Abstract
Listeria monocytogenes is a Gram-positive facultative intracellular pathogen that is highly resistant to lysozyme, a ubiquitous enzyme of the innate immune system that degrades cell wall peptidoglycan. Two peptidoglycan-modifying enzymes, PgdA and OatA, confer lysozyme resistance on L. monocytogenes; however, these enzymes are also conserved among lysozyme-sensitive nonpathogens. We sought to identify additional factors responsible for lysozyme resistance in L. monocytogenes. A forward genetic screen for lysozyme-sensitive mutants led to the identification of 174 transposon insertion mutations that mapped to 13 individual genes. Four mutants were killed exclusively by lysozyme and not other cell wall-targeting molecules, including the peptidoglycan deacetylase encoded by pgdA, the putative carboxypeptidase encoded by pbpX, the orphan response regulator encoded by degU, and the highly abundant noncoding RNA encoded by rli31. Both degU and rli31 mutants had reduced expression of pbpX and pgdA, yet DegU and Rli31 did not regulate each other. Since pbpX and pgdA are also present in lysozyme-sensitive bacteria, this suggested that the acquisition of novel enzymes was not responsible for lysozyme resistance, but rather, the regulation of conserved enzymes by DegU and Rli31 conferred high lysozyme resistance. Each lysozyme-sensitive mutant exhibited attenuated virulence in mice, and a time course of infection revealed that the most lysozyme-sensitive strain was killed within 30 min of intravenous infection, a phenotype that was recapitulated in purified blood. Collectively, these data indicate that the genes required for lysozyme resistance are highly upregulated determinants of L. monocytogenes pathogenesis that are required for avoiding the enzymatic activity of lysozyme in the blood.
INTRODUCTION
Lysozyme is a ubiquitous bactericidal enzyme found in the blood, bodily secretions, and phagocytic cells of all animals (1–3). Lysozyme degrades the bacterial cell wall by hydrolyzing the β1-4 linkage between the N-acetylglucosamine (NAG) and N-acetylmuramic acid (NAM) residues that comprise the peptidoglycan backbone, often resulting in bacteriolysis (4). Not surprisingly, many pathogens have evolved mechanisms of lysozyme resistance (5, 6). The best-characterized mechanisms of lysozyme resistance involve the acetylation state of the peptidoglycan, catalyzed by the deacetylase PgdA and/or the acetyltransferase OatA. PgdA deacetylates the amino group of NAG, while OatA O-acetylates NAM, converting the sugar backbone into a poor lysozyme substrate (7, 8). pgdA mutants of Streptococcus pneumoniae and oatA mutants of Staphylococcus aureus are lysozyme sensitive, and in each case, lysozyme-sensitive mutants are attenuated in animal models of infection (7–10).
Listeria monocytogenes is a facultative intracellular pathogen of animals and humans that causes severe disease in pregnant women and immunocompromised individuals (11). Both PgdA and OatA contribute to lysozyme resistance in L. monocytogenes, and pgdA mutants are attenuated during oral and intravenous (i.v.) infections of mice (9, 10, 12). Indeed, pgdA oatA double mutants are extremely lysozyme sensitive and more than 1,000-fold less virulent in mice (10). In vitro, pgdA oatA double mutants show increased killing upon phagocytosis by macrophages and undergo bacteriolysis in the macrophage cytosol, which leads to induction of macrophage cell death by pyroptosis (10).
Gram-positive pathogens such as L. monocytogenes, Bacillus anthracis, S. aureus, and S. pneumoniae are extremely lysozyme resistant, while many related but nonpathogenic species are lysozyme sensitive (13, 14). One possible explanation for the evolution of lysozyme resistance is the acquisition of enzymes such as PgdA and OatA (14). However, this is probably not the case, since pgdA and oatA are conserved in many nonpathogenic bacilli and staphylococci (13). Why, then, are L. monocytogenes and other pathogens so lysozyme resistant? In this study, we sought to answer this question by performing a forward genetic screen for lysozyme-sensitive mutants of L. monocytogenes. Rather than identify novel cell wall-modifying enzymes, this screening led to the identification of the transcription factor DegU and the abundant noncoding RNA Rli31, which both contributed to lysozyme resistance and L. monocytogenes pathogenesis.
MATERIALS AND METHODS
Bacterial strains.
All of the strains of L. monocytogenes used in this study were in the 10403S background and were cultured in brain heart infusion medium (BHI). Construction of the Δrli31 mutant was performed by amplifying the regions neighboring rli31 with primer pairs JZ298 and JZ299 and JZ300 and JZ301 and then combining them via splice overlap extension PCR. The product was then introduced into L. monocytogenes by allelic exchange with pKSV7 (15). rli31 complement strains were constructed by amplifying rli31 and the rli31 promoter with TB140 and TB141. This fragment was introduced into L. monocytogenes by using pPL2 or pAM401 as described previously (16, 17). For lists of all of the strains and primers used in this study, see Tables S2 and S3 in the supplemental material, respectively.
Transductions were performed by using U153 phage as previously described (18). Briefly, 107 phage were grown in the donor strain, incubated with 108 recipient bacteria, and selected for on BHI-erythromycin plates. For construction of the pgdA pbpX rli31 triple mutant, the rli31::Tn917 transposon was transduced into ΔpgdA pbpX::Tn and selected for on BHI-chloramphenicol plates.
For disk diffusion assays, strains were grown overnight while shaking at 37°C in liquid BHI and 108 bacteria were plated onto BHI agar. A sterile filter disk containing 1 mg of lysozyme was then added to the plate, and bacteria were allowed to grow overnight at 37°C. Zones of inhibition were then measured.
Ethics statement.
This study was carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. All of the protocols were reviewed and approved by the Animal Care and Use Committee at the University of California, Berkeley (Master Animal Use Protocol R235-0813B).
Mouse strains and in vivo infections.
All of the in vivo infections in this study were performed with Crl:CD1(ICR) (CD-1) or C57BL/6J (B6) mice from Charles River and The Jackson Laboratory, respectively. Mice were infected i.v. with 105 logarithmically growing bacteria, and the organs indicated were harvested at 48 h or 30 min postinfection. Organs were homogenized with 0.1% NP-40, and the dilutions indicated were plated onto LB agar. For competitive-index experiments, 105 total bacteria at a 1:1 ratio of each mutant was used for infection and organ homogenates were plated onto both LB and LB containing erythromycin to determine the ratios of the two strains.
For assays in blood, strains were grown to mid-exponential phase in BHI, washed with phosphate-buffered saline (PBS), and diluted 1:100 into defibrinated sheep or horse blood (Hemostat). For assays using bentonite-treated blood, 5 mg of bentonite (Sigma) was added to 1 ml of blood and incubated at 4°C for 30 min. The bentonite was then removed from the blood by two centrifugation steps at 5,000 and then 12,000 rpm.
Northern blot assays and qPCR.
RNA was purified from 20 ml of logarithmically grown bacteria by phenol-chloroform extraction and ethanol precipitation. A 20-μg sample was then loaded onto a 6% urea-polyacrylamide gel and separated by electrophoresis. Gels were stained with a 1:10,000 dilution of SYBR gold (Invitrogen) and imaged by Typhoon (GE). For Northern analysis, nucleotides were transferred to a nylon membrane, probed with 32P-labeled oligonucleotides, and imaged by Typhoon. For quantitative PCR (qPCR) analysis, 4.4 μg of RNA was DNase treated and reverse transcribed with iScript (Bio-Rad). cDNA levels were measured by using SYBR FAST (Kapa Biosystems) and primers (see Table S2 in the supplemental material) specific for the target gene.
Sequencing of transposon insertions.
Flanking regions of transposon insertions were amplified by two rounds of PCR with hot-start polymerase (TaKaRa). For each colony, a small amount of bacteria was resuspended in 100 μl water and 1 μl was used for the first PCR. This reaction amplified the regions flanking the Himar1 transposon with the TN1 primer, which was specific to the transposon, and the Arb1 primer, which contained random nucleotides (see Table S2 in the supplemental material). The second reaction used 1.5 μl of the previous PCR and primers TN2 and Arb2, which were specific to the previously amplified PCR product. The product of this reaction was then treated with 5 μl of ExoSAP-IT (Affymetrix) and sequenced with the primer TNSEQ.
