Abstract
Enzymes embedded into the RNA editing core complex (RECC) catalyze the U-insertion/deletion editing cascade to generate open reading frames in trypanosomal mitochondrial mRNAs. The sequential reactions of mRNA cleavage, U-addition or removal, and ligation are directed by guide RNAs (gRNAs). We combined proteomic, genetic, and functional studies with sequencing of total and complex-bound RNAs to define a protein particle responsible for the recognition of gRNAs and pre-mRNA substrates, editing intermediates, and products. This approximately 23-polypeptide tripartite assembly, termed the RNA editing substrate binding complex (RESC), also functions as the interface between mRNA editing, polyadenylation, and translation. Furthermore, we found that gRNAs represent only a subset of small mitochondrial RNAs, and yet an inexplicably high fraction of them possess 3′ U-tails, which correlates with gRNA's enrichment in the RESC. Although both gRNAs and mRNAs are associated with the RESC, their metabolic fates are distinct: gRNAs are degraded in an editing-dependent process, whereas edited mRNAs undergo 3′ adenylation/uridylation prior to translation. Our results demonstrate that the well-characterized editing core complex (RECC) and the RNA binding particle defined in this study (RESC) typify enzymatic and substrate binding macromolecular constituents, respectively, of the ∼40S RNA editing holoenzyme, the editosome.
INTRODUCTION
Kinetoplastids are a group of unicellular flagellated protozoans that cause parasitic diseases such as African sleeping sickness, Chagas disease, and leishmaniasis. A representative organism, Trypanosoma brucei, is characterized by the presence of a “kinetoplast,” a disc-shaped DNA-containing body located in the mitochondrion adjacent to the flagellum base. The kinetoplast DNA (kDNA) is composed of catenated maxicircles and minicircles: approximately 50 maxicircles encode typical mitochondrial proteins, such as subunits of respiratory complexes, while thousands of minicircles carry guide RNA (gRNA) genes. Because 12 of 18 protein-coding genes are encrypted, their transcripts must undergo U-insertion/deletion editing to create open reading frames (1). The elementary reactions of mRNA cleavage, U-insertion or U-deletion, and ligation are catalyzed by enzymes embedded into the ∼15-polypeptide RNA editing core complex (RECC) (2, 3) and are directed by the gRNA-mRNA hybrid structure (4, 5). Thus, functional mitochondrial mRNAs are created from maxicircle transcripts in a process involving nucleus-encoded proteins and minicircle-encoded gRNAs (6).
The extensively investigated core complex exists in at least three isoforms that share most subunits, including U-insertion TUTase RET2, U-deletion exonucleases REX1 and REX2, RNA ligases REL1 and REL2, and structural proteins. These isoforms are distinct in composition and function due to mutually exclusive associations with endonucleases specific for insertion or deletion sites (7, 8). Notwithstanding the differences among isoforms, RECC was purified as a homogenous ∼20S complex, which enabled a single-particle reconstitution analysis (9, 10). However, in mitochondrial extracts, core complex components were traced into undefined ribonucleoprotein complexes (RNPs) with an apparent sedimentation rate exceeding 40S (11–13). Furthermore, RECC subunits were detected in complexes implicated in gRNA-mRNA annealing (MRP1/2) (11, 14), gRNA uridylation (RET1 TUTase) (15), mRNA kinetoplast polyadenylation [KPAP1 poly(A) polymerase] (16), and translation (17). An additional level of complexity emerged from studies of accessory proteins that are essential for the editing process but that do not belong to the core complex. Most prominent among these are homologous gRNA binding complex subunits 1 and 2 (GRBC1 and GRBC2 [GRBC1/2]) that form a stable a2β2 heterotetramer and are required for gRNA stabilization (14, 18–20). In mitochondria, the GRBC1/2 tetramer associates with several proteins that have been detected in mitochondrial RNA binding complex 1 (21), such as the RGG2 RNA binding protein (22) and polypeptides lacking discernible motifs (23). In contrast, GRBC1 and GRBC2 were found in the purified polyadenylation complex (16, 20) and associated with an RNA binding protein, RGG1 (24), and with the large ribosomal subunit (LSU) (17). RNA interference (RNAi) knockdowns of several GRBC1/2-associated proteins (22, 24–27) decreased the output of edited mRNAs but provided limited insights into how the mRNA transitions between different processing complexes. Indeed, the recent influx of new editing factors and often-disparate information regarding their biological roles and interactions created an exceedingly convoluted picture of the mitochondrial mRNA processing apparatus.
In this work, we have combined proteomic, genetic, and biochemical approaches with sequencing of complex-bound RNAs to decipher the mRNA editing machinery's global architecture, the functionality of its parts, and interactions underlying functional relationships between editing, polyadenylation, and translation. We show that the GRBC1/2 tetramer is the only element within the 10-polypeptide gRNA binding complex (GRBC) which is required for gRNA stabilization. GRBC is connected to the editing core complex via an eight-member module termed the RNA editing mediator complex (REMC) and to the polyadenylation complex via a defined set of five proteins, the polyadenylation mediator complex (PAMC). Held together by protein-protein and RNA-stabilized interactions, GRBC, REMC, and PAMC form a trimodular platform designated the RNA editing substrate binding complex (RESC). This assembly is responsible for binding of RNA editing substrates (preedited mRNAs and gRNAs), intermediates (partially edited mRNAs), and products (edited mRNAs) and, most likely, for delivering edited mRNA to the last processing step, 3′-end adenylation/uridylation. Overall, RESC-mediated interactions provide a rationale for the previously established functional coupling between the completion of editing and postediting mRNA polyadenylation/uridylation and translation (17). We also show that gRNAs, as defined by complementarity to edited mRNAs, represent only a subset among small mitochondrial RNAs and yet their absolute majority possess posttranscriptionally added 3′ U-tails exceeding five nucleotides in length. Remarkably, this bias is reflected by gRNA enrichment in the RESC. Although RESC contains the gRNA-stabilizing GRBC module, we found that gRNAs are degraded during the active editing process whereas the non-gRNAs (ngRNAs) remain unaffected. Collectively, our results demonstrate that the well-characterized editing core complex (RECC) and the RNA binding particle defined in this study (RESC) constitute enzymatic and RNA binding components, respectively, of the U-insertion/deletion RNA editing holoenzyme (editosome).
MATERIALS AND METHODS
Trypanosome culture, RNAi, and protein expression.
The RNAi plasmids were generated by cloning ∼500-bp gene fragments into p2T7-177 vector for tetracycline-inducible expression (28). The constructs were transfected into the procyclic 29-13 T. brucei strain (29). The RNAi knockdowns were performed as previously described (20) and verified by quantitative reverse transcription-PCR (qRT-PCR). For inducible protein expression experiments, full-length genes were cloned into pLEW-MHTAP vector (30).
RNA analysis.
Northern blotting and qRT-PCR were performed as previously described (17). For small-RNA sequencing (RNA-Seq), RNA was extracted from a Renografin density gradient-purified mitochondrial fraction or from affinity-purified complexes and separated on 10% PAGE with 8 M urea. RNA was excised from areas corresponding to 35 to 75 nucleotides (nt), eluted, and processed with a ScriptMiner small RNA-Seq library preparation kit (Epicentre) to generate Illumina-compatible libraries. Single-ended 75-nt stranded sequencing and raw data extraction were performed at the University of California (UC) Irvine Genomics High-Throughput Facility. To perform the gRNA-mRNA alignments and to analyze complex-bound RNAs, we developed a custom python script that generates alternative sequences of the original assembled transcripts by considering GU matches and mapped them to edited mRNAs by the use of the BWA program using default settings (31). GU matches were considered to be equivalent to the canonical matches; we allowed one gap and up to three mismatches in our candidate gRNAs. (See the supplemental material for details.)
Liquid chromatography tandem mass spectrometry (LC MS/MS) analysis, protein identification, and calculation of distributed normalized spectral abundance factors (dNSAF).
Proteomic analysis and interaction network building were performed as described previously (17, 32, 33) with modifications noted in the supplemental material.
RESULTS
RNA editing core and gRNA binding complexes interact via RNA.
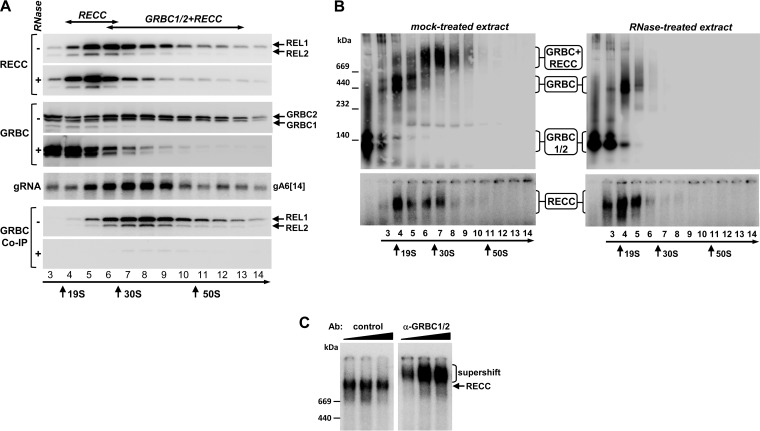
We previously identified guide RNA binding complex proteins 1 and 2 (GRBC1 and GRBC2) in Leishmania tarentolae (14, 20). Since this organism lacks RNAi, T. brucei represents a more attractive system for comprehensive structure/function analysis of RNPs. To gain insight into higher-order interactions of editing complexes, mitochondrial extracts from procyclic (insect) parasites were treated with RNases A and T1 and were fractionated on glycerol gradients. Individual fractions were incubated with [α-32P]ATP to detect RNA editing ligases 1 and 2 (REL1 and REL2) as markers for the editing core complex and were probed with antibodies against GRBC1/2. Representative gRNA was analyzed in each fraction, and immunoprecipitation (IP) was performed with same antibodies to detect GRBC-RECC interactions (Fig. 1A). Under separation conditions, RECC migrated as a broad peak centered at 25S to 30S and extending into the 50S-to-70S region whereas GRBC1/2-containing complexes and gRNA displayed wide-ranging sedimentation profiles peaking in the region from 30S to 40S. The RECC-GRBC co-IP profile mirrored the 30S-to-40S peak but was markedly absent in the 20S-to-25S region. Pretreatment of mitochondrial extract with RNases compacted the RECC into the 20S-to-25S zone and GRBC1/2 into the 10S-to-25S zone and virtually eliminated the RECC-GRBC interaction. To determine the apparent molecular masses of the respective complexes, glycerol gradient fractions were separated on gradient native gels and subjected to immunoblotting (Fig. 1B). Three major GRBC1/2-containing complexes were detected in the mock-treated extract: an ∼200-kDa particle (fractions 3 and 4) resembling the GRBC1/2 tetramer (18), two closely migrating particles (fractions 5 and 6) in the ∼550-kDa range, and a >1.2-MDa complex in fractions 6 to 9. RNase treatment eliminated the largest GRBC1/2-containing complex and compressed the ∼550-kDa complexes into a single band but had only a minor effect on the 200-kDa particle. To conclude, in mitochondrial extracts, GRBC1/2 exists as unassociated tetramer and participates in a stable protein complex of ∼500 kDa, which interacts with RECC via an RNA component, thereby forming an approximately 40S (∼1.2-MDa) RNP.
