Abstract
Septin proteins are conserved structural proteins that often demarcate regions of cell division. The essential nature of the septin ring, composed of several septin proteins, complicates investigation of the functions of the ring, although careful analysis in the model yeast Saccharomyces cerevisiae has elucidated the role that septins play in the cell cycle. Mutation analysis of nonessential septins in the pathogenic fungus Candida albicans has shown that septins also have vital roles in cell wall regulation (CWR), hyphal formation, and pathogenesis. While mutations in nonessential septins have been useful in establishing phenotypes, the septin defect is so slight that identifying causative associations between septins and downstream effectors has been difficult. In this work, we describe decreased abundance by mRNA perturbation (DAmP) alleles of essential septins, which display a septin defect more severe than the defect observed in deletions of nonessential septins. The septin DAmP alleles have allowed us to genetically separate the roles of septins in hyphal growth and CWR and to identify the cyclic AMP pathway as a pathway that likely acts in a parallel manner with septins in hyphal morphogenesis.
INTRODUCTION
Candida albicans is a commensal fungus of the human gastrointestinal and genitourinary tracts. It causes serious systemic disease in patients with compromised immune systems, as well as in immunocompetent patients taking broad-spectrum antibiotics or with indwelling medical devices. Despite therapy, systemic infections can have mortality rates as high as 40% in some immunocompromised populations (1). The ability to switch between yeast and hyphal morphogenetic forms is vital for the pathogenesis of the organism (2, 3). Our focus here is a protein family, called septins, whose members have roles in both antifungal drug sensitivity and hyphal growth of C. albicans.
Septins are highly conserved structural components of animal and fungal cells that have roles in the polarization of cells and cell division. These GTP-binding proteins are stable proteins whose localization and activity is regulated by posttranslational modifications (4). Their essential role in the cell cycle is best understood in the model yeast Saccharomyces cerevisiae. In this fungus, septins localize to the presumptive bud site and remain in the bud neck between mother and daughter cells as the cell cycle progresses, serving as a scaffold to direct protein localization to sites of cell separation (5). Following the conclusion of the cell cycle, these essential proteins rapidly disperse from the bud neck and are recycled to the next presumptive bud (6). Septins appear to behave similarly in the yeast-like cells of the pathogenic fungus C. albicans and are also essential for cell survival (7). Septins, however, behave differently in the hyphal cells of C. albicans. In this growth form, septin rings remain stable for several cell generations, and the behavior of individual septins is altered (8). While these observations strongly support a role for septins during hyphal growth, we know very little about why their continued presence is important in hyphal cells of C. albicans.
Five septins have been characterized in C. albicans that correspond to the five mitotic septins in S. cerevisiae, and they are encoded by CDC3, CDC10, CDC11, CDC12, and SEP7 (an ortholog of S. cerevisiae SSH1). These septins have conserved localization at the bud neck in C. albicans yeast cells, and the septin ring is thought to play a conserved role in the cell cycle during yeast-like growth. CDC3 and CDC12 are essential in both S. cerevisiae and C. albicans and are thought to be critical components in stabilizing the septin ring. CDC10 and CDC11 are also important components of the septin ring. However, cells can survive without CDC10 or CDC11 under ideal growth conditions, a fact that has been utilized when searching for septin functions. In C. albicans, mutations in CDC10 and CDC11 have been used to uncover roles for septins in hyphal growth and cell wall integrity and/or biogenesis (collectively called cell wall regulation [CWR]). C. albicans cells lacking CDC10 or CDC11 are unable to form hyphae on solid surfaces, have defects in hyphal formation in liquid media, and are hypersensitive to the cell wall-targeting antifungal drug caspofungin (7, 9). A recent report has also shown that a temperature-sensitive mutation in CDC12, encoding an essential septin, caused severe hyphal defects in liquid media (10).
Septins are highly regulated proteins. Phosphorylation, sumoylation, acetylation, methylation, and GTP binding are all thought to play some role in regulating septin localization and the stability of the septin filament structures in eukaryotes (4). However, little is known about how septins are regulated or what signals may lie downstream of septins in C. albicans. The cyclin-dependent kinase Cdc28 and the Nim1 kinase Gin4 have demonstrated roles in regulating the septin ring during yeast-like and hyphal growth (11, 12), but it is quite likely that further elements act upstream of septins to regulate their localization and stability. No pathways working downstream of septins have yet been identified in C. albicans, although the Hsl1 and Swe1 protein kinases, which coordinate responses to septins in S. cerevisiae, are present in C. albicans. Identification of the pathways that operate upstream and downstream of septins will provide insight into the contribution of septins to CWR and hyphal morphogenesis.
We have found that a decrease in the level of the essential septins in C. albicans reduces accumulation of other septins at the neck. Reduced septin levels also lead to an increase in susceptibility to cell wall stress, defects in hyphal formation, and reduced virulence in a murine tail vein injection model. In addition, decreasing essential septins in the cell leads to gene expression changes that we used to identify a pathway that may act downstream of or in parallel with septins in hyphal morphogenesis. Overexpression of the protein kinase A (PKA) TPK1 subunit restored hyphal growth in the septin mutant cells, but not cell wall stress resistance, suggesting that distinct pathways exist downstream of septins to mediate cell wall stress and hyphal signals.
MATERIALS AND METHODS
Strains and media.
Strains were grown on yeast extract-peptone-dextrose (YPD) rich medium (10 g yeast extract [RPI], 20 g peptone [RPI], and 2% glucose [Sigma] in 1 liter of H2O) and spider medium (10 g d-mannitol [Sigma], 10 g nutrient broth [BD Difco], 2 g K2HPO4 [Sigma] in 1 liter of H2O). Caspofungin (Merck) and nourseothricin (Werner Bioagents) were added to media at the concentrations described. Primer sequences are listed in Table S1 in the supplemental material.
Mutant strains were made in the BWP17 background (13) (Table 1). Decreased abundance by mRNA perturbation (DAmP) mutant strains were made by replacing the 3′ untranslated region (UTR) of the respective gene with a plasmid sequence (Fig. 1). Two primers approximately 120 bp in length were designed to amplify the plasmid sequence surrounding the URA3 marker in the pGEM-URA plasmid (13). The 3′-most regions of the primers were designed to amplify the plasmid sequence, while the bulk of the primer sequence was specific to the 3′ region of the gene of interest. Primers jrb599 and jrb600 were used to generate the initial CDC3-DAmP allele. Primers jrb602 and jrb603 were used to generate the initial CDC12-DAmP allele. These insertions were selected for on synthetic complete (SC)-Ura agar plates and checked by colony PCR. Primers jrb493 and jrb513 were used to detect the DAmP insertion for CDC3, and primers jrb496 and jrb513 were used to detect the DAmP insertion for CDC12. We replaced the URA3 gene in the DAmP allele with a UAU1 cassette, which contains two overlapping fragments of the URA3 gene surrounding an intact ARG4 gene (Fig. 1B). This construct was used to render the DAmP alleles homozygous by low-frequency mitotic-recombination events. Construction of the homozygous DAmP strains followed previously established methods (14). Briefly, putative Arg+ transformants were selected on SC–Arg-plus-uridine media and checked by colony PCR. Positive colonies were grown for 24 h in YPD liquid medium and patched onto SC-Arg-Ura plates. Putative homozygous DAmP strains were tested by colony PCR to ensure the absence of a wild-type (WT) band and the presence of transformed alleles. Primers jrb493/jrb601 (CDC3) and jrb496/jrb604 (CDC12) were used to detect the wild-type bands for the respective genes. The primer sequences are listed in Table S1 in the supplemental material.
