Abstract
Marine algae of the genus Nannochloropsis are promising producers of biofuel precursors and nutraceuticals and are also harvested commercially for aquaculture feed. We have used quick-freeze, deep-etch electron microscopy, Fourier transform infrared spectroscopy, and carbohydrate analyses to characterize the architecture of the Nannochloropsis gaditana (strain CCMP 526) cell wall, whose recalcitrance presents a significant barrier to biocommodity extraction. The data indicate a bilayer structure consisting of a cellulosic inner wall (∼75% of the mass balance) protected by an outer hydrophobic algaenan layer. Cellulase treatment of walls purified after cell lysis generates highly enriched algaenan preparations without using the harsh chemical treatments typically used in algaenan isolation and characterization. Nannochloropsis algaenan was determined to comprise long, straight-chain, saturated aliphatics with ether cross-links, which closely resembles the cutan of vascular plants. Chemical identification of >85% of the isolated cell wall mass is detailed, and genome analysis is used to identify candidate biosynthetic enzymes.
INTRODUCTION
The genus Nannochloropsis comprises at least six photoautotrophic algal species in the Eustigmatophyceae stramenopile lineage that are found in fresh, brackish, and ocean waters (1). Nannochloropsis cells reproduce asexually, dividing to yield two daughter cells that then shed their mother cell wall (2, 3). Several Nannochloropsis species have been studied as candidate production strains in large-scale biofuel facilities because of their hardy outdoor growth profiles and high lipid yields (4–9). They are also producers of valuable pigments (10) and nutritive oils (11, 12) and are commonly used as an aquaculture feed (13).
Algae are frequently grown in large outdoor ponds until being harvested, dewatered, and extracted for biocommodities. The efficacy of each of these steps—growth, harvesting, dewatering, and extraction—depends upon the composition and architecture of the cell wall. The wall creates a buffer between the external environment and the living protoplast, protecting the cell from environmental pressures. The outer surface of the wall interacts with flocculants (14), and its rigidity helps determine the viscoelastic parameters that characterize algal slurry bulk flow (15). Finally, the cell wall erects mass transfer barriers against dewatering and extraction and may itself contain extractable commodities (16, 17).
Despite the importance of algal cell wall properties in biotechnological applications, little structural information is available for the majority of species. The Chlamydomonas reinhardtii cell wall is the most extensively characterized and appears to be constructed entirely from a suite of hydroxyproline-rich glycoproteins arranged in six distinct layers (18–20). However, algal cell walls display great diversity, varying in molecular components, intra- and intermolecular linkages, and overall structure (21). Wall constituents may include carbohydrates (22), proteins (23, 24), lipids (25, 26), carotenoids (27), tannins (28), and even lignin (29, 30). Much remains to be learned regarding how these constituents cross-link into the networks that form discrete layers around the cell and how they reconfigure in response to physiological and environmental cues.
Among the most extensively studied polymers of the algal cell wall are polysaccharides. These include cellulose (31), chitin-/chitosan-like molecules (32), hemicelluloses (33), pectins (34), fucans (35), alginates (24), ulvans (36), carrageenans (37), and lichenins (38). The polysaccharides in marine algae are frequently sulfated (22).
The composition and architecture of Nannochloropsis cell walls have been assessed in several studies. Brown reported that the polysaccharides of Nannochloropsis oculata contained ∼68% glucose along with about 4 to 8% each rhamnose, mannose, ribose, xylose, fucose, and galactose (39). Recently, Vieler et al. characterized the neutral carbohydrates in the alcohol-insoluble residue (AIR) of Nannochloropsis oceanica (strain CCMP 1779) cell extracts (40). This residue, enriched for cell wall material, was hydrolyzed with trifluoroacetic acid (TFA) followed by Saeman hydrolysis. The authors observed that ∼9% of the AIR was carbohydrate, 90% of which was glucose, ∼3% mannose, and the rest traces of rhamnose, fucose, arabinose, xylose, and galactose. Treatment of the residue with endoglucanase II (EGII), a hydrolyzing enzyme specific for β-1,4-linked glucans, liberated 85% of the glucose, while laminarinase, an enzyme that hydrolyzes β-1,3-glucans, liberated 20%. Bioinformatic analysis of the CCMP 1779 genome yielded two proteins annotated as cellulose synthases, similar to those found in cyanobacteria, and nine proteins that the authors describe as highly similar to plant endoglucanases.
Nannochloropsis cell walls also contain algaenans, a term that likely encompasses several lipid-related species (41, 42). Algaenans are highly resistant to alkali/acid hydrolysis and aqueous/organic solubilization, and their biochemical characterization has been considered tentative since isolation procedures may have induced chemical alterations (43, 44). Published studies indicate that Nannochloropsis algaenan comprises long-chain aliphatic hydrocarbons that are subject to ether cross-linking reactions (41), a description that also applies to the cutan of several species of drought-resistant plants (45). The biosynthetic pathways that produce algaenans and cutans are not presently known.
In this study, ∼86% of the isolated cell wall material of N. gaditana has been positively identified. A new method for isolating Nannochloropsis algaenans was developed, allowing an analysis of native algaenan structure, and this material was characterized by attenuated total reflectance Fourier transform infrared spectroscopy (ATR-FTIR). Furthermore, quick-freeze, deep-etch electron microscopy (QFDE-EM) was used to visualize native and isolated wall components.
MATERIALS AND METHODS
Cell culture.
Nannochloropsis strain CCMP 526 was from the National Center for Marine Algae and Microbiota (formerly CCMP). CCMP 526 was grown at 23°C in f/2 medium (125) with 1.0 g/liter nitrate in a 2-foot by 2-foot flat-panel photobioreactor with a 40-mm width (14-liter capacity). One percent CO2 was bubbled through the cultures, and 70 μmol m−2 s−1 of photosynthetically active radiation (PAR) from cool white fluorescent light illuminated one side. The initial cell density was 4 × 107 cells per ml, and cells were harvested on day 8, during mid-linear phase (4 g/liter). For comparative purposes, CCMP 526 was also grown semicontinuously at 23°C in Roux flasks in Tris-buffered (pH 7.9) artificial seawater (ASW [0.125 M NaCl, 7 mM KNO3, 0.44 mM KH2PO4, 0.24 mM NaHCO3, 0.018 mM FeCl3·6H2O, 0.049 mM MnCl2·4H2O, 1.7 mM CaCl2·2H2O, 13.4 mM MgSO4·7H2O, 13.8 mM MgCl2·6H2O, 0.1 mM Na2EDTA, 0.42 μM CoCl2·6H2O, 0.14 μM Na2MoO4·2H2O, 0.89 μM ZnSO4·7H2O, 0.4 μM CuSO4·5H2O]). These cultures were bubbled with supplemental CO2 (1%) in air; 600 μmol m−2 s−1 of 680-nm light was provided by an LED light bank on one side and 150 μmol m−2 s−1 PAR from cool white fluorescent light was supplied to the other side. Cells grown under the conditions outlined above have been designated “f/2 preparations” or “ASW preparations” throughout the text.
Preparation of cell walls.
Cells pellets were harvested by centrifugation at 4,000 × g for 10 min. Shed cell walls, derived from wall shedding during cell division and from dead cells, formed a red-orange layer on top of the green cell pellet and were carefully removed by pipette. The green whole-cell pellet was resuspended in 50 ml of deionized water (dH2O) and repelleted a total of 3 times to remove traces of medium.
Whole cells were then lyophilized and resuspended to ∼10% solids in dH2O. To every 5 ml of suspended solids, 15 μl of protease inhibitor cocktail (product number P9599; Sigma, St. Louis, MO) and 1 μl of 2,000 U/ml DNase I (NEB, Ipswich, MA) were added. Cells were drawn into the 30-ml chamber of a French press pressure cell (Thermo, Waltham, MA). Seven passes through the French press at a working pressure of 18,000 lb/in2 were required for nearly complete cell lysis, although a small percentage of the cells (∼5%) remained intact even after 7 passages. After every 2 passages, cells were centrifuged at 10,000 × g for 20 min to separate the soluble lysate, which was discarded, from residual cells and cell walls. The latter were resuspended in dH2O as before, the DNase and protease inhibitor cocktail were replenished, and the next passages were performed. Cell walls were separated from residual whole cells by multiple centrifugations at 5,000 × g, each time removing the walls from the green pellet at the bottom of the tube until no green pellet was observed. The walls isolated in this manner are referred to hereinafter as “pressed” cell walls.
Pressed cell walls were separated from residual debris and shed cell walls by layering them upon sucrose gradients of 20, 30, 40, and 60% and centrifuging for 30 to 60 min at 10,000 × g. Shed cell walls and other debris typically remain in the upper strata and were separated from pressed cell walls, which migrated to the bottom of the tube, stuck to the side of the tube, or floated in the 60% sucrose layer. Pressed walls were then washed three times by resuspending the pellet in 50 ml of water and centrifuging as before. After the final wash, the recovered walls were lyophilized.
Every 20 mg of lyophilized cell walls was extracted successively with 10 ml of 80% ethanol, n-hexane–acetone (1:1, vol/vol), and n-hexane for 15 min in a sonicating water bath (model number 91957; Harbor Freight Tools, Calabasas, CA) to remove any loosely associated proteins, lipids, carotenoids, and other soluble matter. Additional washes with n-hexane were performed until the color of the residual material was white or off-white. The organic supernatants from these washes were collected into a preweighed glass beaker and evaporated. The dry residue was weighed. Light microscopy visualizations after these extractions were used to verify that the insoluble material remaining after extraction was morphologically consistent with purified cell walls. The extracted cell walls were lyophilized in preweighed glass vials and weighed. In the sections that follow, “isolated” or “pressed” cell walls refers to walls purified and subjected to organic extraction as described above.
Enzymatic degradation of walls.
For enzymatic digestions of isolated cell walls, approximately 10 mg of walls were used in each 1-ml digestion mixture. Cellulase (product number C0615 [cellulase 1]), chitinase (product number C8241), chitosanase (product number C9830, 25.9 U/ml), lysozyme (product number L6876), lyticase (product number L4025), protease 2 (product number P5147), and sulfatase (product number S1629, 3.37 mg/ml) were purchased from Sigma, while cellulase Onozuka R10 (catalog number 16419.02 [cellulase 2]) was purchased from Serva (Heidelberg, Germany). Stock concentrations of enzymes were 20 mg/ml unless otherwise noted, and enzymes were dissolved in 20 mM sodium phosphate buffer (pH 7) for all digestions. Triplicate samples were digested at room temperature for 24 h on a shaker table. The final enzyme concentrations were 1 mg/ml, and 50 μg/ml of ampicillin was added to each reaction mixture. For one set of controls, enzymes were heat denatured for at least 10 min at 100°C before being added to the cell wall suspensions. As a second set of controls, active enzymes were incubated in reaction mixtures without cell walls. After digestion, the reaction mixtures were centrifuged at 10,000 × g for 5 min. Supernatants were collected, and residual material was washed twice by resuspension in 1 ml of dH2O, followed by centrifugation and supernatant removal. The residual material was then lyophilized and weighed.
Acid hydrolysis of cell walls.
Cell walls isolated from f/2 medium-grown cells were digested in 72% sulfuric acid as described in an analysis protocol from the National Renewable Energy Laboratory (46).
Characterization of monosaccharides.
Supernatants from enzyme digests were assayed for released carbohydrates. For each sample, 490 μl of supernatant was saved for direct analysis by high-performance anion-exchange chromatography (ion chromatography [IC]) with pulsed amperometric detection (HPAEC-PAD) and 10 μl was used for anthrone assays. To the other half of the sample, an equal volume of 4 N TFA was added, and the sample was hydrolyzed to monosaccharides by incubation at 100°C for 6 h. The samples were then evaporated at 65°C under nitrogen. Samples were immediately resuspended in water for analysis.
Monosaccharides were analyzed by HPAEC-PAD on an ICS 5000 equipped with a 2- by 250-mm Carbopac PA-1 analytical column and a 2- by 50-mm Aminotrap guard column (Dionex Corporation, Sunnyvale, CA). The PAD detector used a carbohydrate-certified disposable gold electrode (Thermo) with the gold standard PAD waveform. Neutral sugars and amino sugars were eluted at 31°C and 0.25 ml/min with a 1 mN KOH eluent produced from an eluent generator. Uronic acids were analyzed separately at 40°C and 0.25 ml/min using 100 mN NaOH for 5 min, followed by a 60 to 300 mM gradient of sodium acetate over 10 min to elute the uronic acids. The mobile-phase solutions were degassed and kept under a bed of helium. Individual standards were used to measure the retention times of each sugar and determine carbohydrate concentrations. All major peaks in the IC traces were identified; however, a few very small peaks remained unidentified. Data collection and analysis were done using Chromeleon 7.1.0 software (Dionex).
To validate the IC results, aliquots of 10 μl of supernatants from cellulase digests (cellulase 1) of cell walls and wall-free controls were brought up to 100 μl in dH2O for anthrone quantitation of carbohydrate content (47). A stock solution containing 2 g/liter anthrone reagent in 26.6 N sulfuric acid was prepared, as was a set of 100-μl glucose standards ranging in concentration from 100 to 400 μg/ml. To 100 μl of sample, 900 μl of the anthrone stock solution was added. The samples were placed in boiling water for 12 min and then transferred to an ice bath. Three hundred-microliter aliquots of each were transferred to individual wells of a 96-well plate, and the absorbance at 625 nm was determined using a plate reader (Synergy 2; Biotek, Winooski, VT). The absorbance values were regressed against glucose standards (R2 > 0.999), and sample carbohydrate concentrations were inferred from the resulting regression equations.
Glycosyl linkage analysis.
