Abstract
Competence for genetic transformation in the genus Streptococcus depends on an alternative sigma factor, σX, for coordinated synthesis of 23 proteins, which together establish the X state by permitting lysis of incompetent streptococci, uptake of DNA fragments, and integration of strands of that DNA into the resident genome. Initiation of transient accumulation of high levels of σX is coordinated between cells by transcription factors linked to peptide pheromone signals. In Streptococcus pneumoniae, elevated σX is insufficient for development of full competence without coexpression of a second competence-specific protein, ComW. ComW, shared by eight species in the Streptococcus mitis and Streptococcus anginosus groups, is regulated by the same pheromone circuit that controls σX, but its role in expression of the σX regulon is unknown. Using the strong, but not absolute, dependence of transformation on comW as a selective tool, we collected 27 independent comW bypass mutations and mapped them to 10 single-base transitions, all within rpoD, encoding the primary sigma factor subunit of RNA polymerase, σA. Eight mapped to sites in rpoD region 4 that are implicated in interaction with the core β subunit, indicating that ComW may act to facilitate competition of the alternative sigma factor σX for access to core polymerase.
INTRODUCTION
Streptococcus pneumoniae is a common human pathogen, usually carried asymptomatically in the nasopharynx. A prominent characteristic of the bacterium is a natural ability to take up exogenous DNA from its environment. If the exogenous DNA is integrated into the genome, the bacterium is said to be transformed. Such transformation depends on development of a specialized physiological state, termed competence, development of which is coordinated within a culture by a quorum-sensing (QS) mechanism encoded by two genetic loci, comAB (1) and comCDE (2). Both loci are transcribed at a basal level by the “housekeeping” sigma factor, σA (3, 4). The competence-stimulating peptide (CSP), a product of the comC gene, is secreted by an ABC transporter/protease encoded by comA and comB (4, 5). CSP is sensed by a histidine kinase receptor, ComD, which phosphorylates a cognate response regulator, ComE (1, 4, 6, 7). Phosphorylated ComE activates the promoters of eight operons comprising 13 genes transcribed specifically at competence by binding a direct repeat centered at −40, the ComE box (4, 8, 9). They are designated early competence genes (9) and include both the comAB and comCDE operons (10–13). This organization creates a positive-feedback loop, ensuring a rapid increase in the level of CSP that can cause all the cells in a culture to become competent simultaneously (7, 13). One additional early gene essential for competence is comX, encoding the alternative sigma factor σX, which accumulates to high levels in response to CSP (14), forming a holoenzyme with RNA polymerase (RNAP) and enabling recognition of a noncanonical promoter sequence termed the ComX box, or cinbox, upstream of 23 genes, termed the late competence genes (13–16).
Streptococci have only one known alternative sigma factor, which in all species is linked to expression of the late competence genes. In the Streptococcus mitis and Streptococcus anginosus groups of species, upstream regulators of comX include CSP/ComABCDE QS systems orthologous to the pneumococcal one (17). In all other groups of streptococci, sigX is regulated by a different class of peptide QS circuit, using genes named comS and comR, encoding a peptide pheromone and intracellular receptor, respectively (17). Regulation of σX activity is important, as competence genes encode one or more killer proteins and cognate immunity genes. Within the S. mitis and S. anginosus groups, eight related species (S. anginosus, Streptococcus cristatus, Streptococcus infantis, S. mitis, Streptococcus oligofermentans, Streptococcus oralis, S. pneumoniae, and Streptococcus pseudopneumoniae) add another layer of control of σX activity in the form of a small protein, ComW, which plays an important role during competence development. Since its identification in 2004 in S. pneumoniae (18), attempts to identify the role of ComW have been reported (18–21), but its function remains unclear. comW mutants have several phenotypes that suggest different possible roles for the protein. First, comW mutants are ∼104-fold deficient in transformants (18). Second, in comW mutants, transcription of late competence genes is strongly reduced, to ∼10% of wild-type (WT) levels, and accumulation of σX protein is similarly reduced (19). Third, comW clpE (where ClpEP is a protease that degrades σX) double mutants produce restored amounts of σX protein but are still deficient in transformation (18). Fourth, both the N and C termini of ComW are necessary for σX stability, while the N terminus is necessary for σX function (18). Since none of the roles of ComW are well characterized and available data suggest several alternative roles during transformation, ComW's function in supporting σX activity remains unknown.
To investigate the role of ComW in regulation of competence in an unbiased manner, we designed and carried out a suppressor screen for rare mutations that could bypass the comW requirement and restore the competence lost in ΔcomW strains. Here, we describe a strategy for recovering rare comW bypass mutations and show that they map exclusively to rpoD.
MATERIALS AND METHODS
Bacterial strains and culture media.
The pneumococcal strains used in the study are listed in Table 1. CP2137, a cps ΔcomA derivative of strain Rx1, was used as the WT standard for transformation assays and as a transformation recipient for the ΔcomW::kan deletion. All strains were cultured in CAT medium, supplemented as needed with 1.5% agar. CAT medium was prepared from 5 g of tryptone (Difco Laboratories), 10 g of enzymatic casein hydrolysate (ICN Nutritional Biochemicals), 1 g of yeast extract (Difco), and 5 g of NaCl in 1 liter of H2O; sterilized for 40 min at 121°C; and then supplemented to 0.2% glucose and 0.016 M K2HPO4 before use. Antibiotics were used at the following concentrations: kanamycin (Kn), 200 μg/ml; novobiocin (Nv), 2.5 μg/ml; erythromycin (Em) 0.075 μg/ml; tetracycline (Tc), 0.25 μg/ml; spectinomycin (Spc), 100 μg/ml; and trimethoprim, 100 μg/ml. CSP1 (22) was obtained from NeoBioSci (Cambridge, MA) as a custom synthetic peptide at 80% purity and stored as sterile 0.025% solution in water at −20°C.
TABLE 1.
