Abstract
Ribosome biogenesis is a multistep cellular pathway that involves more than 200 regulatory components to ultimately generate translation-competent 80S ribosomes. The initial steps of this process, particularly rRNA processing, take place in the nucleolus, while later stages occur in the nucleoplasm and cytoplasm. One critical factor of 28S rRNA maturation is the SUMO-isopeptidase SENP3. SENP3 tightly interacts with the nucleolar scaffold protein NPM1 and is associated with nucleolar 60S preribosomes. A central question is how changes in energy supply feed into the regulation of ribosome maturation. Here, we show that the nutrient-sensing mTOR kinase pathway controls the nucleolar targeting of SENP3 by regulating its interaction with NPM1. We define an N-terminal domain in SENP3 as the critical NPM1 binding region and provide evidence that mTOR-mediated phosphorylation of serine/threonine residues within this region fosters the interaction of SENP3 with NPM1. The inhibition of mTOR triggers the nucleolar release of SENP3, thereby likely compromising its activity in rRNA processing. Since mTOR activity is tightly coupled to nutrient availability, we propose that this pathway contributes to the adaptation of ribosome maturation in response to the cellular energy status.
INTRODUCTION
Ribosomes are large ribonucleoprotein complexes functioning as molecular machines in protein synthesis. Mammalian 80S ribosomes are assembled from the small 40S and the large 60S subunit (1). The 60S subunit is composed of the 28S, the 5.8S, and the 5S rRNA and contains at least 46 ribosomal proteins, while the 40S subunit consists of around 30 ribosomal proteins and the 18S rRNA. The maturation of ribosomes is a highly regulated pathway that involves more than 200 nonribosomal proteins, commonly termed trans-acting factors (2, 3). Ribosome biogenesis is initiated in the nucleolus by RNA polymerase I (Pol I), transcribing a 47S precursor rRNA. This precursor is incorporated into a nucleolar 90S preribosomal particle, where specific bases in the rRNA are covalently modified and initial pre-RNA processing steps take place (4, 5). The 40S and 60S ribosomal subunits subsequently take a separate maturation pathway, which proceeds by a number of processing and remodelling events in the nucleolus and the nucleoplasm. Following export to the cytoplasm, both subunits are assembled to translation-competent 80S particles.
As an integral part of protein synthesis, ribosome biogenesis must be tightly coupled to the energy status and nutrient levels of the cell. One key pathway that coordinates energy supply with protein translation is the mammalian target of rapamycin (mTOR) signaling network (6, 7). mTOR is an evolutionarily conserved serine/threonine kinase. In mammalian cells, mTOR is found in two distinct complexes, mTORC1 and mTORC2, that contain specific regulatory subunits and adaptor proteins. For example, Raptor and Rictor are the protein components involved in substrate recruitment and complex assembly of mTORC1 and mTORC2, respectively. mTORC1 positively regulates many anabolic processes, with the role of mTORC1 signaling in translational initiation being the best understood. More recently a function of mTOR signaling in ribosome biogenesis has also been proposed (8–12). In particular, mTOR has been implicated in the control of RNA polymerase I transcription and rRNA processing. However, the molecular basis and the targets of mTOR in the process of ribosome maturation are not well understood.
Here, we show that mTOR signaling is required for the nucleolar targeting of the SUMO-specific isopeptidase SENP3. SENP3 belongs to the SENP family of cysteine proteases, which is comprised of SENP1, SENP2, SENP3, SENP5, SENP6, and SENP7 (13, 14). Members of this family catalyze the removal of the ubiquitin-like SUMO modifier from target proteins. In humans, three related SUMO variants (SUMO1, SUMO2, and SUMO3) act as modifiers that are covalently attached to lysine residues. The conjugation of SUMO to target proteins provides a platform for protein-protein interactions and has a central role in a variety of cellular signaling pathways (15–17). We and others could demonstrate that balanced SUMO conjugation/deconjugation is an important regulatory mechanism for ribosome maturation in higher eukaryotes (18). A key factor in this process is SENP3, which associates with nucleolar pre-60S particles (19, 20). Importantly, the depletion of SENP3 strongly inhibits the processing of the 32S rRNA to the 28S rRNA and precludes the subsequent steps of pre-60S maturation (21, 22). SENP3 associates with the nucleolar scaffold protein NPM1 and a complex comprised of the 60S maturation factors PELP1, TEX10, WDR18, and Las1L. Several subunits of this complex, including NPM1, PELP1, and Las1L, were identified as critical targets of SENP3-mediated deSUMOylation (19, 20). Importantly, the loss of SENP3 leads to the nucleolar release of the PELP1-TEX10-WDR18 complex, suggesting that SENP3-mediated removal of SUMO2/3 from the PELP1 complex is critical for ribosome maturation.
Here, we provide evidence that the mTOR pathway regulates the nucleolar targeting of SENP3. mTOR-mediated phosphorylation of SENP3 facilitates the interaction with its nucleolar binding partner NPM1, thereby promoting nucleolar targeting. Our data suggest that this is a prerequisite for SENP3-dependent 28S rRNA maturation.
MATERIALS AND METHODS
Cell culture and transfection.
HeLa, HEK293T, and U2OS cells were cultured under standard conditions. Plasmid transfections were carried out using the calcium phosphate method for HEK293T cells and FuGeneHD (Roche) for HeLa and U2OS cells according to the manufacturers' instructions. When indicated, the cells were treated with 10 μM Ku-0063794 (Selleck Chemicals) for 6 h or with 2 μM rapamycin (Selleck Chemicals) for 6 h. Amino acid starvation was done in EBSS (Earle's balanced salt solution; Gibco). To inhibit the mTOR/Raptor/MSLT8 complex in the in vitro kinase assay, the complex was preincubated with 1 mM Torin1 (Selleck Chemicals) for 1 h.
For short interfering RNA (siRNA) knockdown experiments, HeLa cells were transfected with the respective siRNAs (120 pmol/well of a six-well plate, 3.5-cm diameter) using Oligofectamine (Invitrogen) according to the manufacturer's protocol. The following siRNA sequences (sense) were used: control, CGUACGCGGAAUACUUCGAdTdT; SENP3, CUGGCCCUGUCUCAGCCAUdTdT; NPM1, GGAAGUCUCUUUAAGAAAAdTdT; Rictor, ACUUGUGAAGAAUCGUAUCdTdT; Raptor, GATGAGGCTGATCTTACAGdTdT.
Cloning and mutagenesis.
Transient transfections of Flag-tagged expression constructs were done with the respective cDNAs cloned into pCI vector (Invitrogen). For bacterial purification of glutathione S-transferase (GST) fusion proteins, pGEX-4T-1 (GE Healthcare) vector was used. All other plasmids are described elsewhere (20, 21). For site-directed mutagenesis, a QuikChange mutagenesis kit (Stratagene) was used according to the manufacturer's instructions.
Expression of recombinant proteins, in vitro transcription/translation, and GST pulldown assays.
GST fusion proteins were expressed in Escherichia coli BL21 as described previously (23). For in vitro transcription/translation, the TnT quick coupled transcription/translation system from Promega was used. One microgram of pCI vector encoding the respective proteins was translated with [35S]methionine (Hartmann Analytic) according to the manufacturer's instructions. GST pulldown experiments were carried out with the in vitro-transcribed/translated products as described previously (24). Phosphatase treatment was done with 200 U of lambda protein phosphatase (New England BioLabs) for 1 h at 30°C according to the standard protocol.
Immunoprecipitation and Western blotting.
For immunoprecipitations of proteins from HeLa, HEK293T, or U2OS cells, cells were lysed in 50 mM HEPES (pH 7.4), 150 mM NaCl, 2 mM EDTA, 0.5% NP-40 containing protease inhibitor cocktail (Complete; Roche) and phosphatase inhibitor cocktail (PhosphoSTOP; Roche). The cell lysates were cleared and incubated overnight with anti-Flag beads (Sigma-Aldrich) for Flag-tagged proteins or with anti-NPM1 (Invitrogen) antibody for precipitating endogenous NPM1, followed by capture on Dynabeads (Life Technologies). Beads then were washed in lysis buffer, collected by centrifugation, and boiled with SDS-PAGE loading buffer for the elution of the bead-bound proteins. After separation by SDS-PAGE, Western blotting was done using ECL detection reagents (Millipore).
