Abstract
The eukaryotic reverse transcriptase telomerase copies its internal RNA template to synthesize telomeric DNA repeats at chromosome ends in balance with sequence loss during cell proliferation. Previous work has established several factors involved in telomerase recruitment to telomeres in yeast and mammalian cells; however, it remains unclear what determines the association of telomerase with telomeres in other organisms. Here we investigate the cell cycle dependence of telomere binding by each of the seven Tetrahymena thermophila telomerase holoenzyme proteins TERT, p65, Teb1, p50, p75, p45, and p19. We observed coordinate cell cycle-regulated recruitment and release of all of the subunits, including the telomeric-repeat DNA-binding subunit Teb1. Using domain truncation and mutagenesis approaches, we investigated which subunits govern the interaction of telomerase holoenzyme with telomeres. Our results show that Teb1 is critical for telomere interaction of other holoenzyme subunits and demonstrate that high-affinity Teb1 DNA-binding activity is necessary and sufficient for cell cycle-regulated telomere association. Overall, these and additional findings indicate that in the ciliate Tetrahymena, telomerase recruitment to telomeres requires direct binding to single-stranded DNA, unlike the indirect DNA recognition through telomere-bound proteins essential in yeast and mammalian cells.
INTRODUCTION
The ends of linear chromosomes are subjected to an onslaught of illicitly activated double-stranded-DNA (dsDNA) repair mechanisms and sequence loss due to incomplete replication by DNA polymerases (1). These biological challenges are in part overcome by the presence of tandem telomeric DNA repeats at the termini of eukaryotic nuclear chromosomes (for example, the sequence TTAGGG in humans and TTGGGG in the ciliate Tetrahymena), which extend to form an overhang on the 3′-OH strand (2–4). Each terminal repeat array is maintained in a dynamic equilibrium of telomeric DNA attrition from genome replication and de novo synthesis by the enzyme telomerase (5–7). The telomerase ribonucleoprotein (RNP) is minimally composed of the catalytic reverse transcriptase (TERT) and an RNA with an internal template (TER) responsible for RNA-dependent extension of the 3′ chromosome end (8, 9). While in vitro DNA synthesis activity can be reconstituted by expression of only TERT and TER, additional subunits of the telomerase complex are required for high activity and processivity in vitro and for telomere elongation by telomerase recruitment to telomeres in vivo (10, 11).
Studies in yeasts and mammalian cells have yielded significant insights into how telomere proteins recruit telomerase by protein-protein interactions (12, 13). In mammalian cells, telomere-bound protein complexes termed shelterin (1, 14) include the dsDNA telomere-binding proteins TRF1 and TRF2 and the single-stranded-DNA (ssDNA) telomere-binding protein POT1 (14, 15). Together with RAP1 and the TIN2 and TPP1 proteins, which bridge TRF1 and TRF2 to POT1, the telomeric-DNA-binding proteins create a network of complexes that block DNA damage response activation (14). Interestingly, TPP1 also interacts with telomerase as an essential step of recruitment in a manner physiologically restricted to S phase of the cell cycle (12, 16–19). In fission yeast, the dsDNA-binding protein Taz1 takes the place of TRF1 and TRF2, Pot1 binds to the single-stranded DNA overhang, and Rap1, Poz1, and Tpz1 function as bridging proteins that link Taz1 to Pot1 (20, 21). Like mammalian TPP1, Tpz1 and another telomere protein, Ccq1, recruit telomerase to telomeres only in S phase of the cell cycle (20, 22).
Ciliates provide yet another model system for studies of telomere and telomerase biology. Their unusual genomic organization of a germ line micronucleus and a polyploid, fragmented-chromosome macronucleus requires tens of thousands of telomeres and an abundance of telomerase (23). In the model organism Tetrahymena, macronuclear telomeres are bound by a POT1 ortholog, Pot1a, which associates with the TPP1 ortholog Tpt1 to functionally cap telomeres and negatively regulate telomerase access (24, 25). Two additional proteins, Pat1 and Pat2, interact with Pot1a-Tpt1, but their biological role is not well understood (25, 26). While Pat1 and Pat2 are not required for telomere end protection, they are essential for end elongation by telomerase. The Tetrahymena telomerase holoenzyme subunits TERT, TER, and p65 (which form the physiological RNP catalytic core) and Teb1, p75, p50, p45, and p19 (subunits necessary for telomerase function at telomeres) are coassembled in both dividing and nondividing cells (27–29). This potentially constitutive assembly of holoenzyme is different from the paradigm set by yeast telomerase holoenzyme subunit regulation by the cell cycle (11, 12, 21, 30). Nonetheless, constitutive Tetrahymena telomerase holoenzyme assembly would be consistent with the dramatic elongation of telomeres in nondividing cells depleted of the Pot1a-Tpt1-Pat1-Pat2 complex (24).
Reconstitution assays have enabled the dissection of the biochemical roles of individual telomerase holoenzyme subunits, providing an important foundation for investigating the mechanism and regulation of telomere elongation in vivo. Extensive studies have demonstrated that within the telomerase catalytic core, both TERT and TER interact with ssDNA. The TERT N-terminal (TEN) domain stimulates active-site use of a short primer-template hybrid, while the remainder of TERT provides some functional recognition and physical protection of ssDNA (31). Within TER, the template binds to the ssDNA 3′ end by hybridization, and other RNA motifs contribute to template placement in the active site (9, 31). Additional telomerase holoenzyme proteins augment the activity of the catalytic core in vitro, but the relation of these in vitro activities to their roles in vivo remains unclear. Among the Tetrahymena telomerase holoenzyme accessory subunits, the p50 N-terminal 30-kDa region (p50N30) confers high repeat addition processivity (RAP) and is sufficient to bind p75 and the catalytic core in vitro, with the C-terminal region of p50 possibly acting to restrain holoenzyme activity (32, 33). Addition of Teb1 to the p50-bound RNP catalytic core increases the elongation rate of tandem repeat synthesis under conditions typical for telomerase assays in vitro. Of the accessory subunits, only Teb1 has domains readily detectable by sequence homology, with a composition of four oligonucleotide/oligosaccharide-binding (OB)-fold domains (27, 34). The domain architecture of Teb1 is paralogous to that of the largest subunit, Rpa1, of the general ssDNA binding factor replication protein A (RPA) (27, 34). Three of the four predicted Teb1 OB-fold domains are confirmed by high-resolution structures, including a cocrystal structure of the highest-affinity Teb1 DNA-binding domain with telomeric ssDNA (35).
To gain additional insight into how these telomerase holoenzyme subunits are assembled in vivo and engaged at the telomere, we utilized chromatin immunoprecipitation (ChIP) assays to investigate cell cycle-regulated changes in the association of Tetrahymena telomerase holoenzyme subunits with telomeres. Here we show that all of the Tetrahymena telomerase proteins have telomere interaction that is restricted in the cell cycle, despite ubiquitous, cell cycle-independent assembly of the high-RAP holoenzyme complex. Furthermore, using numerous domain and sequence variants of p50 and Teb1, we defined Teb1 as a critical subunit in the recruitment of the telomerase holoenzyme to the telomere. By creating a panel of full-length Teb1 proteins defective specifically in holoenzyme assembly or individual sites of DNA interaction, we showed that the affinity of DNA binding influences Teb1 association with telomeres. Together these results suggest a direct DNA interaction mechanism for Tetrahymena telomerase recruitment to telomeres that is distinct from the recruitment mechanisms proposed in other organisms. Overall, our findings provide new structural insights about the Tetrahymena telomerase holoenzyme, contribute to understanding telomerase enzyme mechanism, and illuminate a new level of detail for the cellular process of telomerase recruitment to telomeres.
MATERIALS AND METHODS
Telomerase reconstitutions.
Telomerase reconstitution assays used codon-optimized open reading frames for TERT, p50, and p75 expression in rabbit reticulocyte lysate (RRL) and for Teb1 and p65 expression in Escherichia coli as previously described (27, 33). For RNP catalytic core assembly, recombinant p65 and in vitro-transcribed TER were added to the TERT RRL expression reaction mixture during protein synthesis at 25 nM each. Holoenzyme reconstitution and affinity purification from RRL synthesis reactions were performed as described in previously optimized protocols (32–34). Briefly, telomerase complexes were bound to anti-FLAG M2 affinity resin (Sigma) and washed in T2MG (20 mM Tris-HCl [pH 8.0], 1 mM MgCl2, 10% glycerol, and 2 mM dithiothreitol [DTT]). Recombinant Teb1 was purified using nickel-nitrilotriacetic acid (Ni-NTA)–agarose and added to a final concentration of 200 nM for 20 min at room temperature prior to the activity assay.
Telomerase activity and DNA binding assays.
