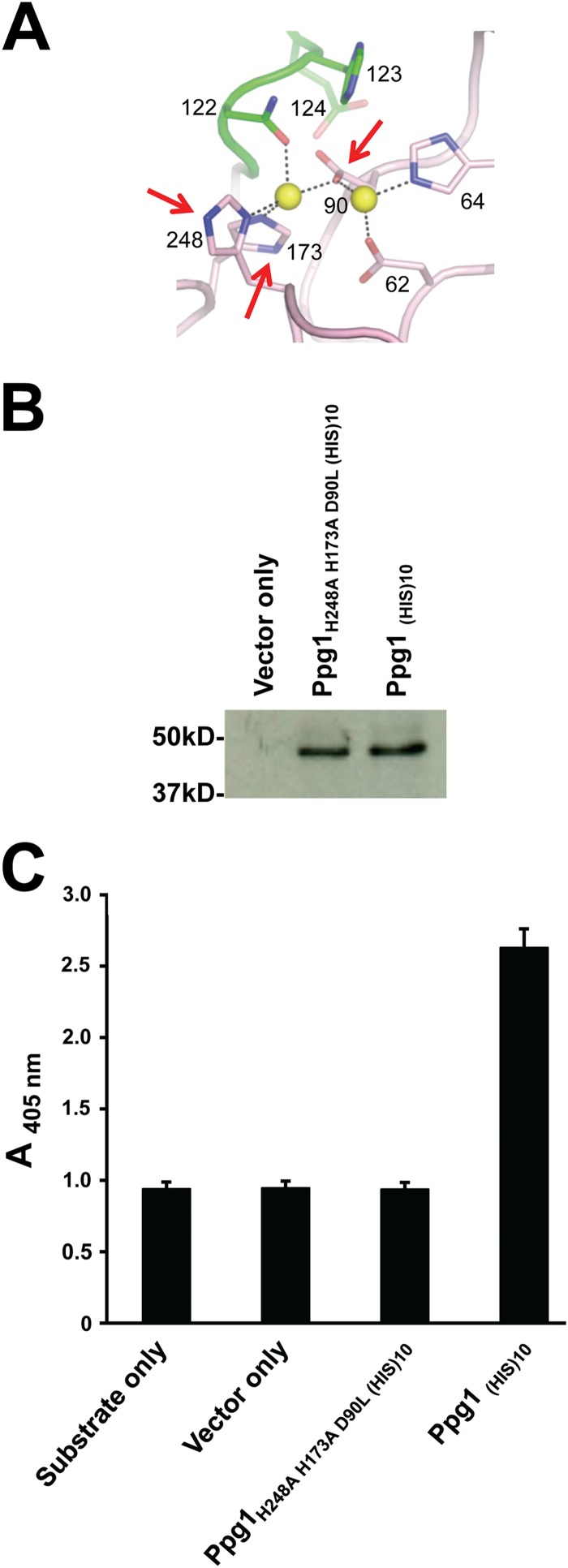
Ppg1, a putative serine/threonine protein phosphatase type 2A, was first identified in C. albicans as part of an amino acid sequence-based screen for orthologs of previously annotated S. cerevisiae protein phosphatases (the identity was 60.8%) (27). A model generated using homology detection and structure prediction HMM-HMM (where HMM stands for hidden Markov model) comparison (HHpred) software (http://toolkit.tuebingen.mpg.de/hhpred) predicted that Ppg1 possesses a conserved putative catalytically active binuclear center (Fig. 1A). In order to confirm that C. albicans PPG1 encodes a phosphatase, recombinant His-tagged Ppg1 was expressed in bacteria and purified. As shown in Fig. 1B and C, Ppg1(HIS)10 was stably expressed and showed significant phosphatase activity compared to vector-only and substrate-only controls. We also observed that Ppg1(HIS)10 phosphatase activity was abolished upon treatment with okadaic acid (see Fig. S1 in the supplemental material), a specific inhibitor of serine/threonine phosphatases of types 1, 2A, and 2B (52). We observed that mutation of three highly conserved amino acid residues in the conserved binuclear center (H248, H173, D90) completely abolished Ppg1(HIS)10 phosphatase activity (Fig. 1C). Importantly, phosphatase activity was not abolished as a consequence of reduced protein stability since the catalytic mutant was expressed at a level equivalent to that of wild-type Ppg1(HIS)10 (Fig. 1B).
FIG 1.
Mutation of the putative Ppg1 catalytic site abolishes phosphatase activity. (A) A model for the putative Ppg1 catalytically active binuclear center generated using homology detection and structure prediction HMM-HMM comparison (HHpred) software (http://toolkit.tuebingen.mpg.de/hhpred). Numbered amino acids represent highly conserved residues in the putative catalytic site. Arrows indicate mutated residues. (B) His-tagged WT and mutated Ppg1 were expressed in an E. coli Rosetta 2(DE3) strain and purified from the supernatant of bacterial extracts using Ni-NTA agarose (Qiagen) affinity chromatography. Western analysis was performed using 20 μg of recombinant purified proteins and an anti-His primary antibody and goat anti-rabbit IgG secondary antibody. (C) Phosphatase activity was determined using 20 μg of the indicated proteins and p-nitrophenyl phosphate (pNPP) as a substrate.
Ppg1 phosphatase activity plays an important role in controlling C. albicans colony morphology, filament extension, and agar invasion.
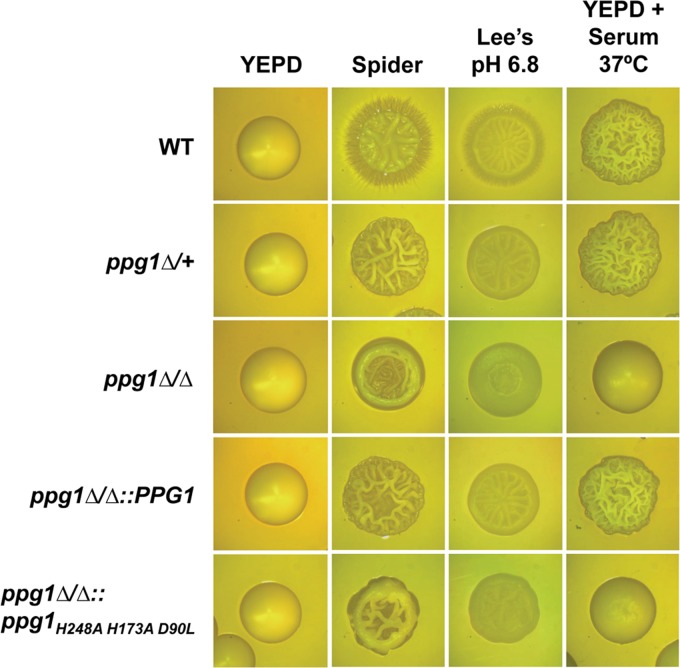
As previously indicated, a large-scale systematic screen of a C. albicans homozygous deletion library demonstrated that the ppg1Δ/Δ mutant strain is defective for morphogenesis and shows reduced abundance in the kidney during infection when combined with a large pool of bar-coded mutants (34). In order to further examine and better characterize these defects, we generated both heterozygous and homozygous ppg1 deletion mutants as well as a ppg1Δ/Δ::PPG1 add-back strain. Initially, all strains were examined for colony morphology on solid filament-inducing media. As indicated in Fig. 2, the ppg1Δ/+ strain showed a clear reduction in fuzziness (filamentous fringes) at the edges of the colonies compared to that of the wild-type (WT) strain on both Spider (nitrogen and carbon starvation) and Lee's pH 6.8 media. However, the ppg1Δ/+ mutant did not appear to show a defect in colony morphology when grown on solid medium in the presence of serum at 37°C. In contrast, the homozygous ppg1Δ/Δ mutant showed more significant colony morphology defects than both WT and ppg1Δ/+ strains on Spider, Lee's pH 6.8, and particularly YEPD plus serum at 37°C media as indicated by significantly reduced wrinkling in addition to an absence of fuzziness at the edges (for Spider and Lee's pH 6.8 media). As expected, colonies of the ppg1Δ/Δ::PPG1 add-back strain showed phenotypes equivalent to those observed with the ppg1Δ/+ strain, and all strains grew as smooth colonies under non-filament-inducing conditions (YEPD medium at 30°C). Interestingly, colonies of a C. albicans ppg1H248A H173A D90L catalytic mutant appeared nearly identical to those of the ppg1Δ/Δ strain but showed slightly greater wrinkling under all solid-medium filament-inducing conditions (Fig. 2). These results indicate that the ppg1Δ/Δ colony morphology defects can be largely attributed to a defect in Ppg1 phosphatase activity.
