Abstract
Both chemotaxis and phagocytosis depend upon actin-driven cell protrusions and cell membrane remodeling. While chemoattractant receptors rely upon canonical G-protein signaling to activate downstream effectors, whether such signaling pathways affect phagocytosis is contentious. Here, we report that Gαi nucleotide exchange and signaling helps macrophages coordinate the recognition, capture, and engulfment of zymosan bioparticles. We show that zymosan exposure recruits F-actin, Gαi proteins, and Elmo1 to phagocytic cups and early phagosomes. Zymosan triggered an increase in intracellular Ca2+ that was partially sensitive to Gαi nucleotide exchange inhibition and expression of GTP-bound Gαi recruited Elmo1 to the plasma membrane. Reducing GDP-Gαi nucleotide exchange, decreasing Gαi expression, pharmacologically interrupting Gβγ signaling, or reducing Elmo1 expression all impaired phagocytosis, while favoring the duration that Gαi remained GTP bound promoted it. Our studies demonstrate that targeting heterotrimeric G-protein signaling offers opportunities to enhance or retard macrophage engulfment of phagocytic targets such as zymosan.
INTRODUCTION
Innate cells such as macrophages can sense pathogens from afar by recognizing chemoattractant stimuli, or nearby, by direct physical contact via cell surface receptors which recognize pathogens or opsonized pathogens (1, 2). Although largely studied independently, phagocytosis and chemotaxis share morphological attributes (3). Both processes converge on signaling to the cytoskeleton, require cell membrane remodeling, and depend upon actin-driven cell protrusions. However, chemotaxis and phagocytosis employ distinct classes of cell membrane receptors. Chemotaxis largely depends upon chemoattractant receptors, which are members of the G-protein coupled receptor (GPCR) family and use heterotrimeric G-proteins (Gαβγ) as signal transducers (4, 5). Ligand engagement of chemoattractant receptors typically results in receptor/heterotrimeric G-protein coupling, Gαi subunit GDP-GTP exchange, Gαi dissociation from Gβγ subunits, downstream effector activation, and directed migration. During chemotaxis, Gβγ subunits have a major role in triggering effectors that control actin assembly and cell migration. In Dictyostelium, a direct link between Gβγ and actin polymerization has been established. The Gβγ effector ElmoE targets Dock-like proteins to activate the small GTPase Rac, which regulates the Arp2/3 complex and F-actin formation (6). Arguing for a direct role for GTP-Gαi2 in breast cancer cell migration, a recent study showed that the chemoattractant CXCL12 stimulates the association of GTP-Gαi2 with Elmo1, promoting the translocation of the Elmo1/Dock180 complex to the cell membrane and leading to Rac activation (7).
Phagocytosis employs several different types of receptors not known to use heterotrimeric G-proteins as signal transducers (2). Neutrophils and macrophages employ Fcγ receptors to mediate the uptake of immunoglobulin G opsonized targets. A complex array of signaling molecules downstream of Fcγ receptors have been implicated in the control of actin polymerization during phagocytosis (2). Similar to chemoattractant signaling, proximal in the phagocytic signaling pathways are Rho GTPases such as Rac1, RhoG, and Cdc42. For example, the phagocytosis of antibody-coated erythrocytes requires Cdc42 signaling to Wiskott-Aldrich syndrome protein (WASP) and neural WASP (NWASP), members of a family of actin nucleation-promoting factors (8). Another phagocytic receptor, dectin-1, detects β-glucans in fungal cell walls and helps coordinate immune defenses against a variety of fungi in both mice and humans (9). Although less well studied, the signaling pathways downstream of dectin-1 overlap those employed by Fcγ receptors. Like Fcγ-mediated phagocytosis, the engagement of dectin-1 results in receptor oligomerization, the activation of Src family kinases and Syk, and the triggering of downstream signaling molecules required for target internalization (9). These include phosphatidylinositol 3-kinase and, as mentioned previously, small GTPases, which help coordinate the actin cytoskeleton rearrangements necessary for membrane extensions and particle engulfment. While G-proteins are not known to regulate dectin-1 signaling, their expression levels are partially controlled by signaling through the GPCRs, such as leukotriene B4 receptor 1 (10).
Several previous studies implicated heterotrimeric G-protein signaling in the control of phagocytosis. Dictyostelium mutants that lacked their solitary G-protein β-subunit exhibited impaired phagocytosis due to improper actin cytoskeleton regulation (11). In another study, pertussis toxin (PTX) inhibited the uptake of antibody-coated Staphylococcus aureus by J774 E-clone cells, a murine macrophage cell line (12). More recently, Gβ1 and Gβ2 knockdowns in primary mouse neutrophils reduced both neutrophil phagocytosis and chemotaxis (13). However, reducing the levels of two Gβ subunits by RNA interference in the J774A.1 mouse macrophage cell line inhibited chemotaxis without affecting phagocytosis (14). To help resolve these differences, we visualized the spatial-temporal dynamics of Gαi, Gβγ, and F-actin in live macrophages during zymosan bioparticle (zymosan) triggered phagocytosis. By combining these data with pharmacologic data, the use of macrophages from gene-targeted mice, and cell line knockdown data, we showed that heterotrimeric G-protein signaling helps regulate actin polymerization to optimize macrophage phagocytosis. During the course of this study, we found that noncanonical G-protein signaling can also impact phagocytosis. In noncanonical G-protein signaling, the guanine exchange factor (GEF) activity exerted by the GPCR is replaced or supplemented by the action of intracellular GEFs, such as Ric-8A. Ric-8A has a dual function in cells. It serves as a molecular chaperone required for the initial association of nascent Gα subunits with cellular membranes (15), and it acts on a complex of Gα protein bound to a G-protein regulatory (GPR)/GoLoco motif-containing protein that releases GTP-Gα and the GPR protein (16). Our study indicates that Ric-8A, Gαi, Gαi nucleotide exchange, and Gβγ signaling contribute to macrophage membrane dynamics and phagocytosis.
MATERIALS AND METHODS
Animals.
C57BL/6 and B6.SJL-Ptprca Pepcb/BoyJ mice were obtained from Jackson Laboratory. Gnai2−/−, Gnai3−/−, and Gnai2G184S/G184S mice have been described elsewhere (17–19). All three strains were backcrossed a minimum of 11 times onto the C57BL/6 background. C57BL/6 GFP-Lifeact mice were kindly provided by Roland Wedlich-Soldne. For bone marrow reconstitution, 7-week-old B6.SJL-Ptprca Pepcb/BoyJ (CD45.1) mice were irradiated twice with 550 rads for total of 1,100 rads and received bone marrow from Gnai2−/− C57BL/6 CD45.2 mice or C57BL/6 CD45.2 mice. The engraftment was monitored by sampling the blood 28 days later. The mice were used 6 to 8 weeks after reconstitution. All mice used in this study were 6 to 14 weeks of age. Mice were housed under specific-pathogen-free conditions. All the animal experiments and protocols used in the study were approved by the NIAID Animal Care and Use Committee (ACUC) at the National Institutes of Health.
Cell culture, transfection, and RNA interference.
RAW 264.7 mouse macrophage cells were obtained from the American Type Culture Collection, and the cells were maintained as directed. Mouse bone marrow-derived macrophages (BMDM) were obtained from the indicated mouse lines and purified as previously described (20, 21). RAW 264.7 cells were transiently transfected with constructs expressing yellow fluorescent protein (YFP)-Gαi2, YFP-Gαi2QL, or YFP-Gαi3 (kindly provided by Alfred Gilman). The PTX-resistant YFP-Gαi2 C352I construct was created by site-directed mutagenesis using standard methods. Red fluorescent protein (RFP)-Lifeact and green fluorescent protein (GFP)-Lifeact were kindly provided by Michael Sixt. For CXCR4-YFP, YFP was fused to the C-terminus of human CXCR. Elmo1-GFP (kindly provided by Kodi S. Ravichandran), Ags3-RFP (22), AGS4-GFP and LGN-GFP (kindly provided by Joe Blumer), ½ YFP-Gβ1 and ½ YFP-Gγ2 (kindly provided by Catherine Berlot), Rgs14-GFP (23), and Ric-8A–GFP and Ric-8A–mCherry (24) have been described elsewhere. RAW 264.7 cells were also permanently transfected (via electroporation) with Elmo1-GFP- and EE-tagged Gαi2QL (Missouri S&T cDNA Resource Center). Following transfection, the cells were maintained in neomycin (400 μg/ml) and analyzed 4 weeks later. For transient RIC-8a knockdown, cells were transfected with a dsRed-shRNA control or dsRed-shRNA–RIC-8A with YFP-Gαi2 as specified. For shRNAs, the following hairpin primers for Ric-8A or a control were subcloned into pSIREN-DsRed vector: Ric-8A (5′-GATCCGCTCAAATGCCTGTGTAATCTTCAAGAGAGATTACACAGGCATTTGAGTTTTTTACGCGTG) and control (5′-GATCCGTGGACAGCTTCATCAACTATTCAAGAGATAGTTGATGAAGCTGTCCATTTTTTACGCGTG). The knockdown constructs have been previously described (25). Transfections were performed using FuGene HD transfection reagent (Roche) according to the manufacturer's protocol. Routinely, a total of 1 to 2 μg of plasmid DNA with 2 μl of transfection reagent was added to each well of a 6-well tissue culture plate and incubated for 18 to 24 h before use. For the small intefering RNA (siRNA) knockdowns, RAW 264.7 cells were transfected with the Transit-tko transfection reagent (Mirus) according to the manufacturer's instructions with mouse Elmo1 or control siRNAs. The Elmo1 siRNA pool consisted of three to five target-specific 19- to 25-nucleotide (nt) siRNAs, and the control siRNA pool was composed of scrambled sequences (Santa Cruz Biotechnology).
Western blot assays.
RAW 264.7 cells or BMDM were lysed in 50 mM HEPES (pH 7.4), 250 mM NaCl, 2 mM EDTA, 1% NP-40, and 10% glycerol containing Complete protease inhibitors (Roche) and supplemented with 100 μM orthovanadate sodium. Proteins (20 μg) were resolved on a 4 to 20% Tris-glycine gels. After transfer to nitrocellulose, the blots were probed with the following antibodies: anti-Ric-8A rabbit polyclonal (1:20,000; G. Tall); anti-Gαi2 (1:1,000; Santa Cruz Biotechnology); antiactin-horseradish peroxidase conjugate (1:20,000; Sigma); and anti-Elmo1 (1:1,000; Santa Cruz Biotechnology). The signal was revealed by enhanced chemiluminescence (Amersham Bioscience).
Phagocytosis.
