Abstract
Plakophilins 1 and 3 (PKP1/3) are members of the arm repeat family of catenin proteins and serve as structural components of desmosomes, which are important for cell-cell-adhesion. In addition, PKP1/3 occur as soluble proteins outside desmosomes, yet their role in the cytoplasm is not known. We found that cytoplasmic PKP1/3 coprecipitated with the RNA-binding proteins FXR1, G3BP, PABPC1, and UPF1, and these PKP1/3 complexes also comprised desmoplakin and PKP2 mRNAs. Moreover, we showed that the interaction of PKP1/3 with G3BP, PABPC1, and UPF1 but not with FXR1 was RNase sensitive. To address the cytoplasmic function of PKP1/3, we performed gain-and-loss-of-function studies. Both PKP1 and PKP3 knockdown cell lines showed reduced protein and mRNA levels for desmoplakin and PKP2. Whereas global rates of translation were unaffected, desmoplakin and PKP2 mRNA were destabilized. Furthermore, binding of PKP1/3 to FXR1 was RNA independent, and both PKP3 and FXR1 stabilized PKP2 mRNA. Our results demonstrate that cytoplasmic PKP1/3 are components of mRNA ribonucleoprotein particles and act as posttranscriptional regulators of gene expression.
INTRODUCTION
The regulation of intercellular adhesion is critical for normal development of all multicellular organisms and for tissue homeostasis. Thereby, cell-cell contacts play an important role, and cadherin-catenin complexes mediate the link to the dynamic forces of the cytoskeleton. In addition to their structural role, catenins are key components of signaling pathways that regulate morphogenesis and tissue homoeostasis. The best-studied member of the catenin family is β-catenin, a central component of the Wnt signaling pathway that triggers transcription of Wnt-specific genes through its interaction with transcription factors (1).
Numerous members of the armadillo (arm) repeat-containing family of catenin proteins are located at cell junctions: β-catenin, plakoglobin, and members of the p120-catenin subfamily, such as p120-catenin itself, p0071-catenin (also known as PKP4), ARVCF (armadillo repeat gene deleted in velocardiofacial syndrome), δ-catenin (also known as neurojungin or neural plakophilin-related protein [NPRAP]), and the plakophilins (PKPs) (2, 3). While the three PKP family members PKP1, -2, and -3 are expressed in a cell-type-specific manner, they are all located at cell borders in desmosomal structures, where they support desmosome assembly and stability (4). PKPs act as desmosomal cross-linkers as they interact with all major desmosomal components, such as desmosomal transmembrane proteins, the desmogleins and desmocollins, cytoplasmic plaque proteins desmoplakin and plakoglobin, and cytoskeletal structures, e.g., the keratins (3, 5). All reported desmosomal interactions are mediated via the N termini of PKPs (6). The lack of PKPs results in a reduction of the size and number of desmosomes and leads to an increase in migration (7–9), underscoring the importance of PKPs as scaffolding proteins.
In addition to their role as structural components of cell-cell contacts, members of the p120-catenin family regulate junctional stability by influencing the endocytosis of cadherins, which modulate the cytoskeleton by interacting with small GTPases and interfere with gene expression through their interaction with transcription factors (10–15). PKP1 localizes both in the nucleus and the cytoplasm (16) and acts as a regulator of mRNA translation by promoting eukaryotic initiation factor 4A1 (eIF4A1) activity (17). Similarly, PKP2 was shown to translocate to the nucleus, where it associates with components of the polymerase III transcription complex (18, 19). In contrast, the functions of PKP3 in cell adhesion and signaling are poorly understood.
Through affinity purification, we previously discovered that the cytoplasmic nonjunctional forms of PKP1 and PKP3 are associated with three RNA-binding proteins (RBPs): FXR1 (fragile X mental retardation syndrome-related protein 1), G3BP (Ras-GTPase-activating protein SH3 domain-binding protein), and PABPC1 [cytoplasmic poly(A)-binding protein 1] (20, 21). In addition, upon environmental stress, PKP1 and PKP3, but not PKP2, were recruited to stress granules, transient cytoplasmic aggregates of translationally stalled mRNAs (22). FXR1 interacts with the 60S ribosomal subunit and influences the translation and stability of bound mRNAs, possibly through interaction with Argonaute 2 (Ago-2) (23–26). G3BP is a binding partner of RasGAP and may thus influence mitogen-activated protein kinase signaling (27). G3BP has been proposed to have endoribonuclease activity involved in mRNA decay (28). In addition, G3BP inhibits translation initiation of certain mRNAs by interacting with their 3′ untranslated regions (UTRs) (29). As the major poly(A)-binding protein, PABPC1 plays a role in mRNA 3′-end processing (30) and promotes cap-dependent translation through its interaction with the translation initiation factor eIF4G (31–34). PABPC1 also enhances mRNA stability in general and has a specific role in microRNA (miRNA)-mediated mRNA decay via its interaction with GW182 (35–38). Moreover, PABPC1 promotes the termination of translation by binding to the eukaryotic release factor 3 (eRF3) (39). Interestingly, this interaction is strongly dependent on the structure of the mRNP, as exemplified by mRNAs with long 3′-UTRs. Such mRNAs are prone to nonsense-mediated mRNA decay (NMD) as a result of the competition between PABPC1 and the central NMD factor up-frameshift 1 (UPF1) for binding to eRF3 (40–43). NMD is not only a translation-dependent quality control mechanism to degrade mRNAs with premature stop codons, as it also regulates the stability of many physiological transcripts (42, 44, 45).
Numerous examples illustrate that RBPs control the processing, translation, stability, transport, and localization of mRNAs (46, 47). Thus, RBPs function as posttranscriptional regulators of gene expression (48). Given that all three RBPs associated with PKP1 or PKP3 affect the turnover and translation of mRNAs in the cytoplasm, it is conceivable that both PKP1 and PKP3 influence gene expression at the posttranscriptional level.
To explore the cytoplasmic functions of PKP1 and PKP3, we generated stable cell lines via knockdown or overexpression, and we found that the knockdown of PKP1 or PKP3 led to reduced protein and mRNA levels of desmoplakin and PKP2. PKP1- and PKP3-associated protein-RNA complexes contained desmoplakin and PKP2 mRNA. Moreover, both PKP1 and PKP3 bound directly to FXR1, in contrast to RNA-mediated interactions with G3BP, PABPC1, and UPF1. Lower mRNA levels were caused by enhanced degradation of desmosomal mRNAs in PKP1- and PKP3-deficient cells. Our findings demonstrate for the first time that cytoplasmic forms of PKP1 and PKP3 influence the stability of specific mRNAs and thus act as posttranscriptional regulators of gene expression.
MATERIALS AND METHODS
Antibodies.
Primary antibodies to PKP1 (PP1 B6-4; Progen Biotechnik), PKP2 (clones PP2/62, PP2/86, PP2/150, and GP-PP2-hCT; Progen Biotechnik), PKP3 (clone PKP3-270.6.2 [Progen Biotechnik] and xPKP3 [20]), desmoplakin (clones DP1&2-2.15, DP1-2.17, DP1&2-2.20, and DP 495; Progen Biotechnik), G3BP (BD Biosciences), FXR1 (clone HPA018246; Sigma-Aldrich), PABPC1 (clone 10E10; ImmuQuest), UPF1/Rent1 (clone A301-902A; Bethyl Laboratories), β-actin (clone AC-15; Novus Biological), anti-myc tag (clone 9106 [Abcam] and clone 1-9E10.2 [ATCC]) were used. For immunoblot analysis, horseradish peroxidase-conjugated secondary antibodies (Jackson ImmunoResearch, Laboratories) were applied.
Constructs.
For generation of stable PKP1 knockdown cells, short hairpin RNA (shRNA) against PKP1a mRNA was designed with Acc65I and HindIII overhangs and cloned into shRNA expression vector psiRNA-h7SKZeo (InvivoGen). As a negative control, an shRNA construct against β-galactosidase (shLac) was used (InvivoGen). The clone with PKP1a target site 2411 (forward, 5′-GTACCTCGAACTTCACCTCCCGATTCTATCAAGAGTAGAATCGGGAGGTGAAGTTCTTTTTGGAA-3′, and reverse, 5′-AGCTTTTCCAAAAAGAACTTCACCTCCCGATTCTACTCTTGATAGAATCGGGAGGTGAAGTTCGA-3′) was selected for further studies as it showed the most efficient protein and mRNA reduction. Generation of PKP3-pEGFP-N1, PKP3-myc-N1, shPKP3-9, shPKP3-1350, and shLuc constructs have been described elsewhere (49). For generation of PKP3 constructs containing a C-terminal myc tag DNA fragment coding for PKP3, domains were amplified by reverse transcription-PCR (RT-PCR) and cloned into the plasmid PKP3-myc-N1 by using EcoRI/KpnI restriction sites. For generation of PKP3 constructs, the following primer sets were used: for PKP3 amino acids (aa) 1 to 230 (PKP3 aa1-230), forward, 5′-TCGAATTCTATGCAGGACGGTAACTT-3′, and reverse, 5′-ATGGTACCGCGTCCAGCCCCCCTGC-3′; for PKP3 aa1-294, forward, 5′-TCGAATTCTATGCAGGACGGTAACTT-3′, and reverse, 5′-ATGGTACCGCGTGGCCCGAGTCAGCCAG-3′; for PKP3 aa1-369, forward, 5′-TCGAATTCTATGCAGGACGGTAACTT-3′, and reverse, 5′-ATGGTACCCGGAGCTTCACCAGCCT-3′; for PKP3 aa370-515, forward, 5′-TCGAATTCATGTTCAACCACGCCAAC-3′, and reverse, 5′-ATGGTACCGCCTCCAGGGCGTGGTT-3′; for PKP3 aa516-797, forward, 5′-TCGAATTCATGGCGGGCAAATGCGAG-3′, and reverse, 5′-ATGGTACCTGTGGGCCCA-3′; for PKP3 aa295-797, forward, 5′-TCGAATTCATGCTGCCGGACGTGCAT-3′, and reverse, 5′-ATGGTACCTGTGGGCCCA-3′. For expression of the myc-tagged bacterial alkaline phosphatase, the vector pc-Myc-CMV-2-BAP was used (immunoprecipitation vector kit; Sigma-Aldrich). Plasmids used for in vitro protein translation were generated by using the Gateway LR-Clonase enzyme mix (Invitrogen) reaction of the entry vectors PKP3-pENTR221 (clone 195039295), PABPC1-pENTR221 (clone 141114054), and FXR1-pENTR221 (clone 105650163) and the destination vectors pDEST 14 (identification no. [ID] V000139) and pDEST17 (ID V000055), which both contain a T7 promoter. Vectors were obtained from the proteomics and genomics core facility of the German Cancer Research Center (DKFZ). For in vitro protein expression of myc-tagged PKP3 full length and domains, the corresponding plasmids were cloned into the vector pcDNA3.1(-) by using EcoRI/AflII restriction sites. For rescue experiments, an RNA interference (RNAi)-resistant PKP3 mutant was produced by using the QuikChange Lightning site-directed mutagenesis kit (Agilent Technologies) and the plasmid PKP3-myc-N1 with the following primers for the rescue construct: forward, 5′-GCTTCGCAAAAATGTCACAGGTATATTGTGGAACCTTTCATCCAGCG-3′, and reverse, 5′-CGCTGGATGAAAGGTTCCACAATATACCTGTGACATTTTTGCGAAGC-3′. The modified PKP3 sequence contained three point mutations, compared to the target sequence of shPKP3-1350.