FIG 2.
Ppg1 phosphatase activity controls C. albicans colony morphology under multiple filament-inducing conditions. Colonies of the indicated strains were grown for 2 days on solid medium under non-filament-inducing conditions (YEPD at 30°C) as well as on the indicated filament-inducing conditions (colonies on YEPD medium plus 10% serum at 37°C were grown for 2 days, and colonies on Spider medium and Lee's pH 6.8 medium were grown at 30°C for 4 days). All colonies were visualized using light microscopy.
To determine the effect of constitutive high-level PPG1 expression on C. albicans colony morphology, we generated a strain in which a single allele of PPG1 was under the control of an E. coli tet operator. In the absence of doxycycline (Dox), a tetracycline derivative, a strong transactivator protein binds as a dimer to the tet operator and directs transcriptional activation. As shown in Fig. S2A in the supplemental material, Northern analysis confirmed that PPG1 is expressed only in the absence of Dox and only when the tet operator is present. Under solid-medium non-filament-inducing conditions (YEPD medium at 30°C), constitutive high-level PPG1 expression did not affect C. albicans colony morphology (see Fig. S2B in the supplemental material). Interestingly, however, PPG1 expression resulted in a noticeable decrease in wrinkling when colonies were grown on Spider medium and YEPD + serum at 37°C. We did not observe any effect of PPG1 expression on filamentation under a variety of liquid-medium inducing conditions (data not shown). These findings indicate that constitutive high-level PPG1 expression results in increased colony morphology defects under certain filament-inducing conditions.
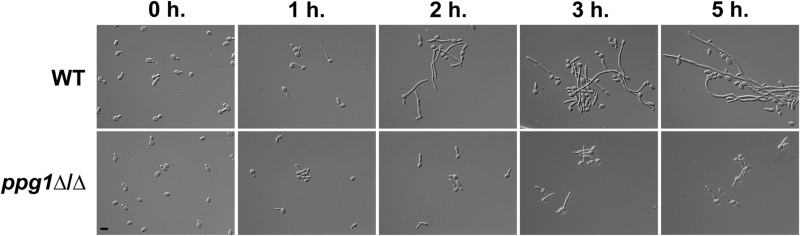
We next sought to determine the effect of the ppg1Δ/Δ mutation on C. albicans cellular morphology under strong liquid-medium filament-inducing conditions. Both WT and ppg1Δ/Δ strains were induced to form filaments by growth in liquid YEPD plus serum at 37°C, and morphology was monitored at specific postinduction time points (Fig. 3). As expected, all cells grew as yeast at the zero-hour time point, just prior to induction. In addition, as observed previously, the WT strain formed short germ tubes at the 1-hour time point and much longer extended filaments (mostly hyphae) over the remainder of the time course. Similar to the wild-type strain, the ppg1Δ/Δ mutant was able to form short germ tubes at the 1-hour time point. However, at the 2-, 3-, and 5-hour time points, cells of the ppg1Δ/Δ mutant showed significantly reduced filament lengths compared to those of the WT strain. In order to determine whether the ppg1Δ/Δ filament extension defect was the result of a delay in filament formation, we allowed the induction time course to proceed for additional time points past 5 h as well as overnight. However, no increase in filamentation or filament length by the ppg1Δ/Δ mutant was observed (data not shown). As expected, both the WT and ppg1Δ/Δ strains grew as yeast cells under non-filament-inducing conditions (YEPD at 30°C) (data not shown). The ppg1H248A H173A D90L catalytic mutant showed a filament extension defect equivalent to that of the ppg1Δ/Δ strain in liquid medium in response to induction by serum at 37°C (see Fig. S3 in the supplemental material). The ppg1Δ/Δ filament extension defect was also rescued in the ppg1Δ/Δ::PPG1 add-back control strain. Altogether, our results indicate that PPG1, and more specifically Ppg1 phosphatase activity, plays an important role in controlling C. albicans filament extension under strong liquid filament-inducing conditions.
FIG 3.
PPG1 is important for filament extension in response to growth under strong filament-inducing conditions. The indicated strains were grown overnight in YEPD medium at 30°C (non-filament-inducing conditions, 0 h time point) and diluted 1:10 into prewarmed YEPD medium plus 10% serum at 37°C (strong filament-inducing conditions). Cells were harvested at the indicated time points (hours), fixed using 4.5% formaldehyde, and washed twice with 1× phosphate-buffered saline (PBS). Images were taken using differential interference contrast (DIC) microscopy. Bar, 10 μm.
During infection, C. albicans filamentous growth is known to play an important role in promoting the invasion of both mucosal cell layers and a variety of host tissues. We performed a simple in vitro assay to determine whether the ppg1Δ/Δ mutation also affects this important virulence trait by examining agar invasion under both non-filament-inducing and a variety of filament-inducing conditions. As shown in Fig. 4, we observed that the ppg1Δ/Δ mutant is highly defective for agar invasion under multiple filament-inducing conditions. Similar to the ppg1Δ/Δ strain, the ppg1H248A H173A D90L catalytic mutant also showed a significant defect in agar invasion. However, this defect appeared to be slightly less severe than that of the ppg1Δ/Δ mutant on Lee's pH 6.8 medium as well as YEPD medium plus serum at 37°C. Interestingly, both the ppg1Δ/+ and the ppg1Δ/Δ::PPG1 add-back strains did not appear to show any agar invasion defect, indicating that expression of a single PPG1 allele is sufficient for normal invasion. As expected, none of the strains were able to invade the agar under non-filament-inducing conditions (YEPD at 30°C). Overall, these results indicate that in addition to filament extension, PPG1 and more specifically Ppg1 phosphatase activity also play an important role in controlling the ability of C. albicans to invade surfaces.