Activity assays of native and reconstituted holoenzyme were performed at room temperature using a standard Tetrahymena telomerase reaction buffer containing 50 mM Tris-acetate (pH 8.0), 2 mM MgCl2, 10 mM spermidine, and 5 mM β-mercaptoethanol. Product synthesis reactions additionally contained 24 nM [α-32P]dGTP, 300 nM unlabeled dGTP, 200 μM unlabeled dTTP, and 200 nM DNA primer (GT2G3)3. Reactions were allowed to proceed for 5 min for purified endogenous holoenzyme and 10 min for recombinant holoenzyme. Tetrahymena cell lysate was assayed at a final dilution of 1:200 for 10 min. A 5′-labeled oligonucleotide DNA recovery control (RC) was added to telomerase products before precipitation. Products were resolved by denaturing gel electrophoresis and detected by phosphorimager analysis using a Typhoon Trio imager.
Expression constructs for sequence-modified Teb1BC and full-length Teb1 were generated using PCR-based mutagenesis. N-terminally six-histidine (His6)-tagged proteins were bacterially expressed and isolated by single-step nickel-agarose purification. Extensive washing and subsequent elution resulted in soluble protein purified to near homogeneity as visualized by SDS-PAGE. Electrophoretic mobility shift assays (EMSAs) with recombinant Teb1 were performed as described previously (34) using the 32P 5′-end-labeled oligonucleotide 5′-GTTGGGGTTGGGGTTGGG-3′ as the probe. Binding affinities were calculated based on free probe signal using ImageQuant software.
Tetrahymena strain construction, cell growth, and enzyme purification from extract.
Tetrahymena endogenous-locus replacement strains TERT-FZZ, Teb1-FZZ, p75-FZZ, p65-FZZ, p50-FZZ, p45-FZZ, and p19-FZZ and transgene strains p50N30-FZZ, p50N25-FZZ, ZZF-p50, Teb1-FZZ, F-Teb1BC, and F-Teb1C were previously described (27, 32, 33). F-Teb1BC and F-Teb1C were expressed in the genetic background of TERT with a C-terminal ZZ tag (33). New strains Teb1 F590A F648A-FZZ, Teb1 Δ555–581 GSGSG-FZZ (ΔZn), Teb1 Δ660–666 AGSSG-FZZ (ΔL45), Teb1 Δ687–701-FZZ (ΔCTαH), Teb1 F293A-FZZ, Teb1 K300A-FZZ, Teb1 F423A-FZZ, Teb1 Y450A-FZZ, Teb1 F603A-FZZ, and Teb1 K660A-FZZ were made by targeting transgene integration at the β-tubulin 1 locus (BTU1) under the control of the metallothionein 1 gene (MTT1) promoter using selection for blasticidin resistance conferred by the bsr2 cassette (36). Strains expressing p50N25-FZZ, ZZF-p50, F-Teb1BC, F-Teb1C, and all of the newly generated Teb1-FZZ strains retain endogenous subunit expression for viability. Genotypes were verified by Southern blotting and Western blotting for the transgene-encoded protein.
Cells were grown in modified Neff medium (0.25% proteose peptone, 0.25% yeast extract, 0.5% dextrose, and 30 μM FeCl3) to mid-log phase (3 × 105 cells/ml). Cells were starved in 10 mM Tris (pH 8.0) for 16 h and refed with modified Neff medium to synchronize the cell cycle at G1. For transgene expression, transgene induction was achieved by addition of CdCl2 to a final concentration of 0.5 μg/ml upon refeeding. Cell counts were measured by fixation in 0.4% formaldehyde. Cell counting was completed in triplicate independent growth cultures.
Cell extract preparation and affinity purification were performed as described previously (27). For Western blots, cell pellets were lysed by heating at 95°C for 5 min in 100 μl of 2× SDS loading buffer (4% SDS, 160 mM Tris-HCl [pH 6.8], 20% glycerol, 0.0025% bromophenol blue, 10% β-mercaptoethanol) to avoid sample proteolysis. Proteins were separated on a 10% SDS-polyacrylamide gel and transferred to Hybond N-XL nitrocellulose membrane (Amersham Biosciences). Blots were blocked in 5% milk and 1× Tris-buffered saline (TBS; 50 mM Tris-HCl [pH 7.5], 150 mM NaCl) then incubated with anti-FLAG M2 mouse monoclonal antibody (Sigma) and antitubulin DM1A mouse monoclonal antibody (EMD Millipore). For TER Northern blot analysis, RNA was spotted onto nitrocellulose membrane, UV-cross-linked, and hybridized using a 32P end-labeled oligonucleotide (5′-AGGTTCAAATAAGTGGTAATGCGGGACAAAAGACTATCG-3′).
ChIP analysis.
For each ChIP assay, 2 × 107 cells per immunoprecipitation were fixed with 0.75% formaldehyde at room temperature for 10 min, quenched with 125 mM Tris (pH 7.5), and washed twice with TBS. Nuclei were isolated as described by Jacob et al. (37). Briefly, pelleted cells were lysed in 10 ml of 1× TMS (10 mM Tris-HCl [pH 7.5], 10 mM MgCl2, 3 mM CaCl2, 250 mM sucrose, and 1 mM DTT), 0.16% Igepal CA-630 for 20 min at 4°C with end-over-end rotation. Sucrose was added to a concentration of 0.816 g/ml, and the lysate was centrifuged at 9,000 × g for 30 min at 4°C. The pelleted nuclei were washed once with TMS before micrococcal nuclease (MNase) digestion. For MNase digestion, nuclei were resuspended in 500 μl 50 mM Tris-HCl (pH 7.5), 60 mM KCl, 15 mM NaCl, 2 mM CaCl2, 0.05% spermidine phosphate, and 1 mM DTT and treated with 6 U MNase (New England BioLabs) for 30 min on ice (generated fragment size of ∼500 bp dsDNA; data not shown). The reaction was stopped by adding EDTA to a final concentration of 50 mM and the nuclei were lysed in the presence of 2% Triton X-100, 250 mM NaCl, and protease inhibitors (0.1 mM phenylmethanesulfonyl fluoride and 1× Sigma protease inhibitor cocktail) followed by incubation at room temperature for 30 min.
The lysate was centrifuged at 16,000 × g for 5 min at 4°C, and the supernatant was immunoprecipitated overnight at 4°C with anti-FLAG M2 affinity resin (Sigma). Precipitates were washed sequentially with 1 ml each buffer A (50 mM Tris-HCl [pH 7.5], 250 mM NaCl, 2% Triton X-100, and 5 mM EDTA), buffer B (50 mM Tris-HCl [pH 7.5], 250 mM NaCl, 1.5% Triton X-100, and 5 mM EDTA), CHAPS buffer (50 mM Tris-HCl [pH 7.5], 250 mM NaCl, 1% Triton X-100, 5 mM EDTA, 0.1% SDS, and 0.05% 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate), and LiCl buffer (50 mM Tris-HCl [pH 7.5], 250 mM NaCl, 1% Triton X-100, 5 mM EDTA, and 150 mM LiCl). DNA cross-links were reversed, and the bound material was eluted by resuspending the resin in 400 μl elution buffer (50 mM Tris-HCl [pH 7.5], 200 mM NaCl, 1% SDS, and 1 mM EDTA) and incubating at 65°C for 16 h.
The eluted DNA was purified by phenol-chloroform-isoamyl alcohol (25:24:1) extraction and ethanol precipitation. The resulting product was spotted on nitrocellulose membranes, UV cross-linked, and hybridized with 5′-end-labeled probe against the telomeric DNA (5′-CCCCAACCCCAACCCCAA-3′) or the subtelomeric ribosomal DNA (rDNA) (5′-TGATAAATAACCAAAAATCAAAGTATTACATCAATAAATAACTTTTACTCAATGTCAAAGAAATTATTGGGG-3′). Dot blots were hybridized in 30 mM NaCl, 3 mM Na3C6H5O7, and 0.1% SDS at 55°C for 12 to 16 h, washed, and then quantified by phosphorimager analysis. Relative signal was quantified using ImageQuant software. The resulting data were normalized to input rDNA signal following subtraction of background, to calculate signal from telomerase per chromosome end rather than per length of telomeric repeat DNA tract.
RESULTS
Tetrahymena telomerase association with telomeres is cell cycle regulated.
To address how telomere interaction by telomerase is coordinated with genome replication in Tetrahymena, we exploited a G1 cell cycle phase enrichment protocol involving nutrient starvation to establish synchrony. Following starvation, cells were refed and samples were collected for ChIP as a time course of cell cycle progression. The micronucleus replicates and divides rapidly, but only in subsequent macronuclear replication does the vast majority of telomere synthesis occur. Macronuclear DNA synthesis occurs broadly over an interval from approximately 2 to 4 h postfeeding, followed by morphological changes that ultimately pinch apart the macronucleus and the cell (Fig. 1A). To confirm the synchrony of cell cycle entry, cell counts were taken in triplicate at several time points. There was no major change in cell number over a 6-h period after feeding, but at approximately 8 h the cell count doubled (Fig. 1B).