TABLE 1.
Strains used in this study
| Strain | Genotype | Source or reference |
|---|---|---|
| BWP17 | ura3/ura3 arg4/arg4 his1/his1 | 13 |
| CW412 | ura3/ura3 arg4/arg4 CDC12-HIS1/his1 CDC12-tUAU1/CDC12-tURA3 | This study |
| DAY185 | ura3/ura3::URA3 arg4/arg4::ARG4 his1/his1::HIS1 | 23 |
| JRB217 | ura3/ura3/URA3 arg4/arg4/UAU1 his1/his1 SEP7-GFP::HIS1/SEP7 | This study |
| JRB271 | ura3/ura3 arg4/arg4 his1/his1 cdc10::ARG4/cdc10::URA3 | This study |
| JRB363 | ura3/ura3 arg4/arg4 his1/his1 cdc10::ARG4/cdc10::URA3 SEP7/SEP7-GFP-HIS1 | This study |
| JRB412 | ura3/ura3 arg4/arg4 his1/HIS1 CDC3-tUAU1/CDC3-tURA3 | This study |
| JRB419 | ura3/ura3 arg4/arg4 HIS1/his1 CDC12-tUAU1/CDC12-tURA3 | This study |
| JRB421 | ura3/ura3 arg4/arg4 CDC3-HIS1/his1 CDC3-tUAU1/CDC3-tURA3 | This study |
| JRB461 | ura3/ura3 arg4/arg4 SEP7-GFP::HIS1-his1/his1 CDC3-tUAU1/CDC3-tURA3 | This study |
| JRB467 | ura3/ura3 arg4/arg4 SEP7-GFP::HIS1-his1/his1 CDC12-tUAU1/CDC12-tURA3 | This study |
| JRB470 | pTDH3-TPK1 ura3/ura3::URA3 arg4/arg4::ARG4 his1/his1::HIS1 | This study |
| JRB473 | pTDH3-TPK1 ura3/ura3 arg4/arg4 his1/HIS1 CDC3-tUAU1/CDC3-tURA3 | This study |
| JRB483 | pTDH3-TPK1 ura3/ura3 arg4/arg4 his1/HIS1 CDC12-tUAU1/CDC12-tURA3 | This study |
| JRB485 | pTDH3-EFG1 ura3/ura3::URA3 arg4/arg4::ARG4 his1/his1::HIS1 | This study |
| JRB490 | pTDH3-EFG1 ura3/ura3 arg4/arg4 his1/HIS1 CDC3-tUAU1/CDC3-tURA3 | This study |
FIG 1.
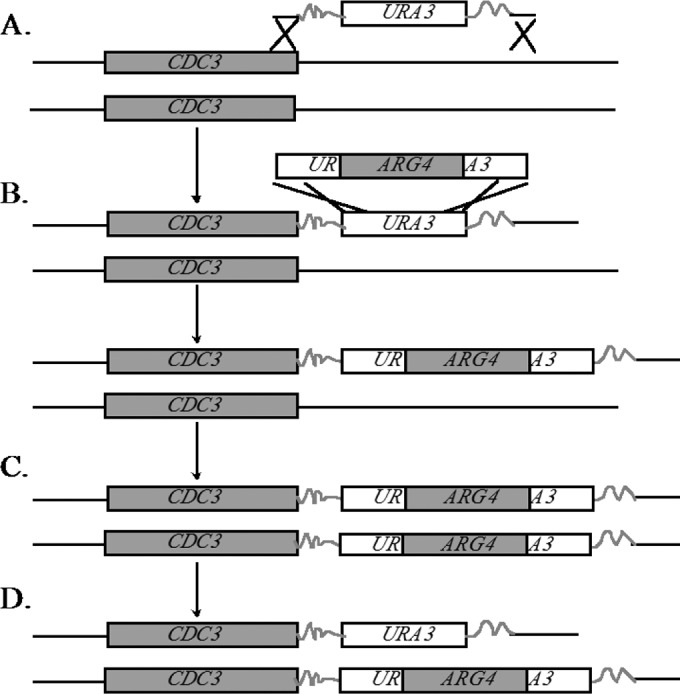
DAmP mutational strategy. (A) A URA3 construct was amplified with primers containing a sequence homologous to the 3′ end of CDC3. This construct was transformed into C. albicans strain BWP17, and positive transformants were selected on SC-Ura agar plates and confirmed by PCR. (B) The URA3 marker was replaced with a UAU1 cassette containing a fully functional ARG4 marker flanked by overlapping portions of the URA3 gene, yielding an Arg+ Ura− strain. (C) Via low-frequency mitotic recombination, homozygosis of the mutation can occur. Because the markers are the same, this event cannot be selected for. (D) Another mitotic-recombination event in one CDC3 allele between the overlapping regions of URA3 loops out the ARG4 marker and yields a strain that can be selected for on SC-Arg-Ura medium. The resulting strains were confirmed by PCR.
To complement the mutant strains, a fragment of DNA 800 to 1,000 bp upstream and 200 to 350 bp downstream of the respective gene was amplified from BWP17 genomic DNA. Primers for CDC3 complementation (jrb471/jrb472) were 60 to 70 bp in length, and the 3′ 20 to 25 bp was gene specific. A 40-mer sequence was tacked onto the 5′ end of each primer sequence to guide in vivo recombination into the pDDB78 plasmid (15). The amplified complementation fragment was cotransformed into the S. cerevisiae BY4741 Δtrp1 strain (from the yeast deletion library [16]) with an EcoRI/NotI-linearized pDDB78 plasmid, and the resulting complementation clone was amplified in Escherichia coli. The primers for CDC12 complementation (jrb666/jrb667) had restriction sites ClaI (jrb666) and NotI (jrb667) added to the 5′ end. The CDC12 sequence was amplified by PCR, digested with ClaI and NotI, and ligated to a ClaI/NotI-linearized pDDB78 plasmid. Each complement clone was digested with NruI to target insertion to the HIS1 locus and transformed into the appropriate mutant strains, complementing the mutation and rendering the strains His+, as well. Complementation was checked by colony PCR using primers jrb493/jrb601 (CDC3) and jrb496/jrb604 (CDC12). pDDB78, digested with NruI, was transformed into the same mutant strains to generate prototrophic, marker-matched strains for comparison with the complemented strains.