RAW 264.7 cells were stimulated with unlabeled, Alexa Fluor 488- or 594-labled zymosan bioparticles (Invitrogen). These bioparticles are fixed Saccharomyces cerevisiae cells covalently labeled with different fluorophores or not and are referred to as zymosan. The zymosan was added at a 20-μg/ml concentration unless otherwise indicated. The engagement and internalization of the particles were confirmed by differential focusing. For flow cytometry, the cells were allowed to internalize the particles for 10 to 30 min at 37°C, and noninternalized particles were removed by washing four times with cold phosphate-buffered saline (PBS). Adherent BMDM cells were recovered with trypsin-EDTA for 5 min at 37°C. Harvested cells were washed again and fixed in 2% paraformaldehyde to prevent quenching of internalized fluorescent particles. The flow cytometry data were acquired using FACSCanto II (Becton Dickson) and analyzed using FlowJo software (Tree Star). Similar cell populations were analyzed by using an ImageStream instrument (Aminis) equipped with 488-nm, 658-nm, and 405-nm lasers. The images obtained at the highest resolution setting had a pixel size of 0.5 μm, with a 16-μm depth of focus. The digital images of cells in flow were captured at 250 cells/second, and green fluorescence along with bright field images were collected. The acquired data were analyzed with the Aminis image data exploration and analysis software (IDEAS). The existing validated algorithm for measuring internalization was applied to analyze the engulfment of zymosan in macrophages. In some experiments, cells were treated with 200 to 500 ng/ml PTX (Calbiochem) for 2 h, 1 nM Pasteurella multocida toxin (PMT; Sigma) for 2 h, or 20 μM gallein (M119; Tocris Bioscience) for 15 min prior to exposure to the phagocytic targets.
For complement- and Fcγ-mediated phagocytosis, 10% (vol/vol) sheep red blood cells (sRBC; Lonza) were stained with the green fluorescent dye CMFDA (5-chloromethylfluorescein diacetate; Molecular Probes) and opsonized with C3b and inactivated C3b (iC3b) or IgG. To coat the sRBC with C3b/iC3b the cells were incubated with anti-sRBC IgM (1:5,000; Cedarlane) for 1 h at 37°C and then with C5-deficient serum (1:10; Sigma) for 20 min at 37°C. For IgG coating, the sRBC were incubated with rabbit anti-sRBC IgG (1:5,000; Rockland Immunochemical) for 1 h at 37°C. For each study, nonopsonized sRBC were included as a control. For the complement phagocytosis assay, the BMDM were treated with 100 nM phorbol myristate acetate for 20 min before addition of the complement-opsonized sRBC. The opsonized (or nonopsonized) sRBC in RPMI medium were added to the primary macrophages at 10:1 and 5:1 ratios and incubated for 30 min at 37°C. After the indicated time periods, the cell mixtures were washed with cold PBS followed by ACK lysing buffer (0.15 M NH4Cl, 1.0 M KHCO3, and 0.01 M Na2-EDTA; pH 7.2) to remove noninternalized sRBC and then fixed with 4% paraformaldehyde–PBS. Phagocytosis was measured by flow cytometric analysis. Macrophage phagocytosis with opsonized S. aureus bioparticles (Invitrogen) was the same protocol as that used with zymosan.
Immunofluorescence.
RAW 264.7 cells stimulated with zymosan were washed and fixed in 4% (wt/vol) paraformaldehyde for 15 min. Cells were then permeabilized with 0.1% Triton X-100 in PBS for 10 min and blocked with 1% bovine serum albumin–PBS for 15 min. For immunostaining, cells were incubated for 4 h at room temperature or overnight at 4°C with antibodies diluted in 1% bovine serum albumin–PBS. Primary antibodies used in these studies included the following: rabbit anti-Gαi3 (1:300; Calbiochem), mouse monoclonal anti-Gαi2 (1:300; Santa Cruz Biotechnology), and mouse monoclonal anti-active αi (1:100; NewEast Biosciences). Secondary antibodies were species-specific antibodies conjugated with Alexa Fluor 488, 568, or 647 (Invitrogen). F-actin was stained using Alexa Fluor 568 or 647 conjugated to phalloidin (Invitrogen). For the F-actin depolymerization experiment, RAW 264.7 cells were treated with 2 nM latrunculin B (Enzo Life Sciences) for 15 min prior to imaging and washout. For imaging Fcγ-dependent phagocytosis, sRBC were incubated with rabbit anti-sRBC and, following cell fixation, the sRBC were detected by immunostaining with Alexa Fluor 568–anti-rabbit Ig.
Cell imaging and data analysis.
For microscopic imaging of phagocytosis, macrophages were plated on collagen-coated glass-bottom dishes (MatTek). All fluorescent images were collected on inverted confocal microscopes. A Leica TCS-SP5-X confocal microscope equipped with an argon and white light laser (Leica Microsystems) and a 100× objective or a 63× oil immersion objective (numerical aperture, 1.4) was used for imaging. The samples were excited with 488- or 561-nm laser lines. For live cell studies, CO2 and temperature were maintained at 5% and 37°C, respectively, using an environmental control system (PeCon GmbH). The dynamics of phagocytosis were imaged as x,y,z stacks over time (four dimensions) with z step sizes ranging from 0.3 to 1 μm. Image analysis was performed using Imaris 7.3.0 (Bitplane AG), ImageJ, and Adobe Photoshop CS3 (Adobe Systems).
Intracellular calcium.
BMDM were seeded at 70,000 cells per 100 μl loading medium (RPMI 1640, 10% fetal bovine serum) into poly-d-lysine-coated 96-well plates (Nalgene Nunc) the day prior to assay. An equal volume of loading buffer (FLIPR Calcium 3 assay kit; Molecular Devices) was added the following day. Cells were incubated for 1 h at 37°C before adding zymosan, and then the calcium flux peak was measured using a FlexStation 3 (Molecular Devices). Data are shown as fluorescent counts, and the y axes show Lm1. In some instances, the cells were treated with PTX at 500 ng/ml for 4 h prior to the assay.
Statistics.
All experiments were performed a minimum of three time. For those experiments where individual cells were counted, a minimum of 100 cells per condition were enumerated. Data are shown as means ± standard errors of the means (SEM). Experimental results were compared by 2-way analysis of variance (ANOVA) or t test. The distribution of phagocytic cells with various numbers of bioparticles were compared by chi square test. All statistical tests were performed using Prism software (GraphPad Software).
RESULTS
Gαi is enriched at cell protrusions, phagocytic sites, and on early phagosomes.
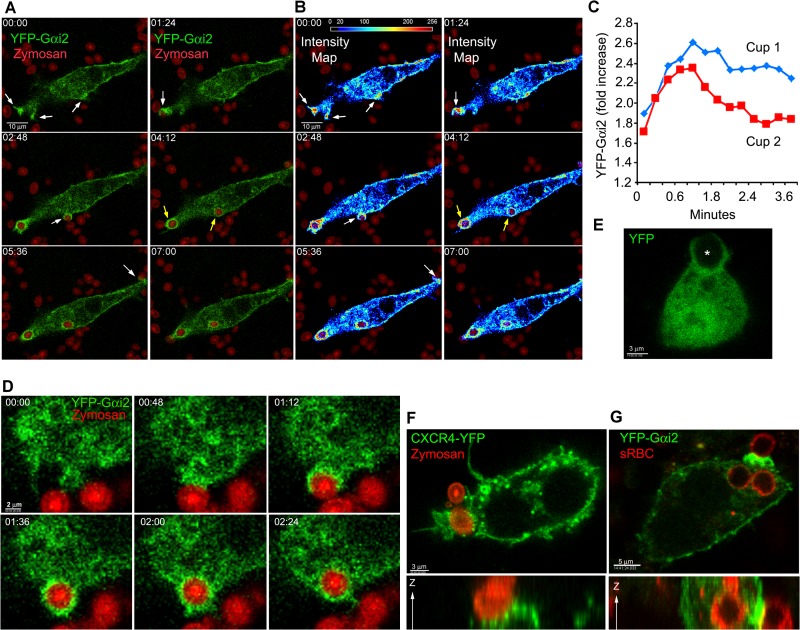
For these studies, we first examined the uptake of fluorescent zymosan by the murine macrophage cell line RAW 264.7. To test the localization of Gαi during phagocytosis, we expressed YFP-Gαi2 proteins (26) in these cells and added fluorescently labeled zymosan. Prior to zymosan capture, cell membrane microspikes and ruffles probed the cell's environment, many of which had enriched expression of YFP-Gαi2. Zymosan capture occurred at these cell protrusion sites (Fig. 1A; see also Video S1 in the supplemental material). Rapidly after the cells engaged zymosan, we observed further recruitment of Gαi proteins to the phagocytic site. The enrichment coincided spatially and temporally with phagocytic cup formation and occurred 1 to 2 min after zymosan engagement. It persisted as the phagocytic cups expanded to engulf the zymosan particle, and YFP-Gαi2 remained associated with the nascent phagosomes. The substitution of YFP-Gαi3 for YFP-Gαi2 led to similar results (N.-N. Huang, unpublished results). An intensity map for YFP-Gαi2 recruitment to phagocytic sites is shown in Fig. 1B. Focusing on the initial contact and engulfment of zymosan revealed an approximately 2-fold increase in YFP-Gαi2 expression at phagocytic cups relative to the average plasma membrane expression level (Fig. 1C). A closer examination showed the enrichment of Gαi2 on the initial and expanding phagocytic cup. Following engulfment, but prior to protrusion of the particle into the cytosol, the level of YFP-Gαi2 declined at the base of the phagosome (Fig. 1D). An examination of the localization of YFP revealed no enrichment at cell protrusion sites or on the phagosome (Fig. 1E). To compare the behavior of a plasma membrane protein, we examined the localization of the chemokine receptor CXCR4 fused to YFP. CXCR4-YFP localized at the plasma membrane and in intracellular vesicle-like structures. Some of the vesicle-like structures accumulated near the developing phagosome. CXCR4-YFP nominally accumulated on the phagocytic cup and the nascent phagosome, less than what was observed with Gαi2-YFP (Fig. 1F and data not shown). To determine whether Gαi2 accumulated at the phagocytic site during Fcγ receptor-mediated phagocytosis, we incubated RAW 264.7 cells expressing YFP-Gαi2 with sRBC previously exposed to sRBC-specific antibody. Again, we found substantial enrichment of Gαi2 on the phagocytic cup following the engagement of the target particle (Fig. 1G). These initial findings showed enhanced expression of Gαi proteins at cell protrusions and at phagocytic sites following exposure to zymosan or antibody-coated sRBC.
FIG 1.
