Abstract
Humoral and cell-mediated immune correlates of protection (COP) for inhalation anthrax in a rhesus macaque (Macaca mulatta) model were determined. The immunological and survival data were from 114 vaccinated and 23 control animals exposed to Bacillus anthracis spores at 12, 30, or 52 months after the first vaccination. The vaccinated animals received a 3-dose intramuscular priming series (3-i.m.) of anthrax vaccine adsorbed (AVA) (BioThrax) at 0, 1, and 6 months. The immune responses were modulated by administering a range of vaccine dilutions. Together with the vaccine dilution dose and interval between the first vaccination and challenge, each of 80 immune response variables to anthrax toxin protective antigen (PA) at every available study time point was analyzed as a potential COP by logistic regression penalized by least absolute shrinkage and selection operator (LASSO) or elastic net. The anti-PA IgG level at the last available time point before challenge (last) and lymphocyte stimulation index (SI) at months 2 and 6 were identified consistently as a COP. Anti-PA IgG levels and lethal toxin neutralization activity (TNA) at months 6 and 7 (peak) and the frequency of gamma interferon (IFN-γ)-secreting cells at month 6 also had statistically significant positive correlations with survival. The ratio of interleukin 4 (IL-4) mRNA to IFN-γ mRNA at month 6 also had a statistically significant negative correlation with survival. TNA had lower accuracy as a COP than did anti-PA IgG response. Following the 3-i.m. priming with AVA, the anti-PA IgG responses at the time of exposure or at month 7 were practicable and accurate metrics for correlating vaccine-induced immunity with protection against inhalation anthrax.
INTRODUCTION
To date, there has not been a systematic evaluation of the relationship between anthrax vaccine-stimulated humoral and cell-mediated immune responses, their relative contributions to protection, or their comparative importance when used singly or in combination to predict the probability of survival in animal models or in humans.
Anthrax toxin protective antigen (PA) is the primary immunogen in licensed anthrax vaccines in the United States and the European Union, as well as in many of the second-generation anthrax vaccines in current development (1). Consequently, the quantitative analysis of anti-PA IgG antibody levels and lethal toxin neutralization activity (TNA) in serum are generally accepted as immunological correlates of protection (COP) for vaccine efficacy in animal models (2). Anti-PA IgG levels and TNA are also considered pivotal for cross-species predictions of anthrax vaccine efficacy in humans, for whom clinical efficacy studies are either impractical or ethically infeasible (3, 4) (http://www.fda.gov/AdvisoryCommittees/CommitteesMeetingMaterials/BloodVaccinesandOtherBiologics/VaccinesandRelatedBiologicalProductsAdvisoryCommittee/ucm239733.htm). Anti-PA IgG and TNA levels, however, are but one part of the spectrum of humoral and cell-mediated immune responses that may contribute to protection. The COP for anthrax may differ depending on vaccine formulations, schedules, and routes of administration (5–10).
The U.S.-licensed anthrax vaccine adsorbed (AVA) (BioThrax) was approved in 1970 for the prevention of anthrax in humans (11–14). The original regimen for AVA was a subcutaneous (s.c.) six-dose primary schedule at 0, 0.5, 1, 6, 12, and 18 months, with subsequent annual boosters. In May 2012, the U.S. Food and Drug Administration (FDA) approved the AVA regimen as an intramuscular (i.m.) three-dose priming schedule at 0, 1, and 6 months, with boosters at 12 and 18 months and annually thereafter (http://www.fda.gov/BiologicsBloodVaccines/Vaccines/ApprovedProducts/ucm304758.htm). These recent changes in the schedule and administration route were based on data from the Centers for Disease Control and Prevention Anthrax Vaccine Research Program (AVRP) (12, 13). The goals of the AVRP were to improve the AVA safety profile and ensure efficacy while minimizing the number of doses required. The study objectives included determining immunological correlates of protection, documenting vaccine efficacy, and optimizing the vaccination schedule and route of administration (14). Due to the low prevalence of inhalation anthrax and the ethical concerns of conducting an efficacy trial in humans, vaccine efficacy and duration of protection were evaluated in rhesus macaques (Macaca mulatta) (15).
The AVRP nonhuman primate (NHP) study used the 0-, 1-, and 6-month intramuscular priming series (3-i.m.) with a full human dose or saline dilutions of AVA to modulate the immune response. The NHP were challenged with high-dose (median, 504× the 50% lethal dose [LD50]) aerosolized Bacillus anthracis spores at month 12, 30, or 52. The PA-specific humoral and PA-stimulated cellular immune response variables were examined during and after the 3-i.m. schedule. In an earlier analysis, the vaccine-induced immune responses were characterized by analysis of variance (ANOVA) and logistic regression. These models were individually fitted to each immunological variable to determine if a variable could predict survival at a specific time point subsequent to the completion of the 3-i.m. priming (15).
In the present analysis, we adopted an alternative strategy to more comprehensively interrogate the AVRP rhesus macaque immunological data to select the best available COP variables. A representative set of 18 immunological responses to PA and 3 response ratios representing Th1/Th2 bias (16, 17), interleukin-4 (IL-4) protein to gamma interferon (IFN-γ) protein, IL-4 mRNA to IFN-γ mRNA, and IL-4-secreting cells to IFN-γ-secreting cells was used to generate a data set of 80 response variables, each of which was considered individually at every available study time point. Together with the AVA dose and time interval between scheduled first vaccination and challenge, we performed variable selection using penalized logistic regressions by two complementing statistical approaches, the stringency of least absolute shrinkage and selection operator (LASSO) and the correlation tolerance of elastic net. Three R software packages, Glmnet (18), Elasticnet (19), and Pensim (20), as well as the C++ software Bayesian binary regression (BBR) (21), which differ in their optimization algorithms and penalty parameter tuning, were used to avoid having to exclude important predictors and to ensure the selection of a reliable set of COP. The simplest plausible sets of variables were derived (22). The selected variables were evaluated for their correlation with survival, adjusting for variable multicollinearity. The receiver operator characteristic (ROC) area under the curve (AUC) was used as the criterion for testing the accuracy of the prediction model. This comprehensive approach utilized all the available information from the NHP study to select the most practicable and accurate COP for AVA-vaccinated NHP.
MATERIALS AND METHODS
NHP study design, vaccination schedule, and challenge.
The rhesus macaque (Macaca mulatta) study design, vaccination, sample schedules, and challenge outcomes have been described in detail (15). Due to the number of vaccinated NHP (n = 114), long duration, and multisite nature of the study, the NHP were organized into 12 groups of 8 to 10 animals plus 2 controls per group, for a total of 137 animals. The control animals were given saline injections instead of AVA but were otherwise treated identically to the vaccinated animals (Table 1). Different groups were vaccinated with the full human dose of AVA (HuAVA), saline-diluted AVA at a 1:5, 1:10, 1:20, or 1:40 dilution, or a saline placebo. The injections were 0.5-ml intramuscular (i.m.) injections administered at 0, 1, and 6 months, followed by aerosol challenge with spores of B. anthracis Ames strain at month 12, 30, or 52.
