Abstract
The circadian clock is finely regulated by posttranslational modifications of clock components. Mouse CRY2, a critical player in the mammalian clock, is phosphorylated at Ser557 for proteasome-mediated degradation, but its in vivo role in circadian organization was not revealed. Here, we generated CRY2(S557A) mutant mice, in which Ser557 phosphorylation is specifically abolished. The mutation lengthened free-running periods of the behavioral rhythms and PER2::LUC bioluminescence rhythms of cultured liver. In livers from mutant mice, the nuclear CRY2 level was elevated, with enhanced PER2 nuclear occupancy and suppression of E-box-regulated genes. Thus, Ser557 phosphorylation-dependent regulation of CRY2 is essential for proper clock oscillation in vivo.
INTRODUCTION
Transcription- and translation-based negative-feedback loops play an important role in circadian clock regulation (1, 2). In mammals, a heterodimer of positive factors, CLOCK and BMAL1, activates the transcription of genes encoding negative factors such as PERIOD (PER1 to -3) and cryptochrome (CRY1 and -2) through binding to E-box enhancer elements in their promoter regions (3). Translated PERs and CRYs associate with each other, enter into cell nuclei, and inhibit their own transcription by interacting with the CLOCK-BMAL1 heterodimer (4). In addition to transcriptional and translational regulation, posttranslational modifications of the clock proteins play critical roles in the clockwork (5–8).
Among the negative factors, CRY1 and CRY2 play major roles for repression of E-box-dependent gene expression (9). They share highly conserved N-terminal and central regions while having unique C-terminal tails. Although the roles of the diverged C-terminal regions remain unclear, both CRY1 and CRY2 have strong repressor activities (9), and therefore, their accumulation and decline are the major period-determining steps for circadian molecular oscillation. Previous studies reported that FBXL3, an F-box-type E3 ubiquitin ligase, enhances proteasomal degradation of CRY1 and CRY2 (10–12). Recently, we and other groups demonstrated that ubiquitination of CRY1 and CRY2 by FBXL21, the closest paralog of FBXL3, stabilizes CRYs (13, 14). It has been reported that FBXL3-dependent CRY1 degradation is regulated by CRY1 phosphorylation (15, 16). In contrast, we previously demonstrated that priming phosphorylation of CRY2 at Ser557 in the C-terminal region by DYRK1A is required for secondary phosphorylation at Ser553 by glycogen synthase kinase 3β (GSK-3β) (17, 18). The dually phosphorylated CRY2 is led to proteasomal degradation (18), which is independent of FBXL3 action (17, 18). So far, the functions of CRY regulators, such as protein kinases and ubiquitin ligases, have been examined by using their inhibitors and/or gene knockout/knockdown (17–19). On the other hand, site-directed mutagenesis in CRY proteins will be the most specific strategy because the upstream regulators would have multiple targets in the clockwork. In the present study, we generated knock-in mice carrying a mutation at the priming phosphorylation site of Ser557 in order to uncover the physiological importance of the sequential phosphorylation of CRY2. We found that Ser557 phosphorylation of CRY2 regulates the nuclear accumulation of CRY2-PER2, a functional repressor complex, and controls the circadian period in both the central and the peripheral clocks in vivo.
MATERIALS AND METHODS
Animal maintenance.
Animal experiments were conducted in accordance with guidelines of The University of Tokyo. Mice were housed at 23°C ± 1°C in cages with commercial chow (CLEA Japan, Inc.) and tap water available ad libitum. Cry1 and Cy2 knockout mice were generated as described in a previous study (20). CAG promoter-driven flippase transgenic mice in the C57BL/6J genetic background were generated by Francis A. Stewart (Dresden University of Technology) (21). PER2::LUC knock-in mice in the C57BL/6J genetic background (22) were obtained from The Jackson Laboratory.
Generation of the CRY2(S557A) targeting construct.
A fragment of the murine Cry2 gene, which includes exons 6 to 11, was amplified from a bacterial artificial chromosome (BAC) clone (RP24-86P20; BACPAC) and subcloned into the pSP72 vector (Promega). The S557A mutation together with a PvuII restriction site were introduced into the targeting construct by PCR-based site-directed mutagenesis. A loxP- and FLP recombination target (FRT)-flanked neo gene under the control of the phosphoglycerate kinase gene (PGK) promoter was inserted into the middle of the intron between exons 10 and 11 as a positive selection marker. At the 3′ end of the targeting vector, the diphtheria toxin A fragment gene (DT-A) driven by the MC1 promoter was added as a negative selection marker.
Generation of CRY2(S557A) knock-in mice.
CRY2(S557A) knock-in mice (CDB accession no. CDB0492K [http://www.cdb.riken.jp/arg/mutant%20mice%20list.html]) were generated according to standard methods (http://www.cdb.riken.jp/arg/Methods.html). The targeting construct (Fig. 1A) was linearized with NotI and electroporated into TT2 cells (23). The targeted embryonic stem (ES) cell clones were isolated and then microinjected into ICR 8-cell-stage embryos to generate chimeric mice, and the generated chimeric males were then mated with C57BL/6J females. Germ line transmission was verified by PCR as well as Southern blotting of tail DNA. For deletion of the neo gene, the obtained heterozygous animals were crossed with CAG-FLPe Tg mice (21). For behavioral analysis and molecular characterization, F0 chimeric mice were backcrossed on the C57BL/6J background for more than eight generations. Heterozygous offspring were intercrossed to produce homozygous mutant animals and their littermate controls. Primers used for genotyping were forward primer 5′-CTCAC CTTCA GAGCA AGGAC-3′ and reverse primer 5′-AAGTA GACAT GGTGG CTCGT ATC-3′. The length of DNA fragments after PvuII digestion is shown in Fig. 1D.
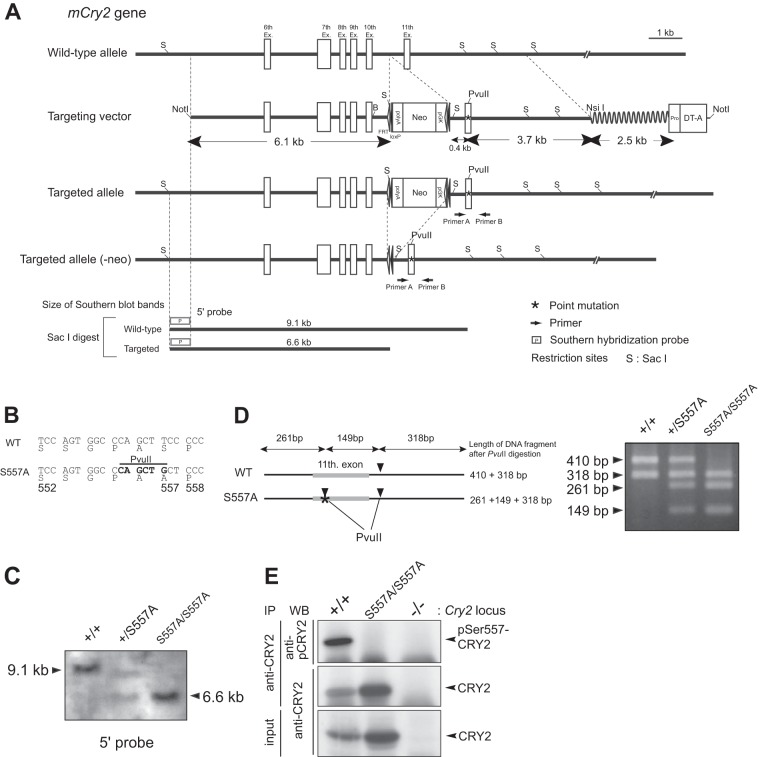
FIG 1.