FIG 1.
Tetrahymena telomerase holoenzyme components have cell cycle-regulated telomere association. (A) Vegetative growth of Tetrahymena schematized to highlight nuclear events starting from starvation-synchronized G1 to the formation of daughter cells. The diploid micronucleus (containing 20 telomeres) duplicates and divides by mitosis, following which the somatic macronucleus (containing ∼30,000 telomeres) replicates and is partitioned by amitotic fission. (B) Cell counts performed after release from starvation to monitor cell division synchronization. A representative cell count is shown from growth of the parental CU522 strain without tagged protein and strains expressing TERT-FZZ, p50-FZZ, p50N30-FZZ, and ZZF-p50 counted in triplicate as independent cultures. Cell cultures for the other expression strains were counted similarly with no difference from the parental strain. (C) Western blot of cells from strains with each holoenzyme component tagged at its endogenous locus by C-terminal fusion to FZZ. (D) Representative activity assays of extracts from wild-type and tagged-subunit strains following release from synchronization. (E) ChIP of each FZZ-tagged telomerase protein expressed from its endogenous locus following release from synchronization. Error bars correspond to the standard errors of the means from multiple independent experiments.
The method of choice for detecting and quantifying chromosome association of telomere-recruited factors like telomerase is telomere ChIP. This technique is typically limited by variable affinity and specificity of antibodies against individual telomere-associated proteins. To overcome this hurdle, we combined endogenous-locus tagging of telomerase holoenzyme proteins with FLAG monoclonal antibody ChIP. Previously generated tagged-subunit strains, each expressing a holoenzyme protein with a C-terminal triple FLAG peptide (F) and tandem protein A domains (ZZ) in place of the untagged native protein from the endogenous locus, were used to ensure physiological subunit expression level and biological function. Consistent with their cellular assembly as a holoenzyme complex, comparable levels of tagged TERT-FZZ, Teb1-FZZ, p75-FZZ, p65-FZZ, p50-FZZ, and p45-FZZ were detected in whole-cell extract by FLAG antibody Western blotting (Fig. 1C). The slightly lower accumulation level of p19-FZZ is a consequence of protein tagging (27).
Telomerase catalytic activity assayed in cell extracts was similar for cells from parental and tagged-subunit-expression strains at different stages of the cell cycle (Fig. 1D). From each strain at each time point, fragmented chromatin was prepared from formaldehyde-cross-linked cells and used for immunoprecipitation of the tagged protein subunit. Because of the relative abundance of endogenous telomerase in Tetrahymena, we were able to quantify protein-bound telomeric repeat DNA directly by hybridization of cross-link-reversed DNA with an end-labeled oligonucleotide complementary to the G-rich telomeric repeats. To control for input extract variations in the preparation of total chromatin, we normalized telomere ChIP signal to rDNA chromosome signal in the input extract (see Materials and Methods). Only low subtelomeric or internal rDNA chromosome hybridization signal was detected in bound ChIP samples (data not shown).
We found that all seven telomerase holoenzyme proteins dramatically increased in telomere association with cell cycle progression to S phase, after which telomere association for all proteins except p50-FZZ decreased (Fig. 1E). Notably, the peripheral holoenzyme subunit Teb1 had a similar telomere ChIP profile compared to the RNP catalytic core subunits TERT and p65 (Fig. 1E), despite indirect interaction of Teb1 with the RNP catalytic core through p50 (33). The delay in loss of telomere ChIP at 6 h unique to p50-FZZ may be a difference imposed by epitope tagging, since previous studies indicated that C-terminal tagging of p50 affects enzyme catalytic activity and increases the length at which telomeres are maintained in cells (27, 32). It is also possible that telomerase holoenzyme architecture changes following telomere recruitment in a manner that increases p50 cross-linking efficiency prior to holoenzyme dissociation, possibly by bringing p50 into closer proximity to product DNA. The alternate explanation that prolonged ChIP signal reflects telomere binding by holoenzyme-free as well as holoenzyme-bound p50 is doubtful, because the biological stability of p50 depends on the other holoenzyme subunits, and thus, p50 is unlikely to be present as a holoenzyme-free protein (27). Overall, we conclude that telomerase holoenzyme subunits are not constitutively bound to telomeres and that all holoenzyme proteins show coordinate telomere recruitment in the cell cycle interval of macronuclear genome replication.
Telomere recruitment is independent of the p50 C-terminal domain but requires p50 interaction with Teb1.
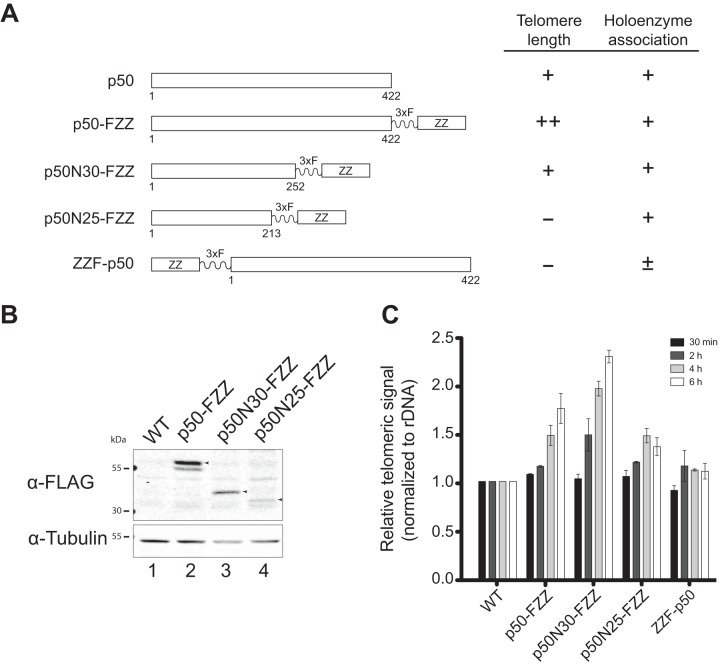
Previous functional analyses demonstrated that the N-terminal 252 residues of p50 (p50N30) support all biochemically identified roles of p50 (32). C-terminal truncation of an additional 39 residues (p50N25) compromised holoenzyme assembly and stability, but the holoenzyme that did assemble with p50N25 retained normal catalytic activity in vivo and in vitro (32). Furthermore, previous structural analyses indicate that all of the p50 subunit density in the holoenzyme structure determined by electron microscopy (EM) derives from p50N30 (32, 33). The C-terminal 20 kDa of p50 appear at least partially disordered in structure, as they are highly sensitive to proteolysis in native cell extract (32, 33). However, because C-terminal tagging of p50 increases telomere length, the p50 C-terminal domain seems likely to have a biological role in holoenzyme regulation (32). In contrast to C-terminal tagging, N-terminal tagging of p50 inhibits the holoenzyme association of Teb1 and disrupts telomerase biological function (33). To investigate how these phenotypes relate to changes in telomerase-telomere interaction, we examined telomere ChIP by N- or C-terminally tagged full-length or truncated p50 proteins (Fig. 2A) in strains with either complete or partial disruption of the endogenous p50 locus (see Materials and Methods).
FIG 2.
Sites of p50 tagging and truncation influence holoenzyme assembly and telomere association. (A) Schematic representation of the tagged p50 proteins used in this study with a summary of their relative function for telomere maintenance and holoenzyme association in vivo (27, 32). (B) Western blot of tagged p50 proteins in cells. The migration of each intact tagged protein is indicated by the arrowhead to the right of the lane. Nonspecific cross-reacting proteins are present in all lanes, including the no-tag protein control (lane 1), whereas proteolysis products of the tagged p50 are lane specific. (C) ChIP performed for p50 proteins as described in the legend to Fig. 1.
We used ChIP to assay C-terminally tagged full-length p50 and p50N30, each expressed in place of the endogenous untagged protein, which gave the tagged proteins similar accumulation levels in cells (Fig. 2B, lanes 1 to 3). Expression of p50N25 was at a lower cellular level (Fig. 2B, lane 4) due to incomplete genetic substitution for the endogenous protein (32). Because ChIP uses denaturing cell lysis and protein purification conditions, it should reduce the differences in p50 purification recovery imposed by native extract proteolysis between the p50-FZZ C-terminal tag and the N-terminal domain (33). Indeed, ChIP by p50-FZZ and p50N30-FZZ was similarly robust, while ChIP by p50N25-FZZ had reduced signal (Fig. 2C). Like p50-FZZ, ChIP of p50N30-FZZ showed the same increase in telomeric signal with the onset of macronuclear replication and the same persistence of ChIP signal at 6 h. As described above for p50-FZZ, the p50N30-FZZ increase in ChIP telomere signal from 4 to 6 h could reflect a change in holoenzyme regulation imposed by elimination of the p50 C-terminal domain. Truncation of p50N30 to p50N25 decreased but did not eliminate telomeric signal association (Fig. 2C), consistent with some p50N25 holoenzyme assembly in vivo. In contrast, ChIP signal was minimal if at all detectable for the N-terminally tagged full-length ZZF-p50 (Fig. 2C). Because Teb1 is the only holoenzyme subunit dissociated by p50 N-terminal tagging (32, 33), this finding suggests that Teb1 is required for telomere association of the other holoenzyme subunits assayed by ZZF-p50 ChIP.
