Abstract
For bacteria, many studies have focused on the role of respiratory enzymes in energy conservation; however, their effect on cell behavior is poorly understood. Pseudomonas aeruginosa can perform both aerobic respiration and denitrification. Previous studies demonstrated that cbb3-type cytochrome c oxidases that support aerobic respiration are more highly expressed in P. aeruginosa under anoxic conditions than are other aerobic respiratory enzymes. However, little is known about their role under such conditions. In this study, it was shown that cbb3 oxidases of P. aeruginosa PAO1 alter anaerobic growth, the denitrification process, and cell morphology under anoxic conditions. Furthermore, biofilm formation was promoted by the cbb3 oxidases under anoxic conditions. cbb3 oxidases led to the accumulation of nitric oxide (NO), which is produced during denitrification. Cell elongation induced by NO accumulation was reported to be required for robust biofilm formation of P. aeruginosa PAO1 under anoxic conditions. Our data show that cbb3 oxidases promote cell elongation by inducing NO accumulation during the denitrification process, which further leads to robust biofilms. Our findings show that cbb3 oxidases, which have been well studied as aerobic respiratory enzymes, are also involved in denitrification and influence the lifestyle of P. aeruginosa PAO1 under anoxic conditions.
INTRODUCTION
Respiration is a fundamental process for energy conservation. In eukaryotic mitochondria, aa3-type cytochrome c oxidase (terminal oxidase) catalyzes the terminal reaction of electron transport in aerobic respiration through oxygen reduction. Furthermore, the generation of reactive oxygen species (ROS) and the oxidation of cytochrome c by aa3 oxidase can induce apoptosis; thus, aa3 oxidase also plays a role in cell fate determination (1, 2). In bacteria, multiple terminal oxidases have been reported to catalyze energy conservation; however, few studies have reported their influence on cell behavior.
Pseudomonas aeruginosa is a ubiquitous bacterium with versatile aerobic and anaerobic respiratory systems (3). Under oxic environments, five terminal oxidases (Cyo, CIO, aa3, cbb3-1, and cbb3-2) reduce oxygen as terminal electron acceptors. The terminal oxidases Cyo and CIO are quinol oxidases, while the terminal oxidases aa3 and cbb3 are cytochrome c oxidases. Compared with other terminal oxidases, cbb3 oxidases possess a high affinity for oxygen. cbb3-1 and cbb3-2 oxidases are encoded by the tetracistronic ccoNOQP-1 and ccoNOQP-2 operons (4). The ccoNOQP-1 operon is constitutively transcribed under various oxygen concentrations (5). On the other hand, the ccoNOQP-2 operon is highly transcribed under low oxygen concentrations because the oxygen-responsive transcriptional regulator ANR upregulates its promoter activity. These enzymatic and transcriptional characteristics of cbb3 oxidases enable P. aeruginosa to grow in low-oxygen environments. In addition, under anoxic conditions, where oxygen is depleted, P. aeruginosa can use N-oxides as alternative electron acceptors in a process known as denitrification. During this process, nitrate (NO3−) is reduced subsequently to nitrite (NO2−), nitric oxide (NO), nitrous oxide (N2O), and, finally, dinitrogen (N2). Each reduction step is performed by NO3− reductase (NAR), NO2− reductase (NIR), NO reductase (NOR), and N2O reductase (NOS). The denitrification process is triggered by physicochemical factors, such as oxygen and N-oxides, via ANR and other transcriptional regulators, such as NarXL and DNR (6, 7). Moreover, quorum sensing (QS) molecules, such as N-acyl-l-homoserine lactone (AHL) and Pseudomonas quinolone signal (PQS), fine-tune the metabolic activity to control the NO concentration (8, 9).
Interestingly, Kawakami et al. reported that cbb3 oxidases of P. aeruginosa are more highly expressed under anoxic denitrifying conditions than are other terminal oxidases, suggesting that cbb3 oxidases may have a unique role under anoxic conditions (5). However, their role has not yet been elucidated. In this study, we showed that cbb3 oxidases of P. aeruginosa PAO1 lead to NO accumulation during the denitrification process under anoxic conditions. This effect resulted in the promotion of biofilm formation via cell elongation. Such a morphological change and complex formation are helpful to survival under many environmental stresses (10, 11). Thus, it is expected that cbb3 oxidases are key enzymes involved in regulating these cell behaviors in response to anoxic environments. Our findings show a new role of cbb3 oxidases, emphasizing their importance in bacterial physiology.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains used in this study are listed in Table 1. For routine cultures, bacterial strains were grown at 37°C in lysogeny broth (LB) medium (Lennox; Nacalai, Kyoto, Japan) or on LB agar plates. When necessary, gentamicin was added at the concentration of 10 μg ml−1 for Escherichia coli and 100 μg ml−1 for P. aeruginosa. Anoxic planktonic cultures were prepared using a method described previously (9). P. aeruginosa was grown oxically in 24-ml test tubes containing 4 ml of LB medium for 10 h and inoculated into butyl-rubber-sealed 17-ml Hungate tubes containing 5 ml of LB medium supplemented with 100 mM KNO3 (LBN medium) at a starting optical density at 660 nm (OD660) of 0.01. The air of the Hungate tubes was replaced with argon by flushing gas through a needle. Anoxic planktonic incubation was performed at 37°C with shaking at 200 rpm. By using a negative-control denitrification mutant (PS1700) (12), it was confirmed that this experimental condition is anoxic (see Fig. S1 in the supplemental material). Hypoxic incubation was carried out by using butyl-rubber-sealed 17-ml Hungate tubes containing 5 ml of LB medium without replacing the air with argon.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristicsa | Source or reference |
|---|---|---|
| Strains | ||
| E. coli | ||
| JM109 | E. coli strain for transformation | TaKaRa |
| DH5α | E. coli strain for transformation | TaKaRa |
| S17-1 | Mobilizer strain | 14 |
| P. aeruginosa | ||
| PAO1 | Wild type | 36 |
| ΔccoN1 mutant | PAO1 with a markerless deletion of ccoN1 gene | This study |
| ΔccoN2 mutant | PAO1 with a markerless deletion of ccoN2 gene | This study |
| ΔccoN1 ΔccoN2 mutant | PAO1 with markerless deletions of ccoN1 and ccoN2 genes | This study |
| Δnir mutant | PAO1 with markerless deletions of nirS to nirN genes | 15 |
| ΔccoN1 ΔccoN2 Δnir mutant | PAO1 with markerless deletions of ccoN1, ccoN2, and nirS to nirN genes | This study |
| PS1700 | narG; 3,835 bp deleted from PA3874 to PA3876 | 12 |
| Plasmids | ||
| pHSG398 | Cloning vector; Cpr | TaKaRa |
| pG19II | pK19mobsac derived suicide vector; sacB Gmr | 13 |
| pG19ccoN1 | ccoN1 deletion cassette in pG19II | This study |
| pG19ccoN2 | ccoN2 deletion cassette in pG19II | This study |
| pG19nir | nirS-to-nirN deletion cassette in pG19II | 15 |
| pMEX9 | pME4510 derived promoter-probe vector; xylE Gmr | 8 |
| pMEXccoN1 | ccoN1 promoter region in pMEX9 | This study |
| pMEXccoN2 | ccoN2 promoter region in pMEX9 | This study |
Cpr, chloramphenicol resistant; Gmr, gentamicin resistant.