PPG1 is important for NRG1 downregulation and affects the expression of multiple filament-specific transcripts in response to serum at 37°C.
Previous studies have shown that downregulation of NRG1, a key transcriptional repressor of filament-specific genes, represents a critical event important for both C. albicans filamentation and expression of the filamentous growth program (22, 53, 54). In order to determine whether PPG1 plays a role in controlling this regulatory event, aliquots of cells from the liquid serum and temperature induction time course experiment described in Fig. 3 (including additional 15- and 30-min time points) were harvested for total RNA preparation, and a Northern analysis was performed to examine NRG1 transcript levels. As shown in Fig. 5, in the WT strain the NRG1 transcript level decreased by the 30-min postinduction time point, remained low through much of the time course, and increased again by the 5-hour time point. In contrast, very little, if any, reduction in NRG1 transcript level was observed in the ppg1Δ/Δ strain, indicating that PPG1 is important for NRG1 downregulation in response to serum at 37°C.
FIG 5.
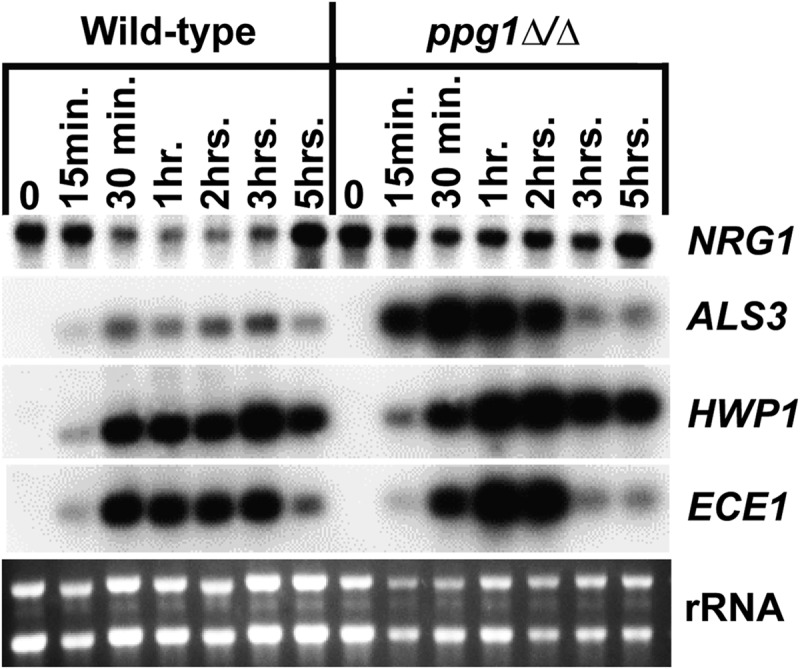
PPG1 is important for NRG1 downregulation and controls filament-specific gene expression in response to serum at 37°C. Cells were harvested at the indicated time points from the strains grown as described for Fig. 3 and used to prepare total RNA for Northern analysis. Three micrograms of total RNA was loaded in each lane, and Northern blots were probed for the indicated transcripts. rRNA is included as a loading control.
We also examined the expression of several filament-specific target genes (ALS3, HWP1, ECE1) in WT versus ppg1Δ/Δ strains in response to induction by serum at 37°C (Fig. 5). Surprisingly, all of these genes showed somewhat increased expression during the early serum and temperature induction time points (up until 2 h). In addition, two filament-specific transcripts (ALS3 and ECE1) also showed a sharp reduction at the later time points (3 and 5 hours postinduction). These results indicate that PPG1 affects both the transcript level and induction kinetics of multiple filament-specific genes under strong filament-inducing conditions.
Functional relationship between PPG1 and the cAMP-PKA filamentous growth signaling pathway.
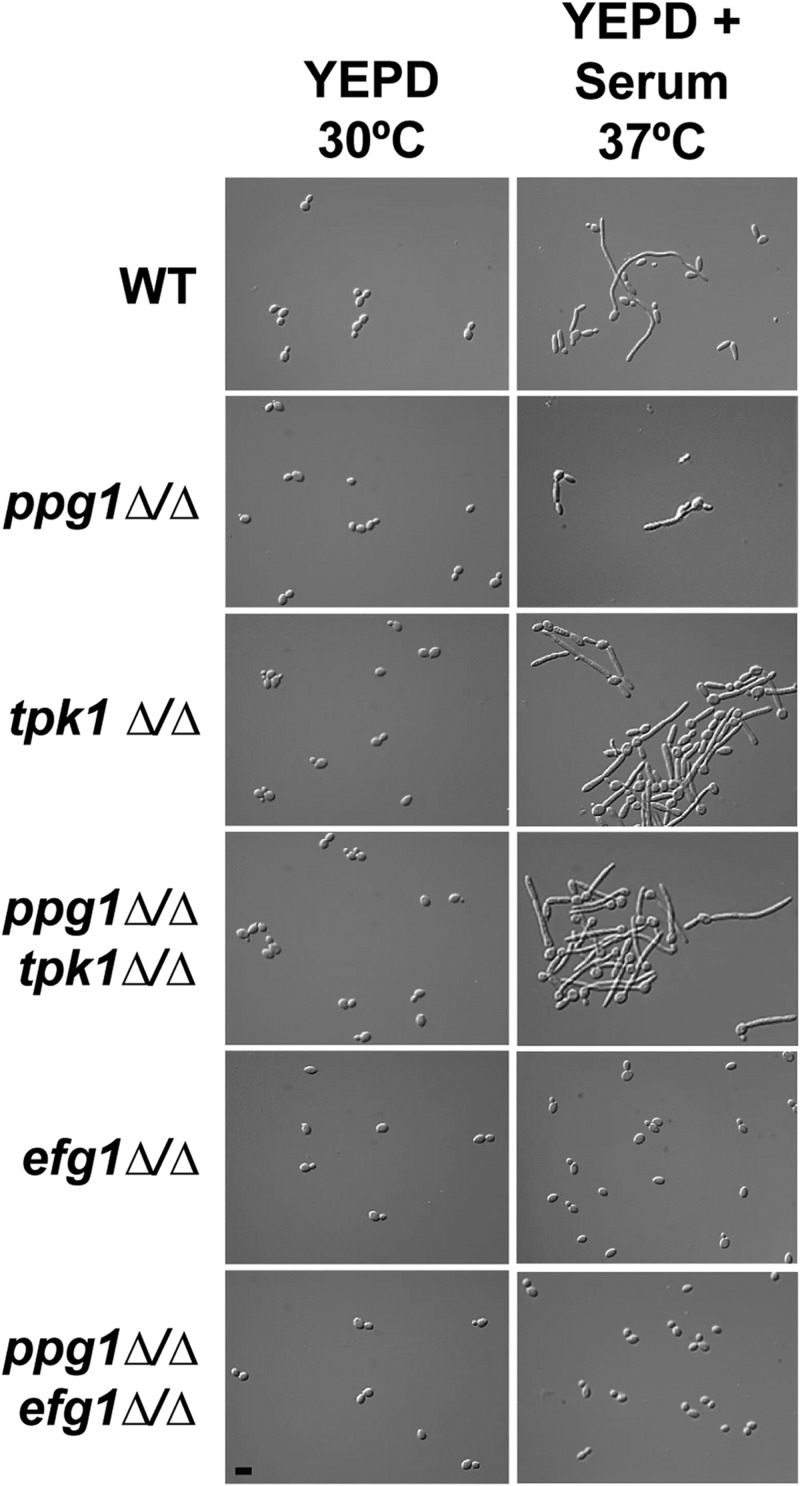
Previous studies have shown that the cAMP-PKA filamentous growth signaling pathway is required for transient removal of the Nrg1 protein from the promoters of filament-specific genes and that a downstream target transcription factor of this pathway, Efg1, is required for downregulation of the NRG1 transcript (54, 55). Our finding that PPG1 is also important for NRG1 downregulation suggested a possible functional relationship between PPG1 and the cAMP-PKA pathway. In order to test this hypothesis, we generated ppg1Δ/Δ tpk1Δ/Δ and ppg1Δ/Δ efg1Δ/Δ double mutants. TPK1 encodes a catalytic subunit of cAMP-dependent protein kinase (56). TPK1 rather than TPK2, which encodes the other cAMP-dependent protein kinase catalytic subunit, was chosen for epistasis analysis because the tpk1Δ/Δ mutant shows the greatest phenotypic difference compared to the ppg1Δ/Δ mutant under liquid-medium filament-inducing conditions. The epistasis analysis was performed using ppg1Δ/Δ, tpk1Δ/Δ, efg1Δ/Δ, ppg1Δ/Δ tpk1Δ/Δ, and ppg1Δ/Δ efg1Δ/Δ mutants. All of these strains, in addition to a wild-type control strain, were induced to form filaments by growth in liquid YEPD medium plus serum at 37°C, and aliquots of cells were fixed for microscopy (Fig. 6). As observed previously, the ppg1Δ/Δ mutant showed a