Abstract
The TIS11/tristetraprolin (TTP) CCCH tandem zinc finger proteins are major effectors in the destabilization of mRNAs bearing AU-rich elements (ARE) in their 3′ untranslated regions. In this report, we demonstrate that the Drosophila melanogaster dTIS11 protein is short-lived due to its rapid ubiquitin-independent degradation by the proteasome. Our data indicate that this mechanism is tightly associated with the intrinsically unstructured, disordered N- and C-terminal domains of the protein. Furthermore, we show that TTP, the mammalian TIS11/TTP protein prototype, shares the same three-dimensional characteristics and is degraded by the same proteolytic pathway as dTIS11, thereby indicating that this mechanism has been conserved across evolution. Finally, we observed a phosphorylation-dependent inhibition of dTIS11 and TTP degradation by the proteasome in vitro, raising the possibility that such modifications directly affect proteasomal recognition for these proteins. As a group, RNA-binding proteins (RNA-BPs) have been described as enriched in intrinsically disordered regions, thus raising the possibility that the mechanism that we uncovered for TIS11/TTP turnover is widespread among other RNA-BPs.
INTRODUCTION
Posttranscriptional regulation plays a central role in the control of gene expression in eukaryotic cells. cis regulatory elements, often located in the 3′ untranslated region (UTR), influence the localization, translation, and degradation status of mRNAs (1, 2) by binding trans-acting microRNAs or RNA-binding proteins (see references 3 and 4 for reviews). AU-rich elements (AREs) are the most common cis-acting determinants found in metazoan mRNAs (5, 6). A large variety of AU-rich element-binding proteins (ARE-BPs) affect either positively or negatively the expression of their target ARE-containing mRNAs (reviewed in reference 7). For example, proteins of the TIS11/tristetraprolin (TTP) and AUF-1 families most frequently promote the degradation of their target mRNA (reviewed in references 8 and 9); TIA-1/R proteins inhibit mRNA translation (10), while HuR binding to AREs generally induces stabilization of the RNA messengers (11).
Redundant, additive, or antagonist effects for ARE-BPs have been reported when these factors are expressed in the same cells. Therefore, the outcome of ARE-mediated gene regulation is largely dependent on the functional availability of each ARE-BP (12–14). In this context, the degradation rate for each ARE-BP is an important parameter for determining the output of ARE control.
The proteasome is an essential proteolytic complex whose main function is to degrade damaged or unnecessary proteins. Proteasome substrates are generally marked by covalent attachment of a Lys-48-type polyubiquitin chain to one of the target's internal lysines. The process requires the combined activity of three classes of enzymes: ubiquitin-activating enzymes (E1), ubiquitin-conjugating enzymes (E2), and ubiquitin ligases that ensure specific targeting for degradation (E3). The ubiquitin-proteasome system (UPS) has been described as essential for the degradation of several ARE-BPs and the regulation of ARE mRNAs. A positive correlation was established between AUF1 degradation by the proteasome and ARE mRNA turnover (15). More recent data have also demonstrated that ubiquitination of AUF1 by the E3 ligase b-Trcp leads to its destruction by the proteasome and to the stabilization of its target mRNAs (16). HuR ubiquitination and subsequent proteasomal degradation upon heat shock has also been described (17). Upon proteasome inhibition, HuR is stabilized, which in turn prevents degradation of the TRAIL receptor DR5 mRNA in an ARE-dependent manner (18). Interestingly, HuR is also subject to nondegradative ubiquitination by the p97-UBXD8 complex, which in turn induces the remodeling of mRNAs that contain ARE-bound HuR protein (19).
The mechanism controlling the degradation of the TTP/TIS11 protein family is not fully understood. In mammals, this family is composed of three members, TTP (ZFP36), BRF1 (ZFP36L1), and BRF2 (ZFP36L2), which all participate in ARE mRNA decay (reviewed in reference 8). The stability of TIS11/TTP proteins is known to be defined by their phosphorylation status, which in turn determines their degradation rate (20–22). Deregulation of this control can have dramatic effects on cell fate, leading to defects in the inflammatory response (23), increased radiation-induced lung toxicity (24), and tumorigenesis (25). Previous studies established the proteasome as the proteolytic complex responsible for the degradation of several TIS11/TTP proteins, as its inhibition stabilizes TTP (20) and BRF1 (21). PEST domains, which are susceptible to target proteins to the proteasome, are also predicted in human and mouse TTP, but mutation of these domains did not alter the level of the corresponding proteins (26). An association of TIS11/TTP proteins with the ubiquitination machinery was also described, as the Cul4B ubiquitin ligase was identified in TTP-containing messenger ribonucleoprotein particles (mRNPs). However, this enzyme did not target TTP for degradation (27). In addition, nondegradative Lys-63-type polyubiquitination of TTP was reported in response to tumor necrosis factor stimulation (in mouse embryonic fibroblast and human umbilical vein endothelial cell lines), thereby reassigning the biological function of TTP from ARE mRNA degradation to modulation of JNK signaling (28).
The molecular mechanism responsible for TIS11/TTP protein decay has remained elusive, in part due to the fact that proteasome inhibitors can affect the expression of this protein family at several levels. The transcriptional activation of the Zfp36 gene encoding TTP depends on NF-κB signaling, whose activity is regulated by the proteasome (20, 29). Moreover, proteasome inhibitors suppress the AKT pathway (30), which plays a central role in regulating BRF1 activity (31, 32). Proper marking of TIS11/TTP proteins as proteasomal substrates by Lys-48-type polyubiquitination has not been described, raising the question of how these proteins are targeted for proteolysis.
In order to get a better insight into the molecular mechanisms controlling the degradation of the TIS11/TTP family of ARE-BPs, we have investigated the mechanisms responsible for the degradation of the unique member of this family found in Drosophila melanogaster, dTIS11. We and others have shown that dTIS11 is able to induce the destabilization of ARE mRNAs (6, 33, 34). Our previous data also implicated the proteasome in the regulation of dTIS11, as pharmacological inhibition of this degradative complex resulted in the accumulation of dTIS11. We report here that dTIS11 is an unstable protein, constitutively produced but rapidly degraded by the proteasome. Recognition of dTIS11 by the proteosomal complex does not require prior ubiquitination, as a nonubiquitinable mutant is efficiently processed by the proteasome. In addition, a proteasome deprived of its ubiquitin-binding domains directly degrades in vitro-translated dTIS11 protein. These results establish dTIS11 as a substrate for the proteasome-dependent ubiquitin-independent proteolysis named “degradation by default.” Many proteins targeted to this degradative pathway possess unstructured, disordered regions that are thought to be important for proteasomal recognition (35, 36). dTIS11 may be an intrinsically disordered protein (IDP), as predicted by bioinformatics analysis, as we found that dTIS11 shares the main characteristics of IDPs, namely, high sensibility to proteolysis and heat resistance. Finally, we report that the mammalian family founding member TTP undergoes the same degradation process, establishing a novel and conserved regulatory mechanism for TIS11/TTP proteins.
MATERIALS AND METHODS
Reagents.
Clasto-lactacystin beta-lactone, epoxomicin, thapsigargin, bafilomycin A1, and NH4Cl were purchased from Sigma-Aldrich. Okadaic acid was purchased from LC Laboratories. Puromycin was purchased from Invivogen. MG132 was purchased from UBPbio.
Plasmids.
Expression vectors for dTIS11V5/HIS, dDREDDV5/HIS, and GFP-DCP1V5/HIS were obtained via reverse transcription-PCR (RT-PCR; for dTIS11 and dDREDD) or amplification from the previously described green fluorescent protein (GFP)-DCP1 construct (37) (kindly provided by E. Izaurralde, Max Planck Institute for Developmental Biology) and then insertion into the pMT/V5-His vector (Life Technologies). GFP-DCP1-CdTIS11V5/HIS was generated by fusion of the coding sequence for GFP-DCP1 with that of the dTIS11 C terminus (amino acids 214 to 436) and insertion into the pMT/V5-His vector. Expression vectors for hemagglutinin (HA)-tagged ubiquitin (Ub) and FLAG-tagged dTIS11 were obtained by replacement of the start codon by an HA or FLAG tag via RT-PCR and insertion into the pAc5.1/V5-His vector (Life Technologies) for HA-ubiquitin and into the pMT/V5-His vector for FLAG-dTIS11. A lysine-less dTIS11KR mutant was designed by replacement of all 14 lysines with arginines. The corresponding DNA sequence was synthesized by Genscript and subcloned into the pMT/V5-His vector (Life Technologies). The UbG76V-GFPV5/HIS coding sequence was generated by PCR from the previously described pcDNA3-UbG76V-eGFP-V5His_20aa construct (38) (Addgene) and inserted into the pMT/V5-His vector. The expression vector for PARP12HIS was kindly provided by D. Hutin (Laboratoire d'Immunobiologie, ULB). The lysine-less TTP mutant form was obtained by replacement of all lysines with arginines, synthesized by Genscript, and subcloned into the pcDNA3.1/Myc-His vector (Life Technologies). All constructs were verified by sequencing. The primers used for the cloning procedures are available on request.
Cell culture, treatments, RNA interference (RNAi), and transfection.
Drosophila S2 cells (Invitrogen) were maintained in serum-free medium (Express Five; Life Technologies) at 24°C. Cells were transfected with FuGENE HD (Promega) according to the manufacturer's instructions. For pMT constructs, transcription driven by the metallothionein promoter was induced by overnight incubation with CuSO4 (0.5 mM). RAW 264.7 and HEK293T cell lines were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum. RAW 264.7 cells were activated with lipopolysaccharide (100 ng/ml; Sigma-Aldrich) for 4 h. HEK293T cells were transfected with Lipofectamine 2000 (Life Technologies), following the manufacturer's instructions.
For RNA interference, double-stranded RNA (dsRNA) was produced with a MEGAscript RNAi kit (Life Technologies), following the manufacturer's instructions. S2 cells were incubated with 10 μg/ml dsRNA targeting β5 for 4 days or gfp or dtis11 for 7 days.
For mRNA half-life measurements, transcription was blocked by using actinomycin D (5 μg/ml; Sigma-Aldrich), and the cells were harvested after 30 min, 1 h, 2 h, and 3 h. Proteasome inhibitors were used in 4-h treatments at final concentrations of 5 μM for clasto-lactacystin-beta-lactone, 40 μM for MG132, and 10 μM for epoxomicin. Lysosome function inhibitors were used in 16-h treatments at final concentrations of 10 mM for NH4Cl, 1 μM for thapsigargin, and 150 nM for bafilomycin A1. Puromycin was used at 50 μg/ml.
For pulse-chase analyses, S2 cells maintained in serum-free medium (ESF 921; Expression Systems) were washed in labeling medium lacking methionine for 1 h, then labeled for 2 h in medium supplemented with 0.1 mCi/ml [35S]methionine (43.48 TBq/mmol; PerkinElmer).
Western blotting and antibodies.
