Abstract
Most freshwater flagellates use contractile vacuoles (CVs) to expel excess water. We have used Chlamydomonas reinhardtii as a green model system to investigate CV function during adaptation to osmotic changes in culture medium. We show that the contractile vacuole in Chlamydomonas is regulated in two different ways. The size of the contractile vacuoles increases during cell growth, with the contraction interval strongly depending on the osmotic strength of the medium. In contrast, there are only small fluctuations in cytosolic osmolarity and plasma membrane permeability. Modeling of the CV membrane permeability indicates that only a small osmotic gradient is necessary for water flux into the CV, which most likely is facilitated by the aquaporin major intrinsic protein 1 (MIP1). We show that MIP1 is localized to the contractile vacuole, and that the expression rate and protein level of MIP1 exhibit only minor fluctuations under different osmotic conditions. In contrast, SEC6, a protein of the exocyst complex that is required for the water expulsion step, and a dynamin-like protein are upregulated under strong hypotonic conditions. The overexpression of a CreMIP1-GFP construct did not change the physiology of the CV. The functional implications of these results are discussed.
INTRODUCTION
In hypotonic media, cells/organisms take up water by osmosis. Water uptake occurs only if the water potential inside the cell is more negative than that outside the cell. The amount of water taken up in a given time depends on (i) the water potential gradient between the medium and the cell interior, (ii) the surface area of the plasma membrane (PM), and (iii) the extent to which the PM is permeable to water. Cells can use three strategies to prevent bursting caused by the osmotic influx of water in a hypotonic environment. First, organisms can modify the composition of its interior, its surface area, and/or its PM permeability to limit water uptake. Second, organisms can use a cell wall to generate a cell wall pressure potential (turgor) to balance the osmotic water potential, as many algae, plants, and fungi do. However, many unicellular protists do not possess a rigid cell wall. Instead, these protists use a third strategy: they employ water pumps called contractile vacuoles (CVs) to remove excess water. CVs are specialized vacuoles that slowly accumulate water during diastole and periodically expel the liquid rapidly into the medium (systole) (1–4).
Although CV morphologies and behaviors differ among various organisms (see Komsic-Buchman and Becker [5] for a recent comparison of the contractile vacuoles of a variety of protists), the basic mechanism (water uptake into the CV by osmosis) appears to be conserved between different eukaryotes. The same proteins/cellular processes have been implicated in CV function in Amoeba, Dictyostelium, Paramecium, Trypanosoma, and green algae (e.g., proton pumps and aquaporins) (3, 4, 6). However, we still do not know anything about what other factors (ion transport systems) are needed to build up the necessary osmotic gradient in any system.
Water channels (known as aquaporins) often facilitate osmotic water flow across membranes. Aquaporins can increase the permeability of the osmotic membrane to water approximately 100-fold compared to that of a pure phospholipid bilayer (7, 8). Aquaporins can be regulated, for example, by phosphorylation at the C terminus (9). Clear evidence has been presented for the presence of aquaporins in the CV membrane of Amoeba proteus (10), Dictyostelium discoideum (11), Leishmania major (12), and Trypanosoma cruzi (13). The genome of Chlamydomonas reportedly contains two putative aquaporins: major intrinsic proteins 1 and 2 (MIP1 and MIP2) (14). Moreover, Anderca et al. (15) reported the ability of MIP1 to facilitate the transport of glycerol, but not water, when expressed in yeast. In spite of this result, the authors suggested in the same paper that MIP1 functions in vivo as a water channel in CVs. In support of this suggestion, we demonstrated previously that CreMIP1 is localized in the CV membrane in Chlamydomonas when tagged with green fluorescent protein (GFP) as a reporter protein (3).
Chlamydomonas is an established green algal model system (16–18). Cells contain two CVs, which are located at the anterior end of the cell close to the basal bodies (19). The ultrastructure of the Chlamydomonas CV has been investigated in some detail (3, 19, 20). At the end of diastole, the CV has a spherical shape approximately 2 μm in diameter. During systole, the CV collapses and fragments into smaller vesicles/vacuoles that vary in size (100 to 200 nm). These vacuoles then fuse with each other to form a new large, spherical CV at the end of diastole. As in other systems, proton pumps have been implicated in CV function in Chlamydomonas (20). We recently isolated Chlamydomonas CV mutants through insertional mutagenesis and demonstrated that the exocyst component SEC6 is required for CV function (3). However, not much is known about the basic physiology of CV function/osmoregulation in Chlamydomonas. Here, we describe the basic regulation of CV activity in Chlamydomonas and its adaptation to media of different osmotic strengths. We also discuss the role of aquaporins in osmoregulation in Chlamydomonas.
MATERIALS AND METHODS
Strains used.
Chlamydomonas sp. strains CC-3395 (arg7-8 cwd mt+) (21) and UVM4 (cwd mt+ arg7) (22) and four individual UVM4-MIP1GFP strains were used (UVM4-MIP1GFP-1 was published previously [3]; UVM4-MIP1GFP-2 to UVM4-MIP1GFP-4 were obtained according to the previously published methods). Culture conditions were as described previously (3), except that fresh cultures were inoculated every 7 days to keep the cell growth in log phase (see Fig. S2 in the supplemental material). For all experiments, 5-day-old cultures (cell density of 106 to 107 cells/ml) were exposed for 2 days to media with different osmotic strengths, i.e., Tris-acetate-phosphate (TAP) either diluted with distilled water (TKA X-CAD; Thermo Electron LED GmbH, Niederelbert, Germany) or augmented with sucrose. The osmolarity of all media was determined using a freezing-point depression osmometer (Osmomat 010; Gonotec, Berlin, Germany).
Light microscopy.
Light and fluorescence microscopy were performed as described by Komsic-Buchmann et al. (3), with 5 to 7 μl cell suspension on each slide. We performed a titration analysis to determine the cytosolic osmolarity of Chlamydomonas sp. strain CC-3395. Cells were incubated for 2 days in media of increasing osmolarity and observed for the presence of CVs.
Calculation of the water permeability coefficient, Pf.
We used the following equations, adapted from references 23 and 24, to determine the water permeability coefficient (Pf) of the cell and CV membrane, respectively: for osmotic pressure, π = RT × C = 2.436 liter MPa mol−1, where C is the osmotic potential of the solution and RT is room temperature (20°C); for osmotic pressure difference, Δπ = π1 − π2; for water flux (J*; in μm3 s−1), J* = efflux per cell per min/60; for the permeability of the membrane to water (LP; in μm s−1 MPa−1), LP = J*/(Δπ × A), where A is surface area; for water permeability (Pf; in μm s−1), Pf = (LP × RT)/VW, where VW is the partial molar volume of water, 0.000018 m3 mol−1.
Subcellular localization of CreMIP1.
A peptide consisting of the 15 C-terminal amino acids of MIP1 was used to generate the α-CreMIP1 antibody in rabbits generated in cooperation with Agrisera (Vännäs, Sweden). The immune serum was purified by affinity chromatography.
For Western blotting, the proteins were extracted from 15 ml of a Chlamydomonas culture. The cells were lysed with SDS buffer supplemented with cOmplete Ultra tablets (2×, EDTA free; Roche, Mannheim, Germany) and heated for 3 min at 95°C. Dithiothreitol (DTT) was added to the supernatant after centrifugation (final concentration, 100 mM). Ten micrograms of protein per sample was separated on a 12% SDS-PAGE gel (25) and transferred to a polyvinylidene difluoride (PVDF) membrane (Immobilon-P; 0.45 μm; Millipore, Darmstadt, Germany). Antibodies were diluted in 2% and 5% milk powder in Tris-buffered saline (TBS). The following primary antibodies were used: α-CreMIP1 (rabbit; 1:10,000; Agrisera, Vännäs, Sweden), α-GFP (mouse; 1:5,000; Roche, Mannheim, Germany), α-Arf1 (rabbit; 1:5,000; Agrisera, Vännäs, Sweden). The following secondary antibodies were used: α-mouse (1:50,000) and α-rabbit (1:80,000) conjugated to horseradish peroxidase (HRP; Sigma, St. Louis, MO). Proteins were detected with chemiluminescence (SuperSignal West Femto maximum sensitivity substrate; Thermo Scientific, Waltham, MA) using the LAS-4000 mini luminescent image analyzer (Fujifilm Europe GmbH).
