Abstract
In many organisms, the geometry of encounter of haploid germ cells is arbitrary. In Saccharomyces cerevisiae, the resulting zygotes have been seen to bud asymmetrically in several directions as they produce diploid progeny. What mechanisms account for the choice of direction, and do the mechanisms directing polarity change over time? Distinct subgroups of cortical “landmark” proteins guide budding by haploid versus diploid cells, both of which require the Bud1/Rsr1 GTPase to link landmarks to actin. We observed that as mating pairs of haploid cells form zygotes, bud site specification progresses through three phases. The first phase follows disassembly and limited scattering of proteins that concentrated at the zone of cell contact, followed by their reassembly to produce a large medial bud. Bud1 is not required for medial placement of the initial bud. The second phase produces a contiguous bud(s) and depends on axial landmarks. As the titer of the Axl1 landmark diminishes, the third phase ultimately redirects budding toward terminal sites and is promoted by bipolar landmarks. Thus, following the initial random encounter that specifies medial budding, sequential spatial choices are orchestrated by the titer of a single cortical determinant that determines whether successive buds will be contiguous to their predecessors.
INTRODUCTION
The relative orientation of each cell in a Saccharomyces cerevisiae mating pair is arbitrary, in the sense that the cells' axes of polarization as they extend toward each other to produce a zygote do not bear a fixed relation to their previous budding. When the resulting zygotes then bud, they produce medial, lateral, or terminal buds (Fig. 1A). The causes of this spatial variability represent a longstanding puzzle. Some, but not all, previous studies have concluded that most initial buds are medial (1–4). In mammalian fertilization, the arbitrary site of sperm entry into the oocyte guides the polarity of initial cleavage. Moreover, in higher organisms, early development often can proceed even if individual cleavage events occur with less than perfect geometry (5).
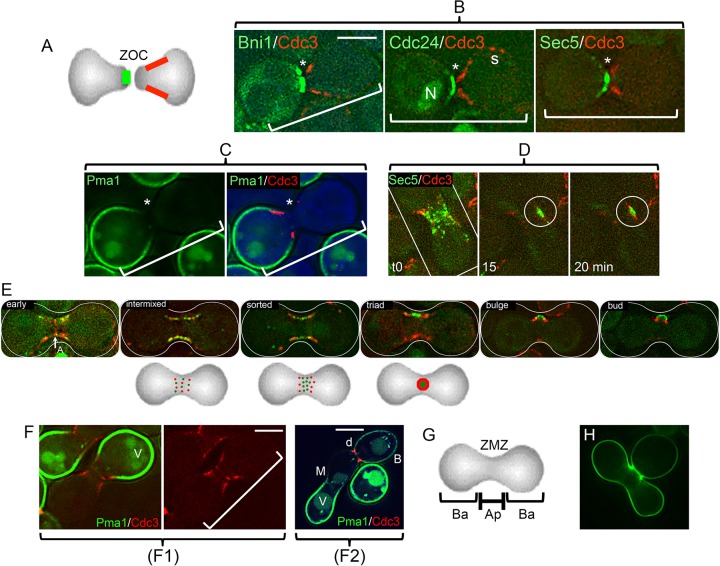
FIG 1.
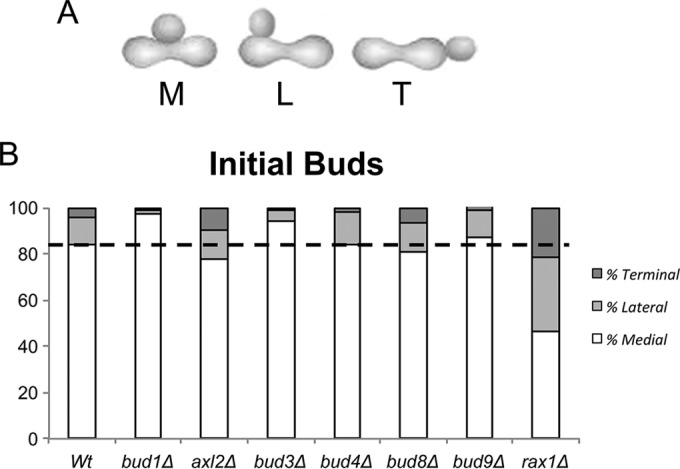
Sites of bud emergence. (A) Zygotes can bud either from the midzone (medial [M]), along the flanking surfaces (lateral [L]), or toward their ends (terminal [T]). The sum of L and T is referred to as “nonmedial.” (B) Landmarks needed for initial bud site specification. Homotypic crosses were conducted, fixed, and counted. Averages of the bud distributions (medial, lateral, and terminal) from at least three independent experiments are plotted in each case. The dashed horizontal line indicates the control value for medial budding as a point of reference. Quantitation is given in Table S2 in the supplemental material. Note that most budding is medial unless Rax1 has been deleted. (We were unable to examine axl1Δ crosses because Axl1 is required for mating. Since rax2Δ strains are prohibitively petite and therefore likely to be genetically unstable, we have not explored the possible significance of the protein.)
Haploid and diploid cells of S. cerevisiae exhibit spatial memory for bud placement. Thus, in the “axial” budding of haploid cells, landmark proteins mark sites of bud emergence and successive bud scars are contiguous with each other, forming unbranched chains. The history of their budding is stably indicated by their circular chitin-rich scars, each of which normally contacts the scar of the preceding and subsequent bud. In contrast, diploid cells usually exhibit “bipolar” budding, with each bud initiating at a polar patch of “landmark” proteins, near which successive bud scars cluster without contacting each other. Daughter cells first bud at the pole that is distal to the mother and then alternate between poles, while the mother cell rebuds at either pole (6–9).
Bud site specification depends on landmark proteins of the cell cortex. Deletion of axial landmarks (Axl1, Axl2/Bud10, Bud3, and Bud4) from haploid cells results in bipolar budding but does not affect budding by diploids. Deletion of bipolar landmarks (Bud8 and Bud9) affects the budding pattern of a/α diploid, but not haploid, cells, causing a “unipolar” appearance, in which scars/buds are clustered at one pole but do not contact each other. Despite their apparently distinct roles, seven of these landmarks are present in both haploid and diploid cells. The exception is Axl1, which is absent from diploid cells. It is also required for production of mating factor a and for cell fusion (10–16) (see Table S1 in the supplemental material). Expression of Axl1 in diploid cells allows them to bud axially (17).
Orientation of actin toward a cortical landmark results in polarized transport of secretory vesicles containing new surface proteins and cell wall components. Such orientation occurs when a landmark protein interacts with the guanine nucleotide exchange factor (GEF) of the transducer GTPase, Bud1/Rsr1. The activated form of Bud1 then interacts with the GEF (Cdc24) of a second GTPase, Cdc42, that guides actin in conjunction with the formin Bni1. Deletion of Bud1 randomizes successive budding in cycling cells (6, 7, 18–21). Related events linking surface determinants to actin are characteristic of many eukaryotes (22).
Like haploid yeast, yeast zygotes can bud at least 20 times (2, 3). As haploid cells bud repeatedly, they accumulate extrachromosomal rDNA circles in the nucleus and exhibit genetic instability, dysfunctional mitochondria, increased vacuolar pH, and accumulation of aggregated and oxidized proteins in the cytoplasm. According to most studies, such molecular burdens are asymmetrically retained by the mother. Age thus can be reset in daughters (23–26). It is not known whether polarity options and mechanisms change during the early stages of development or during replicative aging.
MATERIALS AND METHODS
Cell culture.
Cells were pregrown at room temperature in synthetic medium with glycerol as a carbon source to ensure respiratory competence. Prior to crossing, the cells were shaken for 3 h in fresh glucose-containing medium. The one exception was for experiments in which excess Axl1 was induced. In this case, the cells were pregrown in 2% raffinose and then shifted to 1% raffinose-1% galactose for 3 h before the cross; 1% raffinose-1% galactose was then present during the entire period of zygote formation and budding. Cell growth and experimentation were at room temperature. For experiments in which bud scars were stained, cells were precultured at low density to ensure a minimum of preexisting bud scars.
Almost all experiments were done with cells of S288C background. We have not studied rax2Δ strains because we find that they do not grow on glycerol.
