Abstract
Synergistic advances in optical physics, probe design, molecular biology, labeling techniques and computational analysis have propelled fluorescence imaging into new realms of spatiotemporal resolution and sensitivity. This review aims to discuss advances in fluorescent probes and live-cell labeling strategies, two areas that remain pivotal for future advances in imaging technology. Fluorescent protein– and bio-orthogonal–based methods for protein and RNA imaging are discussed as well as emerging bioengineering techniques that enable their expression at specific genomic loci (for example, CRISPR and TALENs). Important attributes that contribute to the success of each technique are emphasized, providing a guideline for future advances in dynamic live-cell imaging.
Cells are complex machines that integrate intracellular and extracellular signals through the coupled spatiotemporal dynamics of proteins, lipids, metabolites, nucleic acids and glycans. Multicomponent cellular signaling scaffolds are structured in three dimensions with organization on the nanoscale, and signaling pathways are encoded in frequency- and waveform-specific modes1–3. Furthermore, cells are heterogeneous and exhibit phenotypic plasticity, necessitating longitudinal single-cell analyses. Deciphering how this complex and often interdependent symphony of cellular constituents defines healthy and diseased states and how these dynamics propagate from the cellular to the organismal level is one of the great challenges in modern biology. Today, fewer methods provide greater insight into subcellular spatiotemporal dynamics than noninvasive, real-time, specific, sensitive and multiplexed molecular imaging4.
The most widely applied technique for molecular imaging of live cells is the use of fluorescent proteins (FPs) to light up cellular structures such as organelles or biomolecules such as proteins. To identify and track biomolecules in the complex environment of the cell, molecular specificity is essential. FPs generate a fluorescent moiety autocatalytically and, when genetically fused to a protein of interest, offer exquisite labeling specificity. FP fusions can be expressed ectopically, virally and, through recent advances in genome engineering, endogenously under the protein’s native promoter (for example, TALENs and CRISPR)5,6, facilitating long-term imaging throughout organismal development with sensitivity that routinely reaches single molecules. Extensive protein engineering efforts coupled with a focus on the discovery of new FPs have resulted in a powerful palette of fluorescent probes. One of the intriguing things about this field is that engineering efforts not only have been highly successful at targeting some properties, such as brightness, but also have revealed complexity in photophysical properties (for example, photoswitching, kindling and dark-state conversion) that are often confounding. Though these properties may be exploited for specialized microscopy applications, for traditional imaging, they often limit photon output. Such efforts emphasize the need to better understand the photophysical properties of FPs and how such properties influence imaging applications.
Fueled by the obvious benefits that FPs provide for cellular imaging, there has also been a focus on developing methods for labeling biomolecules with small-molecule probes, enabling greater labeling sophistication, and for extending fluorescent tagging to more diverse biomolecules, such as RNA. One such effort includes bio-orthogonal labeling, which is the use of diverse methodology for labeling cellular constituents in vitro and in vivo with unique chemical probes (for example, fluorophores, cross-linking reagents, biotin and so on). Such chemistry must be compatible with the cellular milieu, and fluorophores must be bright and photostable as well as nontoxic and permeable across cellular and organellar membranes. To eliminate nonspecific background, fluorophores should preferably be nonfluorescent (for example, via photoinduced electron transfer, pKa modulation, static quenching and so on) until labeling has occurred and remain nonfluorescent in low-dielectric environments (for example, biological membranes)7. Alternatively, unlabeled fluorophores, if sufficiently permeable, may be removed through iterative rinsing to achieve high imaging contrast. Importantly, the modular nature of many bio-orthogonal labeling systems enables substitution or alteration of the chemical probe, permitting biological questions to be addressed with the molecular diversity provided by modern organic synthesis.
In this review, we provide an overview of two major strategies for tagging biomolecules with fluorescent probes. We briefly describe the demands that microscopy techniques place upon fluorescent probes and emphasize recent developments in both FP engineering and bio-orthogonal labeling to meet these demands. There are a number of important characteristics that all probes must have. In addition to being compatible with the cellular milieu, nontoxic and minimally perturbing, probes need to provide sufficient contrast between the object of interest and background noise. Yet contrast in fluorescence imaging is often influenced by environmental (pH, quenchers, redox agents and O2 concentration) and experimental (excitation wavelength, intensity, duty-cycle and presence of unbound probe) conditions, necessitating careful attention to detail when selecting a probe for a given application8. Additional properties that influence how a probe might be used include photophysical processes such as intersystem crossing, reactive-oxygen sensitization, dark states, quenching and photochromism9. Given the advances in optical imaging (p. 524), improvements to chemical probes and labeling strategies should make possible many of the longstanding goals in fluorescence microscopy (Box 1). Each imaging technique not only provides distinct information content but also places unique demands upon the fluorescent probe (Box 2). Nevertheless, integration of different imaging technologies provides a more complete analysis of biological phenomena.
Box 1 | Longstanding goals of fluorescence microscopy.
Three-dimensional imaging with high spatial resolution
High temporal resolution (<10−3 s)
Low phototoxicity for prolonged imaging (>106 s) of single cells and biological tissues
Single-molecule sensitivity
Simultaneous observation of multiple molecular targets
Nonperturbing labeling strategies
Imaging of proteins under control of native genomic promoters.
Box 2 | Emerging microscopy techniques and fluorescent probe demands.
Wide-field microscopy: Diffraction-limited lateral resolution (~250 nm) and diminished contrast due to illumination and detection of probes above and below the optical focus. Fluorescent probes should be bright and photostable.
Laser-scanning and spinning-disk confocal microscopy: Improved axial resolution (~800 nm) due to rejection of out-of-focus fluorescence emission. Moderately intense and periodic illumination increases phototoxicity and requires particularly photostable bright probes.
Total internal reflection fluorescence microscopy (TIRF): An exponentially decaying evanescent wave propagates ~200 nm into the sample, enabling single-molecule imaging/tracking. Probes should be bright, photostable and resistant to ‘dark-state’ photoconversion for prolonged particle tracking.
Structured illumination: Wide-field technique that computationally recombines images with varying illumination patterns, eliminating out-of-focus blur and improving resolution approximately twofold. Given the increased image acquisition necessary, probes need to be especially bright to increase image acquisition frequency and be resistant to photobleaching.
Super-resolution optical fluctuation imaging: Wide-field technique that uses higher-order spatiotemporal statistical analysis to improve resolution approximately threefold. Fluorescent probes should rapidly fluctuate between emitting and nonemitting states.
PALM and STORM: Sparse single-molecule point-spread functions are analyzed to provide statistical determination of molecule location. PALM and STORM require photoactivatable and photoswitchable probes, respectively, and both methods require bright fluorophores with high contrast between their on and off states.
Stimulated emission depletion (STED) and reversibly saturable optical fluorescence transitions (RESOLFT): Fluorophores are deterministically switched between emitting and nonemitting states, and illumination intensity necessary depends upon the kinetics of the molecular states. Fluorescent probes must be particularly bright for high-resolution imaging, photostable and have robust optical transitions with high contrast between on and off states.
Selective plane illumination microscopy (SPIM): A multiobjective illumination/detection geometry that provides beam-waist limited axial resolution yet diffraction-limited lateral resolution. Fluorescent probes should be bright and photostable.
Genetically encoded FPs
FPs are arguably the most widely used probes in cell biology. The ever-increasing number of FPs with a dizzying array of properties can make it difficult for the cell biologist to identify the best FP for a given application. As has been suggested in numerous other reviews, the key is to pair the FP to the particular application. Core properties to consider are (i) spectral characteristics (i.e., wavelength of excitation and emission); (ii) the brightness and contrast that can be obtained over background fluorescence; (iii) photostability, particularly for long-term time lapse imaging; (iv) thermodynamic stability and residual tendency to oligomerize; and (v) application-tailored FPs with specialized photophysical properties. As microscopy becomes more sophisticated, new FPs with tailored properties continue to be developed, such as reversibly photoswitchable FPs for stochastic optical reconstruction microscopy (STORM) and reversibly saturable optical fluorescence transitions (RESOLFT) imaging, photoactivatable FPs for photoactivation-location microscopy (PALM), rapidly blinking FPs for superresolution optical fluctuation imaging (SOFI) and near-infrared–emitting FPs for in vivo imaging. Furthermore, FPs from unrelated organisms have been developed that rely on sequestration of endogenous cofactors (for example, flavin mononucleotide, biliverdin and bilirubin), expanding the spectral and chemical properties available for FP engineering10,11. Although we still do not have a complete mechanistic understanding of how photophysical properties are tuned by molecular structure, some insights have emerged from recent studies, and these will be highlighted below. In this review, we summarize recent advances in FP engineering based on the categories highlighted above. Select citations are provided in the text, and a comprehensive treatment of the citations can be found in several excellent reviews12,13.
Spectral characteristics of FPs
The chemical composition of the chromophore has an important role in tuning the spectral attributes of the FP (Fig. 1). The chromophore-forming tripeptide can tolerate substitution within the first two positions but not the third because of its role in backbone cyclization. The principle determinant of the excitation and emission wavelengths is the second amino acid in the tripeptide, which in the GFP context contains phenylalanine in UV-excitable FPs14, histidine in blue FPs15, tryptophan in cyan FPs16 and tyrosine in green and yellow FPs17. Elongation of the chromophore conjugation through formation of an acylimine moiety results in red fluorescence (for example, mRuby2)18, and intramolecular cyclization and reduction of the acylimine carbonyl results in orange fluorescence (for example, mOrange2)19. Formation of an acylimine without oxidation of the tyrosine β-carbon results in a UV-excitable blue-fluorescent mutant (for example, TagBFP2)20.
