Abstract
Animal germ cells are totipotent because they maintain a highly unique and specialized epigenetic state for its genome. To accomplish this, germ cells express a rich repertoire of specialized RNA binding protein complexes such as the Piwi proteins and Piwi-interacting RNAs (piRNAs): a germ-cell enriched pathway of the RNA interference (RNAi) phenomenon which includes microRNA and endogenous small interfering RNA pathways. Piwi proteins and piRNAs are deeply conserved in animal evolution and play essential roles in fertility and regeneration. Molecular mechanisms for how these specific ribonucleoproteins act upon the transcriptome and genome are only now coming to light with the application of new systems-wide approaches in both invertebrates and vertebrates. Systems biology studies on invertebrates have revealed that transcriptional and heritable silencing is a main mechanism driven by Piwi proteins and piRNA complexes. In vertebrates, Piwi targeting mechanisms and piRNA biogenesis have progressed, while the discovery that the nuclease activity of Piwi protein is essential for vertebrate germ cell development but not completely required in invertebrates highlights the many complexities of this pathway in different animals. This review recounts how recent systems-wide approaches have a rapidly accelerated our new appreciation for the broad reach of the Piwi pathway on germline genome regulation and what questions facing the field await to be unraveled.
Introduction: RNA interference (RNAi) pathways in animal germ cells
Gene expression control is the sum of both gene activation and gene repression, and in nearly all animal cells, RNAi is a premier pathway for cells to execute broad and rapid gene silencing at the transcriptional and post-transcriptional level. Each cell type expresses specific repertoires of genome-encoded small regulatory RNAs that become incorporated into ribonucleoprotein (RNP) complexes. These small RNAs then serve as guides to direct the RNP complexes to search out target transcripts and genomic loci, thereby providing a dynamic closed-circuit for gene regulation. In essence, the activation of a small RNA-producing gene leads to the repression of a target gene with base-pairing homology to the small RNA.
In animal cells, the most ubiquitous arm of RNAi is the microRNA (miRNA) pathway. The 20–23nt long miRNAs are incorporated into Argonaute (AGO) proteins and have evolved to search for messenger RNA (mRNA) targets using the complementarity of the first 2 – 9 base pairs in the 5' end of the miRNA to ‘seed’ an interaction before locking the interaction in through a combination of mismatches and pairings with the rest of the miRNA (Bartel 2009). The AGO-miRNA RNP forms the core of a larger, less defined RNA Induced Silencing Complex (RISC) that typically seeks the 3' UnTranslated Regions of target mRNAs and can induce inhibition of mRNA translation as well as mRNA destabilization. Although animal genomes encode several hundreds of different individual miRNA sequences, different cell types can express specific sets of miRNAs because each miRNA derives from a single small hairpin structured precursor (~60–100 bp) that typically sits in the middle of an intron or a longer non-coding transcript made by RNA Polymerase II (Pol II) (Carthew and Sontheimer 2009). Despite being short, some miRNAs have remarkably deep conservation through their entire mature miRNA sequence, such as miR-1 and the miR-Let-7, which may be attributed to how each of these miRNAs can regulate a broad number of mRNA targets that are absolutely essential for general animal development (Ambros 2011).
A second arm of RNAi is the endogenous small interfering RNA (endo-siRNA) pathway which is found in invertebrate somatic cells and only mammalian oocytes, cells which do not express vertebrate innate immunity factors that drive cellular shutdown in the presence of long double-stranded RNA (dsRNA) (Okamura and Lai 2008). Although endo-siRNAs are generally ~21nt long, they are different from miRNAs because they are thought to derive from a longer (>100bp) dsRNA precursors forming from either very long fold-back structures; from two RNAs from different loci interacting in trans, or from the direct conversion of an mRNA into dsRNA by an RNA dependent RNA Polymerase (RdRP). In flies, endo-siRNAs preferentially load into Ago2 as opposed to miRNAs tending to load into Ago1, and in nematodes endo-siRNAs partner with a myriad of AGO homologs, however in mammals the distinction between miRNA- and endo-siRNA- AGOs is unclear. The target selection mechanisms for endo-siRNAs are presumed to entail mainly complete complementarity towards genes, repetitive elements such as transposons, and viral transcripts (Ghildiyal and Zamore 2009). The physiological role for endo-siRNAs in animal development remains unclear because mutants that specific disrupt endo-siRNA accumulation in Drosophila have subtle phenotypes, whereas in mammals there is only one Dicer enzyme that processes both miRNA and endo-siRNAS, thus complicating the analysis of endo-siRNAs alone. However, endo-siRNAs generated via RdRPs are likely involved in important gene regulatory effects such in nematode dauer formation (Hall et al. 2013), or in antiviral responses in flies (Goic et al. 2013).
The Piwi pathway: a germ cell innovation
The third arm of RNAi is the Piwi-interacting RNA (piRNA) pathway, which is distinct from miRNAs and endo-siRNAs because piRNAs are mainly enriched in animal germ cells; piRNA biogenesis does not depend on Dicer, the key enzyme that matures miRNAs and endo-siRNAs; and Piwi proteins form a distinct subclade of the AGO protein family (Ishizu et al. 2012). Furthermore, piRNAs appear to derive from single-stranded transcript precursors that either are non-coding with no annotated features, are transcripts that correspond to the 3'UTR of certain protein coding genes, or are transcripts that bear an unusually high concentration of transposable element (TE) sequences which are biased in their orientation so that mature piRNAs are complementarity to the TE’s coding sequence. Somehow, these transcripts are selected and processed into a diverse array of piRNAs for which the pattern of piRNAs sometimes appears erratic yet non-random (Betel et al. 2007), such as some piRNAs that repeatedly accumulate more abundantly and many preferentially begin with a 5' Uridine or having an Adenine at position 10 (see below about the “ping-pong” cycle, and (Siomi et al. 2011).
Despite the characteristics mentioned above, our understanding of piRNA biogenesis is still in its infancy. We know that piRNAs are longer in length than miRNAs and endo-siRNAs (at 25–31 nt), the 3' terminal 2' oxygen is methylated by the Hen1 enzyme, and there are many putative helicases, putative endonucleases, and tudor-domain containing proteins genetically required to generate and/or stabilize piRNAs (Ishizu et al. 2012). However, our mechanistic picture of the functions for these Piwi pathway genes remains murky: the only in vitro piRNA biogenesis activity known is a “Trimmer” activity observed in Bombyx gonadal cell extracts which can trim a long 5' phosphorylated transcript bound by SIWI into a mature piRNA (Kawaoka et al. 2012). And even though genetic advances that place artificial sequences like GFP into a piRNA precursor can give rise to artificial piRNAs (Kawaoka et al. 2012; Muerdter et al. 2012), we still cannot predict exactly which piRNAs would be generated most abundantly from these piRNA-generating transgenes.
However, piRNAs are clearly essential for the proper development of germ cells and fertility, because mutations that ablate Piwi protein function or piRNA biogenesis in both invertebrates and vertebrates result in germ cell death, gonadal atrophy, and ultimately sterility. One molecular consequence of losing piRNAs is the volatile expression of TE transcripts several fold above the negligible level in wild-type animals, and TE mobilization is thought to be damaging germ cell DNA and to cause de novo mutations that result in germ cell apoptosis or failed fertilization. Since a major proportion of metazoan piRNAs are perfectly complementary to the coding sequence of TEs, the role of piRNAs in taming TEs is obvious. However, many animals have large complements of piRNAs that lack homology to TEs, and given the extraordinary diversity of piRNA populations in germ cells compared to miRNAs, the challenge is to determine the full breadth of targets that are regulated by piRNAs.
In this chapter, we cover the monumental progress in recent years towards dissecting the functions and mechanisms of Piwi protein complexes and piRNA biogenesis factors, due in large part to the application of systems-wide approaches to this biomedically and biologically important RNA binding protein pathway. These systems-wide approaches have been built around the extensions of biochemical techniques that recover RNAs and genomic DNAs and characterizing the nucleic acids extremely thoroughly with high-throughput deep sequencing technologies (Figure 1). Supporting these approaches have been heavy dependence on available sequenced and assembled animal genomes and the application of new bioinformatics tools and infrastructure to handle large datasets.
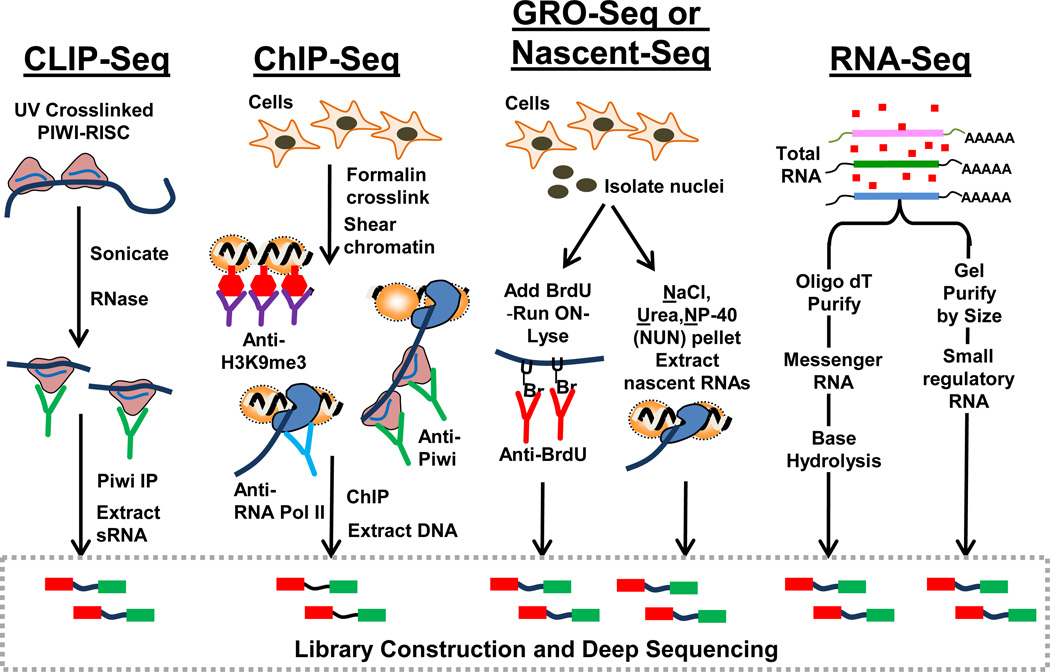
Figure 1. Systems biology approaches applied to study Piwi pathways.
Four main biochemical techniques now enable system-wide analysis of the Piwi-piRNA pathway. Cross-Linking Immuno Preciptiation (CLIP) enriches transcripts associated with Piwi proteins. Chromatin ImmunoPrecipitation (ChIP) enriches genomic DNA regulated by Piwi complexes. Global Run On (GRO) and Nascent RNA analyses reinforced a transcriptional gene silencing role by Piwi, while messenger RNA and piRNA analyses were facilitated by high-throughput deep sequencing of cDNA libraries.
In this review, we will first focus on piRNAs from the nematode Caenorhabditis elegans, which are quite distinct from other metazoan piRNAs, but appear to now influence epigenetic inheritance. Next, we will discuss remarkable new insight into piRNA biogenesis and targeting mechanisms uncovered in Drosophila mutants, ovarian cell line studies and genome-wide knockdown screens. Finally, we will examine the latest findings of vertebrate Piwi protein function and biogenesis that highlight what may be specific distinctions of the Piwi pathway that differ between vertebrates and invertebrates. Because we will be covering mainly the latest progress on how systems biology approaches have propelled the Piwi pathway field, we refer readers to several recent reviews that provide additional detailed and historical coverage of the Piwi field, from early genetics and cell biology to the advent of deep sequencing technology that gave the first comprehensive views of piRNA diversity (Lau 2010; Juliano et al. 2011; Siomi et al. 2011; Ishizu et al. 2012).
Astounding Argonaute diversity in C.elegans
The nematode C.elegans has the distinction of being the first animal where RNAi was describe (Fire et al. 1998), where the first two miRNAs were cloned (Lee et al. 1993; Reinhart et al. 2000), and where its small RNAs were first deeply sequenced by next generation sequencing technology (Ruby et al. 2006). The conservation of Piwi protein function in animal germline development was also first demonstrated in C.elegans via RNAi knockdown more than a decade ago (Cox et al. 1998). However, thorough understanding of C.elegans Piwi protein mutations has only recently progressed significantly due to the need to first investigate the greatly expanded family of AGO genes in C.elegans: 21 Worm-specific AGOs (WAGOs and others) accompany the metazoan AGO-related alg-1, 2 and the Piwi-related genes prg-1, prg-2 and ergo-1. As such, C.elegans is also endowed with a smorgasbord of different small RNAs: miRNAs, exogenous siRNAs, endogenous siRNAs that come in two flavors (22G RNAs and 26G RNAs named by their length and preferred first nucleotide) and piRNAs that are also known as 21U RNAs (Fischer 2010; Ketting 2011). Although miRNAs partition into ALG-1 and ALG-2 complexes, and 21U RNAs partition into Prg-1 and Prg-2 complexes, the line is more blurry amongst the assorted worm AGO proteins for what binds and specifies the biogenesis for 22G and 26G endo-siRNAs (Lau 2010; Claycomb 2012).
Before the systems biology revolution swept over C.elegans, it was assumed at first that the miRNA, piRNA and endo-siRNA pathways were somewhat distinct pathways with just a few shared enzyme factors. Although Dicer is required for the biogenesis of miRNAs and primary exogenous siRNAs that feed into RDE-1 and the 26G endo-siRNAs, there seemed to be a separated generation of secondary endo-siRNAs by different RdRPs: RRF-3 was required for 26G RNA biogenesis whereas RRF-1 and EGO-1 were required for 22G RNA biogenesis. Mutations in prg-1/prg-2 that affected 21U RNA biogenesis did not affect overall endo-siRNA function (Batista et al. 2008; Das et al. 2008), while mutations that affected 22G RNA and 26G RNA accumulations did not impact 21U RNA production (Han et al. 2009; Pavelec et al. 2009).
In fact, most mutations that affect endo-siRNA formation have not displayed overt phenotypes other than an enhanced RNAi (ERI) response when exogenous dsRNAs are used to target endogenous genes. However, notable exceptions are csr-1 and ego-1 mutations that perturb 22G siRNA production in the germline; and eri-3 and alg-3/alg-4 mutations that affect sperm-dependent 26G siRNA (Pavelec et al. 2009; Conine et al. 2010). The csr-1 and ego-1 mutant embryos have severely compromised cell division cycles so that they arrest in development (Claycomb et al. 2009; She et al. 2009), whereas eri-3 and alg3/4 mutants display decreased sperm count at elevated temperatures compared to wild-type (Pavelec et al. 2009; Conine et al. 2010). Gonadal development defects were also observed in prg-1 mutants particularly at elevated temperatures, but at below the standard rearing temperature of 20°C, these mutants of have rescued gonadogenesis despite lacking 21U and 26G small RNAs (Batista et al. 2008; Das et al. 2008). In particular, ergo-1 is a PIWI-subclade AGO gene whose mutants fail to generate 26G siRNAs yet appear nearly normal in development (Vasale et al. 2010). These mild phenotypes seen in the absence of endo-siRNA and piRNA populations in nematodes underscores the mystery of how nematodes can compensate so well when other metazoan germ cells are so dependent on the Piwi pathway.
C.elegans piRNAs are different from other metazoan piRNAs
Although the ERGO-1 and PRG-1/PRG-2 proteins have sequence homology to other metazoan Piwi proteins, we believe that C.elegans piRNAs are highly distinct from metazoan piRNAs, contrary to the impressions of similarity given from phylogenetic trees. First, the 21U RNAs bound by PRG-1/PRG-2 are quite sharply restricted to 21 nt long and shorter than the 24–31 nt length of other animal piRNAs. Second, although the 26G RNAs bound by ERGO-1 are more similar in length to other animal piRNAs, they require Dicer and the RdRP RRF-3 for biogenesis (Vasale et al. 2010), whereas general animal piRNA biogenesis in germ cells is independent of Dicer and RdRP activity (Vagin et al. 2006; Houwing et al. 2007). Third, many other animal piRNAs mature as a complex cluster of overlapping small RNAs derived from a long intergenic transcript (see sections below), but 21U RNA and 26G RNA genomic loci are also strikingly different in their configuration.
Rather than a single long transcript that gives rise to thousands of overlapping piRNAs in other animals, 21U RNAs arise from thousands of individual miniature loci that consist of a small GTTTC-containing motif and other signatures existing upstream of the sequence that is putatively transcribed as a short transcript (Ruby et al. 2006). Transcription of 21U RNAs may triggered by worm specific Forkhead box transcription factors binding at or near this motif to recruit RNA Pol II (Cecere et al. 2012), and then a ~26 nt long capped transcript is then somehow processed at both the 5’ and 3’ ends to yield a mono-phosphorylated 21U RNA (Gu et al. 2012). In the genomes of C.elegans and the related species C.briggsae, the 21U RNA loci are all closely clustered together in two main regions of chromosome IV, suggesting an evolutionary requirement of this arrangement perhaps to facilitate epigenetic control (Ruby et al. 2006; Shi et al. 2013). However, considering how close proximity of each 21U RNA locus is to another, and a recent demonstration that an autonomous transgene with a single 21U RNA locus can efficiently produce an exogenous 21U RNA (Billi et al. 2012), this indicates that we do not fully understand the transcription termination control and the genetic requirements for the concentration of 21U RNA loci onto the single fourth chromosome.