Generation of stable cell lines and experiments.
Stable DU145 cell lines were generated and cultured as described in detail previously (49). The same procedure was used to generate stable PKP1 or PKP3 knockdowns in BPH-1 cells.
For transient knockdown experiments, cells were plated in a 12-well plate at a density of 85,000 cells per well. After incubation for 24 h, cells were washed and transfected with 10 nM control small interfering RNA (siRNA; siGENOME nontargeting siRNA no. 3; Dharmacon), UPF1 siRNA (Silencer Select; Ambion), or FXR1 siRNA (Silencer Select; Ambion) with 1 μl Dharmafect 1 (Dharmacon) according to the manufacturers' instructions. The following siRNA duplexes were used: for UPF1, forward, 5′-GAUGCAGUUCCGCUCCAUU-3′, and reverse, 5′-AAUGGAGCGGAACUGCAUC-3′ (50), and for FXR1, forward, 5′-CGAGCUGAGUGAUUGGUCA-3′, and reverse, 5′-UGACCAAUCACUCAGCUCG-3′ (siRNA ID s15612). At 48 h after transfection, RNA was isolated by using the RNeasy minikit (Qiagen), and RNA was reverse transcribed into cDNA followed by quantitative RT-PCR. Alternatively, cells were lysed after 48 h of siRNA transfection and processed for immunoprecipitation of protein-associated RNA (RIP).
For rescue experiments, cells were cotransfected with plasmids containing a puromycin-resistant gene (pCI-puro [51]) and an RNAi-resistant PKP3 mutant. As a control, an empty vector was processed in parallel. Transfected cells were cultivated for 4 h in serum-free medium and then medium containing 10% fetal bovine serum (Biocrom) and 5% glutamine (Gibco) was added. After 24 h, the cells were selected with 1 μg/ml puromycin (Invitrogen) for another 24 h.
Gel electrophoresis and immunoblotting.
Cells were lysed by adding Laemmli sample buffer that included with Benzonase (Merck). For immunoblotting, proteins decorated with specific primary and secondary antibodies were detected by using a chemiluminescence system (Pierce ECL, Western blotting substrate; Pierce, Rockford, IL).
RNA isolation and quantitative RT-PCR.
Total RNA was isolated from cultured cells by using the RNA isolation kit (RNeasy minikit; Qiagen). Total RNA was transcribed into cDNA (high capacity cDNA kit; Applied Biosystems), and relative gene expression was measured by quantitative RT-PCR with Power-SYBR green master mix (Applied Biosystems) on an ABI StepOnePlus cycler. Primers included those against PKP1 (forward, 5′-GACCAGGACAACTCCACGTT-3′, and reverse, 5′-CTGCTGGTGGTCCCATGTT-3′), PKP2 (forward, 5′-GCAAATGGTTTGCTCGATTT-3′, and reverse, 5′-CTGCTGGTGGTCCCATAGTT-3′), PKP3 (forward, 5′-TGATGAGCTTCGCAAAAATG-3′, and reverse, 5′-CTGAGAGGCTGAGCTGAGGT-3′), desmoplakin (forward, 5′-GCCCTGAGGCGGCAGTTACTC-3′, and reverse, 5′-AGTTCCAGGGTCCGGTTGTTGC-3′), and 18S rRNA (forward, 5′-GTTGGTGGAGCGATTTGTCTG-3′, and reverse, 5′-AGGGCAGGGACTTAATCAACG-3′. In addition, the following primers were used: for UPF1, forward, 5′-AGGCCGACTACGACAAGAAG-3′, and reverse, 5′-ACCGCAGGCATATCTCATCC-3′; for FXR1, forward, 5′-GAAAGCATTGGAAATGTGCAGG-3′, and reverse, 5′-CAGAGGGGTTAGACAGCTCA-3′. For SMG1, primer sequences described in reference 52 were used. 18S rRNA levels were measured as internal references to evaluate RNA recovery and to exclude variations. Ribosomal 18S rRNA was also used for normalization. At least three technical and biological replicates were obtained as indicated. Error bars are presented as means ± standard deviations (SD) or ± standard errors of the means (SEM). Further details have been described elsewhere (49).
Polysome profiling.
For polysome profiling, sucrose block gradients with concentrations of sucrose from 17.5% to 50% in gradient buffer (15 mM Tris-HCl [pH 7.4], 15 mM MgCl2, 300 mM NaCl, 1% Triton X-100) were used. The cells were cultivated for 24 h and then treated with 100 μg/ml cycloheximide (Carl Roth) for 10 min at room temperature. The cells were washed with phosphate-buffered saline (PBS) plus 100 μg/ml cycloheximide. All of the following steps were performed at 4°C. The cells were lysed with gradient buffer containing 100 μg/ml cycloheximide, 500 μg/ml heparin, 0.2 U/ml RNasin Super (Promega), EDTA-free protease inhibitor cocktail (Roche), and 0.1% 2-mercaptoethanol and rotated for 10 min. After centrifugation for 10 min at 10,000 rpm, 250-μl aliquots of the supernatants were fractionated over 17.5%-to-50% sucrose gradients via ultracentrifugation (4°C, 35,000 rpm, 150 min) in a Sorvall Discovery 90SE ultracentrifuge (Thermo Scientific) with an SW60 rotor. Gradients were pumped out with 50% sucrose, and polysome profiles were recorded by measuring the absorbance at 254 nm via a Teledyne ISCO gradient elution system. To quantify global translation rates in cells, the area under the curve of monosomal and polysomal ribosomes was determined and the translation index was calculated by dividing the polysomal area under the curve through the total (polysomes plus monosomes) area under the curve.
Metabolic labeling.
For metabolic labeling, cells were cultured without methionine and cysteine for 1 h. A mixture of [35S]methionine-cysteine (PerkinElmer) was then added to a final concentration of 55 μCi/ml and incubated for 2 h. After labeling, cells were washed with PBS and lysed with 150 μl lysis buffer (15 mM Tris-HCl [pH 7.4], 15 mM MgCl2, 300 mM NaCl, 1% Triton X-100) followed by a centrifugation step (9,000 rpm for 1 min). Twenty-microliter aliquots of the supernatants were spotted on filter papers, air dried, and precipitated in 5% trichloracetate followed by washing with ice-cold 5% trichloracetate and acetone. Incorporation of [35S]methionine-cysteine was determined in an Econofluor-2 system (PerkinElmer) via a scintillation counter (Beckman LS 6000IC). Counts per minute (cpm) were obtained for each sample.
RNA stability assay.
For mRNA stability measurements, the cells were treated with 5 μg/ml actinomycin D (Serva Electrophoresis) and incubated between 1 h and 6 h in an incubator at 37°C. The RNA was then isolated with the RNeasy minikit (Qiagen) and transcribed into cDNA (high-capacity cDNA kit; Applied Biosystems), and mRNA was detected by quantitative RT-PCR, as described before. mRNA half-lives were calculated by assuming a first-order decay rate. mRNA levels were normalized to 18S rRNA and plotted against time. Curves were fitted by linear regression, and mRNA half-lives were calculated as follows: t½ = ln(2)/k; t is time, and k is the rate constant.
IP of proteins and RIP.
For immunoprecipitation (IP) analysis magnetic beads conjugated with secondary pan-mouse IgG antibody or protein A beads (Dynal magnetic beads for both; Invitrogen) were used. IP of proteins has been described in detail before (20). Incubation of lysates with beads was used as a negative control for unspecific binding. For immunoprecipitation of protein-associated RNA (RIP), the incubation time was extended to 2.5 h and an additional washing step (10 min at 4°C) was included. RNA was isolated with TRIzol (Ambion), transcribed into cDNA (high-capacity cDNA kit; Applied Biosystems), and then further used for RT-PCR. To control for unspecific binding, RIP was also performed with antibodies specific for desmoplakin. When using myc rabbit antibody, the IP buffer was supplemented with 20 μg/ml yeast tRNA (Acris), 10 μg/μl IgG-free albumin (Roth), and 3% glycerol, and an additional blocking step (2 h, 4°C) was included. In addition, in some experiments the lysates used for IP were treated with RNase A (2 μl/ml RNase A solution [Qiagen] or 0.2 μg/μl final concentration [Roche]) to analyze RNA-dependent interactions.
In vitro RNA-binding assay.
The PKP2 coding sequence (19) was amplified by RT-PCR with Pfu polymerase (Bioron) with the following primers: (for), 5′-TAATACGACTCACTATAGGGATGGCAGCCCCCGGC-3′, and (rev), 5′-TCAGTCTTTAAGGGAGTGGTAGGC-3′. The RT-PCR product was in vitro transcribed according to the manufacturer's instructions (RiboMAX large-scale RNA production systems -SP6, T7 polymerase; Promega), the RNA was purified (NucAway spin columns; Life Technologies, Ambion), and terminally labeled with poly(AMPs) [Escherichia coli poly(A) polymerase l; Ambion]. In parallel, in vitro protein translation of PKP3 or PABPC1 with PKP3-DEST or PABPC1-DEST plasmids was performed according to the manufacturer's instructions (TNT T7 coupled reticulocyte lysate system; Promega).
For the in vitro RNA-binding assay, 30 μl magnetic beads conjugated with pan-mouse IgG antibodies was used. Samples of in vitro-translated proteins were incubated in physiological salt buffer (140 mM NaCl, 20 mM HEPES, 5 mM EDTA, and 1% NP-40; pH 7.5) with beads coated with primary antibody for 90 min at 4°C. After several washing steps, 1 μg of in vitro-transcribed PKP2 mRNA or 9 μg of total RNA isolated from HEK 293 cells was added and incubated overnight at 4°C in physiological salt buffer supplemented with 0.05% RNasin (RNasin Super; Promega). After several washing steps and transfer into a new tube, RNA was isolated with TRIzol and the total RNA amount was transcribed into cDNA (high-capacity cDNA reverse transcription kit; Applied Biosystems). PKP2 mRNA was amplified by RT-PCR with the primers (for), 5′-TTCTGGGTGGCCTGAAGGAGACT-3′, and (rev), 5′-ACTTCCGGCCGTGAGGTTC-3′, with 25 cycles and detected qualitatively on an agarose gel. To control for unspecific RNA binding, the binding assay was performed by incubation of the RNA applied with beads only.
Protein domain analysis.
For PKP3 domain analysis corresponding constructs (full-length PKP3 or partial PKP3 domains containing a C-terminal myc tag) were transiently transfected into HEK 293 cells by using the transfection reagent X-treme Gene HP (Roche). After 48 h, the cells were used for immunoprecipitation by applying a myc tag-specific antibody. The interaction of PKP3 domains with RNA-binding proteins (IP) or PKP2 mRNA (RIP) was examined by immunoblotting or RT-PCR.
In vitro protein-protein binding assay.