Strain construction.
Standard yeast genetic procedures were used throughout (27). For tagging the N terminus of Rax1 with green fluorescent protein (GFP), we used the integrative plasmid pHP1109, obtained from Hay-Oak Park. For tagging the C terminus of Cdc3 with mCherry, we used the integrative plasmid Yip128-CDC3-mCherry from E. Bi. Landmark deletion strains and tagged strains were verified either by colony PCR or by bioassay, i.e., confirmation of the expected budding phenotype by observation under bright-field microscopy and/or calcofluor white staining of bud scars. Colony PCR was performed using a High-Fidelity PCR kit (Roche).
The diagnostic primers used were as follows. For RAX1, Fwd (5′-CAGAGCAATGCTGGTTATGTAA-3′) and Rev (5′-AAGCCTGCACTTAACGGC-3′). For BUD1, Fwd (5′-TATCATCGCTTAGAAATATTTGGCT-3′) and Rev (5′-AACGCAGCATCTACCGTAAA-3′). To delete RAX1, we copied the hygromycin resistance cassette from pFA6a-hphNT1 (28), using primer Fwd (5′-GTGCACAACGACCTCTAACAATTTCTGCCAAAAAGAAAGTTCAAGAGCGTCCCATTCATCATGCGTACGCTGCAGGTCGAC-3′) and primer Rev (5′-CTGTTTCTTGTACTTAGCGTCACGCGCTATGGAAATATGCGGTGCACAGGTGTTTTTATAATCGATGAATTCGAGCTCG-3′). The product was gel purified and used for transformation. Candidate colonies were then characterized by colony PCR, using the diagnostic primers indicated above.
Strains are listed in Table S4 in the supplemental material. Plasmids are listed in Table S5 in the supplemental material.
Staining and counting of bud scars.
Living cells were stained with calcofluor white (1 μg/ml) (29). After sedimentation and washing, 1-μl samples of the pellet were applied to 1.5% agarose pads, including complete synthetic-glucose growth medium. After overlaying a coverslip and sealing it with Vaseline, they were examined using DAPI (4′,6-diamidino-2-phenylindole) filters with a Deltavision RT epifluorescence microscope and an automated stage (Applied Precision, Inc.). The microscope was equipped with a 100× oil immersion objective (Olympus UPlanApo, 100×/1.40; ∞/0.17/FN26.5), and images were recorded without binning. The microscope uses an Insight Solid State Illuminator 7-color combined unit to deliver the specified excitation bandwidth, combined with an optimized polychroic bandpass filter (Semrock 34-100608-001 Rev. A) and appropriate emission filters. The relevant filters have the following characteristics: DAPI (390/18, 435/48), GFP (475/28, 523/36), and tetramethyl rhodamine isocyanate (TRITC) (542/27, 594/45). Z-stacks of 14 images were captured at 0.2- to 1-μm intervals using a charge-coupled-device (CCD) digital camera (Photometrics CoolSnap HQ). Out-of-focus light was digitally removed using the Softworks deconvolution software (Applied Precision, Inc.). Images were exported as TIF files, and figures were composed using Adobe Photoshop CS (Adobe, Inc.). They were then systematically analyzed. Terminal buds emerge from the external hemispheres of the parental domains. Lateral buds are those that are not terminal and do not emerge from the zygote midzone (ZMZ).
Mating assays.
Equal volumes of actively growing cultures (MATa and MATα) at an optical density at 600 nm (OD600) of ∼0.5 were mixed, sedimented, resuspended, and incubated in stationary plastic microtiter wells without shaking at room temperature. After settling, the cell layer was 50 to 80% confluent, allowing cell-cell contact for the following 4 to 5 h. The cells were then either (i) fixed for 5 min by adding 0.1 volume of 29% formaldehyde and then washed and stored in water, (ii) washed twice in doubly selective medium and then reincubated in the medium with shaking, or (iii) stained with calcofluor white. The composition of the selective medium was designed to limit growth of haploid parental cells (e.g., if one parent was Leu+ ura3 and the other was leu2 Ura+, the selective medium lacked both leucine and uracil). Samples were sonicated before counting.
Note that crosses were not conducted on the surfaces of filters or on solid culture medium to avoid possible changes of surface tension and drying. The often complex shapes of axl2Δ haploid cells and zygotes made it necessary to count only the subpopulation that had conventional shapes.
Data analysis and statistics.
All experiments were conducted three or more times. For each experiment listed in Table S2 in the supplemental material, 200 zygotes were examined to quantitate the relative numbers of zygotes with medial, lateral, or terminal buds. Averages and standard deviations (SD) were calculated. The results from zygotic budding patterns were also analyzed for statistical significance by two-tailed, homoscedastic t tests.
Imaging.
For microscope studies, cells carried chromosomal integrants or centromeric plasmids in which expression of functional copies of FP-tagged proteins was driven from their own promoters. These strains were generated in our laboratory or obtained from other laboratories that have studied them or from Invitrogen.
For microscopy, samples of mating mixtures were applied to agarose pads and imaged as described above. At least 20 cells were observed for each condition, and the selected illustrations are representative of the large majority (>80%). Bright-field images, when present, are in blue. Complete through-focal series were examined in all cases.
RESULTS
Phase I, de novo budding. (i) Most initial budding occurs at the midzone.
In isogenic 4-h crosses of wild-type (wt) haploid cells of several genetic backgrounds, ∼85% of initial buds are medial, judging from inspection of fixed preparations (Fig. 1B; see Table S2, crosses 1 and 2, in the supplemental material). Bud scars stained with calcofluor white that are inherited from haploid parents are seldom adjacent to the zone of contact (ZOC) (30), as we have confirmed (data not shown). This independence of previous budding history motivated our investigation of the nature of initial buds and the contribution of classical budding equipment. We began by localizing tagged proteins that participate in the construction of buds of haploid cells.
(ii) Proteins needed for bud emergence concentrate at the ZOC.
When pairs of cells in mating mixtures establish a synapse-like ZOC prior to cell fusion, cortical proteins redistribute to polarize the cell surface into three regions (Fig. 2A to C and G; see Fig. S1A in the supplemental material). They are characterized by (i) a group of “apical” proteins at the ZOC, as at the tip of the mating projection (31–34); (ii) a “collar” of septins that encircles mating projections and likely serves as a diffusional barrier (35–37); and (iii) a distinct group of proteins that occupies the more distal portion of the cell surface.
FIG 2.