Generation of CRY2(S557A) knock-in mice. (A) Gene-targeting strategy. Shown are schematic representations of the mouse WT Cry2 gene (exons 6 to 11), the targeting vector, the homologously recombined allele, and the homologously recombined allele followed by neo cassette deletion. (B) S557A mutation in the mouse genome. An additional PvuII site at the mutation site was created to identify the genotype. (C) Southern blotting of mouse genomic DNA to confirm homologous recombination. Mouse genomic DNA extracted from ES cells was digested by SacI and hybridized with a 5′ probe whose position is indicated in panel A. (D) Length of fragments digested by PvuII after PCR using primers A and B as shown in panel A. Mice were genotyped by the differences in fragment length. (E) Phosphorylation level of CRY2-Ser557 in mouse liver at ZT18. The nuclear extract of the liver lysate was immunoprecipitated with anti-CRY2 antibody, followed by immunoblot analysis with phosho-Ser557-specific antibody (anti-pCRY2). WB, Western blot.
Southern blot analysis.
Approximately 10 μg of genomic DNA was digested with SacI, electrophoresed on a 0.7% agarose gel, and transferred onto a Hybond+ nylon membrane (Amersham). The membrane was hybridized with a digoxigenin-dUTP-labeled probe and amplified from genomic DNA by using forward primer 5′-GAGCT CCAAG TTCTG GCAGT-3′ and reverse primer 5′-TGGAC TCTTT ACAGA GAAGG-3′. The probe was detected by an alkaline phosphatase-labeled antidigoxigenin Fab fragment (Roche). The positive signals were visualized with chloro-5-substituted adamantyl-1,2-dioxetane phosphate (CSPD)-enhanced chemiluminescence (Roche).
Wheel-running activity.
Five- to ten-week-old mice were housed individually in cages equipped with running wheels. The animals were maintained in a light-tight chamber at constant temperature (23°C ± 1°C) and humidity (65% ± 10%). Mice were entrained to a light-dark (LD) cycle for at least 5 weeks and released into constant darkness (DD). The wheel revolutions were recorded in 5-min bins and analyzed with ClockLab analysis software (Actimetrics). The circadian period of the activity rhythms in DD was determined by using a chi-square periodogram procedure with ClockLab. Mice were exposed to a 30-min light pulse (white light; 150 lx) at circadian time 14 (CT14) or CT22. Activity onsets were calculated by ClockLab to quantify the light-induced phase shift.
Real-time monitoring of cellular rhythms in mouse liver.
CRY2(S557A) knock-in mice were crossed with PER2::LUC knock-in mice (The Jackson Laboratory) (22). Mouse liver tissue was chopped with a razor and cultured in recording medium (phenol red-free Dulbecco's modified Eagle's medium [DMEM; Sigma-Aldrich] supplemented with 10% fetal bovine serum [Equitech Bio, Inc.], 3.5 mg/ml glucose, 25 U/ml penicillin, 25 μg/ml streptomycin, 0.1 mM luciferin, and 10 mM HEPES-NaOH [pH 7.0]) on a Millicell cell culture insert (Millipore). Bioluminescence emitted from the liver slice was recorded by using a dish-type photomultiplier (LumiCycle [Actimetrics] or Kronos [ATTO]). Data analyses were performed by using LumiCycle analysis software (Actimetrics).
Immunoblotting.
Proteins separated by SDS-PAGE were transferred onto a polyvinylidene difluoride membrane (Millipore). The blots were blocked in blocking solution (1% [wt/vol] skim milk in TBS [Tris-buffered saline] [50 mM Tris-HCl, 140 mM NaCl, 1 mM MgCl2 {pH 7.4}]) for 1 h at 37°C and then incubated overnight at 4°C with a primary antibody in blocking solution. The signals were visualized by an enhanced chemiluminescence detection system (PerkinElmer Life Science). The blot membrane was subjected to densitometric scanning, and the band intensities were quantified by using Image Gauge Ver.4.0 software (Fujifilm Science Lab). Primary antibodies used were anti-β-actin, anti-Flag, anti-normal mouse IgG (Sigma), anti-TATA binding protein (TBP), anti-myc, anti-green fluorescent protein (anti-GFP), anti-Raf1 (Santa Cruz Biotechnology), anti-PER2 (Alpha Diagnostic Int.), anti-CLOCK, anti-BMAL1 (24), anti-pSer557-CRY2 antibody (18), anti-CRY1, and anti-CRY2 (13). The primary antibodies were detected by horseradish peroxidase-conjugated anti-rabbit or anti-mouse IgG (Kirkegaard and Perry Laboratories).
Isolation of nuclear and cytosolic fractions of mouse liver.
To analyze the temporal profiles of the expression of clock proteins and genes, mice were entrained to an LD cycle for at least 14 days and transferred to DD conditions. On the second day in DD, mice were sacrificed under dim red light (>640 nm). Mouse liver was homogenized for total protein extraction with buffer A (20 mM Tris-HCl, 1% [vol/vol] Triton X-100, 10% [vol/vol] glycerol, 137 mM NaCl, 1 mM dithiothreitol [DTT], 2 mM EDTA, 4 μg/ml aprotinin, 4 μg/ml leupeptin, 50 mM NaF, 1 mM Na3VO4, 1 mM phenylmethylsulfonyl fluoride [PMSF] [pH 7.8 at 4°C]). Cytosolic and nuclear fractions were isolated from liver and cultured cells as described previously (24).
Immunoprecipitation.
For immunoprecipitation (IP), mouse liver nuclear protein at zeitgeber time 18 (ZT18) or liver cytosolic protein at CT18 was dissolved in IP buffer (20 mM HEPES-NaOH, 137 mM NaCl, 2 mM EDTA, 10% glycerol, 1% Triton X-100, 1 mM DTT, 4 μg/ml aprotinin, 4 μg/ml leupeptin, 50 mM NaF, 1 mM Na3VO4, 1 mM PMSF [pH 7.8 at 4°C]). The extract was incubated with a precipitating antibody at 4°C for 2 h or overnight, followed by incubation with 20 μl protein G-Sepharose at 4°C for 2 h. The beads were washed three times with IP buffer, and the final precipitates were subjected to immunoblotting analysis.
Degradation assay.
For degradation assays of endogenous CRY2, mouse embryonic fibroblasts (MEFs) prepared from wild-type (WT) mice and S557A knock-in mice were treated with 10 μM MG132 (Calbiochem) for 6 h. The culture medium was exchanged with fresh medium containing 100 μg/ml cycloheximide (CHX; Nakalai Tesque), and cells were cultured for 4 or 7 h. The cells were harvested, followed by immunoblot analysis. For assays of overexpressed Flag-CRY2 and myc-PER2, transfected NIH 3T3 cells were treated with 100 μg/ml CHX (Nakalai Tesque), and the cells were cultured for 3 or 6 h. The cells were harvested, followed by immunoblot analysis.