Evolutionary-based comparisons of small RNA populations between the soil nematodes of the Caenorhabditis genus (Shi et al. 2013) and other parasitic nematodes such as Ascaris further highlight the conundrum of the functional role of nematode piRNAs (21U RNAs). The total number of 21U RNAs in C.elegans may still be in flux between ~10,000 – ~24,000 individual sequences as different approaches to library construction, greater sequencing depth and bioinformatics predictions sort out the final tally (Gu et al. 2012; Shi et al. 2013). However, a recent study reported that the 21U RNA gene number may be doubled in C.remanei and C.brenneri compared to C.elegans and C.briggsae. The Shi et al study suggested that 21U RNA diversification may be more important for germline development in gonochoristic nematodes (male and female sexes, C.remanei and C.brenneri) compared to androdioecious nematodes (male and hermaphrodite sexes, C.elegans and C.briggsae). However, the parasitic nematode Ascaris suum is also gonochoristic yet appears to completely lack PRG-1/PRG-2 homologs and 21U RNAs all together, but Ascaris do retain miRNAs and 22G and 26G endo-siRNAs in the genomes (Wang et al. 2011). Gonocyte production is extremely prolific in Ascaris, and there is also a chromosome diminution process in somatic cells of the embryos which eliminates DNA sequences that are typically only expressed in the germline. Although chromosome diminution and maintenance of the germline in protozoans appears to be dependent on a small RNA pathway sharing similarity to animal Piwi pathways (see Conclusion), it is not clear if Ascaris endo-siRNAs are involved in eliminating DNA during chromosome diminution. The miRNA pathway seems to unwavering, therefore we conjecture that nematodes have run the piRNA and endo-siRNA arms of the RNAi pathways through a different “evolutionary gauntlet” compared to other animals with regards to germ cell development, and this principle may even apply when comparing the Piwi pathways of mammals to other vertebrates and invertebrates.
Worm Piwi pathways: more interconnected than we thought
Although we propose that C.elegans piRNAs are different from other animal piRNAs, the field has searched for commonality between C.elegans piRNAs and general animal piRNAs. Because the Piwi pathway clearly suppresses TEs in fly and mammalian germ cells, TEs were examined in the prg-1 and prg-2 null mutants, and only a limited number of TEs such as Tc3 showed a robust up-regulation of its transcript in mutants despite little evidence of TE mobilization (Batista et al. 2008; Das et al. 2008). Another shared feature with general animal piRNAs is that 21U and 26G siRNAs are specifically methylated at the 3' terminal 2' hydroxyl by the HENN1 RNA methylase, similar to fly, fish and mammalian piRNAs and endo-siRNAs being methylated by HEN1 orthologs (Billi et al. 2012; Kamminga et al. 2012; Montgomery et al. 2012). Despite its evolutionary conservation, the functional importance of Hen-1 for general animal piRNAs and endo-siRNAs is not yet clear because in the fly hen-1 mutant there is only a modest decrease in piRNA and endo-siRNA levels and few obvious developmental defects (Horwich et al. 2007; Saito et al. 2007). However, C.elegans 21U piRNAs and 26G endo-siRNAs levels are clearly perturbed and diminished, respectively, in henn-1 null mutants (Billi et al. 2012; Kamminga et al. 2012; Montgomery et al. 2012). Although there are indications that secondary endo-siRNAs like 22G RNAs are not modified with a 2'-O-methyl mark (Montgomery et al. 2012), the gene-silencing function of the 22G RNAs is affected in henn-1 mutants due to the genetic connection between 26G siRNAs and 22G siRNAs.
The genetic connection between 22G and 26G endo-siRNAs has been previously appreciated with the isolation and characterization several mutants that display consistent loss of 22G RNA accumulation and function whenever 26G RNAs were also affected (Conine et al. 2010; Gent et al. 2010; Vasale et al. 2010; Zhang et al. 2011a). As mentioned earlier, the expanded variety of worm AGO proteins mirrors the diversification of 26G and 22G endo-siRNAs: some small RNAs are germline or soma specific (Yigit et al. 2006; Conine et al. 2010; Vasale et al. 2010), sperm or oocyte specific (Han et al. 2009; Gent et al. 2010), perhaps even specific to developmental stages and environmental responses (Hall et al. 2013). The model that emerged was that 26G RNAs were the primary endo-siRNA trigger that could pair to a target transcript, which then subsequently stimulated 22G RNA production and amplification of the silencing process through worm-specific AGO proteins.
Although this 26G to 22G RNA link was apparent, earlier Northern blots indicated 21U RNA accumulation was not linked to bulk 26G RNA nor 22G RNA biogenesis, which pre-dated our understanding of different cohorts of 22G endo-siRNAs, such as CSR-1-specific 22G RNAs versus WAGO specific 22G RNAs (Claycomb et al. 2009; Gu et al. 2009). By constructing C.elegans strains containing integrated transgene reporters that could be targeted and silenced by a 21U RNA and PRG-1, three groups performing small RNA deep sequencing discovered that exogenous, transgene-specific 22G siRNAs were generated in a prg-1 dependent manner (Bagijn et al. 2012; Lee et al. 2012; Luteijn et al. 2012). The most abundant sets of these exogenous 22G RNAs accumulated proximally from the 21U piRNA binding site. Interestingly, exogenous 22G RNAs and transgene silencing were maintained even with 2 mismatches between the 21U RNA and the binding site (Bagijn et al. 2012). Since few perfectly complementary targets to 21U RNAs had been detected, the search was broadened to 21U RNA binding site with up to 3 mismatches and a strong correlation could now be seen between some populations of endogenous 22G RNAs and predicted 21U RNA binding sites on the genome (Bagijn et al. 2012; Lee et al. 2012). Two genes detected as being strongly mis-regulated in the prg-1 mutant were bath-45 and F54F2.2b which lost almost all 22G siRNAs in the prg-1 mutant. However, other genes dependent on prg-1 to produce antisense 22G siRNAs were rather modestly up-regulated while many other genes and TEs that have associated 22G siRNAs retain them regardless of prg-1. Why did prg-1 have such a strong influence on a subset of 22G siRNAs against certain genes and not have much effect on other 22G siRNAs, sometimes against the same gene or elements like Tc3?
To resolve this conundrum, two groups tracked de-repression of their 21U RNA fluorescent reporter transgene in various mutant backgrounds, and discovered that prg-1-mediated silencing in the germline was mainly linked genetically with WAGO-9/HRDE-1, a nuclear-localized worm-specific AGO (WAGO) protein that binds 22G RNAs generated from upstream factors like rrf-1, mut-7, drh-3 and rde-2 (Bagijn et al. 2012; Lee et al. 2012). These WAGO bound 22G RNAs are distinct from CSR-1 bound 22G RNAs even though biogenesis factors like drh-3 may be shared and both CSR-1 and WAGO-9/HRDE-1 are abundantly expressed in the germline. Whereas WAGO-9/HRDE-1 is primarily nuclear, CSR-1 is mostly concentrated in the cytoplasm and perinuclear organelles called P-granules where some RNA processing events are speculated to occur. Nevertheless, CSR-1 may also be exerting effects in the nucleus because during mitosis CSR-1 can be seen on the metaphase plate (Claycomb et al. 2009). Additionally in the C.elegans germline, there are ERGO-1 and Alg-3/Alg-4 AGO proteins binding 26G siRNAs that trigger downstream 22G siRNAs which load into other worm AGO proteins like WAGO-1 (Vasale et al. 2010), (Conine et al. 2010). Thus, the PRG-1/21URNA complex specifies only the downstream generation of WAGO-9/HRDE-1/22GRNA complex in a separate but analogous fashion to how ERGO-1 and ALG-3/ALG-4 direct downstream formation of the WAGO-1/22GRNA and other WAGO complexes. The interplay of the WAGO-9/HRDE-1/22GRNA complex with the CSR-1/22GRNA complex is discussed next.
Germline “everlasting”: small RNAs and the chromatin connection
For such a simple animal with a highly streamlined genome, the C.elegans germline has a staggering and mind-boggling number of different small regulatory RNAs and AGO proteins (Figure 2). In the oocyte alone, the current tally is now four distinct main pathways: the PRG-1::WAGO-9/HRDE-1 pathway, the NRDE-3 pathway, the ERGO-1::WAGO-1 pathway, and the CSR-1 pathway. Other articles have reviewed gene expression control and gene silencing in the C.elegans germline, which also includes the mechanism of silencing unpaired chromosomes during meiosis (Fischer 2010; Lau 2010; Ketting 2011; Claycomb 2012). The salient points for this review are that RNAi pathways are essential for proper germline development to generate gametes that form the totipotent zygote, and C.elegans embryogenesis is extracorporeal, so maternal stores of mRNAs, proteins and fats much be deposited and organized in the oocytes.
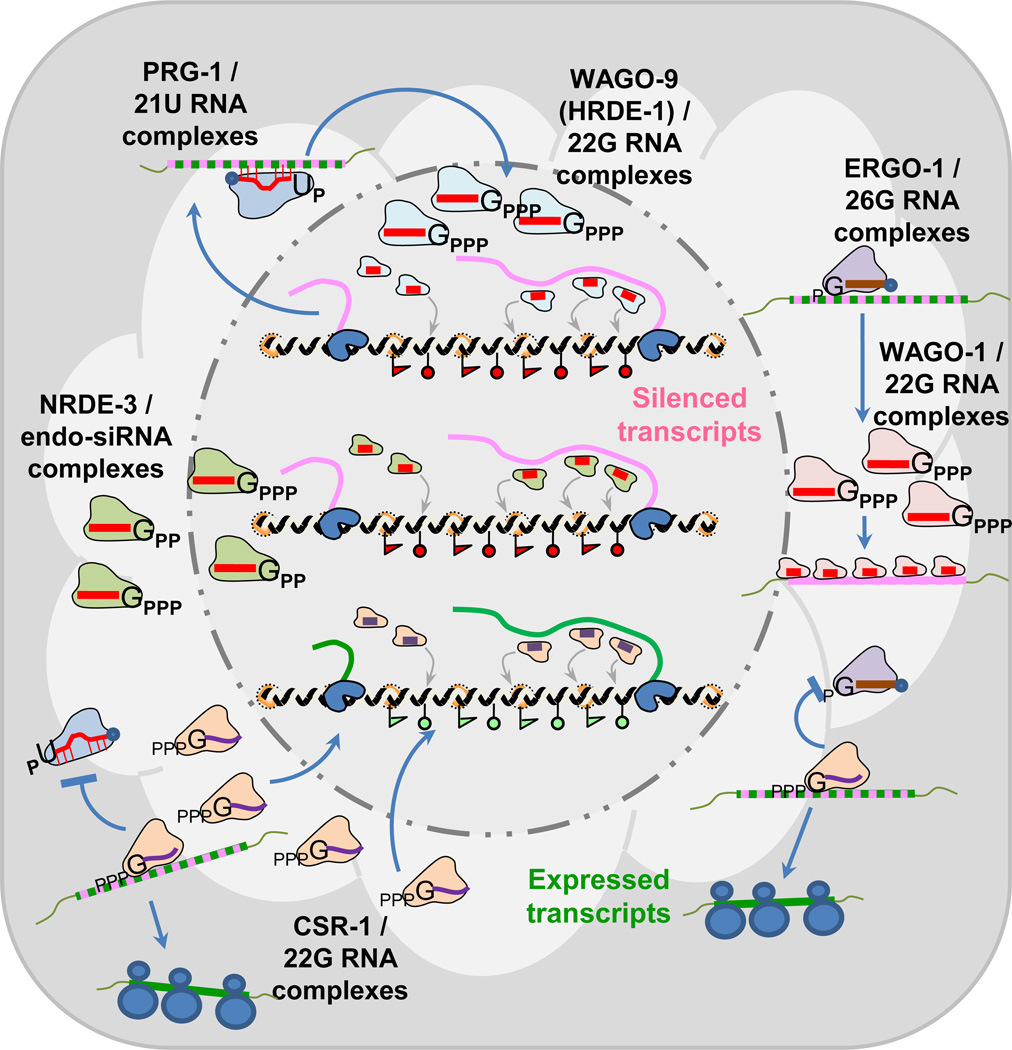
Figure 2. The C.elegans Piwi pathway is genetically connected to other endogenous RNAi pathways.
This diagram represents a developing germ cell in C.elegans with 6 different small RNA pathways operating concurrently, some in the nucleus like the PRG-1 and WAGO-9 complexes and the NRDE-3 and CSR-1 complexes, while ERGO-1 and WAGO-1 are speculated to work in the cytoplasm. Displayed are current models for how these small RNA complexes either silence or maintain gene expression in the C.elegans germline. See text for details.
To highlight one essential maternal factor, the CSR-1 complex is absolutely required for proper embryonic cell division and chromosome segregation events, and molecular studies suggest 22G RNAs guide CSR-1 to specific chromatin domains in order to help establish the boundaries needed to segregate the holocentric chromosomes of C.elegans (Claycomb et al. 2009; van Wolfswinkel et al. 2009). Thus, CSR-1 represents one important layer of epigenetic control in the germline.
However, the future challenge will be to understand why there are so many additional concurrent small RNA- AGO complexes in the C.elegans germ cell that are acting seemingly redundantly or antagonistically. For example, one group has proposed that CSR-1 may serve an anti-silencing role in antagonizing the gene-silencing activities of the WAGO complexes, but the details are obscure for how these two complexes bind similar 22G siRNAs yet partition and regulate different genes ((Shirayama et al. 2012) Fig. 2). Furthermore, in addition to the nucleus-directed PRG-1::WAGO-9/HRDE-1 pathway, there is also the NRDE-3 pathway that is triggered by exogenous siRNAs and also leads to gene silencing at the chromatin level in the nucleus (Guang et al. 2010). Although the nrde-1, nrde-2, and nrde-4 genes are required for gene silencing by the NRDE-3/endo-siRNA complex and the WAGO-9/HRDE-1/22G RNA complex, NRDE-3 is not required for the inherited silencing by WAGO-9/HRDE-1 (Ashe et al. 2012; Buckley et al. 2012). We summarize that there is a crosstalk in common genetic factor as well as partitioning of targeting functions and primary triggers in C.elegans germline RNAi pathways, and much further study is needed to sort all these pathways out.
A new paradigm proposed in these recent studies is that multiple germline RNAi processes establish a mechanism for controlling and discriminating Non-Self versus Self transcripts (Ashe et al. 2012; Shirayama et al. 2012). This paradigm argues that when PRG-1 and 21U RNA fortuitously binds to a foreign transgenes or TEs (an event that accommodates base mismatches), this licenses WAGO-9/HRDE-1 to mount a seemingly permanent silencing of the transgene. This model may be compelling for artificial transgenes, but is less able to explain why PRG-1and WAGO-9/HRDE-1 regulate endogenous protein-coding genes like bath-45 which lack typical features of Non-Self invaders like TEs and viruses as genomic invaders. In fact, most C.elegans TEs and viruses are not massively mis-regulated in prg-1 and wago-9/hrde-1 mutants, perhaps because these Non-Self elements are redundantly controlled by other WAGO/endo-siRNA complexes like NRDE-3 or WAGO-1.
While prg-1 and wago-9/hrde-1 mutant germlines appear as fertile as wild-type, perhaps reflecting the modest expression changes in limited numbers of genes (Batista et al. 2008; Das et al. 2008; Bagijn et al. 2012; Lee et al. 2012), a possible explanation to this conundrum is that the wago-9/hrde-1 mutants lose germline immortality. In other words, although the brood sizes of these mutants are similar to wild-type at the F1 progeny (the first descendants of a mutant in-bred cross), the F2 brood size begins to drop and reaches near complete sterility by the F5 progeny (Buckley et al. 2012). This outcome suggests that the germline may be highly sensitive to subtle yet compounding gene expression perturbations from multiple generations.
Perhaps germline immortality requires redundant levels of small RNAs for regulating key protein expression to levels that we cannot yet pinpoint as important for fertility. In fact, prg-1 mutants only display their brood size deficiencies at elevated temperatures (the phenotype is masked at cooler temperatures), where perhaps germline transcriptional enzyme activity or other gene regulatory processes may be more unbridled. Therefore, we propose that another natural function of the prg-1/wago-9/hrde-1 pathway may be to keep the Self transcriptome and proteome in the germline in check, perhaps to regulate endogenous soma-expressed genes that may be promiscuously expressed in the totipotent epigenetic state of the germline. This idea may also be related to the fact that the general Piwi/piRNA pathways in all other animals from flies to humans are primarily enriched in rapidly dividing germ cells, which display the widest diversity in its transcriptome (Yeo et al. 2004; Pao et al. 2006; Ravasi et al. 2006).
Flies push Piwi / piRNA insights to new heights
Drosophila germline genetics pioneered the discovery of the Piwi pathway (Wilson et al. 1996; Lin and Spradling 1997), (Schupbach and Wieschaus 1989; Schupbach and Wieschaus 1991) and the Drosophila female germline has continued to be the most fruitful system to study this pathway. Basic features of Drosophila piRNA biogenesis steps was previously mentioned in the introduction and are detailed extensively in other reviews (Juliano et al. 2011; Siomi et al. 2011; Ishizu et al. 2012), therefore this section will discuss how recent advances in our recent understanding of piRNA biogenesis and Piwi regulation mechanisms has benefited from systems biology approaches now being applied on Drosophila Piwi pathway mutants and Drosophila gonadal cell lines.
Starting with the beginning of the pathway, there has been recent progress in understanding the regulation of piRNA precursor transcription from the large intergenic piRNA clusters within pericentromeric heterochromatin. The two prototypical major piRNA clusters in Drosophila are the Flamenco locus on the X chromosome and the 42AB piRNA cluster locus on chromosome 2R (Figure 3). In addition to location, these clusters are differentiated because Flamenco generates transcripts from one genomic strand and is mainly expressed in the somatic follicle cells that surround the egg chamber, whereas 42AB generates transcripts from both genomic strands and is mainly expressed in the nurse cells of the female germline and these transcripts are then deposited into the oocyte (Juliano et al. 2011; Siomi et al. 2011). Both clusters, however, are highly concentrated in genomic sequences corresponding to TE relics, mutated copies of TEs that have landed in these clusters to become templates for the piRNAs which can then silence the active TE copies elsewhere in the genome.