Glycosyl linkage analysis was performed by the Complex Carbohydrate Research Center at the University of Georgia. Samples were permethylated, depolymerized, reduced, and acetylated. The resultant partially methylated alditol acetates (PMAAs) were analyzed by gas chromatography-mass spectrometry (GC-MS) as described previously (48).
Amino acid analysis.
Amino acid analysis was conducted by the Molecular Structure Facility at the University of California at Davis. Cell walls were hydrolyzed for 48 h in 6 N HCl–1% phenol at 110°C. Samples (∼2.5 mg) were analyzed for amino acids and related compounds using a lithium citrate buffer system, a Beckman 6300 amino acid analyzer (Beckman, Brea, CA), and an amino acid standard (product number A9906; Sigma).
Fourier transform infrared spectroscopy characterization.
Isolated walls were progressively treated with cellulase 1 and then protease in sodium phosphate buffer at pH 7.0 for 18 h at room temperature before treatment with 6 N HCl for 18 h at 100°C. Four dH2O washes followed after each step, and aliquots from each step were preserved. For ATR-FTIR analysis, ∼1-mg amounts of lyophilized samples of cell walls and algaenan preparations were loaded onto the germanium crystal of a Nexus 470 FTIR spectrophotometer Smart Performer platform (Nicolet, Madison, WI). Spectra were collected using OMNIC software (Nicolet) but were also further analyzed using LabSpec 6 software (Horiba Scientific, Edison, NJ). All spectra were collected across the 4,000 to 600 cm−1 range at a 1 cm−1 spectral resolution. The spectra presented for each sample are the averages of between 200 and 400 spectra. All data were collected at Washington University at St. Louis in the Jens Environmental Molecular and Nanoscale Analysis Laboratory.
Quick-freeze deep-etch electron microscopy.
Samples were rinsed twice in dH2O by pellet centrifugation, pipetted onto a cushioning material, and dropped onto a liquid helium-cooled copper block; the frozen material was transferred to liquid nitrogen and fractured, etched at 80°C for 2 min, and platinum replicated using Pt-C rotary replication as described previously (49). The replicas were examined with a JEOL electron microscope, model JEM 1400, equipped with an AMTV601 digital camera.
Elemental analysis.
Screw-cap Teflon vials (VWR, Randor, PA) were washed thoroughly with dH2O and then pretreated by soaking in trace-metal-grade 6 N nitric acid for 18 h at 100°C. The nitric acid in each vial was replaced with 10 ml of fresh 6 N nitric acid to which 10 to 25 mg of sample was added. A blank with nitric acid but no sample was also prepared as a control. Vials were heated for 18 h at 100°C, and then the nitric acid was evaporated using a hot plate. The insides of the Teflon vials were each washed with 20 ml of dH2O, which was then filtered and used for elemental analysis by inductively coupled-plasma atomic emission spectroscopy (ICP-AES) (Optima 5300 DV; PerkinElmer, Waltham, MA). Cadmium was run as an internal standard and used to correct the values obtained during the analysis. The system uses a nonbaffled cyclonic spray chamber with a type A Meinhard nebulizer. The rates of argon gas flow were 16 liters/min for the plasma, 0.65 liters/min for the nebulizer, and 0.5 liters/min for the auxiliary flow. A custom-made multispectral fitting file was used to correct for spectral interferences between elements. Argon gas was used to purge the spectrometer and introduction system. Dried compressed air was used as the shear gas. Standards were prepared from the AccuTrace reference standard (AccuStandard, New Haven, CT), from analytical standards QCS-7-M and QCS-1 (High Purity Standards, Charleston, SC), and from EM standard ICP-126-5 (Ultra Scientific, North Kingstown, RI). The standards for metals and nonmetals were CCV-1A and -B, respectively (High Purity Standards). Elemental concentrations observed in the blank were subtracted from sample values.
Bioinformatics.
Cell wall-related protein sequences were obtained from the Cell Wall Navigator database (50). Additional cell wall-related protein sequences were extracted from the CAZy database (www.cazy.org) (51). These protein sequences were compared against protein models from the CCMP 526 genome (NCBI BioProject accession number PRJNA73791), genome assembly, and transcriptome shotgun assembly (NCBI BioProject accession number PRJNA157493) using BLAST (52). Combined, these searches yielded a list of protein and nucleotide sequences that were curated to identify putative cell wall-related proteins. Matching protein sequences (E value, <1 × 10−20) were blasted against nonredundant protein sequences in the NCBI database using the DELTA-BLAST algorithm. Matching nucleotide sequences (E value, <10−10) were blasted (blastx) against the same database. In both cases, a match was declared when a protein matched a protein domain with known cell wall-related activity with an E value of less than 10−20 (typically less than 10−100). The resulting list of protein models was mapped back to the genome to prevent redundancy.
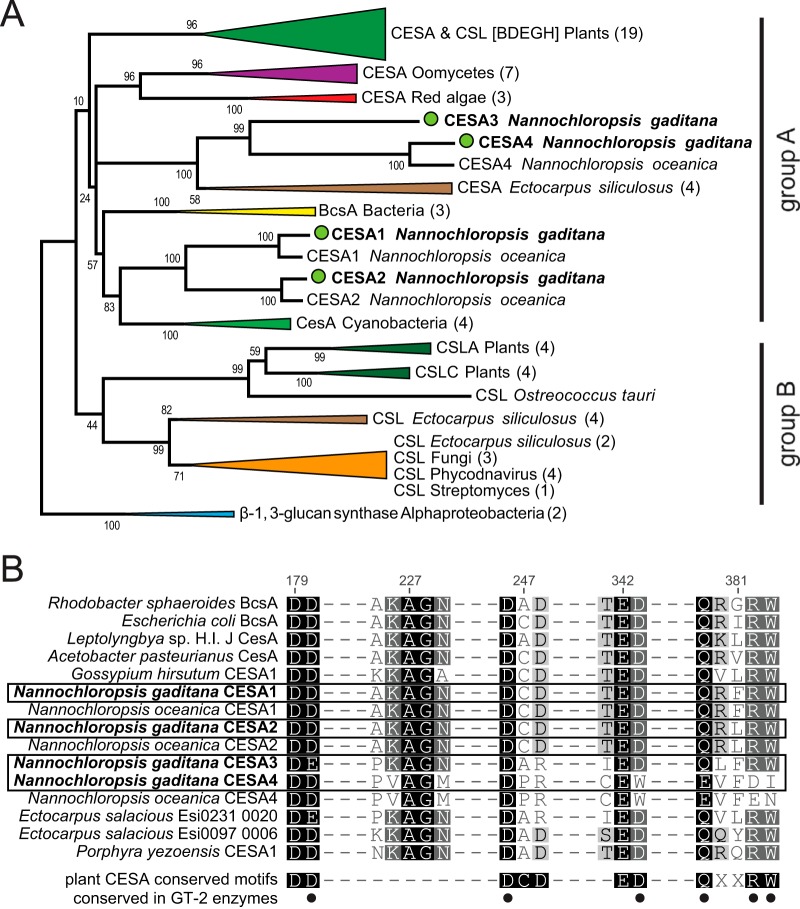
All protein alignments were conducted with MUSCLE (53). A phylogenetic tree of cellulose synthases (CESAs) and cellulose synthase-like (CSL) proteins was inferred by using the maximum-likelihood method based on the Poisson correction model (see Fig. 1) (54). The bootstrap consensus tree inferred from 100 replicates is taken to represent the evolutionary history of the taxa analyzed (55). Branches corresponding to partitions reproduced in less than 10% of bootstrap replicates are collapsed. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (100 replicates) is shown next to the branches (55). Initial trees for the heuristic search were obtained by applying the neighbor-joining method to a matrix of pairwise distances estimated using a JTT (Jones, Taylor, Thornton) model. The analysis involved 71 CESA and CSL sequences from a wide variety of organisms, including plants (56), cyanobacteria (57), bacteria (58), stramenopiles (24), red algae, green algae, and fungi. The accession numbers for these sequences can be found in Table S1 in the supplemental material. All ambiguous positions were removed for each sequence pair. There were a total of 2,826 positions in the final data set. Evolutionary analyses were conducted in MEGA5 (59).
FIG 1.
(A) Phylogenetic tree of cellulose synthases (CESA) and cellulose synthase-like proteins (CSL). The tree was constructed using the maximum-likelihood (ML) approach. N. gaditana sequences are marked with green circles. Numbers indicate bootstrap values in the ML analysis. Numbers in parentheses indicate the numbers of sequences compressed in subtrees. Groups A and B are described in the text. The full list of accession numbers from the proteins used can be found in Table S1 in the supplemental material. (B) Multiple sequence alignments of the motifs common to cellulose synthases. Numbers indicate the amino acid positions in the Rhodobacter sphaeroides BcsA protein (58). Dashes do not represent gaps in the alignment, and not all aligned amino acids are shown. Dots indicate the conserved residues (D, D, D, and QXXRW) found in characterized cellulose synthases within the glycosyl transferase 2 (GT-2) family, as cataloged by the Carbohydrate-Active enZymes (CAZy) database.
All CCMP 526 accession numbers are available at the National Center for Biotechnology Information website (http://www.ncbi.nlm.nih.gov/) and are part of the genome BioProject with accession number PRJNA73791 or the transcriptome shotgun assembly project with accession number PRJNA157493.
RESULTS
The N. gaditana cell wall biomass is primarily cellulose.
In preliminary experiments, we ascertained that intact N. gaditana cells were insensitive to incubation in exogenous cellulase, as determined by their retention of full resistance to chlorophyll release with subsequent exposure to 1% Triton X-100. Given published reports that the outer cell wall layer is composed of algaenan (2), which is expected to block enzyme access, we conducted our subsequent analyses using preparations of isolated cell walls (“pressed” walls; see Materials and Methods).
As an initial screen to characterize the composition of the N. gaditana cell wall, several enzymes were tested for their ability to deconstruct pressed walls from cultures grown in f/2 medium (Table 1). Two cellulase formulations, cellulase 1 (product number C0615; Sigma) and cellulase 2 (Onozuka R10, catalog number 16419.02; Serva), were the most effective, hydrolyzing up to 76% of the cell wall mass. Cocktails containing additional enzymes (chitinase and lyticase) in combination with these cellulases failed to liberate additional biomass (see Fig. S1 in the supplemental material).
TABLE 1.
Monosaccharide composition of N. gaditana cell walls determined after enzymatic digestion
| Enzyme | % of wall mass solubilized (mean ± 1 SD; n = 3)a | % of mass (mean ± 1 SD; n = 3) determined to be carbohydrates by: |
% of glucose in carbohydratesb,c | Other carbohydrates detected | |
|---|---|---|---|---|---|
| HPAEC-PADb | Anthrone | ||||
| Cellulase 1 | 76.0 ± 2.5d | 81.4 ± 8.3d | 78.3 ± 5.6d | 98.9d | Fuc, Man, Rha, GalA |
| 83.5 ± 2.2e | 82.9 ± 3.7e | 78.9 ± 9.3e | 98.0e | ||
| 77.4 ± 2.5f | 73.3 ± 2.6f | 68.7 ± 1.4f | 99.0f | ||
| Cellulase 2 | 71.5 ± 2.0 | 70.1 ± 2.0 | NDg | 97.2 | Fuc, Gal, GalA |
| Chitinase | 69.6 ± 6.3 | 64.3 ± 1.7 | ND | 98.9 | Fuc, GlcN, Rha, GalA |
| Chitosanase | 11.3 ± 1.3 | 11.6 ± 0.5 | ND | 100.0 | None |
| Lyticase | 68.9 ± 3.5 | 65.4 ± 4.1 | ND | 99.2 | Fuc, Rha, GalA |
| Protease | 9.9 ± 6.2 | 1.1 ± 0.2 | ND | 28.5 | Fuc, GlcN, Man, Rha, GalA |
| Zymolyase | 71.0 ± 2.2 | 71.7 ± 3.5 | ND | 99.9 | Fuc, GlcN |
The percentage of the cell wall mass that was solubilized was determined gravimetrically.
Carbohydrates as the percentage of total cell wall mass and glucose as the percentage of total carbohydrate were determined by HPAEC-PAD after digestion of the enzyme hydrolysate with 2 M TFA at 100°C for 6 h. Carbohydrate values reflect the amounts after subtraction of carbohydrates measured in wall-free controls.
Standard deviations were all <0.05%.
Grown in f/2 medium: this sample was used for all digestions using alternate enzymes.
Grown in f/2 medium.
Grown in SW medium.
ND, not determined.
The most effective cellulase (cellulase 1; see Materials and Methods) was used to digest pressed cell walls obtained from two independent cultures grown in f/2 medium and one grown in ASW medium. Supernatants from cellulase digests were then hydrolyzed with 2 N trifluoroacetic acid (TFA) for 6 h at 100°C and analyzed for monosaccharides by both high-performance anion-exchange chromatography with pulsed amperometric detection (HPAEC-PAD) and the anthrone assay. Sample masses were recorded before and after digestion (Table 1). Seventy-three to 83% of the wall mass from each of the three preparations was positively identified as carbohydrate by HPAEC-PAD (69 to 79% by anthrone), of which 98 to 99% of the monomeric saccharides were glucose. The carbohydrate masses were higher for the average of the results for the two f/2 preparations (∼82%, n = 6) than for the single ASW preparation (∼73%, n = 3), a difference that was statistically significant (P < 0.02, Student's two-tailed t test). The results for wall digests of one f/2 cell wall preparation with cellulase 2, chitinase, chitosanase, protease, lyticase, and zymolyase are also given in Table 1. With the exception of the protease treatment, 98 to 99% of the sugar released by each of these enzymes was glucose; however, as shown in Table S2 in the supplemental material, lyticase and zymolase liberated the majority of the glucose in polymeric form, indicating that these enzymes were acting on cross-links and not directly on the cellulose polymer. Hydrolysis with 72% sulfuric acid using an aliquot of the same sample liberated 69% ± 7% (mean ± standard deviation) of the total mass as carbohydrate, as determined by HPAEC-PAD, 98% of which was glucose (data not shown). Small amounts of rhamnose, fucose, galactose, galacturonic acid, mannose, and glucosamine were also detected in all the aforementioned digestion products, as were two unidentified compounds that eluted near the other neutral sugars with concentrations above the background (data not shown). Significantly, no fructose was detected, which indicates that any residual sucrose from the gradient separation step of the wall isolation protocol was successfully removed.