Cell culture and plasmids for transfection.
MEFs were prepared as previously described (13). MEFs, NIH 3T3 cells (RIKEN Cell Bank), and HEK293T cells were cultured and passaged under 5% CO2 in DMEM (Nissui) containing 1.8 mg/ml NaHCO3, 4.5 mg/ml glucose, 100 U/ml penicillin, 100 μg/ml streptomycin, and 10% fetal bovine serum (Equitech Bio, Inc.). NIH 3T3 and HEK293T cells were transiently transfected by using Lipofectamine Plus reagent (Invitrogen) and Lipofectamine 2000 reagent (Invitrogen), respectively, according to the manufacturer's protocols. Mammalian expression vectors encoding Flag-CRY2(WT) and Flag-CRY2(S557A) were described previously (17). Plasmid pEGFP-C1 (Clontech) was used as the transfection control. A vector encoding 6×Myc epitope-tagged PER2 (termed myc-PER2 in the figures) was kindly provided by Louis Ptacek (University of California, San Francisco).
RT-PCR.
Total RNA from mouse liver and MEFs was prepared by using TRIzol reagent (Invitrogen) according to the manufacturer's protocol. Total RNA was reverse transcribed with Superscript II (Invitrogen) and an anchored (dT)16 primer, and the reaction mixture was treated with RNase H (TaKaRa, Japan). The reverse transcripts from liver samples were subjected to real-time PCR (Applied Biosystems) by using GoTaq master mix (Promega) and gene-specific primers. The primers used were G3pdh-Fw (5′-ACGGG AAGCT CACTG GCATG GCCTT-3′), G3pdh-Rv (5′-CATGA GGTCC ACCAC CCTGT TGCTG-3′), Per1-Fw (5′-CAGGC TAACC AGGAA TATTA CCAGC-3′), Per1-Rv (5′-CACAG CCACA GAGAA GGTGT CCTGG-3′), Per2-Fw (5′-GGCTT CACCA TGCCT GTTGT-3′), Per2-Rv (5′-GGAGT TATTT CGGAG GCAAG TGT-3′), Cry1-Fw (5′-CCCAG GCTTT TCAAG GAATG GAACA-3′), Cry1-Rv (5′-TCTCA TCATG GTCAT CAGAC AGAGG-3′), Cry2-Fw (5′-GGGAC TCTGT CTATT GGCAT CTG-3′), Cry2-Rv (5′-GTCAC TCTAG CCCGC TTGGT-3′), Dbp-Fw (5′-AATGA CCTTT GAACC TGATC CCGCT-3′), Dbp-Rv (5′-GCTCC AGTAC TTCTC ATCCT TCTGT-3′), Bmal1-Fw (5′-GCAGT GCCAC TGACT ACCAA GA-3′), Bmal1-Rv (5′-TCCTG GACAT TGCAT TGCAT-3′), Dec1-Fw (5′-ATCAG CCTCC TTTTT GCCTT C-3′), Dec1-Rv (5′-AGCAT TTCTC CAGCA TAGGC AG-3′), Cyp51-Fw (5′-ATACA ACAAT GATCC ACACC CC-3′), and Cyp51-Rv (5′-TCAGA ACCAC ACTCT TCAAC CC-3′). mRNAs from MEFs were quantified by quantitative reverse transcription-PCR (RT-PCR) as described previously (25).
RESULTS
Generation of CRY2(S557A) knock-in mouse.
We previously demonstrated that Ala mutation of Ser557 (S557A mutation), the priming phosphorylation site of CRY2, completely blocked phosphorylation-dependent CRY2 degradation (18). On the other hand, Glu mutation of Ser557 (S557E mutation) did not mimic the phospho-Ser557 state because the S557E mutation failed to enhance GSK-3β-mediated phosphorylation of CRY2. Thus, we employed the S557A mutation to investigate the role of Ser557 phosphorylation of CRY2 in the clockwork in vivo. We designed mutant mice carrying the Ser-to-Ala mutation at amino acid 557 of CRY2. We constructed a targeting vector with a nucleotide replacement for the S557A mutation in the Cry2 gene (Fig. 1A and B). Homologous recombination and the induced point mutation were confirmed by Southern blot analysis and PCR (Fig. 1C and D). CRY2 protein expressed in the liver of homozygous mutant mice showed no immunoreactivity with the antibody recognizing Ser557-phosphorylated CRY2 (18) (Fig. 1E), validating the S557A mutation in vivo. Homozygous CRY2(S557A) knock-in mice were normal in appearance, and the genotypic distribution of the offspring followed Mendelian inheritance.
The S557A mutation lengthened the period of circadian rhythms.
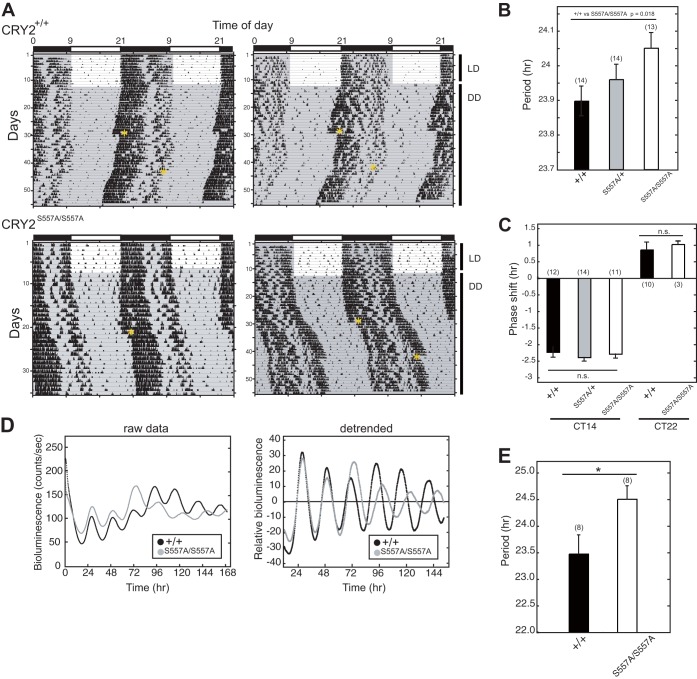
In constant darkness (DD), CRY2(S557A) knock-in mice exhibited a significantly longer free-running period of the wheel-running activity rhythm than their wild-type (WT) littermates (Fig. 2A and B). Both WT and mutant mice, on the other hand, exhibited a similar phase delay or advance of the activity rhythms when exposed to a 30-min light pulse at CT14 or CT22, respectively. No significant alterations in phase shifts were observed between genotypes (Fig. 2C).
FIG 2.
Wheel-running activities and cellular rhythms of CRY2(S557A) knock-in mice. (A) Wheel-running activity records for WT (CRY2+/+) and homozygous mutant (CRY2S557A/S557A) mice are shown in a double-plot format. Animals entrained to LD cycles were transferred to DD. Asterisks in the actogram indicate the time point when a 30-min light pulse was given. (B) Periodogram estimates of the period for each genotype. Data for the 7th to 21st days in DD were used to calculate the circadian period. The circadian periods were plotted as means with standard errors of the means. The number of animals is indicated in parentheses (*, P < 0.05 by Student's t test). (C) Phase shifts in response to a 30-min light pulse at CT14 or CT22 (n.s., nonsignificant difference). (D) PER2::LUC bioluminescence rhythms of mouse liver slices. For detrended data, we subtracted the averaged bioluminescence signals for 24 h. (E) Circadian periods of the liver from the 4th peak to the 6th peak (means with standard errors of the means; n = 8) (*, P < 0.05 by Student's t test).