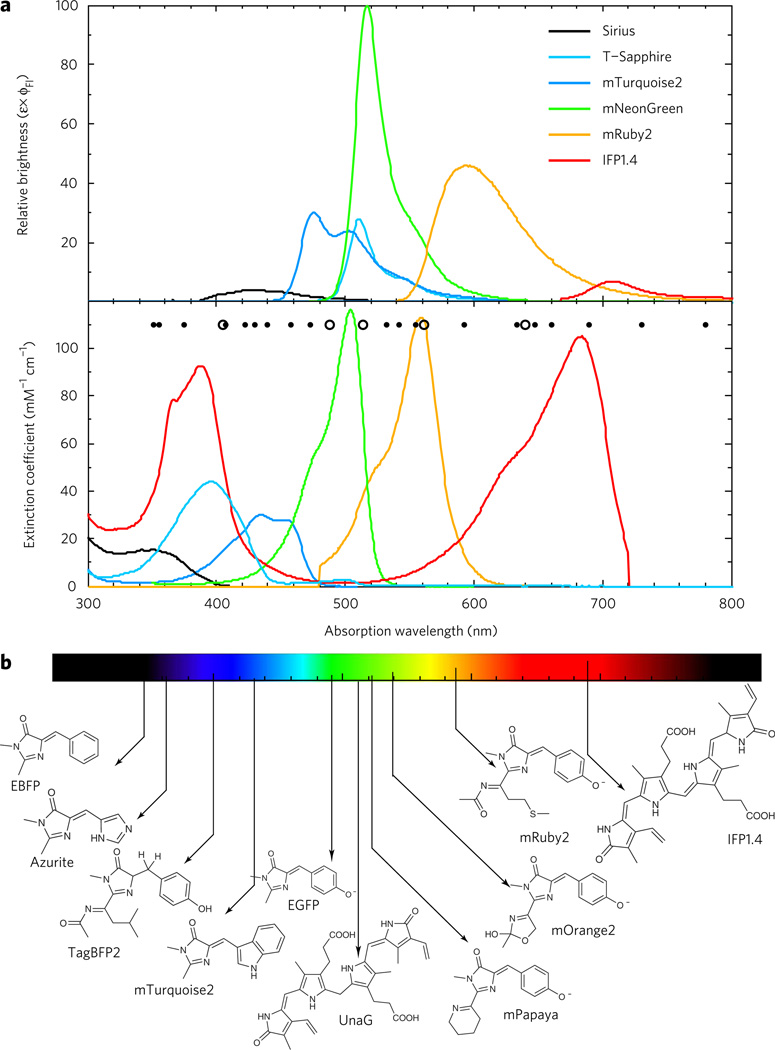
Figure 1. Spectral properties of chromophore classes found in FPs.
(a) Top, fluorescence emission spectra represented in terms of brightness (ε × ϕFl), normalized to the brightest FP available (mNeonGreen). Bottom, absorption spectra of select FPs. Minor dots represent a cross section of solid-state laser sources that are commercially available and major dots are those typically equipped on microscopy equipment. Colors approximate the emission wavelength of the FP. (b) Chromophores and cofactors responsible for fluorescence in select constitutive FPs shown.
FPs with specialized spectral properties include those with large Stokes shifts (>100 nm). Such FPs can be particularly useful for spectral multiplexing, where a single excitation wavelength can excite multiple FPs. For example, LSSmOrange, T-Sapphire and mTagBFP2 can be imaged with a single excitation laser (for example, 405 nm). Additionally, four-color fluorescence cross-correlation spectroscopy using mTagBFP2, T-Sapphire, LSSmKate1 and LSSmOrange could potentially be used for the detection of transient macromolecular complex formation within cells21. The most useful variants include T-Sapphire (ε = 44,000 M−1 cm−1, ϕQY = 0.6, AEx/Em = 399 nm/511 nm) and LSSmOrange (ε = 52,000 M−1 cm−1, ϕQY = 0.45, AEx/Em = 437 nm/572 nm)21. The molecular mechanism leading to the long Stokes shifts involves excitation of a protonated ground-state chromophore that subsequently undergoes excited-state proton transfer.
Brightness and contrast
The overall brightness of an FP is often one of the most important criteria for high-quality imaging. EGFP has long been the gold standard to which other proteins are compared, and for many years other color variants fell far short of the EGFP mark. Recent advances have altered the playing field as molecular evolution efforts have resulted in a new gold standard that supercedes EGFP, and improvements in the blue, cyan and red FP variants have made them more viable candidates. Table 1 summarizes the FP performance leaders for each field, and below we highlight notable recent additions to the FP palette.
Table 1.
Photophysical properties of some leading FPs.
| FP | ε (M−1 cm−1) | ϕFl | Brightness (nm)a | λPA/PS (nm)b | λEx (nm)b | λEm (nm)b | pKAc |
|---|---|---|---|---|---|---|---|
| Constitutively fluorescent | |||||||
| Sirius | 15,000 | 0.24 | 4 | – | 355 | 424 | <3.0 |
| TagBFP2 | 50,600 | 0.64 | 35 | – | 399 | 454 | 2.7 |
| mCerulean3 | 40,000 | 0.87 | 34 | – | 434 | 474 | 3.2 |
| mTurquoise2 | 30,000 | 0.93 | 30 | – | 434 | 474 | 4.5 |
| ECFP | 32,500 | 0.4 | 14 | – | 434 | 477 | 4.7 |
| EGFP | 55,000 | 0.6 | 36 | – | 489 | 509 | 5.9 |
| T-Sapphire | 44,000 | 0.6 | 28 | – | 399 | 511 | 4.9 |
| Clover | 111,000 | 0.76 | 91 | – | 505 | 515 | 6.2 |
| mNeonGreen | 116,000 | 0.8 | 100 | – | 506 | 517 | 5.7 |
| UnaG | 77,300 | 0.51 | 42 | – | 498 | 527 | <4 |
| Venus | 92,200 | 0.57 | 57 | – | 515 | 528 | 6 |
| Citrine | 77,000 | 0.76 | 63 | – | 516 | 529 | 5.7 |
| mPapaya1 | 43,000 | 0.83 | 38 | – | 530 | 541 | 6.8 |
| mKO2 | 63,800 | 0.57 | 39 | – | 551 | 565 | 5.5 |
| mOrange2 | 58,000 | 0.6 | 38 | – | 549 | 565 | 6.5 |
| LSSmOrange | 52,000 | 0.45 | 25 | – | 437 | 572 | 5.7 |
| TagRFP-T | 81,000 | 0.41 | 36 | – | 555 | 584 | 4.6 |
| mRuby2 | 113,000 | 0.38 | 46 | – | 559 | 600 | 5.3 |
| FusionRed | 94,500 | 0.19 | 19 | – | 580 | 608 | 4.6 |
| mCherry | 72,000 | 0.22 | 17 | – | 587 | 610 | <4.5 |
| LSSmKate1 | 31,200 | 0.08 | 3 | – | 463 | 624 | 3.2 |
| mKate2 | 62,500 | 0.4 | 27 | – | 588 | 633 | 5.4 |
| mCardinal2 | 87,000 | 0.19 | 18 | – | 604 | 659 | 5.3 |
| IFP1.4 | 90,000 | 0.07 | 7 | – | 685 | 708 | 4.6 |
| iRFP | 105,000 | 0.059 | 7 | – | 692 | 713 | 4 |
| Irreversibily photoactivatable | |||||||
| PA-GFP | 17,400 | 0.79 | 15 | 405 | 504 | 517 | 4.5 |
| PAmCherry1 | 18,000 | 0.46 | 9 | 405 | 564 | 595 | 6.3 |
| PATagRFP | 66,000 | 0.38 | 27 | 405 | 562 | 595 | 5.3 |
| Irreversibly photoswitchable | |||||||
| PS-CFP2 | 47,000 | 0.23 | 12 | 405 | 490 | 511 | 6.1 |
| mGEos-M | 51,609 | 0.85 | 47 | 405–488 | 503 | 514 | 4.5 |
| Dendra2 | 35,000 | 0.55 | 21 | 405–488 | 553 | 573 | 7.5 |
| mEos3.2 | 32,200 | 0.55 | 19 | 405 | 572 | 580 | 5.8 |
| Kaede | 60,400 | 0.33 | 21 | 405 | 572 | 580 | 5.6 |
| tdEos | 33,000 | 0.6 | 21 | 405 | 569 | 581 | 5.7 |
| mKikGR | 28,000 | 0.63 | 19 | 405 | 580 | 591 | 6.6 |
| Reversibly photoswitchable | |||||||
| rsEGFP2 | 61,000 | 0.3 | 20 | 405–488 | 478 | 503 | 4 |
| mIrisFP | 47,000 | 0.54 | 27 | 405–488 | 486 | 516 | 5.7 |
| Dronpa | 95,000 | 0.85 | 87 | 405 | 503 | 518 | 5 |
| rsFastLime | 39,094 | 0.77 | 32 | 405–488 | 496 | 518 | – |
| Dreiklang | 83,000 | 0.41 | 37 | 365–405 | 511 | 529 | 7.2 |
To assist in comparison of different FPs, brightness is normalized to the brightest FP (mNeonGreen). The molar extinction coefficient ε indicates the absorptivity of the FP and is solely provided for the absorption maxima. The fluorescence quantum yield, ϕFl, or the ratio of emitted photons over absorbed photons, is indicative of the fluorescence efficiency. Brightness is the product of the extinction coefficient and the fluorescence quantum yield.
The excitation (λEx) and emission (λEm) wavelengths are constant for constitutively fluorescent FPs and, in the case of photomodulatable FPs, are provided only for their final fluorescent state following illumination with the activation wavelength (λPA/PS).
The apparent pKA of fluorescence is provided and refers to the pH at which half of the fluorescence remains.
The brightest BFP is TagBFP2. Relative to its precursor, TagBFP, it shows resistance to acylimine hydrolysis and improved brightness. TagBFP2 has been used as a near-UV excitable fusion protein, as a FRET donor for green-emitting FPs and for spectrally multiplexed fluorescence cross-correlation spectroscopy measurements. Cyan FPs are often used as a FRET partner with yellow FPs but have long suffered from low brightness. The recently developed mTurquoise2 is twice as bright as ECFP (ε = 30,000 M−1 cm−1, ϕQY = 0.93)16, with nearly identical spectral properties, making it an attractive replacement for ECFP in FRET sensors. mTurquoise2 was developed using a fluorescence lifetime screen, which provides information on the radiative and non-radiative excited state decay rates, as well as through X-ray crystallography and molecular modeling to identify conformationally dynamic amino acid side chains. Notably, mTurquoise2 is photostable and exhibits monoexponential fluorescence decay, facilitating use in quantitative fluorescence lifetime imaging. The improvements in photophysical performance may be attributed to global rigidification of the chromophore, increased chromophore planarity and improved inter-β-strand hydrogen bonding16. Recently engineered green variants now substantially surpass EGFP in brightness, making them the new FP gold standard. Clover (ε = 111,000 M−1 cm−1 ϕQY = 0.76) and mNeonGreen (ε = 116,000 M−1 cm−1, ϕQY = 0.80), derived from Aequoria victoria and Branchiostoma lanceolatum, respectively, are ~2.5-fold brighter than EGFP17,18. Clover, evolved specifically for FRET analysis with mRuby2 (ε = 113,000 M−1 cm−1, ϕQY = 0.38), an improved red FP, has been used to image kinase activity, membrane voltage and GTPase activity18. The mechanisms by which mNeonGreen and Clover achieve such large extinction coefficients relative to spectrally and chemically similar FPs remain unknown. The class of monomeric RFPs has consistently suffered from diminished brightness, but the recent development of mRuby2, which is 2.7 times brighter than mCherry, is a much-needed advance. Although the molecular origin of the improved extinction coefficient is unknown, deeper biophysical studies on this variant might provide insight to inform future protein engineering efforts.