Cells were lysed in EBC buffer (120 mM NaCl, 0.5% NP-40, 50 mM Tris-HCl [pH 8]) supplemented with phosphatase inhibitors NaF (100 mM), Na3VO4 (0.2 mM), and beta-glycerophosphate (10 mM), and also Complete EDTA-free protease inhibitor cocktail (Roche Applied Science). For ubiquitination detection experiments, cells were lysed in denaturing buffer (6 M urea, 10% glycerol, 20 mM Tris-HCl [pH 8], 1 M NaCl, 1% Triton X-100) supplemented with 15 mM N-ethylmaleimide (Sigma-Aldrich) and 25 μM clasto-lactacystin-beta-lactone. Protein extracts were then incubated on ice for 10 min and cleared by centrifugation. For nickel affinity chromatography purification, protein extracts were incubated with equilibrated Ni-nitrilotriacetic acid (NTA) Superflow beads (Qiagen) at 4°C for 1 h in the corresponding lysis buffer supplemented with 10 mM imidazole, washed in lysis buffer with 25 μM imidazole, and eluted with 150 μM imidazole lysis buffer.
Western blotting was performed with the following primary antibodies: anti-V5, anti-FLAG, anti-GFP, anti-Myc, and antiactin (Sigma-Aldrich) and anti-glyceraldehyde 3-phosphate dehydrogenase (anti-GAPDH; Santa Cruz Biotechnology). The detection of dTIS11 protein by Western blotting was performed with a previously described anti-dTIS11 antibody (39). The antiubiquitin specific antibody (FK2 clone; Enzo Life science) was used as previously described (40). The anti-TTP (Carp-3) and anti-PARP12 antibodies were kindly provided by W. Rigby (Dartmouth Hitchcock Medical Center) and D. Hutin, respectively. Quantifications were performed with the ImageJ software.
Northern blot assays and quantification.
Total RNA was purified using Tri reagent (MBI Fermentas) and analyzed as described previously (33). Quantification of the radioactive signals was performed with a PhosphorImager (GE Healthcare).
In vitro translation and 20S proteasome degradation assay.
In vitro translation was performed in rabbit reticulocyte lysate (Promega) in the presence of [35S]methionine (0.4 mCi/ml; 43.48 TBq/mmol) according to the manufacturer's instructions. In vitro-translated or cell lysate purified proteins were incubated with 1 μg or 0.5 μg purified human 20S proteasome (Enzo Life Sciences), respectively, in 20S buffer (20 mM Tris-HCl [pH 7.2], 1 mM EDTA, 1 mM dithiothreitol [DTT]) at 37°C for the indicated times. 35S-labeled proteins were revealed by SDS-PAGE followed by sodium salicylate gel staining and fluorography.
FastPP assay.
The fast pulse proteolysis (FastPP) assay was performed as described previously (41). Briefly, protein extracts were incubated with 1-mg/liter or 10-mg/liter thermolysin from Bacillus thermoproteolyticus rokko (Sigma-Aldrich) in TL buffer (10 mM CaCl2, 20 mM Na3PO4 [pH 7.2], 150 mM NaCl, 5 mM DTT) for 1 min at the indicated temperatures in a thermocycler. After 1 min, proteolysis was stopped by addition of 17 mM EDTA.
Heat resistance assay.
Protein extracts were heated at 95°C for 10 min and ultracentrifuged for 10 min at 100,000 × g. The supernatant was retrieved, and the pellet was resuspended in 1× Laemmli buffer.
Bioinformatics analysis.
Disorder prediction was performed using default settings with DISOPRED2 (42), IUpred (43), Pondr-fit (44), and DynaMine (45). Protein sequence alignment was performed with ClustalW (46).
RESULTS
dTIS11 is rapidly degraded in Drosophila S2 cells.
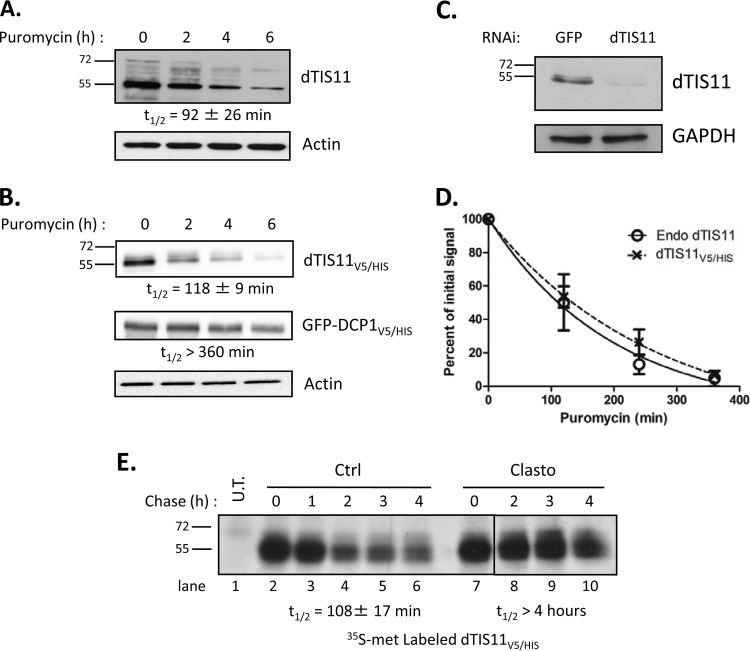
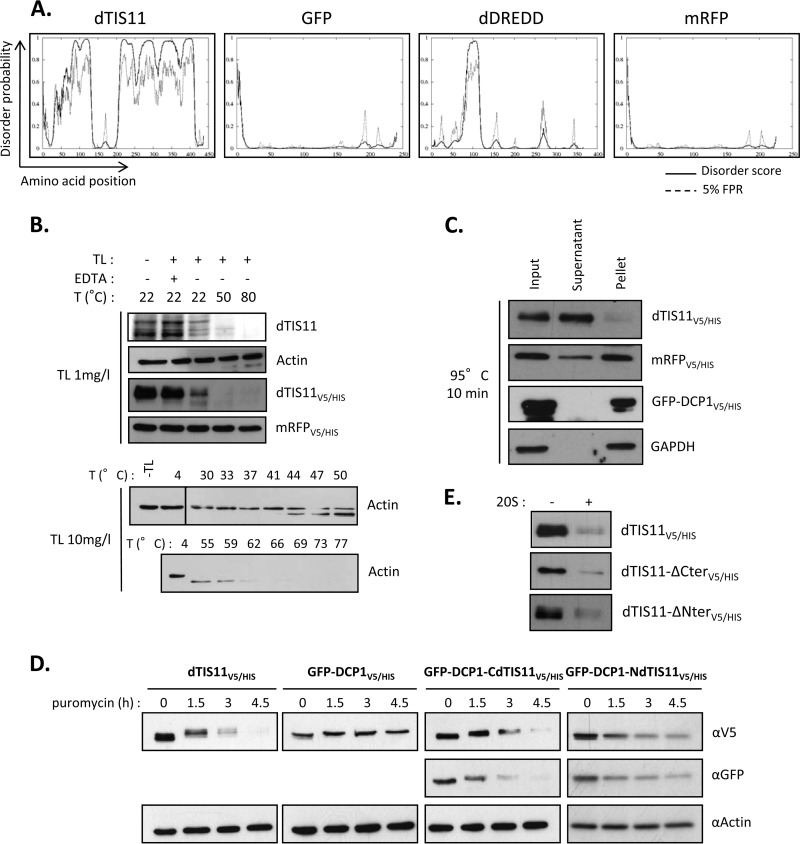
Protein half-lives are highly variable, ranging from a few minutes to several days in eukaryotes (47, 48). Protein decay is thus a key regulatory step, as its balance with protein production establishes a dynamic equilibrium that defines the steady-state level of expression. As a central effector in AU-rich element-mediated mRNA decay (33), the availability and activity of dTIS11 can affect the expression of many target messengers (6, 34). We sought to assess the stability of dTIS11 by measuring its half-life. Drosophila S2 cells were treated with the translation inhibitor puromycin for 0 to 6 h, and resulting levels of dTIS11 were evaluated by Western blotting. Specific detection of dTIS11 by Western blotting was verified upon RNAi inhibition of the corresponding gene (Fig. 1C). Figure 1A shows that dTIS11 was rapidly degraded, with a half-life of approximately 1.5 h (t1/2 = 92 ± 26 min [mean ± standard deviation, or SD]). Slower-migrating bands may correspond to phosphorylated dTIS11, as in vitro dephosphorylation by alkaline phosphatase results in comigration as a single 55-kDa band (data not shown). In parallel, we assessed the stability of a fusion dTIS11 protein, doubly tagged with a V5 epitope and a 6×His motif (dTIS11V5/HIS), in S2 cells. Figure 1B shows that dTIS11V5/HIS has a half-life similar to that of endogenous dTIS11 protein (t1/2 = 118 ± 9 min). In contrast, the control GFP-DCP1V5/HIS produced from the same plasmid backbone and the endogenous actin protein were stable, with half-lives longer than 6 h. Decay curves calculated from four independent experiments confirmed that endogenous and V5/His-tagged dTIS11 proteins are degraded with similar kinetics (Fig. 1D), which indicates that dTIS11V5/HIS may serve as a model for the analysis of the dTIS11 degradation process.
FIG 1.
Rapid degradation of dTIS11. (A and B) S2 cells untransfected (A) or transfected with expression vectors for dTIS11V5/HIS and GFP-DCP1V5/HIS (B) were treated with puromycin for the indicated times. Endogenous dTIS11 was detected by Western blotting with a dTIS11-specific antibody (see Materials and Methods). dTIS11V5/HIS and GFP-DCP1V5/HIS were detected by Western blotting with an anti-V5 antibody. (C) Specificity of the anti-dTIS11 antibody. S2 cells were treated with 10 μg/ml double-stranded RNA targeting GFP or dTIS11 for 7 days. Western blot analysis was performed with anti-dTIS11 and anti-GAPDH antibodies. (D) Quantification of dTIS11 and dTIS11V5/HIS half-lives. Results of four independent experiments performed as described for panels A and B were quantified with the ImageJ software, and the ratios of dTIS11 or dTIS11V5/HIS and actin signals were plotted. The amount of dTIS11 or dTIS11V5/HIS at time zero of puromycin treatment was set to 100% in each experiment and plotted on a linear graph. Half-lives were determined by exponential regression. Error bars represent SD for each experimental condition. (E) Pulse-chase analysis of dTIS11 half-life and effect of proteasome inhibition. Untransfected (U.T.) or dTIS11V5/HIS-expressing S2 cells were labeled with [35S]methionine-containing medium for 90 min, then chased with unlabeled medium supplemented with Clasto or dimethyl sulfoxide (DMSO) as a control, for the indicated times. dTIS11V5/HIS was precipitated on nickel-coupled agarose beads, and precipitates were resolved by SDS-PAGE. Acrylamide gels were analyzed by fluorography. Half-lives were determined by exponential regression after quantification of results of three independent experiments. Vertical bars indicate cuts between separated parts of the same blot.
Half-life measurement using translation inhibitors such as puromycin is indirect and can be affected by side effects of the drug. We therefore sought to confirm the dTIS11 half-life measurement by pulse-chase analysis. In the absence of immunoprecipitaion (IP)-grade antibodies for dTIS11, the experiment was performed on the tagged protein dTIS11V5/HIS. S2 cells expressing dTIS11V5/HIS were cultured in medium containing [35S]methionine and then chased with label-free medium and harvested at several time points. Total protein extracts were submitted to Ni2+-affinity chromatography, and purified dTIS11V5/HIS was analyzed by fluorography after SDS-PAGE separation. The stability of dTIS11V5/HIS directly measured by pulse-chase analysis (t1/2 = 108 ± 17 min) (Fig. 1E, lanes 1 to 6) was similar to that obtained in translation inhibition assays. Both experiments established dTIS11 as an unstable short-lived protein.
dTIS11 degradation requires proteasome activity.
