Abstract
Nearly 5% of membrane proteins are guided to nuclear, endoplasmic reticulum (ER), mitochondrial, Golgi, or peroxisome membranes by their C-terminal transmembrane domain and are classified as tail-anchored (TA) membrane proteins. In Saccharomyces cerevisiae, the guided entry of TA protein (GET) pathway has been shown to function in the delivery of TA proteins to the ER. The sorting complex for this pathway is comprised of Sgt2, Get4, and Get5 and facilitates the loading of nascent tail-anchored proteins onto the Get3 ATPase. Multiple pulldown assays also indicated that Ybr137wp associates with this complex in vivo. Here, we report a 2.8-Å-resolution crystal structure for Ybr137wp from Saccharomyces cerevisiae. The protein is a decamer in the crystal and also in solution, as observed by size exclusion chromatography and analytical ultracentrifugation. In addition, isothermal titration calorimetry indicated that the C-terminal acidic motif of Ybr137wp interacts with the tetratricopeptide repeat (TPR) domain of Sgt2. Moreover, an in vivo study demonstrated that Ybr137wp is induced in yeast exiting the log phase and ameliorates the defect of TA protein delivery and cell viability derived by the impaired GET system under starvation conditions. Therefore, this study suggests a possible role for Ybr137wp related to targeting of tail-anchored proteins.
INTRODUCTION
Eukaryotic membrane-bound proteins have to be targeted to the membranes. Approximately 5% of membrane proteins are tail anchored (TA) to the membrane, and this class of integral membrane proteins can be grouped into the TA family (1–6). TA membrane proteins have a C-terminal trans-membrane domain (TMD). The TMD is inserted into the membrane, while the functional domain(s) is oriented toward the cytosolic side of the cell. TA proteins can be found in nuclear, endoplasmic reticulum (ER), mitochondrial, Golgi, and peroxisome membranes (7, 8). More than 400 human TA proteins have been identified and implicated in membrane fusion, protein translocation, and apoptosis. It was recently shown that TA proteins are delivered to the ER membrane via the guided entry of TA protein (GET) pathway (9–14). Unlike proteins containing an N-terminal signal peptide(s) that is recognized by the signal recognition particle and targeted to the ER in a cotranslational manner, TA proteins expose their targeting signal only after the complete biosynthesis of the C-terminal TMD and are released from the ribosome (15).
In Saccharomyces cerevisiae, Get1 to Get5 and Sgt2 are the major components of the GET pathway (9, 13, 16–18). The first step in the GET pathway is the formation of the Sgt2/Get4/Get5 sorting complex, which recognizes and loads the newly formed TA protein onto the Get3 ATPase (19–21). Get3 then delivers the TA protein to the ER membrane, where the TA protein is released for insertion with the aid of the Get1/Get2 membrane-bound complex (13, 22). Get2 acts as a receptor that recruits the Get3-TA protein complex onto the surface of the ER membrane, while Get1 triggers the release of the TMD of the TA protein from Get3 (9, 12, 23, 24).
Sgt2 is a 38-kDa protein containing a domain with three tetratricopeptide repeats (TPRs) and is well conserved across eukaryotes (25). Starting from the N terminus, Sgt2 has a dimerization domain, a TPR domain, and a Glu/Met-rich C-terminal domain (26–28). Recently, Sgt2 has been shown to interact with Ybr137wp, Hsp104, and Hsp70 via its TPR domain (13, 20). Ybr137wp is the subject of this work, while Hsp104 and Hsp70 are chaperone proteins. A previous immunoprecipitation study indicated that Arg171 and Arg175 of the Sgt2 TPR domain interact with Ybr137wp and the above-mentioned chaperones. Simultaneous mutation of these two arginine residues abolishes the association of Sgt2 with Ybr137wp or the chaperone proteins (13). Notably, the association of Hsp104 or Hsp70 with Sgt2 increases in a yeast strain with a ybr137w deletion (13). The results imply that Ybr137wp and the chaperones compete for the same binding site on the Sgt2 TPR domain. Ybr137wp may regulate TA protein targeting by mediating interactions between Sgt2 and the chaperones. Interestingly, Ybr137wp can form a cytosolic complex with Kap142p (29). Kap142p is a karyopherin that mediates both nuclear export and import (29). However, the role of Ybr137wp in this complex is unclear. We also noted that ybr137w is not conserved outside fungi, even though TA protein targeting and sgt2 are universal to all eukaryotes.
Although many studies have focused on the GET pathway and the role of Sgt2 in the pathway, the function of Ybr137wp remains to be elucidated. To investigate the potential function of Ybr137wp, we first determined the crystal structure of Ybr137wp, which has a homodecameric quaternary structure in both crystal and solution forms. Structural homology analysis suggests that Ybr137wp belongs to the GlcG-like superfamily domain, although they shared low sequence identity. We used isothermal titration calorimetry (ITC) to quantify the binding constant for binding of Sgt2 to Ybr137wp during complex formation, and with the use of in vivo assays, we suggest a possible role for Ybr137wp in the GET pathway.
MATERIALS AND METHODS
Construction of plasmids.
ybr137w encoding Ybr137wp (residues 1 to 179) was PCR amplified by using S. cerevisiae genomic DNA as the template, an upstream primer containing an NdeI site, and a downstream primer containing an XhoI site. The amplified fragment was digested with NdeI and XhoI and then subcloned into the NdeI/XhoI site of pET15b (Novagen) to yield a construct encoding N-terminally His6-tagged Ybr137wp. PCR amplification of the genes encoding full-length Sgt2 (residues 1 to 347) and the TPR domain of Sgt2 (Sgt2TPR) (residues 95 to 227) was performed in a manner similar to that for Ybr137wp. Sgt2ΔC, lacking the Sgt2 C-terminal domain (residues 1 to 227), and Ybr137wpΔC, lacking the Ybr137wp C-terminal domain (residues 1 to 170), were generated by introducing a stop codon using QuikChange site-directed mutagenesis kit reagent (Stratagene) after the codon for position 227 or 170, respectively.
Protein expression and purification.
Plasmids for protein expression were transformed into Escherichia coli BL21(DE3) cells. Transformed cells were grown at 37°C in LB medium supplemented with ampicillin at 100 μg/ml until the optical density at 600 nm (OD600) reached 0.6. Overexpression of His6-Ybr137wp was induced by the addition isopropyl-thio-β-d-galactopyranoside (final concentration, 1.0 mM), and cell growth was continued at 37°C for an additional 3 h. Cells were harvested by centrifugation at 5,000 × g for 20 min at 4°C. Harvested cells were suspended in a solution containing 20 mM Tris-HCl (pH 8.0), 500 mM NaCl, and 5 mM imidazole and then lysed with a Microfluidizer (Microfluidics). The lysate was centrifuged at 30,000 × g for 50 min and then loaded onto a HisTrap HP column equilibrated with the same buffer solution. The column was washed with 10 column volumes of a solution containing 20 mM Tris-HCl (pH 8.0), 500 mM NaCl, and 50 mM imidazole. The His6-tagged proteins were eluted with a solution containing 20 mM Tris-HCl (pH 8.0), 500 mM NaCl, and 200 mM imidazole. Protein fractions were further purified by Superdex 200 gel filtration chromatography using buffer containing 20 mM Tris-HCl (pH 8.0) and 100 mM NaCl. Column fractions were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and those fractions found to contain His6-Ybr137wp were concentrated to ∼10 mg/ml and stored at −80°C. Selenomethionine (SeMet)/His6-Ybr137wp was expressed in cells grown in Overnight Express Autoinduction System 2 medium (Novagen) and was purified in a manner similar to that for Ybr137wp. Sgt2ΔC and Sgt2TPR were also purified in a similar manner.
Size exclusion chromatography.
Size exclusion chromatography (SEC) was performed at 4°C by using an Akta purifier system (GE Healthcare). Briefly, 5-ml samples (100 μM) were individually applied onto a Superdex 200 16/60 PG column (GE Healthcare) that had been equilibrated with a solution containing 20 mM Tris-HCl (pH 8.0) and 100 mM NaCl. The flow rate was 1.0 ml/min, and 2-ml fractions were collected. Peak fractions were then subjected to SDS-PAGE (20% [wt/vol] acrylamide). The column was calibrated by using protein markers (GE Healthcare), including thyroglobulin (669 kDa), ferritin (440 kDa), aldolase (158 kDa), and conalbumin (75 kDa).
Analytical ultracentrifugation.
Analytical ultracentrifugation (AUC) experiments were conducted with an XL-A analytical ultracentrifuge (Beckman-Coulter, Brea, CA) and a Ti An60 rotor. Ybr137wp, Sgt2ΔC, and Ybr137wp/Sgt2ΔC samples in a solution containing 20 mM Tris-HCl (pH 8.0) and 100 mM NaCl were used for all centrifugation experiments. The buffer density and viscosity were calculated by using the SEDNTERP program (30) based on buffer composition. The partial specific volumes were calculated based on the amino acid compositions of Ybr137wp and Sgt2ΔC by using the same program. The sedimentation velocity profiles were collected at 20°C at 280 nm with rotor speeds of 60,000 rpm (Sgt2ΔC) and 35,000 rpm (Ybr137wp). Data were analyzed with the SEDFIT program (31) to fit a continuous distribution model, c(s), according to the Lamm equation. For sedimentation equilibrium experiments, the Ybr137wp/Sgt2ΔC complex was obtained by SEC, and data collection was performed at rotor speeds of 3,200, 3,800, and 7,000 rpm. The resulting absorbance-versus-radial profile was measured at 250 nm for each speed until equilibrium was reached. Sedimentation equilibrium profiles at three rotor speeds were globally fitted to a single discrete species model by using SEDPHAT (32).