Single-residue substitutions of OB-fold DNA binding surfaces impose a DNA binding or telomerase activation defect in full-length Teb1.
Paralogous to Rpa1, Teb1 has an N-terminal domain and three additional OB-fold domains sequentially designated A, B, and C (34). Rpa1 A, B, and C domains interact with ssDNA with the affinity contribution A > B > C (38). Teb1 A and B domains specifically bind the Tetrahymena telomeric repeat G-rich ssDNA with the affinity A > B (34). Teb1C can enhance Teb1B DNA binding activity, but whether Teb1C interacts with DNA directly is unknown. Teb1C does interact directly with the p50 RNP catalytic core and alone can stimulate its catalytic activity. These findings provided the groundwork for understanding what biochemical properties of Teb1 are required for telomerase-telomere interaction.
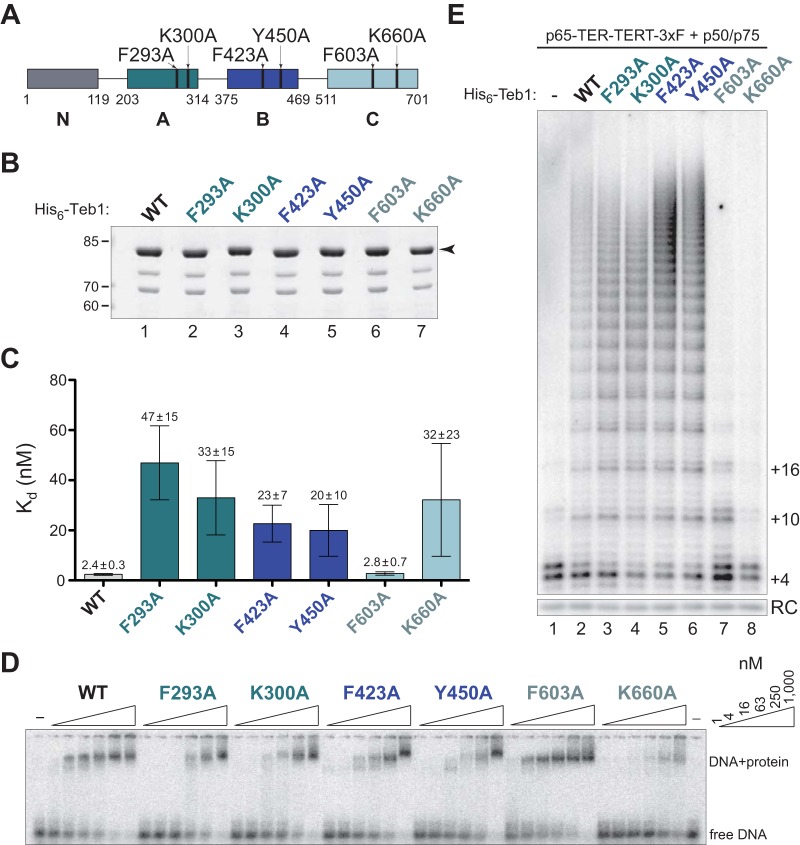
To better characterize the contributions of each Teb1 OB-fold domain to overall Teb1 function, we constructed a panel of full-length Teb1 proteins with single amino acid substitutions on the typical DNA binding surfaces of the A, B, and C domains. Single residue substitutions of alanine were previously assayed in single-domain Teb1A, single-domain Teb1B, and Teb1BC (35). Here we generated two different substitutions in each domain in the full-length-protein context (Fig. 3A), using substitutions that eliminated Teb1A-DNA interaction by EMSA (F293A and K300A), reduced Teb1B-DNA interaction ∼5-fold assayed by filter binding (F423A or Y450A), or reduced Teb1BC DNA binding affinity to near that of Teb1B alone (F603A or K660A). Both Teb1C substitutions also greatly inhibited telomerase catalytic activation.
FIG 3.
Substitutions in canonical DNA binding surfaces of the A, B, and C domains of full-length Teb1 alter DNA binding or telomerase activity in vitro. (A) Schematic of full-length Teb1 single-residue substitutions used in this study. (B) SDS-PAGE of bacterially expressed and purified His6-Teb1 proteins. The arrowhead to the right of the gel indicates full-length Teb1; minor amounts of ∼75- and ∼65-kDa proteins are a copurifying contaminant and Teb1 truncated by proteolysis of the linker between the Teb1 B and C domains, respectively. (C and D) Calculated Kd for each of the Teb1 proteins as determined by EMSA in triplicate (C), with a representative EMSA (D). (E) Activity assay of recombinant full-length Teb1 reconstitution of holoenzyme catalytic activity.
Bacterial expression and affinity purification of N-terminally tagged full-length Teb1 proteins gave predominantly full-length protein by single-step affinity purification (Fig. 3B). We used the purified proteins for ssDNA interaction EMSAs (Fig. 3C and D). The two substitutions in the Teb1A domain had the greatest impact on DNA binding, decreasing affinity ∼20- and ∼15-fold for F293A and K300A, respectively. Substitutions in the Teb1B domain decreased DNA binding affinity less severely, at most ∼10-fold. The Teb1C domain substitution F603A did not affect full-length Teb1 DNA binding affinity, while the K660A substitution imposed a loss of DNA binding activity that was variable between replicate assays, suggestive of a loss of overall protein folding stability.
Next we tested the panel of Teb1 proteins for in vitro reconstitution of high-RAP telomerase activity assayed by direct primer extension with dTTP and radiolabeled dGTP (Fig. 3E). The RNP catalytic core containing bacterially expressed p65, in vitro transcribed TER, and RRL-expressed C-terminally FLAG-tagged TERT (TERT-F) was assembled in RRL and combined with separately RRL-expressed p50 and p75. Telomerase complexes were affinity purified using FLAG antibody resin and then combined with bacterially expressed Teb1 in the activity assay prior to addition of the DNA primer. Full-length Teb1 with a F293A, K300A, F423A, or Y450A substitution in the DNA binding domain showed little if any difference from wild-type Teb1 in high-RAP activity reconstitution (Fig. 3E, lanes 1 to 6). This is consistent with the previously noted lack of correlation between Teb1 DNA binding affinity and processivity stimulation (34). In contrast, full-length Teb1 with a Teb1C F603A or K660A substitution had reduced stimulation of high-RAP DNA product synthesis (Fig. 3E, lanes 7 and 8). Thus, as predicted by domain truncation studies (34), Teb1 biochemical roles in DNA binding and telomerase catalytic activation can be separated in the full-length-protein context.
Distinct surfaces of Teb1C contribute to Teb1BC DNA binding and telomerase catalytic activation.