TABLE 1.
Basic characteristics for nonhuman primates included in the studya
| Group no. | No. in group (no. survived) |
Challenge time (mo) | Vaccine dilution | |
|---|---|---|---|---|
| Treated | Control | |||
| 1 | 10 (8) | 2 (0) | 52 | Undiluted |
| 2 | 9 (9) | 1 (0) | 52 | 1:5 |
| 3 | 10 (6) | 2 (0) | 52 | 1:10 |
| 4 | 10 (5) | 2 (1) | 12 | 1:20 |
| 5 | 10 (4) | 2 (1) | 12 | 1:40 |
| 6 | 10 (8) | 2 (0) | 12 | 1:10 |
| 7 | 10 (6) | 2 (1) | 12 | 1:20 |
| 8 | 10 (9) | 2 (1) | 12 | 1:40 |
| 9 | 9 (6) | 2 (1) | 30 | 1:10 |
| 10 | 10 (10) | 2 (0) | 30 | Undiluted |
| 11 | 8 (8) | 2 (1) | 30 | 1:5 |
| 12 | 8 (7) | 2 (1) | 30 | 1:20 |
| Total | 114 (86) | 23 (7) | ||
A total of 137 rhesus macaques (Macaca mulatta) were included in the study. The animals were injected intramuscularly with a 0.5-ml human dose of AVA or saline-diluted AVA (treated) or saline (control) at 0, 1, and 6 months, followed by aerosol challenge with spores of B. anthracis Ames strain at month 12, 30, or 52.
Humoral and cellular immune responses to AVA.
The immunological variables used in the analysis are listed in Table 2. The methods for determining the total anti-PA IgG concentrations (μg/ml), TNA titers, anti-PA IgG avidity index (AI), lymphocyte (peripheral blood mononuclear cell [PBMC]) stimulation index (SI), and frequencies of PA IgG-specific B cells, IFN-γ-secreting cells, and IL-4-secreting cells are described in detail elsewhere (15) and are reproduced in brief in the supplemental material. The methods for evaluating the PA-specific induction of gene transcription (mRNA) and translation (protein secretion) for cytokines associated with Th1 (IFN-γ and IL-2), Th2 (IL-4 and IL-6), and acute-phase (IL-1β and tumor necrosis alpha [TNF-α]) immune responses in PBMC are provided in Tables S1 to S3 and the supplemental methods in the supplemental material).
TABLE 2.
Assay variables used in the analysise
| Assay typea | Variable name | Target | Time points (mo) |
|---|---|---|---|
| ELISA | IgG | Anti-PA IgG protein | 1, 2, 6, 7, last |
| IL1Be | IL-1β proteinb | 1, 2, 6, 7 | |
| IL2e | IL-2 proteinb | 1, 2, 7 | |
| IL4e | IL-4 proteinb | 1, 2, 7 | |
| IL6e | IL-6 proteinb | 1, 2, 6, 7 | |
| IFNe | IFN-γ proteinb | 1, 2, 7 | |
| TNFe | TNF-α proteinb | 1, 2, 6, 7 | |
| R_IL4IFNe | Ratio of IL-4 protein to IFN-γ protein | 1, 2, 7 | |
| RT-PCR | IL-1Bm | IL-1β mRNAb | 1, 2, 6, 7 |
| IL-2m | IL-2 mRNAb | 1, 2, 6, 7 | |
| IL-4m | IL-4 mRNAb | 1, 2, 6, 7 | |
| IL-6m | IL-6 mRNAb | 1, 2, 6, 7 | |
| IFNm | IFN-γ mRNAb | 1, 2, 6, 7 | |
| TNFm | TNF-α mRNAb | 1, 2, 6, 7 | |
| R_IL4IFNm | Ratio of IL-4 mRNA to IFN-γ mRNA | 1, 2, 6, 7 | |
| Toxin neutralization | TNA | Toxin neutralization activity ED50c | 1, 2, 6, 7, last |
| Lymphocyte stimulation | SI | Lymphocyte stimulation indexb | 1, 2, 6, 7, last |
| Avidity | AI | Anti-PA IgG avidity | 1, 2, 7 |
| ELISpotd | INFeLi | Frequency of IFN-γ-secreting cellsb | 6, 7, last |
| IL4eLi | Frequency of IL-4-secreting cellsb | 1, 6, 7, last | |
| R_IL4IFNeLi | Ratio of frequency of IL-4-secreting cells to frequency of IFN-γ-secreting cells | 6, 7, last |
Eighty assay variables were determined by 6 types of assays.
Determined from in vitro PA-stimulated peripheral blood mononuclear cells (PBMC).
ED50, 50% effective dose.
ELISpot, enzyme-linked immunosorbent spot assay.
The study variables for vaccine dilution and time of challenge were also included in the model, for a total of 82 variables.
Data set construction.
The construction of data sets is detailed in the supplemental material. Briefly, the data were from control and vaccinated animals that completed the study (Table 1). Except for vaccine dose and the interval between first vaccination and aerosol challenge, the primary data set was constructed, with each variable corresponding to an assay, with measurements taken immediately before each injection, 4 weeks after each injection, and the last available time point prior to aerosol challenge. For an assessment of the relative contributions of humoral and cellular immune responses, ratio variables were generated by dividing the Th2 response-related variables by the Th1 response-related variables. The ratio variables were the ratio of IL-4 mRNA to IFN-γ mRNA (R_IL4IFNm), the ratio of secreted IL-4 protein to secreted IFN-γ protein (R_IL4IFNe), and the ratio of the frequency of IL-4-secreting cells to that of IFN-γ-secreting cells (R_IL4IFNeLi). The measurement at each time point was then converted to an individual variable (e.g., anti-PA IgG at month 6 is one variable [IgG_6], and anti-PA IgG at month 7 is a separate variable [IgG_7]). The month-7 time point, which is 1 month after the priming series, was designated “peak,” and the last available time point prior to challenge was designated “last” for all NHP. There was a broad range of missing rates for different variables (see Table S4 in the supplemental material). Multiple imputations were used to minimize the bias from missing data (see Table S5 and methods in the supplemental material). Some variables were not imputed due to the high frequency of missing data and therefore were removed from the data set. The final assay variables (n = 80) used in the analysis are listed in Table 2, in addition to the 2 study variables used for vaccine dilution and the time of challenge.
Variable selection by logistic regression penalized by LASSO and elastic net.