To express a SEP7-GFP-tagged strain, plasmid pJRB103 (9) was digested with BclI (New England BioLabs), and the resulting linearized vector was transformed into his1 auxotrophic strains as indicated in the strain table (Table 1). Transformants were selected on media lacking histidine, and putative insertion mutant strains were checked by colony PCR using primers jrb388/jrb391.
Overexpression of TPK1 and EFG1 was accomplished by replacing the native promoter of one allele of each gene with a TDH3 promoter. A NAT-marked TDH3 promoter construct was amplified from pCJN542 using ∼120-mer oligonucleotides. Twenty base pairs of the 3′ ends of these oligonucleotides was designed to amplify the construct, while the remaining 100 bp was gene specific, designed to guide in vivo recombination in C. albicans. Primers jrb656/jrb657 were used to direct the insertion of the TDH3 promoter upstream of TPK1. Primers SF1501/SF1502 were used to direct the insertion of the TDH3 promoter upstream of EFG1. These promoter replacement transformants were selected on YPD agar with 400 μg/ml nourseothricin. Putative positive transformants were tested by colony PCR (TPK1 primers jrb662/jrb19 and EFG1 primers jrb670/jrb19) for the insertion of the overexpression construct upstream of the intended gene.
Sensitivity and hyphal-growth assays.
For testing sensitivity to caspofungin, strains were grown overnight in liquid YPD medium at 30°C and then diluted to an optical density at 600 nm (OD600) of 3 in H2O. Five-fold dilutions of this stock were plated onto YPD or YPD plus 125 ng/ml caspofungin agar. The plates were incubated for 48 h at 30°C and digitally photographed.
For hyphal-growth assays, 20 μl of the final (least dense) dilution from the sensitivity dilution stocks mentioned above was plated onto spider agar plates. The plates were incubated at 37°C for 4 to 5 days. Colony edges were imaged using a Nikon Eclipse TS100 with a 10× 0.25-numerical-aperture (NA) Ph1 objective. Representative images are shown.
Septin fluorescence microscopy.
SEP7-GFP cells were grown overnight and diluted to an OD600 of ∼0.5 in YPD medium. Two hundred microliters of this culture was pipetted onto glass bottom dishes (MatTek Corporation, Ashland, MA) coated with concanavalin A. The dishes were washed twice with phosphate-buffered saline (PBS) and resuspended with PBS for immediate visualization. The cells were visualized with a Zeiss Axio Observer Z.1 fluorescence microscope and either a 100× 1.4-NA or a 63× objective. Fluorescent images were acquired with an exposure time of 0.5 s on a Coolsnap HQ2 (Photometrics) camera using AxioVision software (Zeiss) or on an AxioCam digital monochrome camera (Zeiss) using Zen pro software (Zeiss). Fluorescence measurements were calculated using ImageJ software (NIH). The average fluorescence intensities within the mother and daughter cells (roughly equivalent measurements) were compared to peak intensity values within the bud neck. Mother/daughter cell averages and bud neck values were subtracted from the average background intensity for each image. P values for fluorescence differences between CDC3-DAmP, CDC12-DAmP, and cdc10Δ/Δ cells and the wild type were <1 × 10−13.
RNA extraction.
Overnight cultures of cells were diluted to an OD600 of 0.2 in 50 ml fresh YPD medium. The cultures were allowed to grow at 30°C with shaking until the culture density reached an OD600 of ∼1. Cells were then harvested by vacuum filtration and flash frozen in a dry-ice–ethyl alcohol (EtOH) bath. The cells were kept frozen on filters at −80°C until RNA extraction. RNA was extracted using a Qiagen RNeasy kit (Qiagen) following the manufacturer's instructions with the following modifications. The cells were resuspended from filters with 1.5 ml ice-cold distilled H2O (dH2O), followed by 20 s of vigorous vortexing. The resuspended cells were transferred to a 1.5-ml tube and spun down following the manufacturer's protocol. Furthermore, during the cell disruption step, the cells were lysed with a bead beater for 3 min at 4°C to maximize cell lysis.
Gene expression analysis.
The expression of genes in the CDC3-DAmP and CDC12-DAmP mutant strains were analyzed by NanoString and quantitative reverse transcription-PCR (qRT-PCR) analysis (described below). They were compared to the expression of protein kinase mutant strains exhibiting hypersensitivity to caspofungin (cla4, hsl1, kin3, swe1, tpk1, yck2, yck3, prk1, ssn3, and hst7) that were analyzed by microarray (described below).
NanoString.
NanoString analysis is a sensitive method for analyzing gene expression of 100 to 800 targets at a time. For our analysis, 295 targets associated with the cell wall and with biofilm formation were selected for analysis. Approximately 100 ng of C. albicans total RNA was mixed with the NanoString probe set (NanoString Technologies, Seattle, WA) and incubated at 65°C overnight. The reaction mixture was then loaded on the NanoString nCounter Prep Station for binding and washing, using the default program. The resultant cartridge was then transferred to the NanoString nCounter digital analyzer for scanning and data collection. A total of 600 fields were captured per sample. The raw data were first adjusted for binding efficiency and background subtraction, following nCounter data analysis guidelines. Five control genes, ACT1, TDH3, YRA1, ARP3, and orf19.7235, were used to normalize gene expression.
qRT-PCR.
Ten micrograms of total RNA was treated with a DNA-free kit (Ambion), followed by first-strand cDNA synthesis from half of the DNA-free RNA using iScript Reverse Transcription Supermix (Bio-Rad). Absence of DNA contamination was confirmed using control sets for which reverse transcriptase was omitted from the cDNA reaction.
Primer3 software (http://frodo.wi.mit.edu/) was used to design primers for CDC3, CDC12, orf19.1862, orf19.251, orf19.7310, and HSP12. Additional primer sets used in these studies were generated in previous studies. The primers are listed in Table S1 in the supplemental material. iQ SYBR green Supermix (2×; Bio-Rad), 1 μl of first-strand cDNA reaction mixture, and 0.1 μM primers were mixed in a total volume of 50 μl per reaction, and real-time PCR was performed in triplicate for each sample on an iCycler iQ real-time PCR detection system (Bio-Rad). The program for amplification had an initial denaturation step at 95°C for 5 min, followed by 40 cycles of 95°C for 45 s and 58°C for 30 s. Product amplification was detected using SYBR green fluorescence during the 58°C step, and the specificity of the reaction was monitored by melting curve analysis following the real-time program. TDH3 was used as a reference gene for normalization of gene expression, which was determined using Bio-Rad iQ5 software (the ΔΔCT method).
Microarray analysis.
Microarray analysis was performed on C. albicans Affymetrix NimbleExpress arrays (CAN04a530004N) as previously described (17). Strains containing mutations in protein kinases were compared to a wild-type marker-matched strain (DAY286). The strains were generated previously (9) and are identified in Table S4 in the supplemental material.