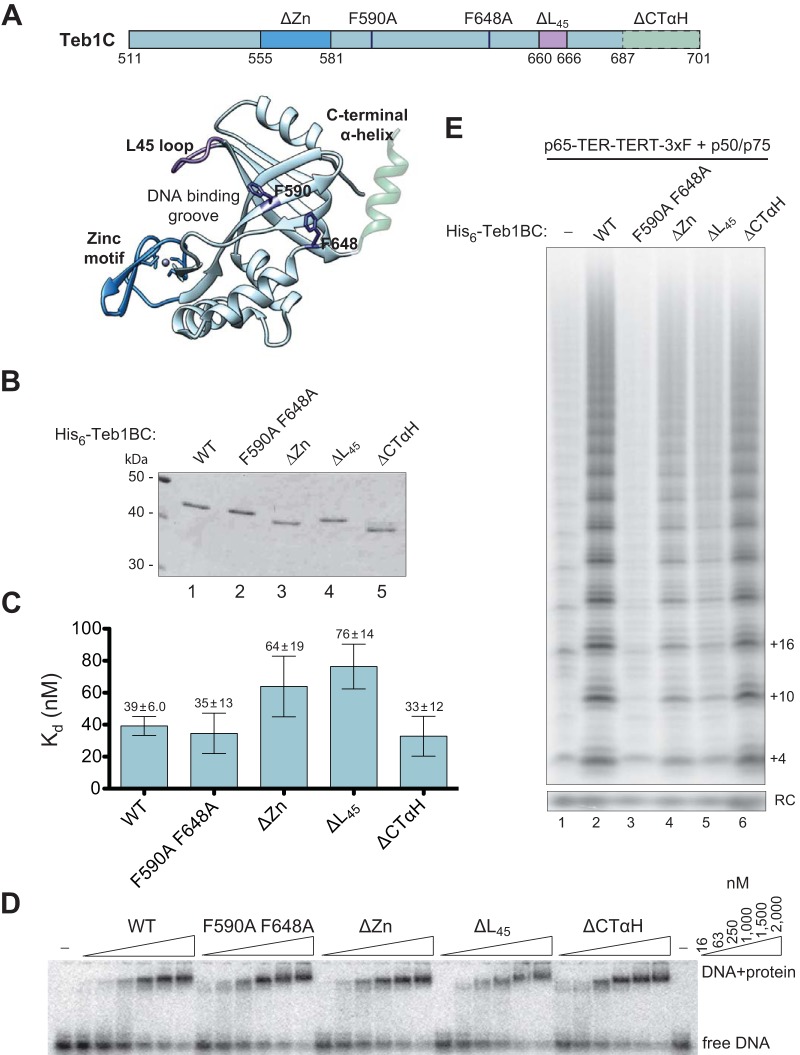
Structural analysis of Teb1C revealed surface features divergent from a canonical OB-fold DNA binding domain (35). Despite distinct structure and lack of high-affinity DNA binding, Teb1C could contact ssDNA to guide its threading from the telomerase active site to Teb1AB in the elongating high-RAP holoenzyme conformation. In addition, with or without a role in DNA contact, Teb1C interaction with the p50-assembled RNP catalytic core stimulates the rate of high-RAP product synthesis (32). Teb1C amino acid substitution can compromise Teb1BC telomerase activation without impact on DNA interaction, and Teb1C alone can stimulate some high RAP activity despite undetectable DNA binding (34). Thus, we sought to additionally resolve the biochemical activities of Teb1C in order to discern how its properties affect telomerase-telomere interaction. To this end, we screened substitutions of bulky side chain residues on the Teb1C surface, informed by the high-resolution Teb1C structure (35) and known determinants of Rpa1 protein-protein and protein-DNA interactions (39–41). Based on previous studies (34), we assayed for potential changes in Teb1C contribution to Teb1BC DNA binding affinity and telomerase catalytic activation.
Teb1C sequence changes of interest (Fig. 4A) were assayed in purified Teb1BC proteins (Fig. 4B) for DNA binding affinity by EMSA (Fig. 4C and D) and for high-RAP activity stimulation by direct primer extension (Fig. 4E). Of particular interest, alanine substitutions of residues F590 and F648 distant from the canonical OB-fold DNA binding surface (Fig. 4A) did not affect the affinity of Teb1BC DNA binding by EMSA (Fig. 4C and D) but entirely eliminated telomerase activation (Fig. 4E). In comparison, replacement of the entire zinc ribbon lobe by a short linker (i.e., residues 555 to 581 changed to GSGSG) (35) only partially inhibited telomerase activation (Fig. 4E). A newly constructed replacement of the unstructured loop connecting OB-fold β-strands 4 and 5 (residues 660 to 666 to AGSSG [ΔL45]) on the potential DNA binding surface (39) had some influence on the DNA binding affinity of Teb1BC and also partially inhibited telomerase activation (Fig. 4C, D, and E). As previously noted (34), deletion of the predicted Teb1 C-terminal α-helix (residues 687 to 701 [ΔCTαH]) that was disordered in the Teb1C structure (35) but has potentially close proximity to the F590/F648 surface (Fig. 4A) had no discernible consequence (Fig. 4C, D, and E). The lack of requirement for the Teb1 ΔCTαH was proposed to reflect evolutionary divergence from Rpa1 to avoid CTαH-mediated heterotrimer association with other RPA subunits (34). Taken together, the results above suggest that instead of the CTαH, a Teb1C surface involving F590 and F648 could be the major determinant for the association of Teb1C with p50 and thus the entire holoenzyme. However, due to the compromised stability of recombinant Teb1 association with other reconstituted holoenzyme components in vitro (33), it was not possible to test this hypothesis directly.
FIG 4.
Teb1C requirements for DNA binding and telomerase activation involve distinct domain surfaces. (A) Schematic of Teb1C variants used in this study. The L45 loop and α-helix at the C terminus were added to the crystal structure (35) by modeling using Chimera (49). (B) SDS-PAGE of bacterially expressed and purified His6-Teb1BC proteins. (C and D) Calculated Kd values for each of the Teb1BC proteins as determined by EMSA in triplicate (C), with a representative EMSA (D). (E) Activity assay of recombinant Teb1BC reconstitution of holoenzyme catalytic activity.
In vivo expression of full-length Teb1 variants reveals structural requirements for Teb1 holoenzyme assembly.
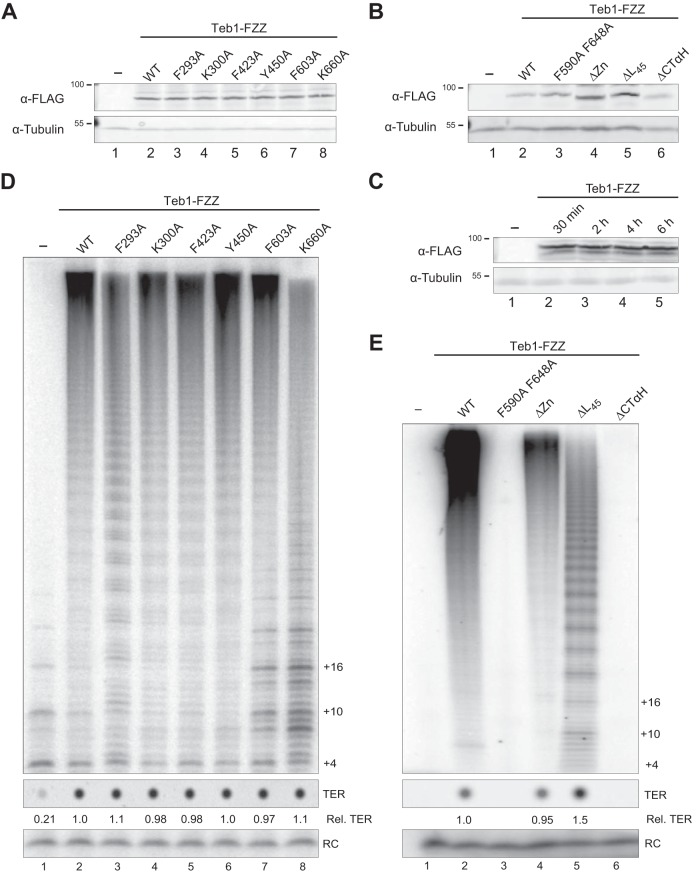
Separation-of-function full-length Teb1 variants provided an opportunity to test the Teb1 biochemical requirements for holoenzyme assembly, catalytic activity, and telomere interaction in vivo. We generated Tetrahymena strains expressing full-length Teb1 proteins with different defects for DNA binding and/or telomerase activation in vitro (Fig. 3 and 4). For consistent comparison independent of the ability of the Teb1 variant to support cell viability in replacement of endogenous Teb1, we integrated expression transgenes driven by the cadmium-inducible MTT1 promoter at the BTU1 locus. Each Teb1 variant or wild-type Teb1 was C-terminally FZZ tagged. We confirmed by Western blotting that equivalent levels of each Teb1-FZZ protein were expressed in cells starved and refed to initiate cell cycle progression, induced for transgene Teb1 expression, and sampled at the postfeeding 4-h time point of macronuclear DNA replication (Fig. 5A and B). Levels of induced Teb1-FZZ protein overexpression remained constant across the time course of cell cycle analysis (Fig. 5C).
FIG 5.
Inducible expression and holoenzyme assembly of FZZ-tagged Teb1 variants reveal requirements for Teb1 holoenzyme assembly. (A and B) Western blots of cells from full-length Teb1-FZZ transgene expression strains with single-residue substitutions of the A, B, or C domain (A) or other Teb1C domain substitutions or deletions (B). (C) Western blot of representative Teb1-FZZ expression across the time course of cell cycle analysis. (D and E) Telomerase activity assay following FLAG antibody purification of Teb1-FZZ complexes from cell extracts.
We first examined the cellular assembly of each tagged Teb1 protein as a telomerase holoenzyme. Cell extracts were used to perform an established F-tag affinity purification (27). Purified complexes associated with tagged Teb1 were assayed in parallel for telomerase activity using the direct primer extension assay and for quantification of TER by dot blot hybridization. To control for nonspecific purification background, cell extract lacking a tagged protein was used in parallel (Fig. 5D and E, lanes 1). Each of the Teb1-FZZ proteins with a single-residue substitution on the typical OB-fold domain DNA binding surface assembled telomerase holoenzyme comparably to wild-type Teb1, as judged by copurification of TER (Fig. 5D). The Teb1A or Teb1B domain variants also had little difference in the telomerase activity product profile compared to wild-type Teb1 (Fig. 5D, lanes 2 to 5), mirroring the in vitro reconstitution results (Fig. 3E). In comparison, in vivo assembly of Teb1C domain variants F603A and K660A revealed that these substitutions did affect assembled holoenzyme catalytic activity (Fig. 5D, lanes 7 and 8). Product DNAs had a pronounced low-RAP profile as well as some of the high-RAP profile of wild-type enzyme. The Teb1 K660A holoenzyme also had reduced catalytic activity per TER relative to wild-type holoenzyme (Fig. 5D, compare lanes 2 and 8), which would be consistent with an overall protein folding problem as well as an activity defect incurred by the K660A substitution.