Whole-genome sequencing.
Genomic DNA was isolated from stationary-phase cultures of L. monocytogenes grown in BHI broth by using the MasterPure DNA purification kit (Epicentre). DNA was then fragmented by using Covaris S22 (Covaris Inc.). Libraries were constructed by using Apollo 324 (IntegenX Inc.), PCR amplified, and multiplexed at the QB3 Functional Genomics Laboratory at the University of California, Berkeley (http://qb3.berkeley.edu/qb3/fgl/). The resulting libraries were sequenced by using single-end reads with a Hiseq 2000 Illumina platform. Sequence data were aligned with the L. monocytogenes 10403S reference genome (GenBank accession no. CP002002.1) by using Bowtie 2 (19), and single nucleotide polymorphisms (SNPs) were identified by using SAMtools (20). Approximately 93% of the reads aligned with the reference, resulting in an average of greater than 50-fold coverage of the genome.
Cell wall purification and muropeptide analysis.
Peptidoglycan was purified from mid-exponential-phase cultures as described previously (21). For extracted cell walls, incubation in 48% hydrofluoric acid was omitted to retain the wall teichoic acids. One milligram of extracted cell walls or peptidoglycan was digested with 100 μg mutanolysin from Streptomyces globulosporus (Sigma) in 12.5 mM sodium phosphate buffer (pH 5.6) for 16 h at 37°C. The reaction was stopped by boiling for 3 min, and the soluble muropeptide fraction was reduced with 10 mg sodium borohydride for 30 min. The pH was lowered to 2 with orthophosphoric acid. Muropeptides were analyzed by high-performance liquid chromatography (HPLC) as described previously (22) with a Hypersil reversed-phase octyldecyl silane column (4.6 by 250 mm, 5-μm particle size; ThermoHypersil-Keystone) at 52°C. Chromatograms corresponded to previously published data (9); however, muropeptide identity assignments were further confirmed by mass spectrometry (MS). MS analysis was carried out by the Unité de Spectrométrie de Masse Structurale et Protéomique at the Institut Pasteur. Fully automated, chip-based nanoelectrospray MS was performed on a NanoMate robot incorporating ESI 400 Chip technology (Advion BioSciences, Ithaca, NY) controlled and manipulated by ChipSoft 8.1.0 software operating under the Windows system while coupled to an Orbitrap Velos mass spectrometer equipped with an electron transfer dissociation module (Thermo Fisher Scientific, Bremen, Germany) by using the off-line nanospray source in the positive-ion mode. A full set of automated positive-ion calibrations was performed immediately prior to mass measurement. The transfer capillary temperature was lowered to 100°C, the sheath and auxiliary gases were switched off, and the source transfer parameters were optimized by using the auto-tune feature. The Fourier transform (FT) automatic gain control was set at 1 × 106 for MS. Spectra were acquired by FT MS over 30 s with one MicroScan and a resolution of 30,000 at m/z 400 before being summed by Qualbrowser in Thermo Xcalibur 2.1. Summed spectra were then deconvoluted by Xtract.
RESULTS
Screen to identify lysozyme-sensitive mutants.
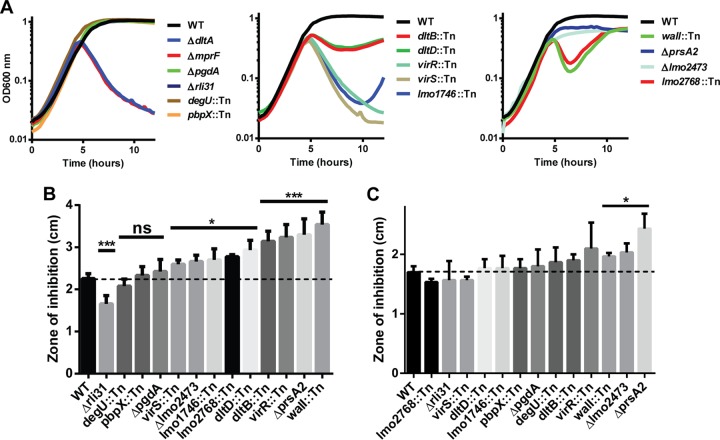
To identify genes required for lysozyme resistance in L. monocytogenes, a library of approximately 40,000 distinct transposon mutants was generated by using a Himar1 transposon delivery vector in wild-type (WT) L. monocytogenes 10403S (23). Bacteria were plated on BHI agar at a concentration of approximately 200 CFU/plate and replica plated onto BHI agar plates containing 1 mg/ml lysozyme. This concentration of lysozyme was chosen because it was sufficient to completely restrict the growth of the ΔpgdA mutant, had no effect on WT bacteria, and was relevant during infection (3, 24). BHI plates were visually compared to BHI-lysozyme plates for the loss of individual colonies (Fig. 1A). Screening was repeated by using a previously described Tn917 library (25), for a total of >50,000 screened colonies. A total of 174 mutants were identified, and the insertion sites were defined by a nested-PCR-based sequencing method (see Materials and Methods). For each gene identified, transposon insertions were transduced into the WT background and rescreened with a lysozyme disk diffusion assay. Transposon insertions mapped to 13 individual genes, and most of the genes were identified by multiple independent transposon insertions (Table 1).
FIG 1.
Screen to identify lysozyme-sensitive mutants in L. monocytogenes. (A) L. monocytogenes transposon mutants were replica plated from BHI (left panels) onto BHI-lysozyme plates (right panels) containing 1 mg/ml chicken egg white lysozyme (Sigma). Arrows indicate colonies defective on BHI-lysozyme plates. (B, C, D) Strains were grown in 2 ml BHI overnight while shaking at 37°C, and 30 μl was spread onto BHI agar. Filter disks containing 1 mg of lysozyme were placed onto the agar and incubated overnight at 37°C, and zones of clearance were measured. Means and standard deviations from at least three separate experiments are presented. ***, P < 0.001. The dotted line indicates the zone of inhibition of WT bacteria by lysozyme.
TABLE 1.
Mutants that cause lysozyme sensitivity in L. monocytogenes
| No. | Locus tag | Gene name | Description | No. of independent insertions | Killinga by: |
|
|---|---|---|---|---|---|---|
| CAMPs | Antibiotics | |||||
| 1 | lmo0415 | pgdA | Peptidoglycan deacetylase | 2 | − | − |
| 2 | lmo0540 | pbpX | Putative carboxypeptidase | 4 | − | − |
| 3 | Intergenic | rli31 | Noncoding RNA | 1 | − | − |
| 4 | lmo2515 | degU | Orphan response regulator | 4 | − | − |
| 5 | lmo0290 | walI | Regulator of WalRK TCSb | 2 | ++ | ++ |
| 6 | lmo0971 | dltD | Modifies teichoic acid with d-Ala | 4 | + | + |
| 7 | lmo0973 | dltB | Modifies teichoic acid with d-Ala | 10 | + | + |
| 8 | lmo1741 | virS | TCS kinase | 3 | ++ | ++ |
| 9 | lmo1745 | virR | TCS response regulator | 6 | ++ | + |
| 10 | lmo1746 | ABC family transporter, vir operon | 9 | ++ | + | |
| 11 | lmo2219 | prsA2 | Extracellular chaperone | 1 | + | ++ |
| 12 | lmo2473 | Unknown | 3 | + | ++ | |
| 13 | lmo2768 | Unknown | 2 | ++ | + | |
−, no killing; +, moderate killing; ++, significant killing of the strains indicated (as observed in the data presented in Fig. 2).
TCS, two-component system.
Mutants were tested for susceptibility to lysozyme by disk diffusion (Fig. 1B and C), confirming that all of the mutants identified were significantly more susceptible to lysozyme than WT L. monocytogenes. The screening results were validated by the identification of mutations of pgdA and prsA2, both of which are required for lysozyme resistance in L. monocytogenes (10, 26). Insertions in oatA were not identified in the screen; however, this was not surprising, as the oatA phenotype is significant only when paired with ΔpgdA (Fig. 1D) (10) and oatA mutants were able to grow on lysozyme plates (data not shown). All of the mutants grew normally in BHI broth, other than the prsA2 and walI mutants, which had slightly delayed growth kinetics (see Fig. S1 in the supplemental material).
