Abstract
The Iho670 fibers of the hyperthermophilic crenarchaeon of Ignicoccus hospitalis were shown to contain several features that indicate them as type IV pilus-like structures. The application of different visualization methods, including electron tomography and the reconstruction of a three-dimensional model, enabled a detailed description of a hitherto undescribed anchoring structure of the cell appendages. It could be identified as a spherical structure beneath the inner membrane. Furthermore, pools of the fiber protein Iho670 could be localized in the inner as well as the outer cellular membrane of I. hospitalis cells and in the tubes/vesicles in the intermembrane compartment by immunological methods.
INTRODUCTION
The motility of microorganisms has already been the focus of interest since van Leeuwenhoek discovered the first organisms under his self-constructed microscopes. In a letter to the Royal Society in London, he described these little animalcules as “very prettily a-moving” and stated that “the biggest sort had a very strong and swift motion, and shot through the water (or spite) like a pike does through water” (1). It took around 300 years until the bacterial flagellum, at present the best-studied prokaryotic motility organelle, could be understood in terms of structure and function. Current data show that the bacterial flagellum not only plays a role in locomotion but also is implied in the type III secretion pathway, colonization of surfaces, or the maintenance of symbiosis between prokaryotes (2–4). In addition, various other types of cell appendages and motility organelles in prokaryotes were studied under structural and functional aspects, like type IV pili, periplasmic flagella of spirochaetes, the junctional pore complex in Myxobacteria and Cyanobacteria, the ratchet structure in Flavobacteria (5), or the terminal organelle involved in adhesion in Mycoplasma pneumoniae (6, 7).
With the discovery of the Archaea as a third domain of life in the 1990s by Carl Woese, archaeal cell appendages received greater attention (8). In particular, the archaeal flagellum was analyzed in detail, and it was shown for Halobacterium some time ago (9, 10) and more recently for Sulfolobus (with movies taken on a thermomicroscope in our labs) that it is able to generate force by rotation (11–14). Moreover, the archaeal flagellum shares some key properties with bacterial type IV pili (15–17): the heterogeneous structure of its filaments, composed of different pilin/flagellin subunits; the existence of homologous genes; a short leader peptide at the pilins/flagellins; and their cleavage by homologous signal peptidases. Like in Bacteria, the overall function of archaeal flagella is motility, although it could be shown for Bacteria as well as for Archaea that flagella also play an important role in adhesion and the formation of cell-cell contacts (11, 12, 18–20).
In addition to the archaeal flagellum, several other archaeal cell appendages, like fimbriae or pili, were described for a multitude of Archaea (21, 22). The cannulae of Pyrodictium, composed of at least three homologous glycoproteins, which form hollow tubes interconnecting the cells on the level of the periplasm after cell division (23, 24), are a unique example. The structurally most astonishing archaeal cell appendages are the hami, 2-μm-long filaments present on the cell surface of the euryarchaeon SM1 (25). They show an uncommon base-to-top organization, with three prickles emanating from the filament at periodic distances, and a distal end with a tripartite, barbed grappling hook, reminiscent of a fishing hook. Functionally, the SM1 cells use these hami for adhesion to different biotic and abiotic surfaces and formation of cell-cell contacts. Another appendage, different from the archaeal flagellum, was observed on cells of the hyperthermophilic crenarchaeon Ignicoccus hospitalis, described as the Iho670 fibers (26). These fibers are built by multiple copies of a 33-kDa protein (Iho670 [product of gene igni_0670]); its primary structure does not show any homology to other cell appendage proteins, like archaeal pili/fimbriae, hami, or cannulae. In the major fiber protein Iho670, a short type IV pilin-like signal peptide of seven amino acids was predicted by FlaFind (27). Edman degradation experiments confirmed that this signal peptide is not present in the mature fiber protein (26); thus, the signal peptide is likely to be removed by a specific peptidase. Cryoelectron microscopy of purified I. hospitalis fibers and three-dimensional (3D) modeling were used to generate a model of the native Iho670 filament with a resolution of 7.5 Å; this model showed fiber filaments to be type IV pilus-like structures, featuring a central core, which is built by α-helices of the hydrophobic N-terminal domain of Iho670 (28). In this study, we present the results of our attempts to localize the Iho670 protein throughout the cell using specific antibodies. Our results show not only that the protein is the essential part of the fiber but also that pools of this protein exist in the inner as well as the outer cellular membrane of I. hospitalis. Moreover, the anchoring structure of the Iho670 fibers is localized in the cell as a structure beneath the inner membrane.
MATERIALS AND METHODS
Mass cultivation and preparation of fibers.
I. hospitalis cells were cultivated anaerobically in half-concentrated synthetic sea water medium (1/2 SME) at 90°C in serum bottles. Mass cultivation was carried out in a 300-liter enamel-protected fermenter as described by Küper et al. (29) by the addition of 0.1% yeast extract. Fibers were obtained from the culture supernatant by 10.5% polyethylene glycol (PEG) and 5.8% NaCl-induced precipitation overnight at 4°C. Fibers were concentrated by centrifugation for 60 min at 16,000 × g in a flowthrough centrifuge overnight, and the pellet was resuspended in a small volume of morpholineethanesulfonic acid (MES) buffer (pH 6). Further purification was achieved with a CsCl gradient (0.55 g/ml) centrifugation for 48 h (SW60-Ti rotor, 250,000 × g, 4°C, Beckman Optima LE-80K ultracentrifuge), collection of the fiber-containing band, and dialysis against MES dialysis buffer (pH 6).
Biochemical approaches.
Protein samples were separated by sodium dodecyl sulfate-polyacrylamide (10%) gel electrophoresis (SDS-PAGE) as described previously (30). Proteins were stained with Coomassie brilliant blue G250 or silver stained as described previously (30, 31). Polyclonal antibodies directed against purified Iho670 fibers were raised in rabbits and purified from serum by protein G affinity chromatography. For Western blot analyses, this primary antibody (rabbit anti-Iho670) was detected by a secondary antibody coupled to alkaline phosphatase (Sigma).
Cultivation of I. hospitalis in cellulose capillary tubes and on carbon-coated gold grids.
Cellulose capillary tubes were filled with an exponentially growing culture of I. hospitalis under anaerobic conditions and were closed at both ends via superglue. Filled cellulose capillary tubes were anaerobically transferred to 20 ml of fresh 1/2 SME and cultivated at 90°C with gentle shaking. Growth of the organisms was controlled by phase-contrast light microscopy before high-pressure freezing and freeze-substitution.