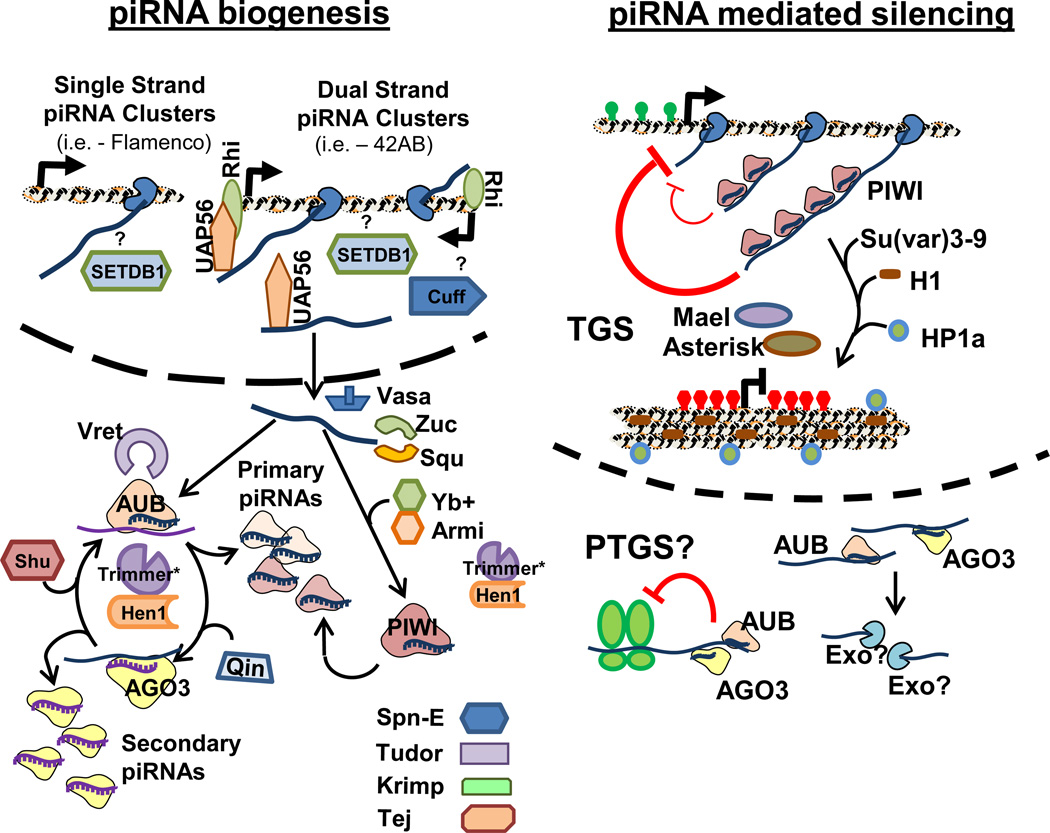
Figure 3. Current models of piRNA biogenesis and piRNA mediated gene silencing mechanisms in Drosophila.
Left) Depiction of known roles for factors genetically implicated in Drosophila piRNA biogenesis. More factors are known to regulate dual strand than single strand piRNA clusters. The piRNA precursor transcripts either are directly processed into primary piRNAs bound by AUB and PIWI, or engage in a subsequent secondary piRNA “ping-pong” amplification by AUB and AGO3. Right) Since AUB and AGO3 are in perinuclear nuage and cytoplasm; they may mediate post-transcriptional gene silencing. The nuclear PIWI binds to nascent transposon transcripts, where it can recruit chromatin modifying factors to induce a repressive chromatin state. Many other factors not shown in this diagram that may affect TE silencing have been identified in knockdown screens, but placement into the model is unclear, as is the role for tudor-domain containing factors like Spn-E, Tudor, Krimp, and Tej.
For piRNAs to be expressed abundantly in gonadal cells there is exquisite regulation of the piRNA cluster loci chromatin. The histone methyltransferase Eggless (also referred to as dSETDB1) may broadly impact transcription of both 42AB and Flamenco clusters by specifically tri-methylating histone H3 on lysine 9 (leaving a H3K9me3 mark) (Rangan et al. 2011). One protein that binds the H3K9Me3 mark is Heterochromatin Protein-1 (HP1, also known as Su(Var)2-05) which is genetically linked to the Piwi pathway (see below). However, a gonad-specific HP1 homolog called Rhino (Rhi) is enriched at and is required for transcription of the 42AB piRNA cluster (Klattenhoff et al. 2009). Associated with Rhi at the 42AB cluster is Cutoff (Cuff), a germ cell specific protein homologous to the Rai1 nuclease, and Cuff is required for expression of both 42AB expression and a single-strand piRNA cluster called the 20A cluster (Pane et al. 2011). Despite appearing as general chromatin-associated factors, Rhi or Cuff are required only for 42AB piRNA expression but not Flamenco piRNA expression, and questions remains as to what specifies Rhi and Cuff to only localize at one piRNA cluster rather than being a general factor like Eggless.
After transcription, RNA processing and transport is likely to play a special role in handling piRNA precursor transcripts. Vasa is one well known germ cell specific RNA-binding protein with putative helicase activity because it contains a DEAD-box domain, and mutations in this gene abrogate 42AB piRNA expression (Malone et al. 2009). Recently, a second DEAD-box containing protein, UAP56, was found to be required for piRNA expression, is localized near Rhi foci on nurse cell chromatin, and appears to bind newly transcribed piRNA-precursor transcripts from 42AB (Zhang et al. 2012). The model from this study is that UAP56 may shuttle a piRNA precursor transcript to the nuclear membrane for a hand off to Vasa for further processing at the nuclear periphery (Zhang et al. 2012).
Pulling back the layers of piRNA biogenesis
Some hints to the puzzle of piRNA biogenesis have come from understanding how the front and back termini of piRNAs are formulated. Despite the immense diversity of piRNA sequences, two common features define the termini of piRNAs – a preference for U at the 5' end, and a highly conserved 2'O-methylation modification added to the 3' end of piRNAs by Hen1 (Horwich et al. 2007; Saito et al. 2007). Recently, a biochemical extract from silkworm worm cells overexpressing the Piwi protein SIWI was able to recapitulate the activity of piRNA 3' end formation and SIWI loading in vitro when given a ~50 bp 5' radioactive phosphate labeled RNA (Kawaoka et al. 2011). The Tomari group also demonstrated that the 3' end trimming was coordinated with Hen-1directed 2'O- methylation, however efforts to purify and identify the putative ribonuclease for ‘Trimmer’ was stymied by the Trimmer activity being restricted to the insoluble fraction (Kawaoka et al. 2011).
Nevertheless, one factor, Zucchini (Zuc), is a tempting candidate for being a Trimmer enzyme because it is absolutely required for piRNA biogenesis and has homology to bacterial endonucleases (Pane et al. 2007). In addition, Zuc’s metazoan homologs are mitochondrial phospholipase D (also known as Pld6 in mouse) (Huang et al. 2011a; Watanabe et al. 2011; Anand and Kai 2012) and is localized to the surface of mitochondrial membranes that might exhibit an insoluble characteristic in extracts. Two recent studies solved the structures of recombinant forms of Zuc and have confirmed that Zuc has endoribonuclease activity in vitro, but the nature of the phospholipase D function remains unclear (Ipsaro et al. 2012; Nishimasu et al. 2012). Structure and biochemical data indicate that Zuc cleaves single-stranded DNA and RNA, and does so without requiring magnesium, a typical ion required by many ribonucleoprotein enzymatic activities including Argonaute’s Slicer activity. The catalytic activity of Zuc was not enhanced against a 5' Uridine (5'U), the preferred starting base of many piRNAs, however its RNase activity was only enhanced with the absence of sodium chloride, which is an interesting reflection of perhaps lower ionic strengths in a membraneous, less soluble microenvironment where Zuc may act to process piRNA precursors.
The “ping-pong” model of piRNA biogenesis is one potential mechanism for specifying the 5' end of many piRNAs. In brief, one Piwi-family protein like Aubergine (AUB) prefers to bind 5'U-starting piRNAs derived from piRNA cluster loci and are antisense to the coding strand of TEs, while Argonaute-3 (AGO3) prefers to bind piRNAs that have a preference of A at position 10 and are derived from TE coding strands (Brennecke et al. 2007; Gunawardane et al. 2007). In this model, AUB and its piRNA binds perfectly complementary to a TE transcript and slices the transcript to yield a cleavage product with a free 5' phosphate. The 5' end of this cleavage product is then bound by AGO3 and “trimming” of this transcript leads to a mature piRNAs. Vice versa in fashion, AGO3 and its piRNA specifies the 5' end and loading the next AUB and piRNA complex (Fig. 3). While the ‘ping-pong’ model is not yet proven biochemically, there is genetic support from ago3 mutations that cause the collapse of AUB piRNAs (Li et al. 2009), and signatures of the ping-pong cycle appear to be conserved in vertebrates as well (Houwing et al. 2007; Aravin et al. 2008; Lau et al. 2009). Recently, a newly identified factor called Qin, which has E3 ligase and TUDOR domains, was shown to modulate the biased loading of specific ping-pong piRNAs into AUB or AGO3. Although qin mutants (also known as kumo, (Anand and Kai 2012)) still produce piRNAs, qin mutations perturb normal AGO3 piRNA loading and affect the protein-protien interactions between AUB and AGO3 (Zhang et al. 2011b).
The piRNA “ping-pong” cycle was originally hypothesized to be hierarchical such that primary piRNAs primed the amplification loop for secondary piRNAs (Brennecke et al. 2007), (Gunawardane et al. 2007), and this connection is supported by mutations in protein factors that abolish primary piRNAs from Flamenco nearly always also ablate the secondary piRNAs exemplified from the 42AB cluster. The mutations of processing factors that destroy primary and secondary piRNA biogenesis include: armi, zuc, vert, shu, and gasz (Saito et al. 2009; Haase et al. 2010; Zamparini et al. 2011; Olivieri et al. 2012; Preall et al. 2012), (Handler et al. 2011; Handler et al. 2013). In contrast are mutations that only affect 42AB but leave Flamenco relatively unaffected, such as spn-E, Krim, vasa, rhi, UAP56, cuff, Kumo and tejas (Lim and Kai 2007; Klattenhoff et al. 2009; Malone et al. 2009; Olivieri et al. 2010; Patil and Kai 2010; Anand and Kai 2012; Zhang et al. 2012). Yb is an exceptional factor because its loss of function mainly affects only the Flamenco piRNAs without much effect on 42AB piRNAs, analogous to the Flamenco promoter mutations that only affect Flamenco piRNAs (Malone et al. 2009; Olivieri et al. 2010; Handler et al. 2011). Although this list of factors can be divided based on their roles in either primary or secondary piRNA biogenesis, it is still unclear how 5' ends of primary piRNAs are defined and why the 3'UTR of certain genic transcripts are selected for primary piRNA biogenesis. Perhaps this gap is attributed to an incompleteness of the list of genes that do impinge upon the Piwi pathway.
Addressing this issue are recent reports by the Hannon and Brennecke labs on genome-wide knockdown screens in flies and cultured follicle cell lines have now greatly expanded this gene list (Czech et al. 2013; Handler et al. 2013; Muerdter et al. 2013). Many previously identified genetic factors implicated in piRNA biogenesis were recovered and validated in these powerful screens, and gratifyingly several new factors have been identified to impact piRNA biogenesis, such as the Drosophila GasZ homolog CG2183, nuclear pore component Nxt1, and the sumolyation E1 ligase Uba2, just to name a few. These screens provided between ~50 – ~90 strongly validated new factors that impact the piRNA pathway in the Drosophila female germline, either at the biogenesis or the gene silencing effector level. Many future studies will ensue to place each of these factors in more discreet positions in the pathway.
Multi-talented Piwi proteins do TGS (and PTGS?)
Insights into how Piwi proteins carry out their effecter role in gene silencing have also dramatically increased recently due to systems approaches in Drosophila. Besides the Drosophila Piwi proteins themselves (PIWI, AUB, AGO3), a couple of Piwi-pathway factors have been defined as effectors of Piwi-mediated gene silencing because their loss of function did not affect piRNA accumulation but did allow for TE transcript up-regulation. Squash (squ) and Maelstrom (mael) were two of the first known effectors because mutations or knockdowns greatly impacted fertility yet most piRNAs were sustained in these animals or gonadal cells (Pane et al. 2007; Malone et al. 2009; Haase et al. 2010). Squ has some homology to an RNAse HII protein, but its biochemical activity is still not well understood, whereas Mael can associates with the microtubule organizing center and serves a role in specifying the polarity of Drosophila oocytes (Sato et al. 2011). A third effecter named Asterix was recently uncovered in the genome-wide RNAi screens as crucial for TE repression by the Piwi pathway despite little change in piRNA production (Muerdter et al. 2013). We can expect more effecters to emerge as the scrutiny turns to how the PIWI complex with piRNAs regulates targets for silencing.
With Drosophila PIWI being the founding member of this protein sub-clade, earlier studies had pointed to a potential transcriptional gene silencing (TGS) role for PIWI. PIWI was known to be nuclear, genetically and biochemically interacted with HP1, and was localized on polytene chromosomes in proximity to HP1 (Cox et al. 2000; Pal Bhadra et al. 2006; Brower-Toland et al. 2007). In addition, Piwi pathway mutations genetically interacted with Polycomb complex genes (Grimaud et al. 2006) and have been shown to modulate heterochromatin formation on transgenic loci harboring TE sequences as well as telomeres and insulated elements (Haynes et al. 2006; Yin and Lin 2007; Moshkovich and Lei 2010; Sentmanat and Elgin 2012). But the question remained as to how directly PIWI was involved in instigating TGS because these studies examined somatic cell phenotypes that were thought to be established by piwi in the germline.
Several recent systems-wide studies interrogating chromatin and nascent RNAs following perturbation of piwi function now clearly indicate that PIWI intimately directs TGS in gonadal cells. These studies corroborate that the loss of PIWI results in strong reduction of HP1 deposition, histone H3 and lysine 9 tri-methylation (H3K9me3), and concomitant increases in RNA Pol II occupancy at TE loci (Sienski et al. 2012; Huang et al. 2013; Le Thomas et al. 2013; Rozhkov et al. 2013). These signatures were consistent with the response of increased levels of steady state and nascent TE transcripts across the Drosophila genome (Sienski et al. 2012; Sytnikova et al. 2013). These data suggests that PIWI association at TE chromatin helps recruit H3K9me3 and HP1 marks that prevent the transcription machinery from engaging. However, the answer to whether H3K9me3 and HP1 marks are the cause or the consequence of gene silencing may be complicated because the Brennecke group observed that the loss of the effecter, Mael, which allows dramatic TE upregulation like the loss of PIWI, had very little reduction in H3K9me3 and HP1 (Sienski et al. 2012). Further studies will hopefully clarify other notable exceptions to the link between HP1 and PIWI, such as continued HP1 deposition at certain piRNA-targeted loci in piwi mutants (Moshkovich and Lei 2010), and PIWI transgenes that delete putative HP1 binding motifs yet are still able to rescue TE silencing (Wang and Elgin 2011).
Another open question regarding PIWI triggering TGS is whether PIWI binds chromatin directly or through nascent transcripts. Although one study was able to achieve a chromatin immunoprecipitation of PIWI (Huang et al. 2013), another study that performed CLIP-Seq on PIWI instead revealed that TE transcripts are the most heavily associated transcripts bound by PIWI, dominating a smaller fraction of genic transcripts (Sytnikova et al. 2013). Although coding gene expression changes have not been detected in fly ovaries deficient of piwi (Le Thomas et al. 2013), several genes did become strongly up-regulated in OSS and OSC cells upon piwi knockdown on the account of a de novo TE insertion in close proximity to the gene (Sienski et al. 2012; Sytnikova et al. 2013). By using the NUN-pellet protocol to obtain nascent transcripts, the Lau group was able to detect nascent TE transcripts up-regulated during PIWI knockdown that were independently transcribed from the nearby affected gene, and this TE transcript was complementary to the TE piRNAs. Furthermore, they showed with crafted reporter genes that PIWI and piRNAs must pair with the nascent transcript and not the DNA, and a threshold number of perfectly complementary piRNAs to the target transcript are required in order to initiate the TGS mechanism (Sytnikova et al. 2013).
Defining how far PIWI-mediated TGS can spread and what chromatin modulators are required for this spread will be an important next question to address. The reporter gene data point to a nascent transcript association requirement and is in agreement with mass spectrometry analysis of alternative splicing and nuclear RNA processing factors associated with PIWI (Le Thomas et al. 2013), indicating that PIWI/piRNA complex can rapidly interact with a TE nascent transcripts and promote a transcriptionally repressed state that can spread at least a few kilobases away. As the scrutiny turns to other chromatin factors, one recent report suggests that depletion of the linker histone H1 can alter H3K9 methylation and in turn unleash TE expression as well as strangely increase the accumulation of piRNA-like species (Lu et al. 2013).
Although the recent attention of TGS mechanisms has been focused on PIWI, other diverse mechanisms of gene regulation may exist for the other Piwi family members. For instance, Aub can regulate Nanos (also known as Nos) mRNA expression by genetically determining its localization to the posterior pole of the Drosophila oocyte where it helps direct the formation of the pole plasm in the embryo (Harris and Macdonald 2001; Megosh et al. 2006). The biased transport of Nanos mRNA is required for Anterior-Posterior patterning and primordial germ cell specification during embryogenesis (Lehmann and Nusslein-Volhard 1991; Wang et al. 1994). To achieve this, AUB can bind the 3'UTR of Nanos mRNA directly with possible help from the Rump protein (Becalska et al. 2011). However, four hours after fertilization, Nanos mRNA is also degraded; and recently Aub was implicated in this mechanism through demonstrations that AUB could associate with the general mRNA degradation factors like Smg and the CCR4 de-adenylase complex (Rouget et al. 2010). Furthermore, it was suggested that the Nanos 3’UTR contained imperfect complementary to some TE piRNAs to specifically recruit AUB (Rouget et al. 2010), however this targeting model does not yet resolve how Nanos gets discriminately selected over other transcripts and directly contrasts with the PIWI-mediated targeting requirements (Sytnikova et al. 2013).