Protein crystallization.
Crystallization trials were set up by using 10 mg/ml Ybr137wp in a solution containing 20 mM Tris-HCl (pH 8.0) and 100 mM NaCl. Initial screening was performed by using a Phoenix robot platform (Rigaku) with commercially available screen reagents (Hampton Research, Molecular Dimension, and Qiagen Ltd.) at 25°C. Crystals (0.06 mm by 0.06 mm by 0.02 mm) that diffracted to a ∼4.5-Å resolution were obtained for the 30% (vol/vol) 2-methyl-2,4-pentandiol (MPD)–20 mM Tris-HCl (pH 8.0) trial. Crystals (0.15 mm by 0.15 mm by 0.02 mm) suitable for X-ray diffraction were obtained by the hanging-drop vapor diffusion method after adjusting the protein concentration to 15 mg/ml at pH ∼8.2 to 8.4 and adding pentaerythritol propoxylate (Aldrich) to ∼2 to 4% (vol/vol). SeMet-Ybr137wp crystals were obtained in a similar manner. The crystals belong to the space group P3121 with cell dimensions of a = b = 135.25 Å and c = 121.13 Å and were diffracted to 2.8- and 2.9-Å resolutions for native and SeMet-labeled crystals, respectively. Native and single-wavelength anomalous diffraction data were generated by using synchrotron X-ray radiation at Beamline 13B1 of the National Synchrotron Radiation Research Center, Taiwan, and collected with a Quantum 310 charge-coupled device (CCD) detector (Area Detector Systems Corp.) at 100 K. The Ybr137wp structure was determined by using selenium single-wavelength anomalous dispersion (Se-SAD) phasing of the SeMet-Ybr137wp data acquired at a peak wavelength of 0.9786 Å. All data were processed by using DENZO and SCALEPACK in HKL2000 (33, 34).
Structure solution and refinement.
The identification of the selenium positions and the generation of initial SAD phases at a 3.0-Å resolution were performed by using PHENIX (35, 36). After identifying the 15 Se positions, the initial phases were further improved to a 2.8-Å resolution by using the maximum likelihood density modification algorithm in PHENIX. Owing to the limited resolution, automatic model building using PHENIX was not successful. Therefore, Coot (37) was used for model building. The native structure (2.8-Å resolution) was energy minimized and then annealed by using PHENIX and CCP4 (35, 36, 38). Sixty-four water molecules were found in the structure by using the water-picking routine in PHENIX. The final model has an R factor of 20.7% for all reflections of >2 σ and between 30.0- and 2.8-Å resolutions and an Rfree value of 26.8% for 5% randomly chosen reflections. A PHENIX-generated Ramachandran plot for the Ybr137wp ϕ-φ angles indicates that 94% of them are in the most favored regions and that 6% are in allowed regions. Solvent-accessible and interface areas were calculated by PISA (39). PyMol and Chimera (40, 41) were used to generate the figures. The Phyre2 server (42) predicted that residues 172 to 177 of Ybr137wp are α-helical. However, due to the lack of electron density at the region occupied by residues 174 to 179, we cannot validate the prediction experimentally.
Isothermal titration calorimetry.
ITC was performed by using a MicroCal iTC200 calorimeter (GE Healthcare) at 25°C. For all experiments, the sample cell contained 60 μM Sgt2ΔC, 20 mM Tris-HCl (pH 8.0), and 100 mM NaCl in a volume of 200 μl. An injection syringe was loaded with 700 μM Ybr137wp or a similar concentration of Ybr137wpΔC in the same buffer. Each experiment consisted of 20 injections. The volumes of the first and subsequent injections were 1 μl and 2 μl, respectively. The time interval between each injection was 240 s. The stirring speed and reference power were 1,000 rpm and 5 μcal/s, respectively. The binding isotherms were integrated and analyzed by using Origin v7.0 (MicroCal).
Yeast strains and plasmids.
All yeast strains used in this study were derived from S. cerevisiae in the BY4741 genetic background. Single-deletion strains were obtained from the Saccharomyces Genome Deletion Consortium (43). All yeast strains with a ybr137w deletion were prepared by PCR replacement of ybr137w with URA3 in the corresponding single-deletion strain. To restore Ybr137wp expression in the deletion strains, cells were transformed with a pRS313 construct containing ybr137w with an endogenous promoter. The gene encoding Flag-SCS2 was constructed by PCR fusion of synthetic DNA encoding the Flag tag and the SCS2 fragment amplified from genomic DNA and then cloned into pYCplac33 between the TEF1 promoter and terminator. The enhanced green fluorescent protein (eGFP)-Sec22 fusion gene was linked with the SacII restriction site sequence and cloned into pYCplac111 between the Gal1 promoter and terminator.
Viability assay.
A single colony of yeast was cultured in yeast extract-peptone-dextrose (YPD) and subcultured daily in fresh YPD for 3 days. Prior to the viability assay, equal amounts of cells cultured overnight were subcultured in YPD and grown for 6 h to log phase at an OD600 of ∼0.5 to 1.0. The cells were washed twice with synthetic complete (SC) medium and then resuspended in the same medium. In the viability assay using galactose, SC medium without glucose (SC-D medium) was used to wash the cells. After measurement of the cell density at 600 nm, cells were resuspended in 5 ml of SC medium or SC-D medium containing galactose to a final OD600 of 0.05 and cultured at 30°C. To assess cell viability, cells were serially diluted with YPD, and a 2-μl sample was dropped onto a YPD plate for each spot. The images were taken after incubation of the plates at 30°C for 36 to 48 h.
Western blotting.
For the quantification of endogenous Ybr137wp, yeast cells were cultured in YPD overnight and subcultured into YPD or SC medium. Two milliliters of cells was harvested at the indicated times. Following OD600 measurements, an equal amount of cells was resuspended in 0.1 N NaOH on ice for 15 min. The treated cells were resuspended in 1× SDS sample buffer and heated at 95°C for 5 min. The proteins were separated by electrophoresis with Bis-Tris 4 to 12% acrylamide gels (Invitrogen) and transferred onto a polyvinylidene difluoride (PVDF) membrane (Merck Millipore). The membrane was blocked in phosphate-buffered saline–Tween (PBST) containing 3% bovine serum albumin (BSA) and incubated with rabbit polyclonal antibodies against Ybr137wp or mouse anti-glucose-6-phosphate dehydrogenase (anti-G6PDH) at room temperature for 1 h, followed by incubation with appropriate secondary antibodies (Abcam).
Blue native gel electrophoresis.
For blue native PAGE analysis, wild-type (WT) and ybr137w deletion yeast strains were cultured in 20 ml YPD overnight. The cells were harvested, resuspended in 0.1 N NaOH, and incubated at room temperature (RT) for 30 min. The treated cells were resuspended in water and sonicated on ice for 2 min to lyse the cells. After centrifugation, the cell lysates were stored at 4°C before use. Ybr137wp with the His tag removed was used as a control. The His tag was removed from the purified recombinant protein by digesting 1 mg of His6-Ybr137wp with 1 unit of thrombin (Novagen) in 1× digestion buffer (20 mM Tris-HCl [pH 8.4], 150 mM NaCl, 2.5 mM CaCl2) at room temperature for 2 days. The His6 tag and undigested recombinant proteins were removed from Ybr137wp by passing the digestion mixture through a Ni column. Blue native gel electrophoresis was conducted with Bis-Tris 4 to 12% acrylamide gels (Invitrogen) according to the NativePAGE gel system protocol. Cell lysates (15 μl at 4 mg/ml) were mixed with 5 μl of 4× NativePAGE loading dye (Invitrogen). The HMW Native Marker kit (GE Healthcare) was used for markers. Electrophoresis was performed with NativePAGE running buffer (50 mM Bis-Tris, 50 mM Tricine [pH 6.8]) and blue cathode buffer (0.02% G-250, 50 mM Bis-Tris, 50 mM Tricine [pH 6.8]) at 150 V for 2.5 h. The markers were cut and stained with Coomassie blue dye. Proteins on the acrylamide gels were transferred onto PVDF membranes and visualized by Western blotting as described above.
Fluorescence microscopy.
Yeast cells harboring pYCplac33 plasmids carrying the Flag-SCS gene were cultured at 30°C for 2 days in SC medium. Cells were fixed with formaldehyde (4% [vol/vol]) for 15 min. Fixed cells were collected by centrifugation (200 × g for 10 min) and then washed with 0.1 M potassium phosphate (pH 6.5) and suspended in 0.1 M potassium phosphate (pH 6.5) with 1.2 M sorbitol. Cells were then treated with Zymolyase (20 g/ml) in the same buffer containing 143 mM β-mercaptoethanol. The resulting spheroplasts were seeded onto poly-l-lysine-coated slides and permeabilized with 100% methanol for 3 min, followed by 100% acetone for 1 s at −20°C. The spheroplasts were next treated with 2% (wt/vol) bovine serum albumin in PBS for 30 min and then with anti-Flag antibody (1:500 dilution; Abcam) and 1% (wt/vol) bovine serum albumin in PBS. After washing with PBS, the spheroplasts were treated with Cy3-conjugated anti-mouse antibodies (1:800 dilution; Rockland) in PBS and 1% (wt/vol) bovine serum albumin for 2 h. The slides were stained with 4′,6-diamidino-2-phenylindole dihydrochloride and then mounted on Vectashield mounting medium (Vector Laboratories) and imaged. For live-cell fluorescence microscopy imaging, yeast cells containing pYCplac111 with the eGFP-Sec22 gene were cultured at 30°C for 2 days in SC medium. After washing with PBS, cells were seeded directly onto slides coated with poly-l-lysine (Sigma) and then visualized with a Zeiss LSM 780 confocal microscope. For the quantification of cells containing puncta, images were taken by using an Olympus IX71 fluorescence microscope mounted with a 100× 1.4-numerical-aperture lens and a Delta Vision Restoration Imaging system. Several hundred cells in 50 random fields from 3 independent experiments were examined.