For electron microscopy (EM), Chlamydomonas cells were transferred into high-salt medium (HSM) augmented with sucrose to reach an osmotic strength of 64 mosM, and HEPES was added (1 mM final concentration). Cell processing for Epon embedding was performed as described previously (3). Cell fixation for LR-Gold embedding was performed simultaneously for 30 min at 4°C with formaldehyde, glutaraldehyde, and aqueous osmium tetroxide simultaneously (final concentrations were 2%, 0.5%, and 0.05%, respectively). For easier handling, the dense cell suspensions were injected after fixation into 1% Difco agar, noble (B&D, Heidelberg, Germany). Samples were washed and dehydrated in an ethanol series and embedded in LR-Gold. Ultrathin sections (60 nm) were cut with a Leica microtome EM UC7 and a diamante knife (45° angle; Diatome). Sections of LR-Gold-embedded cells were processed for immunogold localization according to Geimer (26). Antibodies were used at the following dilutions: α-CreMIP1, 1:200; α-tubulin, 1:200 (positive control; mouse IgG, clone 6 to 11b-1; Sigma, St. Louis, MO, USA); and α-rabbit and α-mouse conjugated with 10-nm gold particles (1:25; Sigma, St. Louis, MO, USA). All micrographs were taken with a transmission electron microscope (CM 10; Phillips, Eindhoven, The Netherlands) and a digital camera (Orius SC200W 1; Gatan, Pleasanton, CA). Images were analyzed using Digital Micrograph (Gatan, Pleasanton, CA) and Adobe Photoshop CS4.
qPCR.
Total RNA was isolated from 15 ml of each Chlamydomonas culture using the peqGOLD plant RNA kit (Peqlab, Erlangen, Germany). DNA was removed from the RNA with DNase I (Fermentas, Burlington, Canada), and cDNA synthesis (RevertAid first-strand cDNA synthesis kit; Fermentas) was conducted using 1 μg total RNA. Quantitative real-time PCR (qPCR) primers were generated using either QuantPrime (27) or Primer3 (28) or were generated manually and tested for an amplification efficiency (E) of 0.9 ≤ E ≤ 1.1 and for specificity (melting curves with a single peak and only one PCR product on a 2 to 4% agarose gel; primer sequences are listed in Table S1 in the supplemental material). qPCR was performed using the Applied Biosystems 7300 cycler (Foster City, CA, USA) and SYBR green (KAPA SYBR fast qPCR mastermix for ABI Prism; Peqlab, Erlangen, Germany) or Maxima SYBR green (Fermentas, Burlington, Canada) with 2 μl 10-fold diluted cDNA (corresponding to 100 ng of total RNA). Nontemplate controls were included each time, and every real-time PCR assay was performed in duplicate. Quantification experiments were repeated three to five times using RNA isolated from independent cultures. Fold changes were calculated based on the relative 2−ΔΔCT method, using RPL34 as an internal standard.
RESULTS
Osmoregulation in Chlamydomonas sp. strain CC-3395.
Initial experiments showed that cellular water homeostasis and CV activity are influenced by the growth status of the cells (see Fig. S1 and S2 in the supplemental material). Cells from cultures, which were kept regularly in the exponential growth phase, exhibit less variability in their cell and CV physiology. In contrast, cells that were exposed to stationary phases (≥7 days before subculturing) exhibited, on average, a larger cell size and smaller CVs, with much greater individual variability (see Fig. S2). Therefore, Chlamydomonas sp. strain CC-3395 was maintained in the exponential growth phase for several weeks prior to all experiments. Cell size, CV size, and CV contraction interval were determined and used to calculate the water flux through the cell, the PM surface area, the CV membrane surface area, and the CV volume (Fig. 1; see Materials and Methods for details). Cell size and CV size (Table 1) were not correlated with the osmotic strength of the medium. In contrast, the CV contraction interval increased with increasing osmotic strength (less osmotic pressure to the cells). The water flux through the cell and the osmotic strength of the medium exhibited a strong negative correlation (linear regression; r2 = 0.898, P = 0.0041) (Fig. 2). Doubling the medium osmolarity nearly halved the water flux through the cells. Thus, cells mainly use changes in CV activity to adapt to media of different osmotic strengths rather than varying cytosolic osmolarity and/or the water permeability of the plasma membrane. The extrapolation of the straight line obtained by linear regression analysis indicates that the average cytosolic osmolarity is approximately 170 mosM.
FIG 1.
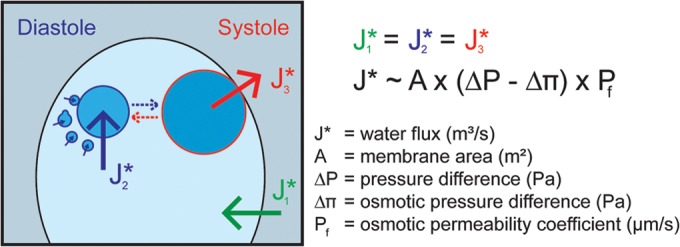
Osmoregulation in Chlamydomonas reinhardtii. Water fluxes across plasma and CV membranes are indicated. The CV on the left is representative of early diastole. At the end of diastole, the large CV (right CV; approximately 2 μm in diameter) develops from small (diameter, 100 to 200 nm) vacuoles (broken blue arrow). The CV on the right depicts the transition from diastole to systole phase. The contractile vacuole associates closely with the PM and expels liquid into the medium. During this process, the CV collapses and fragments into numerous small vacuoles (broken red arrow). To sustain cell size, water fluxes during the three water transport steps must be in equilibrium (J1* = J2* = J3*). The comparison of the CV volume at the end of diastole to the total volume of small vesicles (same amount of membrane material; diameter, 100 to 200 nm) at the beginning of diastole indicates that at least 90% to 95% of the CV volume is discharged during a single CV cycle (assuming ideal spheres for vesicles and the large CV). As the collapsed membrane material of the CV rarely forms ideal spheres, the real percentage of discharge is even higher. For simplicity, the CV volume at the end of diastole is used in all calculations, which gives rise to a small overestimation (less than 5%) of water flux and the calculated membrane permeability coefficient, Pf.
TABLE 1.
Osmoregulation in Chlamydomonasa
| Osmolarity of the medium (mosM) | No. of cells investigated | Cell surface (μm2) | CV vol (μm3) | CV contraction interval (s) | Water efflux (μm3/min cell) | Osmotic PM Pf (μm/s) |
|---|---|---|---|---|---|---|
| 32 | 16 | 248.9 ± 19.9 | 3.4 ± 0.4 | 15.9 ± 1.3 | 25.8 ± 2.3 | 0.69 |
| 64 | 36 | 179.0 ± 8.4 | 2.7 ± 0.2 | 20.4 ± 0.6 | 16.2 ± 1.1 | 0.78 |
| 84 | 21 | 180.7 ± 10.7 | 2.6 ± 0.2 | 25.5 ± 1.1 | 12.8 ± 1.1 | 0.76 |
| 104 | 21 | 168.4 ± 8.0 | 2.2 ± 0.1 | 24.4 ± 0.9 | 10.7 ± 0.7 | 0.89 |
| 124 | 21 | 227.9 ± 13.4 | 2.7 ± 0.2 | 29.0 ± 1.7 | 11.6 ± 1.1 | 1.01 |
| 144 | 20 | 287.0 ± 13.6 | 3.0 ± 0.2 | 107.7 ± 4.7 | 3.5 ± 0.3 | 0.42 |
Mean values and standard errors are given.