The screening for lysozyme-sensitive mutants identified seven gene products previously shown to regulate cell wall and membrane architecture. Of these, the enzyme PrsA2 is a posttranslocation chaperone required for the activity of numerous secreted proteins (26, 27). WalI is a negative regulator of the essential two-component system WalRK, which is required for the expression of autolysins and other cell wall-related enzymes (28–32). Lastly, multiple genes were identified in the vir operon, which regulates the dlt operon and is the only two-component system required for L. monocytogenes virulence (33). The dlt operon is required for d-alanylation of teichoic acid (34), and mutants deficient in dltD show increased autolysis (35).
Apart from pgdA, the contribution of the remaining six genes to lysozyme resistance or to the cell wall was unknown. lmo2473 encodes an uncharacterized protein that has been hypothesized to function in the synthesis of peptidoglycan precursors (36), and lmo2768 encodes an uncharacterized membrane protein with an ABC transporter domain. lmo0540 (here referred to as pbpX) is the homolog of pbpX in B. subtilis, where it contributes to lysozyme resistance (37). pbpX encodes a β-lactamase domain; however, it has been reported that in Mycobacterium smegmatis, PbpX does not contribute to β-lactam antibiotic resistance. Rather, it was proposed to function as a d,d-carboxypeptidase; however, this has never been confirmed biochemically (38). DegU is an orphan response regulator that regulates flagellar and chemotaxis genes in L. monocytogenes and is severely attenuated during the infection of mice (39–43). The phenotype of the degU mutant, however, has remained unexplained since flagellar and chemotaxis genes are not required for virulence (40, 41, 44). Lastly, rli31 encodes a noncoding RNA, and rli31 mutants are attenuated 5-fold in the spleens and livers of infected mice (45, 46).
Because the rli31 mutant was identified from only one unique transposon insertion, we sought to confirm that the rli31 transposon (position 578052 in the L. monocytogenes genome [GenBank accession no. CP002002.1]) disrupted the function of the small noncoding RNA (sRNA) and not neighboring genes. An rli31 deletion mutant was constructed (see Materials and Methods) and was sensitive to lysozyme (Fig. 1C). rli31 was then integrated with its native promoter at a unique locus of the chromosome by using plasmid pPL2 (16), which restored complete lysozyme resistance to the Δrli31 mutant (Fig. 1D). The lysozyme resistance of the Δrli31 mutant was also complemented by the nonintegrative, high-copy-number plasmid pAM401 containing the rli31 gene and the native promoter (Fig. 1D). The Δrli31 mutant had lysozyme susceptibility identical to that of the rli31::Tn mutant, as shown by disk diffusion (Fig. 1D), and these strains behaved identically in all assays.
Nine lysozyme-sensitive mutants are killed by cationic peptides and display increased sensitivity to β-lactam antibiotics.
Seven genes identified in the screen are known to broadly affect cell wall homeostasis, and their phenotypes were likely not specific to lysozyme resistance. To determine if the phenotypes of the remaining mutants were specific to lysozyme, each mutant was treated with a cathelin-related antimicrobial peptide (CRAMP), a cationic antimicrobial peptide (CAMP) (Fig. 2A), and cefuroxime and penicillin G, two β-lactam antibiotics (Fig. 2B and C). dltA and mprF mutants served as positive controls for susceptibility to CAMPs, and both were killed in this assay. DltA is required for d-alanylation of teichoic acid (34), and mprF is required for the transfer of l-lysine onto phospholipids (47), both of which confer a positive charge on the cell surface leading to CAMP resistance. The pgdA mutant served as a control that is killed only by lysozyme and was not killed in either assay (Fig. 2). As predicted, the seven genes known to be involved with cell wall homeostasis were killed by CRAMP and had increased susceptibility to β-lactam antibiotics. It was consistently observed that lmo2473 and prsA2 mutants were killed by CRAMP; however, these mutants were the least susceptible and exhibited only a small amount of additional killing compared to WT bacteria. These two mutants were the most susceptible to penicillin, suggesting that their role in lysozyme susceptibility is due primarily to cell wall and not membrane integrity.
FIG 2.
Treatment of lysozyme-sensitive strains with CRAMP and cell wall-acting antibiotics. (A) A final concentration of 10 μg/ml of purified mouse CRAMP (Anaspec) was added to mid-exponential-phase cells (at 37°C in BHI) of the L. monocytogenes strains indicated, and turbidity was monitored at 10-min intervals. Data are representative of at least three separate experiments and divided into three panels for clarity. (B, C) Thirty-microliter volumes of overnight L. monocytogenes cultures were plated onto BHI, and disks containing 700 ng of cefuroxime (B) or 500 ng of penicillin G (C) were added. The plates were incubated overnight at 37°C, and zones of inhibition were measured. The data shown are the means of at least three separate experiments, and error bars represent the standard deviations of the means. *, P < 0.05; ***, P < 0.001; ns, not statistically significantly different (determined by unpaired two-tailed t test).
The pgdA, rli31, degU, and pbpX mutants were as resistant as WT bacteria to both antibiotics and CAMP treatment. To further determine if these mutants were killed by antimicrobial peptides, the pgdA, degU, pbpX, and rli31 mutants were also treated with high concentrations of a cationic peptide derived from human (RAWVAWRNR) and chicken (NAWVAWRNR) lysozymes (see Fig. S2 in the supplemental material). Identical phenotypes were observed in each mutant and the WT bacteria, while the dltA and mprF mutants were killed by both peptides (see Fig. S2A). Together, these data led to the conclusion that four L. monocytogenes genes were specifically involved with lysozyme resistance. Two encode cell wall-acting enzymes (pgdA and pbpX) and two encode regulators of gene expression (rli31 and degU). Rli31 and DegU were completely uncharacterized with regard to lysozyme sensitivity, and we chose to focus on characterization of the lysozyme susceptibility phenotype of the rli31 mutant.
Rli31 is a constitutively expressed, abundant RNA.
To better characterize the rli31 mutant phenotype, the relative expression of Rli31 was analyzed at various stages of growth in broth by Northern analysis by using a probe specific for Rli31. Rli31 was constitutively expressed during all of the growth phases, from the early exponential phase (optical density at 600 nm [OD600] of 0.3) to mid-exponential (OD600s of 1.3 and 2.3) and stationary (OD600 of 3.9) phases (Fig. 3A). Rli31 migrated at the predicted size of 144 nucleotides (nt) (46) and was not processed into smaller fragments. In addition, Rli31 was observed to be strikingly abundant. Upon the staining of total RNA from L. monocytogenes lysates with a nonspecific nucleotide dye, Rli31 was observable in WT L. monocytogenes lysates alongside other abundant RNAs, such as the 5S rRNA and the 6S sRNA (Fig. 3B). This band was confirmed as Rli31 by Northern analysis with a probe specific for Rli31 (Fig. 3C). The appearance of Rli31 as the only RNA between 100 and 170 nt was surprising, as >40 noncoding RNAs corresponding to this length are predicted to exist in L. monocytogenes (46).
FIG 3.

Rli31 is highly abundant and expressed in all growth phases. (A) Twenty-microgram samples of total RNA collected in the phases of growth in BHI indicated were separated on 6% polyacrylamide. Nucleotides were transferred to a nylon membrane, probed with a 32P-labeled TB13 primer, and imaged by Typhoon. (B) Twenty-microgram samples of total RNA collected in the mid-exponential phase in BHI at 37°C from the WT (lane 1), the Δrli31 mutant (lane 2), and the Δrli31/pAM401:rli31 mutant (lane 3) were separated on 6% polyacrylamide and stained with SYBR gold (Invitrogen). The arrow indicates Rli31, and lane L contains a molecular size ladder. (C) RNA from panel B was transferred to a nylon membrane, probed with a 32P-labeled TB13 primer, and imaged by Typhoon.
Characterization of the rli31 mutant phenotype.