For growth on transmission electron microscopy (TEM) grids, carbon-coated gold grids were transferred into small Teflon carriers and added anaerobically to serum bottles with 20 ml fresh 1/2 SME. After growth of microorganisms directly on this surface, grids were stained for 45 s with 2% uranyl acetate or immunogold labeled.
High-pressure freezing, freeze-substitution, and epon embedding.
Cryopreparation of cells was performed as originally described earlier (32), with modifications described more recently (33). Cellulose capillary tubes containing a sufficient amount of cells were transferred to 20% bovine serum albumin (BSA), cut into small pieces, and high-pressure frozen on gold specimen carriers with an EM PACT 2 high-pressure freezer (Leica Microsystems, Vienna, Austria). Freeze substitution was performed in the EM AFS 2 system (Leica Microsystems, Vienna, Austria) with a substitution medium containing 93% ethanol, 0.5% glutaraldehyde, 1% formaldehyde, and 0.5% uranyl acetate by using the following automatic program: 30 h at −90°C, 8 h at −60°C, 8 h at −30°C, and 3 h at 0°C. Samples were washed three times with ice-cold acetone and infiltrated in epon-acetone (1:1) for 2 h and epon-acetone (2:1) overnight. The cells were embedded in epon and polymerized for 2 days at 60°C. Samples were cut with an EM UC6 ultramicrotome (Leica Microsystems, Vienna, Austria) to sections of 50-nm thickness and transferred to Pioloform-coated copper or nickel slot grids. Sections were stained for 20 min with 2% uranyl acetate and for 1 min with 0.5% lead citrate, except when used for immunogold labeling.
Immunogold labeling on ultrathin sections, carbon-coated gold grids covered with cells, and fibers in solution.
For immunogold labeling, the same rabbit anti-Iho670 antibody that was used in Western blot analyses was utilized. For immunogold labeling on ultrathin sections, cells of I. hospitalis were used which were high-pressure frozen, freeze-substituted, and embedded in epon. The direct immunolabeling of Iho670 fibers in buffer solution/medium was performed by two modified procedures. (i) Carbon-coated grids covered with cells of I. hospitalis were blotted with filter paper and air dried. After heat denaturation at 120°C for 20 min, grids with samples were incubated with primary and secondary antibodies as in other experiments. (ii) Purified Iho670 fibers were incubated with primary and secondary antibodies directly in solution, with brief washing steps in between (centrifugation at 12,000 × g for 5 min), and then applied onto carbon-coated grids. In both experiments, samples were negatively stained using uranyl acetate. In all cases, the primary antibody was detected using secondary antibodies with 6-nm gold or ultrasmall gold, followed by silver enhancement (34, 35). TEM micrographs were recorded at 120 kV, using a 1,024- by 1,024-pixel slow-scan charge-coupled-device (CCD) camera (TVIPS, Gauting, Germany) attached to a CM 12 transmission electron microscope (FEI, Eindhoven, The Netherlands).
Electron tomography and image processing.
Electron tomography was performed using a CM 12 transmission electron microscope (FEI, Eindhoven, The Netherlands) at an accelerating voltage of 120 kV. Tilt series ranged from 50.41° to −46.46° using ultrathin sections and from 68.23° to −60.51° using overgrown carbon-coated gold grids with tilt increments according to the Saxton scheme (36). Data acquisition was carried out using a slow-scan CCD camera (TVIPS). The images were recorded at nominal magnifications of ×22,000 (overgrown gold grids, pixel size of 0.75 nm) or ×17,000 (ultrathin sections, pixel size of 0.93 nm) with equal defocus settings. The images of each individual tilt series were aligned using 15-nm colloidal gold beads as fiducial markers, distributed on the backside of the grids, using TOM Toolbox (37) or IMOD (38, 39). Reconstruction was performed by means of the weighted-back projection method and visualized with AMIRA (Visage Imaging, Richmond, VA).
RESULTS
Identification of Iho670 pools in the membranes of I. hospitalis.
Cells of I. hospitalis were grown in cellulose capillaries to avoid centrifugation steps that had been shown to be destructive for the cell appendages in a previous report (26). After high-pressure freezing, freeze-substitution, and epon embedding, ultrathin sections of the cells were analyzed by transmission electron microscopy. The cells showed good structural preservation, indicated by a tightly packed cytoplasm, undamaged membrane structures, and the existence of fibers which extended above the cell surface. Therefore, these cells were immunolabeled with rabbit anti-Iho670 antibody, a polyclonal antibody raised against purified I. hospitalis fibers, which only reacted with the 33-kDa fiber protein Iho670 in Western blot analyses (see Fig. S1 in the supplemental material). Labeling was done either on cells with Iho670 fibers in solution (Fig. 1) or after cells were grown directly on carbon-coated gold grids (see Fig. S2 in the supplemental material). Electron micrographs showed the antibodies to preferentially bind to the ends of the fiber filament, while binding to the central part of the fiber was rare, in less than 5% out of >100 fibers visualized (40). After a strong heat denaturation step of fibers attached to the carbon film on grids, labeling along the full length of the fiber filament was observed (see Fig. S2). For localization on ultrathin sections, a secondary antibody, coupled to 6-nm gold particles, allowed the subcellular detection of the Iho670 protein. The cells showed clear signals in the inner membrane, the outer cellular membrane, and the tubes/vesicles in the intermembrane compartment and on Iho670 fibers that extended from the surface (Fig. 2). In the cytoplasm itself, as well as in the intermembrane compartment, no signals could be detected. Controls using the preimmune serum instead of the Iho670 antibody, as well as a control without primary antibodies, showed no specific labeling on I. hospitalis cells (data not shown). Therefore, we concluded that I. hospitalis possesses a pool of Iho670 proteins in both membranes. In addition, the tubes/vesicles in the intermembrane compartment (41) were specifically labeled with these antibodies (Fig. 2); one possible explanation is that Iho670 proteins are transported, via these tubes/vesicles, from the inner membrane to the outer cellular membrane, or possibly also vice versa.