FIG 2.
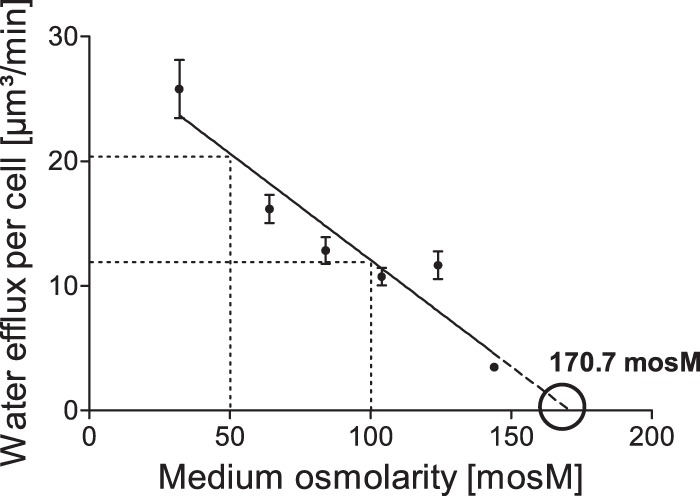
Alteration of the water efflux per cell of Chlamydomonas reinhardtii in response to media with different osmotic strengths. Error bars indicate standard errors of the means (SEM). The SEM of the data point (144/3.5) is too small to be depicted in this diagram.
Under isotonic conditions, no CVs at all were observed in Chlamydomonas. As shown in Fig. S3 in the supplemental material, the titration of CV activity yielded a cytosolic osmolarity of approximately 173 mosM, which is in good agreement with the value obtained by extrapolating the CV efflux to zero in Fig. 2.
Regulation of CV activity.
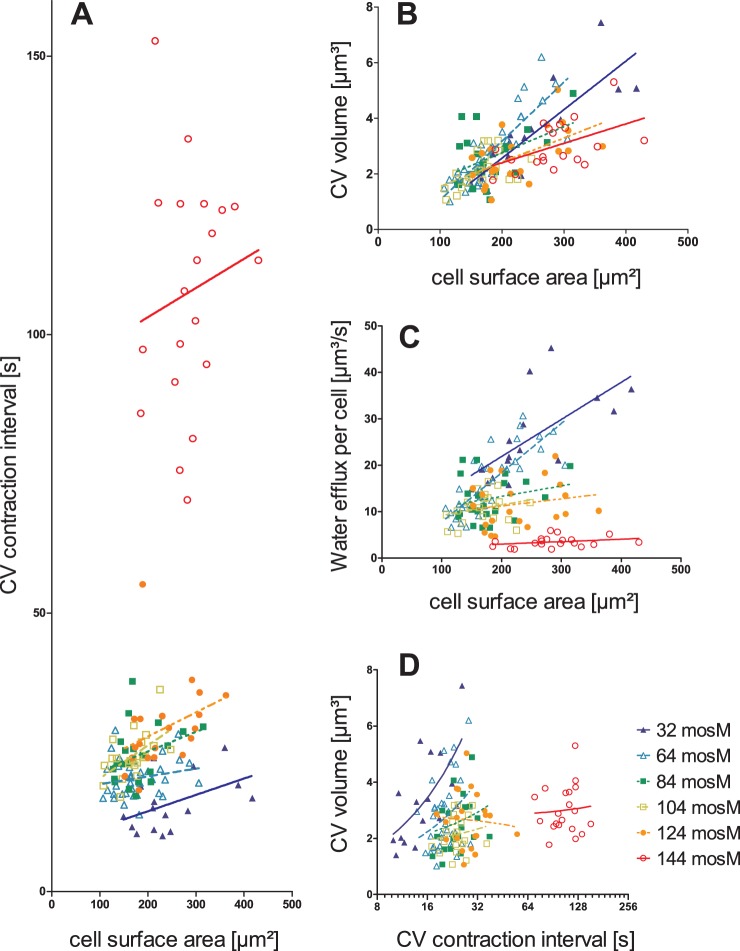
The analysis thus far used population averages and revealed a very good correlation between water transport through the cell and the osmotic strength of the medium. However, as is evident from the standard errors of the means (SEM) depicted for the different cell populations shown in Fig. 2, we observed considerable variation of the water flux within a population of cells. Figure 3 shows scatter plots for individual cells in media of different osmotic strengths (32 mosM to 144 mosM) and depicts the relationship between CV properties and cell size (expressed as cell surface area) as well as the mutual dependence of CV volume and contraction interval. Because our Chlamydomonas cultures are not synchronized, the cells vary greatly in size (100 to 450 μm2 PM surface area). Interestingly, only the CV volume correlated well with the PM surface area, while the duration of the contraction interval strongly depended on the osmolarity of the medium (Fig. 3). These observations indicate that Chlamydomonas cells use CV size to compensate for the increased water influx during cell growth and adjust the contraction interval to adapt the cellular water flux to the medium osmolarity. Our results indicate that there is no correlation between CV size and contraction interval, each is regulated independently. Different combinations of CV size and contraction interval yield the same water flow through the cell, which explains in part the variability of both parameters. However, it is noteworthy that the correlation between CV size and contraction period improves in strong hypotonic media, which indicates that the system is approaching its limits (Fig. 3).
FIG 3.
Relationship of CV properties to cell surface and their mutual dependence in Chlamydomonas reinhardtii. (A) CV contraction intervals versus cell surface area. (B) CV size versus cell surface area. (C) Water efflux per cell versus cell surface area. (D) Relationship between CV volume and CV contraction interval.
Permeability of the plasma membrane.
Given the osmolarities of cytosol and medium, the PM surface area, and the water fluxes through the cell, it is possible to calculate the PM's osmotic permeability coefficient for water, Pf, in all tested media (Table 1). The Pf values of the PM were very low (0.419 to 1.01 μm/s) in all experimental conditions (Table 1). These very low values are in the range of those for membranes reported to lack aquaporins (29), indicating that no aquaporins are present in the PM of Chlamydomonas.
Water uptake into the CV.
Current models regarding CV function suggest that the uptake of water into the CV is mediated by osmosis. Unfortunately, currently it is not possible to determine the osmolarity of the CV lumen in Chlamydomonas; nevertheless, it is possible to model the situation at and in the CV using the determined water fluxes and CV membrane surface areas (calculated from the size of the CV at late diastole). Figure 4 shows combinations of osmotic gradient at the CV membrane and the CV membrane permeability, which yield the determined water fluxes for each osmotic condition. Based on the assumption that the presence of an aquaporin increases the membrane permeability by a factor of 100 (7), the Pf of the CV membrane is between 41.9 and 101 μm/s (Fig. 4, black striated box). As is evident from Fig. 4, only small osmotic gradients (15 to 25 mosM) are needed at the CV membrane to achieve the required water flow.
FIG 4.
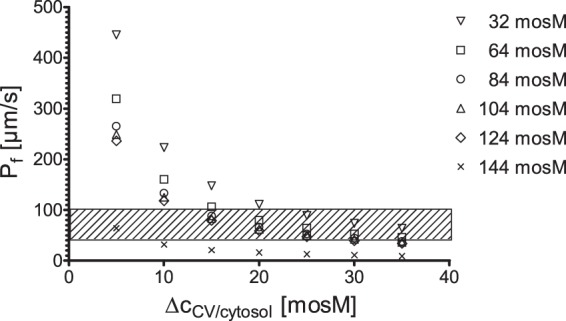
Mutual interdependence between the osmotic water permeability coefficient, Pf, and the osmotic gradient, ΔcCV/cytosol, at the CV membrane in Chlamydomonas reinhardtii. The Pf values corresponding to several selected osmotic gradients. ΔcCV/cytosol values were calculated based on the performance of CVs in Chlamydomonas in different osmotic media. The striated box indicates the suggested range for the Pf for the CV membrane (based on the Pf of the PM [Table 1]).
