Abstract
Integrative and conjugating elements (ICE) are self-transferable DNAs widely present in bacterial genomes, which often carry a variety of auxiliary genes of potential adaptive benefit. One of the model ICE is ICEclc, an element originally found in Pseudomonas knackmussii B13 and known for its propensity to provide its host with the capacity to metabolize chlorocatechols and 2-aminophenol. In this work, we studied the mechanism and target of regulation of MfsR, a TetR-type repressor previously found to exert global control on ICEclc horizontal transfer. By using a combination of ICEclc mutant and transcriptome analysis, gene reporter fusions, and DNA binding assays, we found that MfsR is a repressor of both its own expression and that of a gene cluster putatively coding for a major facilitator superfamily efflux system on ICEclc (named mfsABC). Phylogenetic analysis suggests that mfsR was originally located immediately adjacent to the efflux pump genes but became displaced from its original cis target DNA by a gene insertion. This resulted in divergence of the original bidirectional promoters into two separated individual regulatory units. Deletion of mfsABC did not result in a strong phenotype, and despite screening a large number of compounds and conditions, we were unable to define the precise current function or target of the putative efflux pump. Our data reconstruct how the separation of an ancestor mfsR-mfsABC system led to global control of ICEclc transfer by MfsR.
INTRODUCTION
Efflux systems play an important role in mitigating the effects of toxic substances on bacteria (1). A variety of different efflux systems have been recognized, which have been divided into five main categories: (i) the resistance-nodulation-cell division type (RND), (ii) the major facilitator superfamily (MFS), (iii) small multidrug resistance pumps (SMR), (iv) multidrug and toxic compound extrusion systems (MATE), and (v) ATP-binding cassette (ABC) systems (2). Many of the known efflux systems play a role in antibiotic resistance; therefore, their distribution and propagation in clinically relevant strains pose a major health concern (3). Although mostly located on the bacterial chromosome (1), genes for efflux systems can be transmitted via mobile genetic elements like plasmids or integrative and conjugative elements (ICE) (4–6). Recently, we described a class of widespread ICE among beta- and gammaproteobacteria, which, through a region permissive for gene insertions, can carry a wide range of auxiliary gene functions (7–9). The currently best-studied model of this family, ICEclc, was originally found in Pseudomonas knackmussii B13 (9, 10) and confers the ability to degrade 3-chlorocatechol and 2-aminophenol (7). Homologs to ICEclc, identifiable by a conserved “core” gene region, occur in a range of proteobacterial species, such as Ralstonia sp. strain JS705 (11), Burkholderia xenovorans LB400 (12), Achromobacter, Pseudomonas aeruginosa, Acidovorax sp. strain JS42, Bordetella petrii, Xanthomonas campestris, and Herminiimonas arsenicoxydans (9).
We previously noted that ICEclc carries a gene cluster for a putative MFS efflux system (7). Serendipitously, when studying the effect of mutations in a tetR-type regulator gene named mfsR on ICEclc transfer, we discovered that such mutants also display increased expression of the genes for the putative MFS efflux system (13). Control of MFS efflux is frequently achieved by dedicated transcriptional regulators that can react to chemical stimuli and trigger expression of genes coding for the efflux pumps (14). In particular, tetracycline-type regulators (TetR-type) are often associated with efflux control of a wide range of antibiotics and organic compounds (15), although they have also been shown to regulate other cellular processes (16, 17). TetR family members are typically transcriptional repressors expressed at a low but constitutive level and need the interaction with a ligand (“inducer”) to derepress their target promoters (18, 19).
Given the presence of genes for a putative MFS system on ICEclc (mfs genes) and the link to the TetR-type MfsR protein, we were interested to study their possible regulatory interaction. In order to study the possible implication of MfsR, we used microarray analysis of ICEclc transcription in wild-type and mfsR deletion mutants and measured expression of mCherry in Pseudomonas putida derivatives from transcriptional fusions with a variety of differently sized fragments upstream of mfsR and of the mfs efflux genes. Electrophoretic mobility shift assays (EMSA) with purified C-terminal His-tagged MfsR repressor protein were conducted to further provide evidence for MfsR binding to operator-like sequences present in both promoters. In order to study the effect of having or expressing the genes for the efflux system, we constructed P. putida and P. aeruginosa carrying wild-type ICEclc, with mfsR or mfs efflux pump gene deletions on ICEclc, and examined chemical sensitivities to a range of compounds. Finally, we compared analogous systems to reconstruct the possible origin of the ICEclc-located mfs genes. Our analysis suggests how mfsR on ICEclc became separated from its adjacent target mfs operon as a consequence of a large gene insertion, by which it accidentally acquired a new role as a global regulator of ICEclc transfer initiation.
MATERIALS AND METHODS
Bacterial strains and plasmids.
Escherichia coli DH5α (Gibco Life Technologies, Gaithersburg, MD), DH5α λpir, and S17-1 λpir were routinely used for clonings and plasmid propagation. E. coli BL21(DE3) was used for protein overexpression (10). Strains used during this study are listed in Table S1 in the supplemental material, with details of their construction in Table S2. Wild-type and mutant ICEclc (ΔmfsR ΔmfsABC) were conjugated from their P. putida UWC1 host into P. aeruginosa PAO509, a strain devoid of its five mex efflux pumps (20). Four independent transconjugant colonies of PAO509 from each mating were purified and kept at −80°C for subsequent tests.
Media and growth conditions.