FIG 1.
Cocomplex interactions between editing core and gRNA binding complexes. (A) The impact of RNase treatment on RECC and GRBC1/2 cocomplex interactions. Mock-treated extracts (−) and extracts preincubated with RNase A (0.1 mg/ml) and RNase T1 (1,000 U/ml) (+) were separated on 10% to 30% glycerol gradients for 4 h at 178,000 × g. Sedimentation patterns of the RNA editing core complex were visualized by self-adenylation of RNA editing ligases REL1 and REL2 in the presence of [α-32P]ATP (58). Following immunoprecipitation, anti-GRBC1/2 antibody-coated beads were incubated with [α-32P]ATP to label REL1 and REL2 RNA ligases as a means of detecting the editing core complex. Bound proteins were eluted with SDS gel loading buffer. Alternatively, RNA was extracted from each fraction and separated on a 10% denaturing polyacrylamide gel (PAGE) for hybridization with a gRNA A6[14]-specific probe. (B) RNase treatment eliminates the ∼40S particle. The gradient fractions described for panel A were separated on native 3% to 12% PAGE to estimate the molecular mass of untreated and RNase-resistant RECC and GRBC1/2-containing complexes. (C) “Supershift” of RECC with α-GRBC1/2 antibody. Preadenylated fraction 8 was incubated with 0.1, 0.3, and 1 μg of antigen-purified control and α-GRBC1/2 antibodies and separated on 3% to 12% native PAGE.
To assess the interaction stoichiometry of editing core and GRBC1/2-containing complexes within the ∼40S particle, we performed a “supershift” experiment (Fig. 1C). Fraction 8 from the glycerol gradient was subjected to self-adenylation, incubated with increasing concentrations of antigen-purified control antibodies against nuclear noncanonical poly(A) polymerase 1 (T. brucei ncPAP1 [TbncPAP1] [34]) and antigen-purified α-GRBC1/2 antibodies, and separated on the native gel. The mobility of the RECC was unaffected by the control antibody, while binding of the GRBC1/2 antibody shifted the editing core complex into a slower-migrating band. We concluded that virtually all of the editing core complex within the ∼40S particle is bound to the gRNA binding complex.
A tripartite RESC connects editing core and polyadenylation complexes with the ribosome.
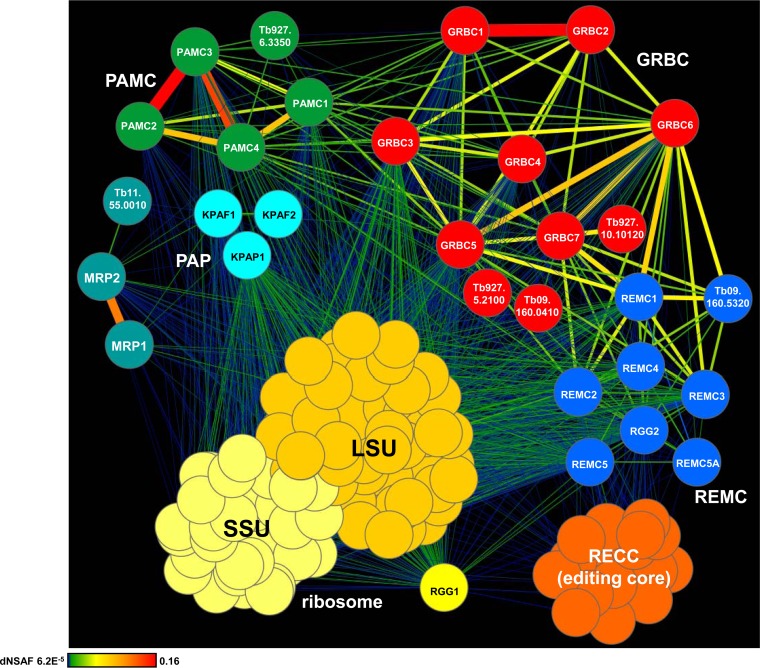
To determine the composition of GRBC1/2-associated complexes and to dissect their RNA-mediated and protein-protein contacts, we took an iterative approach to build the interaction network for mitochondrial RNA processing complexes (Fig. 2; see also Fig. S1 in the supplemental material). The initial data set was obtained by LC-MS/MS analysis of affinity-purified GRBC1 and GRBC2, which was followed by multiple rounds of cross-tagging and LC-MS/MS analysis. In a converging approach, the editing core complex, KPAP1 poly(A) polymerase (16), kinetoplast polyadenylation/uridylation factor 1 (KPAF1) and KPAF2 (KPAF1/2) polyadenylation factors (17), MRP1/2 RNA chaperones (14, 35), RGG1 RNA binding protein (24, 36), and both ribosomal subunits were likewise investigated (see Table S1). All purifications were performed under uniform conditions using mock- and RNase-treated mitochondrial extracts. A label-free quantitative MS strategy was applied to calculate the relative abundance of a given protein in each complex and to quantify interactions based on the distributed normalized spectral abundance factor (dNSAF) (32, 37).
FIG 2.
Interaction network of mitochondrial RNA processing complexes. The network of RNase-resistant interactions was generated in Cytoscape software from bait-prey pairs in which the prey protein was identified with at least 4 unique peptides (see Table S1 in the supplemental material). The edge thickness and color intensity correlate with dNSAF values for bait-prey interactions. For clarity, reciprocal contacts (i.e., bait-bait interactions captured in both purifications) are depicted by a single edge as the sum of two dNSAF values. Color-coded complexes were grouped based on the dNSAF values for each bait-prey pair. The highest dNSAF value corresponds to a GRBC1-GRBC2 direct interaction, which was recapitulated in vitro (18). Gene identifiers (ID) (http://tritrypdb.org) are shown for the proteins detected in the respective complexes with high confidence but not investigated further in this study. SSU, small subunit.
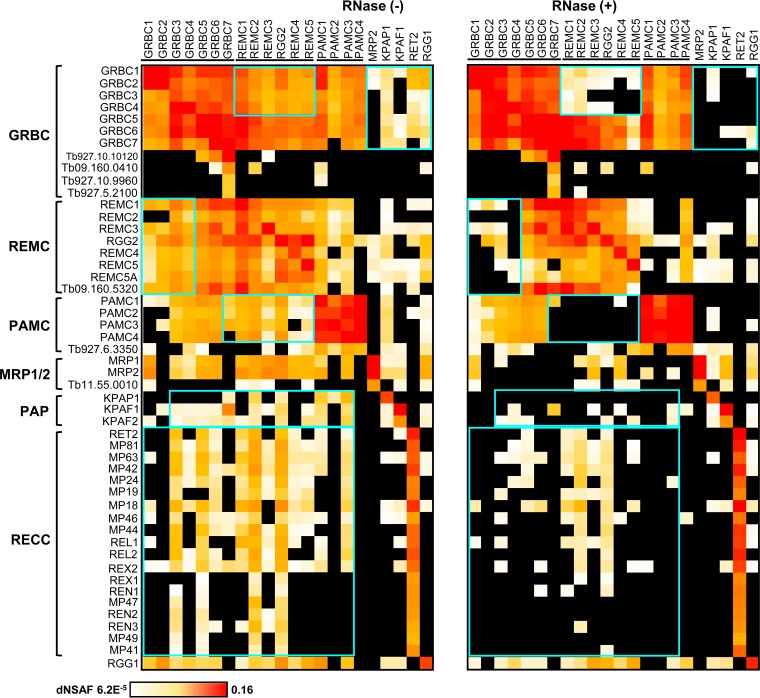
Quantitative clustering of the RNase-resistant protein interaction network revealed the existence of three intrinsically stable modules annotated as follows: (i) the GRBC1/2-containing 10-polypeptide gRNA binding complex (GRBC); (ii) the eight-member RNA editing mediator complex (REMC); and (iii) the set of five proteins that constitute the polyadenylation mediator complex (PAMC) (Fig. 2). These three modules form a platform, here designated the RNA editing substrate binding complex (RESC), that participates in extensive contacts with the RNA editing core and polyadenylation complexes and with the large ribosomal subunit. In agreement with earlier studies (36), RGG1 RNA binding protein was found to be stably associated with the ribosome whereas virtually all interactions of the MRP1/2 complex were mediated by RNA (38). Some relations predicted here were consonant with pairwise yeast two-hybrid (Y2H) mapping of the MRB1 complex (23), such as GRBC2-GRBC6, GRBC4-GRBC6, and GRBC3-GRBC6, while other contacts, such as GRBC6-KPAF1 and REMC5-KPAF1, were not supported by our data. Comparison of RNase-sensitive and -resistant interaction networks indicated that mitochondrial RNAs stabilize intramodular contacts between GRBC, REMC, and PAMC within the RESC assembly and are critical for the recruitment of editing core and polyadenylation complexes (Fig. 3). Identification of three large ribosomal subunit proteins (TbLSU-600, TbLSU-2340, and TbLSU-4710) (see Table S1 in the supplemental material) in the RESC was consistent with the results of an earlier immunological study (17), thus confirming extensive association of the editing apparatus with translation machinery.