FP photostability
As imaging is performed more rapidly on low-abundance molecular targets and for extended periods of time, the photostability of an FP is an increasingly important parameter in FP performance. However, photostability is highly context dependent and depends upon the excitation intensity, excitation source (laser or lamp), excitation duty-cycle (laser-scanning or wide-field) and excitation duration (microseconds in confocal, tens to hundreds of milliseconds in wide-field). Additionally, FPs convert to dark states, including the triplet and isomerized states, which can be either photoprotective or photoreactive8. These experimental difficulties, as well as differences in analysis, render interlaboratory comparisons of photostability challenging. Nevertheless, some progress has been made. Using photobleaching as the sole selection criterion, mOrange2 and TagRFP-T were identified and found to be more photostable than the respective parent proteins (mOrange and TagRFP) under both wide-field and confocal illumination. Likewise, microfluidic platforms have been developed for rapidly measuring the rate of irreversible photobleaching and subsequently sorting single mammalian cells22,23. Mechanistically, photobleaching in FPs may result in part from the low dielectric environment of the protein interior (~4) and large electric fields (10–100 MV cm−1). Strong electric fields could promote photoinduced ionization of the chromophore24, and electron transfer is a principal pathway for photobleaching in small-molecule fluorophores25. Furthermore, electron transfer has been evoked to explain decarboxylation in PA-GFP and DsRed26 as well as conjugation disruption in IrisFP following X-ray illumination27. An improved understanding of how mutations modulate electron transfer in FPs and how to suppress them could provide a desirable increase in FP photon output.
The immediate vicinity surrounding the chromophore also alters FP photophysics. In the absence of protein matrix, the chromophore methylene bridge undergoes rapid cis-trans isomerization and is thus nonfluorescent. Dynamic hydrogen bonding between the protein matrix and the chromophore can stabilize the excited state, resulting in increased Stokes shifts28. With ideal chromophore packing and rigidification, through both Van der Waals and electrostatic interactions, FPs may achieve near-unity fluorescence quantum yields16. Furthermore, the local electric field within the protein interior seems to tune the excitation wavelength of RFPs by the quadratic Stark effect, and misalignment of the electric field with the transition dipole moment may explain their diminished two-photon excitation cross-sections29.
Thermodynamic stability and tendency to oligomerize
In addition to spectral characteristics, overall brightness and photostability, other considerations may be important for guiding FP selection. For example, improved thermodynamic stability seems to permit labeling of a greater repertoire of proteins and improves tolerance in chemically challenging environments (for example, the secretory pathway). In addition, the tendency of FPs to form dimers, tetramers or other aggregates is problematic when the FP is to be fused to another protein. Although major engineering efforts enabled the development of monomeric variants, the experience of many researchers is that some FPs retain a residual tendency to oligomerize30. This has been particularly problematic for the RFPs. The recent development of FusionRed (ε = 94,500 M−1 cm−1, ϕQY = 0.19), which was specifically engineered to be tolerant of a wide range of fusion proteins, is an important step in the right direction, but the brightness of FusionRed is substantially lower than that of mRuby2 (ref. 31).
Application-tailored FPs
In recent years, there has been extensive development of FPs with specialized photophysical properties that can be exploited in advanced imaging techniques (Box 2). Multiple classes of photomodulatable FPs exist. Irreversibly photoactivatable FPs are nonfluorescent until activated with high-energy light (for example, 405 nm) and, following photoactivation, are constitutively fluorescent until irreversible photobleaching of the chromophore. Today, PAGFP and PAmCherry are frequently used in super-resolution imaging, and both form their ‘bright’ state through photoinduced radical reactions and intramolecular decarboxylation32. Irreversibly photoswitchable FPs switch fluorescence from one wavelength to another upon excitation with high-energy light. The most commonly used photoswitchable FPs include PS-CFP2, mEos3, tdEos, Kaede and Dendra2 (ref. 13). A recent addition to the irreversibly photoswitchable FPs is PSmOrange, which was developed to photoconvert to a far-red FP with an excitation maximum that coincides nicely with HeNe lasers (λEx/Em = 636 nm/662 nm). A common mechanism for irreversible photoswitching involves a histidine-containing chromophore tripeptide, which, when excited with ultraviolet light, triggers β-elimination of the protein backbone and glutamate decarboxylation. Both photoactivatable and irreversibly photoswitchable FPs highlight the prevalence of electron transfer in FPs, generating a functional chromophore species as opposed to irreversibly photobleaching. Such probes have been instrumental in super-resolution imaging and pulse-chase experiments and for selectively labeling subpopulations of proteins.
The final class of photomodulatable FPs are reversibly photoswitchable. These FPs rapidly convert to a nonfluorescent state upon excitation. Equilibration back to a bright state can occur spontaneously or can be accelerated via excitation with high-energy laser illumination. Examples include Dronpa, Dreiklang, rsFastLime, rsGFP and mIrisFP12,13. Important attributes include high contrast between the two molecular states, large extinction coefficients and fluorescence quantum yields in the bright state; high photon output before photobleaching; and the ability to withstand a large number of photoswitching cycles. As with most imaging experiments, no single photomodulatable FP is ideal for every application (single-particle tracking, super-resolution structure determination, stoichiometry determination and so on), and careful analysis will be necessary to determine which probe is best for the experiment at hand13.
The identification and subsequent engineering of noncanonical FPs that rely upon sequestration of endogenous cofactors was instrumental in extending the wavelength range of FPs into the infrared. For example, an engineered biliverdin IXα-based bacterial phytochrome named IFP1.4 has excitation and emission maxima of 684 nm and 708 nm, respectively11. Later variants had improved dissociation constants, eliminating the need for exogenous biliverdin addition, and could be engineered to undergo photoactivation for optical lock-in imaging as well as bimolecular fluorescence complementation33–35. Near-infrared FPs (IRFPs) offer many opportunities for further development; in particular, efforts should be focused on improving their fluorescence quantum yield (ϕQY ~ 0.07) and photostability, reducing oligomerization and increasing tolerance of genetic fusions. Another cofactor FP named UnaG was recently discovered in the vertebrate Anguilla japonica that sequesters bilirubin and becomes brightly green fluorescent (ε = 77,300 M−1 cm−1, ϕQY = 0.51)36. Addition of bilirubin to apoUnaG results in immediate and titratable fluorescence and can be used for the determination of bilirubin concentrations in human serum and diagnosis of kernicterus and jaundice36. As UnaG and biliverdin-based IRFPs do not rely upon O2 for chromophore formation (unlike canonical FPs), they provide an opportunity to image obligate and facultative anaerobes. Another cofactor FP, miniSOG, binds flavin mononucleotide and upon photoexcitation rapidly generates singlet oxygen (1O2, ϕROS = 0.47) and has been used for genetically encoded diaminobenzidine oxidation and EM contrast formation10.
Bio-orthogonal methods for protein labeling
Methods of bio-orthogonal labeling of proteins
A growing number of methods for labeling biomolecules with fluorescent probes and other chemical handles for imaging have been reported in the literature. In this review we focus on techniques that are currently capable of labeling intracellular proteins or RNA. Thus, we do not cover the many reported systems that remain poorly compatible with live-cell labeling of intracellular targets. One of the distinct advantages of bio-orthogonal labeling is the freedom and flexibility to use a wide repertoire of small-molecule probes with diverse properties as well as the ability to couple innovative probes such as indicators and sensors to proteins of interest. A requirement of these probes is that they must be cell permeable or be delivered by microinjection into a cell. Labeling in situ can be covalent (chemical and enzymatic) or noncovalent (binding equilibria). Covalent methods can provide improved imaging contrast, particularly when the unconjugated fluorophore is nonfluorescent or can be readily washed out of the cell. Covalent methods are also compatible with pulse-chase studies, permitting analysis of intracellular trafficking, degradation and biogenesis. However, noncovalent labeling methods also offer some unique advantages. If the dissociation rate (koff) is not negligible, recycling of the fluorophore decreases the apparent rate of photobleaching and thus may enable longer-term imaging. Additionally, through careful control of the ligand concentration relative to the KD and optimized (kon) and (koff) rates, binding-induced ‘blinking’ of the fluorophore could be used for super-resolution imaging.
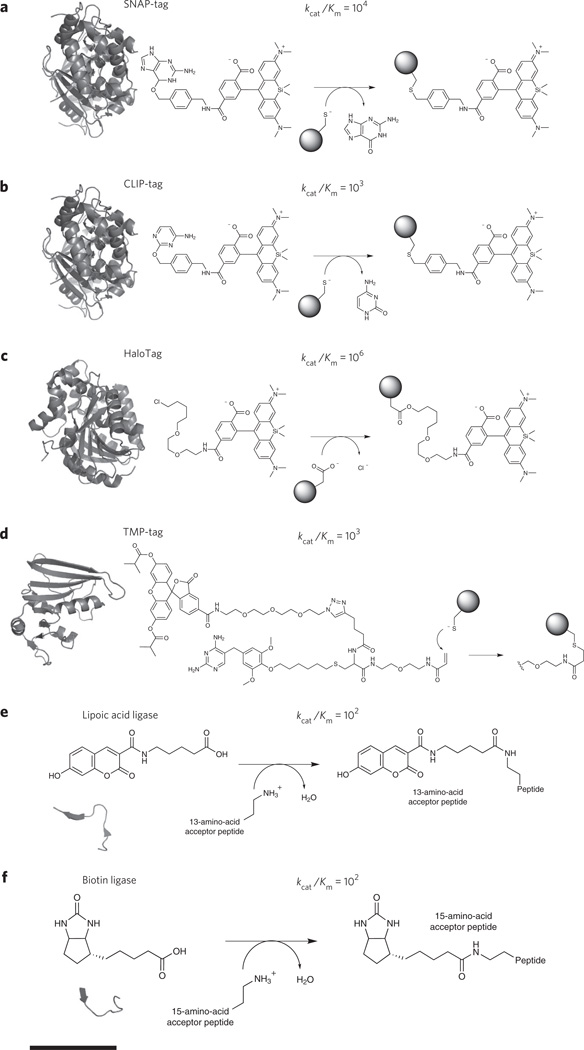
Broadly, there are two types of enzymatic methods for covalent labeling of proteins: those that undergo self-labeling via reaction with a suicide inhibitor and those that recognize and transfer a fluorescent label to a peptide motif that is appended to a protein of interest. For enzymatic labeling reactions, the kinetics should be fast (kcat/KM >106) under physiological conditions, with a KM sensitive enough to label low-abundance targets, and labeling should be stoichiometric and occur to near completion. The fluorescent probe should either be nonfluorescent when unligated, or the unreacted probe should be readily washed out of cells. Self-labeling technologies include SNAP-tag, CLIP-tag, HaloTag and TMP-Tag7,37–39. Labeling with SNAP-tag is performed by fusing the protein of interest to O6-alkylguanine transferase (AGT; Fig. 2a). Incubation of benzylguanine-tethered fluorophores to SNAP-tag–expressing cells results in site-specific AGT cysteine alkylation, covalently tethering the fluorophore to the AGT fusion protein. CLIP-tag is another AGT variant that self-labels via reactions with fluorescent derivatives of O2-benzylcytosine (Fig. 2b)38. Despite the similarities between SNAP-tag and CLIP-tag, both methods offer substrate specificity and can thus be used simultaneously to image multiple molecular species38. AGT has been engineered to eliminate native DNA binding and improve the kinetics and thermodynamics of its reactivity. HaloTag, derived from an engineered variant of haloalkane dehalogenase, reacts with fluorophore-conjugated chloroalkanes (Fig. 2c)37. Each system offers rapid protein labeling in vivo and in vitro (103–106 M−1 s−1). Fluorophores in these systems are typically fluorescent throughout the labeling procedure. Consequently, thorough removal of unlabeled probes is necessary to achieve high imaging contrast. Nevertheless, substrates with excellent photophysical properties and cell permeability are widely available, eliminating the need for expertise in organic chemistry. A hybrid system, referred to as TMP-tag, involves fusion of a protein of interest to Escherichia coli dihydrofolate reductase (molecular weight ~18 kDa). Binding of trimethroprim (TMP) is preferentially, but not exclusively, limited to prokaryotic dihydrofolate reductase and triggers a proximity-induced reaction between a cysteine and an acrylimide-TMP-fluorophore conjugate (Fig. 2d)39.