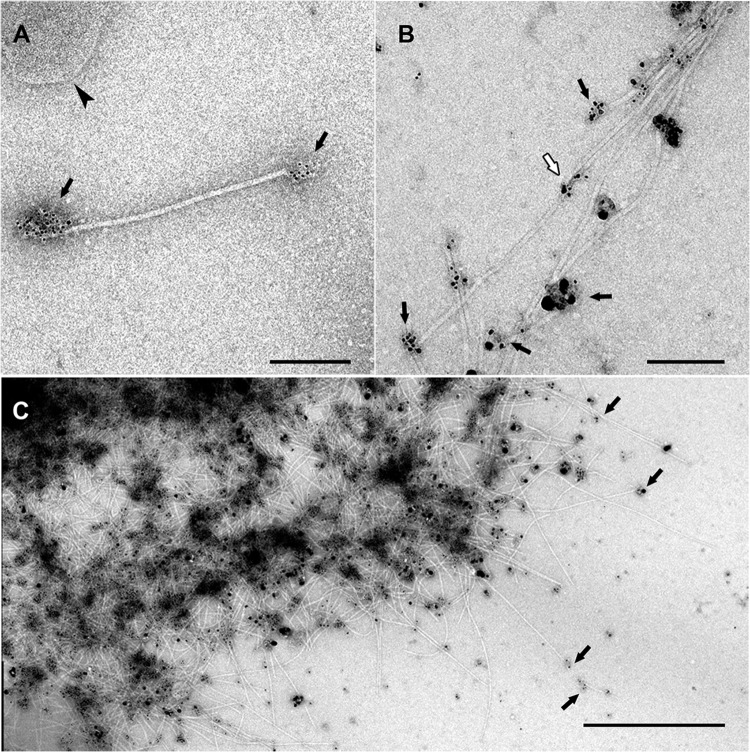
FIG 1.
Immunogold labeling of Iho670 fibers in solution. The labeling of purified fiber proteins was performed using the rabbit anti-Iho670 primary antibody (dilution of 1:10) and the goat anti-rabbit secondary antibody (6-nm gold, dilution of 1:5 [A]; ultrasmall gold, dilution of 1:5 [B]) with subsequent silver enhancement for 6 min. The images show single filaments (A) or aggregates of filaments (B, C). In most cases, the antibodies bind to the ends of the filaments where epitopes can be accessed (black arrows). Binding to intermediate regions is rare (white arrow). Sometimes, small pieces of membranes can be found after the purification (arrowhead in panel A). Scale bars, 200 nm (A and B) and 1 μm (C).
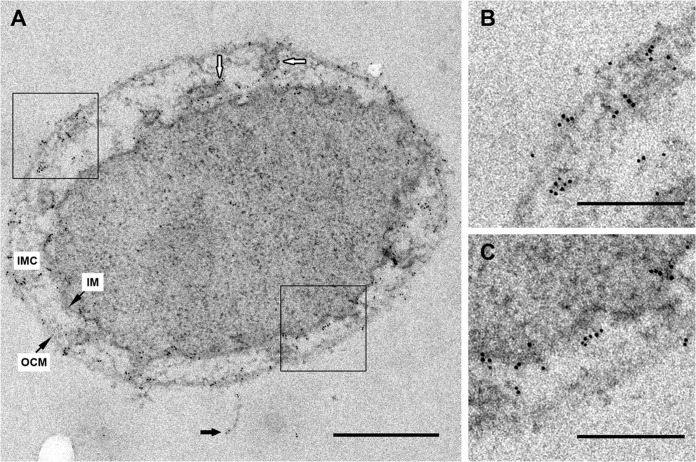
FIG 2.
Immunogold localization of Iho670 on ultrathin sections. Labeling was performed on sections of high-pressure frozen, freeze-substituted, and epon-embedded I. hospitalis cells. (A) Ultrathin section of an I. hospitalis cell with a fiber, indicated by black arrows. Primary antibody, rabbit anti-Iho670 (dilution of 1:100); secondary antibody, goat anti-rabbit antibody and 6-nm gold (dilution of 1:50). White arrows indicate the labeling of tubes/vesicles in the intermembrane compartment. At higher magnification, the selected areas in panel A exhibit signals in the outer cellular membrane (B) and the inner membrane (C). Scale bars, 500 nm (A) and 200 nm (B, C).
Search for similar proteins.
A BLASTP search was conducted in the nonredundant NCBI protein database (April 2014), using the Iho670 protein sequence, aiming to identify similar proteins from other archaeal species which have been isolated and described. It revealed only proteins with low similarity scores (<45), with the similarity restricted to the first 50 amino acids, from the following crenarchaea: Hyperthermus butylicus, Ignisphaera aggregans, Vulcanisaeta distributa, Caldisphaera lagunensis, and Caldivirga maquilingensis. The next hit was another putative protein of Ignicoccus hospitalis, Igni_0668. It showed a distinct similarity to Igni_0670 in the hydrophobic N-terminal region, up to amino acid 25, and, in contrast to the other crenarchaeal proteins, a weak overall sequence similarity (query cover, 69%; identity, 23%; score, 36.6; E value, 0.22). In the KEGG database, it is listed as a paralog of the fiber protein Igni_0670. The corresponding protein, Igni_0668, was heterologously expressed in Rosetta(DE3)pLysS using the expression vector pET29b, and highly specific antibodies were raised in rabbits (verified by Western blotting). On ultrathin sections, these antibodies distinctly labeled both the inner and the outer cellular membranes (see Fig. S3 in the supplemental material, arrows); in most cases, labeling on both membranes was observed on opposing sites (white arrows).
Localization of the anchoring structure of the Iho670 fibers.
The localization and visualization of anchoring structures of archaeal cell appendages are quite challenging, with only few successful examples. The unique anchoring structures that could be visualized so far are restricted to Halobacteria and were described as a “polar cap” or a “discoid-like lamellar structure” (DLS) (42, 43). In both cases, these flagellum anchors were thought to be located in a membrane-like structure beneath the inner membrane, but a detailed description of these structures is still absent. In order to localize the anchoring structure of the fibers of I. hospitalis, a huge number of ultrathin sections (far more than 100) were analyzed via transmission electron microscopy. For that purpose, cells were grown in cellulose capillaries to avoid centrifugation, high-pressure frozen, freeze-substituted, and embedded in epon to guarantee optimal structural preservation. In ultrathin sections, those regions that showed fibers in a longitudinal cut and those which were still in contact with the cell body of I. hospitalis were of particular interest. Different electron microscopic images were taken that clearly revealed the fibers crossing the outer cellular membrane and extending into the intermembrane compartment (Fig. 3). Because of the dynamic 3D structure of the intermembrane compartment, it was not unambiguously clear if the fibers also crossed the inner membrane. To clarify this question, electron tomography and image processing were performed on selected ultrathin sections. The resulting tomogram clearly displayed that the fiber crosses the inner membrane and is anchored in a roundish structure at about 65 nm beneath the inner membrane (Fig. 4). A membrane-like plateau, as seen by Speranskii et al. (43), could not be detected, however. Based on these results, we deductively locate the anchoring structure of the fibers of I. hospitalis to a place beneath the inner membrane.