significant defect in filament extension. Also consistent with previous observations (56), the tpk1Δ/Δ mutant showed a level of filamentation similar to that of a WT strain. Interestingly, the ppg1Δ/Δ tpk1Δ/Δ double mutant showed significant filamentation with a phenotype identical to that of the tpk1Δ/Δ single mutant. In addition, both the efg1Δ/Δ and ppg1Δ/Δ efg1Δ/Δ mutants showed identical phenotypes and grew entirely as yeast cells (consistent with a previous report for the efg1Δ/Δ mutant [12]). These results indicate that both efg1Δ/Δ and tpk1Δ/Δ mutations are epistatic to the ppg1Δ/Δ mutation during liquid-medium filament induction in response to serum at 37°C and suggest that PPG1 functions upstream of TPK1 and EFG1 to affect the C. albicans cAMP-PKA signaling pathway. Similar epistatic relationships were observed when colony morphologies were examined on solid Spider medium (see Fig. S4 in the supplemental material). However, the tpk1Δ/Δ and ppg1Δ/Δ tpk1Δ/Δ mutants showed reduced fuzziness (filamentation) at the colony fringes compared to the WT strain (as previously reported for the tpk1Δ/Δ mutant [56]) and the ppg1Δ/Δ efg1Δ/Δ mutant showed mild colony wrinkling, suggesting that Efg1 and Ppg1 may perform partially independent functions under this condition. A similar epistasis analysis has suggested that PPG1 does not function via the C. albicans MAP kinase signaling pathway (data not shown).
FIG 6.
The tpk1Δ/Δ and efg1Δ/Δ mutations are epistatic to the ppg1Δ/Δ mutation with respect to cellular morphology under strong liquid-medium filament-inducing conditions. The indicated strains were grown overnight in YEPD at 30°C, saturated cultures were diluted under the indicated filament-inducing (YEPD + serum at 37°C) and -noninducing (YEPD at 30°C) conditions, and cells were harvested at the 3-h postinduction time point. After fixing in 4.5% formaldehyde, cells were washed twice with 1× PBS and visualized using DIC microscopy. Bar, 10 μm.
Ppg1 phosphatase activity is important for C. albicans filament extension and virulence in a mouse model of systemic candidiasis.
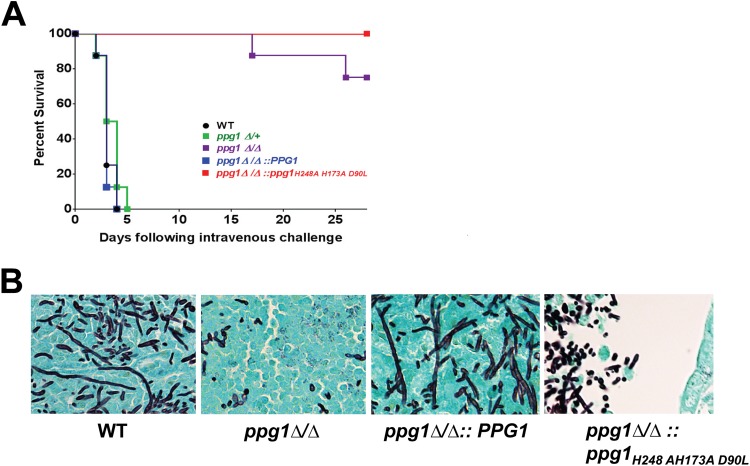
In order to determine the requirement of PPG1 and Ppg1 phosphatase activity for virulence in a mouse model of systemic candidiasis, mice were inoculated by tail vein with WT, ppg1Δ/+, ppg1Δ/Δ, ppg1Δ/Δ::PPG1 add-back, and ppg1Δ/Δ:: ppg1H248A H173A D90L mutant strains. None of these strains showed a significant growth defect at 37°C as determined by a growth curve. As indicated in Fig. 7A, all mice inoculated with wild-type, ppg1Δ/+, and ppg1Δ/Δ::PPG1 strains died within 5 days. In contrast, all mice inoculated with the ppg1Δ/Δ::ppg1H248A H173A D90L catalytic mutant and 75% of mice inoculated with the ppg1Δ/Δ strain were alive at 28 days postinfection. These results indicate that Ppg1, and more specifically Ppg1 phosphatase activity, is important for virulence in a mouse model of systemic candidiasis. Interestingly, strains bearing a single allele of PPG1 were not defective for virulence. A histological analysis indicated, as expected, that long extended filaments were present in tissues infected with WT and ppg1Δ/Δ::PPG1 add-back strains (Fig. 7B). In contrast, both ppg1Δ/Δ and ppg1Δ/Δ::ppg1H248A H173A D90L catalytic mutant strains generally showed a mixture of yeast and short filaments in the kidneys during infection in vivo, also associated with reduced tissue penetration and lack of invasive growth. Overall, these results indicate that Ppg1 and Ppg1 phosphatase activity are important for filament extension, tissue invasion, and virulence during infection in vivo and are consistent with our previous in vitro findings.
FIG 7.