Interestingly, Nanos mRNA posterior pole localization also depends on the function of protein chaperones like Hsp90 (Song et al. 2007), and genetic phenomenon like canalization has been proposed to connect the role of Hsp90 chaperones and the piRNA pathway (Sato and Siomi 2010). The term canalization describes how organisms maintain developmental robustness despite so much variability from environmental pressures and genetic variation. Hsp90 chaperones are thought to be one level of this canalization because they help mutated or misfolded proteins still fold into functional enzymes, thereby “buffering” phenotypes to remain similar across a population of varying genotypes. However, to explain how Hsp90 perturbations allowed genetic variation to magnify, the connection to piRNAs was only recently made in a study showing that genetic and chemical disruptions of Hsp90 activity can negatively affect piRNA populations and therefore yield de-repression of TEs (Specchia et al. 2010). This was further supported by data showing that Hsp90 may foster post-translational modifications of PIWI necessary for TE repression (Gangaraju et al. 2011), and a new linking of the Piwi pathway to Shu/FKBP6 as additional chaperones that may have functions related to Hsp90 (Olivieri et al. 2012; Preall et al. 2012; Xiol et al. 2012). These findings reflect how beautifully complex and intertwined these genetic and biochemical pathways to are ensure proper animal germ cell development.
The role piRNAs play in hybrid dysgenesis and species evolution
Ultimately, a key evolutionary pressure for the Piwi and piRNA pathway is to ensure fertility within a species, but it seems the Piwi pathway is also a potent licensing system that opposes the formation of certain hybrids. In the hybrid dysgenesis phenomenon, daughter animals resulting from a mating of two hybrids seem to suffer poorer health compared to parents, such as near sterility despite normal development to adulthood. For intraspecies hybrid dysgenesis of D. melanogaster strains (Shpiz and Kalmykova 2009), dysgenesis has a pattern that suggests a maternally deposited epigenetic factor to the embryo is a key determinant to ensure fertility, while the lack of the maternal factor meant the embryo could not respond to some harmful paternal factor. A breakthrough in understanding hybrid dysgenesis systems was placing piRNAs as the maternally deposited factor required to respond to a particular TE imparted by the paternal chromosomes (Brennecke et al. 2008; Malone et al. 2009). Compellingly, in a Drosophila mother with an entirely intact Piwi pathway and diverse piRNA populations, lacking just the small complement of piRNAs against a novel TE that invades the father’s germline can spell trouble in the progeny.
Interestingly, with time dysgenic daughters can begin to regain some fertility, but by what mechanism? By carefully tracking piRNAs and re-sequencing of progeny genomes in the P-element system of hybrid dysgenesis, the Theurkauf group showed that P-element TEs were indeed mobilizing in the dysgenic progeny, and likely causing genomic damage in the germ cells that result in sterility (Khurana et al. 2011). But when the mobile TE lands back into a piRNA cluster, new P-element piRNAs begin to emerge in aging dysgenic progeny that also begin to regain some (but not all) fertility. This observation was bolstered by other studies demonstrating an analogous recovery from complete infertility in dysgenic progeny from an I-R element hybrid system (Grentzinger et al. 2012) and in a paramutation-based genetic system for P-elements (de Vanssay et al. 2012). Together, these studies indicate that primary piRNA biogenesis systems are insufficient to handle novel TEs contributed by the paternal genome. Instead, the gain of TEs landing into specific piRNA clusters that yield maternally deposited piRNAs are needed to stimulate some aspect of “ping-pong cycle” for sustaining piRNA function through embryogenesis and into larval gonad development.
Although the study of the Piwi pathway in dysgenic strains of D. melanogaster has been extremely insightful, these strains are non-natural products of careful laboratory rearing practices, whereas in the wild the selection pressures are stronger and more rapid on the Piwi pathway to maintain fertility. One evolutionary study has found that the piRNA compositions in D. melanogaster populations certainly provide evidence for a positive selection signal (Lu and Clark 2010), however there also seems to be a tolerance for Drosophila to allow TEs more leeway to expand in the genome, perhaps due to the canalization roles that Piwi proteins and piRNAs play. The role of piRNA incompatibility with general species barriers has also been proposed (Shpiz and Kalmykova 2009), but the first study to examine piRNAs in an actual interspecies hybrid was in a special cross of a D. melanogaster Hmr mutant with the closest relative D. simulans, in which hybrid progeny development can proceed to adulthood (Kelleher et al. 2012). Although these interspecies hybrid progeny eventually share similar phenotypic deficiencies with intraspecies dysgenic hybrids, such as distorted populations of maternally deposited piRNAs, sterility, and loss of repression for a broad number of TEs, the Barbash group could not detect a clear correlation between TEs de-repressed highly in the hybrid and decreases or divergences in sequences between parental piRNAs and hybrid progeny piRNAs. Furthermore, by showing that a transgene bearing the D. simulans Aub was unable to rescue the D. melanogaster aub mutant, the authors suggest there may be underlying deficiencies or incompatibilities between the Piwi pathway protein components of different species rather than just differences in piRNAs may account for dysgenesis in interspecies hybrids (Kelleher et al. 2012). These fly experiments are initial forays of evolutionary studies applied to the Piwi pathway; however, a greater diversity of model systems like vertebrates must be brought to bear to see how pervasive these genetic and evolutionary phenomena extend into animal evolution.
Regulating the vertebrate piRNA pathway
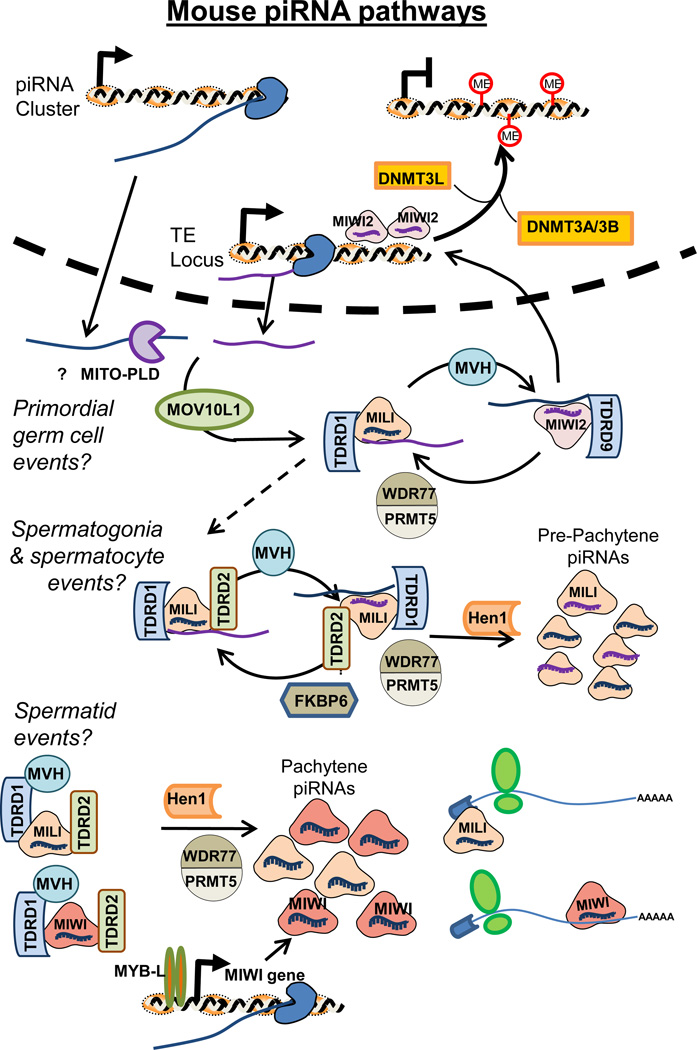
Similar to flies, mammalian genomes encode at least three clear Piwi homologs, and nomenclature for vertebrate Piwi homologs is a cruel joke of history whereby the first letter of the animal “helps” denote the homologs’ origin, hence the mouse encodes Miwi, Miwi2, and Mili (Miwi-like), while humans encode Hiwi, Hiwi2, and Hili. A Miwi knockout mouse was one of the first mutants demonstrating the conserved function of Piwi pathway in vertebrates for germ cell development (Deng and Lin 2002). However, as the Piwi pathway in mammals and other model vertebrates come into better focus, it is apparent that there are important differences between vertebrates and flies in the function and requirement of Piwi pathway components. For example, although knockout mutations in any of the loci of Miwi, Mili or Miwi2, render male mice completely infertile because of spermatogenic arrest and atrophy of the testes; all female homozygous mutants are superficially as fertile as heterozygous littermates. This contrasts with flies where severe Piwi pathway mutations impairs fertility in males and females (Cox et al. 2000), and contrasts with zebrafish where Ziwi and Zili null mutants in both genders cannot form rudimentary gonads and become sterile masculinized fish (Houwing et al. 2007; Houwing et al. 2008). Although Mili and some piRNAs are expressed in mouse oocytes, the proportion of piRNAs is comparably lower with respect to abundant endo-siRNAs in mouse eggs (Tam et al. 2008), (Watanabe et al. 2008), and mammalian oogenesis is typically restricted to very few mitotic divisions after birth while fish and fly ovaries continuously engage in mitotic cell divisions of oogonia. We speculate that the Piwi pathway may be most essential in germ cells that must engage in continuous rounds of mitosis during gametogenesis, but formally testing this hypothesis is challenging.
Mouse Piwi protein expression is temporally regulated during spermatogenesis, such that Miwi2 is expressed earliest and only temporarily in the embryonic gonad, whereas Miwi is expressed latest after 10 days postpartum (dpp) when the first synchronized wave of spermatogenesis approaches the pachytene stage of meiosis. Mili’s expression is most pervasive, starting at 12.5 days post coitus in male primordial germ cells and extending through adulthood. The temporal regulation of the Piwi proteins themselves coincide with three rough categories of piRNA expression as well, pre-natal piRNAs, pre-pachytene piRNAs, and post-pachytene piRNAs, which differ on the level of how many TE sequences and how strong a ping-pong cycle signature is present. In pre-natal piRNAs, a ping-pong cycle signature exists between Miwi2 and Mili piRNAs which tend to be more enriched in TE sequences such as LINE1, the most prevalent TE class in mammalian genomes. After birth but before 12 dpp, when only Mili dominates, these pre-pachytene piRNAs have some enrichment of TE sequences suggesting Mili can perform the ping-pong cycle with itself as well as a strong enrichment of 3'UTR genic piRNAs (Robine et al. 2009). Finally, post-pachytene piRNAs are incorporated into obht Miwi and Mili, are depleted in TE sequences, almost devoid of ping-pong cycle signatures (Beyret et al. 2012), and derive from over a hundred intergenic and genic clusters that presumably express the piRNA precursor as single-stranded RNA (Robine et al. 2009).
Although reasons are still unknown for the temporal expression of mammalian Piwi pathway genes, an important transcription factor has recently been discovered to play a role in this temporal expression of Miwi and post-pachytene piRNAs. A mouse mutagenesis screen had previously pinpointed the transcription factor A-MYB being required for spermatogenesis as well as breast tissue development (Toscani et al. 1997). Recently, A-MYB was implicated in promoting pachytene piRNA and Miwi transcript expression in spermatocytes and spermatids (Li et al. 2013). ChIP-Seq of A-MYB found it at the promoters of Miwi and other piRNA biogenesis factor genes like MitoPLD, Mov10L, and Tdrd6, the promoters of pachytene piRNA clusters, and its own promoter, therefore it may engage in a feed forward circuit that allows A-MYB to ramp-up transcription of both piRNA-precursors and piRNA biogenesis machinery. However, the presence of A-MYB in breast tissue, which lack mouse Piwi protein and piRNA expression, also suggests that A-MYB itself is not sufficient to drive the expression of the entire Piwi pathway. Perhaps the Piwi pathway demands a combination of transcription factors, such as the USF and NF-Y factors recently suggested to also affect Miwi expression (Hou et al. 2012). Finally, although Miwi and piRNAs are most highly expressed in round spermatids, they are absent in mature sperm, and this turnover may either be part of the standard process of cytoplasmic shedding during the final stages of spermatogenesis, or it may be part of an active process of turnover by the proteasome (Zhao et al. 2013).
Not quite the same: Piwi pathway nuances in vertebrates
With so many genetic factors implicated in the Piwi pathways in Drosophila, we might expect that the larger mammalian genomes would encode as many if not more Piwi pathway factors. However, there are only as many mouse Piwi protein effectors as there are in Drosophila, but it remains a challenge to designate which mouse Piwi gene is the true fly ortholog. MIWI2 has the same nuclear localization and chromatin regulation characteristics with PIWI, while MILI and MIWI are cytoplasmic like AUB and AGO3. However, PIWI and MIWI mainly engage in primary piRNA biogenesis and not the ping-pong cycle, while MILI and MIWI2 can engage in the cycle in the mouse. Finally, MILI and MIWI2 have been genetically connected to directing de novo DNA methylation of TE elements during embryogenesis (see below), whereas in flies there is only minor evidence that DNA methylation has an influence on Drosophila development and no evidence yet for a link to PIWI, AUB or AGO3.
Amongst the mammalian Piwi pathway factors that can be clearly assigned to orthologs in Drosophila, there appears to be a difference in the degree that mutations in mouse genes affect piRNA biogenesis and gametogenesis compared to the fly genes. Many mutations in the Drosophila Piwi pathways are extremely severe, causing complete sterility and major piRNA loss. However, other than the mouse Piwi genes themselves, the only other gene mutation to cause complete loss of mouse piRNAs in the testes is the putative helicase Mov10-Like (Mov10L, which is distinct from the Mov10 paralog that is required for miRNA maturation into the RISC (Meister et al. 2005; Frost et al. 2010). Mov10L is the mammalian ortholog of Drosophila armi, and the Mov10L mutant male is infertile with shrunken testes that are devoid of functional sperm due to changes in spermatogonia and spermatocyte organelle morphology and an increased buildup of mitochondria (Zheng et al. 2010).
In contrast are several mouse Piwi factors where the mutations in mouse still cause spermatogenic arrest and male infertility, but their effect on global piRNA production during the initial waves of spermatogenesis are surprisingly mild. For example, whereas Drosophila vasa mutant ovaries are deficient of most piRNAs (Malone et al. 2009), mutations in Mouse Vasa Homolog (MVH) seem to only cause loss of Miwi2 piRNAs in the fetal testes, while Mili piRNAs can still accumulate (Kuramochi-Miyagawa et al. 2010). Mouse Maelstom (ortholog of Drosophila mael) resides most frequently in the piP (piRNA Processing) bodies, associates with the Tdrd9 and Tdrd1 factors, and influences Miwi2 piRNAs but not Mili piRNAs (Soper et al. 2008; Aravin et al. 2009). Since the suppression of mouse TEs is affected in mouse mael mutants without loss of piRNAs, the effector role of silencing rather than piRNA biogenesis may be conserved between mouse and fly mael orthologs. Although mutations and knockdowns of fly Zuc severely compromise Drosophila piRNAs, knockout mutants in the mouse ortholog MitoPLD seems to affect the production of some piRNAs, but not others, particularly SINE element piRNAs bound by Mili (Huang et al. 2011a; Watanabe et al. 2011). Mouse GasZ knockout mutants (Ma et al. 2009) were created before knockdowns of fly GasZ (CG2183) were studied (Czech et al. 2013; Handler et al. 2013), and whereas TEs are de-represssed in both fly and mouse GasZ mutants, piRNA biogenesis is much more severely impacted in the fly GasZ mutant. Knockdowns and mutations of fly Shutdown (Shu) strongly impair piRNA biogenesis (Olivieri et al. 2012; Preall et al. 2012), but knockouts of the mouse ortholog, FKBP6, only causes some loss of Miwi2 piRNAs but effects are quite modest against Mili piRNAs (Xiol et al. 2012). Finally, knockdown of the glycerol-3-phosphate acyltransferase 2 (GPAT2), a mitochondrial outer membrane protein, in a germinal stem cell mouse line was shown to affect piRNA loading into Mili (Shiromoto et al. 2013) and its fly homolog CG5508 also recovered in one of the RNAi screens for being required in TE silencing (Czech et al. 2013) but role for CG5508 in piRNA biogenesis not yet known.
A large group of genes linked to the Piwi pathway and also conserved throughout animal evolution are the tudor-domain-containing proteins which are specifically expressed in germ cells. The tudor domain was named after the founding member, the Drosophila Tudor (Tud) gene whose closest mammalian homolog is TuDoR-Domain 6 (TDRD6). Tud and TDRD6 are the largest Tudor proteins with 11 and 8 repeated tudor domain in the fly and mouse genes, respectively. Tud mutants display the grandchildless phenotype where the daughters develop infertility, similar to hybrid dysgenesis and to the PRMT5 arginine methylase mutants that deposits symmetrical dimethyl arginines onto Aub and AGO3 to facilitate its interaction with Tud (Kirino et al. 2009; Nishida et al. 2009). Some amount of TE de-repression and piRNA decrease is observed in these mutants, but these phenotypes are mild compared to more severe phenotypes of Piwi protein mutations themselves or other biogenesis factors. Like Tud in flies, Tdrd6 is also required for spermatogenesis in mouse and associates with Miwi and Mili (Vasileva et al. 2009). The tudor domain is hypothesized to foster protein-protein docking interactions, so the presence of just multiple tudor domains in Tud and Tdrd6 suggests their main function is to act as scaffolds.