For protein-binding studies, PKP3 full-length and deletion constructs and FXR1 were produced in vitro according to the manufacturer's instructions (TNT T7 coupled reticulocyte lysate system; Promega). Aliquots of in vitro-translated PKP3 full-length and PKP3 deletion constructs were incubated with primary antibodies specific for myc tag. After binding for 2 h at 4°C, the beads were washed and incubated with in vitro-translated FXR1 protein for 2 h at 4°C. After several washing steps, bound proteins were eluted and separated via SDS-PAGE followed by Western blotting. Incubation of in vitro-translated FXR1 with beads loaded with myc tag antibody only was used as a negative control for unspecific binding. All incubations were carried out in IP buffer supplemented with RNase A.
Statistical analysis.
In independent assays, each experiment was repeated at least three times, including technical replicates. Data were analyzed with the unpaired Student's t test, and a P value of <0.05 was considered indicative of a statistically significant result. For quantification of PKP2 protein expression levels, ImageJ software was used.
RESULTS
PKP1 and PKP3 associate with FXR1 in an RNA-independent manner.
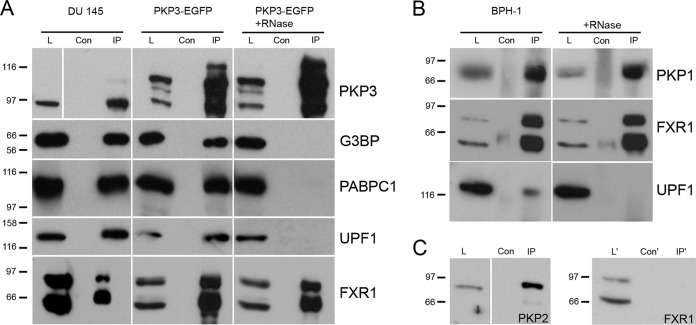
We previously identified G3BP, PABPC1, and FXR1 as interaction partners of PKP1/3 (20, 21). While reanalyzing our mass spectrometry data, we noticed that UPF1 also copurified with PKP3. UPF1 is an RNA helicase required for NMD (41, 53, 54). To further explore these interactions, we immunoprecipitated endogenous PKP3 from lysates of DU 145 cells and confirmed the association of PKP3 with G3BP, PABPC1, FXR1, and UPF1 (Fig. 1A). We also observed these interactions with overexpressed PKP3-EGFP (enhanced green fluorescent protein). Until then, an RNase inhibitor was included in all experiments to prevent disruption of protein-RNA complexes. To determine whether these interactions were dependent on the presence of RNA, we added RNase A to the lysates prior to immunoprecipitation. Whereas the efficiency of the PKP3 immunoprecipitation was not affected by RNase A treatment, the interactions with G3BP, PABPC1, and UPF1 were lost (Fig. 1A). In contrast, FXR1 remained associated with PKP3 even after RNase A treatment. Our earlier observations on recruitment of PKP1 and PKP3 into stress granules (20) suggested that PKP1, like PKP3, is also involved in RNA metabolism. Indeed, we found that endogenous PKP1 also coimmunoprecipitated FXR1 and UPF1 (Fig. 1B). Upon RNase A treatment, UPF1 was no longer associated with PKP1, whereas FXR1 remained bound to PKP1 (Fig. 1B). Interestingly, we did not detect an interaction between PKP2 and FXR1 (Fig. 1C).
FIG 1.
PKP1 and PKP3 are associated with RNA-binding proteins. (A) PKP3 immunoprecipitation with lysates from DU 145 wild-type and PKP3-overexpressing (PKP3-EGFP) cell lines. By immunoprecipitation using PKP3-specific antibodies, PKP3 complexes were enriched; for detection of PKP3 in DU 145 lysates, results from a longer exposure time are shown. The interactions of PKP3 with RNA-binding proteins G3BP, PABPC1, UPF1, and FXR1 were detected by immunoblotting. FXR1 occurred in two major bands representing short, more widely expressed splice variants (89, 90). RNA-mediated protein-protein interactions were analyzed by RNase A treatment of lysates from DU 145-PKP3-EGFP cells. (B) PKP1 immunoprecipitation with lysates from BPH-1 cells. By immunoprecipitation using PKP1-specific antibodies, PKP1 complexes were enriched. The interactions with RNA-binding proteins FXR1 and UPF1 were detected by immunoblotting. (C) PKP2 immunoprecipitation with lysates from DU 145 cells. By using immunoprecipitation with PKP2-specific antibodies, PKP2 complexes were enriched. The interaction with FXR1 was checked by immunoblotting. Note the portion of FXR1 in the PKP1/PKP3 IP sample after RNase A treatment, while G3BP, PABPC1, and UPF1 were completely undetectable. Lanes L and L′, total lysate; Con and Con′, negative control for unspecific binding; IP and IP′, immunoprecipitation results. For all immunoblots, molecular masses are given in kDa on the left.
Taken together, we found that UPF1 is part of PKP3- and PKP1-containing complexes, and we demonstrated that the interaction of PKP3/PKP1 with G3BP, PABPC1, and UPF1 is RNA dependent. In contrast, FXR1 interacts in an RNA-independent fashion with PKP3 and PKP1 but not with PKP2, indicative of a closer connection between PKP1/PKP3 and FXR1.
PKP1- and PKP3-associated complexes contain mRNA of desmosomal proteins.
As it was recently shown that FXR1 binds the mRNA of the desmosomal protein desmoplakin (55), we wondered whether PKP-FXR1 complexes bound mRNAs of desmosomal mRNAs. To look into this, we immunoprecipitated PKP3 from DU 145 cell lysates and tested for the presence of mRNA by RT-PCR. As a control, we used antibodies against another desmosomal protein, desmoplakin, which also interacts with PKP3 and PKP1 (56). Indeed, PKP2 and desmoplakin mRNAs were detected in the PKP3 immunoprecipitate but not in the desmoplakin immunoprecipitate (Fig. 2A). Similarly, PKP1 was found to associate with PKP2 and desmoplakin mRNAs (Fig. 2B). In contrast, the two mRNAs were not detected after PKP2 immunoprecipitation (Fig. 2C).
FIG 2.
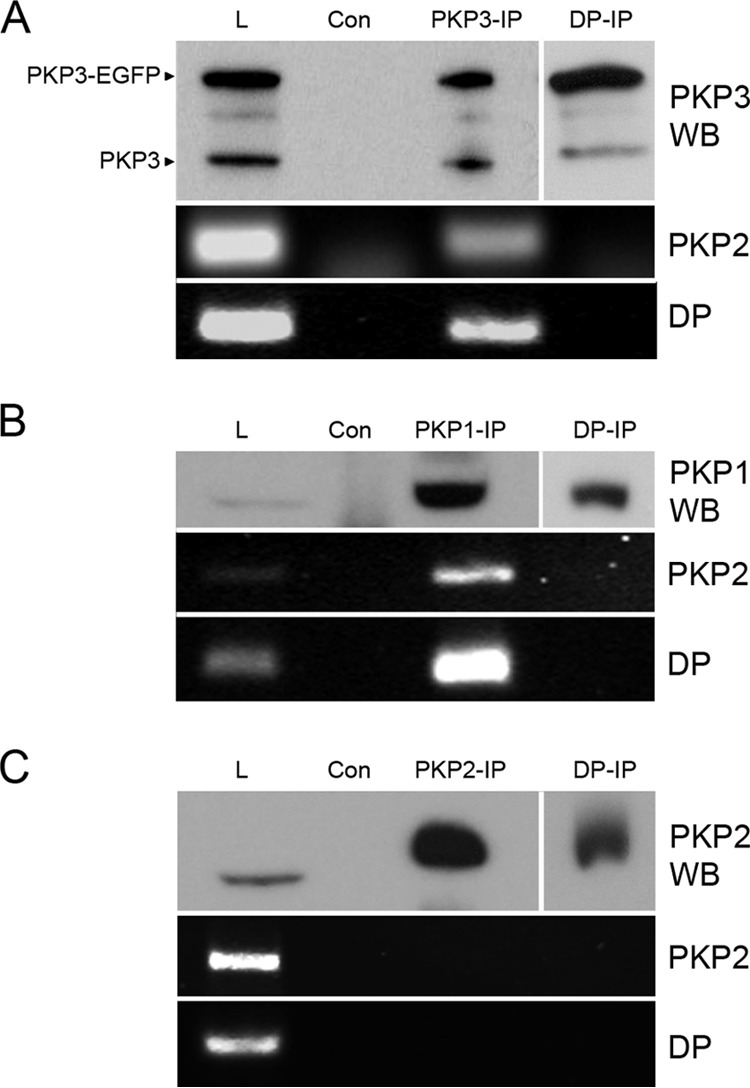
PKP2 and desmoplakin mRNAs were detected in PKP1 and PKP3 complexes. Detection of PKP2 and desmoplakin mRNA is shown in PKP3-EGFP-containing complexes (A), in PKP1 complexes from BPH-1 cells (B), and in PKP2 complexes from DU 145 cells (C). By immunoprecitiation using PKP3-, PKP1-, or PKP2-specific antibody complexes were enriched (compare total lysate [L], negative control for unspecific binding [Con], and immunoprecipitate [PKP3-IP/PKP1-IP/PKP2-IP]). In parallel, desmoplakin-specific antibodies were used (DP-IP). The distribution of PKP3 or PKP1 in different samples was detected by immunoblotting (PKP3 WB, PKP1 WB, and PKP2 WB); for DP-IP, results from a longer exposure time are shown. In corresponding samples, the occurrence of PKP2 and desmoplakin (DP) mRNA was determined after RNA isolation by RT-PCR. Note that PKP2 and desmoplakin mRNAs could be detected in total lysates and PKP3 and PKP1 immunoprecipitates.
PKP1 and PKP3 knockdown reduces mRNA and protein levels of desmosomal proteins.