E. coli and P. putida were grown at 37°C and 30°C, respectively. E. coli was cultivated on Luria-Bertani medium (LB), whereas P. putida was grown on LB and 21C minimal medium (MM) (21, 22) complemented with either 10 mM 3-chlorobenzoate (3-CBA) or 15 mM succinate as the sole carbon and energy source. P. aeruginosa PAO509 and its derivatives containing wild-type or mutant ICEclc were grown at 30°C and 37°C in MM with 5 mM 3-CBA. When necessary, the following antibiotics were added to culture media at the indicated concentrations: kanamycin (Km) at 25 μg/ml, gentamicin (Gm) at 20 μg/ml, and ampicillin (Ap) at 100 (for E. coli) or 500 μg/ml (for P. putida). Chromium(III) chloride was added at concentrations between 1 and 200 mg/liter.
DNA manipulations.
Isolation of both chromosomal and plasmid DNA, PCR, restriction enzyme digestion, ligation, and electroporation were performed by standard procedures (22). Gene deletions were created by double recombination and marker selection as described elsewhere (23). Complementations of mutations were tested in strains with a single inserted gene copy delivered as mini-Tn7 through the pUC18-miniTn7_Gm system (24). Nucleotide positions are given according to GenBank accession number AJ617740.2 (ICEclc).
MIC assays.
The MICs of a set of 20 antimicrobial compounds (see Table S3 in the supplemental material) were determined according to standard procedures (25) for three strains of P. putida UWC1 (carrying wild-type ICEclc [strain 2737], ICEclc-ΔmfsABC [strain 4165], and ICEclc-ΔmfsR [strain 3543]). MICs were further determined for triclosan (in a concentration range from 0.004 to 1,024 mg/liter of triclosan, in 2-fold increments), and chromium(III) chloride hexahydrate (0.5 to 5,000 mg/liter in 2-fold increments) with P. aeruginosa PAO509 strains (carrying ICEclc, ICEclc-ΔmfsABC, and ICEclc-ΔmfsR). The MIC was taken as the lowest concentration for and above which no growth was visible by naked eye after incubation for 16 h at 30°C (P. putida) or 37°C (P. aeruginosa).
Reporter gene fusions.
Fragments from upstream mfsR or mfsA were produced by PCR (see Table S2 in the supplemental material), cloned in front of a promoterless mcherry gene in E. coli, and delivered in single copy into the chromosomes of various P. putida UWC1 strains (see Table S1) by using the mini-Tn5 delivery system of plasmid pCK218 (26). Transformants were selected on Km-containing plates and verified by PCR. For each reporter fusion, at least three independent clones with different insertion positions of the transposed fragment were used as replicates in fluorometry measurements. P. putida reporter strains were grown overnight with rotary shaking in MM with 15 mM succinate supplemented with 25 μg/ml of Km. An aliquot of 5 μl of each culture was then inoculated into 195 μl of fresh MM with 15 mM succinate and Km in 96-well black polystyrene microtiter plates with flat transparent bottoms (Greiner Bio-One VACUETTE Schweiz GmbH). The plates were incubated at 30°C with orbital shaking (300 rpm), and the culture turbidities (optical densities at 600 nm [OD600]) and mCherry fluorescence (excitation, 590 nm; emission, 620 nm) were recorded at regular intervals. Fluorescence intensities were normalized by culture turbidity and subtracted by the normalized fluorescence of P. putida UWC1 carrying a single-copy promoterless mcherry mini-Tn5 insertion.
Gene reporter induction assay.
P. putida UWC1 (ICEclc) carrying the PmfsA-6-mcherry promoter fusion was cultured on MM with 15 mM succinate and 25 μg/ml of Km in Erlenmeyer flasks. At mid-exponential phase, one of the following compounds was added to the cultures: 10 mM 3-CBA, 0.5 mM 3-chlorocatechol, 5 mM 2-aminophenol, and 4% or 0.4% ethanol. The culture turbidity and the mCherry fluorescence were monitored for the next two and half hours and compared to those for the uninduced control. The same strain was also grown on MM with 15 mM succinate or 10 mM 3-CBA and 25 μg/ml of Km for 35 h, during which the culture absorbance and the mCherry fluorescence were measured.
Biolog phenotypic microarray.
Chemical sensitivities to 960 compounds or conditions of strains of P. putida UWC1 (carrying ICEclc and ICEclc-ΔmfsABC) were determined by Biolog phenotypic microarray PM09-20 service for cells growing on nutrient broth (Biolog Inc., Hayward, CA).
Microarray analysis.
Transcriptome profiling of ICEclc-specific genes in P. putida UWC1 strains carrying ICEclc (wild type) and ICEclc-ΔmfsR (strain 3543) was performed as described previously (27). Strains were grown on MM with 10 mM 3-CBA as the sole carbon and energy source. Cells were harvested at mid-exponential phase and at 48 h after the entry into stationary phase. Total RNA was extracted; Cy3-labeled cDNA was synthesized by reverse transcription, purified, and hybridized to Agilent 8x15K custom ICEclc microarray slides as previously described (27). After a washing, slides were scanned and data extracted and analyzed using GeneSpring GX (version 12; Agilent Technologies, Santa Clara, CA).
Protein purification.
The mfsR gene was amplified by PCR under inclusion of NdeI and XhoI restriction sites (see Table S2 in the supplemental material). After digestion by NdeI and XhoI, the fragment was ligated into pET-22b(+) (Novagen; catalog no. 69744-3) predigested with the same enzymes, thus adding a hexahistidine tag at the C-terminal end. The plasmid [pET22b(+)-mfsR-His6] was transformed into E. coli BL21(DE3), which was subsequently grown in LB with Ap at 25°C with rotary shaking at 220 rpm. At mid-exponential phase, cells were induced with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for 1 h. Cells were collected by centrifugation and lysed using a French press. His-tagged protein (MfsR-His6) was purified by nickel-nitrilotriacetic acid (Ni-NTA) resin according to procedures outlined by the supplier (Qiagen AG, Hilden, Germany) and dialyzed overnight in N-cyclohexyl-3-aminopropanesulfonic acid (CAPS)-based DNA binding buffer (containing 10 mM CAPS at pH 10.5, 250 mM NaCl, 10 mM magnesium acetate, 10 mM KCl, 0.1 mM EDTA, 1 mM dithiothreitol [DTT], and 5% [vol/vol] glycerol) using a Slide-A-Lyzer cassette (Thermo Fisher Scientific, Inc., Waltham, MA).