Gαi proteins are recruited to phagocytic cups and phagosomes. (A and B) Time-lapse confocal microscopy images of RAW 264.7 macrophages expressing YFP-Gαi2 proteins (green) and exposed to fluorescent zymosan (red). White arrows indicate Gαi proteins at cell protrusions near zymosan and sites of Gαi2 proteins accumulated at phagocytic cups; yellow arrows indicate internalized phagosomes coated by Gαi proteins. Numbers represent elapsed time (in minutes) after zymosan had settled onto cells. Bar, 10 μm. Panel B shows an intensity map of Gαi2 localization during phagocytosis. The Gαi2 signal is shown as a pseudocolor intensity map, where yellow and red indicate 1.5- to 3-fold intensity increases over light blue regions. Images began at initial zymosan contact. (C) Time course of YFP-Gαi2 recruitment to phagocytic cups and nascent phagosomes over 3 min. Intensity is shown relative to average plasma membrane expression level. (D) Time-lapse confocal microscopy of RAW 264.7 macrophages expressing YFP-Gαi2 (green) and exposed to zymosan (red). Images were acquired every 24 s. Numbers are the elapsed time following initial contact with zymosan. Bar, 2 μm. (E) Confocal microscopy of RAW 264.7 cells expressing YFP and exposed to zymosan. Locations of zymosan particles are indicated with an asterisk. (F) Confocal microscopy of RAW 264.7 cells expressing CXCR4-YFP and exposed to fluorescent zymosan. Following fixation, the cells were imaged. Both x-y and z-x projections are shown. Bar, 3 μm. (G) Confocal microscopy of RAW 264.7 cells expressing YFP-Gαi2 and exposed to sRBC previously incubated with a rabbit anti-sRBC antibody. Following fixation, the cells were immunostained for rabbit Ig (red) to outline the sRBC and imaged. Both x-y and z-y projections are shown.
Gαi and F-actin are coordinately expressed during phagocytosis.
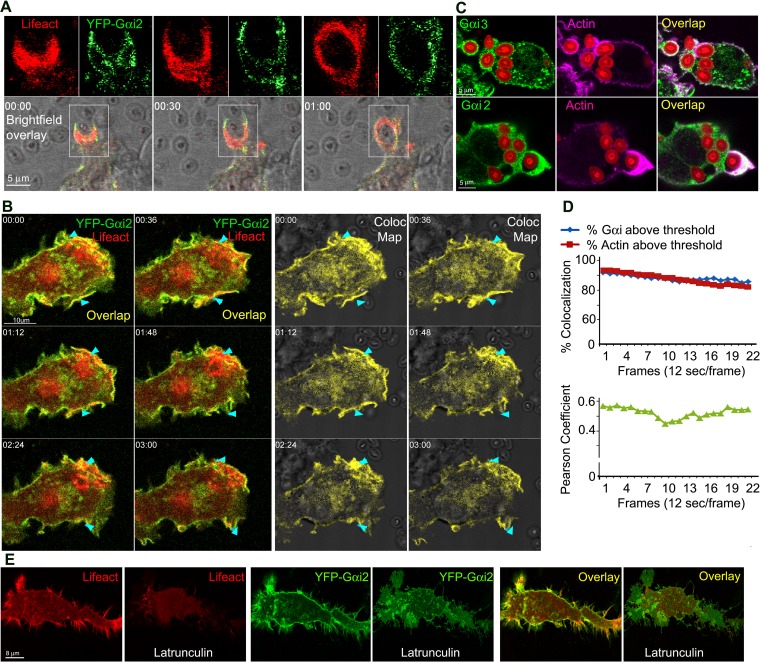
To delineate the relationship between actin polymerization and Gαi protein localization during phagocytosis, we imaged YFP-Gαi2 and RFP-Lifeact, a marker for F-actin (27), using RAW 264.7 cells exposed to unlabeled zymosan. YFP-Gαi2 proteins dynamically coordinated with RFP-Lifeact in cell membrane protrusions, ruffles, and during the initial stages of phagocytosis (Fig. 2A). Occasionally both proteins condensed into a filamentous-like structure surrounding the phagocytic cup prior to particle engulfment (Fig. 2B; see also Video S2 in the supplemental material). Next, we verified that the distribution of Gαi2-YFP faithfully represented endogenous Gαi proteins by immunostaining mouse BMDM for Gαi proteins and with Alexa Fluor 647-phalloidin after exposure of the cells to fluorescently labeled zymosan. Endogenous Gαi proteins were enriched at cell protrusions and at the phagocytic cup and partially colocalized with F-actin at those sites (Fig. 2C). Using RAW 264.7 cells to examine the spatial-temporal distribution of YFP-Gαi2 and RFP-Lifeact during phagocytosis, we found a highly significant level of Gαi2 and F-actin colocalization (Fig. 2D). These results demonstrated a close spatial-temporal distribution of Gαi and F-actin at cell protrusion sites and during the early steps in the phagocytosis of zymosan.
FIG 2.
Gαi proteins colocalize with F-actin at cell protrusions and at phagocytic cups. (A) Time-lapse confocal microscopy of RAW 264.7 cells expressing YFP-Gαi2 (green) and RFP-Lifeact (red) and exposed to zymosan focusing on a phagocytic cup. Numbers represent elapsed time (in minutes). For each time point, a bright-field overlay image and two fluorescent images electronically zoomed 2× are shown. Bar, 5 μm. (B) Time-lapse confocal microscopy images of RAW 264.7 cells expressing YFP-Gαi2 (green) and RFP-Lifeact (red) and exposed to zymosan. Arrowheads indicate Gαi together with actin condensed at membrane protrusions and ruffles targeting and engulfing zymosan. An overlap and corresponding colocalization map is shown. Imaging began 5 min after zymosan addition. Bar, 10 μm. (C) Confocal microscopy of BMDM exposed to fluorescent zymosan and immunostained for Gαi2, Gαi3, and F-actin (phalloidin). Bar, 5 μm. (D) Quantification of YFP-Gαi2 and RFP-Lifeact colocalization over time. The percent colocalization of RFP-Lifeact and YFP-Gαi2 above threshold (above) and Pearson's colocalization coefficient (below) are shown. Images acquired at 12 s/frame. (E) Confocal microscopy of RAW 264.7 cells expressing RFP-Lifeact (red) and YFP-Gαi2 (green) before or after a 15-min exposure to latrunculin B. Individual and overlap images are shown. Bar, 8 μm.
To determine whether the enrichment of Gαi at cell protrusions depended on the presence of F-actin, we treated cells with latrunculin B (28), an agent that interferes with actin polymerization, and examined how the disruption of F-actin assembly affected Gαi localization. To limit extreme changes in shape and detachment of the cells from the substratum, short latrunculin B treatments were applied to yield incomplete F-actin depolymerization. The treated cells rapidly underwent a visible redistribution of Gαi proteins as the actin depolymerized (Fig. 2E). Cell protrusions that contacted phagocytic targets rapidly disappeared. Next, we washed out the latrunculin B and observed how F-actin and YFP-Gαi2 reappeared in the cells. Over time, we found F-actin at new cell protrusions and filopodia. The reappearance of F-actin was often accompanied by an increasing enrichment of Gαi2 at the same sites (see Video S3 in the supplemental material). These results are supported by 3D image reconstructions and colocalization measurements (data not shown), and they implicate Gαi proteins in filopodium formation and the recognition of zymosan, and they support the previously observed spatial and temporal coordination between F-actin and Gαi localization in cell protrusions and during phagocytosis.
Inhibiting Gαi nucleotide exchange reduces membrane dynamics, phagocytosis, F-actin levels, and zymosan-induced increases in intracellular Ca2+.
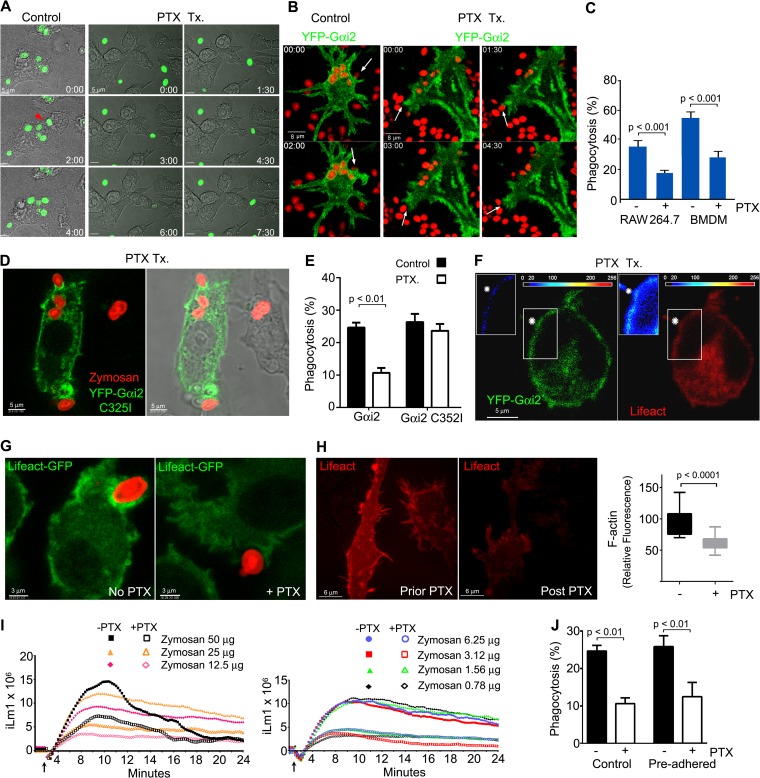
GPCR-mediated Gαi nucleotide exchange and chemotaxis can be blocked by exposing cells to PTX, which catalyzes the ADP-ribosylation of Gαi proteins, inhibiting the release of GDP and the Gβγ dimer. To ascertain whether PTX affected zymosan-triggered phagocytosis, we treated RAW 264.7 cells and then exposed the cells to labeled zymosan. We found that the PTX treatment reduced membrane ruffling and the number of cells that engulfed zymosan (Fig. 3A). To visualize the impact of PTX on Gαi2 dynamics during phagocytosis, we treated RAW 264.7 cells expressing YFP-Gαi2 and exposed them to fluorescent zymosan. PTX treatment hampered cell protrusions, lamellipodium formation, and zymosan capture (Fig. 3B and C; see also Video S4 in the supplemental material). To determine whether we could rescue the phagocytic defect in the PTX-treated RAW 264.7 cells, we expressed YFP-Gαi2 C352I, treated the cells with PTX, and exposed the cells to fluorescent zymosan. The C352I mutation renders Gαi2 insensitive to PTX-mediated ADP-ribosylation and can rescue signaling in PTX-treated cells (29). We found that the PTX-resistant YFP-Gαi2 construct accumulated at phagocytic sites in PTX-treated cells, in contradistinction to the wild-type Gαi2 (Fig. 3D). In addition, the construct nearly completely rescued the phagocytic defect in the PTX-treated RAW 264.7 cells (Fig. 3E).
FIG 3.