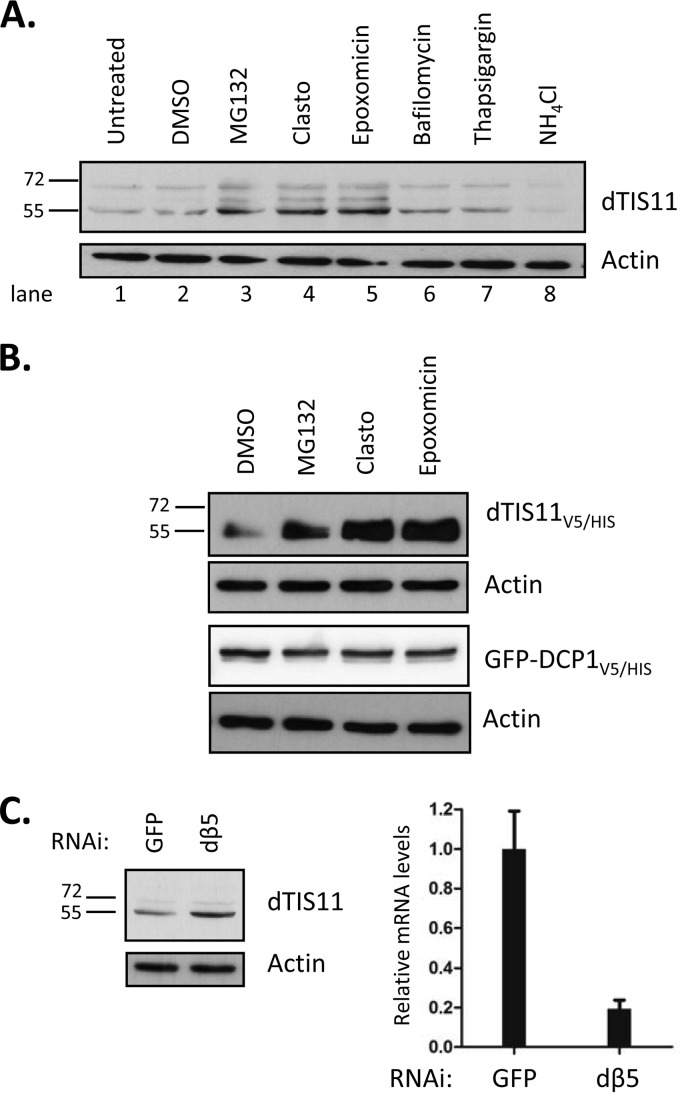
In eukaryotic cells, two pathways are responsible for most proteolytic events: slow selective digestion in the lysosome and targeted degradation by the proteasome. As dTIS11 appears to be efficiently and specifically degraded, we investigated the implications for the proteasome in this process. We previously showed that the proteasome inhibitor clasto-lactacystin-beta-lactone (Clasto) increases the level of dTIS11 protein detectable in S2 cells (33). Pulse-chase analysis of dTIS11V5/HIS degradation performed in S2 cells treated with Clasto suggested that this protein is indeed degraded in a proteasome-dependent manner, as its half-life increased from 108 min to more than 4 h in the presence of Clasto (Fig. 1E, lanes 7 to 10). To confirm this result and to assess the potential role of lysosomal function in endogenous dTIS11 proteolysis, S2 cells were incubated in the presence of inhibitors of both pathways. Treatment with MG132, Clasto, or epoxomicin, proteasome inhibitors of three different classes (49), induced a marked accumulation of dTIS11 in S2 cells (Fig. 2A, compare lanes 1 and 2 with lanes 3 to 5). In contrast, dTIS11 did not accumulate in response to treatment with bafilomycin and NH4Cl (Fig. 2A, compare lanes 1 and 2 with lanes 6 and 8) at doses where these inhibitors of lysosomal function block the accumulation of Lysotracker in S2 cells (data not shown). Inhibition of the lysosomal function by thapsigargin treatment provided similar results (Fig. 2A).
FIG 2.
dTIS11 degradation requires proteasome activity. (A and B) Effects of proteasome and lysosomal activity inhibitors on dTIS11 accumulation. S2 cells untransfected (A) or transfected with an expression vector encoding dTIS11V5/HIS or GFP-DCP1V5/HIS (B) were treated with the indicated inhibitors for 4 h (proteasome inhibitors) or 16 h (lysosomal activity inhibitors) before harvesting and analysis by Western blotting. (C) RNAi inhibition of the proteasome leads to dTIS11 accumulation. S2 cells were treated with double-stranded RNA for GFP or dβ5. (Left panel) Endogeneous dTIS11 and actin levels were assessed by Western blotting as described for Fig. 1. (Right panel) dβ5 mRNA levels were measured by quantitative RT-PCR and normalized with RPL32 mRNA levels.
As observed for endogenous dTIS11, the three classes of proteasome inhibitors increased the accumulation of dTIS11V5/HIS in transfected S2 cells (Fig. 2B, upper lane). In contrast, the accumulation levels of the GFP-DCP1V5/HIS control and of the endogenous actin protein were not modified by these treatments (Fig. 2B, lower lanes). Moreover, efficient inhibition of proteasome subunit β5 by RNA interference led to endogenous dTIS11 accumulation (Fig. 2C). Together, our results thus indicate that dTIS11 is highly unstable in Drosophila S2 cells because of its continuous degradation by the proteasome.
Ubiquitin-independent degradation of dTIS11 by the proteasome.
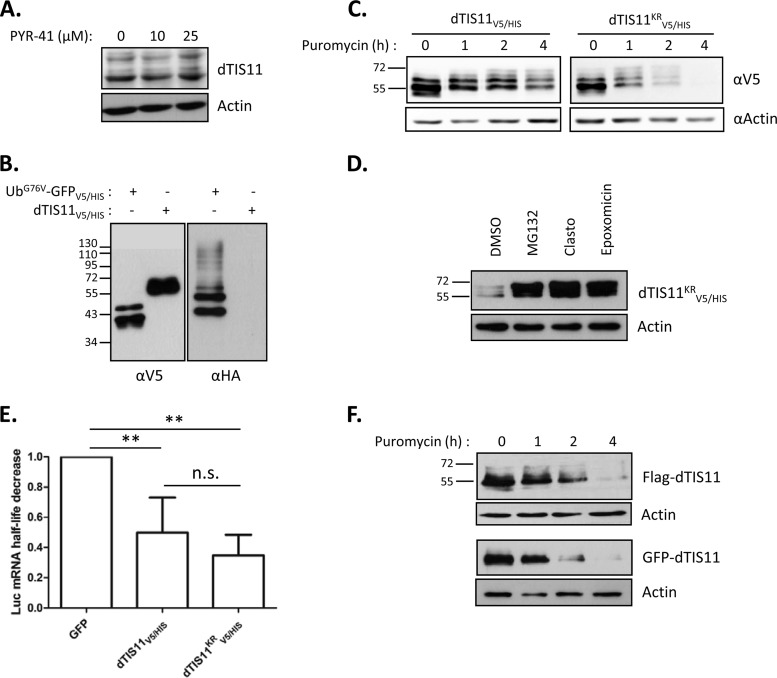
Because proteasomal activity is required for dTIS11 degradation, we investigated the implication of the ubiquitination system in dTIS11 processing. First, we sought to directly detect dTIS11 ubiquitination. This can be achieved by observing coprecipitation of ubiquitin with the protein of interest. Polyubiquitinated proteins were immunoprecipitated from S2 cell extracts by using an antiubiquitin specific antibody as previously described (40). Under these conditions, we failed to detect the endogenous dTIS11 protein in the immunoprecipitated fraction (data not shown). To assess the implication of the ubiquitin system in the degradation of endogenous dTIS11, S2 cells were treated with the E1 ubiquitin-activating enzyme inhibitor PYR-41 (50). Although this compound is active in S2 cells (51), we did not detect any increase of dTIS11 accumulation when S2 cells were treated with sufficient levels of PYR-41 (Fig. 3A).
FIG 3.
Ubiquitin-independent degradation of dTIS11 by the proteasome. (A) dTIS11 accumulation is insensitive to PYR-41 treatment. Cells were treated with PYR-41 at 10 μM for 16 h or at 25 μM for 2 h. Protein extracts were analyzed by Western blotting as described in the text. (B) Ubiquitination of dTIS11V5/HIS was not detectable. Protein extracts from Drosophila S2 cells coexpressing HA-ubiquitin with either UbG76VGPFV5/HIS or dTIS11V5/HIS were subjected to IMAC. Western blotting of Ni2+-NTA-retained proteins with anti-V5 and anti-HA antibodies revealed the presence of V5-tagged proteins and HA-ubiquitin in the His-enriched extract. (C and D) The lysine-less dTIS11KRV5/HIS mutant is unstable and degraded by the proteasome. S2 cells transfected with expression vectors encoding wild-type dTIS11V5/HIS or lysine-less dTIS11KRV5/HIS were treated with puromycin for the indicated times (C) or with the proteasome inhibitors MG132, Clasto, epoxomicin, or dimethyl sulfoxide (DMSO) as a control for 4 h (D). Protein extracts were analyzed by Western blotting by using the indicated antibodies. (E) dTIS11KRV5/HIS was functional and destabilized ARE-bearing mRNA. S2 cells expressing an ARE-fused luciferase reporter with GFPV5/HIS, dTIS11V5/HIS or dTIS11KRV5/HIS were treated with actinomycin D for up to 3 h. Luciferase mRNA levels were quantified by Northern blotting, and the associated half-lives were determined by exponential regression. The luciferase half-life decrease was calculated by reporting the half-life obtained under each condition and comparing it to that calculated for the GFP control. A Mann-Whitney U test was used for the statistical analysis. n.s., nonsignificant; *, P < 0.05; **, P < 0/01. (F) The context of the N-terminal amino acid does not control dTIS11 degradation. Flag-dTIS11- or GFP-dTIS11-expressing cells were treated with puromycin for the indicated times. Protein extracts were analyzed by Western blotting using anti-FLAG, anti-GFP, or antiactin antibodies.
In order to potentiate the ubiquitination process and facilitate its detection, S2 cells were cotransfected with expression vectors coding for dTIS11V5/HIS and for a HA-tagged ubiquitin (HA-ubiquitin). As a positive control, we also transfected cells with an expression vector for UbG76V-GFPV5/HIS, since this fusion is known to undergo efficient polyubiquitination and subsequent proteasomal degradation (38). The cells were then treated with MG132 to prevent degradation of the ubiquitinated moieties. Proteins were extracted in highly denaturing urea buffer supplemented with the deubiquitinase inhibitor N-ethylmaleimide to prevent hydrolysis of the ubiquitin-substrate bonds in the extract. Affinity chromatography was then performed on Ni2+-coupled beads for isolation of dTIS11V5/HIS or UbG76V-GFPV5/HIS, followed by separation via SDS-PAGE. Western blotting with an anti-V5 antibody showed that precipitation of both proteins occurred with similar efficiency (Fig. 3B, left panel). Interestingly, UbG76V-GFPV5/HIS was seen as two bands that were separated by a distance that matched the attachment of a single ubiquitin particle (8 kDa). Blotting of Ni2+-purified extracts with anti-HA antibodies showed the expected ladder/smear, which corresponded to polyubiquitinated UbG76V-GFPV5/HIS, but no HA-ubiquitin signal associated with dTIS11V5/HIS (Fig. 3B, right panel), which suggests that dTIS11V5/HIS is not predominantly ubiquitinated under these conditions.