Aquaporins in Chlamydomonas.
The published genome of Chlamydomonas (30) contains three putative aquaporins: MIP1 (Cre12.g549300.t1.2), MIP2 (Cre17.g711250.t1.2), and MIP3 (Cre01.g038800.t1.2). MIP1 and MIP2 have been characterized in silico by Anderberg et al. (14); they are members of the algal MIP-D subfamily (associated with the X intrinsic protein [XIP] subfamily), and it has been suggested that they are glycerol (MIP1) or water and glycerol (MIP2) transporters. MIP3 exhibits the greatest similarity to plant small basic intrinsic proteins (SIPs) (31), which generally are localized in the plant endoplasmic reticulum (ER) membrane (32). MIP1 and MIP3 were readily detected using reverse transcription-PCR, indicating that MIP2 is expressed only weakly, if at all, in our culture conditions. In keeping with this result, there are no expression data available for MIP2 in the databases.
MIP1 is localized to the CV.
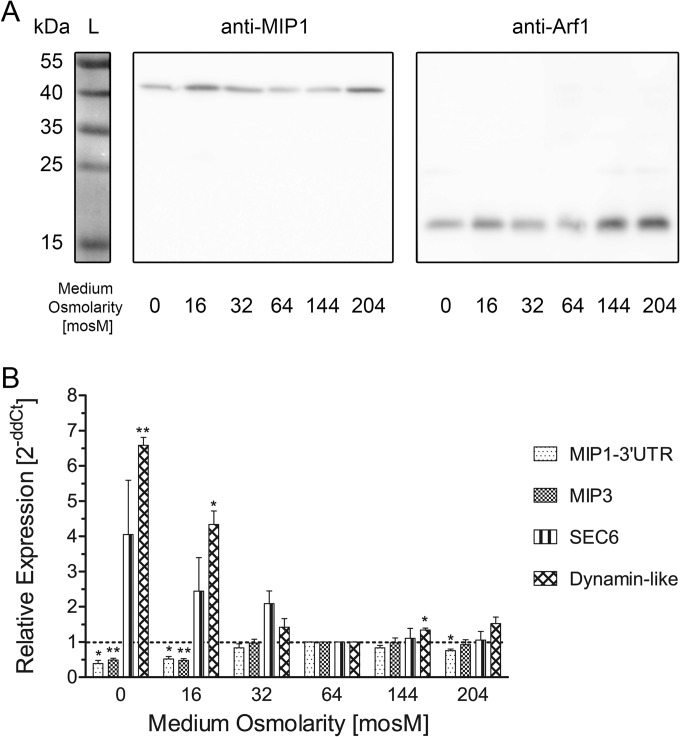
We raised a new rabbit polyclonal antibody against a C-terminal, 15-amino-acid peptide (anti-CreMIP1). As shown in Fig. 5A, the antibody specifically recognizes a single band of approximately 43 kDa on Western blots using whole-cell extracts from Chlamydomonas sp. strain CC-3395 for SDS-PAGE and blotting, which is significantly larger than the expected size (31.54 kDa; based on the sequence of the published gene model [30]). Pretreatment of the protein samples with N-glycosidase F did not increase the mobility of the protein detected by the antibody, suggesting that the decreased mobility is not due to N-glycosylation (not shown; see Discussion).
FIG 5.
Expression of the aquaporin CreMIP1 in response to different osmotic media in Chlamydomonas reinhardtii. (A) Western blot. Whole-cell extracts of Chlamydomonas were probed with an α-CreMIP1 antibody (expected molecular mass, 31.54 kDa) and an α-Arf1 antibody (loading control; expected molecular mass, 20.59 kDa). (B) qPCR. **, P < 0.01; *, P < 0.05. Error bars indicate SEM.
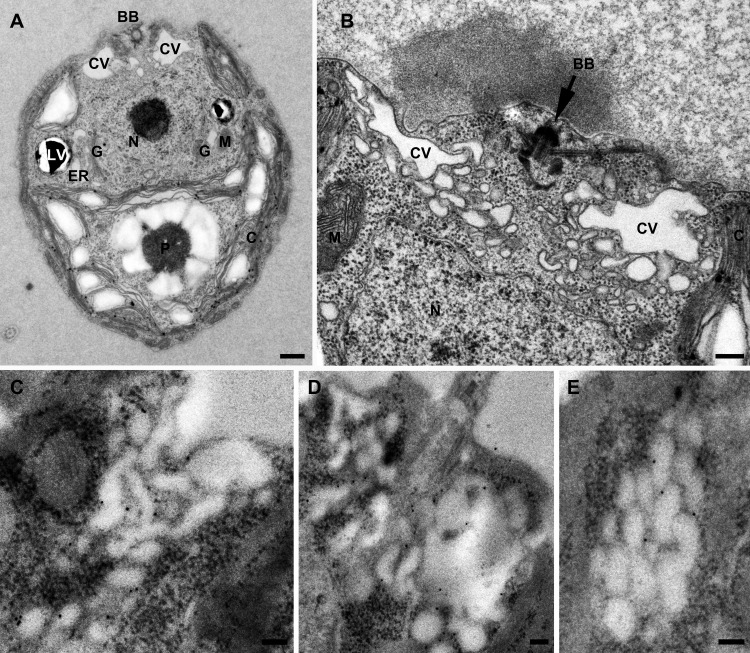
Cells were chemically fixed and LR-Gold embedded in order to prepare them for immunogold electron microscopy using anti-CreMIP1 (Fig. 6). Specific gold labeling was observed only at the membranes of the CV. The labeling of the CV membrane was homogenous, and the number of gold particles increased linearly with the length of the CV membrane profile examined (not shown). This finding indicated that MIP1 most likely is evenly distributed on all membranes of the CV complex, whether it consists of 100-nm to 200-nm vesicles or a large vacuole. A few gold particles sometimes were observed on the Golgi complex (possibly representing newly biosynthesized MIP1 en route to the CV), lytic vacuoles, plastids, mitochondria, nucleus, and PM. However, the labeling density was much lower for these compartments than on the CV membrane (not shown). Controls using the secondary antibody alone showed only weak, nonspecific labeling (not shown).
FIG 6.
CreMIP1 is localized at the CV membrane in Chlamydomonas reinhardtii. Ultrastructure of Chlamydomonas reinhardtii. (A) Longitudinal section; (B) cross-section through the two CV regions of a cell. (C to E) Immunogold localization of CreMIP1 at the CV membranes using 10-nm gold particles. BB, basal body; C, chloroplast; CV, contractile vacuole; ER, endoplasmic reticulum; G, Golgi complex; LV, lytic vacuole; M, mitochondrion; N, nucleus; P, pyrenoid. Scale bars are 500 nm (A), 200 nm (B), and 100 nm (C to E).
The level of aquaporin expression does not differ in media of different osmotic conditions.
We investigated MIP1 protein levels using Western blotting (Fig. 5A) and the level of MIP1 transcript using qPCR (Fig. 5B). No significant differences were observed regarding MIP1 protein levels in cells exposed to different osmotic conditions, even when we lowered the osmotic potential to 0 mosM (i.e., distilled water; amazingly, cells survived for 2 days in distilled water). The protein level fluctuated somewhat but exhibited no clear pattern. However, our qPCR results indicated a small but significant ∼2-fold downregulation of MIP1 (and MIP3) in strong hypotonic media at 0 and 16 mosM. In contrast, the SEC6 (Cre17.g744847.t1.1) transcript, known to be required for CV function (3), exhibited a trend toward upregulation in strong hypotonic media. In addition, a transcript coding for a dynamin-like protein (Cre13g.569000.t1.1) was significantly upregulated. This protein is present in the same expression cluster as MIP1 in Ning et al. (33); therefore, it might be involved in the CV cycle, possibly during systole.