Construction of P. aeruginosa mutants.
The plasmids used in this study are listed in Table 1. The PCR primers used in this study are listed in Table S1 in the supplemental material. pG19ccoN1 and pG19ccoN2 plasmids carrying deletion cassettes of ccoN1 and ccoN2 were constructed based on a previously described method (13). The pG19II-derived plasmids (pG19ccoN1 and pG19ccoN2) were transferred into P. aeruginosa PAO1 wild-type (WT) and ΔccoN1 strains by conjugation with the mobilizer E. coli S17-1 (14), followed by homologous recombination as previously described (13). Target gene deletions were confirmed by PCR analysis. In addition, to construct the ΔccoN1 ΔccoN2 Δnir mutant, which is devoid of NIR and does not produce NO, pG19nir (15) was transferred into the ΔccoN1 ΔccoN2 strain by conjugation with E. coli S17-1, followed by homologous recombination.
C23O specific activity assay.
The ccoN1 and ccoN2 transcriptional fusion plasmids were constructed by cloning the promoter regions of ccoN1 and ccoN2 (4, 5) into the pMEX9 reporter plasmid upstream of xylE. These plasmids were introduced into the P. aeruginosa PAO1 WT (16). The specific activities of catechol 2,3-dioxygenase (C23O) (the xylE gene product) in the strains carrying reporter plasmids were determined by monitoring the absorbance at 375 nm (A375) by following a method described previously (13).
DIC and bright-field observation.
For differential interference contrast (DIC) observation of cells in planktonic shaking cultures, 5-μl quantities of the cultures were mounted onto micro-cover glasses (VWR International, USA) by following a method described previously (17). For the bright-field observation of cells in biofilms, biofilms formed in 6-well plates were washed with phosphate-buffered saline (PBS). After scraping of the biofilms with PBS, 10-μl quantities of the suspensions were mounted onto micro-cover glasses. A Carl Zeiss laser scanning microscope (LSM 710) equipped with a 63×/1.40 numerical aperture Plan-Apochromat objective (Carl Zeiss, Oberkochen, Germany) was used to acquire DIC and bright-field images. The images were captured using an AxioCam MRm digital camera and analyzed with the AxioVision software (version 4.8; Carl Zeiss).
Denitrification activity assay.
Denitrification activities were measured by analytical methods described previously (9, 12). NO3− concentrations were determined by the brucine-sulfanilic acid method (18). NO2− concentrations were determined by the sulfanilamide-naphthylethylene diamine method (18). N2O and N2 concentrations were measured with a gas chromatograph (model GC-8AIT; Shimadzu).
DPA assay.
Quantification of the cellular levels of deoxynucleoside triphosphates (dNTPs) in cell extracts was performed based on a previously described method (19). Anoxic incubation of P. aeruginosa was performed using butyl-rubber-sealed 500-ml Erlenmeyer flasks containing 80 ml of LBN medium at 37°C with shaking at 200 rpm. Anoxically grown cells were washed in 100 mM potassium phosphate buffer (pH 7.5) and sonicated. Lysed cell extracts containing equal amounts of protein were mixed with diphenylamine (DPA) reagent, consisting of 2 g of DPA (Wako, Osaka, Japan) dissolved in 100 ml of pure acetic acid and 2.75 ml of concentrated sulfuric acid. After incubation at 37°C for 4 h, the A595 was measured.
NO detection.
Cellular NO levels were measured based on a previously described method (20) with a NO detection reagent, diaminofluorescein-2 diacetate (DAF-2 DA) (Sekisui Medical, Tokyo, Japan). One-milliliter quantities of planktonic shaking cultures grown under anoxic conditions were incubated with 10 μM DAF-2 DA. After incubation at 37°C for 1 h and washing with PBS, the green fluorescence of a reaction product, DAF-2 T, was measured using a Varioskan flash fluorometer (Thermo Scientific, Waltham, MA) at an excitation wavelength of 495 nm and an emission wavelength of 515 nm. The fluorescent intensity was normalized to OD660.
Biofilm formation assay.
Overnight cultures of P. aeruginosa grown under oxic conditions were diluted to an OD660 of 0.1 in LBN medium. Aliquots (100 μl) of the cultures were inoculated into 900 μl of LBN medium in 24-well polystyrene plates to achieve an OD660 of 0.01. The plates were incubated at 37°C under anoxic conditions. For anoxic incubation, an AnaeroPack system (Mitsubishi Gas Chemical, Tokyo, Japan) was used (17). After incubation, quantification of biofilms was performed with the crystal violet staining method, as previously described (17, 21).
Biofilm visualization with COCRM.
For biofilm visualization, 6-well polystyrene plates were used. Overnight cultures of P. aeruginosa grown under oxic conditions were diluted to an OD660 of 0.5 in LBN medium. Aliquots (0.1 ml) of the cultures were inoculated into 4.9 ml of LBN medium in 6-well plates to achieve an OD660 of 0.01. The plates were anoxically incubated at 37°C with an AnaeroPack system. After incubation for 24 h, biofilms that had formed at the bottom of the 6-well plates were washed with PBS, and the structures were visualized with continuous-optimizing confocal reflection microscopy (COCRM) (22, 23). A Carl Zeiss LSM 5 PASCAL equipped with a 63×/0.9 numerical aperture W N-Achroplan water immersion objective was used to acquire COCRM images. Biofilms were illuminated with a 514-nm argon laser, and the reflected light was collected. COCRM images were analyzed with the LSM 5 PASCAL software (version 3.5; Carl Zeiss).
LIVE/DEAD bacterial viability assay.
The bacterial viability in planktonic shaking cultures was determined by using a Baclight LIVE/DEAD bacterial viability staining kit (Molecular Probes, Eugene, OR), by following a previously described method (24). A Carl Zeiss LSM 5 PASCAL equipped with a 63×/1.4 numerical aperture Plan-Apochromat objective was used to acquire LIVE/DEAD images. Live SYTO9-stained cells and dead propidium iodide-stained cells were excited with an argon laser and a helium neon laser, respectively, and detected with a 560-nm long-pass filter and a 505- to 530-nm band-pass filter, respectively. LIVE/DEAD images were analyzed with the LSM 5 PASCAL software (version 3.5; Carl Zeiss).
RESULTS
cbb3 oxidases alter cell morphology under anaerobic denitrifying growth.