The glycosyl linkages of a pressed cell wall preparation obtained from cells grown in ASW medium were also determined after the polysaccharides were permethylated, depolymerized, reduced, and acetylated. The resulting partially methylated alditol acetates (PMAAs) were analyzed by gas chromatography-mass spectrometry (GC-MS); the results are presented in Table 2. The vast majority of the residues were common to cellulose, namely, 1,4-linked glucopyranosyl (78%) and terminal glucopyranosyl (8%), the latter included both reducing and nonreducing terminal residues. Small amounts of 1,6-linked (3%), 1,3-linked (1.2%), 1,4,6-linked (0.6%), and 1,3,4-linked (0.5%) glucopyranosyl residues were also found; glucose accounted for 91.4% of the total residues detected. Also found were traces of fucopyranosyl, rhamnopyranosyl, mannopyranosyl, galactopyranosyl, xylopyranosyl, and N-acetylglucopyranosyl carbohydrates. Of these, only xylose was not detected in the monomeric composition analysis (Table 1). α and β stereoisomers were not distinguishable using this method; it should also be noted that the relatively mild acid hydrolysis method used likely did not release all of the carbohydrate monomers.
TABLE 2.
Glycosyl linkage analysis of N. gaditana cell wall polysaccharides
| Glycosyl linkage | % Present |
|---|---|
| 1,4-Linked glucopyranosyl residue (4-Glc) | 78.1 |
| Terminal glucopyranosyl residue (t-Glc)a | 7.9 |
| 1,6-Linked glucopyranosyl residue (6-Glc) | 3.1 |
| Terminal fucopyranosyl residue (t-Fuc)a | 1.9 |
| 1,2-Linked rhamnopyranosyl residue (2-Rha) | 1.5 |
| Terminal N-acetylglucosamine residue (t-GlcNAc)a | 1.3 |
| 1,3-Linked glucopyranosyl residue (3-Glc) | 1.2 |
| 1,4-Linked N-acetylglucosamine residue (4-GlcNAc) | 1.0 |
| Terminal mannopyranosyl residue (t-Man)a | 0.7 |
| Terminal fucopyranosyl residue (t-Gal)a | 0.6 |
| 1,4,6-Linked glucopyranosyl residue (4,6-Glc) | 0.6 |
| 1,3,4-Linked glucopyranosyl residue (3,4-Glc) | 0.5 |
| 1,4-Linked mannopyranosyl residue (4-Man) | 0.4 |
| Terminal xylopyranosyl residue (t-Xyl)a | 0.3 |
| 1,2-Linked mannopyranosyl residue (2-Man) | 0.3 |
| Terminal rhamnopyranosyl residue (t-Rha)a | 0.2 |
| 1,2-Linked fucopyranosyl residue (2-Fuc) | 0.2 |
| 1,6-Linked fucopyranosyl residue (6-Gal) | 0.1 |
Terminal residues may come from the reducing or nonreducing end of a polysaccharide.
Genomic analysis of cellulose-related sequences.
The evidence for a predominance of cellulose in the N. gaditana cell wall prompted a query of the CCMP 526 genome for the presence of genes encoding putative cellulose biosynthetic and degradative enzymes. Several genes encoding homologs of characterized cellulose synthases (Fig. 1) and β-glycoside hydrolases were identified (Table 3). To simplify the presentation, the modifiers “candidate” and “putative” are not used below, but it is by definition the case that until gene products are purified and assayed, all gene sequences identified by homology encode candidate enzymes.
TABLE 3.
β-Glycoside hydrolase enzymes encoded in Nannochloropsis gaditanaa
| GH family | Description | Identifier or GenBank accession no. of gene model |
|---|---|---|
| GH1 | β-Glucosidase (cel1b) | Nga02538 |
| GH1 | β-Glucosidase (cel1b) | Nga04378.01 |
| GH1 | β-Glucosidase (cel1b) | Nga00952 |
| GH3 | β-Glucosidase (b7;bgla;bghg1;ao090009000356) | Nga06223 |
| GH3 | β-Glucosidase (b7;bgla;bghg1;ao090009000356) | Nga06304 |
| GH3 | β-Glucosidase (b7;bgla;bghg1;ao090009000356) | Nga00051 |
| GH3 | β-Glucosidase (b7;bgla;bghg1;ao090009000356) | Nga03687 |
| GH6 | Cellobiohydrolase ii (cbhii;cbh2;cbh-ii;trcel6a) | Nga31009 |
| GH8 | Endo-β-1,4-glucanase a (cela;cthe_0269) | Nga30029 |
| GH8 | Endo-β-1,4-glucanase a (cela;cthe_0269) | Nga01206 |
| GH8 | Endo-β-1,4-glucanase a (cela;cthe_0269) | Nga03336 |
| GH8 | Endo-β-1,4-glucanase a (cela;cthe_0269) | Nga10019 |
| GH9 | Endo-β-1,4-glucanase 8/cellulase 1 (atcel1;egase) | Nga05528 |
| GH9 | Endo-β-1,4-glucanase 8/cellulase 1 (atcel1;egase) | Nga30804 |
| GH9 | Endo-β-1,4-glucanase 8/cellulase 1 (atcel1;egase) | Nga06309 |
| GH9 | Endo-β-1,4-glucanase 8/cellulase 1 (atcel1;egase) | Nga30060 |
| GH9 | Endo-β-1,4-glucanase 8/cellulase 1 (atcel1;egase) | Nga31020 |
| GH9 | Endo-β-1,4-glucanase 8/cellulase 1 (atcel1;egase) | Nga00928 |
| GH9 | Endo-β-1,4-glucanase 8/cellulase 1 (atcel1;egase) | Nga10014 |
| GH10 | Endo-β-1,4-glucanase/exo-β-1,4-glucanase (celb;egb) | Nga06757 |
| GH10 | Endo-β-1,4-glucanase/exo-β-1,4-glucanase (celb;egb) | Nga06788 |
| GH10 | Endo-β-1,4-glucanase/exo-β-1,4-glucanase (celb;egb) | Nga02660 |
| GH10 | Endo-β-1,4-glucanase/exo-β-1,4-glucanase (celb;egb) | Nga03399 |
| GH10 | Endo-β-1,4-glucanase/exo-β-1,4-glucanase (celb;egb) | Nga02713 |
| GH10 | Endo-β-1,4-glucanase/exo-β-1,4-glucanase (celb;egb) | Nga10015 |
| GH10 | Endo-β-1,4-glucanase/exo-β-1,4-glucanase (celb;egb) | Nga10016 |
| GH10 | Endo-β-1,4-glucanase/exo-β-1,4-glucanase (celb;egb) | Nga10017 |
| GH10 | Endo-β-1,4-glucanase/exo-β-1,4-glucanase (celb;egb) | Nga10018 |
| GH16 | Xyloglucan:xyloglucosyltransferase-like | JU963924.1b |
| CBM1c | Fungal cellulose binding domain | JU970214.1b |
Glycoside hydrolase enzymes found in N. gaditana thought to act on cellulose. Other GH enzymes are present in N. gaditana.
GenBank accession number for associated transcript.
Not a GH enzyme but involved in cellulose metabolism.
Among the multiple glycosyltransferases and glycosylhydrolases identified, four displayed homology to cellulose synthase and seven to glycoside hydrolase family 9 (GH9) cellulases. Additionally, genes encoding multiple isoforms of other β-hydrolytic enzymes are present in the genome, including genes for enzymes in the GH1 and GH3 (e.g., β-glucosidase), GH6, GH8, and GH10 families, which in sum could potentially deconstruct cellulose to glucose. Also found were two homologs of genes encoding UDP-glucose pyrophosphorylase, which is required for glucose activation in the synthesis of cellulose (and other glucose polymers), where one of these homologs is fused to a phosphoglucomutase gene (GenBank accession number JU963155).
Cellulose synthase protein models derived from N. gaditana transcripts CESA1 (GenBank accession number JU963065), CESA2 (GenBank accession number JU963444), CESA3 (GenBank accession numberJU981242), and CESA4 (GenBank accession numberJU963368) were aligned to the sequences of plant, algal, bacterial, and fungal cellulose synthases (CESA) and cellulose synthase-like (CSL) proteins (Fig. 1). Using the maximum-likelihood method, a phylogeny was inferred (Fig. 1A). The four CESA-like proteins found in N. gaditana cluster into two clades: clade I with CESA1 and CESA2 and clade II with CESA3 and CESA4. Plant CESA and CSL proteins are divided into two groups, one directly related to cyanobacteria (group A in Fig. 1A) and the other to cyanobacterial-derived sequences in green algae (group B in Fig. 1A) (60). Both of the Nannochloropsis CESA clades are located within group A, which also includes CESA from red algae, Oomycetes, and Ectocarpus salacious and CESA/CSL from land plants. Within this group, clade I clusters closely with cyanobacterial CesA and bacterial BcsA proteins, while clade II clusters with E. siliculosus CESA. Both bacterial BcsA and cyanobacterial CesA enzymes have been experimentally verified to produce cellulose, including biochemically characterized enzymes from Rhodobacter sphaeroides (58, 61), Gluconacetobacter xylinus (62), Nostoc punctiforme, Anabaena species (63), and Thermosynechococcus vulcanus (64). No matches were found with the poorly defined bacterial BcsB to -G or the BcsQ and BcsZ protein sequences that are associated with cellulose synthesis in some bacterial systems (65, 66). The four E. siliculosus CESA proteins that cluster with CESA3 and CESA4 are likely genuine cellulose synthases (24). E. siliculosus produces cellulose microfibrils in its cell wall and has nine CESA/CSL proteins. Five are similar to group B CSL proteins from Actinobacteria and fungi, while the remaining four are similar to group A CESA proteins (24).
A number of the characteristic motifs of cellulose synthases are present in the N. gaditana homologs. The glycosyltransferase 2 (GT-2) enzyme family includes processive cellulose synthases that have the conserved amino acids D, D, D, and QXXRW that are critical for activity (56, 67). The third Asp is proposed to form a catalytic base, while the other motifs are predicted to be involved in substrate and/or product interactions, with the conserved Trp of the QXXRW motif interacting with the newly polymerized cellulose (58, 67). These motifs are found in CESA1, CESA2, and CESA3, but only the first two aspartic acids are found in CESA4 (Fig. 1B). In plant cellulose synthases, these conserved motifs have been further defined to DD, DCD, ED, and QXXRW (56). CESA1 and CESA2 have perfect matches for these sequence motifs, while CESA3 substitutes DE and DAR for the DD and DCD motifs, respectively. CESA1 and CESA2 both contain the sequences AKAGN and TED, which bookend a pocket that is reported to bind UDP-glucose (58). CESA3 substitutes a proline for the first alanine in the AKAGN motif and an isoleucine for threonine in the TED motif (Fig. 1B). These motifs are the least conserved in CES4, which substitutes PVAGM for the AKAGN motif and CEW for the TED motif. Interestingly, an E. siliculosus cellulose synthase carries all of the same substitutions that are found in CES3 (DE, DAR, and PKAGN).
All four of the cellulose synthases in N. gaditana CCMP 526 have homologs in the N. oceanica CCMP 1779 genome and a second sequenced N. gaditana (B-31) genome (9, 40, 68). Near-exact matches of CESA1, CESA2, and CESA4 are found in the N. gaditana B-31 genome, with amino acid identities of 99.7% or greater, whereas matches with 80.3, 86, and 73.3% amino acid identity, respectively, are found in the N. oceanica CCMP 1779 genome (see Table S3 in the supplemental material). Although homology to CESA3 was found in the other Nannochloropsis genomes, assembly in this region of the respective genomes is poor, yielding only partial sequences that could not be assembled into a contiguous section of DNA. A complete CESA3 sequence was only recoverable from the N. gaditana CCMP 526 transcriptome shotgun assembly (5). A pairwise identity analysis (see Table S3) indicates that the CESA1, CESA2, and CESA4 orthologs between Nannochloropsis species share higher percent identities than do paralogs (CESA1 and CESA2; CESA3 and CESA4), which suggests that the CESA genes were duplicated prior to speciation of N. gaditana and N. oceanica (Fig. 1A).
Transcript levels of the CESA genes were queried from CCMP 526 transcriptome data sets (5, 9). In linear growth, the CESA1, -2, -3, and -4 genes had transcript expression levels of 451, 802, 295, and 57 reads per kilobase per million mapped reads (RPKM), respectively. These levels of expression put CESA2 as the 113th most highly expressed gene under these conditions, with CESA1 and CESA3 among the top 525 expressed genes and CES4 as approximately the 3,000th most expressed gene, out of ∼9,000 genes. Under nitrogen deprivation, the expression levels did not change appreciably at the time assessed (16 h).