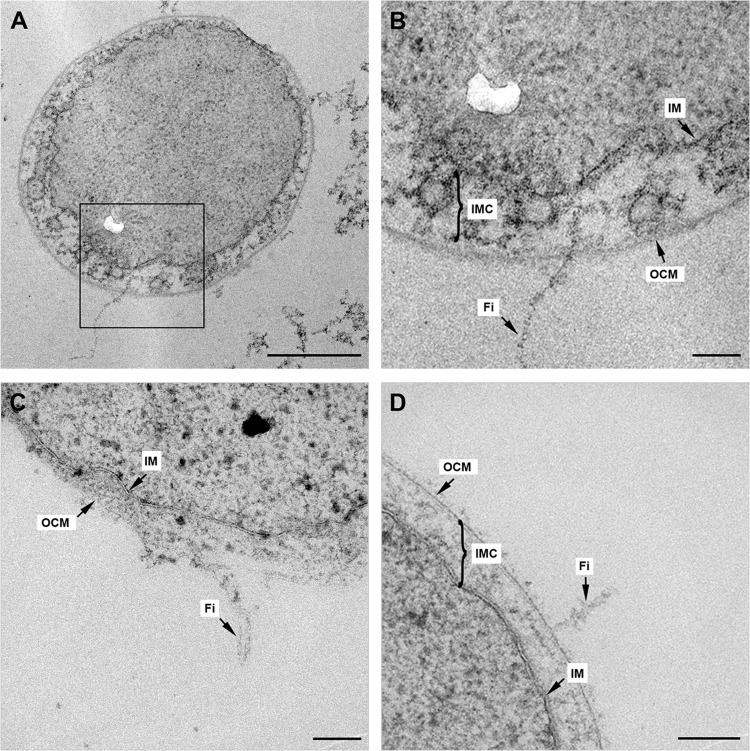
FIG 3.
Illustration of I. hospitalis fibers in ultrathin sections. (A to D) Ultrathin sections of I. hospitalis cells exhibiting longitudinal cuts, including fibers. (B) At higher magnification of the selected area in panel A, the connection of the fiber with the cell is visible. IM, inner membrane; IMC, intermembrane compartment; Fi, fiber; OCM, outer cellular membrane. Scale bars, 500 nm (A) and 100 nm (B, C, D). The white kidney-shaped area in panels A and B is a defect in the epon section.
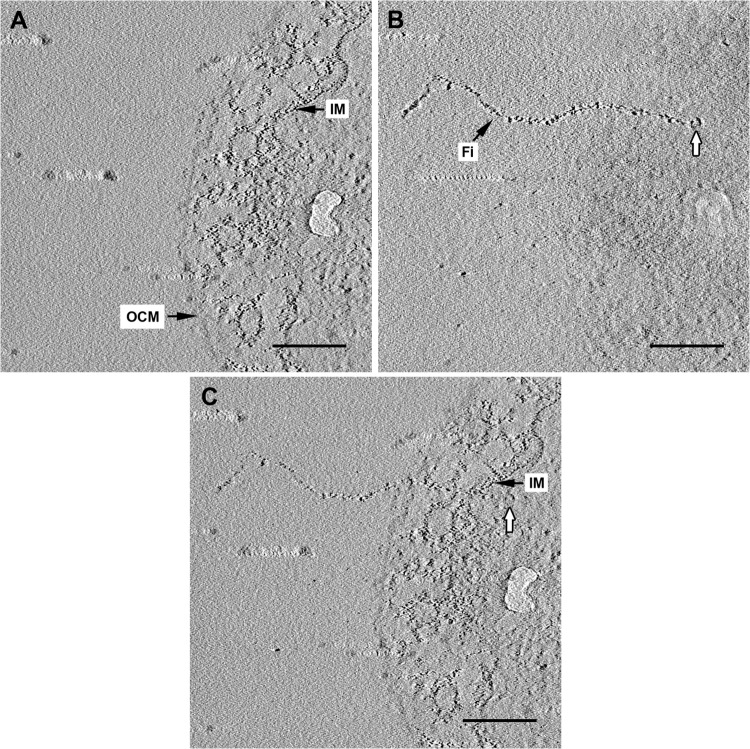
FIG 4.
Electron tomography of an ultrathin section of I. hospitalis. (A, B) Selected slices of the final tomogram after reconstruction of a tilt series of an I. hospitalis cell displaying the membranes (A) and a single fiber, with a knob-like structure at its end inside the cytoplasm (white arrows in panels B and C). (C) After merging both images, the localization of the fiber is more obvious, passing both membranes and being anchored within the cytoplasm. IM, inner membrane; OCM, outer cellular membrane; Fi, fiber. Scale bars, 200 nm.