Variable selection was performed on 20 imputed data sets. Variables that were selected ≥10 times out of the 20 imputed data sets were included in the final variable set for each selection method (see the supplemental material) (23, 24). To have the highest confidence that the best correlates were identified, we selected software packages that employed two statistical approaches and that differed in their optimization algorithms and penalty parameter tuning. LASSO and elastic net variable selections were performed in three R packages: Glmnet (18), Elasticnet (19), and Pensim (20), and LASSO variable selection was performed in the C++ software package BBR (21) (see Table S6 in the supplemental material). LASSO may undergo too-stringent shrinkage and thus ignore important predictors, while elastic net has a grouping effect, selecting important predictors even if they are correlated with each other. Elastic net may consequently select too many predictors, resulting in overfitting in the prediction model (18, 19). Repeated (60 times) 10-fold cross-validations were performed to select the best penalty parameters. The optimal sets of variables were selected, with the penalty parameters being where the cross-validation error is minimal or the cross-validated likelihood is maximal. The simplest plausible (parsimonious) sets of variables were selected by applying the “1-standard error rule” (24), choosing the variables with the penalty parameters for which the cross-validation error reached the sum of the minimum cross-validation error and one standard error. The correlation between each selected variable and survival was examined by simple logistic regression in SAS version 9.3 (SAS Institute, Cary, NC) with the original unimputed data set.
Collinearity or multicollinearity diagnosis and evaluation of model performance.
Although variables with high rates of missing data are not useful for building practicable models, these variables are still worthy of examination. If the performances of the models that include these variables are compared with those of the models that exclude these variables, the importance of these variables will be known. If these variables do not add much to the performances of the models, these variables can be safely excluded from the models. Otherwise, these variables should be taken into consideration in the future generation of prediction models. Therefore, the model evaluation was performed with the imputed data sets including variables that were heavily imputed.
The emphasis on PA as the primary immunogen and the interrelatedness of the many facets of acquired immunity may result in data from two or more immunoassays being highly correlated with each other. In addition, variables from the same immunoassay but that are measured at different time points may also be highly correlated. Therefore, collinearity or multicollinearity may occur if these variables are present in the same regression models. Logistic regression does not work properly on models containing collinear or multicollinear variables. Therefore, before an evaluation of each variable set for the potential to be a prediction model with good performance, a collinearity or multicollinearity diagnosis was completed (see the supplemental material). To overcome collinearity or multicollinearity, principal component logistic regression (PCLR) was performed to evaluate the performances of the models built with each selected variable set that had collinearity or multicollinearity. For performing PCLR, principal component analysis (PCA) was done using Prcomp in R to generate an eigenvector and score matrix, which were used for logistic regression using glm.fit in R. For variable sets that did not have collinearity or multicollinearity, logistic regression was done using glm.fit in R. The PCLR and logistic regression models are described in detail in the supplemental material. To compare the AUCs between the models, paired permutation tests were performed (25, 26), with a Bonferroni-corrected significance level of 0.0025 for multiple comparisons.
Practicable and accurate correlates of protection.
For further performance evaluation as a COP, PCLR models with multiple variables that had multicollinearity, and logistic models with one variable or two variables that did not have collinearity, were built on the original unimputed data set. PCLR and logistic regressions were performed with the original unimputed data set by glm.fit in R or proc logistic in SAS. To compare the prediction accuracy of the logistic regression models that were built with different variables but on the same original data set, the Roccontrast statement in proc logistic in SAS was used to perform dependent AUC comparisons. To compare the prediction accuracy of the PCLR models with that of the logistic regression models, with both built on the same original data set, paired permutation tests (25, 26) were performed.
RESULTS
Variable selection by logistic regression penalized by LASSO or elastic net.
Parsimonious variable selections are summarized in Table 3. Additionally, optimal variable selections were performed with Glmnet (18), Elasticnet (19), the R package Pensim (20), and BBR (21) by choosing the minimum cross-validation error or the maximum cross-validated likelihood, thus minimizing prediction error (see Table S7 in the supplemental material). Additional details for parsimonious and optimal variable selections are included in the supplemental material (see Tables S7 to S13 in the supplemental material). The three variables last anti-PA IgG and SI at months 2 (SI_2) and 6 (SI_6) were selected by all the methods. Among these packages, all the parsimonious variable selections by LASSO (Par_LASSO) chose only these three variables. Parsimonious selection by elastic net with the Elasticnet package (Par_elastic_Elasticnet) additionally selected both anti-PA IgG and TNA at month 7 (peak). Parsimonious selection by elastic net with the Glmnet package (Par_elastic_Glmnet) further selected IL4e and TNFe at month 1, anti-PA IgG, TNA, and IFNeLi at month 6, and R_IL4IFNeLi and R_IL4IFNm at month 7. The optimal selections chose additional variables IL4eLi at month 1, IL1Be at month 2, IL1Be, TNFe, and R_IL4IFNeLi at month 6, IL1Bm and IL4eLi at month 7, and IL4eLi and R_IL4IFNeLi at the last time point (see Table S7).
TABLE 3.
Summary of parsimonious variable selectionsb
| Variable name | Time point (mo) | Simple logistic regression results |
BBR LASSO | Elasticnet |
Glmnet |
||||
|---|---|---|---|---|---|---|---|---|---|
| Intercept (P value) | Parameter (P value) | AUC (95% Cl) | LASSO | Elastic net | LASSO | Elastic net | |||
| IgG | 6 | 0.8952 (<0.0001) | 1.7332 (<0.0001)a | 0.7724 (0.6956–0.8491) | X | ||||
| 7 | −0.6569 (0.0582) | 1.0009 (<0.0001)a | 0.7956 (0.7208–0.8703) | X | X | ||||
| Last | 0.7105 (0.0015) | 2.1628 (<0.0001)a | 0.8214 (0.7514–0.8914) | • | • | • | • | • | |
| TNA | 6 | −1.346 (0.0060) | 1.8551 (<0.0001)a | 0.7416 (0.6689–0.8143) | X | ||||
| 7 | −1.9889 (0.0006) | 1.0808 (<0.0001)a | 0.7918 (0.7158–0.8678) | X | X | ||||
| SI | 2 | −4.9416 (<0.0001) | 1.4900 (<0.0001)a | 0.7860 (0.7086–0.8633) | • | • | • | • | • |
| 6 | −3.9370 (0.0376) | 1.4369 (0.0111)a | 0.7095 (0.5809–0.8381) | • | • | • | • | • | |
| IFNeLi | 6 | −0.3719 (0.3090) | 0.7881 (0.0004)a | 0.7073 (0.6077–0.8068) | X | ||||
| R_IL4IFNeLi | 7 | 0.6808 (0.0020) | −0.1508 (0.3684) | 0.5428 (0.4195–0.6661) | X | ||||
| IL4e | 1 | −2.0654 (0.5856) | 2.0972 (0.4594) | 0.5149 (0.4790–0.5508) | X | ||||
| TNFe | 1 | 3.2168 (0.0787) | −1.4247 (0.1622) | 0.6627 (0.5485–0.7769) | X | ||||
| R_IL4IFNm | 7 | 0.6608 (0.0004) | −0.7249 (0.0508) | 0.5829 (0.4844–0.6815) | X | ||||
| No. of variables | 3 | 3 | 5 | 3 | 12 | ||||
| Variable set identifier | Par_LASSOc | Par_LASSOc | Par_elastic_Elasticnet | Par_LASSOc | Par_elastic_Glmnet | ||||
P value of <0.05 from Wald chi-square test of the parameter estimate.