Genes that are involved with cellulose degradation or remodeling were also queried in the N. gaditana genome, including those encoding cellulases, xyloglucan endotransglycosylases/hydrolases (XTH, glycoside hydrolase family 16 [GH16]), expansins, and swollenins. Representative members of glycoside hydrolase families 1, 3, 5, 6, 7, 8, 9, 10, 12, 16, 44, 45, 48, 61, and 124, as curated by the Carbohydrate-Active enZymes (CAZy) database (51), as well as expansins and swollenins (69), were used as search queries. Homologs to GH1, -3, -6, -8, -9, -10, and -16 members are present (Table 3), but homologs to expansins and swollenins were not identified. A total of 21 endo-β-1,4-glucanases and exo-β-1,4-glucanases were found in the N. gaditana genome, including members from the GH6, -8, -9, and -10 families. GH9 enzymes have been found in both eukaryotes and bacteria, and in plants, some members have been proposed to contribute to cellulose remodeling during growth (70). The GH9 enzymes found in N. gaditana are all highly similar to the biochemically characterized cellulase of Clostridium thermocellum, which has activity against both cellulose and lichenin but little against laminarin or xylan (71, 72). Alignment of the putative CCMP 526 cellulases with the C. thermocellum cellulase indicates that the proteins share signature motifs (see Fig. S2 in the supplemental material). These include the DLXGGXXDAGD, HRR, and DXXXXXXXXE motifs that contain amino acids necessary for catalytic activity (73, 74). Our analysis also indicates that regions with the sequences LFXEXQRXG and WRXD are conserved among the C. thermocellum and CCMP 526 cellulases. Interestingly, one GH6 family cellobiohydrolase II (CBHII)-like enzyme was found in the N. gaditana genome. Typically, this family of enzymes is found in fungi and other cellulolytic organisms, where the enzyme acts processively from the nonreducing ends of cellulose chains to generate cellobiose (75). A GH6 homolog is also found in the stramenopile Aureococcus anophagefferens but appears to be absent in oomycetes, which include plant pathogens. Additionally, another protein with possible fungal origin, a fungal-type cellulose-binding domain (CBM1), was also found in N. gaditana. A search for plant-type xyloglucan endotransglycosylases/hydrolases (GH16) yielded one match in the transcriptome assembly. Three GH1 and four GH3 enzymes were also present. The enzymes in these families have β-glucosidase, β-galactosidase, or β-mannosidase activity (51). In the case of β-glucosidases, these enzymes hydrolyze the exocellulase product, typically cellobiose, into individual monosaccharides (76), so these enzymes may also play a role in N. gaditana cellulose reorganization/metabolism.
The cell wall preparations contain amino acids and minerals.
Amino acid analysis was performed on samples of pressed walls harvested from cells grown on both f/2 and ASW medium (Table 4). The amino acid ratios were consistent among triplicate biological samplings. Amino acid content as a percentage of total biomass was also consistent, averaging 6.2% ± 1.7%. The total amino acid mass is likely underreported because sulfur-containing residues (cysteine and methionine) and tryptophan were not quantitatively recovered by the method used.
TABLE 4.
Amino acid analysis of cell wall materiala
| Residue | % of total proteinb |
|---|---|
| Ala | 7.9 ± 0.3 |
| Arg | 7.7 ± 1.4 |
| Asx | 10.8 ± 0.7 |
| Glx | 9.7 ± 0.8 |
| Gly | 6.7 ± 0.2 |
| His | 2.0 ± 0.2 |
| Ile | 4.7 ± 0.1 |
| Leu | 8.5 ± 0.2 |
| Lys | 6.6 ± 0.2 |
| Phe | 5.5 ± 0.3 |
| Pro | 5.3 ± 0.5 |
| Ser | 5.4 ± 0.1 |
| Thr | 7.4 ± 0.2 |
| Tyr | 3.1 ± 0.4 |
| Val | 8.0 ± 0.4 |
Cysteine and methionine were not quantitatively recovered by the analysis, and tryptophan was not recovered at all.
The protein content as a percentage of the total biomass was 6.2% ± 1.7% (n = 4; mean ± 1 standard deviation).
To further characterize the balance of the cell wall mass, elemental analysis of the wall material was performed using ICP-AES. Four biological replicates were tested, two from cells grown in f/2 medium and two from cells grown in ASW medium. An average of 1.1% ± 0.6% of the cell wall mass could be attributed to the 31 elements assayed, which did not include C, H, N, or O (see Table S4 in the supplemental material). The concentrations of Mg, Ca, Na, and S were several times higher in the ASW-grown samples than in those grown in f/2, though in each case, these were small fractions of the total biomass. The median sulfur content for these samples was 0.3% of the total biomass; however, sulfate could not be detected by ion chromatography (IC) following sulfatase digestion of cell walls. Measurements of the amino acids and select elements of the carbohydrates identified are combined in Table 5. In total, 86.5% ± 7.2% of the mass of the cell wall material has been identified. The remainder of the mass balance is likely comprised of algaenan, as described below.
TABLE 5.
Mass balance of the N. gaditana cell wall preparations
| Component | % of total biomass |
|---|---|
| Carbohydrate (n = 3) | 79.2 ± 4.9 |
| Protein (n = 4) | 6.2 ± 1.7 |
| Mineral (n = 4) | 1.1 ± 0.6 |
| Total | 86.5 ± 7.2 |
The wall comprises an outer algaenan layer protecting an inner cellulose layer.
Many algae produce algaenan-containing cell walls, including vegetative cells of Scenedesmus, some Chlorella species, and all Nannochloropsis species tested (25, 77–81), cysts of Chlamydomonas, Haematococcus, and Polytomella (49, 82–89), and Botryococcus colonies (126). In thin sections of osmicated, dehydrated specimens, the presence of algaenan usually (43) correlates with the presence of a layer called the trilaminar sheath (TLS), a narrow dark-light-dark domain at the wall periphery (2, 77, 80, 81, 83, 88, 90). An excellent thin-section image of the TLS from N. occulata is found in Fig. 3H of Murakami and Hashimoto (2), beneath which is a second wall layer and then the plasma membrane.
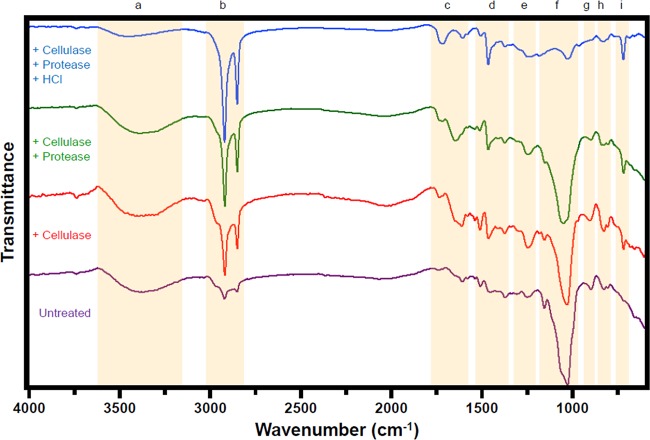
FIG 3.
FTIR spectra of CCMP 526 pressed walls. Spectra are provided for pressed walls and following stepwise cellulase, protease, and HCl digestion. Nine notable wavenumber regions (a to i) have been highlighted.
Figure 2A shows a model of the Nannochloropsis wall based on quick-freeze deep-etch EM (QFDE-EM), wherein living cells or wall preparations are snap-frozen at liquid-helium temperatures without fixation and then fractured and etched (surface ice sublimated under vacuum) and platinum replicated (49). Layer 1, which adopts the TLS configuration in thin section (2) but forms a single layer in replicas, is underlain by a thicker layer 2 (Fig. 2B). Struts often connect layer 2 to the plasma membrane (Fig. 2C), and extensions of unknown composition protrude from the outer surface of layer 1 (Fig. 2C). Consistent with our observations using Triton X-100, no morphological changes in wall structure were observed when intact cells were exposed to cellulases (not shown).
FIG 2.
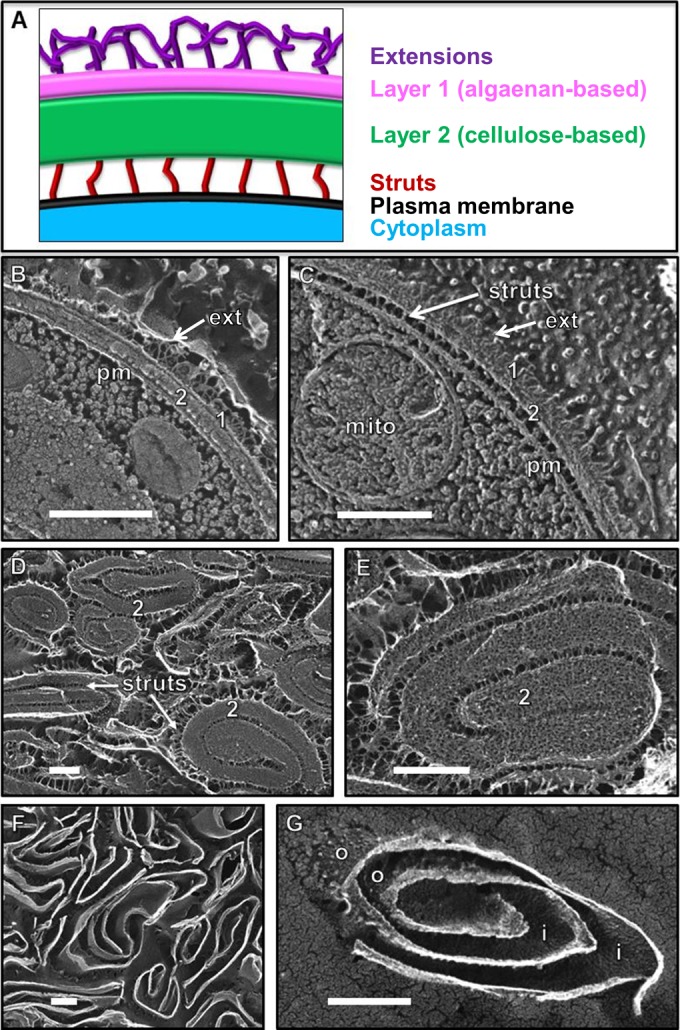
(A) Model of the Nannochloropsis wall based on quick-freeze deep-etch EM (QFDE-EM) images. The native layer 1 adopts a TLS configuration in thin section (2) but forms a single layer in replicas and is overlain with fibrous extensions. Layer 2 is thicker and often associates with the plasma membrane via narrow struts. (B) Cross fracture through the native wall showing its two layers (1, 2), distal extensions (ext), and close association with the plasma membrane (pm). (C) Tangential fracture through the two native wall layers showing distal extensions (ext) and proximal struts attaching to the plasma membrane (pm). mito, mitochondrion. (D and E) Pressed cell walls, with the prominent fibrillar layer 2 masking the presence of layer 1. (F and G) Same pressed wall preparation as shown in panels D and E after 24 h of incubation in cellulase. Fibrillar layer 2 and struts were digested, exposing layer 1, whose outer (o) and inner (i) faces are granular. All scale bars, 250 nm.
Images of pressed cell walls are shown in Fig. 2D and E. Each wall curves in on itself to form a concentric whorl interconnected by struts. Layer 2 has a fluffy, often porous appearance with a delicate fibrous substructure. These fibers have a far narrower caliber than the cellulose fibrils found in red algae and land plants that are assembled under the aegis of linear rows or rosettes of cellulose synthases, and no linear rows or rosettes are evident in the plasma membrane of N. gaditana.
Figure 2F and G show this same pressed wall preparation after digestion with cellulase 1 as described in Materials and Methods. Layer 2 and the struts have been completely removed, generating an apparently pure preparation of layer 1 whose outer surface (Fig. 2G, o) retains the extensions and whose newly exposed inner layer (Fig. 2G, i) is granular.
Spectroscopy of intact and enzyme-digested wall preparations.
The presence of lipid-derived, nonhydrolyzable algaenan is well established in N. salina and other species of Nannochloropsis, where it is described as comprising highly saturated, long-chain aliphatic material that is cross-linked by ether bonds (41, 91). We found a similar material in the residual material after cellulase digestion of N. gaditana cell walls. Whereas algaeanan has heretofore been distinguished from other biomass constituents as an insoluble residue following severe acid and base hydrolysis, our enzymatic approach has allowed the study of material unlikely to be chemically altered or contaminated with artifacts such as Maillard condensation or caramelization products (92).
Aliquots of pressed cell walls, sequentially exposed to a cellulase and then a protease and then to HCl acid hydrolysis, were analyzed by attenuated total reflectance Fourier transform infrared (ATR-FTIR) spectroscopy (Fig. 3; Table 6). Spectra of nondigested walls are dominated by chemical groups associated with carbohydrates, such as the broad band centered around 3,380 cm−1 (hydroxyl, H-bonded, O-H stretch), the intense band maximizing at 1,029 cm−1 (hydroxyl/ether, C-O stretch), and the narrow band at 1,157 cm−1 (glycosidic ether, C-O-C stretch). Treatment with cellulase greatly diminishes these bands, although it does not eliminate them entirely, and allows vibrational frequencies associated with the long aliphatic chains of algaenan to become more apparent, such as intense bands at 2,920 cm−1 (methylene, C-H symmetric stretch) and 2,852 cm−1 (methyl/methylene, C-H asymmetric stretch), plus narrow bands at 1,465 cm−1 (methylene, C-H scissoring), 1,376 cm−1 (methyl, C-H rock), and 720 cm−1 [methylene, −(CH2)n when n ≥ 3, C-H rock]. Notably, low-intensity bands, typically indicative of protein, are also visible at 1,646 cm−1 (amide I, C=O stretch), 1,540 cm−1 (amide II, N-H/C-N stretch), and 1,307 cm−1 (amide III, C-N stretch/N-H bend).
TABLE 6.