Initial bud formation and reorganization of proteins from the ZOC. (A) Diagram of typical apical and basal protein distributions. The two opposed cells have established a ZOC. They have not yet fused. Selected cortical proteins assume highly polarized distributions at this point. Two distributions are indicated: apical (green) and a subapical “collar” (red). Proteins implicated in membrane fusion, actin polarization, and secretion are in the apical group (panel B; see Fig. S2A in the supplemental material). Septins form the collar. A further group has a basal orientation (see below). (B) Examples of distributions of GFP-tagged proteins that are adjacent to the ZOC (*). In each case, a cell expressing the tagged protein in question (the formin Bni1; the guanine exchange factor for Cdc42, Cdc24; or the exocyst protein Sec5) was crossed with a cell expressing a septin (Cdc3-mCherry). Note that the septin forms a collar subapical to the ZOC (35). The example for Cdc24-GFP—some of which is in the nucleus (N)—shows that septins can also persist at cortical sites (s) that mark sites of previous cytokinesis. The brackets mark the position and orientation of the apposed pairs of cells. The strains were ATY5176 (Bni1), ATY4980 (Cdc24), ATY4312 (Sec5), and ATY5545 (Cdc3). (See Fig. S1A in the supplemental material.) (C) Example of a protein that has a basal distribution. A cell expressing both the GFP-tagged plasma membrane proton ATPase, Pma1, and the septin, Cdc3-mCherry, was crossed with a nonfluorescent cell. The left image shows only GFP. The right image shows both signals, as well as a bright-field image in blue. The strains were ATY5297 × ATY5545. (D) Scattering of an exocyst protein along the ZMZ cortex. Time lapse series of a cell expressing Sec5-GFP crossed with a cell expressing Cdc3-mCherry. Note that at time zero many foci of Sec5-GFP are beginning to be flanked by the septin. At the 15-min time point, Sec5-GFP forms a patch (circled) before completion of the septin ring to form a triad at 20 min. The strains were ATY4312 × ATY5545. (E) Stages of redistribution of the apical protein, Cdc24-GFP, and Cdc3-mCherry. After cell fusion, the nuclear pool of Cdc24-GFP vanishes, the septin annulus (A) is transiently visible, and Cdc3-mCherry and Cdc24-GFP resolve into foci that then scatter and intermix throughout the cortex of the ZMZ. Over the following 10 to 20 min, they sort so that the red foci (septin) progressively flank the green signal. This culminates in the formation of a characteristic “triad,” which subsequently bends and can be recognized as a small bud, which will further enlarge. The three diagrams beneath the micrographs illustrate the progressive morphogenesis that gives rise to the triad. To avoid photobleaching, a composite, not time lapse, series is shown. The strains were ATY4980 × ATY5545. (F) Domain separation persists after cell fusion. (F1) A cell expressing Pma1-GFP and Cdc3-mCherry was crossed with an unlabeled cell, and the two cells have fused. The red septin signal has redistributed symmetrically and shows initial stages of formation of the medial annulus. Note that septins continue to abut on Pma1-GFP, which does not invade the ZMZ. For clarity, the right-hand image includes only the red. V, vacuole. The strains were ATY5896 × ATY4373. (F2) Crosses were conducted between cells expressing Pma1-GFP and cells expressing Cdc3-mCherry. After 5 h, note the sharp discontinuity (d) of distribution of Pma1-GFP at the bud neck. Also note the lack of transfer beyond the ZMZ (M) of preexisting Pma1-GFP from the parent that contributed the lower-left portion of the zygote. B, bud. The strains were ATY5297 × ATY5545. (G) Diagram of apical (Ap) and basal (Ba) domains of early zygotes. The ZMZ is labeled. (H) Overview of the initial bud. Note the size of the nearly mature medial bud of this calcofluor white-stained zygote. It is not unusual to encounter initial buds that are even larger relative to the size of the body of the zygote. The calcofluor white signal is pseudocolored green. The strains were ATY3852 × ATY4373. Scale bars, 5 μm.
The apical group includes proteins that are of central importance for polarized growth and bud construction (Bni1, Cdc24, and Sec5). Moreover, the a-factor transporter, and components of the mitogen-activated protein kinase (MAPK) cascade that responds to mating factors, concentrate at the ZOC, where they surely contribute to intense signaling. Several landmark proteins are also present. Among them is Rax1, which has a particular significance for initial budding (see below). The ZOC is the site of frequent exocytosis and endocytosis, judging from the presence of exocyst proteins, proteins implicated in endocytosis, and evidence of new cell wall synthesis (38) (see Fig. S2A and S3 in the supplemental material). Hypotonicity sensors and proteins that signal to Pkc1 are also present, presumably regulating cell fusion and remodeling of the cell wall. The basal group of proteins includes the proton ATPase Pma1, as well as the glycerol transporter Fps1, which promotes cell fusion (39). Upon cell-cell fusion, the apical proteins resolve into foci that scatter along the cortex of the zygote midzone (ZMZ) without accumulating at more terminal regions of the zygote, where Pma1 remains. Furthermore, the septin collar is supplanted by a medial annulus (35) that gives rise to foci of septins that are interspersed among the foci of apical proteins along the ZMZ (Fig. 2D to F; see Fig. S2B in the supplemental material).
The medial bud then forms de novo. In this process, the field of scattered apical proteins (e.g., Bni1, Cdc24, Sec5, and also the scaffold protein Bem1 [data not shown]) reorganizes so that foci of the septin (red) preferentially flank the apical proteins (green). They then further reorganize to form cortical “triads” that mature to become buds (Fig. 2D and E; see Fig. S1B and C in the supplemental material). As in haploid cells, this progressive reorganization of septins likely results from the balance of local exocytosis and endocytosis (18, 40–42).
The distribution of apical proteins after cell fusion has mechanistic implications for bud site specification. This is because many yeast plasma membrane proteins have only limited lateral mobility and because septin assemblies are likely to further curtail their dispersion (18, 33, 40, 41, 43). Thus, since the ZOC furnishes an enriched group of key proteins—some of which already functioned in polarized growth before cell fusion—it can be thought of as priming the ZMZ for de novo budding. During these events, cortical sites may continually associate with actin filaments that converged at the ZOC prior to cell fusion. These filaments do appear to lose uniformity of orientation upon cell fusion; however, this would be expected to result from the splaying out from the ZOC of fixed sites at which they contact the cortex (30, 35, 44).
(iii) Distinctive features of de novo buds.
Calcofluor white staining of zygotes formed over a 4-h period shows that the large majority have a single medial bud scar. We refer to this as a “de novo” bud, since it forms at a site where previous budding had not occurred. The diameter of this scar was generally at least 1.5 times greater than the diameter of other zygotic bud scars and even larger than the scars of cycling haploid cells and diploid cells (Fig. 3A to D). Indeed, the buds themselves are disproportionately large in comparison to the size of the parental lobes of the zygote (Fig. 2H).
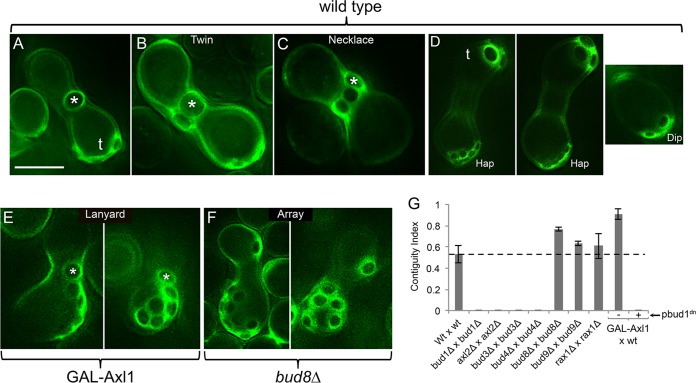
FIG 3.
Localization of bud scars stained with calcofluor white. In each case, zygotes were formed over 5 h and then allowed to bud for an additional 15 h. The calcofluor white signal is pseudocolored green. (A) Note the large medial scar (*), as well as smaller terminal scars (t). The strains were ATY4307 × ATY4303. (B) Note the twin medial scars. The strains were ATY4307 × ATY4303. (C) Note the necklace of medial scars. As many as six encircling scars can be found. The strains were ATY4307 × ATY4303. (D) (Left two images) Size difference between the small preexisting contiguous haploid scars (Hap) and the terminal scars (t) formed by the zygote. (Right image) Diploid bud scars (Dip) imaged at the same magnification. The strains were ATY4307 × ATY4303. (E) Cross between a wt strain and a strain in which excess Axl1 can be induced. The cells were shifted from raffinose to galactose medium for 3 h before mixing. They were then crossed for 5 h and reincubated for 15 h in the presence of galactose. Note the lanyards that extend from the ZMZ. The strains were ATY3328 × ATY6125. (F) Cross between two strains that lack Bud8. Note the arrays of scars that lie adjacent to each other but are not generally contiguous. Comparable observations were made in crosses between strains that lack Bud9. The strains were ATY5884 × ATY6284. (G) Quantitation of contiguity. Pairs of cells were crossed for 5 h and reincubated for 15 h, as described in the text. The mating mixtures were then stained with calcofluor white and examined. To calculate the contiguity index, the number of zygotes that showed medial contiguity was divided by the total number of zygotes that had a medial scar. The penultimate bar describes a cross (in galactose) between a wt cell and a cell carrying galactose-inducible Axl1. The final bar pertains to a cross identical to wt × GAL-Axl1, with the inclusion of [pbud1dn] in both partners. The data are presented as averages ± standard deviations. The dashed horizontal line indicates the control value for medial budding. In comparison to the wild-type cross, the data for crosses of bud8Δ strains and bud9Δ strains and the GAL-Axl1 cross are significant, with P values of <0.05. The strains were wt × wt (ATY3207 × ATY4303), bud1Δ × bud1Δ (ATY402 × ATY4907), axl2Δ × axl2Δ (ATY5881 × ATY5860), bud3Δ × bud3Δ (ATY5882 × ATY5861), bud4Δ × bud4Δ (ATY5883 × ATY5862), bud8 Δ × bud8Δ (ATY5884 × ATY5863), bud9Δ × bud9Δ (ATY5885 × ATY5864), rax1Δ × rax1Δ (ATY5052 × ATY5477), wt × GAL-Axl1 (ATY3328 × ATY6125), and wt [pbud1dn] × GAL-Axl1 [pbud1dn] (ATY5056 × ATY6179). All crosses were in the presence of glucose, except for the last two, which were in galactose-inducing medium. Scale bar, 5 μm.