Detection of polyubiquitination may be limited by the low sensitivity of the assay, rapid deubiquitination in the protein extracts, and/or specific degradation of the modified moieties. Therefore, we could not exclude that dTIS11 ubiquitination might still occur in S2 cells. We thus sought to determine whether cryptic ubiquitination is necessary for dTIS11 recognition by the proteasome. Because degradative ubiquitination is generally initiated by the covalent binding of a ubiquitin molecule to an internal lysine of the substrate (52), we analyzed the behavior of dTIS11 when mutated for all 14 lysines conservatively replaced with equally positively charged arginine residues (dTIS11KRV5/HIS). S2 cells were transfected with expression vectors encoding dTIS11V5/HIS or the dTIS11KRV5/HIS mutant and were treated or not with puromycin for 4 h. Western blot analysis showed that simultaneous mutation of all ubiquitin-accepting sites did not stabilize dTIS11 (Fig. 3C), while the mutant remained a substrate for the proteasome, as shown by the dTIS11KRV5/HIS accumulation upon MG132, Clasto, or epoxomicin treatment (Fig. 3D). Importantly, simultaneous replacement of as many as 14 residues may extensively alter the protein structure, leading to the production of a polypeptide recognized as aberrant and targeted for degradation, regardless of its natural degradative process. We thus verified dTIS11KRV5/HIS functionality in promoting ARE mRNA decay. To do this, we compared the abilities of dTIS11V5/HIS and dTIS11KRV5/HIS to destabilize a reporter mRNA construct (Luc-ARE; described in a previous study [33]). S2 cells expressing the reporter mRNA were cotransfected with expression vectors for dTIS11V5/HIS, dTIS11KRV5/HIS, or GFPV5/HIS as a control. We observed by Northern blotting and subsequent quantification of the Luc-ARE mRNA half-life that, in contrast to GFP, wild-type and lysine-less dTIS11were able to destabilize the target reporter mRNA to a similar extent (Fig. 3E), arguing in favor of the production of dTIS11KRV5/HIS as a properly folded and active protein.
While the vast majority of ubiquitination processes occur on internal lysines of the substrate, examples of alternative ubiquitination have been reported in which the N-terminal amino acid is used as the ubiquitination accepting site (see reference 53 for a review). We thus verified that modifying the environment of dTIS11 at its N terminus by addition of either a FLAG or a GFP coding sequence upstream of the dtis11 start codon AUG (FLAG-dTIS11 and GFP-dTIS11) did not alter its degradation pattern. As for the lysine-less mutant, this modification did not stabilize the protein (Fig. 3F).
Taken together, our results suggest that while dTIS11 is a proteasome substrate in Drosophila S2 cells, its degradation does not require prior marking by the ubiquitination machinery.
Direct degradation of dTIS11 by the 20S proteasome in the absence of ubiquitin and ATP.
Ubiquitin-independent proteasome-dependent degradation has been reported for a limited but growing number of proteins. It has been described to involve different forms of the proteasome: the 20S proteasome, the 26S proteasome, which has been most extensively studied, or other proteasome variants (see reference 36 for a recent review). The 20S core is formed by four heptameric rings that are stacked as a barrel-shaped complex, defining a narrow 53-Å degradation chamber shut by the N termini of its outer subunits. As such, and while bearing all three proteolytic activities of the proteasome, it alone stands gated, unable to recognize ubiquitin or to process large folded proteins. In contrast, association of the 20S core with 19S caps into the 26S proteasome allows for the degradation of ubiquitinated proteins, as the 19S provides opening of the 20S gated entrance, ubiquitin recognition, and ATP-dependent substrate unfolding (reviewed in reference 54).
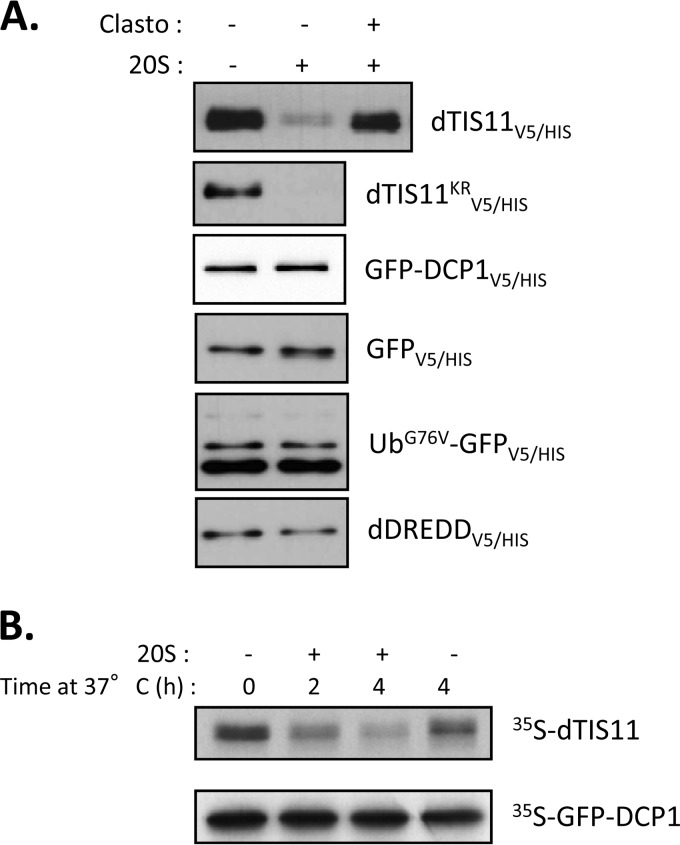
As we established dTIS11 as a bona fide substrate for the proteasome with no requirement for prior ubiquitination, we investigated the ability of the 20S proteasome to directly recognize and process dTIS11. First, we tested whether purified 20S could degrade dTIS11 extracted from Drosophila S2 cells. For this, S2 cells were transfected with expression vectors for dTIS11V5/HIS, GFP-dTIS11KRV5/HIS, or several negative controls (GFPV5/HIS, GFP-DCP1V5/HIS, UbG76V-GFPV5/HIS, and dDREDDV5/HIS). The corresponding proteins were purified by Ni2+-affinity chromatography, incubated with commercially available purified 20S proteasome in the absence of ATP, and submitted to Western blot analysis. Figure 4A shows that the 20S degradation assay for dTIS11V5/HIS and dTIS11KRV5/HIS resulted in thorough degradation of these proteins, while all negative controls, including ubiquitinated UbG76V-GFPV5/HIS, remained unaffected. Coincubation with clasto-lactacystin-beta-lactone reverted dTIS11V5/HIS degradation, confirming the dependence on proteasomal activity in this assay. These results showed that dTIS11 can be directly degraded by the 20S proteasome, in the absence of the ATP necessary for the unfolding processes. To test whether dTIS11 was also sensitive to degradation by the 20S proteasome when produced in a heterologous, acellular system, in vitro translation of dTIS11 mRNA was programmed in reticulocyte lysate and the translation products were subsequently exposed to purified 20S proteasome. Figure 4B shows efficient and 20S-dependent degradation of in vitro-translated dTIS11, while the negative-control GFP-DCP1 remained stable. Altogether, these results demonstrate that dTIS11 can be directly recognized and processed by the 20S proteasome in the absence of ATP and ubiquitin.
FIG 4.
Direct recognition of dTIS11 by the 20S proteasome. (A) ATP-independent degradation of dTIS11 by purified 20S proteasome. The indicated proteins were purified using IMAC from S2 cells transfected with corresponding expression vectors and incubated for 1 h in the presence of 0.5 μg of purified 20S proteasome with or without Clasto. Western blot analysis was performed with anti-V5 antibodies. (B) 20S proteasome-mediated degradation of dTIS11 produced by in vitro translation. dTIS11 and GFP-DCP1 were translated in vitro in the presence of [35S]methionine and then incubated with 1 μg 20S proteasome for the indicated times. Labeled proteins were revealed by SDS-PAGE followed by sodium salicylate gel staining and fluorography.
dTIS11 behaves as an intrinsically disordered protein.
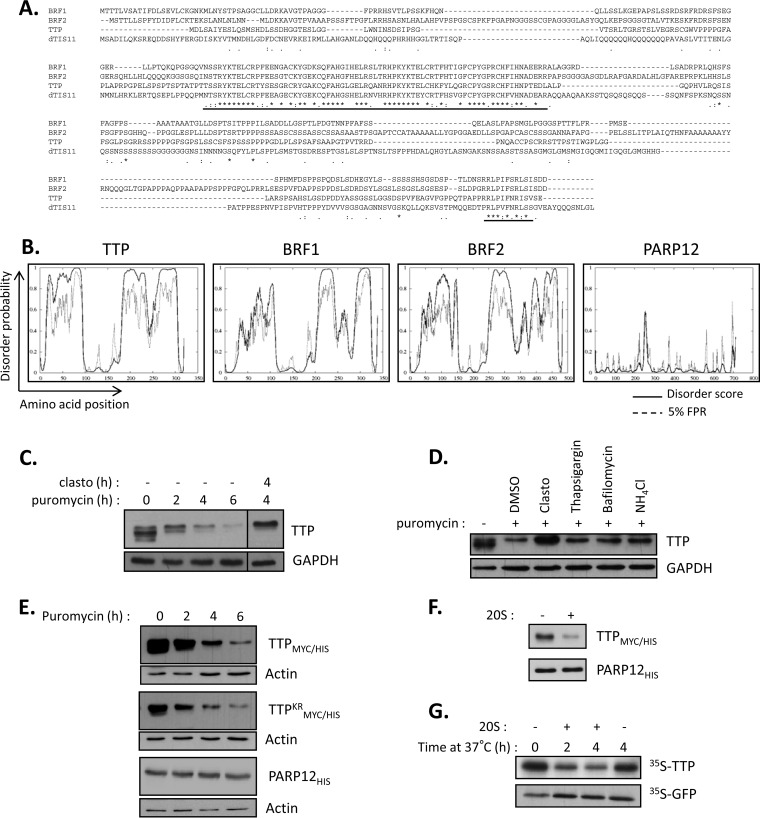
The ubiquitin-independent proteasome-dependent degradation, also known as “degradation by default” (55), is of increasing prevalence as the number of proteins described to undergo such degradation grows. Strikingly, many of these proteins present large disordered domains that lack a stable tertiary structure; hence, their recent classification as IDP (56). Moreover, protein disorder normalized for protein length was shown as the major feature in determining a short half-life for proteins (57). As dTIS11 is involved in this “by default” pathway, we examined the possibility that dTIS11 protein would present one or more IDRs. We first performed bioinformatic analysis on the dTIS11 amino acid sequence, looking for predicted IDRs. Figure 5A shows the results returned by DISOPRED2, an algorithm trained on high-resolution X-ray crystal data to calculate a probability of disorder for each amino acid of the query protein. The predicted disorder for dTIS11 appeared very high throughout the whole sequence, with the notable exceptions of the tandem zinc finger domains (amino acids 135 to 163 and 173 to 201), as well as short stretches at the N and C termini. In contrast, the disorder profiles for GFP and Drosophila DREDD were much flatter, which correlated with the inability of the 20S proteasome to process these proteins in vitro (Fig. 4A). Similar results were obtained when using Pondr-fit, IUpred, and DynaMine, three additional disorder prediction algorithms (data not shown).