A dramatic difference was observed for Teb1-FZZ with the F590A and F648A substitutions, which did not recover any associated telomerase activity or TER (Fig. 5E, lanes 2 and 3). Thus, these substitutions on a Teb1C surface far from the canonical DNA binding interface abrogated full-length Teb1 holoenzyme assembly in vivo. Teb1-FZZ with Teb1C zinc ribbon or L45 loop deletion efficiently assembled telomerase holoenzyme based on the level of copurified TER, but these holoenzymes had compromised catalytic activity (Fig. 5E, lanes 4 and 5). Curiously, RAP as well as overall activity was affected for the holoenzyme with Teb1ΔL45-FZZ (Fig. 5E, lane 5), suggestive of a possible role for the L45 loop in clamping product DNA. Intriguingly, Teb1ΔCTαH did not support holoenzyme assembly in vivo (Fig. 5E, lane 6). This finding was surprising given that deletion of the CTαH was inconsequential for robust high-RAP-activity reconstitution in vitro (Fig. 4E, lane 6). We suggest that the need for Teb1 CTαH function could be obviated in vitro by the high concentration of Teb1 required in the activity assay (see Discussion).
Teb1 DNA binding activity independent of holoenzyme association is sufficient for telomere interaction.
We next used the full-length Teb1-FZZ transgene expression strains to determine the biochemical requirements for Teb1-mediated telomerase recruitment to telomeres by ChIP. Overexpressed wild-type Teb1-FZZ gave almost 20-fold telomeric DNA signal enrichment over background binding assayed using cell extract lacking tagged protein (Fig. 6A), which is an ∼5-fold increase in the amount of telomeric ChIP signal compared to ChIP of Teb1-FZZ tagged at its endogenous locus (Fig. 1E). This increase in telomere association is consistent with the finding that Teb1 overexpression induces telomere loss and growth arrest (27), which could result from telomere structure changes imposed by the additional Teb1 binding. Remarkably, even overexpressed Teb1 retained cell cycle specificity of telomere interaction, evident as the sharp peak of telomere interaction at 4 h after release from starvation (Fig. 6A). This result highlights the tight regulation of Teb1-telomere interaction during S phase of the cell cycle and suggests that the cell cycle regulation of Teb1-telomere interaction is controlled by a mechanism autonomous to Teb1.
FIG 6.
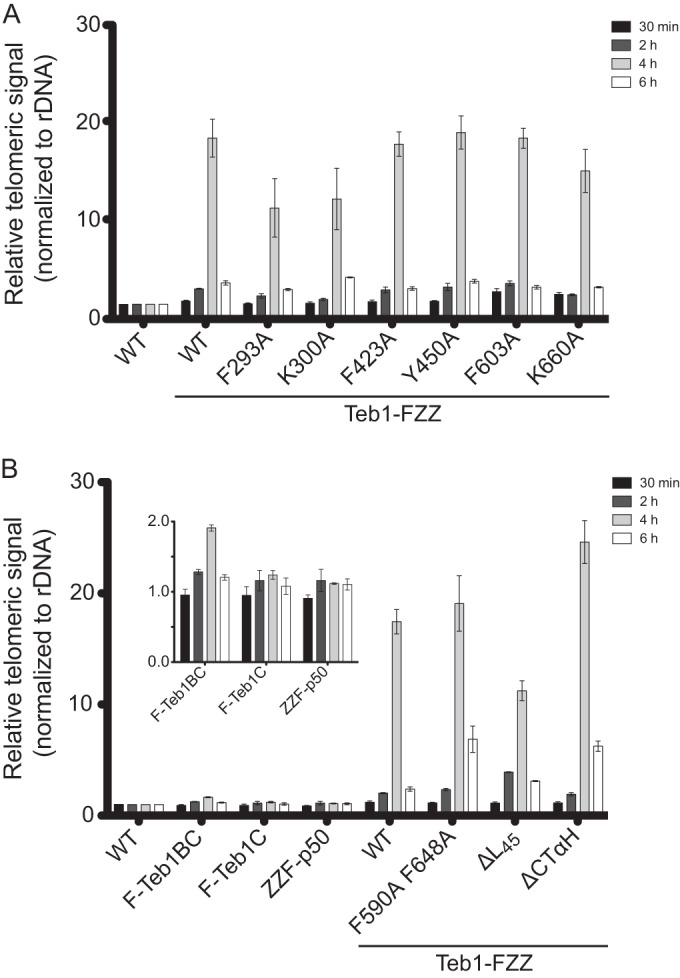
DNA binding mediates Teb1-telomere interaction. ChIP was performed as described in the legend to Fig. 1 for the Teb1 transgene expression strains with (A) single amino acid substitutions and (B) domain truncations and additional Teb1C variants. The inset shows a subset of the same results plotted with a different vertical scale.
We compared ChIP by the transgene-expressed wild-type Teb1-FZZ and the Teb1-FZZ single-residue substitutions on the canonical OB-fold DNA binding surfaces, which for Teb1 A and B domains compromised full-length Teb1 DNA binding in vitro (Fig. 3C and D). The F293A and K300A substitutions within Teb1A reduced the telomere ChIP signal, whereas the F423A and Y450A substitutions in Teb1B and the F603A and K660A substitutions in Teb1C did not substantially change telomere ChIP in comparison to wild-type Teb1 (Fig. 6A). No single-residue substitution eliminated telomere interaction by full-length Teb1-FZZ in vivo, consistent with the results for full-length Teb1 DNA binding in vitro. Nonetheless, among the single-residue Teb1 variants, the substitutions that did reduce telomere interaction were those with the greatest impact on Teb1 DNA binding affinity in vitro. To extend this connection, we compared ChIP by full-length Teb1-FZZ to ChIP by F-Teb1BC or F-Teb1C also expressed from the MTT1 promoter of a transgene integrated at the BTU1 locus (33). Compared to full-length Teb1, Teb1BC showed an ∼10-fold decrease in telomeric ChIP signal and Teb1C an ∼20-fold decrease in telomeric ChIP signal (Fig. 6A). We conclude that Teb1BC catalytic activation of telomerase to high-RAP product synthesis is insufficient for telomere recruitment, at least under conditions of competition with the coexpressed endogenous Teb1 required for cell viability (33).
Finally, we compared ChIP by the transgene-expressed wild-type Teb1-FZZ to that of the Teb1-FZZ proteins with Teb1C deletions or substitutions beyond the canonical OB-fold DNA binding surface. Teb1ΔL45 showed a decrease in telomere interaction but retained a ChIP signal within ∼2-fold of the wild-type signal (Fig. 6B). Of particular interest were the two Teb1 variants lacking any biochemically detectable holoenzyme assembly. Both the F590A F648A and ΔCTαH Teb1-FZZ proteins gave telomeric ChIP signals equaling or exceeding that of the wild-type Teb1-FZZ (Fig. 6B). These findings indicate that Teb1 associates with telomeres without a requirement for coassembly with the other telomerase holoenzyme subunits. Overall, the results above suggest the conclusion that in Tetrahymena, an integral subunit of a constitutively assembled telomerase holoenzyme mediates telomerase recruitment to telomeres by direct sequence-specific but cell cycle-regulated association with ssDNA (Fig. 7). These studies of Tetrahymena telomerase reveal a mechanism for telomerase-telomere interaction that is distinct from telomerase recruitment mechanisms proposed for yeast or mammalian cell model systems.
FIG 7.
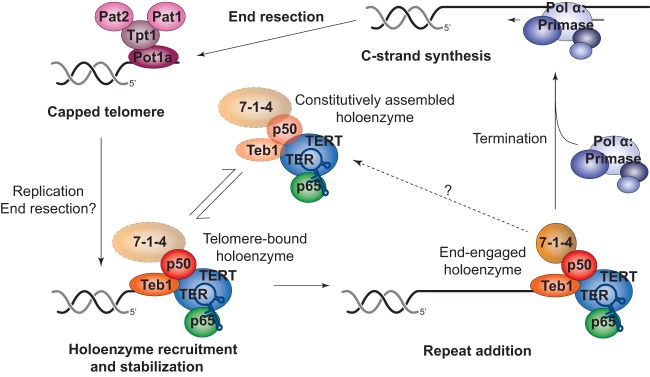
Working model of Tetrahymena telomerase recruitment to and regulation at telomeres. During G1, telomeres are capped by the four-protein complex consisting of Pot1a, Tpt1, Pat1, and Pat2. S phase marks the start of DNA replication and synthesis, which initiates a series of events to orchestrate a handoff between Pot1a and Teb1. After holoenzyme recruitment to the telomeres, telomerase engages in productive repeat addition. Termination of this step may be coupled to disassembly and/or displacement by Polα-primase to extend the C-strand and favor reestablishment of the Tetrahymena telomere protein complex.