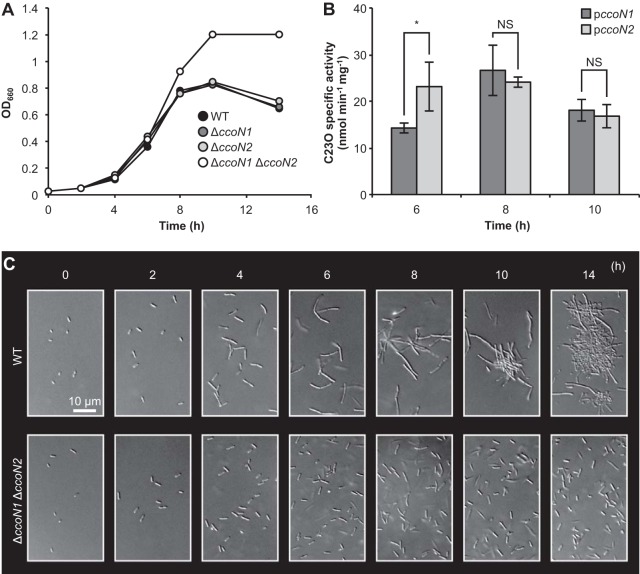
To investigate the effect of two cbb3 oxidases on cell growth, P. aeruginosa PAO1 WT and ΔccoN deletion mutants that are deficient in cbb3-1 and/or cbb3-2 oxidases were incubated oxically in LB medium or anoxically in LBN medium. Under anoxic denitrifying planktonic conditions, the ΔccoN1 ΔccoN2 mutant grew better than the WT (Fig. 1A). Growth of ΔccoN single mutants was similar to that of the WT. The CFU value of the ΔccoN1 ΔccoN2 mutant was higher than those of the WT and the ΔccoN single mutants at 14 h (see Fig. S2 in the supplemental material). These results show that cbb3-1 and cbb3-2 oxidases alter anaerobic denitrifying growth. cbb3-1 subunits of P. aeruginosa have a high degree of amino acid similarity to cbb3-2 subunits (4). Therefore, two cbb3 oxidases may complement each other under anoxic denitrifying conditions. On the other hand, aerobic planktonic growth of ΔccoN1 and ΔccoN1 ΔccoN2 mutants was slightly delayed compared with that of the WT and the ΔccoN2 mutant (see Fig. S3A). In addition, the ΔccoN1 ΔccoN2 mutant showed less growth than the WT under hypoxic conditions in the absence of nitrate (see Fig. S4), which was consistent with a previous report (25).
FIG 1.
Effect of cbb3 oxidases on anaerobic growth and cell morphology. Anoxic incubation of P. aeruginosa was performed using Hungate tubes containing argon gas and LBN medium at 37°C with shaking at 200 rpm. (A) Time course of OD measurements. Three independent experiments were carried out, and representative data are shown. (B) Transcriptional activities of two cbb3 oxidases. ccoN promoter-dependent C23O activities in cells possessing pMEXccoN1 or pMEXccoN2 were measured and normalized to the C23O activities in cells possessing pMEX9. The values represent the means ± standard deviations from three independent experiments. NS, not significant. *, P < 0.05 (unpaired two-tailed Student's t test). (C) Time course of DIC images. Three independent experiments were carried out, and representative images are shown.
Kawakami et al. examined the expression levels of cbb3 oxidases under different oxygen concentrations (5). To monitor whether cbb3 oxidases were expressed under our experimental conditions, promoter fusion plasmids with the xylE gene fused with promoter regions of the ccoN1 and ccoN2 genes were used. The specific activities of C23O, the xylE gene product, were determined by a previously described method (13). As expected, promoter expression of cbb3-1 and cbb3-2 oxidases in the P. aeruginosa PAO1 WT was confirmed during the logarithmic to stationary phase (Fig. 1B), suggesting that these respiratory enzymes have a physiological role under anoxic denitrifying conditions. In a study by Yoon et al., it was demonstrated that P. aeruginosa PAO1 elongates and aggregates in anoxic planktonic shaking cultures (26). Similar phenomena were observed in the WT by DIC observation. In contrast, the ΔccoN1 ΔccoN2 mutant did not elongate under anoxic denitrifying conditions (Fig. 1C). Both strains were rod shaped under oxic conditions (see Fig. S3B). These results show that cbb3 oxidases alter cell morphology under anaerobic denitrifying growth.
cbb3 oxidases alter the denitrification process.
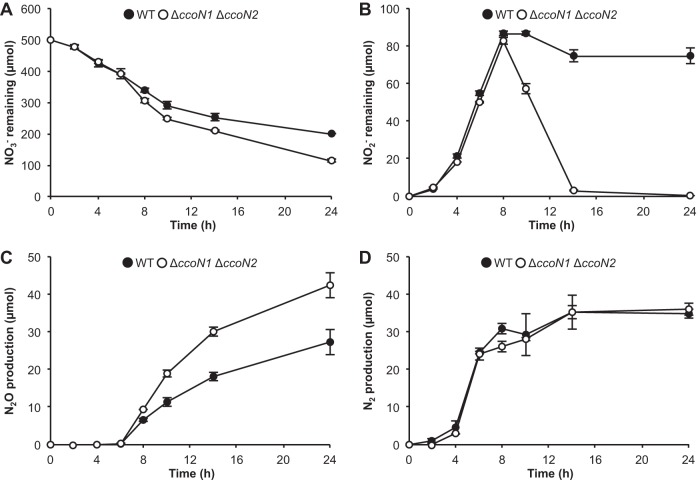
Under anoxic conditions where NO3− is present, P. aeruginosa can grow through denitrification, in which NO3− is reduced subsequently to NO2−, NO, N2O, and, finally, to N2 (3). Cell elongation under anoxic conditions is attributed to the accumulation of NO that is produced during such a process (26). Our data showed that cbb3 oxidases affect cell morphology; hence, the effect of cbb3 oxidases on the denitrification process was investigated. As shown in Fig. 2A and B, the ΔccoN1 ΔccoN2 mutant reduced more NO3− and NO2− than the WT. N2O production was higher in the ΔccoN1 ΔccoN2 mutant than in the WT (Fig. 2C). N2 production of the ΔccoN1 ΔccoN2 mutant was similar to that of the WT (Fig. 2D). These results show that cbb3 oxidases alter the denitrification process. As shown by our data, the amounts of NO2− and N2O were most affected by the cbb3 oxidases, suggesting that cbb3 oxidases alter the denitrification process in between the reduction of NO2− to N2O.
FIG 2.
Effect of cbb3 oxidases on the denitrification process. The amounts of NO3− remaining (A), NO2− remaining (B), N2O production (C), and N2 production (D) were measured in planktonic shaking cultures. The values represent the means ± standard deviations from three independent experiments.
cbb3 oxidases alter cell morphology through NO.