Ppg1 phosphatase activity is important for C. albicans virulence and filament extension during infection in vivo. Eight (n = 8) female BALB/c mice (6 to 8 weeks old) were inoculated by tail vein with 3.5 × 105 CFU of the indicated strains, and survival was monitored for 28 days. A Kaplan-Meier test indicated that ppg1Δ/Δ and ppg1Δ/Δ::ppg1H248A H173A D90L strains showed a statistically significant difference in virulence compared to the wild-type strain (P < 0.0001). (B) Kidneys from mice infected with the indicated strains were fixed, paraffin embedded, sectioned, and stained with Grocott-Gomori methenamine silver (GMS).
DISCUSSION
Protein phosphatases are known to function as key regulatory molecules in controlling a wide variety of biological processes. Previous studies in yeast and higher eukaryotes have shown that PP2A-type phosphatases, in particular, control many cellular processes, including actin cytoskeleton organization, mitosis, DNA replication, cell wall integrity, cellular proliferation, apoptosis, gene expression, and signal transduction (35, 37, 57–59). In the major human fungal pathogen Candida albicans, protein phosphatases are known to regulate cell cycle progression, cation homeostasis, cell wall integrity, macrophage killing, antifungal drug resistance (e.g., as mediated by calcineurin), osmotolerance, and the response to oxidative stress and DNA-damaging agents (33, 60–66). However, considerably less is known about the ability of phosphatases to control morphology, one of the most important C. albicans virulence traits.
Here, we report the characterization of PPG1, which encodes a PP2A-type serine/threonine phosphatase that plays a specific important role in controlling C. albicans morphology, invasion, and virulence. The ppg1Δ/Δ mutant was highly defective for colony morphology under a variety of solid-medium filament-inducing conditions, suggesting that PPG1 is important for the ability of C. albicans to respond to multiple environmental cues. Interestingly, this mutant was capable of forming germ tubes during liquid serum and temperature filament induction but showed a significant defect in filament extension. Epistasis analysis has suggested that PPG1 does not appear to function in the UME6-HGC1 pathway, which has previously been shown to control hyphal filament extension (49, 67) (data not shown); PPG1 is also unlikely to function through EED1, another component of this pathway (68). Consistent with the observed defect in filament extension, the ppg1Δ/Δ mutant was also defective for both agar invasion in vitro and tissue invasion during infection in vivo, which may at least partially account for the significantly reduced virulence of this strain. However, Ppg1 may also control additional virulence-related properties.
Our demonstration that the ppg1H248A H173A D90L catalytic mutant shows nearly equivalent defects in colony morphology, filamentation, invasion, and virulence to those of the ppg1Δ/Δ strain strongly suggests that these defects can be attributed primarily to a loss in Ppg1 phosphatase activity. Interestingly, however, we did observe several minor differences in the phenotypes of ppg1H248A H173A D90L and ppg1Δ/Δ strains. On solid media, the ppg1H248A H173A D90L catalytic mutant showed slightly greater colony wrinkling and invasion than the ppg1Δ/Δ strain. In addition, while all mice inoculated with the ppg1H248A H173A D90L catalytic mutant survived within 28 days postinfection, one-quarter of the mice inoculated with the ppg1Δ/Δ strain succumbed to infection. These minor differences suggest that in addition to phosphatase activity, Ppg1 may possess other activities or perform other functions that make a very small contribution to C. albicans virulence and virulence-related properties.
Does PPG1 function to promote C. albicans filament extension and virulence by targeting known filamentous growth signaling pathways and transcriptional regulators? Multiple lines of evidence suggest that Ppg1 specifically targets the cAMP-PKA filamentation pathway. We have shown that mutations in Tpk1, a catalytic subunit of the cAMP-dependent protein kinase, and Efg1, the downstream transcription factor target of the cAMP-PKA pathway, are epistatic to the ppg1Δ/Δ mutation with respect to filamentation in liquid YEPD plus serum at 37°C, conditions that are known to activate the cAMP-PKA pathway. These results suggest that Ppg1 functions upstream of both Tpk1 and Efg1. Consistent with this finding, we have also demonstrated that PPG1 is specifically important for downregulation of NRG1, a key transcriptional repressor of filament-specific genes, in response to growth in serum at 37°C. Downregulation of NRG1 requires Efg1, and recent work has shown that the cAMP-PKA pathway also directs a transient displacement of the Nrg1 protein from promoters of filament-specific genes (54, 55). Because Efg1 is known to functionally and/or physically interact with other transcriptional regulators that respond to different environmental cues, including Flo8, Sfl1, Sfl2, and Czf1 (69–71), Ppg1 may also modulate the function of these factors as well.
Our demonstration that PPG1 is important for downregulation of NRG1 may to some extent explain the observed ppg1Δ/Δ mutant defects in morphology and virulence. According to this hypothesis, in the absence of NRG1 downregulation in the ppg1Δ/Δ mutant, filament-specific genes would not be induced (or would show reduced expression), leading to yeast phase growth. Surprisingly, we observed increased, rather than decreased, expression of three filament-specific transcripts during the early time points of serum and temperature induction in the ppg1Δ/Δ versus wild-type strains. However, in the case of two of these genes, there was a reduction in transcript level at the later time points. These results suggest a more complex relationship between PPG1 and filament-specific gene expression. During the early stages of filamentation, filament-specific gene expression may show a transient increase in the ppg1Δ/Δ strain as a result of loss of inhibitory dephosphorylation of filamentous growth signaling pathway components that respond to environmental cues. However, in later stages of filament development Ppg1 may play a more important role in maintaining filament-specific gene expression and hyphal growth. Consistent with this hypothesis, ppg1 mutants can form normal germ tubes but are specifically defective for filament extension in the later time points of a serum and temperature induction time course. In addition, a previous study has suggested that filament-specific genes are differentially regulated in a temporal manner during hyphal initiation versus maintenance phases (55).
How exactly is the Ppg1 phosphatase controlled in response to filament-inducing conditions? PPG1 is not a filament-induced gene and appears to be expressed at very low constitutive levels (our unpublished observations). In addition, we have observed that high-level PPG1 expression causes a slight reduction in colony wrinkling only in the presence of solid medium filament-inducing conditions. These results suggest that Ppg1, similar to other phosphatases, may function to modulate and carefully fine-tune the activity of major phosphorylation pathways (such as cAMP-PKA). While an epistasis analysis has suggested that Ppg1 functions independently of the MAP kinase pathway (data not shown), we cannot exclude the possibility that Ppg1 targets additional filamentous growth signaling pathways. Most likely, Ppg1 itself is regulated at the posttranslational level, possibly by phosphorylation. Our observation that the ppg1Δ/Δ mutant is defective for filamentation during infection in vivo and in response to a wide variety of environmental conditions in vitro suggests that Ppg1 phosphatase activity is controlled by multiple filament-inducing signals. Consistent with this hypothesis, a recent study in S. cerevisiae has indicated that phosphatases in the same PP2A subfamily as Ppg1 are activated by glucose at the posttranslational level via the cAMP-PKA pathway (72). At this point, the exact mechanism(s) by which C. albicans Ppg1 phosphatase activity is controlled by environmental cues that induce filamentation and promote virulence in the host remains unclear. It is hoped that future studies will shed more light in this area and also identify Ppg1-interacting partners and substrates. Ultimately, gaining a better understanding of how phosphatases, such as Ppg1, function to regulate and fine-tune the activity of C. albicans filamentous growth signaling pathways should improve our knowledge of the complex regulatory circuits that control morphology and virulence in fungal pathogens and could provide information leading to the development of more effective antifungal strategies.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Brian Wickes and P. John Hart for useful advice and assistance during the course of the experiments. We thank Brian Wickes for comments on the manuscript. We also thank Alexander Johnson, P. John Hart, and Peter Dube for strains, plasmids, and antibodies (respectively).