Figure 2. Covalent bio-orthogonal labeling mechanisms.
Each panel provides the approximate size of the fusion protein, shown as a crystal structure. Chemical reactions have been enlarged for clarity, and all of the rates are measured in M−1 s−1. For all panels, scale bar (at bottom) represents 1.5 nm. (a) Genetic fusion of SNAP-tag to a protein of interest and subsequent suicide inhibition with fluorescent O6-benzylguanine derivatives results in a fluorescently labeled protein (Protein Data Bank (PDB) code 3KYZ). The fluorophore shown is SiR-SNAP, a silicon-based near-infrared fluorophore. (b) In the same way as SNAP-tag, CLIP-tag reacts with fluorescent O2-benzylcytosine derivatives (PDB code 3KYZ). (c) HaloTag fusion proteins react with chlorinated haloalkanes to form an irreversible fluorophore-alkyl-enzyme tether (PDB code 4KAA). (d) TMP-tag binds trimethoprim with nanomolar affinity and accelerates thiol reactivity with an acrylamide appendage (right) through proximity effects, irreversibly labeling the protein of interest (PDB code 1RD7). (e) Lipoic acid ligase, expressed in trans to the protein of interest labeled with a 13-amino-acid peptide, catalyzes the site-specific incorporation of coumarin. (f) Biotin ligase, which is analogous to lipoic acid ligase, catalyzes the site-specific incorporation of biotin or biotin isomeres, which can be chemically modified or used as an antigen for subsequent detection.
SNAP-tag (20 kDa), CLIP-tag (20 kDa), HaloTag (33 kDa) and TMP-Tag (18 kDa) are similar in size to FPs (30 kDa) and therefore do not alleviate concerns that the larger label size could, in principle, lead to perturbation of protein function. However, a wide range of diverse probes have been developed, particularly for the SNAP-tag system, enabling studies that would not be possible with an FP tag. For example, the SNAP-tag has been used to couple small-molecule fluorescent indicators for Ca2+, Zn2+ and H2O2 to proteins of interest and thus, in principle, enables measurement of these signaling species in the immediate vicinity of a protein of interest40–42; the solvatochromic dye Nile Red has been used with the SNAP-tag fusion system to couple this ‘turn-on’ dye to membrane proteins43; and, finally, chemical labeling can be used to measure protein turnover and half-life in cells and animals44.
To address size concerns and potentially allow for labeling at internal sites in a protein structure as opposed to just the N or C termini, enzymatic peptide labeling techniques have been developed in which a small-molecule probe is coupled to a small (~12–15 amino acids, ~1.7 kDa) peptide tag. Such systems include lipoic acid ligase, fluorophore ligase and biotin ligase and comprise three components: an enzyme that must be expressed in the cell, a protein of interest with a genetically encoded engineered peptide sequence and a cell-permeable small-molecule probe. Lipoic acid ligase from E. coli was engineered to ligate a variety of functionalized substrates to a 13-amino-acid peptide. Referred to as probe incorporation mediated by enzymes (PRIME), functionalized substrates were used for two-step labeling via azide-driven ‘click’ chemistry with cyclooctyne-conjugated fluorophores. More recently, a fluorophore ligase was generated, enabling direct incorporation of 7-hydroxycoumarin, 7-aminocoumarin and Pacific Blue (Fig. 2e)45,46. Steric collisions within the active site of lipoic acid ligase seem to necessitate small fluorophores, traditionally limiting imaging to the near-UV. Additionally, incorporation of different fluorophores requires reengineering of enzyme substrate specificity and tolerance. An analogous method using biotin ligase, which recognizes a 15-amino-acid peptide, has been used to label proteins with biotin isomeres functionalized with alkyne and azide chemistry (Fig. 2f)47. Biotin ligase, however, suffers from incorporation of endogneous biotin when expressed intracellularly but is particularly well suited for extracellular labeling via biotin-streptavidin binding or secondary click chemistry–driven labeling. Compared to the aforementioned self-labeling techniques (for example, SNAP-tag), biotin and lipoic acid also suffer from ~104-fold slower enzyme kinetics. Nevertheless, the small size of the peptide tag (<1.7 kDa) and small distance between the label and its target should facilitate improved image interpretation as super-resolution imaging advances toward the molecular scale.
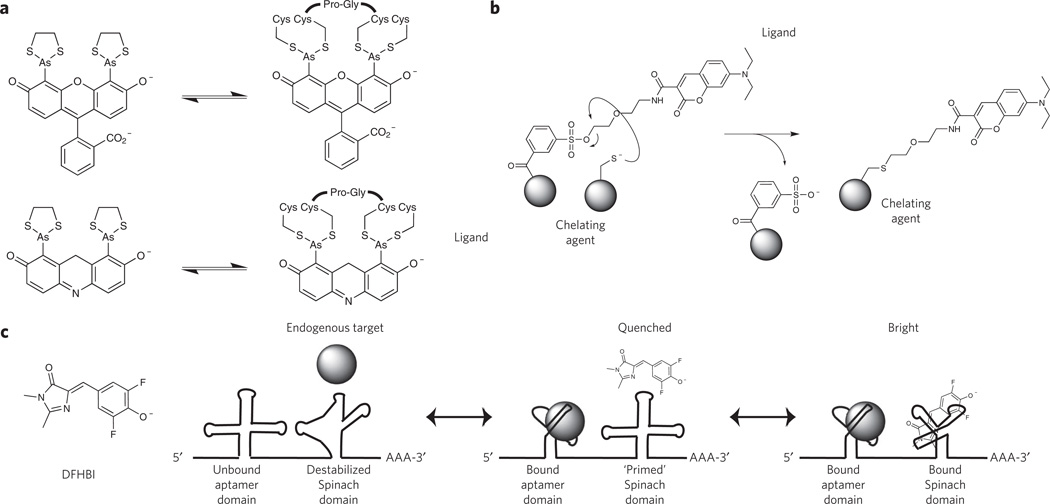
Noncovalent peptide-based methods based on reversible binding of a probe to a peptide target have also been developed for protein tagging. Here, the probe should have a high affinity, i.e., a low KD (for example, <10−8 M), which allows labeling of low-abundance targets. A limitation of this approach is that the probe must be present throughout the experiment and thus should be weakly fluorescent or nonfluorescent in the unbound state to minimize background fluorescence. Ideally, large binding-induced changes in fluorescence intensity (ΔF/F >100) permit visualization of bound probes over freely diffusing probes. Continued presence of the probe also necessitates careful measurements of toxicity and perturbation of cellular processes. FlAsH and ReAsH are respectively biarsenical fluorescein and resorufin derivatives that reversibly bind a tetracysteine peptide motif with high affinity (KD ~10−11 M) and a large increase in fluorescence intensity (ΔF/F ~ 50,000; Fig. 3a)48. These probes are limited to imaging in reducing environments and suffer from diminished contrast with low-abundance targets owing to nonspecific partitioning and fluorescence in biological membranes. Although these probes served as early examples of the power of conjugating chemical fluorophores and fluorescent indicators to proteins of interest, more recent engineering efforts seem to have focused on covalent labeling approaches.
Figure 3. Bio-orthogonal labeling mechanisms based on reversible binding equilibria.
(a) FlAsH (top) and ReAsH (bottom) recognize a specific peptide motif (Cys-Cys-Pro-Gly-Cys-Cys) with high affinity (KD ~4 pM) and undergo a large increase in fluorescence intensity upon binding. (b) Ligand-specific binding by a protein, RNA or generic chelating agent triggers proximity-induced reactivity between a nucleophile located on the chelating group and the reactive tosyl-conjugated fluorophore. (c) A multidomain RNA construct with a target-specific aptamer domain and a target-dependent ′Spinach′ domain. An endogenous protein serves as the ligand for the aptamer domain, and binding of the protein triggers a conformational rearrangement in the Spinach domain, allowing productive binding of DFHBI to yield a fluorescent state. In the absence of the antigen, Spinach cannot bind DFHBI and remains nonfluorescent.