Virulence assays.
Seven-week-old male ICR mice (Harlan-Sprague) were inoculated by intravenous injection in the lateral tail vein with 1 × 106 cells of C. albicans strains in 0.2 ml of normal saline. The mice were followed until they were moribund, at which point they were sacrificed. Survival curves were calculated according to the Kaplan-Meier method using the PRISM program (GraphPad Software) and compared using the log-rank (Mantel-Cox) test. A P value of <0.05 was considered significant. This murine study was approved by the University of Pittsburgh Institutional Animal Care and Use Committee and performed according to its guidelines.
RESULTS
DAmP alleles of essential septins cause reduced septin accumulation at the bud neck.
Studies with mutants with mutations in the nonessential septin genes CDC10 and CDC11 in C. albicans have demonstrated that the septin ring is vital for CWR (9), hyphal morphogenesis (7), and pathogenesis (18). It has been assumed that the septin ring is weakened in cdc10Δ/Δ and cdc11Δ/Δ mutant cells and that deletions of these septins exhibit a mild version of a complete loss of the septin ring. While we believe that this may be true for certain functions, we believe that the essential septins have additional roles in CWR and morphogenesis that can be uncovered only by studying mutations in these septins.
Septin proteins are stable over multiple cell generations (6), with localization cues driven by posttranslational modifications. This stability renders traditional approaches to study C. albicans essential gene function, such as promoter substitutions, difficult to assess. Therefore, a DAmP strategy was used to mutate CDC3 and CDC12 in C. albicans. In this approach, the 3′ UTRs of both alleles of CDC3 and CDC12 were replaced with foreign non-UTR sequence, removing the polyadenylation addition sites. Replacement of the 3′ UTR with vector sequence should destabilize the RNA by preventing polyadenylation. Accordingly, we found that removal of this sequence leads to a decrease in RNA accumulation of the targeted septin (Fig. 2A). The RNA levels of CDC3 were not altered in a CDC12-DAmP strain, and the RNA levels of CDC12 were not altered in a CDC3-DAmP mutant strain (see Fig. S1A in the supplemental material), nor did the septin DAmP mutations significantly affect the RNA levels of other mitotic septins (see Fig. S1B in the supplemental material). This demonstrates that reduction of expression of a single septin does not affect the regulation of other septins and that the phenotypes we observe are due to the reduced expression of the targeted septin.
FIG 2.
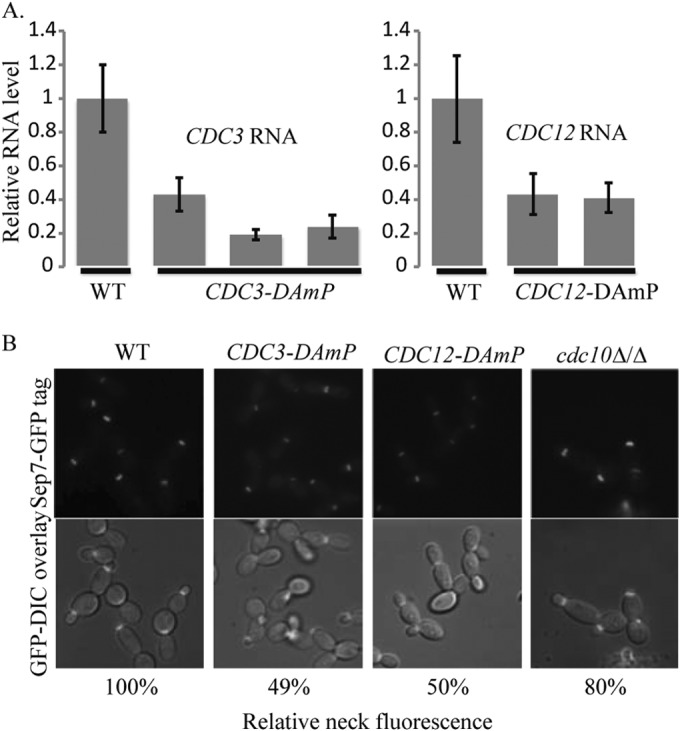
Septin DAmP alleles reduce septin gene expression and septin accumulation at the bud neck. (A) The expression of CDC3 or CDC12 in yeast cells was tested in three independently isolated CDC3-DAmP strains (JRB412, JRB413, and JRB415, from left to right in the left graph) and two independently isolated CDC12-DAmP mutant strains (JRB418 and JRB419, from left to right in the right graph). Expression in the mutant strains was normalized to expression in a marker-matched wild-type control, DAY185 (WT). (B) The localization and fluorescence intensity of Sep7-GFP were measured in CDC3-DAmP (JRB461; 159 cells), CDC12-DAmP (JRB467; 131 cells), and cdc10Δ/Δ (JRB363; 152 cells) mutant yeast cells. Fluorescence levels from a Sep7-GFP-tagged marker-matched wild-type strain (JRB217; 78 cells) (WT) were used to normalize mutant values. P values between the marker-matched control and the mutant cells were highly significantly different (P < 1 × 10−17 for CDC3-DAmP, P < 1 × 10−16 for CDC12-DAmP, and P < 1 × 10−14 for the cdc10Δ/Δ strain), as were the values between the CDC3-DAmP/CDC12-DAmP and the cdc10Δ/Δ mutant cells (P < 1 × 10−61 and P < 1 × 10−36, respectively).
A SEP7-GFP fusion gene was introduced into the DAmP septin mutant strains in order to assess septin ring formation. We compared the new DAmP mutant strains to SEP7-GFP-containing cdc10Δ/Δ and WT strains. Localization of the septin ring in yeast cells was not affected by the deletion of CDC10 or by the DAmP mutations in CDC3 and CDC12 (Fig. 2B). However, the fluorescence intensity of Sep7-GFP at the bud neck was reduced by approximately 50% in both the CDC3-DAmP and the CDC12-DAmP strains compared to the wild type, suggesting that the septin ring was physically affected by reduced levels of essential septins (Fig. 2B; see Table S2 in the supplemental material). This reduction of fluorescence was not as severe in the cdc10Δ/Δ strain (∼37% reduction in fluorescence), suggesting that the DAmP alleles of the essential septin genes cause a more severe defect and might allow us to uncover novel septin functions.
DAmP septins have hyphal-growth and CWR defects.
Septins are important for both hyphal growth and CWR, although their specific roles in these processes are poorly defined. Based on the reduction of septin accumulation at the neck observed in the septin DAmP mutant strains, we anticipated that these strains would also be defective in hyphal growth and CWR. To test for CWR defects in the septin DAmP mutant strains, we utilized a drug, caspofungin, that targets the enzyme responsible for producing the β-1,3-glucan, a major component of the cell wall. This drug has devastating effects on the cell wall of the fungus and is currently used as a clinical antifungal drug. Mutant and complemented strains were tested for the ability to grow in the presence of 125 ng/ml caspofungin. Compared to the wild-type and complemented strains, the DAmP mutant strains displayed hypersensitivity to caspofungin, indicating that the reduction in septins at the bud neck does indeed have an impact on CWR (Fig. 3A).
