Abstract
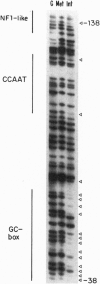
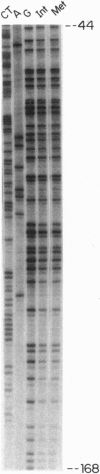
We report that ligation-mediated PCR (LMPCR) can be used for high-resolution study of metaphase chromosomes, and we discuss the role of metaphase chromatin structure in the preservation of differentiated cell states. The X chromosome-linked human PGK1 (phosphoglycerate kinase 1) promoter region was investigated, and euchromatic active X chromosome (Xa) metaphase chromatin was compared with interphase Xa chromatin and to heterochromatic inactive X chromosome (Xi) metaphase and interphase chromatin. We find that (i) good-quality data at single-nucleotide resolution can be obtained by LMPCR analysis of dimethyl sulfate-treated intact metaphase cells; (ii) transcription factors present on the Xa promoter of interphase chromatin are not present on metaphase chromatin, establishing that the transcription complex on the PGK1 promoter must form de novo each cell generation; and (iii) the dimethyl sulfate reactivity pattern of Xa and Xi chromatin at metaphase is very similar to that of naked DNA. These results are discussed in the context of models for heritable chromatin structure and epigenetic mechanisms for cell memory, and they are also relevant to more general aspects of chromatin structure and differences between euchromatin and heterochromatin.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bauer W. R., Hayes J. J., White J. H., Wolffe A. P. Nucleosome structural changes due to acetylation. J Mol Biol. 1994 Feb 25;236(3):685–690. doi: 10.1006/jmbi.1994.1180. [DOI] [PubMed] [Google Scholar]
- Brown D. D. The role of stable complexes that repress and activate eucaryotic genes. Cell. 1984 Jun;37(2):359–365. doi: 10.1016/0092-8674(84)90366-0. [DOI] [PubMed] [Google Scholar]
- Firth J. D., Ebert B. L., Pugh C. W., Ratcliffe P. J. Oxygen-regulated control elements in the phosphoglycerate kinase 1 and lactate dehydrogenase A genes: similarities with the erythropoietin 3' enhancer. Proc Natl Acad Sci U S A. 1994 Jul 5;91(14):6496–6500. doi: 10.1073/pnas.91.14.6496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazit B., Cedar H., Lerer I., Voss R. Active genes are sensitive to deoxyribonuclease I during metaphase. Science. 1982 Aug 13;217(4560):648–650. doi: 10.1126/science.6283640. [DOI] [PubMed] [Google Scholar]
- Gottesfeld J. M., Wolf V. J., Dang T., Forbes D. J., Hartl P. Mitotic repression of RNA polymerase III transcription in vitro mediated by phosphorylation of a TFIIIB component. Science. 1994 Jan 7;263(5143):81–84. doi: 10.1126/science.8272869. [DOI] [PubMed] [Google Scholar]
- Groudine M., Conkin K. F. Chromatin structure and de novo methylation of sperm DNA: implications for activation of the paternal genome. Science. 1985 May 31;228(4703):1061–1068. doi: 10.1126/science.2986289. [DOI] [PubMed] [Google Scholar]
- Hansen R. S., Ellis N. A., Gartler S. M. Demethylation of specific sites in the 5' region of the inactive X-linked human phosphoglycerate kinase gene correlates with the appearance of nuclease sensitivity and gene expression. Mol Cell Biol. 1988 Nov;8(11):4692–4699. doi: 10.1128/mcb.8.11.4692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornstra I. K., Nelson D. L., Warren S. T., Yang T. P. High resolution methylation analysis of the FMR1 gene trinucleotide repeat region in fragile X syndrome. Hum Mol Genet. 1993 Oct;2(10):1659–1665. doi: 10.1093/hmg/2.10.1659. [DOI] [PubMed] [Google Scholar]
- Iverson B. L., Dervan P. B. Adenine specific DNA chemical sequencing reaction. Nucleic Acids Res. 1987 Oct 12;15(19):7823–7830. doi: 10.1093/nar/15.19.7823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeppesen P., Turner B. M. The inactive X chromosome in female mammals is distinguished by a lack of histone H4 acetylation, a cytogenetic marker for gene expression. Cell. 1993 Jul 30;74(2):281–289. doi: 10.1016/0092-8674(93)90419-q. [DOI] [PubMed] [Google Scholar]
- Kerem B. S., Goitein R., Richler C., Marcus M., Cedar H. In situ nick-translation distinguishes between active and inactive X chromosomes. Nature. 1983 Jul 7;304(5921):88–90. doi: 10.1038/304088a0. [DOI] [PubMed] [Google Scholar]
- Klevecz R. R. Automated cell cycle analysis. Methods Cell Biol. 1975;10:157–172. doi: 10.1016/s0091-679x(08)60735-9. [DOI] [PubMed] [Google Scholar]
