Abstract
Caldicellulosiruptor bescii efficiently degrades cellulose, xylan, and native grasses at high temperatures above 70°C under anaerobic conditions. C. bescii extracellularly secretes multidomain glycoside hydrolases along with proteins of unknown function. In this study, we analyzed the C. bescii proteins that bind to the cell walls of timothy grass by using mass spectrometry, and we identified four noncatalytic plant cell wall-binding proteins (PWBPs) with high pI values (9.2 to 9.6). A search of a conserved domain database showed that these proteins possess a common domain related to solute-binding proteins. In addition, 12 genes encoding PWBP-like proteins were detected in the C. bescii genomic sequence. To analyze the binding properties of PWBPs, recombinant PWBP57 and PWBP65, expressed in Escherichia coli, were prepared. The PWBPs displayed a wide range of binding specificities: they bound to cellulose, lichenan, xylan, arabinoxylan, glucuronoxylan, mannan, glucomannan, pectin, oligosaccharides, and the cell walls of timothy grass. The proteins showed the highest binding affinity for the plant cell wall, with association constant (Ka) values of 5.2 × 106 to 44 × 106 M−1 among the insoluble polysaccharides tested, as measured using depletion binding isotherms. Affinity gel electrophoresis demonstrated that the proteins bound to the acidic polymer pectin most strongly among the soluble polysaccharides tested. Fluorescence microscopic analysis showed that the proteins bound preferentially to the cell wall in a section of grass leaf. Binding of noncatalytic PWBPs with high pI values might be necessary for efficient utilization of polysaccharides by C. bescii at high temperatures.
INTRODUCTION
The plant cell wall is the most abundant bioresource on Earth (1), and efficient conversion of the cellulosic biomass to fermentable sugars is a crucial process for cost-effective production of lignocellulosic biofuels (2). The plant cell wall is composed of polysaccharides (cellulose, hemicellulose, and pectin) and lignin, a heterogeneous aromatic polymer. Cellulose, a β-1,4-linked glucose polymer, is the main structural fiber, surrounded by hemicellulose and pectin, and has a tightly packed crystalline structure. Hemicellulose contains a variety of polysaccharides, including xylan (a β-1,4-linked xylose polymer), xyloglucan (a β-1,4-linked glucose polymer with xylose side chains), glucuronoxylan (a xylan with side chains of glucuronic acid and glucuronic acid methyl ester), mannan (a β-1,4-linked mannose polymer), and β-1,3/1,4-linked glucose polymers (3). Pectin is an acidic polymer composed mainly of galacturonic acids with partial esterification.
Microbial degradation of the plant cell wall plays a primary role in the organic carbon cycle on Earth. Many hydrolases that degrade plant cell walls consist of a single glycoside hydrolase (GH) domain and a single carbohydrate-binding module (CBM). CBMs are defined as discrete folds with carbohydrate-binding activity within a carbohydrate-active enzyme, such as glycoside hydrolase, glycosyltransferase, polysaccharide lyase, or carbohydrate esterase (4). CBMs are classified into families based on similarities in their amino acid sequences; currently, more than 60 CBM families have been identified (5). CBMs present in carbohydrate-active enzymes have been well characterized (6), but little is known about the biochemical properties and microbiological functions of plant cell wall-binding proteins (PWBPs) that lack a catalytic domain.
Caldicellulosiruptor bescii efficiently degrades crystalline cellulose, xylan, and nonpretreated plant biomass, such as switchgrass and hardwood poplar (7). C. bescii is an anaerobic, Gram-positive, nonmotile, non-spore-forming, and extremely thermophilic bacterium with a growth temperature range of 40 to 90°C (optimum of 72 to 80°C) (8, 9), and it is the most thermophilic bacterium capable of efficiently degrading microcrystalline cellulose (10). A crude extract prepared from C. bescii cells shows stronger degradation activity in relation to microcrystalline cellulose and nonpretreated plant biomass compared to the filamentous fungus Trichoderma reesei (11). Some cellulolytic bacteria (e.g., Clostridium thermocellum) produce a large cellulase complex called a cellulosome (12), but Caldicellulosiruptor species do not (10). To degrade the plant cell wall, Caldicellulosiruptor species secrete a diverse set of free cellulases and hemicellulases along with noncatalytic proteins (13–15). The major enzymes are composed of multidomain structures, two GHs, and one to three CBMs (13, 14, 16, 17). Several multidomain enzymes have been biochemically characterized (18–23). Nevertheless, the function and biochemical properties of noncatalytic proteins secreted by Caldicellulosiruptor species are unknown. Therefore, we tested the hypothesis that the proteins secreted by C. bescii include a noncatalytic protein(s) that preferentially binds to polysaccharides present in the plant cell wall. Here, we identify the proteins with high pI values that possess a common domain.
MATERIALS AND METHODS
Bacterial culture.
The C. bescii DSM 6725 strain was obtained from the German Collection of Microorganisms and Cell Cultures (Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH, Braunschweig, Germany). Timothy grass (Phleum pratense), which is used as a feed crop for cattle, was dried at 60°C for 2 days and milled with a ZM100 rotor mill (Retsch, Haan, Germany). Milled powder passed through a mesh with 0.08-mm trapezium holes was used as the substrate. C. bescii was anaerobically cultured at 75°C in a basal medium (pH 7.6 to 7.8) consisting of 2 g/liter yeast extract, 20 mM HEPES, 1 g/liter urea, 0.2 g/liter NaCl, 0.2 g/liter KCl, 0.2 g/liter NH4Cl, 0.18 g/liter MgCl2·6H2O, 72 mg/liter Na2HPO4·12H2O, 20 mg/liter (NH4)2SO4, 50 mg/liter CaCl2·2H2O, 3 mg/liter FeSO4·7H2O, 0.5 mg/liter MnCl2·4H2O, 1 ml/liter each of modified trace mineral and vitamin solutions (11), 1 mg/liter resazurin, and 1 g/liter cysteine-HCl. Timothy grass (5 g/liter) was added to the basal medium as a carbon source.
Preparation of the plant cell wall-binding protein fraction.
C. bescii was cultured in the medium containing timothy grass until it reached the middle of the exponential growth phase (approximately 2 days). The culture supernatant was decanted, and then the sedimented timothy grass was packed into a column (0.5-cm diameter, 5-cm height). The packed timothy grass was thoroughly washed with deionized water and transferred to a 1.5-ml microtube. The proteins that bound to timothy grass were eluted by boiling in a 2× sodium dodecyl sulfate (SDS) sample buffer containing 100 mM Tris-HCl (pH 6.8), 4% (wt/vol) SDS, 20% glycerol, and 1 mg/liter bromophenol blue. The eluted solution served as the plant cell wall-binding protein fraction.
Zymography.
The plant cell wall-binding fraction was separated using SDS-polyacrylamide gel electrophoresis (PAGE) at 10 mA with an 8% (wt/vol) gel containing 2 g/liter carboxymethylcellulose (CMC). The gel was washed with deionized water once, followed by washing three times with 20 mM 2-(N-morpholino)ethanesulfonic acid (MES) buffer (pH 6.0) for 15 min. The gel was incubated in buffer containing 20 mM MES (pH 6.0), 1 mM CaCl2, and 1 mM MgCl2 at 75°C overnight. The gel was stained with a dye solution (1 g/liter Congo red in 1% ethanol) for 30 min and was then washed with 1 M NaCl until positive signals appeared as clear bands.
Mass spectrometry.
Proteins were identified using mass spectrometry as described previously (11). In brief, the plant cell wall-binding fraction was separated by SDS-PAGE using an 8 to 16% gradient gel and was stained with Coomassie brilliant blue (CBB) dye. The proteins were excised from the gel and then digested with trypsin. The digested peptides were analyzed on a 4800 Plus matrix-assisted laser desorption/ionization time of flight/time of flight (MALDI-TOF/TOF) analyzer (AB Sciex, MA). Genes encoding the binding proteins were identified by using MASCOT software (24).