Approximate wavenumber assignments of major bands in ATR-FTIR results
| Region | Wavenumber range (cm−1) | Major bands (cm−1) | Functional groups | Comment |
|---|---|---|---|---|
| a | 3,170–3,630 | 3,380 or 3,445 | ν(O-H)/ν(N-H) | Very broad; change of center after HCl treatment |
| b | 2,800–3,020 | 2,920, 2,852 | ν(CH3)/ν(CH2) | Alkane |
| c | 1,570–1,770 | 1,610, 1,720 | νC = C/νC = O | Alkene/ester |
| d | 1,380–1,520 | 1,380, 1,465, 1,512 | δ(CH3)/δ(CH2)/δ(C = C) | CH3 and CH2 scissoring; aromatic C=C ring breathing |
| e | 1,230–1,290 | 1,240 | ν(C-O-C) | Ether |
| f | 980–1,200 | 1,029, 1,157 | ν(C-O)/δ(O-H) | 1,157 cm−1 eliminated by HCl treatment |
| g | 880–930 | 900 | ωo(C-H) | CH wag (out of plane) |
| h | 770–870 | 827 | δ(C-H)/τ(C-O-C) | Trisubstituted alkene/epoxide |
| i | 670–750 | 720 | ρ(CH2)n | Long-chain (CH2) rocking; n > 3 |
Interestingly, protease treatment does not alter these protein-indicative bands—in fact, no spectral changes are evident—whereas they are abolished with HCl treatment. Such treatment also elicits a distinct shift of the highest-wavenumber band to a new wavenumber, centered near 3,445 cm−1, indicating that the 3,380 cm−1-centered bands in pre-HCl samples may also obscure an amide N-H stretching contribution (secondary amides are generally assigned near 3,300 cm−1). Although aliphatic wavenumbers are obviously pronounced following HCl treatment and dominate the spectra, additional, often weak, bands indicative of oxygen moieties are also abundant.
Our spectra generally agree with the conclusions of Gelin et al. (78) without being potentially compromised by preparative artifacts. However, our HCl treatment of enzyme-hydrolyzed pressed walls also reveals a distinct band at 1,719 cm−1 (C=O stretch) which may indicate carboxyl, aldehyde, ketone, or ester functional groups. Close observation of many wavenumbers over the entire spectrum strongly suggests the presence of carboxyl groups, but this does not preclude other oxygen-containing moieties.
DISCUSSION
Notes on the cell wall preparations.
Nannochloropsis gaditana cells were grown in either f/2 or ASW medium, and their cell walls isolated from freeze-dried biomass after successive steps, including (i) bursting the cells in a French press, (ii) separating walls from other cellular components on a sucrose gradient, and (iii) extracting the wall fraction with solvents of decreasing polarity to remove contaminants (e.g., sucrose, adsorbing salts, proteins, lipids, and carotenoids). Because material was lost during the French press and sucrose separation steps, we were unable to quantify cell wall mass as a percentage of cell dry weight; however, the final yields (after sample losses) were ∼2% of the cell dry weight. Upon inspection with electron microscopy, the wall preparations appeared highly homogeneous and uncontaminated by other materials (Fig. 2D and E).
The pressed wall preparation is a variation of that published by Takeda (93), with the use of a sucrose rather than a Percoll gradient being dictated by concerns that residual Percoll might complicate glycosyl linkage analysis. No trace of sucrose or fructose could be found in the resulting preparations when digested and analyzed by IC. The Takeda method also includes an α-amylase digestion to remove starch, but this step was deemed unnecessary because (i) Nannochloropsis species are not believed to produce starch, as is also the case for other stramenopiles (94), (ii) no starch grains are evident by QFDE-EM of intact cells, and (iii) genomic evidence of starch biosynthetic enzymes (i.e., starch synthase and isoamylase) is lacking (5, 9, 40, 68, 93, 95–97). Furthermore, the low abundance of 1,3-linked glucose detected, combined with the absence of detectable mannitol, suggested minimal contamination with laminarin, a storage product common in related brown algae (98). It should be noted that the β-1,3-linked storage polysaccharide presumed to be laminarin has not been verified in Nannochloropsis, nor has chrysolaminarin (99) or mycolaminarin (100), which are found in more distantly related diatoms and oomycetes, respectively.
We are aware that the use of chemical treatments to remove adventitious material may inadvertently remove constituent wall molecules as well. That said, during the organic extraction steps, only ∼5% of the sucrose gradient-separated cell wall mass was removed (data not shown).
Cellulose in the N. gaditana cell wall.
The bulk (∼75%) of N. gaditana cell wall preparations was determined to be cellulose, which we show by QFDE-EM to form the inner layer of a bilayered wall. Monosaccharide quantification after both chemical and enzymatic digestion of the cell wall preparations demonstrates that glucose is by far the dominant carbohydrate (Table 1). Linkage analysis confirmed the predominance of 1,4-linked glucose (Table 2); however, as this methodology uses relatively mild hydrolysis conditions, only a fraction of all carbohydrate linkages was determined. The pressed wall material was highly susceptible to cellulase digestion, as evidenced by both HPAEC-PAD and EM data (Table 1; Fig. 2F and G). The lyticase and zymolyase enzymes liberated little free glucose, but TFA digestion of the enzyme hydrolysates produced predominantly glucose (see Table S2 in the supplemental material); we therefore suggest that they are not acting directly on the major wall polysaccharide but, rather, act on critical cross-links to release the wall polymers. All of these treatments liberated glucose and only traces of other saccharides, consistent with a predominately cellulosic material.
Cellulose synthesis is performed in the plasma membrane by the action of cellulose synthase complexes, also called terminal complexes (101–103). The primary substrate for cellulose synthase, UDP-glucose, is synthesized either anabolically from glucose by UDP-glucose pyrophosphorylase or catabolically from sucrose by sucrose synthase (102). We have identified in silico homologs to three cellulose synthases containing core amino acids central to activity, two UDP-glucose pyrophosphorylases, and seven cellulases, demonstrating that N. gaditana at least putatively possesses the metabolic machinery for cellulose synthesis and catabolism. A search for sucrose synthase sequences derived from 18 plant species yielded no homologues in the two N. gaditana genomes and one N. oceanica genome queried, suggesting that Nannochloropsis utilizes the anabolic pathway to UDP-glucose formation.
Cellulose synthase terminal complexes assemble in geometrical configurations, often as linear rows or hexagonal rosettes, and cellulose morphology is directly related to these configurations (103). Commonly found in land plants and some algae, rosettes tend to produce the crystalline cellulose Iβ allomorph; for example, the rosette terminal complexes of Micrasterias and Spyrogyra are thought to correlate with the perpendicular orientation of the cellulose Iβ fibrils (104). Linear terminal complexes have also been characterized in algae, such as Valonia, Oocystis, and Boergesneia, and are associated with cellulose Iα production (103, 105–107). The configuration of Nannochloropsis terminal complexes and the degree of cellulose crystallinity are currently unknown. CESA3 is similar to a cellulose synthase from the brown alga E. siliculosus that forms long, linear terminal complexes (108). No such configurations have been visualized in the Nannochloropsis cell membrane in our QFDE-EM studies. CESA1 and CESA2 are highly similar to the bacterial BcsA enzymes, which in the case of Escherichia coli have been suggested to produce amorphous cellulose (65).
Processive β-glycosyltransferases contain the characteristic motifs D, D, D, and QXXRW located in two protein domains, A (D, D) and B (D, QXXRW). Both domain A and B are conserved in processive cellulase enzymes, while nonprocessive enzymes typically lack domain B (109). CESA1, -2, and -3 have both the A and B domain, whereas CESA4 is lacking key residues that coordinate UDP (DCD, R in QXXRW), act as the catalytic base (ED), and stabilize the acceptor glucan (W in QXXRW), indicating that despite homology, CESA4 likely does not encode a genuine cellulose synthase.
Three of the E. siliceous cellulose synthases are highly expressed in female gametes just after fertilization, which correlates with rapid primary cell wall biogenesis (110). These three CESA genes are members of the E. siliceous CESA clade that forms a cluster with Nannochloropsis CESA3 (Fig. 1A, group A). Despite some differences in the E. siliceous enzyme motifs relative to the sequences of prototypical plant cellulose synthases (Fig. 1B), it seems likely that these enzymes are responsible for producing the microfibrils of cellulose found in the cell wall of E. siliceous. Nannochloropsis CESA3 shares these distinctive motifs with the E. siliceous CESA enzymes, which include DE instead of DD and DAR instead of DCD.
There is a surprising lack of diversity in the monosaccharide composition of the cell wall preparations, although other saccharides and uronic acids, including 1,2-linked and terminal rhamnose and fucose residues, 1,4-linked and terminal mannose and N-acetylglucosamine residues, and 1,6-linked and terminal galactose residues (Table 2), were present in trace amounts. Treatment with cellulase 2 yielded 2.7% of the polysaccharide content as galactose (data not shown), but in all other enzymatic treatments, the masses of nonglucose monosaccharides and uronic acids were less than 1% of the sample mass (Table 1).
Cell wall amino acids.
Amino acid analysis indicated that ∼6% of the cell wall mass is comprised of amino acids. The individual amino acid ratios were highly reproducible between runs, and cell wall preparations were rigorously washed with both polar and nonpolar solvents, suggesting that the amino acids were not adventitiously associated with cell walls but, rather, represented an integral cell wall constituent. Attempts to isolate purified proteins from cell wall preparations incubated with denaturing buffers failed to reveal well resolved protein bands using SDS gel electrophoresis. This may be due to the proteins being embedded within the cell wall and unable to diffuse from this matrix or to proteins being covalently linked to insoluble algaenan (see below) or a cell wall carbohydrate. Additionally, the current cell wall preparations yield relatively small amounts of biomass, and the proteins may be below the limit of detection, particularly if there is heterogeneity due to differing degrees of posttranslational modification (i.e., glycosylation). BLAST searches for homologs of other algal cell wall proteins (e.g., hydroxyproline-rich glycoproteins from C. reinhardtii) failed to reveal any significant matches. Despite our inability to identify cell wall proteins, the amino acid analysis suggests the presence of tightly associated proteins (perhaps covalently linked to other wall constituents or encapsulated within the cell wall). Additional investigation is required to ascertain whether the amino acids correspond to specific polypeptides encoded in the genome or whether they are assembled by nonribosomal pathways (111, 112) that are core components of the cell wall.
We compared the cell wall composition of N. gaditana cells grown in ASW and f/2 media. While subtle differences are noted with respect to some cell wall components, including minerals and minor carbohydrates, the main component of the cell walls (glucose) does not change drastically in response to varying the composition of the medium, demonstrating the dominant structural role of cellulose in the N. gaditana wall. It remains to be determined whether stresses like nutrient limitation and high light may affect the composition of Nannochloropsis cell walls.
The algaenan outer wall.
The FTIR and EM data presented in this paper clearly demonstrate that the outer wall of N. gaditana contains algaenan (Fig. 2 and 3), which we propose to be primarily responsible for the wall's recalcitrance to breakage. The term algaenan encompasses disparate types of enzymatically and chemically resistant aliphatic material that may derive from distinct biochemical pathways in different organisms. The algaenan of Nannochloropsis salina and an unspecified Nannochloropsis has been proposed to comprise straight-chain (∼C30), highly saturated aliphatic compounds joined by ether bonds at terminal and one or two midchain positions (41). There is some debate about the structure of algaenan in Tetraedron minimum and Scenedesmus communis, but it appears to consist of very-long-chain (up to C120) monomeric (di)carboxylic acids and not the ester- and ether-linked polymers that were originally proposed (43, 113). Meanwhile, the algaenan found in the colonial matrix of three races of Botryococcus braunii comprises polyacetals that are either cross-linked by terpene epoxides (B and L race) or not (A race) (114, 115).
The composition of algaenan has been ascertained using diverse methodologies that have the potential to affect structural determinations (43, 44). To date, isolation has depended on severe hydrolysis reactions at high temperature to degrade any reactive polysaccharides, lipids, and proteins (41, 44, 113). Such harsh treatments have invited the criticism that the fundamental algaenan composition may be altered or contaminated with by-products, such as Maillard condensation products (44). In the present study, a 24-h digestion of pressed cell walls from N. gaditana at room temperature, first with cellulase and then protease, left behind what appeared to be a homogeneous preparation of algaenan-containing outer cell walls (Fig. 2F and G), albeit with some remaining carbohydrate and protein signatures (Fig. 3). This enzymatic approach permitted the analysis of a highly enriched algaenan preparation without exposure to high temperature or strongly oxidizing/reducing conditions. Subsequent acid hydrolysis reduced, if not eliminated, the carbohydrate and protein signatures in the FTIR but may have created artifacts (discussed below). However, comparison of the FTIR spectra of the material before and after acid hydrolysis afforded us the opportunity to separate artifactual features from genuine ones.
The results after cellulase treatment demonstrate the removal of carbohydrate (and potentially protein) by the sizable reduction of related FTIR bands, such as those associated with glycosidic ethers (1,157 cm−1) and amides (1,646 cm−1). Saponification is often performed to remove ester-linked fatty acids from crude algaenan (41, 44, 113); however, saponification followed by acid-catalyzed methyl esterification (fatty acid methyl ester [FAME] analysis) on a subset of algaenan preparations did not yield detectable fatty acid methyl esters by GC-flame ionization detection analysis (data not shown). Since there was a clear lack of ester functionalities in our material, as demonstrated by the FTIR data and FAME analysis, this obviated the need for saponification in subsequent sample analysis. The lack of esters is perhaps attributable to the preparatory steps employed, which may have yielded a material less contaminated (i.e., by membranes) than in previous work.
Our FTIR data generally support the proposed Nannochloropsis algaenan model of Gelin et al. (78) by confirming the predominance of long-chain methylenic stretches with ether linkages and few sites of unsaturation. However, there are notable differences between our two sets of results. First, our data suggest that alcohol moieties are present and that some of these may be midchain. The latter conclusion is drawn from comparing the FTIR spectra before and after acid hydrolysis of the enzymatically isolated algaenan. After acid hydrolysis, alcohol-related bands at 3,380 cm−1and 1,052 cm−1 are diminished and trans alkenes become more apparent with the band at 965 cm−1. The disappearance of the alcohol bands can be attributed, at least in part, to removal of residual carbohydrates, but the emergence of trans unsaturation intimates the dehydration of secondary alcohols to form alkenes. Alternatively, the removal of carbohydrate –OH bands may have simply unmasked preexisting carbon double bonds. Alcohol groups were either not observed or were of low abundance in the algaenan of other studies, but sites of unsaturation were observed (41, 43, 113). Sites of unsaturation may be derived from dehydration reactions, since the algaenan in those studies was isolated only after severe acid hydrolysis. In addition to finding alcohol groups in the algaenan material, we observed indications of carboxyl and/or aldehyde functionalities after acid hydrolysis, neither of which was identified in the prior study of Nannochloropsis algaenan, although the former was found in the algaenans of freshwater algae (41, 43).