Because NO accumulation induces cell elongation (26), it can be hypothesized that cbb3 oxidases affect NO accumulation, which leads to cell elongation. To test this hypothesis, NIR mutants that do not produce NO were used. Compared with the growth difference between the WT and the ΔccoN1 ΔccoN2 mutant, Δnir and ΔccoN1 ΔccoN2 Δnir mutants had similar but not identical growth (Fig. 1A and Fig. 3A and B; see also Fig. 4B), suggesting that NO is one of the main causes of the growth difference. Consistent with other reports (26), the Δnir mutant did not elongate as the WT and was rod shaped, similar to the ΔccoN1 ΔccoN2 mutant (Fig. 1C and 3C). When planktonic shaking cultures were observed by LIVE/DEAD staining, the WT culture was found to comprise dead or membrane-damaged cells, while the NIR mutant culture comprised intact cells (see Fig. S5 in the supplemental material). This result was consistent with previous observations (27, 28). The ΔccoN1 ΔccoN2 mutant culture comprised intact cells, similar to NIR mutants. These results support the hypothesis that cbb3 oxidases affect NO accumulation during denitrification.
FIG 3.
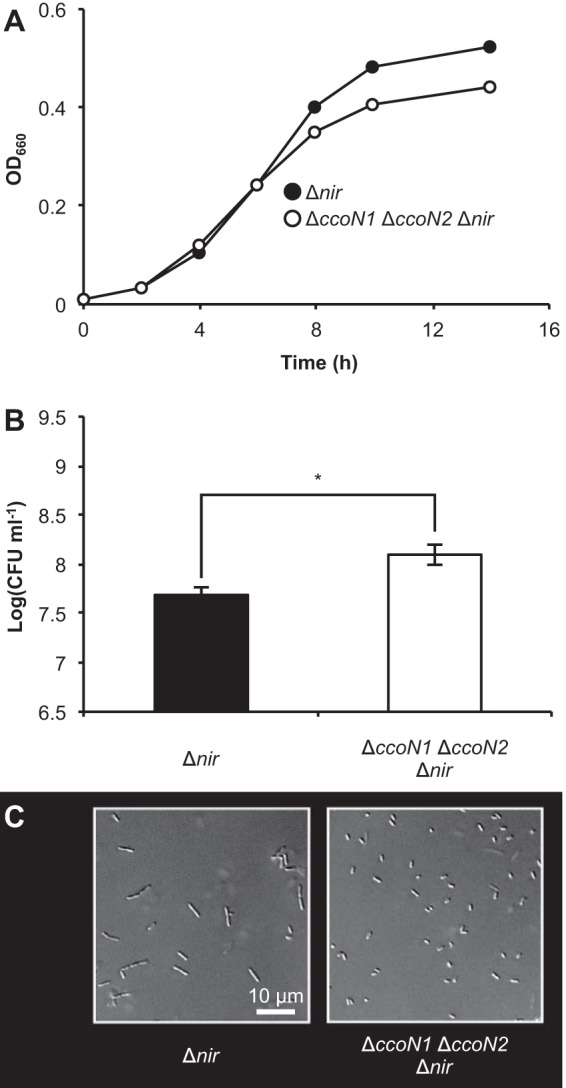
Anaerobic growth and cell morphology of NIR mutants. (A) Time course of OD measurements. Three independent experiments were carried out, and representative data are shown. (B) CFU at 14 h. The values represent the means ± standard deviations from three independent experiments. *, P < 0.01 (unpaired two-tailed Student's t test). (C) DIC images of NIR mutants at 14 h. Three independent experiments were carried out, and representative images are shown.
FIG 4.
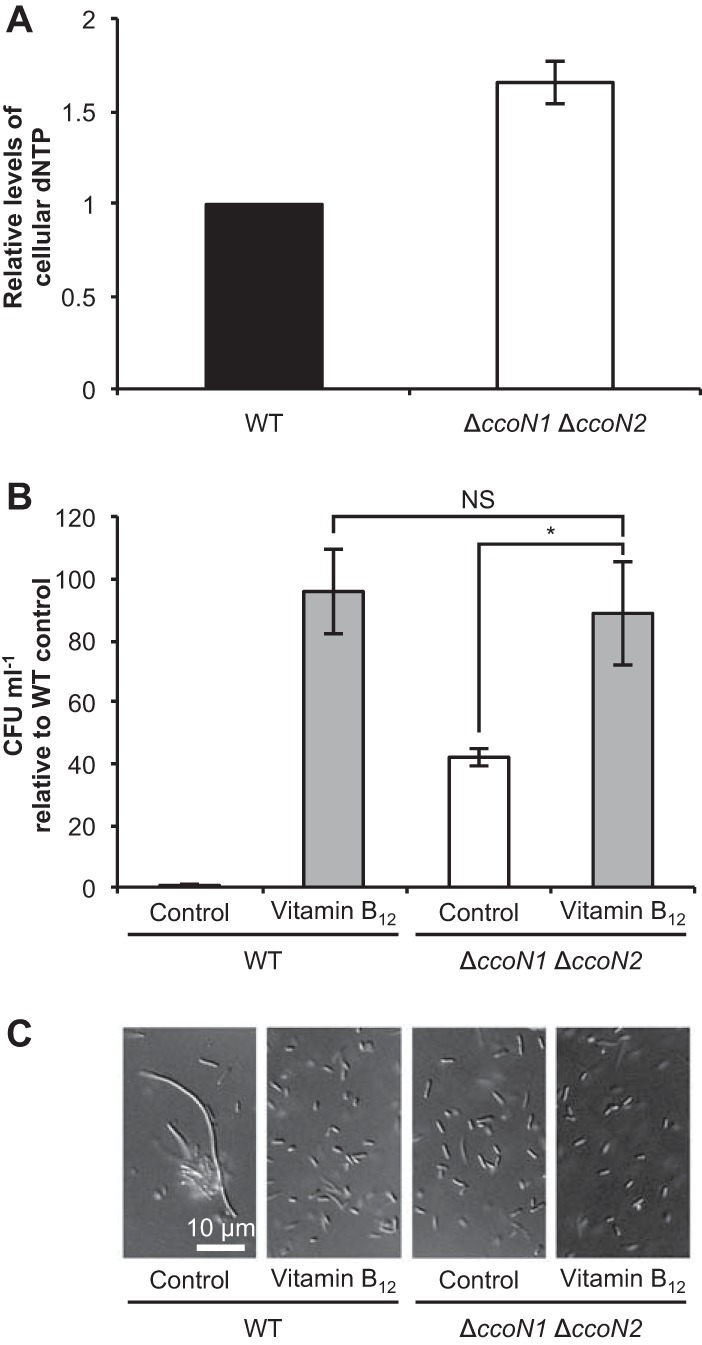
Effect of cbb3 oxidases on DNA synthesis. (A) Relative cellular dNTP levels at 14 h as determined by a DPA assay. Anoxic incubation of P. aeruginosa was performed using Erlenmeyer flasks containing argon gas and LBN medium at 37°C with shaking at 200 rpm. Cell extracts containing equal amounts of protein were used in this assay. The values represent the means ± standard deviations from three independent experiments. (B) Relative CFU at 24 h in LBN medium supplemented with 100 nM vitamin B12. The values represent the means ± standard deviations from three independent experiments. NS, not significant. *, P < 0.05 (unpaired two-tailed Student's t test). (C) DIC images at 24 h. Three independent experiments were carried out, and representative images are shown.