FIG 3.
Global comparison of RNase-resistant and RNase-sensitive interactions. Heat maps were generated from the data presented in Table S1 in the supplemental material (ribosomal subunits are omitted). Highlighted boxes indicate interactions that were susceptible to RNase treatment.
Guide RNAs are critical for the RESC-editing core complex interaction.
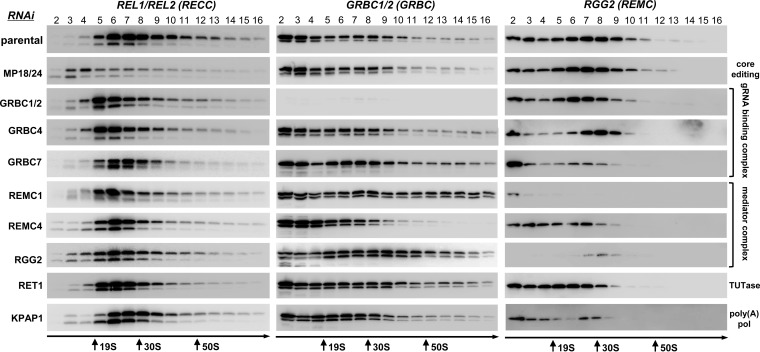
As documented by studies of the core editing complex, loss of an individual subunit may lead to degradation of an entire complex or of a subcomplex or may exert negligible effects on complex integrity. In many cases, such reductions reflect the extent of protein-protein interactions and provide cues for identification of direct binding partners (39). We further reasoned that disruptive effects of RNase treatment on cocomplex interactions may be narrowed to a particular RNA class by eliminating a key enzyme responsible for its biogenesis. Inducible RNAi cell lines have been generated to target most RESC subunits, and mitochondrial extracts were analyzed by sedimentation in glycerol gradients under highly reproducible conditions (Fig. 4; see also Fig. S2 and S3 in the supplemental material). Sedimentation patterns of the editing core complex and RESC's gRNA binding and editing mediator modules have been visualized in representative knockdown cell lines. In addition to RESC knockdowns, repression of MP18/24 was used to destabilize the editing core complex (40, 41). Furthermore, RET1 TUTase was depleted to eliminate gRNAs, hence, edited mRNAs, while preserving unedited and preedited mRNAs (18, 42). Finally, edited and unedited mRNAs but not preedited mRNAs have been downregulated by knocking down the KPAP1 poly(A) polymerase (16).
FIG 4.
Effects of knockdowns of individual subunits on editing core complexes, GRBCs, and REMCs. Mitochondrial fractions from RNAi cell lines induced for 60 h were extracted with detergent, and soluble contents were separated for 5 h at 178,000 ×g in a 10% to 30% glycerol gradient. The RNA editing core complex was detected by self-adenylation of RNA ligases; GRBC and REMC modules were visualized with antibodies against GRBC1/2 and RGG2, respectively. Thyroglobulin (19S) and bacterial ribosomal subunits were separated in parallel experiments as apparent S-value markers.
Although neither knockdown destabilized the entire RESC assembly, RNAi of specific subunits triggered elimination of potential interacting partners, e.g., loss of RGG2 in REMC1 RNAi, which is consistent with Y2H mapping (23) and strong network interaction (Fig. 2). Likewise, downregulation of GRBC1 but not GRBC2 was observed in GRBC7 RNAi. Remarkably, repression of REMC4, which was predicted to interact with GRBC1/2, GRBC5, and GRBC6 by both Y2H and protein-protein interaction network analysis (Fig. 2), reduced the abundance of both GRBC and REMC modules in the 50S-to-70S range and caused their redistribution to a lighter region. Disassembly of the editing core complex in MP18/24 RNAi led to insignificant changes in GRBC and REMC patterns, as would be expected from the transient nature of RESC-RECC interaction. In contrast, GRBC1/2 RNAi, which eliminates mature gRNAs (20), caused the core complex to sediment in lighter fractions. Downregulation of gRNA abundance by means of the use of RET1 TUTase RNAi had similar effects on the core complex sedimentation, suggesting that gRNAs play a key role in RESC contacts with the RECC. To that end, elimination of edited mRNAs by KPAP1 knockdown (16) did not alter the sedimentation patterns of GRBC, REMC, or the core complex. Overall, our data suggest that, in contrast to the stable and interdependent organization of the editing core complex, RESC represents a more dynamic assembly held together by protein-protein interactions which are facilitated by RNA-mediated contacts. Importantly, gRNAs play a key role in RESC interaction with the editing core complex.
GRBC and REMC modules participate in distinct phases of mRNA editing.
The compositional complexity of RESC exceeds that of the editing core complex (Fig. 2), but none of the components, except RGG2, display discernible motifs that would suggest a function. We have previously established that GRBC1/2 knockdowns abolish gRNAs and, therefore, the editing process (18, 20). Interestingly, the RGG1 RNA binding protein, which according to our data interacts predominantly with the ribosome (Fig. 2), was shown to be essential for RNA editing (24). To reconcile the subunit distribution among complexes with their participation in the editing process, we next analyzed cell growth phenotypes, mRNAs, and ribosomal RNAs in respective RNAi knockdowns (Table 1; see also Fig. S4 in the supplemental material). Inducible repression of all genes but GRBC3 and ribosome-associated LSU-4710 triggered moderate to severe growth inhibition phenotypes, suggesting functions essential to cell viability. It must be noted that RNAi knockdowns in T. brucei are typically reflected by a moderate (20% to 80%) loss of targeted mRNA as measured by qRT-PCR but result in deeper (80% to 90%) protein ablation (Fig. 4; see also reference 44). To establish a possible connection between the observed phenotypes and the impeded editing process, the relative abundances of unedited, cis-edited (CO2), moderately edited (Cyb and Murf2), and panedited transcripts and ribosomal RNAs have been determined by quantitative RT-PCR (Table 1; see also Fig. S5 in the supplemental material). In the case of cis-edited CO2 mRNA, the guiding sequence is embedded in the 3′ untranslated region of the same molecule whereas almost all other known editing sites can be aligned with trans-acting gRNAs (45). Remarkably, knockdowns of all GRBC subunits except GRBC7 had minor effects on CO2 mRNA whereas REMC subunits 1, 3, and 4 were essential for cis editing (see Fig. S5). In contrast, with the exception of GRBC3, GRBC knockdowns induced virtually uniform downregulation of edited RNAs whereas the requirements for REMC components were transcript specific: in REMC2-depleted cells, the effects ranged from neutral to minor to virtual elimination of edited mRNAs, e.g., Murf2 versus RPS12 versus ND8. In contrast to a published report (24), RGG1 knockdown did not inhibit editing. In agreement with LSU-2340 association with the large ribosomal subunit, the corresponding knockdown reduced levels of 12S rRNA. To conclude, all GRBC components, except subunit 3, are universally essential for editing directed by trans-acting gRNAs whereas REMC subunits are required for editing of a single mRNA (ND7 mRNA in REMC5 RNAi), a few mRNAs (Murf 2, RPS12, CO3, ND3, ND7, and ND8 mRNAs in REMC4 RNAi), or all mRNAs (REMC1).
TABLE 1.