Overexpression of MIP1-GFP does not change the CV physiology.
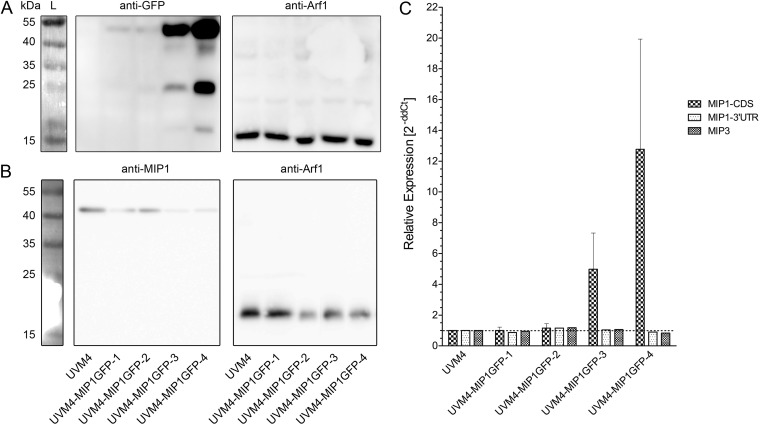
To investigate the role of MIP1 in more detail, we characterized four independent MIP1-GFP-expressing strains. Western blotting (Fig. 7A and B) and qPCR analysis (Fig. 7C) confirmed that all strains expressed CreMIP1-GFP in addition to CreMIP1 (compare MIP1-CDS with MIP1–3′UTR in the qPCR analysis, where CDS is coding sequence and UTR is untranslated region). The strains expressed CreMIP1-GFP at different levels; it was strongly overexpressed in two strains (UVM4-MIP1GFP-3 and UVM4-MIP1GFP-4) (Fig. 7C).
FIG 7.
Expression of the aquaporin CreMIP1 in different strains of Chlamydomonas reinhardtii UVM4 expressing CreMIP1-GFP fusion protein. (A) Western blots. Whole-cell extracts of Chlamydomonas reinhardtii were probed with an α-GFP antibody and an α-Arf1 antibody (loading control; expected molecular mass, 20.59 kDa). L, protein ladder. (B) Endogenous CreMIP1 (expected molecular mass, 31.54 kDa) is present in cell extracts of all cell lines. (C) qPCR. The primer pair MIP1-CDS (CDS = coding sequence) detects all transcripts of MIP1 and MIP1-GFP, while the primer pair MIP1–3′UTR (UTR = untranslated region) detects only transcripts of the endogenous MIP1. Error bars indicate SEM.
The anti-GFP antibody detected two bands with apparent molecular sizes of approximately 54 kDa and 25 kDa. While the latter is in good agreement with the known molecular mass of GFP (26.5 kDa), the upper band runs faster than expected for the MIP1-GFP fusion construct (calculated size, 58.04 kDa) (see Discussion). Using the GFP antibody, overexpression of CreMIP1-GFP was easily detected in two of the four strains at the protein level. We also detected free GFP in the CreMIP1-GFP-overexpressing strains, which indicates some cleavage of the CreMIP1-GFP construct in these strains. Interestingly, the level of endogenous CreMIP1 protein (Fig. 7B) appears to decrease as CreMIP1-GFP levels increase (Fig. 7A), although the expression level of endogenous CreMIP1 mRNA (Fig. 7B, MIP1–3′UTR) remained stable in all strains.
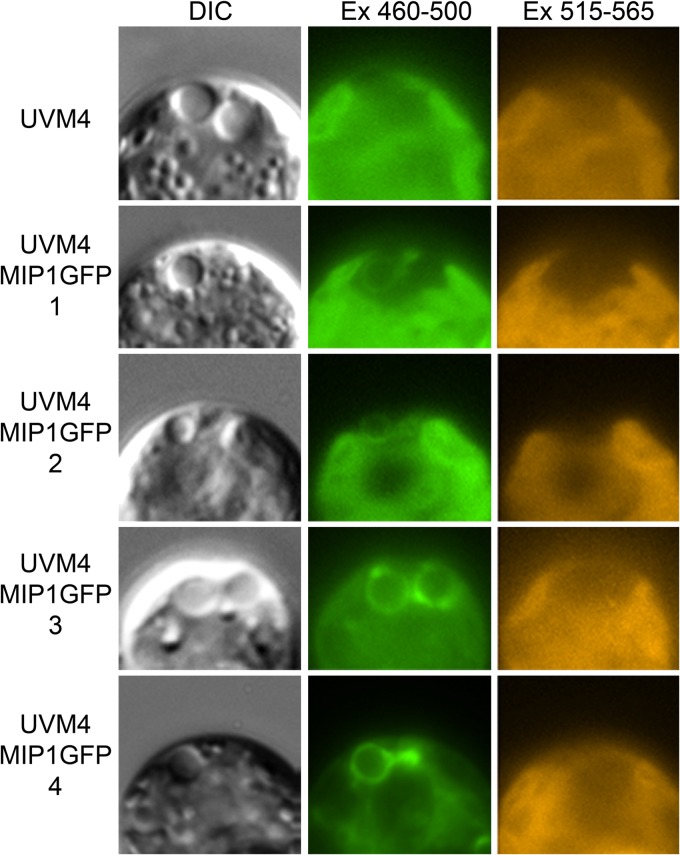
All CreMIP1-GFP-expressing strains showed a GFP signal at the CV (Fig. 8), indicating that all strains contained MIP1-GFP in the CV membrane. As expected from the Western blotting results, we detected additional GFP fluorescence (most likely in the lytic vacuoles and the cytoplasm) in UVM4-MIP1GFP-4 and, to a lesser extent, in UVM4-MIP1GFP-3. Video microscopy showed that the large vacuole shrinks asymmetrically (collapses toward the PM) at the end of diastole. Moreover, CreMIP1-GFP is never incorporated into the PM during systole (see Video S1 in the supplemental material).
FIG 8.
In vivo localization of CreMIP1 tagged with GFP in four different cell lines of Chlamydomonas reinhardtii UVM4-MIP1GFP. Each row displays the anterior region of the same cell in three different channels, as indicated above the rows. DIC, differential interference contrast; Ex, excitation wave length in nm.
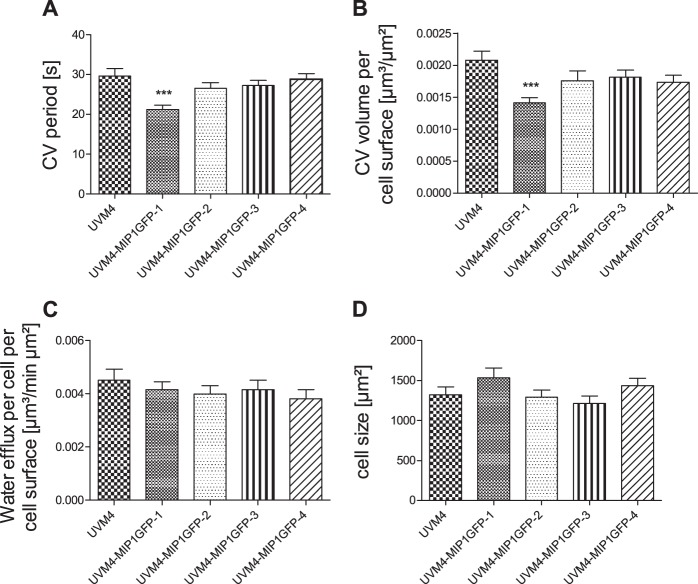
Despite the different expression levels of CreMIP1-GFP, three of the four MIP-GFP-expressing strains did not differ in CV physiology compared with the parental UVM4 strain (Fig. 9). We observed a significant change in CV pumping rate and size in only one strain (UVM4-MIP1GFP-1, the strain with the lowest CreMIP1-GFP expression rate), which compensated for each other and yielded the same water flow.