NO derived from denitrification inhibits DNA synthesis by ribonucleotide reductase (RNR), especially class II-type RNR (RNR II), during anaerobic growth of P. aeruginosa PAO1 (19). Because RNR catalyzes the formation of deoxyribonucleotides from ribonucleotides (29), cellular dNTP levels in the WT and the ΔccoN1 ΔccoN2 mutant were quantified by a DPA assay. The dNTP level of the ΔccoN1 ΔccoN2 mutant was higher than that of the WT (Fig. 4A), suggesting that cbb3 oxidases inhibit RNR II activity, presumably through NO. Vitamin B12, a coenzyme of RNR II, has been reported to restore defective anaerobic growth caused by NO (19). In the absence of vitamin B12, the ΔccoN1 ΔccoN2 mutant grew better than the WT. When supplemented with vitamin B12, growth of the ΔccoN1 ΔccoN2 mutant was similar to that of the WT (Fig. 4B). Furthermore, cell morphology of the ΔccoN1 ΔccoN2 mutant was similar to that of the WT in the presence of vitamin B12 (Fig. 4C). Growth of the ΔccoN1 ΔccoN2 mutant supplemented with vitamin B12 was slightly higher and the cell length was shorter than for the control, suggesting that NO also influences anaerobic growth and cell morphology of the ΔccoN1 ΔccoN2 mutant. Taken together, these results indicate that cbb3 oxidases promote cell elongation under anoxic denitrifying conditions by affecting NO accumulation.
cbb3 oxidases alter the cellular NO level.
To provide further evidence that cbb3 oxidases induce NO accumulation, cellular NO levels of the WT and the ΔccoN1 ΔccoN2 mutant were measured with a NO detection reagent, DAF-2 DA. As expected, the NO levels of the ΔccoN1 ΔccoN2 mutant were lower than for the WT (Fig. 5A). Furthermore, the WT phenotype was restored in the ΔccoN1 ΔccoN2 mutant when a NO donor, sodium nitroprusside (SNP; Sigma-Aldrich, St. Louis, MO), was added in the culture. Addition of SNP resulted in low growth yield of the ΔccoN1 ΔccoN2 mutant and induced the accumulation of NO2−, as observed in the WT (Fig. 1A and 2B), compared with those of the control (Fig. 5B). In addition, elongated cells and cell aggregates observed in the WT (Fig. 1C) were observed in the ΔccoN1 ΔccoN2 mutant by the addition of SNP (Fig. 5C). The influence of SNP was diminished by the addition of a NO scavenger, 2-phenyl-4,4,5,5-tetramethylimidazoline-1-oxyl-3-oxide (PTIO; Wako), confirming that NO but not iron or cyanide released from SNP affected the cell physiology of the ΔccoN1 ΔccoN2 mutant. These results show that cbb3 oxidases alter the cellular NO level during the denitrification process.
FIG 5.
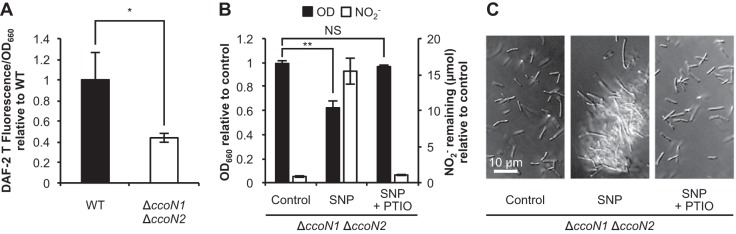
Effect of cbb3 oxidases on the cellular NO level. (A) Relative cellular NO levels at 6 h as determined by a NO detection reagent, DAF-2 DA. The values represent the means ± standard deviations from three independent experiments. *, P < 0.05 (unpaired two-tailed Student's t test). (B) Relative ODs and the amounts of NO2− remaining for the ΔccoN1 ΔccoN2 mutant at 14 h in LBN medium supplemented with 1 μM NO donor SNP and/or 2 mM NO scavenger PTIO. The values represent the means ± standard deviations from three independent experiments. NS, not significant. **, P < 0.01 (unpaired two-tailed Student's t test). (C) DIC images at 14 h. Three independent experiments were carried out, and representative images are shown.
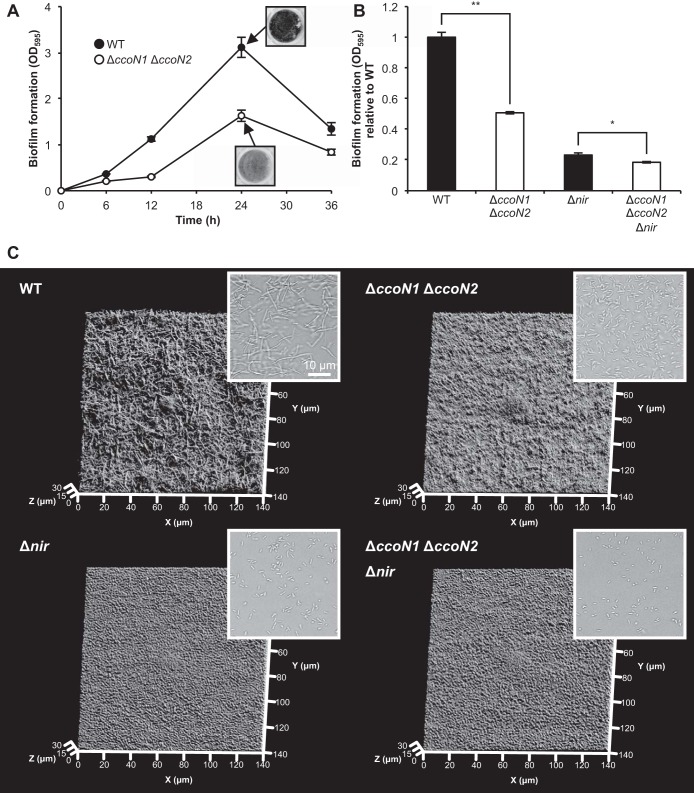
cbb3 oxidases promote the formation of biofilms composed of elongated cells.