Effects of RNAi knockdowns on cell viability and mitochondrial RNAsa
| Subunit | MRB1(s) | Motif(s) | Gene ID(s) | Cell growth | Guide RNA change | Edited mRNA change |
Preedited mRNA change |
Never-edited mRNA change |
rRNA change |
||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| General | Specific | General | Specific | General | Specific | 9S | 12S | ||||||
| Guide RNA binding complex (GRBC) | |||||||||||||
| TbGRBC1/2 | GAP1/GAP2 | None | Tb927.7.2570/Tb927.2.3800 | +++ | D | D | I | None | D | None | |||
| TbGRBC3 | MRB8620 | None | Tb11.01.8620 | None | None | None | None | I | None | None | None | ||
| TbGRBC4 | MRB5390 | None | Tb11.02.5390 | +++ | I | D | None | None | None | None | None | ||
| TbGRBC5 | MRB11870 | None | Tb927.10.11870 | +++ | I | D | I | None | None | None | |||
| TbGRBC6 | MRB3010 | None | Tb927.5.3010 | +++ | I | D | I | None | None | None | |||
| TbGRBC7 | None | Tb11.01.0880 | +++ | I | D | I | None | None | None | ||||
| RNA editing mediator complex (REMC) | |||||||||||||
| TbREMC1 | MRB10130 | None | Tb927.10.10130 | +++ | I | D | None | None | D | D | None | ||
| TbREMC2 | MRB1860 | None | Tb927.2.1860 | +++ | None | D | None | None | D | None | None | ||
| TbREMC3 | MRB800 | None | Tb927.7.800 | ++ | None | D | None | None | D | D | D | ||
| TbREMC4 | MRB8180 | None | Tb927.8.8180 | + | None | D | None | None | None | None | None | None | |
| TbREMC5/5A | MRB4160 | None | Tb927.4.4160 | +++ | I | D | None | None | I | None | None | ||
| MRB8170 | None | Tb927.8.8170 | +++ | I | D | None | None | I | None | None | |||
| TbRGG2 | TbRGG2 | RGG/RRM | Tb927.10.10830 | +++ | None | None | D/I | None | I/None | None | I/None | None | None |
| Polyadenylation mediator complex (PAMC) | |||||||||||||
| TbPAMC1 | None | Tb927.1.1730 | +++ | I | |||||||||
| TbPAMC2 | None | Tb927.6.1200 | + | None | |||||||||
| TbPAMC3 | None | Tb927.10.1730 | ++ | None | |||||||||
| TbPAMC4 | None | Tb927.1.3010 | + | None | D | I | I | None | None | ||||
| RNA editing core complex (RECC) | |||||||||||||
| TbMP18/24 | OB fold | Tb927.10.5120/Tb427.10.5110 | +++ | I | D | I | D | None | None | ||||
| TbRET2 | TUTase | Tb427.07.1550 | +++ | I | D | I | D | None | None | ||||
| RGG1 RNA binding protein (RGG1) | |||||||||||||
| TbRGG1 | RGG | Tb927.6.2230 | +++ | None | D | None | None | None | D | ||||
| Large ribosomal subunit (LSU) | |||||||||||||
| TbLSU-600 | None | Tb927.10.600 | ++ | n/d | D | None | D | None | None | ||||
| TbLSU-2340 | None | Tb11.01.2340 | + | n/d | D | None | None | None | D | ||||
| TbLSU-4710 | NUDIX | Tb927.7.4710 | None | n/d | I | None | None | D | None | None | |||
Growth inhibition phenotypes were ranked none, weak (+), moderate (++), and severe (+++). ID, identifier; D, decrease by more than 20% over 96 h of RNAi induction; I, increase by more than 20% over 96 h of RNAi induction; n/d, not determined; OB fold, oligonucleotide/oligosaccharide binding fold; NUDIX, nucleoside diphosphates linked to some moiety X. Proteins detected in various accounts of the MRB1 complex are indicated (19, 21, 23, 27, 43).
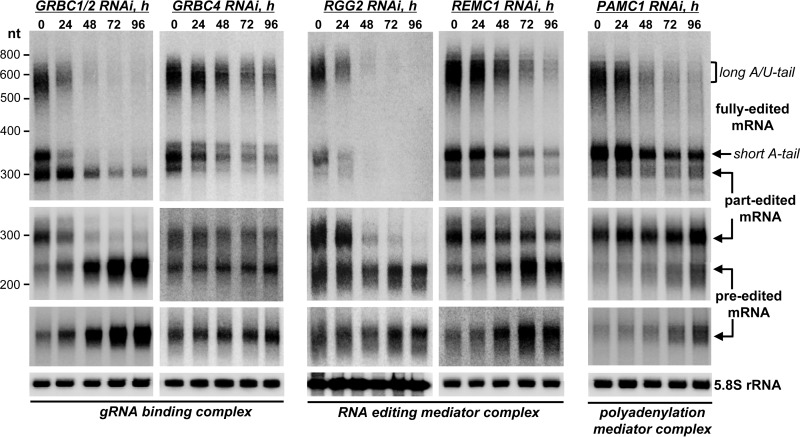
Inhibition of editing by a particular knockdown, as conventionally measured by qRT-PCR at a single RNAi induction time point, attests to a decreased output of a multicomponent system but provides limited information about specific steps that are disrupted. To integrate complex associations with the impact of representative knockdowns on specific editing steps, we next investigated RNAi-triggered changes in preedited, partially edited (∼70% completed), and fully edited mRNAs (Fig. 5). The ribosomal protein RPS12 mRNA was chosen as a model because it represents a single editing domain; i.e., multiple overlapping gRNAs direct editing in a strict 3′-to-5′ hierarchical order (45, 46). It has been also established that pre- and partially edited mRNAs possess short 3′ A-tails whereas fully edited molecules undergo the postediting addition of a long A/U-tail (16, 17). As would be expected from gRNA ablation in the GRBC1/2 RNAi background, inactivation of editing led to a loss of all edited forms and to an approximately 10-fold accumulation of preedited mRNA. In contrast, GRBC4 RNAi did not significantly alter the levels of pre-edited and partially edited forms but caused the gradual loss of both short- and long-tailed fully edited mRNAs. Although this protein engages in direct interactions with GRBC1/2, -5, and -6 subunits (Fig. 2) (23), the GRBC integrity was not compromised. On the other hand, GRBC's interaction with REMC was affected, as suggested by the loss of RGG2 in the 10S-to-20S region (Fig. 4). In agreement with RGG2's proposed role in the processivity of editing (22), its knockdown eliminated partially and fully edited mRNAs but did not affect the preedited mRNA. To further test whether a compromised REMC module would inhibit the processivity of editing, we analyzed REMC1 knockdown and found effects that virtually mirror those of RGG2's RNAi. Collectively, these findings suggest that the intact editing core complex and the gRNA binding module (GRBC) are sufficient for initiation but not for the processive editing of the entire domain. Apparently, the REMC module is chiefly responsible for GRBC-RECC interaction, thereby acting as an essential component in a process involving multiple overlapping gRNAs.
FIG 5.
Accumulation of editing intermediates in RNAi knockdowns correlates with placement of respective subunits within RESC assemblies. Data represent the results of Northern blotting of preedited, partially edited (46 editing events at the 5′ region were not completed and 107 events in the preceding region were completed), and fully edited RPS12 mRNAs. Total RNA was separated on a 5% polyacrylamide–8 M urea gel and hybridized with single-stranded DNA probes. Cytosolic rRNA (5.8S) was used as a loading control.
The PAMC module is required for postediting adenylation/uridylation.
Functional coupling between mRNA editing and polyadenylation is manifested by preediting addition of the short 3′ A-tail, which serves to stabilize mRNAs during editing (16), and the postediting adenylation/uridylation, which enables mRNA binding to the ribosome (17). The former processing event is accomplished by KPAP1 poly(A) polymerase, while the latter requires both KPAP1 and RET1 TUTase activities. Since knockdowns of the polyadenylation mediator complex components (Fig. 2) did not significantly affect the levels of edited mRNAs (see Fig. S5 in the supplemental material), we next tested whether the PAMC module is required for either pre- or postediting 3′-end modifications. The PAMC1 subunit was selected for in-depth analysis because KPAP1, RET1, and kinetoplast polyadenylation/uridylation factors 1 and 2 (KPAF1/2) were detected with high confidence in the affinity-purified PAMC1 complex (see Table S1). In agreement with the qRT-PCR data, PAMC1 RNAi did not significantly alter the abundance of short A-tailed preedited, partially edited, or fully edited forms of RPS12 mRNA (Fig. 5). These minor changes indicate that the preediting adenylation and editing processes were largely uncompromised. However, the postediting A/U-tailing was effectively abolished. Considering that only fully edited mRNAs undergo 3′ A/U-tailing (16) in a reaction that requires KPAP1 poly(A)polymerase, RET1 TUTase, and KPAF1/2 factors (17), these findings indicate that the PAMC module is essential for coordinating the completion of mRNA editing and the 3′ A/U-tailing, which in turn activates mitochondrial translation. Collectively, these experiments establish RESC as the trident platform in which specific modules are responsible for gRNA stabilization (GRBC), recruitment of the RNA editing core complex (REMC), and mRNA transition between editing and 3′ polyadenylation/uridylation (PAMC).
Guide RNAs are selectively degraded during the active editing process.
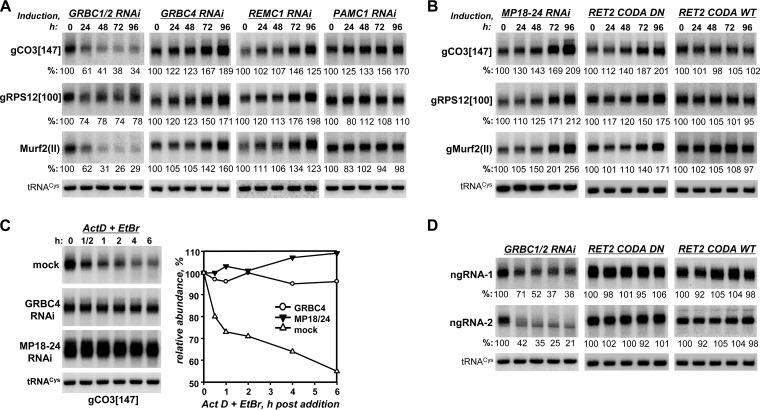
Because knockdowns of many GRBC and REMC subunits obstructed editing, we next inquired whether knockdown of any protein investigated in this work would affect gRNA levels. The relative abundances of representative maxicircle-encoded [gMurf2(II)] and minicircle-encoded (gCO3[147] and gRPS12[100]) gRNAs were analyzed by Northern blotting at 24-h intervals after RNAi induction (Fig. 6A and Table 1; see also Fig. S6 in the supplemental material). In agreement with a previous study of individual GRBC1 and GRBC2 proteins (20), simultaneous repression of the two proteins triggered a rapid decline in gRNA steady-state levels. However, depletion of most other subunits in GRBC and REMC modules led to gRNA accumulation whereas knockdowns of PAMC subunits left gRNAs unaffected. Such outcomes could be anticipated for PAMC since the module is chiefly responsible for interaction with the polyadenylation complex, but the gRNA upregulation concomitant with inhibition of editing was puzzling.
FIG 6.