Protein structure accession number.
The atomic coordinates for Ybr137wp have been deposited in the Protein Data Bank (PDB) under accession number 4CLC.
RESULTS
Structure of S. cerevisiae Ybr137wp.
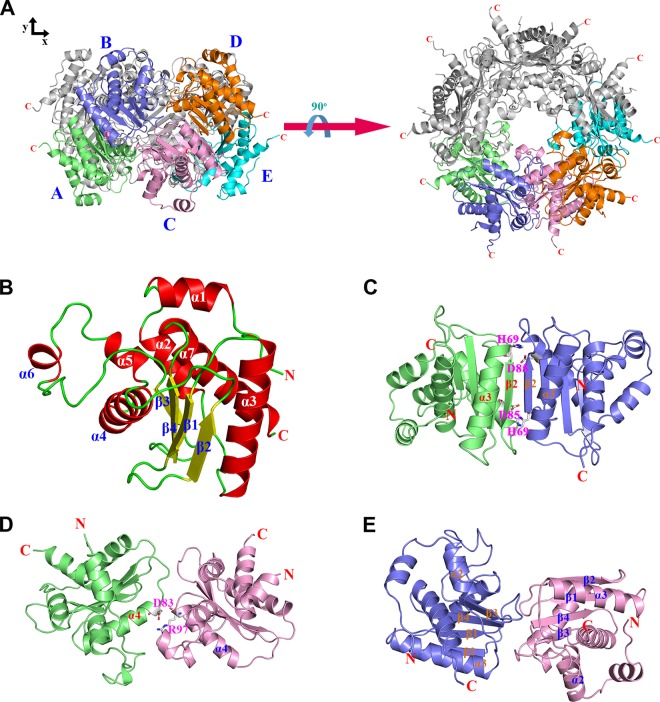
The crystal structure of full-length S. cerevisiae Ybr137wp was determined at a 2.8-Å resolution by using the single-wavelength anomalous dispersion (SAD) phases from an SeMet-derivative crystal. The statistics for data collection and refinement are summarized in Tables 1 and 2. The final model contains five Ybr137wp molecules in the asymmetric unit. Surprisingly, these five Ybr137wp molecules can form a decamer in the crystal (Fig. 1A). The Ybr137wp monomer contains seven α-helices (α1 to α7) and four β-strands (β1 to β4) (Fig. 1B). The four β-strands form an antiparallel β-sheet constituting the core of Ybr137wp surrounded by α-helices.
TABLE 1.
X-ray data collection statistics
| Parameter | Valuea |
|
|---|---|---|
| Native Ybr137wp | SeMet Ybr137wp | |
| Cell statistics | ||
| Space group | P3121 | P3121 |
| Unit cell parameters (Å) | a = b = 135.25, c = 121.50 | a = b = 135.97, c = 122.25 |
| Resolution range (Å) | 50.0-2.8 | 50.0-2.9 |
| Mosaicity (°) | 0.4 | 0.3 |
| Data processing statistics | ||
| Total no. of reflections | 235,271 | 322,258 |
| No. of unique reflections | 30,600 | 29,247 |
| Multiplicity | 7.7 (7.7) | 11.0 (11.2) |
| Completeness (%) | 95.7 (96.3) | 99.8 (100.0) |
| I/σ (I) | 33.8 (6.8) | 36.4 (4.2) |
| Rmerge (%)b | 5.9 (41.2) | 6.0 (54.4) |
Values in the parentheses are for the highest-resolution shell.
Rmerge = ΣhklΣi |Ii(hkl) − [I(hkl)]|/ΣhklΣi Ii(hkl), where Ii(hkl) is the observed intensity and [I(hkl)] is the average intensity from multiple observations of symmetry-related reflections.
TABLE 2.
Refinement statistics
| Refinement parameter | Value for Ybr137wp |
|---|---|
| Resolution range (Å) | 25.0–2.8 |
| No. of molecules per asymmetric unit | 5 |
| R/Rfree (%)a | 20.7/26.8 |
| No. of residues per asymmetric unit | 856 |
| No. of atoms per asymmetric unit | 6,602 |
| No. of water molecules per asymmetric unit | 64 |
| Ramachandran plot (% core/allowed/disallowed) | 94/6/0 |
| RMSD from ideality | |
| Bond lengths (Å) | 0.009 |
| Bond angles (°) | 1.4 |
| B-factor (Å2) | |
| Avg | 42.9 |
| Macromolecules | 43.0 |
| Solvent | 35.2 |
R = Σhkl ‖Fobs| − |Fcalc‖/Σhkl |Fobs|, where |Fobs| and |Fcalc| are the observed and calculated structure factor amplitudes, respectively. Rfree was calculated on the basis of 5% of the total number of reflections that were randomly omitted from the refinement. RMSD, root mean square deviation.
FIG 1.
Structural features of Ybr137wp. (A) Decamer conformation of Ybr137wp. The five Ybr137wp molecules in an asymmetric unit are designated Mol A to Mol E and are shown in green, blue, pink, orange, and cyan, respectively. The left and right panels are the side view and top view of the decamer, respectively. (B) Ribbon diagram of the Ybr137wp monomer. The four β-strands (β1 to β4), seven α-helices (α1 to α7), and loops in the protein are shown in yellow, red, and green, respectively. (C to E) Three types of interface interactions between subunits, the AB-type interface (C), the AC-type interface (D), and the BC-type interface (E). The corresponding molecules are colored as described above for panel A. The residues involved in salt bridge formation are depicted as a stick model.
The decamer structure is formed mainly by a dimer designed as an AB dimer through a pseudo-5-fold symmetry (Fig. 1A). In addition to the AB dimer, there are two other types of protein-protein interactions, denoted AC and BC, in the decamer, as shown in Fig. 1A. To determine which interaction represented the major structural element to form the decamer, we used the PISA server (39) to measure the surface areas of these interfaces. The surface area buried by the AB dimer (Fig. 1C) is 929.9 Å2, whereas those of the AC (Fig. 1D) and BC (Fig. 1E) interfaces are 446.8 Å2 and 620.2 Å2, respectively. Therefore, the AB dimer most likely plays the major elemental dimer to form the Ybr137wp decamer, and AC and BC interactions contribute to assemble and stabilize the decamer conformation. The AB dimer interface mainly involves β2 from one chain arranged in an antiparallel orientation with the β2 of the neighboring molecule through 2-fold symmetry (Fig. 1C). The intermolecular interactions stabilizing the interface involve electrostatic interactions, hydrogen bond networks, and van der Waals interactions. Specifically, His69 from one chain and Asp85 from the other chain form reciprocal salt bridges (Fig. 1C).
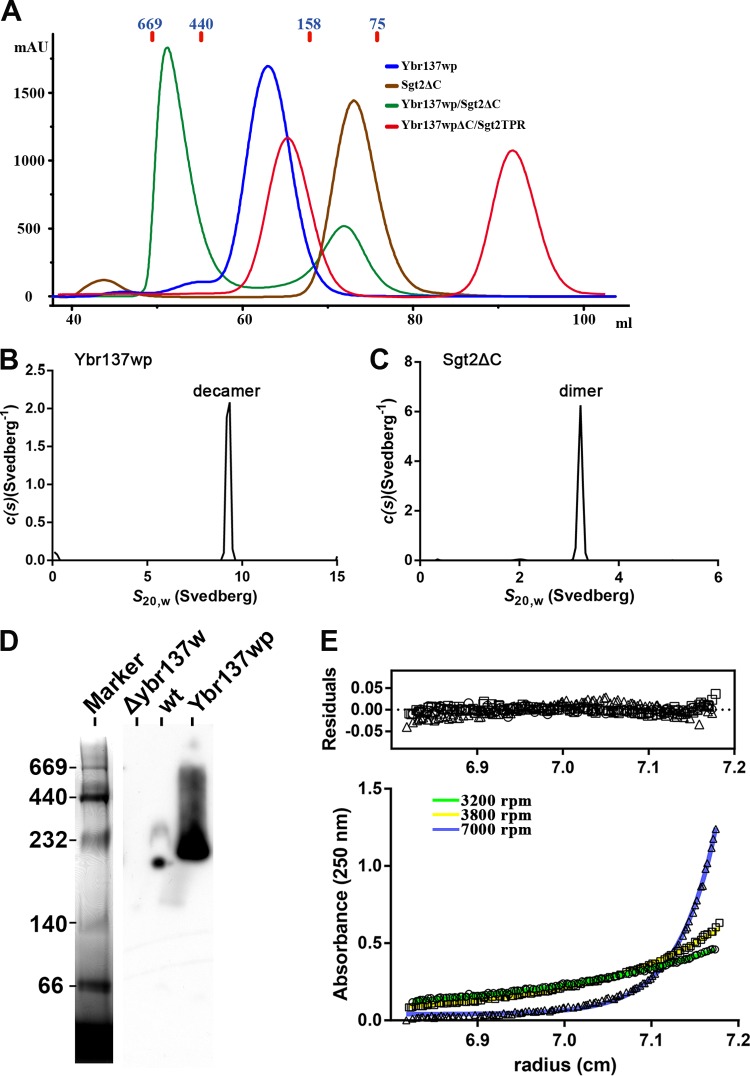
To determine if Ybr137wp is a decamer in solution, we performed size exclusion chromatography (SEC) analysis. Ybr137wp oligomerizes in solution with the major peak migrating at an elution position of ∼250 kDa, a mass consistent with a Ybr137wp decamer, since the theoretical molecular mass of the His6-tagged Ybr137wp monomer is 22.6 kDa (Fig. 2A). We also performed sedimentation velocity studies by analytical ultracentrifugation (AUC). The sedimentation velocity results revealed that Ybr137wp has a sedimentation coefficient of 9.2 S at a concentration of 46 μM (Fig. 2B). The corresponding c(s) distribution yielded a molecular mass of ∼230 kDa, which is consistent with the SEC result and matches the theoretical mass of the Ybr137wp decamer.