•, variables that were selected by all five selection methods. X, variables that were selected by 1 or 2 methods.
Parsimonious selections by LASSO in all the packages selected the same variables that were considered one variable set, Par_LASSO, for subsequent analyses.
Simple logistic regression with each of the selected variables showed statistically significant (P < 0.05) positive correlations between survival and anti-PA IgG at months 6 and 7 (peak), as well as at the last time point, TNA at months 6 and 7, SI at months 2 and 6, and IFN-γ-secreting cell frequency at month 6 (Table 3). The ratio of the IL-4 to IFN-γ mRNA levels (R_IL4IFNm) at month 6, which was chosen only in the optimal selections, also had a statistically significant correlation with survival (see Table S7 in the supplemental material).
Evaluation of survival prediction models with selected sets of variables.
The selected variable sets were used to build regression models to test their survival prediction performance. The PCLR models were built with variable sets that had multicollinearity. Logistic regression was applied to the three-variable set (Par_LASSO) that did not have multicollinearity. When the models with the three parsimonious variable sets Par_LASSO, Par_elastic_Elasticnet, and Par_elastic_Glmnet were applied to the imputed data sets, the mean AUCs were 0.8492, 0.8494, and 0.9022, respectively (Table 4; see also Table S14 in the supplemental material). Therefore, the model with variable set Par_LASSO was the most parsimonious among the three. However, this model was limited by the fact that in the unimputed original data, there were 71 out of 137 animals (51.82%) that did not have an SI measurement at month 6. Using the variable SI at month 6 to predict survival with the unimputed data will therefore have low statistical power and may create bias. To test the impact of deleting SI at month 6 on the model performance, a model with SI at month 6 excluded from Par_LASSO was applied to the 20 imputed data sets, and its performance was compared with that of other models. This model had an AUC of 0.8409, which is not statistically significantly different from that of the model with variable set Par_LASSO, suggesting that SI at month 6 did not play a significant role in model performance (Table 4; see also Table S14 in the supplemental material). Therefore, SI at month 6 was excluded in further evaluations.
TABLE 4.
Summary of performance of PCLR and logistic regression models containing parsimoniously selected variables
| Regressiona | Variable set identifier | No. of variables | Mean AUC | Paired multiple comparisonsb |
|---|---|---|---|---|
| PCLR | Par_elastic_Elasticnet | 5 | 0.8494 | NS |
| Par_elastic_Glmnet | 12 | 0.9022 | S | |
| Logistic regression | Par_LASSOc | 3 | 0.8492 | NS |
| Last anti-PA IgG + SI_2 | 2 | 0.8409 | Reference model |
PCLR or logistic regression was performed for each selected variable set with the imputed data sets.
Multiple comparisons of AUCs between a model and the model with the variables last anti-PA IgG and SI_2 were performed by a paired permutation test with a Bonferroni-corrected significance level of 0.0025; NS, P > 0.0025; S, P ≤ 0.0025.
Parsimonious selections by LASSO in all the packages selected the same variables that were considered one variable set, Par_LASSO.
All together, the model with last anti-PA IgG and SI at month 2 was the most parsimonious. Together with their low missing rates in the unimputed data set, these two variables are worthy of consideration in the generation of practicable prediction models.
Practicable and accurate correlates of protection.
The last anti-PA IgG measurement concurrent with aerosol exposure was consistently selected by all the variable selection methods. For situations where a preexposure sample was not available, however, the peak measurement was found to be the best practicable alternative for predicting protection against future exposure. The peak response variables were therefore included in the regression model evaluations.
TNA was statistically significantly correlated with survival but did not provide the same good level of accuracy as anti-PA IgG. However, TNA is considered to be species neutral since it does not rely on a species-specific conjugate antibody, allowing a direct comparison in the same assay of serum antibody functional activity across animal species and genera. TNA features prominently in the literature as an immunological bridge to extrapolate data in animals in order to predict anthrax vaccine effectiveness in humans (4, 27). Furthermore, anti-PA IgG and TNA levels in NHP were highly correlated (15), and corresponding data in humans are available (12, 13, 15). Similar to peak anti-PA IgG, peak TNA was included in the regression model evaluations as a potential alternative to last anti-PA IgG.
Consequently, the parsimonious variable selection was expanded to include the following five variables: last anti-PA IgG, last TNA, peak anti-PA IgG, peak TNA, and SI at month 2. These variables were evaluated both singly and in combinations in regression models of predicted survival probability in NHP. A collinearity or multicollinearity diagnosis was used to detect collinearity or multicollinearity and generate the combinations of these variables for the models. Multicollinearity was present in all the variable combinations composed of three or more variables. Among the two-variable combinations, collinearity was not present between SI at month 2 and any of the other four variables, nor between last TNA and peak anti-PA IgG or peak TNA (data not shown). Last anti-PA IgG had only mild collinearity with peak TNA (variance inflation factor [VIF], 2.57; tolerance, 0.39 [the cutoff value is 2.5 for VIF and 0.4 for tolerance]; details are in the supplemental methods and references 12 and 13 in the supplemental material) and peak anti-PA IgG (VIF, 2.68; tolerance, 0.37) (28, 29). Because PCLR can be done only with more than three variables, the two-variable logistic regression models containing either last anti-PA IgG and peak TNA or last anti-PA IgG and peak anti-PA IgG were fitted. A PCLR model containing all five variables or the Par_elastic_Elasticnet variable set except SI at month 6 was fitted for comparison with the single-variable and two-variable logistic regression models (Table 5). The variable set Par_elastic_Glmnet that contains 12 variables was not examined at this step, due to the high frequency of missing data within some of these variables in the original unimputed data set.
TABLE 5.