We then examined the circadian period of the liver clock as a representative of peripheral clocks. CRY2(S557A) knock-in mice were crossed with PER2::LUC homozygous reporter mice, in which PER2 is expressed in a circadian manner as a fusion protein with luciferase (LUC) (22). The bioluminescence rhythms recorded from cultured liver slices indicated that the S557A mutation significantly lengthened the circadian period of the liver clock (Fig. 2D and E). Together, our results revealed that Ser557 phosphorylation of CRY2 plays a pivotal role in controlling the oscillation speed of behavioral circadian rhythms in vivo and the peripheral clock in the liver.
Expression profile of clock proteins in CRY2(S557A) knock-in mice.
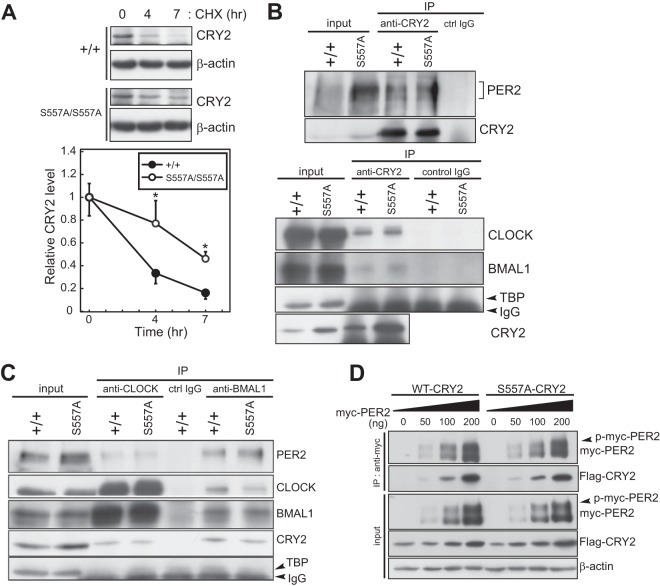
We next investigated the in vivo effect of the S557A mutation on the molecular property of CRY2. We first determined the stability of endogenous CRY2 protein in mouse embryonic fibroblasts (MEFs) prepared from the knock-in and WT mice. In the presence of CHX, a protein synthesis inhibitor, CRY2(S557A) was more stabilized than CRY2(WT) (Fig. 3A), indicating that Ser557-dependent degradation of endogenous CRY2 is attenuated in mutant mice. Coimmunoprecipitation experiments using liver lysates showed that CRY2(S557A) formed a complex with PER2, BMAL1, and CLOCK similarly to CRY2(WT) (Fig. 3B and C). The activity of CRY2 binding to PER2 expressed in HEK293T cells was also largely unaffected by the S557A mutation (Fig. 3D).
FIG 3.
Molecular properties of CRY2(S557A) in mutant mice. (A) Degradation assay of endogenous CRY2. MEFs prepared from WT or CRY2(S557A) mutant mice were treated with MG132 for 6 h and then treated with CHX. The cells were collected at the indicated time points. Quantified intensities were plotted (means with standard errors of the means; n = 3) by setting the peak value to 1 (*, P < 0.05 by Student's t test). (B) Immunoprecipitation experiments examining the interaction of CRY2 with PER2, CLOCK, and BMAL1 in mouse liver at ZT18. Proteins in liver nuclear extracts were immunoprecipitated with anti-CRY2. (C) Proteins in liver nuclear extracts at ZT18 were immunoprecipitated with anti-CLOCK or anti-BMAL1, and the immunoprecipitated product was probed with anti-CRY2, anti-CLOCK, anti-BMAL1, and anti-PER2 antibodies. Normal mouse IgG was used as the control. (D) Interaction of overexpressed CRY2(S557A) with myc-PER2 in HEK293T cells. myc-PER2 was expressed together with Flag-CRY2(WT) or its mutant protein, Flag-CRY2(S557A), in HEK293T cells. Flag-CRY2(WT) or Flag-CRY2(S557A) was coimmunoprecipitated with anti-myc antibody. p-myc-PER2 indicates the phosphorylated form of myc-PER2.
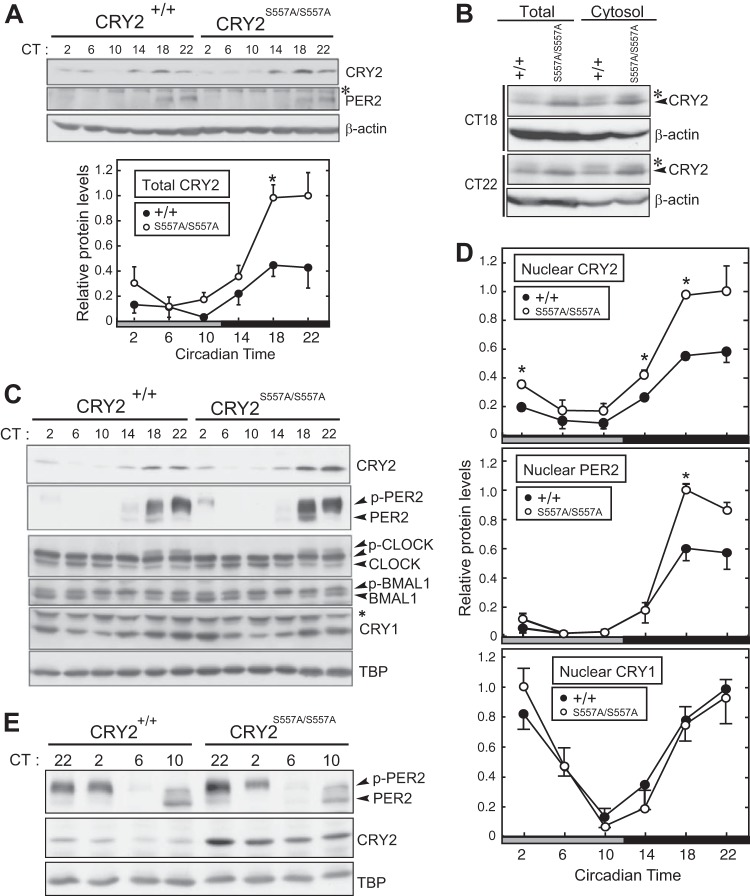
We then analyzed the temporal expression profiles of the clock proteins in the liver. CRY2 levels in total liver lysates of mutant mice, compared to WT mice, were markedly elevated during the subjective night (CT14 to -22) (Fig. 4A). On the other hand, no detectable change in total PER2 levels between mutant and WT mice was observed (Fig. 4A). Upregulations of CRY2 protein by the S557A mutation were observed in both cytosolic (Fig. 4B) and nuclear (Fig. 4C and D) fractions during the subjective night. We found that nuclear PER2 protein was also upregulated at CT18 and CT22 in the mutant liver (Fig. 4C and D). On the other hand, nuclear CRY1 levels were almost unaffected by the mutation (Fig. 4C and D). We also observed no significant alteration between the WT and the S557A mutant in not only the nuclear CLOCK and BMAL1 levels but also the temporal profiles of their protein band shifts (Fig. 4C), which represent their phosphorylation states (24). Importantly, nuclear CRY2 protein remained at a higher level in mutant liver, even during the subjective day of the subsequent cycle, when CRY2 levels in WT liver were kept very low in the nucleus (Fig. 4E). The excess CRY2-PER2 complex in the nucleus is most likely attributed to the period-lengthening effect of the S557A mutation (Fig. 2B and E). It is evident that Ser557-dependent regulation of CRY2 contributes to maintaining both CRY2 and PER2 protein levels in the appropriate range in cell nuclei.