FIG 2.
Oligomer and complex formation of Ybr137wp and Sgt2 in solution. (A) Size exclusion chromatograms of Ybr137wp, Sgt2ΔC, Sgt2ΔC/Ybr137wp, and Ybr137wpΔC/Sgt2TPR. The molecular masses (kDa) of four markers are indicated in blue. mAU, milli-absorbance units. (B and C) Sedimentation velocity analyses of Ybr137wp and Sgt2ΔC. c(s) distribution analysis of sedimentation velocity data is shown for Ybr137wp (B) and Sgt2ΔC (C) at 20°C. (D) Blue native PAGE analysis of cell lysates from wild-type and Δybr137w cells after being cultured in YPD for 16 h. The proteins were resolved by blue native PAGE on 4-to-12% gradient gels. Recombinant Ybr137w with the His6 tag removed was used as the control. The native PAGE marker with the indicated molecular masses (kDa) was stained with Coomassie blue dye, and Ybr137wp was visualized by Western blotting with anti-Ybr137wp antibodies. (E) Sedimentation equilibrium data for the Sgt2ΔC/Ybr137wp complex. Shown are the absorbance-versus-radius profiles for the Sgt2ΔC/Ybr137wp complex after attaining sedimentation equilibrium at 3,200 rpm (○), 3,800 rpm (□), and 7,000 rpm (△) scanned at 250 nm. Best-fit distributions are shown as a solid line. Residuals versus the radial position were plotted under the resulting best-fit analysis at three rotor speeds. Data were extracted with SEDFIT and analyzed with SEDPHAT.
Given these results, we concluded that the decameric structure is probably essential for Ybr137wp function. To further support this hypothesis, we performed blue native PAGE using proteins from yeast crude extracts, and the results clearly indicated that endogenous Ybr137wp forms an oligomer with a molecular weight equivalent to that of a decamer in vivo (Fig. 2D). Therefore, the decameric quaternary structure is detectable in vivo and in vitro.
Comparison of Ybr137wp and proteins with similar structures.
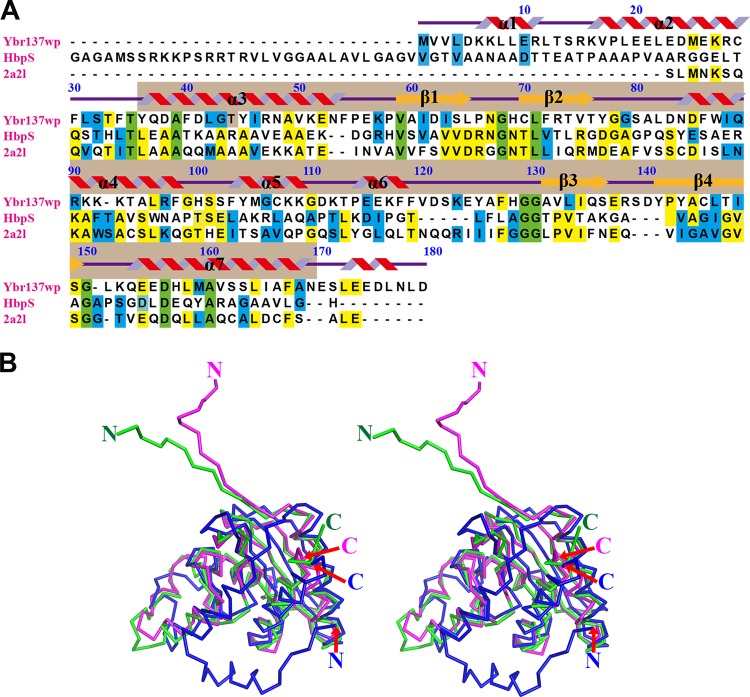
Although the function of Ybr137wp is not clear, our crystallography study shows that its tertiary structure is similar to those of a heme-degrading protein, HbpS from Streptomyces reticuli (PDB accession number 3FPV) (44), and a protein of unknown function from Klebsiella pneumoniae (PDB accession number 2A2L). However, Ybr137wp has only 14 and 16% sequence identity to HbpS and the K. pneumoniae protein, respectively (Fig. 3A). Structural homology analysis using the Phyre2 server (42) suggests that Ybr137wp belongs to the GlcG-like superfamily domain observed in bacteria, archaea, and eukaryotes. In general, members of the GlcG-like superfamily have 5 α-helices and 4 β-strands arranged in an α-β(2)-α(3)-β(2)-α configuration (Fig. 3A). We used a SCOP database search (45) and found >4,100 proteins containing a GlcG-like domain. Although the GlcG-like superfamily is large, there are only two representative structures deposited in the Protein Data Bank. We therefore compared the Ybr137wp structure with those of HbpS and the K. pneumoniae protein. The most significant structural differences were found in the N-terminal regions of the proteins (Fig. 3B). Residues 1 to 30 of Ybr137wp form two extra α-helices, whereas the N-terminal regions of HbpS and the K. pneumoniae protein form random coils and pointed in a direction opposite to that of Ybr137wp. Although the GlcG-like portion (residues 36 to 169) of Ybr137wp does not display striking conformational disparity for other GlcG-like proteins, there are several minor structure differences in the loop regions.
FIG 3.
Sequence alignment and structural comparison. (A) Sequence of Ybr137wp compared with those of HbpS and a K. pneumoniae protein of unknown function (PDB accession number 2A2L). Numbering of residue is based on Ybr137wp. Secondary structural elements of helices and strands are shown in red and orange, respectively. α1 and α2 helices are observed only Ybr137wp. Identical amino acid residues are highlighted in green and yellow when the sequences are identical among all three and between two proteins, respectively. Sequences in blue are amino acids that have similarity between any two proteins. Sequence alignment was performed by using the Biology Work Bench from the San Diego Supercomputer Center (SDSC) and displayed with the program BOXSHADE. The light brown color below the secondary structure pattern indicates the GlcG-like domain. The secondary structures are designated according to the crystal structure of Ybr137wp; however, residues 171 to 179 representing α-helices are the prediction result from Phyre2. (B) Stereo view for structural comparison of Ybr137wp (blue), HbpS (magenta), and the protein reported under PDB accession number 2A2L (green).
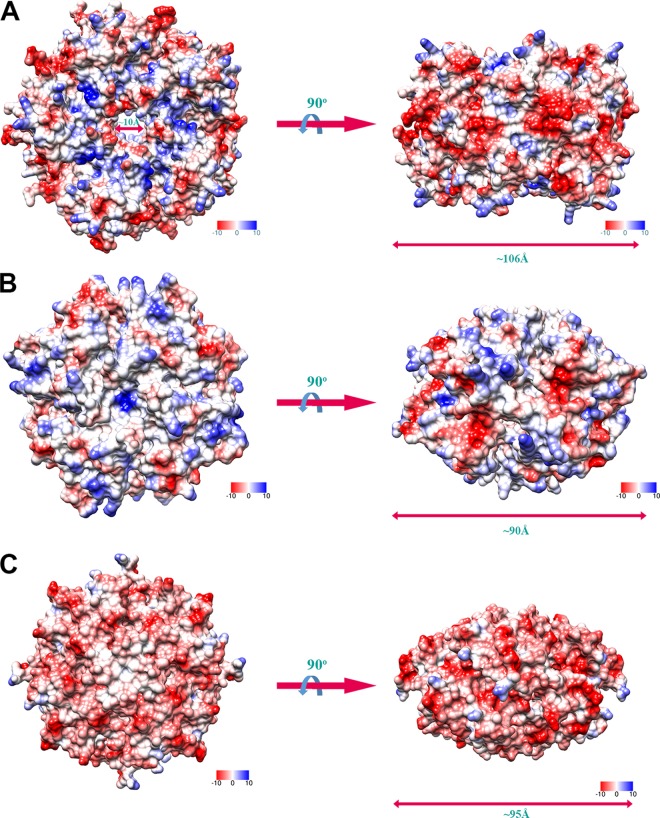
We also noted that Ybr137wp has a decameric structure, while HbpS and the K. pneumoniae protein form octamers in crystals. The Ybr137wp decamer is ∼106 Å in diameter and contains a pore through its middle with a diameter of ∼10 Å (Fig. 4). However, both HbpS and the K. pneumoniae protein did not form a pore, and the diameter is smaller than that of the Ybr137wp decamer (Fig. 4B and C). We compared the surface potentials of these three oligomers. As shown in Fig. 4, the negatively and positively charged residues are almost equally distributed on the surface of the Ybr137wp decamer, while the HbpS octamer has more positive charge potential on the protein surface than Ybr137wp and the K. pneumoniae protein. Surprisingly, the surface potential of the K. pneumoniae protein is mainly negatively charged. These results indicated that these three proteins may be involved in different pathways with different functions.