(iv) Genetic requirements for initial bud site specification.
To learn whether initial bud site specification relies on Bud1 and landmarks, we evaluated the sites of budding in crosses of corresponding deletion strains. Strikingly, most initial buds emerge at the ZMZ regardless of whether Bud1 is present. Indeed, in its absence, medial budding becomes even more frequent than in the wt. Moreover, single deletion of Axl2, Bud3, Bud4, Bud8, or Bud9 does not reduce the predominance of medial budding (Fig. 1B; see Table S2, crosses 3 to 8, in the supplemental material). Axl1 deletions were not studied, since the protein contributes to production of the MATa mating factor (17). Although tagged Axl1 was detected at the ZOC and Axl2 was detected at the site of initial bud emergence, Bud3 and Bud4 were not detected at these sites (data not shown; see Fig. S1A in the supplemental material).
(v) A role for Rax1, Far1, and the position of the nucleus?
Unlike Axl and Bud landmarks, Rax1 is implicated in polarity specification in both haploid and diploid cells. When deleted from haploid cells, its importance is evident if Axl1, Bud3, or Bud4 is also missing. Each of these deletion strains shows increased axial budding in a rax1Δ background (45, 46). When Rax1 is deleted from diploid cells, daughter cells bud indiscriminately at both poles, perhaps due to delocalization of Bud8 or Bud9 (13, 14, 45).
In rax1Δ zygotes, initial nonmedial budding increases to ∼50% (Fig. 1B; see Table S2, cross 9, in the supplemental material). This increase does not appear to reflect extensive relocation of proteins responsible for actin guidance. For example, there is no indication that fusion of rax1Δ cells quickly allows Bni1 or Cdc24 to spread along the length of early zygotes (see Fig. S1G and H in the supplemental material). Deletion of a second protein (Sst2) with an RGS domain that can regulate G-protein signaling (similar to the RGS domain of Rax1) (47) does not increase nonmedial budding (see Table S2, cross 10, in the supplemental material).
Interestingly, the increase in nonmedial budding in rax1Δ zygotes requires Bud1 (see Fig. S1F and Table S2, cross 15, in the supplemental material). Thus, a latent Bud1-dependent bud site specification pathway does exist in early zygotes. To learn whether this latent pathway depends on known landmarks, we deleted single landmarks from the rax1Δ strains used to generate zygotes and then evaluated the sites of budding in homotypic crosses. None of the classical landmarks is strongly implicated (see Fig. S1F and Table S2, crosses 14 and 16 to 19, in the supplemental material).
Rax1 concentrates at the ZOC and appears at the midpoint of triads upon definition of the site of bud emergence. It is also irregularly distributed along much of the zygote surface (see Fig. S1A to E in the supplemental material), where it generally forms patches that can have a characteristic rough or “floret” substructure in face view (see Fig. S1D in the supplemental material). Interestingly, upon fusion of a cell expressing GFP-Rax1 with a nonfluorescent cell, GFP-Rax1 distributes symmetrically across the ZMZ (see Fig. S1E in the supplemental material) but does not further invade the other parental domain (see Fig. S1E in the supplemental material).
If indeed the ZMZ concentrates factors that direct medial budding, circumstances that destabilize the ZMZ could promote nonmedial budding. To test this idea, we inactivated Cdc12, one of the septins that concentrates at the ZMZ. When cdc12-6 strains were crossed at a semirestrictive temperature, there was a modest but distinct decrease in initial medial budding to ∼60% (see Table S2, cross 13, in the supplemental material). Temperatures above 26°C could not be studied because of the bizarre shapes of zygotes and buds that form under these conditions.
Other candidate participants, the Far1 protein and the position of the nucleus, appear not to be relevant to the predominance of medial budding. (i) Far1 is required for the polarization of mating pairs toward each other and does not require Bud1 to do so (30, 48–52). We observe that homotypic crosses of far1-H7 strains still show ∼85% medial budding (see Table S2, cross 11, in the supplemental material). Moreover, Far1-GFP, which was present at the ZOC, vanishes well before bud emergence (see Fig. S2C in the supplemental material). (ii) Regarding the possible importance of the position of its cytoplasmic microtubules (1, 8, 53), initial buds remain predominantly medial in kar1 × wt crosses in which nuclear congression is inhibited (54) (see Table S2, cross 12, in the supplemental material).
Phase II, contiguous budding: a transient haploid-like phase.
Initial buds are largely medial. When zygotes are allowed to form for 4 h and then are incubated for an additional 2 h under conditions that oppose further zygote formation (see Materials and Methods), an adjacent medial scar is present on ∼30% of them (n = 144). Figure 3B illustrates this situation in a cross that included further incubation. We therefore consider these “twins” to be the first and second buds that form. The margins of these scars contact each other, as in the axial budding of haploid cells. Strikingly, when the reincubation is extended to 15 h, about half of the zygotes have either twins or “necklaces” of scars that encircle much of the ZMZ (Fig. 3C). To provide a simple measure of the frequency of contiguous budding, we calculated a contiguity index. This is the number of zygotes that show contiguity (twins, necklaces, or lanyards [see below]) divided by that number plus the number that have a solitary medial bud. In 5-h-plus-15-h protocols, the index for wt crosses is 0.53 ± 0.08 (Fig. 3G; see Table S3 in the supplemental material). At this time, a range of budding histories is evident: 14% have no medial scar, 42% have a single medial scar, 31% have a twin, and 13% have a necklace (n = 138).
Although de novo medial buds form regardless of whether Bud1 and axial landmarks are present, formation of twins requires both Bud1 and the axial landmarks: Axl2, Bud3, and Bud4. Thus, in each case, homotypic 5-h-plus-15-h crosses of corresponding deletion strains show an index of <0.01 (Fig. 3G; see Table S3 in the supplemental material). Correspondingly, at cytokinesis, as for haploid and diploid cells, the necks of de novo buds are encircled by double rings of Axl2, Bud3, and Bud4 (see Table S1 in the supplemental material). Both the genetic evidence and these localizations argue that these proteins guide the formation of the second bud of the twin.
Since deletion of Axl1 drastically reduces zygote formation, to investigate its possible involvement in contiguity, we induced Axl1 during zygote formation and studied the distribution of scars using 2-step protocols with a 15-h reincubation. The resulting zygotes showed a dramatic increase in contiguity (index = 0.91 ± 0.05). Moreover, in addition to having twins and necklaces, lanyards of up to nine contiguous scars extended away from the ZMZ (Fig. 3E). In these cases, almost all the scars were included in the lanyards. The contiguity that results from excess Axl1 requires Bud1, judging from the impact of coexpression of a dominant-negative form of Bud1 (20) (Fig. 3G, far-right bar; see Table S3 in the supplemental material). In contrast to the impact of deleting axial landmarks, deletion of bipolar landmarks (especially Bud8) modestly increases the contiguity index (Fig. 3G; see Table S3 in the supplemental material). Since, as we explain below, at least Bud8 and Bud9 promote terminal budding, the impact of their deletion on medial contiguity could reflect competition between these pathways.
Thus, haploid bud site specification equipment is active in early zygotes, and although there are diminishing amounts of Axl1, haploid determinants often cause buds to be contiguous to the de novo bud or to each other.
These observations also provide a novel explanation of why Axl1 is no longer transcribed after cell fusion: were Axl1 to continue to be synthesized in zygotes, lanyards would be commonplace and lateral, and especially terminal, budding would be reduced. Dispersal of progeny would therefore be at least somewhat limited.