Cloning, expression, and purification of recombinant proteins.
The DNA fragments encoding amino acid residues 22 to 500 of PWBP57 (GenBank accession number ACM59723) and residues 29 to 557 of PWBP65 (GenBank accession number ACM59962) were amplified from the genomic DNA of C. bescii DSM 6725 by PCR using the primers listed in Table S1 of the supplemental material. Truncated N-terminal regions of these proteins contained a putative secretion signal (25). The amplified DNA fragments encoding PWBP57 and PWBP65 were inserted into the pCold I (TaKaRa Bio, Inc., Shiga, Japan) and pET28b (Novagen, WI) vectors, respectively, such that the six-histidine tag sequences were fused to the N termini of the protein products. The vectors were introduced into the Escherichia coli strain Rosetta 2 (Novagen), and the recombinant proteins were overexpressed via induction with 0.2 mM isopropyl-1-thio-β-d-galactopyranoside. The PWBP57 and PWBP65 cells were cultured overnight at 16°C and 18°C, respectively. The two recombinant proteins were purified using the same procedure. Cells producing the recombinant proteins were resuspended in binding buffer (20 mM HEPES [pH 7.2], 20 mM imidazole, 0.3 M NaCl, 1 mM dithiothreitol, 0.025% Tween 20). After cell disruption by sonication, the cell debris was removed by centrifugation at 14,000 × g for 20 min. The soluble fraction was heat treated at 75°C for 20 min and then recentrifuged to remove insoluble material. The soluble fraction was applied to a nickel resin HisTrap HP column (GE Healthcare Biosciences, NJ). After unbound proteins were washed out with binding buffer containing 50 mM imidazole, the recombinant proteins were eluted using binding buffer containing 0.5 M imidazole. Dialysis and removal of the six-histidine tag by protease treatment were performed simultaneously. Factor Xa for PWBP57 or thrombin for PWBP65 was added to the eluted solution, and the solutions were immediately dialyzed against 20 mM HEPES (pH 7.5) buffer containing 0.1 M NaCl and 2 mM 2-mercaptoethanol (2ME) at 4°C overnight. Tag removal was checked after dialysis; tagged proteins were not detected by SDS-PAGE. Tag-removed proteins were further purified using a HisTrap SP HP ion-exchange resin (GE Healthcare Biosciences) with a linear NaCl gradient elution from 0.1 M to 1.0 M in 20 mM HEPES (pH 7.5) containing 2 mM 2ME. Purified proteins were dialyzed against buffer M (pH 6.0) containing 20 mM MES and 2 mM 2ME. Protein concentrations were determined with protein assay CBB solution (Nacalai Tesque, Inc., Kyoto, Japan), using bovine serum albumin (BSA) as a standard.
Fluorescence microscopy.
The PWBP57 and PWBP65 proteins were fluorescently labeled with the HiLyte Fluor 647 and HiLyte Fluor 555 dyes (Dojindo Molecular Technologies, Inc., Gaithersburg, MD) according to the manufacturer's instructions. Leaves of Italian ryegrass (Lolium multiflorum) were fixed with a solution containing 5% formalin, 5% acetate, and 45% ethanol. The leaves were embedded in paraffin and sliced at 5 to 6 μm. After deparaffinization, the sections were incubated with 1 mg/ml BSA in buffer M for 10 min at room temperature for suppression of nonspecific binding. The sections were washed with buffer M and then incubated with the PWBPs (0.1 mg/ml each) in buffer M for 10 min. Localization of the proteins was observed under a laser scanning microscope (LSM 700; Carl Zeiss Japan, Tokyo, Japan).
Insoluble polysaccharide-binding assays.
Each protein (7 μg) was mixed with insoluble polysaccharides (0.5 to 1 mg), Avicel (Merck KGaA, Darmstadt, Germany), α-cellulose (Nacalai Tesque), phosphoric acid-swollen cellulose (PASC), insoluble mannan from the ivory nut (Megazyme, Wicklow, Ireland), insoluble xylan from oat spelts (Sigma-Aldrich, MO), insoluble arabinoxylan (Megazyme), wheat starch (Nacalai Tesque), chitin from crab (Nacalai Tesque), timothy grass, or rice straw in buffer M. PASC was prepared from Avicel as described previously (26). Rice straw was prepared using the same procedure as that used for timothy grass. The mixtures (200 μl) were incubated with end-over-end rotation at 25°C for 30 min and then centrifuged at 16,000 × g for 15 min. After removal of the supernatant, the precipitates were extensively washed three times with buffer M. The bound proteins were eluted by boiling in 2× SDS sample buffer and were then analyzed using SDS-PAGE in a 10% (wt/vol) gel. The proteins were visualized with CBB staining, and the intensities of bands were quantified using ImageJ software.
Affinity gel electrophoresis.
Each protein (3 μg) was analyzed by electrophoresis under native conditions in a 16% (wt/vol) acrylamide gel (pH 5.4 to 5.6) containing 120 mM potassium acetate and 0.1 mg/ml of one of the following: lichenan, a soluble polysaccharide from Cetraria islandica (MP Biomedicals, Inc., CA); xyloglucan from tamarind (Megazyme); soluble xylan from beechwood (Sigma-Aldrich); glucuronoxylan (4-O-methyl-d-glucurono-d-xylan; Sigma-Aldrich); locust bean gum (galactomannan; Wako, Osaka, Japan); glucomannan from konjac (Megazyme); pectin from citrus (Nacalai Tesque). The electrophoresis was conducted at 10 mA and 25°C for 2.5 to 3 h using running buffer (pH 3.8 to 4.0) consisting of 27 mM acetic acid and 70 mM alanine. Methyl green dye was loaded into empty wells, and electrophoresis was stopped according to migration of the dye. The proteins were visualized using CBB staining.
Depletion binding isotherm.
Increasing concentrations of the proteins (1 to 10 μM) were mixed with insoluble polysaccharides, PASC (2 mg/ml), xylan from oat spelts (3 mg/ml), insoluble mannan (3 mg/ml), or timothy grass (3 mg/ml) in buffer M. The mixtures (200 μl) were incubated with end-over-end rotation at 25°C for 30 min and then centrifuged at 16,000 × g for 15 min. The concentration of free protein present in the supernatant was determined using protein assay CBB solution. The amount of bound protein was calculated by subtracting free protein from total protein. The binding data were analyzed by nonlinear regression using the single-site binding model (a Langmuir-type isotherm) (27), which yields the association constant (Ka) and Bmax (the amount of bound protein at a saturation concentration).
Isothermal titration calorimetry.
To suppress dilution heat, the recombinant proteins were extensively dialyzed against buffer M, and ligands were dissolved in the same buffer. Cello-oligosaccharides with degrees of polymerization (DP) values of 2 to 6, xylo-oligosaccharides with DPs of 2 to 6, and mannohexaose, laminarihexaose, and xyloglucan heptasaccharide (Xyl3Gluc4) were used as ligands. The protein (50 μM) was titrated at 25°C by using successive injections of ligands (0.5 mM) at 120-s intervals with continuous stirring at 1,000 rpm with a MicroCal iTC200 instrument (GE Healthcare Biosciences). Nonlinear regression with the single-site binding model was used to analyze the binding heat data. Thermodynamic parameters were calculated using the Gibbs free energy equation (ΔG = ΔH − TΔS = −RTlnKa), where H is the enthalpy (in joules), T is the temperature (in kelvin), S is the entropy (in joules per kelvin), and R is the ideal gas constant (8.314 J/mol-K). All oligosaccharides were purchased from Megazyme.
RESULTS
Identification of plant cell wall-binding proteins.