FIG 4.
Clock protein profiles for mutant mouse liver. (A) Protein rhythms of CRY2 and PER2 in total lysates of mouse liver. The liver tissues were isolated at the indicated times on the second day in DD. The bottom panel shows the quantification of CRY2 (means with standard errors of the means; n = 3). The asterisk in the blot indicates a nonspecific band. (B) Expression profiles of CRY2 protein in whole-liver lysates and the cytosolic fraction at CT18 and CT22. Asterisks in the blot indicate nonspecific bands. (C) Circadian profiles of expression levels of CRY2, PER2, CRY1, CLOCK, and BMAL1 proteins in liver nuclear extract. TBP was used as a loading control and a nuclear marker. The asterisk indicates a nonspecific band. p-PER2, p-CLOCK, and p-BMAL1 indicate the phosphorylated forms of these proteins. (D) Quantification of CRY2, PER2, and CRY1 protein levels (means with standard errors of the means; n = 3). (*, P < 0.05 by Student's t test). (E) Circadian profiles of CRY2 and PER2 protein expression in liver nuclear extract in the declining phase of CRY2 of the next circadian day. The liver tissues were isolated at the indicated times on the second day (CT22) and the third day (CT2, -6, and -10) in DD. p-PER2 indicates the phosphorylated form of PER2.
Temporal mRNA expression profiles of clock genes.
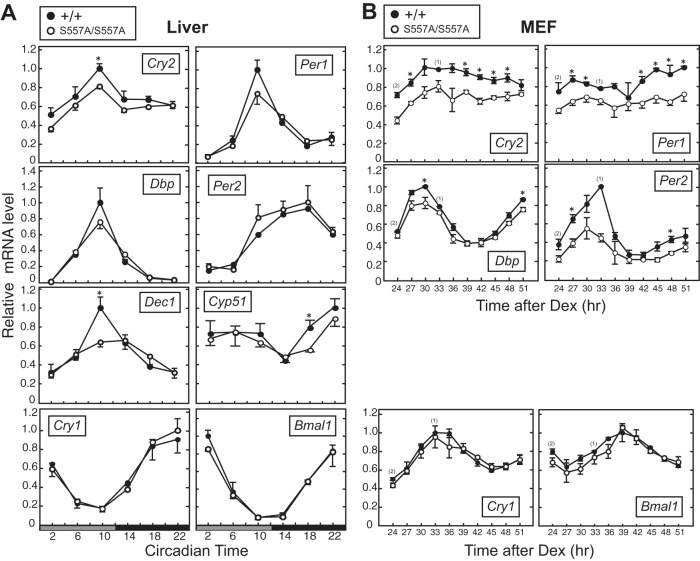
Because CRY2 is a strong repressor of E-box-dependent transcription (9), the S557A mutation-induced increase in the level of the CRY2 protein was expected to alter E-box-dependent gene transcriptions. In the liver of CRY2(S557A) knock-in mice, mRNA levels of the representative E-box-regulated genes Cry2, Per1, Dbp, and Dec1 all showed trends toward downregulation compared to those in the liver of WT mice (Fig. 5A). In MEFs, the S557A mutation caused significant reductions in the mRNA abundances of Cry2, Per1, Per2, and Dbp as well (Fig. 5B). On the other hand, expression levels of Cry1 and Bmal1, whose transcriptions are regulated mainly through the RORE cis element (26, 27), were intact in mutant mouse liver and MEFs (Fig. 5A and B). The mRNA levels of Cyp51, known to be regulated by CRY (28), were also decreased by the S557A mutation (Fig. 5A). These results indicate that Ser557 of CRY2 is required for normal expression of a subset of the clock genes as well as proper outputs of the circadian clock.
FIG 5.
Temporal expression profiles of clock genes in CRY2(S557A) knock-in mice. (A) Expression rhythms of clock genes in mouse liver. Mouse liver was isolated at the indicated time points on the second day in DD. mRNA levels of the clock genes were quantified by real-time quantitative PCR (means with standard errors of the means; n = 3) (*, P < 0.05 by Student's t test). (B) mRNA levels of clock genes in MEFs synchronized by dexamethasone (Dex) pulse treatment. Twenty-four hours after the dexamethasone pulse, cells were harvested at intervals of 3 h. mRNA abundance was quantified by quantitative RT-PCR (means with standard errors of the means; n = 3 [n = 2 for data for the WT at 24 h and n = 1 for data for the WT at 33 h]).
CRY2 regulates the cellular distribution of PER2.