DISCUSSION
This study provides mechanistic insight into how Tetrahymena telomerase is governed in its action at telomeres. We show that Teb1 is necessary for telomerase-telomere interaction and also sufficient for telomere interaction as a holoenzyme subunit dissociated from the RNP catalytic core. Furthermore, using mutagenesis for selective disruption of different Teb1 biochemical properties, we determined that telomere association by Teb1 depends on its high affinity of DNA binding. Together these results support a model in which telomere recruitment of telomerase occurs through direct interaction of Teb1-containing holoenzyme with DNA (Fig. 7), rather than by telomerase interaction with a telomere-bound protein, as in yeasts and mammalian cells (16, 20, 30, 42–44). Activity assays here and in previous studies (27–29, 32, 33) suggest that a Tetrahymena telomerase holoenzyme is largely assembled off the telomere, although dynamic exchange of Teb1 and/or 7-1-4 could occur (Fig. 7). Constitutive assembly of telomerase holoenzyme makes sense in light of the offset micronuclear and macronuclear S phase in Tetrahymena, which require telomerase function over an extended interval of cell growth. Despite cell extract evidence for apparently constitutive telomerase holoenzyme assembly, the ChIP assays of every telomerase protein subunit across a synchronized cell cycle demonstrate conclusively that Tetrahymena telomerase-telomere interaction is cell cycle regulated. Therefore, after S phase is complete, telomerase holoenzyme must be excluded from telomeric DNA interaction.
We suggest that because a Tetrahymena telomerase holoenzyme is recruited to ssDNA, the cell cycle regulation of telomerase-telomere interaction can derive from direct DNA binding competition for the telomere 3′ overhang (Fig. 7). Tetrahymena telomeres are capped by Pot1a in complex with Tpt1, Pat1, and Pat2 (25, 26), bound to an overhang of 14 to 15 or 20 to 21 nucleotides with a TGGGGT-3′OH end permutation (45, 46). This length of overhang is insufficient for binding both Pot1a and Teb1 (34). Because Pot1a depletion triggers runaway telomere elongation even in nondividing cells (24), telomerase exclusion from chromosome termini may depend on a low off-rate of Pot1a from bound DNA achieved in part by Pot1a interactions with other telomere proteins (25, 47). Alternately, Teb1 binding to telomeres could be competed by RPA, with subsequent RPA replacement by Pot1a complexes. After Pot1a binding, genome replication or potentially C-strand resection would then be required to displace Pot1a and allow telomerase recruitment (Fig. 7). However, simple binding competition predicts that the extended ssDNA length of telomeres elongated by telomerase would be a highly favorable substrate for additional elongation, which is not consistent with telomere length homeostasis. Thus, we suggest that new repeat synthesis by telomerase is coupled to C-strand synthesis in a Teb1-dependent manner that is disadvantageous to another Teb1 engagement of the same telomere (Fig. 7). Synthesis-dependent telomerase recruitment of its own displacement factors provides a compelling model for the function of Rpa1-like domain architecture in Teb1, because Rpa1 recruits second-strand synthesis activity by conformational change induced upon DNA binding (48).
The DNA interaction affinity of Teb1 derives from the central Teb1 A and B domains, which combined have higher affinity for telomeric repeat sequence than does full-length Tetrahymena Rpa1 (34). Removal of the entire Teb1A domain in the Teb1BC protein and Teb1A amino acid substitution in full-length Teb1 each reduced ChIP, but the amounts of ChIP loss are not directly comparable due to potential differences in cross-linking efficiency with the change in DNA binding site length and potential differences in competition with endogenous Teb1. Teb1C interaction with DNA is not detectable directly by EMSA, but a contribution of Teb1C to DNA interaction would be consistent with the reduced telomerase RAP and telomere interaction imposed by the Teb1C ΔL45 substitution. In crystallography studies, the predicted Teb1C terminal α-helix peptide was not ordered in position relative to the Teb1C OB fold. However, based on the position of the C-terminal residue of the OB fold, the peptide extension could be near the F590/F648 protein face away from the canonical DNA binding cleft (Fig. 4A). The importance of F590 and F648 for Teb1 assembly with the p50 RNP catalytic core is supported by both in vitro and in vivo telomerase reconstitutions, whereas the significance of the α-helix at the C terminus was evident only with in vivo reconstitution. This terminal α-helix could interact with as-yet unidentified Rpa2 and Rpa3 homologs that chaperone telomerase holoenzyme assembly in vivo. A more likely explanation lies in the failure of recombinant versus endogenous Teb1 to support high-affinity p50 interaction, compensated by the use of a high concentration of Teb1 in the reconstituted-enzyme activity assays. The biochemical challenge to in vitro folding of a physiological Teb1-p50-RNP catalytic core holoenzyme conformation parallels by the inefficient reconstitution of purified human TPP1 with recombinant human telomerase RNP in vitro (18). Studies of Tetrahymena telomerase holoenzyme conformational change upon ssDNA binding, elongation, and termination (Fig. 7) will provide informative comparison for general insights about telomerase mechanism and regulation at telomeres.
ACKNOWLEDGMENTS
Funding for this work was provided by an NSF Graduate Research Fellowship under grant DGE-1106400 to H.E.U. and NIH R01 GM054198 to K.C. Molecular graphics and analyses were performed with the UCSF Chimera package. Chimera is developed by the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco (supported by NIGMS P41-GM103311).
Footnotes
Published ahead of print 15 September 2014
REFERENCES
- 1.O'Sullivan RJ, Karlseder J. 2010. Telomeres: protecting chromosomes against genome instability. Nat. Rev. Mol. Cell Biol. 11:171–181. 10.1038/nrm2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Moyzis RK, Buckingham JM, Cram LS, Dani M, Deaven LL, Jones MD, Meyne J, Ratliff RL, Wu J-R. 1988. A highly conserved repetitive DNA sequence, (TTAGGG)n, present at the telomeres of human chromosomes. Proc. Natl. Acad. Sci. U. S. A. 85:6622–6626. 10.1073/pnas.85.18.6622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blackburn EH, Greider CW, Szostak JW. 2006. Telomeres and telomerase: the path from maize, Tetrahymena and yeast to human cancer and aging. Nat. Med. 12:1133–1138. 10.1038/nm1006-1133. [DOI] [PubMed] [Google Scholar]
- 4.Blackburn EH, Gall JG. 1978. A tandemly repeated sequence at the termini of the extrachromosomal ribosomal RNA genes in Tetrahymena. J. Mol. Biol. 120:33–53. 10.1016/0022-2836(78)90294-2. [DOI] [PubMed] [Google Scholar]
- 5.Greider CW. 1996. Telomere length regulation. Annu. Rev. Biochem. 65:337–365. 10.1146/annurev.bi.65.070196.002005. [DOI] [PubMed] [Google Scholar]
- 6.Pfeiffer V, Lingner J. 2013. Replication of telomeres and the regulation of telomerase. Cold Spring Harb. Perspect. Biol. 5:a010405. 10.1101/cshperspect.a010405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Lange T. 2009. How telomeres solve the end-protection problem. Science 326:948–952. 10.1126/science.1170633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hengesbach M, Akiyama BM, Stone MD. 2011. Single-molecule analysis of telomerase structure and function. Curr. Opin. Chem. Biol. 15:845–852. 10.1016/j.cbpa.2011.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blackburn EH, Collins K. 2011. Telomerase: an RNP enzyme synthesizes DNA. Cold Spring Harb. Perspect. Biol. 3:205–213. 10.1101/cshperspect.a003558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Podlevsky JD, Chen JJ. 2012. It all comes together at the ends: telomerase structure, function, and biogenesis. Mutat. Res. 730:3–11. 10.1016/j.mrfmmm.2011.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Egan ED, Collins K. 2012. Biogenesis of telomerase ribonucleoproteins. RNA 18:1747–1759. 10.1261/rna.034629.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nandakumar J, Cech TR. 2013. Finding the end: recruitment of telomerase to telomeres. Nat. Rev. Mol. Cell Biol. 14:69–82. 10.1038/nrm3505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stewart JA, Chaiken MF, Wang F, Price CM. 2012. Maintaining the end: roles of telomere proteins in end-protection, telomere replication and length regulation. Mutat. Res. 730:12–19. 10.1016/j.mrfmmm.2011.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Palm W, de Lange T. 2008. How shelterin protects mammalian telomeres. Annu. Rev. Genet. 42:301–334. 10.1146/annurev.genet.41.110306.130350. [DOI] [PubMed] [Google Scholar]