PTX treatment affects membrane dynamics, reduces phagocytosis, and decreases zymosan-triggered intracellular Ca2+ increases. (A) Time-lapse microscopy (differential interference contrast optics) of RAW 264.7 cells untreated (left) or treated with PTX (right) and exposed to zymosan (green). The cells were treated for 2 h with PTX (200 ng/ml) prior the addition of zymosan. Bar, 5 μm. (B) Time-lapse confocal microscopy of YFP-Gαi2-expressing RAW 264.7 cells, untreated (left) or treated with PTX (right) following exposure to zymosan (red). Arrows indicate phagocytic sites. Bar, 8 μm. (C) Effects of PTX treatment on macrophage phagocytosis. RAW 264.7 or BMDM, PTX treated or not, were exposed to Alexa Fluor 594-labeled zymosan, and zymosan capture was quantitated by flow cytometry. Data were compared by using a paired t test (right panel). (D) Confocal and bright-field microscopy of RAW 264.7 cell expressing YFP-Gαi2 C235I, treated with PTX for 2 h, and exposed to fluorescent zymosan. A representative cell shows enrichment of YFP-Gαi2 C235I at phagocytic sites despite PTX pretreatment. (E) Effect of PTX on zymosan phagocytosis by RAW 264.7 cells expressing either YFP-Gαi2 or YFP-Gαi2 C325I. Statistical analysis of the data was performed with a t test. (F) Confocal microscope images of a PTX-treated RAW 264.7 cell expressing YFP-Gαi2 (green) and RFP-Lifeact (red) and exposed to zymosan. The insets show intensity maps of YFP-Gαi2 or RFP-Lifeact, as indicated. The zymosan engagement site is shown by an asterisk. Bar, 5 μm. (G) Confocal microscope images of BMDM from Lifeact mice treated with PTX or not and exposed to fluorescent zymosan. Bar, 3 μm. (H) Confocal microscopy of RAW 264.7 cells expressing RFP-Lifeact prior to and after a 3-h PTX exposure. The same microscope field is shown from before and 3 h after the addition of PTX (400 ng/ml). Bar, 8 μm. Immunofluorescence levels measured before and after PTX treatment. Statistics were performed with the t test. (I) Intracellular Ca2+ responses of BMDM to zymosan. Cells were pretreated with PTX (open symbols) or not (closed symbols). The arrow is at the time of zymosan addition. (J) Phagocytosis assay using RAW 264.7 cells expressing YFP-Gαi2 treated with PTX, or not, and allowed to preadhere to fluorescent zymosan, or not. To adhere the bioparticles, RAW 264.7 cells were cooled to 4°C (30 min), after which zymosan was added. Thirty minutes later, the cells were warmed to 37°C for 20 min to allow phagocytosis to proceed before fixation. Data were analyzed with the t test.
To examine the impact of PTX on actin dynamics, we imaged RAW 264.7 cells expressing YFP-Gαi2 and RFP-Lifeact following exposure of the cells to unlabeled zymosan. PTX treatment reduced the recruitment of F-actin to cell protrusions and phagocytic sites (Fig. 3F). Imaging BMDM prepared from Lifeact mice that had been exposed or not to PTX gave similar results as we observed with RAW 264.7 cells. PTX treatment reduced the number of phagocytic events and decreased F-actin at phagocytic sites (Fig. 3G). To examine the effect of PTX on F-actin levels at sites of cell protrusion, we imaged the same cells prior to and after a 3-h PTX treatment. We found a 30 to 50% reduction of F-actin at cell adhesion and protrusion sites (Fig. 3H). Next, as heterotrimeric G-protein signaling often leads to increases in intracellular Ca2+, we tested whether zymosan elicited an intracellular Ca2+ response and if so whether it was sensitive to PTX. Previously, zymosan had been shown to elicit an increase in intracellular Ca2+ in human neutrophils (30). We found that zymosan elicited a strong rise in intracellular Ca2+ in BMDM that was partially sensitive to PTX treatment. At the highest zymosan concentrations, PTX reduced the rise in intracellular Ca2+ by 50%, and at the lower concentrations the intracellular Ca2+ rise was reduced by 70% (Fig. 3I). To test whether the reduced phagocytosis noted in the PTX-treated cells arose from impaired localization of the phagocytic targets (i.e., defective chemotaxis), we exposed RAW 264.7 cells that had been pretreated with PTX, or not, to fluorescent zymosan at 4°C to allow the cells to preadhere prior to raising the temperature to 37°C, to allow phagocytosis to proceed. Arguing that target localization was not impacted by the PTX treatment in these experiments, we found that the PTX pretreatment remained effective in inhibiting phagocytosis even when the target cells directly contacted the zymosan beads (Fig. 3J). These data indicated that Gαi nucleotide exchange is associated with the increased levels of F-actin observed during the early stages of phagocytosis, and it helps maintains the constitutive levels of F-actin found in macrophages.
GTP-Gαi2 enhances phagocytosis and F-actin formation.
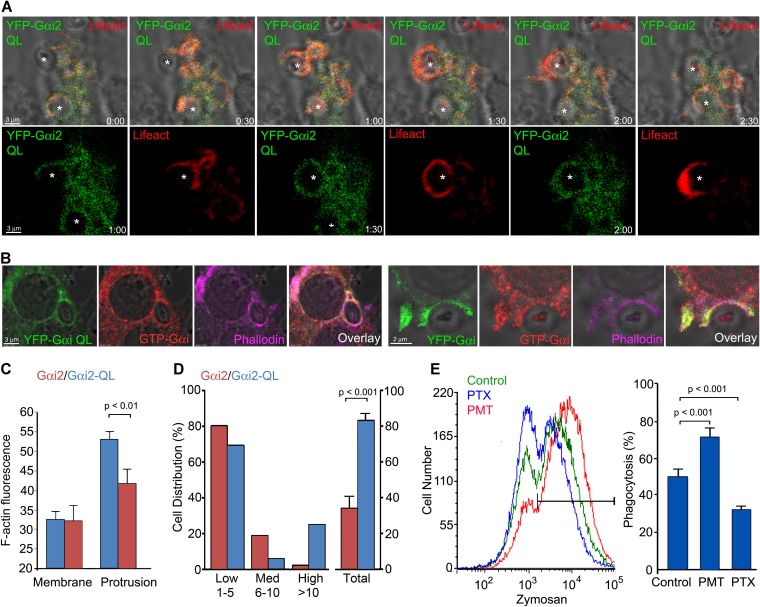
Next, we tested whether colocalization of Gαi with F-actin depended upon its nucleotide binding status, which in turn controls its association with guanine dissociation inhibitors (GDIs), such as Gβγ subunits. We expressed in RAW 264.7 cells YFP-Gαi2Q205L (QL), a constitutively active Gαi2 protein that lacks GTPase activity (31). Hence, it is largely GTP bound and unassociated with a GDI. We found that YFP-Gαi2 QL localized somewhat differently than the wild-type (WT) protein, as less YFP-Gαi2QL localized at the plasma membrane. Although present at phagocytic sites, it was not as enriched as the WT protein (Fig. 4A; see also Video S5 in the supplemental material). To verify that GTP-Gαi was present at phagocytic sites, we made use of an antibody that recognizes Gαi only when it is GTP bound. First, we transfected RAW 264.7 cells with YFP-Gαi2 QL, and then we exposed the cells to zymosan and immunostained for GTP-Gαi and used Alexa Fluor 647-phalloidin to detect F-actin. As expected, we observed good colocalization between YFP-Gαi2 QL, the immunostained GTP-Gαi, and F-actin on an early phagosome (Fig. 4B, left panels). Next, we analyzed RAW 264.7 cells expressing the wild-type YFP-Gαi2. Again, we detected an overlap between YFP-Gαi2, GTP-bound Gαi, and F-actin at cell protrusions and at the phagocytic cup (Fig. 4B, right panels).
FIG 4.
Localization of GTP-Gαi2 at phagocytic cups and iamges showing that increasing the level of GTP-Gαi2 in macrophages enhances zymosan phagocytosis. (A) Time-lapse confocal microscopy of RAW 264.7 cells expressing YFP-Gαi2 QL (green) and RFP-Lifeact (red) exposed to zymosan. Sequential images, including a bright-field overlay, are shown. Individual images corresponding to 3 time points are shown below. Zymosan is indicated by asterisks. Bar, 3 μm. (B) Confocal microcopy images of RAW 264.7 cells expressing YFP-Gαi2QL or YFP-Gαi2 immunostained for GTP-Gαi and F-actin delineated by phalloidin-Alexa Fluor 647 staining. Individual and overlay images are shown. Bars, 3 μm (left); 2 μm (right). (C) F-actin levels at cell protrusions in RFP-Lifeact-expressing RAW 264.7 cells cotransfected with YFP-Gαi2 QL or YFP-Gαi2. Shown is a ratio between RFP-Lifeact expression at cell protrusions and that at unperturbed plasma membrane. Data were analyzed by using the t test. (D) Phagocytosis of zymosan by YFP-positive RAW 264.7 cells expressing either YFP-Gαi2QL or YFP-Gαi2. The percentages of YFP-positive cells that had engulfed either 1 to 5, 6 to 10, or more than 10 zymosan particles were enumerated (first 6 columns). The last 2 column are the percentage of YFP-positive cells that engulfed zymosan. Data were analyzed by paired t test. (E) Flow cytometry of RAW 264.7 cells either untreated or treated with PTX or PMT prior to exposure to Alexa Fluor 594-labeled zymosan for 30 min. PTX and PMT treatment was for 2 h prior to zymosan addition. The histogram shows untreated RAW 264.7 macrophages (green) and PTX-treated (blue) or PMT-treated (red) cells. Data were analyzed with the t test.
Despite the failure of YFP-Gαi2 QL to accumulate at phagocytic sites to the same extent as the wild-type protein, F-actin levels at cell protrusion sites in those cells expressing the mutant protein were high (Fig. 4C), and phagocytic engulfment of zymosan occurred more rapidly than in the cells expressing the WT protein (∼45 s versus ∼80 s). Comparison of the uptake of fluorescent zymosan by YFP-positive RAW 264.7 cells expressing YFP-Gαi2 or YFP-Gαi2 QL revealed that the constitutively active Gαi protein increased the number of particles engulfed per cell and enhanced the percentage of RAW 264.7 cells that captured particles (Fig. 4D).
Another way to activate Gαi as well as Gαq and Gα13 is to treat cells with a toxin made by the bacterium Pasteurella multocida. This toxin (PMT) deamidates a conserved glutamine residue in the α-subunit of heterotrimeric G proteins that is required for GTP hydrolysis, thereby locking Gα in a GTP-bound state (32). To assess the effect of PMT on macrophage phagocytosis, we treated RAW 264.7 cells with PMT for 2 h prior to incubating the cells with fluorescent zymosan. The PMT pretreatment resulted in a 40% increase in the number of macrophages that engulfed zymosan over a 30-min span, while a similar-duration pretreatment with PTX caused an approximately 40% reduction compared to untreated cells (Fig. 4E). These data indicate that the increased levels of GTP-bound Gαi2 elevate F-actin levels at cell protrusions and facilitate zymosan phagocytosis.
Inhibiting Gβγ signaling impairs zymosan phagocytosis.