Statistics of regression models for survival predictions
| Model (mo) by type | Variable (mo)a | Intercept (P value)b | Parameter (P value)b | AUC (95% CI)c | P value for AUC comparison |
|---|---|---|---|---|---|
| Logistic | |||||
| Peak anti-PA IgG | IgG_7 | −0.6569 (0.0582) | 1.0009 (<0.0001) | 0.7956 (0.7208–0.8703) | 0.2712 |
| Last anti-PA IgG | IgG_last | 0.7105 (0.0015) | 2.1628 (<0.0001) | 0.8214 (0.7514–0.8914) | Reference model |
| Last anti-PA IgG (12) | IgG_last (12) | 0.5416 (0.0707) | 1.4605 (0.0049) | 0.7240 (0.5970–0.8509) | NAd |
| Last anti-PA IgG (30) | IgG_last (30) | 1.0465 (0.0269) | 2.3380 (0.0036) | 0.8758 (0.7721–0.9796) | NA |
| Last anti-PA IgG (52) | IgG_last (52) | 0.3651 (0.5105) | 4.4283 (0.0026) | 0.9071 (0.8014–1.000) | NA |
| Peak TNA | TNA_7 | −1.9889 (0.0006) | 1.0808 (<0.0001) | 0.7918 (0.7158–0.8678) | 0.2651 |
| Last TNA | TNA_last | −1.1918 (0.0086) | 1.6011 (<0.0001) | 0.7270 (0.6526–0.8014) | 0.0009e |
| SI_2 | SI_2 | −4.9416 (<0.0001) | 1.4900 (<0.0001) | 0.7860 (0.7086–0.8633) | 0.3873 |
| SI_2 + peak anti-PA IgG | SI_2 | −2.8355 (0.0553) | 0.6652 (0.1390) | 0.8121 (0.7412–0.8831) | 0.7435 |
| IgG_7 | 0.7267 (0.0136) | ||||
| SI_2 + last anti-PA IgG | SI_2 | −1.6413 (0.2724) | 0.6155 (0.1140) | 0.8424 (0.7758–0.9090) | 0.0777 |
| IgG_last | 1.7901 (0.0001) | ||||
| SI_2 + peak TNA | SI_2 | −3.9897 (0.0024) | 0.7455 (0.0918) | 0.8100 (0.7384–0.8817) | 0.7115 |
| TNA_7 | 0.7368 (0.0197) | ||||
| SI_2 + last TNA | SI_2 | −4.3354 (0.0006) | 1.0212 (0.0057) | 0.7927 (0.7182–0.8672) | 0.3462 |
| TNA_last | 0.9827 (0.0224) | ||||
| Peak anti-PA IgG + last anti-PA IgG | IgG_7 | 0.4400 (0.3400) | 0.1991 (0.5027) | 0.8247 (0.7546–0.8947) | 0.5252 |
| IgG_last | 1.8750 (0.0012) | ||||
| Peak TNA + last anti-PA IgG | TNA_7 | 0.2292 (0.7834) | 0.1928 (0.5497) | 0.8266 (0.7570–0.8963) | 0.3859 |
| IgG_last | 1.9140 (0.0008) | ||||
| Peak anti-PA IgG + last TNA | IgG_7 | −1.1664 (0.0116) | 0.7297 (0.0037) | 0.7942 (0.7202–0.8682) | 0.2058 |
| TNA_last | 0.7490 (0.1054) | ||||
| Peak TNA + last TNA | TNA_7 | −2.1130 (0.0003) | 0.7725 (0.0058) | 0.7956 (0.7217–0.8695) | 0.2639 |
| TNA_last | 0.7589 (0.1059) | ||||
| PCLR | |||||
| SI_2 + peak anti-PA IgG + peak TNA + last anti-PA IgG | PC1 | 1.1927 (<0.0001) | −0.0356 (<0.0001) | 0.8403 (0.7744–0.9062) | 0.1780 |
| PC2 | 0.5438 (0.0076) | ||||
| PC3 | 1.7489 (0.0046) | ||||
| PC4 | −0.0977 (09300) | ||||
| SI_2 + peak anti-PA IgG + last TNA + peak TNA + last anti-PA IgG | PC1 | 1.1510 (<0.0001) | −0.0346 (<0.0001) | 0.8432 (0.7768–0.9097) | 0.6185 |
| PC2 | 0.5254 (0.0091) | ||||
| PC3 | 0.7204 (0.1153) | ||||
| PC4 | 2.1960 (0.0148) | ||||
| PC5 | 0.9948 (0.4113) |
PC1, PC2, PC3, PC4, and PC5 are principal components.
Intercept and parameter values are for the logistic regression for the indicated variable or component. Note that models with multiple variables have only a single intercept.
AUC is given as a measure of model accuracy.
NA, not applicable, due to dependent ROC curve comparisons being performed in this analysis, and thus ROC curve comparisons here should be within exactly the same sample.
P value with a significance level of <0.05 for AUC comparison with that of the logistic regression model with last anti-PA IgG only.
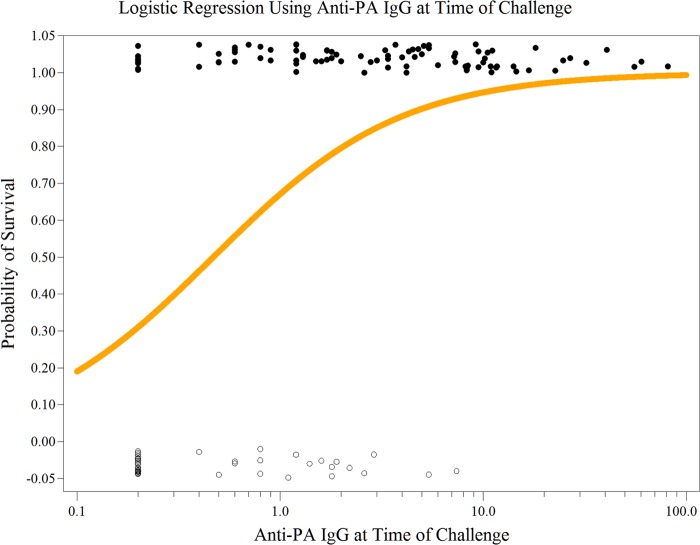
Of the single-variable models (Table 5), last anti-PA IgG had the highest AUC (0.8214; 95% confidence interval [CI], 0.7514 to 0.8914), peak anti-PA IgG, peak TNA, and SI at month 2 had lower but not statistically significantly lower AUCs (0.7956, 95% CI, 0.7208 to 0.8703; 0.7918, 95% CI, 0.7158 to 0.8678; and 0.7860, 95% CI, 0.7086 to 0.8633, respectively), and last TNA had statistically significantly lower AUC (0.7270; 95% CI, 0.6526 to 0.8014). Logistic regression using last anti-PA IgG at specific challenge times generated different parameter estimates but similar significant P values compared with those for overall last anti-PA IgG (Table 5). The different parameter estimates were due to differences in the magnitude of the anti-PA IgG responses at these time points. Last anti-PA IgG had a parameter estimate of 2.1628 (Table 5), indicating that a 1-log10 unit (10-fold) increase in last anti-PA IgG can make an 8.70-fold increase in the odds of survival (probability of survival/probability of death). The logistic regression model for last anti-PA IgG is shown in Fig. 1.
FIG 1.
Logistic regression of last anti-PA IgG concentration versus survival. NHP survivors (●) (1.0 on the y axis) and nonsurvivors (○) (0 on the y axis) are plotted with a slight vertical y axis displacement so that overlapping points may be seen. The logistic regression line (orange line) is the predicted survival based on the NHP last anti-PA IgG (μg/ml) measurements.