Immunofluorescence.
For immunofluorescence, HeLa cells were fixed in either 3.4% paraformaldehyde at room temperature or in methanol (MeOH) at −20°C for 10 min, permeabilized with 0.5% Triton X-100, and processed using standard protocols. Images were acquired with an LSM 510 Meta (Zeiss) and Leica TCS SP8. Primary antibodies used for detecting the respective proteins are listed below. The following secondary antibodies were used: Alexa Fluor 488–goat anti-rabbit (Invitrogen), Cy3–goat anti-mouse (Jackson ImmunoResearch), Alexa Fluor 555–goat anti-rabbit (Invitrogen), fluorescein isothiocyanate (FITC)–donkey anti-mouse (Jackson ImmunoResearch), and Alexa Fluor 488–goat anti-rat (Invitrogen) antibodies.
Antibodies used for Western blotting and immunofluorescence.
The following antibodies were used for Western blotting, immunoprecipitation, and immunofluorescence: anti-Flag (clone M2; Sigma-Aldrich), anti-Flag (F7425; Sigma-Aldrich), antivinculin (clone hVIN-1; Sigma-Aldrich), anti-PELP1 (A300-180A; Bethyl Laboratories), anti-SENP3 (D20A10 XP) (Cell Signaling), anti-NPM/B23 (clone NA24; Santa Cruz), anti-NPM (32-5200; Invitrogen), PES1 (rat monoclonal antibody; gift from Dirk Eick, Helmholtz Zentrum, Munich), anti-β-tubulin (clone E7; Developmental Studies Hybridoma Bank), anti-Rictor (2140S; Cell Signaling), anti-Raptor (2280S; Cell Signaling), and anti-p70S6K (T389) (9234S; Cell Signaling).
Metabolic labeling and analysis of rRNA.
Pulse-chase labeling was carried out in HeLa cells. Cells first were incubated in phosphate-free medium (Gibco) for 1 h and pulsed with 20 μCi of [32P]orthophosphoric acid (Hartmann Analytic) for 1 h at 37°C. The cells then were washed with phosphate-buffered saline (PBS) and chased in regular Dulbecco's modified Eagle medium (DMEM) for 1 h. RNA was extracted from cells using an RNA purification kit (Roche). Finally, 1 μg of RNA was run on a denaturing agarose gel and stained with ethidium bromide, and the different rRNA species were visualized by autoradiography. Quantification of the rRNA was performed with a Bio-Rad phosphorimager.
In vitro kinase assay.
HEK293T or HeLa cells were plated on a 15-cm dish and grown to 70% confluence. The cells then were transfected with 30 μg of the Flag-tagged constructs and grown for an additional 48 h. Flag-tagged proteins were immunoprecipitated from cells on anti-Flag–agarose beads and washed three times with kinase buffer (10 mM HEPES, pH 7.5, 50 mM NaCl, 50 mM β-glycerophosphate, 1 mM dithiothreitol [DTT], 10 mM MgCl2, 4 mM MnCl2). The bead-bound proteins then were incubated with 300 ng of a catalytic fragment (spanning amino acids [aa] 1362 to 2549) of human mTOR kinase (40061; BPS Bioscience) in the presence of [32P]ATP (0.1 μCi/μl) for 1 h at 30°C. In the experiment shown in Fig. 3B, Flag-SENP3 was incubated with 300 ng of the mTOR complex (40300; BPS Biosciences) consisting of mTOR (aa 1362 to 2549), Raptor, and mLST8 for 5 min at 30°C. After incubation, the beads were washed with kinase buffer and boiled with sample buffer to elute the proteins. The eluted proteins then were run on an SDS-PAGE gel and dried, and phosphorylation was detected by autoradiography.
FIG 3.
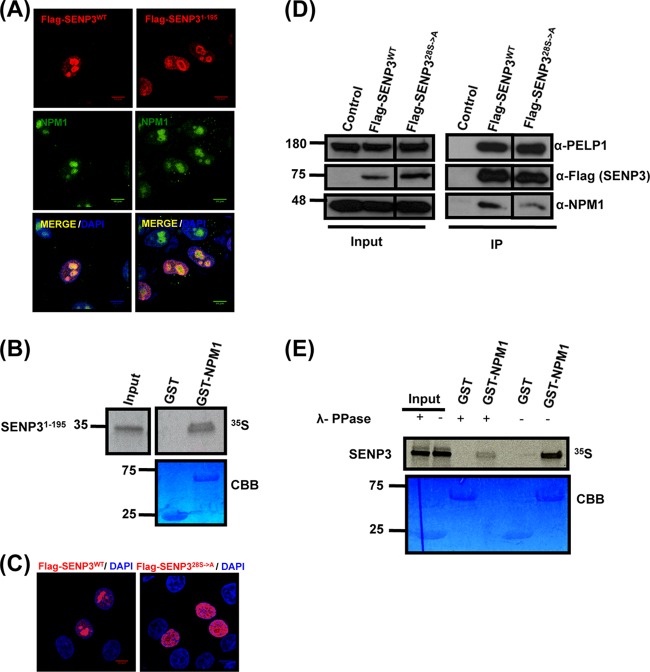
Localization and NPM1 interaction of SENP3 is determined by its N-terminal serine/threonine residues. (A) Full-length Flag-SENP3 and Flag-SENP31-195 were transiently expressed in HeLa cells, and their colocalization with NPM1 was determined by indirect immunofluorescence using anti-Flag and anti-NPM1 antibodies, respectively. (B) [35S]methionine-labeled SENP31-195 was generated by in vitro transcription/translation and tested for NPM1 interaction by GST pulldown using GST-NPM1 as the bait. CBB, Coomassie brilliant blue. (C) HeLa cells were transiently transfected with plasmids expressing wild-type Flag-SENP3 or Flag-SENP328S→A and stained with anti-Flag antibody for indirect immunofluorescence. (D) Interaction of wild-type Flag-SENP3 and Flag-SENP328S→A with endogenous NPM1 and PELP1 was monitored by Western blotting after immunoprecipitation of the transiently expressed Flag-tagged proteins in HeLa cells. The different lanes shown originate from the same blot taken at the same exposure times. (E) [35S]methionine-labeled SENP3 was generated by in vitro transcription/translation and either mock treated or treated with lambda phosphatase (λ-PPase). The proteins were used for a pulldown assay with either GST or GST-NPM1, and the interaction of SENP3 with NPM1 was detected by autoradiography.
MS/MS.
Flag-tagged SENP3 was immunoprecipitated from cells and in vitro phosphorylated using recombinant mTOR kinase. The eluted protein then was separated on an SDS-PAGE gel, and the bands at the size corresponding to SENP3 were excised and trypsin digested. The digested peptide mixture, at a concentration of 12.5 ng/μl, was desalted, purified by a C18 stage tip method, separated by online nano-liquid chromatography (LC), and analyzed using electrospray tandem mass spectrometry (MS/MS). The experiments were performed on an Easy-nLCII (Thermo Scientific) system connected to an LTQ Orbitrap elite mass spectrometer (Thermo Scientific) equipped with a nanoelectrospray ion source (Proxeon Biosystems, Odense, Denmark). MS/MS spectra were acquired in the ion trap by using the collision-induced dissociation (CID) mode. The peptides were separated with 60-min gradients from 5 to 35% acetonitrile in 0.5% acetic acid. The Orbitrap machine was operated in the data-dependent mode. A resolution value of 120,000 was set to acquire an MS scan. Up to 10 of the most intense ions prescan were selected for CID fragmentation. Selected target ions for MS/MS were dynamically excluded for 90 s.
Data analysis was carried out using MaxQuant software (version 1.2.2.5) supported by Mascot as the database search engine for peptide identifications. All data were filtered with a 1% false discovery rate (FDR) at the peptide level. MS/MS peak lists were filtered to contain, at most, six peaks per 100-Da interval searched by Andromeda against a concentrated forward and reverse version of UniProt and containing frequently observed contaminants, like human keratins. Initial precursor mass tolerance was set to 7 ppm, and the MS/MS mass tolerance was 0.5 Da. Phosphorylation (serine, threonine, tyrosine), N-acetyl protein, and oxidized methionine were searched as variable modifications, whereas cysteine carbamidomethylation was searched as a fixed modification.