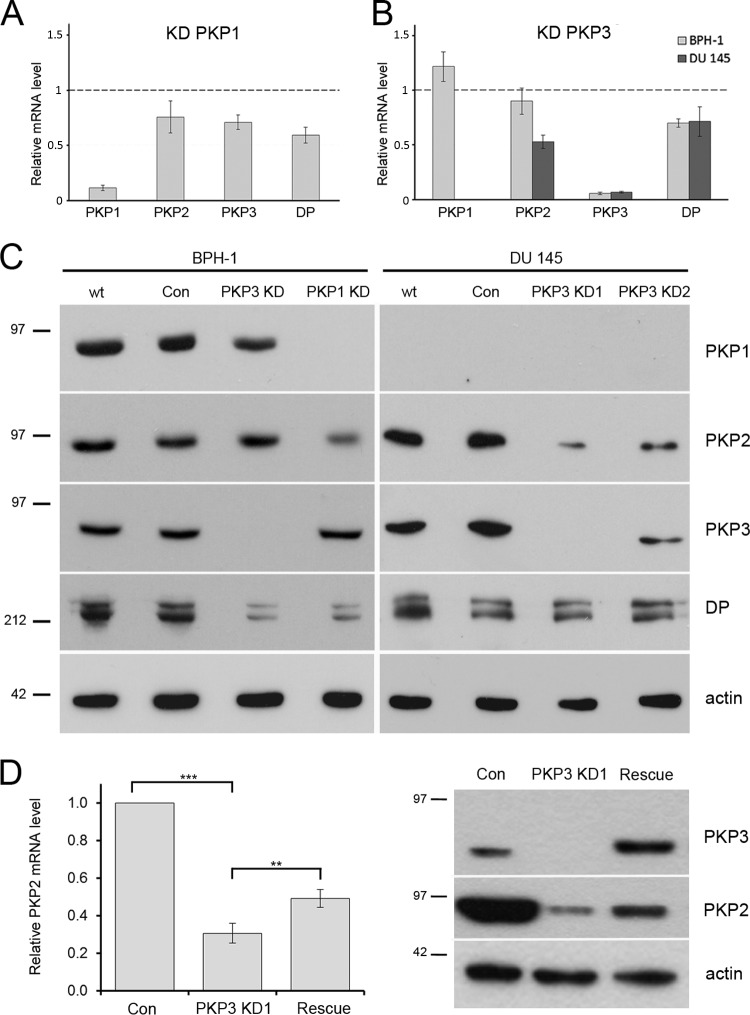
Having shown that PKP2 and desmoplakin mRNAs are specifically associated with PKP1 and PKP3 particles, we hypothesized that this might have an impact on their expression. On one hand, we used the benign prostatic cell line BPH-1, which expresses all three PKPs, to knock down either PKP1 or PKP3 (Fig. 3). Furthermore, the malignant prostatic cell line DU 145, which expresses only PKP2 and PKP3 (49), was used to knock down PKP3. We stably transfected shRNA constructs that target different sequences of PKP1 or PKP3 into these cell lines. Measurement of PKP1 and PKP3 mRNA and protein levels confirmed specific knockdown in the stable shRNA cell lines (Fig. 3A to C).
FIG 3.
Comparison of RNA and protein levels of desmosomal proteins in cells with different PKP1 and PKP3 levels. (A) Relative mRNA expression levels of PKP1, PKP2, and PKP3 and desmoplakin (DP) were measured by quantitative RT-PCR in BPH1-shPKP1 cells. (B) Relative mRNA expression levels of PKP1, PKP2, PKP3, and desmoplakin (DP) were measured in BPH1-shPKP3-1350 and DU145-shPKP3-1350 cells. The results in panels A and B were normalized to 18S rRNA. The means ± SEM of 3 independent experiments are shown relative to the corresponding control cell lines. Note that both PKP1 and PKP3 knockdown leads to decreased PKP2 or desmoplakin mRNA expression levels. (C) The protein levels in cell lines with PKP1 or PKP3 knockdowns (KD) were compared to wild-type and negative-control cell lines by immunodetection. Note that PKP1 and PKP3 knockdown leads to a decreased PKP2 or desmoplakin protein level. Immunoblot on the left, wt, BPH-1; Con, BPH1-shLac; PKP3 KD, BPH1-shPKP3-1350; PKP1 KD, BPH1-shPKP1. Immunoblots on the right: wt, DU 145; Con, DU145-shLuc; PKP3 KD1, DU145-shPKP3-1350; PKP3 KD2, DU145-shPKP3-9. (D) Upon expression of an RNAi-resistant PKP3 cDNA in DU 145-shPKP3-1350 cells, the RNA and protein levels of PKP2 were rescued (biological replicates n = 4). The detection of β-actin is provided as a loading control. Error bars show SD. **, P < 0.01; ***, P < 0.001. Molecular mass markers (in kDa) are indicated on the left.
To analyze the effect of altered PKP1 and PKP3 levels on the expression of desmosomal proteins, we determined PKP2 and desmoplakin mRNA levels in the different cell lines by quantitative RT-PCR. In PKP1-depleted cells, the expression levels of PKP2, PKP3, and desmoplakin were diminished (Fig. 3A). Furthermore, in PKP3-depleted cell lines, expression levels of PKP2 and desmoplakin were decreased; they were strongest for PKP2 in DU145-shPKP3-1350 cells (Fig. 3B). By immunoblot analysis, we confirmed that PKP2 protein expression was reduced in PKP1- and PKP3-depleted cells. In addition, a reduction of desmoplakin was also noticed in both BPH-1 cell lines depleted for PKP1 or PKP3 (Fig. 3C). To rule out off-target effects, we performed a rescue experiment. Upon transient transfection of an RNAi-resistant PKP3-cDNA construct in DU 145 PKP3-deficient cells, PKP3 was reexpressed. This led to a significant upregulation of PKP2 mRNA levels, and also to an increase in protein (Fig. 3D).
Taken together, knockdown of PKP1 and PKP3 led to reduced expression of PKP2 and desmoplakin mRNAs. Importantly, lower expression of levels PKP2 and desmoplakin were also observed at the protein level, demonstrating that PKP1 and PKP3 are physiologically important to sustain the expression of other desmosomal proteins.
PKP3 does not affect global translation.
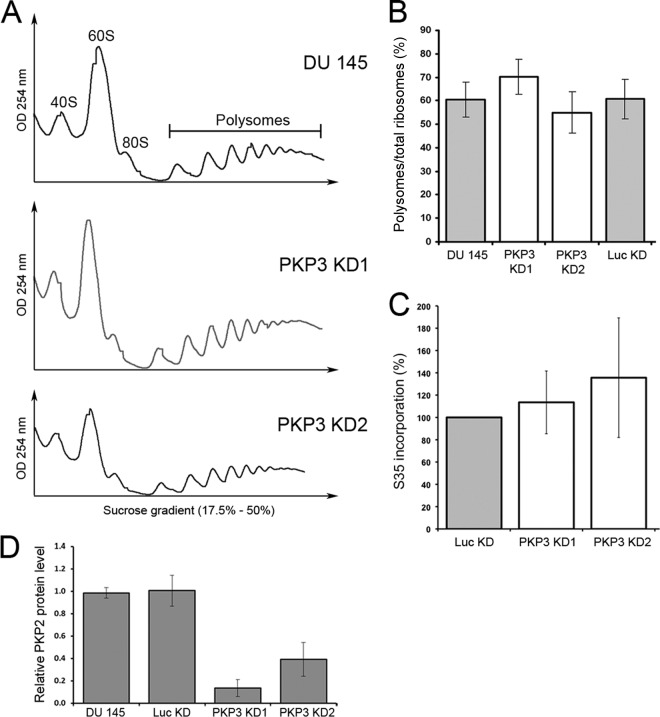
PKP1/3 is found in complexes with RBPs (20, 21) that play important roles in translation regulation (23, 29, 31, 57). In addition, PKP1 has been reported to act as a regulator of mRNA translation by promoting eIF4A1 activity (17). We therefore wondered if the global translation rate was changed in PKP3 knockdown cells. To this end, we recorded polysome profiles by sucrose gradient centrifugation of wild-type DU 145 and two corresponding PKP3 knockdown cell lines (Fig. 4A). However, no obvious differences were detected between the polysome profiles of the different cell lines. For a quantitative assessment of the profiles, the percentage of polysomal ribosomes was calculated by dividing the area under the polysomal part of the curve by the area under the entire curve. The percentage of polysomal ribosomes, which serves as a measure for the overall translation activity, did not differ consistently between the cell lines (Fig. 4B). As an alternative approach, we measured the rate of protein synthesis by labeling with [35S]methionine-cysteine (Fig. 4C). Incorporation of [35S]methionine-cysteine into precipitable protein did not show any major changes between the cell lines. Taken together, the polysome profiles and metabolic labeling experiments provided no evidence for a role of PKP3 in regulating global translation.
FIG 4.
Influence of PKP3 levels on the overall translation rate. (A) Polysome profiling analyses of DU 145 cell lines with different PKP3 levels compared to wild type (DU 145). The panels show profiles obtained by UV recording (254 nm). Peaks corresponding to 40S, 60S, and 80S ribosomes as well as polysomes are labeled in the profile of DU 145 cells. (B) Determination of the translation index by calculation of the ratio of polysomes to total ribosomes of profiles obtained from PKP3 knockdown DU 145 cell lines and control cells. (C) Analysis of translation rate by [35S]methionine-cysteine labeling of DU 145 cell lines with different PKP3 levels. The results are shown as percentages in relation to results with the negative-control cell line. Note that the results indicate that reduced PKP3 levels did not show a significant influence on the overall translation rate. (D) Quantification of PKP2 protein levels determined by Western blotting using ImageJ software. The relative PKP2 protein levels in DU 145 cells are compared to Luc KD, PKP3 KD1, and PKP3 KD2 cells (biological replicates: n = 3). PKP2 protein expression was normalized to β-actin expression. The results are means ± SD. Abbreviations used correspond to the following cell lines: PKP3 KD1, DU145-shPKP3-1350; PKP3 KD2, DU145-shPKP3-9; Luc KD, DU145-shLuc.
Although PKP3 obviously does not have a general role in global translation, it may influence the translation of specific genes. We therefore quantified the protein expression level of PKP2 in DU 145 cells and compared it with that in PKP3-deficient cell lines (Fig. 4D). We found that the relative PKP2 protein level was reduced by up to 80%, an observation that was in line with earlier reports on the RBPs being part of PKP3 complexes (23, 29, 31, 57).
PKP1/3-mediated stabilization of mRNAs encoding desmosomal proteins depends on FXR1.
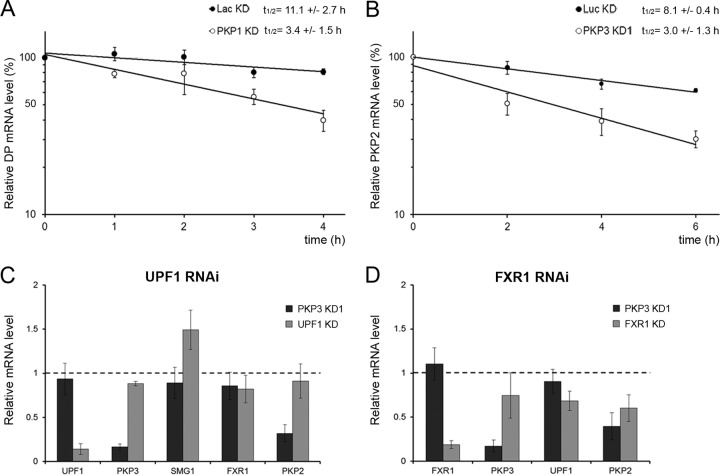
In addition to their functions in translation, all four RBPs found in PKP1/3 complexes are involved in regulating mRNA stability (26, 28, 38, 40, 54, 58). Given that PKP1 and PKP3 knockdown leads to reduced mRNA levels of desmosomal proteins (Fig. 3A and B), we investigated the influence of PKP1 and PKP3 on the mRNA stability of desmoplakin and PKP2. Transcription was blocked with actinomycin D, and RNA was isolated after 0, 1, 2, 3, 4, or 6 h. The amount of desmoplakin and PKP2 mRNA over time was then measured by quantitative RT-PCR (Fig. 5A and B). While desmoplakin mRNA was relatively stable in the BPH-1 control cells (half-life, 11.1 ± 2.7 h), its degradation was accelerated in the PKP1 knockdown cell line (half-life, 3.4 ± 1.5 h). Accordingly, PKP2 mRNA was relatively stable in the DU 145 control cells (half-life, 8.1 ± 0.4 h), and its degradation was accelerated in the PKP3 knockdown cell line (half-life, 3.0 ± 1.3 h). The differences in mRNA half-lives were significant (P = 0.005 for both).