To determine why the rli31 mutant was killed by lysozyme, cell walls from WT L. monocytogenes and the rli31 mutant were purified and the muropeptide compositions of these strains were analyzed by reverse-phase HPLC (Fig. 4A). O-acetylation and covalently attached modifications were also analyzed by omitting the hydrofluoric acid treatment during cell wall purification (Fig. 4B). Muropeptide composition was confirmed by MS, and the abundance of eluted molecules was measured as a percentage of the total absorbance at 206 nm (see Table S1 in the supplemental material). In these analyses, the cell walls of the rli31 mutant contained an abundance of O-acetylated muropeptides similar to that of WT bacteria, while multiple species of N-deacetylated glycans were less abundant (for example, 8.49 to 5.72% GlcNMTriPDAPNH2; see Table S1 in the supplemental material). The analysis also identified a significant increase in muramyl-tripeptides in the rli31 mutant (3.85 to 5.27%) and an increase in 3-4 (d-ala–diaminopimelic acid [DAP]) linkages (3.62 to 5.65%), indicating a difference in peptide cross-linking. The difference in acetylation suggested that pgdA was misregulated in the rli31 mutant. The alterations in cross-linking suggested that pbpX may also contribute to the rli31 mutant phenotype.
FIG 4.
Characterization of the rli31 mutant phenotype. (A, B) HPLC analysis of the muropeptide composition of WT and Δrli31 mutant L. monocytogenes. Deacetylated muropeptides are red, and O-acetylated muropeptides are blue. Muropeptide abbreviations: GlcNAc, NAG; GlcN, glucosamine; M, NAM; TriPDAPNH2, l-alanyl-γ-d-glutamyl-amidated meso-diaminopimelic acid; TetraPDAPGlcNAc, l-alanyl-γ-d-glutamyl-amidated meso-diaminopimelyl-d-alanine; OAc, O-acetylated. (A) Samples were treated with hydrofluoric acid. (B) Samples were not treated with hydrofluoric acid to retain covalent modifications. (C) Multiple rli31 and pgdA mutants were independently passaged with increasing concentrations of lysozyme (50, 100, 200, 500, 1,000 μg/ml) in BHI broth while shaking at 37°C. The resulting strains were grown to mid-exponential phase and treated with 1 mg/ml lysozyme along with the Δrli31 mutant parent strain. Turbidity was monitored at 10-min intervals. sup., suppressor. (D) PgdA and PbpX transcripts of the strains indicated were measured by qPCR, normalized to BglA, and compared to the transcript levels in WT L. monocytogenes. Error bars represent standard deviations of the means. Statistical analyses were performed by using a two-tailed t test assuming a null hypothesis of 1. **, P < 0.01.
A genetic approach was then undertaken in which suppressor mutations were generated in the Δrli31 background that restored lysozyme resistance to this mutant. Individual rli31 colonies were subcultured in broth with increasing concentrations of lysozyme (50, 100, 200, 500, and 1,000 μg/ml) until these mutants were resistant to 1 mg/ml lysozyme (Fig. 4C). Three of these strains were then deep sequenced, and SNP analysis revealed that all three uniquely derived strains had a single thymidine insertion 16 nt upstream of the pgdA transcriptional start site, at position 436736 on the chromosome (Table 2). qPCR analysis of the pgdA transcript levels in these strains revealed 3.1-fold ± 0.39-fold upregulation of pgdA. These data suggested that the rli31 phenotype could be complemented by overexpression of pgdA.
TABLE 2.
Variants identified in rli31 mutant suppressor strainsa
| Δrli31 suppressor strain | Location | Reference | Alteration | Description |
|---|---|---|---|---|
| 1 | 436736 | T | 16 nt upstream of pgdA transcriptional start site | |
| 2 | 436736 | T | 16 nt upstream of pgdA transcriptional start site | |
| 3 | 436736 | T | 16 nt upstream of pgdA transcriptional start site | |
| 2418019 | A | Intergenic region 5′ of lmo2387 |
Individually derived Δrli31 mutants were serially passaged with increasing concentrations of lysozyme until the resulting mutants were resistant to 1 mg/ml lysozyme. Whole-genome sequencing identified variants as described in Materials and Methods.
Together, the biochemical and genetic analyses suggested that the rli31 mutant phenotype was due to misregulation of pgdA and possibly pbpX. To determine if Rli31 regulated pgdA or pbpX, RNA was purified from WT bacteria and the rli31 mutant and real-time qPCRs were performed with primers specific for these two genes. These data showed that pgdA was significantly downregulated (8-fold below the WT) and pbpX was slightly downregulated in the rli31 mutant (3-fold below the WT, Fig. 4D). These qPCR data matched the biochemical and genetic analyses and provided an explanation for the lysozyme sensitivity of the rli31 mutant.
Rli31 functions independently of DegU.
It remained unclear why the degU mutant was lysozyme sensitive and if this phenotype involved Rli31. RNA was purified from the WT and the degU mutant, and qPCRs were performed with primers specific for transcripts of pgdA, pbpX, and rli31. Significant downregulation of pgdA (18-fold below the WT) and pbpX (8-fold below the WT) was observed in the degU mutant (Fig. 4D). However, no difference in Rli31 abundance was observed in the degU mutant and no change was observed in the amount of degU transcript in the rli31 mutant.
To determine if the Rli31 and DegU phenotypes were epistatic, the degU mutation was transduced into the Δrli31 mutant and the resulting double mutant was treated with 10 μg/ml lysozyme (Fig. 5A). The rli31 degU double mutant was killed significantly more than either single mutant by treatment with lysozyme. Because DegU regulates chemotaxis and motility genes, any regulation of DegU by Rli31 would result in a motility phenotype. Upon inoculation into soft agar, neither the rli31 mutant nor the rli31 overexpression construct displayed a motility defect, while the degU mutant was not motile, as expected (see Fig. S3 in the supplemental material). Together, these data suggested that Rli31 and DegU both regulate pgdA and pbpX but by independent mechanisms.
FIG 5.
The rli31 mutant phenotype is due principally to the regulation of pgdA and pbpX. Strains were grown to an OD600 of 0.5, and the indicated concentrations of lysozyme were added. Bacteria were plated for CFU counting at the intervals shown. The data are averages of at least three independent experiments, and error bars specify standard errors. Two-tailed t tests indicate statistically significant difference between the degU::Tn and Δrli31 degU::Tn (A), ΔpgdA and ΔpgdA rli31::Tn (B), ΔpbpX and ΔpbpX rli31::Tn (C), ΔoatA and ΔoatA rli31::Tn (D), ΔpgdA ΔoatA and ΔpgdA ΔoatA rli31::Tn (E), and ΔpgdA pbpX::Tn and ΔpgdA pbpX::Tn rli31::Tn (F) mutant strains. **, P < 0.01; *, P < 0.05; ns, no significant difference.
The rli31 mutant phenotype is due mostly to regulation of pgdA and pbpX.
To determine if the lysozyme sensitivity of the rli31 mutant is due exclusively to the regulation of pgdA and pbpX, the rli31 transposon was transduced into the ΔpgdA mutant and the resulting double mutant strain was treated with a concentration of lysozyme that did not fully kill either single mutant (167 μg/ml). In this assay, the double mutant was killed significantly more than either the Δrli31 or the ΔpgdA mutant (Fig. 5B). A similar phenotype was also observed with the pbpX rli31 double mutant (Fig. 5C), the oatA rli31 double mutant (Fig. 5D), and the oatA pgdA rli31 triple mutant (Fig. 5E). The pgdA pbpX rli31 triple mutant was then compared with the pgdA pbpX double mutant (Fig. 5F). The differences between these strains were smaller than those in the other experiments, where only a 2- to 10-fold difference was observed between these strains at 1 and 2 h and no statistically significant difference existed at 4 h. These data suggested that most of the rli31 mutant phenotype was attributable to the regulation of pgdA and pbpX. However, because the rli31 phenotype was somewhat additive when paired with the pgdA pbpX double mutant at 1 and 2 h, this suggested that the target of Rli31 must have a small additional function distinct from the regulation of pgdA and pbpX, which are required for lysozyme resistance in L. monocytogenes.
The rli31 phenotype was additive with the pgdA and oatA phenotypes in vitro, yet it remained unclear how the rli31 phenotype affected the pgdA and oatA phenotypes during infection. Upon the i.v. infection of CD-1 mice, the oatA rli31 double mutant was significantly more attenuated than the oatA mutant (see Fig. S4A in the supplemental material). When mice were infected with strains harboring pgdA mutations, however, CFU were barely recoverable. The dynamic range was small, and no statistically significant difference between the pgdA and pgdA rli31 mutants was observed when a total of 10 mice were used. Despite this, the difference between the pgdA oatA double mutant and the pgdA oatA rli31 triple mutant was significant (see Fig. S4B). These data provided in vivo evidence that Rli31 functions independently of PgdA and OatA.