RESULTS
Nucleolar localization of SENP3 is crucial for ribosome maturation.
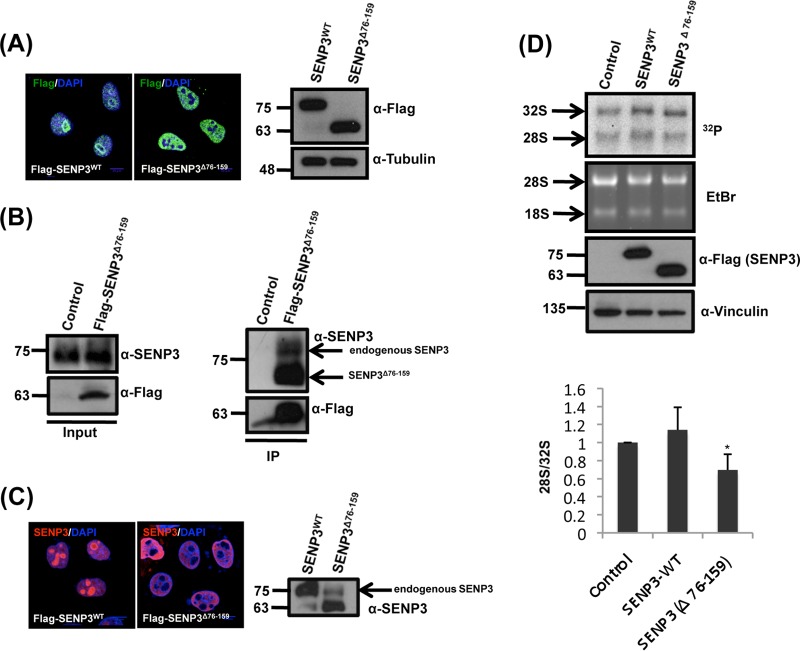
We and others have shown that SENP3 is central for ribosome maturation (21, 22). However, it has remained unclear whether this function depends on nucleolar localization of the protein. To address this point, we generated a deletion mutant of SENP3 (SENP3Δ76-159) that is excluded from the nucleolus and exhibits a strictly nucleoplasmic distribution when expressed as a Flag-tagged version in HeLa cells (Fig. 1A) (14, 25). In anti-Flag immunoprecipitation experiments, this variant is capable of binding the endogenous full-length SENP3 protein (Fig. 1B). Accordingly, due to its elevated protein level, the ectopic expression of SENP3Δ76-159 drags the endogenous SENP3 protein out of the nucleolus (Fig. 1C). Thus, the expression of SENP3Δ76-159 allowed us to investigate the requirement of nucleolar SENP3 for 28S rRNA maturation and 60S ribosomal subunit biogenesis. 28S rRNA processing from the 32S precursor was studied by pulse-chase labeling of nascent rRNA with [32P]orthophosphate. In cells transfected with wild-type SENP3, the mature 28S rRNA species is typically at least as abundant as its 32S rRNA precursor. In contrast, in the presence of SENP3Δ76-159, the formation of the mature 28S rRNA is significantly compromised compared to that of the cells expressing wild-type SENP3 (Fig. 1D, upper). The quantification of the 28S to 32S ratio showed that in cells expressing SENP3Δ76-159, the 28S/32S rRNA ratio was reduced to around 60% compared to that of wild-type cells (Fig. 1D, lower). This reduction in 28S maturation by SENP3Δ76-159 is comparable to the effect observed upon RNA interference (RNAi)-mediated depletion of the protein, indicating that SENP3Δ76-159 acts as a dominant-negative mutant (21, 22) (also see Fig. 6B).
FIG 1.
Nucleolar SENP3 is required for ribosome biogenesis. (A, left) HeLa cells transiently expressing Flag-SENP3 or Flag-SENP3Δ76-159 were stained with anti-Flag antibody to determine their localization. (Right) Immunoblotting of the transfected samples was performed with anti-Flag antibody to test for their expression levels, and antitubulin was used as a loading control. (B) Flag-tagged SENP3Δ76-159 was expressed in HEK293T cells and purified by immunoprecipitation (IP) with anti-Flag beads. Flag-SENP3Δ76-159 along with the interacting proteins was eluted from the beads with SDS sample buffer, separated by SDS-PAGE, and detected using anti-Flag or anti-SENP3 antibody, respectively. (C) As described for panel A, except the cells were stained with anti-SENP3 antibody to simultaneously determine the localization of exogenous and endogenous SENP3 (left), and the level of the endogenous SENP3 was compared with the exogenous SENP3 levels by immunoblotting using anti-SENP3 antibody (right). (D) HeLa cells were transiently transfected with wild-type SENP3 or the SENP3Δ76-159 variant. After 48 h, the cells were pulse labeled with [32P]orthophosphate for 1 h and chased for 1 h with unlabeled medium. RNA was purified from cells, separated under denaturing conditions on an agarose gel, and subjected to autoradiography. (Upper) The steady-state RNA levels were visualized by ethidium bromide (EtBr) staining of the gel. Transfection efficiency was monitored by immunoblotting using an anti-Flag antibody. Antivinculin staining served as a loading control. (Lower) The 28/32S rRNA ratio then was quantified using a phosphorimager. Values represent averages from three independent experiments, with error bars indicating standard deviations (SD), and a P value of <0.05.
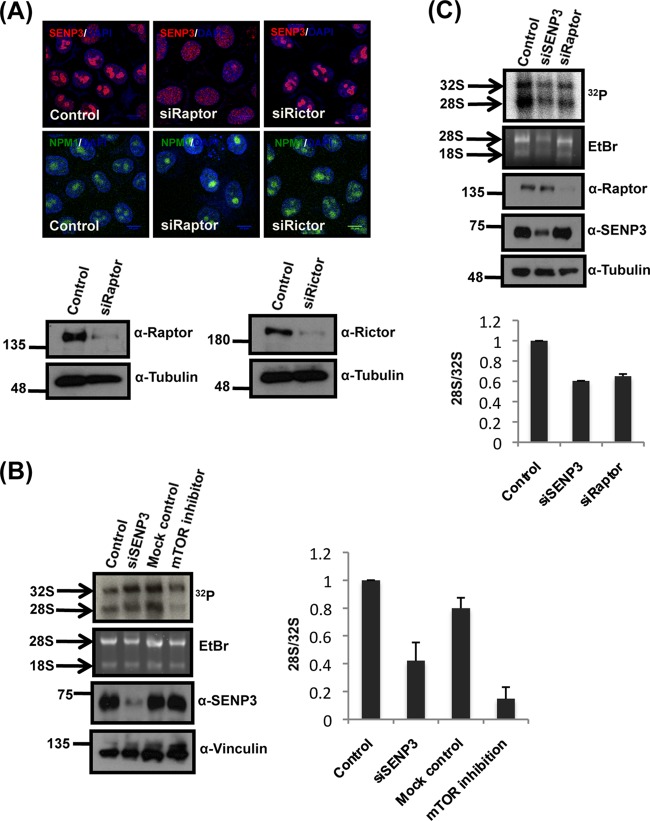
FIG 6.
Inhibition of mTOR signaling reduces 28S maturation. (A, upper) The localization of SENP3 and NPM1 was monitored by immunofluorescence in HeLa cells after the siRNA-mediated depletion of Rictor or Raptor. (Lower) Efficient depletion of the respective proteins was verified by immunoblotting with anti-Raptor or anti-Rictor antibody with antitubulin serving as a loading control. (B) A pulse-chase rRNA processing assay, as described in the legend to Fig. 1D, was performed in HeLa cells after siRNA-mediated depletion of SENP3 or treatment of cells with Ku-0063794. After extraction, the RNA was run on a denaturing agarose gel, stained with EtBr, and subjected to autoradiography for the detection of the various rRNA species. The efficiency of depletion and levels of SENP3 was controlled by Western blotting with anti-SENP3 antibody. Vinculin was used as a loading control. The 28/32S rRNA ratio was quantified using a phosphorimager. The values represent averages from 3 independent experiments, and the error bars indicate standard errors of the means. (C) As described for panel B, except the cells first were transfected with control siRNA or siRNA directed against SENP3 or Raptor before the pulse-chase assay. The efficient depletion of the respective proteins was monitored by immunoblotting with anti-SENP3 and anti-Raptor antibodies, using tubulin as a loading control. Phosphorimager quantifications are as shown in the lower panel.