The GRBC1/2 tetramer is responsible for gRNA and gRNA-like small RNA stabilization. (A) Steady-state gRNA levels in GRBC, REMC, and PAMC subunit knockdowns. Total RNA was isolated at the indicated time points after RNAi induction and analyzed by Northern blotting. Relative levels of gRNA abundance were calculated in reference to the nucleus-encoded but mitochondrion-localized tRNACys(GCA). (B) Steady-state gRNA levels in cells with perturbed core editing complex (MP18-24 RNAi) and eliminated (RET2 CODA DN) or restored (RET2 CODA WT) U-insertion editing activity. RNA was analyzed as described for panel A. (C) Guide RNA decay in T. brucei cells depleted of GRBC4 and TbMP18/TbMP24. After 48 h of RNAi induction, actinomycin D (Act D) and ethidium bromide (EtBr) were added to inhibit transcription. Total RNA was isolated from cells collected at indicated time points, separated on 10% polyacrylamide–8 M urea gel, transferred onto a membrane, and sequentially probed for gRNAs and tRNACys(GCA). (D) Steady-state levels of abundant uridylated small RNAs that did not fulfill the gRNA prediction criteria used in this study or by Koslowsky et al. (45) were tested in GRBC1/2 knockdown cells and RET2 knock-in cell lines with inhibited editing (RET2 CODA DN) or restored editing (RET2 CODA WT). ngRNA1 sequence, 5′-AGTTTATGTCTAATTTCACTGATCTACCTAGTATAAAATATCATACACGTATTGTATATTTTTTTTTTTTTTT-3′; ngRNA2 sequence, 5′-ATACTAGGTAGATCAGTGAAATTAGACATAAACTGTATTATAATAAGATATATATTTTTTTTTTTTTT-3′.
The inverse correlation between edited mRNA and gRNA levels in multiple GRBC and REMC knockdowns (Fig. 5 and 6A and Table 1) led us to hypothesize that gRNA accumulation may be an outcome of the inhibited editing process. This would suggest that the editing-dependent gRNA degradation takes precedence over gRNA recycling under normal conditions. To test this hypothesis, we performed simultaneous knockdown of editing core complex structural subunits TbMP18 and TbMP24 and knock-ins with catalytically inactive and active variants of RET2 TUTase (47, 48). In these genetic backgrounds, the RNA editing core complex was virtually eliminated (Fig. 4) and an essential editing enzyme was inactivated by a point mutation, respectively. As a result, gRNA abundance increased in TbMP18/TbMP24 knockdown or knock-in with the inactive RET2 but remained constant in cells with active RET2 knock-in (Fig. 6B). The editing-dependent gRNA degradation was confirmed by in vivo decay assays in cells lacking TbMP18/24 and GRBC4. Following transcription blockade performed with actinomycin D and ethidium bromide, gRNA levels rapidly declined in mock-treated cells but remained virtually unaffected in induced RNAi cell lines for the duration of the decay assay (Fig. 6C).
These surprising observations raised the issue of whether the phenomenon of editing-dependent degradation is specific for gRNAs. Indeed, earlier reports indicated the presence of gRNA-like molecules that could not be readily aligned with edited mRNAs (49, 50) and small RNAs of polarity opposite that of gRNAs (18). To address the specificity of editing-dependent guide RNA degradation, we next isolated 35-to-75-nt-long RNAs from highly enriched mitochondrial fractions and, after filtering out nucleus-encoded contaminations, obtained ∼15 × 10 (6) raw read counts by strand-specific RNA-Seq (Table 2). To define gRNA species, assembled transcripts (∼6.8 × 106) were mapped to edited mRNAs, allowing G-U base pairs but no more than one gap and three mismatches. The edited mRNA coverage was compared to that obtained from the same data set by scripts utilized by Koslowsky et al. (45); the latter approach also allowed internal mismatches but provided more flexibility at gRNA's 3′ and 5′ extremities. Both algorithms generated virtually complete coverage of all known editing sites; the inclusive method of Koslowsky et al. allowed more sequences to be assigned as gRNAs, while the restrictive method used in this study identified fewer RNAs that map to nonedited regions, such as cis-edited CO2 mRNA (see Fig. S7 in the supplemental material). Candidate gRNAs and non-guide RNAs (ngRNA) were then clustered based on the length of the oligo(U) tail. Using our mapping parameters, the numbers of ngRNA sequence classes and read counts significantly exceeded those of gRNAs (Table 2). Remarkably, 96% of predicted gRNAs possessed U-tails of six nucleotides or longer whereas substantial fractions of ngRNA had either no uridylate residues (∼25%) or short U-tails (26%) at their 3′ ends. Guide RNAs bearing long U-tails accounted for more than 95% of the known editing sites, although the extent of coverage per nucleotide of mRNA, and hence the relative abundances of the corresponding guide RNAs, varied dramatically (see Fig. S7 in the supplemental material).
TABLE 2.
Oligo(U) tail length distribution in non-guide RNAs and candidate guide RNAs
| Parameter | Non-guide RNA value for indicated subtype category |
Candidate guide RNA value for indicated subtype category |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No U-tail |
1–5 U's |
6+ U's |
No U-tail |
1–5 U's |
6+ U's |
|||||||
| − | + | − | + | − | + | − | + | − | + | − | + | |
| Sequence count | 6,559 | 6,946 | 6,945 | 7,398 | 12,416 | 12,691 | 65 | 64 | 89 | 90 | 3,373 | 3,449 |
| Sequence % | 25.3 | 25.7 | 26.8 | 27.4 | 47.9 | 46.9 | 1.8 | 1.8 | 2.5 | 2.5 | 95.7 | 95.7 |
| Read count | 1,239,452 | 1,600,083 | 764,293 | 827,715 | 10,676,762 | 10,899,794 | 2,726 | 3,260 | 3,285 | 5,563 | 281,167 | 364,566 |
| Read % | 9.8 | 12.0 | 6.0 | 6.2 | 84.2 | 81.8 | 0.9 | 0.9 | 1.1 | 1.5 | 97.9 | 97.6 |
Small mitochondrial RNAs were mock treated (−) or incubated with 5′-to-3′ Terminator exonuclease (+) prior to construction of stranded RNA-Seq libraries.
To determine the changes in ngRNA levels under conditions that either destabilize gRNAs (loss of GRBC1/2) or stabilize gRNAs (inhibition of editing), we selected two abundant 3′ uridylated species for Northern blotting (ngRNA-1 and ngRNA-2) (Fig. 6D). The effective downregulation of ngRNAs upon loss of GRBC1/2 indicated that small RNAs that do not fulfill the definition of gRNAs are nonetheless stabilized via binding to GRBC. In contrast, steady levels of these molecules in genetic backgrounds with inhibited (Fig. 6D, RET2 CODA DN) or restored (Fig. 6D, RET2 CODA WT) editing demonstrated the specificity of editing-dependent gRNA degradation. Thus, an active editing process accelerates gRNA decay whereas GRBC1 and GRBC2 represent protein factors essential for the stability of small mitochondrial RNAs irrespective of their participation in editing.
RESC functions as the main RNA binding platform within the editosome.
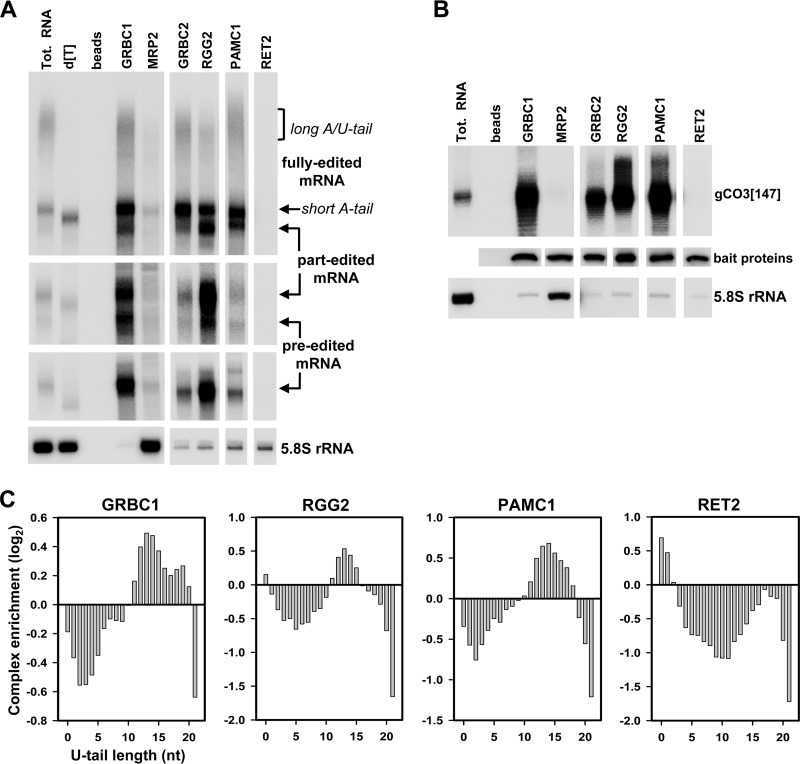
RNA binding properties have been ascribed to subunits of the editing core and GRBC complexes, and MRP1/2 RNA chaperones some other proteins (reviewed in reference 6). The RNA's stabilizing role in interactions among editing complexes has also been established (Fig. 1, 3, and 4). Although we have identified gRNAs as essential for RESC-core complex contacts, the nature of the main RNA binding platform in the editing apparatus remained unclear. To address this issue, we applied ultrarapid affinity purification (17) to isolate RESC modules and the editing core complex; the MRP1/2 complex, which nonspecifically binds single- and double-stranded RNAs with high affinity (14, 51), was used as a control. Northern blotting of bound RNAs revealed a consistent association of the preedited and edited mRNAs with all three RESC modules. In addition, partially edited mRNA was enriched in RGG2-containing REMC (Fig. 7A; see also Fig. S8 in the supplemental material). Surprisingly, within the dynamic range afforded by hybridization techniques, we were unable to detect either editing substrates or products bound to the editing core complex (Fig. 7A, RET2). Distribution of the four mRNA forms distinguished by our assay (preedited, partially edited, fully edited with a short tail, and fully edited with a long tail) between complexes that transiently interact with RESC (Fig. 2) was also instructive: only trace mRNA amounts were detected in MRP1/2 (Fig. 7A) and in polyadenylation factor KPAF1 (see Fig. S8 in the supplemental material).