Labeling of proteins with the aforementioned methods requires modification of the protein itself, either through genetic fusions with another protein (for example, FP and SNAP-tag) or a peptide tag (for example, biotin ligase and fluorophore ligase). To enable native protein tracking, several methods have been developed. Initially, ligands that could bind the active site of a specific protein were synthetically coupled to reactive functional groups such that protein-specific binding of the ligand accelerated SN2 reactivity between the functional group and nearby amino acids via proximity effects (similar to TMP-Tag)39. Unfortunately, irreversible conjugation of the ligand near the active site results in constitutive binding, rendering the protein nonfunctional, hence perturbing the cellular system. Although useful for pharmacological inhibition of oncogenic proteins49, this strategy remains nonideal for functional imaging of active protein targets. To overcome this limitation, next-generation affinity labeling techniques use a phenylsulfonate ester moiety that concomitantly releases the ligand upon nucleophilic attack (Fig. 3b)50. This has been shown to chemically label proteins with high specificity in living cells and intact animals50 and was recently used for FRET imaging of rapamycin-induced intermolecular complexation in living cells51. Another recently developed approach used an RNA aptamer to image bacterial protein expression52. In this system, binding of a protein to an RNA domain allosterically modulated binding and fluorescence of a GFP chromophore analog (3,5-difluoro-4-hydroxybenzylidene imidazolinone (DFHBI)) in a secondary ‘Spinach’ aptamer domain (Fig. 3c)53. Given that aptamers have been developed for diverse cellular constituents using systematic evolution of ligands by exponential enrichment (SELEX), this method could be applied to any other cellular target. Nevertheless, Spinach suffers from diminished contrast and will most likely require further development for widespread use (further details below). A final alternative, recently used to detect the formation of transient signaling complexes of internalized G protein–coupled receptors, are camelid-derived single-domain ‘nanobodies’ (molecular weight ~15 kDa) fused to GFP54. Nanobodies have been developed for a variety of targets, can be expressed in eukaryotic cells and are also capable of directly modulating FP photophysics55.
Methods for labeling of RNA
RNA elements are now widely recognized for their ability to modulate specific cellular outcomes well beyond the role of mRNA in coding for proteins. For example, ablation of long noncoding RNA elements alters cellular differentiation and development and can result in embryonic lethality or growth defects56. Furthermore, functional mRNA transcripts are actively sequestered and released from subcellular structures, altering mRNA storage, microRNA-mediated gene silencing and neuronal degeneration57,58. Given these and many other observations, it has become evident that RNA imaging methods are necessary to decipher the spatiotemporal dynamics of coding and noncoding RNA in vivo.
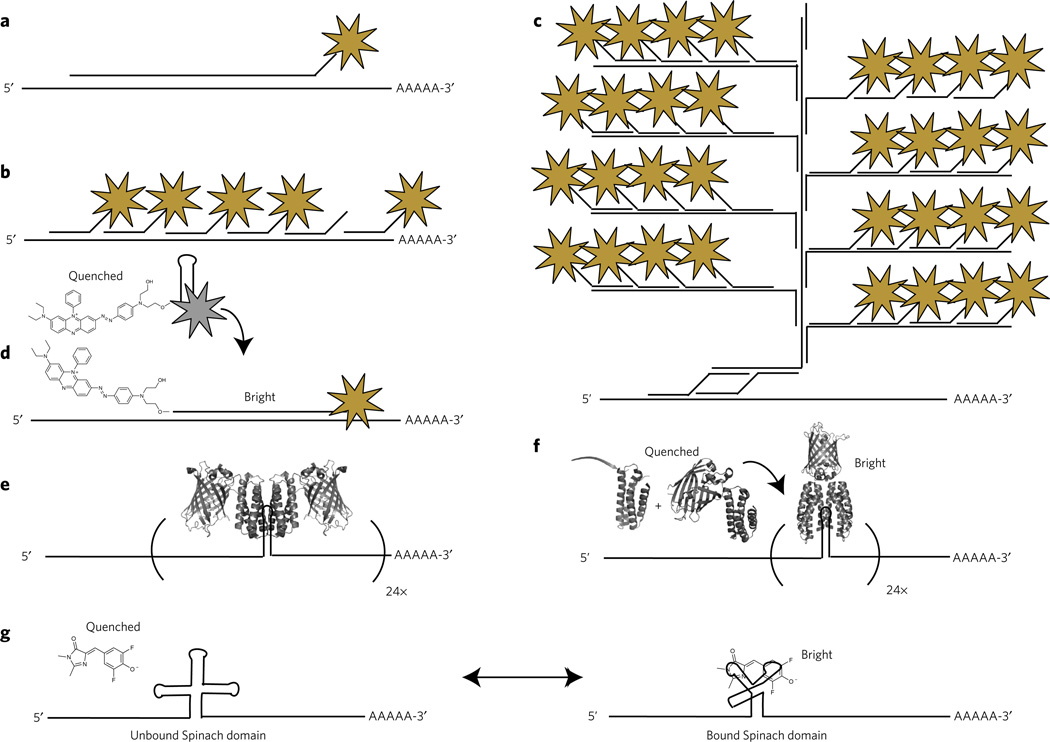
Early efforts to label endogenous RNA in fixed cells, referred to as fluorescence in situ hybridization (FISH), relied upon long antisense RNA or DNA oligonucleotides that could be chemically modified and probed by immunofluorescence (Fig. 4a)59. As DNA synthesis technology improved, antisense oligonucleotides were truncated (~20–50 nucleotides), directly labeled with fluorophores and multiplexed throughout the RNA target (Fig. 4b) and most recently via branched DNA structures that result in thousands of fluorophores per RNA transcript (Fig. 4c). Impressively, branched DNA labeling, coupled with robotics and image analysis, provides single-transcript sensitivity and has been used to analyze transcriptome heterogeneity at the single-cell level60. Although rare, multivalent FISH techniques have also been applied to live-cell imaging with single-transcript sensitivity61. Dense fluorophore labeling is necessary to detect single RNA transcripts over a background of nonhybridized fluorescent oligonucleotides. Alternatively, decreased labeling densities can be achieved with a fluorophore-quencher pair that reduces fluorescence until hybridization (Fig. 4d)62. Although these approaches are powerful means to label endogenous RNA, they require introduction of labeled oligonucleotides into the cellular milieu. Thus, the cells must either be fixed and permeabilized or the probes must be directly microinjected into cells61.
Figure 4. Methods for labeling RNA biomolecules.
(a,b) Initial efforts to label RNAs in vivo were performed on chemically fixed samples and included long antisense DNA and RNA oligonucleotides labeled with a single fluorophore (shown as a star) (a) or multiple shorter oligonucleotides, each labeled with a single fluorophore (b). (c) Branched DNA structures provide increased specificity through the use of two adaptor oligonucleotides that nucleate the formation of a branched and multiply labeled oligonucleotides analogous to DNA origami. (d) A ‘molecular beacon’ fluorophore-quencher conjugate remains nonfluorescent until hybridization with the sequence of interest. (e) For live-cell imaging, 24 tandem repeats of a hairpin structure are inserted into the 3′ untranslated region of a transcript. Two GFP-MS2 (or equivalent orthogonal phage protein) fusion proteins identify each hairpin motif and bind with high affinity, generating a transcript with 48 GFP molecules and enabling routine single-molecule imaging of RNA in live cells. (f) To eliminate nonspecific background, a multicomponent system (for example, Pum-HD) oligomerizes on a unique RNA hairpin structure, triggering bimolecular complementation of GFP. (g) DFHBI, which is a GFP chromophore analog that is nonfluorescent when free in solution, is recognized by a specific RNA aptamer sequence, ′Spinach′, that turns on fluorescence upon binding.
Genetically encoded RNA imaging methods have also been developed, circumventing the challenges involved in transport of fluorescently labeled oligonucleotides across the cellular membrane. The most widely used approach makes use of RNA binding proteins from bacteriophage that recognize specific RNA sequences and structures. The RNA motif can be genetically incorporated onto the 3′ UTR of an RNA of interest and the RNA binding protein fused to an FP. Because of their sequence specificity, bacteriophage-adopted systems can be spectrally multiplexed, and examples of bacteriophage systems include MS2 (ref. 63), boxB64, PP7 (ref. 65) and pumilio homology domain (Pum-HD)66. Owing to a large fluorescence background, many of these systems require tandem repeats of the RNA hairpin motif. For example, to achieve single-transcript sensitivity with MS2, 24 tandem repeats of the 19 nucleotide stem-loop motif must be inserted into 3′ UTR of the transcript, ultimately recruiting 48 MS2-GFP proteins and resulting in a complex >1 MDa (Fig. 4e). To improve contrast and circumvent the dense labeling necessary to achieve single-transcript sensitivity, an alternative approach of using two independently optimized Pum-HD fragments for adjacent RNA sequences, coupled with bimolecular fluorescence complementation, was reported (Fig. 4f). When coupled with total internal reflection microscopy, this method provided sufficient contrast to observe singly labeled mRNA translocation on molecular motors66.
A third approach involves the use of RNA aptamers capable of modulating the fluorescent properties of a small-molecule probe. As early as 1998, an aptamer capable of oxidizing nonfluorescent dihydrotetramethylrosamine to the fluorescent tetramethylrosamine was reported67. More recently, borrowing from the observation that GFP’s chromophore is nonfluorescent until rigidification, an aptamer (referred to as Spinach) that is capable of activating fluorescence in the cell-permeable GFP-analog DFHBI was used to image 5S rRNA in living cells (Fig. 4g)53. Subsequent studies suggested that Spinach is thermodynamically sensitive to RNA context and unfolding at cellular temperatures68. Further mutagenesis identified Spinach2, an aptamer that remained folded in a greater diversity of RNA contexts at physiological temperatures, resulting in a ~3.2-fold increase in fluorescence brightness in vivo68. Other fluorescence-modulating aptamers have been developed, including aptamer-induced disruption of molecular beacon–like fluorescence quenching69,70 and rigidification of malachite green analogs71. Although these methods show promise, several challenges will need to be addressed for widespread usage. As binding is reversible, cells must be continuously bathed in the fluorogenic compound, and cytotoxicity should be closely monitored. Continuous bathing also requires little to no background fluorescence for standard wide-field imaging techniques. Rapid excitation of the fluorescent complexes can result in isomerization, photoinduced unbinding of the fluorogenic probe and diminished signals, although fluorophore recycling is a useful property for minimizing photobleaching72. Lastly, lower dissociation constants (KD) will need to be demonstrated for low-copy RNA detection, and fluorophores with improved photophysics should enable truly single-molecule imaging.
Approaches for incorporation of fluorescent tags in cells
Prior to 2008, fluorescence imaging experiments relied upon ectopic or virally transduced expression of FP-tagged proteins under constitutively active (for example, cytomegalovirus) or chemically inducible promoters (for example, Tet-On). Overexpression must be carefully monitored, and control experiments are necessary to demonstrate that the exogenous expression of a tagged signaling element (for example, a kinase, phosphatase, GTPase and so on) does not perturb network dynamics4. However, genomic labeling of proteins under endogenous promoters eliminates these concerns, providing native, cell type– and cell state–specific expression profiles and dynamics. Fortunately, genome engineering technology has advanced rapidly, and scientists from diverse backgrounds are using these emerging techniques to genetically manipulate cells with high fidelity.