As shown in Fig. 1C, cbb3 oxidases promote bacterial aggregation under anoxic planktonic conditions. Cell aggregation often shares common mechanisms with biofilm formation (30, 31). Therefore, cbb3 oxidases may also influence bacterial biofilm formation under anoxic static conditions. The insets in Fig. 6A show crystal violet-stained biofilms formed at the bottom of 24-well plates. From the initial attachment stage (0 to 12 h), the amount of biofilm formed by the ΔccoN1 ΔccoN2 mutant was less than that formed by the WT (Fig. 6A). At 24 h, the ΔccoN1 ΔccoN2 mutant formed half as much biofilm as the WT. Biofilm formation of ΔccoN single mutants was similar to that of the WT (see Fig. S6 in the supplemental material). These results show that cbb3-1 and cbb3-2 oxidases alter anaerobic biofilm formation. As shown in Fig. S7A in the supplemental material, the ΔccoN1 ΔccoN2 mutant reduced NO3− earlier than the WT under this condition. NO2− was less accumulated in the culture of the ΔccoN1 ΔccoN2 mutant than in that of the WT (see Fig. S7B), supporting our idea that cbb3 oxidases alter anaerobic biofilm formation through denitrification. A previous study demonstrated that cell elongation promoted by NO is required for robust biofilm formation of P. aeruginosa PAO1 under anoxic denitrifying conditions (26). In contrast with the difference in biofilm formation between the WT and the ΔccoN1 ΔccoN2 mutant, Δnir and ΔccoN1 ΔccoN2 Δnir mutants, which do not produce NO, formed similar amounts of biofilms (Fig. 6A and B), suggesting that NO is one of the main factors responsible for the difference in biofilm formation. Cell morphological changes in biofilms were confirmed by the bright-field observation of cells scraped from biofilms and the observation of biofilm structures using COCRM (22, 23). The WT biofilms were composed of elongated cells, while ΔccoN1 ΔccoN2 and NIR mutant biofilms were composed of rod-shaped cells (Fig. 6C). These results show that cbb3 oxidases promote mature biofilm formation via cell elongation under anoxic denitrifying conditions. Because of NO accumulation by the cbb3 oxidases (Fig. 5A), cell elongation and the formation of biofilms composed of elongated cells are triggered.
FIG 6.
Effect of cbb3 oxidases on biofilm formation. Anoxic incubation of P. aeruginosa was performed using 24-well (A and B) or 6-well (C) plates containing LBN medium incubated at 37°C. An AnaeroPack system was used for anoxic static incubation. (A) Time course of the amounts of biofilm formation. The values represent the means ± standard deviations from three independent experiments. The insets show crystal violet-stained biofilms formed at the bottom of 24-well plates. (B) Relative amounts of biofilm formation of NIR mutants at 24 h. The values represent the means ± standard deviations from three independent experiments. *, P < 0.01; **, P < 0.0001 (unpaired two-tailed Student's t test). (C) Three-dimensional images of biofilm structures at 24 h visualized by COCRM. Each projection shows fields of 140 by 140 μm (x-y), as indicated. The insets show the bright-field images of cells scraped from biofilms. Three independent experiments were carried out, and representative images are shown.
DISCUSSION
In this study, we uncovered a novel role for cbb3 oxidases in anoxic denitrifying environments in P. aeruginosa PAO1. It was shown that cbb3 oxidases induce NO accumulation during the denitrification process and promote the maturation of the anaerobic biofilm structure via cell elongation.
Impact of cbb3 oxidases on cell behaviors under anoxic conditions.
cbb3 oxidases of P. aeruginosa are more highly expressed under anoxic denitrifying conditions (oxygen concentration of 0%) than under oxic conditions (5). Similarly, Rosenbaum et al. demonstrated transcriptional expression of cbb3 oxidase during biofilm formation of Shewanella oneidensis MR-1 under anoxic conditions (20% CO2–80% N2) when an electrode was used as a terminal electron acceptor; however, the role of cbb3 oxidase is not well understood (32). In P. aeruginosa, under oxic conditions, cbb3 oxidases support energy conservation by catalyzing oxygen reduction in aerobic respiration. Consistent with its role in conserving energy under low-oxygen conditions, the deletion of cbb3 oxidases resulted in less biofilm formation in the oxic flow cell chamber than for the WT, where oxygen becomes limited (25). Our current study was carried out under anoxic conditions, and cbb3 oxidases were shown to promote biofilm formation (Fig. 6A). Rather than through energy conservation, we hypothesize that cbb3 oxidases affect biofilm formation through NO stress. Growth of the ΔccoN1 ΔccoN2 mutant was similar to that of the WT in the presence of vitamin B12, which alleviates NO stress (Fig. 4B). Moreover, cbb3 oxidases led to the accumulation of NO (Fig. 5A). These results support our hypothesis that NO stress rather than energy conservation is a key factor that determines the effect of cbb3 oxidases under anoxic conditions. Consistent with this idea, Yoon et al. demonstrated that NO stress derived from denitrification induces cell elongation, leading to anaerobic biofilm formation (26). Taken together, our data show that cbb3 oxidases have different roles depending on the environment (Fig. 7).
FIG 7.
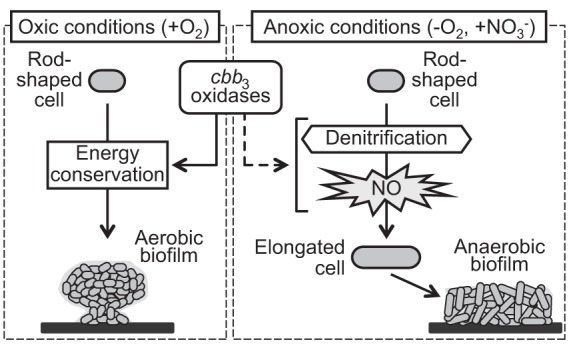
Proposed model depicting how the roles of cbb3 oxidases differ depending on the environment. Under oxic conditions, cbb3 oxidases support energy conservation through oxygen reduction, resulting in aerobic biofilm formation. On the other hand, under anoxic denitrifying conditions, cbb3 oxidases lead to NO accumulation during the denitrification process. NO promotes cell elongation and subsequent anaerobic biofilm formation. It is unclear how cbb3 oxidases induce NO accumulation.
NO is involved in regulating several phenotypes related to cell-cell interactions and social behaviors in P. aeruginosa. For instance, NO can promote membrane vesicle (MV) production and type III secretion system (T3SS) expression, which are involved in host infection (15, 33). NO not only promotes biofilm formation under anoxic conditions (26) but also functions as a signal to induce biofilm dispersal under certain conditions (28). Although NO plays important roles in cell physiology, how NO accumulation is controlled is poorly understood. QS has been indicated to balance the activities of NIR and NOR enzymes, thus controlling NO accumulation, while the precise mechanism is unknown (8, 9, 27). Our results show that cbb3 oxidases are also involved in the control of NO accumulation, adding new factors that are involved in the control of this important molecule and further implying the impact of the cbb3 oxidases in regulating cell behaviors.
In P. aeruginosa PAO1, cell elongation through NO stress is required for mature biofilm formation under anoxic conditions (26). Under anoxic environments, specific mat-like biofilm structures composed of elongated cells are formed, which are different from the mushroom-like biofilm structures composed of rod-shaped cells that form under oxic environments. It is suggested that cell elongation and mature biofilm formation can provide an advantage under stressful environmental conditions (10, 11). However, some clinical isolates of P. aeruginosa remain rod shaped and form mature biofilms under anoxic conditions (17). This result indicates the presence of unknown factors leading to mature biofilm formation in these clinical isolates. It will be interesting to determine whether cbb3 oxidases are involved in the variation among strains under anoxic conditions.