Lysozyme-sensitive mutants are attenuated in vivo within 30 min and are killed upon inoculation into blood in vitro.
To evaluate the contribution of the four genes required for resistance to the enzymatic activity of lysozyme, the phenotypes of these mutants were analyzed in vivo. Six- to 8-week-old CD-1 female mice were infected i.v., and the CFU counts in their spleens and livers were determined after 48 h. The pgdA and degU mutants were attenuated 3 to 5 logs, while the rli31 and pbpX mutants were attenuated 5-fold (Fig. 6A).
FIG 6.
Serum kills lysozyme-sensitive L. monocytogenes. (A) CFU counts in organs of CD-1 mice infected i.v. for 48 h. Shown are combined data from two separate experiments with at least eight mice per group. A two-tailed Mann-Whitney t test was used for the statistical comparison of each group with the WT. *, P < 0.05; **, P < 0.01; ***, P < 0.0001. (B) CFU counts in organs of CD-1 mice infected i.v. for 30 min with the strains of bacteria indicated. Shown are combined data from at least two separate experiments with at least eight mice per group. A two-tailed P value is reported for the comparison of each group with the WT. *, P < 0.05; **, P < 0.01; ***, P < 0.0001. (C, D) Strains were grown to mid-exponential phase in BHI, washed with PBS, and diluted 1:100 in defibrinated sheep or horse blood (Hemostat). Bacteria were plated for CFU counting at the times indicated. Data represent means and standard errors from at least three separate experiments. (D) Blood was treated with 5 mg of bentonite (Sigma) for 30 min at 4°C immediately prior to inoculation. A two-tailed P value is reported for the comparison of the pgdA oatA pbpX mutant strain (no bentonite) with the WT. A two-tailed P value is also reported for the comparison of the values obtained with the pgdA oatA pbpX mutant strain with bentonite with those obtained without bentonite. ***, P < 0.0001.
We sought to explain the severe loss of virulence of lysozyme-sensitive L. monocytogenes observed in vivo. The lysozyme concentration varies from organ to organ (48, 49); therefore, the defect observed in vivo may be due to killing in a specific organ. However, the rli31 mutant phenotypes in various organs were similar, ranging from 5- to 20-fold (see Fig. S5A in the supplemental material). We next reasoned that the major bactericidal factor could be macrophages, neutrophils, or serum, since all have been previously implicated in the killing of lysozyme-sensitive bacteria (9, 50, 51). Upon the infection of bone marrow-derived macrophages, only a small defect was observed when the pgdA oatA pbpX triple mutant, the most lysozyme-sensitive strain, was used (see Fig. S5B). Infection of macrophages with other lysozyme-sensitive strains showed even smaller differences between the mutant and WT bacteria (data not shown).
Next, to test if serum could kill lysozyme-sensitive mutants, i.v. infections were performed and mice were sacrificed at 30 min postinfection, a time point when L. monocytogenes has encountered the blood but neutrophils have not yet migrated to areas of infection (52). For these infections, we chose three mutants that differ in lysozyme susceptibility, the rli31 mutant, the pgdA mutant, and the pgdA oatA pbpX triple mutant. Surprisingly, the pgdA oatA pbpX triple mutant lost 97% of its viability after 30 min, the pgdA mutant lost 92.5%, and the rli31 mutant lost a small but significant number of CFU (Fig. 6B). These data suggest that serum is a major factor responsible for the killing of lysozyme-sensitive L. monocytogenes during infection.
To directly test if serum killed lysozyme-sensitive bacteria, the four strains specifically killed by lysozyme were inoculated into purified blood and plated for CFU over time. The pgdA mutant lost >104 CFU and the degU mutant lost >40-fold CFU after 4 h (Fig. 6C). The rli31 mutant was killed 5-fold more than the WT, and the pbpX mutant was similar to the WT. Most strikingly, the pgdA oatA pbpX triple mutant lost 103 CFU within 30 min and CFU were barely recoverable after 2 h (Fig. 6D). The loss of CFU of various lysozyme-sensitive strains observed in blood correlated precisely with their lysozyme susceptibility and the defect observed in vivo. To directly test if lysozyme was the bactericidal factor responsible, the blood was treated with bentonite, which removes lysozyme activity from serum but leaves complement intact (53, 54). Upon bentonite treatment, the pgdA oatA pbpX triple mutant behaved identically to WT bacteria at all of the time points examined (Fig. 6D). Bentonite treatment also fully rescued the killing of the pgdA mutant in blood (data not shown). To ensure that bentonite treatment did not remove a bactericidal factor other than lysozyme, bentonite-treated blood was supplemented with 10 and 25 μg/ml of chicken egg white lysozyme, which are physiological concentrations found in blood (3, 55). Lysozyme addition completely restored the bactericidal activity of blood (see Fig. S6 in the supplemental material).
DISCUSSION
The results of this study show that L. monocytogenes uses three enzymes (PgdA, PbpX, and OatA) and two regulators of gene expression (DegU and Rli31) to resist the bactericidal activity of lysozyme. The two regulators and one of the enzymes, PbpX, were previously not associated with L. monocytogenes lysozyme resistance. The degU and rli31 mutants were extremely susceptible to lysozyme and were attenuated during in vivo infection. Both the degU and rli31 mutants displayed a reduced abundance of pbpX and pgdA mRNAs, and lysozyme-resistant rli31 suppressor mutants upregulated the expression of pgdA. These data suggested that DegU and Rli31 are the major regulators of lysozyme resistance in L. monocytogenes that act by increasing the expression of pgdA and pbpX. All of these factors contributed to L. monocytogenes pathogenesis in mice and were required for survival in blood.
Rli31 is a 144-nt noncoding RNA that contains a Rho-independent transcriptional terminator and is predicted to be transcribed by the essential housekeeping transcription factor SigA (46). A previous study using a different strain of L. monocytogenes showed that rli31 abundance was relatively low during growth in broth but upregulated during infection and that rli31 mutants were attenuated 5-fold in the spleens and livers of infected mice (46). In agreement, we also observed that rli31 mutants were 5-fold attenuated in the spleens and livers of infected mice, but in contrast to the previous study, we found that Rli31 was among the most abundant RNAs in L. monocytogenes. pgdA and pbpX were downregulated in the rli31 mutant, and upregulation of pgdA restored lysozyme resistance to the rli31 mutant. Lastly, the pgdA pbpX double mutant was killed similarly to the pgdA pbpX rli31 triple mutant by lysozyme treatment, suggesting that the rli31 mutant phenotype was due to regulation of the two major lysozyme resistance enzyme-encoding genes, pgdA and pbpX. sRNAs have diverse functions in bacteria, including regulation of translation, targeting of mRNA for degradation, and modulation of protein activity (56). It was therefore possible that Rli31 interacted directly with the pgdA and pbpX transcripts, leading to mRNA degradation or to translational repression. However, both genes contain relatively small 5′ untranslated regions (44 and 26 nt, respectively [57]) and neither gene contains any homology to Rli31 as determined by BLAST searches, by target prediction programs (TargetRNA [58], RNApredator [59]), or by any other method of measuring homology. In the future, we aim to identify the target(s) of Rli31 to explain how Rli31 regulates pgdA and pbpX.
The results of this and three other studies show that L. monocytogenes degU mutants are considerably attenuated in mice (40, 41, 44). DegU is responsible for the expression of chemotaxis and flagellar genes, but chemotaxis genes are not required for L. monocytogenes virulence (40, 41, 44, 60). The data presented here suggest that the attenuation of the degU mutant in vivo is due to lysozyme sensitivity caused by the downregulation of pbpX and pgdA. In B. subtilis, DegU is cotranscribed with its cognate kinase, which is encoded by degS, but L. monocytogenes lacks degS and it remains unclear how DegU is regulated posttranslationally. Further studies are required to determine if DegU regulates pgdA and pbpX directly or indirectly and to determine whether this regulation is affected by phosphorylation of DegU.