- 15.Baumann P, Cech TR. 2001. Pot1, the putative telomere end-binding protein in fission yeast and humans. Science 292:1171–1175. 10.1126/science.1060036. [DOI] [PubMed] [Google Scholar]
- 16.Xin H, Liu D, Wan M, Safari A, Kim H, O'Connor MS, Songyang Z. 2007. TPP1 is a homologue of ciliate TEBP-beta and interacts with POT1 to recruit telomerase. Nature 445:559–562. 10.1038/nature05469. [DOI] [PubMed] [Google Scholar]
- 17.Nandakumar J, Bell CF, Weidenfeld I, Zaug AJ, Leinwand LA, Cech TR. 2012. The TEL patch of telomere protein TPP1 mediates telomerase recruitment and processivity. Nature 492:285–259. 10.1038/nature11648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sexton AN, Youmans DT, Collins K. 2012. Specificity requirements for human telomere protein interaction with telomerase holoenzyme. J. Biol. Chem. 287:34455–34464. 10.1074/jbc.M112.394767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhong FL, Batista LF, Freund A, Pech MF, Venteicher AS, Artandi SE. 2012. TPP1 OB-fold domain controls telomere maintenance by recruiting telomerase to chromosome ends. Cell 150:481–494. 10.1016/j.cell.2012.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miyoshi T, Kanoh J, Saito M, Ishikawa F. 2008. Fission yeast Pot1-Tpp1 protects telomeres and regulates telomere length. Science 320:1341–1344. 10.1126/science.1154819. [DOI] [PubMed] [Google Scholar]
- 21.Moser BA, Nakamura TM. 2009. Protection and replication of telomeres in fission yeast. Biochem. Cell Biol. 87:747–758. 10.1139/O09-037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tomita K, Cooper JP. 2008. Fission yeast Ccq1 is telomerase recruiter and local checkpoint controller. Genes Dev. 22:3461–3474. 10.1101/gad.498608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jahn CL, Klobutcher LA. 2002. Genome remodeling in ciliated protozoa. Annu. Rev. Microbiol. 56:489–520. 10.1146/annurev.micro.56.012302.160916. [DOI] [PubMed] [Google Scholar]
- 24.Jacob NK, Lescasse R, Linger BR, Price CM. 2007. Tetrahymena POT1a regulates telomere length and prevents activation of a cell cycle checkpoint. Mol. Cell. Biol. 27:1592–1601. 10.1128/MCB.01975-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Linger BR, Morin GB, Price CM. 2011. The Pot1a-associated proteins Tpt1 and Pat1 coordinate telomere protection and length regulation in Tetrahymena. Mol. Biol. Cell 22:4161–4170. 10.1091/mbc.E11-06-0551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Premkumar VL, Cranert S, Linger BR, Morin GB, Minium S, Price C. 2014. The 3′ overhangs at Tetrahymena thermophila telomeres are packaged by four proteins, Pot1a, Tpt1, Pat1, and Pat2. Eukaryot. Cell 13:240–245. 10.1128/EC.00275-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Min B, Collins K. 2009. An RPA-related sequence-specific DNA-binding subunit of telomerase holoenzyme is required for elongation processivity and telomere maintenance. Mol. Cell 36:609–619. 10.1016/j.molcel.2009.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Witkin KL, Collins K. 2004. Holoenzyme proteins required for the physiological assembly and activity of telomerase. Genes Dev. 18:1107–1118. 10.1101/gad.1201704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Witkin KL, Prathapam R, Collins K. 2007. Positive and negative regulation of Tetrahymena telomerase holoenzyme. Mol. Cell. Biol. 27:2074–2083. 10.1128/MCB.02105-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moser BA, Chang YT, Kosti J, Nakamura TM. 2011. Tel1ATM and Rad3ATR kinases promote Ccq1-Est1 interaction to maintain telomeres in fission yeast. Nat. Struct. Mol. Biol. 18:1408–1413. 10.1038/nsmb.2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu RA, Collins K. 2014. Human telomerase specialization for repeat synthesis by unique handling of primer-template duplex. EMBO J. 33:921–935. 10.1002/embj.201387205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hong K, Upton H, Miracco EJ, Jiang J, Zhou ZH, Feigon J, Collins K. 2013. Tetrahymena telomerase holoenzyme assembly, activation, and inhibition by domains of the p50 central hub. Mol. Cell. Biol. 33:3962–3971. 10.1128/MCB.00792-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jiang J, Miracco EJ, Hong K, Eckert B, Chan H, Cash DD, Min B, Zhou ZH, Collins K, Feigon J. 2013. The architecture of Tetrahymena telomerase holoenzyme. Nature 496:187–192. 10.1038/nature12062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Min B, Collins K. 2010. Multiple mechanisms for elongation processivity within the reconstituted Tetrahymena telomerase holoenzyme. J. Biol. Chem. 285:16434–16443. 10.1074/jbc.M110.119172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zeng Z, Min B, Huang J, Hong K, Yang Y, Collins K, Lei M. 2011. Structural basis for Tetrahymena telomerase processivity factor Teb1 binding to single-stranded telomeric-repeat DNA. Proc. Natl. Acad. Sci. U. S. A. 108:20357–20361. 10.1073/pnas.1113624108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Couvillion MT, Collins K. 2012. Biochemical approaches including the design and use of strains expressing epitope-tagged proteins. Methods Cell Biol. 109:347–355. 10.1016/B978-0-12-385967-9.00012-8. [DOI] [PubMed] [Google Scholar]
- 37.Jacob NK, Stout AR, Price CM. 2004. Modulation of telomere length dynamics by the subtelomeric region of Tetrahymena telomeres. Mol. Biol. Cell 15:3719–3728. 10.1091/mbc.E04-03-0237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wold MS. 1997. Replication protein A: a heterotrimeric, single-stranded DNA-binding protein required for eukaryotic DNA metabolism. Annu. Rev. Biochem. 66:61–92. 10.1146/annurev.biochem.66.1.61. [DOI] [PubMed] [Google Scholar]
- 39.Bochkareva E, Belegu V, Korolev S, Bochkarev A. 2001. Structure of the major single-stranded DNA-binding domain of replication protein A suggests a dynamic mechanism for DNA binding. EMBO J. 20:612–618. 10.1093/emboj/20.3.612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lao Y, Lee CG, Wold MS. 1999. Replication protein A interactions with DNA. 2. Characterization of double-stranded DNA-binding/helix-destabilization activities and the role of the zinc-finger domain in DNA interactions. Biochemistry 38:3974–3984. [DOI] [PubMed] [Google Scholar]
- 41.Bochkareva E, Korolev S, Lees-Miller SP, Bochkarev A. 2002. Structure of the RPA trimerization core and its role in the multistep DNA-binding mechanism of RPA. EMBO J. 21:1855–1863. 10.1093/emboj/21.7.1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yamazaki H, Tarumoto Y, Ishikawa F. 2012. Tel1(ATM) and Rad3(ATR) phosphorylate the telomere protein Ccq1 to recruit telomerase and elongate telomeres in fission yeast. Genes Dev. 26:241–246. 10.1101/gad.177873.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Abreu E, Aritonovska E, Reichenbach P, Cristofari G, Culp B, Terns RM, Lingner J, Terns MP. 2010. TIN2-tethered TPP1 recruits human telomerase to telomeres in vivo. Mol. Cell. Biol. 30:2971–2982. 10.1128/MCB.00240-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tejera AM, Stagno d'Alcontres M, Thanasoula M, Marion RM, Martinez P, Liao C, Flores JM, Tarsounas M, Blasco MA. 2010. TPP1 is required for TERT recruitment, telomere elongation during nuclear reprogramming, and normal skin development in mice. Dev. Cell 18:775–789. 10.1016/j.devcel.2010.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jacob NK, Kirk KE, Price CM. 2003. Generation of telomeric G strand overhangs involves both G and C strand cleavage. Mol. Cell 11:1021–1032. 10.1016/S1097-2765(03)00131-X. [DOI] [PubMed] [Google Scholar]
- 46.Jacob NK, Skopp R, Price CM. 2001. G-overhang dynamics at Tetrahymena telomeres. EMBO J. 20:4299–4308. 10.1093/emboj/20.15.4299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang F, Podell ER, Zaug AJ, Yang Y, Baciu P, Cech TR, Lei M. 2007. The POT1-TPP1 telomere complex is a telomerase processivity factor. Nature 445:506–510. 10.1038/nature05454. [DOI] [PubMed] [Google Scholar]
- 48.Fan J, Pavletich NP. 2012. Structure and conformational change of a replication protein A heterotrimer bound to ssDNA. Genes Dev. 26:2337–2347. 10.1101/gad.194787.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. 2004. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25:1605–1612. 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]