Although our data show that NO stress is the main cause for cell elongation, the cells of the ΔccoN1 ΔccoN2 Δnir mutant were shorter than those of the Δnir mutant under planktonic conditions (Fig. 3C), suggesting that other mechanisms that affect cell morphology may exist. Under static conditions, the cells of the ΔccoN1 ΔccoN2 Δnir mutant were shorter than those of the Δnir mutant, and biofilm formation of the ΔccoN1 ΔccoN2 Δnir mutant was decreased compared with that of the Δnir mutant (Fig. 6B and C). Thus, cell morphological changes affect anaerobic biofilm formation via unknown mechanisms. It has been reported that the mutation or overexpression of the terminal oxidase CIO results in a cell division defect in P. aeruginosa (34). cbb3 oxidases may also affect cell morphology via a common unknown mechanism under anoxic conditions.
Conclusions.
Collectively, our data show that cbb3 oxidases of P. aeruginosa PAO1 lead to NO accumulation during the denitrification process and promote biofilm formation via cell elongation in anoxic denitrifying environments. It is unique that cbb3 oxidases are strongly involved in biofilm formation in anoxic environments where oxygen-dependent respiratory activity is not dominant. A few studies have reported an additional role for bacterial respiratory enzymes. For example, mutations in the cyo operon encoding cytochrome o ubiquinol oxidase influence carbon catabolite repression of phenol degradation by Pseudomonas putida (35). Our data reveal a new aspect of cbb3 oxidases that depends on environmental conditions and further imply the multifunctionality of bacterial respiratory enzymes.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by a Grant-in-Aid for Scientific Research (60292520) to N.N. from the Ministry of Education, Culture, Sports, Science and Technology in Japan (MEXT). M.T. was supported by a Grant-in-Aid for Scientific Research (25701012) from MEXT. The Japan Science and Technology Agency, CREST, and ALCA also provided financial support for this study.
Footnotes
Published ahead of print 2 September 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.01978-14.
REFERENCES
- 1.Brown GC, Borutaite V. 2008. Regulation of apoptosis by the redox state of cytochrome c. Biochim. Biophys. Acta 1777:877–881. 10.1016/j.bbabio.2008.03.024. [DOI] [PubMed] [Google Scholar]
- 2.Hüttemann M, Helling S, Sanderson TH, Sinkler C, Samavati L, Mahapatra G, Varughese A, Lu G, Liu J, Ramzan R, Vogt S, Grossman LI, Doan JW, Marcus K, Lee I. 2012. Regulation of mitochondrial respiration and apoptosis through cell signaling: cytochrome c oxidase and cytochrome c in ischemia/reperfusion injury and inflammation. Biochim. Biophys. Acta 1817:598–609. 10.1016/j.bbabio.2011.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arai H. 2011. Regulation and function of versatile aerobic and anaerobic respiratory metabolism in Pseudomonas aeruginosa. Front. Microbiol. 2:103. 10.3389/fmicb.2011.00103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Comolli JC, Donohue TJ. 2004. Differences in two Pseudomonas aeruginosa cbb3 cytochrome oxidases. Mol. Microbiol. 51:1193–1203. 10.1046/j.1365-2958.2003.03904.x. [DOI] [PubMed] [Google Scholar]
- 5.Kawakami T, Kuroki M, Ishii M, Igarashi Y, Arai H. 2010. Differential expression of multiple terminal oxidases for aerobic respiration in Pseudomonas aeruginosa. Environ. Microbiol. 12:1399–1412. 10.1111/j.1462-2920.2009.02109.x. [DOI] [PubMed] [Google Scholar]
- 6.Arai H, Kodama T, Igarashi Y. 1997. Cascade regulation of the two CRP/FNR-related transcriptional regulators (ANR and DNR) and the denitrification enzymes in Pseudomonas aeruginosa. Mol. Microbiol. 25:1141–1148. 10.1046/j.1365-2958.1997.5431906.x. [DOI] [PubMed] [Google Scholar]
- 7.Schreiber K, Krieger R, Benkert B, Eschbach M, Arai H, Schobert M, Jahn D. 2007. The anaerobic regulatory network required for Pseudomonas aeruginosa nitrate respiration. J. Bacteriol. 189:4310–4314. 10.1128/JB.00240-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Toyofuku M, Nomura N, Fujii T, Takaya N, Maseda H, Sawada I, Nakajima T, Uchiyama H. 2007. Quorum sensing regulates denitrification in Pseudomonas aeruginosa PAO1. J. Bacteriol. 189:4969–4972. 10.1128/JB.00289-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Toyofuku M, Nomura N, Kuno E, Tashiro Y, Nakajima T, Uchiyama H. 2008. Influence of the Pseudomonas quinolone signal on denitrification in Pseudomonas aeruginosa. J. Bacteriol. 190:7947–7956. 10.1128/JB.00968-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davey ME, O'Toole GA. 2000. Microbial biofilms: from ecology to molecular genetics. Microbiol. Mol. Biol. Rev. 64:847–867. 10.1128/MMBR.64.4.847-867.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Justice SS, Hunstad DA, Cegelski L, Hultgren SJ. 2008. Morphological plasticity as a bacterial survival strategy. Nat. Rev. Microbiol. 6:162–168. 10.1038/nrmicro1820. [DOI] [PubMed] [Google Scholar]
- 12.Yawata Y, Nomura N, Uchiyama H. 2008. Development of a novel biofilm continuous culture method for simultaneous assessment of architecture and gaseous metabolite production. Appl. Environ. Microbiol. 74:5429–5435. 10.1128/AEM.00801-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maseda H, Sawada I, Saito K, Uchiyama H, Nakae T, Nomura N. 2004. Enhancement of the mexAB-oprM efflux pump expression by a quorum-sensing autoinducer and its cancellation by a regulator, MexT, of the mexEF-oprN efflux pump operon in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 48:1320–1328. 10.1128/AAC.48.4.1320-1328.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Simon R, O'Connell M, Labes M, Pühler A. 1986. Plasmid vectors for the genetic analysis and manipulation of rhizobia and other gram-negative bacteria. Methods Enzymol. 118:640–659. 10.1016/0076-6879(86)18106-7. [DOI] [PubMed] [Google Scholar]
- 15.Toyofuku M, Zhou S, Sawada I, Takaya N, Uchiyama H, Nomura N. 24 September 2013. Membrane vesicle formation is associated with pyocin production under denitrifying conditions in Pseudomonas aeruginosa PAO1. Environ. Microbiol. 10.1111/1462-2920.12260. [DOI] [PubMed] [Google Scholar]