FIG 5.
PKP1 and PKP3 influence mRNA stability of desmoplakin and PKP2 together with FXR1. (A) Desmoplakin mRNA stability measurements in BPH-1 PKP1 knockdown (PKP1 KD; open circles) and negative-control BPH-1 cells (Lac KD; closed circles) after actinomycin D treatment. (B) PKP2 mRNA stability measurements in DU 145 PKP3 knockdown cells (PKP3 KD1; open circles) and negative-control DU 145 cells (Luc KD; closed circles) after actinomycin D treatment. RNA was isolated after 0, 1, 2, 3, 4, or 6 h, and the amount of mRNA was detected by quantitative RT-PCR. The data of four independent biological replicates were normalized to 0-h time point data, and means ± SEM were determined. Note that the differences in mRNA half-lives were significantly different (P = 0.005 for desmoplakin and PKP2). (C and D) Quantitative RT-PCR analyses of endogenous transcripts in DU 145-shPKP3-1350 (PKP3 KD1) cells, in DU 145-shLuc cells depleted of UPF1 (C), or FXR1 (D) via RNAi. The values shown in panels C and D are the average fold changes (mean ± SD) from three independent experiments relative to control DU 145-shLuc cells. A value of 1 indicates no change. All mRNA levels were normalized to 18S rRNA levels.
Because the NMD core component UPF1 copurified with PKP1/3-associated complexes that also contained PKP2 mRNA, we wondered whether the stability of PKP2 mRNA was regulated by UPF1. Depletion of UPF1 in DU 145 cells by siRNA transfection reduced the UPF1 mRNA levels by more than 70% (Fig. 5C). While NMD target transcripts such as SMG1 (52) were upregulated by UPF1 knockdown, the mRNA level of PKP2 was unchanged in both control and PKP3 knockdown DU 145 cells, indicating that PKP2 is not a direct target of UPF1-mediated mRNA degradation. In contrast, depletion of FXR1 in DU 145 cells reduced the PKP2 mRNA level similarly to the level observed in PKP3 knockdown cells (Fig. 5D). Therefore, we concluded that both proteins, PKP3 and FXR1, stabilize PKP2 mRNA.
PKP3 binds PKP2 mRNA indirectly via C-terminal arm repeats.
Since PKP3 forms a complex with RBPs and PKP2 mRNA (Fig. 1 and 2), we examined if PKP3 binds directly to PKP2 mRNA. To this end, we established a solid-phase assay where in vitro-translated PKP3 protein was immobilized on beads and mixed with in vitro-transcribed, polyadenylated PKP2 mRNA. Immobilization of in vitro-translated protein was verified by immunoblotting (Fig. 6A). Binding of PKP2 mRNA to the immobilized protein was then tested by RT-PCR, yet direct association of PKP2 mRNA with PKP3 was not detected (Fig. 6B). PABPC1, which was immobilized as a positive control, did bind PKP2 mRNA. Since the cDNA used for in vitro transcription of PKP2 mRNA lacked 1,678 nucleotides (nt) of the 3′-UTR, we performed the same assay with total RNA isolated from HEK 293 cells, which express PKP2 mRNA. Again, no binding of PKP2 mRNA to PKP3 could be detected, whereas binding to PABPC1 was observed (Fig. 6B). Having shown that both PKP3 and FXR1 stabilize PKP2 mRNA (Fig. 5D), we wondered whether FXR1 might mediate the recruitment of PKP2 mRNA to PKP3-associated complexes. FXR1 contains sequence motifs characteristic of RNA-binding proteins, including two KH domains and an RGG box (59). Therefore, we knocked down FXR1 in DU 145 cells and checked if PKP3-containing complexes still bound PKP2 mRNA. Indeed, in these cells PKP2 mRNA was no longer detectable in PKP3-containing complexes (Fig. 6C). Therefore, we concluded that RNA recruitment to PKP3-containing complexes requires FXR1.
FIG 6.
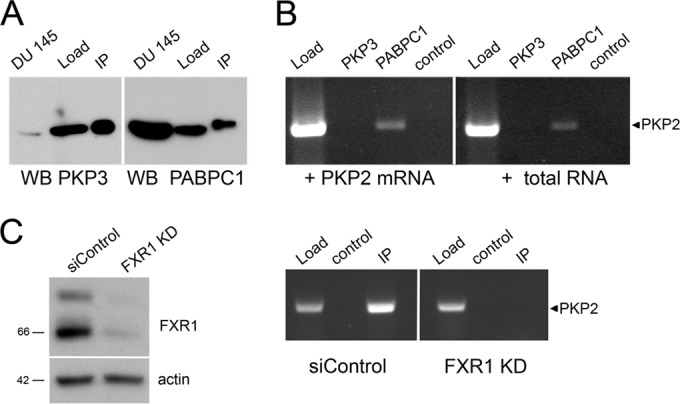
The RNA-binding capacity of PKP3. (A and B) In vitro RNA-binding assay to analyze the RNA-binding capacity of PKP3. (A) The proteins PKP3 and PABPC1 were synthesized by in vitro translation and immobilized on beads using specific antibodies. The efficiencies of protein synthesis and loading were verified by immunoblotting using specific antibodies (PKP3 and PABPC1). The sizes of the in vitro-synthesized proteins were compared with total lysates of DU 145 cells. Load, in vitro-translated protein; IP, immunoprecipitation results. (B) Beads loaded with PKP3 or PABPC1 were incubated with in vitro-transcribed PKP2 mRNA that was synthetically polyadenylated or total mRNA isolated from HEK 293 cells. Binding of PKP2 mRNA was verified by RT-PCR with a PKP2-specific primer. Note that PKP2 mRNA was detectable only in beads loaded with PABPC1. Load, RNA sample; PKP3, beads loaded with PKP3; PABPC1, beads loaded with PABPC1; control, beads incubated with RNA only. (C) Upon FXR1 knockdown in DU 145 cells overexpressing PKP3-EGFP, PKP2 mRNA was no longer detectable in PKP3 complexes. By immunoprecipitation using specific antibodies, PKP3-containing complexes were enriched from DU 145-PKP3-EGFP cells transfected with unrelated siRNAs (siControl) or FXR1 knockdown cells (FXR1 KD). Compare total lysate (Load), negative control for unspecific binding (control), and immunopreciptiate (IP) results. The occurrence of PKP2 mRNA was determined after RNA isolation by RT-PCR.
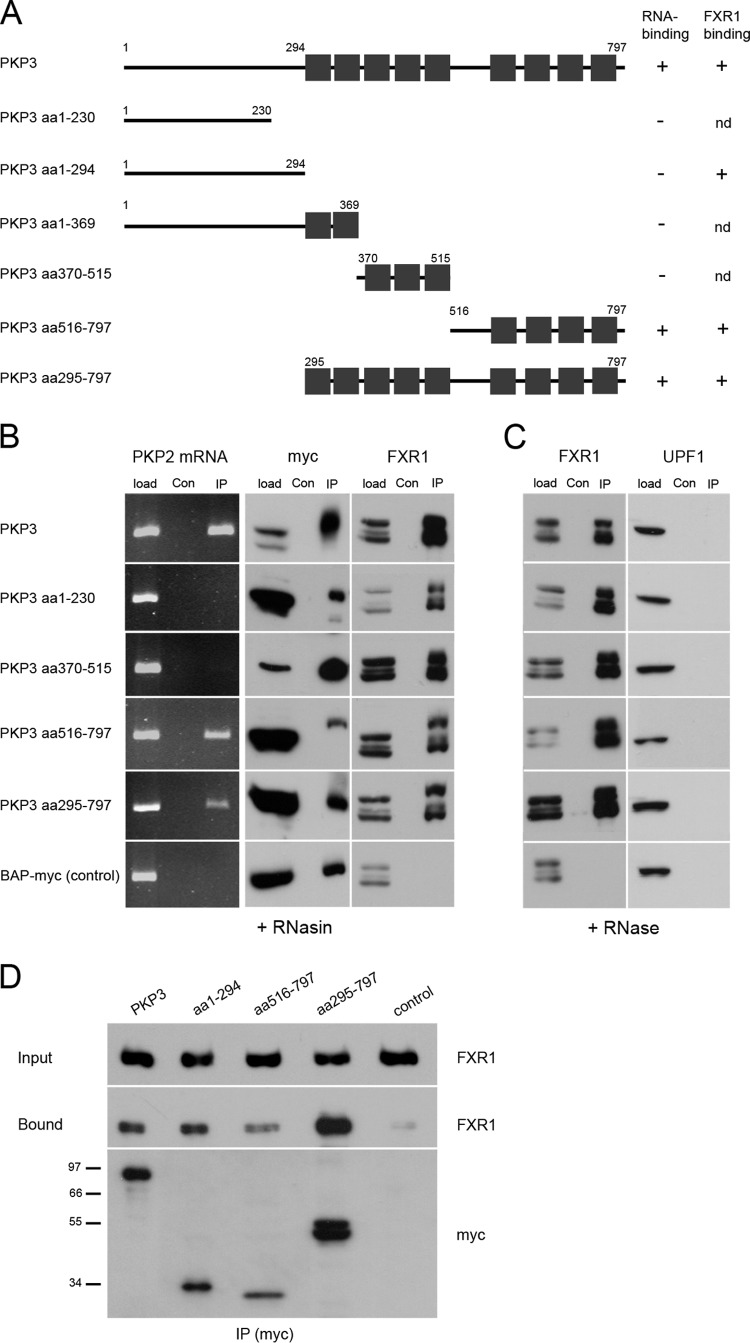
Finally, we wanted to identify PKP3 domains that mediate the interaction with FXR1. A series of myc-tagged PKP3 fragments were expressed in HEK 293 cells, which contain endogenous PKP2 mRNA. Three PKP3 fragments comprised different portions of the N terminus, and three other fragments contained the C-terminal arm repeat domain or parts thereof (Fig. 7A). All six fragments, full-length PKP3-myc and, as a negative control, myc-tagged bacterial alkaline phosphatase, were examined for binding of PKP2 mRNA after immunoprecipitation. PKP3 fragments containing the C-terminal arm repeat domains 6 to 9 (PKP3, PKP3 aa295-797, and PKP3 aa516-797) were found to coimmunoprecipitate PKP2 mRNA (Fig. 7B). In contrast, all six PKP3 fragments were able to bind to FXR1, and this interaction occurred independently of RNA (Fig. 7C). To verify which PKP3 fragment binds to FXR1 directly, an in vitro approach was applied. PKP3 fragments were in vitro translated and affinity-purified via their myc tag, followed by incubation with in vitro-translated FXR1. Again, all PKP3 fragments bound FXR1 (Fig. 7D). Therefore, we conclude that both the N-terminal and C-terminal domains of PKP3 interact with FXR1.
FIG 7.