M.T.A. was supported by a Ruth L. Kirschstein National Research Service Award for Individual Postdoctoral Fellows from the National Institute of Dental and Craniofacial Research (F32DE023471). D.K. was supported by National Institute of Allergy and Infectious Diseases grant 5RO1AI083344 in addition to a Voelcker Young Investigator Award from the Max and Minnie Tomerlin Voelcker Fund.
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health, National Institute of Allergy and Infectious Diseases, or National Institute of Dental and Craniofacial Research.
Footnotes
Published ahead of print 17 October 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/EC.00199-14.
REFERENCES
- 1.Edmond MB, Wallace SE, McClish DK, Pfaller MA, Jones RN, Wenzel RP. 1999. Nosocomial bloodstream infections in United States hospitals: a three-year analysis. Clin. Infect. Dis. 29:239–244. 10.1086/520192. [DOI] [PubMed] [Google Scholar]
- 2.Wisplinghoff H, Bischoff T, Tallent SM, Seifert H, Wenzel RP, Edmond MB. 2004. Nosocomial bloodstream infections in US hospitals: analysis of 24,179 cases from a prospective nationwide surveillance study. Clin. Infect. Dis. 39:309–317. 10.1086/421946. [DOI] [PubMed] [Google Scholar]
- 3.Calderone RA, Clancy CJ. 2012. Candida and candidiasis, 2nd ed. ASM Press, Washington, DC. [Google Scholar]
- 4.Dupont PF. 1995. Candida albicans, the opportunist. A cellular and molecular perspective. J. Am. Podiatr. Med Assoc. 85:104–115. 10.7547/87507315-85-2-104. [DOI] [PubMed] [Google Scholar]
- 5.Weig M, Gross U, Muhlschlegel F. 1998. Clinical aspects and pathogenesis of Candida infection. Trends Microbiol. 6:468–470. 10.1016/S0966-842X(98)01407-3. [DOI] [PubMed] [Google Scholar]
- 6.Klepser ME. 2001. Antifungal resistance among Candida species. Pharmacotherapy 21(8 Part 2):124S–132S. [DOI] [PubMed] [Google Scholar]
- 7.Maubon D, Garnaud C, Calandra T, Sanglard D, Cornet M. 2014. Resistance of Candida spp. to antifungal drugs in the ICU: where are we now? Intensive Care Med. 40:1241–1255. 10.1007/s00134-014-3404-7. [DOI] [PubMed] [Google Scholar]
- 8.Odds FC. 1988. Candida and candidosis, 2nd ed. Baillière Tindall, London, United Kingdom. [Google Scholar]
- 9.Brown AJ. 2002. Expression of growth form-specific factors during morphogenesis in Candida albicans, p 87–93 In Calderone RA. (ed), Candida and candidiasis. ASM Press, Washington, DC. [Google Scholar]
- 10.Kumamoto CA, Vinces MD. 2005. Contributions of hyphae and hypha-co-regulated genes to Candida albicans virulence. Cell. Microbiol. 7:1546–1554. 10.1111/j.1462-5822.2005.00616.x. [DOI] [PubMed] [Google Scholar]
- 11.Korting HC, Hube B, Oberbauer S, Januschke E, Hamm G, Albrecht A, Borelli C, Schaller M. 2003. Reduced expression of the hyphal-independent Candida albicans proteinase genes SAP1 and SAP3 in the efg1 mutant is associated with attenuated virulence during infection of oral epithelium. J. Med. Microbiol. 52:623–632. 10.1099/jmm.0.05125-0. [DOI] [PubMed] [Google Scholar]
- 12.Lo HJ, Kohler JR, DiDomenico B, Loebenberg D, Cacciapuoti A, Fink GR. 1997. Nonfilamentous C. albicans mutants are avirulent. Cell 90:939–949. 10.1016/S0092-8674(00)80358-X. [DOI] [PubMed] [Google Scholar]
- 13.Dalle F, Wachtler B, L'Ollivier C, Holland G, Bannert N, Wilson D, Labruere C, Bonnin A, Hube B. 2010. Cellular interactions of Candida albicans with human oral epithelial cells and enterocytes. Cell. Microbiol. 12:248–271. 10.1111/j.1462-5822.2009.01394.x. [DOI] [PubMed] [Google Scholar]
- 14.Lorenz MC, Bender JA, Fink GR. 2004. Transcriptional response of Candida albicans upon internalization by macrophages. Eukaryot. Cell 3:1076–1087. 10.1128/EC.3.5.1076-1087.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Saville SP, Lazzell AL, Monteagudo C, Lopez-Ribot JL. 2003. Engineered control of cell morphology in vivo reveals distinct roles for yeast and filamentous forms of Candida albicans during infection. Eukaryot. Cell 2:1053–1060. 10.1128/EC.2.5.1053-1060.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Braun BR, Johnson AD. 1997. Control of filament formation in Candida albicans by the transcriptional repressor TUP1. Science 277:105–109. 10.1126/science.277.5322.105. [DOI] [PubMed] [Google Scholar]
- 17.Mitchell AP. 1998. Dimorphism and virulence in Candida albicans. Curr. Opin. Microbiol. 1:687–692. 10.1016/S1369-5274(98)80116-1. [DOI] [PubMed] [Google Scholar]
- 18.Brown AJ. 2002. Morphogenetic signaling pathways in Candida albicans, p 95–106 In Calderone RA. (ed), Candida and candidiasis. ASM Press, Washington, DC. [Google Scholar]
- 19.Brown AJ, Gow NA. 1999. Regulatory networks controlling Candida albicans morphogenesis. Trends Microbiol. 7:333–338. 10.1016/S0966-842X(99)01556-5. [DOI] [PubMed] [Google Scholar]
- 20.Biswas S, Van Dijck P, Datta A. 2007. Environmental sensing and signal transduction pathways regulating morphopathogenic determinants of Candida albicans. Microbiol. Mol. Biol. Rev. 71:348–376. 10.1128/MMBR.00009-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sudbery PE. 2011. Growth of Candida albicans hyphae. Nat. Rev. Microbiol. 9:737–748. 10.1038/nrmicro2636. [DOI] [PubMed] [Google Scholar]
- 22.Kadosh D, Johnson AD. 2005. Induction of the Candida albicans filamentous growth program by relief of transcriptional repression: a genome-wide analysis. Mol. Biol. Cell 16:2903–2912. 10.1091/mbc.E05-01-0073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nantel A, Dignard D, Bachewich C, Harcus D, Marcil A, Bouin AP, Sensen CW, Hogues H, van het Hoog M, Gordon P, Rigby T, Benoit F, Tessier DC, Thomas DY, Whiteway M. 2002. Transcription profiling of Candida albicans cells undergoing the yeast-to-hyphal transition. Mol. Biol. Cell 13:3452–3465. 10.1091/mbc.E02-05-0272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu H, Kohler J, Fink GR. 1994. Suppression of hyphal formation in Candida albicans by mutation of a STE12 homolog. Science 266:1723–1726. 10.1126/science.7992058. [DOI] [PubMed] [Google Scholar]
- 25.Tebarth B, Doedt T, Krishnamurthy S, Weide M, Monterola F, Dominguez A, Ernst JF. 2003. Adaptation of the Efg1p morphogenetic pathway in Candida albicans by negative autoregulation and PKA-dependent repression of the EFG1 gene. J. Mol. Biol. 329:949–962. 10.1016/S0022-2836(03)00505-9. [DOI] [PubMed] [Google Scholar]