When C. bescii was cultured in medium containing microcrystalline cellulose (Avicel), the Avicel appeared as slimy particles (see Fig. S1 in the supplemental material), suggesting that the cells secreted viscous substances. To identify noncatalytic proteins that bind to plant cell walls, we cultured the cells in medium containing timothy grass instead of Avicel, because native plant biomass is a more realistic substrate for C. bescii. Bound proteins were extracted from grass cell walls and analyzed by SDS-PAGE (Fig. 1A). CMCase activity was detected for bands corresponding to molecular masses of >110 kDa (Fig. 1B), which probably resulted from hydrolysis by multidomain enzymes. Mass spectrometry identified 12 proteins, including five multidomain enzymes, in the binding fraction (Table 1). Four plant cell wall-binding proteins (Athe_0181, Athe_0847, Athe_0597, and Athe_0614) with pI values of 9.2 to 9.6 were identified in bands H to K, which were negative for CMC activity (Fig. 1B). These proteins were named PWBPs to designate them as plant cell wall-binding proteins, with numbers corresponding to their molecular weights.
FIG 1.
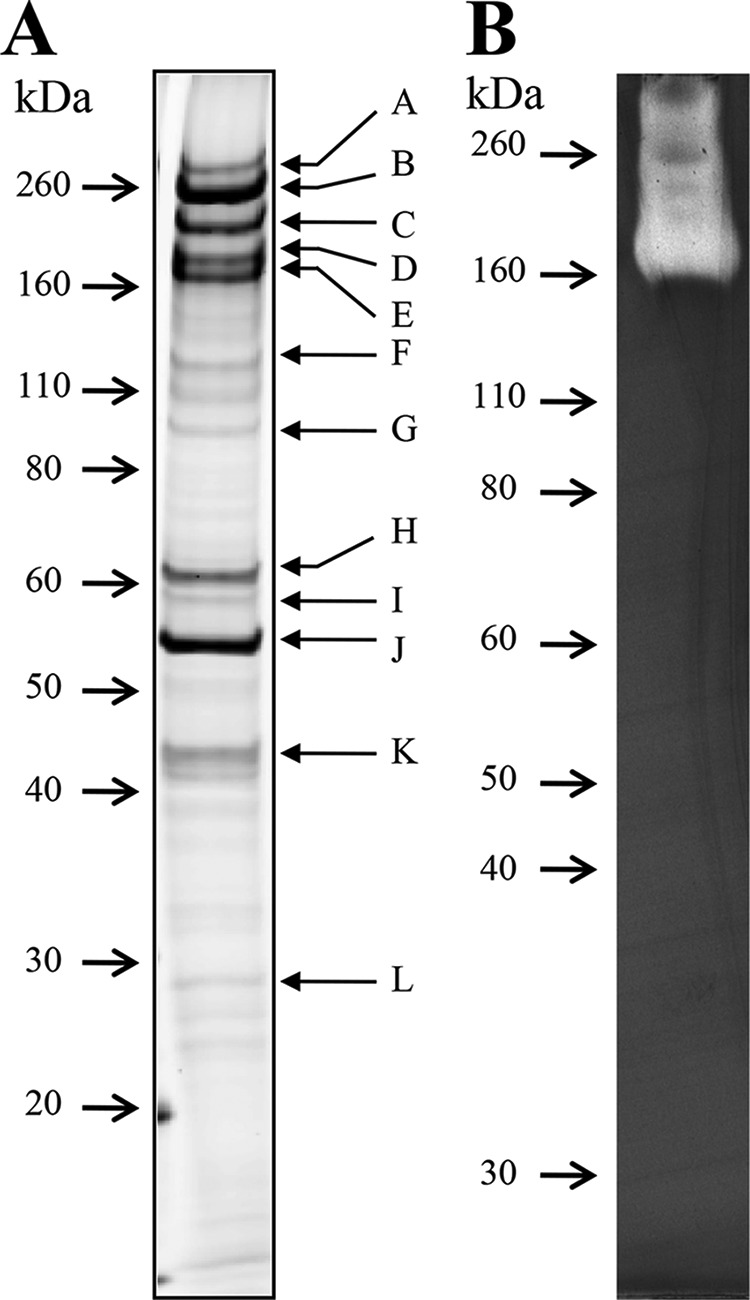
Profiles of protein binding to the plant cell wall of timothy grass. (A) The protein fraction was analyzed by SDS-PAGE with an 8 to 16% (wt/vol) gradient gel and stained with Coomassie brilliant blue. (B) CMC zymogram. The protein fraction was separated by SDS-PAGE with an 8% (wt/vol) gel containing CMC and then negatively stained with Congo red. Arrows indicate the positions of molecular mass markers.
TABLE 1.
Mass spectrometry analysis results for proteins bound to cell walls of timothy grass
| Banda | Locus tag | GenBank accession no. | Classification by annotation programs | Molecular mass (kDa) | pI |
|---|---|---|---|---|---|
| A | Athe_1860 | ACM60948 | Putative glucanase with GH74 and GH48 domains | 209 | 5.6 |
| B | Athe_1867 | ACM60955 | CelA (Cel9A/Cel48A) | 195 | 5.9 |
| C | Athe_1857 | ACM60945 | Putative xylanase/cellulase with GH10 and 48 domains | 165 | 6.1 |
| D | Athe_1865 | ACM60953 | Cel9B/Man5A | 151 | 5.6 |
| E | Athe_1859 | ACM60947 | Man5B/Cel44A | 142 | 5.9 |
| F | Athe_2303 | ACM61373 | S-layer homology domain protein | 109 | 4.9 |
| G | Athe_0460 | ACM59592 | Putative cellobiose phosphorylase | 93 | 5.7 |
| H | Athe_0181 | ACM59333 | PWBP66 | 66 | 9.2 |
| I | Athe_0847 | ACM59962 | PWBP65 | 65 | 9.3 |
| J | Athe_0597 | ACM59723 | PWBP57 | 57 | 9.3 |
| K | Athe_0614 | ACM59738 | PWBP49 | 49 | 9.6 |
| L | Athe_0297 | ACM59437 | Protein with unknown function | 32 | 8.6 |
These SDS-PAGE bands are shown in Fig. 1A.
Domain structure.
The four PWPBs did not show similarity (>40% amino acid sequence identity) to proteins with known functions in BLAST searches and did not contain a CBM or catalytic domain, according to a Conserved Domain Database (CDD) (28). Interestingly, the CDD search showed that these proteins possess a common domain related to the solute-binding protein (SBP) superfamily (Fig. 2). PWBP49, PWBP65, and PWBP66 contain an SBP bacterial family 1 (SBP bac 1) domain. PWBP57 has a two-domain structure containing an N-terminal UgpB domain and a C-terminal conserved domain of unknown function (DUF3502). The UgpB protein, a member of the SBP superfamily, binds to sn-glycerol-3-phosphate and is structurally similar to SBPs (29). Sequence identity (in amino acid residues) was 13 to 32% among the SBP domains of PWBPs.
FIG 2.
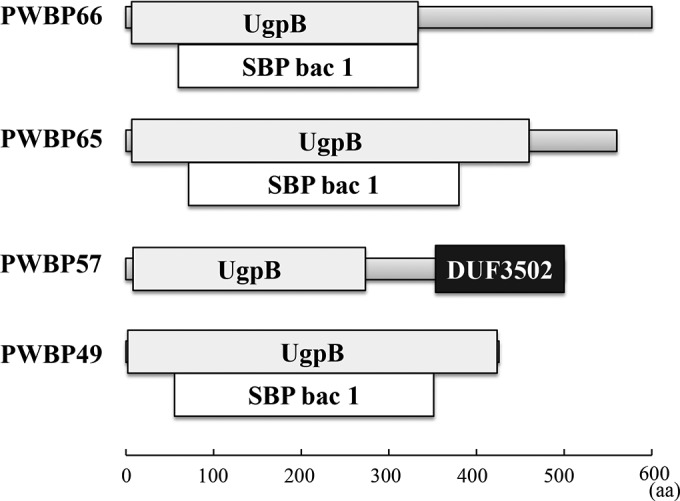
Domain structure of the plant cell wall-binding proteins. Structures predicted by searches in the Conserved Domain Database are shown.