FIG 3.
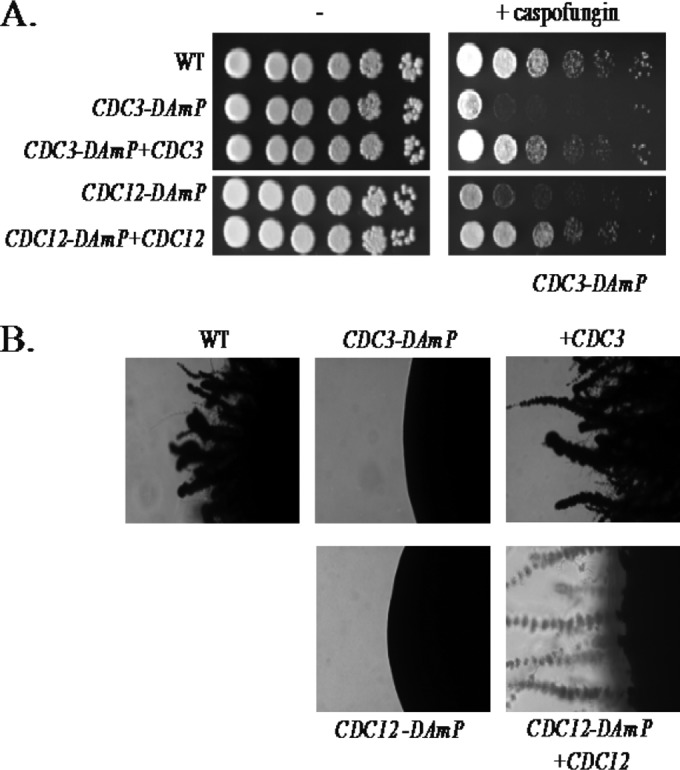
Septin DAmP alleles are hypersensitive to cell wall stress and defective for hyphal development on solid surfaces. (A) Growth of a marker-matched wild-type strain (DAY185) was compared to that of CDC3-DAmP (JRB412), CDC3-DAmP+CDC3 (JRB421), CDC12-DAmP (JRB419), and CDC12-DAmP+CDC12 (CW412) mutant strains in the presence (+ caspofungin) and absence (−) of 125 ng/ml caspofungin. The strains were allowed to grow for 2 days at 30°C and imaged. The spots represent 5-fold dilutions from left to right. (B) The strains indicated for panel A were spotted onto spider agar plates and incubated at 37°C for 6 days. Images of the edges of representative single colonies are shown.
There are a number of signals that induce C. albicans to switch from yeast-like growth to hyphal growth. High temperatures, low pH, nutrient starvation, contact with a surface, and the presence of serum can all induce hyphal growth. Mutants with deletions of nonessential septins display a modest hyphal defect under liquid induction conditions but are very defective in hyphal formation on a solid surface (7). DAmP mutants of essential septins also displayed a moderate hyphal defect in liquid media (data not shown) and were very defective in hyphal formation on a solid surface (Fig. 3B; see Fig. S2 in the supplemental material). Complementation restored normal hyphal growth to the septin DAmP mutants.
Transcriptional profiles of DAmP septins reveal a connection with the cyclic AMP (cAMP) pathway.
Transcriptional profiles of 300 genes in CDC3-DAmP, CDC12-DAmP, and cdc10Δ/Δ strains were measured with NanoString technology in the absence of stress. The 300 genes tested were selected largely for putative roles in cell wall biogenesis or for their presence in the cell wall, along with several control genes. Profiles were performed in triplicate for each mutation with independent mutant strains when available. While there was significant overlap between CDC3-DAmP and CDC12-DAmP homozygote mutant profiles, the cdc10Δ/Δ mutation had little to no impact on gene expression (see Table S3 in the supplemental material), consistent with preliminary microarray and qRT-PCR analyses of the strain (data not shown). Complementation of the CDC3-DAmP mutation with a wild-type copy of CDC3 at the HIS1 locus restored wild-type levels of gene expression to the subset of interesting genes from the NanoString data set that we tested by RT-PCR (Table 2).
TABLE 2.
Complementation of a DAmP mutant restores gene expression
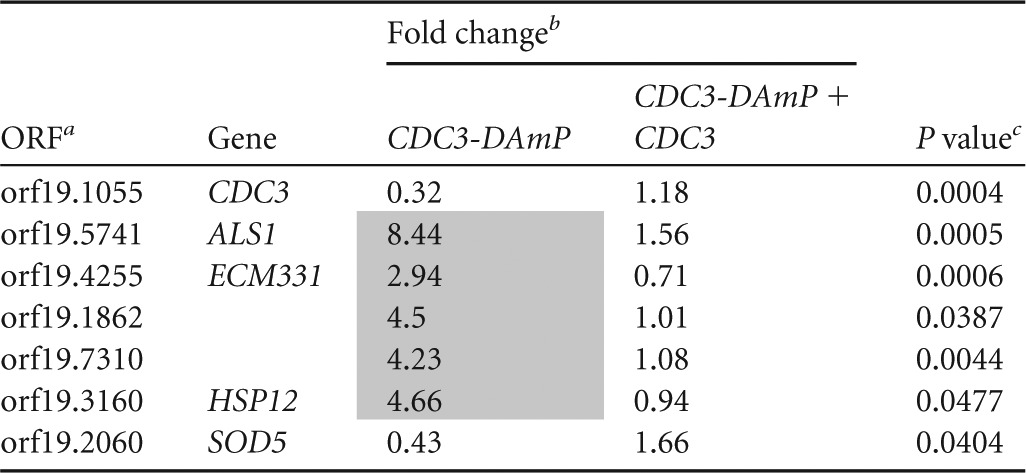
ORF, open reading frame.
The data represent the fold change normalized to a marker-matched wild-type control strain (DAY185). Upregulated genes are shaded.
P values were measured by t test comparisons of mutant and complemented strains.
The transcriptional profile of the CDC3-DAmP and CDC12-DAmP homozygote mutant strains was compared to preliminary microarray data for 11 protein kinase mutant strains (see Table S4 in the supplemental material). The protein kinase mutations were homozygous insertion mutations, and these mutant strains, like the septin mutant strains, were also hypersensitive to caspofungin (9). Eleven mutant strains were tested, i.e., those with mutations in cla4, hsl1, ire1, kin3, swe1, tpk1, yck2, yck3, prk1, ssn3, and hst7. Of the mutant strains tested, only the tpk1−/− strain showed a sizeable overlap with the septin DAmP mutants. Like the CDC3-DAmP and CDC12-DAmP mutations, few genes were impacted by the tpk1−/− mutation, but the few genes that were affected generally overlapped. In all, approximately 58% of the genes upregulated in the tpk1−/− mutant were also upregulated in the CDC3-DAmP and CDC12-DAmP mutant strains (Table 3), suggesting some overlap in function between septins and the cAMP pathway.