Strains and primers used in this study
| Strain or primer | Description or sequence (5′–3′) | Sourcea and/or reference | Location |
|---|---|---|---|
| Strains | |||
| CPM7 | CP1250 SsbB−::pEVP3::SsbB+; SsbB+; Smr Cmr | 13 | |
| CP1250 | Rx1 hex malM511 str-1 bgl-1; low-α-galactosidase background; Smr | 1 | |
| CP1344 | CP1250 ΔclpC::PcTet; Smr Tcr | 18 | |
| CP1376 | CP1250 comW::kan; Smr Knr | 9 | |
| CP1500 | Rx1 hex nov-r1 bry-r str-1 ery-r1 ery-r2; Nvr Emr Smr | 21 | |
| CP1759 | CP1250 comW::Spc; Spcr Smr | 12 | |
| CP2000 | CP1250 Δcps; Smr | 21 | |
| CP2107 | CP2000 Δcps SsbB−::pEVP3::SsbB+; SsbB+; Smr Cmr | CP2000 × CPM7 (21) | |
| CP2136 | CP2107 comA::Cheshire; Smr Cmr Emr | 35 | |
| CP2137 | CP2000 ΔCheshire comA mutant; Smr Cmr | 35 | |
| CP2451 | CP2137 rpoD-L363F; Smr Cmr | CP2137 × NYT1 {rpoD} | |
| CP2452 | CP2137 rpoD-A171V; Smr Cmr | CP2137 × ALT4 {rpoD} | |
| CP2453 | CP2137 rpoD-R355H; Smr Cmr | CP2137 × FLT4 {rpoD} | |
| CP2454 | CP2137 rpoD-R316H; Smr Cmr | CP2137 × ILT1 {rpoD} | |
| CP2455 | CP2451 ΔcomW::kan; Smr Cmr Knr | CP2451 × CP1376 {ΔcomW::kan} | |
| CP2456 | CP2452 ΔcomW::kan; Smr Cmr Knr | CP2452 × CP1376 {ΔcomW::kan} | |
| CP2457 | CP2453 ΔcomW::kan; Smr Cmr Knr | CP2453 × CP1376 {ΔcomW::kan} | |
| CP2458 | CP2454 ΔcomW::kan; Smr Cmr Knr | CP2454 × CP1376 {ΔcomW::kan} | |
| CP2459 | ALT4 rpoD-V171; Smr Cmr Knr Nvr Emr Tcr | ALT4 × CP2137 {rpoD} | |
| CP2460 | FLT4 rpoD-H355; Smr Cmr Knr Nvr Emr Tcr | FLT4 × CP2137 {rpoD} | |
| CP2461 | ILT1 rpoD-H316; Smr Cmr Knr Nvr Emr Tcr | ILT4 × CP2137 {rpoD} | |
| CP2462 | NYT1 rpoD-F363; Smr Cmr Knr Nvr Emr Tcr | NYT1 × CP2137 {rpoD} | |
| CP2463 | CP2137 ΔcomW::kan; Smr Cmr Knr | CP2137 × CP1376 {ΔcomW::kan} | |
| ALT4 | CP2463 rpoD-A171V; Smr Cmr Nvr Emr Tcr | Spontaneous rpoD mutationb | |
| FLT4 | CP2463 rpoD-R355H; Smr Cmr Nvr Emr Tcr | Spontaneous rpoD mutationb | |
| ILT1 | CP2463 rpoD-R316H; Smr Cmr Nvr Emr Tcr | Spontaneous rpoD mutationb | |
| NYT1 | CP2463 rpoD-L363F; Smr Cmr Nvr Emr Tcr | Spontaneous rpoD mutationb | |
| Primers | |||
| DAM497 | CAATTGACTATATTAGAGGCGAGACA | spr0016 | |
| DAM500 | TATCAAGCGCATCATTCAAGATAACAG | purA | |
| YT18 | GCCCTAGAAGAATTGGAACG | dnaG | |
| YT20 | CAGGGTCGATGACTTCTTCC | spr0980 | |
| YT30 | GACAGGCTTTGAGTCTCTTGATGG | spr0977 | |
| YT31 | CGGACGCTCAAACTTGGCTAATTC | spr0982 | |
| YT34 | CCATTGCCAAACGCTATGTC | rpoD | |
| YT36 | GTCAACCGCCTTCATCAAGCC | rpoD | |
| YT40 | TATGGGCTTGATGAAGGC | rpoD | |
| YT41 | TATGGGCTTGATGAAGGT | rpoD | |
| YT42 | ATACGCTCACGAGTTACG | rpoD | |
| YT43 | CGTGAAGAAAATGTTCTGCG | rpoD | |
| YT44 | CGTGAAGAAAATGTTCTGCA | rpoD | |
| YT45 | TCACATCTGCCTCGATTG | cpoA | |
| YT46 | TTGCTACGACTTGGTTGGC | rpoD | |
| YT47 | TTGCTACGACTTGGTTGGT | rpoD | |
| YT48 | GTCAATGACCCTGTCCGTATG | cpoA | |
| YT49 | CAAGTCGTAGCAAACCGC | rpoD | |
| YT50 | CAAGTCGTAGCAAACCGT | rpoD | |
| YT51 | CACGGTAAGCACCTGAAAC | cpoA | |
| YT76 | AGCGCCGACAGGGATTGGGA | dinG | |
| YT77 | ACATTGGCCTTTTGACGTGCAT | ezrA |
Braces indicate transfer of the gene only via the PCR amplicon. ×, transformation cross as recipient X donor.
Followed by transformation with nov-r1, ery-r1 ery-r2, and ΔclpC::PcTet donors.
Preparation of donor DNA.
The primers used for PCR amplification of donor DNA are listed in Table 1. The comW::kan deletion fragment was amplified from strain CP1376 using primers DAM497 and DAM500. rpoD fragments (5.5 kb) were amplified from strain CP2137 using primers YT30 and YT31. Amplification was performed in 50-μl reaction mixtures with 1 μl Phire HotStart II polymerase (Thermo Scientific) and Phire Reaction Buffer, 10 ng template DNA, 200 μmol/liter of each deoxynucleoside triphosphate (dNTP), and 0.5 μmol/liter of each primer. The amplification conditions were 98°C for 90 s and then 98°C for 15 s, 56°C for 15 s, and 72°C for 105 s for 30 cycles, followed by a 5-min 72°C final extension. The gyrB Nvr marker was prepared as a 7.4-kb amplicon using primers YT76 and YT77 and CP1500 DNA as the template. Amplification was performed as described above, except with 65°C as the annealing temperature. The PCR products were purified using a DNA Clean and Concentrate kit (Zymo Research). Genomic donor DNA was extracted from strains CP1500, CP1344, and CP1759 (Table 1).
Transformation assays.
The standard assay for transformation was done essentially as previously described (19). Log-phase culture at an optical density at 550 nm (OD550) of 0.05 at 37°C was incubated with 0.1 μg/ml DNA, 250 ng/ml CSP, 0.5 mM CaCl2, and 0.04% bovine serum albumin for 80 min at 37°C. Portions of the culture were then embedded (in 1.5 ml of CAT mixed with 1.5 ml of CAT agar) in sandwich plates and overlaid with the relevant antibiotic, as previously described (19). After 15 h at 37°C, colonies were counted. The transformation efficiency was expressed as CFU per ml of cells transformed at an OD550 of 0.05. To determine the transformation efficiency relative to the WT, the transformant yield of a strain was divided by that for a parallel WT culture. In the WT, typical yields for the PCR-amplified Nvr donor were 50% to 90% of cells transformed, while typical yields for the genomic DNA preparations were ∼2%, ∼1%, and 0.1 to 0.3% for Emr, Tcr, and Spcr markers, respectively.
Estimation of spontaneous mutation frequency.
An indication of the number of spontaneous mutations present in a mutant library was obtained by determining the number of trimethoprim-resistant mutants, which arise by inactivation of any of several proteins, including thymidine synthetase and the Ami transporters. Mutants insensitive to 100 to 200 μg/ml trimethoprim occurred at a frequency of ∼10−4.
Transformation and recovery of enrichment libraries.
Mutant libraries were prepared by serial dilutions of cells from a single isolated colony of CP2463 for 35 doublings in culture volumes of 10 ml. To obtain an enriched suppressor mutant library, 108 CFU in 1 ml CAT were exposed to CSP and DNA as described for the standard transformation assay. The entire culture was spread onto 20 100-mm selective CAT agar plates. After 15 h at 37°C in 5% CO2, 2 × 104 of the resulting transformant colonies were resuspended in 10 ml CAT. During outgrowth at 37°C for several hours, viable cells increased from ∼105 to 108 CFU/ml. This pool of transformant clones was stored at −80°C with 10% glycerol. The recovered transformant library pool is estimated to contain 20,000 transformant clones, each at 5,000 cells/ml.
Whole-genome sequencing (WGS) and analysis.
Genomic DNA for sequencing was prepared from 109 CFU by 0.2% Triton lysis and purified using a Genomic DNA Clean and Concentrate kit (Zymo Research). For sequencing on the Ion Torrent Proton sequencer, genomic DNA extracts were sheared to an average fragment size of 200 bp using a Covaris S220 focused ultrasonicator and purified using AMPure beads (Beckman-Coulter, Brea, CA). Library preparation followed the Wafergen Prep PGM 200 DNA Library protocol, using an Apollo 324 system (Wafergen, Fremont, CA) according to the manufacturer's instructions. DNA libraries containing Ion Xpress Barcode Adapters (Life Technologies, Grand Island, NY) were quantified in a Qubit 2.0 fluorometer (Life Technologies) and pooled in approximately equimolar ratio. The pool was quantified using the Library Quantification kit for Ion Torrent (KAPA Biosystems, Woburn, MA) and diluted to 26 nM prior to emulsion PCR, which was performed on the OneTouch2 instrument (Life Technologies) according to the manufacturer's instructions. Subsequently, the emulsion was broken, and Ion sphere particles (ISPs) containing amplified DNA fragments were recovered using the OneTouchES instrument (Life Technologies). The ISPs were loaded onto a Proton I chip and analyzed on the Ion Torrent Proton sequencer using 200-bp chemistry (V2) according to the manufacturer's instructions. Sequence data were demultiplexed on the Proton server and exported as FASTQ files. Using the CLC genomics workbench (CLC Bio), sequencing reads from each strain were mapped to the reference genome sequence of strain R6 (23), a laboratory ancestor of Rx1 (24). Variants that indicated a single nucleotide polymorphisms (SNP) in any of the strains relative to the reference were compiled (see Data set S1 in the supplemental material) and filtered as described in Results (see Table S1 in the supplemental material).