B. subtilis also contains pbpX, oatA, and the genes for multiple peptidoglycan deacetylases (13, 37, 61), yet B. subtilis is not a pathogen and is significantly more sensitive to lysozyme than L. monocytogenes. The MIC of lysozyme is 6 μg/ml for WT B. subtilis (13) and 2,000 μg/ml for L. monocytogenes (data not shown). Therefore, there is a >300-fold difference between these two species in lysozyme sensitivity although both encode highly related enzymes. Why, then, is L. monocytogenes so lysozyme resistant? Our data suggest that L. monocytogenes has not acquired novel enzymes to mediate lysozyme resistance but rather utilizes two regulators of gene expression, DegU and Rli31, to upregulate the expression of common cell wall-modifying enzymes. Therefore, it is the upregulation of lysozyme resistance genes by DegU and Rli31 that accounts for the difference in lysozyme resistance between L. monocytogenes and B. subtilis. Microarray analysis of the degU regulon in B. subtilis did not identify pbpX or a deacetylase (62), suggesting that L. monocytogenes has evolved lysozyme resistance by modifying the DegU regulon to include pgdA and pbpX. The conservation of these enzymes across bacterial species suggests that these enzymes are also required in nonpathogens for growth and/or for surviving exposure to low concentrations of antibacterial molecules found in the environment. However, overexpression of these enzymes can convert them into factors essential for pathogenesis.
Pathogenic staphylococci are lysozyme resistant because of the expression of oatA, while nonpathogenic species are often lysozyme sensitive (8, 14). It was previously suggested that these nonpathogens lack oatA (14), but simple BLAST searches indicate that genes with high homology to S. aureus oatA exist in many nonpathogenic, lysozyme-sensitive staphylococci, including S. carnosus (76% identity), S. xylosus (77% identity), and S. equorum (62% identity). On the basis of the results of our study, we hypothesize that oatA is likely expressed at very low levels in these organisms and is probably upregulated in pathogenic staphylococci by uncharacterized regulators. Indeed, inducible expression of S. aureus oatA in lysozyme-sensitive S. carnosus led to increased lysozyme resistance (14).
Lysozyme resistance clearly represents a common mechanism of bacterial pathogenesis, as lysozyme-sensitive mutants are severely attenuated in multiple models of infection. However, it has been difficult to assign a direct role to lysozyme in vivo since mice contain two lysozyme genes, LysM and LysP. LysM LysP double mutant mice do not exist, to our knowledge, and LysM−/− mice still have nearly WT levels of lysozyme in their blood (10, 64). However, bone marrow-derived macrophages from LysM−/− mice lack lysozyme and are therefore appropriate models to examine the roles of lysozyme during infection (10). Lysozyme-sensitive L. monocytogenes bacteria are more susceptible to killing in bone marrow-derived macrophages from B6 mice, leading to hyperinduction of multiple inflammatory pathways, but show no differences in LysM−/− macrophages (10). Therefore, pgdA mutants are clearly more susceptible to killing by lysozyme in macrophages, resulting in increased inflammation. However, these phenotypes do not result in a significant loss of CFU during macrophage infection (10) (see Fig. S5B in the supplemental material). The most striking bactericidal activity observed in vivo occurred during the first 30 min after i.v. infection, where lysozyme-sensitive bacteria lost >90% of their CFU, a phenotype that can be faithfully recapitulated in vitro in blood (Fig. 6). These data suggest that serum is a major host factor responsible for the killing of lysozyme-sensitive bacteria in vivo, at least upon i.v. infection. During oral infection, L. monocytogenes disseminates to the liver in blood via the portal vein (65), and a large number of bacteria in the blood is required to cause meningitis (11, 66). Therefore, exposure to blood is likely to occur during human listeriosis, where L. monocytogenes causes meningitis, abortion in pregnant women, and sepsis (11). The results of this and other studies suggest that the role of lysozyme in vivo is multifactorial (9, 10, 12, 50, 67) but that serum lysozyme is the major factor that protects against lysozyme-sensitive L. monocytogenes during i.v. infection. The robust killing of lysozyme-sensitive bacteria in vivo illustrates why pathogens have evolved multiple and novel regulatory strategies to rewire the expression of essential cell wall processes to become lysozyme resistant.
Supplementary Material
ACKNOWLEDGMENTS
We thank Kathleen Ryan and members of the Portnoy lab for helpful discussions and critical reading of the manuscript.
Daniel A. Portnoy has a consulting relationship with and a financial interest in Aduro Biotech, and both he and the company stand to benefit from the commercialization of the results of this research.
This work was supported by National Institutes of Health grant 1PO1 AI63302 and National Institutes of Health grant 1R01 AI27655 to D.A.P. (http://www.nih.gov). Work done in the lab of I.G.B. was supported by an ERC starting grant (PGNfromSHAPEtoVIR no. 202283).
Footnotes
Published ahead of print 25 August 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.02053-14.
REFERENCES
- 1.Callewaert L, Michiels CW. 2010. Lysozymes in the animal kingdom. J. Biosci. 35:127–160. 10.1007/s12038-010-0015-5. [DOI] [PubMed] [Google Scholar]
- 2.Gordon S, Todd J, Cohn ZA. 1974. In vitro synthesis and secretion of lysozyme by mononuclear phagocytes. J. Exp. Med. 139:1228–1248. 10.1084/jem.139.5.1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hankiewicz J, Swierczek E. 1974. Lysozyme in human body fluids. Clin. Chim. Acta 57:205–209. 10.1016/0009-8981(74)90398-2. [DOI] [PubMed] [Google Scholar]
- 4.Vollmer W, Joris B, Charlier P, Foster S. 2008. Bacterial peptidoglycan (murein) hydrolases. FEMS Microbiol. Rev. 32:259–286. 10.1111/j.1574-6976.2007.00099.x. [DOI] [PubMed] [Google Scholar]
- 5.Davis KM, Weiser JN. 2011. Modifications to the peptidoglycan backbone help bacteria to establish infection. Infect. Immun. 79:562–570. 10.1128/IAI.00651-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vollmer W. 2008. Structural variation in the glycan strands of bacterial peptidoglycan. FEMS Microbiol. Rev. 32:287–306. 10.1111/j.1574-6976.2007.00088.x. [DOI] [PubMed] [Google Scholar]
- 7.Vollmer W, Tomasz A. 2000. The pgdA gene encodes for a peptidoglycan N-acetylglucosamine deacetylase in Streptococcus pneumoniae. J. Biol. Chem. 275:20496–20501. 10.1074/jbc.M910189199. [DOI] [PubMed] [Google Scholar]
- 8.Bera A, Herbert S, Jakob A, Vollmer W, Gotz F. 2005. Why are pathogenic staphylococci so lysozyme-resistant? The peptidoglycan O-acetyltransferase OatA is the major determinant for lysozyme resistance of Staphylococcus aureus. Mol. Microbiol. 55:778–787. 10.1111/j.1365-2958.2004.04446.x. [DOI] [PubMed] [Google Scholar]