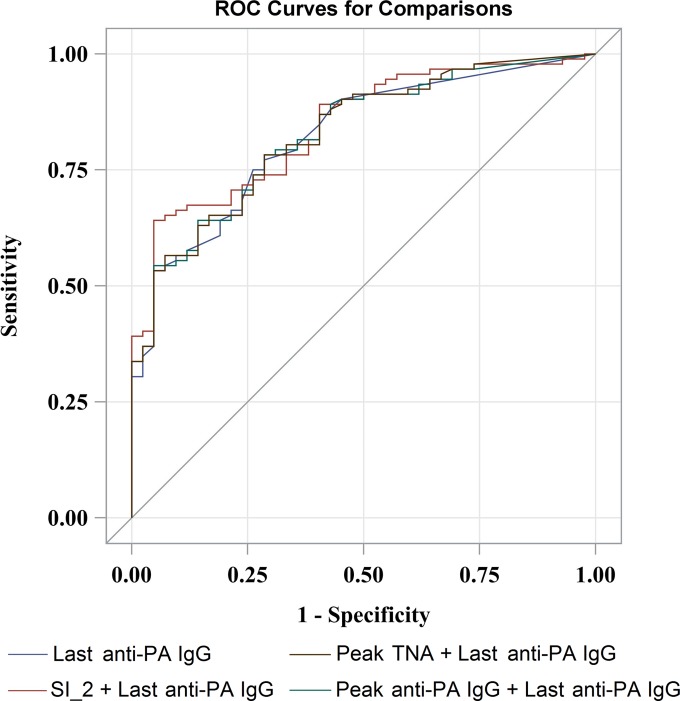
In each two-variable model, there was always one variable with a nonsignificant parameter estimate (Table 5). The 2 two-variable models that contain last anti-PA IgG had AUCs of 0.8247 (95% CI, 0.7546 to 0.8947) and 0.8266 (95% CI, 0.7570 to 0.8963), which were very close to the AUC of the model with last anti-PA IgG only. The model with SI_2 and last anti-PA IgG had an AUC of 0.8424 (95% CI, 0.7758 to 0.9090), the highest accuracy of all the one-variable and two-variable models (Table 5). The PCLR model containing all five variables had an AUC of 0.8432 (95% CI, 0.7768 to 0.9097), and the PCLR model containing the Par_elastic_Elasticnet variable set not including SI at month 6 had an AUC of 0.8403 (95% CI, 0.7744 to 0.9062). Permutation tests showed that compared with the single-variable model containing last anti-PA IgG, only the single-variable model containing last TNA had a statistically significantly lower AUC (0.7270; 95% CI, 0.6526 to 0.8014; P = 0.0009) (Table 5). The ROC curves for the two-variable models that had a higher AUC than that of the last anti-PA IgG model roughly overlap the ROC curve of the last anti-PA IgG model (Fig. 2).
FIG 2.
Comparisons of ROC curves among logistic regression models. The ROC curves were from logistic regressions containing the single variable last anti-PA IgG and two variables with combinations of SI_2, last anti-PA IgG, and peak anti-PA IgG and TNA.
Overall, anti-PA IgG at the last time point and SI at months 2 and 6 were the three best correlates of protection in rhesus macaques. Of these, the single-variable last anti-PA IgG model had the highest prediction accuracy in this genus and thus is the most appropriate single-variable model for survival predictions (Table 5).
DISCUSSION
Anti-PA antibody levels have been an accepted serological indicator for protection against anthrax since acellular vaccines for human use were first developed (1, 30–34). More recently, serum TNA has been the focal point for correlates of protection in animal models and estimates of survival probability in humans (4, 27, 35). Despite this focus, there has been no systematic evaluation of the relationship between PA-stimulated humoral and cell-mediated immune responses, their relative contributions to protection, or their comparative importance when used singly or in combination to predict the probability of survival.
We applied a comprehensive strategy using logistic regression penalized by LASSO or elastic net in four different software applications with dissimilar algorithms, methods, and criteria for penalty parameter tuning. The strategy selected the optimal and parsimonious COP variable combinations and time points that correlated with survival.
The two statistical approaches of LASSO and elastic net were applied because LASSO may perform shrinkage too stringently, and the resulting prediction model may exclude important predictors. Alternatively, elastic net may be too lenient in its variable selection, with subsequent overfitting in the prediction model, and consequently, variations within the sample may be disproportionately interpreted as variations in the study population. In order to avoid overfitting of the prediction model, parsimonious sets of variables were derived. The selected variables were then evaluated for correlation with survival. Models without collinearity or multicollinearity issues were evaluated using logistic regression, and models containing variables with multicollinearity were evaluated by PCLR. The AUC was used as the criterion for testing the accuracy of the prediction model. The AUC measures the ability of the model to correctly identify the survival status. AUCs between 0.90 and 1, 0.80 and 0.90, 0.70 and 0.80, and 0.50 and 0.70 represent high, good, moderate, and low discrimination performance of the model, respectively (36). This comprehensive approach utilized all the available information from the study to select the most plausible and practicable COP for AVA-vaccinated rhesus macaques.
Statistically significant positive correlations with survival were identified for anti-PA IgG at months 6 and 7 and the last time point before infectious challenge (last), for TNA at months 6 and 7 (peak), SI at months 2 and 6, and the frequency of IFN-γ-secreting cells at month 6. The ratio of IL-4 mRNA to IFN-γ mRNA transcription levels at month 6 had a statistically significant negative correlation with survival. The different software applications consistently selected anti-PA IgG responses at challenge (last) and SI at months 2 and 6, thus confirming their importance as immune correlates of protection in rhesus macaques. The frequency of missing data for SI at month 6 was 71 out of 137 (51.82%), causing potential issues of bias and low statistical power. Deleting SI at month 6 from Par_LASSO did not lower the prediction accuracy much. The suitable variables were then used in logistic regression models for survival predictions. Although their performance in the analysis was lower, the peak and last TNA responses were also included in the models, primarily due to their positive correlation with survival and the cross-species utility of TNA for predicting survival probabilities in humans. Therefore, the final selected single-correlate variables were peak anti-PA IgG, peak TNA, last anti-PA IgG, last TNA, and SI at month 2. The final dual-correlate variables were combinations of peak anti-PA IgG, last anti-PA IgG, peak TNA, last TNA, and SI at month 2.
The single-correlate logistic regression models using peak TNA and peak anti-PA IgG responses described the immune status on completion of the 3-i.m. priming series and were valid predictors of survival at later time points. The model with only last anti-PA IgG had good predictive accuracy (AUC, 0.8214; 95% CI, 0.7514 to 0.8914). The predictive accuracy of last TNA was moderate, with an AUC of 0.7270 (95% CI, 0.6526 to 0.8014), which was statistically significantly less than the last anti-PA IgG AUC (P = 0.0009). This difference is likely due to the lower analytical sensitivity of the TNA assay. At the last time point, there were 104 values that were above the limit of detection (LOD) of the anti-PA IgG assay but only 69 that were above the LOD of the TNA assay. Peak anti-PA IgG, peak TNA, and SI at month 2 were moderately accurate predictors of survival, with an AUC of >0.78.
In the dual-correlate logistic regression models, there was always one parameter estimate that was not statistically significant, indicating that it was not making a significant contribution to the predictive power of the model. Compared with the single-correlate model with last anti-PA IgG only, the predictive accuracy was slightly improved but not statistically significantly different using the dual-correlate models containing last anti-PA IgG or PCLR models containing all five variables or the Par_elastic_Elasticnet variable set except SI at month 6, and it was slightly decreased but not statistically significantly different using all other dual-correlate models. Given the lower technical complexity, higher sample stability, and higher throughput of an enzyme-linked immunosorbent assay (ELISA) than those of a lymphocyte proliferation assay, together with the ability to accurately calibrate ELISA standards and quantify the IgG analyte for each species (15, 37), these data confirm that last anti-PA IgG provides an appropriate correlate of protection for cross-species survival predictions (4).