Targeted gene sequencing at the rpoD locus.
To sequence DNA near the rpoD gene, a 5.5-kb amplicon was prepared using primers YT30 and YT31 as described above, followed by sequencing reads using primers YT18, YT20, YT34, and YT36 at the University of Illinois at Chicago (UIC) Research Resources Center (RRC) sequencing facility, providing >3-fold coverage of 100% of the rpoD gene.
Allele exchange crosses.
To avoid possible disruption of the dnaG-rpoD-spr0980 operon, candidate suppressor alleles were transferred directly by transformation, without any linked selective marker gene, by taking advantage of the high replacement frequency achieved during natural genetic transformation with large, pure donor gene fragments. To replace one rpoD allele with another, a 5.5-kb amplicon encompassing rpoD and flanking genes, prepared by PCR from cells from a donor strain template with primers YT30 and YT31, as described above, was used as a donor at 100 ng/ml to transform a competent culture of the recipient strain. Single colonies were scanned for the replacement allele by SNP determination as described below. Overall, the frequencies of allele replacement under these conditions were 60% for the WT and 12% for suppressed comW mutants. A purified subclone of each positive colony, verified by sequencing the entire rpoD gene, was retained for further analysis.
SNP determination by mismatched-primer assay.
rpoD alleles were distinguished by a mismatched-primer PCR assay. Primers were designed to match ∼20 bases, including each SNP as the 3′-terminal base of the primer: YT41, YT44, YT47, and YT50. Identical primers, except for the 3′-terminal base, which matched the WT sequence, were also created: YT40, YT43, YT46, and YT49. They were paired with a second primer, YT42, YT45, YT48, or YT51, to amplify a 500-bp diagnostic fragment. Thus, primer set YT40, YT41, and YT42 was used to detect the base change corresponding to the A171V amino acid substitution. Using YT40 or YT41 as the forward primer and YT42 is the reverse primer, two separate reactions were performed. The two reaction products were compared on a gel for the presence or absence of a band. The same was done with primer sets YT43, YT44, and YT45 for R316H; YT46, YT47, and YT48 for R355H; and YT49, YT50, and YT51 for L363F. Detection of a product with the WT primers indicated a WT rpoD sequence, while a product with only the primers matching the mutant allele indicated a mutant rpoD allele. Amplification was performed in 10-μl reaction mixtures, with 0.2 μl Phire HotStart II polymerase (Thermo Scientific) and Phire Reaction Buffer, 1 ng template DNA, 200 μmol/liter of each dNTP, and 0.5 μmol/liter of each primer. The amplification conditions were 98°C for 90 s and then 98°C for 15 s, 69°C for 15 s, and 72°C for 15 s for 20 cycles, followed by a 5-min 72°C final extension.
RESULTS
Rare ΔcomW suppressors are recovered by serial enrichment.
The expression of genes regulated by an alternative sigma factor typically depends on numerous additional factors affecting the entire life cycle of the sigma factor itself, including its synthesis, its availability, its stability, its success in competing for occupancy of limiting core polymerase, and interaction of the resulting holoenzyme with specific promoter sites. As existing data indicate that ComW might affect various levels in the life cycle of σX, including synthesis, stability, and activity, but no molecular mechanism has yet been identified, we wished to obtain an unbiased genetic indication of the most important site of action of ComW by collecting and mapping suppressor mutations that could partially bypass the requirement for comW in competence development.
Although the severe transformation deficiency of comW mutants (10,000-fold reduced) offers the possibility of strong enrichment for such suppressors in a single step of transformation, a preliminary screen of a transposon library (provided by P. Burghout, Nijmegen, The Netherlands) did not yield any such suppressors; because transposon insertions produce predominantly null mutations, this may indicate that important players might be essential for viability (data not shown). To allow recovery of suppressors that were not null mutations, we sought conditions that would provide a sufficiently rich array of base substitutions to allow recovery of more subtle bypass mutations but not such a high density of base changes as to make mapping by WGS cumbersome. Because strain Rx1 carries a mutation inactivating the HexAB mismatch repair system, which creates a mild mutator phenotype (24), we conducted a pilot enrichment in the Rx1 derivative CP2463, in which comW was replaced by a Knr cassette and a comA deletion made development of competence absolutely dependent on exogenous CSP.
A single comW mutant colony of strain CP2463 was grown for ∼35 doublings to create a library of mixed, potentially mutant subclones (Fig. 1). The total frequency of trimethoprim-resistant mutants in a similar culture was estimated as 0.01% (data not shown), indicating the presence of numerous spontaneous base pair changes in the pool. Subjecting this library to transformation could preferentially retrieve mutations that suppress the loss of transformation caused by ComW deficiency, but as the transformation efficiency of a ΔcomW mutant is nonzero (10−4), transformants of rare-suppressor-bearing mutants would not be expected to dominate the pool of transformants obtained from a single round of transformation. However, we anticipated that the enrichment achieved in this pool would be compounded by a second round of transformation of the entire transformant pool and that suppressor mutants might predominate after further repetition of the enrichment.
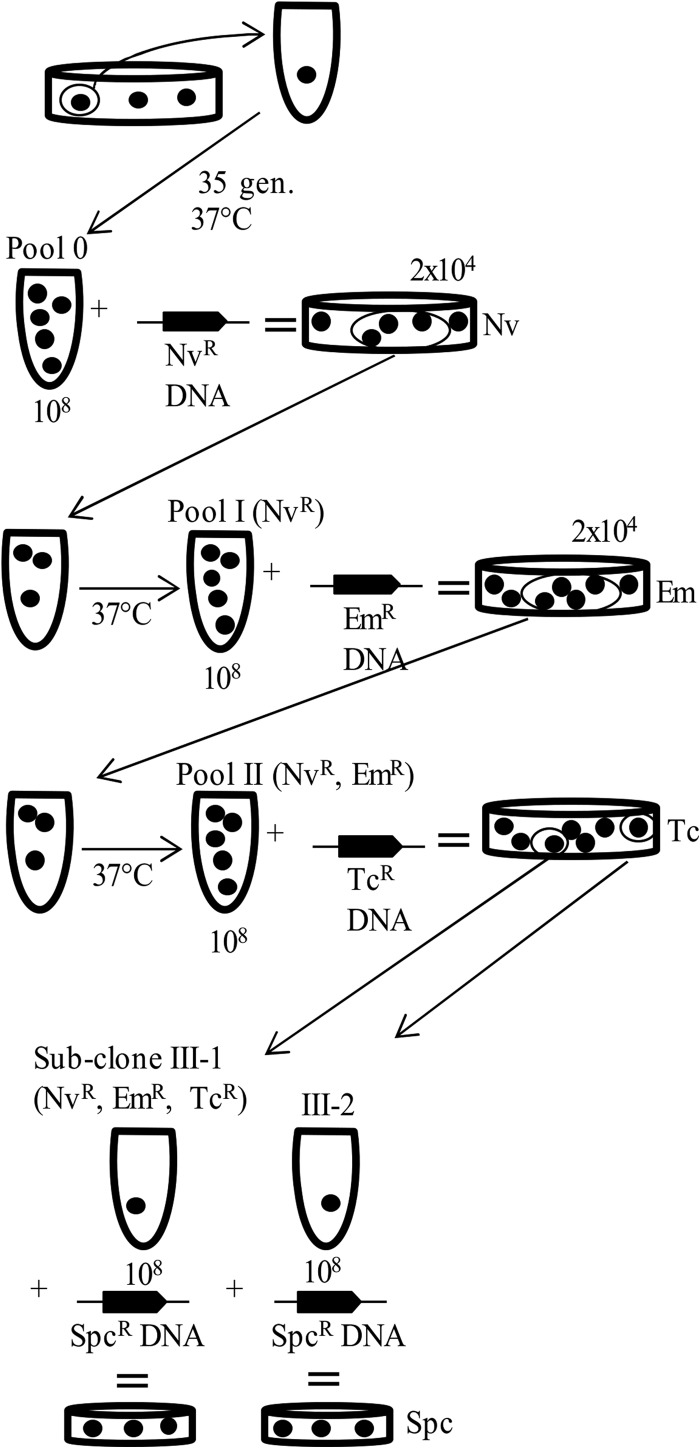
FIG 1.