Although the mass spectrometric analysis identified 12 proteins, many proteins in the plant cell wall-binding fraction were unidentified, suggesting that additional SBP-related proteins were present in that fraction. We searched for genes encoding SBP-related proteins that lacked a catalytic domain in the C. bescii genomic sequence, and we detected 12 protein-coding genes (locus tag numbers 0105, 0175, 0849, 1388, 1896, 1913, 1942, 2091, 2264, 2310, 2554, and 2574 [see Fig. S2 in the supplemental material]). Each of these putative PWBPs possessed a putative secretion signal; their pI values ranged from 8.7 to 9.6 (average, 9.4).
Phylogenetic analysis.
Proteins homologous to PWBPs were retrieved from databases through BLAST searches. All Caldicellulosiruptor species possess PWBP homologs (see Fig. S3 in the supplemental material). Outside the genus Caldicellulosiruptor, proteins showing the highest identity to PWBP49 (51 to 52% amino acid sequence similarity) belonged to Deinococcus geothermalis, Clostridium termitidis, and Clostridium cellulolyticum (see Fig. S3A). Proteins from Cellulosilyticum lentocellum, C. termitidis, and Clostridium stercorarium showed the highest identity to PWBP57 (46 to 50%). Other proteins with 35 to 41% identity to PWBP57 belonged to various genera in the classes Clostridia and Bacilli (see Fig. S3B). Proteins with the highest identity to PWBP65 (43 to 46%) belonged to Thermoanaerobacterium thermosaccharolyticum, Thermoanaerobacterium xylanolyticum, and Mahella australiensis. Similar to PWBP57, most proteins related to PWBP65 belonged to microbes in the classes Clostridia and Bacilli (see Fig. S3C). No proteins with >35% identity to PWBP66 were found.
Binding to the plant cell wall.
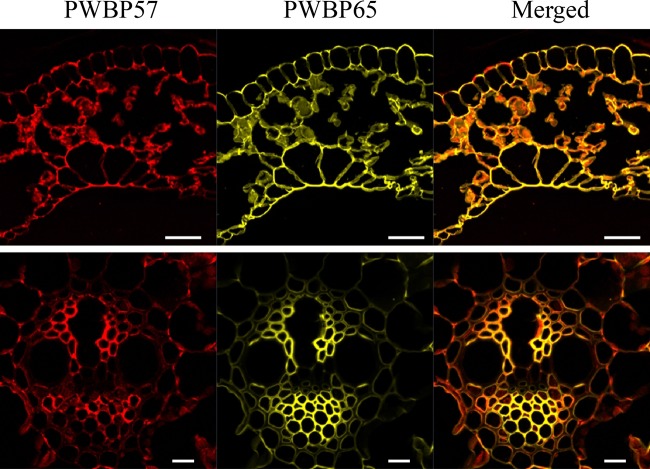
To evaluate the biochemical properties of the PWBP proteins, we attempted to express the genes encoding PWBP49, PWBP57, PWBP65, and PWBP66 in E. coli. PWBP49, PWBP57, and PWBP65 were successfully expressed (30), but PWBP66 was not, probably as a result of its cytotoxicity. Here, we report the biochemical properties of PWBP57 and PWBP65; PWBP49 is currently under examination. The recombinant PWBP57 and PWBP65 proteins were purified to near homogeneity by using two-step column chromatography (see Fig. S4 in the supplemental material). To analyze the protein localization on plant tissue, the recombinant proteins labeled with fluorescent dyes were incubated with a section of a grass leaf and observed under a fluorescence microscope. PWBP57 and PWBP65 bound preferentially to the cell wall (Fig. 3). Most stained areas overlapped; however, PWBP57 appeared to bind to xylem surrounding vessels in vascular tissue, whereas PWBP65 appeared to bind to both phloem and xylem. PWBP65 bound more strongly to primary cell walls, while PWBP57 bound to both primary and secondary cell walls.
FIG 3.
Preferential binding to the plant cell wall. PWBP57 (red) and PWBP65 (yellow) labeled with fluorescent dyes were incubated with a section of a grass leaf, and localization of the proteins was examined under a fluorescence microscope. Upper panels show the overall structure of the leaf cells. Lower panels show close-up images of vascular bundle cells. Bars in upper and lower panels are 50 and 10 μm, respectively.
Binding to insoluble polysaccharides.
The binding specificities of the proteins to insoluble polysaccharides were examined in a pulldown assay using SDS-PAGE. PWBP57 and PWBP65 bound to plant cell walls of timothy grass and rice straw and to a wide variety of polysaccharides (Fig. 4), including Avicel, α-cellulose, amorphous cellulose (PASC), mannan, xylan from oat spelts, and arabinoxylan. PWBP57 showed the strongest binding to PASC (9.4 μg/mg of substrate). The amount of PWBP65 bound to PASC was at levels similar to those of α-cellulose, mannan, xylan, and the plant cell wall (5.5 to 7.2 μg/mg of substrate). For both proteins, binding to Avicel and arabinoxylan was weak (1.4 to 1.8 μg/mg of substrate) relative to the other substrates. Binding to wheat starch and chitin from crab by both proteins was negligible.
FIG 4.
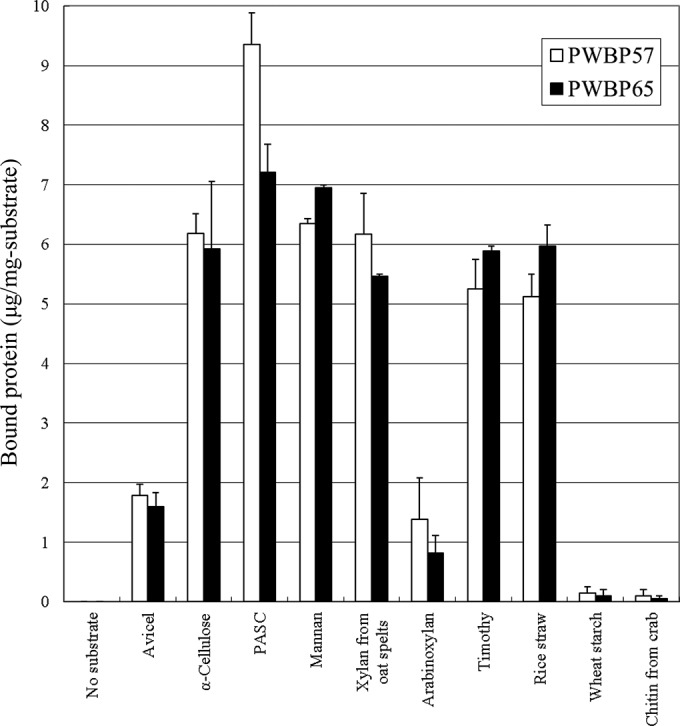
Binding of plant cell wall-binding proteins to insoluble polysaccharides as measured in the pulldown assay. The proteins were mixed with insoluble polysaccharides, and the bound proteins were eluted by boiling in 4% SDS. The eluted protein fraction was analyzed using SDS-polyacrylamide gel electrophoresis, and the intensity of bands was quantified using ImageJ software. Data are means ± standard deviations.
Binding to soluble polysaccharides.
Binding specificities to soluble polysaccharides were examined using affinity gel electrophoresis (31). The gel profiles are shown in Fig. 5 and summarized in Table S2 in the supplemental material. In the absence of a substrate, PWBP57 and PWBP65 migrated in the gel because of their positive charges. The presence of pectin in the gel decreased the mobility of the PWBPs most severely, suggesting that these proteins have a strong affinity for pectin. Xylan moderately decreased the mobilities of both proteins. The mobility of PWBP57 was decreased in the presence of lichenan (a β-1,3/1,4-linked glucan), but the effect of lichenan was weaker for PWBP65. Both proteins interacted weakly with glucuronoxylan and glucomannan. PWBP65 also interacted weakly with xyloglucan, but the interaction of PWBP57 with xyloglucan was undetectable. Interaction with galactomannan was not detected for either protein. Aprotinin, a trypsin inhibitor with a pI of 10.6, was analyzed as a control protein, using xylan and pectin as substrates. The migration of protein in the presence of xylan or pectin was almost identical to that in the absence of polysaccharide (see Fig. S5 in the supplemental material). This result verified that not all proteins interacted with the polysaccharides in the gels and that changes in migration reflected properties of the protein.