- 26.Bockmuhl DP, Ernst JF. 2001. A potential phosphorylation site for an A-type kinase in the Efg1 regulator protein contributes to hyphal morphogenesis of Candida albicans. Genetics 157:1523–1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hanaoka N, Takano Y, Shibuya K, Fugo H, Uehara Y, Niimi M. 2008. Identification of the putative protein phosphatase gene PTC1 as a virulence-related gene using a silkworm model of Candida albicans infection. Eukaryot. Cell 7:1640–1648. 10.1128/EC.00129-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Csank C, Makris C, Meloche S, Schröppel K, Röllinghoff M, Dignard D, Thomas DY, Whiteway M. 1997. Derepressed hyphal growth and reduced virulence in a VH1 family-related protein phosphatase mutant of the human pathogen Candida albicans. Mol. Biol. Cell 8:2539–2551. 10.1091/mbc.8.12.2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fan J, Wu M, Jiang L, Shen SH. 2009. A serine/threonine protein phosphatase-like protein, CaPTC8, from Candida albicans defines a new PPM subfamily. Gene 430:64–76. 10.1016/j.gene.2008.10.022. [DOI] [PubMed] [Google Scholar]
- 30.Lee CM, Nantel A, Jiang L, Whiteway M, Shen SH. 2004. The serine/threonine protein phosphatase SIT4 modulates yeast-to-hypha morphogenesis and virulence in Candida albicans. Mol. Microbiol. 51:691–709. 10.1111/j.1365-2958.2003.03879.x. [DOI] [PubMed] [Google Scholar]
- 31.Bader T, Bodendorfer B, Schroppel K, Morschhauser J. 2003. Calcineurin is essential for virulence in Candida albicans. Infect. Immun. 71:5344–5354. 10.1128/IAI.71.9.5344-5354.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Blankenship JR, Wormley FL, Boyce MK, Schell WA, Filler SG, Perfect JR, Heitman J. 2003. Calcineurin is essential for Candida albicans survival in serum and virulence. Eukaryot. Cell 2:422–430. 10.1128/EC.2.3.422-430.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sanglard D, Ischer F, Marchetti O, Entenza J, Bille J. 2003. Calcineurin A of Candida albicans: involvement in antifungal tolerance, cell morphogenesis and virulence. Mol. Microbiol. 48:959–976. 10.1046/j.1365-2958.2003.03495.x. [DOI] [PubMed] [Google Scholar]
- 34.Noble SM, French S, Kohn LA, Chen V, Johnson AD. 2010. Systematic screens of a Candida albicans homozygous deletion library decouple morphogenetic switching and pathogenicity. Nat. Genet. 42:590–598. 10.1038/ng.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Evans DR, Stark MJ. 1997. Mutations in the Saccharomyces cerevisiae type 2A protein phosphatase catalytic subunit reveal roles in cell wall integrity, actin cytoskeleton organization and mitosis. Genetics 145:227–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Garbers C, DeLong A, Deruere J, Bernasconi P, Soll D. 1996. A mutation in protein phosphatase 2A regulatory subunit A affects auxin transport in Arabidopsis. EMBO J. 15:2115–2124. [PMC free article] [PubMed] [Google Scholar]
- 37.Lin FC, Arndt KT. 1995. The role of Saccharomyces cerevisiae type 2A phosphatase in the actin cytoskeleton and in entry into mitosis. EMBO J. 14:2745–2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Posas F, Clotet J, Muns MT, Corominas J, Casamayor A, Arino J. 1993. The gene PPG encodes a novel yeast protein phosphatase involved in glycogen accumulation. J. Biol Chem. 268:1349–1354. [PubMed] [Google Scholar]
- 39.Banerjee M, Thompson DS, Lazzell A, Carlisle PL, Pierce C, Monteagudo C, Lopez-Ribot JL, Kadosh D. 2008. UME6, a novel filament-specific regulator of Candida albicans hyphal extension and virulence. Mol. Biol. Cell 19:1354–1365. 10.1091/mbc.E07-11-1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Noble SM, Johnson AD. 2005. Strains and strategies for large-scale gene deletion studies of the diploid human fungal pathogen Candida albicans. Eukaryot. Cell 4:298–309. 10.1128/EC.4.2.298-309.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fu J, Hettler E, Wickes BL. 2006. Split marker transformation increases homologous integration frequency in Cryptococcus neoformans. Fungal Genet. Biol. 43:200–212. 10.1016/j.fgb.2005.09.007. [DOI] [PubMed] [Google Scholar]
- 42.Reuss O, Vik A, Kolter R, Morschhauser J. 2004. The SAT1 flipper, an optimized tool for gene disruption in Candida albicans. Gene 341:119–127. 10.1016/j.gene.2004.06.021. [DOI] [PubMed] [Google Scholar]
- 43.Banerjee M, Uppuluri P, Zhao XR, Carlisle PL, Vipulanandan G, Villar CC, Lopez-Ribot JL, Kadosh D. 2013. Expression of UME6, a key regulator of Candida albicans hyphal development, enhances biofilm formation via Hgc1- and Sun41-dependent mechanisms. Eukaryot. Cell 12:224–232. 10.1128/EC.00163-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lackey E, Vipulanandan G, Childers DS, Kadosh D. 2013. Comparative evolution of morphological regulatory functions in Candida species. Eukaryot. Cell 12:1356–1368. 10.1128/EC.00164-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Seetharaman SV, Taylor AB, Holloway S, Hart PJ. 2010. Structures of mouse SOD1 and human/mouse SOD1 chimeras. Arch. Biochem. Biophys. 503:183–190. 10.1016/j.abb.2010.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Guthrie C, Fink GR. 1991. Guide to yeast genetics and molecular biology. Academic Press, San Diego, CA. [Google Scholar]
- 47.Lee KL, Buckley HR, Campbell CC. 1975. An amino acid liquid synthetic medium for the development of mycelial and yeast forms of Candida albicans. Sabouraudia 13:148–153. 10.1080/00362177585190271. [DOI] [PubMed] [Google Scholar]
- 48.Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. 1992. Current protocols in molecular biology. Greene Publishing Associates and Wiley-Interscience, New York, NY. [Google Scholar]