Dimerization assay.
Sixty-two micrograms of freshly purified MfsR-His6 protein fraction was incubated with 0.4% glutaraldehyde for 30 min at room temperature. The reaction mixture was then boiled and loaded on a 12% sodium dodecyl sulfate-polyacrylamide gel and run for 2 h at 250 V (28).
Fluorescence-based electrophoretic mobility shift assays.
DNA fragments encompassing the PmfsR and PmfsA promoters were produced by PCR (see Table S2 in the supplemental material) and purified from gel. Then, they were ligated into pGEM-T Easy (Promega) and transformed into E. coli DH5α for propagation. The plasmids were extracted by using a Genomed's Jetstar Midiprep and linearized by ScaI digestion. Linear plasmids were used as templates in the PCR using M13 primers with attached Dyomics681 dye at their 5′ ends (Microsynth AG, Switzerland). The resulting Dyomics681-labeled promoter fragments were purified on a gel and used in subsequent EMSA at a 0.5 nM final concentration. Each EMSA reaction was carried out in a total volume of 20 μl of CAPS-based DNA-binding buffer, which further contained 0.2 μg of salmon sperm DNA, 1 μg of bovine serum albumin (BSA), and 0.5 μM freshly purified MfsR-His6 protein (or no MfsR, in the case of negative controls). EMSA reaction mixtures were incubated for 30 min at 22 to 24°C and protected from direct sunlight. Following incubation, the EMSA reaction mixtures were loaded on a CAPS-based native 4% polyacrylamide gel and run for 4 to 6 h at 50 V in CAPS buffer. Gels were subsequently incubated for 1 h in gel drying solution (containing 20% ethanol, 10% methanol, 10% isopropanol, 2% glycerol, and 58% H2O, all vol/vol) and directly scanned on a fluorescence bed scanner at 680 and 700 nm (Li-Cor Odyssey; Li-Cor Biosciences).
Homology searches and synteny comparisons.
BLASTN and BLASTP analyses were carried out using ICEclc mfs gene sequences as a query and ICEberg (29) and NCBI nucleotide collection (nr/nt) or nonredundant protein sequences (nr) as databases. COBALT alignments and Clustal Omega phylogenetic treeing were performed using online services on the NCBI and EBI servers, respectively. Synteny analysis between ICEclc and related genomes was performed with the WebACT tool (30).
Microarray data accession number.
Microarray data have been deposited and are accessible from the GEO database under accession number GSE51391.
RESULTS
A putative major facilitator system encoded on ICEclc.
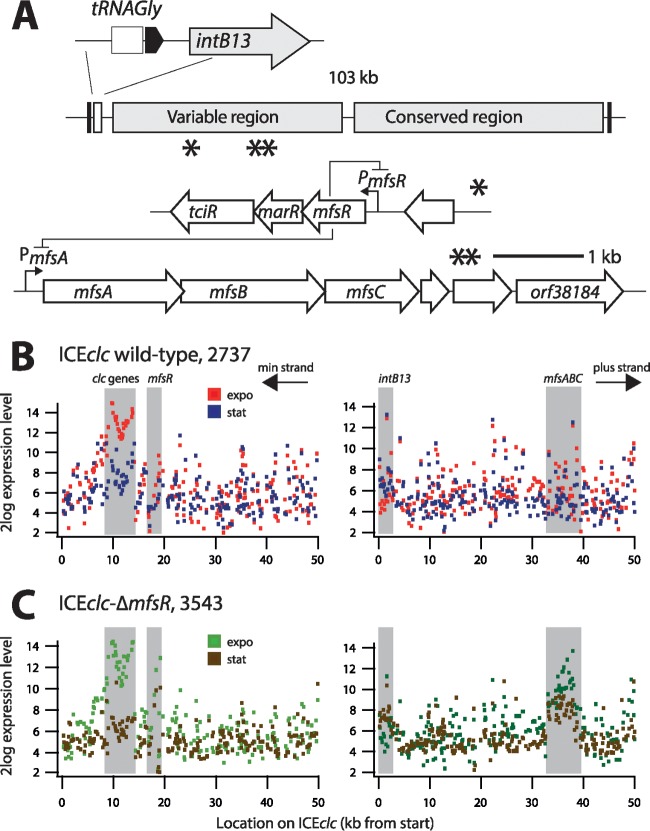
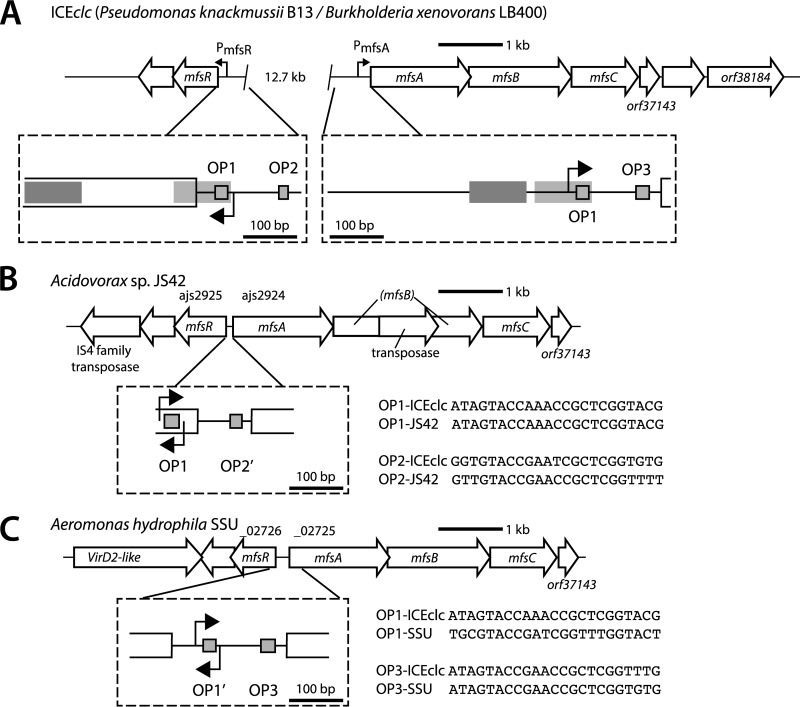