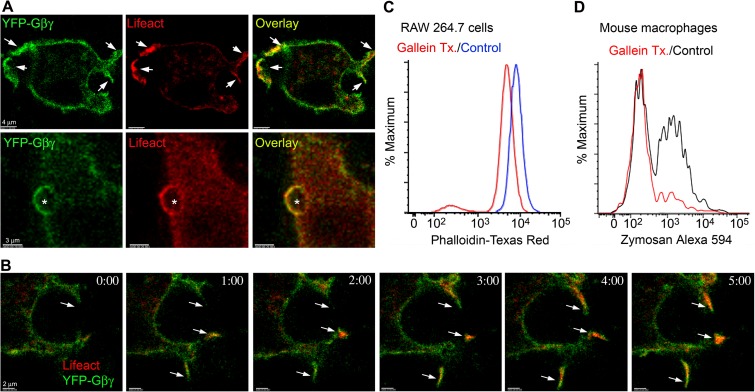
The different localization patterns of the WT and the GTPase-deficient Gαi prompted us to examine the localization of Gβγ subunits at phagocytic sites. To do so, we expressed an amino-terminal fragment of YFP fused to Gβ1 and a carboxyl-terminal YFP fragment fused to Gγ2 in RAW 264.7 cells. When associated, the proteins create a functional YFP-tagged Gβ1γ2 dimer (33). Much like YFP-Gαi, YFP-Gβ1γ2 expression was enriched at the plasma membrane, more so at F-actin-rich cell protrusions, and at phagocytic cups (Fig. 5A). In addition, we often observed a coordinated enrichment of RFP-Lifeact at cell protrusion sites (Fig. 5B). To determine if Gβγ signaling contributed to F-actin levels, we treated RAW 264.7 cells with gallein, a known pharmacologic inhibitor of the binding of Gβγ to effectors such as phospholipase Cβ2/3, pREX guanine nucleotide exchange factor, G-protein receptor kinase 2, and phosphoinositide 3-kinase γ (34). Overnight treatment reduced constitutive F-actin levels in RAW 264.7 cells, as assessed by phalloidin staining (Fig. 5C). Next, we checked whether acute inhibition of Gβγ signaling affected phagocytosis. We incubated BMDM with gallein for 15 min prior to adding fluorescent zymosan. Fifteen minutes later, we assessed the percentage of phagocytic cells by flow cytometry. We found that the gallein preincubation reduced BMDM phagocytosis of zymosan by approximately 60% (Fig. 5D). These experiments showed that Gβγ accumulates at cell protrusions and at phagocytic sites, as does Gαi, and that Gβγ signaling promotes F-actin formation and facilitates zymosan phagocytosis.
FIG 5.
Involvement of Gβγ subunits in macrophage phagocytosis. (A) Confocal microscopy images of RAW 264.7 cells expressing RFP-Lifeact (red), ½ YFP-Gβ1, and ½ YFP-Gγ2 exposed (bottom) or not (top) to zymosan. Sites of strong overlap are indicated with arrows (top). Bar, 4 μm. Location of zymosan is indicated with an asterisk (bottom). Bar, 3 μm. (B) Time-lapse confocal microscopy of RAW 264.7 cells expressing RFP-Lifeact (red), ½ YFP-Gβ1, and ½ YFP-Gγ2. Arrows show cell protrusions where YFP-Gβ1γ2 and RFP-Lifeact eventually colocalized. Bar, 2 μm. (C) Flow cytometry of RAW 264.7 cells treated with gallein, or not, and stained for phalloidin. RAW 264.7 cells were treated overnight with 10 μM gallein prior to phalloidin staining and flow cytometry. (D) Flow cytometry of mouse BMDM treated with gallein (20 μM, 15 min) or not and then exposed to fluorescent zymosan (10 μg) for an additional 30 min.
Genetic evidence that mouse Gαi proteins regulate phagocytosis.
Macrophages express two Gαi isoforms, Gαi2 and Gαi3 (http://www.immgen.org/databrowser/index.html). We first tested the impact of the loss of Gαi3 by comparing BMDM prepared from WT or Gnai3−/− mice (17). We noted a reduction in the number of phagocytic macrophages and a decrease in the number of particles engulfed per cell (Fig. 6A; see also Video S6 in the supplemental material). We quantitated these results by standard flow cytometry and found in three separate experiments an approximately 25% reduction in the number of phagocytic cells and greater than 50% reduction in the number of particles engulfed per cell (data not shown). A similar analysis was performed using ImageStream cytometry, which combines flow cytometry with high-resolution microscopy. Initially, we tested our ability to discriminate different numbers of phagocytized beads per cell by using WT macrophages exposed to fluorescent zymosan (Fig. 6B). We found that we could readily distinguish cells that had engulfed <2 particles, 2 to 4 particles, or greater than 4 particles. We then compared WT and Gnai3−/− macrophages exposed to fluorescent zymosan. This revealed a reduction in the number of phagocytic Gnai3−/− macrophages similar to what we had noted with standard flow cytometry but also a sharp reduction in the number of Gnai3−/− macrophages that engulfed multiple particles (Fig. 6C). Next, we evaluated BMDM prepared from WT mice reconstituted with WT or Gnai2−/− bone marrow cells. We used bone marrow-reconstituted mice because of the difficulty in obtaining Gnai2−/− mice. They must be derived from the mating of heterozygotes and there is considerable perinatal and postnatal loss of the mice with the desired genotype. In addition, bone marrow reconstitution avoids the impact of the loss of Gαi2 in nonhematopoietic cell types. When tested, the Gnai2−/− BMDM also had a reduced uptake of zymosan (Fig. 6D).
FIG 6.
Altered phagocytosis in BMDM prepared from Gnai3−/−, Gnai2−/−, and Gnai2G184S/G18S mice. (A) Confocal microscopy of WT and Gnai3−/− BMDM exposed to fluorescent zymosan (green) for 15 min prior to fixation and staining with phalloidin (red). Bar, 20 μm. (B) ImageStream analysis of WT macrophages exposed to fluorescent zymosan. Parameters were set to segregate WT macrophages, based on the number of fluorescent beads that they acquired. Representative images are shown. (C) ImageStream analysis of WT and Gnai3−/− BMDM exposed to fluorescent zymosan. The percent phagocytosis is shown on the right. The percentage of macrophages that captured 2 or less, 3 to 4, or 5 or more particles are shown in the pie charts. Data were analyzed using the chi-square test. (D) Flow cytometry results, comparing phagocytosis of WT and Gnai2−/− BMDM exposed to various concentrations of fluorescent zymosan for 20 min. Data were analyzed by using a paired t test. (E) Flow cytometry results comparing WT and Gnai2G184S/G184S BMDM exposed to fluorescent zymosan for 20 min. Data were analyzed by using a 2-way ANOVA. (F) Flow cytometry results from the analysis of WT, Gnai2−/−, and Gnai3−/− BMDM. Isotype controls and dectin-1 expression levels are shown. (G) Flow cytometry results from the analysis of WT, PTX-treated WT, Gnai2−/−, and Gnai3−/− BMDM exposed to fluorescent S. aureus bioparticles, anti-sRBC-coated sRBC, or complement-coated sRBC. Data were analyzed by using a 2-way ANOVA.
A genetic gain-of-function approach can also be used to assess the role of Gαi signaling in a signal transduction pathway. The introduction of a serine in Gαi2 at glycine 184 (G184S) blocks the interaction of Regulator of G-protein signaling (RGS) proteins from binding Gαi2 and accelerating the intrinsic GTPase activity of Gαi2 (18). Normally, RGS proteins limit the duration that Gαi remains GTP bound, thereby decreasing Gαi- and Gβγ-mediated effector activation. Knock-in of such a mutation into mice causes neutrophil bone marrow retention, poor recruitment of neutrophils to inflammatory sites, and abnormal chemoattractant receptor signaling (20). We purified BMDM from wild-type and Gnai2G184S/G184S bone marrow-reconstituted mice and assessed their ability to engulf fluorescent zymosan beads by using flow cytometry. We tried several concentrations of zymosan, and in each instance the Gnai2G184S/G184S bone marrow-reconstituted mice BMDM engulfed more zymosan than those prepared from wild-type bone marrow-reconstituted mice (Fig. 6E).
Since dectin-1 expression is sensitive to Gαi signaling (10), we checked the levels of dectin-1 expression on wild-type, Gnai2−/−, and Gnai3−/− macrophages. We found that while all of them expressed high levels of dectin-1, the Gnai2−/− macrophages had an approximately 30% reduction compared to wild-type cells (Fig. 6F). In contrast, the levels of F4/80, CD11b, and CD11c were similar on the macrophages from the different mice. Furthermore, a short-term PTX treatment did not affect dectin-1 expression (data not shown). Although modest, the reduction in dectin-1 noted with the Gnai2−/− macrophages may contribute to the phagocytic defect in these cells; however, a reduction of dectin-1 cannot account for the decreased phagocytic activity of the Gnai3−/− macrophages.
To test the impact of inhibiting Gαi2/3 nucleotide exchange on the phagocytosis of other targets by primary macrophage, we used Staphylococcus aureus bioparticles, sRBC coated with antibody, or sRBC coated with complement. We found that the PTX treatment did not affect the phagocytosis of the S. aureus bioparticles, but it reduced both Fcγ- and complement-mediated phagocytosis of sRBC (approximately 30 and 50%, respectively). The Gnai2−/− macrophages were similarly impaired, although the Gnai3−/− macrophage showed no significant defect compared to wild-type cells (Fig. 6G). Together these data indicate that while not essential for macrophage phagocytosis, Gαi signaling has a significant regulatory role.
Recruitment of Elmo1 to the phagocytic cup depends upon Gαi nucleotide exchange.