Physical interaction with NPM1 determines the nucleolar localization of SENP3.
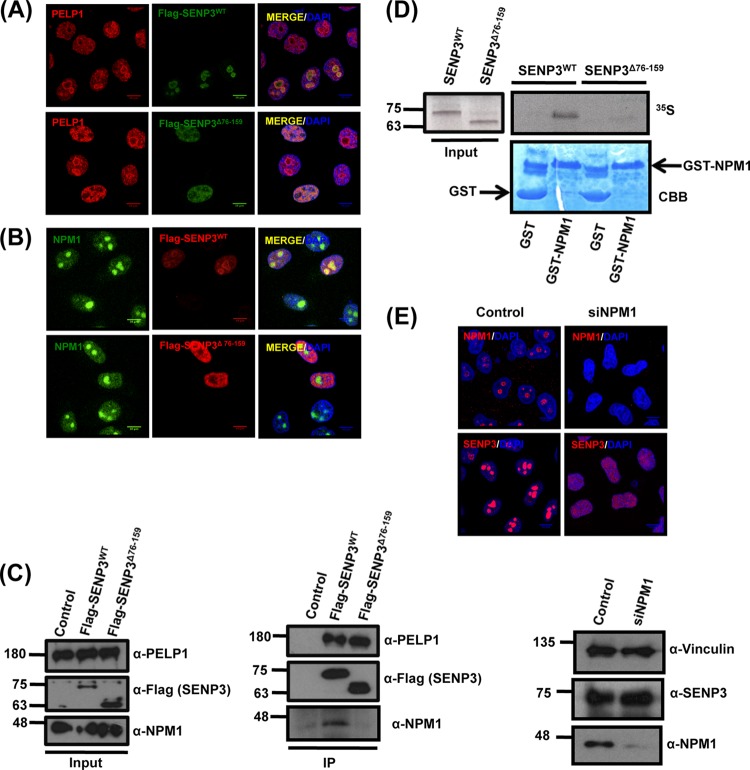
Previous work by our group and others has demonstrated that SENP3 is associated with NPM1 and the PELP1-Tex10-WDR18 complex (19–22, 26, 27). Therefore, we set out to monitor whether SENP3Δ76-159 affects the localization of PELP1 and NPM1. In cells expressing SENP3Δ76-159, PELP1 was largely depleted from the nucleolus and distributed to the nucleoplasm (Fig. 2A), while NPM1 remained in the nucleolus under these conditions (Fig. 2B). The most straightforward interpretation of these data is that SENP3Δ76-159 still is capable of binding PELP1 but has lost its ability to bind NPM1. Consistent with this assumption, in anti-Flag immunoprecipitation experiments endogenous PELP1 copurifies with Flag-SENP3Δ76-159 as well as wild-type Flag-SENP3 (Fig. 2C). In contrast, only wild-type SENP3, but not SENP3Δ76-159, is associated with endogenous NPM1 (Fig. 2C). These data suggest that the NPM1 binding domain in SENP3 overlaps its nucleolar targeting region. To further substantiate this view, we performed in vitro binding assays. Recombinantly expressed GST-NPM1 was used as the bait in GST pulldown assays and tested for interaction with full-length SENP3 or SENP3Δ76-159, which was generated by in vitro transcription/translation. In this setup, full-length SENP3, but not SENP3Δ76-159, bound to NPM1, further supporting the view that binding to NPM1 is critical for the nucleolar compartmentalization of SENP3 (Fig. 2D). In agreement with this interpretation, SENP3 acquires diffuse staining in the nucleoplasm upon RNAi-mediated depletion of NPM1 (Fig. 2E). Collectively, our data suggest that an N-terminal region, which is needed for the physical interaction with NPM1, directly mediates nucleolar targeting of SENP3.
FIG 2.
NPM1 interaction is required for the nucleolar targeting of SENP3. (A) HeLa cells transiently expressing Flag-SENP3 or Flag-SENP3Δ76-159 were stained with anti-Flag and anti-PELP1 antibodies to determine their localization. (B) As described for panel A, except the cells were stained with anti-Flag and anti-NPM1 antibodies after transfection. (C) HeLa cells were transiently transfected with Flag-SENP3 or Flag-SENP3Δ76-159, and their interaction with NPM1 and PELP1 was determined after immunoprecipitation on Flag beads by immunoblotting using anti-Flag, anti-NPM1, and anti-PELP1 antibodies. (D) [35S]methionine-labeled SENP3 and SENP3Δ76-159 were generated by in vitro transcription/translation and used for GST pulldown assays using GST or GST-NPM1 bound to glutathione-Sepharose beads. After separation by SDS-PAGE, interactions were detected via autoradiography. (Upper) Autoradiography showing 35S-labeled SENP3 variants and their interaction with GST-NPM1. (Lower) Coomassie staining of the SDS-PAGE gel showing GST and GST-NPM1. (E) HeLa cells were either mock transfected or transfected with siRNA directed against NPM1. Seventy-two hours posttransfection, the cells were fixed and stained with anti-SENP3 or anti-NPM1 antibody in order to visualize their respective localizations through indirect immunofluorescence (top). Western blotting was performed with anti-SENP3 and anti-NPM1 antibodies to check for their proteins levels, and antivinculin was used as a loading control (bottom).
N-terminal serine/threonine residues are required for nucleolar targeting and NPM1 binding of SENP3.
We next aimed to determine whether the N-terminal region of SENP3 not only is required but also is sufficient for NPM1 binding and nucleolar targeting. To this end, we generated an N-terminal fragment of SENP3 ranging from amino acid 1 to 195. When expressed as a Flag-tagged protein in HeLa cells, this fragment was targeted to the nucleolus (Fig. 3A). Moreover, it bound GST-NPM1 in an in vitro binding experiment (Fig. 3B), further demonstrating that nucleolar targeting of SENP3 coincides with binding to NPM1. With respect to the importance of the NPM1-SENP3 interaction for nucleolar targeting, we aimed to understand how this interaction is regulated. Since we had previously shown that SENP3 is phosphorylated, we hypothesized that phosphorylation affected the association of SENP3 with NPM1 (28). To address this point generally, we performed an alanine scan and generated a Flag-tagged version of full-length SENP3 in which all 28 serine/threonine residues in the N-terminal region (aa 1 to 195) were replaced by alanine. We found that, unlike wild-type Flag-SENP3, Flag-SENP328S→A is not properly targeted to the nucleolus and exhibits a predominantly nucleoplasmic distribution (Fig. 3C). Further, we tested the ability of this mutant to bind NPM1. When comparing anti-Flag immunoprecipitates from wild-type Flag-SENP3 and Flag-SENP328S→A expressing cells, a drastic reduction of NPM1 binding was observed for the Flag-SENP328S→A mutant (Fig. 3D). However, Flag-SENP328S→A still properly interacts with PELP1 (Fig. 3D). To further support the idea that it is indeed the phosphorylation of SENP3 that controls the interaction with NPM1, we performed an in vitro binding experiment. To this end, we generated SENP3 by in vitro transcription/translation using rabbit reticulocyte lysate, which typically recapitulates cellular phosphorylation events. Subsequently, the protein was dephosphorylated through the addition of lambda phosphatase (λ-PPase), and its binding to GST-NPM1 was compared to that of the mock-treated phosphorylated SENP3. As shown above, SENP3 exhibits a strong and specific interaction with GST-NPM1. Intriguingly, however, this binding is strongly impaired upon dephosphorylating SENP3, suggesting that the phosphorylation status of SENP3 determines its binding to NPM1 (Fig. 3E).
The N-terminal region of SENP3 is phosphorylated by mTOR.