FIG 5.
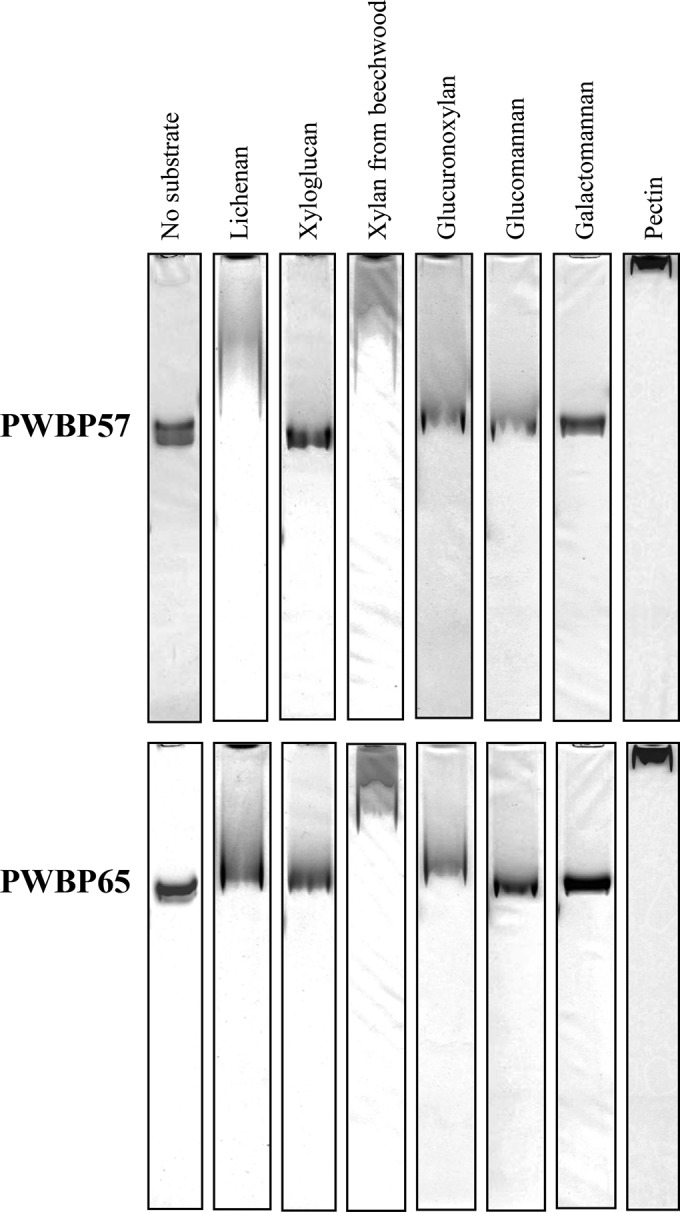
Binding to soluble polysaccharides according to affinity gel electrophoresis. The plant cell wall-binding proteins were analyzed by electrophoresis under native conditions using gels that contained the indicated soluble polysaccharides and then stained with Coomassie brilliant blue. The resulting profiles are shown.
Binding capacity and affinity for insoluble polysaccharides.
Binding to insoluble polysaccharides was further analyzed by depletion binding isotherms (31) using four substrates: PASC, mannan, xylan from oat spelts, and cell walls of timothy grass. The raw data are shown in Fig. S6A in the supplemental material and are summarized in Table 2. Both proteins showed the highest affinity for plant cell walls, with Ka values of 5.2 × 106 to 44 × 106 M−1. Xylan was the second-best binding partner for both proteins. As for binding capacity, PASC was the best (4.5 μmol/g of substrate) for PWBP57, followed by mannan. The binding capacity of PWBP65 was similar for the four substrates (0.8 to 1.3 μmol/g of substrate).
TABLE 2.
Binding capacities and affinities for insoluble polysaccharidesa
| Substrate | PWBP57 |
PWBP65 |
||
|---|---|---|---|---|
| Bmax (μmol/g of substrate) | Ka (106 M−1) | Bmax (μmol/g of substrate) | Ka (106 M−1) | |
| Timothy grass cell wall | 1.11 ± 0.04 | 43.5 ± 13.2 | 0.79 ± 0.02 | 5.17 ± 1.13 |
| Xylan from oat spelts | 1.36 ± 0.09 | 20.4 ± 9.9 | 1.25 ± 0.05 | 2.06 ± 0.36 |
| PASC | 4.48 ± 0.63 | 0.59 ± 0.02 | 1.2 ± 0.13 | 1.02 ± 0.31 |
| Mannan | 2.30 ± 0.94 | 0.14 ± 0.09 | 0.90 ± 0.08 | 1.22 ± 0.30 |
Determined using depletion binding isotherms. Data are means ± standard deviations.
Binding to oligosaccharides.
Calorimetric analyses with isothermal titration were performed to reveal the binding preference of PWBPs to oligosaccharides (raw data are shown in Fig. S6B in the supplemental material; the data are summarized in Table 3). PWBP57 bound to cello-oligosaccharides from trimers to hexamers, with Ka values of 1.1 × 107 to 4.1 × 107 M−1, while heat binding was not detected for the dimer cellobiose. Heat binding of PWBP57 to xylohexaose, mannohexaose, or laminarihexaose (a β-1,3-linked glucohexaose) was not detected. PWBP65 bound to both cello- and xylo-oligosaccharides, and its binding affinities for tetramers and pentamers were higher than those for trimers and hexamers. Both proteins bound weakly to the branched oligosaccharide Xyl3Gluc4, which consists of a cellotetraose main chain with three xylose side chains. Although thermodynamic parameters were not determined precisely, weak heat binding of PWBP65 to cellobiose, xylobiose, mannohexaose, and laminarihexaose was observed, with apparent Ka values at 104 to 105 M−1 (data not shown).
TABLE 3.
PWBP affinities for oligosaccharidesa
| Protein | Ligand | Ka (106 M−1) | ΔH (kJ/mol) | TΔS (kJ/mol) |
|---|---|---|---|---|
| PWBP57 | Cellotriose | 33.5 ± 11.8 | 23.6 ± 0.3 | 66.5 |
| Cellotetraose | 41.1 ± 27.2 | 30.0 ± 0.3 | 73.5 | |
| Cellopentaose | 14.5 ± 2.69 | 35.1 ± 0.3 | 76.0 | |
| Cellohexaose | 11.1 ± 2.48 | 33.1 ± 0.4 | 73.4 | |
| Xyl3Gluc4 | 0.0124 ± 0.0070 | −51.7 ± 29.2 | −27.8 | |
| PWBP65 | Cellotriose | 0.248 ± 0.006 | −47.0 ± 1.0 | −14.8 |
| Cellotetraose | 6.71 ± 2.42 | −54.7 ± 1.1 | −15.7 | |
| Cellopentaose | 2.92 ± 1.31 | −37.4 ± 1.2 | −0.5 | |
| Cellohexaose | 0.466 ± 0.152 | −25.7 ± 1.2 | 6.7 | |
| Xylotriose | 1.89 ± 0.41 | −62.1 ± 1.2 | −26.3 | |
| Xylotetraose | 6.35 ± 1.26 | −70.8 ± 0.8 | −31.9 | |
| Xylopentaose | 8.43 ± 2.53 | −59.3 ± 0.9 | −19.7 | |
| Xylohexaose | 3.88 ± 1.10 | −52.6 ± 1.0 | −15.0 | |
| Xyl3Gluc4 | 0.206 ± 0.029 | −41.3 ± 0.1 | −11.0 |
Determined by isothermal titration calorimetry. Data are means ± standard deviations.