- 9.Boneca IG, Dussurget O, Cabanes D, Nahori MA, Sousa S, Lecuit M, Psylinakis E, Bouriotis V, Hugot JP, Giovannini M, Coyle A, Bertin J, Namane A, Rousselle JC, Cayet N, Prevost MC, Balloy V, Chignard M, Philpott DJ, Cossart P, Girardin SE. 2007. A critical role for peptidoglycan N-deacetylation in Listeria evasion from the host innate immune system. Proc. Natl. Acad. Sci. U. S. A. 104:997–1002. 10.1073/pnas.0609672104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rae CS, Geissler A, Adamson PC, Portnoy DA. 2011. Mutations of the Listeria monocytogenes peptidoglycan N-deacetylase and O-acetylase result in enhanced lysozyme sensitivity, bacteriolysis, and hyperinduction of innate immune pathways. Infect. Immun. 79:3596–3606. 10.1128/IAI.00077-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vázquez-Boland JA, Kuhn M, Berche P, Chakraborty T, Dominguez-Bernal G, Goebel W, Gonzalez-Zorn B, Wehland J, Kreft J. 2001. Listeria pathogenesis and molecular virulence determinants. Clin. Microbiol. Rev. 14:584–640. 10.1128/CMR.14.3.584-640.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aubry C, Goulard C, Nahori MA, Cayet N, Decalf J, Sachse M, Boneca IG, Cossart P, Dussurget O. 2011. OatA, a peptidoglycan O-acetyltransferase involved in Listeria monocytogenes immune escape, is critical for virulence. J. Infect. Dis. 204:731–740. 10.1093/infdis/jir396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guariglia-Oropeza V, Helmann JD. 2011. Bacillus subtilis σ(V) confers lysozyme resistance by activation of two cell wall modification pathways, peptidoglycan O-acetylation and d-alanylation of teichoic acids. J. Bacteriol. 193:6223–6232. 10.1128/JB.06023-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bera A, Biswas R, Herbert S, Gotz F. 2006. The presence of peptidoglycan O-acetyltransferase in various staphylococcal species correlates with lysozyme resistance and pathogenicity. Infect. Immun. 74:4598–4604. 10.1128/IAI.00301-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smith K, Youngman P. 1992. Use of a new integrational vector to investigate compartment-specific expression of the Bacillus subtilis spoIIM gene. Biochimie 74:705–711. 10.1016/0300-9084(92)90143-3. [DOI] [PubMed] [Google Scholar]
- 16.Lauer P, Chow MY, Loessner MJ, Portnoy DA, Calendar R. 2002. Construction, characterization, and use of two Listeria monocytogenes site-specific phage integration vectors. J. Bacteriol. 184:4177–4186. 10.1128/JB.184.15.4177-4186.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Camilli A, Tilney LG, Portnoy DA. 1993. Dual roles of plcA in Listeria monocytogenes pathogenesis. Mol. Microbiol. 8:143–157. 10.1111/j.1365-2958.1993.tb01211.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hodgson DA. 2000. Generalized transduction of serotype 1/2 and serotype 4b strains of Listeria monocytogenes. Mol. Microbiol. 35:312–323. 10.1046/j.1365-2958.2000.01643.x. [DOI] [PubMed] [Google Scholar]
- 19.Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9:357–359. 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25:2078–2079. 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Girardin SE, Boneca IG, Viala J, Chamaillard M, Labigne A, Thomas G, Philpott DJ, Sansonetti PJ. 2003. Nod2 is a general sensor of peptidoglycan through muramyl dipeptide (MDP) detection. J. Biol. Chem. 278:8869–8872. 10.1074/jbc.C200651200. [DOI] [PubMed] [Google Scholar]
- 22.Antignac A, Rousselle JC, Namane A, Labigne A, Taha MK, Boneca IG. 2003. Detailed structural analysis of the peptidoglycan of the human pathogen Neisseria meningitidis. J. Biol. Chem. 278:31521–31528. 10.1074/jbc.M304749200. [DOI] [PubMed] [Google Scholar]
- 23.Zemansky J, Kline BC, Woodward JJ, Leber JH, Marquis H, Portnoy DA. 2009. Development of a mariner-based transposon and identification of Listeria monocytogenes determinants, including the peptidyl-prolyl isomerase PrsA2, that contribute to its hemolytic phenotype. J. Bacteriol. 191:3950–3964. 10.1128/JB.00016-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aine E, Morsky P. 1984. Lysozyme concentration in tears—assessment of reference values in normal subjects. Acta Ophthalmol. (Copenh.) 62:932–938. [DOI] [PubMed] [Google Scholar]
- 25.Camilli A, Portnoy A, Youngman P. 1990. Insertional mutagenesis of Listeria monocytogenes with a novel Tn917 derivative that allows direct cloning of DNA flanking transposon insertions. J. Bacteriol. 172:3738–3744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Forster BM, Zemansky J, Portnoy DA, Marquis H. 2011. Posttranslocation chaperone PrsA2 regulates the maturation and secretion of Listeria monocytogenes proprotein virulence factors. J. Bacteriol. 193:5961–5970. 10.1128/JB.05307-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alonzo F, III, Xayarath B, Whisstock JC, Freitag NE. 2011. Functional analysis of the Listeria monocytogenes secretion chaperone PrsA2 and its multiple contributions to bacterial virulence. Mol. Microbiol. 80:1530–1548. 10.1111/j.1365-2958.2011.07665.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dubrac S, Bisicchia P, Devine KM, Msadek T. 2008. A matter of life and death: cell wall homeostasis and the WalKR (YycGF) essential signal transduction pathway. Mol. Microbiol. 70:1307–1322. 10.1111/j.1365-2958.2008.06483.x. [DOI] [PubMed] [Google Scholar]
- 29.Dubrac S, Msadek T. 2008. Tearing down the wall: peptidoglycan metabolism and the WalK/WalR (YycG/YycF) essential two-component system. Adv. Exp. Med. Biol. 631:214–228. 10.1007/978-0-387-78885-2_15. [DOI] [PubMed] [Google Scholar]
- 30.Bisicchia P, Noone D, Lioliou E, Howell A, Quigley S, Jensen T, Jarmer H, Devine KM. 2007. The essential YycFG two-component system controls cell wall metabolism in Bacillus subtilis. Mol. Microbiol. 65:180–200. 10.1111/j.1365-2958.2007.05782.x. [DOI] [PubMed] [Google Scholar]
- 31.Howell A, Dubrac S, Andersen KK, Noone D, Fert J, Msadek T, Devine K. 2003. Genes controlled by the essential YycG/YycF two-component system of Bacillus subtilis revealed through a novel hybrid regulator approach. Mol. Microbiol. 49:1639–1655. 10.1046/j.1365-2958.2003.03661.x. [DOI] [PubMed] [Google Scholar]
- 32.Ng WL, Kazmierczak KM, Winkler ME. 2004. Defective cell wall synthesis in Streptococcus pneumoniae R6 depleted for the essential PcsB putative murein hydrolase or the VicR (YycF) response regulator. Mol. Microbiol. 53:1161–1175. 10.1111/j.1365-2958.2004.04196.x. [DOI] [PubMed] [Google Scholar]
- 33.Mandin P, Fsihi H, Dussurget O, Vergassola M, Milohanic E, Toledo-Arana A, Lasa I, Johansson J, Cossart P. 2005. VirR, a response regulator critical for Listeria monocytogenes virulence. Mol. Microbiol. 57:1367–1380. 10.1111/j.1365-2958.2005.04776.x. [DOI] [PubMed] [Google Scholar]
- 34.Abachin E, Poyart C, Pellegrini E, Milohanic E, Fiedler F, Berche P, Trieu-Cuot P. 2002. Formation of d-alanyl-lipoteichoic acid is required for adhesion and virulence of Listeria monocytogenes. Mol. Microbiol. 43:1–14. 10.1046/j.1365-2958.2002.02723.x. [DOI] [PubMed] [Google Scholar]
- 35.Steen A, Palumbo E, Deghorain M, Cocconcelli PS, Delcour J, Kuipers OP, Kok J, Buist G, Hols P. 2005. Autolysis of Lactococcus lactis is increased upon d-alanine depletion of peptidoglycan and lipoteichoic acids. J. Bacteriol. 187:114–124. 10.1128/JB.187.1.114-124.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sauer JD, Witte CE, Zemansky J, Hanson B, Lauer P, Portnoy DA. 2010. Listeria monocytogenes triggers AIM2-mediated pyroptosis upon infrequent bacteriolysis in the macrophage cytosol. Cell Host Microbe 7:412–419. 10.1016/j.chom.2010.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ho TD, Hastie JL, Intile PJ, Ellermeier CD. 2011. The Bacillus subtilis extracytoplasmic function σ factor σ(V) is induced by lysozyme and provides resistance to lysozyme. J. Bacteriol. 193:6215–6222. 10.1128/JB.05467-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Flores AR, Parsons LM, Pavelka MS., Jr 2005. Characterization of novel Mycobacterium tuberculosis and Mycobacterium smegmatis mutants hypersusceptible to beta-lactam antibiotics. J. Bacteriol. 187:1892–1900. 10.1128/JB.187.6.1892-1900.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gueriri I, Bay S, Dubrac S, Cyncynatus C, Msadek T. 2008. The Pta-AckA pathway controlling acetyl phosphate levels and the phosphorylation state of the DegU orphan response regulator both play a role in regulating Listeria monocytogenes motility and chemotaxis. Mol. Microbiol. 70:1342–1357. 10.1111/j.1365-2958.2008.06496.x. [DOI] [PubMed] [Google Scholar]