Connected to the Piwi pathway are three other tudor domain-containing genes with clear orthologs between flies and mice: papi, whose mouse homolog is Tdrd2 (also called Tdrkh); tejas (tej), whose mouse homolog is Tdrd5; and spindle-E (Spn-E), whose mouse homolog is Tdrd9. These genes differ from Tud because they only contain a single tudor domain, but Papi has transmembrane domains, Tej has a LOTUS domain, and Spn-E has helicase and RNA binding domains (Handler et al. 2011). Papi associates with Ago3 and its mutant may display higher TE expression (Liu et al. 2011), but RNAi knockdown of Papi has not replicated this phenotype (Czech et al. 2013; Handler et al. 2013; Muerdter et al. 2013), and Papi’s influence on piRNA production is not clear. Tdrd2 interacts strongly with Miwi (Chen et al. 2009), and a recent knockout mouse of Tdrd2 shows LINE1 de-repression, spermatogenic arrest before pachytene is reached, and a very interesting lengthening of Mili-bound piRNAs to ~6–8 nt longer (Saxe et al. 2013). Although Tejas and Tdrd5 are essential for female and male germline development, respectively, only Tejas has been shown to be important for piRNA biogenesis and TE silencing (Patil and Kai 2010; Yabuta et al. 2011). Classic female sterile fly mutations of Spn-E suffer severe loss of piRNAs (Vagin et al. 2006; Malone et al. 2009), but while TDRD9 knockout mouse are male sterile, they have overall similar levels of piRNAs in pre-pachytene stage testis to wild type, with just minor differences in a few specific piRNA sequences (Chuma et al. 2006; Shoji et al. 2009). Although TDRD9 associates with Miwi2 and its localization in spermatogonia is influenced by Mili (Shoji et al. 2009), it is enigmatic why in contrast to Spn-E that mouse Tdrd9 is not critical for piRNA biogenesis.
For other tudor domain-containing genes in fly and mouse, the homologs have diverged such that either the obvious ortholog is not apparent, or that designating the ortholog bioinformatically may not be obvious. For example, Tdrd1 has been proposed to the closest homolog to Drosophila Qin in one study (Siomi et al. 2010) or to CG9925 in another study which suggested Qin was more related to Tdrd4 (Handler et al. 2011). Tdrd1 is essential for male germ cell development and TE silencing via promoting efficient piRNA biogenesis in Mili, but whether Tdrd1 has a role in the ping-pong cycle like Qin remains open because there are still significant piRNAs present in the Tdrd1 −/− mutant (Reuter et al. 2009; Vagin et al. 2009; Wang et al. 2009). A purification of zebrafish Tdrd1 identified longer transcripts which may be putative piRNA precursors (Huang et al. 2011b), perhaps analogous to the putative piRNA precursors found associated in an ARMI purification from Drosophila OSC cells (Saito et al. 2010). Although the Tdrd4 knockout mouse has not been reported yet, knockdown of CG9925 did not elicit TE de-repression (Handler et al. 2011), thus it remains open which is the clear Drosophila ortholog of Tdrd1. Then there is the example of Drosophila Krimper, which lacks any mammalian ortholog (Siomi et al. 2010; Handler et al. 2011), but clearly has a strong effect on the Piwi pathway in flies when mutated or knocked down (Lim and Kai 2007; Malone et al. 2009; Handler et al. 2011).
Mammalian Piwi proteins need ‘slicing’ activity to do their jobs
Amongst the commonalities between the Piwi pathways of flies and mice, the silencing mechanisms of the Piwi proteins themselves have been puzzlingly complex. Like certain AGO proteins, the Piwi-group proteins retain the key amino acid residues required for the endonucleolytic slicing activity that occurs upon a target transcript substrate that is selected by the guide small RNA in the protein. The slicing activity is crucial to the ping-pong cycle model (Brennecke et al. 2007; Gunawardane et al. 2007; Li et al. 2009), however it has not been reported whether AUB and AGO3 transgenes mutated in catalytic residues can rescue the aub and ago3 mutants. Although PIWI has slicing activity in vitro (Saito et al. 2006), two studies using a Piwi transgene mutated in the catalytic residues indicate that slicing activity is dispensable for TE silencing in Drosophila OSC cells and in the female germline (Saito et al. 2010; Darricarrere et al. 2013). With the revelation that Drosophila Piwi mediates TGS, the dispensability of the slicing activity is surprising because Ago1 from fission yeast absolutely requires slicing activity in order to manifest its TGS on heterochromatic loci (Irvine et al. 2006).
Two mouse Piwi proteins, Mili and Miwi2, have also been implicated to promote TGS upon TEs in the early development phases of fetal and pre-pubertal sperm. In Mili and Miwi2 knockout mice, there is a substantial loss of DNA methylation upon chromatin containing LINE1 sequences (Aravin et al. 2007; Carmell et al. 2007; Aravin et al. 2008; Kuramochi-Miyagawa et al. 2008), mirroring the phenotype of dnmt3l mutants, the gene responsible for de novo re-establishment of this silencing mark on TEs during mammalian embryogenesis after parental epigenetic imprints are erased and resorted (Lane et al. 2003). The genetic function of Mili and Miwi2 in specifying DNA methylation of TEs is compelling, and may even extended to imprinted loci like Rasgrf1 (Watanabe et al. 2011). However, the biochemical mechanism and questions of how directly do Mili and Miwi2 direct DNA methylation remain obscure because dnmt3l mutants have no effect on piRNA biogenesis (Aravin et al. 2008), and proteomic analysis of Mili and Miwi2 complexes have not identified associated DNA methyltransferases (Vagin et al. 2009). In addition, not all TEs are equally repressed at the TGS level, such as the IAP TE which is not affected in Miwi2 mutants (Kuramochi-Miyagawa et al. 2008).
In contrast to Miwi2 and Mili being genetically linked to TGS, Miwi has mainly been thought to regulate target transcripts post-transcriptionally, although demonstrating this mechanism has not been fully addressed. It is clear that Miwi is mainly cytoplasmic and concentrated in the perinuclear organelle called the chromatoid body, a dense structure believed to be a transit hub for mRNAs (Kotaja and Sassone-Corsi 2007). Miwi can also bind and perhaps stabilize some mRNAs (Deng and Lin 2002) as well as pachytene piRNA precursor transcripts (Vourekas et al. 2012), but whether Miwi has sequence binding specificity or how mechanistically Miwi can select to preferentially bind certain transcripts is unclear (Robine et al. 2009). A translational regulation role for Miwi and Mili has mainly been portrayed from density gradient fractionations of adult testis extracts that show Miwi and Mili co-fractionating with poly-ribosomes (Grivna et al. 2006; Unhavaithaya et al. 2009). It has been challenging to formally test the translational regulatory activity of Miwi and Mili upon synthetic mRNAs and transgenes in testes extracts competent for in vitro translation. However the recent report of a mouse gonadal cell culture line (Germline Stem Cells, or GSCs; (Shiromoto et al. 2013) with endogenous Mili and piRNAs might possibly open a door to more rigorous biochemistry if these cells can be cultured in bulk.
Mammalian Piwi proteins were first demonstrated to exhibit Slicer activity in vitro (Lau et al. 2006), and this activity was thought to sustain a ping-pong cycle signature between Miwi2 and Mili (Aravin et al. 2008). However, only mammalian Ago2 retains slicer activity to form the RISC capable for target RNA cleavage, while Ago1, -3, and -4 are incapable of slicing activity (Liu et al. 2004) but can regulate targets through binding and recruiting translation repressing factors. Although Ago2’s catalytic residues are critical for mouse hematopoiesis because Ago2 slicing is required to process the essential miR-451 miRNA (Cheloufi et al. 2010; Cifuentes et al. 2010), a mutated Ago2 transgene lacking slicing activity could rescue hematopoiesis in Ago2 −/− mice, suggesting that the Ago2 slicing activity is highly specialized (O'Carroll et al. 2007).
To address the importance of the slicing activity in mouse Piwi proteins, point mutations that disrupted the catalytic residues of Miwi (Reuter et al. 2011), Mili and Miwi2 (De Fazio et al. 2011) were generated and revealed some surprising and perplexing genetic effects. Slicing-inactive Mili mutant males (MiliDAH) were infertile and displayed up-regulated LINE1 TE expression and decreased DNA methylation, but interestingly overall piRNA production within Mili was comparable to wild-type with mainly a mild reduction of piRNAs complementary to LINE1 sequences (De Fazio et al. 2011). Interestingly, it was Miwi2 piRNA loading that was perturbed in MiliDAH mutants, suggesting Mili slicing was required for specifying a ping-pong cycle mechanism to load piRNAs into Miwi2. Perplexingly, inactivation of slicing residues in Miwi2 failed to reveal a reciprocal defect in piRNA production neither in Mili nor in Miwi2 itself, with TE silencing also intact (De Fazio et al. 2011). Whereas catalytic residue mutations in Mili and Miwi2 are genetically recessive, the catalytic residue mutation in Miwi (MiwiADH) turned out to be a dominant negative in causing male sterility in the heterozygous state (Reuter et al. 2011). If one speculates that Miwi proteins form dimers as an explanation of the dominant negative phenotype, then it was very mysterious that pachytene piRNA accumulation and general testes transcriptome profiles were largely unaffected in the MiwiADH mutant. Indeed, this study displayed LINE1 TE upregulation in MiwiADH mutants and some evidence that Miwi may use certain piRNAs to slice LINE1 TE transcripts post-transcriptionally as a means to suppress TE mobilization (Reuter et al. 2011). Recently, conditional Mili knockout and conditional slicing MiliDAH mutants were shown to still exhibit mouse male sterility despite functional Mili capable of establishing proper DNA methylation of TE loci (Di Giacomo et al. 2013), thus leading to the model that LINE1 TEs reanimate during pachytene stages of meiosis but require both Mili and Miwi protein to prevent germ cell damage. Since we still do not know how highly efficient mechanisms for primary piRNA biogenesis in mouse spermatocytes can proceed, apparently even when Slicing activity is compromised, these genetic studies have raised more open questions and new conundrums to the Piwi pathway that await future novel approaches and additional model organisms and germ cell systems to fully dissect this pathway.
Conclusion – Piwi “on the brain” and going beyond the germline?
The Piwi pathway genes and piRNAs are clearly most influential and abundantly expressed in animal gonads, but the detection and genetic influence of this pathway in other somatic animal tissues continues to be debated. It is possible that Piwi-mediated chromatin modifications determined in the germ cells and early embryo are later perpetuated in a Piwi-independent fashion via a “cellular memory” to somatic tissues like eye pigmentation or salivary gland polytene chromomsomes (Pal-Bhadra et al. 2002; Brower-Toland et al. 2007; Moshkovich and Lei 2010). However, caution should also be heeded in determining whether bonafide piRNAs are indeed expressed in somatic cells, because several abundant and stable structural RNA fragments have been misclassified as potential piRNAs in somatic cells without the confirmation that they are indeed loaded into a Piwi protein complex (Janic et al. 2010; Lee et al. 2011; Yan et al. 2011).
However, there has been a convergence of data implicating that some invertebrate animal neurons may harbor its own repository of piRNAs and Piwi machinery. The sea slug Aplysia is a classic model system for neurophysiology because of its exceptionally large, characterized ganglion comprised of many cell types including neurons (Bailey et al. 2004). In addition to miRNAs, it was recently shown that the Aplysia ganglions contain putative piRNAs (Rajasethupathy et al. 2012), but characterization of the Aplysia piRNAs, whether they are derived from large intergenic cluster or target TEs, is hampered by the rough draft stage of the Aplysia genome (L. et al. 2009; Rajasethupathy et al. 2012). Injection of mimics and inhibitors against a single putative Aplysia piRNA was somehow able to instill neurophysiological responses and DNA methylation changes in these ganglions (Rajasethupathy et al. 2012). A focused scrutiny of specific neurons in the fly brain has also suggested that Piwi proteins may be expressed and playing a role in controlling TE mobilization in the fly brain (Perrat et al. 2013). As a possible mechanism for fly neuroplasticity, this study hypothesized that certain neurons lacking AUB and AGO3 would allow TE activity to yield heterogeneity within the Drosophila brain that could be useful for learning and memory (Perrat et al. 2013). Biochemical analyses for the small amount of tissue from specific fly neurons is technically challenging, however genetic and chemical ablation of miRNAs and endo-siRNAs had previously suggested the existence of putative piRNAs in fly heads (Ghildiyal and Zamore 2009). So as technology in this arena advances, including more efficient and cost-effective systems biology approaches, we expect to uncover new avenues where the Piwi pathway and piRNAs may influence animal behavior and other processes beyond the animal germline.
Clearly, flies and mice have dominated the recent Piwi pathway scene because system approaches have succeeded due in part to the ease of genetically manipulating flies and the large knowledgebase for mice as a model mammal of humans. As such, both organisms have the greatest foundation of genomics resources and largest community of researchers. But to understand the Piwi pathway fully will demand going beyond the fly and mouse and making use of other vertebrates and invertebrate models. For example, to ask if vertebrates are susceptible to hybrid dysgenesis like in flies where a mother lacking just one class of piRNAs will produce sterile daughters, mice may not be an optimal system because mammalian oogenesis does not appear to depend on the Piwi pathway and mammalian sperm do not transmit piRNAs as epigenetic factors like oocytes can. However, the frog Xenopus tropicalis has well characterized piRNA clusters configured in an analogous fashion to mammalian clusters (Armisen et al. 2009; Lau et al. 2009), as well as highly abundant levels of Xiwi in the oocyte that appears to be concentrated assymetrically in the germ plasm (Lau et al. 2009), specialized cytoplasm that can specify the formation of primordial germ cells (Houston and King 2000).
In addition to gametogenesis, the Piwi pathway may also be connected to regeneration of lost body parts in certain animals. Planarians and cnidarians contain a large and dispersed population of special stem cells called neoblasts that express Piwi proteins and piRNAs (Reddien et al. 2007; Palakodeti et al. 2008). These neoblasts are the key cells endowing these animals with extraordinary regenerative capabilities such as restoring complete body plans from nearly any piece of severed tissue (Petersen and Reddien 2009). In fact, the major small RNA populations in these animals are not miRNAs but rather piRNAs (Grimson et al. 2008; Palakodeti et al. 2008). Amongst vertebrates, the axolotl salamander is prominently one of a few vertebrates capable of regenerating a severed limb during adulthood, and recently it was suggested that axolotl Piwi transcripts might be triggered for expression in the regenerating blastema (Zhu et al. 2012). Perhaps somatic cells in the blastema are reanimating the Piwi pathway as a form of induced de-differentiation similar to induced pluriopotency, and indeed mutations of a tumor suppressor gene in flies may possibly be one route to achieve this de-differentiation state (Janic et al. 2010). We speculate that the majority of other vertebrates where somatic cells lack the expression of the Piwi pathway may explain our limited regenerative potential.
Finally, we discuss lastly but not the least that the Piwi pathway is not the sole purview of multicellular organisms: the single celled protozoans have creatively co-opted the Piwi pathway for reproductive and growth functions in fantastic fashion. Tetrahymena encodes at least eight genes that are more similar to metazoan Piwi genes than Ago genes, and the Tetrahymena Piwi genes (TWI) have very essential roles in growth (Couvillion Collins 2010) and nuclear RNA metabolism (Couvillion et al. 2010). Notably, Tetrahymena express scnRNAs that are just one form of protozoan piRNAs, which are required for meiosis, conjugation, and a complex process of DNA elimination from the macronucleus (Mochizuki and Gorovsky 2004a; Schoeberl et al. 2012). The Tetrahymena scnRNAs appear to direct chromatin modification events that lead to the removal of large swaths of DNA to yield a slimmed macronucleus perhaps optimized for genic transcription (Mochizuki et al. 2002; Mochizuki and Gorovsky 2004b). But in opposite fashion, another protozoan, Oxytricha, uses its piRNAs to instead specify the retention of DNA elements and preventing genes from being lost when its genome rearranges into thousands of tiny chromosomes (Fang et al. 2012; Swart et al. 2013). These quirky protozoan Piwi pathways are a testament to the rich diversity of natural biological processes, and should motivate us to continue casting a wide net of systems biology approaches across multiple model organisms.
Figure 4. The mouse spermatogenic piRNA pathway is tightly regulated throughout development.
During early spermatogenesis, primary pre-pachytene piRNAs are produced from transposon and genic transcripts. These MILI-bound, sense-oriented piRNAs can then direct “Ping-pong” cycle production of MIWI2-bound or MILI-bound antisense piRNAs in primordial germ cells and spermatogonia, respectively. MIWI2-piRNA complexes are then able to translocate to the nucleus to initiate transcriptional gene silencing by promoting de novo DNA methylation. After meiosis and during spermatid maturation, non-repetitive pachytene piRNAs begin to dominate and load into both MILI and MIWI with the assistance of tudor-domain containing proteins. The post-pachytene MIWI and MILI complexes are hypothesized to regulate target transcripts post-transcriptionally.
Acknowledgments
We are grateful to members of the Lau lab, Julie Claycomb and Dianne Schwarz for discussions and critical reading of this manuscript. If a late-breaking report was missed, it was because the field has progressed so rapidly that we could not cite it after this article was submitted. This work was supported by the Searle Scholars foundation to NCL.