Strategy for enrichment of comW bypass mutants. A single colony was picked and grown for 35 generations to create pool 0, a library of potential suppressor mutations. Then, 108 cells of pool 0 were transformed with Nvr DNA and plated, and 2 × 104 Nvr transformant colonies were collected to create pool I. Next, 108 cells of pool I were transformed with Emr DNA, and 2 × 104 Nvr Emr transformant colonies were collected to create pool II. After 108 cells of pool II were transformed with Tcr DNA, individual Nvr Emr Tcr transformant colonies were picked and transformed with Spcr DNA to determine the transformation efficiency. The dots in tubes represent cells; the dots on plates represent colonies.
To maximize recovery of rare suppressors in the first cycle of enrichment, a pure 7.4-kb Nvr DNA amplicon, which routinely transforms the WT with 80 to 90% efficiency, was chosen to transform a portion of the mutant library (100 million cells). Transformant colonies (2 × 104) were collected as a pool of Nvr cells and outgrown to produce a working pool stock. This pool was not rich in suppressors (Table 2); indeed, the recovered Nvr library did not transform detectably more than the ΔcomW background rate of 10−4 compared to the WT strain. The pool of Nvr transformants was next transformed with a genomic Emr donor DNA. Among 20 resulting Emr transformants, none transformed better than the ΔcomW mutant (data not shown). Since suppressors were thus clearly not dominant among the resulting Nvr Emr transformants, a second enrichment library was collected as a pool of 20,000 of the Nvr Emr transformants (Fig. 1) and subjected to a third cycle of transformation using genomic Tcr donor DNA (Fig. 1). Among six resulting Nvr Emr Tcr transformants, four transformed at a much higher frequency (400- to 2,000-fold) than the comW mutant parent (Table 2). This indicates that suppressors with increased transformation efficiencies dominated the resulting Nvr Emr Tcr transformant collection and established that three cycles of enrichment by this procedure could suffice to recover rare stable comW bypass mutants.
TABLE 2.
Recovery of suppressor mutants through serial transformational enrichment
| Culturea | Transformation efficiencyb |
Relative efficiencye | |
|---|---|---|---|
| WTc | Mutantd | ||
| Spontaneous-mutation library (pool 0) | 5 × 107 | 2 × 104 | 0.0004 |
| Collected Nvr library (pool I) | 5 × 105 | 1 × 102 | 0.0002 |
| Collected Nvr Emr library (pool II) | 5 × 105 | 5 × 102 | 0.001 |
| Nvr Emr Tcr clone 1 | 1 × 105 | 3 × 102 | 0.003 |
| Nvr Emr Tcr clone 2 | 1 × 105 | 9 × 101 | 0.0009 |
| Nvr Emr Tcr clone 3 | 1 × 105 | 4 × 103 | 0.04 |
| Nvr Emr Tcr clone 4 | 1 × 105 | 1 × 104 | 0.1 |
| Nvr Emr Tcr clone 5 | 1 × 105 | 1 × 104 | 0.1 |
| Nvr Emr Tcr clone 6 | 1 × 105 | 2 × 104 | 0.2 |
Pool or subclone in enrichment series PRT.
Transformation efficiency, number of drug-resistant CFU/ml, determined in quadruplicate. A PCR marker was used for pool 0, and genomic markers were used for pools I and II and clones 1 to 6. SD values were below 20%.
WT control.
Mutant pool or clone.
Relative efficiency, transformation efficiency of the strain or pool relative to that of the WT.
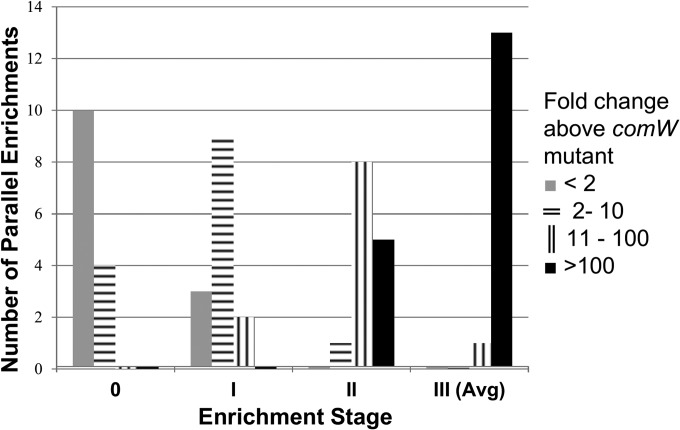
To obtain additional independent bypass mutants, this compound enrichment process was repeated in parallel for 13 additional separate single-colony subclones of CP2463, following the same series of successive enrichment cycles. After the first enrichments, 9 of the 14 Nvr pools transformed at slightly elevated rates, while two transformed 10 times as well as the comW parent, indicating that suppressor mutants were present in the pools, but at a low frequency (Fig. 2). After the second series of enrichment cycles, 5 of the 14 Nvr Emr pools transformed at rates at least 100-fold above that of the comW parent. After the third enrichment, among five Nvr Emr Tcr transformant subclones tested from each enrichment, at least one transformed at a rate at least 20-fold higher than that of the ΔcomW parent (Fig. 2 and Table 3), indicating that suppressor mutants dominated in all 14 cases.
FIG 2.
Serial enrichment of suppressor mutants from 14 independent libraries. Shown are counts of pools with transformation efficiencies in four ranges, for the initial mutant library (stage 0) and 3 successive cycles of enrichment. Values for enrichment stage III reflect averages (Avg) of subclone efficiencies (Table 3).
TABLE 3.
σA substitutions in bypass mutants
| Independent enrichmenta | Individualb | Relative efficiencyc | σA substitutiond |
|---|---|---|---|
| ALT | 5 | 0.002 | R297C |
| 2 | 0.007 | R316C | |
| 1 | 0.01 | P287S | |
| 3 | 0.02 | A171V | |
| 4 | 0.05 | A171V | |
| DET | 1 | 0.0003 | − |
| 3 | 0.006 | A171V | |
| 4 | 0.009 | − | |
| 5 | 0.05 | A171V | |
| FLT | 3 | 0.004 | R355H |
| 5 | 0.006 | R355H | |
| 1 | 0.03 | R316H | |
| 4 | 0.08 | R355H | |
| GAT | 5 | 0.00008 | − |
| 4 | 0.02 | R316H | |
| 2 | 0.06 | R316H | |
| 3 | 0.08 | A171V | |
| 1 | 0.09 | R316H | |
| ILT | 3 | 0.0004 | − |
| 4 | 0.1 | R316H | |
| 2 | 0.26 | R316H | |
| 1 | 0.3 | R316H | |
| 5 | 0.31 | R316H | |
| MAT | 2 | 0.002 | − |
| 1 | 0.003 | − | |
| 4 | 0.003 | Y290C | |
| 3 | 0.01 | R316H | |
| 5 | 0.02 | A171V | |
| MTT | 4 | 0.008 | R316H |
| 3 | 0.03 | A171T | |
| 2 | 0.06 | A171V | |
| 1 | 0.07 | R316H | |
| 5 | 0.07 | R316H | |
| NCT | 1 | 0.001 | − |
| 3 | 0.02 | A171V | |
| 5 | 0.02 | A171V | |
| 2 | 0.07 | A171V | |
| 4 | 0.07 | A171V | |
| NVT | 2 | 0.003 | V314A |
| 4 | 0.02 | A171V | |
| 5 | 0.02 | A171V | |
| 3 | 0.07 | V314A | |
| 1 | 0.1 | R316H | |
| NYT | 5 | 0.00008 | − |
| 4 | 0.008 | L363F | |
| 1 | 0.18 | L363F | |
| 3 | 0.28 | R316H | |
| 2 | 0.41 | R316H | |
| OHT | 1 | 0.05 | R355H |
| PRT | 2 | 0.0009 | − |
| 1 | 0.003 | − | |
| 3 | 0.04 | R355H | |
| 4 | 0.1 | R316H | |
| 5 | 0.1 | R316H | |
| 6 | 0.2 | R316H | |
| SCT | 5 | 0.08 | R316H |
| 4 | 0.1 | R316H | |
| 2 | 0.13 | R316H | |
| TXT | 2 | 0.06 | R316H |
| 5 | 0.1 | R316H | |
| 4 | 0.11 | R316H | |
| WT | 1 | WT | |
| ΔcomW | 0.0001 | WT |
Name of enrichment series.