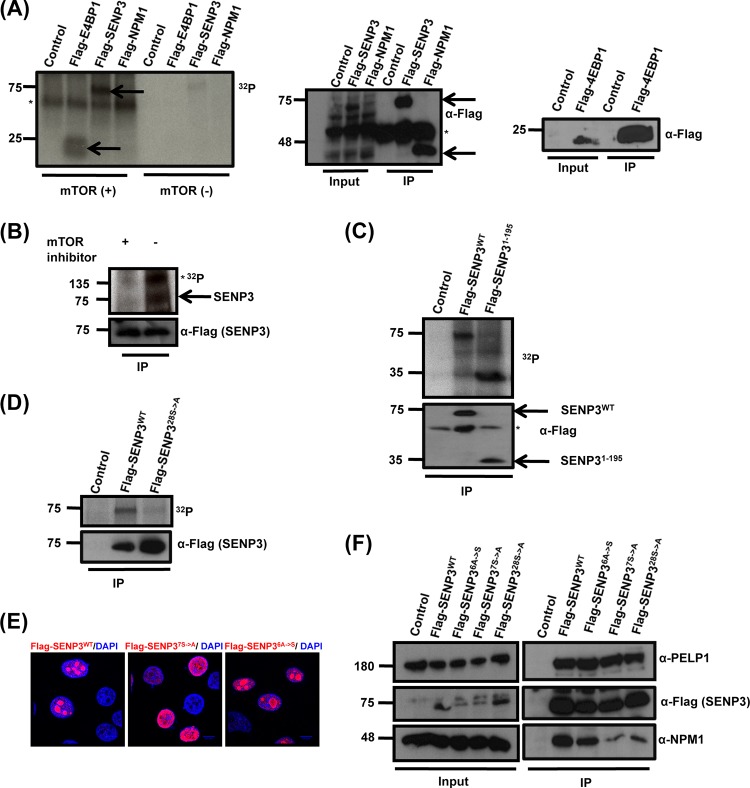
Based on the findings described above, we asked which kinase pathway could be involved in phosphorylation-mediated binding of SENP3 to NPM1. In light of the recently described role of mTOR signaling in ribosome biogenesis, we considered mTOR as a candidate kinase (8, 9, 11). Therefore, we asked whether SENP3 is a direct target of mTOR-mediated phosphorylation. Because SENP3 cannot be purified in bacteria, we tackled this question by immunoprecipitating Flag-tagged SENP3 from HEK293 cells and, after stringent washing, used the purified protein for an in vitro phosphorylation assay with recombinant mTOR in the presence of [32P]ATP (Fig. 4A). In the initial experiment, we used a catalytic mTOR fragment (ranging from amino acid 1362 to the end), which was expressed in a baculovirus-infected Sf9 cell system. Interestingly, the autoradiography revealed that SENP3 is strongly phosphorylated by mTOR with efficiency comparable to that of the known mTOR target 4EBP1 (Fig. 4A). The phosphorylation of SENP3 was strictly dependent on the addition of recombinant mTOR to immunopurified SENP3, excluding the contribution of a copurified kinase in the phosphorylation process. Also of note, no phosphorylation of NPM1 by mTOR could be observed in this experimental setup, demonstrating at least some specificity for mTOR-mediated phosphorylation (Fig. 4A). The phosphorylation of SENP3 also was detected after only 5 min of incubation with the recombinant mTOR/Raptor/MLST8 complex purified from baculovirus-infected Sf9 cells (Fig. 4B). The specificity of the reaction was further demonstrated by the loss of SENP3 phosphorylation as well as mTOR autophosphorylation upon the addition of the mTOR inhibitor Torin1 (Fig. 4B, left lane).
FIG 4.
mTOR phosphorylates SENP3 at its N terminus. (A) HEK293T cells were transfected with plasmids expressing Flag-SENP3, Flag-NPM1, Flag-4EBP1, or empty vector, and the respective proteins were immunoprecipitated using Flag-agarose beads. The proteins bound to the beads then were used for in vitro phosphorylation assay in the presence of [32P]ATP with or without the catalytic fragment of mTOR (aa 1362 to 2549). (Left) Subsequently, the samples were separated by SDS-PAGE and phosphorylation was detected by autoradiography. IgGs are indicated by an asterisk. (Right) The expression and immunoprecipitation of the Flag-tagged constructs was monitored by Western blotting with anti-Flag antibody. (B) Flag-tagged SENP3 was immunoprecipitated from HeLa cells and used for in vitro phosphorylation assay with [32P]ATP with either the mock-treated or Torin1-treated mTOR/Raptor/MLST8 complex. (Upper) The samples then were subjected to SDS-PAGE, followed by autoradiography. mTOR autophosphorylation is indicated by an asterisk. (Lower) Immunopurified SENP3 was monitored by Western blotting with anti-Flag antibody. (C, upper) mTOR-mediated phosphorylation of Flag-SENP3 or Flag-SENP31-195 was analyzed in an in vitro phosphorylation assay as described for panel A. (Lower) IP was verified by immunoblotting with anti-Flag antibody. (D) mTOR-mediated phosphorylation of wild-type Flag-SENP3 and Flag-SENP328S→A was done as described for panels A and B. (Lower) IPs were verified by immunoblotting with anti-Flag antibody. (E) HeLa cells were transiently transfected with plasmids expressing wild-type Flag-SENP3, Flag-SENP37S→A, or Flag-SENP36A→S and stained with anti-Flag antibody for indirect immunofluorescence. (F) Interaction of wild-type Flag-SENP3, Flag-SENP37S→A, Flag-SENP36A→S, or Flag-SENP328S→A with endogenous NPM1 and PELP1 was monitored by Western blotting after immunoprecipitation of the transiently expressed Flag-tagged proteins in HeLa cells.
Based on the requirement of the N-terminal region of SENP3 in nucleolar targeting and NPM1 binding, we next tested the immunopurified N-terminal fragment of SENP3 (SENP31-195) for mTOR-mediated phosphorylation. Like full-length SENP3, SENP31-195 was efficiently phosphorylated by mTOR, suggesting that major mTOR phosphorylation sites reside in this region (Fig. 4C). In line with this idea, the Flag-SENP328S→A variant described above, in which all serine/threonine residues in the N-terminal region (aa 1 to 195) were replaced by alanine, did not undergo significant mTOR-mediated phosphorylation (Fig. 4D). To further pinpoint the mTOR-dependent phosphosites in SENP3, we performed mass spectrometric analysis of SENP31-195 after in vitro phosphorylation. The analysis covered 23 out of 28 S/T residues in this region and identified five potential phosphosites (S25, S26, S141, T142, and T145) that were unphosphorylated in the mock-treated sample (Table 1). Based on these data, we generated a variant of full-length SENP3 (Flag-SENP37S→A) where these residues, together with two more serine residues directly adjacent to these sites (S139 and S143), were changed to alanine. The two additional serine mutations were included to avoid their use as phosphosites in the absence of the canonical sites. We then were interested to see whether the removal of these 7 S/T residues in the context of full-length SENP3 affects its subnuclear localization and NPM1 binding. Importantly, unlike wild-type Flag-SENP3, Flag-SENP37S→A does not properly localize to the nucleolus and exhibits a predominantly nucleoplasmic distribution in the majority of cells (Fig. 4E). Moreover, like Flag-SENP328S→A, Flag-SENP37S→A did not efficiently bind NPM1, as evident from anti-Flag coimmunoprecipitation experiments (Fig. 4F). Also of note, the interaction of Flag-SENP37S→A with PELP1 was not compromised, suggesting that the N-terminal phosphosites in SENP3 are critical for NPM1 binding but not the association with PELP1. To further understand if N-terminal phosphosites are necessary and sufficient for nucleolar targeting of SENP3 and NPM1 binding, we reintroduced serine 25 and the five S/T residues in the region covering amino acids 139 to 145 in the SENP328S→A variant. This mutant (Flag-SENP36A→S) exhibits normal binding of SENP3 to NPM1 and shows nucleolar localization (Fig. 4E and F), underscoring the importance of these residues for nucleolar targeting and NPM1 binding.
TABLE 1.
Mass spectrometric analysis of the in vitro-phosphorylated SENP31-195 fragmenta
| mTOR treatment | Peptide sequence | Phosphosite(s) | m/z | Mass | PEPb | Scorec | Delta scored |
|---|---|---|---|---|---|---|---|
| mTOR+ | ETIQGTGSWGPEPPGPGIPPAYSSPR | S25, S26 | 905.753 | 2714.24 | 2.3E−09 | 142.23 | 120.56 |
| STSLTFHWK | S141, T142, T145 | 633.751 | 1265.49 | 0.07933 | 88.681 | 70.872 | |
| mTOR− | ETIQGTGSWGPEPPGPGIPPAYSSPR | 879.098 | 2634.27 | 6.8E−07 | 123.12 | 93.645 | |
| STSLTFHWK | 553.785 | 1105.56 | 0.05124 | 95.099 | 51.329 |
Parameters from control samples and mTOR-treated samples are shown for the peptides containing S25 and S26 and S141, T142, and T145.