REFERENCES
- Ambros V. MicroRNAs and developmental timing. Current opinion in genetics & development. 2011;21:511–517. doi: 10.1016/j.gde.2011.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anand A, Kai T. The tudor domain protein kumo is required to assemble the nuage and to generate germline piRNAs in Drosophila. The EMBO journal. 2012;31:870–882. doi: 10.1038/emboj.2011.449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravin AA, Sachidanandam R, Bourc'his D, Schaefer C, Pezic D, Toth KF, Bestor T, Hannon GJ. A piRNA pathway primed by individual transposons is linked to de novo DNA methylation in mice. Molecular cell. 2008;31:785–799. doi: 10.1016/j.molcel.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravin AA, Sachidanandam R, Girard A, Fejes-Toth K, Hannon GJ. Developmentally regulated piRNA clusters implicate MILI in transposon control. Science. 2007;316:744–747. doi: 10.1126/science.1142612. [DOI] [PubMed] [Google Scholar]
- Aravin AA, van der Heijden GW, Castaneda J, Vagin VV, Hannon GJ, Bortvin A. Cytoplasmic compartmentalization of the fetal piRNA pathway in mice. PLoS Genet. 2009;5:e1000764. doi: 10.1371/journal.pgen.1000764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armisen J, Gilchrist MJ, Wilczynska A, Standart N, Miska EA. Abundant and dynamically expressed miRNAs, piRNAs, and other small RNAs in the vertebrate Xenopus tropicalis. Genome research. 2009;19:1766–1775. doi: 10.1101/gr.093054.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashe A, Sapetschnig A, Weick EM, Mitchell J, Bagijn MP, Cording AC, Doebley AL, Goldstein LD, Lehrbach NJ, Le Pen J, et al. piRNAs can trigger a multigenerational epigenetic memory in the germline of C. elegans. Cell. 2012;150:88–99. doi: 10.1016/j.cell.2012.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagijn MP, Goldstein LD, Sapetschnig A, Weick EM, Bouasker S, Lehrbach NJ, Simard MJ, Miska EA. Function, targets, and evolution of Caenorhabditis elegans piRNAs. Science. 2012;337:574–578. doi: 10.1126/science.1220952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey CH, Kandel ER, Si K. The persistence of long-term memory: a molecular approach to self-sustaining changes in learning-induced synaptic growth. Neuron. 2004;44:49–57. doi: 10.1016/j.neuron.2004.09.017. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batista PJ, Ruby JG, Claycomb JM, Chiang R, Fahlgren N, Kasschau KD, Chaves DA, Gu W, Vasale JJ, Duan S, et al. PRG-1 and 21U-RNAs interact to form the piRNA complex required for fertility in C. elegans. Molecular cell. 2008;31:67–78. doi: 10.1016/j.molcel.2008.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becalska AN, Kim YR, Belletier NG, Lerit DA, Sinsimer KS, Gavis ER. Aubergine is a component of a nanos mRNA localization complex. Developmental biology. 2011;349:46–52. doi: 10.1016/j.ydbio.2010.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betel D, Sheridan R, Marks DS, Sander C. Computational analysis of mouse piRNA sequence and biogenesis. PLoS Comput Biol. 2007;3:e222. doi: 10.1371/journal.pcbi.0030222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beyret E, Liu N, Lin H. piRNA biogenesis during adult spermatogenesis in mice is independent of the ping-pong mechanism. Cell research. 2012;22:1429–1439. doi: 10.1038/cr.2012.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billi AC, Alessi AF, Khivansara V, Han T, Freeberg M, Mitani S, Kim JK. The Caenorhabditis elegans HEN1 ortholog, HENN-1, methylates and stabilizes select subclasses of germline small RNAs. PLoS Genet. 2012;8:e1002617. doi: 10.1371/journal.pgen.1002617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R, Hannon GJ. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell. 2007;128:1089–1103. doi: 10.1016/j.cell.2007.01.043. [DOI] [PubMed] [Google Scholar]
- Brennecke J, Malone CD, Aravin AA, Sachidanandam R, Stark A, Hannon GJ. An epigenetic role for maternally inherited piRNAs in transposon silencing. Science. 2008;322:1387–1392. doi: 10.1126/science.1165171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brower-Toland B, Findley SD, Jiang L, Liu L, Yin H, Dus M, Zhou P, Elgin SC, Lin H. Drosophila PIWI associates with chromatin and interacts directly with HP1a. Genes & development. 2007;21:2300–2311. doi: 10.1101/gad.1564307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckley BA, Burkhart KB, Gu SG, Spracklin G, Kershner A, Fritz H, Kimble J, Fire A, Kennedy S. A nuclear Argonaute promotes multigenerational epigenetic inheritance and germline immortality. Nature. 2012;489:447–451. doi: 10.1038/nature11352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmell MA, Girard A, van de Kant HJ, Bourc'his D, Bestor TH, de Rooij DG, Hannon GJ. MIWI2 Is Essential for Spermatogenesis and Repression of Transposons in the Mouse Male Germline. Developmental cell. 2007 doi: 10.1016/j.devcel.2007.03.001. [DOI] [PubMed] [Google Scholar]
- Carthew RW, Sontheimer EJ. Origins and Mechanisms of miRNAs and siRNAs. Cell. 2009;136:642–655. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cecere G, Zheng GX, Mansisidor AR, Klymko KE, Grishok A. Promoters Recognized by Forkhead Proteins Exist for Individual 21U-RNAs. Molecular cell. 2012;47:734–745. doi: 10.1016/j.molcel.2012.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheloufi S, Dos Santos CO, Chong MM, Hannon GJ. A dicer-independent miRNA biogenesis pathway that requires Ago catalysis. Nature. 2010;465:584–589. doi: 10.1038/nature09092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Jin J, James DA, Adams-Cioaba MA, Park JG, Guo Y, Tenaglia E, Xu C, Gish G, Min J, et al. Mouse Piwi interactome identifies binding mechanism of Tdrkh Tudor domain to arginine methylated Miwi. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:20336–20341. doi: 10.1073/pnas.0911640106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuma S, Hosokawa M, Kitamura K, Kasai S, Fujioka M, Hiyoshi M, Takamune K, Noce T, Nakatsuji N. Tdrd1/Mtr-1, a tudor-related gene, is essential for male germ-cell differentiation and nuage/germinal granule formation in mice. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:15894–15899. doi: 10.1073/pnas.0601878103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cifuentes D, Xue H, Taylor DW, Patnode H, Mishima Y, Cheloufi S, Ma E, Mane S, Hannon GJ, Lawson ND, et al. A novel miRNA processing pathway independent of Dicer requires Argonaute2 catalytic activity. Science. 2010;328:1694–1698. doi: 10.1126/science.1190809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claycomb JM. Caenorhabditis elegans small RNA pathways make their mark on chromatin. DNA and cell biology. 2012;31(Suppl 1):S17–S33. doi: 10.1089/dna.2012.1611. [DOI] [PubMed] [Google Scholar]
- Claycomb JM, Batista PJ, Pang KM, Gu W, Vasale JJ, van Wolfswinkel JC, Chaves DA, Shirayama M, Mitani S, Ketting RF, et al. The Argonaute CSR-1 and its 22G-RNA cofactors are required for holocentric chromosome segregation. Cell. 2009;139:123–134. doi: 10.1016/j.cell.2009.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conine CC, Batista PJ, Gu W, Claycomb JM, Chaves DA, Shirayama M, Mello CC. Argonautes ALG-3 and ALG-4 are required for spermatogenesis-specific 26G-RNAs and thermotolerant sperm in Caenorhabditis elegans. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:3588–3593. doi: 10.1073/pnas.0911685107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couvillion MT, Sachidanandam R, Collins K. A growth-essential Tetrahymena Piwi protein carries tRNA fragment cargo. Genes & development. 2010;24:2742–2747. doi: 10.1101/gad.1996210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox DN, Chao A, Baker J, Chang L, Qiao D, Lin H. A novel class of evolutionarily conserved genes defined by piwi are essential for stem cell self-renewal. Genes & development. 1998;12:3715–3727. doi: 10.1101/gad.12.23.3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox DN, Chao A, Lin H. piwi encodes a nucleoplasmic factor whose activity modulates the number and division rate of germline stem cells. Development. 2000;127:503–514. doi: 10.1242/dev.127.3.503. [DOI] [PubMed] [Google Scholar]
- Czech B, Preall JB, McGinn J, Hannon GJ. A Transcriptome-wide RNAi Screen in the Drosophila Ovary Reveals Factors of the Germline piRNA Pathway. Molecular cell. 2013 doi: 10.1016/j.molcel.2013.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darricarrere N, Liu N, Watanabe T, Lin H. Function of Piwi, a nuclear Piwi/Argonaute protein, is independent of its slicer activity. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:1297–1302. doi: 10.1073/pnas.1213283110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das PP, Bagijn MP, Goldstein LD, Woolford JR, Lehrbach NJ, Sapetschnig A, Buhecha HR, Gilchrist MJ, Howe KL, Stark R, et al. Piwi and piRNAs act upstream of an endogenous siRNA pathway to suppress Tc3 transposon mobility in the Caenorhabditis elegans germline. Molecular cell. 2008;31:79–90. doi: 10.1016/j.molcel.2008.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Fazio S, Bartonicek N, Di Giacomo M, Abreu-Goodger C, Sankar A, Funaya C, Antony C, Moreira PN, Enright AJ, O'Carroll D. The endonuclease activity of Mili fuels piRNA amplification that silences LINE1 elements. Nature. 2011;480:259–263. doi: 10.1038/nature10547. [DOI] [PubMed] [Google Scholar]
- de Vanssay A, Bouge AL, Boivin A, Hermant C, Teysset L, Delmarre V, Antoniewski C, Ronsseray S. Paramutation in Drosophila linked to emergence of a piRNA-producing locus. Nature. 2012;490:112–115. doi: 10.1038/nature11416. [DOI] [PubMed] [Google Scholar]
- Deng W, Lin H. miwi, a murine homolog of piwi, encodes a cytoplasmic protein essential for spermatogenesis. Developmental cell. 2002;2:819–830. doi: 10.1016/s1534-5807(02)00165-x. [DOI] [PubMed] [Google Scholar]
- Di Giacomo M, Comazzetto S, Saini H, De Fazio S, Carrieri C, Morgan M, Vasiliauskaite L, Benes V, Enright AJ, O'Carroll D. Multiple Epigenetic Mechanisms and the piRNA Pathway Enforce LINE1 Silencing during Adult Spermatogenesis. Molecular cell. 2013;50:601–608. doi: 10.1016/j.molcel.2013.04.026. [DOI] [PubMed] [Google Scholar]
- Fang W, Wang X, Bracht JR, Nowacki M, Landweber LF. Piwi-interacting RNAs protect DNA against loss during Oxytricha genome rearrangement. Cell. 2012;151:1243–1255. doi: 10.1016/j.cell.2012.10.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- Fischer SE. Small RNA-mediated gene silencing pathways in C. elegans. The international journal of biochemistry & cell biology. 2010;42:1306–1315. doi: 10.1016/j.biocel.2010.03.006. [DOI] [PubMed] [Google Scholar]
- Frost RJ, Hamra FK, Richardson JA, Qi X, Bassel-Duby R, Olson EN. MOV10L1 is necessary for protection of spermatocytes against retrotransposons by Piwi-interacting RNAs. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:11847–11852. doi: 10.1073/pnas.1007158107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangaraju VK, Yin H, Weiner MM, Wang J, Huang XA, Lin H. Drosophila Piwi functions in Hsp90-mediated suppression of phenotypic variation. Nature genetics. 2011;43:153–158. doi: 10.1038/ng.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gent JI, Lamm AT, Pavelec DM, Maniar JM, Parameswaran P, Tao L, Kennedy S, Fire AZ. Distinct phases of siRNA synthesis in an endogenous RNAi pathway in C. elegans soma. Molecular cell. 2010;37:679–689. doi: 10.1016/j.molcel.2010.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghildiyal M, Zamore PD. Small silencing RNAs: an expanding universe. Nat Rev Genet. 2009;10:94–108. doi: 10.1038/nrg2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goic B, Vodovar N, Mondotte JA, Monot C, Frangeul L, Blanc H, Gausson V, Vera-Otarola J, Cristofari G, Saleh MC. RNA-mediated interference and reverse transcription control the persistence of RNA viruses in the insect model Drosophila. Nature immunology. 2013;14:396–403. doi: 10.1038/ni.2542. [DOI] [PubMed] [Google Scholar]
- Grentzinger T, Armenise C, Brun C, Mugat B, Serrano V, Pelisson A, Chambeyron S. piRNA-mediated transgenerational inheritance of an acquired trait. Genome research. 2012;22:1877–1888. doi: 10.1101/gr.136614.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimaud C, Bantignies F, Pal-Bhadra M, Ghana P, Bhadra U, Cavalli G. RNAi components are required for nuclear clustering of Polycomb group response elements. Cell. 2006;124:957–971. doi: 10.1016/j.cell.2006.01.036. [DOI] [PubMed] [Google Scholar]
- Grimson A, Srivastava M, Fahey B, Woodcroft BJ, Chiang HR, King N, Degnan BM, Rokhsar DS, Bartel DP. Early origins and evolution of microRNAs and Piwi-interacting RNAs in animals. Nature. 2008;455:1193–1197. doi: 10.1038/nature07415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grivna ST, Pyhtila B, Lin H. MIWI associates with translational machinery and PIWI-interacting RNAs (piRNAs) in regulating spermatogenesis. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:13415–13420. doi: 10.1073/pnas.0605506103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu W, Lee HC, Chaves D, Youngman EM, Pazour GJ, Conte D, Jr, Mello CC. CapSeq and CIP-TAP identify Pol II start sites and reveal capped small RNAs as C. elegans piRNA precursors. Cell. 2012;151:1488–1500. doi: 10.1016/j.cell.2012.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu W, Shirayama M, Conte D, Jr, Vasale J, Batista PJ, Claycomb JM, Moresco JJ, Youngman EM, Keys J, Stoltz MJ, et al. Distinct argonaute-mediated 22G-RNA pathways direct genome surveillance in the C. elegans germline. Molecular cell. 2009;36:231–244. doi: 10.1016/j.molcel.2009.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guang S, Bochner AF, Burkhart KB, Burton N, Pavelec DM, Kennedy S. Small regulatory RNAs inhibit RNA polymerase II during the elongation phase of transcription. Nature. 2010;465:1097–1101. doi: 10.1038/nature09095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunawardane LS, Saito K, Nishida KM, Miyoshi K, Kawamura Y, Nagami T, Siomi H, Siomi MC. A slicer-mediated mechanism for repeat-associated siRNA 5' end formation in Drosophila. Science. 2007;315:1587–1590. doi: 10.1126/science.1140494. [DOI] [PubMed] [Google Scholar]
- Haase AD, Fenoglio S, Muerdter F, Guzzardo PM, Czech B, Pappin DJ, Chen C, Gordon A, Hannon GJ. Probing the initiation and effector phases of the somatic piRNA pathway in Drosophila. Genes & development. 2010;24:2499–2504. doi: 10.1101/gad.1968110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall SE, Chirn GW, Lau NC, Sengupta P. RNAi pathways contribute to developmental history-dependent phenotypic plasticity in C. elegans. RNA. 2013 doi: 10.1261/rna.036418.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han T, Manoharan AP, Harkins TT, Bouffard P, Fitzpatrick C, Chu DS, Thierry-Mieg D, Thierry-Mieg J, Kim JK. 26G endo-siRNAs regulate spermatogenic and zygotic gene expression in Caenorhabditis elegans. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:18674–18679. doi: 10.1073/pnas.0906378106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handler D, Meixner K, Pizka M, Lauss K, Schmied C, Gruber FS, Brennecke J. The Genetic Makeup of the Drosophila piRNA Pathway. Molecular cell. 2013 doi: 10.1016/j.molcel.2013.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handler D, Olivieri D, Novatchkova M, Gruber FS, Meixner K, Mechtler K, Stark A, Sachidanandam R, Brennecke J. A systematic analysis of Drosophila TUDOR domain-containing proteins identifies Vreteno and the Tdrd12 family as essential primary piRNA pathway factors. The EMBO journal. 2011;30:3977–3993. doi: 10.1038/emboj.2011.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris AN, Macdonald PM. Aubergine encodes a Drosophila polar granule component required for pole cell formation and related to eIF2C. Development. 2001;128:2823–2832. doi: 10.1242/dev.128.14.2823. [DOI] [PubMed] [Google Scholar]
- Haynes KA, Caudy AA, Collins L, Elgin SC. Element 1360 and RNAi components contribute to HP1-dependent silencing of a pericentric reporter. Current biology : CB. 2006;16:2222–2227. doi: 10.1016/j.cub.2006.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horwich MD, Li C, Matranga C, Vagin V, Farley G, Wang P, Zamore PD. The Drosophila RNA methyltransferase, DmHen1, modifies germline piRNAs and single-stranded siRNAs in RISC. Current biology : CB. 2007;17:1265–1272. doi: 10.1016/j.cub.2007.06.030. [DOI] [PubMed] [Google Scholar]
- Hou Y, Yuan J, Zhou X, Fu X, Cheng H, Zhou R. DNA demethylation and USF regulate the meiosis-specific expression of the mouse Miwi. PLoS Genet. 2012;8:e1002716. doi: 10.1371/journal.pgen.1002716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houston DW, King ML. Germ plasm and molecular determinants of germ cell fate. Current topics in developmental biology. 2000;50:155–181. doi: 10.1016/s0070-2153(00)50008-8. [DOI] [PubMed] [Google Scholar]
- Houwing S, Berezikov E, Ketting RF. Zili is required for germ cell differentiation and meiosis in zebrafish. The EMBO journal. 2008;27:2702–2711. doi: 10.1038/emboj.2008.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houwing S, Kamminga LM, Berezikov E, Cronembold D, Girard A, van den Elst H, Filippov DV, Blaser H, Raz E, Moens CB, et al. A Role for Piwi and piRNAs in Germ Cell Maintenance and Transposon Silencing in Zebrafish. Cell. 2007;129:69–82. doi: 10.1016/j.cell.2007.03.026. [DOI] [PubMed] [Google Scholar]