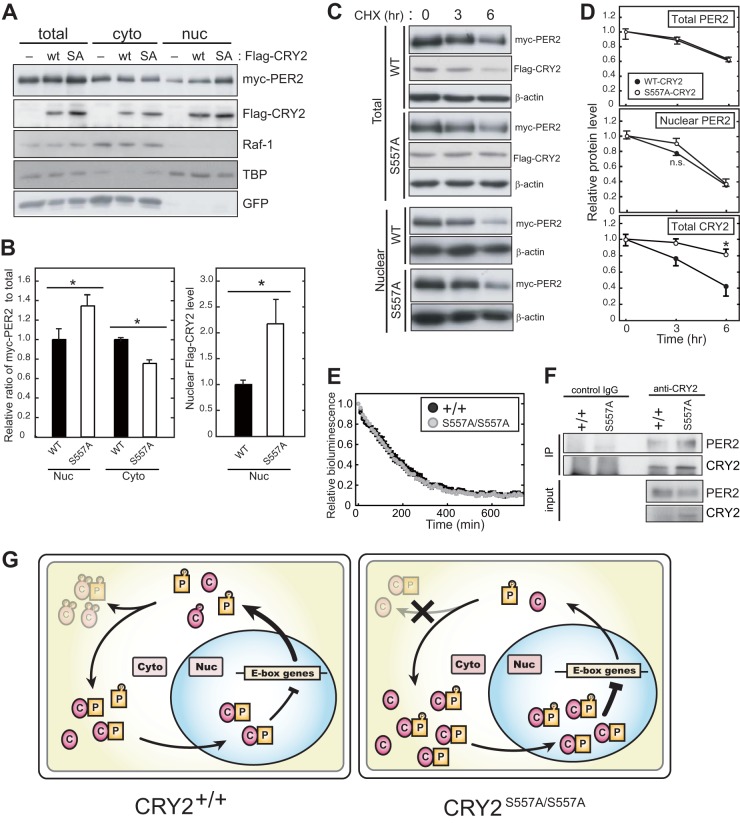
It is well established that PER proteins accumulate in the nucleus during the nighttime when E-box-mediated transcription is inhibited (29, 30), and its nuclear transport is a critical period-determining step (31). In the liver of CRY2(S557A) knock-in mice, nuclear PER2 levels were remarkably increased, whereas total PER2 levels were largely unaffected (Fig. 4A and D). These observations suggest that CRY2 phosphorylation regulates the intracellular distribution of the CRY2-PER2 complex. To investigate whether the S557A mutation affects the nuclear accumulation of the CRY2-PER2 complex in cultured cells, we examined the intracellular distribution of myc-PER2 coexpressed with CRY2(WT) or CRY2(S557A). The S557A mutation allowed CRY2 to accumulate more abundantly in both the cytosolic and nuclear fractions (Fig. 6A and B). Although Ser557 is located near the nuclear localization signal (NLS) (32), the intracellular distribution of CRY2 between the cytosol and nucleus was not affected by the S557A mutation (Fig. 4B and C and 6A). On the other hand, nuclear occupancy of myc-PER2 was increased in CRY2(S557A)-expressing cells (Fig. 6A and B). Of note, no significant change in the decay rate of the nuclear myc-PER2 levels was observed following CRY2(S557A) expression (Fig. 6C, bottom, and D). These results exclude the possibility that the nuclear accumulation of PER2 observed in mutant mice is due to the stabilization of nuclear PER2. In fact, in the presence of CHX, the bioluminescence signals of endogenous PER2::LUC in S557A mutant and WT MEFs showed decay rates indistinguishable from each other (Fig. 6E). Coimmunoprecipitation assays showed that the level of PER2 bound with CRY2 in the liver cytosol was increased in the knock-in mice, whereas the cytosolic PER2 level was not increased in the mutant mice (Fig. 6F). It is most likely that overaccumulation of CRY2(S557A) in the cytosol enhances the formation of the CRY2-PER2 complex and facilitates the nuclear transport of (CRY2-)PER2. In summary, we propose that the disruption of the phosphorylation-dependent regulation of CRY2 at Ser557 leads to the excessive accumulation of the negative factors, resulting in an abnormal periodicity of the circadian clock.
FIG 6.
Intracellular distribution of PER2 regulated by CRY2 protein. (A) Effect of CRY2(WT) or CRY2(S557A) on the intracellular distribution of PER2 in NIH 3T3 cells. myc-PER2 together with Flag-CRY2(WT) or Flag-CRY2(S557A) was transiently expressed in NIH 3T3 cells. The cells were fractionated into the cytosolic and nuclear fractions, and the protein levels of myc-PER2 and Flag-CRY2 were quantified by Western blotting. Raf-1 and TBP were used as the cytosol and nucleus markers, respectively. SA, S557A. (B) Ratios of nuclear and cytosolic myc-PER2 to total myc-PER2 (means with standard errors of the means; n = 3) (*, P < 0.05 by Student's t test). (C) Effect of CRY2(WT) or CRY2(S557A) on PER2 stability in NIH 3T3 cells. myc-PER2 was transiently expressed together with CRY2(WT) or CRY2(S557A) in NIH 3T3 cells. Forty-two hours after transfection, transfected cells were treated with 100 μg/ml CHX for 3 or 6 h. Temporal changes of myc-PER2 and Flag-CRY2 protein levels were analyzed by Western blotting. (D) Quantification of data from panel C. Relative protein levels were plotted by setting the peak value to 1.0 (means with standard errors of the means; n = 3) (*, P < 0.05 by Student's t test). (E) Decay rates of bioluminescence signals of PER2::LUC in MEFs in the presence of CHX treatment. MEFs were prepared from CRY2+/+ PER2LUC/LUC mice or CRY2S557A/S557A PER2LUC/LUC double mutant mice. The bioluminescence signals derived from PER2::LUC were monitored in recording medium containing 100 μg/ml CHX. Relative bioluminescence signals were plotted by setting the peak value to 1.0 (means with standard errors of the means; n = 6). (F) Interaction of CRY2 and PER2 in the cytosolic fraction. Proteins in the liver cytosolic fraction prepared at ZT18 were immunoprecipitated with anti-CRY2 antibody and the immunoprecipitated product were probed with anti-CRY2 and anti-PER2 antibodies. Normal mouse IgG was used as the control. (G) Molecular model for the role of Ser557 phosphorylation in the circadian clock. P and C indicate PER2 proteins and CRY2 proteins, respectively.
DISCUSSION
We propose a molecular model for Ser557 phosphorylation-dependent degradation of CRY2 in the mammalian circadian clock (Fig. 6G). In WT mouse liver, phosphorylation at Ser557 of translated CRY2 allows subsequent phosphorylation at Ser553 by GSK-3β, leading to proteasome-mediated CRY2 degradation (17). This degradation mechanism suppresses the overaccumulation of CRY2 in both the cytosol and, as a consequence, the nucleus. We previously found that the phosphorylation of CRY2 by GSK-3β is dependent on phospho-Ser557, and therefore, the S557A mutation completely blocks the GSK-3β-mediated degradation of CRY2 and upshift of the CRY2 protein band representing Ser553 phosphorylation (17, 18). Thus, in mutant mouse liver, it is most likely that the mutation of Ser557 attenuates the second phosphorylation at Ser553 and that CRY2(S557A) evades the sequential phosphorylation-dependent degradation (Fig. 3A). As a result, CRY2 expression is maintained at a higher level in mutant mice (Fig. 4C and E). Importantly, the S557A mutation of CRY2 enhanced the nuclear accumulation of PER2 as well as CRY2 (Fig. 4C and D). Because overaccumulated CRY2 forms a complex with PER2 more abundantly (Fig. 6F), it is suggested that elevated levels of CRY2(S557A) proteins facilitate the nuclear translocation of PER2 protein (Fig. 4C and 6B). This upregulation of the nuclear CRY2-PER2 complex should underlie the inhibition of E-box-mediated transcription of the clock genes (Fig. 5A), because the repressor activity of CRY2(S557A) is comparable to that of CRY2(WT) (33). The excessive nuclear accumulation of CRY2-PER2 should delay the restart of next-cycle transcriptional activation mediated by CLOCK-BMAL1, resulting in a lengthening of the circadian period in mutant mice (Fig. 2B and E).