Nannochloropsis algaenans appear to be much the same biopolymer as the cutan found in drought-resistant land plants, such as Agave and Clivia (45, 113, 116–118). Isolation of cutan from plant cuticular material is performed in much the same way as classical algaenan isolation, namely, through solvent extraction and the application of strong reducing/oxidizing reagents to remove carbohydrates, proteins, and free and ester-bound lipids (45, 116–119). As with the characterization of algaenan, structural determinations of cutan are uncertain because of study-to-study variation in isolation procedures and the potential for artifact generation introduced by aggressive isolation approaches (45, 118, 120). However, it appears that this material comprises long-chain (∼C30) alkanes and alkenes joined by ether linkages and is, thus, at least an analogue of Nannochloropsis algaenan (45, 121).
Given the structural similarities between cutan and Nannochloropsis algaenan, it is tempting to speculate that similar biosynthetic machinery produces the two compounds. Radiolabeled linoleic acid (C18:2) fed to Clivia miniata is incorporated into the aliphatic residue (i.e., cutan-rich material) of its leaves (118). Intriguingly, Nannochloropsis produces C18:1, C18:2, and C18:3 fatty acids, predominantly C18:1, and its algaenan contains long-chain aliphatics with substitutions at the ω18 position (9, 41, 122). A number of possible algaenan precursors functionalized or unsaturated at what would be the ω18 position (the nomenclature changes according to functionalization) have been found in Nannochloropsis, including C32:1 alcohols, C32 and C32:1 diols, C32 hydroxy fatty acids, C32 dihydroxy fatty acids, and C32 hydoxy ketones (41, 123, 124). This suggests that a C18 fatty acid may either be elongated or condensed with a similar fatty molecule to produce C28-C34 algaenan constituents, perhaps by the action of polyketide synthase(s).
Six polyketide synthases have been identified in silico in the CCMP 526 genome. Although the functions of these enzymes are not yet known, we consider these promising targets for future interrogation in algaenan biosynthesis. Two lack dehydratase and enoyl reductase modules, and three have fatty acyl-reductase domains, consistent with the synthesis of predominately methylenic monomers with alcohols/aldehydes for cross-linking. The FTIR spectra of algaenan indicate few CH3 moieties relative to CH2 moieties, suggesting that a predominately terpenoid pathway to algaenan biosynthesis is unlikely. Although extensive work remains to characterize the composition and biosynthetic routes to algaenan and its cross-linking, the methods for algaenan purification described here and the availability of multiple Nannochloropsis genomes will greatly facilitate such efforts.
Supplementary Material
ACKNOWLEDGMENTS
This material is based upon work supported by the Sustainable Algal Biofuels Consortium funded at CSM by the state of Colorado Energy Collaboratory in support of award DE-EE0003372 from the U.S. Department of Energy, Bioenergy Technology Office. H.G.G. was funded by the New American University Fellowship at Arizona State University. Additional support was provided to M.J.S. and M.C.P. by the Air Force Office of Scientific Research (grant FA9550-11-1-0211). U.G. was funded by contract DE-EE0003046 awarded to the National Alliance for Advanced Biofuels and Bioproducts (NAABB) from the U.S. Department of Energy and by grant SC0006873 from the DOE Office of Biological and Environmental Research.
Footnotes
Published ahead of print 19 September 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/EC.00183-14.
REFERENCES
- 1.Andersen RA, Brett RW, Potter D, Sexton JP. 1998. Phylogeny of the Eustigmatophyceae based upon 18S rDNA, with emphasis on Nannochloropsis. Protist 149:61–74. 10.1016/S1434-4610(98)70010-0. [DOI] [PubMed] [Google Scholar]
- 2.Murakami R, Hashimoto H. 2009. Unusual nuclear division in Nannochloropsis oculata (Eustigmatophyceae, Heterokonta) which may ensure faithful transmission of secondary plastids. Protist 160:41–49. 10.1016/j.protis.2008.09.002. [DOI] [PubMed] [Google Scholar]
- 3.Rodolfi L, Zittelli GC, Barsanti L, Rosati G, Tredici MR. 2003. Growth medium recycling in Nannochloropsis sp mass cultivation. Biomol. Eng. 20:243–248. 10.1016/S1389-0344(03)00063-7. [DOI] [PubMed] [Google Scholar]
- 4.Bondioli P, Della Bella L, Rivolta G, Chini Zittelli G, Bassi N, Rodolfi L, Casini D, Prussi M, Chiaramonti D, Tredici MR. 2012. Oil production by the marine microalgae Nannochloropsis sp. F&M-M24 and Tetraselmis suecica F&M-M33. Bioresour. Technol. 114:567–572. 10.1016/j.biortech.2012.02.123. [DOI] [PubMed] [Google Scholar]
- 5.Jinkerson RE, Radakovits R, Posewitz MC. 2013. Genomic insights from the oleaginous model alga Nannochloropsis gaditana. Bioengineered 4:37–43. 10.4161/bioe.21880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moazami N, Ashori A, Ranjbar R, Tangestani M, Eghtesadi R, Nejad AS. 2012. Large-scale biodiesel production using microalgae biomass of Nannochloropsis. Biomass Bioenerg. 39:449–453. 10.1016/j.biombioe.2012.01.046. [DOI] [Google Scholar]
- 7.Quinn JC, Yates T, Douglas N, Weyer K, Butler J, Bradley TH, Lammers PJ. 2012. Nannochloropsis production metrics in a scalable outdoor photobioreactor for commercial applications. Bioresour. Technol. 117:164–171. 10.1016/j.biortech.2012.04.073. [DOI] [PubMed] [Google Scholar]
- 8.Radakovits R, Jinkerson RE, Darzins A, Posewitz MC. 2010. Genetic engineering of algae for enhanced biofuel production. Eukaryot. Cell 9:486–501. 10.1128/EC.00364-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Radakovits R, Jinkerson RE, Fuerstenberg SI, Tae H, Settlage RE, Boore JL, Posewitz MC. 2012. Draft genome sequence and genetic transformation of the oleaginous alga Nannochloropsis gaditana. Nat. Comm. 3:686. 10.1038/ncomms1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lubián L, Montero O, Moreno-Garrido I, Huertas IE, Sobrino C, González-del Valle M, Parés G. 2000. Nannochloropsis (Eustigmatophyceae) as source of commercially valuable pigments. J. Appl. Phycol. 12:249–255. 10.1023/A:1008170915932. [DOI] [Google Scholar]
- 11.Krienitz L, Wirth M. 2006. The high content of polyunsaturated fatty acids in Nannochloropsis limnetica (Eustigmatophyceae) and its implication for food web interactions, freshwater aquaculture and biotechnology. Limnologica 36:204–210. 10.1016/j.limno.2006.05.002. [DOI] [Google Scholar]
- 12.Sukenik A. 1991. Ecophysiological considerations in the optimization of eicosapentaenoic acid production by Nannochloropsis sp. (Eustigmatophyceae). Bioresour. Technol. 35:263–269. 10.1016/0960-8524(91)90123-2. [DOI] [Google Scholar]
- 13.Ferreira M, Coutinho P, Seixas P, Fábregas J, Otero A. 2009. Enriching rotifers with “premium” microalgae. Nannochloropsis gaditana. Mar. Biotechnol. (N. Y.) 11:585–595. 10.1007/s10126-008-9174-x. [DOI] [PubMed] [Google Scholar]
- 14.Cheng Y-S, Zheng Y, Labavitch JM, VanderGheynst JS. 2011. The impact of cell wall carbohydrate composition on the chitosan flocculation of Chlorella. Process Biochem. 46:1927–1933. 10.1016/j.procbio.2011.06.021. [DOI] [Google Scholar]
- 15.Adesanya VO, Vadillo DC, Mackley MR. 2012. The rheological characterization of algae suspensions for the production of biofuels. J. Rheol. (N. Y. N. Y.) 56:925–939. 10.1122/1.4717494. [DOI] [Google Scholar]
- 16.Eggers R, Sievers U, Stein W. 1985. High pressure extraction of oil seed. J. Am. Oil Chem. Soc. 62:1222–1230. 10.1007/BF02541832. [DOI] [Google Scholar]
- 17.Sowbhagya H, Sushma SB, Rastogi N, Naidu MM. 2013. Effect of pretreatments on extraction of pigment from marigold flower. J. Food Sci. Technol. 50:122–128. 10.1007/s13197-011-0313-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Imam SH, Buchanan MJ, Shin HC, Snell WJ. 1985. The Chlamydomonas cell wall: characterization of the wall framework. J. Cell Biol. 101:1599–1607. 10.1083/jcb.101.4.1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goodenough UW, Heuser JE. 1985. The Chlamydomonas cell wall and its constituent glycoproteins analyzed by the quick-freeze, deep-etch technique. J. Cell Biol. 101:1550–1568. 10.1083/jcb.101.4.1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee J-H, Waffenschmidt S, Small L, Goodenough U. 2007. Between-species analysis of short-repeat modules in cell wall and sex-related hydroxyproline-rich glycoproteins of Chlamydomonas. Plant Phys. 144:1813–1826. 10.1104/pp.107.100891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Work VH, Bentley FK, Scholz MJ, D'Adamo S, Gu HY, Vogler BW, Franks DT, Stanish LF, Jinkerson RE, Posewitz MC. 2013. Biocommodities from photosynthetic microorganisms. Environ. Prog. Sustain. Energy 32:989–1001. 10.1002/ep.11849. [DOI] [Google Scholar]
- 22.Kloareg B, Quatrano RS. 1988. Structure of the cell walls of marine algae and ecophysiological functions of the matrix polysaccharides. Oceanogr. Mar. Biol. Annu. Rev. 26:259–315. [Google Scholar]
- 23.Wang S-B, Hu Q, Sommerfeld M, Chen F. 2004. Cell wall proteomics of the green alga Haematococcus pluvialis (Chlorophyceae). Proteomics 4:692–708. 10.1002/pmic.200300634. [DOI] [PubMed] [Google Scholar]
- 24.Michel G, Tonon T, Scornet D, Cock JM, Kloareg B. 2010. The cell wall polysaccharide metabolism of the brown alga Ectocarpus siliculosus. Insights into the evolution of extracellular matrix polysaccharides in Eukaryotes. New Phytol. 188:82–97. 10.1111/j.1469-8137.2010.03374.x. [DOI] [PubMed] [Google Scholar]
- 25.Gelin F, Volkman JK, Largeau C, Derenne S, Damste JSS, De Leeuw JW. 1999. Distribution of aliphatic, nonhydrolyzable biopolymers in marine microalgae. Org Geochem. 30:147–159. 10.1016/S0146-6380(98)00206-X. [DOI] [Google Scholar]
- 26.Kodner RB, Surnmons RE, Knoll AH. 2009. Phylogenetic investigation of the aliphatic, non-hydrolyzable biopolymer algaenan, with a focus on green algae. Org. Geochem. 40:854–862. 10.1016/j.orggeochem.2009.05.003. [DOI] [Google Scholar]
- 27.Burczyk J, Szkawran H, Zontek I, Czygan FC. 1981. Carotenoids in the outer cell wall layer of Scenedesmus (Chlorophyceae). Planta 151:247–250. 10.1007/BF00395176. [DOI] [PubMed] [Google Scholar]
- 28.Koivikko R, Loponen J, Honkanen T, Jormalainen V. 2005. Contents of soluble, cell wall-bound AND exuded phlorotannins in the brown alga Fucus vesiculosus, with implications on their ecological functions. J. Chem. Ecol. 31:195–212. 10.1007/s10886-005-0984-2. [DOI] [PubMed] [Google Scholar]
- 29.Martone PT, Estevez JM, Lu FC, Ruel K, Denny MW, Somerville C, Ralph J. 2009. Discovery of lignin in seaweed reveals convergent evolution of cell-wall architecture. Curr. Biol. 19:169–175. 10.1016/j.cub.2008.12.031. [DOI] [PubMed] [Google Scholar]
- 30.Sorensen I, Pettolino FA, Bacic A, Ralph J, Lu FC, O'Neill MA, Fei ZZ, Rose JKC, Domozych DS, Willats WGT. 2011. The charophycean green algae provide insights into the early origins of plant cell walls. Plant J. 68:201–211. 10.1111/j.1365-313X.2011.04686.x. [DOI] [PubMed] [Google Scholar]
- 31.Tamura H, Mine I, Okuda K. 1996. Cellulose-synthesizing terminal complexes and microfibril structure in the brown alga Sphacelaria rigidula (Sphacelariales, Phaeophyceae). Phycol. Res. 44:63–68. 10.1111/j.1440-1835.1996.tb00039.x. [DOI] [Google Scholar]
- 32.Kapaun E, Reisser W. 1995. A chitin-like glycan in the cell wall of a Chlorella sp. (Chlorococcales, Chlorophyceae). Planta 197:577–582. [Google Scholar]
- 33.Domozych D, Stewart K, Mattox K. 1980. The comparative aspects of cell wall chemistry in the green algae (chlorophyta). J. Mol. Evol. 15:1–12. 10.1007/BF01732578. [DOI] [PubMed] [Google Scholar]