- Huang H, Gao Q, Peng X, Choi SY, Sarma K, Ren H, Morris AJ, Frohman MA. piRNA-associated germline nuage formation and spermatogenesis require MitoPLD profusogenic mitochondrial-surface lipid signaling. Developmental cell. 2011a;20:376–387. doi: 10.1016/j.devcel.2011.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang HY, Houwing S, Kaaij LJ, Meppelink A, Redl S, Gauci S, Vos H, Draper BW, Moens CB, Burgering BM, et al. Tdrd1 acts as a molecular scaffold for Piwi proteins and piRNA targets in zebrafish. The EMBO journal. 2011b;30:3298–3308. doi: 10.1038/emboj.2011.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang XA, Yin H, Sweeney S, Raha D, Snyder M, Lin H. A major epigenetic programming mechanism guided by piRNAs. Developmental cell. 2013;24:502–516. doi: 10.1016/j.devcel.2013.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ipsaro JJ, Haase AD, Knott SR, Joshua-Tor L, Hannon GJ. The structural biochemistry of Zucchini implicates it as a nuclease in piRNA biogenesis. Nature. 2012;491:279–283. doi: 10.1038/nature11502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irvine DV, Zaratiegui M, Tolia NH, Goto DB, Chitwood DH, Vaughn MW, Joshua-Tor L, Martienssen RA. Argonaute slicing is required for heterochromatic silencing and spreading. Science. 2006;313:1134–1137. doi: 10.1126/science.1128813. [DOI] [PubMed] [Google Scholar]
- Ishizu H, Siomi H, Siomi MC. Biology of PIWI-interacting RNAs: new insights into biogenesis and function inside and outside of germlines. Genes & development. 2012;26:2361–2373. doi: 10.1101/gad.203786.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janic A, Mendizabal L, Llamazares S, Rossell D, Gonzalez C. Ectopic expression of germline genes drives malignant brain tumor growth in Drosophila. Science. 2010;330:1824–1827. doi: 10.1126/science.1195481. [DOI] [PubMed] [Google Scholar]
- Juliano C, Wang J, Lin H. Uniting germline and stem cells: the function of Piwi proteins and the piRNA pathway in diverse organisms. Annual review of genetics. 2011;45:447–469. doi: 10.1146/annurev-genet-110410-132541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamminga LM, van Wolfswinkel JC, Luteijn MJ, Kaaij LJ, Bagijn MP, Sapetschnig A, Miska EA, Berezikov E, Ketting RF. Differential impact of the HEN1 homolog HENN-1 on 21U and 26G RNAs in the germline of Caenorhabditis elegans. PLoS Genet. 2012;8:e1002702. doi: 10.1371/journal.pgen.1002702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaoka S, Izumi N, Katsuma S, Tomari Y. 3' end formation of PIWI-interacting RNAs in vitro. Molecular cell. 2011;43:1015–1022. doi: 10.1016/j.molcel.2011.07.029. [DOI] [PubMed] [Google Scholar]
- Kawaoka S, Mitsutake H, Kiuchi T, Kobayashi M, Yoshikawa M, Suzuki Y, Sugano S, Shimada T, Kobayashi J, Tomari Y, et al. A role for transcription from a piRNA cluster in de novo piRNA production. RNA. 2012;18:265–273. doi: 10.1261/rna.029777.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelleher ES, Edelman NB, Barbash DA. Drosophila interspecific hybrids phenocopy piRNA-pathway mutants. PLoS biology. 2012;10:e1001428. doi: 10.1371/journal.pbio.1001428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ketting RF. The many faces of RNAi. Developmental cell. 2011;20:148–161. doi: 10.1016/j.devcel.2011.01.012. [DOI] [PubMed] [Google Scholar]
- Khurana JS, Wang J, Xu J, Koppetsch BS, Thomson TC, Nowosielska A, Li C, Zamore PD, Weng Z, Theurkauf WE. Adaptation to P element transposon invasion in Drosophila melanogaster. Cell. 2011;147:1551–1563. doi: 10.1016/j.cell.2011.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirino Y, Kim N, de Planell-Saguer M, Khandros E, Chiorean S, Klein PS, Rigoutsos I, Jongens TA, Mourelatos Z. Arginine methylation of Piwi proteins catalysed by dPRMT5 is required for Ago3 and Aub stability. Nat Cell Biol. 2009;11:652–658. doi: 10.1038/ncb1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klattenhoff C, Xi H, Li C, Lee S, Xu J, Khurana JS, Zhang F, Schultz N, Koppetsch BS, Nowosielska A, et al. The Drosophila HP1 homolog Rhino is required for transposon silencing and piRNA production by dual-strand clusters. Cell. 2009;138:1137–1149. doi: 10.1016/j.cell.2009.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotaja N, Sassone-Corsi P. The chromatoid body: a germ-cell-specific RNA-processing centre. Nat Rev Mol Cell Biol. 2007;8:85–90. doi: 10.1038/nrm2081. [DOI] [PubMed] [Google Scholar]
- Kuramochi-Miyagawa S, Watanabe T, Gotoh K, Takamatsu K, Chuma S, Kojima-Kita K, Shiromoto Y, Asada N, Toyoda A, Fujiyama A, et al. MVH in piRNA processing and gene silencing of retrotransposons. Genes & development. 2010;24:887–892. doi: 10.1101/gad.1902110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuramochi-Miyagawa S, Watanabe T, Gotoh K, Totoki Y, Toyoda A, Ikawa M, Asada N, Kojima K, Yamaguchi Y, Ijiri TW, et al. DNA methylation of retrotransposon genes is regulated by Piwi family members MILI and MIWI2 in murine fetal testes. Genes & development. 2008;22:908–917. doi: 10.1101/gad.1640708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- L ML, J J, J RJ, S P, A K, M M, J WP, B B, S LE, R KE. Aplysia Genome Project. p. 11× coverage sequencing of Aplysia californica genome. Broad Institute. 2009 [Google Scholar]
- Lane N, Dean W, Erhardt S, Hajkova P, Surani A, Walter J, Reik W. Resistance of IAPs to methylation reprogramming may provide a mechanism for epigenetic inheritance in the mouse. Genesis. 2003;35:88–93. doi: 10.1002/gene.10168. [DOI] [PubMed] [Google Scholar]
- Lau NC. Small RNAs in the animal gonad: guarding genomes and guiding development. The international journal of biochemistry & cell biology. 2010;42:1334–1347. doi: 10.1016/j.biocel.2010.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau NC, Ohsumi T, Borowsky M, Kingston RE, Blower MD. Systematic and single cell analysis of Xenopus Piwi-interacting RNAs and Xiwi. The EMBO journal. 2009;28:2945–2958. doi: 10.1038/emboj.2009.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau NC, Seto AG, Kim J, Kuramochi-Miyagawa S, Nakano T, Bartel DP, Kingston RE. Characterization of the piRNA complex from rat testes. Science. 2006;313:363–367. doi: 10.1126/science.1130164. [DOI] [PubMed] [Google Scholar]
- Le Thomas A, Rogers AK, Webster A, Marinov GK, Liao SE, Perkins EM, Hur JK, Aravin AA, Toth KF. Piwi induces piRNA-guided transcriptional silencing and establishment of a repressive chromatin state. Genes & development. 2013;27:390–399. doi: 10.1101/gad.209841.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee EJ, Banerjee S, Zhou H, Jammalamadaka A, Arcila M, Manjunath BS, Kosik KS. Identification of piRNAs in the central nervous system. RNA. 2011;17:1090–1099. doi: 10.1261/rna.2565011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HC, Gu W, Shirayama M, Youngman E, Conte D, Jr, Mello CC. C. elegans piRNAs mediate the genome-wide surveillance of germline transcripts. Cell. 2012;150:78–87. doi: 10.1016/j.cell.2012.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee RC, Feinbaum RL, Ambros V. The C elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- Lehmann R, Nusslein-Volhard C. The maternal gene nanos has a central role in posterior pattern formation of the Drosophila embryo. Development. 1991;112:679–691. doi: 10.1242/dev.112.3.679. [DOI] [PubMed] [Google Scholar]
- Li C, Vagin VV, Lee S, Xu J, Ma S, Xi H, Seitz H, Horwich MD, Syrzycka M, Honda BM, et al. Collapse of germline piRNAs in the absence of Argonaute3 reveals somatic piRNAs in flies. Cell. 2009;137:509–521. doi: 10.1016/j.cell.2009.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XZ, Roy CK, Dong X, Bolcun-Filas E, Wang J, Han BW, Xu J, Moore MJ, Schimenti JC, Weng Z, et al. An ancient transcription factor initiates the burst of piRNA production during early meiosis in mouse testes. Molecular cell. 2013;50:67–81. doi: 10.1016/j.molcel.2013.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim AK, Kai T. Unique germ-line organelle, nuage, functions to repress selfish genetic elements in Drosophila melanogaster. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:6714–6719. doi: 10.1073/pnas.0701920104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H, Spradling AC. A novel group of pumilio mutations affects the asymmetric division of germline stem cells in the Drosophila ovary. Development. 1997;124:2463–2476. doi: 10.1242/dev.124.12.2463. [DOI] [PubMed] [Google Scholar]
- Liu J, Carmell MA, Rivas FV, Marsden CG, Thomson JM, Song JJ, Hammond SM, Joshua-Tor L, Hannon GJ. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004;305:1437–1441. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- Liu L, Qi H, Wang J, Lin H. PAPI, a novel TUDOR-domain protein, complexes with AGO3, ME31B and TRAL in the nuage to silence transposition. Development. 2011;138:1863–1873. doi: 10.1242/dev.059287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Clark AG. Population dynamics of PIWI-interacting RNAs (piRNAs) and their targets in Drosophila. Genome research. 2010;20:212–227. doi: 10.1101/gr.095406.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X, Wontakal SN, Kavi H, Kim BJ, Guzzardo PM, Emelyanov AV, Xu N, Hannon GJ, Zavadil J, Fyodorov DV, et al. Drosophila H1 regulates the genetic activity of heterochromatin by recruitment of Su(var)3–9. Science. 2013;340:78–81. doi: 10.1126/science.1234654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luteijn MJ, van Bergeijk P, Kaaij LJ, Almeida MV, Roovers EF, Berezikov E, Ketting RF. Extremely stable Piwi-induced gene silencing in Caenorhabditis elegans. The EMBO journal. 2012;31:3422–3430. doi: 10.1038/emboj.2012.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, Buchold GM, Greenbaum MP, Roy A, Burns KH, Zhu H, Han DY, Harris RA, Coarfa C, Gunaratne PH, et al. GASZ is essential for male meiosis and suppression of retrotransposon expression in the male germline. PLoS Genet. 2009;5:e1000635. doi: 10.1371/journal.pgen.1000635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone CD, Brennecke J, Dus M, Stark A, McCombie WR, Sachidanandam R, Hannon GJ. Specialized piRNA pathways act in germline and somatic tissues of the Drosophila ovary. Cell. 2009;137:522–535. doi: 10.1016/j.cell.2009.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megosh HB, Cox DN, Campbell C, Lin H. The role of PIWI and the miRNA machinery in Drosophila germline determination. Current biology : CB. 2006;16:1884–1894. doi: 10.1016/j.cub.2006.08.051. [DOI] [PubMed] [Google Scholar]
- Meister G, Landthaler M, Peters L, Chen PY, Urlaub H, Luhrmann R, Tuschl T. Identification of novel argonaute-associated proteins. Current biology : CB. 2005;15:2149–2155. doi: 10.1016/j.cub.2005.10.048. [DOI] [PubMed] [Google Scholar]
- Mochizuki K, Fine NA, Fujisawa T, Gorovsky MA. Analysis of a piwi-related gene implicates small RNAs in genome rearrangement in tetrahymena. Cell. 2002;110:689–699. doi: 10.1016/s0092-8674(02)00909-1. [DOI] [PubMed] [Google Scholar]
- Mochizuki K, Gorovsky MA. Conjugation-specific small RNAs in Tetrahymena have predicted properties of scan (scn) RNAs involved in genome rearrangement. Genes & development. 2004a;18:2068–2073. doi: 10.1101/gad.1219904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochizuki K, Gorovsky MA. RNA polymerase II localizes in Tetrahymena thermophila meiotic micronuclei when micronuclear transcription associated with genome rearrangement occurs. Eukaryotic cell. 2004b;3:1233–1240. doi: 10.1128/EC.3.5.1233-1240.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery TA, Rim YS, Zhang C, Dowen RH, Phillips CM, Fischer SE, Ruvkun G. PIWI associated siRNAs and piRNAs specifically require the Caenorhabditis elegans HEN1 ortholog henn-1. PLoS Genet. 2012;8:e1002616. doi: 10.1371/journal.pgen.1002616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moshkovich N, Lei EP. HP1 recruitment in the absence of argonaute proteins in Drosophila. PLoS Genet. 2010;6:e1000880. doi: 10.1371/journal.pgen.1000880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muerdter F, Guzzardo PM, Gillis J, Luo Y, Yu Y, Chen C, Fekete R, Hannon GJ. A Genome-wide RNAi Screen Draws a Genetic Framework for Transposon Control and Primary piRNA Biogenesis in Drosophila. Molecular cell. 2013 doi: 10.1016/j.molcel.2013.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muerdter F, Olovnikov I, Molaro A, Rozhkov NV, Czech B, Gordon A, Hannon GJ, Aravin AA. Production of artificial piRNAs in flies and mice. RNA. 2012;18:42–52. doi: 10.1261/rna.029769.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida KM, Okada TN, Kawamura T, Mituyama T, Kawamura Y, Inagaki S, Huang H, Chen D, Kodama T, Siomi H, et al. Functional involvement of Tudor and dPRMT5 in the piRNA processing pathway in Drosophila germlines. The EMBO journal. 2009;28:3820–3831. doi: 10.1038/emboj.2009.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimasu H, Ishizu H, Saito K, Fukuhara S, Kamatani MK, Bonnefond L, Matsumoto N, Nishizawa T, Nakanaga K, Aoki J, et al. Structure and function of Zucchini endoribonuclease in piRNA biogenesis. Nature. 2012;491:284–287. doi: 10.1038/nature11509. [DOI] [PubMed] [Google Scholar]
- O'Carroll D, Mecklenbrauker I, Das PP, Santana A, Koenig U, Enright AJ, Miska EA, Tarakhovsky A. A Slicer-independent role for Argonaute 2 in hematopoiesis and the microRNA pathway. Genes & development. 2007;21:1999–2004. doi: 10.1101/gad.1565607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura K, Lai EC. Endogenous small interfering RNAs in animals. Nat Rev Mol Cell Biol. 2008;9:673–678. doi: 10.1038/nrm2479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivieri D, Senti KA, Subramanian S, Sachidanandam R, Brennecke J. The cochaperone shutdown defines a group of biogenesis factors essential for all piRNA populations in Drosophila. Molecular cell. 2012;47:954–969. doi: 10.1016/j.molcel.2012.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivieri D, Sykora MM, Sachidanandam R, Mechtler K, Brennecke J. An in vivo RNAi assay identifies major genetic and cellular requirements for primary piRNA biogenesis in Drosophila. The EMBO journal. 2010;29:3301–3317. doi: 10.1038/emboj.2010.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal-Bhadra M, Bhadra U, Birchler JA. RNAi related mechanisms affect both transcriptional and posttranscriptional transgene silencing in Drosophila. Molecular cell. 2002;9:315–327. doi: 10.1016/s1097-2765(02)00440-9. [DOI] [PubMed] [Google Scholar]
- Pal Bhadra M, Bhadra U, Birchler JA. Misregulation of sex-lethal and disruption of male-specific lethal complex localization in Drosophila species hybrids. Genetics. 2006;174:1151–1159. doi: 10.1534/genetics.106.060541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palakodeti D, Smielewska M, Lu YC, Yeo GW, Graveley BR. The PIWI proteins SMEDWI-2 and SMEDWI-3 are required for stem cell function and piRNA expression in planarians. RNA. 2008;14:1174–1186. doi: 10.1261/rna.1085008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pane A, Jiang P, Zhao DY, Singh M, Schupbach T. The Cutoff protein regulates piRNA cluster expression and piRNA production in the Drosophila germline. The EMBO journal. 2011;30:4601–4615. doi: 10.1038/emboj.2011.334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pane A, Wehr K, Schupbach T. zucchini and squash encode two putative nucleases required for rasiRNA production in the Drosophila germline. Developmental cell. 2007;12:851–862. doi: 10.1016/j.devcel.2007.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pao SY, Lin WL, Hwang MJ. In silico identification and comparative analysis of differentially expressed genes in human and mouse tissues. BMC genomics. 2006;7:86. doi: 10.1186/1471-2164-7-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patil VS, Kai T. Repression of Retroelements in Drosophila Germline via piRNA Pathway by the Tudor Domain Protein Tejas. Current biology : CB. 2010 doi: 10.1016/j.cub.2010.02.046. [DOI] [PubMed] [Google Scholar]
- Pavelec DM, Lachowiec J, Duchaine TF, Smith HE, Kennedy S. Requirement for the ERI/DICER complex in endogenous RNA interference and sperm development in Caenorhabditis elegans. Genetics. 2009;183:1283–1295. doi: 10.1534/genetics.109.108134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrat PN, DasGupta S, Wang J, Theurkauf W, Weng Z, Rosbash M, Waddell S. Transposition-driven genomic heterogeneity in the Drosophila brain. Science. 2013;340:91–95. doi: 10.1126/science.1231965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen CP, Reddien PW. Wnt signaling and the polarity of the primary body axis. Cell. 2009;139:1056–1068. doi: 10.1016/j.cell.2009.11.035. [DOI] [PubMed] [Google Scholar]
- Preall JB, Czech B, Guzzardo PM, Muerdter F, Hannon GJ. shutdown is a component of the Drosophila piRNA biogenesis machinery. RNA. 2012;18:1446–1457. doi: 10.1261/rna.034405.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajasethupathy P, Antonov I, Sheridan R, Frey S, Sander C, Tuschl T, Kandel ER. A role for neuronal piRNAs in the epigenetic control of memory-related synaptic plasticity. Cell. 2012;149:693–707. doi: 10.1016/j.cell.2012.02.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangan P, Malone CD, Navarro C, Newbold SP, Hayes PS, Sachidanandam R, Hannon GJ, Lehmann R. piRNA production requires heterochromatin formation in Drosophila. Current biology : CB. 2011;21:1373–1379. doi: 10.1016/j.cub.2011.06.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravasi T, Suzuki H, Pang KC, Katayama S, Furuno M, Okunishi R, Fukuda S, Ru K, Frith MC, Gongora MM, et al. Experimental validation of the regulated expression of large numbers of non-coding RNAs from the mouse genome. Genome research. 2006;16:11–19. doi: 10.1101/gr.4200206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddien PW, Bermange AL, Kicza AM, Sanchez Alvarado A. BMP signaling regulates the dorsal planarian midline and is needed for asymmetric regeneration. Development. 2007;134:4043–4051. doi: 10.1242/dev.007138. [DOI] [PubMed] [Google Scholar]
- Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR, Ruvkun G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- Reuter M, Berninger P, Chuma S, Shah H, Hosokawa M, Funaya C, Antony C, Sachidanandam R, Pillai RS. Miwi catalysis is required for piRNA amplification-independent LINE1 transposon silencing. Nature. 2011;480:264–267. doi: 10.1038/nature10672. [DOI] [PubMed] [Google Scholar]
- Reuter M, Chuma S, Tanaka T, Franz T, Stark A, Pillai RS. Loss of the Mili-interacting Tudor domain-containing protein-1 activates transposons and alters the Mili-associated small RNA profile. Nat Struct Mol Biol. 2009;16:639–646. doi: 10.1038/nsmb.1615. [DOI] [PubMed] [Google Scholar]
- Robine N, Lau NC, Balla S, Jin Z, Okamura K, Kuramochi-Miyagawa S, Blower MD, Lai EC. A broadly conserved pathway generates 3'UTR-directed primary piRNAs. Current biology : CB. 2009;19:2066–2076. doi: 10.1016/j.cub.2009.11.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouget C, Papin C, Boureux A, Meunier AC, Franco B, Robine N, Lai EC, Pelisson A, Simonelig M. Maternal mRNA deadenylation and decay by the piRNA pathway in the early Drosophila embryo. Nature. 2010;467:1128–1132. doi: 10.1038/nature09465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozhkov NV, Hammell M, Hannon GJ. Multiple roles for Piwi in silencing Drosophila transposons. Genes & development. 2013;27:400–412. doi: 10.1101/gad.209767.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruby JG, Jan C, Player C, Axtell MJ, Lee W, Nusbaum C, Ge H, Bartel DP. Large-scale sequencing reveals 21U-RNAs and additional microRNAs and endogenous siRNAs in C. elegans. Cell. 2006;127:1193–1207. doi: 10.1016/j.cell.2006.10.040. [DOI] [PubMed] [Google Scholar]
- Saito K, Inagaki S, Mituyama T, Kawamura Y, Ono Y, Sakota E, Kotani H, Asai K, Siomi H, Siomi MC. A regulatory circuit for piwi by the large Maf gene traffic jam in Drosophila. Nature. 2009;461:1296–1299. doi: 10.1038/nature08501. [DOI] [PubMed] [Google Scholar]
- Saito K, Ishizu H, Komai M, Kotani H, Kawamura Y, Nishida KM, Siomi H, Siomi MC. Roles for the Yb body components Armitage and Yb in primary piRNA biogenesis in Drosophila. Genes & development. 2010;24:2493–2498. doi: 10.1101/gad.1989510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K, Nishida KM, Mori T, Kawamura Y, Miyoshi K, Nagami T, Siomi H, Siomi MC. Specific association of Piwi with rasiRNAs derived from retrotransposon and heterochromatic regions in the Drosophila genome. Genes & development. 2006;20:2214–2222. doi: 10.1101/gad.1454806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K, Sakaguchi Y, Suzuki T, Siomi H, Siomi MC. Pimet, the Drosophila homolog of HEN1, mediates 2'-O-methylation of Piwi- interacting RNAs at their 3' ends. Genes & development. 2007;21:1603–1608. doi: 10.1101/gad.1563607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato K, Nishida KM, Shibuya A, Siomi MC, Siomi H. Maelstrom coordinates microtubule organization during Drosophila oogenesis through interaction with components of the MTOC. Genes & development. 2011;25:2361–2373. doi: 10.1101/gad.174110.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato K, Siomi H. Is canalization more than just a beautiful idea? Genome biology. 2010;11:109. doi: 10.1186/gb-2010-11-3-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxe JP, Chen M, Zhao H, Lin H. Tdrkh is essential for spermatogenesis and participates in primary piRNA biogenesis in the germline. The EMBO journal. 2013 doi: 10.1038/emboj.2013.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoeberl UE, Kurth HM, Noto T, Mochizuki K. Biased transcription and selective degradation of small RNAs shape the pattern of DNA elimination in Tetrahymena. Genes & development. 2012;26:1729–1742. doi: 10.1101/gad.196493.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schupbach T, Wieschaus E. Female sterile mutations on the second chromosome of Drosophila melanogaster. I. Maternal effect mutations. Genetics. 1989;121:101–117. doi: 10.1093/genetics/121.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schupbach T, Wieschaus E. Female sterile mutations on the second chromosome of Drosophila melanogaster. II. Mutations blocking oogenesis or altering egg morphology. Genetics. 1991;129:1119–1136. doi: 10.1093/genetics/129.4.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sentmanat MF, Elgin SC. Ectopic assembly of heterochromatin in Drosophila melanogaster triggered by transposable elements. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:14104–14109. doi: 10.1073/pnas.1207036109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- She X, Xu X, Fedotov A, Kelly WG, Maine EM. Regulation of heterochromatin assembly on unpaired chromosomes during caenorhabditis elegans meiosis by components of a small RNA-mediated pathway. PLoS Genet. 2009;5:e1000624. doi: 10.1371/journal.pgen.1000624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Z, Montgomery TA, Qi Y, Ruvkun G. High-throughput sequencing reveals extraordinary fluidity of miRNA, piRNA, and siRNA pathways in nematodes. Genome research. 2013;23:497–508. doi: 10.1101/gr.149112.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirayama M, Seth M, Lee HC, Gu W, Ishidate T, Conte D, Jr, Mello CC. piRNAs initiate an epigenetic memory of nonself RNA in the C. elegans germline. Cell. 2012;150:65–77. doi: 10.1016/j.cell.2012.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiromoto Y, Kuramochi-Miyagawa S, Daiba A, Chuma S, Katanaya A, Katsumata A, Nishimura K, Ohtaka M, Nakanishi M, Nakamura T, et al. GPAT2, a mitochondrial outer membrane protein, in piRNA biogenesis in germline stem cells. RNA. 2013;19:803–810. doi: 10.1261/rna.038521.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoji M, Tanaka T, Hosokawa M, Reuter M, Stark A, Kato Y, Kondoh G, Okawa K, Chujo T, Suzuki T, et al. The TDRD9-MIWI2 complex is essential for piRNA-mediated retrotransposon silencing in the mouse male germline. Developmental cell. 2009;17:775–787. doi: 10.1016/j.devcel.2009.10.012. [DOI] [PubMed] [Google Scholar]
- Shpiz S, Kalmykova A. Epigenetic transmission of piRNAs through the female germline. Genome biology. 2009;10:208. doi: 10.1186/gb-2009-10-2-208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sienski G, Donertas D, Brennecke J. Transcriptional silencing of transposons by Piwi and maelstrom and its impact on chromatin state and gene expression. Cell. 2012;151:964–980. doi: 10.1016/j.cell.2012.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siomi MC, Mannen T, Siomi H. How does the royal family of Tudor rule the PIWI-interacting RNA pathway? Genes & development. 2010;24:636–646. doi: 10.1101/gad.1899210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siomi MC, Sato K, Pezic D, Aravin AA. PIWI-interacting small RNAs: the vanguard of genome defence. Nat Rev Mol Cell Biol. 2011;12:246–258. doi: 10.1038/nrm3089. [DOI] [PubMed] [Google Scholar]
- Song Y, Fee L, Lee TH, Wharton RP. The molecular chaperone Hsp90 is required for mRNA localization in Drosophila melanogaster embryos. Genetics. 2007;176:2213–2222. doi: 10.1534/genetics.107.071472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soper SF, van der Heijden GW, Hardiman TC, Goodheart M, Martin SL, de Boer P, Bortvin A. Mouse maelstrom, a component of nuage, is essential for spermatogenesis and transposon repression in meiosis. Developmental cell. 2008;15:285–297. doi: 10.1016/j.devcel.2008.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Specchia V, Piacentini L, Tritto P, Fanti L, D'Alessandro R, Palumbo G, Pimpinelli S, Bozzetti MP. Hsp90 prevents phenotypic variation by suppressing the mutagenic activity of transposons. Nature. 2010;463:662–665. doi: 10.1038/nature08739. [DOI] [PubMed] [Google Scholar]
- Swart EC, Bracht JR, Magrini V, Minx P, Chen X, Zhou Y, Khurana JS, Goldman AD, Nowacki M, Schotanus K, et al. The Oxytricha trifallax macronuclear genome: a complex eukaryotic genome with 16,000 tiny chromosomes. PLoS biology. 2013;11:e1001473. doi: 10.1371/journal.pbio.1001473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sytnikova YA, Post C, Chirn GW, Clark J, Lau NC. Target binding and silencing capacity of the Drosophila PIWI protein and piRNAs. Submitted. 2013 doi: 10.1261/rna.046300.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam OH, Aravin AA, Stein P, Girard A, Murchison EP, Cheloufi S, Hodges E, Anger M, Sachidanandam R, Schultz RM, et al. Pseudogene-derived small interfering RNAs regulate gene expression in mouse oocytes. Nature. 2008;453:534–538. doi: 10.1038/nature06904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toscani A, Mettus RV, Coupland R, Simpkins H, Litvin J, Orth J, Hatton KS, Reddy EP. Arrest of spermatogenesis and defective breast development in mice lacking A-myb. Nature. 1997;386:713–717. doi: 10.1038/386713a0. [DOI] [PubMed] [Google Scholar]
- Unhavaithaya Y, Hao Y, Beyret E, Yin H, Kuramochi-Miyagawa S, Nakano T, Lin H. MILI, a PIWI-interacting RNA-binding protein, is required for germ line stem cell self-renewal and appears to positively regulate translation. The Journal of biological chemistry. 2009;284:6507–6519. doi: 10.1074/jbc.M809104200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagin VV, Sigova A, Li C, Seitz H, Gvozdev V, Zamore PD. A distinct small RNA pathway silences selfish genetic elements in the germline. Science. 2006;313:320–324. doi: 10.1126/science.1129333. [DOI] [PubMed] [Google Scholar]
- Vagin VV, Wohlschlegel J, Qu J, Jonsson Z, Huang X, Chuma S, Girard A, Sachidanandam R, Hannon GJ, Aravin AA. Proteomic analysis of murine Piwi proteins reveals a role for arginine methylation in specifying interaction with Tudor family members. Genes & development. 2009;23:1749–1762. doi: 10.1101/gad.1814809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Wolfswinkel JC, Claycomb JM, Batista PJ, Mello CC, Berezikov E, Ketting RF. CDE-1 affects chromosome segregation through uridylation of CSR-1-bound siRNAs. Cell. 2009;139:135–148. doi: 10.1016/j.cell.2009.09.012. [DOI] [PubMed] [Google Scholar]
- Vasale JJ, Gu W, Thivierge C, Batista PJ, Claycomb JM, Youngman EM, Duchaine TF, Mello CC, Conte D., Jr Sequential rounds of RNA-dependent RNA transcription drive endogenous small-RNA biogenesis in the ERGO-1/Argonaute pathway. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:3582–3587. doi: 10.1073/pnas.0911908107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasileva A, Tiedau D, Firooznia A, Muller-Reichert T, Jessberger R. Tdrd6 is required for spermiogenesis, chromatoid body architecture, and regulation of miRNA expression. Current biology : CB. 2009;19:630–639. doi: 10.1016/j.cub.2009.02.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vourekas A, Zheng Q, Alexiou P, Maragkakis M, Kirino Y, Gregory BD, Mourelatos Z. Mili and Miwi target RNA repertoire reveals piRNA biogenesis and function of Miwi in spermiogenesis. Nat Struct Mol Biol. 2012;19:773–781. doi: 10.1038/nsmb.2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Dickinson LK, Lehmann R. Genetics of nanos localization in Drosophila. Developmental dynamics : an official publication of the American Association of Anatomists. 1994;199:103–115. doi: 10.1002/aja.1001990204. [DOI] [PubMed] [Google Scholar]
- Wang J, Czech B, Crunk A, Wallace A, Mitreva M, Hannon GJ, Davis RE. Deep small RNA sequencing from the nematode Ascaris reveals conservation, functional diversification, and novel developmental profiles. Genome research. 2011;21:1462–1477. doi: 10.1101/gr.121426.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Saxe JP, Tanaka T, Chuma S, Lin H. Mili interacts with tudor domain-containing protein 1 in regulating spermatogenesis. Current biology : CB. 2009;19:640–644. doi: 10.1016/j.cub.2009.02.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang SH, Elgin SC. Drosophila Piwi functions downstream of piRNA production mediating a chromatin-based transposon silencing mechanism in female germ line. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:21164–21169. doi: 10.1073/pnas.1107892109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T, Chuma S, Yamamoto Y, Kuramochi-Miyagawa S, Totoki Y, Toyoda A, Hoki Y, Fujiyama A, Shibata T, Sado T, et al. MITOPLD is a mitochondrial protein essential for nuage formation and piRNA biogenesis in the mouse germline. Developmental cell. 2011;20:364–375. doi: 10.1016/j.devcel.2011.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T, Totoki Y, Toyoda A, Kaneda M, Kuramochi-Miyagawa S, Obata Y, Chiba H, Kohara Y, Kono T, Nakano T, et al. Endogenous siRNAs from naturally formed dsRNAs regulate transcripts in mouse oocytes. Nature. 2008;453:539–543. doi: 10.1038/nature06908. [DOI] [PubMed] [Google Scholar]
- Wilson JE, Connell JE, Macdonald PM. aubergine enhances oskar translation in the Drosophila ovary. Development. 1996;122:1631–1639. doi: 10.1242/dev.122.5.1631. [DOI] [PubMed] [Google Scholar]
- Xiol J, Cora E, Koglgruber R, Chuma S, Subramanian S, Hosokawa M, Reuter M, Yang Z, Berninger P, Palencia A, et al. A role for Fkbp6 and the chaperone machinery in piRNA amplification and transposon silencing. Molecular cell. 2012;47:970–979. doi: 10.1016/j.molcel.2012.07.019. [DOI] [PubMed] [Google Scholar]
- Yabuta Y, Ohta H, Abe T, Kurimoto K, Chuma S, Saitou M. TDRD5 is required for retrotransposon silencing, chromatoid body assembly, and spermiogenesis in mice. The Journal of cell biology. 2011;192:781–795. doi: 10.1083/jcb.201009043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Z, Hu HY, Jiang X, Maierhofer V, Neb E, He L, Hu Y, Hu H, Li N, Chen W, et al. Widespread expression of piRNA-like molecules in somatic tissues. Nucleic acids research. 2011;39:6596–6607. doi: 10.1093/nar/gkr298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo G, Holste D, Kreiman G, Burge CB. Variation in alternative splicing across human tissues. Genome biology. 2004;5:R74. doi: 10.1186/gb-2004-5-10-r74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yigit E, Batista PJ, Bei Y, Pang KM, Chen CC, Tolia NH, Joshua-Tor L, Mitani S, Simard MJ, Mello CC. Analysis of the C. elegans Argonaute family reveals that distinct Argonautes act sequentially during RNAi. Cell. 2006;127:747–757. doi: 10.1016/j.cell.2006.09.033. [DOI] [PubMed] [Google Scholar]
- Yin H, Lin H. An epigenetic activation role of Piwi and a Piwi-associated piRNA in Drosophila melanogaster. Nature. 2007;450:304–308. doi: 10.1038/nature06263. [DOI] [PubMed] [Google Scholar]
- Zamparini AL, Davis MY, Malone CD, Vieira E, Zavadil J, Sachidanandam R, Hannon GJ, Lehmann R. Vreteno, a gonad-specific protein, is essential for germline development and primary piRNA biogenesis in Drosophila. Development. 2011;138:4039–4050. doi: 10.1242/dev.069187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Montgomery TA, Gabel HW, Fischer SE, Phillips CM, Fahlgren N, Sullivan CM, Carrington JC, Ruvkun G. mut-16 and other mutator class genes modulate 22G and 26G siRNA pathways in Caenorhabditis elegans. Proceedings of the National Academy of Sciences of the United States of America. 2011a;108:1201–1208. doi: 10.1073/pnas.1018695108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Wang J, Xu J, Zhang Z, Koppetsch BS, Schultz N, Vreven T, Meignin C, Davis I, Zamore PD, et al. UAP56 couples piRNA clusters to the perinuclear transposon silencing machinery. Cell. 2012;151:871–884. doi: 10.1016/j.cell.2012.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Xu J, Koppetsch BS, Wang J, Tipping C, Ma S, Weng Z, Theurkauf WE, Zamore PD. Heterotypic piRNA Ping-Pong requires qin a protein with both E3 ligase and Tudor domains. Molecular cell. 2011b;44:572–584. doi: 10.1016/j.molcel.2011.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S, Gou LT, Zhang M, Zu LD, Hua MM, Hua Y, Shi HJ, Li Y, Li J, Li D, et al. piRNA-triggered MIWI ubiquitination and removal by APC/C in late spermatogenesis. Developmental cell. 2013;24:13–25. doi: 10.1016/j.devcel.2012.12.006. [DOI] [PubMed] [Google Scholar]
- Zheng K, Xiol J, Reuter M, Eckardt S, Leu NA, McLaughlin KJ, Stark A, Sachidanandam R, Pillai RS, Wang PJ. Mouse MOV10L1 associates with Piwi proteins and is an essential component of the Piwi-interacting RNA (piRNA) pathway. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:11841–11846. doi: 10.1073/pnas.1003953107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu W, Pao GM, Satoh A, Cummings G, Monaghan JR, Harkins TT, Bryant SV, Randal Voss S, Gardiner DM, Hunter T. Activation of germline-specific genes is required for limb regeneration in the Mexican axolotl. Developmental biology. 2012;370:42–51. doi: 10.1016/j.ydbio.2012.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]