Phase III, lateral and terminal bud emergence.
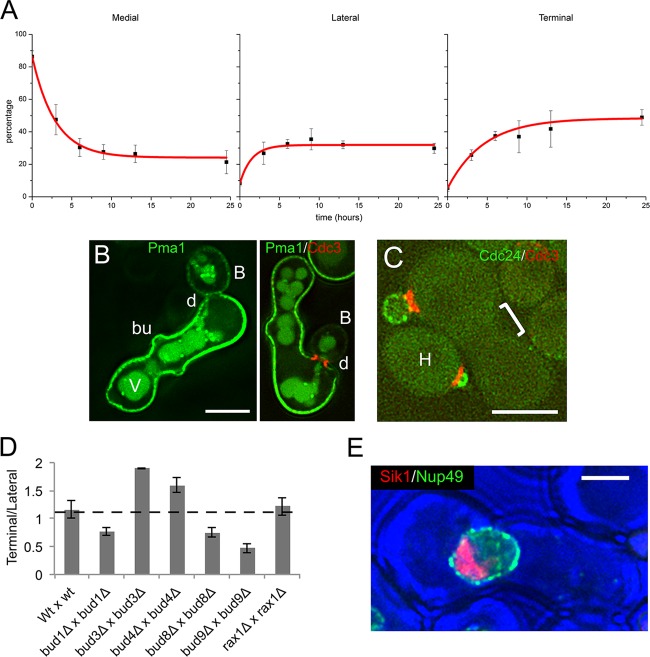
Later budding must account for most of the diploid progeny of zygotes. In fact, bud sites become widely distributed and progressively more terminal with time. Thus, examination of fixed preparations showed that ∼75% of buds were nonmedial at the +15-h time point and that more buds were emerging at terminal sites than at lateral sites (Fig. 4A). Calcofluor white staining showed that the cumulative ratio of terminal to lateral scars (the T/L ratio) was 1.16 ± 0.16 (n = 138) (see Table S3 in the supplemental material). At this time, the average number of scars per zygote was 5.2 ± 0.56 (SD), and no systematic clustering or contiguity of scars occurred outside the midzone. Scars were found in both parental domains of ∼90% of zygotes (n = 138).
FIG 4.
Impact of repeated budding. (A) Population-based time course of budding. After 4 h, crosses were transferred to doubly selective medium and then were sampled at a succession of time points before fixation and counting to tabulate the abundance of medial, lateral, and terminal buds. Note the progressive decline in the frequency of medial budding with time and the reciprocal increase in lateral and terminal buds. Shown are averages of three experiments ± SD. The strains were ATY4307 × ATY4303. (B) Shapes of zygotes and their buds. After allowing zygote formation for 5 h between cells that express Pma1-GFP and cells that express Cdc3-mCherry, the mating mixture was transferred to doubly selective medium for 15 h. A variety of unusually shaped zygotes is seen, often with a medial bulge (bu) or a terminal curve, as on the right. The discontinuity (d) of distribution of Pma1-GFP at the bud neck is evident. B, bud; V, vacuole. The strains were ATY5297 × ATY5545. (C) The zygote midzone after overnight culture. Crosses of cells expressing Cdc24-GFP and cells expressing Cdc3-mCherry were transferred to doubly selective medium after 5 h, cultured overnight, and then examined. Note that the midzone of the zygotes (bracket) shows no sign of persistence of the two labeled proteins. When no bud is present, Cdc24-GFP is not obviously localized. H, haploid. The strains were ATY5001 × ATY5545. (D) Lateral versus terminal distribution of bud scars. Isogenic pairs of cells were crossed for 5 h and then reincubated for 15 h before staining with calcofluor white to localize bud scars and calculate the ratio of terminal to lateral scars. The data are presented as averages ± standard deviations. The dashed horizontal line indicates the control value for medial budding. In comparison to the wild-type cross, the P values for the other crosses (bud1Δ, bud3Δ, bud4Δ, bud8Δ, and bud9Δ) are significant, with P values of <0.05. The strains were as in the legend to Fig. 3G. (E) Lack of disorganization of the nucleolus with age. In the illustrated zygote, one of the parental cells expresses a tagged nucleolar protein, Sik1-mRFP, and the tagged nucleoporin, Nup49-GFP. The cross was allowed to proceed for 5 h and was then cultured for an additional 15 h in medium that allows zygotes to bud but limits growth of the parental haploid cells. Note the compact appearance of the large nucleolus, as in our previous studies of initial budding (64). The strains were ATY4066 × ATY4373. Scale bars, 5 μm.
Despite the modest progression toward increasingly terminal sites with time, there was no absolutely constant order of bud emergence. Thus, inspection of fixed preparations showed that some terminal buds were present at the end of the initial 5-h cross (see Table S2 in the supplemental material) and that the relative frequency of lateral-bud emergence increased to a plateau value rather than peaking at intermediate times and then diminishing. Moreover, although terminal buds accounted for more than half of all buds being formed after 15 h, lateral and medial buds were still forming at this time (Fig. 4A; see Table S2, crosses 1 and 20, in the supplemental material). Flexibility in the order of placement of successive buds is also indicated, after calcofluor white staining, by the presence of zygotes (∼14% of the total) that lacked medial scars and yet had lateral and/or terminal scars.
Zygotes that have budded over 15 h can generally be recognized by deformations of their shape that reflect the presence of bud scars (Fig. 4B). Bud emergence per se nevertheless appears normal at this time and therefore seems to provide a visible counterpart of the phenotypic resetting of age that is characteristic of the progeny of older mothers (24, 55). Thus, Bni1, Cdc3, and Cdc24 constituted typical triads during formation of their nonmedial buds, and both the bud neck septin ring and the shape of daughter cells appeared pristine (Fig. 4B and C and data not shown). Also, as for early zygotes, Axl2, Bud3, and Bud4 formed double rings at the site of cytokinesis (see Table S2 in the supplemental material), and a sharp discontinuity of composition was characteristic of the cell surface at the level of the bud neck (Fig. 4B).
Genetic requirements for later bud site selection.
Calcofluor white staining of 5-h-plus-15-h crosses of corresponding deletion strains showed that Bud1 and landmarks are not required to reorient budding toward nonmedial sites (see Fig. S4A in the supplemental material). Nevertheless, Bud1 and certain landmarks do affect the balance between lateral and terminal events. Thus, crosses of Bud1 deletion strains showed somewhat less terminal budding than wt crosses (Fig. 4D; see Table S3 in the supplemental material), suggesting that some landmark(s) normally promotes reorientation of actin toward terminal regions of the cortex. Consistent with this hypothesis, deletion of Bud8 or Bud9 also decreased the T/L ratio. Moreover, upon deletion of Bud8 or Bud9 (but not Rax1), a minor proportion of zygotes (∼15%) accumulated striking lateral “arrays” of bud scars in which no systematic contiguity was obvious (Fig. 3F). As expected for a functional landmark, a limited amount of tagged Bud8 could be detected at the cell cortex, including terminal regions (see Fig. S4B in the supplemental material).
The impact of deleting axial landmarks is altogether distinct from that of deleting bipolar landmarks. Thus, deletion of Bud3 or Bud4 actually promoted bud emergence at terminal sites (Fig. 4D). (Deletion of Axl2 has a similar effect, but it cannot be accurately quantitated because of the often bizarre shapes of axl2Δ zygotes.) This effect again provides evidence of competition between the participation of haploid and bipolar landmarks.
The nonmedial shift that normally occurs over 15 h does not depend on events that are characteristic of replicative aging. Thus, deletion of Fob1 (56, 57) did not affect the proportion of medial versus nonmedial buds (see Table S2, cross 21, in the supplemental material). Furthermore, unlike haploid cells that have undergone replicative aging (58), we did not observe enlargement or fragmentation of the nucleolus in 15-h zygotes (Fig. 4E).