- 16.Farinha MA, Kropinski AM. 1990. High efficiency electroporation of Pseudomonas aeruginosa using frozen cell suspensions. FEMS Microbiol. Lett. 58:221–225. [DOI] [PubMed] [Google Scholar]
- 17.Fang H, Toyofuku M, Kiyokawa T, Ichihashi A, Tateda K, Nomura N. 2013. The impact of anaerobiosis on strain-dependent phenotypic variations in Pseudomonas aeruginosa. Biosci. Biotechnol. Biochem. 77:1747–1752. 10.1271/bbb.130309. [DOI] [PubMed] [Google Scholar]
- 18.Nicholas DJD, Nason A. 1957. Determination of nitrate and nitrite. Methods Enzymol. 3:981–984. 10.1016/S0076-6879(57)03489-8. [DOI] [Google Scholar]
- 19.Lee KM, Go J, Yoon MY, Park Y, Kim SC, Yong DE, Yoon SS. 2012. Vitamin B12-mediated restoration of defective anaerobic growth leads to reduced biofilm formation in Pseudomonas aeruginosa. Infect. Immun. 80:1639–1649. 10.1128/IAI.06161-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Su S, Panmanee W, Wilson JJ, Mahtani HK, Li Q, Vanderwielen BD, Makris TM, Rogers M, McDaniel C, Lipscomb JD, Irvin RT, Schurr MJ, Lancaster JR, Kovall RA, Hassett DJ. 2014. Catalase (KatA) plays a role in protection against anaerobic nitric oxide in Pseudomonas aeruginosa. PLoS One 9:e91813. 10.1371/journal.pone.0091813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Merritt JH, Kadouri DE, O'Toole GA. 2005. Growing and analyzing static biofilms. Curr. Protoc. Microbiol. 00:B:1B.1:1B.1.1–1B.1.17. 10.1002/9780471729259.mc01b01s00. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yawata Y, Toda K, Setoyama E, Fukuda J, Suzuki H, Uchiyama H, Nomura N. 2010. Monitoring biofilm development in a microfluidic device using modified confocal reflection microscopy. J. Biosci. Bioeng. 110:377–380. 10.1016/j.jbiosc.2010.04.002. [DOI] [PubMed] [Google Scholar]
- 23.Inaba T, Ichihara T, Yawata Y, Toyofuku M, Uchiyama H, Nomura N. 2013. Three-dimensional visualization of mixed species biofilm formation together with its substratum. Microbiol. Immunol. 57:589–593. 10.1111/1348-0421.12064. [DOI] [PubMed] [Google Scholar]
- 24.Tashiro Y, Nomura N, Nakao R, Senpuku H, Kariyama R, Kumon H, Kosono S, Watanabe H, Nakajima T, Uchiyama H. 2008. Opr86 is essential for viability and is a potential candidate for a protective antigen against biofilm formation by Pseudomonas aeruginosa. J. Bacteriol. 190:3969–3978. 10.1128/JB.02004-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alvarez-Ortega C, Harwood CS. 2007. Responses of Pseudomonas aeruginosa to low oxygen indicate that growth in the cystic fibrosis lung is by aerobic respiration. Mol. Microbiol. 65:153–165. 10.1111/j.1365-2958.2007.05772.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yoon MY, Lee KM, Park Y, Yoon SS. 2011. Contribution of cell elongation to the biofilm formation of Pseudomonas aeruginosa during anaerobic respiration. PLoS One 6:e16105. 10.1371/journal.pone.0016105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yoon SS, Hennigan RF, Hilliard GM, Ochsner UA, Parvatiyar K, Kamani MC, Allen HL, DeKievit TR, Gardner PR, Schwab U, Rowe JJ, Iglewski BH, McDermott TR, Mason RP, Wozniak DJ, Hancock RE, Parsek MR, Noah TL, Boucher RC, Hassett DJ. 2002. Pseudomonas aeruginosa anaerobic respiration in biofilms: relationships to cystic fibrosis pathogenesis. Dev. Cell 3:593–603. 10.1016/S1534-5807(02)00295-2. [DOI] [PubMed] [Google Scholar]
- 28.Barraud N, Hassett DJ, Hwang SH, Rice SA, Kjelleberg S, Webb JS. 2006. Involvement of nitric oxide in biofilm dispersal of Pseudomonas aeruginosa. J. Bacteriol. 188:7344–7353. 10.1128/JB.00779-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jordan A, Torrents E, Sala I, Hellman U, Gibert I, Reichard P. 1999. Ribonucleotide reduction in Pseudomonas species: simultaneous presence of active enzymes from different classes. J. Bacteriol. 181:3974–3980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Allesen-Holm M, Barken KB, Yang L, Klausen M, Webb JS, Kjelleberg S, Molin S, Givskov M, Tolker-Nielsen T. 2006. A characterization of DNA release in Pseudomonas aeruginosa cultures and biofilms. Mol. Microbiol. 59:1114–1128. 10.1111/j.1365-2958.2005.05008.x. [DOI] [PubMed] [Google Scholar]
- 31.Alhede M, Kragh KN, Qvortrup K, Allesen-Holm M, van Gennip M, Christensen LD, Jensen P, Nielsen AK, Parsek M, Wozniak D, Molin S, Tolker-Nielsen T, Høiby N, Givskov M, Bjarnsholt T. 2011. Phenotypes of non-attached Pseudomonas aeruginosa aggregates resemble surface attached biofilm. PLoS One 6:e27943. 10.1371/journal.pone.0027943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rosenbaum MA, Bar HY, Beg QK, Segrè D, Booth J, Cotta MA, Angenent LT. 2012. Transcriptional analysis of Shewanella oneidensis MR-1 with an electrode compared to Fe(III)citrate or oxygen as terminal electron acceptor. PLoS One 7:e30827. 10.1371/journal.pone.0030827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Van Alst NE, Wellington M, Clark VL, Haidaris CG, Iglewski BH. 2009. Nitrite reductase NirS is required for type III secretion system expression and virulence in the human monocyte cell line THP-1 by Pseudomonas aeruginosa. Infect. Immun. 77:4446–4454. 10.1128/IAI.00822-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tavankar GR, Mossialos D, Williams HD. 2003. Mutation or overexpression of a terminal oxidase leads to a cell division defect and multiple antibiotic sensitivity in Pseudomonas aeruginosa. J. Biol. Chem. 278:4524–4530. 10.1074/jbc.M210355200. [DOI] [PubMed] [Google Scholar]
- 35.Petruschka L, Burchhardt G, Müller C, Weihe C, Herrmann H. 2001. The cyo operon of Pseudomonas putida is involved in carbon catabolite repression of phenol degradation. Mol. Genet. Genomics 266:199–206. 10.1007/s004380100539. [DOI] [PubMed] [Google Scholar]
- 36.Holloway BW, Krishnapillai V, Morgan AF. 1979. Chromosomal genetics of Pseudomonas. Microbiol. Rev. 43:73–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.