Tcr clone number.
Ratio of mutant transformation efficiency to that of the WT. The SD values of quadruplicate measurements were below 20%.
Boldface, mutation identified by WGS; −, isolate not sequenced.
We conclude that rare, spontaneously arising suppressors of the comW transformation deficiency can be recovered repeatedly by three cycles of compounded enrichments.
All suppressor mutants contain an SNP in rpoD.
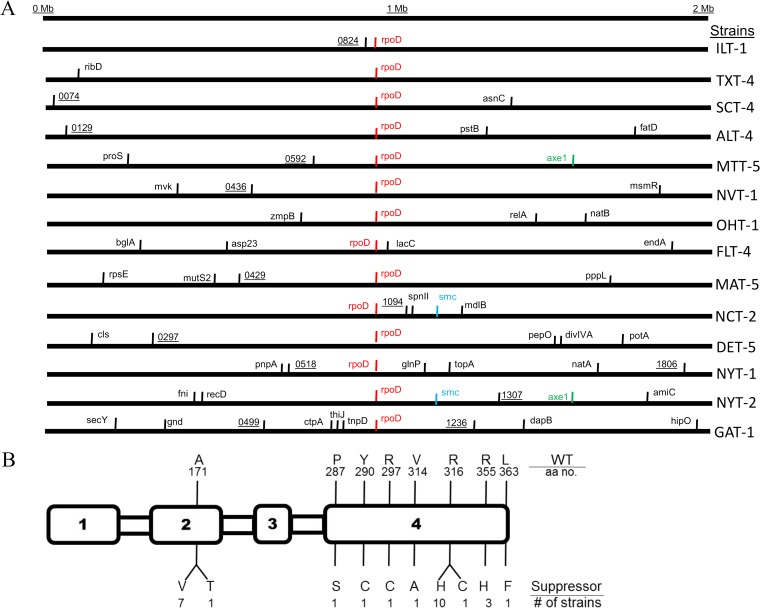
Since competence was substantially restored in each of at least 14 independently derived ΔcomW mutants, we sought to learn whether suppression was caused by single-gene mutations and to identify mutations that might be linked to the suppressor phenotype. To identify candidate unmarked mutations, WGS was carried out on a total of 16 strains, including 14 suppressor mutants, the ΔcomW parent (CP2463), and the corresponding comW+ wild type (CP2137). The sequencing reads were aligned to an annotated reference sequence of S. pneumoniae strain R6 (23), which is derived, like Rx1, from the clinical isolate D39 (25). Approximately 9,000 SNPs were identified in total (∼600 per sequenced strain) (Table 4; see Data set S1 in the supplemental material). After removing from consideration SNPs in regions with less than 40-fold coverage, ∼6,600 SNPs mapped to coding regions as identified in the R6 genome annotation. Of these, ∼2,700 cause synonymous amino acid changes. Among the remaining 3,900 SNPs, 3,768 reflected divergence of the Rx1 and R6 lineages (24), i.e., differed from the R6 sequence but were present in all of our sequenced strains (14 suppressors, the WT, and the ΔcomW parent). Among the remaining 132 SNPs, 64 were mapped to donor drug markers introduced during enrichment cycles. The remaining 68 SNPs of interest were distributed among 53 unique genes among the sequenced suppressor strains, ranging from 2 to 10 SNPs per strain. However, only the gene rpoD contained an SNP in each of the 14 sequenced suppressor strains (Table 4; see Table S1 in the supplemental material). As the remaining 54 SNPs were scattered among 52 other genes (Fig. 3A), we hypothesized that the single gene hit in every suppressor strain, rpoD, was the site of the effective bypass mutations.
TABLE 4.
Filtering of SNPs detected by WGS in 14 suppressor strains
| Stage of analysis | Total no. of SNPs in 14 strains | No. of SNPs per strain |
|---|---|---|
| All SNPs detected vs. R6a | 9,000 | 640 |
| High-quality sequence coverageb | 7,500 | 540 |
| Changes in ORFsc | 6,600 | 470 |
| Nonsynonymous | 3,900 | 280 |
| Absent from parentd | 132 | 9 |
| Outside introduced markerse | 68 | 5 |
| In rpoD | 14 | 1 |
Total SNPs after alignment to accession no. NC_003098.1.
SNPs remaining after removal of SNPs with poor sequence coverage.
Open reading frames (ORFs) as annotated in accession number NC_003098.1.
SNPs absent from the ΔcomW parent.
SNPs not introduced by Nvr, Emr, or Tcr transformation.
FIG 3.
Locations of 68 amino acid changes identified by whole-genome and rpoD-targeted sequencing. (A) Mapping of base changes identified by WGS in 14 independent comW bypass mutants. Nonsynonymous substitutions are organized by relative genome positions, and gene designations are as annotated in accession number NC_003098.1. Black, unique gene hit; color, 2 or more hits in the same gene; black horizontal line, entire S. pneumoniae genome. The names of the bypass strains sequenced by WGS are on the right. (B) Map of predicted amino acid residue changes in RpoD (σA) among 27 suppressor mutants (Table 3). Boxes, four conserved regions of σA, 369 amino acids (aa), as assigned by Vassylyev et al. (32). The WT residues and positions are shown above the protein, and the suppressor residues and numbers of cases are shown below.
Using Sanger sequencing of PCR products, we confirmed the presence of each rpoD mutation in the 14 suppressor strains used for WGS and the absence of any rpoD mutation from the WT, the ΔcomW parent, and the Nvr, Tcr, Spcr, and Emr donor strains, indicating that the mutations arose during the mutagenesis and enrichment cycles. When rpoD was similarly sequenced in the remaining 37 suppressor mutants identified among cycle 3 Nvr Emr Tcr transformants (Table 3), several more independent instances of the four rpoD mutations identified by WGS were revealed, as well as six additional rpoD nucleotide changes, as shown in Fig. 3B.
Altogether, the mapping and sequence data supported an inference that bypass of the comW requirement is available solely or principally by specific modification of σA.
Four mutant rpoD alleles substantially rescue transformation in ΔcomW strains and account for most or all of the suppression phenotype.