The binding of PWBP57 to cello-oligosaccharides was an endothermic reaction with increased entropy. In contrast, the binding of PWBP65 to cello- and xylo-oligosaccharides was exothermic with reduced entropy. Binding to Xyl3Gluc4 was exothermic for both proteins.
Circular dichroism analysis was conducted to examine the effects of oligosaccharide binding on protein thermostability (see Fig. S7A in the supplemental material). Binding to cellotetraose increased the melting point (Tm) values of PWBP57 and PWBP65 by 5.5°C and 2.1°C, respectively (see Table S3 in the supplemental material). Competitive binding experiments were performed to compare binding affinities between an oligosaccharide and an insoluble polysaccharide. The binding of PWBP57 and PWBP65 to cell walls of timothy grass was not affected by the presence of cellotetraose, even when the oligosaccharide concentration was increased up to a 10-fold excess over that of the proteins (see Fig. S7B).
DISCUSSION
We identified four noncatalytic PWBPs with high pI values (9.2 to 9.6) that are secreted by C. bescii. According to database searches, the amino acid sequences of PWBPs do not show significant similarities to those of proteins with experimentally verified functions, and they do not contain a CBM or catalytic domain. Fluorescence microscopy demonstrated that PWBP57 and PWBP65 preferentially bind to plant cell walls. PWBPs possess a common domain related to SBPs, as predicted by the database searches. All Caldicellulosiruptor species have the PWBP-homologous genes. PWBPs are currently annotated with extracellular solute-binding proteins in databases, but biochemical experiments that demonstrate their function have not yet been reported. In addition to the four PWBPs, 12 genes encoding putative PWBPs are present in the C. bescii genome (see Fig. S2 in the supplemental material); extracellular secretion of each of these putative PWBPs has been experimentally confirmed (13, 14). C. bescii secretes a wide variety of PWBPs, including the 12 putative PWBPs; this observation points to the importance of noncatalytic polysaccharide-binding proteins in the utilization of polysaccharides. Proteins showing similarity to PWBPs are found in various bacteria, including cellulolytic or hemicellulolytic bacteria, such as C. lentocellum, C. termitidis, and C. stercorarium (32–34). Nevertheless, proteins homologous to PWBPs were not detected in cellulosome-producing bacteria.
On the basis of database searches, PWBP57 is predicted to contain two domains, UgpB and DUF3502. The UgpB domain belongs to cluster B in the SBP superfamily and is structurally similar to SBPs (29). Most SBPs with specific binding to saccharides are classified in cluster B and are a component of ATP-binding cassette (ABC) transporters for substrate uptake. Despite little sequence similarity, the overall three-dimensional structure of SBPs is highly conserved (35). SBPs consist of two globular domains connected by a hinge region, and the substrate-binding site is located at the interface between the two domains. Substrate binding leads to a conformational change from an open to a closed form in which the two domains are tightly packed, stabilizing the substrate-protein complex. The binding of PWBP57 and PWBP65 to cellotetraose increases their Tms by 2 to 6°C. This result implies that the SBP domains of PWBP57 and PWBP65 are stabilized by substrate binding in a manner similar to that found in typical SBPs.
Thus far, most SBPs have been reported to bind to soluble substrates with low molecular weights, such as metal ions, inorganic ions, amino acids, peptides (3 to 35 amino acids), and oligosaccharides (35). SBPs normally display high binding specificity, but PWBP57 and PWBP65 are capable of binding to a variety of plant cell wall saccharides, from soluble oligosaccharides to insoluble macromolecules. These proteins bind to microcrystalline cellulose, amorphous cellulose, lichenan, xyloglucan, xylan, arabinoxylan, glucuronoxylan, mannan, and oligosaccharides, including the branched molecule Xyl3Gluc4. This wide range of substrate specificity suggests that the predicted globular SBP domains of PWBP57 and PWBP65 may open more widely than those of conventional SBPs because of a more flexible or longer hinge region.
The Ka value for PWBP57 and PWBP65 for oligosaccharides is 0.2 × 106 to 44 × 106 M−1. This value is within the range (105 to 107 M−1) reported for most oligosaccharide-binding SBPs (35). Typically, binding of a protein to a soluble carbohydrate is enthalpically driven with entropic compensation; this mechanism is consistent with the thermodynamics of PWBP65. On the other hand, the binding of PWBP57 to cello-oligosaccharides is an endothermic reaction driven by an increase in entropy. Increased entropy in protein-ligand binding is thought to result from dehydration at the contact surface (27). Thus, the conformation of the contact surface and amino acid residues responsible for substrate binding probably differs between PWBP57 and PWBP65. Both proteins bind weakly to the branched oligosaccharide Xyl3Gluc4 rather than to its straight form, cellotetraose, indicating that they show a preference for straight-chain saccharides. The branched side chains of Xyl3Gluc4 may apply steric constraints to the predicted globular domains to induce the closed conformation. Binding of PWBP57 to xylohexaose, mannohexaose, or laminarihexaose is not detectable, but this protein can bind to their longer forms—xylan, mannan, and lichenan, respectively. Binding of PWBP57 to these saccharides appears to require sugar chains with lengths greater than a hexamer.
PWBP65 displays wider substrate specificity than PWBP57 in relation to oligosaccharides; the former binds to xylo-oligosaccharides and weakly to mannohexaose and laminarihexaose. Although they have similar characteristics, these proteins also show differences in polysaccharide binding. In particular, the binding capacity of PWBP57 for PASC is 4-fold greater than that of PWBP65, and the affinity of PWBP57 for insoluble xylan is 10-fold higher than that of PWBP65. Fluorescence microscopic analysis revealed slight differences between the profiles of leaf cells to which PWBP57 and PWBP65 bind. The walls of xylem cells are larger and are more lignified than phloem cell walls, which are more cellulosic. Plant cell walls consist of a variety of polysaccharides, and the surface structures formed by these polysaccharides differ among plant species. C. bescii may require many types of PWBPs to recognize the different surface structures of plant cell walls.
Among the insoluble polysaccharides tested, the binding affinities of PWBP57 and PWBP65 are highest for the plant cell wall. Binding of the proteins to the cell wall is not affected even when cellotetraose is present (see Fig. S7B in the supplemental material), suggesting that the proteins exhibit a preference for polysaccharides over oligosaccharides. Affinity gel electrophoresis shows that the proteins bind most strongly to acidic soluble polysaccharides (pectin). Hemicellulose has negatively charged residues, such as glucuronic acid in glucuronoxylan (3). Because PWBPs are highly basic proteins (pI 9.2 to 9.6), it is possible that the high affinity for the plant cell wall results from interactions with the negatively charged residues in hemicellulose and pectin.
CBMs present in glycoside hydrolases have been well characterized, but the role of noncatalytic binding proteins secreted by plant cell wall-degrading bacteria is poorly understood. Enzyme-substrate binding is a key step for efficient degradation of insoluble polysaccharides, including cellulose. CBMs facilitate binding of enzymes to substrates and enhance catalytic activity (6). Removal of a CBM leads to a reduction or a complete loss of catalytic activity toward insoluble polysaccharides, whereas the activity toward soluble substrates is often not affected (36, 37). Swollenin of T. reesei and EXLX1 of Bacillus subtilis are noncatalytic cellulose-binding proteins (38, 39) that are related to plant expansins (40). These proteins loosen filter paper by disrupting hydrogen bonds between cellulose microfibrils. EXLX1 is a basic protein (pI 9.2) that reportedly enhances cellulase activity (41). We tested the filter paper-loosening activities of PWBP57 and PWBP65 by using an extensometer, but we did not detect activity (data not shown). In addition, we tested the enhancement of cellulase activity by the PWBPs by using a truncated cellulase consisting of GH9 and a CBM3 from C. bescii CelA (18); no enhancement was observed (data not shown).