FIG 7.
RNA editing substrates, intermediates, and products are held by the RESC. (A) Distribution of preedited, partially edited, and fully edited mRNAs among RESC modules and core editing complexes. Following rapid affinity pulldown, RNA was extracted from magnetic beads, separated on a 5% polyacrylamide–8 M urea gel, and probed for the respective mRNA species. [dT], total (Tot.) RNA was treated with RNase H in the presence of 18-mer (d[T]) to remove poly(A) tails; beads, IgG-coated magnetic beads were incubated with extract from the parental cell line. (B) Distribution of guide RNAs among complexes. The same RNA samples as described for panel A were probed for a specific gRNA. RNA amounts were normalized to tandem affinity purification (TAP)-tagged bait proteins detected by quantitative Western blotting with antibodies against calmodulin binding peptide. (C) The profile of the distribution of U-tail lengths in complex-bound transcripts. Bars above and below zero indicate relative enrichment and absence, respectively, of small RNAs with corresponding U-tails. A similarly sized fraction of total mitochondrial RNA was used as the reference.
Analysis of gCO3[147] guide RNA distribution among the same complexes exposed its preferential binding to RESC but not to the editing core complex (Fig. 7B; see also Fig. S8 in the supplemental material). Although these data further implicate RESC as the main binding platform for editing substrates, both gRNAs and ngRNAs require the same subunits (GRBC1 and GRBC2) for stabilization (Fig. 6A and D). These considerations warranted further investigation into features that may distinguish gRNAs from the more abundant and diverse ngRNAs (Table 2) in terms of selective association with RESC. We next isolated 35-to-75-nt-long molecules from affinity-purified complexes and performed strand-specific RNA-Seq. Because gRNAs possess 5′ triphosphates, all samples were mock treated or were incubated with the 5′-3′ exonuclease (Terminator) that digests RNA with 5′ monophosphate to remove potential cytosolic RNA contamination. To that end, a high degree of similarity between treated and untreated data sets in total mitochondrial and complex-bound RNAs suggests that most mitochondrial RNAs in this size range are protected against degradation (Table 2; see also Fig. S9 in the supplemental material). To assess gRNA enrichment in affinity-purified complexes, we summed the read counts that mapped to all edited transcripts for each sample and performed a Fisher exact test to estimate the percentage of significantly enriched RNAs versus the percentage of the same-sized fraction in mitochondrial RNA (see Table S2). These studies demonstrated that gRNAs are significantly enriched in all RESC modules (GRBC, REMC, and PAMC) but not in the editing core complex. Finally, the profile of the distribution of the lengths of U-tails in complex-enriched RNAs revealed a strong correlation between the presence of 10 to 18 uridines at the 3′ end and an association of specific RNAs with all three RESC modules (Fig. 7C). Unexpectedly, predominantly nonuridylated and monouridylated RNAs were found in the editing core complex. Considering that RNAs with 0 to 5 U's constitute ∼50% of the ngRNA population but are nearly absent among gRNAs (Table 2) and the fact that no significant gRNA enrichment was observed in the core complex (see Table S2), we concluded that the presence of the 10-to-18-nucleotide-long oligo(U) tail represents a defining feature of functional gRNAs, which enables their recruitment by the RESC.
DISCUSSION
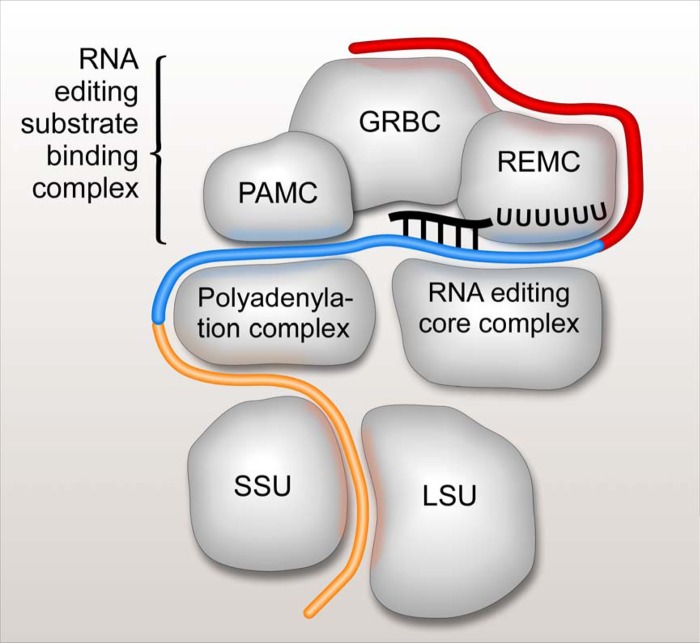
The concept of the U-insertion/deletion RNA editing holoenzyme, or the editosome, has evolved from purification of the seven-polypeptide complex (52) to the definition of a stable complex of ∼15 proteins (2, 3) (the ∼20S editosome, or the RNA editing core complex [RECC]) to the understanding that RECC functions as part of a still larger assembly (reviewed in references 6, 53, 54, and 55). The discoveries of functional coupling between mRNA editing and 3′ polyadenylation and translation added even more complexity to the process of generating translationally competent mitochondrial mRNAs (16, 17, 56). In this study, we applied a unified strategy involving biochemical fractionations, quantitative proteomics, RNAi knockdowns, and analysis of editing substrates, intermediates, and products to define the global composition of the U-insertion/deletion editosome and the functions of its main components. We show that the editosome consists of two specialized elements: a modular RNA editing substrate binding complex (RESC; ∼23 polypeptides) that is responsible for binding of RNA editing substrates (preedited mRNA and gRNA), intermediates (partially edited mRNAs), and products (fully edited mRNAs) and the catalytic RNA editing core complex (RECC). In light of the observed RNA distribution between RESC and RECC, we propose a general model in which gRNAs and preedited mRNAs, intermediates, and products remain bound to RESC during the editing process whereas the enzymatic core complex interacts transiently with this assembly to target individual editing sites (Fig. 8).
FIG 8.
Schematic representation of mRNA interactions with RNA editing substrate binding, RNA editing core and polyadenylation complexes, and the ribosome. The RNA editing substrate binding complex (RESC) consists of three modules: a guide RNA binding complex (GRBC), an RNA editing mediator complex (REMC), and a polyadenylation mediator complex (PAMC). These modules are responsible for gRNA stabilization, recruitment of RNA editing core complexes, and recruitment of polyadenylation complexes, respectively. Preedited mRNA is depicted in red, edited mRNA is shown in blue, and edited and polyadenylated/uridylated (translation-competent) mRNA is colored in orange.
Although gRNAs are chiefly responsible for RESC-RECC interactions, the RNase-resistant contacts and outcomes of genetic knockdowns pointed to the existence of a specialized module within RESC that accommodates contacts with the core editing complex. All subunits of this module, termed the RNA editing mediator complex (REMC), were essential for normal cell growth but exerted transcript-specific effects. For example, REMC1 was critical for all trans- and cis-editing events whereas REMC2 and RGG2 were dispensable for editing of moderately edited mRNAs (Cyb and Murf2). In contrast, REMC2 was required for editing of a single panedited mRNA (ND7) while repression of RGG2 negatively affected all panedited mRNAs (see Fig. S5 in the supplemental material). Accumulation of editing intermediates in REMC knockdowns (Fig. 5) and analysis of bound RNAs (Fig. 7) further implicated this module in both initiation and progression of editing. Cross-tagging experiments provided compelling evidence that, within the RESC superassembly, REMC is engaged in extensive contacts with another module, the guide RNA binding complex (GRBC). Initially defined by the presence of a GRBC1/2 heterotetramer (18, 20), the GRBC module consists of at least 10 polypeptides lacking detectable motifs. From the functional standpoint, GRBC1 and GRBC2 remain the only mitochondrial proteins essential for gRNA stability; knockdowns of GRBC subunits 4 to 7 triggered gRNA upregulation concomitant with inhibition of RNA editing.
The network of protein-protein and RNA-stabilized interactions places GRBC at the interface between REMC and the third distinct RESC module, the polyadenylation mediator complex (PAMC). The definition of PAMC and its contacts with KPAP1 poly(A) polymerase, RET1 TUTase, and KPAF1/2 polyadenylation/uridylation factors provide a physical basis for the functional connection between polyadenylation and editing processes. The KPAP1-catalyzed preediting polyadenylation was not impacted by PAMC1 RNAi, while the postediting addition of a long A/U-tail, which enables mRNA binding to the small ribosomal subunit and also requires KPAP1, RET1 TUTase, and KPAF1/2 factors, was severely compromised (Fig. 5). It is noteworthy that long-tailed and short-tailed mRNAs are present in similar proportions in total RNA whereas long-tailed mRNAs are highly (∼90%) enriched in the ribosome-bound population (17). In contrast, approximately 90% of fully edited mRNAs bound to all RESC modules possessed short A-tails (Fig. 7A; see also Fig. S8 in the supplemental material). Thus, it appears that PAMC channels fully edited mRNAs into the postedited adenylation/uridylation pathway by interacting with the polyadenylation complex (Fig. 8).