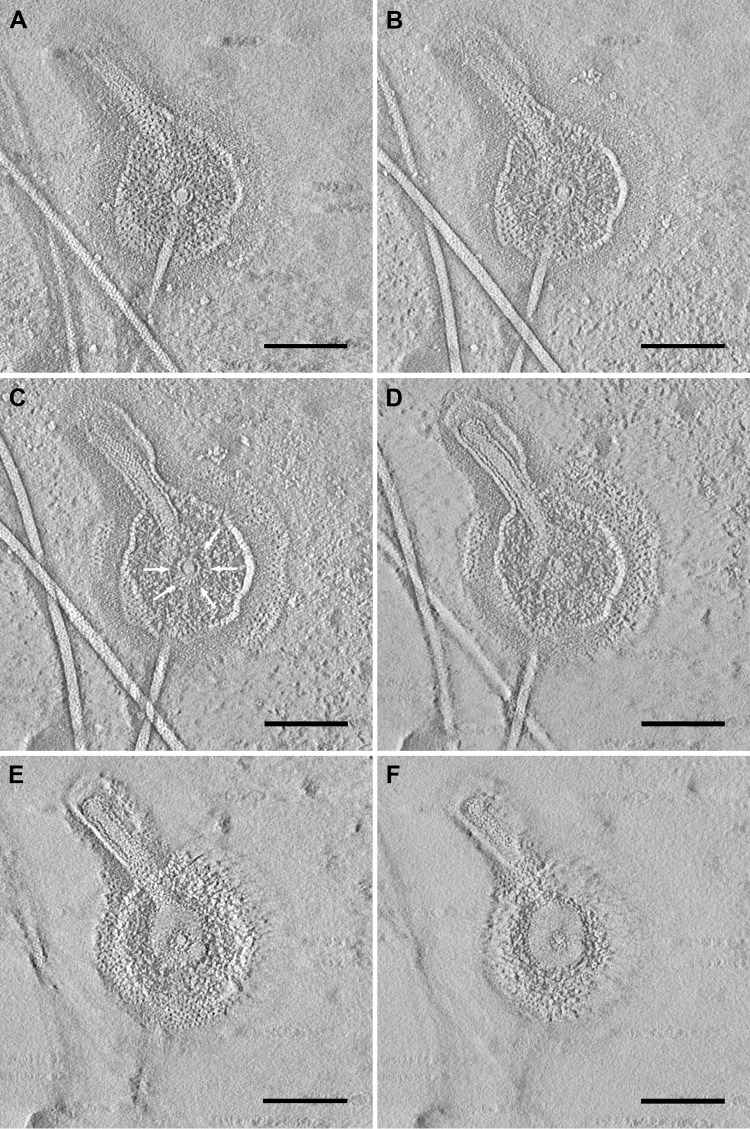
Electron tomography on structures showing fiber-associated complexes supports this assumption. For this experiment, I. hospitalis cells were grown directly on carbon-coated gold grids for subsequent electron microscopy of adherent cells and smoothly lysed (by incubation for 2 h in 1/2 SME with a modified pH of 6.75). In fact, not all cells lysed, but about 20 to 50% did (estimated); there were still “intact” cells visible after this treatment (Fig. 5). Nevertheless, this incubation led to the unraveling of a significant number of fiber-associated complexes (arrows in Fig. 5A). The complexes could be seen either without any cell contact or still in association with cells/cell remnants of I. hospitalis (Fig. 5). In all cases, they exhibited a spherical structure surrounded by two different membranes, from which the fiber arises. Immunogold labeling with an antibody directed against the main protein of the outer cellular membrane (44) assigned one of the membranes associated with the complexes to be fragments of the outer cellular membrane of I. hospitalis (data not shown; see reference 40). For that reason, we interpreted the other membrane to be fragments of the inner membrane. Electron tomography showed that the spherical structure encases the fiber; the central knob is surrounded by a cage-like structure (Fig. 6C, arrows; see also Fig. S4 in the supplemental material). That aggregate is once more coated by the inner membrane and additionally by the outer cellular membrane as the outermost sheath. We therefore conclude that the fiber is anchored in a spherical structure beneath the inner membrane.
FIG 5.
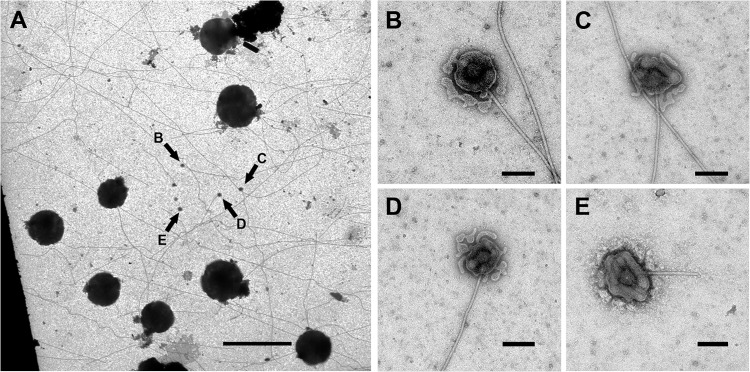
Fiber-associated complexes on overgrown carbon-coated gold grids. (A) General overview of an overgrown carbon-coated gold grid after negative staining with uranyl acetate, displaying several coccoid I. hospitalis cells. The fiber-associated complexes are indicated by arrows in panel A and are shown enlarged in panels B to E. Scale bars, 5 μm (A) and 100 nm (B to E).
FIG 6.
Electron tomography of a fiber-associated complex. (A to F) Selected sections of the final tomogram after reconstruction of the tilt series of a fiber-associated complex of I. hospitalis. The spherical complex encases the fiber; the central knob is surrounded by a cage-like structure (C, arrows). Scale bars, 100 nm.
DISCUSSION
In previous studies, we showed that the fibers of Ignicoccus hospitalis represent structures sharing similarities to bacterial type IV pili and archaeal flagella, with implications for supercoiling and polymorphism (26, 28, 45). Our current results enabled us to gain insight into the anchoring mechanism of these unique cell surface appendages.
Localization of Iho670 proteins in I. hospitalis.
Different immunogold labeling experiments showed clear signals with the rabbit anti-Iho670 antibody over the whole length of the fiber, so we concluded that this antibody is highly specific. The detection of significant amounts of fiber proteins in both membranes and, in addition, in the tubes/vesicles in the intermembrane compartment supports the hypothesis that the tubes/vesicles might be involved in transporting proteins from the cytoplasm and inner membrane to the outer cellular membrane, or possibly also vice versa, a function already previously postulated for this highly dynamic membrane system (41, 46). In organisms expressing type IV pili, an inner membrane pool of pilins could also be found, where they are thought to be used for assembly of new type IV pili (47). At least to a certain extent, these pilins could be the result of a degradation process of type IV pili, so a recycling of pilin proteins is also possible (48, 49). At least the N-terminal portion of the I. hospitalis fiber protein shows similarities to bacterial type IV pili, although with a different supercoiling mechanism (28). Thus, the protein pools in the I. hospitalis membranes could also be seen as a reservoir for fiber proteins. It has to be mentioned here that I. hospitalis fibers do not show any homology to archaeal flagella or fimbriae known today and only weak similarity to a few “hypothetical proteins” from other crenarchaea; hence, the I. hospitalis fibers represent an independent variant of cell appendages (28). With relation to the Ignicoccus habitat, a region with extremely high rates of water flow (50), and the brittle structure of these fibers (26), a reservoir of fiber proteins is highly expedient, enabling a fast reassembly in the case of damage. So far, there is no evidence that the fibers are used for motility, as demonstrated for flagella in other archaeal species (26, 51). Thus, an adhesive function for these structures is highly supposable, strengthened by the fact that growing I. hospitalis cells on surfaces results in a high abundance of fibers on these surfaces (see Fig. 2 in reference 28). In this context, the fiber proteins in the I. hospitalis outer cellular membrane could also be used as adhesins for a first attachment onto surfaces, intensified by the production of fibers at a later time point. In fact, adhesion proteins in the outer membranes of organisms are quite common (52–54); see, e.g., CBP-12, a type IV pilin-like protein in the Fibrobacter succinogenes outer membrane (55).
Since its first description in 2009, the Iho670 fiber protein of I. hospitalis remains a unique protein; searches for a similar protein among the Archaea are still unsuccessful. A single protein in I. hospitalis itself, Igni_0668, exhibits some weak but distinct similarity; inside the cell, it is also located in both membranes. It remains to be shown in future studies, investigating protein-protein interactions, or using Northern blot analysis, whether this protein, and possibly the Igni_0669 “hypothetical” protein, is involved in fiber assembly or in building up the anchor structure.