FIG 4.
Electrostatic surface potential of Ybr137wp in a decameric conformation (A), HbpS from Streptomyces reticuli in an octameric conformation (B), and a protein from Klebsiella pneumoniae with unknown function in an octameric conformation (PDB accession number 2A2L) (C). Positively and negatively charged residues are shown in blue and red, respectively.
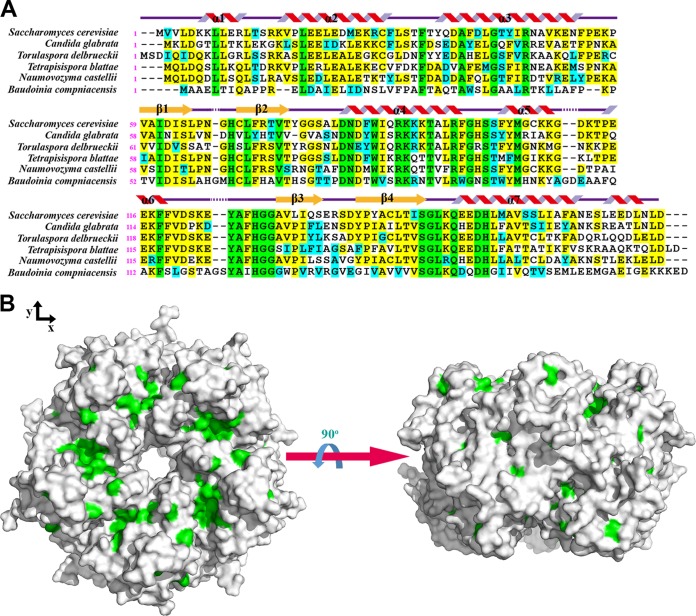
We also compared several species containing Ybr137wp-like proteins by sequence alignment. Ybr137wp can be found only in fungi. We selected Candida albicans, Torulaspora delbrueckii, Tetrapisispora phaffii, Naumovozyma castellii, and Baudoinia compniacensis for comparison with Saccharomyces cerevisiae. Except for Baudoinia compniacensis, which belongs to the Teratosphaeriaceae family and is distantly related to Saccharomyces cerevisiae, all species are members of the Saccharomycetaceae family. As shown in Fig. 5, there are only 32 conserved residues in this comparison. Most of these residues are located near the protein-protein-interacting interfaces, such as H69, D83, and R97, as mentioned above. Due to the lack of high sequence similarity to any protein with a known function and quaternary structure, the current structural information does not allow us to predict a possible function for Ybr137wp.
FIG 5.
(A) Sequence alignment of Ybr137wp with other Ybr137wp-like proteins from the indicated species. Fully conserved residues are shown in green when the sequences are identical in all species, in yellow when the sequences are identical, and in blue when similarity existed. Sequence alignment was performed by using the Biology Work Bench from SDSC and was displayed with the program BOXSHADE. The secondary structure is based on the crystal structure of Ybr137wp. Since we did not observe the C-terminal structure (residue 171 to 179) of Ybr137wp, the α-helix represented here is from the prediction result by Phyre2. (B) Fully conserved residues are shown in green on the decamer surface of Ybr137wp, generated by PyMol.
Interaction between Sgt2 and Ybr137wp.
Coimmunoprecipitation studies have shown that Sgt2 associates with the Get3, Get4, and Get5 proteins and chaperone proteins such as Hsp104 and Hsp70 (20). Interestingly, a then-novel protein encoded by ybr137w was also found to associate with the Sgt2 complex (13, 17, 20). To characterize the interaction between Ybr137wp and Sgt2, we performed an in vitro binding assay using purified recombined proteins. We initially constructed His-tagged full-length Sgt2 containing the dimerization (residues 1 to 72), TPR (residues 95 to 227), and C-terminal TA protein-binding (residues 228 to 347) domains. However, during the course of purification, full-length Sgt2 was rapidly degraded, producing fragments with a molecular mass of ∼25 to 35 kDa. Therefore, we reconstructed a truncated Sgt2 lacking the unstable C-terminal domain (Sgt2ΔC) (residues 1 to 227) for binding assays. As shown in Fig. 2A, SEC results for Sgt2ΔC revealed a molecular mass matching the dimeric state of the protein, consistent with data from previous studies (20, 46). We also performed sedimentation velocity analysis by AUC, and the results revealed that Sgt2ΔC has a sedimentation coefficient of 3.2 S (Fig. 2C). The c(s) distribution analysis revealed that the molecular mass is 54 kDa and closely matches that of the dimeric conformation.
We next verified the association between Sgt2 and Ybr137wp by SEC. When Ybr137wp and Sgt2ΔC were mixed in a 1:1.5 molar ratio and then subjected to SEC, the Ybr137wp/Sgt2ΔC complex eluted between the 440- and 669-kDa markers in the presence of 100 mM NaCl (Fig. 2A). Because Ybr137wp is a decamer, the SEC results may indicate that the stoichiometry of the Ybr137wp/Sgt2ΔC complex trends toward 1:1. To further confirm the molecular mass of the Ybr137wp/Sgt2ΔC complex, sedimentation equilibrium experiments were performed. The resulting absorbance-versus-radial position plots at three rotor speeds are shown in Fig. 2E. All plots were globally fitted to a single discrete species model and represented best-fit distributions. The molecular mass of Ybr137wp/Sgt2ΔC in solution was determined to be 499,587 Da. This result is consistent with SEC results and again suggests that the molar ratio of Ybr137wp/Sgt2ΔC in the complex can be 1:1.
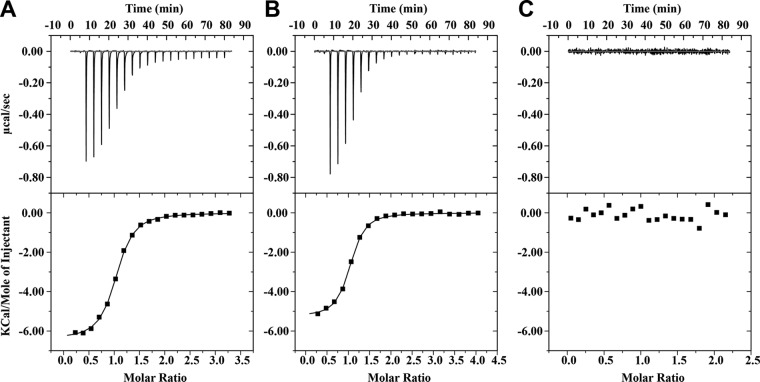
We next used isothermal titration calorimetry (ITC) to evaluate the affinity of binding between Sgt2 and Ybr137wp. Integration of the binding isotherms indicated that the dissociation constant (Kd) for Ybr137wp/Sgt2ΔC is 1.36 ± 0.09 μM in assay buffer containing 20 mM Tris-HCl (pH 8.0) and 100 mM NaCl (Fig. 6A). The resulting stoichiometry of 0.99 indicated that one Ybr137w decamer is capable of interacting with five Sgt2ΔC dimers. To directly assess the role of the Sgt2 dimerization domain in Ybr137wp/Sgt2ΔC complex formation, we performed ITC using Sgt2TPR and Ybr137wp and found that the binding affinity and stoichiometry of this complex were similar to those of the Ybr137wp/Sgt2ΔC complex and that the Kd is 1.38 ± 0.09 μM (Fig. 6B). This result indicated that the Sgt2 dimerization domain does not interact directly with Ybr137wp.
FIG 6.
Isothermal titration calorimetry data reveal Sgt2 binding to Ybr137wp. (A) Sgt2ΔC titrated into Ybr137wp; (B) Sgt2TPR titrated into Ybr137wp; (C) Sgt2TPR titrated into Ybr137wpΔC.
Previous studies indicated that the acidic C-terminal DDLD motif of Hsp104 is responsible for its association with other proteins (17). We noted that Ybr137wp also contains an acidic motif, EEDL, in its C-terminal region. To assess whether Ybr137wp uses a similar motif to bind the TPR domain of Sgt2, we generated a Ybr137wp construct lacking the C-terminal EEDL motif (residues 1 to 170) (Ybr137wpΔC) and used it for binding assays with ITC detection. The ITC results indicated that Ybr137ΔC and Sgt2 do not associate (Fig. 6C). We also carried out SEC to examine if Ybr137ΔC can form a complex with Sgt2TPR. We mixed Ybr137ΔC with Sgt2TPR at a molar ratio of 1:1. The SEC results indicate that no peak higher than the Ybr137wΔC decamer molecular mass can be detected (Fig. 2A) and are consistent with the ITC results. Taken together, the results presented here strongly suggest that Ybr137wp interacts with the TPR domain of Sgt2 via its C-terminal acidic motif. This result is consistent with our structure, since the C-terminal region of Ybr137wp protrudes from the surface of the decamer. However, these results do not delineate the function of Ybr137wp. Therefore, we performed in vivo experiments to determine whether Ybr137wp participates in the GET pathway.