Complementarity to edited mRNA remains the formal criterion for designating a small mitochondrial RNA a “guide RNA” (57). We show in this work that, irrespective of the computational parameters for gRNA prediction, a substantial fraction of small mitochondrial RNAs cannot be considered gRNAs, and yet the GRBC1/2 proteins are critical for stabilizing both gRNAs and ngRNAs. This finding indicates that the GRBC1/2 tetramer represents a small RNA binding interface of the RESC. Although the functions of ngRNAs remain to be established, we noticed that many such sequences form double-stranded hybrids with gRNAs. Furthermore, we posit that editing-dependent selective degradation may serve as the criterion for designating a specific RNA a gRNA. In any event, the editing-dependent gRNA degradation provides initial clues for resolution of a long-standing conundrum concerning how fully edited mRNAs are cleared from bound gRNAs prior to translation.
The abundance and sequence complexity of ngRNAs exceed those of putative gRNAs, thus raising the issue of the selectivity of gRNA recruitment by RESC (Fig. 7C and Table 2; see also Table S2 in the supplemental material). Our results demonstrate that both gRNAs and ngRNAs maintain 5′ triphosphates at the sequence level (see Fig. S9 in the supplemental material) but their 3′ uridylation states differ dramatically: nearly 50% of non-gRNAs had U-tails ranging from zero to five U's in length, while virtually all gRNAs possess oligo(U) tails more than six nucleotides in length. In terms of distribution among editing complexes, gRNAs with 10 to 18 U's were significantly enriched in affinity-purified RESC modules but not in the editing core complex. This finding underscores the presence of an elongated U-tail as the main recognition element that enables gRNA recruitment by RESC. Relative enrichment of non- or monouridylated gRNAs in RECC (Fig. 7), on the other hand, may reflect the presence of 3′-5′ U-specific exonucleases REX1 and REX2 in the core complex or may indicate that deuridylation precedes editing-dependent gRNA degradation (Fig. 6). The essential issues for further investigation include the mechanism of small mitochondrial RNA processing that favors uridylation of guide RNAs and the identity of the GRBC subunit(s) responsible for binding of uridylated RNAs. In this work, we show that GRBC1 and GRBC2 remain the only proteins within the GRBC complex that are required for gRNA stability. In contrast, a previous study demonstrated that the recombinant GRBC1/2 complex binds uridylated and nonuridylated short RNAs equally well (18), which points to another protein within GRBC as responsible for enhancing the binding of uridylated RNAs. The surprising finding of GRBC1/2 being essential for the maintenance of both guide RNAs and non-guide RNA species has far-reaching ramifications, as it suggests that there is a common mechanism for all small mitochondrial RNA processing and stabilization. In sum, our studies defined the U-insertion/deletion editosome as the ∼40S particle composed of the catalytic core complex (RECC) and the trimodular RNA substrate binding complex (RESC).
Supplementary Material
ACKNOWLEDGMENTS
We thank Dona Koslowsky and Yanni Sun for sharing a gRNA prediction script, Laurie Read for the kind gift of RGG2 antibodies, and members of our laboratories for discussions. We also thank the UC Irvine Genomics High-Throughput Facility staff for technical assistance.
This work was supported by NIH grants R01AI091914 and R01AI101057 to R.A. and R01GM074830 and R21CA161807 to L.H., a Biomedical Informatics Training predoctoral fellowship to R.M.K. (5T15LM007443-10; principal investigator, P. Baldi), and an MGHPCC Seed Fund program grant to S.M.
Footnotes
Published ahead of print 15 September 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MCB.01075-14.
REFERENCES
- 1.Benne R, Van den Burg J, Brakenhoff J, Sloof P, Van Boom J, Tromp M. 1986. Major transcript of the frameshifted coxII gene from trypanosome mitochondria contains four nucleotides that are not encoded in the DNA. Cell 46:819–826. 10.1016/0092-8674(86)90063-2. [DOI] [PubMed] [Google Scholar]
- 2.Aphasizhev R, Aphasizheva I, Nelson RE, Gao G, Simpson AM, Kang X, Falick AM, Sbicego S, Simpson L. 2003. Isolation of a U-insertion/deletion editing complex from Leishmania tarentolae mitochondria. EMBO J. 22:913–924. 10.1093/emboj/cdg083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Panigrahi AK, Schnaufer A, Ernst NL, Wang B, Carmean N, Salavati R, Stuart K. 2003. Identification of novel components of Trypanosoma brucei editosomes. RNA 9:484–492. 10.1261/rna.2194603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blum B, Bakalara N, Simpson L. 1990. A model for RNA editing in kinetoplastid mitochondria: “guide” RNA molecules transcribed from maxicircle DNA provide the edited information. Cell 60:189–198. 10.1016/0092-8674(90)90735-W. [DOI] [PubMed] [Google Scholar]
- 5.Seiwert SD, Heidmann S, Stuart K. 1996. Direct visualization of uridylate deletion in vitro suggests a mechanism for kinetoplastid RNA editing. Cell 84:831–841. 10.1016/S0092-8674(00)81062-4. [DOI] [PubMed] [Google Scholar]
- 6.Aphasizhev R, Aphasizheva I. 2011. Uridine insertion/deletion mRNA editing in trypanosomes: a playground for RNA-guided information transfer. Wiley Interdiscip. Rev. RNA 2:669–685. 10.1002/wrna.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carnes J, Trotter JR, Peltan A, Fleck M, Stuart K. 2008. RNA editing in Trypanosoma brucei requires three different editosomes. Mol. Cell. Biol. 28:122–130. 10.1128/MCB.01374-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Panigrahi AK, Ernst NL, Domingo GJ, Fleck M, Salavati R, Stuart KD. 2006. Compositionally and functionally distinct editosomes in Trypanosoma brucei. RNA 12:1038–1049. 10.1261/rna.45506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Golas MM, Bohm C, Sander B, Effenberger K, Brecht M, Stark H, Goringer HU. 2009. Snapshots of the RNA editing machine in trypanosomes captured at different assembly stages in vivo. EMBO J. 28:766–778. 10.1038/emboj.2009.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li F, Ge P, Hui WH, Atanasov I, Rogers K, Guo Q, Osato D, Falick AM, Zhou ZH, Simpson L. 2009. Structure of the core editing complex (L-complex) involved in uridine insertion/deletion RNA editing in trypanosomatid mitochondria. Proc. Natl. Acad. Sci. U. S. A. 106:12306–12310. 10.1073/pnas.0901754106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Corell RA, Read LK, Riley GR, Nellissery JK, Allen TE, Kable ML, Wachal MD, Seiwert SD, Myler PJ, Stuart KD. 1996. Complexes from Trypanosoma brucei that exhibit deletion editing and other editing-associated properties. Mol. Cell. Biol. 16:1410–1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Osato D, Rogers K, Guo Q, Li F, Richmond G, Klug F, Simpson L. 2009. Uridine insertion/deletion RNA editing in trypanosomatid mitochondria: in search of the editosome. RNA 15:1338–1344. 10.1261/rna.1642809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pollard VW, Harris ME, Hajduk SL. 1992. Native mRNA editing complexes from Trypanosoma brucei mitochondria. EMBO J. 11:4429–4438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aphasizhev R, Aphasizheva I, Nelson RE, Simpson L. 2003. A 100-kD complex of two RNA-binding proteins from mitochondria of Leishmania tarentolae catalyzes RNA annealing and interacts with several RNA editing components. RNA 9:62–76. 10.1261/rna.2134303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aphasizhev R, Sbicego S, Peris M, Jang SH, Aphasizheva I, Simpson AM, Rivlin A, Simpson L. 2002. Trypanosome mitochondrial 3′ terminal uridylyl transferase (TUTase): the key enzyme in U-insertion/deletion RNA editing. Cell 108:637–648. 10.1016/S0092-8674(02)00647-5. [DOI] [PubMed] [Google Scholar]
- 16.Etheridge RD, Aphasizheva I, Gershon PD, Aphasizhev R. 2008. 3′ adenylation determines mRNA abundance and monitors completion of RNA editing in T. brucei mitochondria. EMBO J. 27:1596–1608. 10.1038/emboj.2008.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aphasizheva I, Maslov D, Wang X, Huang L, Aphasizhev R. 2011. pentatricopeptide repeat proteins stimulate mRNA adenylation/uridylation to activate mitochondrial translation in trypanosomes. Mol. Cell 42:106–117. 10.1016/j.molcel.2011.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aphasizheva I, Aphasizhev R. 2010. RET1-catalyzed uridylylation shapes the mitochondrial transcriptome in Trypanosoma brucei. Mol. Cell. Biol. 30:1555–1567. 10.1128/MCB.01281-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hashimi H, Cicova Z, Novotna L, Wen YZ, Lukes J. 2009. Kinetoplastid guide RNA biogenesis is dependent on subunits of the mitochondrial RNA binding complex 1 and mitochondrial RNA polymerase. RNA 15:588–599. 10.1261/rna.1411809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Weng J, Aphasizheva I, Etheridge RD, Huang L, Wang X, Falick AM, Aphasizhev R. 2008. Guide RNA-binding complex from mitochondria of trypanosomatids. Mol. Cell 32:198–209. 10.1016/j.molcel.2008.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Panigrahi AK, Zikova A, Dalley RA, Acestor N, Ogata Y, Anupama A, Myler PJ, Stuart KD. 2008. Mitochondrial complexes in trypanosoma brucei: a novel complex and a unique oxidoreductase complex. Mol. Cell Proteomics 7:534–545. 10.1074/mcp.M700430-MCP200. [DOI] [PubMed] [Google Scholar]