The anchoring structure of Iho670 fibers.
By using electron tomography of ultrathin sections and of fiber complexes, it was possible to localize the anchoring structure of the fibers to a place underneath the inner membrane, showing the fiber arising from a spherical structure. As a consequence, an anchoring structure in the outer cellular membrane or directly in the inner membrane can be excluded. That is in contrast to the anchoring structure supposed for bacterial type IV pili, which is assumed to be located in or directly underneath the inner membrane (56); however, a definite localization of this structure is absent. In addition, hardly any information is reported concerning the anchoring structure of archaeal flagella, with the notable exception of Sulfolobus: here, some biochemical data exist (57). Direct structural data obtained by electron microscopy are poor or missing, however. That might be due to inadequate isolation methods or labile anchoring structures, which cannot be conserved by the techniques used so far. By now, only knob-like structures could be found by chance in flagellum preparations of some methanogenic species, in addition to the so-called “polar cap” or “discoid-like lamellar structure” (DLS) in halophilic Archaea (42, 58, 59; our unpublished data). A similarity of the anchoring structure of the I. hospitalis fibers to this discoid lamellar structure is unlikely, because there is no evidence for a membrane-like structure beneath the inner membrane, as it is proposed by Metlina (58) for the DLS. Although the DLS was described by the use of ultrathin sections, one has to admit that the cells presented in these studies (58, 60) exhibit a poor structural preservation (due to inferior preparation methods) and therefore most likely do not reflect the native state. Similarities to the polar cap of Halobacterium are difficult to discuss, because a definite description for this structure does not exist. Kupper et al. (42) were solely able to describe round patches from which numerous flagella emerged. The anchoring structure presented here was obtained from ultrathin sections of I. hospitalis cells which were grown in cellulose capillaries and high-pressure frozen, freeze-substituted, and epon embedded. Therefore, we conclude that the anchor of the Iho670 fiber described here represents a “close-to-native” structure. To our knowledge, this is the first definite localization of the anchoring structure of an archaeal cell appendage.
The theory of an anchoring structure underneath the inner membrane as a spherical or roundish structure is strengthened by tomograms recorded from fiber-associated complexes found on overgrown carbon-coated gold grids, indicating the fiber to arise from a spherical structure. Also in these complexes, the basal point of the fiber can be localized to an area which is at some distance from the inner membrane. The complexes originated from lysed cells, due to osmotic effects and micelle formation of the inner and the outer cellular membrane. Isolation and biochemical analyses of these complexes are highly promising to gain more information on proteins that are involved in this structure.
The data presented here are in line with our current hypothesis that the fibers of I. hospitalis are type IV pilus-like structures, according to obvious similarities in structure and biochemical context. Finally, a definite localization of an anchoring structure of an archaeal cell appendage could be demonstrated for the first time. The results of the subcellular localization experiments demonstrate the main fiber protein to be present in both cellular membranes and in the tubes/vesicles in the intermembrane compartment, strongly indicating a transport activity from one membrane to the other.
Supplementary Material
ACKNOWLEDGMENTS
We thank E. Papst, T. Hader, and K. Eichinger for technical assistance and A. Zenker for expert artwork.
This work was supported by DFG grant WI 731/10-1 to R.R. and R.W., DFG grant HU 703/2-1/2 to Harald Huber and R.R., and the LOEWE program of the state of Hessen (Synmikro) to A.K.
Footnotes
Published ahead of print 25 August 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.01861-14.
REFERENCES
- 1.Leewenhoeck A. 1684. An abstract of a letter from Mr. Anthony Leewenhoeck at Delft, dated Sept. 17. 1683. Containing some microscopical observations, about animals in the scurf of the teeth, the substance call'd worms in the nose, the cuticula consisting of scales. Philos. Trans. 14:568–574. 10.1098/rstl.1684.0030. [DOI] [Google Scholar]
- 2.Aizawa SI. 2001. Bacterial flagella and type III secretion systems. FEMS Microbiol. Lett. 21:157–164. 10.1016/S0378-1097(01)00301-9. [DOI] [PubMed] [Google Scholar]
- 3.Nachamkin I, Yang X, Stern NJ. 1993. Role of Campylobacter jejuni flagella as colonization factors for three-day-old chicks: analysis with flagella mutants. Appl. Environ. Microbiol. 95:1269–1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shimoyama T, Kato S, Ishii S, Watanabe K. 2009. Flagellum mediates symbiosis. Science 323:1574. 10.1126/science.1170086. [DOI] [PubMed] [Google Scholar]
- 5.Bardy SL, Ng SYM, Jarrell KF. 2003. Prokaryotic motility structures. Microbiology 149:295–304. 10.1099/mic.0.25948-0. [DOI] [PubMed] [Google Scholar]
- 6.Seto S, Layh-Schmitt G, Kenri T, Miyata M. 2001. Visualization of the attachment organelle and cytadherence proteins of Mycoplasma pneumonia by immunofluorescence microscopy. J. Bacteriol. 183:1621–1630. 10.1128/JB.183.5.1621-1630.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hasselbring BM, Jordan JL, Krause RW, Krause DC. 2006. Terminal organelle development in the cell wall-less bacterium Mycoplasma pneumoniae. Proc. Natl. Acad. Sci. U. S. A. 103:16478–16483. 10.1073/pnas.0608051103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Woese CR, Kandler O, Wheelis ML. 1990. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc. Natl. Acad. Sci. U. S. A. 87:4576–4579. 10.1073/pnas.87.12.4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marwan W, Alam M, Oesterhelt D. 1991. Rotation and switching of the flagellar motor assembly in Halobacterium halobium. J. Bacteriol. 173:1971–1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alam M, Claviez M, Oesterhelt D, Kessel M. 1984. Flagella and motility behaviour of square bacteria. EMBO J. 3:2899–2903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thomas NA, Bardy SL, Jarrell KF. 2001. The archaeal flagellum: a different kind of prokaryotic motility structure. FEMS Microbiol. Rev. 25:147–174. 10.1111/j.1574-6976.2001.tb00575.x. [DOI] [PubMed] [Google Scholar]