One such genome engineering technology is the type II CRISPR-Cas system, an RNA-guided, dsDNA blunt endonuclease73. Originally evolved in archaea and bacteria as a form of adaptive immunity to foreign DNA epitopes, CRISPR-Cas has been successfully used in the eukaryotic context for genomic knockouts and knock-ins as well as for transcription activation and repression74,75. Genome engineering can be performed through both homologous and nonhomologous end-joining mechanisms. Cas9, the RNA-guided dsDNA endonuclease, can be fluorescently tagged with an FP and, through mutagenesis, converted into a nickase or a nuclease-null variant (nnCas9). The RNA guide (crRNA) targets specific genomic sequences and, when coupled with fluorescently tagged nnCas9, can be used for dynamic DNA imaging in living specimens76. Furthermore, the crRNA can be genetically fused to MS2 tandem repeats, providing yet another interesting mechanism for imaging dsDNA–crRNA–Cas9 triplex formation.
Unlike the CRISPR-Cas system, where DNA specificity is provided by crRNA, Xanthomonas transcription-activator like (TAL) effectors encode specificity through direct interactions between the protein side chains and the dsDNA77. TAL effectors have a repetitive architecture, consisting of 13–28 tandem repeats of an ~34-amino-acid domain. Each domain contains two amino acids, commonly referred to as the repeat variable diresidue, that are mutated to preferentially bind one nucleotide (A, T, C or G)5. Although each domain and its repeat variable diresidue has nucleotide promiscuity, the overall TAL effector specificity remains high because 13–28 sequential nucleotides are recognized by the full-length protein. Like CRISPR-Cas, TAL effectors can be fused to FPs, transcription activators, repressors and potentially epigenetic modifiers. Genome engineering is accomplished by generating two TAL effectors fused to FokI that target DNA sequences ~6–40 base pairs apart. Coincident binding of the TAL effectors enables FokI dimerization, activation and cleavage of the DNA followed by homologous or nonhomologous end-joining DNA repair mechanisms. Other mechanisms for genome engineering have been reported, including zinc-finger nucleases78. Nevertheless, genome engineering requires absolute sequence specificity and cleavage precision, particularly for human applications79. Intriguing possibilities continue to emerge from advances in genetic engineering, and it will be interesting to see how these developments affect not only imaging but also our understanding of transcription regulation, genome dynamics and epigenetics75.
Conclusions and outlook
Fluorescence imaging has steadily progressed throughout the last decade, breaking through experimental and conceptual limitations and shedding light upon previously unobservable biological phenomena1,80. The tools highlighted in this review have been instrumental in these advances. Yet, as the characteristics of probes and our ability to label biomolecules have improved, so too have the complexity of questions the biological community seeks to address. This sets up an iterative cycle, where, as we push the boundaries of what we can measure by fluorescence imaging, we demand probes and technologies with increasingly stringent or specialized properties.
FP discovery and engineering have resulted in a broad color palette of genetically encodable fluorescent probes. Looking forward, we must ask what performance metrics will be necessary for further imaging advances and what are fundamental avenues for improvement of these probes? Here, we highlight three potential directions that could substantially improve FP-based imaging experiments. First, brighter probes will become increasingly important as researchers adopt low-copy expression platforms, such as expression of a tagged protein from its endogenous locus using genome editing technology. The best blue, yellow and red FPs are roughly half as bright as the best green FP, suggesting substantial room for improvement. However, the development of improved systems alone is insufficient unless the scientific community understands mechanistically what governs the improved performance. For example, spectroscopic studies detailing how Clover and mNeonGreen achieve large extinction coefficients and how these properties can be further improved and translated to other FPs could help guide efforts for improving the blue, yellow and red variants. Second, we need a deeper mechanistic understanding of complex photophysical properties such as dark-state conversion and photobleaching. It is clear that when structural and spectroscopic studies are coupled with protein engineering efforts, it is possible to gain molecular insight into factors that influence essential characteristics such as brightness and photoswitching. Although our understanding of some properties (for example, quantum yield) is increasing and engineering efforts targeting these properties have been successful, our understanding of other properties such as dark-state conversion and photobleaching remains very limited24. As the biological community demands increasingly photostable proteins, such as those for single-particle tracking of low-copy proteins, or increasingly sophisticated properties, such as those for modulation of dark-state conversion rates for SOFI, a deeper understanding of the mechanism (or mechanisms) of these processes and how these mechanisms are influenced by the protein structure would substantially aid in engineering efforts. Third, FP engineers should continue to identify and mutate new autofluorescent proteins, particularly those with a noncanonical fold that may provide improved contrast in the blue and near-infrared spectral regions. A bright and photostable FP excitable at 640 nm, with sufficient contrast for subcellular imaging, would be particularly useful. All engineering efforts would be greatly aided by high-throughput selection platforms for the directed evolution of new FP mutants or identification of small-molecule scaffolds with improved photophysics22,23.
Bioorthogonal labeling methods have made great strides in the last decade, enabling labeling of proteins with smaller tags, internal labeling (as opposed to just N- and C-terminal fusions), tagging of proteins with chemical probes for a greater diversity of applications and extension of imaging technology to nonproteinaceous cellular constituents (for example, nucleotides, glycans and metabolites). Some technologies, such as SNAP-tag, are relatively advanced and have seen widespread application, whereas others, such as PRIME, show great promise but have not yet been widely tested. How might these technologies evolve in the future? The protein-based systems (SNAP-tag, CLIP-tag, HaloTag and TMP-Tag) are comparable in size to FPs, so their main advantage over the simpler FP system is their ability to conjugate specialized small molecules, permitting experiments that cannot be done with FP technology. The SNAP-tag system has been increasingly exploited for sophisticated applications, and presents a huge opportunity for further development of these systems. Possible ways to develop these systems include, for example, using them to couple fluorescent probes for cellular analytes, metabolites or signaling molecules so that such species can be tracked in defined locations, such as at the mouth of channels or pores; coupling of chemical probes that are brighter and more photostable than FPs for super-resolution microscopy; and, finally, labeling biomolecules with probes that could be used in multimodal imaging, such as correlative fluorescence and electron microscopy. The peptide-based labeling systems enable attachment of a much smaller tag and thus will be instrumental for labeling tricky proteins that are easily perturbed by the size of the fusion moiety. However, these systems typically only accept a limited number of substrates, and hence efforts to expand the substrates and particularly to develop fluorophore targeting schemes using bright, photostable, red-shifted fluorophores would be valuable.
Looking beyond the requirements of probes themselves, it is interesting to speculate just how far imaging can take us. Given the complexity of intracellular signal dynamics as well as tissue architecture, a commonly cited limitation of microscopy is that it is restricted to simultaneous analysis of only one to four fluorescently labeled targets. Consequently, methods for increasing the number of targets will be necessary to reveal the true complexity of biological systems. Several approaches have been developed to address this concern, including spectral unmixing, spectral deconvolution and spectral painting (for example, Brianbow)81,82. If applied to fixed specimens, iterative labeling and imaging of specific epitopes can increase the dimensionality of the data83–85. For example, in array tomography, thin (~30–100 nm) specimens are dehydrated, preserved and subjected to multiple rounds of immunofluorescence labeling, imaging and antibody removal. In one demonstration, 17 neuronal markers were imaged with single-synapse resolution83, an approach that in theory could be applied to ~100 antigens84. This method is also compatible with EM, providing unique opportunities for three-dimensional correlative imaging with broad-spectrum or genetically encoded (for example, Mini-SOG) EM contrast mechanisms and may be subjected to deconvolution methods that provide sub-diffraction resolution10,86. Likewise, advancements in high-resolution image-based MS could circumvent the diffraction limit of light altogether while also providing images based on ~100 antigens87. Nevertheless, one of the defining features of light microscopy is its compatibility with living specimens, and the biological community would greatly benefit from innovations in live-cell multiplexing. Alternative biocompatible contrast agents with narrow resonances (for example, luminescence, Raman and so on88) and methods for labeling biomolecules with them would be useful and accelerate emerging forms of cellular bioimaging89.
Fueled by advances in camera technology and high-speed optics, microscopists today can collect data at a rate comparable to that of particle physicists90,91. Like other aspects of ‘big data’, this phenomenon is accelerating, and nascent imaging technologies will demand increasingly complex and multidimensional computational analysis to extract biologically meaningful and quantitative information4. Sophisticated image analysis could have a profound effect on our knowledge of the spatiotemporal organization of biology, elucidating molecular stoichiometry92 and subcellular RNA identity93. Furthermore, given that the local microenvironment is a principal determinant of disease94, democratization of biomimetic tissue substrates (for example, tissue on a chip) and improvements to in vivo imaging (for example, adaptive optics and far-red FPs with large two-photon absorption cross-sections) are also necessary. The next decade will probably be as exciting as the last, especially as established imaging technologies become more widely applied to biological processes and new technologies (for example, negative refractive index imaging with metamaterials95 and increasingly sensitive micro- and nanoscale nitrogen-vacancy NMR) continue to emerge96. Improved probes and labeling strategies provide the bedrock for such advances.
Footnotes
Competing financial interests
The authors declare no competing financial interests.
References
- 1. Kanchanawong P, et al. Nanoscale architecture of integrin-based cell adhesions. Nature. 2010;468:580–584. doi: 10.1038/nature09621. An elegant study on the three-dimensional nanostructure of mechanochemical signaling domains using interferometry-based super-resolution imaging.
- 2. Toettcher JE, Weiner OD, Lim WA. Using optogenetics to interrogate the dynamic control of signal transmission by the Ras/Erk module. Cell. 2013;155:1422–1434. doi: 10.1016/j.cell.2013.11.004. This article demonstrates the power of fluorescence imaging coupled with optogenetics and proteomics for interrogation of frequency-dependent signal transduction.
- 3.Hao N, Budnik BA, Gunawardena J, O’Shea EK. Tunable signal processing through modular control of transcription factor translocation. Science. 2013;339:460–464. doi: 10.1126/science.1227299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Machacek M, et al. Coordination of rho GTPase activities during cell protrusion. Nature. 2009;461:99–103. doi: 10.1038/nature08242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boch J, et al. Breaking the code of DNA binding specificity of TAL-type III effectors. Science. 2009;326:1509–1512. doi: 10.1126/science.1178811. [DOI] [PubMed] [Google Scholar]
- 6.Cong L, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lukinavičius G, et al. A near-infrared fluorophore for live-cell super-resolution microscopy of cellular proteins. Nat. Chem. 2013;5:132–139. doi: 10.1038/nchem.1546. A bio-orthogonal–compatible silicon-based small-molecule fluorophore provides excellent contrast in the near-infrared and its use in vivo. This paper demonstrates how optimal probe photophysical properties can enable imaging from live-cell super-resolution to thick and scattering tissue samples.