Ybr137wp mediates GET-dependent defects.
Numerous studies have demonstrated that deletions of GET proteins directly affect TA protein delivery to membranes. The mutants form puncta containing the TA proteins or have a decreased efficiency of inserting the TA protein into the ER membrane (9, 47). Due to the various functions of TA proteins, deletion of GET proteins can result in a wide range of phenotypic changes, such as increased sensitivity to temperature and antibiotics and secretion of ER-resident proteins (9, 48, 49). To understand the involvement of Ybr137wp in the GET system, we assessed the viability of yeast cells with a knockout of ybr137w or a gene encoding a GET protein at both 30°C and 40°C. As shown in Fig. 7A, strains with a get3, get5, or sgt2 deletion have decreased yeast cell viability at 40°C, although normal viability was maintained at 30°C. Conversely, cells of a strain with a ybr137w deletion behave as wild-type yeast cells at elevated temperatures. Interestingly, the severity of the viability defect for each strain was dependent on the missing protein, as follows: Get3 > Get5 > Sgt2. This order was also observed for GFP-Sed5 mislocalization and hygromycin B sensitivity in GET protein-deleted cell lines (17).
FIG 7.
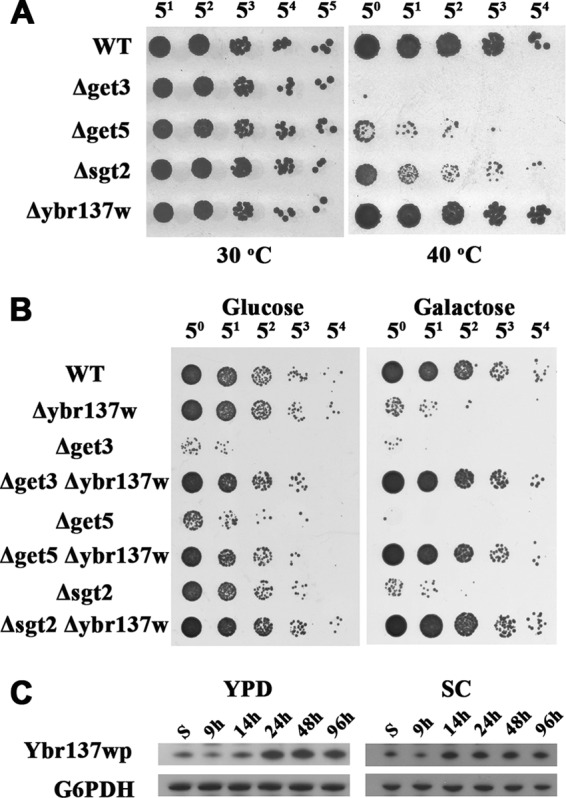
Genetic interactions of ybr137w and get genes. (A) S. cerevisiae wild-type or mutant cells, as indicated, were incubated at 30°C or 40°C on YPD plates. (B) After 5 days of incubation in SC medium with glucose or galactose, cells were serially diluted and incubated on YPD plates. (C) Time course of Ybr137wp expression in yeast. The expression levels of endogenous Ybr137wp in wild-type S. cerevisiae incubated in YPD (left) and SC (right) media were detected by using rabbit anti-Ybr137w antiserum. S indicates cell lysate after subculture.
Since the viability of each GET protein deletion strain was different at 40°C, the temperature sensitivity may have obscured the involvement of Ybr137wp in the GET pathway. Thus, we assessed the influence of ybr137w on other GET pathway-dependent phenotypes. A previous study showed that deletions of two members of the GET pathway can further enhance the temperature and antibiotic sensitivities of the affected yeast (9). We next examined the growth defect using cells with a ybr137w and GET protein double deletion. We found that yeast cells with a get3, get5, or sgt2 deletion have low viability at day 5 in synthetic complete (SC) medium at 30°C (Fig. 7B, left). In contrast, the viability of cells of the ybr137w deletion strain was comparable to that of WT cells. Surprisingly, cells with GET gene and ybr137w double deletions behaved similarly to WT cells (Fig. 7B, left).
We also noted that the amount of Ybr137wp was small in log-phase cultures but increased significantly after culturing for 14 h and reached a maximal level at 24 h in YPD medium, whereas it increased to a maximal level at 14 h in SC medium (Fig. 7C). The time point of increasing Ybr137w expression levels matches the diauxic shift of yeast, at which point glucose is mostly consumed. This phenomenon suggests that Ybr137wp might modulate the function of proteins in the GET pathway when the cells exhaust the nutrients in the medium. To test this hypothesis, we assessed the viability of ybr137w and GET pathway protein double-deletion mutants in SC medium with galactose as the carbon source. As shown in Fig. 7B (right), all single-deletion strains, including the Δybr137w strain, had lower cell viability in SC medium containing galactose instead of glucose. Nevertheless, the ybr137w deletion can rescue Get3, Get5, or Sgt2 deletion strains in galactose-containing medium (Fig. 7B, right). These results suggest that Ybr137wp in conjunction with the GET pathway may also function as a response to starvation.
Recently, Powis et al. demonstrated that all cytosolic components of the GET system are translocated into a distinct focus containing TA protein in the absence of glucose (50). Thus, we investigated whether Ybr137wp affects the formation of TA protein-containing foci. The ER membrane proteins Sec22 and SCS2 have been used as model substrates to study the delivery of TA proteins by the GET system (9, 13). We established yeast strains expressing Flag-tagged Sec22 or GFP-tagged SCS2 in WT and Δget3 yeast cells. As shown in Fig. 8A, the tagged Sec22 and SCS2 proteins localized mainly at the perinuclear ER and cortical ER in the wild-type strain. In contrast, noticeable fractions of puncta containing Sec22 or SCS2 were observed in the Δget3 strain, implying a defect in TA protein delivery (Fig. 8A and B). However, deletion of ybr137w alone did not affect the distribution of Sec22 and SCS2. Interestingly, deletion of both ybr137w and get3 decreased the formation of Sec22 and SCS2 puncta significantly compared to that in cells with only a get3 deletion (Fig. 8A and B). This rescue effect of the ybr137w deletion on punctum formation is consistent with our findings of viability defects, as shown in Fig. 7. To further confirm the effect of Ybr137wp on this phenotype, we transformed a plasmid bearing the ybr137w gene driven by an endogenous promoter. Ybr137wp expressed in Δget3 Δybr137w double-deletion cells restored the formation of TA protein puncta (Fig. 8A). Taken together, it appears that Ybr137wp expression is induced in yeast cells exiting the log phase and mediates the functions of the GET system in terms of TA protein delivery and cell viability.
FIG 8.
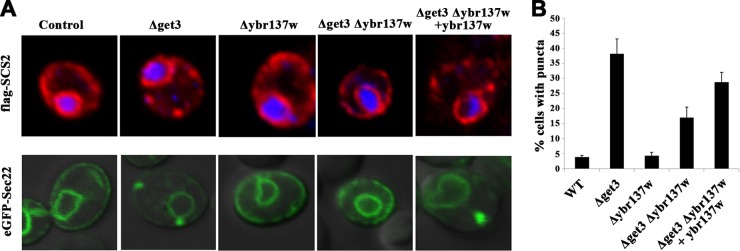
Localization of TA proteins in S. cerevisiae. (A) Subcellular localization of Flag-SCS2 (top) and eGFP-Sec22 (bottom) expressed in wild-type S. cerevisiae or the indicated strains after 2 days in SC medium. (B) Quantitative analysis of the formation of Sec22 puncta. Error bars represent the means ± standard deviations from several hundred cells in three independent experiments.
DISCUSSION
TRC40/Asna1 was first discovered in mammalian cells in 2007 as an ATPase responsible for efficient delivery of TA proteins to ER membrane. Its yeast homolog, Get3, and related proteins (Get1, Get2, Get4, Get5, and Sgt2) in the GET system were subsequently identified (9, 11, 13, 20, 51). Substantial evidence has demonstrated that Sgt2 acts upstream of the GET pathway (13, 17). Sgt2 is complexed with Get4 and Get5. The C-terminal methionine-rich domain of Sgt2 binds the TA protein and then transfers it to Get3 for ER membrane targeting with the aid of the Get4/Get5 complex. Moreover, the TPR domain of Sgt2 binds chaperones such as Hsp104 and Hsp70 (SSA1 in yeast) (13). It has been shown that Hsp70 can stimulate TA protein integration in vitro (9). Thus, it is speculated that Sgt2 can also recruit chaperones to prevent the aggregation of newly synthesized TA proteins before Get3 binding.