In this study, we showed that CRY2 knock-in mice exhibited longer circadian periods. However, we should mention that the period-lengthening effect of the mutation on the central clock was modest compared to the large effect on the peripheral liver clock. In general, the central clock in the suprachiasmatic nucleus (SCN) is more resistant to genetic perturbation than peripheral clocks because of the tight coupling among SCN neurons (34). In a previous study, we showed that the knockdown of Dyrk1A, a protein kinase responsible for Ser557 phosphorylation and overexpression of CRY2(S557A) in cultured cells, shortened the circadian period of the cellular rhythm (17). These effects of kinase knockdown and overexpression of CRY2(S557A) in the presence of endogenous CRY2(WT) on the circadian period appear opposite to those of the S557A mutation in vivo demonstrated in the present study (Fig. 2B and E). Such a difference is probably attributable to the fact that the S557A mutation of endogenous CRY2 in mice (this study) caused a more severe effect on the nuclear abundance of the CRY2-PER2 complex than in previous studies (17). Indeed, Dyrk1A knockdown advanced the timing of nuclear CRY2 accumulation, whereas it had no significant effect on CRY2 levels at peak timing (17). Also, for S557A mutant-overexpressing cells, it is predicted that the nuclear CRY2 level is not as high as that in CRY2(S557A) knock-in cells for the following reasons. First, in the previous overexpression study, we used NIH 3T3 cells stably expressing CRY2(S557A), in which the mutant CRY2 protein level was approximately half that of endogenous wild-type CRY2 (17), and therefore, accumulation of nuclear CRY2 could be far more modest than that observed in the present study. Second, in previous studies, CRY2(S557A) overexpression was driven by a constitutive promoter, which may fail to regulate the proper time-of-day-dependent nuclear accumulation of the CRY2-PER2 complex (17). In the knock-in cells, on the other hand, nuclear CRY2 protein levels were dramatically increased during the accumulating phase. Importantly, CRY2 accumulation was accompanied by increased nuclear PER2 transport (Fig. 4C and D). The overaccumulated CRY2-PER2 complex should need a longer time to be cleared from the nucleus, leading to the lengthening of the circadian period. This model is supported by a previous simulation-based study indicating that an increase of PER2 nuclear transport rather than its stability has a period-lengthening effect (31). Here, we demonstrate that Ser557 phosphorylation of CRY2 is critical not only for the slow accumulation of nuclear CRY2 protein in the increasing phase (ZT10 to -18) but also for suppressing the overaccumulation of CRY2 protein at peak timing (ZT22) (Fig. 4C and D).
Collectively, we demonstrate the in vivo roles of the C-terminal phosphorylation signal of CRY2 in the circadian clockwork. Ser557 phosphorylation of CRY2 promotes CRY2 degradation and inhibits the overaccumulation of the CRY2-PER2 complex in the nucleus, which is critical for period determination of the circadian clock (Fig. 6G). To date, a variety of posttranslational modification sites in clock proteins have been identified, but the site-specific physiological functions of these modifications remained mostly undetermined in the central clock. The present study clearly demonstrated that site-specific mutagenesis of a gene is a strong strategy for understanding the molecular mechanism of the circadian clock.
ACKNOWLEDGMENTS
We thank Francis A. Stewart (Dresden University of Technology) for CAG-FLPe transgenic mice.
This work was supported in part by grants-in-aid for scientific research and by the Global COE program (Integrative Life Science Based on the Study of Biosignaling Mechanisms) from MEXT, Japan. A.H. and N.K. were supported by JSPS research fellowships for young scientists.
Footnotes
Published ahead of print 6 October 2014
REFERENCES
- 1.Takahashi JS. 1995. Molecular neurobiology and genetics of circadian rhythms in mammals. Annu. Rev. Neurosci. 18:531–553. 10.1146/annurev.ne.18.030195.002531. [DOI] [PubMed] [Google Scholar]
- 2.Dunlap JC. 1999. Molecular bases for circadian clocks. Cell 96:271–290. 10.1016/S0092-8674(00)80566-8. [DOI] [PubMed] [Google Scholar]
- 3.Bunger MK, Wilsbacher LD, Moran SM, Clendenin C, Radcliffe LA, Hogenesch JB, Simon MC, Takahashi JS, Bradfield CA. 2000. Mop3 is an essential component of the master circadian pacemaker in mammals. Cell 103:1009–1017. 10.1016/S0092-8674(00)00205-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shearman LP, Sriram S, Weaver DR, Maywood ES, Chaves I, Zheng B, Kume K, Lee CC, Hastings MH, Reppert SM. 2000. Interacting molecular loops in the mammalian circadian clock. Science 288:1013–1019. 10.1126/science.288.5468.1013. [DOI] [PubMed] [Google Scholar]
- 5.Schibler U. 2006. Circadian time keeping: the daily ups and downs of genes, cells, and organisms. Prog. Brain Res. 153:271–282. 10.1016/S0079-6123(06)53016-X. [DOI] [PubMed] [Google Scholar]
- 6.Gallego M, Virshup DM. 2007. Post-translational modifications regulate the ticking of the circadian clock. Nat. Rev. Mol. Cell Biol. 8:139–148. 10.1038/nrm2106. [DOI] [PubMed] [Google Scholar]
- 7.Grimaldi B, Nakahata Y, Kaluzova M, Masubuchi S, Sassone-Corsi P. 2009. Chromatin remodeling, metabolism and circadian clocks: the interplay of CLOCK and SIRT1. Int. J. Biochem. Cell Biol. 41:81–86. 10.1016/j.biocel.2008.08.035. [DOI] [PubMed] [Google Scholar]
- 8.Reischl S, Kramer A. 2011. Kinases and phosphatases in the mammalian circadian clock. FEBS Lett. 585:1393–1399. 10.1016/j.febslet.2011.02.038. [DOI] [PubMed] [Google Scholar]
- 9.Kume K, Zylka MJ, Sriram S, Shearman LP, Weaver DR, Jin X, Maywood ES, Hastings MH, Reppert SM. 1999. mCRY1 and mCRY2 are essential components of the negative limb of the circadian clock feedback loop. Cell 98:193–205. 10.1016/S0092-8674(00)81014-4. [DOI] [PubMed] [Google Scholar]
- 10.Siepka SM, Yoo S-H, Park J, Song W, Kumar V, Hu Y, Lee C, Takahashi JS. 2007. Circadian mutant Overtime reveals F-box protein FBXL3 regulation of cryptochrome and period gene expression. Cell 129:1011–1023. 10.1016/j.cell.2007.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Busino L, Bassermann F, Maiolica A, Lee C, Nolan PM, Godinho SI, Draetta GF, Pagano M. 2007. SCFFbxl3 controls the oscillation of the circadian clock by directing the degradation of cryptochrome proteins. Science 316:900–904. 10.1126/science.1141194. [DOI] [PubMed] [Google Scholar]