- 8.Dean KM, et al. Analysis of red-fluorescent proteins provides insight into dark-state conversion and photodegradation. Biophys. J. 2011;101:961–969. doi: 10.1016/j.bpj.2011.06.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stennett EMS, Ciuba MA, Levitus M. Photophysical processes in single molecule organic fluorescent probes. Chem. Soc. Rev. 2014;43:1057–1075. doi: 10.1039/c3cs60211g. [DOI] [PubMed] [Google Scholar]
- 10.Shu X, et al. A genetically encoded tag for correlated light and electron microscopy of intact cells, tissues, and organisms. PLoS Biol. 2011;9:e1001041. doi: 10.1371/journal.pbio.1001041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shu X, et al. Mammalian expression of infrared fluorescent proteins engineered from a bacterial phytochrome. Science. 2009;324:804–807. doi: 10.1126/science.1168683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nienhaus K, Nienhaus GU. Fluorescent proteins for live-cell imaging with super-resolution. Chem. Soc. Rev. 2014;43:1088. doi: 10.1039/c3cs60171d. [DOI] [PubMed] [Google Scholar]
- 13.Sengupta P, van Engelenburg SB, Lippincott-Schwartz J. Superresolution imaging of biological systems using photoactivated localization microscopy. Chem. Rev. 2014;114:3189–3202. doi: 10.1021/cr400614m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tomosugi W, et al. An ultramarine fluorescent protein with increased photostability and pH insensitivity. Nat. Methods. 2009;6:351–353. doi: 10.1038/nmeth.1317. [DOI] [PubMed] [Google Scholar]
- 15.Ai HW, Shaner NC, Cheng Z, Tsien RY, Campbell RE. Exploration of new chromophore structures leads to the identification of improved blue fluorescent proteins. Biochemistry. 2007;46:5904–5910. doi: 10.1021/bi700199g. [DOI] [PubMed] [Google Scholar]
- 16. Goedhart J, et al. Structure-guided evolution of cyan fluorescent proteins towards a quantum yield of 93% Nat. Commun. 2012;3:751. doi: 10.1038/ncomms1738. This article demonstrates how the application of careful selection pressures and sophisticated analysis can continue to improve fluorescent protein performance.
- 17. Shaner NC, et al. A bright monomeric green fluorescent protein derived from branchiostoma lanceolatum. Nat. Methods. 2013;10:407–409. doi: 10.1038/nmeth.2413. A unique computational approach results in the brightest green fluorescent protein yet.
- 18.Lam AJ, et al. Improving FRET dynamic range with bright green and red fluorescent proteins. Nat. Methods. 2012;9:1005–1012. doi: 10.1038/nmeth.2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shaner NC, et al. Improving the photostability of bright monomeric orange and red fluorescent proteins. Nat. Methods. 2008;5:545–551. doi: 10.1038/nmeth.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Subach OM, Cranfill PJ, Davidson MW, Verkhusha VV. An enhanced monomeric blue fluorescent protein with the high chemical stability of the chromophore. PLoS ONE. 2011;6:e28674. doi: 10.1371/journal.pone.0028674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shcherbakova DM, Hink MA, Joosen L, Gadella TW, Verkhusha VV. An orange fluorescent protein with a large stokes shift for single-excitation multicolor FCCS and FRET imaging. J. Am. Chem. Soc. 2012;134:7913–7923. doi: 10.1021/ja3018972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Davis LM, Lubbeck JL, Dean KM, Palmer AE, Jimenez R. Microfluidic cell sorter for use in developing red fluorescent proteins with improved photostability. Lab Chip. 2013;13:2320–2327. doi: 10.1039/c3lc50191d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lubbeck JL, Dean KM, Ma H, Palmer AE, Jimenez R. Microfluidic flow cytometer for quantifying photobleaching of fluorescent proteins in cells. Anal. Chem. 2012;84:3929–3937. doi: 10.1021/ac202825z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vegh RB, et al. Chromophore photoreduction in red fluorescent proteins is responsible for bleaching and phototoxicity. J. Phys. Chem. B. 2014;118:4527–4534. doi: 10.1021/jp500919a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vogelsang J, et al. A reducing and oxidizing system minimizes photobleaching and blinking of fluorescent dyes. Angew. Chem. Int. Ed. Engl. 2008;47:5465–5469. doi: 10.1002/anie.200801518. [DOI] [PubMed] [Google Scholar]
- 26.Habuchi S, et al. Evidence for the isomerization and decarboxylation in the photocon-version of the red fluorescent protein DsRed. J. Am. Chem. Soc. 2005;127:8977–8984. doi: 10.1021/ja047023o. [DOI] [PubMed] [Google Scholar]
- 27.Adam V, et al. Structural basis of X-ray–induced transient photobleaching in a photoactivatable green fluorescent protein. J. Am. Chem. Soc. 2009;131:18063–18065. doi: 10.1021/ja907296v. [DOI] [PubMed] [Google Scholar]
- 28.Konold P, Regmi CK, Chapagain PP, Gerstman BS, Jimenez R. Hydrogen bond flexibility correlates with stokes shift in mPlum variants. J. Phys. Chem. B. 2014;118:2940–2948. doi: 10.1021/jp412371y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Drobizhev M, Tillo S, Makarov NS, Hughes TE, Rebane A. Color hues in red fluorescent proteins are due to internal quadratic stark effect. J. Phys. Chem. B. 2009;113:12860–12864. doi: 10.1021/jp907085p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Costantini LM, Snapp EL. Fluorescent proteins in cellular organelles: serious pitfalls and some solutions. DNA Cell Biol. 2013;32:622–627. doi: 10.1089/dna.2013.2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shemiakina II, et al. A monomeric red fluorescent protein with low cytotoxicity. Nat. Commun. 2012;3:1204. doi: 10.1038/ncomms2208. [DOI] [PubMed] [Google Scholar]
- 32.Subach FV, et al. Photoactivation mechanism of PAmCherry based on crystal structures of the protein in the dark and fluorescent states. Proc. Natl. Acad. Sci. USA. 2009;106:21097–21102. doi: 10.1073/pnas.0909204106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Filonov GS, et al. Bright and stable near-infrared fluorescent protein for in vivo imaging. Nat. Biotechnol. 2011;29:757–761. doi: 10.1038/nbt.1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Piatkevich KD, Subach FV, Verkhusha VV. Far-red light photoactivatable near-infrared fluorescent proteins engineered from a bacterial phytochrome. Nat. Commun. 2013;4:2153. doi: 10.1038/ncomms3153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Filonov GS, Verkhusha VV. A near-infrared BiFC reporter for in vivo imaging of protein-protein interactions. Chem. Biol. 2013;20:1078–1086. doi: 10.1016/j.chembiol.2013.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kumagai A, et al. A bilirubin-inducible fluorescent protein from eel muscle. Cell. 2013;153:1602–1611. doi: 10.1016/j.cell.2013.05.038. [DOI] [PubMed] [Google Scholar]
- 37.Los GV, et al. HaloTag: a novel protein labeling technology for cell imaging and protein analysis. ACS Chem. Biol. 2008;3:373–382. doi: 10.1021/cb800025k. [DOI] [PubMed] [Google Scholar]
- 38.Gautier A, et al. An engineered protein tag for multiprotein labeling in living cells. Chem. Biol. 2008;15:128–136. doi: 10.1016/j.chembiol.2008.01.007. [DOI] [PubMed] [Google Scholar]
- 39.Gallagher SS, Sable JE, Sheetz MP, Cornish VW. An in vivo covalent TMP-tag based on proximity-induced reactivity. ACS Chem. Biol. 2009;4:547–556. doi: 10.1021/cb900062k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tomat E, Nolan EM, Jaworski J, Lippard SJ. Organelle-specific zinc detection using zinpyr-labeled fusion proteins in live cells. J. Am. Chem. Soc. 2008;130:15776–15777. doi: 10.1021/ja806634e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Srikun D, Albers AE, Nam CI, Iavarone AT, Chang CJ. Organelle-targetable fluorescent probes for imaging hydrogen peroxide in living cells via SNAP-tag protein labeling. J. Am. Chem. Soc. 2010;132:4455–4465. doi: 10.1021/ja100117u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bannwarth M, et al. Indo-1 derivatives for local calcium sensing. ACS Chem. Biol. 2009;4:179–190. doi: 10.1021/cb800258g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Prifti E, et al. A fluorogenic probe for SNAP-tagged plasma membrane proteins based on the solvatochromic molecule Nile red. ACS Chem. Biol. 2014;9:606–612. doi: 10.1021/cb400819c. [DOI] [PubMed] [Google Scholar]
- 44.Bojkowska K, et al. Measuring in vivo protein half-life. Chem. Biol. 2011;18:805–815. doi: 10.1016/j.chembiol.2011.03.014. [DOI] [PubMed] [Google Scholar]
- 45. Uttamapinant C, et al. A fluorophore ligase for site-specific protein labeling inside living cells. Proc. Natl. Acad. Sci. USA. 2010;107:10914–10919. doi: 10.1073/pnas.0914067107. This article, using an engineered lipoic acid ligase from E. coli that directly couples 7-hydroxycoumarin to a 13-amino-acid peptide, demonstrates intracellular and minimally perturbative labeling of actin, microtubule associated proteins and synaptic membrane proteins in living cells.