Inspection of the ICEclc sequence had indicated the presence of a set of genes for a putative major facilitator system efflux pump (orf32963-orf34495-orf36077) (7), which we propose to rename here in analogy to other systems as mfsABC (see Fig. S1 in the supplemental material). Gene organization and microarray analysis of ICEclc gene expression suggested further that the mfsABC genes are part of a transcriptional unit that continues with three further open reading frames (Fig. 1A). The three subsequent open reading frames, orf37143, orf37489, and orf38184, are predicted to code for a protein with unknown function, a lipid A deacylase (PagL-like protein), and a putative esterase of the alpha-beta hydrolase family, respectively. As we previously noticed by serendipity when selecting for mutants impaired in ICEclc transfer, expression of the mfsABC gene cluster is elevated compared to that in the wild type in transposon insertion mutants in a gene named mfsR (13). Indeed, hybridization of reverse-transcribed Cy3-labeled mRNA from cultures grown on 3-CBA showed around 12.6-fold-higher expression of the mfsABC cluster in a P. putida UWC1 ICEclc derivative in which the gene mfsR was deleted (ICEclc-ΔmfsR; [strain 3543]) (see Table S1 in the supplemental material) than in UWC1 carrying ICEclc (wild type) (Fig. 1B and C). The difference in expression level occurred in mRNA from both exponential and stationary-phase cells (Fig. 1C). This suggested that mfsR was directly or indirectly implicated in regulating expression of the mfsABC cluster, in addition to its previously described control of mfsR-marR-tciR expression (Fig. 1A) (13).
FIG 1.
Genetic map of ICEclc and the relevant regions for microarray analysis. (A) ICEclc schematic representation. Arrows indicate predicted open reading frames and their orientation. Single and double asterisks depict the locations of mfsR-marR-tciR and mfsABC-orf38184, respectively. (B and C) Log 2-fold mean expression level of ICEclc genes in P. putida UWC1 carrying ICEclc (strain 2737) and ICEclc-ΔmfsR (strain 3543), respectively. RNA was harvested from cells in exponential phase growing with 10 mM 3-CBA as the sole carbon and energy source and in subsequent stationary phase. Diagrams display mean expression levels from triplicate microarrays for probes plotted along the length of ICEclc (kb) on both strands (labeled “min” and “plus”). Gray-shaded zones indicate expression in the regions of the intB13 integrase gene, the clc genes for 3-chlorocatechol metabolism, mfsR, and mfsABC.
MfsR represses both its own promoter and that of the mfsABC-orf38184 cluster.
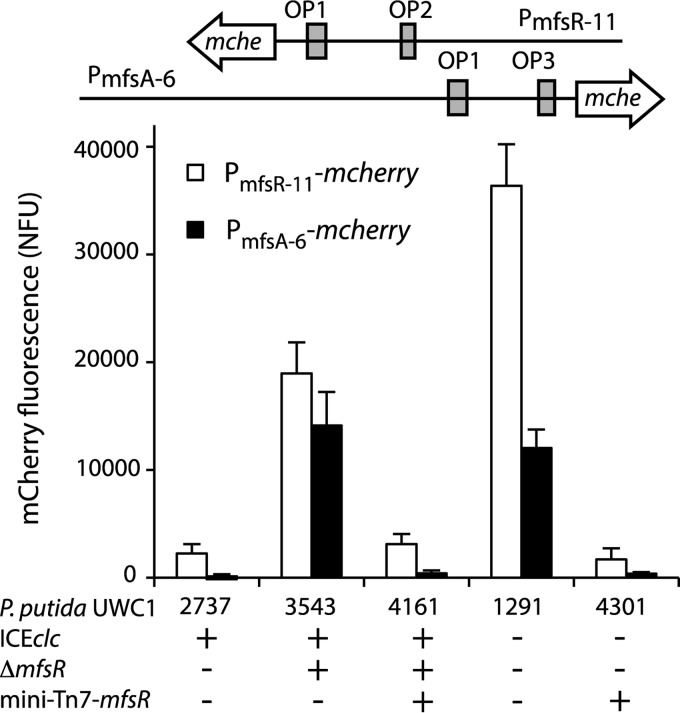
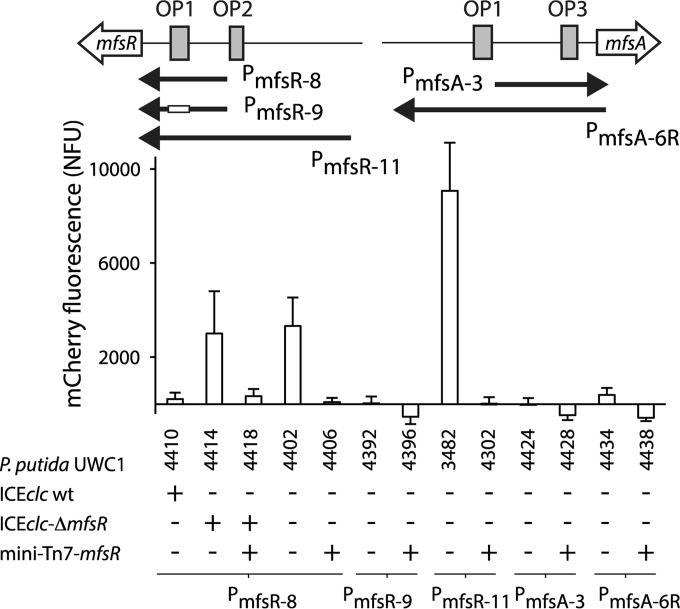