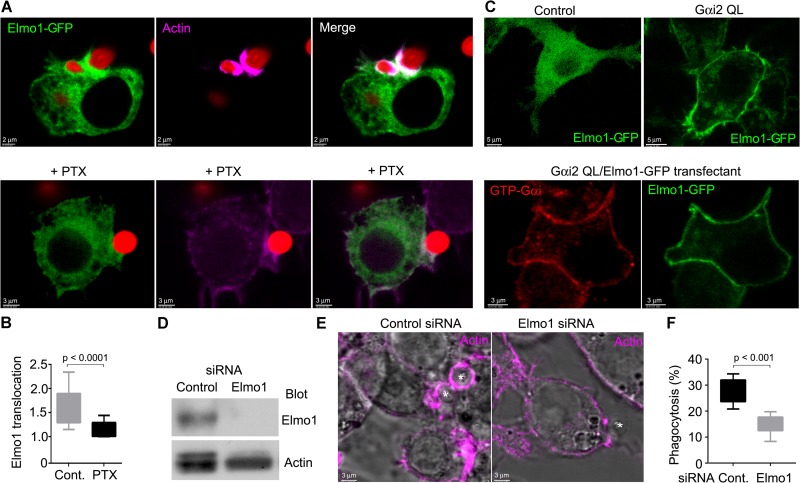
Our observation that Gαi signaling contributed to phagocytosis and the recent observation that CXCL12 stimulation promoted an interaction between Gαi2 and ELMO1 in a human breast cancer cell line prompted us to determine whether Elmo1 was recruited to phagocytic cups. To do so, we introduced Elmo1-GFP into RAW 264.7 cells and exposed the cells to zymosan. In the absence of zymosan, Elmo1-GFP localized predominately in the cytosol with a low level of expression at the plasma membrane; however, zymosan exposure recruited Elmo1-GFP to the phagocytic cup (Fig. 7A and B), while treatment of the cells with pertussis inhibited the recruitment (Fig. 7A and B). Consistent with a role for Gαi signaling in the plasma membrane localization of Elmo1, coexpression of the constitutively active form of Gαi2 shifted much of Elmo1-GFP from the cytosol to the plasma membrane (Fig. 7C). Initial transient coexpression of Elmo1-GFP and a nonlabeled Gαi2QL shifted some Elmo1-GFP to the membrane, but much remained in the cytosol (data not shown). To reduce the impact of transient overexpression and to allow better membrane targeting of the mutant Gαi2 protein, we selected cell lines that coexpressed Gαi2QL and Elmo1-GFP or a control vector with the Elmo1 construct by using RAW264.7 cells. Four weeks later, we examined the intracellular location of Elmo1-GFP. We found in those cells expressing Elmo1-GFP and Gαi2QL that the majority of Elmo1-GFP localized to the plasma membrane. We verified the expression of Gαi2QL by immunostaining the cells with an antibody that recognizes GTP-Gαi (Fig. 7C). To assess the role of Elmo1 in the accumulation of F-actin at phagocytic cups and on phagocytosis, we reduced its expression in RAW 264.7 cells by RNA interference. Immunoblotting cell lysates from control or Elmo1-siRNA-treated cells revealed the successful lowering of Elmo1 expression (Fig. 7D). Less F-actin accumulated at phagocytic sites, and the reduction in Elmo1 expression decreased the phagocytosis of zymosan beads (Fig. 7E and F). These data show that the recruitment of Elmo1 to phagocytic sites helps promote zymosan phagocytosis.
FIG 7.
Gαi signaling recruits Elmo1 to the phagocytic cup to enhance zymosan phagocytosis in RAW 264.7 cells. (A, top) Confocal microscopy of RAW 264.7 cells expressing Elmo1-GFP and exposed to fluorescent zymosan and stained with Alexa Fluor 647-phalloidin. Bar, 2 μm. (Bottom) Confocal microscopy of PTX-treated RAW 264.7 cells expressing Elmo1-GFP and exposed to fluorescent zymosan and stained with Alexa Fluor 647-phalloidin. Bar, 3 μm. (B) Quantification of Elmo1 enrichment at phagocytic cups (relative to plasma membrane levels) in either nontreated or PTX-treated RAW 264.7 cells exposed to zymosan. A whisker plot is shown. Data were analyzed by using a t test. (C, top) Confocal microscopy of RAW264.7 cells permanently transfected with Elmo1-GFP in the presence or absence of Gαi2QL. Bar, 5 μm. (Bottom) Confocal microscopy of RAW 264.7 cells expressing Gαi2QL-Elmo1-GFP and immunostained for GTP-Gαi. Bar, 3 μm. (D) Immunoblot of Elmo1 and actin expression in siRNA-treated cells. (E) Superimposed bright-field and confocal microscopy images of Elmo1 knockdown and control cells stained for F-actin (phalloidin) and exposed to zymosan (asterisks). (F) The percentage of phagocytic cells in Elmo1 knockdown and control cells. Results are shown as a whisker plot. Data were analyzed by using the t test.
Noncanonical G-protein signaling pathways are involved in phagocytosis.
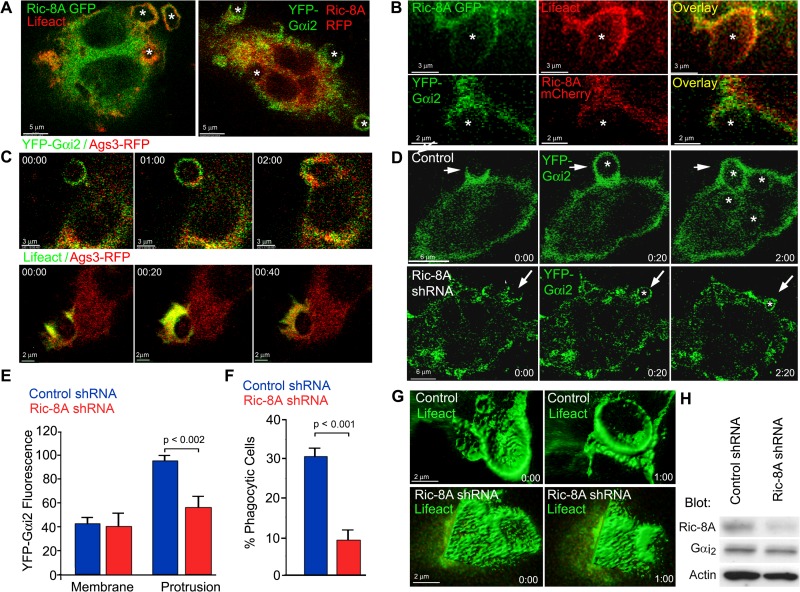
As noted above, PTX blocks the GEF activity of GPCRs for Gαi, but it also blocks the GEF activity of Ric-8A, an intracellular protein implicated in mitotic spindle localization (25). While Ric-8A also functions to chaperone Gα subunits to cell membranes, PTX does not block the chaperone activity of Ric-8A (35). Supporting a role for Ric-8A in phagocytosis, we found that in RAW 264.7 cells cotransfected with Ric-8A–GFP and RFP-Lifeact, or with YFP-Gαi2 and Ric-8A–mCherry, that Ric-8A partially colocalized with both proteins at phagocytic cups and phagosomes (Fig. 8A and B). To assess whether other proteins involved in noncanonical G-protein signaling were enriched at phagocytic sites, we expressed Rgs14-GFP, Ags3-RFP, Ags4-GFP, or LGN-GFP in RAW 264.7 cells and exposed the cells to zymosan. All of the fusion proteins localized predominately in the cytosol (data not shown). Of the four fusion proteins, only the Ags3-RFP fusion protein accumulated at phagocytic sites (Fig. 8C). To test whether Ric-8A might facilitate phagocytosis, we examined the impact of reducing the level of Ric-8A mRNA. We expressed a dsRed-shRNARic-8A or a control construct along with YFP-Gαi2 in RAW 264.7 cells and exposed the transfected cells to zymosan. We found that the cells transfected with the Ric-8A shRNA recruited less YFP-Gαi2 to cell protrusions and engulfed zymosan more slowly than did control cells (Fig. 8D and E). In addition, transfection of the dsRed-shRNA–Ric-8A in RAW 264.7 cells reduced the percentage of phagocytic cells compared to RAW 264.7 cells transfected with a control shRNA (Fig. 8F). When we transfected RAW 264.7 cells with GFP-Lifeact and either a control dsRed-shRNA or a dsRed-shRNA–Ric-8A and exposed the cells to zymosan, we often observed initial phagocytic cup formation but a subsequent engulfment failure in the cells with lowered Ric-8A expression (Fig. 8G). Immunoblotting verified the reduction in Ric-8A following transfection of the Ric-8A shRNA with a minimal effect on Gαi levels (Fig. 8H). These data indicate that lowering Ric-8A impedes phagocytic cup closure and reduces macrophage phagocytosis.
FIG 8.
Ric-8A promotes phagocytosis and phagocytic cup closure. (A) Confocal microscopy of RAW 264.7 cells expressing Ric-8A–GFP and RFP-Lifeact (left) or YFP-Gαi2 and Ric-8A-mCherry (right) following exposure to zymosan. Individual images, fluorescent overlay, and bright-field fluorescent overlay are shown in the region of a phagocytic cup. The locations of zymosan are indicated by asterisks. Bar, 5 μm. (B) Zoomed individual confocal images of RAW 264.7 cell from the image set acquired for panel A. Bars, 3 μm (top), 2 μm (bottom). (C) Time-lapse confocal microscopy of RAW 264.77.4 cells expressing Ags3-RFP and either Gαi2-YFP (top) or GFP-Lifeact (bottom) and exposed to zymosan. Bar, 3 μm (top), 2 μm (bottom). (D) Time-lapse confocal microscopy of RAW 264.7 cells expressing YFP-Gαi2 and either a control shRNA (top) or a Ric-8A shRNA (bottom) and exposed to zymosan. The locations of zymosan are indicated with asterisks. Bars, 6 μm. (E) Recruitment of YFP-Gαi2 to cell protrusions in RAW 264.7 cells expressing either a control shRNA or a Ric-8A shRNA. Fluorescent intensities at the cell membrane and at cell protrusions are shown. Statistics were determined by using the t test. (F) The percentage of RAW 264.7 cells expressing a control or a Ric-8A shRNA that engulfed zymosan over a 15-min period. Statistics were performed using the t test. (G) Time-lapse confocal microscopy of RAW 264.7 cells expressing GFP-Lifeact and either a control (top) or a Ric-8A shRNA (bottom) and exposed to zymosan. The surface rendering of the phagocytic cup was done using Imaris software. Bar, 2 μm. (H) Immunoblot of Ric-8A, Gαi2, and actin levels in cell lysates prepared from control and Ric-8A shRNA-expressing RAW 264.7 cells.
DISCUSSION
The recruitment of macrophages to inflammatory sites, directed cell protrusions, and the engulfment of pathogens/particles to various degrees depend upon heterotrimeric G-protein signaling. Initial macrophage-pathogen encounters are driven by canonical G-protein signaling (3). Cell polarization, chemotaxis, directed cell protrusions, and cell ruffling all depend upon the release of Gβγ subunits from Gαi (5, 36). Autocrine and/or paracrine GPCR signaling, which arises following initial pathogen-macrophage interactions, likely modulate the subsequent capture and engulfment events (37). The direct recognition, engulfment, and propulsion of targets into the interior of cells depends upon signaling from capturing receptors (2). Our study reveals a cross talk between the pathways initiated by capturing receptors and pathways that employ Gi as a signal transducer. Our results indicate that the appropriate activation of Gαi and the release of Gβγ subunits contribute to F-actin dynamics and the overall efficiency of zymosan phagocytosis by macrophages. Activated Gαi likely recruits Elmo1, providing a link to Dock proteins and the activation of small GTPases. We also have provided evidence that Ric-8A, a noncanonical GEF, augments the engulfment of zymosan by macrophages.