TABLE 3.
Comparison of gene expression levels between CDC3-DAmP, CDC12-DAmP, and tpk1−/− strains
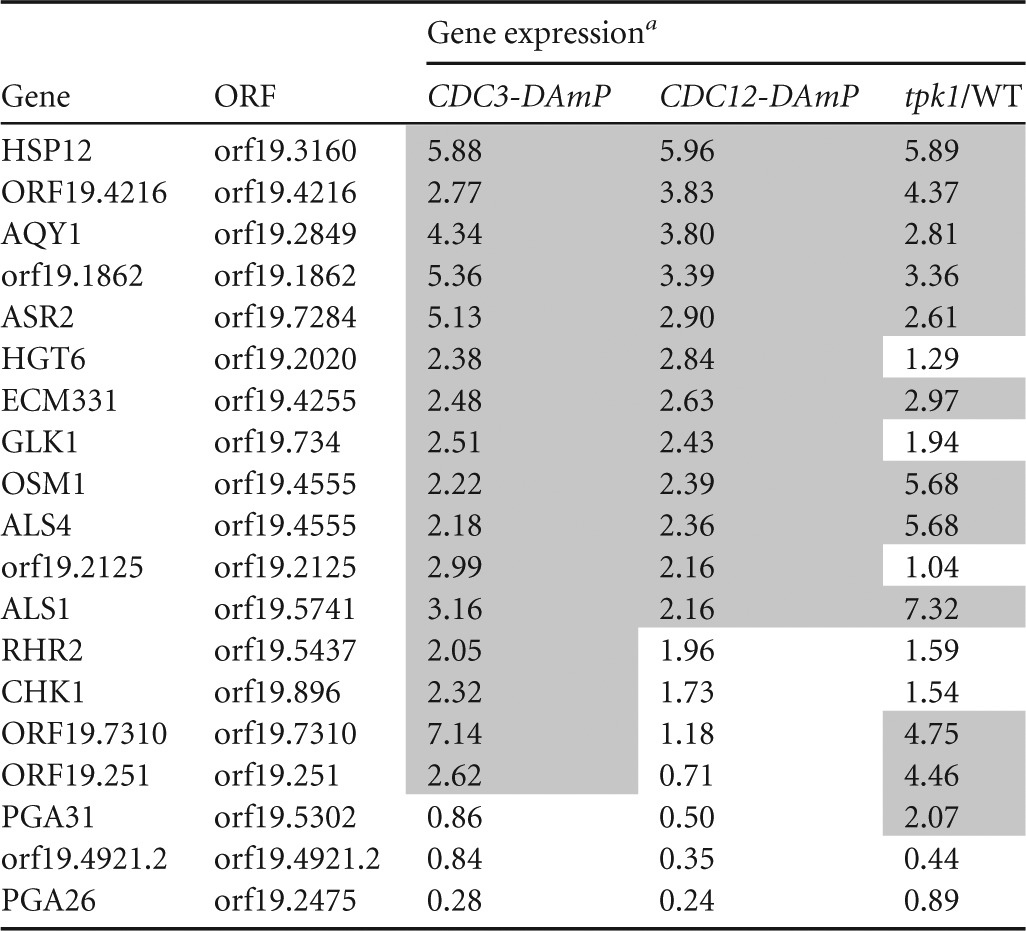
Gene expression is fold change and normalized to a marker-matched wild-type control (DAY185). Upregulated genes are shaded.
Overexpression of TPK1 and EFG1 restores hyphal growth and virulence to septin DAmP strains.
Tpk1 is one of two protein kinase A subunits in the cyclic AMP pathway, a conserved pathway in eukaryotes that generally serves as a signal transduction pathway sensing nutrient conditions. Like septins, Tpk1 has a role in filamentation on solid surfaces and in CWR, while Tpk2 is involved in filamentation in liquid media (9, 19, 20). Our hypothesis, based on our transcriptional-profiling data, was that the cAMP pathway either acts upstream of septins or lies downstream of septin-mediated signaling. We had previously shown that septin localization was unaffected by the tpk1−/− mutation (9), and thus, the latter hypothesis seemed most likely. If true, overexpression of components of the cAMP pathway should restore wild-type hyphal growth and CWR to septin DAmP strains. Therefore, we replaced the promoter of one allele of the protein kinase gene TPK1 or the transcription factor gene EFG1, a downstream target of Tpk1, with the promoter for the highly expressed gene TDH3 in the CDC3-DAmP, CDC12-DAmP, and wild-type strains. These strains were then tested for sensitivity to caspofungin and for the ability to form hyphae on spider agar at 37°C. As hypothesized, overexpression of EFG1 or TPK1 restored hyphal formation to the septin DAmP strains (Fig. 4A and data not shown). However, overexpression of EFG1 or TPK1 did not restore wild-type caspofungin sensitivity to these strains (data not shown). This result is the first indication we have had that the roles of septins in hyphal formation and CWR can be genetically separated.
FIG 4.
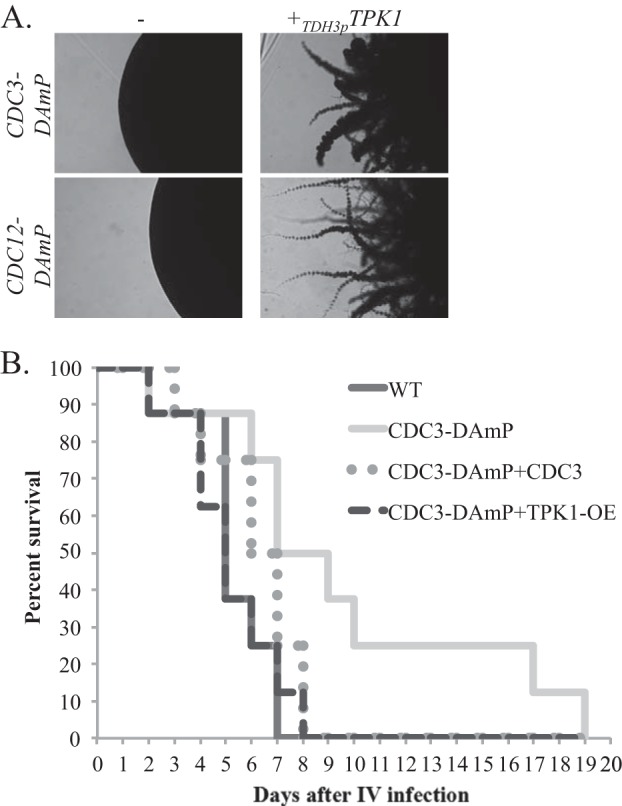
Overexpression of TPK1 and EFG1 restores hyphal growth and virulence defects of a CDC3-DAmP mutant strain. (A) CDC3-DAmP (JRB412), CDC12-DAmP (JRB418), and septin DAmP strains overexpressing TPK1 were spotted onto spider agar plates and incubated at 37°C for 6 days. Images of the edges of representative single colonies are shown. (B) A marker-matched wild-type strain (DAY185) and CDC3-DAmP (JRB412), CDC3-DAmP+CDC3 (JRB421), and CDC3-DAmP+TPK1-OE (JRB473) strains were injected into the tail veins of 8 mice each. Survival of infected strains was recorded daily. The time to death was significantly delayed for mice infected with the CDC3-DAmP strain (P = 0.017) but was not significantly different in the mice infected with the wild-type, CDC3-DAmP+CDC3 (P = 0.18), or CDC3-DAmP+TPK1-OE (P = 1.0) strain.