FIG 5.
dTIS11 is a potential intrinsically disordered protein. (A) dTIS11 is predicted to be highly disordered. Disorder profiles were calculated using DISOPRED2 and are shown for the indicated proteins. Plain lines, disorder score per residue; dashed lines, prediction threshold at a 5% false-positive rate. (B) dTIS11 is a preferred substrate for proteolysis by thermolysin. Whole-protein extracts from untransfected S2 cells (dTIS11 and actin panels) or dTIS11V5/HIS- or mRFPV5/HIS-expressing cells were incubated with 1 or 10 mg/liter TL for 1 min at the indicated temperatures. Western blot analysis was performed with anti-dTIS11, antiactin, or anti-V5 antibodies. Vertical bars indicate cuts between separated parts of the same blot. (C) Heat resistance of dTIS11. Whole-protein extracts from dTIS11V5/HIS-, mRFPV5/HIS-, or GFP-DCP1V5/HIS-expressing cells were boiled at 95°C for 10 min and then centrifuged at 100,000 × g for 10 min. Western blot analysis was performed on samples before centrifugation (input), and the soluble fraction was analyzed after centrifugation (supernatant) or the resuspended insoluble fraction (pellet). (D) dTIS11 N- and C-terminal domains act as signals for degradation. S2 cells expressing dTIS11V5/HIS, GFP-DCP1V5/HIS, GFP-DCP1-CdTIS11V5/HIS, or GFP-DCP1-NdTIS11V5/HIS were treated with puromycin for the indicated times. Western blot analysis was performed using the indicated antibodies. (E) The dTIS11 N terminus and C terminus are dispensable for 20S recognition. The indicated proteins were purified using IMAC from S2 cells transfected with corresponding expression vectors and incubated for 1 h in the presence of 0.5 μg of purified 20S proteasome. Western blot analysis was performed with anti-V5 antibodies.
Because dTIS11 is predicted to have extended disordered regions and because it complies with the main hallmark of IDPs, namely, susceptibility to degradation by the 20S proteasome, we next sought to determine whether dTIS11 also shared biochemical properties with IDPs. FastPP is a pulse proteolysis assay that measures a protein's degree of structuration in relation to temperature (41). This assay takes advantage of the specificity for unfolded states of the heat-resistant protease thermolysin (TL), which cleaves nearby exposed hydrophobic residues Phe, Val, Ile, and Leu. Incubation with TL thus results in degradation of the preferred disordered substrates, sparing folded proteins. In parallel, the incubation temperature can be increased to favor unfolding and thus susceptibility to TL. The amount of TL and the temperature needed for the TL to digest a protein give a measure of its structuration state. Figure 5B shows the results for FastPP analysis for dTIS11. Protein extracts from S2 cells untransfected (panels for dTIS11 and actin), or expressing dTIS11V5/HIS or mRFPV5/HIS as a highly structured control (Fig. 5A, right panel), were incubated with 1 or 10 mg/liter of TL at 4, 22, 50, or 80°C and then analyzed by Western blotting. Endogenous as well as overexpressed dTIS11 were both efficiently degraded, starting at 22°C. Degradation of dTIS11 also required low TL concentrations (1 mg/liter) that proved insufficient for the degradation of the actin and mRFPV5/HIS controls, even at the highest temperature. In fact, actin degradation required a 10-fold-higher concentration of TL and higher temperatures, of 50 to 60°C (Fig. 5B, bottom panel). As we would expect for IDPs, we found that dTIS11 was highly susceptible to this degradation process. Our results suggest that the dTIS11 unfolded state is already achieved at S2 cell culture temperatures of 24°C.
Another feature common in IDPs is their resistance to high temperatures. Because most IDPs have a low hydrophobic content, they do not present a hydrophobic core and are less likely to lose solubility upon heating (see reference 58 for a review). We thus tested dTIS11 heat resistance. Protein extracts from S2 cells expressing dTIS11V5/HIS, mRFPV5/HIS, or GFP-DCP1V5/HIS were boiled at 95°C for 10 min and then ultracentrifuged at 100,000 × g to separate soluble from insoluble fractions. Western blot analysis showed that dTIS11V5/HIS was mainly soluble, in contrast with the insoluble control proteins mRFPV5/HIS, GFP-DCP1V5/HIS, and endogenous GAPDH (Fig. 5C). Together, our data indicate that dTIS11, a protein predicted to be mainly disordered, also behaves like an IDP in several aspects.
As dTIS11 appears to be disordered and because disordered stretches have been shown to signal for proteasomal digestion, we next tested the capacity of regions of dTIS11 that were predicted to be disordered to change the fate of a stable protein. For this, we constructed expression vectors for GFP-DCP1-CdTIS11V5/HIS and GFP-DCP1-NdTIS11V5/HIS, built from the fusion of GFP-DCP1 with dTIS11 C-terminal region (CdTIS11; amino acids 214 to 436) and N-terminal region (NdTIS11; amino acids 1 to 110), respectively. The stability of these fusions was analyzed in S2 cells after translation inhibition. We observed that addition of CdTIS11 or NdTIS11 induced destabilization of the otherwise-stable GFP-DCP1V5/HIS, as shown by Western blotting using an anti-V5 antibody (Fig. 5D). Detection of the fusion protein by the N-terminal GFP moiety revealed that the degradation was complete, thus establishing dTIS11 C- and N-terminal domains as functional signals for degradation. While both domains are sufficient for destabilization, neither is necessary for dTIS11 turnover, as versions of dTIS11V5/HIS lacking the N terminus (dTIS11-ΔNterV5/HIS; amino acids 111 to 436) or most of the C-terminal domain (dTIS11-ΔCterV5/HIS, amino acids 1 to 253) were efficiently degraded by the 20S proteasome (Fig. 5E).
dTIS11 ubiquitin-independent proteasome-dependent degradation is conserved from Drosophila to mammals.
The TIS11 family of proteins is present in eukaryotes from yeast to mammals. However, while the identification of proteins in this family is based on homology of their highly conserved tandem zinc finger domains, conservation for the rest of their sequence is much less conserved. This is easily observed when looking at a sequence alignment between Drosophila dTIS11 and the three mouse homologs, TTP, BRF1, and BRF2 (Fig. 6A). Yet, the implication of the proteasome in the degradation of TIS11/TTP proteins has been broadly established (20, 21). We thus addressed the hypothesis that the mammalian TIS11/TTP proteins would undergo the same degradation process as dTIS11. First, we analyzed the three mouse sequences by using DISOPRED2. We observed that despite sharing low sequence homology, their disorder profiles showed striking similarity with dTIS11, with an overall highly disordered degree, aside from the zinc finger domains and short stretches at their N and C termini (Fig. 6B). We next sought to test our hypothesis experimentally on TTP, the founding member of the family. The corresponding Zfp36 gene is induced upon immunological challenge in macrophages and is then rapidly downregulated. We thus looked at the stability of the TTP protein in LPS-activated mouse macrophage cells, RAW 264.7. Translation inhibition experiments showed that the TTP half-life is very short and is comparable to that of dTIS11 (t1/2 = 114 ± 35 min). In addition, proteasome inhibition by clasto-lactacystin-beta-lactone prevented TTP degradation (Fig. 6C), implicating, as expected, the proteasome in this process. We also verified that TTP degradation did not require lysosomal activity in RAW 264.7 cells (Fig. 6D). To test whether ubiquitination is required for TTP proteasomal degradation, we compared the stability of wild-type TTPMYC/HIS with its lysine-less mutant, TTPKRMYC/HIS. Of note, both proteins were doubly tagged with a Myc epitope for detection and 6×His repetition for purification. HEK293T cells were transfected with expression vectors for TTPMYC/HIS, TTPKRMYC/HIS, or another zinc finger (ZC3H1) RNA-binding protein, PARP12HIS, as a negative control. Translation inhibition experiments allow for the visualization of the degradation patterns. As observed for Drosophila dTIS11, conservative mutation of all 5 lysines into equally positively charged arginines did not prevent TTP efficient degradation, while the PARP12HIS control remained stable (Fig. 6E). We next analyzed the ability of the 20S proteasome to directly recognize and degrade TTPMYC/HIS purified from HEK293T extracts. Figure 6F shows efficient degradation of TTPMYC/HIS when incubated with purified 20S proteasome, compared to the PARP12HIS control. Interestingly, the disorder profile for PARP12 showed a much lower probability for destructuration, again correlating with the protein's insensitivity to 20S degradation (Fig. 6B and F). Finally, in vitro-translated TTP was degraded by purified 20S proteasome, indicating that TTP proteasomal degradation does not require ATP or ubiquitin (Fig. 6G).
FIG 6.
Evolutionary conservation of ubiquitin-independent proteasome-dependent degradation for TIS11/TTP proteins. (A) Sequence alignment based on ClustalW analysis of Drosophila dTIS11 and three mouse TIS11/TTP family proteins. Single letters, amino acids; asterisk, identical amino acid; colon, conserved substitution; period, semiconserved substitution. Highly conserved tandem zinc finger domains and the C-terminal stretch are underlined. (B) Disorder prediction calculated by using DISOPRED2 for the indicated proteins. Plain lines, disorder score per residue; dashed lines, prediction threshold at a 5% false-positive rate. (C) TTP is unstable and degraded by the proteasome. RAW 264.7 cells were stimulated with lipopolysaccharide for 4 h and treated with Clasto and puromycin for the indicated times. TTP was revealed by Western blotting with an anti-TTP antibody. Vertical bars indicate cuts between separated parts of the same blot. (D) Proteasome inhibition leads to TTP accumulation. RAW 264.7 cells were left untreated or cotreated with puromycin and the indicated inhibitors for 4 h for proteasome inhibitors or for 16 h for inhibition of lysosomal activity, before harvesting and analysis by Western blotting. (E) Ubiquitination is not required for TTP degradation. HEK293T cells were transfected with expression vectors for wild-type TTPMYC/HIS, lysine-less mutant TTPKRMYC/HIS, or PARP12HIS and treated with puromycin for the indicated times. TTPMYC/HIS and TTPKRMYC/HIS were detected by using anti-MYC antibodies, and PARP12HIS was detected with a PARP12-specific antibody. (F) ATP-independent degradation of TTPMYC/HIS by purified 20S proteasome. TTPMYC/HIS and PARP12HIS were purified using IMAC on HEK293T cells transfected with the corresponding expression vectors and incubated with 0.5 μg purified 20S proteasome for 45 min. (G) 20S proteasomal degradation of TTP in the absence of ubiquitin. TTP and GFP were translated in vitro in the presence of [35S]methionine and then incubated with 1 μg 20S proteasome for the indicated times. Labeled proteins were revealed by SDS-PAGE followed by sodium salicylate gel staining and fluorography.
Together, these results establish that as with Drosophila dTIS11, mammalian proteins of theTIS11/TTP family are targeted to the “degradation by default” pathway by ubiquitin-independent proteasome-dependent degradation.
Phosphorylation-dependent regulation of TTP and dTIS11 degradation by the 20S proteasome.