Using a combination of genetic, pharmacologic, and cell imaging data, our study provides compelling evidence that Gαi nucleotide exchange occurs at phagocytic sites and that the generation of Gαi-GTP and the release of Gβγ subunits are functionally relevant for macrophages to engage and engulf zymosan and likely other targets. However, the failure of PTX to inhibit S. aureus bioparticle phagocytosis suggests that Gαi signaling is more relevant for larger targets. A major question is how Gαi nucleotide exchange is initiated. Zymosan may engage an unknown GPCR or other cell surface receptor that triggers Gαi nucleotide exchange. The rapid increase in intracellular Ca2+ following exposure of macrophages to zymosan and its inhibition by PTX are consistent with the engagement of a GPCR signaling pathway. Additional support for GPCR involvement came from a study that linked Gα12/13 and Gαi signaling to changes in cyclic AMP levels following zymosan exposure (38). It was postulated that either a direct zymosan-GPCR interaction resulted in G-protein activation or that zymosan via a Toll-like receptor 2 (TLR2) interaction triggered transactivation of a GPCR (38). Rather than a direct ligand-receptor interaction, the engagement of zymosan or another large target may sufficiently deform the cell membrane to trigger a mechanosensor linked to Gαi. Several GPCRs have been linked to mechanoreception (39). A less likely, but alternative, mechanism is that an unknown macrophage receptor triggers the recruitment and activation of Ric-8A, which serves to trigger Gα nucleotide exchange. However, a more likely scenario is that Ric-8A functions to amplify GPCR-initiated signaling. Ric-8A has been shown to amplify signaling through a Gq-linked receptor, and a recent study in Dictyostelium revealed that Ric8 acts downstream in a chemoattractant signaling pathway to augment Gα signaling (40, 41).
Based on the analysis of the Gnai2−/−, Gnai2 G184S+/+, and Gnai3−/− macrophages, both Gαi2 and Gαi3 contributed to zymosan phagocytosis, although only the loss of Gαi2 reduced Fcγ- and complement-mediated phagocytosis of coated sRBC. In hematopoietic cells, the loss of Gαi2 is usually more detrimental than the loss of Gαi3, in part because of the higher expression level of Gαi2 versus Gαi3, but in addition, some signaling pathways are more dependent on one isoform than the other. For example, chemotaxis is much more sensitive to loss of Gαi2 than the loss of Gαi3 (4, 42). Macrophages from Gnai2 G184S+/+ mice, which have potentiated Gαi2 signaling, confirmed the importance of Gαi2 signaling in the phagocytosis of zymosan, yet macrophages from these same mice been shown to have reduced Toll-like receptor-induced inflammatory cytokine production (43). The role of Gαi proteins in TLR signaling pathways remains poorly understood and may contribute to some of the phagocytic defects noted in the Gαi-deficient mice. Further studies are needed to better understand how Gαi nucleotide exchange is regulated by other signaling pathways, such as those triggered by TLRs.
During chemoattractant receptor-triggered chemotaxis, Gαi nucleotide exchange is thought to function predominately to release Gβγ subunits. GTP-Gαi has been ascribed little or no role. In contrast, our data indicate that GTP-Gαi signaling helps regulate phagocytosis. The expression of Gαi2 QL sped up phagocytosis, modulated F-actin dynamics, and increased the percentage of phagocytic cells. In providing a mechanism for how GTP-Gαi augments F-actin formation and phagocytosis, we found that Elmo1 was recruited by GTP-Gαi to the plasma membrane. During phagocytosis, the levels of Elmo1 increased at the phagocytic cup, and reducing Elmo1 expression impaired zymosan chemotaxis. Our results are consistent with the previous demonstration of CXCL12-induced translocation of a trimeric complex between Gαi2, ELMO1, and Dock180 (7). This links GTP-Gαi to Rac activation and F-actin formation. Since Elmo1/Dock180 has been previously connected to phagocytosis, Gαi nucleotide exchange may affect phagocytosis by generating GTP-Gαi.
The recruitment of Gβγ subunits to cell protrusions and phagocytic cups, and the reduction in phagocytosis following the addition of a competitive inhibitor of Gβγ signaling, both support a role for Gβγ in zymosan phagocytosis by BMDM. Both canonical and noncanonical G-protein signaling can release Gβγ subunits from Gαi. In the standard G-protein signaling model, GPCR-triggered Gα guanine nucleotide exchange releases Gβγ subunits from the heterotrimer, while in noncanonical G-protein signaling, a GPR motif-containing protein competes with Gβγ subunits for Gα binding. In the latter scenario, Ric-8A targets GDP-Gα associated with a GPR motif containing protein to release GTP-Gα and the GPR motif protein (16). A GPR motif containing protein Ags3 (Gpsm1) has been shown to act in a GPCR-independent pathway to release Gβγ subunits that promote renal tubule epithelial cell proliferation (44). Furthermore, studies in Dictyostelium have shown that Ric8 can amplify Gα signaling by increasing the availability of Gβγ subunits (40).
The involvement of Ric-8A in macrophage phagocytosis argues that a GPR motif containing protein or proteins also participates. Murine macrophages express several candidates, including Ags3/Gpsm1, LGN/Gpms2/Ags2, Ags4/Gpsm3, Rgs12, and Rgs14 (http://www.immgen.org/databrowser/index.html). However, little information is available about these proteins in macrophage biology. Using fluorescent fusion proteins, we tested the possible recruitment of Ags3, LGN, Ags4, and Rgs14, and only Ags3 was enriched at phagocytic cups. We are currently testing whether reducing the expression of these proteins impacts phagocytosis. Suggesting that they may have functional roles, myeloid cells from Gpsm3−/− mice migrate less efficiently to Ccl2 and Cx3cl1 than do wild-type mouse myeloid cells (45). Importantly, many of these proteins have additional domains that can interact with other relevant proteins. For example, Ags3/Gpsm1 binds LC3 (46). This interaction may affect phagosome recruitment of LC3 or, conversely, the recruitment of Ags3/Gpsm1 to phagosomes.
In this study, we used a macrophage cell line and BMDM to show that both Gαi and Gβγ signaling affect actin dynamics and zymosan bioparticle phagocytosis. Live cell imaging demonstrated that Gαi and Gβγ are enriched with F-actin at cell membrane protrusions and phagocytic sites. Inhibiting Gαi nucleotide exchange, reducing Ric-8A expression, or inhibiting Gβγ signaling all substantially reduced phagocytosis. BMDM that lack Gαi2 or Gαi3 also had a reduced capacity to engulf zymosan. In contrast, Gnai2G184S/G184S BMDM, wild-type BMDM treated with Pasteurella multocida toxin, or macrophages expressing a constitutively active Gαi protein had enhanced capacities to engulf zymosan bioparticles. Together, our findings show that heterotrimeric G-protein signaling pathways contribute to the recognition, capture, and engulfment of zymosan and likely other pathogens. Our studies support the further development of small-molecule inhibitors of Gβγ signaling such as gallein as anti-inflammatory agents.
Supplementary Material
ACKNOWLEDGMENTS
We thank David Stephany and Kevin Holmes (Research Technologies Branch, NIAID) for help with some of the flow cytometry experiments. We gratefully acknowledge Juraj Kabat (Core Imaging Facility, NIAID) for his quantification and modeling of some of the imaging data and Hyeseon Cho (NIAID) for making the PTX-resistant YFP-Gαi2 construct. We also thank Anthony Fauci for his continued support.
This research was supported by the intramural program of the National Institute of Allergy and Infectious Diseases.
Footnotes
Published ahead of print 15 September 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MCB.00325-14.
REFERENCES
- 1.Aderem A, Underhill DM. 1999. Mechanisms of phagocytosis in macrophages. Annu. Rev. Immunol. 17:593–623. 10.1146/annurev.immunol.17.1.593. [DOI] [PubMed] [Google Scholar]
- 2.Flannagan RS, Jaumouille V, Grinstein S. 2012. The cell biology of phagocytosis. Annu. Rev. Pathol. 7:61–98. 10.1146/annurev-pathol-011811-132445. [DOI] [PubMed] [Google Scholar]
- 3.Heinrich V, Lee CY. 2011. Blurred line between chemotactic chase and phagocytic consumption: an immunophysical single-cell perspective. J. Cell Sci. 124:3041–3051. 10.1242/jcs.086413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kehrl JH, Hwang IY, Park C. 2009. Chemoattract receptor signaling and its role in lymphocyte motility and trafficking. Curr. Top. Microbiol. Immunol. 334:107–127. 10.1007/978-3-540-93864-4_5. [DOI] [PubMed] [Google Scholar]
- 5.Van Haastert PJ, Devreotes PN. 2004. Chemotaxis: signalling the way forward. Nat. Rev. Mol. Cell Biol. 5:626–634. 10.1038/nrm1435. [DOI] [PubMed] [Google Scholar]
- 6.Yan J, Mihaylov V, Xu X, Brzostowski JA, Li H, Liu L, Veenstra TD, Parent CA, Jin T. 2012. A Gβγ effector, ElmoE, transduces GPCR signaling to the actin network during chemotaxis. Dev. Cell 22:92–103. 10.1016/j.devcel.2011.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li H, Yang L, Fu H, Yan J, Wang Y, Guo H, Hao X, Xu X, Jin T, Zhang N. 2013. Association between Gαi2 and ELMO1/Dock180 connects chemokine signalling with Rac activation and metastasis. Nat. Commun. 4:1706. 10.1038/ncomms2680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Park H, Cox D. 2009. Cdc42 regulates Fc gamma receptor-mediated phagocytosis through the activation and phosphorylation of Wiskott-Aldrich syndrome protein (WASP) and neural-WASP. Mol. Biol. Cell 20:4500–4508. 10.1091/mbc.E09-03-0230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goodridge HS, Underhill DM, Touret N. 2012. Mechanisms of Fc receptor and dectin-1 activation for phagocytosis. Traffic 13:1062–1071. 10.1111/j.1600-0854.2012.01382.x. [DOI] [PubMed] [Google Scholar]
- 10.Serezani CH, Kane S, Collins L, Morato-Marques M, Osterholzer JJ, Peters-Golden M. 2012. Macrophage dectin-1 expression is controlled by leukotriene B4 via a GM-CSF/PU.1 axis. J. Immunol. 189:906–915. 10.4049/jimmunol.1200257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peracino B, Borleis J, Jin T, Westphal M, Schwartz JM, Wu L, Bracco E, Gerisch G, Devreotes P, Bozzaro S. 1998. G protein beta subunit-null mutants are impaired in phagocytosis and chemotaxis due to inappropriate regulation of the actin cytoskeleton. J. Cell Biol. 141:1529–1537. 10.1083/jcb.141.7.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Damiani MT, Colombo MI. 2001. Involvement of heterotrimeric G proteins in phagocytosis and recycling from the phagosomal compartment. Exp. Cell Res. 271:189–199. 10.1006/excr.2001.5354. [DOI] [PubMed] [Google Scholar]