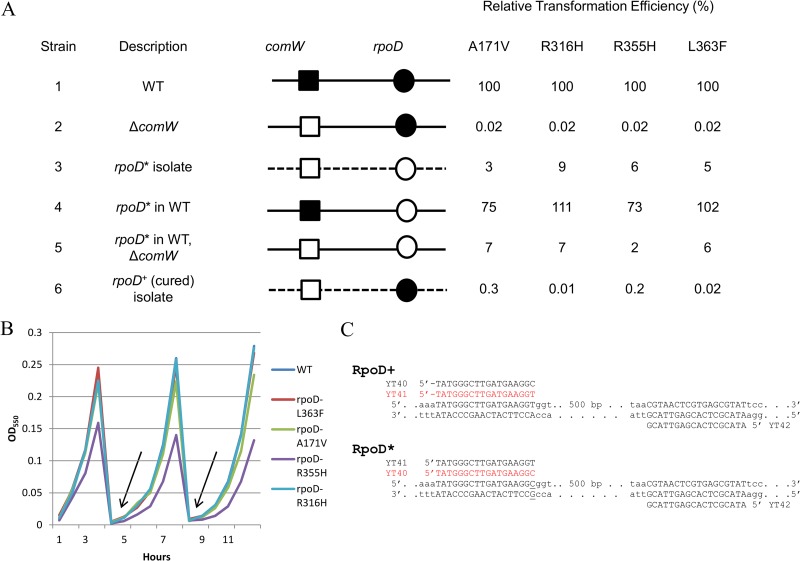
To determine directly whether the mutations identified in rpoD could bypass the loss of competence in comW mutants, two sets of allele replacement crosses were done for four representative rpoD suppressor alleles, including the three that had been recovered most frequently. Each selected rpoD allele was transferred to a WT background, followed by deletion of comW to allow detection of possible suppression. Conversely, the rpoD* allele (where the asterisk indicates any of the four mutant rpoD alleles) in each of the four corresponding suppressor mutants was replaced by the wild-type sequence (Fig. 4A). Since the mutations were not linked to a selective marker, we used a single-nucleotide primer mismatch PCR assay (Fig. 4C) to distinguish rpoD transformants from untransformed progeny. In all four cases, the backcross of mutant rpoD alleles was sufficient to increase transformation in a comW background to the level observed in the original suppressor mutant (Fig. 4A). In each case, introduction of the putative suppressor allele in the WT background did not itself detectably affect the transformation efficiency or growth rate (Fig. 4A and B). However, after the removal of comW, each rpoD* comW double mutant strongly suppressed the transformation deficiency typical of comW mutants. Thus, suppression of the comW transformation defect was indeed linked to the mutant rpoD* alleles and not to any other mutations that may have accumulated during enrichment of suppressor mutants.
FIG 4.
Linkage between rpoD* mutations and the comW bypass phenotype. (A) Backcross analysis of the bypass phenotypes of four rpoD* isolates. Shown is a comparison of the transformation efficiency of the suppressor isolate (line 3) to those of isolates with rpoD* in a WT background (line 4) and with rpoD* in a transformant ΔcomW WT background (line 5) and a suppressor isolate cured of the rpoD* mutation (line 6). Solid lines, WT genome; dashed lines, genome of isolate recovered from enrichment; ■, comW+; □, ΔcomW; ●, rpoD+; ○, rpoD* (the bypass mutant residues are indicated at the tops of the data columns). For mutations A171V, R316H, R355H, and L363F, the suppressor isolates used were ALT4, ILT1, FLT4, and NYT1, respectively; the strains with backcrossed rpoD* alleles in a ΔcomW WT background were CP2456, CP2458, CP2457, and CP2455; and the rpoD+ cured strains were CP2459, CP2461, CP2460, and CP2462. Standard deviation (SD) values were below 20%. (B) Growth of strains CP2451 to -54 containing rpoD* mutations over 17 generations. The arrows indicate 1:100 dilutions of the exponentially growing culture in fresh medium. (C) Allele-specific alternative primer pairs used for mismatch PCR genotyping of segregants after transformation with 5.5-kb donor rpoD amplicons. The SNP in the A171V mutant is underlined. The productive allele-specific primer for each allele is shown in red.
To test the linkage of suppression to the mutant alleles of rpoD further, we cured each of the four suppressor mutant types of its rpoD* SNP by replacement with the WT rpoD (Fig. 4A). For each mutation, one suppressor isolate was selected at random for the cross, although three of the mutations arose more than once. On curing of their rpoD mutations, the four strains lost their suppression phenotype, two completely and two partially (Fig. 4A). In the former cases, this indicates that suppression is entirely explained by the mutations in rpoD. In the latter cases, it appears to be possible that an additional mutation is present in the recovered suppressor mutants that accounts for a modest level of suppression (∼10-fold higher than that of the comW mutant). Nonetheless, as the corresponding backcross strains containing only the mutant allele of rpoD transformed at the level of the original suppressor mutant, it is clear that in both cases the mutant rpoD allele was primarily responsible for the comW bypass phenotype.
We conclude that each of the four substitutions in σA, A171V, R316H, R355H, and L363F, is individually both necessary and sufficient for comW bypass in the suppressor mutants recovered through repeated transformational enrichment.
DISCUSSION
To seek clues to the critical function of ComW during competence development, rare suppressors were retrieved from a library of spontaneous mutants prepared in a comW mutant background. This strategy provided a minimally biased way to identify possible ComW interaction partners, especially by allowing the examination of essential genes. Whole-genome sequencing and linkage analysis identified 10 different spontaneous mutations that substantially increased transformation efficiency in the ComW-deficient background. Remarkably, all 10 suppressors mapped to rpoD, which encodes the essential primary sigma factor subunit of RNA polymerase. This immediately implicates the housekeeping sigma factor in the function of ComW during competence development in S. pneumoniae and may explain why a previous approach using a mariner transposon was not successful at identifying comW suppressors.
Although there is no currently available structure for the S. pneumoniae RNA polymerase holoenzyme, the strong conservation of the sequence and structure of bacterial RpoD proteins suggests it could be informative to inspect the structure of σ70 in the Escherichia coli holoenzyme for clues to the nature of the pneumococcal comW bypass mechanism. To identify homologous residues in E. coli and Thermus aquaticus σ70 that correspond to those affected in the pneumococcal bypass mutants, we aligned regions 2 to 4 of RpoD proteins from several species (Fig. 5). To make comparisons, we used E. coli, T. aquaticus, and Staphylococcus aureus sequences, for which there are available crystal structures, and the Streptococcus mutans sequence, because it is a Streptococcus species that is naturally transformable but does not have ComW. Region 4 of σA, the site of most of the bypass substitutions, forms a protrusion rooted by region 3 in the cleft between the β and β′ subunits (Fig. 6). In the shape of a slightly cupped left hand, it exposes several surfaces with known functions in the initiation of transcription. The base knuckles and thumb are targeted by many transcriptional activator proteins, such as CAP and the λCI repressor, while residues of the little finger interact with PhoB (Fig. 6A) (26). The palm forms a hydrophobic surface that cradles the β flap tip, while the wrist also contacts core residues in the β and β′ subunits. The comW bypass substitutions identified here map exclusively to the palm and wrist, surfaces thought to interact with core subunits but not with either promoter DNA or known DNA-binding regulators (Fig. 6B). However, the latter surfaces do contain residues that are important in E. coli and T. aquaticus for core affinity (27, 28), as well as residues altered by DksA/ppGpp bypass mutations that facilitate activity of the alternative sigma factor σN (Fig. 6C) (29). Colocation of residues altered in comW bypass mutants with DksA bypass residues and core affinity residues suggests that ComW may serve in some way to modulate σA/σX competition in favor of σX and that weakening σA affinity for core renders this ComW activity less critical for competence development. If σA were to have a reduced affinity for core, it is possible that this would manifest in observable changes in growth. We did not observe such changes under standard growth conditions (Fig. 4B), and the literature regarding the residues affecting ppGpp bypass (29) and affinity for core (27, 28) lacks any observations on corresponding changes in the growth rate. However, there are some mutations in rpoD that cause growth defects under certain conditions, such as high temperatures (30) and exposure to ppGpp (31), which we have not yet examined. The alanine residue affected by the remaining two comW bypass mutations (S. pneumoniae A171 or E. coli A415) is buried within the three-dimensional (3D) structure of region 2, where it forms part of the interface between two successive alpha-helices (E399-D417 and K425-Q445). We speculate that disruption of this packing may also weaken the interaction between σA and core.
FIG 5.
Alignment of σA homologs. S. pneumoniae σA amino acid residues 147 to 369 were aligned with σA sequences from accession numbers Q9EZJ8.1 (T. aquaticus), YP_491259.1 (E. coli), EIA15345.1 (S. aureus), NP_721232.1 (S. pneumoniae), and NP_358573.1 (S. mutans), using Clustal Omega with default parameters (33). Asterisks, identical residues; colons, conserved residues; periods, semiconserved residues; red, residues in S. pneumoniae σA replaced by bypass mutations.