Posterior error probability of the identification. This value essentially operates as a P value, where smaller is more significant.
Andromeda score for the best-associated MS/MS spectrum.
Score difference to the second-best identified peptide.
mTOR signaling controls the interaction between SENP3 and NPM1.
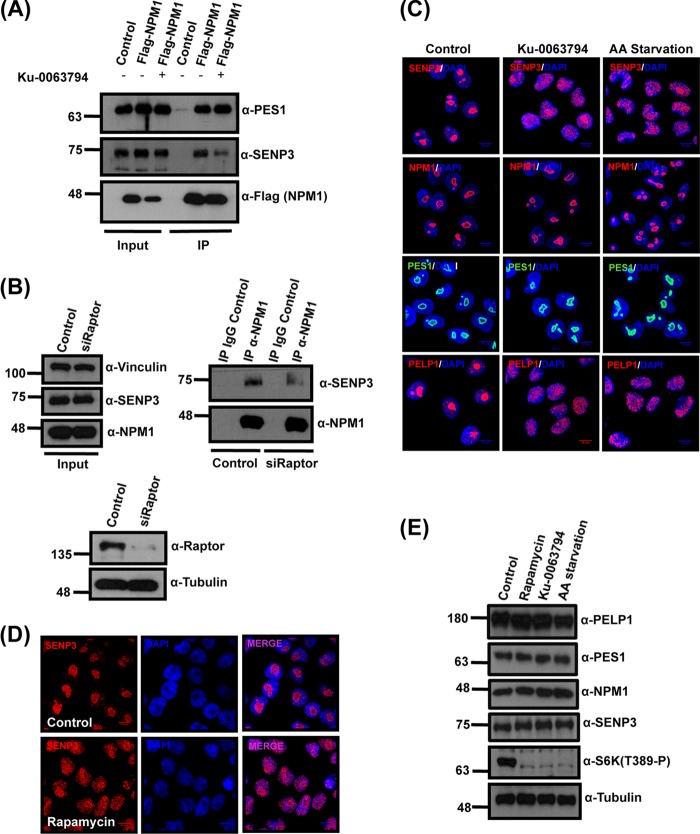
The findings described above led us to investigate whether, in a cellular setting, mTOR activity is involved in controlling nucleolar targeting of SENP3 and binding of the protein to NPM1. First, to see whether mTOR inhibition translates into a release of SENP3 from the nucleolus, we initially choose Ku-0063794, a small-molecule inhibitor of both mTORC1 and mTORC2, to block mTOR signaling (29). The SENP3-NPM1 interaction was investigated by coimmunoprecipitation experiments in a cell line expressing Flag-tagged NPM1 (21). In mock-treated control cells, a significant fraction of endogenous SENP3 is found in anti-Flag immunoprecipitates (Fig. 5A). However, upon mTOR inhibition, this robust interaction is strongly compromised, suggesting that active mTOR signaling is facilitating the interaction between NPM1 and SENP3 (Fig. 5A). Moreover, blocking mTOR signaling by depletion of Raptor weakens the interaction of endogenous NPM1 and SENP3 (Fig. 5B). Of note, the inhibition of mTOR generally does not affect the association of NPM1 with its interaction partners, since binding of the 60S maturation factor PES1 to NPM1 is not impaired upon Ku-0063794 treatment (30). From these data, we conclude that active mTOR signaling specifically controls the dynamic interaction of NPM1 and SENP3. Based on our finding that NPM1 recruits SENP3 to the nucleolus, we investigated whether the impairment of NPM1/SENP3 binding, which is induced by mTOR inhibition, translates into a release of SENP3 from the nucleolus. In immunofluorescence experiments, we indeed observed a striking nucleoplasmic redistribution of SENP3 when mTOR signaling is blocked with Ku-0063794 (Fig. 5C). A similar redistribution of SENP3 was observed upon amino acid starvation or treatment with rapamycin (Fig. 5C and D). It is worth nothing that, under these conditions, phosphorylation of the mTOR target S6-kinase was lost, but the levels of SENP3 as well as PES1 and NPM1 were unchanged, suggesting that the observed effects are not due to alterations in protein abundance (Fig. 5E). Amino acid starvation and rapamycin more specifically inhibit mTORC1 rather than mTORC2, indicating that the nucleolar targeting of SENP3 most likely is controlled through the mTORC1 complex. To further validate this point, we performed siRNA-mediated knockdowns of Raptor and Rictor (Fig. 6A), which are the protein components involved in substrate recruitment and complex assembly of mTORC1 and mTORC2, respectively. The knockdown of Raptor but not Rictor affected the nucleolar accumulation of SENP3 without affecting NPM1 (Fig. 6A). In summary, these data show that mTOR signaling, most likely through the mTORC1 complex, controls the nucleolar targeting of SENP3. Considering the importance of nucleolar SENP3 for proper ribosome maturation, we next asked whether nucleolar release of SENP3 induced by the inhibition of mTOR signaling is accompanied by defects in ribosome formation. Intriguingly, like SENP3 depletion, Ku-0063794 treatment or depletion of Raptor dramatically affects 28S rRNA maturation, as monitored by pulse-chase labeling of rRNA processing (Fig. 6B and C). In summary, these data indicate that active mTOR signaling is required for the nucleolar accumulation of SENP3, which in turn ensures productive 28S rRNA maturation and 60S biogenesis.
FIG 5.
mTOR affects SENP3 localization and SENP3-NPM1 interaction. (A) U2OS cells expressing Flag-NPM1 were either mock treated or treated with the mTOR inhibitor Ku-0063794. Subsequently, Flag-NPM1 was immunoprecipitated from these cells using Flag-agarose beads and separated by SDS-PAGE. Anti-Flag antibody was used to verify the immunoprecipitation of Flag-NPM1, and anti-SENP3 or anti-PES1 antibody was used to check for coimmunoprecipitation. (B) HeLa cells were transfected with either control siRNA or siRNA directed against Raptor, and 72 h later the cells were harvested for immunoprecipitation of the endogenous NPM1. (Upper right) The samples then were probed by immunoblotting with anti-NPM1 or anti-SENP3 antibody to check for NPM1 levels and coimmunoprecipitation of SENP3. (Upper left and lower) The proteins levels in control and knockdown samples were monitored by Western blotting with anti-NPM1, anti-SENP3, and anti-Raptor antibodies with antitubulin and antivinculin antibodies as loading controls. (C) HeLa cells were either mock treated, treated with Ku-0063794, or starved in Earle's balanced salt solution (EBSS) for 6 h and stained with anti-SENP3, anti-NPM1, anti-PES1, and anti-PELP1 antibodies to visualize their respective localization by indirect immunofluorescence. (D) SENP3 localization in HeLa cells was detected by immunofluorescence using anti-SENP3 antibody after treatment with rapamycin. (E) Immunoblotting of the HeLa cell lysates was done to check for changes in protein levels using anti-PELP1, anti-SENP3, anti-PES1, anti-S6K(T389-P), and anti-NPM1 antibodies after EBSS starvation or rapamycin or Ku-0063794 treatment. Antitubulin antibody was used as a loading control.
DISCUSSION
The SUMO-specific isopeptidase SENP3 is critically involved in mammalian ribosome biogenesis by stimulating the maturation of the 28S rRNA. Here, we show that this function requires the nucleolar localization of SENP3, which is determined by its interaction with the nucleolar binding partner NPM1. We demonstrate that the mTOR signaling pathway promotes this interaction by directly targeting the N terminus of SENP3.