- 22.Ammerman ML, Presnyak V, Fisk JC, Foda BM, Read LK. 2010. TbRGG2 facilitates kinetoplastid RNA editing initiation and progression past intrinsic pause sites. RNA 16:2239–2251. 10.1261/rna.2285510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ammerman ML, Downey KM, Hashimi H, Fisk JC, Tomasello DL, Faktorova D, Kafkova L, King T, Lukes J, Read LK. 2012. Architecture of the trypanosome RNA editing accessory complex, MRB1. Nucleic Acids Res. 40:5637–5650. 10.1093/nar/gks211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hashimi H, Zikova A, Panigrahi AK, Stuart KD, Lukes J. 2008. TbRGG1, an essential protein involved in kinetoplastid RNA metabolism that is associated with a novel multiprotein complex. RNA 14:970–980. 10.1261/rna.888808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Acestor N, Panigrahi AK, Carnes J, Zikova A, Stuart KD. 2009. The MRB1 complex functions in kinetoplastid RNA processing. RNA 15:277–286. 10.1261/rna.1353209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fisk JC, Ammerman ML, Presnyak V, Read LK. 2008. TbRGG2, an essential RNA editing accessory factor in two Trypanosoma brucei life cycle stages. J. Biol. Chem. 283:23016–23025. 10.1074/jbc.M801021200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kafková L, Ammerman ML, Faktorová D, Fisk JC, Zimmer SL, Sobotka R, Read LK, Lukes J, Hashimi H. 2012. Functional characterization of two paralogs that are novel RNA binding proteins influencing mitochondrial transcripts of Trypanosoma brucei. RNA 18:1846–1861. 10.1261/rna.033852.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wickstead B, Ersfeld K, Gull K. 2002. Targeting of a tetracycline-inducible expression system to the transcriptionally silent minichromosomes of Trypanosoma brucei. Mol. Biochem. Parasitol. 125:211–216. 10.1016/S0166-6851(02)00238-4. [DOI] [PubMed] [Google Scholar]
- 29.Wirtz E, Leal S, Ochatt C, Cross GA. 1999. A tightly regulated inducible expression system for conditional gene knock-outs and dominant-negative genetics in Trypanosoma brucei. Mol. Biochem. Parasitol. 99:89–101. 10.1016/S0166-6851(99)00002-X. [DOI] [PubMed] [Google Scholar]
- 30.Jensen BC, Kifer CT, Brekken DL, Randall AC, Wang Q, Drees BL, Parsons M. 2007. Characterization of protein kinase CK2 from Trypanosoma brucei. Mol. Biochem. Parasitol. 151:28–40. 10.1016/j.molbiopara.2006.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760. 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fang L, Kaake RM, Patel VR, Yang Y, Baldi P, Huang L. 2012. Mapping the protein interaction network of the human COP9 signalosome complex using a label-free QTAX strategy. Mol. Cell Proteomics 11:138–147. 10.1074/mcp.M111.016352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kaake RM, Milenkovic T, Przulj N, Kaiser P, Huang L. 2010. Characterization of cell cycle specific protein interaction networks of the yeast 26S proteasome complex by the QTAX strategy. J. Proteome. Res. 9:2016–2029. 10.1021/pr1000175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Etheridge RD, Clemens DM, Aphasizhev R. 2009. Identification and characterization of nuclear non-canonical poly(A) polymerases from Trypanosoma brucei. Mol. Biochem. Parasitol. 164:66–73. 10.1016/j.molbiopara.2008.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Köller J, Müller UF, Schmid B, Missel A, Kruft V, Stuart K, Göringer HU. 1997. Trypanosoma brucei gBP21. An arginine-rich mitochondrial protein that binds to guide RNA with high affinity. J. Biol. Chem. 272:3749–3757. [DOI] [PubMed] [Google Scholar]
- 36.Vanhamme L, Perez-Morga D, Marchal C, Speijer D, Lambert L, Geuskens M, Alexandre S, Ismaïli N, Göringer U, Benne R, Pays E. 1998. Trypanosoma brucei TBRGG1, a mitochondrial oligo(U)-binding protein that co-localizes with an in vitro RNA editing activity. J. Biol. Chem. 273:21825–21833. 10.1074/jbc.273.34.21825. [DOI] [PubMed] [Google Scholar]
- 37.Zhang Y, Wen Z, Washburn MP, Florens L. 2010. Refinements to label free proteome quantitation: how to deal with peptides shared by multiple proteins. Anal. Chem. 82:2272–2281. 10.1021/ac9023999. [DOI] [PubMed] [Google Scholar]
- 38.Allen TE, Heidmann S, Reed R, Myler PJ, Goringer HU, Stuart KD. 1998. Association of guide RNA binding protein gBP21 with active RNA editing complexes in Trypanosoma brucei. Mol. Cell. Biol. 18:6014–6022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schnaufer A, Ernst NL, Palazzo SS, O'Rear J, Salavati R, Stuart K. 2003. Separate insertion and deletion subcomplexes of the Trypanosoma brucei RNA editing complex. Mol. Cell 12:307–319. 10.1016/S1097-2765(03)00286-7. [DOI] [PubMed] [Google Scholar]
- 40.Salavati R, Ernst NL, O'Rear J, Gilliam T, Tarun S, Jr, Stuart K. 2006. KREPA4, an RNA binding protein essential for editosome integrity and survival of Trypanosoma brucei. RNA 12:819–831. 10.1261/rna.2244106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tarun SZ, Jr, Schnaufer A, Ernst NL, Proff R, Deng J, Hol W, Stuart K. 2008. KREPA6 is an RNA-binding protein essential for editosome integrity and survival of Trypanosoma brucei. RNA 14:347–358. 10.1261/rna.763308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Aphasizhev R, Aphasizheva I, Simpson L. 2003. A tale of two TUTases. Proc. Natl. Acad. Sci. U. S. A. 100:10617–10622. 10.1073/pnas.1833120100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ammerman ML, Tomasello DL, Faktorova D, Kafkova L, Hashimi H, Lukes J, Read LK. 2013. A core MRB1 complex component is indispensable for RNA editing in insect and human infective stages of Trypanosoma brucei. PLoS One 8:e78015. 10.1371/journal.pone.0078015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pusnik M, Schneider A. 2012. A trypanosomal pentatricopeptide repeat protein stabilizes the mitochondrial mRNAs of cytochrome oxidase subunits 1 and 2. Eukaryot. Cell 11:79–87. 10.1128/EC.05213-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Koslowsky D, Sun Y, Hindenach J, Theisen T, Lucas J. 2014. The insect-phase gRNA transcriptome in Trypanosoma brucei. Nucleic Acids Res. 42:1873–1886. 10.1093/nar/gkt973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Maslov DA, Simpson L. 1992. The polarity of editing within a multiple gRNA-mediated domain is due to formation of anchors for upstream gRNAs by downstream editing. Cell 70:459–467. 10.1016/0092-8674(92)90170-H. [DOI] [PubMed] [Google Scholar]
- 47.Ringpis GE, Aphasizheva I, Wang X, Huang L, Lathrop RH, Hatfield GW, Aphasizhev R. 2010. Mechanism of U insertion RNA editing in trypanosome mitochondria: the bimodal TUTase activity of the core complex. J. Mol. Biol. 399:680–695. 10.1016/j.jmb.2010.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ringpis GE, Stagno J, Aphasizhev R. 2010. Mechanism of U-insertion RNA editing in trypanosome mitochondria: characterization of RET2 functional domains by mutational analysis. J. Mol. Biol. 399:696–706. 10.1016/j.jmb.2010.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Madej MJ, Alfonzo JD, Huttenhofer A. 2007. Small ncRNA transcriptome analysis from kinetoplast mitochondria of Leishmania tarentolae. Nucleic Acids Res. 35:1544–1554. 10.1093/nar/gkm004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Madej MJ, Niemann M, Huttenhofer A, Goringer HU. 2008. Identification of novel guide RNAs from the mitochondria of Trypanosoma brucei. RNA Biol. 5:84–91. 10.4161/rna.5.2.6043. [DOI] [PubMed] [Google Scholar]
- 51.Schumacher MA, Karamooz E, Zikova A, Trantirek L, Lukes J. 2006. Crystal structures of T. brucei MRP1/MRP2 guide-RNA binding complex reveal RNA matchmaking mechanism. Cell 126:701–711. 10.1016/j.cell.2006.06.047. [DOI] [PubMed] [Google Scholar]
- 52.Rusché LN, Cruz-Reyes J, Piller KJ, Sollner-Webb B. 1997. Purification of a functional enzymatic editing complex from Trypanosoma brucei mitochondria. EMBO J. 16:4069–4081. 10.1093/emboj/16.13.4069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Aphasizhev R, Aphasizheva I. 2014. Mitochondrial RNA editing in trypanosomes: small RNAs in control. Biochimie 100:125–131. 10.1016/j.biochi.2014.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Göringer HU. 2012. ‘Gestalt,' composition and function of the Trypanosoma brucei editosome. Annu. Rev. Microbiol. 66:65–82. 10.1146/annurev-micro-092611-150150. [DOI] [PubMed] [Google Scholar]
- 55.Hashimi H, Zimmer SL, Ammerman ML, Read LK, Lukes J. 2013. Dual core processing: MRB1 is an emerging kinetoplast RNA editing complex. Trends Parasitol. 29:91–99. 10.1016/j.pt.2012.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kao CY, Read LK. 2005. Opposing effects of polyadenylation on the stability of edited and unedited mitochondrial RNAs in Trypanosoma brucei. Mol. Cell. Biol. 25:1634–1644. 10.1128/MCB.25.5.1634-1644.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Maslov DA, Simpson L. 2007. Strategies of kinetoplastid cryptogene discovery and analysis. Methods Enzymol. 424:127–139. 10.1016/S0076-6879(07)24006-6. [DOI] [PubMed] [Google Scholar]
- 58.Bakalara N, Simpson AM, Simpson L. 1989. The Leishmania kinetoplast-mitochondrion contains terminal uridylyltransferase and RNA ligase activities. J. Biol. Chem. 264:18679–18686. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.