- 40.Gueriri I, Cyncynatus C, Dubrac S, Arana AT, Dussurget O, Msadek T. 2008. The DegU orphan response regulator of Listeria monocytogenes autorepresses its own synthesis and is required for bacterial motility, virulence and biofilm formation. Microbiology 154:2251–2264. 10.1099/mic.0.2008/017590-0. [DOI] [PubMed] [Google Scholar]
- 41.Knudsen GM, Olsen JE, Dons L. 2004. Characterization of DegU, a response regulator in Listeria monocytogenes, [sic] involved in regulation of motility and contributes to virulence. FEMS Microbiol. Lett. 240:171–179. 10.1016/j.femsle.2004.09.039. [DOI] [PubMed] [Google Scholar]
- 42.Mauder N, Williams T, Fritsch F, Kuhn M, Beier D. 2008. Response regulator DegU of Listeria monocytogenes controls temperature-responsive flagellar gene expression in its unphosphorylated state. J. Bacteriol. 190:4777–4781. 10.1128/JB.00258-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Williams T, Joseph B, Beier D, Goebel W, Kuhn M. 2005. Response regulator DegU of Listeria monocytogenes regulates the expression of flagella-specific genes. FEMS Microbiol. Lett. 252:287–298. 10.1016/j.femsle.2005.09.011. [DOI] [PubMed] [Google Scholar]
- 44.Williams T, Bauer S, Beier D, Kuhn M. 2005. Construction and characterization of Listeria monocytogenes mutants with in-frame deletions in the response regulator genes identified in the genome sequence. Infect. Immun. 73:3152–3159. 10.1128/IAI.73.5.3152-3159.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Toledo-Arana A, Dussurget O, Nikitas G, Sesto N, Guet-Revillet H, Balestrino D, Loh E, Gripenland J, Tiensuu T, Vaitkevicius K, Barthelemy M, Vergassola M, Nahori MA, Soubigou G, Regnault B, Coppee JY, Lecuit M, Johansson J, Cossart P. 2009. The Listeria transcriptional landscape from saprophytism to virulence. Nature 459:950–956. 10.1038/nature08080. [DOI] [PubMed] [Google Scholar]
- 46.Mraheil MA, Billion A, Mohamed W, Mukherjee K, Kuenne C, Pischimarov J, Krawitz C, Retey J, Hartsch T, Chakraborty T, Hain T. 2011. The intracellular sRNA transcriptome of Listeria monocytogenes during growth in macrophages. Nucleic Acids Res. 39:4235–4248. 10.1093/nar/gkr033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thedieck K, Hain T, Mohamed W, Tindall BJ, Nimtz M, Chakraborty T, Wehland J, Jansch L. 2006. The MprF protein is required for lysinylation of phospholipids in listerial membranes and confers resistance to cationic antimicrobial peptides (CAMPs) on Listeria monocytogenes. Mol. Microbiol. 62:1325–1339. 10.1111/j.1365-2958.2006.05452.x. [DOI] [PubMed] [Google Scholar]
- 48.Cross M, Mangelsdorf I, Wedel A, Renkawitz R. 1988. Mouse lysozyme M gene: isolation, characterization, and expression studies. Proc. Natl. Acad. Sci. U. S. A. 85:6232–6236. 10.1073/pnas.85.17.6232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wehkamp J, Chu H, Shen B, Feathers RW, Kays RJ, Lee SK, Bevins CL. 2006. Paneth cell antimicrobial peptides: topographical distribution and quantification in human gastrointestinal tissues. FEBS Lett. 580:5344–5350. 10.1016/j.febslet.2006.08.083. [DOI] [PubMed] [Google Scholar]
- 50.Milani CJ, Aziz RK, Locke JB, Dahesh S, Nizet V, Buchanan JT. 2010. The novel polysaccharide deacetylase homologue Pdi contributes to virulence of the aquatic pathogen Streptococcus iniae. Microbiology 156:543–554. 10.1099/mic.0.028365-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fittipaldi N, Sekizaki T, Takamatsu D, de la Cruz Dominguez-Punaro M, Harel J, Bui NK, Vollmer W, Gottschalk M. 2008. Significant contribution of the pgdA gene to the virulence of Streptococcus suis. Mol. Microbiol. 70:1120–1135. 10.1111/j.1365-2958.2008.06463.x. [DOI] [PubMed] [Google Scholar]
- 52.Conlan JW, North RJ. 1991. Neutrophil-mediated dissolution of infected host cells as a defense strategy against a facultative intracellular bacterium. J. Exp. Med. 174:741–744. 10.1084/jem.174.3.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Glynn AA, Milne CM. 1967. A kinetic study of the bacteriolytic and bactericidal action of human serum. Immunology 12:639–653. [PMC free article] [PubMed] [Google Scholar]
- 54.Feingold DS, Goldman JN, Kuritz HM. 1968. Locus of the action of serum and the role of lysozyme in the serum bactericidal reaction. J. Bacteriol. 96:2118–2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Prixova J. 1975. Serum lysozyme in mice subjected to combined immunosuppression with 6-mercaptopurine and hydrocortisone. Folia Microbiol. (Praha) 20:509–512. 10.1007/BF02891711. [DOI] [PubMed] [Google Scholar]
- 56.Storz G, Vogel J, Wassarman KM. 2011. Regulation by small RNAs in bacteria: expanding frontiers. Mol. Cell 43:880–891. 10.1016/j.molcel.2011.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wurtzel O, Sesto N, Mellin JR, Karunker I, Edelheit S, Becavin C, Archambaud C, Cossart P, Sorek R. 2012. Comparative transcriptomics of pathogenic and non-pathogenic Listeria species. Mol. Syst. Biol. 8:583. 10.1038/msb.2012.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tjaden B. 2012. Computational identification of sRNA targets. Methods Mol. Biol. 905:227–234. 10.1007/978-1-61779-949-5_14. [DOI] [PubMed] [Google Scholar]
- 59.Eggenhofer F, Tafer H, Stadler PF, Hofacker IL. 2011. RNApredator: fast accessibility-based prediction of sRNA targets. Nucleic Acids Res. 39(Web Server Issue):W149–W154. 10.1093/nar/gkr467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Murray EJ, Kiley TB, Stanley-Wall NR. 2009. A pivotal role for the response regulator DegU in controlling multicellular behaviour. Microbiology 155:1–8. 10.1099/mic.0.023903-0. [DOI] [PubMed] [Google Scholar]
- 61.Kobayashi K, Sudiarta IP, Kodama T, Fukushima T, Ara K, Ozaki K, Sekiguchi J. 2012. Identification and characterization of a novel polysaccharide deacetylase C (PdaC) from Bacillus subtilis. J. Biol. Chem. 287:9765–9776. 10.1074/jbc.M111.329490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ogura M, Yamaguchi H, Yoshida K, Fujita Y, Tanaka T. 2001. DNA microarray analysis of Bacillus subtilis DegU, ComA and PhoP regulons: an approach to comprehensive analysis of B.subtilis two-component regulatory systems. Nucleic Acids Res. 29:3804–3813. 10.1093/nar/29.18.3804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Reference deleted.
- 64.Markart P, Faust N, Graf T, Na CL, Weaver TE, Akinbi HT. 2004. Comparison of the microbicidal and muramidase activities of mouse lysozyme M and P. Biochem. J. 380:385–392. 10.1042/BJ20031810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Melton-Witt JA, Rafelski SM, Portnoy DA, Bakardjiev AI. 2012. Oral infection with signature-tagged Listeria monocytogenes reveals organ-specific growth and dissemination routes in guinea pigs. Infect. Immun. 80:720–732. 10.1128/IAI.05958-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Berche P. 1995. Bacteremia is required for invasion of the murine central nervous system by Listeria monocytogenes. Microb. Pathog. 18:323–336. 10.1006/mpat.1995.0029. [DOI] [PubMed] [Google Scholar]
- 67.Kaoukab-Raji A, Biskri L, Bernardini ML, Allaoui A. 2012. Characterization of SfPgdA, a Shigella flexneri peptidoglycan deacetylase required for bacterial persistence within polymorphonuclear neutrophils. Microbes Infect. 14:619–627. 10.1016/j.micinf.2012.01.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.