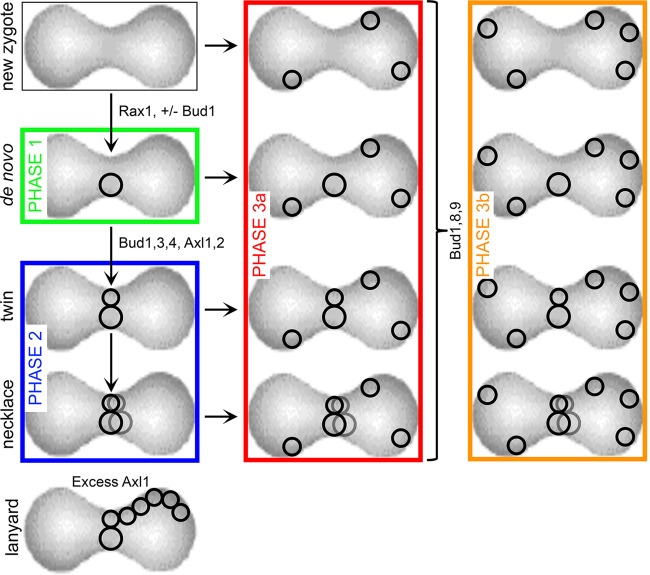
The sequential use of axial and then bipolar landmarks is summarized in Fig. 5 and Table 1.
FIG 5.
Model of sequential-budding options for zygotes. An unbudded zygote (new zygote) is at the upper left. (Phase 1) The first bud (de novo) is medial for ∼ 85% of zygotes. This step does not require Bud1. It becomes less frequent in the absence of Rax1. (Phase 2) After the initial medial bud—if Axl1 is still sufficiently abundant and the other axial landmarks are present—further contiguous budding occurs. These events generate the pattern labeled “twin” and, subsequently, that labeled “necklace.” Judging from the distribution of bud scars after overnight budding, 42% of the initial zygotes have a single medial bud, while 31% have twins and 13% have a necklace. In the presence of excess Axl1, contiguous chains of scars (lanyards) extend from the midzone along the lateral flank of the zygote (bottom left). (Phase 3) Regardless of whether medial buds are present, both lateral and terminal buds can emerge (3a). In the presence of Bud8 and Bud9, the relative amount of terminal budding increases (3b).
TABLE 1.
Bud1 and landmark involvement in successive phases of budding
| Phase | Event | Influence by Bud1 | Landmark participation |
|---|---|---|---|
| I | Medial, de novo | No | Rax1 |
| II | Medial, contiguous | Yes | Axl1, Axl2, Bud3, Bud4 |
| III | Lateral/terminal | Yes | Bud8, Bud9 |
DISCUSSION
The relative orientation of parental yeast cells is arbitrary when they encounter each other. The phase I de novo budding by zygotes nevertheless shows spatial uniformity and bypasses any preexisting landmarks and/or bud scars. Production of these new buds does not depend on the Bud1 GTPase, perhaps because no reorientation of actin is required. Moreover, apart from Rax1, landmark proteins are not required. These observations strongly contrast with the well-established paradigms for bud site specification in cycling cells.
The initial medial preference appears be a manifestation of the process by which the ZOC concentrates proteins that will contribute to bud formation and then gives rise to the ZMZ. Their lack of dispersion along the length of the zygote could reflect (i) restriction of the diffusion of underlying binding sites, which could be linked to the newly synthesized—and perhaps chemically distinct—portion of the cell wall that surrounds the ZMZ; (ii) the same protein-specific features that caused them to concentrate at the ZOC; or (iii) their association with actin filaments that persist from prior to cell fusion.
There is no counterpart of de novo buds during the life cycle of haploid or diploid cells since—apart from the buds that emerge from germinating spores—budding is continuous. The large diameter of the necks of these buds signifies that the size of the cytokinetic ring of septins and contractile proteins is not fixed, reminiscent of consequences of deletion of the bud neck kinase, Cla4, from haploid cells (59).
Rax1 promotes initial medial budding, likely due to its presence at the midpoint of triads. In the absence of Rax1, the pathway that increases nonmedial events depends on Bud1 but is only modestly dependent on classical landmarks. A seemingly independent and novel characteristic of Rax1 is that the consequences of its deletion for later budding are distinct from the consequences of deleting Bud8 or Bud9.
In phase II, buds often form adjacent to the de novo bud. Their contiguity and dependence on Bud1 and axial landmarks are reminiscent of haploid budding. Taken together with the observation that forced expression of Axl1 increases contiguous budding, these events can be considered a manifestation of holdover of the full set of determinants of haploid budding. The later transition to predominant lateral and terminal budding can thus be attributed to the decrease of the titer of Axl1.
Without further guidance, the absence of Axl1 could lead to full randomization of bud site selection. Indeed, buds do emerge at lateral and terminal sites in the absence of single landmarks or Bud1. Nevertheless, deletion of Bud8 or Bud9 diminishes the frequency of terminal budding and leads to the accumulation of lateral “arrays” of scars. These arrays may be a counterpart of the unipolar budding that is characteristic of diploid cells from which either one of these proteins has been deleted. In diploid cells, Bud8 and Bud9 concentrate at the two poles, having been deposited at either the apex (Bud8) or base (Bud9) of new buds (14, 60). Upon expression from high-copy-number plasmids, we detected a faint GFP-Bud8 signal along the cortex and at the termini of late zygotes.
Bud site selection often involves both an “overt” (most frequent) option and a backup or “latent” option(s). For example, in cycling cells, in addition to the well-recognized axial and bipolar budding patterns, modest numbers of buds do form at other sites. Changes of bud site selection also occur (i) upon repeated budding by diploid cells and (ii) in haploid cells after starvation. These changes are attributable to shifting of actin orientation, as occurs upon stress (8, 61, 62).
Bud site specification in zygotes usually begins with a medial bud and then shows hallmarks of involvement of classical landmarks; however, conformity is incomplete—some zygotes lack medial buds, some have a medial bud that is not twinned, some buds emerge at terminal sites even when bipolar landmarks are absent, etc. The functional plasticity of these cells could be essential for their survival.
In brief, for 85% of zygotes, the first bud is medial, apparently because of the local concentration of factors that are essential for bud formation. The importance of Rax1 likely is a reflection of its normally being part of the initial triad. The following one or more buds can also be medial, and if so, they are usually contiguous to the de novo bud. Since this situation is exaggerated upon overexpression of Axl1, and since it requires Bud1 and each of the axial landmarks, we attribute this contiguity to the lingering presence of Axl1, along with the other axial landmarks. Upon escape from contiguous budding, lateral and terminal budding increase. Given the relative timing of lateral versus terminal bud emergence and the impact of bipolar landmarks on terminal budding, we conclude that this phase is a composite, i.e., an early stage that allows both lateral and terminal budding and a later, landmark-dependent stage that is more terminal.
The lack of a geometrically precise pattern of zygotic bud site specification over the period that we have studied should be contrasted with the seemingly more precise patterning of the following (diploid) generation. In this regard, these zygotes lack accurate spatial memory. They may constitute a normally occurring variant of the stochastic “wandering” of polarity guidance proteins in cells exposed to low doses of pheromone and in cells with lesions in signal transmission to actin (19, 52, 63). We suggest that a prerequisite for this increased consistency is that progeny cells emerge from a bud neck that provides a fixed spatial point of reference.
Supplementary Material
ACKNOWLEDGMENTS
We thank D. McDonald for use of the Deltavision microscope and the following for strains and plasmids: R. Arkowitz, E. Bi, A. Camacho, S. Emr, I. Herskowitz, M. Lord, H.-U. Moesch, H.-O. Park, and K. Runge. Thanks are due to Mouki Ayyagari, Ken Farabaugh, Solomon Hwang, Purnima Jaiswal, Richard Lee, Robin Su, Richmond Wong, and Krysta Wyatt for help with accessory experiments and to J. Coller, P. deBoer, D. Manor, M. Snider, and A. Zhu for advice; to M. McMurray for comments on the text; and to S. Rose Kang, C-L. Ni, V. Parmar, and J. Saks for daily camaraderie.
This work was supported by NIH grants R01GM089872 (Institute of General Medical Sciences), P30AI036219, and P30CA043703 and by the Visconsi family.
Footnotes
Published ahead of print 29 August 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/EC.00161-14.