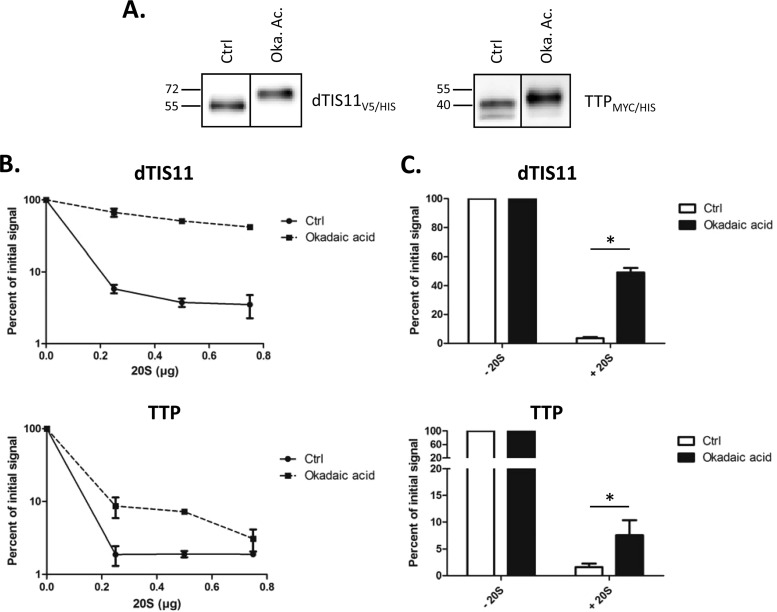
In mammals, TIS11/TTP are substrates for phosphorylation events that result in stabilization of these factors (21). We tested whether phosphorylation affected TTP and dTIS11 degradation by the 20S proteasome. For this, S2 cells or HEK293T cells expressing dTIS11V5/HIS and TTPMYC/HIS, respectively, were treated or not with okadaic acid, a phosphatase inhibitor able to induce phosphorylation of TIS11/TTP proteins in mammalian cells (21, 59). Protein extracts obtained from these cells were then submitted to immobilized-metal affinity chromatography (IMAC) for HIS tag-based protein purification. Figure 7A confirms that okadaic acid treatment induced a shift toward higher apparent molecular weight bands, corresponding to phosphorylated forms for dTIS11V5/HIS and TTPMYC/HIS. Subsequent incubation with increasing amounts of 20S proteasome led to a significant decrease in the degradation efficiency for proteins obtained from okadaic acid-treated cells (Fig. 7B and C). These results suggest that phosphorylation prevents, at least in part, the degradation of both dTIS11 and TTP by the proteasome.
FIG 7.
Phosphorylation regulates degradation of dTIS11 and TTP by the 20S proteasome in vitro. (A) Okadaic acid treatment induced phosphorylation of dTIS11 and TTP. S2 cells or HEK293T cells transfected with expression vectors for dTIS11V5/HIS or TTPMYC/HIS, respectively, were treated or not with okadaic acid (Oka. Ac.). His-tagged proteins were purified by IMAC and detected by Western blotting using anti-V5 or anti-MYC antibodies. Vertical bars indicate cuts between separated parts of the same blot. (B) Phosphorylation-regulated degradation of dTIS11 and TTP. Purified dTIS11V5/HIS or TTPMYC/HIS obtained as described for panel A were incubated for 1 h with the indicated amounts (B) or 0.5 μg (C) of 20S proteasome and then detected by Western blotting. Two (B) and four (C) independent experiments were quantified using the ImageJ software; results are reported relative to the corresponding value in the absence of 20S proteasome and plotted on the graph. Error bars represent SD for each experimental condition. The Mann-Whitney U test was performed for statistical analysis. *, P < 0.05.
Notably, the stabilizing effect observed for TTP is much smaller than that seen for dTIS11. This could reveal intrinsic differences within regulatory aspects for the two proteins, or it could be due to the experimental setting used. Indeed, okadaic acid treatment may not affect dTIS11 and TTP phosphorylation to the same extent. Moreover, the associated hyperphosphorylation state may never even be achieved under normal conditions. Additionally, the proteasome concentration and the molecular environment in this assay do not reflect the physiological conditions of the cell, which could stress the differences observed between the two proteins. Nevertheless, our results clearly establish a phosphorylation-dependent effect on TIS11/TTP degradability by the 20S proteasome in vitro.
DISCUSSION
In order to maintain proper regulation of genetic expression, cells need to control the availability of main regulators, such as ARE-BPs. This can be achieved by adjusting the balance between protein production and degradation. In this study, we showed that, similar to other ARE-BPs, TIS11/TTP proteins possess high turnover rates, which ensure the setting of rapid transitions in protein levels. However, while ARE-BPs such as AUF1 or HuR are targeted to degradation by ubiquitin-dependent mechanisms (16, 17), we showed that TIS11/TTP proteins enter a ubiquitin-independent degradation-by-default pathway. The Drosophila dTIS11 protein and the mammalian TTP protein are both targeted for degradation by the same mechanism. Therefore, our results uncover a new evolutionarily conserved regulation mechanism for TIS11/TTP proteins. Drosophila dTIS11, as the sole member of the family in this organism, might be essential for regulation of a potentially broad spectrum of mRNAs, rather establishing dTIS11 as a general buffering protein that limits the expression of its targets. Evolutionary complexification by gene duplication may have then required fine-tuned and target-specific controls of the TIS11/TTP members by adding layers of regulation on each family member, such as implementing transcriptional and signaling-dependent regulation. Nevertheless, our results indicate that the mechanism controlling TIS11/TTP protein stability has been conserved across evolution from invertebrates to mammals.
Previous studies pointed to the proteasome as the main proteolytic complex responsible for TIS11/TTP proteins. However, this conclusion was most often drawn from pharmacological studies using drugs such as MG132 or lactacystin, which also possess inhibitory effects on lysosome activity (reviewed in reference 49). Thus, the potential implication of autophagy-driven lysosomal degradation, another pathway of cytosolic protein degradation (60), in TIS11/TTP turnover has not been clearly assessed. Here, we showed that in contrast to proteasomal inhibition, disturbance of lysosomal degradation by drugs targeting different steps of this process did not affect dTIS11 and TTP levels. In addition, a lysosome-deficient rabbit reticulocyte lysate programmed with either dtis11 mRNA or TTP mRNA produced proteasome-sensitive dTIS11 and TTP, thus establishing these proteins as bona fide substrates of the proteasome.
Although proteasome dependent, we showed that TIS11/TTP degradation does not require ubiquitination. Rather, our results suggest that the destructuration states of these proteins are essential for proteasomal recognition. Intrinsically disordered regions can be found as localized stretches in the primary sequence, linkers separating two folded domains, or even as extended parts of IDPs. Some of these regions have also been shown to serve as recognition domains for the 20S proteasome, interacting with the gate of the 20S and clearing the entrance to the degradation chamber (56). In addition, IDPs theoretically do not require active unfolding processes, as they do not present a stable 3-dimensional structure, thus allowing their entry in the degradation-by-default process.
While lacking tertiary structure under physiological conditions, many IDPs undergo induced folding upon binding to a partner. This process is thought to allow increased binding kinetics and structural plasticity (reviewed in reference 61). Furthermore, protein stabilization following binding/folding events has been described for several proteasome substrates (62–64). The overall 3-dimensional structure of TIS11/TTP proteins has not been elucidated, with the exception of two conserved regions, both predicted as folded in our study. First, the high-resolution crystal structure of the TTP C-terminal short stretch was crucial in understanding its function for the recruitment of the CCR4-NOT deadenylation complex (65). Second, nuclear magnetic resonance analysis of TTP and BRF1 tandem zinc finger domains showed that while folding of these regions depends on the availability of bivalent cations, their overall structuration is modified in the presence of target RNA sequences (66). In addition, complete structuration of the TTP second zinc finger requires proper binding to a target ARE (67), thus raising the possibility that target RNA availability controls the stability of TIS11/TTP proteins, as this mechanism has been described for the yeast RNA-BP YB-1 (63). Alternatively, other binding partners could prevent TIS11/TTP degradation by precluding the access of the proteasome to their disordered regions.
In mammals, the TIS11/TPP proteins are subject to extensive phosphorylation (21, 31, 68). A dephosphorylation assay revealed that the Drosophila dTIS11 is also heavily phosphorylated, which we then confirmed by mass spectrometry analysis (data not shown). Previous studies have shown that the degradation of TIS11/TTP factors is controlled by their phosphorylation status. In fact, several authors have proposed a model in which binding of phosphorylated forms of TIS11/TTP to the 14-3-3 adaptor protein would prevent their degradation (20, 21, 59). This model is mainly based on the fact that phosphorylation induces both TIS11/TTP stabilization and binding to 14-3-3. However, the causality link between the two events is not clear. In fact, addition of recombinant 14-3-3 in our in vitro degradation assay did not stabilize dTIS11 or TTP (data not shown). Our observations, though limited to our in vitro analysis, thus raise an alternate hypothesis in which phosphorylation would directly affect TIS11/TTP recognition by the proteasome, as such a process has been observed for at least one other protein (69). 14-3-3 adaptors are able to prevent TIS11/TTP dephosphorylation (70). Their stabilizing role could thus be strictly indirect with regard to proteasomal digestion.
The disordered state here associated with a degradation process is also known to provide additional properties to IDPs. Intrinsic disorder allows for an increased interaction surface and better exposure of interaction motifs, facilitating associations with several partners. This could explain the rather high number of interacting partners formally identified for TTP, including 14-3-3 proteins, the kinases p38, MK2, and MEKK4, and also members of the deadenylation machinery (reference 8 and references therein). Two-hybrid analysis also identified 31 potential interactions partners for TTP, 4 of which were later confirmed by coimmunoprecipitation experiments (71). Flexibility and adaptability are other crucial features of disordered regions. These would allow RNA-BPs to bind similar yet different RNA molecules as well as increase their “capture radius.” In addition, the induced folding upon binding, common in IDPs, provides low-affinity/high-specificity binding (72), which is ideal for RNA-BP to target RNA recognition. These considerations may explain the observed enrichment in IDRs for RNA-BPs (73, 74). In accordance with this hypothesis, the disordered linker present in the tandem zinc finger domains of the BRF1 protein was shown to specifically contact AU-rich elements of mRNAs (66). This binding is controlled by an induced folding mechanism in which the tandem zinc finger becomes more rigid and stable upon mRNA contact (75). This mode of RNA recognition is correlated with a large diversity of mRNA targets identified for BRF1 (76).
RNA-BPs represent one of the largest protein families in eukaryotes (77). Although the average stability of RNA-BPs is higher than that of non-RNA-BPs (78), it remains to be determined whether the IDRs found to be enriched in RNA-BPs could serve as functional signals for proteasomal degradation for specific subsets of RNA-BPs.
ACKNOWLEDGMENTS
We thank Vincent Raussens for helpful discussions and William Rigby for providing CARP-3 anti-TTP antibody.
Work in our laboratory is supported by the Fund for Medical Scientific Research (Belgium; grant 2.4556.08), the Fonds Brachet, the Fonds Van Buuren, the Actions de Recherches Concertées (grants AV.06/11-345 and AV.12/17), and the FEDER Hainaut Biomed program. L. V. Ngoc is supported by a doctoral grant of the Fonds pour la Recherche en Industrie et Agriculture (Belgium).