Hyphal development is a key component of C. albicans virulence, and deletion of nonessential septins causes attenuated virulence in the fungus, likely due to the defective hyphal formation in these strains (18). Like the mutations in nonessential septins, a DAmP mutation in the essential CDC3 septin gene led to attenuated virulence in a murine systemic-infection model (Fig. 4B). Complementation with a functional copy of CDC3 restored wild-type virulence to CDC3-DAmP mutant cells. Additionally, overexpression of TPK1 also restores wild-type virulence in CDC3-DAmP mutant cells, strongly suggesting that the virulence defect of septin mutant strains is mediated through targets held in common with the cAMP pathway.
DISCUSSION
Septins have long been considered a structural element of the cell and indeed have been called “the fourth component of the cytoskeleton” (21). Our previous work and work by others suggest that septins are more than simple scaffolding structures for cell separation. Septins are dispersed from the bud neck when the cell wall is stressed, and they appear to pull functional cell wall biosynthesis machinery with them when they move (9, 22). This response may be part of the normal cellular defense against cell wall stress, and at least portions of this response are conserved in S. cerevisiae. The function of septins in C. albicans hyphae appears to be somewhat distinct from their roles in yeast cell division. Septins persist at sites of previous division in hyphae well after a septum has been laid between the filament segments, and their role at this stage is completely unknown (8). We sought to further elucidate the roles of septins in cell wall stress and hyphal development by analyzing the roles of essential septins in these processes.
The septin ring is an essential structure in dividing cells, and the essential nature of the ring has made genetic analysis of septin function difficult. Analysis of nonessential components of the septin ring, encoded by CDC10, CDC11, and SEP7, have been informative in elucidating phenotypes associated with septin ring defects, including hyphal defects, virulence defects, and CWR defects. Expression analysis of these mutant strains, however, demonstrates no evidence of defects affecting gene expression, nor does the septin ring itself appear to be strongly affected by their loss in C. albicans (Fig. 2B). We used a different approach, decreased abundance by mRNA perturbation, to analyze the effect that mutating an essential septin might have on the cellular phenotype and gene expression.
The homozygous CDC3-DAmP and CDC12-DAmP strains shared some phenotypes with deletion mutants of nonessential septins, suggesting that the DAmP approach is a valid method for analysis of essential gene function in C. albicans. Both septin DAmP mutant strains had hyphal-development, CWR, and virulence defects, similar to strains with mutations in nonessential septins. Unlike the nonessential-septin mutants, however, the DAmP mutant strains demonstrated a stark decrease of septin accumulation at the septin ring, as well as a transcriptional response to the mutation. These results are consistent with the rationale that impairment of essential septins might reveal new biological functions not evident from the nonessential-septin mutants.
We used transcriptional analysis to identify pathways that might act in parallel with septins for their roles in CWR or hyphal formation. The transcriptional profiles of CDC3-DAmP and CDC12-DAmP mutant strains were very similar to each other yet distinct from those of other DAmP mutant strains (data not shown), suggesting that this profile is septin specific and not caused by our mutational approach. Furthermore, the profile did not overlap the cdc10Δ/Δ mutant profile, suggesting the DAmP mutations were more severe than a deletion of a nonessential septin (see Table S3 in the supplemental material). Further evidence that this expression profile is septin specific is the observation that complementation of the DAmP strains with a wild-type copy of the mutated septin restores the wild-type expression profile. Using expression data from strains with similar phenotypes, we identified an overlap between the septin DAmP profile and the tpk1−/− profile.
tpk1 encodes one of two PKA proteins in C. albicans that are an integral part of the cAMP signaling pathway. We hypothesized that septins were either being regulated in part by the cAMP pathway or that septins were acting upstream of some component in the cAMP pathway. Our evidence suggests that septin localization is not regulated by the cAMP pathway but that a pathway acting downstream of septins likely acts in parallel with the cAMP pathway. One possibility for this pathway is the Hsl1/Swe1 pathway, which acts downstream of septins to regulate the cell cycle in response to the state of septins at the bud neck. It is the only pathway that has thus far been shown to act downstream of septins. However, neither Hsl1 nor Swe1 mutant strains are defective in hyphal formation (22), suggesting that another, yet to be characterized pathway is acting downstream of septins to trigger hyphal growth in liquid media.
The transcriptional-profile similarities between tpk1−/− and septin DAmP mutant strains suggests that a genetic relationship exists between septins and the cAMP pathway. We tested two hypotheses, that septin stability at the neck is affected by the cAMP pathway and that a septin signal feeds into the cAMP pathway upstream of cAMP, neither of which was supported by the results of our experiments. Septin stability at the bud neck in septin DAmP strains was unaffected by the overexpression of TPK1, and cAMP levels in yeast cells were at wild-type levels in a septin DAmP strain (data not shown). These data suggest that the cAMP pathway and a pathway functioning downstream of septins in hyphal formation act in parallel to regulate hyphal development and virulence in C. albicans.
Our work suggests that distinct pathways act in response to septins during cell wall damage and hyphal growth. Overexpression of TPK1 and EFG1 was sufficient to complement the hyphal-formation defect in septin DAmP mutant strains but did not restore wild-type sensitivity to cell wall damage. This suggests that the cellular responses downstream of septins in these two processes are distinct. Whether these pathways respond to septin changes or are activated by differentially regulated septins themselves remains to be seen. It is clear that the tools we have generated will provide us with the opportunity to identify novel pathways acting downstream of septins in distinct settings.
Supplementary Material
ACKNOWLEDGMENTS
We thank the members of our laboratories for their advice and discussions. We particularly thank Tatyana Aleynikova for her technical support. We are also grateful to Merck Research Laboratories for providing caspofungin.
This work was supported by NIH program grant P20GM103427 (J.R.B.), NIH postdoctoral fellowship F32AI071439 (J.R.B.), a National University of Ireland Traveling Studentship (S.F.), and NIH grant 5R01AI057804 (A.P.M.).
Footnotes
Published ahead of print 12 September 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/EC.00127-14.