Fay et al. (4) noted in their cross-species meta-analysis that vaccine formulation plays a statistically significant role in the quantitative assessment of correlates of protection. In the AVRP study, both the NHP and human cohorts were treated with AVA in experiments designed to match the schedule and time of measurement as closely as possible. The quantitative model generated from the AVRP NHP data therefore should be used only to bridge to data from human cohorts tested with the same vaccine schedule. The anti-PA IgG concentration at the time of challenge should be a suitable correlate for other PA-based vaccines, although any cross-species bridging should be based on data from clinical and nonclinical studies with matching vaccine formulations, schedules, and times of measurement. Anthrax vaccines that do not rely on PA as the primary immunogen may require further study to identify the most suitable correlates.
The anti-PA IgG levels at the time of challenge (last) were the most accurate single measure for determining the probability of survival against inhalation anthrax in rhesus macaques completing a 3-i.m. priming series of AVA. The SI responses at months 2 and 6 and peak anti-PA IgG and TNA were also strong correlates of protection for rhesus macaques. These single correlates of protection were selected from 80 assay and 2 study variables. None of the multivariable models evaluated were statistically significantly better than the single-correlate last anti-PA IgG model. In the absence of a last anti-PA IgG measurement concurrent with aerosol exposure, the month-7 anti-PA IgG and TNA responses to 3-i.m. priming are suitable alternative correlates of protection.
Supplementary Material
ACKNOWLEDGMENTS
The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention.
We thank David Madigan from Columbia University and Nong Shang from the Centers for Disease Control and Prevention for their critical comments and constructive advice.
This study was funded through the Centers for Disease Control and Prevention (Atlanta, GA) with additional support from the Biomedical Advanced Research and Development Authority (BARDA) (Washington, DC). Battelle was funded under DHHS CDC contract 200-2000-10065. L.C. and S.D. were funded by the Atlanta Research and Education Foundation (AREF) through the Department of Veterans Affairs, Veterans Health Administration, Office of Research and Development (Atlanta, GA).
Footnotes
Published ahead of print 3 September 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/CVI.00469-14.
REFERENCES
- 1. Saile E, Quinn CP. 2010. Anthrax vaccines, p 269–293 In Bergman NH. (ed), Bacillus anthracis and anthrax. Wiley-Blackwell, Hoboken, NJ. [Google Scholar]
- 2. Bienek DR, Loomis LJ, Biagini RE. 2009. The anthrax vaccine: no new tricks for an old dog. Hum. Vaccin. 5:184–189. 10.4161/hv.5.3.7308. [DOI] [PubMed] [Google Scholar]
- 3. Gronvall GK, Trent D, Borio L, Brey R, Nagao L, Alliance for Biosecurity 2007. The FDA animal efficacy rule and biodefense. Nat. Biotechnol. 25:1084–1087. 10.1038/nbt1007-1084. [DOI] [PubMed] [Google Scholar]
- 4. Fay MP, Follmann DA, Lynn F, Schiffer JM, Stark GV, Kohberger R, Quinn CP, Nuzum EO. 2012. Anthrax vaccine-induced antibodies provide cross-species prediction of survival to aerosol challenge. Sci. Transl. Med. 4:151ra126. 10.1126/scitranslmed.3004073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. McBride BW, Mogg A, Telfer JL, Lever MS, Miller J, Turnbull PC, Baillie L. 1998. Protective efficacy of a recombinant protective antigen against Bacillus anthracis challenge and assessment of immunological markers. Vaccine 16:810–817. 10.1016/S0264-410X(97)00268-5. [DOI] [PubMed] [Google Scholar]
- 6. Livingston BD, Little SF, Luxembourg A, Ellefsen B, Hannaman D. 2010. Comparative performance of a licensed anthrax vaccine versus electroporation based delivery of a PA encoding DNA vaccine in rhesus macaques. Vaccine 28:1056–1061. 10.1016/j.vaccine.2009.10.111. [DOI] [PubMed] [Google Scholar]
- 7. Turnbull PC, Broster MG, Carman JA, Manchee RJ, Melling J. 1986. Development of antibodies to protective antigen and lethal factor components of anthrax toxin in humans and guinea pigs and their relevance to protective immunity. Infect. Immun. 52:356–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Turnbull PC, Leppla SH, Broster MG, Quinn CP, Melling J. 1988. Antibodies to anthrax toxin in humans and guinea pigs and their relevance to protective immunity. Med. Microbiol. Immunol. 177:293–303. [DOI] [PubMed] [Google Scholar]
- 9. Keitel WA, Treanor JJ, El Sahly HM, Evans TG, Kopper S, Whitlow V, Selinsky C, Kaslow DC, Rolland A, Smith LR, Lalor PA. 2009. Evaluation of a plasmid DNA-based anthrax vaccine in rabbits, nonhuman primates and healthy adults. Hum. Vaccin. 5:536–544. 10.4161/hv.5.8.8725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Luxembourg A, Hannaman D, Nolan E, Ellefsen B, Nakamura G, Chau L, Tellez O, Little S, Bernard R. 2008. Potentiation of an anthrax DNA vaccine with electroporation. Vaccine 26:5216–5222. 10.1016/j.vaccine.2008.03.064. [DOI] [PubMed] [Google Scholar]
- 11. Brachman PS, Gold H, Plotkin SA, Fekety FK, Werrin M, Ingraham NR. 1962. Field evaluation of human anthrax vaccine. Am. J. Public Health Nations Health 52:632–645. 10.2105/AJPH.52.4.632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Marano N, Plikaytis BD, Martin SW, Rose C, Semenova VA, Martin SK, Freeman AE, Li H, Mulligan MJ, Parker SD, Babcock J, Keitel W, El Sahly H, Poland GA, Jacobson RM, Keyserling HL, Soroka SD, Fox SP, Stamper JL, McNeil MM, Perkins BA, Messonnier N, Quinn CP, Anthrax Vaccine Research Program Working Group 2008. Effects of a reduced dose schedule and intramuscular administration of anthrax vaccine adsorbed on immunogenicity and safety at 7 months: a randomized trial. JAMA 300:1532–1543. 10.1001/jama.300.13.1532. [DOI] [PubMed] [Google Scholar]