The maturation of ribosomes is a central cellular process that needs to be coordinated with the cellular energy status. An important regulatory principle of ribosome biogenesis is the separation of specific maturation steps into distinct cellular compartments (4, 5). The RNA polymerase I-mediated transcription of the 47S rRNA precursor takes place in the inner core of the nucleolus, at the interface of fibrillar centers and the dense fibrillar component (DFC) (5). The subsequent processing steps of the precursor typically are subclassified into early and late processing events, with the early steps occurring in the DFC region of the nucleolus and later steps in the granular region and the nucleoplasm. The cleavage of the rRNA precursor within the internal transcribed spacer (ITS2) region, which separates the 5.8S and the 28S regions in the rRNA, takes place in the GC region. In mammals, this processing step generates the pre-28S rRNA species and a 12S intermediate, which is further trimmed to the 5.8S form in the nucleoplasm (2). Several factors involved in ITS2 cleavage are concentrated in the nucleolus. One key factor is NPM1/B23, which serves as a marker for GCs and seems to act as a scaffold for several multiprotein complexes implicated in 28S maturation (31). We and others have identified the SUMO2/3-specific isopeptidase SENP3 as a major interaction partner of NPM1 (19, 20–22).
We have shown that SENP3 also resides in the GC and accordingly is required for proper 28S maturation. Here, we demonstrated that the nucleolar residency of SENP3 is needed for its function in 28S rRNA maturation. Furthermore, we show that the nucleolar recruitment of SENP3 requires direct binding to NPM1 through a defined domain in the N-terminal domain of SENP3. Importantly, our work reveals that serines 25 and 26, together with a stretch of serine/threonine residues spanning residues 139 to 145, can be targeted by mTOR-mediated phosphorylation. An SENP3 version lacking seven serine/threonine residues is severely compromised in nucleolar targeting and interaction with NPM1, even though it shows residual phosphorylation (data not shown). Very similarly, the inhibition of the mTOR kinase strongly affects the binding of SENP3 to NPM1 and induces SENP3's release from the nucleolus. Therefore, our data support the idea that mTOR-dependent phosphorylation of SENP3 promotes its interaction with NPM1 and the recruitment to the nucleolus, which ultimately stimulates ribosome biogenesis. The N-terminal region of SENP3 with the potential phosphorylation sites defined here is highly conserved in mammals. Notably, S26, which is one of the mTOR-targeted residues, is adjacent to a proline residue, a signature that typically is found in mTOR substrates. How the phosphorylation of SENP3 contributes to NPM1 binding currently is unknown. Recent structural work defined an arginine-rich motif in several NPM1 interaction partners as the critical region for NPM1 binding (32). It has been proposed that these arginine-rich stretches bind to an acidic groove in the N-terminal oligomerization domain of NPM1 to acquire its pentameric conformation (32). We have shown previously that the oligomerization domain serves as the SENP3 binding region in NPM1. Intriguingly, the region in SENP3 we now defined as the NPM1 binding domain contains two R-rich stretches (boldface) that are separated by the identified phospho-Ser/Thr cluster (underlined) between residues 139 and 145 (125RRRRAMRAFRMLLYSKSTSLTFHWKLWGRHRGRRR160). One scenario is that the phosphorylation of these residues facilitates the exposure of the arginine motif through intramolecular interactions in the SENP3 N-terminal domain. Alternatively, the negative charges introduced by phosphorylation could directly contribute to NPM1 binding through additional electrostatic interactions. Notably, however, replacing the serine residues in SENP3 with negatively charged amino acids did not lead to enhanced binding to NPM1, suggesting that negative charges alone are not sufficient for this process (data not shown). This is in line with several examples where the introduction of phosphomimetic residues cannot functionally replace the natural phosphoserine residues (33).
Mitrea et al. have shown that posttranslational modifications within the oligomerization domain of NPM1 regulate the transition from the pentameric to the monomeric state and the concomitant binding of NPM1 interactors (32). Although our in vitro experiments support the idea that mTOR directly acts on SENP3, mTOR signaling may have additional indirect roles in this pathway. This also is evident from our observation that TOR inhibition causes a much more severe defect in ribosome maturation than SENP3 depletion. For example, high-throughput proteomic data suggest that the phosphorylation of NPM1 at S70 is sensitive to mTOR inhibitors (34, 35). Thus, S70 phosphorylation by mTOR or a downstream kinase may contribute to the control of NPM1/SENP3 binding and nucleolar recruitment of SENP3. Changes in protein phosphatase activities, like those of PP2A, also might be implicated in this process, since mTOR inhibition is known to stimulate their activities.
Currently it is unclear in which cellular compartment mTOR phosphorylates SENP3. However, components of the mTORC1 complex are found in the nucleolus, making the nucleolar phosphorylation of SENP3 a plausible event (36). Interestingly, previous work has demonstrated that mTOR regulates RNA polymerase I-dependent transcription of rRNA by modulating the activity of the transcription factor TIF-IA (10, 11). Intriguingly, reminiscent of our model, mTOR was proposed to determine the nucleolar localization of TIF-IA by controlling its interaction with RNA Pol I.
Recent work has revealed that mild oxidative stress induces the nucleolar release of SENP3, suggesting that the spatial regulation of SENP3 is relevant as a nutrient-sensing and redox-sensing pathway (37–39). Redox sensing of SENP3 is mediated by oxidative modifications of specific cysteine residues, which goes along with the stabilization of the protein and ultimately leads to the activation of the HIF1 pathway by SENP3-mediated deSUMOylation of p300 in the nucleoplasm (37–39). Furthermore, in a cellular model of endoplasmic reticulum stress, the unfolded protein response kinase PERK was reported to trigger ubiquitin-independent degradation of a cytosolic fraction of SENP3. In summary, these data highlight the need for SENP3 control at multiple levels. Our data reveal a novel pathway which now implicates mTOR signaling in the delicate control of SENP3 function. We propose that this pathway coordinates ribosome biogenesis with nutrient availability.
ACKNOWLEDGMENTS
We thank Ivan Dikic and all members of our institute for support and stimulating discussions.
This work was funded by DFG Priority programs SPP1365, SFB684, SFB815, and LOEWE Ub-Net.
Footnotes
Published ahead of print 6 October 2014
REFERENCES
- 1.Thomson E, Ferreira-Cerca S, Hurt E. 2013. Eukaryotic ribosome biogenesis at a glance. J. Cell Sci. 126:4815–4821. 10.1242/jcs.111948. [DOI] [PubMed] [Google Scholar]
- 2.Henras AK, Soudet J, Gerus M, Lebaron S, Caizergues-Ferrer M, Mougin A, Henry Y. 2008. The post-transcriptional steps of eukaryotic ribosome biogenesis. Cell. Mol. Life Sci. 65:2334–2359. 10.1007/s00018-008-8027-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kressler D, Hurt E, Bassler J. 2010. Driving ribosome assembly. Biochim. Biophys. Acta 1803:673–683. 10.1016/j.bbamcr.2009.10.009. [DOI] [PubMed] [Google Scholar]
- 4.Hernandez-Verdun D, Roussel P, Thiry M, Sirri V, Lafontaine DL. 2010. The nucleolus: structure/function relationship in RNA metabolism. Wiley interdisciplinary reviews. RNA 1:415–431. 10.1002/wrna.39. [DOI] [PubMed] [Google Scholar]
- 5.Boisvert FM, van Koningsbruggen S, Navascues J, Lamond AI. 2007. The multifunctional nucleolus. Nat. Rev. Mol. Cell Biol. 8:574–585. 10.1038/nrm2184. [DOI] [PubMed] [Google Scholar]