REFERENCES
- 1.Pfaller MA, Diekema DJ. 2007. Epidemiology of invasive candidiasis: a persistent public health problem. Clin. Microbiol. Rev. 20:133–163. 10.1128/CMR.00029-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lo H-J, Köhler JR, DiDomenico B, Loebenberg D, Cacciapuoti A, Fink GR. 1997. Nonfilamentous C. albicans mutants are avirulent. Cell 90:939–949. 10.1016/S0092-8674(00)80358-X. [DOI] [PubMed] [Google Scholar]
- 3.Saville SP, Lazzell AL, Monteagudo C, Lopez-Ribot JL. 2003. Engineered control of cell morphology in vivo reveals distinct roles for yeast and filamentous forms of Candida albicans during infection. Eukaryot. Cell 2:1053–1060. 10.1128/EC.2.5.1053-1060.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hernandez-Rodriguez Y, Momany M. 2012. Posttranslational modifications and assembly of septin heteropolymers and higher-order structures. Curr. Opin. Microbiol. 15:660–668. 10.1016/j.mib.2012.09.007. [DOI] [PubMed] [Google Scholar]
- 5.Longtine MS, Bi E. 2003. Regulation of septin organization and function in yeast. Trends Cell Biol. 13:403–409. 10.1016/S0962-8924(03)00151-X. [DOI] [PubMed] [Google Scholar]
- 6.McMurray MA, Thorner J. 2008. Septin stability and recycling during dynamic structural transitions in cell division and development. Curr. Biol. 18:1203–1208. 10.1016/j.cub.2008.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Warenda A, Konopka J. 2002. Septin function in Candida albicans morphogenesis. Mol. Biol. Cell 13:2732–2746. 10.1091/mbc.E02-01-0013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gonzalez-Novo A, Correa-Bordes J, Labrador L, Sanchez M, Vazquez de Aldana CR, Jimenez J. 2008. Sep7 is essential to modify septin ring dynamics and inhibit cell separation during Candida albicans hyphal growth. Mol. Biol. Cell 19:1509–1518. 10.1091/mbc.E07-09-0876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blankenship JR, Fanning S, Hamaker JJ, Mitchell AP. 2010. An extensive circuitry for cell wall regulation in Candida albicans. PLoS Pathog. 6:e1000752. 10.1371/journal.ppat.1000752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li L, Zhang C, Konopka JB. 2012. A Candida albicans temperature-sensitive cdc12-6 mutant identifies roles for septins in selection of sites of germ tube formation and hyphal morphogenesis. Eukaryot. Cell 11:1210–1218. 10.1128/EC.00216-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li CR, Yong JY, Wang YM, Wang Y. 2012. CDK regulates septin organization through cell-cycle-dependent phosphorylation of the Nim1-related kinase Gin4. J. Cell Sci. 125:2533–2543. 10.1242/jcs.104497. [DOI] [PubMed] [Google Scholar]
- 12.Wang Y. 2009. CDKs and the yeast-hyphal decision. Curr. Opin. Microbiol. 12:644–649. 10.1016/j.mib.2009.09.002. [DOI] [PubMed] [Google Scholar]
- 13.Wilson RB, Davis D, Mitchell AP. 1999. Rapid hypothesis testing with Candida albicans through gene disruption with short homology regions. J. Bacteriol. 181:1868–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Enloe B, Diamond A, Mitchell AP. 2000. A single-transformation gene function test in diploid Candida albicans. J. Bacteriol. 182:5730–5736. 10.1128/JB.182.20.5730-5736.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Spreghini E, Davis DA, Subaran R, Kim M, Mitchell AP. 2003. Roles of Candida albicans Dfg5p and Dcw1p cell surface proteins in growth and hypha formation. Eukaryot. Cell 2:746–755. 10.1128/EC.2.4.746-755.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Winzeler EA, Shoemaker DD, Astromoff A, Liang H, Anderson K, Andre B, Bangham R, Benito R, Boeke JD, Bussey H, Chu AM, Connelly C, Davis K, Dietrich F, Dow SW, El Bakkoury M, Foury F, Friend SH, Gentalen E, Giaever G, Hegemann JH, Jones T, Laub M, Liao H, Liebundguth N, Lockhart DJ, Lucau-Danila A, Lussier M, M'Rabet N, Menard P, Mittmann M, Pai C, Rebischung C, Revuelta JL, Riles L, Roberts CJ, Ross-MacDonald P, Scherens B, Snyder M, Sookhai-Mahadeo S, Storms RK, Veronneau S, Voet M, Volckaert G, Ward TR, Wysocki R, Yen GS, Yu K, Zimmermann K, Philippsen P, Johnston M, Davis RW. 1999. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285:901–906. 10.1126/science.285.5429.901. [DOI] [PubMed] [Google Scholar]
- 17.Liu TT, Znaidi S, Barker KS, Xu L, Homayouni R, Saidane S, Morschhauser J, Nantel A, Raymond M, Rogers PD. 2007. Genome-wide expression and location analyses of the Candida albicans Tac1p regulon. Eukaryot. Cell 6:2122–2138. 10.1128/EC.00327-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Warenda AJ, Kauffman S, Sherrill TP, Becker JM, Konopka JB. 2003. Candida albicans septin mutants are defective for invasive growth and virulence. Infect. Immun. 71:4045–4051. 10.1128/IAI.71.7.4045-4051.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bockmuhl DP, Krishnamurthy S, Gerads M, Sonneborn A, Ernst JF. 2001. Distinct and redundant roles of the two protein kinase A isoforms Tpk1p and Tpk2p in morphogenesis and growth of Candida albicans. Mol. Microbiol. 42:1243–1257. 10.1046/j.1365-2958.2001.02688.x. [DOI] [PubMed] [Google Scholar]
- 20.Fanning S, Xu W, Beaurepaire C, Suhan JP, Nantel A, Mitchell AP. 2012. Functional control of the Candida albicans cell wall by catalytic protein kinase A subunit Tpk1. Mol. Microbiol. 86:284–302. 10.1111/j.1365-2958.2012.08193.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mostowy S, Cossart P. 2012. Septins: the fourth component of the cytoskeleton. Nat. Rev. Mol. Cell. Biol. 13:183–194. 10.1038/nrm3284. [DOI] [PubMed] [Google Scholar]
- 22.Badrane H, Nguyen MH, Blankenship JR, Cheng S, Hao B, Mitchell AP, Clancy CJ. 2012. Rapid redistribution of phosphatidylinositol-(4,5)-bisphosphate and septins during the Candida albicans response to caspofungin. Antimicrob. Agents Chemother. 56:4614–4624. 10.1128/AAC.00112-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Davis D, Edwards JE, Jr, Mitchell AP, Ibrahim AS. 2000. Candida albicans RIM101 pH response pathway is required for host-pathogen interactions. Infect. Immun. 68:5953–5959. 10.1128/IAI.68.10.5953-5959.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