Footnotes
Published ahead of print 22 September 2014
REFERENCES
- 1.Zhao W, Pollack JL, Blagev DP, Zaitlen N, McManus MT, Erle DJ. 2014. Massively parallel functional annotation of 3′ untranslated regions. Nat. Biotechnol. 32:387–391. 10.1038/nbt.2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Oikonomou P, Goodarzi H, Tavazoie S. 2014. Systematic identification of regulatory elements in conserved 3′-UTRs of human transcripts. Cell Rep. 7:281–292. 10.1016/j.celrep.2014.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sun W, Julie Li Y-S, Huang H-D, Shyy JY-J, Chien S. 2010. microRNA: a master regulator of cellular processes for bioengineering systems. Annu. Rev. Biomed. Eng. 12:1–27. 10.1146/annurev-bioeng-070909-105314. [DOI] [PubMed] [Google Scholar]
- 4.Vindry C, Vo Ngoc L, Kruys V, Gueydan C. 2014. RNA-binding protein-mediated post-transcriptional controls of gene expression: integration of molecular mechanisms at the 3′ end of mRNAs? Biochem. Pharmacol. 89:431–440. 10.1016/j.bcp.2014.04.003. [DOI] [PubMed] [Google Scholar]
- 5.Xie X, Lu J, Kulbokas EJ, Golub TR, Mootha V, Lindblad-Toh K, Lander ES, Kellis M. 2005. Systematic discovery of regulatory motifs in human promoters and 3′ UTRs by comparison of several mammals. Nat. Cell Biol. 434:338–345. 10.1038/nature03441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Spasic M, Friedel CC, Schott J, Kreth J, Leppek K, Hofmann S, Ozgur S, Stoecklin G. 2012. Genome-wide assessment of AU-rich elements by the AREScore algorithm. PLoS Genet. 8:e1002433. 10.1371/journal.pgen.1002433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barreau C. 2005. AU-rich elements and associated factors: are there unifying principles? Nucleic Acids Res. 33:7138–7150. 10.1093/nar/gki1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brooks SA, Blackshear PJ. 2013. Tristetraprolin (TTP): interactions with mRNA and proteins, and current thoughts on mechanisms of action. Biochim. Biophys. Acta 1829:666–679. 10.1016/j.bbagrm.2013.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.White EJF, Brewer G, Wilson GM. 2013. Post-transcriptional control of gene expression by AUF1: mechanisms, physiological targets, and regulation. Biochim. Biophys. Acta 1829:680–688. 10.1016/j.bbagm.2012.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Piecyk M, Wax S, Beck AR, Kedersha N, Gupta M, Maritim B, Chen S, Gueydan C, Kruys V, Streuli M, Anderson P. 2000. TIA-1 is a translational silencer that selectively regulates the expression of TNF-alpha. EMBO J. 19:4154–4163. 10.1093/emboj/19.15.4154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Simone LE, Keene JD. 2013. Mechanisms coordinating ELAV/Hu mRNA regulons. Curr. Opin. Genet. Dev. 23:35–43. 10.1016/j.gde.2012.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lal A, Mazan Mamczarz K, Kawai T, Yang X, Martindale JL, Gorospe M. 2004. Concurrent versus individual binding of HuR and AUF1 to common labile target mRNAs. EMBO J. 23:3092–3102. 10.1038/sj.emboj.7600305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liao B, Hu Y, Brewer G. 2007. Competitive binding of AUF1 and TIAR to MYC mRNA controls its translation. Nat. Struct. Mol. Biol. 14:511–518. 10.1038/nsmb1249. [DOI] [PubMed] [Google Scholar]
- 14.Tiedje C, Ronkina N, Tehrani M, Dhamija S, Laass K, Holtmann H, Kotlyarov A, Gaestel M. 2012. The p38/MK2-driven exchange between tristetraprolin and HuR regulates AU-rich element-dependent translation. PLoS Genet. 8:e1002977. 10.1371/journal.pgen.1002977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Laroia G. 1999. Control of mRNA decay by heat shock-ubiquitin-proteasome pathway. Science 284:499–502. 10.1126/science.284.5413.499. [DOI] [PubMed] [Google Scholar]
- 16.Li ML, Defren J, Brewer G. 2013. Hsp27 and F-box protein-TrCP promote degradation of mRNA decay factor AUF1. Mol. Cell. Biol. 33:2315–2326. 10.1128/MCB.00931-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Abdelmohsen K, Srikantan S, Yang X, Lal A, Kim HH, Kuwano Y, Galban S, Becker KG, Kamara D, de Cabo R, Gorospe M. 2009. Ubiquitin-mediated proteolysis of HuR by heat shock. EMBO J. 28:1271–1282. 10.1038/emboj.2009.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kandasamy K, Kraft AS. 2008. Proteasome inhibitor PS-341 (VELCADE) induces stabilization of the TRAIL receptor DR5 mRNA through the 3′-untranslated region. Mol. Cancer Ther. 7:1091–1100. 10.1158/1535-7163.MCT-07-2368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhou H-L, Geng C, Luo G, Lou H. 2013. The p97-UBXD8 complex destabilizes mRNA by promoting release of ubiquitinated HuR from mRNP. Genes Dev. 27:1046–1058. 10.1101/gad.215681.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brook M, Tchen CR, Santalucia T, McIlrath J, Arthur JSC, Saklatvala J, Clark AR. 2006. Posttranslational regulation of tristetraprolin subcellular localization and protein stability by p38 mitogen-activated protein kinase and extracellular signal-regulated kinase pathways. Mol. Cell. Biol. 26:2408–2418. 10.1128/MCB.26.6.2408-2418.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Benjamin D, Schmidlin M, Min L, Gross B, Moroni C. 2006. BRF1 protein turnover and mRNA decay activity are regulated by protein kinase B at the same phosphorylation sites. Mol. Cell. Biol. 26:9497–9507. 10.1128/MCB.01099-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hitti E, Iakovleva T, Brook M, Deppenmeier S, Gruber AD, Radzioch D, Clark AR, Blackshear PJ, Kotlyarov A, Gaestel M. 2006. Mitogen-activated protein kinase-activated protein kinase 2 regulates tumor necrosis factor mRNA stability and translation mainly by altering tristetraprolin expression, stability, and binding to adenine/uridine-rich element. Mol. Cell. Biol. 26:2399–2407. 10.1128/MCB.26.6.2399-2407.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jalonen U, Paukkeri EL, Moilanen E. 2008. Compounds that increase or mimic cyclic adenosine monophosphate enhance tristetraprolin degradation in lipopolysaccharide-treated murine J774 macrophages. J. Pharmacol. Exp. Ther. 326:514–522. 10.1124/jpet.107.133702. [DOI] [PubMed] [Google Scholar]
- 24.Ray D, Shukla S, Allam US, Helman A, Ramanand SG, Tran L, Bassetti M, Krishnamurthy PM, Rumschlag M, Paulsen M, Sun L, Shanley TP, Ljungman M, Nyati MK, Zhang M, Lawrence TS. 2013. Tristetraprolin mediates radiation-induced TNF-α production in lung macrophages. PLoS One 8:e57290. 10.1371/journal.pone.0057290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bourcier C, Griseri P, Grepin R, Bertolotto C, Mazure N, Pages G. 2011. Constitutive ERK activity induces downregulation of tristetraprolin, a major protein controlling interleukin8/CXCL8 mRNA stability in melanoma cells. Am. J. Physiol. Cell Physiol. 301:C609–C618. 10.1152/ajpcell.00506.2010. [DOI] [PubMed] [Google Scholar]
- 26.Rigby WFC, Roy K, Collins J, Rigby S, Connolly JE, Bloch DB, Brooks SA. 2005. Structure/function analysis of tristetraprolin (TTP): p38 stress-activated protein kinase and lipopolysaccharide stimulation do not alter TTP function. J. Immunol. 174:7883–7893. 10.4049/jimmunol.174.12.7883. [DOI] [PubMed] [Google Scholar]
- 27.Pfeiffer JR, Brooks SA. 2012. Cullin 4B is recruited to tristetraprolin-containing messenger ribonucleoproteins and regulates TNF-mRNA polysome loading. J. Immunol. 188:1828–1839. 10.4049/jimmunol.1102837. [DOI] [PubMed] [Google Scholar]
- 28.Schichl YM, Resch U, Lemberger CE, Stichlberger D, de Martin R. 2011. Novel phosphorylation-dependent ubiquitination of tristetraprolin by mitogen-activated protein kinase/extracellular signal-regulated kinase kinase kinase 1 (MEKK1) and tumor necrosis factor receptor-associated factor 2 (TRAF2). J. Biol. Chem. 286:38466–38477. 10.1074/jbc.M111.254888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Deleault KM, Skinner SJ, Brooks SA. 2008. Tristetraprolin regulates TNF TNF-alpha mRNA stability via a proteasome dependent mechanism involving the combined action of the ERK and p38 pathways. Mol. Immunol. 45:13–24. 10.1016/j.molimm.2007.05.017. [DOI] [PubMed] [Google Scholar]
- 30.Wu YT, Ouyang W, Lazorchak AS, Liu D, Shen HM, Su B. 2011. mTOR complex 2 targets Akt for proteasomal degradation via phosphorylation at the hydrophobic motif. J. Biol. Chem. 286:14190–14198. 10.1074/jbc.M111.219923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schmidlin M, Lu M, Leuenberger SA, Stoecklin G, Mallaun M, Gross B, Gherzi R, Hess D, Hemmings BA, Moroni C. 2004. The ARE-dependent mRNA-destabilizing activity of BRF1 is regulated by protein kinase B. EMBO J. 23:4760–4769. 10.1038/sj.emboj.7600477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Graham JR, Hendershott MC, Terragni J, Cooper GM. 2010. mRNA degradation plays a significant role in the program of gene expression regulated by phosphatidylinositol 3-kinase signaling. Mol. Cell. Biol. 30:5295–5305. 10.1128/MCB.00303-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lauwers A, Twyffels L, Soin R, Wauquier C, Kruys V, Gueydan C. 2009. Post-transcriptional regulation of genes encoding anti-microbial peptides in Drosophila. J. Biol. Chem. 284:8973–8983. 10.1074/jbc.M806778200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cairrao F, Halees AS, Khabar KSA, Morello D, Vanzo N. 2009. AU-rich elements regulate Drosophila gene expression. Mol. Cell. Biol. 29:2636–2643. 10.1128/MCB.01506-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu C-W, Corboy MJ, DeMartino GN, Thomas PJ. 2003. Endoproteolytic activity of the proteasome. Science 299:408–411. 10.1126/science.1079293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Erales J, Coffino P. 2014. Ubiquitin-independent proteasomal degradation. Biochim. Biophys. Acta 1843:216–221. 10.1016/j.bbamcr.2013.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Eulalio A, Behm-Ansmant I, Schweizer D, Izaurralde E. 2007. P-body formation is a consequence, not the cause, of RNA-mediated gene silencing. Mol. Cell. Biol. 27:3970–3981. 10.1128/MCB.00128-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Beskow A, Grimberg KB, Bott LC, Salomons FA, Dantuma NP, Young P. 2009. A conserved unfoldase activity for the p97 AAA-ATPase in proteasomal degradation. J. Mol. Biol. 394:732–746. 10.1016/j.jmb.2009.09.050. [DOI] [PubMed] [Google Scholar]