In most cases, binding of proteins to insoluble substrates is weakened at high temperatures because of increased kinetic energy. This effect may pose a problem for the extremely thermophilic bacterium C. bescii, which can grow at temperatures up to 90°C (9). Our present data show that when C. bescii is cultured in Avicel-containing medium, the Avicel appears as slimy particles, a phenomenon that is likely to be caused by secreted substances. The viscous coating on the Avicel particles might help the cells or proteins remain bound to the particle surface at high temperatures. The concentration of bacterial cells in high-temperature environments, such as hot springs, is generally much lower than that in medium-temperature environments. Because C. bescii cells are nonmotile, efficient access to the plant cell wall is expected to be crucial to the wall degradation process. We can hypothesize that PWBPs bind to and neutralize negatively charged regions on the plant cell wall and that the basicity of these proteins attracts negatively charged molecules to the binding sites. The C. bescii multidomain enzymes are slightly acidic (pI 5.6 to 6.1 [Table 1]) and are expected to interact nonspecifically with the basic PWBPs. On the basis of this hypothesis, multidomain enzymes might be recruited to regions bound by PWBPs on the plant cell wall. The bacterial cell wall typically has a net negative charge (42). Teichoic acids are negatively charged cell wall polymers in Gram-positive bacteria and they perform important functions in cell adhesion and biofilm formation (43). We speculate that the positively charged residues of PWBPs are involved in bacterial adhesion to the plant cell wall through nonspecific interactions.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported in part by JSPS KAKENHI grant 24580490.
We thank the Instrument Center of the Institute for Molecular Science for assistance with the isothermal titration calorimetric analyses. We thank T. Kiyoshi for providing the grass samples and H. Kurumizaka and N. Horikoshi for support of the circular dichroism experiments.
Footnotes
Published ahead of print 25 August 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.01897-14.
REFERENCES
- 1.Gilbert HJ. 2010. Biochemistry and structural biology of plant cell wall deconstruction. Plant Physiol. 153:444–455. 10.1104/pp.110.156646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jordan DB, Bowman MJ, Braker JD, Dien BS, Hector RE, Lee CC, Mertens JA, Wagschal K. 2012. Plant cell walls to ethanol. Biochem. J. 442:241–252. 10.1042/BJ20111922. [DOI] [PubMed] [Google Scholar]
- 3.Scheller HV, Ulvskov P. 2010. Hemicelluloses. Annu. Rev. Plant Biol. 61:263–289. 10.1146/annurev-arplant-042809-112315. [DOI] [PubMed] [Google Scholar]
- 4.Cantarel BL, Coutinho PM, Rancurel C, Bernard T, Lombard V, Henrissat B. 2009. Carbohydrate-active enzymes database (CAZy): an expert resource for glycogenomics. Nucleic Acids Res. 37:D233–D238. 10.1093/nar/gkn663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lombard V, Ramulu HG, Drula E, Coutinho PM, Henrissat B. 2014. Carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 42:D490–D495. 10.1093/nar/gkt1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bolam DN, Ciruela A, McQueen-Mason S, Simpson P, Williamson MP, Rixon JE, Boraston A, Hazlewood GP, Gilbert HJ. 1998. Pseudomonas cellulose-binding domains mediate their effects by increasing enzyme substrate proximity. Biochem. J. 331:775–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang SJ, Kataeva I, Hamilton-Brehm SD, Engle NL, Tschaplinski TJ, Doeppke C, Davis M, Westpheling J, Adams MWW. 2009. Efficient degradation of lignocellulosic plant biomass without pretreatment by the thermophilic anaerobe “Anaerocellum thermophilum” DSM 6725. Appl. Environ. Microbiol. 75:4762–4769. 10.1128/AEM.00236-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Svetlichnyi VA, Svetlichnaya TP, Chernykh NA, Zavarzin GA. 1990. Anaerocellum thermophilum gen. nov. sp.: an extremely thermophilic cellulolytic eubacterium isolated from hot springs in the valley of geysers. Microbiology 59:598–604. [Google Scholar]
- 9.Yang SJ, Kataeva I, Wiegel J, Yin YB, Dam P, Xu Y, Westpheling J, Adams MWW. 2010. Classification of “Anaerocellum thermophilum” strain DSM 6725 as Caldicellulosiruptor bescii sp. nov. Int. J. Syst. Evol. Microbiol. 60:2011–2015. 10.1099/ijs.0.017731-0. [DOI] [PubMed] [Google Scholar]
- 10.Blumer-Schuette SE, Kataeva I, Westpheling J, Adams MWW, Kelly RM. 2008. Extremely thermophilic microorganisms for biomass conversion: status and prospects. Curr. Opin. Biotechnol. 19:210–217. 10.1016/j.copbio.2008.04.007. [DOI] [PubMed] [Google Scholar]
- 11.Kanafusa-Shinkai S, Wakayama J, Tsukamoto K, Hayashi N, Miyazaki Y, Ohmori H, Tajima K, Yokoyama H. 2013. Degradation of microcrystalline cellulose and non-pretreated plant biomass by a cell-free extracellular cellulase/hemicellulase system from the extreme thermophilic bacterium Caldicellulosiruptor bescii. J. Biosci. Bioeng. 115:64–70. 10.1016/j.jbiosc.2012.07.019. [DOI] [PubMed] [Google Scholar]
- 12.Fontes C, Gilbert HJ. 2010. Cellulosomes: highly efficient nanomachines designed to deconstruct plant cell wall complex carbohydrates. Annu. Rev. Biochem. 79:655–681. 10.1146/annurev-biochem-091208-085603. [DOI] [PubMed] [Google Scholar]
- 13.Lochner A, Giannone RJ, Rodriguez M, Shah MB, Mielenz JR, Keller M, Antranikian G, Graham DE, Hettich RL. 2011. Use of label-free quantitative proteomics to distinguish the secreted cellulolytic systems of Caldicellulosiruptor bescii and Caldicellulosiruptor obsidiansis. Appl. Environ. Microbiol. 77:4042–4054. 10.1128/AEM.02811-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dam P, Kataeva I, Yang SJ, Zhou FF, Yin YB, Chou WC, Poole FL, Westpheling J, Hettich R, Giannone R, Lewis DL, Kelly R, Gilbert HJ, Henrissat B, Xu Y, Adams MWW. 2011. Insights into plant biomass conversion from the genome of the anaerobic thermophilic bacterium Caldicellulosiruptor bescii DSM 6725. Nucleic Acids Res. 39:3240–3254. 10.1093/nar/gkq1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lochner A, Giannone RJ, Keller M, Antranikian G, Graham DE, Hettich RL. 2011. Label-free quantitative proteomics for the extremely thermophilic bacterium Caldicellulosiruptor obsidiansis reveal distinct abundance patterns upon growth on cellobiose, crystalline cellulose, and switchgrass. J. Proteome Res. 10:5302–5314. 10.1021/pr200536j. [DOI] [PubMed] [Google Scholar]
- 16.Andrews G, Lewis D, Notey J, Kelly R, Muddiman D. 2010. Part I: characterization of the extracellular proteome of the extreme thermophile Caldicellulosiruptor saccharolyticus by GeLC-MS2. Anal. Bioanal. Chem. 398:377–389. 10.1007/s00216-010-3955-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Blumer-Schuette SE, Lewis DL, Kelly RM. 2010. Phylogenetic, microbiological, and glycoside hydrolase diversities within the extremely thermophilic, plant biomass-degrading genus Caldicellulosiruptor. Appl. Environ. Microbiol. 76:8084–8092. 10.1128/AEM.01400-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zverlov V, Mahr S, Riedel K, Bronnenmeier K. 1998. Properties and gene structure of a bifunctional cellulolytic enzyme (CelA) from the extreme thermophile “Anaerocellum thermophilum” with separate glycosyl hydrolase family 9 and 48 catalytic domains. Microbiology 144:457–465. 10.1099/00221287-144-2-457. [DOI] [PubMed] [Google Scholar]