FIG 9.
Changes in the expression level of CreMIP1-GFP do not affect the CV physiology. (A) CV period. (B) CV volume. (C) Water efflux. (D) Cell size. ***, P < 0.001; n = 20. Error bars indicate SEM.
DISCUSSION
Osmoregulation.
The contractile vacuoles of Chlamydomonas have been investigated before (19, 20, 34–36). Luykx et al. (19) used Chlamydomonas reinhardtii strain 137c for their studies (stock numbers CC-124 and CC-125). The 137c cells possess a typical Chlamydomonas cell wall. In contrast, the CC-3395 strain used in this study completely lacks a cell wall (21) (Fig. 6A and B). However, the basic characteristics of the CV cycle are very similar between the two strains, which suggests that the presence of a cell wall does not play an important role in osmoregulation in Chlamydomonas. The two strains differ mainly with regard to their cytosolic osmolarity: 173 mosM (CC-3395) versus 125 mosM (137c). The latter is more similar to the cytosolic osmolarities determined for other green algae (Scherffelia dubia, ∼93 mosM [37]; Mesostigma viride, ∼85 mosM [38]). The reason for the increased cytosolic osmolarity of CC-3395 compared to that of 137c currently is not known.
Luykx et al. (19) observed that larger cells tend to have larger CV; however, the authors did not perform a regression analysis, and their results depicted in Fig. 2 (19) demonstrate only weak relationships between CV size and cell size (most likely a saturation curve) and CV interval and cell size. We observed a good linear correlation between CV size and cell size and no relationship between the contraction interval and the cell size. The observed contraction interval is much longer in 137c than in CC-3395 cells (28 s and 20 s, respectively, in TAP medium), which might reflect the different cytosolic osmolarities determined for the two strains. Both studies observed a decrease in the contraction interval when the osmotic strength of the medium is reduced, which suggests that the contraction interval is centrally involved in the adaptation process in all Chlamydomonas strains.
A major finding of this study is the low water permeability of the PM, which also has been observed often for other protists (summarized in reference 39). The low water permeability might be an adaptation to their naturally hypoosmotic environment, as it reduces the water flux into the cell. A low rate of water uptake also is supported by the low cytosolic osmolarities observed for many freshwater protists (e.g., green algae, as described above). Water removal by CVs is energized by proton pumps in all investigated systems (see Introduction); thus, these adaptations probably are important to limit the energy consumed by this process.
A major difference between Chlamydomonas reinhardtii strain CC-3395 and other well-investigated protists is that Chlamydomonas only modifies the CV's contraction interval to adapt to media of different osmotic strengths (constant cytosolic osmolarity and water permeability of the PM). Even after 3 months of cultivation in medium at 204 mosM, no active CVs were observed (not shown). This differs completely from other investigated protists (mainly ciliates) that use CVs for osmoregulation (summarized in reference 39). For example, within 2 days the ciliate Paramecium caudatum modifies its cytosolic osmolarity to reestablish a hypertonic cytosol (39). In addition, many protists apparently modify PM permeability and CV osmolarity while adapting to media of different osmotic strengths (39). Therefore, Chlamydomonas uses a much simpler system than other protists (fewer parameters to adjust) to adapt to media of different osmotic strengths. This might represent a major advantage for using Chlamydomonas to analyze CV function and regulation.
Aquaporins and CV function.
For immunization, we used the C-terminal peptide (15 amino acids), which has no close similar sequences in the predicted Chlamydomonas proteome. The affinity-purified antibody recognized a single band in whole-cell extracts; however, the electrophoretic mobility of the endogenous protein and CreMIP1-GFP were not as expected (see Results). At first glance, the aberrant electrophoretic mobility observed for CreMIP1 and CreMIP1-GFP appear to contradict each other (reduced versus increased mobility). However, it seems possible that the 43-kDa band represents a CreMIP1 dimer. SDS-stable oligomerization of aquaporins also has been observed in other systems (e.g., see reference 40). Therefore, the aberrant size of CreMIP1-GFP (54 kDa versus the expected size of 58.04 kDa) likely represents only an aquaporin monomer tagged with GFP. However, it should be noted that the protein model for CreMIP1 has not been experimentally confirmed. Additional work is needed to clarify this question.
Immunogold localization confirmed that CreMIP1 localizes to the CV in Chlamydomonas, which supports the in vivo localization of the GFP-tagged protein CreMIP1-GFP. Furthermore, we were able to show that CreMIP1 was downregulated approximately 2-fold at the transcriptional level when cells were exposed to strong hypotonic media but was unchanged at the protein level. This indicates that CreMIP1 is regulated mainly at the protein level. The observed decrease of MIP1 protein in the strongly MIP1-GFP-overexpressing strains might support this. However, we cannot rule out that an unknown process biased the Western blot analysis or qPCR results. Additional work is needed to address this question in detail.
The level of CreMIP1-GFP expression had no demonstrable effect on CV physiology in any of the tested strains. The physiology in three of the four strains tested did not change at all, while in one strain the changes in CV size and interval compensated for each other. As the three other strains did not show any modification of the CV cycle, we believe that these changes were caused by the insertion of the expression cassette. This leads us to question the functionality of the aquaporin CreMIP1 when tagged with GFP at the C terminus. The function of aquaporins can be modified by their oligomerization state (e.g., increased water permeability upon oligomerization [41]). If our explanations for the observed aberrant mobilities in SDS-PAGE are correct, then adding GFP at the C terminus might prevent oligomerization and reduce or even abolish water transport through CreMIP1-GFP. Another possibility for the stable CV physiology of the four UVM4-MIP1GFP strains is that the C terminus is involved in the regulation of the aquaporin, for example, by phosphorylation (42, 43). This hypothesis is supported by the presence of several putative phosphorylation sites at the C terminus (not shown). Additional work is needed to address this question.
Functional implications.
Our findings have important implications for CV regulation. The water transport machinery resides in all CV membranes (proton pumps, aquaporins, and other transporters that generate the required gradient) and allows water transport into the lumen of all vesicles/vacuoles. Therefore, the water transport capacity of the CV is proportional to the entire surface area of the CV membrane structures. During cell growth, the size of the CV increases (more CV membrane material) and more water is expelled per contraction interval (i.e., in the same amount of time). However, the total CV membrane surface area scales to the second power with CV size (radius), while the CV volume scales to the third power. Thus, the larger CV cannot be filled in the same amount of time unless the water transport capacity of the membrane is increased, which points to the regulation of the water transport capacity of the CV membrane during adaptation to cell growth. A similar argument can be made for an increase of the water transport capacity of the CV membranes during adaptation to different media. With more hypotonic pressure to the cells, the CV's contraction interval decreases, while the CV size does not change. Therefore, the CV must be filled to the same volume in a shorter period of time, which can only be achieved by an increase of the water transport capacity. To summarize, both adaptation processes involve the regulation of the water transport capacity (i.e., aquaporin regulation and/or alteration of the osmotic gradient). Similarly, more membrane fusion events per unit of time are required to build a larger CV at a constant pumping rate during cell growth or a vacuole of the same size in a shorter contraction interval during adaptation to a stronger hypotonic medium. Because adaptation to media of less osmotic strength requires more systole steps per minute (water expulsion and CV fragmentation), we can expect proteins involved in these steps to be upregulated as well. Indeed, qPCR showed that the Chlamydomonas SEC6 transcript (which is involved in exocytosis) tended toward upregulation in media of low osmotic strength (Fig. 5B). Similarly, the transcript that encodes a dynamin-like protein that might be involved in CV fragmentation also is strongly upregulated (Fig. 5B). This suggests that at least some of the protein machinery indeed is under transcriptional control. Currently, we are performing transcriptome analyses to identify putative members of the CV protein machinery through expression studies using SEC6 and the aquaporin MIP1 as CV markers.