In order to better understand the mechanism of MfsR-regulated expression from the mfsR and mfsA promoter regions, reporter gene fusions were constructed. A 656-bp region upstream of mfsR (PmfsR-11) (Fig. 2A) and a 486-bp region upstream of mfsA (PmfsA-6) (Fig. 2B) were each fused to a promoterless mcherry gene and introduced in single copy on the chromosome of P. putida UWC1 using mini-Tn5 delivery. mCherry expression from both mfsR and mfsA promoters was low in P. putida UWC1 with ICEclc but 10 to 15 times higher in P. putida UWC1 without ICEclc (Fig. 3). mCherry expression was also 7-fold higher in P. putida UWC1 with ICEclc-ΔmfsR (Fig. 3) but was diminished to wild-type ICEclc levels when complementing mfsR in a single copy using mini-Tn7 delivery and expressed from its own promoter (Fig. 3; see also Table S1 in the supplemental material). mCherry expression from both promoters also strongly decreased in P. putida UWC1 without ICEclc but expressing mfsR from the same single-copy mini-Tn7 insertion (strain 4301) (Fig. 3). Note that the deletion of mfsR in strain 3543 expands into the marR reading frame, but since full complementation of function is achieved by adding a single-copy mfsR gene under the control of its own promoter, we conclude that inactivation of a putative MarR protein in strain 3543 and its derivatives has no effect on mfsR or mfsA expression. Taken together, the results demonstrated that both mfsR and mfsA promoters are constitutively expressed in the absence of MfsR and that MfsR alone is sufficient to repress transcription. The 2-fold difference in mCherry expression from the mfsR promoter between P. putida UWC1 without ICEclc and with ICEclc-ΔmfsR (Fig. 3) may indicate that another ICEclc-located factor exerts additional control.
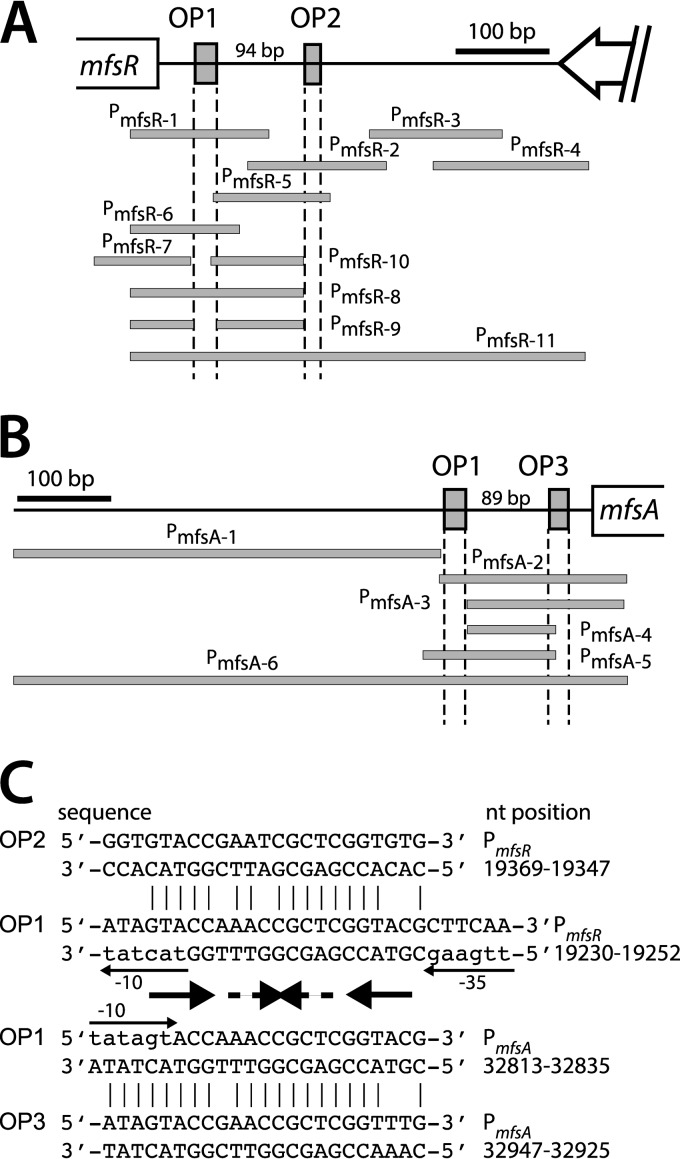
FIG 2.
Overview of the studied mfsR and mfsA promoter regions. (A) PCR-amplified and cloned promoter fragments upstream of mfsR (gray bars). The OP1 sequence was deleted in fragment PmfsR-9. (B) Cloned promoter fragments upstream of mfsA. Gray boxes represent MfsR operator sequences (OP1, OP2, and OP3). (C) Details of the OP sequences in the mfsR and mfsA promoter regions. Palindromic sequences within the OP1 box are indicated by thick arrows (5′-GTACCNANCNGNTNGGTAC, of which OP2 and OP3 share only the first part, 5′-NNACCGAN5TCGGTAC). Thin arrows point to the −35/−10 motifs and transcription direction in the OP1 box of the mfsR promoter and the reverse −10 motif and transcription direction within OP1 in PmfsA. Nucleotide positions refer to numbering of ICEclc in GenBank (accession number AJ617740.2). For the complete sequence, see Fig. S2 in the supplemental material.