Analysis of complex formation of PKP3 fragments. (A) Schematic drawing of myc-tagged full-length (PKP3) and fragments consisting of amino acids (aa) 1 to 230, aa 1 to 294, aa 1 to 369, aa 370 to 515, aa 516 to 597, and aa 295 to 797, as indicated. The numbers indicate amino acid positions, and the boxes represent the arm repeat domains (nine in total). All constructs were C-terminally tagged with a myc epitope. (B) The association of the PKP3 constructs, exogenously expressed in HEK 293 cells, with PKP2 mRNA was analyzed after RNA isolation from immunoprecipitates by RT-PCR. The enrichment of myc-tagged proteins was verified by immunoblotting using a myc-specific antibody. In parallel, coprecipitation of FXR1 was followed. A construct coding for myc-tagged bacterial alkaline phosphatase was used as a negative control (BAP-myc). (C) The direct binding of FXR1 and UPF1 to PKP3 fragments was compared by RNase A treatment. (D) Binding of PKP3 fragments to FXR1 in vitro. FXR1- and PKP3-myc-tagged constructs were in vitro translated, and mixtures of FXR1 and PKP3 fragments were immunoprecipitated with myc tag antibody. Binding of FXR1 to beads loaded with myc tag antibody was used as a specificity control. Molecular masses (in kDa) are indicated on the left.
DISCUSSION
PKP1 and PKP3 are important components of desmosomal cell-cell adhesions in epithelial cells. PKP3-deficient mice develop skin abnormalities and are prone to cutaneous inflammation under stress conditions (60). Counter to earlier views, PKP3 may protect the skin from inflammation, not only because of its structural role in cell adhesion but also because cytoplasmic interaction partners hint at an additional regulatory function of PKP3. Especially, the association of PKP3 with RBPs led us to postulate a role of PKP3 in posttranscriptional gene regulation (20). The present study revealed PKP1 and PKP3 to be components of mRNPs in complex with G3BP, FXR1, PABPC1, and UPF1. Furthermore, PKP1- and PKP3-containing mRNPs showed enrichment of PKP2 and desmoplakin mRNAs, and knockdown experiments suggested that PKP1/3 enhances the mRNA stability of desmosomal proteins.
Our observation that PKP1 and PKP3 influence the expression of both constitutive and cell-type-specific desmosomal plaque components raises the question about the biological role of this finding. It is well documented that changes in the expression of a desmosomal component may influence the expression of other components. The constitutive inactivation of PKP2 in mice leads to a reduced expression of desmoglein 2 (61), and the conditional inactivation of desmoplakin in skin results in a decreased expression of PKP2 and increased expression of PKP3 (62). Diverse mechanisms have been reported that specify the availability of desmosomal components, such as transcriptional regulation (63, 64), protein modification (65), or proteolytic degradation (66). Importantly, members of the p120-catenin subfamily not only play a structural role in cell-cell adhesion but also influence cell signaling in the cytoplasm and the nucleus (10–14, 67). Furthermore, it is known from p120-catenin and β-catenin that changes in cellular localization and interaction partners occur during cancer development and may promote or inhibit malignant transformation (68–71).
The detection of PKP1 and PKP3 in mRNPs points to a role in posttranscriptional gene regulation. All identified PKP1- and PKP3-interacting RBPs contribute to mRNA stability and translation regulation (27, 30, 72, 73). As the protein levels of PKP2 and desmoplakin were found to correlate with PKP1 or PKP3 expression, we wondered whether both PKPs have an impact on translation. Indeed, PKP1 has been reported to stimulate translation through its interaction with eIF4A1 in vitro (17). Although PKP3 was not found to alter the global translation rate, the translation of specific mRNAs, e.g., PKP2, may be regulated by PKP3.
Another mode of posttranscriptional regulation is the modulation of mRNA turnover. We found that PKP1/3 enhances mRNA stability of desmoplakin and PKP2 and thereby sustains their expression. To understand how PKP3 influences PKP2 mRNA stability, we examined whether PKP3 directly binds to mRNA. Close inspection of the PKP3 amino acid sequence did not reveal any known RNA-binding motif (74). However, this does not rule out direct binding to RNA, and indeed systematic purification approaches have recently led to the identification of new RNA-binding proteins that lack canonical RNA-binding domains (75, 76). PKP3 has a very high isoelectric point, above 9, and several basic residues are scattered along the entire amino acid sequence (77). PKP3 contains nine arm repeat domains that consist of an approximately 40-residue motif that forms three helices arranged in a triangular shape (78). All arm-repeats together form a basic grove that might mediate electrostatic interactions to negatively charged molecules such as RNA. While PKP2 mRNA clearly is associated with PKP3-containing complexes, we could not detect a direct interaction of PKP3 with PKP2 mRNA in vitro.
To elucidate how PKP1/3 might influence the mRNA stability of desmosomal proteins, we took a closer look at interacting proteins. By immunoprecipitation, we identified PKP1 and PKP3 as components of mRNPs containing not only G3BP, FXR1, and PABPC1 but also UPF1 as a new binding partner. UPF1 is a central component of NMD, which eliminates aberrant mRNAs harboring a premature termination codon. Moreover, UPF1 is also involved in the regulation of many physiological mRNAs (42, 44, 45). UPF1-mediated mRNA decay is stimulated by long 3′-UTRs of more than 1,000 nt, and indeed PKP2 mRNA has a 3′-UTR of 1,678 nt. However, UPF1 knockdown did not change PKP2 mRNA levels, indicating that PKP2 is not an NMD target.
Our observation that the association of PKP1 and PKP3 with FXR1 was resistant to RNase A treatment suggests that PKP1/3 are bound to mRNPs by protein-protein interactions. The finding that all PKP3 domains expressed in vivo copurify with FXR1 implies that there are different modes for how PKP3 and FXR1 may interact. Within mRNPs, PKP3 may serve as a scaffold, recruiting additional factors. FXR1 may in turn mediate the binding to mRNA, because upon FXR1 depletion PKP3 complexes no longer contain mRNA. The shortest isoform of FXR1 is known to bind to AU-rich elements (AREs) in the 3′-UTR of several mRNAs (24). Indeed, the 3′-UTR of PKP2 mRNA contains putative ARE elements. Moreover, it was recently shown that FXR1 binds to the mRNA of desmoplakin and another cell adhesion molecule, talin2, and alters the expression of these cell adhesion proteins (55). Depending on the cellular context, FXR1 can either repress or activate the translation of targets (23, 25). In addition, it was recently reported that FXR1 regulates the mRNA stability of the cell cycle exit regulator p21 (79). Several studies suggest that these diverse actions may be regulated by miRNAs bound to mRNPs (80, 81).
It is interesting to compare PKP1 and PKP3 with the protein HuR, an RBP that affects target mRNA stability and translation. HuR binds to AU-rich regulatory motifs in the 3′-UTR of, for example, p53, vascular endothelial growth factor, COX-2, β-catenin, cyclin, and tumor necrosis factor alpha mRNAs (82). Thereby, HuR influences cellular processes such as apoptosis, proliferation, cell cycle, and tumor progression (83–87). The stabilizing effect of HuR on mRNA is tightly regulated by its modification, and an imbalance may lead to diseased states like cancer (88). To what extent PKP1 and PKP3 more generally influence posttranscriptional gene regulation through interaction with proteins or RNAs, and whether such interactions promote carcinogenesis, needs to be clarified in future studies. Here we have shown for the first time that the mRNA level of a desmosomal component is regulated by another desmosomal protein, adding posttranscriptional gene regulation to the multitude of mechanisms that fine-tune the desmosomal adhesive strength to local requirements.
ACKNOWLEDGMENTS
This work was supported by grants from the German Cancer Aid (109248) and the Deutsche Forschungsgemeinschaft (HO 2455/3-1).
Footnotes
Published ahead of print 15 September 2014
REFERENCES
- 1.Valenta T, Hausmann G, Basler K. 2012. The many faces and functions of beta-catenin. EMBO J. 31:2714–2736. 10.1038/emboj.2012.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carnahan RH, Rokas A, Gaucher EA, Reynolds AB. 2010. The molecular evolution of the p120-catenin subfamily and its functional associations. PLoS One 5:e15747. 10.1371/journal.pone.0015747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Neuber S, Muhmer M, Wratten D, Koch PJ, Moll R, Schmidt A. 2010. The desmosomal plaque proteins of the plakophilin family. Dermatol. Res. Pract. 2010:101452. 10.1155/2010/101452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bonne S, Gilbert B, Hatzfeld M, Chen X, Green KJ, van Roy F. 2003. Defining desmosomal plakophilin-3 interactions. J. Cell Biol. 161:403–416. 10.1083/jcb.200303036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Holthofer B, Windoffer R, Troyanovsky S, Leube RE. 2007. Structure and function of desmosomes. Int. Rev. Cytol. 264:65–163. 10.1016/S0074-7696(07)64003-0. [DOI] [PubMed] [Google Scholar]
- 6.Hatzfeld M. 2007. Plakophilins: multifunctional proteins or just regulators of desmosomal adhesion? Biochim. Biophys. Acta 1773:69–77. 10.1016/j.bbamcr.2006.04.009. [DOI] [PubMed] [Google Scholar]
- 7.McMillan JR, Haftek M, Akiyama M, South AP, Perrot H, McGrath JA, Eady RA, Shimizu H. 2003. Alterations in desmosome size and number coincide with the loss of keratinocyte cohesion in skin with homozygous and heterozygous defects in the desmosomal protein plakophilin 1. J. Invest. Dermatol. 121:96–103. 10.1046/j.1523-1747.2003.12324.x. [DOI] [PubMed] [Google Scholar]
- 8.South AP, Wan H, Stone MG, Dopping-Hepenstal PJ, Purkis PE, Marshall JF, Leigh IM, Eady RA, Hart IR, McGrath JA. 2003. Lack of plakophilin 1 increases keratinocyte migration and reduces desmosome stability. J. Cell Sci. 116:3303–3314. 10.1242/jcs.00636. [DOI] [PubMed] [Google Scholar]