Our results indicate that Chlamydomonas can independently sense cell size and the osmotic strength of the medium, and this ability allows the cell to adapt CV function to its changing needs. Because the CV represents a built-in cellular sensor for osmoregulation in Chlamydomonas, Chlamydomonas might represent a good model with which to investigate questions related to the control of cell size as well as signal perception and transduction in osmoregulation. These are important questions in modern biology (44) that have yet to be resolved. For example, it still is not clear which parameter cells actually sense (45). In plants, a histidine kinase that compensates for a yeast osmosensor mutant has been implicated as one component (46). BLASTp analysis using this protein has detected several putative histidine kinases of unknown function in Chlamydomonas (data not shown). Thus, it seems possible that the green plants share an ancient osmoregulatory pathway and that Chlamydomonas is a useful model system in which to study plant osmoregulation.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the Deutsche Forschungsgemeinschaft (Be1779/12-3).
We thank Bill Snell (Texas, USA) for sharing transcriptomic data prior to publication.
Footnotes
Published ahead of print 12 September 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/EC.00163-14.
REFERENCES
- 1.Patterson DJ. 1981. Contractile vacuole complex behavior as a diagnostic character for free living amoeba. Protistologica 17:243–248. [Google Scholar]
- 2.Allen RD. 2000. The contractile vacuole and its membrane dynamics. Bioessays 22:1035–1042. . [DOI] [PubMed] [Google Scholar]
- 3.Komsic-Buchmann K, Stephan LM, Becker B. 2012. The SEC6 protein is required for contractile vacuole function in Chlamydomonas reinhardtii. J. Cell Sci. 125:2885–2895. 10.1242/jcs.099184. [DOI] [PubMed] [Google Scholar]
- 4.Plattner H. 2013. Contractile vacuole complex–its expanding protein inventory. Int. Rev. Cell Mol. Biol. 306:371–416. 10.1016/B978-0-12-407694-5.00009-2. [DOI] [PubMed] [Google Scholar]
- 5.Komsic-Buchmann K, Becker B. 2013. Contractile vacuoles in green algae–structure and function, p 123–142 In Katsaros KHC. (ed), Advances in algal cell biology. de Gruyter, Berlin, Germany. [Google Scholar]
- 6.Ulrich PN, Jimenez V, Park M, Martins VP, Atwood J, Moles K, Collins D, Rohloff P, Tarleton R, Moreno SNJ, Orlando R, Docampo R. 2011. Identification of contractile vacuole proteins in Trypanosoma cruzi. PLoS One 6:e18013. 10.1371/journal.pone.0018013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang B, Fukuda N, van Hoek A, Matthay MA, Ma T, Verkman AS. 2000. Carbon dioxide permeability of aquaporin-1 measured in erythrocytes and lung of aquaporin-1 null mice and in reconstituted proteoliposomes. J. Biol. Chem. 275:2686–2692. 10.1074/jbc.275.4.2686. [DOI] [PubMed] [Google Scholar]
- 8.Zeidel ML, Ambudkar SV, Smith BL, Agre P. 1992. Reconstitution of functional water channels in liposomes containing purified red cell CHIP28 protein. Biochemistry 31:7436–7440. 10.1021/bi00148a002. [DOI] [PubMed] [Google Scholar]
- 9.Chaumont F, Tyerman SD. 2014. Aquaporins: highly regulated channels controlling plant water relations. Plant Physiol. 164:1600–1618. 10.1104/pp.113.233791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nishihara E, Yokota E, Tazaki A, Orii H, Katsuhara M, Kataoka K, Igarashi H, Moriyama Y, Shimmen T, Sonobe S. 2008. Presence of aquaporin and V-ATPase on the contractile vacuole of Amoeba proteus. Biol. Cell 100:179–188. 10.1042/BC20070091. [DOI] [PubMed] [Google Scholar]
- 11.Von Bülow J, Müller-Lucks A, Kai L, Bernhard F, Beitz E. 2012. Functional characterization of a novel aquaporin from Dictyostelium discoideum amoebae implies a unique gating mechanism. J. Biol. Chem. 287:7487–7494. 10.1074/jbc.M111.329102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Figarella K, Uzcategui NL, Zhou Y, LeFurgey A, Ouellette M, Bhattacharjee H, Mukhopadhyay R. 2007. Biochemical characterization of Leishmania major aquaglyceroporin LmAQP1: possible role in volume regulation and osmotaxis. Mol. Microbiol. 65:1006–1017. 10.1111/j.1365-2958.2007.05845.x. [DOI] [PubMed] [Google Scholar]
- 13.Montalvetti A, Rohloff P, Docampo R. 2004. A functional aquaporin co-localizes with the vacuolar proton pyrophosphatase to acidocalcisomes and the contractile vacuole complex of Trypanosoma cruzi. J. Biol. Chem. 279:38673–38682. 10.1074/jbc.M406304200. [DOI] [PubMed] [Google Scholar]
- 14.Anderberg HI, Danielson JÅH, Johanson U. 2011. Algal MIPs, high diversity and conserved motifs. BMC Evol. Biol. 11:110. 10.1186/1471-2148-11-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Anderca MI, Suga S, Furuichi T, Shimogawara K, Muto S. 2004. Functional identification of the glycerol transport activity of Chlamydomonas reinhardtii CrMIP1. Plant Cell Physiol. 45:1313–1319. 10.1093/pcp/pch141. [DOI] [PubMed] [Google Scholar]
- 16.Blaby IK, Blaby-Haas CE, Tourasse N, Hom EFY, Lopez D, Aksoy M, Grossman A, Umen J, Dutcher S, Porter M, King S, Witman GB, Stanke M, Harris EH, Goodstein D, Grimwood J, Schmutz J, Vallon O, Merchant SS, Prochnik S. 17 June 2014. The Chlamydomonas genome project: a decade on. Trends Plant Sci. 10.1016/j.tplants.2014.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grossman AR, Harris EE, Hauser C, Lefebvre PA, Martinez D, Rokhsar D, Shrager J, Silflow CD, Stern D, Vallon O, Zhang Z. 2003. Chlamydomonas reinhardtii at the crossroads of genomics. Eukaryot. Cell 2:1137–1150. 10.1128/EC.2.6.1137-1150.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Merchant SS, Prochnik SE, Vallon O, Harris EH, Karpowicz SJ, Witman GB, Terry A, Salamov A, Fritz-Laylin LK, Maréchal-Drouard L, Marshall WF, Qu L-H, Nelson DR, Sanderfoot AA, Spalding MH, Kapitonov VV, Ren Q, Ferris P, Lindquist E, Shapiro H, Lucas SM, Grimwood J, Schmutz J, Cardol P, Cerutti H, Chanfreau G, Chen C-L, Cognat V, Croft MT, Dent R, Dutcher S, Fernández E, Fukuzawa H, González-Ballester D, González-Halphen D, Hallmann A, Hanikenne M, Hippler M, Inwood W, Jabbari K, Kalanon M, Kuras R, Lefebvre PA, Lemaire SD, Lobanov AV, Lohr M, Manuell A, Meier I, Mets L, Mittag M, Mittelmeier T, Moroney JV, Moseley J, Napoli C, Nedelcu AM, Niyogi K, Novoselov SV, Paulsen IT, Pazour G, Purton S, Ral J-P, Riaño-Pachón DM, Riekhof W, Rymarquis L, Schroda M, Stern D, Umen J, Willows R, Wilson N, Zimmer SL, Allmer J, Balk J, Bisova K, Chen C-J, Elias M, Gendler K, Hauser C, Lamb MR, Ledford H, Long JC, Minagawa J, Page MD, Pan J, Pootakham W, Roje S, Rose A, Stahlberg E, Terauchi AM, Yang P, Ball S, Bowler C, Dieckmann CL, Gladyshev VN, Green P, Jorgensen R, Mayfield S, Mueller-Roeber B, Rajamani S, Sayre RT, Brokstein P, Dubchak I, Goodstein D, Hornick L, Huang YW, Jhaveri J, Luo Y, Martínez D, Ngau WCA, Otillar B, Poliakov A, Porter A, Szajkowski L, Werner G, Zhou K, Grigoriev IV, Rokhsar DS, Grossman AR. 2007. The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science 318:245–250. 10.1126/science.1143609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Luykx P, Hoppenrath M, Robinson DG. 1997. Structure and behavior of contractile vacuoles in Chlamydomonas reinhardtii. Protoplasma 198:73–84. 10.1007/BF01282133. [DOI] [Google Scholar]