REFERENCES
- 1.Byers B, Goetsch L. 1974. Duplication of spindle plaques and integration of the yeast cell cycle. Cold Spring Harbor Symp. Quant. Biol. 38:123–131. 10.1101/SQB.1974.038.01.016. [DOI] [PubMed] [Google Scholar]
- 2.Aufderheide KJ. 1975. Cytoplasmic inheritance in Saccharomyces cerevisiae: comparison of first zygotic budsite to mitochondrial inheritance patterns. Mol. Gen. Genet. 140:231–241. 10.1007/BF00334268. [DOI] [PubMed] [Google Scholar]
- 3.Muller I. 1985. Parental age and the life-span of zygotes of Saccharomyces cerevisiae. Antonie Van Leeuwenhoek 51:1–10. 10.1007/BF00444223. [DOI] [PubMed] [Google Scholar]
- 4.Birky CW., Jr 1978. Transmission genetics of mitochondria and chloroplasts. Annu. Rev. Genet. 12:471–512. 10.1146/annurev.ge.12.120178.002351. [DOI] [PubMed] [Google Scholar]
- 5.Zernicka-Goetz M. 2005. Cleavage pattern and emerging asymmetry of the mouse embryo. Nat. Rev. Mol. Cell Biol. 6:919–928. 10.1038/nrm1782. [DOI] [PubMed] [Google Scholar]
- 6.Bi E, Park HO. 2012. Cell polarization and cytokinesis in budding yeast. Genetics 191:347–387. 10.1534/genetics.111.132886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kang PJ, Beven L, Hariharan S, Park HO. 2010. The Rsr1/Bud1 GTPase interacts with itself and the Cdc42 GTPase during bud-site selection and polarity establishment in budding yeast. Mol. Biol. Cell 21:3007–3016. 10.1091/mbc.E10-03-0232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chant J, Pringle JR. 1995. Patterns of bud-site selection in the yeast Saccharomyces cerevisiae. J. Cell Biol. 129:751–765. 10.1083/jcb.129.3.751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hartwell LH. 1973. Synchronization of haploid yeast cell cycles, a prelude to conjugation. Exp. Cell Res. 76:111–117. 10.1016/0014-4827(73)90425-4. [DOI] [PubMed] [Google Scholar]
- 10.Gao XD, Sperber LM, Kane SA, Tong Z, Tong AH, Boone C, Bi E. 2007. Sequential and distinct roles of the cadherin domain-containing protein Axl2p in cell polarization in yeast cell cycle. Mol. Biol. Cell 18:2542–2560. 10.1091/mbc.E06-09-0822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kang PJ, Angerman E, Jung CH, Park HO. 2012. Bud4 mediates the cell-type-specific assembly of the axial landmark in budding yeast. J. Cell Sci. 125:3840–3849. 10.1242/jcs.103697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen T, Hiroko T, Chaudhuri A, Inose F, Lord M, Tanaka S, Chant J, Fujita A. 2000. Multigenerational cortical inheritance of the Rax2 protein in orienting polarity and division in yeast. Science 290:1975–1978. 10.1126/science.290.5498.1975. [DOI] [PubMed] [Google Scholar]
- 13.Kang PJ, Angerman E, Nakashima K, Pringle JR, Park HO. 2004. Interactions among Rax1p, Rax2p, Bud8p, and Bud9p in marking cortical sites for bipolar bud-site selection in yeast. Mol. Biol. Cell 15:5145–5157. 10.1091/mbc.E04-07-0600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Krappmann AB, Taheri N, Heinrich M, Mosch HU. 2007. Distinct domains of yeast cortical tag proteins Bud8p and Bud9p confer polar localization and functionality. Mol. Biol. Cell 18:3323–3339. 10.1091/mbc.E06-10-0899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Galgoczy DJ, Cassidy-Stone A, Llinas M, O'Rourke SM, Herskowitz I, DeRisi JL, Johnson AD. 2004. Genomic dissection of the cell-type-specification circuit in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U. S. A. 101:18069–18074. 10.1073/pnas.0407611102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Elia L, Marsh L. 1998. A role for a protease in morphogenic responses during yeast cell fusion. J. Cell Biol. 142:1473–1485. 10.1083/jcb.142.6.1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fujita A, Oka C, Arikawa Y, Katagai T, Tonouchi A, Kuhara S, Misumi Y. 1994. A yeast gene necessary for bud-site selection encodes a protein similar to insulin-degrading enzymes. Nature 372:567–570. 10.1038/372567a0. [DOI] [PubMed] [Google Scholar]
- 18.Okada S, Leda M, Hanna J, Savage NS, Bi E, Goryachev AB. 2013. Daughter cell identity emerges from the interplay of cdc42, septins, and exocytosis. Dev. Cell 26:148–161. 10.1016/j.devcel.2013.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu CF, Savage NS, Lew DJ. 2013. Interaction between bud-site selection and polarity-establishment machineries in budding yeast. Philos. Trans. R. Soc. Lond. B Biol. Sci. 368:20130006. 10.1098/rstb.2013.0006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bender A, Pringle JR. 1989. Multicopy suppression of the cdc24 budding defect in yeast by CDC42 and three newly identified genes including the ras-related gene RSR1. Proc. Natl. Acad. Sci. U. S. A. 86:9976–9980. 10.1073/pnas.86.24.9976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chant J, Herskowitz I. 1991. Genetic control of bud site selection in yeast by a set of gene products that constitute a morphogenetic pathway. Cell 65:1203–1212. 10.1016/0092-8674(91)90015-Q. [DOI] [PubMed] [Google Scholar]
- 22.Chen B, Brinkmann K, Chen Z, Pak CW, Liao Y, Shi S, Henry L, Grishin NV, Bogdan S, Rosen MK. 2014. The WAVE regulatory complex links diverse receptors to the actin cytoskeleton. Cell 156:195–207. 10.1016/j.cell.2013.11.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hughes AL, Gottschling DE. 2012. An early age increase in vacuolar pH limits mitochondrial function and lifespan in yeast. Nature 492:261–265. 10.1038/nature11654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Steinkraus KA, Kaeberlein M, Kennedy BK. 2008. Replicative aging in yeast: the means to the end. Annu. Rev. Cell Dev. Biol. 24:29–54. 10.1146/annurev.cellbio.23.090506.123509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Spokoini R, Moldavski O, Nahmias Y, England JL, Schuldiner M, Kaganovich D. 2012. Confinement to organelle-associated inclusion structures mediates asymmetric inheritance of aggregated protein in budding yeast. Cell Rep. 2:738–747. 10.1016/j.celrep.2012.08.024. [DOI] [PubMed] [Google Scholar]
- 26.Lopez-Otin C, Blasco MA, Partridge L, Serrano M, Kroemer G. 2013. The hallmarks of aging. Cell 153:1194–1217. 10.1016/j.cell.2013.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Amberg D, Burke D, Strathern D. 2005. Methods in yeast genetics: a Cold Spring Harbor Laboratory course manual, 2005 ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 28.Janke C, Magiera MM, Rathfelder N, Taxis C, Reber S, Maekawa H, Moreno-Borchart A, Doenges G, Schwob E, Schiebel E, Knop M. 2004. A versatile toolbox for PCR-based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast 21:947–962. 10.1002/yea.1142. [DOI] [PubMed] [Google Scholar]
- 29.Pringle JR, Preston RA, Adams AE, Stearns T, Drubin DG, Haarer BK, Jones EW. 1989. Fluorescence microscopy methods for yeast. Methods Cell Biol. 31:357–435. 10.1016/S0091-679X(08)61620-9. [DOI] [PubMed] [Google Scholar]
- 30.Valtz N, Peter M, Herskowitz I. 1995. FAR1 is required for oriented polarization of yeast cells in response to mating pheromones. J. Cell Biol. 131:863–873. 10.1083/jcb.131.4.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Merlini L, Dudin O, Martin SG. 2013. Mate and fuse: how yeast cells do it. Open Biol. 3:130008. 10.1098/rsob.130008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Suchkov DV, DeFlorio R, Draper E, Ismael A, Sukumar M, Arkowitz R, Stone DE. 2010. Polarization of the yeast pheromone receptor requires its internalization but not actin-dependent secretion. Mol. Biol. Cell 21:1737–1752. 10.1091/mbc.E09-08-0706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Valdez-Taubas J, Pelham HR. 2003. Slow diffusion of proteins in the yeast plasma membrane allows polarity to be maintained by endocytic cycling. Curr. Biol. 13:1636–1640. 10.1016/j.cub.2003.09.001. [DOI] [PubMed] [Google Scholar]