- 9.Kundu ST, Gosavi P, Khapare N, Patel R, Hosing AS, Maru GB, Ingle A, Decaprio JA, Dalal SN. 2008. Plakophilin3 downregulation leads to a decrease in cell adhesion and promotes metastasis. Int. J. Cancer 123:2303–2314. 10.1002/ijc.23797. [DOI] [PubMed] [Google Scholar]
- 10.Pieters T, van Roy F, van Hengel J. 2012. Functions of p120ctn isoforms in cell-cell adhesion and intracellular signaling. Front. Biosci. 17:1669–1694. 10.2741/4012. [DOI] [PubMed] [Google Scholar]
- 11.Bass-Zubek AE, Hobbs RP, Amargo EV, Garcia NJ, Hsieh SN, Chen X, Wahl JK, III, Denning MF, Green KJ. 2008. Plakophilin 2: a critical scaffold for PKC alpha that regulates intercellular junction assembly. J. Cell Biol. 181:605–613. 10.1083/jcb.200712133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mariner DJ, Wang J, Reynolds AB. 2000. ARVCF localizes to the nucleus and adherens junction and is mutually exclusive with p120(ctn) in E-cadherin complexes. J. Cell Sci. 113:1481–1490. [DOI] [PubMed] [Google Scholar]
- 13.Bass-Zubek AE, Godsel LM, Delmar M, Green KJ. 2009. Plakophilins: multifunctional scaffolds for adhesion and signaling. Curr. Opin. Cell Biol. 21:708–716. 10.1016/j.ceb.2009.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cho K, Vaught TG, Ji H, Gu D, Papasakelariou-Yared C, Horstmann N, Jennings JM, Lee M, Sevilla LM, Kloc M, Reynolds AB, Watt FM, Brennan RG, Kowalczyk AP, McCrea PD. 2010. Xenopus kazrin interacts with ARVCF-catenin, spectrin and p190B RhoGAP, and modulates RhoA activity and epithelial integrity. J. Cell Sci. 123:4128–4144. 10.1242/jcs.072041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McCrea PD, Gu D. 2010. The catenin family at a glance. J. Cell Sci. 123:637–642. 10.1242/jcs.039842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schmidt A, Langbein L, Rode M, Pratzel S, Zimbelmann R, Franke WW. 1997. Plakophilins 1a and 1b: widespread nuclear proteins recruited in specific epithelial cells as desmosomal plaque components. Cell Tissue Res. 290:481–499. 10.1007/s004410050956. [DOI] [PubMed] [Google Scholar]
- 17.Wolf A, Krause-Gruszczynska M, Birkenmeier O, Ostareck-Lederer A, Huttelmaier S, Hatzfeld M. 2010. Plakophilin 1 stimulates translation by promoting eIF4A1 activity. J. Cell Biol. 188:463–471. 10.1083/jcb.200908135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mertens C, Hofmann I, Wang Z, Teichmann M, Sepehri Chong S, Schnolzer M, Franke WW. 2001. Nuclear particles containing RNA polymerase III complexes associated with the junctional plaque protein plakophilin 2. Proc. Natl. Acad. Sci. U. S. A. 98:7795–7800. 10.1073/pnas.141219498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mertens C, Kuhn C, Franke WW. 1996. Plakophilins 2a and 2b: constitutive proteins of dual location in the karyoplasm and the desmosomal plaque. J. Cell Biol. 135:1009–1025. 10.1083/jcb.135.4.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hofmann I, Casella M, Schnolzer M, Schlechter T, Spring H, Franke WW. 2006. Identification of the junctional plaque protein plakophilin 3 in cytoplasmic particles containing RNA-binding proteins and the recruitment of plakophilins 1 and 3 to stress granules. Mol. Biol. Cell 17:1388–1398. 10.1091/mbc.E05-08-0708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang C, Strobel P, Marx A, Hofmann I. 2013. Plakophilin-associated RNA-binding proteins in prostate cancer and their implications in tumor progression and metastasis. Virchows Arch. 463:379–390. 10.1007/s00428-013-1452-y. [DOI] [PubMed] [Google Scholar]
- 22.Kedersha N, Anderson P. 2007. Mammalian stress granules and processing bodies. Methods Enzymol. 431:61–81. 10.1016/S0076-6879(07)31005-7. [DOI] [PubMed] [Google Scholar]
- 23.Vasudevan S, Steitz JA. 2007. AU-rich-element-mediated upregulation of translation by FXR1 and Argonaute 2. Cell 128:1105–1118. 10.1016/j.cell.2007.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Garnon J, Lachance C, Di Marco S, Hel Z, Marion D, Ruiz MC, Newkirk MM, Khandjian EW, Radzioch D. 2005. Fragile X-related protein FXR1P regulates proinflammatory cytokine tumor necrosis factor expression at the post-transcriptional level. J. Biol. Chem. 280:5750–5763. 10.1074/jbc.M401988200. [DOI] [PubMed] [Google Scholar]
- 25.Lachance C, Thuraisingam T, Garnon J, Roter E, Radzioch D. 2007. Posttranscriptional gene expression regulation in CpG-activated macrophages depends on FXR1P RNA-binding protein. FEMS Immunol. Med. Microbiol. 51:422–430. 10.1111/j.1574-695X.2007.00317.x. [DOI] [PubMed] [Google Scholar]
- 26.Khera TK, Dick AD, Nicholson LB. 2010. Fragile X-related protein FXR1 controls post-transcriptional suppression of lipopolysaccharide-induced tumour necrosis factor-alpha production by transforming growth factor-beta1. FEBS J. 277:2754–2765. 10.1111/j.1742-4658.2010.07692.x. [DOI] [PubMed] [Google Scholar]
- 27.Irvine K, Stirling R, Hume D, Kennedy D. 2004. Rasputin, more promiscuous than ever: a review of G3BP. Int. J. Dev. Biol. 48:1065–1077. 10.1387/ijdb.041893ki. [DOI] [PubMed] [Google Scholar]
- 28.Li WM, Barnes T, Lee CH. 2010. Endoribonucleases–enzymes gaining spotlight in mRNA metabolism. FEBS J. 277:627–641. 10.1111/j.1742-4658.2009.07488.x. [DOI] [PubMed] [Google Scholar]
- 29.Ortega AD, Willers IM, Sala S, Cuezva JM. 2010. Human G3BP1 interacts with beta-F1-ATPase mRNA and inhibits its translation. J. Cell Sci. 123:2685–2696. 10.1242/jcs.065920. [DOI] [PubMed] [Google Scholar]
- 30.Mangus DA, Evans MC, Jacobson A. 2003. Poly(A)-binding proteins: multifunctional scaffolds for the post-transcriptional control of gene expression. Genome Biol. 4:223. 10.1186/gb-2003-4-7-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Amrani N, Ghosh S, Mangus DA, Jacobson A. 2008. Translation factors promote the formation of two states of the closed-loop mRNP. Nature 453:1276–1280. 10.1038/nature06974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Preiss T, Hentze MW. 2003. Starting the protein synthesis machine: eukaryotic translation initiation. Bioessays 25:1201–1211. 10.1002/bies.10362. [DOI] [PubMed] [Google Scholar]
- 33.Martineau Y, Derry MC, Wang X, Yanagiya A, Berlanga JJ, Shyu AB, Imataka H, Gehring K, Sonenberg N. 2008. Poly(A)-binding protein-interacting protein 1 binds to eukaryotic translation initiation factor 3 to stimulate translation. Mol. Cell. Biol. 28:6658–6667. 10.1128/MCB.00738-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gray NK, Coller JM, Dickson KS, Wickens M. 2000. Multiple portions of poly(A)-binding protein stimulate translation in vivo. EMBO J. 19:4723–4733. 10.1093/emboj/19.17.4723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tritschler F, Huntzinger E, Izaurralde E. 2010. Role of GW182 proteins and PABPC1 in the miRNA pathway: a sense of deja vu. Nat. Rev. Mol. Cell Biol. 11:379–384. 10.1038/nrm2885. [DOI] [PubMed] [Google Scholar]
- 36.Fabian MR, Mathonnet G, Sundermeier T, Mathys H, Zipprich JT, Svitkin YV, Rivas F, Jinek M, Wohlschlegel J, Doudna JA, Chen CY, Shyu AB, Yates JR, III, Hannon GJ, Filipowicz W, Duchaine TF, Sonenberg N. 2009. Mammalian miRNA RISC recruits CAF1 and PABP to affect PABP-dependent deadenylation. Mol. Cell 35:868–880. 10.1016/j.molcel.2009.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zekri L, Huntzinger E, Heimstadt S, Izaurralde E. 2009. The silencing domain of GW182 interacts with PABPC1 to promote translational repression and degradation of microRNA targets and is required for target release. Mol. Cell. Biol. 29:6220–6231. 10.1128/MCB.01081-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huntzinger E, Braun JE, Heimstadt S, Zekri L, Izaurralde E. 2010. Two PABPC1-binding sites in GW182 proteins promote miRNA-mediated gene silencing. EMBO J. 29:4146–4160. 10.1038/emboj.2010.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cosson B, Berkova N, Couturier A, Chabelskaya S, Philippe M, Zhouravleva G. 2002. Poly(A)-binding protein and eRF3 are associated in vivo in human and Xenopus cells. Biol. Cell 94:205–216. 10.1016/S0248-4900(02)01194-2. [DOI] [PubMed] [Google Scholar]
- 40.Ivanov PV, Gehring NH, Kunz JB, Hentze MW, Kulozik AE. 2008. Interactions between UPF1, eRFs, PABP and the exon junction complex suggest an integrated model for mammalian NMD pathways. EMBO J. 27:736–747. 10.1038/emboj.2008.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hogg JR, Goff SP. 2010. Upf1 senses 3′UTR length to potentiate mRNA decay. Cell 143:379–389. 10.1016/j.cell.2010.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nicholson P, Yepiskoposyan H, Metze S, Zamudio Orozco R, Kleinschmidt N, Muhlemann O. 2010. Nonsense-mediated mRNA decay in human cells: mechanistic insights, functions beyond quality control and the double-life of NMD factors. Cell. Mol. Life Sci. 67:677–700. 10.1007/s00018-009-0177-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kervestin S, Jacobson A. 2012. NMD: a multifaceted response to premature translational termination. Nat. Rev. Mol. Cell Biol. 13:700–712. 10.1038/nrm3454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Czaplinski K, Weng Y, Hagan KW, Peltz SW. 1995. Purification and characterization of the Upf1 protein: a factor involved in translation and mRNA degradation. RNA 1:610–623. [PMC free article] [PubMed] [Google Scholar]
- 45.Stalder L, Muhlemann O. 2008. The meaning of nonsense. Trends Cell Biol. 18:315–321. 10.1016/j.tcb.2008.04.005. [DOI] [PubMed] [Google Scholar]
- 46.Collins LJ, Kurland CG, Biggs P, Penny D. 2009. The modern RNP world of eukaryotes. J. Hered. 100:597–604. 10.1093/jhered/esp064. [DOI] [PubMed] [Google Scholar]
- 47.Glisovic T, Bachorik JL, Yong J, Dreyfuss G. 2008. RNA-binding proteins and post-transcriptional gene regulation. FEBS Lett. 582:1977–1986. 10.1016/j.febslet.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Keene JD, Tenenbaum SA. 2002. Eukaryotic mRNPs may represent posttranscriptional operons. Mol. Cell 9:1161–1167. 10.1016/S1097-2765(02)00559-2. [DOI] [PubMed] [Google Scholar]