- Klevecz R. R., Braly P. S. Circadian and ultradian cytokinetic rhythms of spontaneous human cancer. Ann N Y Acad Sci. 1991;618:257–276. doi: 10.1111/j.1749-6632.1991.tb27248.x. [DOI] [PubMed] [Google Scholar]
- Kornberg R. D., Lorch Y. Chromatin structure and transcription. Annu Rev Cell Biol. 1992;8:563–587. doi: 10.1146/annurev.cb.08.110192.003023. [DOI] [PubMed] [Google Scholar]
- Li E., Beard C., Jaenisch R. Role for DNA methylation in genomic imprinting. Nature. 1993 Nov 25;366(6453):362–365. doi: 10.1038/366362a0. [DOI] [PubMed] [Google Scholar]
- Manuelidis L. A view of interphase chromosomes. Science. 1990 Dec 14;250(4987):1533–1540. doi: 10.1126/science.2274784. [DOI] [PubMed] [Google Scholar]
- Matsui S. I., Weinfeld H., Sandberg A. A. Quantitative conservation of chromatin-bound RNA polymerases I and II in mitosis. Implications for chromosome structure. J Cell Biol. 1979 Feb;80(2):451–464. doi: 10.1083/jcb.80.2.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- McGhee J. D., Felsenfeld G. Reaction of nucleosome DNA with dimethyl sulfate. Proc Natl Acad Sci U S A. 1979 May;76(5):2133–2137. doi: 10.1073/pnas.76.5.2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PRESCOTT D. M., BENDER M. A. Synthesis of RNA and protein during mitosis in mammalian tissue culture cells. Exp Cell Res. 1962 Mar;26:260–268. doi: 10.1016/0014-4827(62)90176-3. [DOI] [PubMed] [Google Scholar]
- Paro R. Mechanisms of heritable gene repression during development of Drosophila. Curr Opin Cell Biol. 1993 Dec;5(6):999–1005. doi: 10.1016/0955-0674(93)90084-4. [DOI] [PubMed] [Google Scholar]
- Pfeifer G. P., Riggs A. D. Chromatin differences between active and inactive X chromosomes revealed by genomic footprinting of permeabilized cells using DNase I and ligation-mediated PCR. Genes Dev. 1991 Jun;5(6):1102–1113. doi: 10.1101/gad.5.6.1102. [DOI] [PubMed] [Google Scholar]
- Pfeifer G. P., Singer-Sam J., Riggs A. D. Analysis of methylation and chromatin structure. Methods Enzymol. 1993;225:567–583. doi: 10.1016/0076-6879(93)25037-3. [DOI] [PubMed] [Google Scholar]
- Pfeifer G. P., Steigerwald S. D., Hansen R. S., Gartler S. M., Riggs A. D. Polymerase chain reaction-aided genomic sequencing of an X chromosome-linked CpG island: methylation patterns suggest clonal inheritance, CpG site autonomy, and an explanation of activity state stability. Proc Natl Acad Sci U S A. 1990 Nov;87(21):8252–8256. doi: 10.1073/pnas.87.21.8252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeifer G. P., Tanguay R. L., Steigerwald S. D., Riggs A. D. In vivo footprint and methylation analysis by PCR-aided genomic sequencing: comparison of active and inactive X chromosomal DNA at the CpG island and promoter of human PGK-1. Genes Dev. 1990 Aug;4(8):1277–1287. doi: 10.1101/gad.4.8.1277. [DOI] [PubMed] [Google Scholar]
- Riggs A. D. DNA methylation and cell memory. Cell Biophys. 1989 Aug-Oct;15(1-2):1–13. doi: 10.1007/BF02991574. [DOI] [PubMed] [Google Scholar]
- Riggs A. D. DNA methylation and late replication probably aid cell memory, and type I DNA reeling could aid chromosome folding and enhancer function. Philos Trans R Soc Lond B Biol Sci. 1990 Jan 30;326(1235):285–297. doi: 10.1098/rstb.1990.0012. [DOI] [PubMed] [Google Scholar]
- Riggs A. D., Pfeifer G. P. X-chromosome inactivation and cell memory. Trends Genet. 1992 May;8(5):169–174. doi: 10.1016/0168-9525(92)90219-t. [DOI] [PubMed] [Google Scholar]
- Rivier D. H., Rine J. Silencing: the establishment and inheritance of stable, repressed transcription states. Curr Opin Genet Dev. 1992 Apr;2(2):286–292. doi: 10.1016/s0959-437x(05)80286-2. [DOI] [PubMed] [Google Scholar]
- Saitoh Y., Laemmli U. K. Metaphase chromosome structure: bands arise from a differential folding path of the highly AT-rich scaffold. Cell. 1994 Feb 25;76(4):609–622. doi: 10.1016/0092-8674(94)90502-9. [DOI] [PubMed] [Google Scholar]
- Th'ng J. P., Guo X. W., Swank R. A., Crissman H. A., Bradbury E. M. Inhibition of histone phosphorylation by staurosporine leads to chromosome decondensation. J Biol Chem. 1994 Apr 1;269(13):9568–9573. [PubMed] [Google Scholar]
- Tommasi S., LeBon J. M., Riggs A. D., Singer-Sam J. Methylation analysis by genomic sequencing of 5' region of mouse Pgk-1 gene and a cautionary note concerning the method. Somat Cell Mol Genet. 1993 Nov;19(6):529–541. doi: 10.1007/BF01233380. [DOI] [PubMed] [Google Scholar]
- Wolffe A. P. Transcription complexes. Prog Clin Biol Res. 1990;322:171–186. [PubMed] [Google Scholar]