- 12.Albers SV, Meyer B. 2011. The archaeal cell envelope. Nat. Rev. Microbiol. 9:414–426. 10.1038/nrmicro2576. [DOI] [PubMed] [Google Scholar]
- 13.Klingl A, Flechsler J, Heimerl T, Rachel R. 19 September 2013. Archaeal cells. In eLS. John Wiley & Sons Ltd, Chichester, United Kingdom. 10.1002/9780470015902.a0000383.pub2. [DOI] [Google Scholar]
- 14.Shahapure R, Driessen RPC, Haurat MF, Albers SV, Dame RT. 2013. The archaellum: a rotating type IV pilus. Mol. Microbiol. 91:716–723. 10.1111/mmi.12486. [DOI] [PubMed] [Google Scholar]
- 15.Jarrell KF, Bayley DP, Kostyukova AS. 1996. The archaeal flagellum: a unique motility structure. J. Bacteriol. 178:5057–5064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bayley DP, Jarrell KF. 1998. Further evidence to suggest that archaeal flagella are related to bacterial type IV pili. J. Mol. Evol. 46:370–373. [PubMed] [Google Scholar]
- 17.Ng SYM, Chaban B, Jarrell KF. 2006. Archaeal flagella, bacterial flagella and type IV pili: a comparison of genes and posttranslational modifications. J. Mol. Microbiol. Biotechnol. 11:167–191. 10.1159/000094053. [DOI] [PubMed] [Google Scholar]
- 18.Näther DJ, Rachel R, Wanner G, Wirth R. 2006. Flagella of Pyrococcus furiosus: multifunctional organelles, made for swimming, adhesion to various surfaces, and cell-cell contacts. J. Bacteriol. 188:6915–6923. 10.1128/JB.00527-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bellack A, Huber H, Rachel R, Wanner G, Wirth R. 2011. Methanocaldococcus villosus sp. nov., a heavily flagellated archaeon that adheres to surfaces and forms cell-cell contacts. Int. J. Syst. Evol. Microbiol. 61:1239–1245. 10.1099/ijs.0.023663-0. [DOI] [PubMed] [Google Scholar]
- 20.Lassak K, Neiner T, Ghosh A, Klingl A, Wirth R, Albers SV. 2011. Molecular analysis of the crenarchaeal flagellum. Mol. Microbiol. 83:110–124. 10.1111/j.1365-2958.2011.07916.x. [DOI] [PubMed] [Google Scholar]
- 21.Wang YA, Yu X, Ng SY, Jarrell KF, Egelman EH. 2008. The structure of an archaeal pilus. J. Mol. Biol. 29:456–466. 10.1016/j.jmb.2008.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thoma C, Frank M, Rachel R, Schmid S, Näther D, Wanner W, Wirth R. 2008. The Mth60 fimbriae of Methanothermobacter thermoautotrophicus are functional adhesins. Environ. Microbiol. 10:2785–2795. 10.1111/j.1462-2920.2008.01698.x. [DOI] [PubMed] [Google Scholar]
- 23.Rieger G, Rachel R, Hermann R, Stetter KO. 1995. Ultrastructure of the hyperthermophilic archaeon Pyrodictium abyssi. J. Struct. Biol. 115:78–87. 10.1006/jsbi.1995.1032. [DOI] [Google Scholar]
- 24.Nickell S, Hegerl R, Baumeister W, Rachel R. 2003. Pyrodictium cannulae enter the periplasmic space but do not enter the cytoplasm, as revealed by cry-electron tomography. J. Struct. Biol. 141:32–42. 10.1016/S1047-8477(02)00581-6. [DOI] [PubMed] [Google Scholar]
- 25.Moissl C, Rachel R, Briegel A, Engelhardt H, Huber R. 2005. The unique structure of archaeal hami, highly complex cell appendages with nano-grappling hooks. Mol. Microbiol. 56:361–370. 10.1111/j.1365-2958.2005.04294.x. [DOI] [PubMed] [Google Scholar]
- 26.Müller DW, Meyer C, Gürster S, Küper U, Huber H, Rachel R, Wanner G, Wirth R, Bellack A. 2009. The Iho670 fibers of Ignicoccus hospitalis. a new type of archaeal cell surface appendage. J. Bacteriol. 191:6465–6468. 10.1128/JB.00858-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Szabó Z, Stahl AO, Albers SV, Kissinger JC, Driessen AJM, Pohlschroder M. 2007. Identification of diverse archaeal proteins with class III signal peptides cleaved by distinct archaeal prepilin peptidases. J. Bacteriol. 189:772–778. 10.1128/JB.01547-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yu X, Goforth C, Meyer C, Rachel R, Wirth R, Schröder DF, Egelman EH. 2012. Filaments from Ignicoccus hospitalis show diversity of packing in proteins containing N-terminal type IV pilin helices. J. Mol. Biol. 422:274–281. 10.1016/j.jmb.2012.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Küper U, Meyer C, Müller V, Rachel R, Huber H. 2010. Energized outer membrane and spatial separation of metabolic processes in the hyperthermophilic archaeon Ignicoccus hospitalis. Proc. Natl. Acad. Sci. U. S. A. 107:3152–3156. 10.1073/pnas.0911711107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schägger H, von Jagow G. 1987. Tricine-sodium dodecyl sulphate-polyacrylamide gel electrophoresis for separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 166:223–231. [DOI] [PubMed] [Google Scholar]
- 31.Blum H, Beier H, Gross HJ. 1987. Improved silver staining of plant proteins, RNA and DNA in polyacrylamid gels. Electrophoresis 8:93–99. 10.1002/elps.1150080203. [DOI] [Google Scholar]
- 32.Hohenberg H, Mannweiler K, Müller M. 1994. High-pressure freezing of cell suspensions in cellulose capillary tubes. J. Microsc. 175:34–43. 10.1111/j.1365-2818.1994.tb04785.x. [DOI] [PubMed] [Google Scholar]
- 33.Rachel R, Meyer C, Klingl A, Gürster S, Heimerl T, Wasserburger N, Burghardt T, Küper U, Bellack A, Schopf S, Wirth R, Huber H, Wanner G. 2010. Analysis of the ultrastructure of Archaea by electron microscopy. Methods Cell Biol. 96:47–69. 10.1016/S0091-679X(10)96003-2. [DOI] [PubMed] [Google Scholar]
- 34.Danscher G. 1981. Localisation of gold in biological tissue. A photochemical method for light and electron microscopy. Histochemistry 71:81–88. [DOI] [PubMed] [Google Scholar]