- 12.Godinho SIH, Maywood ES, Shaw L, Tucci V, Barnard AR, Busino L, Pagano M, Kendall R, Quwailid MM, Romero MR, O'Neill J, Chesham JE, Brooker D, Lalanne Z, Hastings MH, Nolan PM. 2007. The after-hours mutant reveals a role for Fbxl3 in determining mammalian circadian period. Science 316:897–900. 10.1126/science.1141138. [DOI] [PubMed] [Google Scholar]
- 13.Hirano A, Yumimoto K, Tsunematsu R, Matsumoto M, Oyama M, Kozuka-Hata H, Nakagawa T, Lanjakornsiripan D, Nakayama KI, Fukada Y. 2013. FBXL21 regulates oscillation of the circadian clock through ubiquitination and stabilization of cryptochromes. Cell 152:1106–1118. 10.1016/j.cell.2013.01.054. [DOI] [PubMed] [Google Scholar]
- 14.Yoo S-H, Mohawk JA, Siepka SM, Shan Y, Huh SK, Hong H-K, Kornblum I, Kumar V, Koike N, Xu M, Nussbaum J, Liu X, Chen Z, Chen ZJ, Green CB, Takahashi JS. 2013. Competing E3 ubiquitin ligases govern circadian periodicity by degradation of CRY in nucleus and cytoplasm. Cell 152:1091–1105. 10.1016/j.cell.2013.01.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gao P, Yoo S-H, Lee K-J, Rosensweig C, Takahashi JS, Chen BP, Green CB. 2013. Phosphorylation of the cryptochrome 1 C-terminal tail regulates circadian period length. J. Biol. Chem. 288:35277–35286. 10.1074/jbc.M113.509604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lamia KA, Sachdeva UM, DiTacchio L, Williams EC, Alvarez JG, Egan DF, Vasquez DS, Juguilon H, Panda S, Shaw RJ, Thompson CB, Evans RM. 2009. AMPK regulates the circadian clock by cryptochrome phosphorylation and degradation. Science 326:437–440. 10.1126/science.1172156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kurabayashi N, Hirota T, Sakai M, Sanada K, Fukada Y. 2010. DYRK1A and glycogen synthase kinase 3, a dual-kinase mechanism directing proteasomal degradation of CRY2 for circadian timekeeping. Mol. Cell. Biol. 30:1757–1768. 10.1128/MCB.01047-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harada Y, Sakai M, Kurabayashi N, Hirota T, Fukada Y. 2005. Ser-557-phosphorylated mCRY2 is degraded upon synergistic phosphorylation by glycogen synthase kinase-3 beta. J. Biol. Chem. 280:31714–31721. 10.1074/jbc.M506225200. [DOI] [PubMed] [Google Scholar]
- 19.Hirota T, Lewis WG, Liu AC, Lee JW, Schultz PG, Kay SA. 2008. A chemical biology approach reveals period shortening of the mammalian circadian clock by specific inhibition of GSK-3β. Proc. Natl. Acad. Sci. U. S. A. 105:20746–20751. 10.1073/pnas.0811410106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vitaterna MH, Selby CP, Todo T, Niwa H, Thompson C, Fruechte EM, Hitomi K, Thresher RJ, Ishikawa T, Miyazaki J. 1999. Differential regulation of mammalian period genes and circadian rhythmicity by cryptochromes 1 and 2. Proc. Natl. Acad. Sci. U. S. A. 96:12114–12119. 10.1073/pnas.96.21.12114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schaft J, Ashery-Padan R, van der Hoeven F, Gruss P, Stewart AF. 2001. Efficient FLP recombination in mouse ES cells and oocytes. Genesis 31:6–10. 10.1002/gene.1076. [DOI] [PubMed] [Google Scholar]
- 22.Yoo S-H, Yamazaki S, Lowrey PL, Shimomura K, Ko CH, Buhr ED, Siepka SM, Hong H-K, Oh WJ, Yoo OJ. 2004. PERIOD2::LUCIFERASE real-time reporting of circadian dynamics reveals persistent circadian oscillations in mouse peripheral tissues. Proc. Natl. Acad. Sci. U. S. A. 101:5339–5346. 10.1073/pnas.0308709101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yagi T, Tokunaga T, Furuta Y, Nada S, Yoshida M, Tsukada T, Saga Y, Takeda N, Ikawa Y, Aizawa S. 1993. A novel ES cell line, TT2, with high germline-differentiating potency. Anal. Biochem. 214:70–76. 10.1006/abio.1993.1458. [DOI] [PubMed] [Google Scholar]
- 24.Yoshitane H, Takao T, Satomi Y, Du NH, Okano T, Fukada Y. 2009. Roles of CLOCK phosphorylation in suppression of E-box-dependent transcription. Mol. Cell. Biol. 29:3675–3686. 10.1128/MCB.01864-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kon N, Hirota T, Kawamoto T, Kato Y, Tsubota T, Fukada Y. 2008. Activation of TGF-β/activin signalling resets the circadian clock through rapid induction of Dec1 transcripts. Nat. Cell Biol. 10:1463–1469. 10.1038/ncb1806. [DOI] [PubMed] [Google Scholar]
- 26.Ueda HR, Hayashi S, Chen W, Sano M, Machida M, Shigeyoshi Y, Iino M, Hashimoto S. 2005. System-level identification of transcriptional circuits underlying mammalian circadian clocks. Nat. Genet. 37:187–192. 10.1038/ng1504. [DOI] [PubMed] [Google Scholar]
- 27.Ukai-Tadenuma M, Yamada RG, Xu H, Ripperger JA, Liu AC, Ueda HR. 2011. Delay in feedback repression by cryptochrome 1 is required for circadian clock function. Cell 144:268–281. 10.1016/j.cell.2010.12.019. [DOI] [PubMed] [Google Scholar]
- 28.Oishi K, Miyazaki K, Kadota K, Kikuno R, Nagase T, Atsumi G-I, Ohkura N, Azama T, Mesaki M, Yukimasa S, Kobayashi H, Iitaka C, Umehara T, Horikoshi M, Kudo T, Shimizu Y, Yano M, Monden M, Machida K, Matsuda J, Horie S, Todo T, Ishida N. 2003. Genome-wide expression analysis of mouse liver reveals CLOCK-regulated circadian output genes. J. Biol. Chem. 278:41519–41527. 10.1074/jbc.M304564200. [DOI] [PubMed] [Google Scholar]
- 29.Lee C, Etchegaray JP, Cagampang FR, Loudon AS, Reppert SM. 2001. Posttranslational mechanisms regulate the mammalian circadian clock. Cell 107:855–867. 10.1016/S0092-8674(01)00610-9. [DOI] [PubMed] [Google Scholar]
- 30.Yagita K, Tamanini F, van Der Horst GT, Okamura H. 2001. Molecular mechanisms of the biological clock in cultured fibroblasts. Science 292:278–281. 10.1126/science.1059542. [DOI] [PubMed] [Google Scholar]
- 31.St John PC, Hirota T, Kay SA, Doyle FJ. 2014. Spatiotemporal separation of PER and CRY posttranslational regulation in the mammalian circadian clock. Proc. Natl. Acad. Sci. U. S. A. 111:2040–2045. 10.1073/pnas.1323618111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sakakida Y, Miyamoto Y, Nagoshi E, Akashi M, Nakamura TJ, Mamine T, Kasahara M, Minami Y, Yoneda Y, Takumi T. 2005. Importin alpha/beta mediates nuclear transport of a mammalian circadian clock component, mCRY2, together with mPER2, through a bipartite nuclear localization signal. J. Biol. Chem. 280:13272–13278. 10.1074/jbc.M413236200. [DOI] [PubMed] [Google Scholar]
- 33.Sanada K, Harada Y, Sakai M, Todo T, Fukada Y. 2004. Serine phosphorylation of mCRY1 and mCRY2 by mitogen-activated protein kinase. Genes Cells 9:697–708. 10.1111/j.1356-9597.2004.00758.x. [DOI] [PubMed] [Google Scholar]
- 34.Liu AC, Welsh DK, Ko CH, Tran HG, Zhang EE, Priest AA, Buhr ED, Singer O, Meeker K, Verma IM, Doyle FJ, III, Takahashi JS, Kay SA. 2007. Intercellular coupling confers robustness against mutations in the SCN circadian clock network. Cell 129:605–616. 10.1016/j.cell.2007.02.047. [DOI] [PMC free article] [PubMed] [Google Scholar]