- 13.Zhang Y, Tang W, Jones MC, Xu W, Halene S, Wu D. 2010. Different roles of G protein subunits β1 and β2 in neutrophil function revealed by gene expression silencing in primary mouse neutrophils. J. Biol. Chem. 285:24805–24814. 10.1074/jbc.M110.142885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hwang JI, Choi S, Fraser ID, Chang MS, Simon MI. 2005. Silencing the expression of multiple Gβ-subunits eliminates signaling mediated by all four families of G proteins. Proc. Natl. Acad. Sci. U. S. A. 102:9493–9498. 10.1073/pnas.0503503102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gabay M, Pinter ME, Wright FA, Chan P, Murphy AJ, Valenzuela DM, Yancopoulos GD, Tall GG. 2011. Ric-8 proteins are molecular chaperones that direct nascent G protein alpha subunit membrane association. Sci. Signal. 4:ra79. 10.1126/scisignal.2002223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ahmed SM, Angers S. 2013. Emerging non-canonical functions for heterotrimeric G proteins in cellular signaling. J. Recept. Signal. Transduct. Res. 33:177–183. 10.3109/10799893.2013.795972. [DOI] [PubMed] [Google Scholar]
- 17.Gohla A, Klement K, Piekorz RP, Pexa K, Svom Dahl Spicher K, Dreval V, Haussinger D, Birnbaumer L, Nurnberg B. 2007. An obligatory requirement for the heterotrimeric G protein Gi3 in the antiautophagic action of insulin in the liver. Proc. Natl. Acad. Sci. U. S. A. 104:3003–3008. 10.1073/pnas.0611434104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang X, Fu Y, Charbeneau RA, Saunders TL, Taylor DK, Hankenson KD, Russell MW, D'Alecy LG, Neubig RR. 2006. Pleiotropic phenotype of a genomic knock-in of an RGS-insensitive G184S Gnai2 allele. Mol. Cell. Biol. 26:6870–6879. 10.1128/MCB.00314-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rudolph U, Finegold MJ, Rich SS, Harriman GR, Srinivasan Y, Brabet P, Boulay G, Bradley A, Birnbaumer L. 1995. Ulcerative colitis and adenocarcinoma of the colon in G alpha i2-deficient mice. Nat. Genet. 10:143–150. 10.1038/ng0695-143. [DOI] [PubMed] [Google Scholar]
- 20.Cho H, Kamenyeva O, Yung S, Gao JL, Hwang IY, Park C, Murphy PM, Neubig RR, Kehrl JH. 2012. The loss of RGS protein-Gα(i2) interactions results in markedly impaired mouse neutrophil trafficking to inflammatory sites. Mol. Cell. Biol. 32:4561–4571. 10.1128/MCB.00651-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shi CS, Shenderov K, Huang NN, Kabat J, Abu-Asab M, Fitzgerald KA, Sher A, Kehrl JH. 2012. Activation of autophagy by inflammatory signals limits IL-1β production by targeting ubiquitinated inflammasomes for destruction. Nat. Immunol. 13:255–263. 10.1038/ni.2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vural A, Oner S, An N, Simon V, Ma D, Blumer JB, Lanier SM. 2010. Distribution of activator of G-protein signaling 3 within the aggresomal pathway: role of specific residues in the tetratricopeptide repeat domain and differential regulation by the AGS3 binding partners Gi(alpha) and mammalian inscuteable. Mol. Cell. Biol. 30:1528–1540. 10.1128/MCB.01018-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cho H, Kim DU, Kehrl JH. 2005. RGS14 is a centrosomal and nuclear cytoplasmic shuttling protein that traffics to promyelocytic leukemia nuclear bodies following heat shock. J. Biol. Chem. 280:805–814. 10.1074/jbc.M408163200. [DOI] [PubMed] [Google Scholar]
- 24.Boularan C, Kamenyeva O, Cho H, Kehrl JH. 2014. Resistance to inhibitors of cholinesterase (Ric)-8A and Gαi contribute to cytokinesis abscission by controlling vacuolar protein-sorting (Vps)34 activity. PLoS One 9:e86680. 10.1371/journal.pone.0086680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Woodard GE, Huang NN, Cho H, Miki T, Tall GG, Kehrl JH. 2010. Ric-8A and Gi alpha recruit LGN, NuMA, and dynein to the cell cortex to help orient the mitotic spindle. Mol. Cell. Biol. 30:3519–3530. 10.1128/MCB.00396-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gibson SK, Gilman AG. 2006. Giα and Gβ subunits both define selectivity of G protein activation by α2-adrenergic receptors. Proc. Natl. Acad. Sci. U. S. A. 103:212–217. 10.1073/pnas.0509763102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Riedl J, Crevenna AH, Kessenbrock K, Yu JH, Neukirchen D, Bista M, Bradke F, Jenne D, Holak TA, Werb Z, Sixt M, Wedlich-Soldner R. 2008. Lifeact: a versatile marker to visualize F-actin. Nat. Methods 5:605–607. 10.1038/nmeth.1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Spector I, Shochet NR, Kashman Y, Groweiss A. 1983. Latrunculins: novel marine toxins that disrupt microfilament organization in cultured cells. Science 219:493–495. 10.1126/science.6681676. [DOI] [PubMed] [Google Scholar]
- 29.Leaney JL, Tinker A. 2000. The role of members of the pertussis toxin-sensitive family of G proteins in coupling receptors to the activation of the G protein-gated inwardly rectifying potassium channel. Proc. Natl. Acad. Sci. U. S. A. 97:5651–5656. 10.1073/pnas.080572297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lyman CA, Simons ER, Melnick DA, Diamond RD. 1988. Induction of signal transduction in human neutrophils by Candida albicans hyphae: the role of pertussis toxin-sensitive guanosine triphosphate-binding proteins. J. Infect. Dis. 158:1056–1064. 10.1093/infdis/158.5.1056. [DOI] [PubMed] [Google Scholar]
- 31.Hermouet S, Merendino JJ, Jr, Gutkind JS, Spiegel AM. 1991. Activating and inactivating mutations of the alpha subunit of Gi2 protein have opposite effects on proliferation of NIH 3T3 cells. Proc. Natl. Acad. Sci. U. S. A. 88:10455–10459. 10.1073/pnas.88.23.10455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Orth JH, Fester I, Siegert P, Weise M, Lanner U, Kamitani S, Tachibana T, Wilson BA, Schlosser A, Horiguchi Y, Aktories K. 2013. Substrate specificity of Pasteurella multocida toxin for alpha subunits of heterotrimeric G proteins. FASEB J. 27:832–842. 10.1096/fj.12-213900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hynes TR, Yost E, Mervine S, Berlot CH. 2008. Multicolor BiFC analysis of competition among G protein beta and gamma subunit interactions. Methods 45:207–213. 10.1016/j.ymeth.2008.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smrcka AV, Lehmann DM, Dessal AL. 2008. G protein betagamma subunits as targets for small molecule therapeutic development. Comb. Chem. High Throughput Screen. 11:382–395. 10.2174/138620708784534761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Oner SS, Maher EM, Gabay M, Tall GG, Blumer JB, Lanier SM. 2013. Regulation of the G-protein regulatory-Gαi signaling complex by nonreceptor guanine nucleotide exchange factors. J. Biol. Chem. 288:3003–3015. 10.1074/jbc.M112.418467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bohdanowicz M, Schlam D, Hermansson M, Rizzuti D, Fairn GD, Ueyama T, Somerharju P, Du G, Grinstein S. 2013. Phosphatidic acid is required for the constitutive ruffling and macropinocytosis of phagocytes. Mol. Biol. Cell 24:1700–1712. 10.1091/mbc.E12-11-0789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee SP, Serezani CH, Medeiros AI, Ballinger MN, Peters-Golden M. 2009. Crosstalk between prostaglandin E2 and leukotriene B4 regulates phagocytosis in alveolar macrophages via combinatorial effects on cyclic AMP. J. Immunol. 182:530–537. 10.4049/jimmunol.182.1.530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jiang LI, Sternweis PC, Wang JE. 2013. Zymosan activates protein kinase A via adenylyl cyclase VII to modulate innate immune responses during inflammation. Mol. Immunol. 54:14–22. 10.1016/j.molimm.2012.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Storch U, Mederos y Schnitzler M, Gudermann T. 2012. G protein-mediated stretch reception. Am. J. Physiol. Heart Circ. Physiol. 302:H1241–H1249. 10.1152/ajpheart.00818.2011. [DOI] [PubMed] [Google Scholar]
- 40.Kataria R, Xu X, Fusetti F, Keizer-Gunnink I, Jin T, van Haastert PJ, Kortholt A. 2013. Dictyostelium Ric8 is a nonreceptor guanine exchange factor for heterotrimeric G proteins and is important for development and chemotaxis. Proc. Natl. Acad. Sci. U. S. A. 110:6424–6429. 10.1073/pnas.1301851110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nishimura A, Okamoto M, Sugawara Y, Mizuno N, Yamauchi J, Itoh H. 2006. Ric-8A potentiates Gq-mediated signal transduction by acting downstream of G protein-coupled receptor in intact cells. Genes Cells 11:487–498. 10.1111/j.1365-2443.2006.00959.x. [DOI] [PubMed] [Google Scholar]
- 42.Wiege K, Le DD, Syed SN, Ali SR, Novakovic A, Beer-Hammer S, Piekorz RP, Schmidt RE, Nurnberg B, Gessner JE. 2012. Defective macrophage migration in Gαi2- but not Gαi3-deficient mice. J. Immunol. 189:980–987. 10.4049/jimmunol.1200891. [DOI] [PubMed] [Google Scholar]
- 43.Li P, Neubig RR, Zingarelli B, Borg K, Halushka PV, Cook JA, Fan H. 2012. Toll-like receptor-induced inflammatory cytokines are suppressed by gain of function or overexpression of Gα(i2) protein. Inflammation 35:1611–1617. 10.1007/s10753-012-9476-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Regner KR, Nozu K, Lanier SM, Blumer JB, Avner ED, Sweeney WE, Jr, Park F. 2011. Loss of activator of G-protein signaling 3 impairs renal tubular regeneration following acute kidney injury in rodents. FASEB J. 25:1844–1855. 10.1096/fj.10-169797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Giguere PM, Billard MJ, Laroche G, Buckley BK, Timoshchenko RG, McGinnis MW, Esserman D, Foreman O, Liu P, Siderovski DP, Tarrant TK. 2013. G-protein signaling modulator-3, a gene linked to autoimmune diseases, regulates monocyte function and its deficiency protects from inflammatory arthritis. Mol. Immunol. 54:193–198. 10.1016/j.molimm.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Garcia-Marcos M, Ear J, Farquhar MG, Ghosh P. 2011. A GDI (AGS3) and a GEF (GIV) regulate autophagy by balancing G protein activity and growth factor signals. Mol. Biol. Cell 22:673–686. 10.1091/mbc.E10-08-0738. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.