In this report, we demonstrated that Ybr137wp and the Sgt2 TPR domain interact directly with a remarkable binding affinity (Fig. 6). Given the facts that removal of the TPR domain on Sgt2 has no effect on the distribution of Sed5 (a Golgi membrane-residing TA protein) in yeast (17) and that Sgt2 with a mutation at the TPR domain can still load the TA protein Sec22 onto Get3 in vitro (13), the biological function of the TPR domain of Sgt2 and the role of its association with Ybr137wp in the GET pathway remain elusive. Indeed, the role of Ybr137wp in the delivery of TA proteins was assessed by using the temperature-sensitive phenotype commonly found in yeast when any protein in its GET pathway is deleted (Fig. 7A). The results are consistent with those of a previous study (17) and indicate that Ybr137wp is not essential for routine TA protein targeting under normal conditions. However, we observed that the Ybr137wp expression level is lower in yeast cells grown in log phase under normal conditions but was significantly increased in yeast after 9 h of culturing in SC medium or after 24 h of culturing in YPD medium. These time points of increasing Ybr137wp expression levels coincide with the diauxic shift of yeast. Previous studies indicated that the glucose concentration is dramatically decreased to exhausted levels in minimal defined (MD) limiting medium at 9 h (52), whereas glucose is exhausted in rich YPD medium after 18 h of culturing (53). In both studies, the mRNA levels of ybr137w were found to be transiently and significantly increased during the diauxic shift and reduced after this shift. We furthermore found that the Ybr137wp protein levels during the diauxic shift in limited and rich media are maintained through later phases, suggesting a possible function of Ybr137wp in these later phases.
The TPR domain of Sgt2 is also involved in the homeostasis of other proteins (54, 55). A recent study revealed that Sgt2 interacts with two key chaperones (Ssa1 and Hsp104) involved in prion pathogenesis and affects prion formation in yeast (56). Furthermore, the mammalian homolog of Sgt2, SGTA, antagonized the Bag6-dependent ubiquitination of mislocalized proteins (55). The protein interaction networks and the time when the gene is expressed often provide strong clue to its biological function. Therefore, it is suggested that Sgt2 can recruit or associate with different proteins to participate in or modulate numerous cellular functions, especially under stress conditions. In support of this hypothesis, a previous study showed that Get3 as well as other cytosolic members of the GET system translocated into distinct foci in response to starvation (50). Under nutritional stress, the risk of protein aggregation increases because less ATP is available for chaperone activity (50). Therefore, it was suggested that TA proteins would be actively translocated into Get3-containing foci and held by Get3 to protect them from aggregation and that the activity of the GET system might be regulated in response to starvation. In our study, we found that the formation of aggregated Sec22 or SCS2 in yeast with an impaired GET system depends on the presence of Ybr137wp after 2 days of culture. Deletion of ybr137w and the gene encoding Get3 significantly prevented TA protein aggregation (Fig. 8). This result indicated that Ybr137wp expressed in later phases plays a role in the GET system. Nevertheless, the rescue effect on punctum formation by Ybr137wp also appeared to affect the viability of yeast in long-term (>3-day) cultures, which would be a newly identified phenotype associated with Get3, Get5, and Sgt2. Since Ybr137wp shared the same binding site with Hsp104 and Hsp70 on the Sgt2TPR domain, the deletion of ybr137w might increase the occupancy of chaperones on Sgt2. Therefore, it is possible that more Hsp104 is recruited to Sgt2 to disaggregate TA protein puncta in the absence of Ybr137wp and rescue the viability defect in the Δget3 strain. Taken together, we suggest that Ybr137wp, when bound to Sgt2, modulates the function of the GET system under starvation conditions. However, the detailed molecular mechanism for this function remains to be addressed.
ACKNOWLEDGMENTS
We thank Ming F. Tam for helpful discussion and suggestions. All crystallographic data were collected at Beamline 13B1 at the National Synchrotron Radiation Research Center (NSRRC) in Taiwan. We also acknowledge the use of Analytical Ultracentrifugation XL-A (AUC) in the Biophysics Core Facility, Scientific Instrument Center, Academia Sinica.
This work was supported by research grants from Academia Sinica and the National Science Council (NSC101-2311-B-001-022-MY3 to C.-D.H.), Taiwan.
Footnotes
Published ahead of print 6 October 2014
REFERENCES
- 1.Borgese N, Brambillasca S, Soffientini P, Yabal M, Makarow M. 2003. Biogenesis of tail-anchored proteins. Biochem. Soc. Trans. 31:1238–1242. 10.1042/BST0311238. [DOI] [PubMed] [Google Scholar]
- 2.Chen YA, Scheller RH. 2001. SNARE-mediated membrane fusion. Nat. Rev. Mol. Cell Biol. 2:98–106. 10.1038/35052017. [DOI] [PubMed] [Google Scholar]
- 3.Kutay U, Ahnert-Hilger G, Hartmann E, Wiedenmann B, Rapoport TA. 1995. Transport route for synaptobrevin via a novel pathway of insertion into the endoplasmic reticulum membrane. EMBO J. 14:217–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wattenberg B, Lithgow T. 2001. Targeting of C-terminal (tail)-anchored proteins: understanding how cytoplasmic activities are anchored to intracellular membranes. Traffic 2:66–71. 10.1034/j.1600-0854.2001.20108.x. [DOI] [PubMed] [Google Scholar]
- 5.High S, Abell BM. 2004. Tail-anchored protein biosynthesis at the endoplasmic reticulum: the same but different. Biochem. Soc. Trans. 32:659–662. 10.1042/BST0320659. [DOI] [PubMed] [Google Scholar]
- 6.Borgese N, Fasana E. 2011. Targeting pathways of C-tail-anchored proteins. Biochim. Biophys. Acta 1808:937–946. 10.1016/j.bbamem.2010.07.010. [DOI] [PubMed] [Google Scholar]
- 7.Borgese N, Gazzoni I, Barberi M, Colombo S, Pedrazzini E. 2001. Targeting of a tail-anchored protein to endoplasmic reticulum and mitochondrial outer membrane by independent but competing pathways. Mol. Biol. Cell 12:2482–2496. 10.1091/mbc.12.8.2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Borgese N, Colombo S, Pedrazzini E. 2003. The tale of tail-anchored proteins: coming from the cytosol and looking for a membrane. J. Cell Biol. 161:1013–1019. 10.1083/jcb.200303069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schuldiner M, Metz J, Schmid V, Denic V, Rakwalska M, Schmitt HD, Schwappach B, Weissman JS. 2008. The GET complex mediates insertion of tail-anchored proteins into the ER membrane. Cell 134:634–645. 10.1016/j.cell.2008.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Favaloro V, Spasic M, Schwappach B, Dobberstein B. 2008. Distinct targeting pathways for the membrane insertion of tail-anchored (TA) proteins. J. Cell Sci. 121:1832–1840. 10.1242/jcs.020321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stefanovic S, Hegde RS. 2007. Identification of a targeting factor for posttranslational membrane protein insertion into the ER. Cell 128:1147–1159. 10.1016/j.cell.2007.01.036. [DOI] [PubMed] [Google Scholar]
- 12.Wang F, Whynot A, Tung M, Denic V. 2011. The mechanism of tail-anchored protein insertion into the ER membrane. Mol. Cell 43:738–750. 10.1016/j.molcel.2011.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang F, Brown EC, Mak G, Zhuang J, Denic V. 2010. A chaperone cascade sorts proteins for posttranslational membrane insertion into the endoplasmic reticulum. Mol. Cell 40:159–171. 10.1016/j.molcel.2010.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chartron JW, Clemons WM, Jr, Suloway CJ. 2012. The complex process of GETting tail-anchored membrane proteins to the ER. Curr. Opin. Struct. Biol. 22:217–224. 10.1016/j.sbi.2012.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Egan B, Beilharz T, George R, Isenmann S, Gratzer S, Wattenberg B, Lithgow T. 1999. Targeting of tail-anchored proteins to yeast mitochondria in vivo. FEBS Lett. 451:243–248. 10.1016/S0014-5793(99)00581-5. [DOI] [PubMed] [Google Scholar]
- 16.Jonikas MC, Collins SR, Denic V, Oh E, Quan EM, Schmid V, Weibezahn J, Schwappach B, Walter P, Weissman JS, Schuldiner M. 2009. Comprehensive characterization of genes required for protein folding in the endoplasmic reticulum. Science 323:1693–1697. 10.1126/science.1167983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kohl C, Tessarz P, von der Malsburg K, Zahn R, Bukau B, Mogk A. 2011. Cooperative and independent activities of Sgt2 and Get5 in the targeting of tail-anchored proteins. Biol. Chem. 392:601–608. 10.1515/BC.2011.066. [DOI] [PubMed] [Google Scholar]
- 18.Chartron JW, Gonzalez GM, Clemons WM., Jr 2011. A structural model of the Sgt2 protein and its interactions with chaperones and the Get4/Get5 complex. J. Biol. Chem. 286:34325–34334. 10.1074/jbc.M111.277798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chartron JW, Suloway CJ, Zaslaver M, Clemons WM., Jr 2010. Structural characterization of the Get4/Get5 complex and its interaction with Get3. Proc. Natl. Acad. Sci. U. S. A. 107:12127–12132. 10.1073/pnas.1006036107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chang YW, Chuang YC, Ho YC, Cheng MY, Sun YJ, Hsiao CD, Wang C. 2010. Crystal structure of Get4-Get5 complex and its interactions with Sgt2, Get3, and Ydj1. J. Biol. Chem. 285:9962–9970. 10.1074/jbc.M109.087098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bozkurt G, Wild K, Amlacher S, Hurt E, Dobberstein B, Sinning I. 2010. The structure of Get4 reveals an alpha-solenoid fold adapted for multiple interactions in tail-anchored protein biogenesis. FEBS Lett. 584:1509–1514. 10.1016/j.febslet.2010.02.070. [DOI] [PubMed] [Google Scholar]