- 34.Domozych DS, Serfis A, Kiemle SN, Gretz MR. 2007. The structure and biochemistry of charophycean cell walls. I. Pectins of Penium margaritaceum. Protoplasma 230:99–115. 10.1007/s00709-006-0197-8. [DOI] [PubMed] [Google Scholar]
- 35.Berteau O, Mulloy B. 2003. Sulfated fucans, fresh perspectives: structures, functions, and biological properties of sulfated fucans and an overview of enzymes active toward this class of polysaccharide. Glycobiology 13:29R–40R. 10.1093/glycob/cwg058. [DOI] [PubMed] [Google Scholar]
- 36.Lahaye M, Robic A. 2007. Structure and functional properties of ulvan, a polysaccharide from green seaweeds. Biomacromolecules 8:1765–1774. 10.1021/bm061185q. [DOI] [PubMed] [Google Scholar]
- 37.Michel G, Helbert W, Kahn R, Dideberg O, Kloareg B. 2003. The structural bases of the processive degradation of ι-carrageenan, a main cell wall polysaccharide of red algae. J. Mol. Biol. 334:421–433. 10.1016/j.jmb.2003.09.056. [DOI] [PubMed] [Google Scholar]
- 38.Ford CW, Percival E. 1965. Paper 551. Polysaccharides synthesised by Monodus subterraneus. Part II. The cell wall glucan. J. Chem. Soc. (Resumed) 0:3014–3016. 10.1039/JR9650003014. [DOI] [Google Scholar]
- 39.Brown MR. 1991. The amino-acid and sugar composition of 16 species of microalgae used in mariculture. J. Exp. Mar. Biol. Ecol. 145:79–99. 10.1016/0022-0981(91)90007-J. [DOI] [Google Scholar]
- 40.Vieler A, Wu G, Tsai CH, Bullard B, Cornish AJ, Harvey C, Reca IB, Thornburg C, Achawanantakun R, Buehl CJ, Campbell MS, Cavalier D, Childs KL, Clark TJ, Deshpande R, Erickson E, Armenia Ferguson A, Handee W, Kong Q, Li X, Liu B, Lundback S, Peng C, Roston RL, Sanjaya Simpson JP, Terbush A, Warakanont J, Zauner S, Farre EM, Hegg EL, Jiang N, Kuo MH, Lu Y, Niyogi KK, Ohlrogge J, Osteryoung KW, Shachar-Hill Y, Sears BB, Sun Y, Takahashi H, Yandell M, Shiu SH, Benning C. 2012. Genome, functional gene annotation, and nuclear transformation of the heterokont oleaginous alga Nannochloropsis oceanica CCMP1779. PLoS Genet. 8:e1003064. 10.1371/journal.pgen.1003064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gelin F, Volkman JK, deLeeuw JW, Damste JSS. 1997. Mid-chain hydroxy long-chain fatty acids in microalgae from the genus Nannochloropsis. Phytochemistry 45:641–646. 10.1016/S0031-9422(97)00068-X. [DOI] [Google Scholar]
- 42.Jeffree CE. 2007. The fine structure of the plant cuticle, p 11–125 In Riederer M, Müller C. (ed), Annual plant reviews, vol 23 Blackwell Publishing Ltd., Oxford, United Kingdom. [Google Scholar]
- 43.Allard B, Rager MN, Templier J. 2002. Occurrence of high molecular weight lipids (C80+) in the trilaminar outer cell walls of some freshwater microalgae. A reappraisal of algaenan structure. Org. Geochem. 33:789–801. 10.1016/S0146-6380(02)00029-3. [DOI] [Google Scholar]
- 44.Allard B, Templier J, Largeau C. 1998. An improved method for the isolation of artifact-free algaenans from microalgae. Org. Geochem. 28:543–548. 10.1016/S0146-6380(98)00012-6. [DOI] [Google Scholar]
- 45.Boom A, Damste JSS, de Leeuw JW. 2005. Cutan, a common aliphatic biopolymer in cuticles of drought-adapted plants. Org. Geochem. 36:595–601. 10.1016/j.orggeochem.2004.10.017. [DOI] [Google Scholar]
- 46.Sluiter AD, Hames BR, Ruiz RO, Scarlata C, Sluiter JB, Templeton DW, Crocker D. 2008. Determination of structural carbohydrates and lignin in biomass: laboratory analytical procedure (LAP). National Renewable Energy Laboratory, Golden, CO. [Google Scholar]
- 47.Morris DL. 1948. Quantitative determination of carbohydrates with Dreywood's anthrone reagent. Science 107:254–255. 10.1126/science.107.2775.254. [DOI] [PubMed] [Google Scholar]
- 48.York WS, Darvill AG, McNeil M, Stevenson TT, Albersheim P. 1986. Isolation and characterization of plant cell walls and cell wall components. Methods Enzymol. 118:3–40. 10.1016/0076-6879(86)18062-1. [DOI] [Google Scholar]
- 49.Heuser JE. 2011. The origins and evolution of freeze-etch electron microscopy. J. Electron Microsc. (Tokyo) 60:S3–S29. 10.1093/jmicro/dfr044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Girke T, Lauricha J, Tran H, Keegstra K, Raikhel N. 2004. The Cell Wall Navigator Database. A systems-based approach to organism-unrestricted mining of protein families involved in cell wall metabolism. Plant Physiol. 136:3003–3008. 10.1104/pp.104.049965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cantarel BL, Coutinho PM, Rancurel C, Bernard T, Lombard V, Henrissat B. 2009. The Carbohydrate-Active enZymes database (CAZy): an expert resource for glycogenomics. Nucleic Acids Res. 37:D233–D238. 10.1093/nar/gkn663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402. 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792–1797. 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zuckercandl E, Pauling L. 1965. Evolutionary divergence and convergence in proteins, p 97–166 In Bryson V, Vogel HJ. (ed), Evolving genes and proteins. Academic Press, New York, NY. [Google Scholar]
- 55.Felsenstein J. 1985. Confidence-limits on phylogenies—an approach using the bootstrap. Evolution 39:783–791. 10.2307/2408678. [DOI] [PubMed] [Google Scholar]
- 56.Sethaphong L, Haigler CH, Kubicki JD, Zimmer J, Bonetta D, DeBolt S, Yingling YG. 2013. Tertiary model of a plant cellulose synthase. Proc. Natl. Acad. Sci. U. S. A. 110:7512–7517. 10.1073/pnas.1301027110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Paul R, Jinkerson RE, Buss K, Steel J, Mohr R, Hess WR, Chen M, Fromme P. 2014. Draft genome sequence of the filamentous cyanobacterium Leptolyngbya sp. strain Heron Island J, exhibiting chromatic acclimation. Genome Announc. 2(1):e01166–13. 10.1128/genomeA.01166-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Morgan JL, Strumillo J, Zimmer J. 2013. Crystallographic snapshot of cellulose synthesis and membrane translocation. Nature 493:181–186. 10.1038/nature11744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28:2731–2739. 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yin Y, Huang J, Xu Y. 2009. The cellulose synthase superfamily in fully sequenced plants and algae. BMC Plant Biol. 9:99. 10.1186/1471-2229-9-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Omadjela O, Narahari A, Strumillo J, Melida H, Mazur O, Bulone V, Zimmer J. 2013. BcsA and BcsB form the catalytically active core of bacterial cellulose synthase sufficient for in vitro cellulose synthesis. Proc. Natl. Acad. Sci. U. S. A. 110:17856–17861. 10.1073/pnas.1314063110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nobles D, Brown RM. 2004. The pivotal role of cyanobacteria in the evolution of cellulose synthases and cellulose synthase-like proteins. Cellulose 11:437–448. 10.1023/B:CELL.0000046339.48003.0e. [DOI] [Google Scholar]
- 63.Nobles DR, Romanovicz DK, Brown RM., Jr 2001. Cellulose in cyanobacteria. Origin of vascular plant cellulose synthase? Plant Physiol. 127:529–542. 10.1104/pp.010557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kawano Y, Saotome T, Ochiai Y, Katayama M, Narikawa R, Ikeuchi M. 2011. Cellulose accumulation and a cellulose synthase gene are responsible for cell aggregation in the cyanobacterium Thermosynechococcus vulcanus RKN. Plant Cell Physiol. 52:957–966. 10.1093/pcp/pcr047. [DOI] [PubMed] [Google Scholar]
- 65.Le Quere B, Ghigo JM. 2009. BcsQ is an essential component of the Escherichia coli cellulose biosynthesis apparatus that localizes at the bacterial cell pole. Mol. Microbiol. 72:724–740. 10.1111/j.1365-2958.2009.06678.x. [DOI] [PubMed] [Google Scholar]
- 66.Zogaj X, Nimtz M, Rohde M, Bokranz W, Romling U. 2001. The multicellular morphotypes of Salmonella typhimurium and Escherichia coli produce cellulose as the second component of the extracellular matrix. Mol. Microbiol. 39:1452–1463. 10.1046/j.1365-2958.2001.02337.x. [DOI] [PubMed] [Google Scholar]
- 67.Saxena IM, Brown RM. 1997. Identification of cellulose synthase(s) in higher plants: sequence analysis of processive beta-glycosyltransferases with the common motif 'D, D, D35Q(R,Q)XRW'. Cellulose 4:33–49. 10.1023/A:1018411101036. [DOI] [Google Scholar]
- 68.Corteggiani Carpinelli E, Telatin A, Vitulo N, Forcato C, D'Angelo M, Schiavon R, Vezzi A, Giacometti GM, Morosinotto T, Valle G. 2014. Chromosome scale genome assembly and transcriptome profiling of Nannochloropsis gaditana in nitrogen depletion. Mol. Plant. 7:323–335. 10.1093/mp/sst120. [DOI] [PubMed] [Google Scholar]
- 69.Saloheimo M, Paloheimo M, Hakola S, Pere J, Swanson B, Nyyssonen E, Bhatia A, Ward M, Penttila M. 2002. Swollenin, a Trichoderma reesei protein with sequence similarity to the plant expansins, exhibits disruption activity on cellulosic materials. Eur. J. Biochem. 269:4202–4211. 10.1046/j.1432-1033.2002.03095.x. [DOI] [PubMed] [Google Scholar]
- 70.Cosgrove DJ. 2005. Growth of the plant cell wall. Nat. Rev. Mol. Cell. Biol. 6:850–861. 10.1038/nrm1746. [DOI] [PubMed] [Google Scholar]
- 71.Hazlewood GP, Davidson K, Laurie JI, Huskisson NS, Gilbert HJ. 1993. Gene sequence and properties of Cell, a family E endoglucanase from Clostridium thermocellum. J. Gen. Microbiol. 139:307–316. 10.1099/00221287-139-2-307. [DOI] [PubMed] [Google Scholar]
- 72.Gilad R, Rabinovich L, Yaron S, Bayer EA, Lamed R, Gilbert HJ, Shoham Y. 2003. CelI, a noncellulosomal family 9 enzyme from Clostridium thermocellum, is a processive endoglucanase that degrades crystalline cellulose. J. Bacteriol. 185:391–398. 10.1128/JB.185.2.391-398.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Nishida Y, Suzuki K, Kumagai Y, Tanaka H, Inoue A, Ojima T. 2007. Isolation and primary structure of a cellulase from the Japanese sea urchin Strongylocentrotus nudus. Biochimie 89:1002–1011. 10.1016/j.biochi.2007.03.015. [DOI] [PubMed] [Google Scholar]
- 74.Zhou W, Irwin DC, Escovar-Kousen J, Wilson DB. 2004. Kinetic studies of Thermobifida fusca Cel9A active site mutant enzymes. Biochemistry 43:9655–9663. 10.1021/bi049394n. [DOI] [PubMed] [Google Scholar]
- 75.Koivula A, Ruohonen L, Wohlfahrt G, Reinikainen T, Teeri TT, Piens K, Claeyssens M, Weber M, Vasella A, Becker D, Sinnott ML, Zou JY, Kleywegt GJ, Szardenings M, Stahlberg J, Jones TA. 2002. The active site of cellobiohydrolase Cel6A from Trichoderma reesei: the roles of aspartic acids D221 and D175. J. Am. Chem. Soc. 124:10015–10024. 10.1021/ja012659q. [DOI] [PubMed] [Google Scholar]
- 76.Jeng WY, Wang NC, Lin MH, Lin CT, Liaw YC, Chang WJ, Liu CI, Liang PH, Wang AH. 2011. Structural and functional analysis of three beta-glucosidases from bacterium Clostridium cellulovorans, fungus Trichoderma reesei and termite Neotermes koshunensis. J. Struct. Biol. 173:46–56. 10.1016/j.jsb.2010.07.008. [DOI] [PubMed] [Google Scholar]
- 77.Allard B, Templier J. 2000. Comparison of neutral lipid profile of various trilaminar outer cell wall (TLS)-containing microalgae with emphasis on algaenan occurrence. Phytochemistry 54:369–380. 10.1016/S0031-9422(00)00135-7. [DOI] [PubMed] [Google Scholar]
- 78.Gelin F, Boogers I, Noordeloos AAM, Damste JSS, Riegman R, De Leeuw JW. 1997. Resistant biomacromolecules in marine microalgae of the classes eustigmatophyceae and chlorophyceae: geochemical implications. Org. Geochem. 26:659–675. 10.1016/S0146-6380(97)00035-1. [DOI] [Google Scholar]
- 79.Grossi V, Blokker P, Damste JSS. 2001. Anaerobic biodegradation of lipids of the marine microalga Nannochloropsis salina. Org. Geochem. 32:795–808. 10.1016/S0146-6380(01)00040-7. [DOI] [Google Scholar]
- 80.Pickett-Heaps JD, Staehelin LA. 1975. The ultrastructure of Scenedesmus (Chlorophyceae). II. Cell division and colony formation. J. Phycol. 11:186–202. 10.1111/j.1529-8817.1975.tb02766.x. [DOI] [Google Scholar]