- 34.Proszynski TJ, Klemm R, Bagnat M, Gaus K, Simons K. 2006. Plasma membrane polarization during mating in yeast cells. J. Cell Biol. 173:861–866. 10.1083/jcb.200602007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tartakoff AM, Aylyarov I, Jaiswal P. 2013. Septin-containing barriers control the differential inheritance of cytoplasmic elements. Cell Rep. 3:223–236. 10.1016/j.celrep.2012.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim HB, Haarer BK, Pringle JR. 1991. Cellular morphogenesis in the Saccharomyces cerevisiae cell cycle: localization of the CDC3 gene product and the timing of events at the budding site. J. Cell Biol. 112:535–544. 10.1083/jcb.112.4.535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ford SK, Pringle JR. 1991. Cellular morphogenesis in the Saccharomyces cerevisiae cell cycle: localization of the CDC11 gene product and the timing of events at the budding site. Dev. Genet. 12:281–292. 10.1002/dvg.1020120405. [DOI] [PubMed] [Google Scholar]
- 38.Marsh LR. 1997. The pathways of cell and nuclear fusion in S. cerevisiae. Mol. Cell. Biol. Yeast Saccharomyces 3:827–888. [Google Scholar]
- 39.Philips J, Herskowitz I. 1997. Osmotic balance regulates cell fusion during mating in Saccharomyces cerevisiae. J. Cell Biol. 138:961–974. 10.1083/jcb.138.5.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Iwase M, Luo J, Nagaraj S, Longtine M, Kim HB, Haarer BK, Caruso C, Tong Z, Pringle JR, Bi E. 2006. Role of a Cdc42p effector pathway in recruitment of the yeast septins to the presumptive bud site. Mol. Biol. Cell 17:1110–1125. 10.1091/mbc.E05-08-0793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Orlando K, Sun X, Zhang J, Lu T, Yokomizo L, Wang P, Guo W. 2011. Exo-endocytic trafficking and the septin-based diffusion barrier are required for the maintenance of Cdc42p polarization during budding yeast asymmetric growth. Mol. Biol. Cell 22:624–633. 10.1091/mbc.E10-06-0484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Marco E, Wedlich-Soldner R, Li R, Altschuler SJ, Wu LF. 2007. Endocytosis optimizes the dynamic localization of membrane proteins that regulate cortical polarity. Cell 129:411–422. 10.1016/j.cell.2007.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Caudron F, Barral Y. 2009. Septins and the lateral compartmentalization of eukaryotic membranes. Dev. Cell 16:493–506. 10.1016/j.devcel.2009.04.003. [DOI] [PubMed] [Google Scholar]
- 44.Karpova TS, McNally JG, Moltz SL, Cooper JA. 1998. Assembly and function of the actin cytoskeleton of yeast: relationships between cables and patches. J. Cell Biol. 142:1501–1517. 10.1083/jcb.142.6.1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fujita A, Lord M, Hiroko T, Hiroko F, Chen T, Oka C, Misumi Y, Chant J. 2004. Rax1, a protein required for the establishment of the bipolar budding pattern in yeast. Gene 327:161–169. 10.1016/j.gene.2003.11.021. [DOI] [PubMed] [Google Scholar]
- 46.Lord M, Inose F, Hiroko T, Hata T, Fujita A, Chant J. 2002. Subcellular localization of Axl1, the cell type-specific regulator of polarity. Curr. Biol. 12:1347–1352. 10.1016/S0960-9822(02)01042-4. [DOI] [PubMed] [Google Scholar]
- 47.Chasse SA, Flanary P, Parnell SC, Hao N, Cha JY, Siderovski DP, Dohlman HG. 2006. Genome-scale analysis reveals Sst2 as the principal regulator of mating pheromone signaling in the yeast Saccharomyces cerevisiae. Eukaryot. Cell 5:330–346. 10.1128/EC.5.2.330-346.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tyers M, Futcher B. 1993. Far1 and Fus3 link the mating pheromone signal transduction pathway to three G1-phase Cdc28 kinase complexes. Mol. Cell. Biol. 13:5659–5669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Peter M, Herskowitz I. 1994. Direct inhibition of the yeast cyclin-dependent kinase Cdc28-Cln by Far1. Science 265:1228–1231. 10.1126/science.8066461. [DOI] [PubMed] [Google Scholar]
- 50.Butty AC, Pryciak PM, Huang LS, Herskowitz I, Peter M. 1998. The role of Far1p in linking the heterotrimeric G protein to polarity establishment proteins during yeast mating. Science 282:1511–1516. 10.1126/science.282.5393.1511. [DOI] [PubMed] [Google Scholar]
- 51.Arkowitz RA. 2009. Chemical gradients and chemotropism in yeast. Cold Spring Harb. Perspect. Biol. 1:a001958. 10.1101/cshperspect.a001958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Arkowitz RA. 2013. Cell polarity: wanderful exploration in yeast sex. Curr. Biol. 23:R10–R12. 10.1016/j.cub.2012.11.037. [DOI] [PubMed] [Google Scholar]
- 53.Segal M, Bloom K, Reed SI. 2000. Bud6 directs sequential microtubule interactions with the bud tip and bud neck during spindle morphogenesis in Saccharomyces cerevisiae. Mol. Biol. Cell 11:3689–3702. 10.1091/mbc.11.11.3689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Conde J, Fink GR. 1976. A mutant of Saccharomyces cerevisiae defective for nuclear fusion. Proc. Natl. Acad. Sci. U. S. A. 73:3651–3655. 10.1073/pnas.73.10.3651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Unal E, Kinde B, Amon A. 2011. Gametogenesis eliminates age-induced cellular damage and resets life span in yeast. Science 332:1554–1557. 10.1126/science.1204349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kaeberlein M, McVey M, Guarente L. 1999. The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes Dev. 13:2570–2580. 10.1101/gad.13.19.2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Defossez PA, Prusty R, Kaeberlein M, Lin SJ, Ferrigno P, Silver PA, Keil RL, Guarente L. 1999. Elimination of replication block protein Fob1 extends the life span of yeast mother cells. Mol. Cell 3:447–455. 10.1016/S1097-2765(00)80472-4. [DOI] [PubMed] [Google Scholar]
- 58.Sinclair DA, Mills K, Guarente L. 1997. Accelerated aging and nucleolar fragmentation in yeast sgs1 mutants. Science 277:1313–1316. 10.1126/science.277.5330.1313. [DOI] [PubMed] [Google Scholar]
- 59.Blanco N, Reidy M, Arroyo J, Cabib E. 2012. Crosslinks in the cell wall of budding yeast control morphogenesis at the mother-bud neck. J. Cell Sci. 125:5781–5789. 10.1242/jcs.110460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zahner JE, Harkins HA, Pringle JR. 1996. Genetic analysis of the bipolar pattern of bud site selection in the yeast Saccharomyces cerevisiae. Mol. Cell. Biol. 16:1857–1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Delley PA, Hall MN. 1999. Cell wall stress depolarizes cell growth via hyperactivation of RHO1. J. Cell Biol. 147:163–174. 10.1083/jcb.147.1.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kono K, Saeki Y, Yoshida S, Tanaka K, Pellman D. 2012. Proteasomal degradation resolves competition between cell polarization and cellular wound healing. Cell 150:151–164. 10.1016/j.cell.2012.05.030. [DOI] [PubMed] [Google Scholar]
- 63.Slaughter BD, Smith SE, Li R. 2009. Symmetry breaking in the life cycle of the budding yeast. Cold Spring Harb. Perspect. Biol. 1:a003384. 10.1101/cshperspect.a003384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tartakoff AM, Jaiswal P. 2009. Nuclear fusion and genome encounter during yeast zygote formation. Mol. Biol. Cell 20:2932–2942. 10.1091/mbc.E08-12-1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.