FIG 3.
Reporter gene expression from mfsR and mfsA promoter fragments in the presence or absence of mfsR in P. putida UWC1. P. putida UWC1 host strains (for details, see Table S1 in the supplemental material) with relevant genetic details are indicated below the graph. mCherry fluorescence was measured 20 h after inoculation in MM with 15 mM succinate as the sole carbon and energy source and is normalized by the culture density (NFU, normalized fluorescence units).
Regions upstream of mfsR and mfsABC share similar sequence motifs.
Since mfsR encodes a TetR-type transcriptional regulator, and most members of this family are known to be autorepressors (15), we hypothesized that the upstream regions of mfsABC and mfsR would contain similar DNA motifs acting as MfsR binding sites. Indeed, BLASTN comparison of the sequences upstream of mfsR and of mfsA revealed identical 23-bp regions, which we designated OP1 (Fig. 2). In addition, each region contained a second imperfect repeat of OP1, named OP2 (for the mfsR promoter region) and OP3 (for mfsA). Interestingly, OP2 lies 94 bp upstream of OP1 in the mfsR promoter region (Fig. 2A), whereas OP3 is located 89 bp downstream of OP1 in the mfsA promoter region (Fig. 2B). Both OP1 boxes have an internal palindromic sequence, 5′-GTACCNANCNGNTNGGTAC, of which OP2 and OP3 only share the first part, 5′-NNACCGA(N5)TCGGTAC (Fig. 2C; see also Fig. S2 in the supplemental material). The two OP boxes in both promoter regions are located in the opposite orientation relative to their target gene (Fig. 2C).
MfsR binds to both mfsR and mfsA promoter regions.
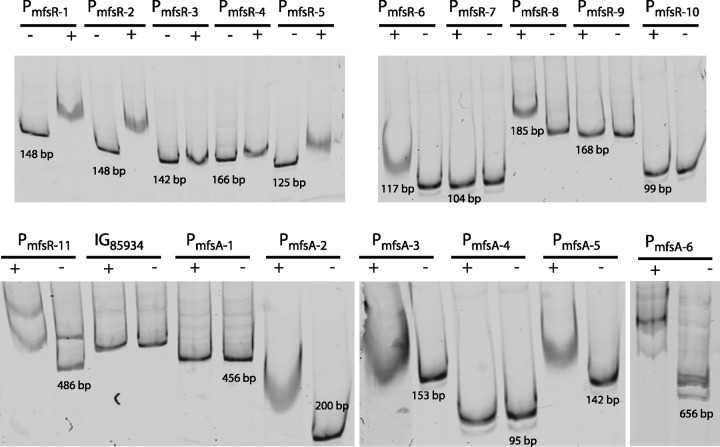
In order to verify whether MfsR directly binds to both PmfsR and PmfsA promoter regions and whether the OP1, OP2, and OP3 boxes are sufficient for binding, EMSA were performed with purified MfsR, tagged with hexahistidine at the C-terminal end (MfsR-His6). Nickel-NTA-purified MfsR-His6 from E. coli has an observed size of 25 kDa (predicted size, 25.8 kDa), whereas glutaraldehyde cross-linking resulted in the formation of a 50-kDa protein band (Fig. 4). This suggests MfsR-His6 to form homodimers in solution, which is in agreement with several observations that TetR family members occur as homodimers (31, 32). Even larger protein bands were seen in cross-linked preparations (e.g., 95 kDa) (Fig. 4), which we believe may have been higher-order multimers or artifacts of the procedure. Purified MfsR-His6 was incubated with a variety of different-length fluorescently end-labeled fragments (depicted in Fig. 2A and B) from both mfsR and mfsA promoters. The resulting DNA-protein complexes were separated on nondenaturing polyacrylamide gels and imaged using fluorometry (Fig. 5). As expected, MfsR-His6 binds to the “complete” PmfsR and PmfsA promoter regions (PmfsR-11 and PmfsA-6, respectively) (Fig. 5) but not to a random other ICEclc intergenic region IG85934 (upstream of orf85934). For some fragments, MfsR-His6-DNA complexes yielded two identifiable bands (e.g., PmfsR-11 and PmfsA-6 in Fig. 5), but these seemed to be artifacts of the labeling procedure (i.e., even present without protein added). Every subfragment containing at least the OP1 box (fragments PmfsR-1, PmfsR-6, PmfsR-8, PmfsR-11, PmfsA-2, PmfsA-5, and PmfsA-6), the OP2 box (fragments PmfsR-2, PmfsR-5, and PmfsR-11), or the OP3 box (fragments PmfsA-2, PmfsA-3, and PmfsA-6) was bound by purified MfsR-His6 (Fig. 5). In contrast, none of the fragments without at least one of the OP boxes (fragments PmfsR-3, PmfsR-4, PmfsR-7, PmfsR-9, PmfsR-10, PmfsA-1, and PmfsA-4) was bound by MfsR-His6. Collectively, these results indicate that MfsR directly binds to fragments encompassing at least one of the OP boxes. The exact positions of MfsR interaction in the operator DNA may lie entirely or partially within the predicted OP box sequence (Fig. 2C).
FIG 4.