- 35.Stierhof YD, Humbel BM, Herrmann R, Otten MT, Schwarz H. 1992. Direct visualization and silver enhancement of ultrasmall antibody-bound gold particles on immunolabeled ultrathin resin sections. Scanning Microsc. 6:1009–1012. [Google Scholar]
- 36.Saxton WO, Baumeister W, Hahn M. 1984. Three-dimensional reconstruction of imperfect two-dimensional crystals. Ultramicroscopy 13:57–70. 10.1016/0304-3991(84)90057-3. [DOI] [PubMed] [Google Scholar]
- 37.Nickell S, Förster F, Linaroudis A, Del Net W, Beck F, Hegerl R, Baumeister W, Plitzko J. 2005. TOM software toolbox: acquisition and analysis for electron tomography. J. Struct. Biol. 149:227–234. 10.1016/j.jsb.2004.10.006. [DOI] [PubMed] [Google Scholar]
- 38.Kremer JM, Mastronade DN, McIntosh JR. 1996. Computer visualization of three-dimensional image data using IMOD. J. Struct. Biol. 116:71–76. 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
- 39.Mastronarde DN. 2006. Tomographic reconstruction with the IMOD software package. Microsc. Microanal. 12:178–179. 10.1017/S1431927606069467.17481355 [DOI] [Google Scholar]
- 40.Meyer C. 2010. Die fibers von Ignicoccus hospitalis: ultrastruktur, verankerung, und molekularbiologische untersuchungen. Ph.D. thesis University of Regensburg, Regensburg, Germany. [Google Scholar]
- 41.Huber H, Küper U, Daxer S, Rachel R. 2012. The unusual cell biology of the hyperthermophilic crenarchaeon Ignicoccus hospitalis. Antonie Van Leeuwenhoek 102:203–219. 10.1007/s10482-012-9748-5. [DOI] [PubMed] [Google Scholar]
- 42.Kupper J, Marwan W, Typke D, Grünberg H, Uwer U, Gluch M, Oesterhelt D. 1994. The flagellar bundle of Halobacterium salinarum is inserted into a distinct polar cap structure. J. Bacteriol. 176:5184–5187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Speranskii VV, Metlina AL, Novikova TM, Bakeyeva LY. 1996. Disk-like lamellar structure as part of the archaeal flagellar apparatus. Biophysics 41:167–173. [Google Scholar]
- 44.Burghardt T, Näther DJ, Junglas B, Huber H, Rachel R. 2007. The dominating outer membrane protein of the hyperthermophilic archaeum Ignicoccus hospitalis: a novel pore-forming complex. Mol. Microbiol. 63:166–176. 10.1111/j.1365-2958.2006.05509.x. [DOI] [PubMed] [Google Scholar]
- 45.Albers SV, Szabó Z, Driessen AJ. 2003. Archaeal homolog of bacterial type IV prepilin signal peptidase with broad substrate specificity. J. Bacteriol. 185:3918–3925. 10.1128/JB.185.13.3918-3925.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rachel R, Wyschkony I, Riehl S, Huber H. 2002. The ultrastructure of Ignicoccus: evidence for a novel outer membrane and for intracellular vesicle budding in an archaeon. Archaea 1:1–9. 10.1155/2002/371325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Parge HE, Forest KT, Hickley MJ, Christensen DA, Getzoff ED, Tainer JA. 1995. Structure of the fiber-forming protein pilin at 2.6 Å resolution. Nature 378:32–38. 10.1038/378032a0. [DOI] [PubMed] [Google Scholar]
- 48.Morand PC, Bille E, Morelle S, Eugène E, Beretti JL, Wolfgang M, Meyer TF, Koomey M, Nassif X. 2004. Type IV pilus retraction in pathogenic Neisseria is regulated by PilC proteins. EMBO J. 23:2009–2017. 10.1038/sj.emboj.7600200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Skerker JM, Berg HC. 2001. Direct observation of extension and retraction of type IV pili. Proc. Natl. Acad. Sci. U. S. A. 98:6901–6904. 10.1073/pnas.121171698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fricke H, Giere O, Stetter K, Alfrdesson GA, Kristjansson JK, Stoffers P, Svavarsson J. 1989. Hydrothermal vent communities at the shallow subpolar Mid-Atlantic ridge. Mar. Biol. 102:425–429. 10.1007/BF00428495. [DOI] [Google Scholar]
- 51.Herzog B, Wirth R. 2012. Swimming behavior of selected species of Archaea. Appl. Environ. Microbiol. 78:1670–1674. 10.1128/AEM.06723-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kaplan CW, Lux R, Haake SK, Shi W. 2009. The Fusobacterium nucleatum outer membrane protein RadD is an arginine-inhibitable adhesion required for inter-species adherence and the structured architecture of multi-species biofilm. Mol. Microbiol. 71:35–47. 10.1111/j.1365-2958.2008.06503.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang P, Chomel BB, Schau MK, Goo JS, Droz S, Kelminson KL, George SS, Lerche NW, Koehler JE. 2004. A family of variably expressed outer-membrane proteins (Vomp) mediates adhesion and autoaggregation in Bartonella quintana. Proc. Natl. Acad. Sci. U. S. A. 101:13630–13635. 10.1073/pnas.0405284101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hoiczyk E, Roggenkamp A, Reichenbecher M, Lupas A, Heesemann J. 2000. Structure and sequence analysis of Yersinia YadA and Moraxella UspAS reveal a novel class of adhesins. EMBO J. 19:5989–5999. 10.1093/emboj/19.22.5989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jun HS, Qi M, Gong J, Egbosimba EE, Forsberg CW. 2007. Outer membrane proteins of Fibrobacter succinogenes with potential roles in adhesion to cellulose and in cellulose digestion. J. Bacteriol. 189:6806–6815. 10.1128/JB.00560-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bardy SL, Ng SY, Jarrell KF. 2004. Recent advances in the structure and assembly of the archaeal flagellum. J. Mol. Microbiol. Biotechnol. 7:41–51. 10.1159/000077868. [DOI] [PubMed] [Google Scholar]
- 57.Banerjee A, Neiner T, Tripp P, Albers S-V. 2013. Insights into subunit interactions in the Sulfolobus acidocaldarius archaellum cytoplasmic complex. FEBS J. 280:6141–6149. 10.1111/febs.12534. [DOI] [PubMed] [Google Scholar]
- 58.Metlina AL. 2004. Bacterial and archaeal flagella as prokaryotic motility organelles. Biochemistry (Moscow) 69:1203–1212. [DOI] [PubMed] [Google Scholar]
- 59.Kalmokoff ML, Jarrell KF, Koval SF. 1988. Isolation of flagella from the archaeabacterium Methanococcus voltae by phase separation with Triton X-114. J. Bacteriol. 170:1752–1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bakeeva LE, Metlina AL, Novikova TM, Speranski VV. 1992. Ul'trastruktura zhgutikovogo apparata Halobacterium salinarium. Dokl. RAN 326:914–915. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.