- 22.Liou ST, Cheng MY, Wang C. 2007. SGT2 and MDY2 interact with molecular chaperone YDJ1 in Saccharomyces cerevisiae. Cell Stress Chaperones 12:59–70. 10.1379/CSC-220R.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stefer S, Reitz S, Wang F, Wild K, Pang YY, Schwarz D, Bomke J, Hein C, Lohr F, Bernhard F, Denic V, Dotsch V, Sinning I. 2011. Structural basis for tail-anchored membrane protein biogenesis by the Get3-receptor complex. Science 333:758–762. 10.1126/science.1207125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mariappan M, Mateja A, Dobosz M, Bove E, Hegde RS, Keenan RJ. 2011. The mechanism of membrane-associated steps in tail-anchored protein insertion. Nature 477:61–66. 10.1038/nature10362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kordes E, Savelyeva L, Schwab M, Rommelaere J, Jauniaux JC, Cziepluch C. 1998. Isolation and characterization of human SGT and identification of homologues in Saccharomyces cerevisiae and Caenorhabditis elegans. Genomics 52:90–94. 10.1006/geno.1998.5385. [DOI] [PubMed] [Google Scholar]
- 26.Tobaben S, Varoqueaux F, Brose N, Stahl B, Meyer G. 2003. A brain-specific isoform of small glutamine-rich tetratricopeptide repeat-containing protein binds to Hsc70 and the cysteine string protein. J. Biol. Chem. 278:38376–38383. 10.1074/jbc.M301558200. [DOI] [PubMed] [Google Scholar]
- 27.Liou ST, Wang C. 2005. Small glutamine-rich tetratricopeptide repeat-containing protein is composed of three structural units with distinct functions. Arch. Biochem. Biophys. 435:253–263. 10.1016/j.abb.2004.12.020. [DOI] [PubMed] [Google Scholar]
- 28.Worrall LJ, Wear MA, Page AP, Walkinshaw MD. 2008. Cloning, purification and characterization of the Caenorhabditis elegans small glutamine-rich tetratricopeptide repeat-containing protein. Biochim. Biophys. Acta 1784:496–503. 10.1016/j.bbapap.2007.12.003. [DOI] [PubMed] [Google Scholar]
- 29.Yoshida K, Blobel G. 2001. The karyopherin Kap142p/Msn5p mediates nuclear import and nuclear export of different cargo proteins. J. Cell Biol. 152:729–740. 10.1083/jcb.152.4.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Laue TM, Shah BD, Ridgeway TM, Pelletier SL. 1992. Computer-aided interpretation of analytical sedimentation data for proteins, p 90–125 In Harding SE, Rowe AJ, Horton JC. (ed), Analytical ultracentrifugation in biochemistry and polymer science. Royal Society of Chemistry, Cambridge, United Kingdom. [Google Scholar]
- 31.Schuck P. 2000. Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and Lamm equation modeling. Biophys. J. 78:1606–1619. 10.1016/S0006-3495(00)76713-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vistica J, Dam J, Balbo A, Yikilmaz E, Mariuzza RA, Rouault TA, Schuck P. 2004. Sedimentation equilibrium analysis of protein interactions with global implicit mass conservation constraints and systematic noise decomposition. Anal. Biochem. 326:234–256. 10.1016/j.ab.2003.12.014. [DOI] [PubMed] [Google Scholar]
- 33.Otwinowski Z, Minor W. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276:307–326. 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 34.Otwinowski Z, Minor W. 2001. Denzo & Scalepack, p 226–235 In Rossman MG, Arnold E. (ed), International tables for crystallography, vol F Macromolecular crystallography Kluwer Academic Publishers, Dordrecht, Netherlands. [Google Scholar]
- 35.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. 2010. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66:213–221. 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Echols N, Headd JJ, Hung LW, Jain S, Kapral GJ, Grosse Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner RD, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. 2011. The Phenix software for automated determination of macromolecular structures. Methods 55:94–106. 10.1016/j.ymeth.2011.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Emsley P, Cowtan K. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60:2126–2132. 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 38.Potterton E, Briggs P, Turkenburg M, Dodson E. 2003. A graphical user interface to the CCP4 program suite. Acta Crystallogr. D Biol. Crystallogr. 59:1131–1137. 10.1107/S0907444903008126. [DOI] [PubMed] [Google Scholar]
- 39.Krissinel E, Henrick K. 2007. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372:774–797. 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 40.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. 2004. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25:1605–1612. 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 41.Schrodinger LLC. 2010. The PyMOL molecular graphics system, version 1.3r1. Schrodinger LLC, New York, NY. [Google Scholar]
- 42.Kelley LA, Sternberg MJ. 2009. Protein structure prediction on the Web: a case study using the Phyre server. Nat. Protoc. 4:363–371. 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- 43.Winzeler EA, Shoemaker DD, Astromoff A, Liang H, Anderson K, Andre B, Bangham R, Benito R, Boeke JD, Bussey H, Chu AM, Connelly C, Davis K, Dietrich F, Dow SW, El Bakkoury M, Foury F, Friend SH, Gentalen E, Giaever G, Hegemann JH, Jones T, Laub M, Liao H, Liebundguth N, Lockhart DJ, Lucau-Danila A, Lussier M, M'Rabet N, Menard P, Mittmann M, Pai C, Rebischung C, Revuelta JL, Riles L, Roberts CJ, Ross-MacDonald P, Scherens B, Snyder M, Sookhai-Mahadeo S, Storms RK, Veronneau S, Voet M, Volckaert G, Ward TR, Wysocki R, Yen GS, Yu K, Zimmermann K, Philippsen P, Johnston M, Davis RW. 1999. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285:901–906. 10.1126/science.285.5429.901. [DOI] [PubMed] [Google Scholar]
- 44.Ortiz de Orue Lucana D, Bogel G, Zou P, Groves MR. 2009. The oligomeric assembly of the novel haem-degrading protein HbpS is essential for interaction with its cognate two-component sensor kinase. J. Mol. Biol. 386:1108–1122. 10.1016/j.jmb.2009.01.017. [DOI] [PubMed] [Google Scholar]
- 45.Andreeva A, Howorth D, Chandonia JM, Brenner SE, Hubbard TJ, Chothia C, Murzin AG. 2008. Data growth and its impact on the SCOP database: new developments. Nucleic Acids Res. 36:D419–D425. 10.1093/nar/gkm993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chang YW, Lin TW, Li YC, Huang YS, Sun YJ, Hsiao CD. 2012. Interaction surface and topology of Get3-Get4-Get5 protein complex, involved in targeting tail-anchored proteins to endoplasmic reticulum. J. Biol. Chem. 287:4783–4789. 10.1074/jbc.M111.318329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yamagata A, Mimura H, Sato Y, Yamashita M, Yoshikawa A, Fukai S. 2010. Structural insight into the membrane insertion of tail-anchored proteins by Get3. Genes Cells 15:29–41. 10.1111/j.1365-2443.2009.01362.x. [DOI] [PubMed] [Google Scholar]
- 48.Schuldiner M, Collins SR, Thompson NJ, Denic V, Bhamidipati A, Punna T, Ihmels J, Andrews B, Boone C, Greenblatt JF, Weissman JS, Krogan NJ. 2005. Exploration of the function and organization of the yeast early secretory pathway through an epistatic miniarray profile. Cell 123:507–519. 10.1016/j.cell.2005.08.031. [DOI] [PubMed] [Google Scholar]
- 49.Hu Z, Potthoff B, Hollenberg CP, Ramezani-Rad M. 2006. Mdy2, a ubiquitin-like (UBL)-domain protein, is required for efficient mating in Saccharomyces cerevisiae. J. Cell Sci. 119:326–338. 10.1242/jcs.02754. [DOI] [PubMed] [Google Scholar]
- 50.Powis K, Schrul B, Tienson H, Gostimskaya I, Breker M, High S, Schuldiner M, Jakob U, Schwappach B. 2013. Get3 is a holdase chaperone and moves to deposition sites for aggregated proteins when membrane targeting is blocked. J. Cell Sci. 126:473–483. 10.1242/jcs.112151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Copic A, Dorrington M, Pagant S, Barry J, Lee MC, Singh I, Hartman JL, IV, Miller EA. 2009. Genomewide analysis reveals novel pathways affecting endoplasmic reticulum homeostasis, protein modification and quality control. Genetics 182:757–769. 10.1534/genetics.109.101105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Brauer MJ, Saldanha AJ, Dolinski K, Botstein D. 2005. Homeostatic adjustment and metabolic remodeling in glucose-limited yeast cultures. Mol. Biol. Cell 16:2503–2517. 10.1091/mbc.E04-11-0968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.DeRisi JL, Iyer VR, Brown PO. 1997. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science 278:680–686. 10.1126/science.278.5338.680. [DOI] [PubMed] [Google Scholar]
- 54.Xu Y, Cai M, Yang Y, Huang L, Ye Y. 2012. SGTA recognizes a noncanonical ubiquitin-like domain in the Bag6-Ubl4A-Trc35 complex to promote endoplasmic reticulum-associated degradation. Cell Rep. 2:1633–1644. 10.1016/j.celrep.2012.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Leznicki P, High S. 2012. SGTA antagonizes BAG6-mediated protein triage. Proc. Natl. Acad. Sci. U. S. A. 109:19214–19219. 10.1073/pnas.1209997109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kiktev DA, Patterson JC, Muller S, Bariar B, Pan T, Chernoff YO. 2012. Regulation of chaperone effects on a yeast prion by cochaperone Sgt2. Mol. Cell. Biol. 32:4960–4970. 10.1128/MCB.00875-12. [DOI] [PMC free article] [PubMed] [Google Scholar]