- 13. Wright JG, Plikaytis BD, Rose CE, Parker SD, Babcock J, Keitel W, El Sahly H, Poland GA, Jacobson RM, Keyserling HL, Semenova VA, Li H, Schiffer J, Dababneh H, Martin SK, Martin SW, Marano N, Messonier NE, Quinn CP. 2014. Non-inferiority and safety of reduced schedules and intramuscular injection of anthrax vaccine adsorbed (AVA). Vaccine 32:1019–1028. 10.1016/j.vaccine.2013.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Institute of Medicine (US) Committee to Review the CDC Anthrax Vaccine Safety and Efficacy Research Program. 2002. An assessment of the CDC anthrax vaccine safety and efficacy research program. National Academies Press, Washington, DC. [PubMed] [Google Scholar]
- 15. Quinn CP, Sabourin CL, Niemuth NA, Li H, Semenova VA, Rudge TL, Mayfield HJ, Schiffer J, Mittler RS, Ibegbu CC, Wrammert J, Ahmed R, Brys AM, Hunt RE, Levesque D, Estep JE, Barnewall RE, Robinson DM, Plikaytis BD, Marano N, AVRP Laboratory Working Group 2012. A three-dose intramuscular injection schedule of anthrax vaccine adsorbed generates sustained humoral and cellular immune responses to protective antigen and provides long-term protection against inhalation anthrax in rhesus macaques. Clin. Vaccine Immunol. 19:1730–1745. 10.1128/CVI.00324-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Scola MP, Thompson SD, Brunner HI, Tsoras MK, Witte D, Van Dijk MA, Grom AA, Passo MH, Glass DN. 2002. Interferon-gamma:interleukin-4 ratios and associated type 1 cytokine expression in juvenile rheumatoid arthritis synovial tissue. J. Rheumatol. 29:369–378. [PubMed] [Google Scholar]
- 17. Shirai A, Conover J, Klinman DM. 1995. Increased activation and altered ratio of interferon-gamma:interleukin-4 secreting cells in MRL-lpr/lpr mice. Autoimmunity 21:107–116. 10.3109/08916939508993357. [DOI] [PubMed] [Google Scholar]
- 18. Friedman J, Hastie T, Tibshirani R. 2010. Regularization paths for generalized linear models via coordinate descent. J. Stat. Softw. 33:1–22. 10.1111/j.1467-9868.2005.00503.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zou H, Hastie T. 2005. Regularization and variable selection via the elastic net. J. R. Stat. Soc. 67:301–320. 10.1111/j.1467-9868.2005.00503.x. [DOI] [Google Scholar]
- 20. Waldron L, Pintilie M, Tsao MS, Shepherd FA, Huttenhower C, Jurisica I. 2011. Optimized application of penalized regression methods to diverse genomic data. Bioinformatics 27:3399–3406. 10.1093/bioinformatics/btr591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Genkin A, Lewis DD, Madigan D. 2007. Large-scale Bayesian logistic regression for text categorization. Technometrics 49:291–304. 10.1198/004017007000000245. [DOI] [Google Scholar]
- 22. Hastie T, Tibshirani R, Friedman J. 2009. The elements of statistical learning: data mining, inference, and prediction, 2nd ed. Springer-Verlag, New York, NY. [Google Scholar]
- 23. Heymans MW, van Buuren S, Knol DL, van Mechelen W, de Vet HCW. 2007. Variable selection under multiple imputation using the bootstrap in a prognostic study. BMC Med. Res. Methodol. 7:33. 10.1186/1471-2288-7-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Austin PC, Tu JV. 2004. Bootstrap methods for developing predictive models. Am. Stat. 58:131–137. 10.1198/0003130043277. [DOI] [Google Scholar]
- 25. Venkatraman ES. 2000. A permutation test to compare receiver operating characteristic curves. Biometrics 56:1134–1138. 10.1111/j.0006-341X.2000.01134.x. [DOI] [PubMed] [Google Scholar]
- 26. Robin X, Turck N, Hainard A, Tiberti N, Lisacek F, Sanchez JC, Müller M. 2011. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics 12:77. 10.1186/1471-2105-12-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ionin B, Hopkins RJ, Pleune B, Sivko GS, Reid FM, Clement KH, Rudge TL, Jr, Stark GV, Innes A, Sari S, Guina T, Howard C, Smith J, Swoboda ML, Vert-Wong E, Johnson V, Nabors GS, Skiadopoulos MH. 2013. Evaluation of immunogenicity and efficacy of anthrax vaccine adsorbed for postexposure prophylaxis. Clin. Vaccine Immunol. 20:1016–1026. 10.1128/CVI.00099-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Allison PD. 1999. Multiple regression: a primer. Pine Forge Press, Thousand Oaks, CA. [Google Scholar]
- 29. Belsley DA, Kuh K, Welsch RE. 1980. Regression diagnostics: identifying influential data and sources of collinearity. John Wiley & Sons, New York, NY. [Google Scholar]
- 30. Thorne CB, Belton FC. 1957. An agar-diffusion method for titrating Bacillus anthracis immunizing antigen and its application to a study of antigen production. J. Gen. Microbiol. 17:505–516. 10.1099/00221287-17-2-505. [DOI] [PubMed] [Google Scholar]
- 31. Little SF, Webster WM, Ivins BE, Fellows PF, Norris SL, Andrews GP. 2004. Development of an in vitro-based potency assay for anthrax vaccine. Vaccine 22:2843–2852. 10.1016/j.vaccine.2003.12.027. [DOI] [PubMed] [Google Scholar]
- 32. Pitt ML, Little SF, Ivins BE, Fellows P, Barth J, Hewetson J, Gibbs P, Dertzbaugh M, Friedlander AM. 2001. In vitro correlate of immunity in a rabbit model of inhalational anthrax. Vaccine 19:4768–4773. 10.1016/S0264-410X(01)00234-1. [DOI] [PubMed] [Google Scholar]
- 33. Reuveny A, White MD, Adar YY, Kafri Y, Altboum YZ, Gozes Y, Kobiler D, Shafferman A, Velan B. 2001. Search for correlates of protective immunity conferred by anthrax vaccine. Infect. Immun. 69:2888–2893. 10.1128/IAI.69.5.2888-2893.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Pittman PR, Gibbs PH, Cannon TL, Friedlander AM. 2002. Anthrax vaccine: short-term safety experience in humans. Vaccine 20:972–978. 10.1016/S0264-410X(01)00387-5. [DOI] [PubMed] [Google Scholar]
- 35. Pittman PR, Fisher D, Quinn X, Schmader T, Barrera-Oro JG. 2013. Effect of delayed anthrax vaccine dose on Bacillus anthracis protective antigen IgG response and lethal toxin neutralization activity. Vaccine 31:5009–5014. 10.1016/j.vaccine.2013.08.086. [DOI] [PubMed] [Google Scholar]
- 36. Swets JA. 1988. Measuring the accuracy of diagnostic systems. Science 240:1285–1293. 10.1126/science.3287615. [DOI] [PubMed] [Google Scholar]
- 37. Semenova VA, Schiffer J, Steward-Clark E, Soroka S, Schmidt DS, Brawner MM, Lyde F, Thompson R, Brown N, Foster L, Fox S, Patel N, Freeman AE, Quinn CP. 2012. Validation and long term performance characteristics of a quantitative enzyme linked immunosorbent assay (ELISA) for human anti-PA IgG. J. Immunol. Methods 376:97–107. 10.1016/j.jim.2011.12.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.