- 49.Breuninger S, Reidenbach S, Sauer CG, Strobel P, Pfitzenmaier J, Trojan L, Hofmann I. 2010. Desmosomal plakophilins in the prostate and prostatic adenocarcinomas: implications for diagnosis and tumor progression. Am. J. Pathol. 176:2509–2519. 10.2353/ajpath.2010.090737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jin H, Suh MR, Han J, Yeom KH, Lee Y, Heo I, Ha M, Hyun S, Kim VN. 2009. Human UPF1 participates in small RNA-induced mRNA downregulation. Mol. Cell. Biol. 29:5789–5799. 10.1128/MCB.00653-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Scott SD, Joiner MC, Marples B. 2002. Optimizing radiation-responsive gene promoters for radiogenetic cancer therapy. Gene Ther. 9:1396–1402. 10.1038/sj.gt.3301822. [DOI] [PubMed] [Google Scholar]
- 52.Huang L, Lou CH, Chan W, Shum EY, Shao A, Stone E, Karam R, Song HW, Wilkinson MF. 2011. RNA homeostasis governed by cell type-specific and branched feedback loops acting on NMD. Mol. Cell 43:950–961. 10.1016/j.molcel.2011.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hwang J, Sato H, Tang Y, Matsuda D, Maquat LE. 2010. UPF1 association with the cap-binding protein, CBP80, promotes nonsense-mediated mRNA decay at two distinct steps. Mol. Cell 39:396–409. 10.1016/j.molcel.2010.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kim YK, Furic L, Desgroseillers L, Maquat LE. 2005. Mammalian Staufen1 recruits Upf1 to specific mRNA 3′UTRs so as to elicit mRNA decay. Cell 120:195–208. 10.1016/j.cell.2004.11.050. [DOI] [PubMed] [Google Scholar]
- 55.Whitman SA, Cover C, Yu L, Nelson DL, Zarnescu DC, Gregorio CC. 2011. Desmoplakin and talin2 are novel mRNA targets of fragile x-related protein-1 in cardiac muscle. Circ. Res. 109:262–271. 10.1161/CIRCRESAHA.111.244244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hofmann I, Mertens C, Brettel M, Nimmrich V, Schnolzer M, Herrmann H. 2000. Interaction of plakophilins with desmoplakin and intermediate filament proteins: an in vitro analysis. J. Cell Sci. 113:2471–2483. [DOI] [PubMed] [Google Scholar]
- 57.Siomi MC, Zhang Y, Siomi H, Dreyfuss G. 1996. Specific sequences in the fragile X syndrome protein FMR1 and the FXR proteins mediate their binding to 60S ribosomal subunits and the interactions among them. Mol. Cell. Biol. 16:3825–3832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhong N, Ju W, Nelson D, Dobkin C, Brown WT. 1999. Reduced mRNA for G3BP in fragile X cells: evidence of FMR1 gene regulation. Am. J. Med. Genet. 84:268–271. . [DOI] [PubMed] [Google Scholar]
- 59.Siomi MC, Siomi H, Sauer WH, Srinivasan S, Nussbaum RL, Dreyfuss G. 1995. FXR1, an autosomal homolog of the fragile X mental retardation gene. EMBO J. 14:2401–2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sklyarova T, Bonne S, D'Hooge P, Denecker G, Goossens S, De Rycke R, Borgonie G, Bosl M, van Roy F, van Hengel J. 2008. Plakophilin-3-deficient mice develop hair coat abnormalities and are prone to cutaneous inflammation. J. Invest. Dermatol. 128:1375–1385. 10.1038/sj.jid.5701189. [DOI] [PubMed] [Google Scholar]
- 61.Grossmann KS, Grund C, Huelsken J, Behrend M, Erdmann B, Franke WW, Birchmeier W. 2004. Requirement of plakophilin 2 for heart morphogenesis and cardiac junction formation. J. Cell Biol. 167:149–160. 10.1083/jcb.200402096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vasioukhin V, Bowers E, Bauer C, Degenstein L, Fuchs E. 2001. Desmoplakin is essential in epidermal sheet formation. Nat. Cell Biol. 3:1076–1085. 10.1038/ncb1201-1076. [DOI] [PubMed] [Google Scholar]
- 63.Savagner P, Yamada KM, Thiery JP. 1997. The zinc-finger protein slug causes desmosome dissociation, an initial and necessary step for growth factor-induced epithelial-mesenchymal transition. J. Cell Biol. 137:1403–1419. 10.1083/jcb.137.6.1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Aigner K, Descovich L, Mikula M, Sultan A, Dampier B, Bonne S, van Roy F, Mikulits W, Schreiber M, Brabletz T, Sommergruber W, Schweifer N, Wernitznig A, Beug H, Foisner R, Eger A. 2007. The transcription factor ZEB1 (δEF1) represses plakophilin 3 during human cancer progression. FEBS Lett. 581:1617–1624. 10.1016/j.febslet.2007.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Muller J, Ritt DA, Copeland TD, Morrison DK. 2003. Functional analysis of C-TAK1 substrate binding and identification of PKP2 as a new C-TAK1 substrate. EMBO J. 22:4431–4442. 10.1093/emboj/cdg426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pasdar M, Nelson WJ. 1988. Kinetics of desmosome assembly in Madin-Darby canine kidney epithelial cells: temporal and spatial regulation of desmoplakin organization and stabilization upon cell-cell contact. I. Biochemical analysis. J. Cell Biol. 106:677–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Reynolds AB. 2007. p120-catenin: past and present. Biochim. Biophys. Acta 1773:2–7. 10.1016/j.bbamcr.2006.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.van Hengel J, van Roy F. 2007. Diverse functions of p120ctn in tumors. Biochim. Biophys. Acta 1773:78–88. 10.1016/j.bbamcr.2006.08.033. [DOI] [PubMed] [Google Scholar]
- 69.Reynolds AB, Roczniak-Ferguson A. 2004. Emerging roles for p120-catenin in cell adhesion and cancer. Oncogene 23:7947–7956. 10.1038/sj.onc.1208161. [DOI] [PubMed] [Google Scholar]
- 70.Clevers H. 2006. Wnt/beta-catenin signaling in development and disease. Cell 127:469–480. 10.1016/j.cell.2006.10.018. [DOI] [PubMed] [Google Scholar]
- 71.Ying Y, Tao Q. 2009. Epigenetic disruption of the WNT/beta-catenin signaling pathway in human cancers. Epigenetics 4:307–312. 10.4161/epi.4.5.9371. [DOI] [PubMed] [Google Scholar]
- 72.Chamieh H, Ballut L, Bonneau F, Le Hir H. 2008. NMD factors UPF2 and UPF3 bridge UPF1 to the exon junction complex and stimulate its RNA helicase activity. Nat. Struct. Mol. Biol. 15:85–93. 10.1038/nsmb1330. [DOI] [PubMed] [Google Scholar]
- 73.Mazroui R, Huot ME, Tremblay S, Filion C, Labelle Y, Khandjian EW. 2002. Trapping of messenger RNA by Fragile X Mental Retardation protein into cytoplasmic granules induces translation repression. Hum. Mol. Genet. 11:3007–3017. 10.1093/hmg/11.24.3007. [DOI] [PubMed] [Google Scholar]
- 74.Lunde BM, Moore C, Varani G. 2007. RNA-binding proteins: modular design for efficient function. Nat. Rev. Mol. Cell Biol. 8:479–490. 10.1038/nrm2178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Baltz AG, Munschauer M, Schwanhausser B, Vasile A, Murakawa Y, Schueler M, Youngs N, Penfold-Brown D, Drew K, Milek M, Wyler E, Bonneau R, Selbach M, Dieterich C, Landthaler M. 2012. The mRNA-bound proteome and its global occupancy profile on protein-coding transcripts. Mol. Cell 46:674–690. 10.1016/j.molcel.2012.05.021. [DOI] [PubMed] [Google Scholar]
- 76.Castello A, Fischer B, Eichelbaum K, Horos R, Beckmann BM, Strein C, Davey NE, Humphreys DT, Preiss T, Steinmetz LM, Krijgsveld J, Hentze MW. 2012. Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell 149:1393–1406. 10.1016/j.cell.2012.04.031. [DOI] [PubMed] [Google Scholar]
- 77.Schmidt A, Langbein L, Pratzel S, Rode M, Rackwitz HR, Franke WW. 1999. Plakophilin 3: a novel cell-type-specific desmosomal plaque protein. Differentiation 64:291–306. 10.1046/j.1432-0436.1999.6450291.x. [DOI] [PubMed] [Google Scholar]
- 78.Choi HJ, Weis WI. 2005. Structure of the armadillo repeat domain of plakophilin 1. J. Mol. Biol. 346:367–376. 10.1016/j.jmb.2004.11.048. [DOI] [PubMed] [Google Scholar]
- 79.Davidovic L, Durand N, Khalfallah O, Tabet R, Barbry P, Mari B, Sacconi S, Moine H, Bardoni B. 2013. A novel role for the RNA-binding protein FXR1P in myoblasts cell-cycle progression by modulating p21/Cdkn1a/Cip1/Waf1 mRNA stability. PLoS Genet. 9:e1003367. 10.1371/journal.pgen.1003367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Pasquinelli AE. 2012. MicroRNAs and their targets: recognition, regulation and an emerging reciprocal relationship. Nat. Rev. Genet. 13:271–282. 10.1038/nrg3162. [DOI] [PubMed] [Google Scholar]
- 81.Palanisamy V, Jakymiw A, Van Tubergen EA, D'Silva NJ, Kirkwood KL. 2012. Control of cytokine mRNA expression by RNA-binding proteins and microRNAs. J. Dent. Res. 91:651–658. 10.1177/0022034512437372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hinman MN, Lou H. 2008. Diverse molecular functions of Hu proteins. Cell. Mol. Life Sci. 65:3168–3181. 10.1007/s00018-008-8252-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Atasoy U, Curry SL, Lopez de Silanes I, Shyu AB, Casolaro V, Gorospe M, Stellato C. 2003. Regulation of eotaxin gene expression by TNF-alpha and IL-4 through mRNA stabilization: involvement of the RNA-binding protein HuR. J. Immunol. 171:4369–4378. 10.4049/jimmunol.171.8.4369. [DOI] [PubMed] [Google Scholar]
- 84.Brennan CM, Steitz JA. 2001. HuR and mRNA stability. Cell. Mol. Life Sci. 58:266–277. 10.1007/PL00000854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Levy NS, Chung S, Furneaux H, Levy AP. 1998. Hypoxic stabilization of vascular endothelial growth factor mRNA by the RNA-binding protein HuR. J. Biol. Chem. 273:6417–6423. 10.1074/jbc.273.11.6417. [DOI] [PubMed] [Google Scholar]
- 86.Mazan-Mamczarz K, Galban S, Lopez de Silanes I, Martindale JL, Atasoy U, Keene JD, Gorospe M. 2003. RNA-binding protein HuR enhances p53 translation in response to ultraviolet light irradiation. Proc. Natl. Acad. Sci. U. S. A. 100:8354–8359. 10.1073/pnas.1432104100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wang W, Furneaux H, Cheng H, Caldwell MC, Hutter D, Liu Y, Holbrook N, Gorospe M. 2000. HuR regulates p21 mRNA stabilization by UV light. Mol. Cell. Biol. 20:760–769. 10.1128/MCB.20.3.760-769.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Abdelmohsen K, Gorospe M. 2010. Posttranscriptional regulation of cancer traits by HuR. Wiley Interdiscip. Rev. RNA 1:214–229. 10.1002/wrna.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kirkpatrick LL, McIlwain KA, Nelson DL. 1999. Alternative splicing in the murine and human FXR1 genes. Genomics 59:193–202. 10.1006/geno.1999.5868. [DOI] [PubMed] [Google Scholar]
- 90.Dube M, Huot ME, Khandjian EW. 2000. Muscle specific fragile X related protein 1 isoforms are sequestered in the nucleus of undifferentiated myoblast. BMC Genet. 1:4. 10.1186/1471-2156-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]