FIG 6.
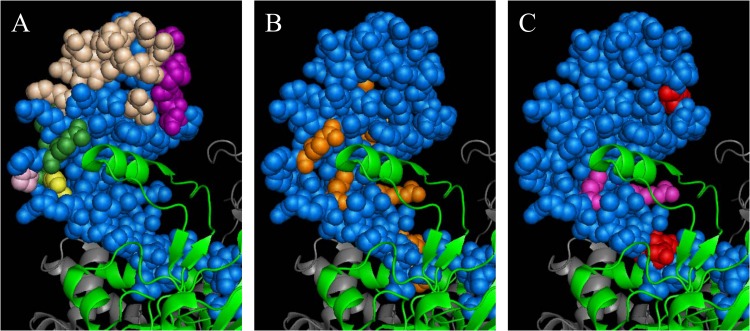
Locations of DNA-contacting and protein interaction residues, comW bypass residues, and bypass and affinity-affecting residues in region 4 of σA in a holoenzyme. Shown is the crystal structure of a holoenzyme from E. coli (34), Protein Data Bank (PDB) ID 4LJZ. σ70, space-filling blue; β-subunit, green ribbon; β′, gray ribbon. (A) Residues contacting DNA or regulatory proteins in region 4. Beige, DNA-binding residues; dark green, CAP-interacting residues; pink, FNR-interacting residues; yellow, λCI-interacting residues; purple, PhoB-interacting residues (all according to Campbell et al. [26]). (B) comW suppressor residues in region 4. Orange, residues corresponding to comW bypass mutations identified in this study. (C) Bypass and affinity residues from E. coli and T. thermophilus in region 4. Red, residues corresponding to bypass mutations that facilitate the activity of an alternative sigma factor according to Laurie et al. (29); magenta, mutations that reduce σA affinity for RNA according to Dove et al. (27) and Nickels et al. (28).
Despite the predominant location of comW bypass mutations in areas of σA that interact with core subunits, neither the mechanism of action of ComW nor the mechanism bypassing the comW requirement is revealed directly by these data. They do not even reveal which of the comW phenotypes are relieved in the bypass mutants. The location of affected residues in bypass mutants simply points to σA/σX competition (Fig. 7), which could be impacted by mechanisms affecting the level of σX or by those affecting more directly its interaction with core polymerase. However, it can be thought to “tip the balance” among known mechanisms affecting alternative sigma factor activity if the absence of other suppressor sites is considered. For example, activity of ComW either as an anti-anti-sigma or as an anti-protease-adapter would imply that these partners could be sites for additional effective suppressor mutations. The absence of such sites from our collection of suppressors suggests they are not important targets of ComW activity. In contrast, exclusive targeting of this part of σA is easily accommodated if ComW acts to exclude σA from polymerase, to promote loading of σX onto core polymerase, or to provide extra free core enzyme.
FIG 7.
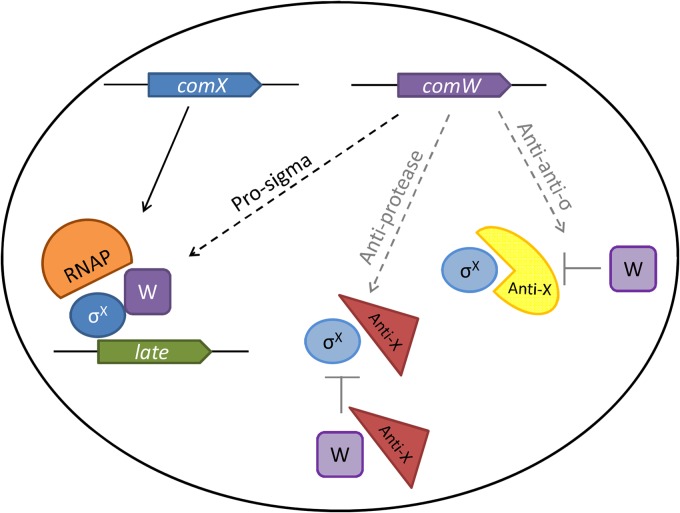
Model of potential ComW functions. Possible functions of ComW as a prosigma, anti-protease-adapter or as an anti-anti-σ. The prosigma function promoting competition with σA is in boldface because it is consistent with recovery of bypass mutations exclusively in rpoD. Anti-protease and anti-anti-σ functions are lighter because no bypass mutations were in protease subunits/adapters or in other proteins orthologous to anti-σ factors. Dashed lines, possible functions; pentagons, genes; orange half-circle, core RNA polymerase; blue circles, σX; purple squares, ComW; yellow, anti-σX; red triangles, protease.
Supplementary Material
ACKNOWLEDGMENTS
We thank Greg A. Harrison, Osamah Hasan, Marlin Amy Halder, Om Bhetuwal, and Haijing Bai for assistance with experiments. We thank the UIC Research Resources Center for sequencing and assistance with sequence analysis and Peter Burghout for the gift of the pneumococcal mariner T7 Transposon Library.
This work was supported in part by the National Science Foundation under grant no. MCB-1020863.
Footnotes
Published ahead of print 11 August 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.01933-14.
REFERENCES
- 1.Pestova EV, Havarstein LS, Morrison DA. 1996. Regulation of competence for genetic transformation in Streptococcus pneumoniae by an auto-induced peptide pheromone and a two-component regulatory system. Mol. Microbiol. 21:853–862. 10.1046/j.1365-2958.1996.501417.x. [DOI] [PubMed] [Google Scholar]
- 2.Hui FM, Morrison DA. 1991. Genetic transformation in Streptococcus pneumoniae: nucleotide sequence analysis shows comA, a gene required for competence induction, to be a member of the bacterial ATP-dependent transport protein family. J. Bacteriol. 173:372–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Morrison DA, Jaurin B. 1990. Streptococcus pneumoniae possesses canonical Escherichia coli (sigma 70) promoters. Mol. Microbiol. 4:1143–1152. 10.1111/j.1365-2958.1990.tb00689.x. [DOI] [PubMed] [Google Scholar]
- 4.Martin B, Soulet AL, Mirouze N, Prudhomme M, Mortier-Barrière I, Granadel C, Noirot-Gros MF, Noirot P, Polard P, Claverys JP. 2013. ComE/ComE∼P interplay dictates activation or extinction status of pneumococcal X-state (competence). Mol. Microbiol. 87:394–411. 10.1111/mmi.12104. [DOI] [PubMed] [Google Scholar]
- 5.Lacks SA. 2004. Transformation, p 89–115 In Mitchell TJ, Morrison DA, Spratt BG, Tuomanen EI. (ed), The pneumococcus. ASM Press, Washington, DC. [Google Scholar]
- 6.Håvarstein LS, Gaustad P, Nes IF, Morrison DA. 1996. Identification of the streptococcal competence-pheromone receptor. Mol. Microbiol. 21:863–869. 10.1046/j.1365-2958.1996.521416.x. [DOI] [PubMed] [Google Scholar]
- 7.Ween O, Gaustad P, Havarstein LS. 1999. Identification of DNA binding sites for ComE, a key regulator of natural competence in Streptococcus pneumoniae. Mol. Microbiol. 33:817–827. 10.1046/j.1365-2958.1999.01528.x. [DOI] [PubMed] [Google Scholar]
- 8.Oggioni MR, Morrison DA. 2008. Cooperative regulation of competence development in Streptococcus pneumoniae: cell-to-cell signaling via a peptide pheromone and an alternative sigma factor, p 345–362 In Winans SC, Bassler BL. (ed), Chemical communication among bacteria. ASM Press, Washington, DC. [Google Scholar]
- 9.Martin B, Granadel C, Campo N, Hénard V, Prudhomme M, Claverys JP. 2010. Expression and maintenance of ComD-ComE, the two-component signal-transduction system that controls competence of Streptococcus pneumoniae. Mol. Microbiol. 75:1513–1528. 10.1111/j.1365-2958.2010.07071.x. [DOI] [PubMed] [Google Scholar]
- 10.Peterson SN, Sung CK, Cline R, Desai BV, Snesrud EC, Luo P, Walling J, Li H, Mintz M, Tsegaye G, Burr PC, Do Y, Ahn S, Gilbert J, Fleischmann RD, Morrison DA. 2004. Identification of competence pheromone responsive genes in Streptococcus pneumoniae by use of DNA microarrays. Mol. Microbiol. 51:1051–1070. 10.1046/j.1365-2958.2003.03907.x. [DOI] [PubMed] [Google Scholar]