- 19.Te'o VSJ, Saul DJ, Bergquist PL. 1995. CelA, another gene coding for a multidomain cellulase from the extreme thermophile Caldocellum saccharolyticum. Appl. Microbiol. Biotechnol. 43:291–296. 10.1007/BF00172827. [DOI] [PubMed] [Google Scholar]
- 20.Su X, Mackie RI, Cann IKO. 2012. Biochemical and mutational analyses of a multi-domain cellulase/mannanase from Caldicellulosiruptor bescii. Appl. Environ. Microbiol. 78:2230–2240. 10.1128/AEM.06814-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ye LB, Su XY, Schmitz GE, Moon YH, Zhang J, Mackie RI, Cann IKO. 2012. Molecular and biochemical analyses of the GH44 module of CbMan5B/Cel44A, a bifunctional enzyme from the hyperthermophilic bacterium Caldicellulosiruptor bescii. Appl. Environ. Microbiol. 78:7048–7059. 10.1128/AEM.02009-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yi ZL, Su XY, Revindran V, Mackie RI, Cann I. 2013. Molecular and biochemical analyses of CbCel9A/Cel48A, a highly secreted multi-modular cellulase by Caldicellulosiruptor bescii during growth on crystalline cellulose. PLoS One 8:e84172. 10.1371/journal.pone.0084172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brunecky R, Alahuhta M, Xu Q, Donohoe BS, Crowley MF, Kataeva IA, Yang SJ, Resch MG, Adams MWW, Lunin VV, Himmel ME, Bomble YJ. 2013. Revealing nature's cellulase diversity: the digestion mechanism of Caldicellulosiruptor bescii CelA. Science 342:1513–1516. 10.1126/science.1244273. [DOI] [PubMed] [Google Scholar]
- 24.Perkins DN, Pappin DJC, Creasy DM, Cottrell JS. 1999. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20:3551–3567. . [DOI] [PubMed] [Google Scholar]
- 25.Emanuelsson O, Brunak S, von Heijne G, Nielsen H. 2007. Locating proteins in the cell using TargetP, SignalP and related tools. Nat. Protoc. 2:953–971. 10.1038/nprot.2007.131. [DOI] [PubMed] [Google Scholar]
- 26.Wood TM. 1988. Preparation of crystalline, amorphous, and dyed cellulase substrates. Methods Enzymol. 160:19–25. 10.1016/0076-6879(88)60103-0. [DOI] [Google Scholar]
- 27.Creagh AL, Ong E, Jervis E, Kilburn DG, Haynes CA. 1996. Binding of the cellulose-binding domain of exoglucanase Cex from Cellulomonas fimi to insoluble microcrystalline cellulose is entropically driven. Proc. Natl. Acad. Sci. U. S. A. 93:12229–12234. 10.1073/pnas.93.22.12229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marchler-Bauer A, Lu SN, Anderson JB, Chitsaz F, Derbyshire MK, DeWeese-Scott C, Fong JH, Geer LY, Geer RC, Gonzales NR, Gwadz M, Hurwitz DI, Jackson JD, Ke ZX, Lanczycki CJ, Lu F, Marchler GH, Mullokandov M, Omelchenko MV, Robertson CL, Song JS, Thanki N, Yamashita RA, Zhang DC, Zhang NG, Zheng CJ, Bryant SH. 2011. CDD: a conserved domain database for the functional annotation of proteins. Nucleic Acids Res. 39:D225–D229. 10.1093/nar/gkq1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wuttge S, Bommer M, Jager F, Martins BM, Jacob S, Licht A, Scheffel F, Dobbek H, Schneider E. 2012. Determinants of substrate specificity and biochemical properties of the sn-glycerol-3-phosphate ATP binding cassette transporter (UgpB-AEC2) of Escherichia coli. Mol. Microbiol. 86:908–920. 10.1111/mmi.12025. [DOI] [PubMed] [Google Scholar]
- 30.Yokoyama H, Yamashita T, Horikoshi N, Kurumizaka H, Kagawa W. 2013. Crystallization and preliminary X-ray diffraction analysis of the secreted protein Athe_0614 from Caldicellulosiruptor bescii. Acta Crystallogr. F Struct. Biol. Cryst. Commun. 69:438–440. 10.1107/S174430911300554X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Abbott DW, Boraston AB. 2012. Quantitative approaches to the analysis of carbohydrate-binding module function. Methods Enzymol. 510:211–231. 10.1016/B978-0-12-415931-0.00011-2. [DOI] [PubMed] [Google Scholar]
- 32.Hethener P, Brauman A, Garcia JL. 1992. Clostridium termitidis sp. nov. a cellulolytic bacterium from the gut of the wood-feeding termite, Nasutitermes lujae. Syst. Appl. Microbiol. 15:52–58. 10.1016/S0723-2020(11)80138-4. [DOI] [Google Scholar]
- 33.Murray WD, Hofmann L, Campbell NL, Madden RH. 1986. Clostridium lentocellum sp. nov., a cellulolytic species from river sediment containing paper-mill waste. Syst. Appl. Microbiol. 8:181–184. 10.1016/S0723-2020(86)80074-1. [DOI] [Google Scholar]
- 34.Madden RH. 1983. Isolation and characterization of Clostridium stercorarium sp. nov., cellulolytic thermophile. Int. J. Syst. Bacteriol. 33:837–840. [Google Scholar]
- 35.Berntsson RPA, Smits SHJ, Schmitt L, Slotboom DJ, Poolman B. 2010. A structural classification of substrate-binding proteins. FEBS Lett. 584:2606–2617. 10.1016/j.febslet.2010.04.043. [DOI] [PubMed] [Google Scholar]
- 36.Kleine J, Liebl W. 2006. Comparative characterization of deletion derivatives of the modular xylanase XynA of Thermotoga maritima. Extremophiles 10:373–381. 10.1007/s00792-006-0509-0. [DOI] [PubMed] [Google Scholar]
- 37.Guillen D, Sanchez S, Rodriguez-Sanoja R. 2010. Carbohydrate-binding domains: multiplicity of biological roles. Appl. Microbiol. Biotechnol. 85:1241–1249. 10.1007/s00253-009-2331-y. [DOI] [PubMed] [Google Scholar]
- 38.Saloheimo M, Paloheimo M, Hakola S, Pere J, Swanson B, Nyyssonen E, Bhatia A, Ward M, Penttila M. 2002. Swollenin, a Trichoderma reesei protein with sequence similarity to the plant expansins, exhibits disruption activity on cellulosic materials. Eur. J. Biochem. 269:4202–4211. 10.1046/j.1432-1033.2002.03095.x. [DOI] [PubMed] [Google Scholar]
- 39.Kerff F, Amoros A, Herman R, Sauvage E, Petrella S, Filee P, Charlier P, Joris B, Tabuchi A, Nikolaidis N, Cosgrove DJ. 2008. Crystal structure and activity of Bacillus subtilis YoaJ (EXLX1), a bacterial expansin that promotes root colonization. Proc. Natl. Acad. Sci. U. S. A. 105:16876–16881. 10.1073/pnas.0809382105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McQueen-Mason S, Durachko DM, Cosgrove DJ. 1992. Two endogenous proteins that induce cell wall extension in plants. Plant Cell 4:1425–1433. 10.1105/tpc.4.11.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lin H, Shen Q, Zhan JM, Wang Q, Zhao YH. 2013. Evaluation of bacterial expansin EXLX1 as a cellulase synergist for the saccharification of lignocellulosic agro-industrial wastes. PLoS One 8:e75022. 10.1371/journal.pone.0075022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dickson JS, Koohmaraie M. 1989. Cell surface charge characteristics and their relationship to bacterial attachment to meat surfaces. Appl. Environ. Microbiol. 55:832–836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brown S, Maria JPS, Walker S. 2013. Wall teichoic acids of gram-positive bacteria. Annu. Rev. Microbiol. 67:313–336. 10.1146/annurev-micro-092412-155620. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.