- 81.Staehelin LA, Pickett-Heaps JD. 1975. The ultrastructure of Scenedesmus (Chlorophyceae). I. Species with the “reticulate” or “arty” type of ornamental layer. J. Phycol. 11:163–185. 10.1111/j.1529-8817.1975.tb02765.x. [DOI] [Google Scholar]
- 82.Blokker P, Schouten S, de Leeuw JW, Damste JSS, van den Ende H. 1999. Molecular structure of the resistant biopolymer in zygospore cell walls of Chlamydomonas monoica. Planta 207:539–543. 10.1007/s004250050515. [DOI] [Google Scholar]
- 83.Damiani MC, Leonardi PI, Pieroni OI, Caceres EJ. 2006. Ultrastructure of the cyst wall of Haematococcus pluvialis (Chlorophyceae): wall development and behaviour during cyst germination. Phycologia 45:616–623. 10.2216/05-27.1. [DOI] [Google Scholar]
- 84.Daniel P, Henley J, VanWinkle-Swift K. 2007. Altered zygospore wall ultrastructure correlates with reduced abiotic stress resistance in a mutant strain of Chlamydomonas monoica (Chlorophyta). J. Phycol. 43:112–119. 10.1111/j.1529-8817.2006.00313.x. [DOI] [Google Scholar]
- 85.Grief C, O'Neill MA, Shaw PJ. 1987. The zygote cell wall of Chlamydomonas reinhardtii: a structural, chemical and immunological approach. Planta 170:433–445. 10.1007/BF00402977. [DOI] [PubMed] [Google Scholar]
- 86.Malmberg AE, VanWinkle-Swift KP. 2001. Zygospore germination in Chlamydomonas monoica (Chlorophyta): timing and pattern of secondary zygospore wall degradation in relation to cytoplasmic events. J. Phycol. 37:86–94. 10.1046/j.1529-8817.2001.037001086.x. [DOI] [Google Scholar]
- 87.Montsant A, Zarka A, Boussiba S. 2001. Presence of a nonhydrolyzable biopolymer in the cell wall of vegetative cells and astaxanthin-rich cysts of Haematococcus pluvialis (Chlorophyceae). Mar. Biotechnol. (N. Y.) 3:515–521. 10.1007/s1012601-0051-0. [DOI] [PubMed] [Google Scholar]
- 88.VanWinkle-Swift KP, Rickoll WL. 1997. The zygospore wall of Chlamydomonas monoica (Chlorophyceae): morphogenesis and evidence for the presence of sporopollenin. J. Phycol. 33:655–665. 10.1111/j.0022-3646.1997.00655.x. [DOI] [Google Scholar]
- 89.Wang SB, Chen F, Sommerfeld M, Hu Q. 2005. Isolation and proteomic analysis of cell wall-deficient Haematococcus pluvialis mutants. Proteomics 5:4839–4851. 10.1002/pmic.200400092. [DOI] [PubMed] [Google Scholar]
- 90.Brown DL, Leppard GG, Massalski A. 1976. Fine structure of encystment of the quadriflagellate alga, Polytomella agilis. Protoplasma 90:139–154. 10.1007/BF01276484. [DOI] [PubMed] [Google Scholar]
- 91.Gelin F, Boogers I, Noordeloos AAM, Damste JSS, Hatcher PG, deLeeuw JW. 1996. Novel, resistant microalgal polyethers: an important sink of organic carbon in the marine environment? Geochim. Cosmochim. Acta 60:1275–1280. 10.1016/0016-7037(96)00038-5. [DOI] [Google Scholar]
- 92.Allard B, Templier J, Largeau C. 1997. Artifactual origin of mycobacterial bacteran. Formation of melanoidin-like artifact macromolecular material during the usual isolation process. Org. Geochem. 26:691–703. [Google Scholar]
- 93.Takeda H. 1991. Sugar composition of the cell wall and the taxonomy of Chlorella (Chlorophyceae). J. Phycol. 27:224–232. 10.1111/j.0022-3646.1991.00224.x. [DOI] [Google Scholar]
- 94.Michel G, Tonon T, Scornet D, Cock JM, Kloareg B. 2010. Central and storage carbon metabolism of the brown alga Ectocarpus siliculosus: insights into the origin and evolution of storage carbohydrates in Eukaryotes. New Phytol. 188:67–81. 10.1111/j.1469-8137.2010.03345.x. [DOI] [PubMed] [Google Scholar]
- 95.Dong HP, Williams E, Wang DZ, Xie ZX, Hsia RC, Jenck A, Halden R, Li J, Chen F, Place AR. 2013. Responses of Nannochloropsis oceanica IMET1 to long-term nitrogen starvation and recovery. Plant Physiol. 162:1110–1126. 10.1104/pp.113.214320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Tomaselli L. 2007. The microalgal cell, p 1–19 In Richmond A. (ed), Handbook of microalgal culture: biotechnology and applied phycology. Blackwell Publishing Ltd., Oxford, United Kingdom. [Google Scholar]
- 97.Wang D, Ning K, Li J, Hu J, Han D, Wang H, Zeng X, Jing X, Zhou Q, Su X, Chang X, Wang A, Wang W, Jia J, Wei L, Xin Y, Qiao Y, Huang R, Chen J, Han B, Yoon K, Hill RT, Zohar Y, Chen F, Hu Q, Xu J. 2014. Nannochloropsis genomes reveal evolution of microalgal oleaginous traits. PLoS Genet. 10:e1004094. 10.1371/journal.pgen.1004094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Percival EGV, Ross AG. 1951. Paper 156. The constitution of laminarin. Part II. The soluble laminarin of Laminaria digitata. J. Chem. Soc. (Resumed) 0:720–726. 10.1039/JR9510000720. [DOI] [Google Scholar]
- 99.Beattie A, Hirst EL, Percival E. 1961. Studies on the metabolism of the Chrysophyceae. Comparative structural investigations on leucosin (chrysolaminarin) separated from diatoms and laminarin from the brown algae. Biochem. J. 79:531–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wang MC, Bartnicki-Garcia S. 1974. Mycolaminarans: storage (1→3)-β-D-glucans from the cytoplasm of the fungus phytophthora palmivora. Carbohydr. Res. 37:331–338. 10.1016/S0008-6215(00)82922-5. [DOI] [Google Scholar]
- 101.Carpita NC. 2011. Update on mechanisms of plant cell wall biosynthesis: how plants make cellulose and other (1->4)-beta-D-glycans. Plant Physiol. 155:171–184. 10.1104/pp.110.163360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Endler A, Persson S. 2011. Cellulose synthases and synthesis in Arabidopsis. Mol. Plant. 4:199–211. 10.1093/mp/ssq079. [DOI] [PubMed] [Google Scholar]
- 103.Tsekos I. 1999. The sites of cellulose synthesis in algae: Diversity and evolution of cellulose-synthesizing enzyme complexes. J. Phycol. 35:635–655. 10.1046/j.1529-8817.1999.3540635.x. [DOI] [Google Scholar]
- 104.Kim N-H, Herth W, Vuong R, Chanzy H. 1996. The cellulose system in the cell wall of Micrasterias. J. Struct. Biol. 117:195–203. 10.1006/jsbi.1996.0083. [DOI] [PubMed] [Google Scholar]
- 105.Brown RM, Jr, Montezinos D. 1976. Cellulose microfibrils: visualization of biosynthetic and orienting complexes in association with the plasma membrane. Proc. Natl. Acad. Sci. U. S. A. 73:143–147. 10.1073/pnas.73.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Itoh T, Brown RM., Jr 1984. The assembly of cellulose microfibrils in Valonia macrophysa Kutz. Planta 160:372–381. 10.1007/BF00393419. [DOI] [PubMed] [Google Scholar]
- 107.Kudlicka K, Wardrop A, Itoh T, Brown RM. 1987. Further evidence from sectioned material in support of the existence of a linear terminal complex in cellulose synthesis. Protoplasma 136:96–103. 10.1007/BF01276358. [DOI] [Google Scholar]
- 108.Katsaros C, Reiss H-D, Schnepf E. 1996. Freeze-fracture studies in brown algae: putative cellulose-synthesizing complexes on the plasma membrane. Eur. J. Phycol. 31:41–48. 10.1080/09670269600651171. [DOI] [Google Scholar]
- 109.Saxena IM, Brown RM, Fevre M, Geremia RA, Henrissat B. 1995. Multidomain architecture of beta-glycosyl transferases: implications for mechanism of action. J. Bacteriol. 177:1419–1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Lipinska AP, D'Hondt S, Van Damme EJ, De Clerck O. 2013. Uncovering the genetic basis for early isogamete differentiation: a case study of Ectocarpus siliculosus. BMC Genomics 14:909. 10.1186/1471-2164-14-909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Freeman R, Geier H, Weigel KM, Do J, Ford TE, Cangelosi GA. 2006. Roles for cell wall glycopeptidolipid in surface adherence and planktonic dispersal of Mycobacterium avium. Appl. Environ. Microbiol. 72:7554–7558. 10.1128/AEM.01633-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Hansen FT, Droce A, Sorensen JL, Fojan P, Giese H, Sondergaard TE. 2012. Overexpression of NRPS4 leads to increased surface hydrophobicity in Fusarium graminearum. Fungal Biol. 116:855–862. 10.1016/j.funbio.2012.04.014. [DOI] [PubMed] [Google Scholar]
- 113.Blokker P, Schouten S, Van den Ende H, De Leeuw JW, Hatcher PG, Damste JSS. 1998. Chemical structure of algaenans from the fresh water algae Tetraedron minimum, Scenedesmus communis and Pediastrum boryanum. Org. Geochem. 29:1453–1468. 10.1016/S0146-6380(98)00111-9. [DOI] [Google Scholar]
- 114.Metzger P, Rager M-N, Largeau C. 2007. Polyacetals based on polymethylsqualene diols, precursors of algaenan in Botryococcus braunii race B. Org. Geochem. 38:566–581. 10.1016/j.orggeochem.2006.12.003. [DOI] [Google Scholar]
- 115.Metzger P, Rager MN, Fosse C. 2008. Braunicetals: acetals from condensation of macrocyclic aldehydes and terpene diols in Botryococcus braunii. Phytochemistry 69:2380–2386. 10.1016/j.phytochem.2008.06.004. [DOI] [PubMed] [Google Scholar]
- 116.Gupta NS, Collinson ME, Briggs DEG, Evershed RP, Pancost RD. 2006. Reinvestigation of the occurrence of cutan in plants: implications for the leaf fossil record. Paleobiology 32:432–449. 10.1666/05038.1. [DOI] [Google Scholar]
- 117.Nip M, Tegelaar EW, Deleeuw JW, Schenck PA, Holloway PJ. 1986. A new non-saponifiable highly aliphatic and resistant bio-polymer in plant cuticles—evidence from pyrolysis and C-13-NMR analysis of present-day and fossil plants. Naturwissenschaften 73:579–585. 10.1007/BF00368768. [DOI] [Google Scholar]
- 118.Villena JF, Dominguez E, Stewart D, Heredia A. 1999. Characterization and biosynthesis of non-degradable polymers in plant cuticles. Planta 208:181–187. 10.1007/s004250050548. [DOI] [PubMed] [Google Scholar]
- 119.Schouten S, Moerkerken P, Gelin F, Baas M, de Leeuw JW, Damste JSS. 1998. Structural characterization of aliphatic, non-hydrolyzable biopolymers in freshwater algae and a leaf cuticle using ruthenium tetroxide degradation. Phytochemistry 49:987–993. 10.1016/S0031-9422(97)00942-4. [DOI] [Google Scholar]
- 120.McKinney DE, Bortiatynski JM, Carson DM, Clifford DJ, DeLeeuw JW, Hatcher PG. 1996. Tetramethylammonium hydroxide (TMAH) thermochemolysis of the aliphatic biopolymer cutan: insights into the chemical structure. Org. Geochem. 24:641–650. 10.1016/0146-6380(96)00055-1. [DOI] [Google Scholar]
- 121.Schmidt HW, Schonherr J. 1982. Development of plant cuticles: occurrence and role of non-ester bonds in cutin of Clivia miniata Reg. leaves. Planta 156:380–384. 10.1007/BF00397478. [DOI] [PubMed] [Google Scholar]
- 122.MacDougall KM, McNichol J, McGinn PJ, O'Leary SJ, Melanson JE. 2011. Triacylglycerol profiling of microalgae strains for biofuel feedstock by liquid chromatography-high-resolution mass spectrometry. Anal. Bioanal. Chem. 401:2609–2616. 10.1007/s00216-011-5376-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Mejanelle L, Sanchez-Gargallo A, Bentaleb I, Grimalt JO. 2003. Long chain n-alkyl diols, hydroxy ketones and sterols in a marine eustigmatophyte, Nannochloropsis gaditana, and in Brachionus plicatilis feeding on the algae. Org. Geochem. 34:527–538. 10.1016/S0146-6380(02)00246-2. [DOI] [Google Scholar]
- 124.Volkman JK, Barrett SM, Dunstan GA, Jeffrey SW. 1992. C-30-C-32 alkyl diols and unsaturated alcohols in microalgae of the class Eustigmatophyceae. Org. Geochem. 18:131–138. 10.1016/0146-6380(92)90150-V. [DOI] [Google Scholar]
- 125.Guillard RRL. 1975. Culture of phytoplankton for feeding marine invertebrates, p 29–60 In Smith WL, Chanley MH. (ed), Culture of marine invertebrate animals. Plenum Press, New York, NY. [Google Scholar]
- 126.Weiss TL, Roth R, Goodson C, Vitha S, Black I, Azadi P, Rusch J, Holzenburg A, Devarenne TP, Goodenough U. 2012. Colony organization in the green alga Botryococcus braunii (race B) is specified by a complex extracellular matrix. Eukaryot. Cell 11:1424–1440. 10.1128/EC.00184-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.