- 49.Carlisle PL, Kadosh D. 2010. Candida albicans Ume6, a filament-specific transcriptional regulator, directs hyphal growth via a pathway involving Hgc1 cyclin-related protein. Eukaryot. Cell 9:1320–1328. 10.1128/EC.00046-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bradford MM. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248–254. 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 51.Childers DS, Mundodi V, Banerjee M, Kadosh D. 2014. A 5′ UTR-mediated translational efficiency mechanism inhibits the Candida albicans morphological transition. Mol. Microbiol. 92:570–585. 10.1111/mmi.12576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Takai A, Murata M, Torigoe K, Isobe M, Mieskes G, Yasumoto T. 1992. Inhibitory effect of okadaic acid derivatives on protein phosphatases. A study on structure-affinity relationship. Biochem. J. 284(Part 2):539–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Murad AMA, Leng P, Straffon M, Wishart J, Macaskill S, MacCallum D, Schnell N, Talibi D, Marechal D, Tekaia F, d'Enfert C, Gaillardin C, Odds FC, Brown AJP. 2001. NRG1 represses yeast-hypha morphogenesis and hypha-specific gene expression in Candida albicans. EMBO J. 20:4742–4752. 10.1093/emboj/20.17.4742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Braun BR, Kadosh D, Johnson AD. 2001. NRG1, a repressor of filamentous growth in C. albicans, is down-regulated during filament induction. EMBO J. 20:4753–4761. 10.1093/emboj/20.17.4753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lu Y, Su C, Wang A, Liu H. 2011. Hyphal development in Candida albicans requires two temporally linked changes in promoter chromatin for initiation and maintenance. PLoS Biol. 9:e1001105. 10.1371/journal.pbio.1001105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bockmuhl DP, Krishnamurthy S, Gerads M, Sonneborn A, Ernst JF. 2001. Distinct and redundant roles of the two protein kinase A isoforms Tpk1p and Tpk2p in morphogenesis and growth of Candida albicans. Mol. Microbiol. 42:1243–1257. [DOI] [PubMed] [Google Scholar]
- 57.Snaith HA, Armstrong CG, Guo Y, Kaiser K, Cohen PT. 1996. Deficiency of protein phosphatase 2A uncouples the nuclear and centrosome cycles and prevents attachment of microtubules to the kinetochore in Drosophila microtubule star (mts) embryos. J. Cell Sci. 109(Part 13):3001–3012. [DOI] [PubMed] [Google Scholar]
- 58.Kinoshita N, Yamano H, Niwa H, Yoshida T, Yanagida M. 1993. Negative regulation of mitosis by the fission yeast protein phosphatase ppa2. Genes Dev. 7:1059–1071. 10.1101/gad.7.6.1059. [DOI] [PubMed] [Google Scholar]
- 59.Seshacharyulu P, Pandey P, Datta K, Batra SK. 2013. Phosphatase: PP2A structural importance, regulation and its aberrant expression in cancer. Cancer Lett. 335:9–18. 10.1016/j.canlet.2013.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Clemente-Blanco A, Gonzalez-Novo A, Machin F, Caballero-Lima D, Aragon L, Sanchez M, de Aldana CR, Jimenez J, Correa-Bordes J. 2006. The Cdc14p phosphatase affects late cell-cycle events and morphogenesis in Candida albicans. J. Cell Sci. 119:1130–1143. 10.1242/jcs.02820. [DOI] [PubMed] [Google Scholar]
- 61.Adam C, Erdei E, Casado C, Kovacs L, Gonzalez A, Majoros L, Petrenyi K, Bagossi P, Farkas I, Molnar M, Pocsi I, Arino J, Dombradi V. 2012. Protein phosphatase CaPpz1 is involved in cation homeostasis, cell wall integrity and virulence of Candida albicans. Microbiology 158:1258–1267. 10.1099/mic.0.057075-0. [DOI] [PubMed] [Google Scholar]
- 62.Martinez-Esparza M, Martinez-Vicente E, Gonzalez-Parraga P, Ros JM, Garcia-Penarrubia P, Arguelles JC. 2009. Role of trehalose-6P phosphatase (TPS2) in stress tolerance and resistance to macrophage killing in Candida albicans. Int. J. Med. Microbiol. 299:453–464. 10.1016/j.ijmm.2008.12.001. [DOI] [PubMed] [Google Scholar]
- 63.Zhao Y, Feng J, Li J, Jiang L. 2012. Mitochondrial type 2C protein phosphatases CaPtc5p, CaPtc6p, and CaPtc7p play vital roles in cellular responses to antifungal drugs and cadmium in Candida albicans. FEMS Yeast Res. 12:897–906. 10.1111/j.1567-1364.2012.00840.x. [DOI] [PubMed] [Google Scholar]
- 64.Leiter E, Gonzalez A, Erdei E, Casado C, Kovacs L, Adam C, Olah J, Miskei M, Molnar M, Farkas I, Hamari Z, Arino J, Pocsi I, Dombradi V. 2012. Protein phosphatase Z modulates oxidative stress response in fungi. Fungal Genet. Biol. 49:708–716. 10.1016/j.fgb.2012.06.010. [DOI] [PubMed] [Google Scholar]
- 65.Wang H, Gao J, Li W, Wong AH, Hu K, Chen K, Wang Y, Sang J. 2012. Pph3 dephosphorylation of Rad53 is required for cell recovery from MMS-induced DNA damage in Candida albicans. PLoS One 7:e37246. 10.1371/journal.pone.0037246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sun LL, Li WJ, Wang HT, Chen J, Deng P, Wang Y, Sang JL. 2011. Protein phosphatase Pph3 and its regulatory subunit Psy2 regulate Rad53 dephosphorylation and cell morphogenesis during recovery from DNA damage in Candida albicans. Eukaryot. Cell 10:1565–1573. 10.1128/EC.05042-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zheng X, Wang Y, Wang Y. 2004. Hgc1, a novel hypha-specific G1 cyclin-related protein regulates Candida albicans hyphal morphogenesis. EMBO J. 23:1845–1856. 10.1038/sj.emboj.7600195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Martin R, Moran GP, Jacobsen ID, Heyken A, Domey J, Sullivan DJ, Kurzai O, Hube B. 2011. The Candida albicans-specific gene EED1 encodes a key regulator of hyphal extension. PLoS One 6:e18394. 10.1371/journal.pone.0018394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cao F, Lane S, Raniga PP, Lu Y, Zhou Z, Ramon K, Chen J, Liu H. 2006. The Flo8 transcription factor is essential for hyphal development and virulence in Candida albicans. Mol. Biol. Cell 17:295–307. 10.1091/mbc.E05-06-0502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Znaidi S, Nesseir A, Chauvel M, Rossignol T, d'Enfert C. 2013. A comprehensive functional portrait of two heat shock factor-type transcriptional regulators involved in Candida albicans morphogenesis and virulence. PLoS Pathog. 9:e1003519. 10.1371/journal.ppat.1003519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Vinces MD, Haas C, Kumamoto CA. 2006. Expression of the Candida albicans morphogenesis regulator gene CZF1 and its regulation by Efg1p and Czf1p. Eukaryot. Cell 5:825–835. 10.1128/EC.5.5.825-835.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Castermans D, Somers I, Kriel J, Louwet W, Wera S, Versele M, Janssens V, Thevelein JM. 2012. Glucose-induced posttranslational activation of protein phosphatases PP2A and PP1 in yeast. Cell Res. 22:1058–1077. 10.1038/cr.2012.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.