- 20.Robinson DG, Hoppenrath M, Oberbeck K, Luykx P, Ratajczak R. 1998. Localization of pyrophosphatase and V-ATPase in Chlamydomonas reinhardtii. Bot. Acta 111:108–122. 10.1111/j.1438-8677.1998.tb00685.x. [DOI] [Google Scholar]
- 21.Shimogawara K, Fujiwara S, Grossman A, Usuda H. 1998. High-efficiency transformation of Chlamydomonas reinhardtii by electroporation. Genetics 148:1821–1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Neupert J, Karcher D, Bock R. 2009. Generation of Chlamydomonas strains that efficiently express nuclear transgenes. Plant J. Cell Mol. Biol. 57:1140–1150. 10.1111/j.1365-313X.2008.03746.x. [DOI] [PubMed] [Google Scholar]
- 23.Buchanan BB, Gruissem W, Jones RL. 2000. Biochemistry and molecular biology of plants. American Society of Plant Physiologists, Rockville, MD. [Google Scholar]
- 24.Taiz L, Zeiger E. 2006. Plant physiology, 4th ed. Sinauer Associate, Inc., Sunderland, MA. [Google Scholar]
- 25.Laemmli UK. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685. 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 26.Geimer S. 2009. Immunogold labeling of flagellar components in situ. Methods in cell biology, 1st ed. Elsevier, New York, NY. [DOI] [PubMed] [Google Scholar]
- 27.Arvidsson S, Kwasniewski M, Riaño-Pachón DM, Mueller-Roeber B. 2008. QuantPrime–a flexible tool for reliable high-throughput primer design for quantitative PCR. BMC Bioinformatics 9:465. 10.1186/1471-2105-9-465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Untergasser A, Cutcutache I, Koressaar T, Ye J, Faircloth BC, Remm M, Rozen SG. 2012. Primer3–new capabilities and interfaces. Nucleic Acids Res. 40:e115. 10.1093/nar/gks596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maurel C. 1997. Aquaporins and water permeability of plant membranes. Annu. Rev. Plant Physiol. Plant Mol. Biol. 48:399–429. 10.1146/annurev.arplant.48.1.399. [DOI] [PubMed] [Google Scholar]
- 30.Goodstein DM, Shu S, Howson R, Neupane R, Hayes RD, Fazo J, Mitros T, Dirks W, Hellsten U, Putnam N, Rokhsar DS. 2012. Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 40:D1178–D1186. 10.1093/nar/gkr944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Abascal F, Irisarri I, Zardoya R. 2014. Diversity and evolution of membrane intrinsic proteins. Biochim. Biophys. Acta 1840:1468–1481. 10.1016/j.bbagen.2013.12.001. [DOI] [PubMed] [Google Scholar]
- 32.Ishikawa F, Suga S, Uemura T, Sato MH, Maeshima M. 2005. Novel type aquaporin SIPs are mainly localized to the ER membrane and show cell-specific expression in Arabidopsis thaliana. FEBS Lett. 579:5814–5820. 10.1016/j.febslet.2005.09.076. [DOI] [PubMed] [Google Scholar]
- 33.Ning J, Otto TD, Pfander C, Schwach F, Brochet M, Bushell E, Goulding D, Sanders M, Lefebvre PA, Pei J, Grishin NV, Vanderlaan G, Billker O, Snell WJ. 2013. Comparative genomics in Chlamydomonas and Plasmodium identifies an ancient nuclear envelope protein family essential for sexual reproduction in protists, fungi, plants, and vertebrates. Genes Dev. 27:1198–1215. 10.1101/gad.212746.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ruiz FA, Marchesini N, Seufferheld M, Govindjee Docampo R. 2001. The polyphosphate bodies of Chlamydomonas reinhardtii possess a proton-pumping pyrophosphatase and are similar to acidocalcisomes. J. Biol. Chem. 276:46196–46203. 10.1074/jbc.M105268200. [DOI] [PubMed] [Google Scholar]
- 35.Weiss RL. 1983. Coated vesicles in the contractile vacuole/mating structure region of Chlamydomonas. J. Ultrastruct. Res. 85:33–44. 10.1016/S0022-5320(83)90114-4. [DOI] [Google Scholar]
- 36.Luykx P, Hopenrath M, Robinson DG. 1997. Osmoregulatory mutants that affect the function of the contractile vacuole in Chlamydomonas reinhardtii. Protoplasma 200:99–111. 10.1007/BF01280738. [DOI] [Google Scholar]
- 37.Becker B, Hickisch A. 2005. Inhibition of contractile vacuole function by brefeldin A. Plant Cell Physiol. 46:201–212. 10.1093/pcp/pci014. [DOI] [PubMed] [Google Scholar]
- 38.Buchmann K, Becker B. 2009. The system of contractile vacuoles in the green alga Mesostigma viride (Streptophyta). Protist 160:427–443. 10.1016/j.protis.2009.01.002. [DOI] [PubMed] [Google Scholar]
- 39.Allen RD, Naitoh Y. 2002. Osmoregulation and contractile vacuoles of protozoa, p 351–394 In Stein W, Zeuthen T. (ed), Molecular mechanisms of water transport across biological membranes. Elsevier, New York, NY. [DOI] [PubMed] [Google Scholar]
- 40.Yasui M, Kwon TH, Knepper MA, Nielsen S, Agre P. 1999. Aquaporin-6: an intracellular vesicle water channel protein in renal epithelia. Proc. Natl. Acad. Sci. U. S. A. 96:5808–5813. 10.1073/pnas.96.10.5808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Otto B, Uehlein N, Sdorra S, Fischer M, Ayaz M, Belastegui-Macadam X, Heckwolf M, Lachnit M, Pede N, Priem N, Reinhard A, Siegfart S, Urban M, Kaldenhoff R. 2010. Aquaporin tetramer composition modifies the function of tobacco aquaporins. J. Biol. Chem. 285:31253–31260. 10.1074/jbc.M110.115881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Johansson I, Karlsson M, Johanson U, Larsson C, Kjellbom P. 2000. The role of aquaporins in cellular and whole plant water balance. Biochim. Biophys. Acta 1465:324–342. 10.1016/S0005-2736(00)00147-4. [DOI] [PubMed] [Google Scholar]
- 43.Verkman AS, Mitra AK. 2000. Structure and function of aquaporin water channels. Am. J. Physiol. Renal Physiol. 278:F13–F28. [DOI] [PubMed] [Google Scholar]
- 44.Marshall WF, Young KD, Swaffer M, Wood E, Nurse P, Kimura A, Frankel J, Wallingford J, Walbot V, Qu X, Roeder AHK. 2012. What determines cell size? BMC Biol. 10:101. 10.1186/1741-7007-10-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pedersen SF, Kapus A, Hoffmann EK. 2011. Osmosensory mechanisms in cellular and systemic volume regulation. J. Am. Soc. Nephrol. 22:1587–1597. 10.1681/ASN.2010121284. [DOI] [PubMed] [Google Scholar]
- 46.Urao T, Yakubov B, Satoh R, Yamaguchi-Shinozaki K, Seki M, Hirayama T, Shinozaki K. 1999. A transmembrane hybrid-type histidine kinase in Arabidopsis functions as an osmosensor. Plant Cell 11:1743–1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.