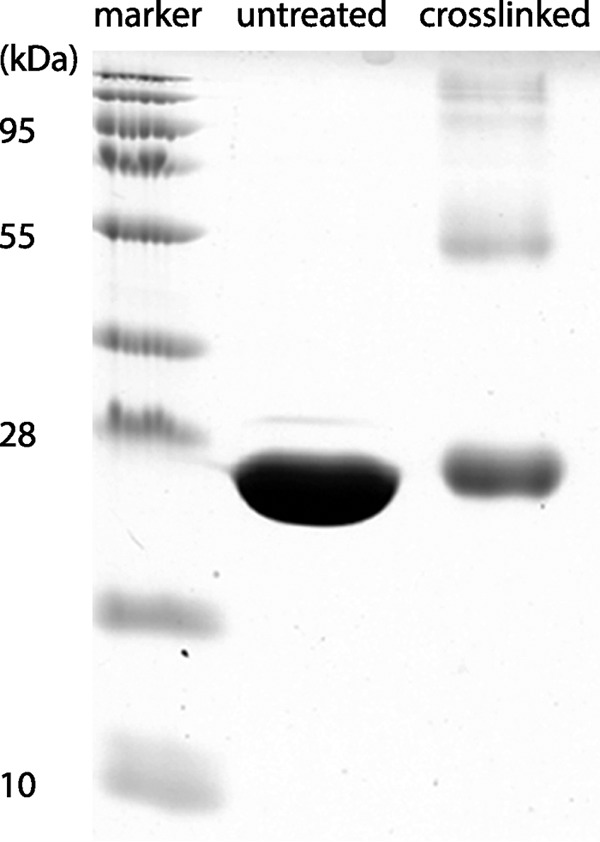
Purification of MfsR-His6 protein. Shown is untreated Ni-NTA-purified MfsR-His6 from E. coli extract and glutaraldehyde-cross-linked protein on a 12% SDS-PAGE gel.
FIG 5.
DNA binding of purified MfsR-His6 to mfsR and mfsA promoter fragments in electrophoretic mobility shift experiments (see Fig. 2A and B for fragment location). Reaction mixtures contain 0.5 nM DNA fragment labeled at the 5′ end with Dyomics681 infrared dye. −, no MfsR-His6 added; +, 0.5 μM MfsR-His6 added. Numbers in the lanes indicate the fragment size.
A single OP1 box is sufficient for MfsR repression.
To determine whether both OP boxes in the upstream regions were necessary for repression, we measured cellular fluorescence from a shorter mfsR promoter fragment (PmfsR-8) (Fig. 2A), fused to a promoterless mcherry and inserted in single copy onto the chromosome of P. putida UWC1. Expression of mCherry from PmfsR-8 in P. putida UWC1 is 3-fold lower than that of PmfsR-11 (Fig. 6, lanes 4 and 8). As before for the longer mfsR promoter fragment (PmfsR-11) (Fig. 6), mCherry expression from PmfsR-8 was high in P. putida UWC1 without ICEclc and in P. putida UWC1 with ICEclc-ΔmfsR (strain 3543) (Fig. 6, lanes 4 and 2). In contrast, mCherry expression from PmfsR-8 was low in P. putida UWC1 with ICEclc, in P. putida UWC1 with ICEclc-ΔmfsR (strain 3543) complemented with a single-copy mini-Tn7 mfsR insertion, and in P. putida UWC1 with mini-Tn7 mfsR (Fig. 6, lanes 1, 3, and 5, respectively). This indicates that the OP1 box is sufficient to exert repression by MfsR, although upstream cis elements may be important for achieving full activity from the mfsR promoter.
FIG 6.
Reporter gene expression of mfsR and mfsA promoter fragments fused to a promoterless mcherry gene in a single copy in P. putida UWC1 with (+) or without (−) ICEclc or ICEclc-ΔmfsR, in the presence (+) or absence (−) of complemented mfsR. Fluorescence was measured 25.5 h after inoculation in MM plus succinate and is normalized by the culture density. For strain numbers, see Table S1 in the supplemental material.
In contrast, deleting the OP1 box in the mfsR promoter resulted in the complete loss of mCherry expression both in P. putida UWC1 without ICEclc and in P. putida UWC1 expressing mfsR (PmfsR-9) (Fig. 6). This suggests not only that interaction of MfsR with OP1 was abolished but also that the promoter itself was damaged. A shorter fragment of the mfsA promoter with only OP3 was not functional in P. putida UWC1 (PmfsA-3) (Fig. 6, lane 9). This also suggests that fragment PmfsA-3 does not encompass the mfsA promoter, which must therefore lie further upstream. Bioinformatic predictions suggested that indeed a −35 and a −10 box may exist upstream and partially overlapping with OP1 (Fig. 2C; see also Fig. S2 in the supplemental material).
Because mfsR is distantly located from mfsABC-orf38184, we finally tested whether the mfsA upstream region had retained the classical promoter configuration of TetR/TetA (i.e., two overlapping but divergent promoters). The fragment PmfsA-6 (Fig. 2B) was cloned in the opposite orientation with respect to the mcherry gene. P. putida UWC1 and P. putida UWC1 miniTn7 mfsR carrying a single copy of the reverse PmfsA-6 promoter fusion, however, did not express any mCherry fluorescence (PmfsA-6R) (Fig. 6, lanes 12 and 13). Therefore, we concluded that there was no promoter in the reverse direction that would help explain the presence of the secondary OP box.
Possible functionality of the MfsABC system.