- 11.Peterson SN, Cline RT, Tettelin H, Sharov V, Morrison DA. 2000. Gene expression analysis of the Streptococcus pneumoniae competence regulon by use of DNA microarrays. J. Bacteriol. 182:6192–6202. 10.1128/JB.182.21.6192-6202.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Håvarstein LS, Coomaraswamy G, Morrison DA. 1995. An unmodified heptadecapeptide pheromone induces competence for genetic transformation in Streptococcus pneumoniae. Proc. Natl. Acad. Sci. U. S. A. 21:11140–11144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee MS, Morrison DA. 1999. Identification of a new regulator in Streptococcus pneumoniae linking quorum sensing to competence for genetic transformation. J. Bacteriol. 181:5004–5016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Luo P, Li H, Morrison DA. 2003. ComX is a unique link between multiple quorum sensing outputs and competence in Streptococcus pneumoniae. Mol. Microbiol. 50:623–633. 10.1046/j.1365-2958.2003.03714.x. [DOI] [PubMed] [Google Scholar]
- 15.Luo P, Morrison DA. 2003. Transient association of an alternative sigma factor, ComX, with RNA polymerase during the period of competence for genetic transformation in Streptococcus pneumoniae. J. Bacteriol. 185:349–358. 10.1128/JB.185.1.349-358.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Morrison DA. 2007. Genetic transformation in Streptococcus pneumoniae: strategies for genome modification, p 3–24 In Hackenbeck R, Chhatwal S. (ed), Molecular biology of streptococci. Horizon Scientific Press, Norfolk, United Kingdom. [Google Scholar]
- 17.Håvarstein LS. 2010. Increasing competence in the genus Streptococcus. Mol. Microbiol. 78:541–544. 10.1111/j.1365-2958.2010.07380.x. [DOI] [PubMed] [Google Scholar]
- 18.Luo P, Li H, Morrison DA. 2004. Identification of ComW as a new component in the regulation of genetic transformation in Streptococcus pneumoniae. Mol. Microbiol. 54:172–183. 10.1111/j.1365-2958.2004.04254.x. [DOI] [PubMed] [Google Scholar]
- 19.Sung CK, Morrison DA. 2005. Two distinct functions of ComW in stabilization and activation of the alternative sigma factor ComX in Streptococcus pneumoniae. J. Bacteriol. 187:3052–3061. 10.1128/JB.187.9.3052-3061.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Piotrowski A, Luo P, Morrison DA. 2009. Competence for genetic transformation in Streptococcus pneumoniae: termination of activity of the alternative sigma factor ComX is independent of proteolysis of ComX and ComW. J. Bacteriol. 191:3359–3366. 10.1128/JB.01750-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weng L, Piotrowski A, Morrison DA. 2013. Exit from competence for genetic transformation in S. pneumoniae is regulated at multiple levels. PLoS One 8:e64197. 10.1371/journal.pone.0064197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pozzi G, Masala L, Iannelli F, Manganelli R, Havarstein LS, Piccoli L, Simon D, Morrison DA. 1996. Competence for genetic transformation in encapsulated strains of Streptococcus pneumoniae: two allelic variants of the peptide pheromone. J. Bacteriol. 178:6087–6090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hoskins J, Alborn WE, Jr, Arnold J, Blaszczak LC, Burgett S, DeHoff BS, Estrem ST, Fritz L, Fu DJ, Fuller W, Geringer C, Gilmour R, Glass JS, Khoja H, Kraft AR, Lagace RE, LeBlanc DJ, Lee LN, Lefkowitz EJ, Lu J, Matsushima P, McAhren SM, McHenney M, McLeaster K, Mundy CW, Nicas TI, Norris FH, O'Gara M, Peery RB, Robertson GT, Rockey P, Sun PM, Winkler ME, Yang Y, Young-Bellido M, Zhao G, Zook CA, Baltz RH, Jaskunas SR, Rosteck PR, Jr, Skatrud PL, Glass JI. 2001. Genome of the bacterium Streptococcus pneumoniae strain R6. J. Bacteriol. 183:5709–5717. 10.1128/JB.183.19.5709-5717.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tiraby JG, Fox MS. 1973. Marker discrimination in transformation and mutation of pneumococcus. Proc. Natl. Acad. Sci. U. S. A. 70:3541–3545. 10.1073/pnas.70.12.3541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tiraby JG, Fox MS, Bernheimer H. 1975. Marker discrimination in deoxyribonucleic acid-mediated transformation of various Pneumococcus strains. J. Bacteriol. 121:608–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Campbell EA, Muzzin O, Chlenov M, Sun JL, Olson A, Weinman O, Trester-Zedlitz ML, Darst SA. 2002. Structure of the bacterial RNA polymerase promoter specificity σ subunit. Mol. Cell 9:527–539. 10.1016/S1097-2765(02)00470-7. [DOI] [PubMed] [Google Scholar]
- 27.Dove SL, Darst SA, Hochschild A. 2003. Region 4 of σ as a target for transcription regulation. Mol. Microbiol. 48:863–874. 10.1046/j.1365-2958.2003.03467.x. [DOI] [PubMed] [Google Scholar]
- 28.Nickels BE, Garrity SJ, Mekler V, Minakhin L, Severinov K, Ebright RH, Hochschild A. 2005. The interaction between σ70 and the β-flap of Escherichia coli RNA polymerase inhibits extension of nascent RNA during early elongation. Proc. Natl. Acad. Sci. U. S. A. 102:4488–4493. 10.1073/pnas.0409850102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Laurie AD, Bernardo LMD, Sze CC, Skarfstad E, Szalewska-Palasz A, Nystrom T, Shingler V. 2003. The role of the alarmone (p)ppGpp in σN competition for core RNA polymerase. J. Biol. Chem. 278:1494–1503. 10.1074/jbc.M209268200. [DOI] [PubMed] [Google Scholar]
- 30.Grossman AD, Zhou YN, Gross C, Heilig J, Christie GE, Calendar R. 1985. Mutations in the rpoH (htpR) gene of Escherichia coli K-12 phenotypically suppresses a temperature-sensitive mutant defective in the σ70 subunit of RNA polymerase. J. Bacteriol. 161:939–943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hernandez JV, Cashel M. 1995. Changes in conserved region 3 of Escherichia coli σ70 mediate ppGpp-dependent functions in vivo. J. Mol. Biol. 252:536–549. 10.1006/jmbi.1995.0518. [DOI] [PubMed] [Google Scholar]
- 32.Vassylyev DG, Sekine SI, Laptenko O, Lee J, Vassylyeva MN, Borukhov S, Yokoyama S. 2002. Crystal structure of a bacterial holoenzyme at 2.6Å resolution. Nature 417:712–719. 10.1038/nature752. [DOI] [PubMed] [Google Scholar]
- 33.Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J, Thompson JD, Higgins DG. 2011. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7:539. 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bae B, Davis E, Brown D, Campbell EA, Wigneshweraraj S, Darst SA. 2013. Phage T7 Gp2 inhibition of Escherichia coli RNA polymerase involved misappropriation of σ70 domain 1.1. Proc. Natl. Acad. Sci. U. S. A. 110:19772–19777. 10.1073/pnas.1314576110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Weng L, Biswas I, Morrison DA. 2009. A self-deleting Cre-lox-ermAM cassette, Cheshire, for markerless gene deletion in Streptococcus pneumoniae. J. Microbiol. Methods 79:353–357. 10.1016/j.mimet.2009.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.