- 46.Liu DS, Phipps WS, Loh KH, Howarth M, Ting AY. Quantum dot targeting with lipoic acid ligase and HaloTag for single-molecule imaging on living cells. ACS Nano. 2012;6:11080–11087. doi: 10.1021/nn304793z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Howarth M, Takao K, Hayashi Y, Ting AY. Targeting quantum dots to surface proteins in living cells with biotin ligase. Proc. Natl. Acad. Sci. USA. 2005;102:7583–7588. doi: 10.1073/pnas.0503125102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Martin BR, Giepmans BNG, Adams SR, Tsien RY. Mammalian cell based optimization of the biarsenical-binding tetracysteine motif for improved fluorescence and affinity. Nat. Biotechnol. 2005;23:1308–1314. doi: 10.1038/nbt1136. [DOI] [PubMed] [Google Scholar]
- 49.Cohen MS, Zhang C, Shokat KM, Taunton J. Structural bioinformatics–based design of selective, irreversible kinase inhibitors. Science. 2005;308:1318–1321. doi: 10.1126/science1108367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tsukiji S, Miyagawa M, Takaoka Y, Tamura T, Hamachi I. Ligand-directed tosyl chemistry for protein labeling in vivo. Nat. Chem. Biol. 2009;5:341–343. doi: 10.1038/nchembio.157. [DOI] [PubMed] [Google Scholar]
- 51.Tamura T, Kioi Y, Miki T, Tsukiji S, Hamachi I. Fluorophore labeling of native FKBP12 by ligand-directed tosyl chemistry allows detection of its molecular interactions in vitro and in living cells. J. Am. Chem. Soc. 2013;135:6782–6785. doi: 10.1021/ja401956b. [DOI] [PubMed] [Google Scholar]
- 52.Song W, Strack RL, Jaffrey SR. Imaging bacterial protein expression using genetically encoded RNA sensors. Nat. Methods. 2013;10:873–875. doi: 10.1038/nmeth.2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Paige JS, Wu KY, Jaffrey SR. RNA mimics of green fluorescent protein. Science. 2011;333:642–646. doi: 10.1126/science.1207339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Irannejad R, et al. Conformational biosensors reveal GPCR signalling from endosomes. Nature. 2013;495:534–538. doi: 10.1038/nature12000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kirchhofer A, et al. Modulation of protein properties in living cells using nanobodies. Nat. Struct. Mol. Biol. 2010;17:133–138. doi: 10.1038/nsmb.1727. [DOI] [PubMed] [Google Scholar]
- 56.Fatica A, Bozzoni I. Long non-coding RNAs: new players in cell differentiation and development. Nat. Rev. Genet. 2014;15:7–21. doi: 10.1038/nrg3606. [DOI] [PubMed] [Google Scholar]
- 57.Brengues M, Teixeira D, Parker R. Movement of eukaryotic mRNAs between polysomes and cytoplasmic processing bodies. Science. 2005;310:486–489. doi: 10.1126/science.1115791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ramaswami M, Taylor JP, Parker R. Altered ribostasis: RNA-protein granules in degenerative disorders. Cell. 2013;154:727–736. doi: 10.1016/j.cell.2013.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rudkin GT, Stollar BD. High resolution detection of DNA-RNA hybrids in situ by direct immunofluorescence. Nature. 1977;265:472–473. doi: 10.1038/265472a0. [DOI] [PubMed] [Google Scholar]
- 60. Battich N, Stoeger T, Pelkmans L. Image-based transcriptomics in thousands of single human cells at single-molecule resolution. Nat. Methods. 2013;10:1127–1133. doi: 10.1038/nmeth.2657. This article, using sophisticated reagent delivery, FISH probes based on branched DNA technology and automated image analysis, demonstrates the power of single-molecule genome-wide RNA imaging.
- 61.Santangelo PJ, et al. Single molecule–sensitive probes for imaging RNA in live cells. Nat. Methods. 2009;6:347–349. doi: 10.1038/nmeth.1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tyagi S, Kramer FR. Molecular beacons: probes that fluoresce upon hybridization. Nat. Biotechnol. 1996;14:303–308. doi: 10.1038/nbt0396-303. [DOI] [PubMed] [Google Scholar]
- 63.Bertrand E, et al. Localization of ash mRNA particles in living yeast. Mol. Cell. 1998;2:437–445. doi: 10.1016/s1097-2765(00)80143-4. [DOI] [PubMed] [Google Scholar]
- 64.Daigle N, Ellenberg J. λN-GFP: an RNA reporter system for live-cell imaging. Nat. Methods. 2007;4:633–636. doi: 10.1038/nmeth1065. [DOI] [PubMed] [Google Scholar]
- 65.Alcid EA, Jurica MS. A protein-based EM label for RNA identifies the location of exons in spliceosomes. Nat. Struct. Mol. Biol. 2008;15:213–215. doi: 10.1038/nsmb.1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ozawa T, Natori Y, Sato M, Umezawa Y. Imaging dynamics of endogenous mitochondrial RNA in single living cells. Nat. Methods. 2007;4:413–419. doi: 10.1038/nmeth1030. [DOI] [PubMed] [Google Scholar]
- 67.Wilson C, Szostak JW. Isolation of a fluorophore-specific DNA aptamer with weak redox activity. Chem. Biol. 1998;5:609–617. doi: 10.1016/s1074-5521(98)90289-7. [DOI] [PubMed] [Google Scholar]
- 68. Strack RL, Disney MD, Jaffrey SR. A superfolding Spinach2 reveals the dynamic nature of trinucleotide repeat containing RNA. Nat. Methods. 2013;10:1219–1224. doi: 10.1038/nmeth.2701. An RNA aptamer with improved folding and thermodynamic stability, Spinach2, enables dynamic live-cell imaging of ‘toxic’ RNAs.
- 69.Lee J, et al. Combining SELEX screening and rational design to develop light-up fluorophore-RNA aptamer pairs for RNA tagging. ACS Chem. Biol. 2010;5:1065–1074. doi: 10.1021/cb1001894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sunbul M, Jäschke A. Contact-mediated quenching for RNA imaging in bacteria with a fluorophore-binding aptamer. Angew. Chem. Int. Ed. Engl. 2013;52:13401–13404. doi: 10.1002/anie.201306622. [DOI] [PubMed] [Google Scholar]
- 71.Lux J, Peña EJ, Bolze F, Heinlein M, Nicoud J-F. Malachite green derivatives for two-photon RNA detection. Chem Bio Chem. 2012;13:1206–1213. doi: 10.1002/cbic.201100747. [DOI] [PubMed] [Google Scholar]
- 72.Han KY, Leslie BJ, Fei J, Zhang J, Ha T. Understanding the photophysics of the Spinach-DFHBI RNA aptamer–fluorogen complex to improve live cell RNA imaging. J. Am. Chem. Soc. 2013;135:19033–19038. doi: 10.1021/ja411060p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jinek M, et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Qi LS, et al. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell. 2013;152:1173–1183. doi: 10.1016/j.cell.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Mali P, Esvelt KM, Church GM. Cas9 as a versatile tool for engineering biology. Nat. Methods. 2013;10:957–963. doi: 10.1038/nmeth.2649. A thought-provoking perspective on Cas9-mediated bioengineering.
- 76.Chen B, et al. Dynamic imaging of genomic loci in living human cells by an optimized CRISPR/Cas system. Cell. 2013;155:1479–1491. doi: 10.1016/j.cell.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bogdanove AJ, Voytas DF. TAL effectors: customizable proteins for DNA targeting. Science. 2011;333:1843–1846. doi: 10.1126/science.1204094. [DOI] [PubMed] [Google Scholar]
- 78.Santiago Y, et al. Targeted gene knockout in mammalian cells by using engineered zinc-finger nucleases. Proc. Natl. Acad. Sci. USA. 2008;105:5809–5814. doi: 10.1073/pnas.0800940105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Pattanayak V, Ramirez CL, Joung JK, Liu DR. Revealing off-target cleavage specificities of zinc-finger nucleases by in vitro selection. Nat. Methods. 2011;8:765–770. doi: 10.1038/nmeth.1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Betzig E, et al. Imaging intracellular fluorescent proteins at nanometer resolution. Science. 2006;313:1642–1645. doi: 10.1126/science.1127344. [DOI] [PubMed] [Google Scholar]
- 81.Tang J, van Panhuys N, Kastenmüller W, Germain R. The future of immunoimaging-deeper, bigger, more precise, and definitively more colorful. Eur. J. Immunol. 2013;43:1407–1412. doi: 10.1002/eji.201243119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Livet J, et al. Transgenic strategies for combinatorial expression of fluorescent proteins in the nervous system. Nature. 2007;450:56–62. doi: 10.1038/nature06293. [DOI] [PubMed] [Google Scholar]
- 83.Micheva KD, Busse B, Weiler NC, O’Rourke N, Smith SJ. Single-synapse analysis of a diverse synapse population: proteomic imaging methods and markers. Neuron. 2010;68:639–653. doi: 10.1016/j.neuron.2010.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zrazhevskiy P, Gao X. Quantum dot imaging platform for single-cell molecular profiling. Nat Commun. 2013;4:1619. doi: 10.1038/ncomms2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Schubert W, et al. Analyzing proteome topology and function by automated multidimensional fluorescence microscopy. Nat. Biotechnol. 2006;24:1270–1278. doi: 10.1038/nbt1250. [DOI] [PubMed] [Google Scholar]
- 86.Wang G, Smith SJ. Sub-diffraction limit localization of proteins in volumetric space using Bayesian restoration of fluorescence images from ultrathin specimens. PLOS Comput. Biol. 2012;8:e1002671. doi: 10.1371/journal.pcbi.1002671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Angelo M, et al. Multiplexed ion beam imaging of human breast tumors. Nat. Med. 2014;20:436–442. doi: 10.1038/nm.3488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Gorris HH, Wolfbeis OS. Photon-upconverting nanoparticles for optical encoding and multiplexing of cells, biomolecules, and microspheres. Angew. Chem. Int. Ed. Engl. 2013;52:3584–3600. doi: 10.1002/anie.201208196. [DOI] [PubMed] [Google Scholar]
- 89.Saar BG, et al. Video-rate molecular imaging in vivo with stimulated Raman scattering. Science. 2010;330:1368–1370. doi: 10.1126/science.1197236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Gao L, et al. Noninvasive imaging beyond the diffraction limit of 3D dynamics in thickly fluorescent specimens. Cell. 2012;151:1370–1385. doi: 10.1016/j.cell.2012.10.008. New selective-plane microscope with improved resolution interrogates single cells and developing embryos with unprecedented sensitivity.
- 91.Ahrens MB, Orger MB, Robson DN, Li JM, Keller PJ. Whole-brain functional imaging at cellular resolution using light-sheet microscopy. Nat. Methods. 2013;10:413–420. doi: 10.1038/nmeth.2434. [DOI] [PubMed] [Google Scholar]
- 92.Puchner EM, Walter JM, Kasper R, Huang B, Lim WA. Counting molecules in single organelles with superresolution microscopy allows tracking of the endosome maturation trajectory. Proc. Natl. Acad. Sci. USA. 2013;110:16015–16020. doi: 10.1073/pnas.1309676110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lee JH, et al. Highly multiplexed subcellular RNA sequencing in situ. Science. 2014;343:1360–1363. doi: 10.1126/science.1250212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Quail DF, Joyce JA. Microenvironmental regulation of tumor progression and metastasis. Nat. Med. 2013;19:1423–1437. doi: 10.1038/nm.3394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Xu T, Agrawal A, Abashin M, Chau KJ, Lezec HJ. All-angle negative refraction and active flat lensing of ultraviolet light. Nature. 2013;497:470–474. doi: 10.1038/nature12158. [DOI] [PubMed] [Google Scholar]
- 96.Mamin HJ, et al. Nanoscale nuclear magnetic resonance with a nitrogen-vacancy spin sensor. Science. 2013;339:557–560. doi: 10.1126/science.1231540. [DOI] [PubMed] [Google Scholar]