- 39.Twyffels L, Wauquier C, Soin R, Decaestecker C, Gueydan C, Kruys V. 2013. A masked PY-NLS in Drosophila TIS11 and its mammalian homolog tristetraprolin. PLoS One 8:e71686. 10.1371/journal.pone.0071686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fujimuro M, Yokosawa H. 2005. Production of antipolyubiquitin monoclonal antibodies and their use for characterization and isolation of polyubiquitinated proteins. Methods Enzymol. 399:75–86. [DOI] [PubMed] [Google Scholar]
- 41.Minde DP, Maurice MM, Rüdiger SGD. 2012. Determining biophysical protein stability in lysates by a fast proteolysis assay, FASTpp. PLoS One 7:e46147. 10.1371/journal.pone.0046147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ward JJ, Sodhi JS, McGuffin LJ, Buxton BF, Jones DT. 2004. Prediction and functional analysis of native disorder in proteins from the three kingdoms of life. J. Mol. Biol. 337:635–645. 10.1016/j.jmb.2004.02.002. [DOI] [PubMed] [Google Scholar]
- 43.Dosztanyi Z, Csizmok V, Tompa P, Simon I. 2005. IUPred: web server for the prediction of intrinsically unstructured regions of proteins based on estimated energy content. Bioinformatics 21:3433–3434. 10.1093/bioinformatics/bti541. [DOI] [PubMed] [Google Scholar]
- 44.Xue B, Dunbrack RL, Williams RW, Dunker AK, Uversky VN. 2010. PONDR-FIT: a meta-predictor of intrinsically disordered amino acids. Biochim. Biophys. Acta 1804:996–1010. 10.1016/j.bbapap.2010.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dinavahi R, Pancsa R, Tompa P, Lenaerts T, Vranken WF. 2014. The DynaMine webserver: predicting protein dynamics from sequence. Nucleic Acids Res.42(Web Server issue):W264–W270. 10.1093/nar/gku270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948. 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 47.Schwanhäusser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, Chen W, Selbach M. 2011. Global quantification of mammalian gene expression control. Nature 473:337–342. 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- 48.Belle A, Tanay A, Bitincka L, Shamir R, O'Shea EK. 2006. Quantification of protein half-lives in the budding yeast proteome. Proc. Natl. Acad. Sci. U. S. A. 103:13004–13009. 10.1073/pnas.0605420103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kisselev AF, van der Linden WA, Overkleeft HS. 2012. Proteasome inhibitors: an expanding army attacking a unique target. Chem. Biol. 19:99–115. 10.1016/j.chembiol.2012.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang Y, Kitagaki J, Dai R-M, Tsai YC, Lorick KL, Ludwig RL, Pierre SA, Jensen JP, Davydov IV, Oberoi P, Li C-CH, Kenten JH, Beutler JA, Vousden KH, Weissman AM. 2007. Inhibitors of ubiquitin-activating enzyme (E1), a new class of potential cancer therapeutics. Cancer Res. 67:9472–9481. 10.1158/0008-5472.CAN-07-0568. [DOI] [PubMed] [Google Scholar]
- 51.Li S, Chen Y, Shi Q, Yue T, Wang B, Jiang J. 2012. Hedgehog-regulated ubiquitination controls smoothened trafficking and cell surface expression in Drosophila. Plos Biol. 10:e1001239. 10.1371/journal.pbio.1001239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Finley D. 2009. Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu. Rev. Biochem. 78:477–513. 10.1146/annurev.biochem.78.081507.101607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ciechanover A, Ben-Saadon R. 2004. N-terminal ubiquitination: more protein substrates join in. Trends Cell Biol. 14:103–106. 10.1016/j.tcb.2004.01.004. [DOI] [PubMed] [Google Scholar]
- 54.Tomko RJ, Jr, Hochstrasser M. 2013. Molecular architecture and assembly of the eukaryotic proteasome. Annu. Rev. Biochem. 82:415–445. 10.1146/annurev-biochem-060410-150257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Asher G, Reuven N, Shaul Y. 2006. 20S proteasomes and protein degradation “by default.” Bioessays 28:844–849. 10.1002/bies.20447. [DOI] [PubMed] [Google Scholar]
- 56.Tsvetkov P, Asher G, Paz A, Reuven N, Sussman JL, Silman I, Shaul Y. 2008. Operational definition of intrinsically unstructured protein sequences based on susceptibility to the 20S proteasome. Proteins 70:1357–1366. 10.1002/prot.21614. [DOI] [PubMed] [Google Scholar]
- 57.Tompa P, Prilusky J, Silman I, Sussman JL. 2008. Structural disorder serves as a weak signal for intracellular protein degradation. Proteins 71:903–909. 10.1002/prot.21773. [DOI] [PubMed] [Google Scholar]
- 58.Tompa P, Fersht A. 2009. Structure and function of intrinsically disordered proteins. Taylor & Francis Group, Boca Raton, FL. [Google Scholar]
- 59.Sun L, Stoecklin G, Van Way S, Hinkovska-Galcheva V, Guo R-F, Anderson P, Shanley TP. 2007. Tristetraprolin (TTP)-14-3-3 complex formation protects TTP from dephosphorylation by protein phosphatase 2a and stabilizes tumor necrosis factor-alpha mRNA. J. Biochem. 282:3766–3777. 10.1074/jbc.M607347200. [DOI] [PubMed] [Google Scholar]
- 60.Rogov V, Dötsch V, Johansen T, Kirkin V. 2014. Interactions between autophagy receptors and ubiquitin-like proteins form the molecular basis for selective autophagy. Mol. Cell 53:167–178. 10.1016/j.molcel.2013.12.014. [DOI] [PubMed] [Google Scholar]
- 61.Kovacs D, Szabo B, Pancsa R, Tompa P. 2013. Intrinsically disordered proteins undergo and assist folding transitions in the proteome. Arch. Biochem. Biophys. 531:80–89. 10.1016/j.abb.2012.09.010. [DOI] [PubMed] [Google Scholar]
- 62.Asher G, Bercovich Z, Tsvetkov P, Shaul Y, Kahana C. 2005. 20S Proteasomal degradation of ornithine decarboxylase is regulated by NQO1. Mol. Cell 17:645–655. 10.1016/j.molcel.2005.01.020. [DOI] [PubMed] [Google Scholar]
- 63.Sorokin AV, Selyutina AA, Skabkin MA, Guryanov SG, Nazimov IV, Richard C, Th'ng J, Yau J, Sorensen PH, Ovchinnikov LP. 2005. Proteasome-mediated cleavage of the Y-box-binding protein 1 is linked to DNA-damage stress response. EMBO J. 24:3602–3612. 10.1038/sj.emboj.7600830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Alvarez-Castelao B, Castaño JG. 2005. Mechanism of direct degradation of IκBα by 20S proteasome. FEBS Lett. 579:4797–4802. 10.1016/j.febslet.2005.07.060. [DOI] [PubMed] [Google Scholar]
- 65.Fabian MR, Frank Rouya F, Siddiqui C, Lai N, Karetnikov WS, Blackshear A, Nagar PJ, Sonenberg B, N. 2013. Structural basis for the recruitment of the human CCR4-NOT deadenylase complex by tristetraprolin. Nat. Struct. Mol. Biol. 20:735–739. 10.1038/nsmb.2572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hudson BP, Martinez-Yamout MA, Dyson HJ, Wright PE. 2004. Recognition of the mRNA AU-rich element by the zinc finger domain of TIS11d. Nat. Struct. Mol. Biol. 11:257–264. 10.1038/nsmb738. [DOI] [PubMed] [Google Scholar]
- 67.Blackshear PJ. 2003. Characteristics of the interaction of a synthetic human tristetraprolin tandem zinc finger peptide with AU-rich element-containing RNA substrates. J. Biol. Chem. 278:19947–19955. 10.1074/jbc.M301290200. [DOI] [PubMed] [Google Scholar]
- 68.Cao H, Deterding LJ, Blackshear PJ. 2007. Phosphorylation site analysis of the anti-inflammatory and mRNA-destabilizing protein tristetraprolin. Expert Rev. Proteomics 4:711–726. 10.1586/14789450.4.6.711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Poppek D, Keck S, Ermak G, Jung T, Stolzing A, Ullrich O, Davies KJA, Grune T. 2006. Phosphorylation inhibits turnover of the tau protein by the proteasome: influence of RCAN1 and oxidative stress. Biochem. J. 400:511. 10.1042/BJ20060463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sandler H, Stoecklin G. 2008. Control of mRNA decay by phosphorylation of tristetraprolin. Biochem. Soc. Trans. 36:491. 10.1042/BST0360491. [DOI] [PubMed] [Google Scholar]
- 71.Kedar VP, Darby MK, Williams JG, Blackshear PJ. 2010. Phosphorylation of human tristetraprolin in response to its interaction with the Cbl interacting protein CIN85. PLoS One 5:e9588. 10.1371/journal.pone.0009588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Schulz GE. 1979. Nucleotide binding proteins. Elsevier/North- Holland Biomedical Press, New York, NY. [Google Scholar]
- 73.Castello A, Fischer B, Eichelbaum K, Horos R, Beckmann BM, Strein C, Davey NE, Humphreys DT, Preiss T, Steinmetz LM, Krijgsveld J, Hentze MW. 2012. Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell 149:1393–1406. 10.1016/j.cell.2012.04.031. [DOI] [PubMed] [Google Scholar]
- 74.Kwon SC, Yi H, Eichelbaum K, Föhr S, Fischer B, You KT, Castello A, Krijgsveld J, Hentze MW, Kim VN. 2013. The RNA-binding protein repertoire of embryonic stem cells. Nat. Struct. Mol. Biol. 20:1122–1130. 10.1038/nsmb.2638. [DOI] [PubMed] [Google Scholar]
- 75.Qin F, Chen Y, Li Y-X, Chen H-F. 2009. Induced fit for mRNA/TIS11d complex. J. Chem. Phys. 131:115103. 10.1063/1.3224126. [DOI] [PubMed] [Google Scholar]
- 76.Zhang L, Prak L, Rayon-Estrada V, Thiru P, Flygare J, Lim B, Lodish HF. 2013. ZFP36L2 is required for self-renewal of early burst-forming unit erythroid progenitors. Nature 499:92–96. 10.1038/nature12215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ray D, Kazan H, Cook KB, Weirauch MT, Najafabadi HS, Li X, Gueroussov S, Albu M, Zheng H, Yang A, Na H, Irimia M, Matzat LH, Dale RK, Smith SA, Yarosh CA, Kelly SM, Nabet B, Mecenas D, Li W, Laishram RS, Qiao M, Lipshitz HD, Piano F, Corbett AH, Carstens RP, Frey BJ, Anderson RA, Lynch KW, Penalva LOF, Lei EP, Fraser AG, Blencowe BJ, Morris QD, Hughes TR. 2013. A compendium of RNA-binding motifs for decoding gene regulation. Nature 499:172–177. 10.1038/nature12311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mittal N, Roy N, Babu MM, Janga SC. 2009. Dissecting the expression dynamics of RNA-binding proteins in posttranscriptional regulatory networks. Proc. Natl. Acad. Sci. U. S. A. 106:20300–20305. 10.1073/pnas.0906940106. [DOI] [PMC free article] [PubMed] [Google Scholar]