- 6.Laplante M, Sabatini DM. 2012. mTOR signaling in growth control and disease. Cell 149:274–293. 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cornu M, Albert V, Hall MN. 2013. mTOR in aging, metabolism, and cancer. Curr. Opin. Genet. Dev. 23:53–62. 10.1016/j.gde.2012.12.005. [DOI] [PubMed] [Google Scholar]
- 8.Iadevaia V, Huo Y, Zhang Z, Foster LJ, Proud CG. 2012. Roles of the mammalian target of rapamycin, mTOR, in controlling ribosome biogenesis and protein synthesis. Biochem. Soc. Trans. 40:168–172. 10.1042/BST20110682. [DOI] [PubMed] [Google Scholar]
- 9.Iadevaia V, Zhang Z, Jan E, Proud CG. 2012. mTOR signaling regulates the processing of pre-rRNA in human cells. Nucleic Acids Res. 40:2527–2539. 10.1093/nar/gkr1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mayer C, Zhao J, Yuan X, Grummt I. 2004. mTOR-dependent activation of the transcription factor TIF-IA links rRNA synthesis to nutrient availability. Genes Dev. 18:423–434. 10.1101/gad.285504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mayer C, Grummt I. 2006. Ribosome biogenesis and cell growth: mTOR coordinates transcription by all three classes of nuclear RNA polymerases. Oncogene 25:6384–6391. 10.1038/sj.onc.1209883. [DOI] [PubMed] [Google Scholar]
- 12.Reiter A, Steinbauer R, Philippi A, Gerber J, Tschochner H, Milkereit P, Griesenbeck J. 2011. Reduction in ribosomal protein synthesis is sufficient to explain major effects on ribosome production after short-term TOR inactivation in Saccharomyces cerevisiae. Mol. Cell. Biol. 31:803–817. 10.1128/MCB.01227-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hickey CM, Wilson NR, Hochstrasser M. 2012. Function and regulation of SUMO proteases. Nat. Rev. Mol. Cell Biol. 13:755–766. 10.1038/nrm3478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mukhopadhyay D, Dasso M. 2007. Modification in reverse: the SUMO proteases. Trends Biochem. Sci. 32:286–295. 10.1016/j.tibs.2007.05.002. [DOI] [PubMed] [Google Scholar]
- 15.Gareau JR, Lima CD. 2010. The SUMO pathway: emerging mechanisms that shape specificity, conjugation and recognition. Nat. Rev. Mol. Cell Biol. 11:861–871. 10.1038/nrm3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wilkinson KA, Henley JM. 2010. Mechanisms, regulation and consequences of protein SUMOylation. Biochem. J. 428:133–145. 10.1042/BJ20100158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van der Veen AG, Ploegh HL. 2012. Ubiquitin-like proteins. Annu. Rev. Biochem. 81:323–357. 10.1146/annurev-biochem-093010-153308. [DOI] [PubMed] [Google Scholar]
- 18.Raman N, Nayak A, Muller S. 2013. The SUMO system: a master organizer of nuclear protein assemblies. Chromosoma 122:475–485. 10.1007/s00412-013-0429-6. [DOI] [PubMed] [Google Scholar]
- 19.Castle CD, Cassimere EK, Denicourt C. 2012. LAS1L interacts with the mammalian Rix1 complex to regulate ribosome biogenesis. Mol. Biol. Cell 23:716–728. 10.1091/mbc.E11-06-0530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Finkbeiner E, Haindl M, Muller S. 2011. The SUMO system controls nucleolar partitioning of a novel mammalian ribosome biogenesis complex. EMBO J. 30:1067–1078. 10.1038/emboj.2011.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haindl M, Harasim T, Eick D, Muller S. 2008. The nucleolar SUMO-specific protease SENP3 reverses SUMO modification of nucleophosmin and is required for rRNA processing. EMBO Rep. 9:273–279. 10.1038/embor.2008.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yun C, Wang Y, Mukhopadhyay D, Backlund P, Kolli N, Yergey A, Wilkinson KD, Dasso M. 2008. Nucleolar protein B23/nucleophosmin regulates the vertebrate SUMO pathway through SENP3 and SENP5 proteases. J. Cell Biol. 183:589–595. 10.1083/jcb.200807185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stehmeier P, Muller S. 2009. Phospho-regulated SUMO interaction modules connect the SUMO system to CK2 signaling. Mol. Cell 33:400–409. 10.1016/j.molcel.2009.01.013. [DOI] [PubMed] [Google Scholar]
- 24.Schmidt D, Muller S. 2002. Members of the PIAS family act as SUMO ligases for c-Jun and p53 and repress p53 activity. Proc. Natl. Acad. Sci. U. S. A. 99:2872–2877. 10.1073/pnas.052559499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nishida T, Tanaka H, Yasuda H. 2000. A novel mammalian Smt3-specific isopeptidase 1 (SMT3IP1) localized in the nucleolus at interphase. Eur. J. Biochem. 267:6423–6427. 10.1046/j.1432-1327.2000.01729.x. [DOI] [PubMed] [Google Scholar]
- 26.Kuo ML, den Besten W, Thomas MC, Sherr CJ. 2008. Arf-induced turnover of the nucleolar nucleophosmin-associated SUMO-2/3 protease Senp3. Cell Cycle 7:3378–3387. 10.4161/cc.7.21.6930. [DOI] [PubMed] [Google Scholar]
- 27.Nishida T, Yamada Y. 2008. SMT3IP1, a nucleolar SUMO-specific protease, deconjugates SUMO-2 from nucleolar and cytoplasmic nucleophosmin. Biochem. Biophys. Res. Commun. 374:382–387. 10.1016/j.bbrc.2008.07.047. [DOI] [PubMed] [Google Scholar]
- 28.Klein UR, Haindl M, Nigg EA, Muller S. 2009. RanBP2 and SENP3 function in a mitotic SUMO2/3 conjugation-deconjugation cycle on Borealin. Mol. Biol. Cell 20:410–418. 10.1091/mbc.E08-05-0511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Garcia-Martinez JM, Moran J, Clarke RG, Gray A, Cosulich SC, Chresta CM, Alessi DR. 2009. Ku-0063794 is a specific inhibitor of the mammalian target of rapamycin (mTOR). Biochem. J. 421:29–42. 10.1042/BJ20090489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang J, Yang Y, Wu J. 2009. B23 interacts with PES1 and is involved in nucleolar localization of PES1. Acta Biochim. Biophys. Sin. 41:991–997. 10.1093/abbs/gmp096. [DOI] [PubMed] [Google Scholar]
- 31.Lindstrom MS. 2011. NPM1/B23: a multifunctional chaperone in ribosome biogenesis and chromatin remodeling. Biochem. Res. Int. 2011:195209. 10.1155/2011/195209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mitrea DM, Grace CR, Buljan M, Yun MK, Pytel NJ, Satumba J, Nourse A, Park CG, Madan Babu M, White SW, Kriwacki RW. 2014. Structural polymorphism in the N-terminal oligomerization domain of NPM1. Proc. Natl. Acad. Sci. U. S. A. 111:4466–4471. 10.1073/pnas.1321007111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dephoure N, Gould KL, Gygi SP, Kellogg DR. 2013. Mapping and analysis of phosphorylation sites: a quick guide for cell biologists. Mol. Biol. Cell 24:535–542. 10.1091/mbc.E12-09-0677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hsu PP, Kang SA, Rameseder J, Zhang Y, Ottina KA, Lim D, Peterson TR, Choi Y, Gray NS, Yaffe MB, Marto JA, Sabatini DM. 2011. The mTOR-regulated phosphoproteome reveals a mechanism of mTORC1-mediated inhibition of growth factor signaling. Science 332:1317–1322. 10.1126/science.1199498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yu Y, Yoon SO, Poulogiannis G, Yang Q, Ma XM, Villen J, Kubica N, Hoffman GR, Cantley LC, Gygi SP, Blenis J. 2011. Phosphoproteomic analysis identifies Grb10 as an mTORC1 substrate that negatively regulates insulin signaling. Science 332:1322–1326. 10.1126/science.1199484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vazquez-Martin A, Cufi S, Oliveras-Ferraros C, Menendez JA. 2011. Raptor, a positive regulatory subunit of mTOR complex 1, is a novel phosphoprotein of the rDNA transcription machinery in nucleoli and chromosomal nucleolus organizer regions (NORs). Cell Cycle 10:3140–3152. 10.4161/cc.10.18.17376. [DOI] [PubMed] [Google Scholar]
- 37.Wang Y, Yang J, Yang K, Cang H, Huang XZ, Li H, Yi J. 2012. The biphasic redox sensing of SENP3 accounts for the HIF-1 transcriptional activity shift by oxidative stress. Acta Pharmacol. Sin. 33:953–963. 10.1038/aps.2012.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang Y, Yang J, Yi J. 2012. Redox sensing by proteins: oxidative modifications on cysteines and the consequent events. Antioxid. Redox Signal. 16:649–657. 10.1089/ars.2011.4313. [DOI] [PubMed] [Google Scholar]
- 39.Yan S, Sun X, Xiang B, Cang H, Kang X, Chen Y, Li H, Shi G, Yeh ET, Wang B, Wang X, Yi J. 2010. Redox regulation of the stability of the SUMO protease SENP3 via interactions with CHIP and Hsp90. EMBO J. 29:3773–3786. 10.1038/emboj.2010.245. [DOI] [PMC free article] [PubMed] [Google Scholar]