To study the possible role of the mfs system of ICEclc in tolerance to antimicrobial compounds, we tested mutants of P. putida UWC1 or P. aeruginosa PAO509 carrying one copy of ICEclc in which mfsR or mfsABC was deleted. MICs of P. putida UWC1 strains for 16 antibiotics and 5 other compounds were in most cases not more than 2-fold different for any of the tested compounds in the same assay, with a weak tendency of P. putida UWC1 with ICEclc-ΔmfsR to display the lowest MICs (see Table S3 in the supplemental material). In addition, MICs were determined for triclosan with P. aeruginosa PAO509, a strain in which all five copies of Mex-type efflux systems (i.e., mexAB-oprM, mexCD-oprJ, mexEF-oprN, mexJK, and mexXY) are deleted, resulting in hypersensitivity to triclosan (20). Whereas P. aeruginosa PAO1 is indeed very resistant to triclosan and strain PAO509 extremely sensitive, MICs for triclosan were nondistinguishable between strain PAO509 and PAO509 carrying wild-type ICEclc and even a little lower in PAO509 carrying ICEclc-ΔmfsABC or ICEclc-ΔmfsR (see Table S4).
In addition, potential differences in sensitivities to 960 chemical compounds and conditions between P. putida UWC1 carrying ICEclc-ΔmfsABC and P. putida UWC1 carrying ICEclc were screened in Biolog phenotypic arrays (see Fig. S3 in the supplemental material). Biolog tests indicated that P. putida lacking the mfsABC genes was more sensitive to chromium(III) chloride and carbamate (see Fig. S3). However, independent experimental repetitions measuring growth rates in liquid cultures, growth yields, and ethidium-bromide partitioning in individual cells using flow cytometry (33) were not able to confirm any increase of chromium(III) chloride sensitivity for P. putida UWC1 carrying ICEclc-ΔmfsABC seen with Biolog compared to that of P. putida UWC1 carrying wild-type ICEclc. Finally, none of the aromatic substrates for which catabolism is encoded on ICEclc (7), such as 3-CBA, 3-chlorocatechol, or 2-aminophenol, led to a derepression of the PmfsA-mcherry fusion in P. putida UWC1 carrying ICEclc when added to the culture media (data not shown).
DISCUSSION
Recently, we showed that mfsR controls expression from a cluster of three consecutive regulatory genes (including itself) (Fig. 1A), exerting a global level of control on ICEclc transfer (13). ICEclc transfer occurs only from a small fraction of cells (3 to 5%) that develop “transfer competence” under stationary-phase conditions (34). Deletion of mfsR, as well as uncontrolled expression of the downstream gene tciR, invokes a massive increase of the number of ICEclc transfer-competent cells in stationary phase and subsequent ICEclc transfer (13). We found in this work that MfsR also controls the genes for a putative efflux system on ICEclc. Promoter reporter fusions in a variety of wild-type and mutant backgrounds confirmed that MfsR is solely responsible for the repression of both the mfsR and the mfsA promoters, which are upstream of the first gene of the putative efflux system. This was further confirmed by in vitro DNA binding studies with purified MfsR-His6 protein. As we argue in more detail below, we speculate that this unusual dual control by MfsR is the result of an ancient gene acquisition into an ICEclc precursor mfs system and subsequent gene rearrangement.
The MfsR repressor mechanism is analogous to that of TetR-type transcriptional regulators more in general. TetR-like regulatory systems often occur in a configuration where the regulator gene is oriented divergently from the target gene it controls, interspaced by some 200 to 300 bp (15). This intergenic region typically contains two relatively close paired operator sites with overlapping and divergent promoters. For example, two 15-bp internal palindromic sequences spaced by 11 bp form the TetR operator in the tetR-tetA system (35), whereas for the ttgR-ttgA system the TtgR operator consists of a 30-bp region containing two 11-bp palindromic sequences interspaced by 5 bp (18, 36). Like other TetR family members, MfsR forms homodimers in solution (Fig. 4) and specifically binds to operator sites in both promoters PmfsR and PmfsA (Fig. 5). Unusual in the MfsR system is that both PmfsR and PmfsA promoters carry two more widely separated operator sites (OP1 and OP2 or OP3, ∼90-bp distance) (see Fig. S2 in the supplemental material). Both contain palindromic sequences, which in analogy to other TetR proteins may be key features for MfsR recognition. One of the operator sites (OP1) is completely conserved in both promoters, whereas the others (OP2 and OP3) are slightly divergent to each other and to OP1 (Fig. 2C; see also Fig. S2). A single operator box (OP1) is enough for MfsR to prevent transcription of PmfsR, but deletion of the OP1 sequence completely abolished expression (Fig. 6, PmfsR-9), suggesting that it carries (part of) the promoter sequence itself. Indeed, a possible −35 motif (TTGAAG) is located just upstream of OP1 and a putative −10 motif (TACTAT; 17-bp spacing) is part of OP1 (Fig. 2C; see also Fig. S2). Hence, the role of the second and more upstream OP box, OP2, is not immediately clear. It is reasonable to assume that a second repressor-binding box in the immediate vicinity (94 bp) of the acting repressor (OP1) can increase the local concentration of repressor protein, which may enhance the efficiency of repression (37). Alternatively, OP2 is a now-defunct relic inherited from a past configuration in which mfsR was directly upstream and divergently oriented from mfsABC (see further below for why such a scenario may be likely).
The order of the OP1 and OP3 boxes in the mfsA promoter region is reversed compared to the mfsR promoter (Fig. 2; see also Fig. S2 in the supplemental material). A shorter version of PmfsA (including OP3 but not OP1) did not invoke expression of the fused reporter gene in the absence of MfsR (Fig. 6, PmfsA-3). This suggests that similar to the case for PmfsR, the promoter lies most probably within (at least partly) OP1. Indeed, a putative −10 motif overlaps with OP1 in the mfsA promoter (TATAGT), with a putative −35 motif 17 bp upstream (TTGACA), but these lie in reverse orientation to the −35/−10 motifs around OP1 in the PmfsR context (see Fig. S2). Despite the presence of the OP1 box, the reverse orientation of the mfsA promoter fragment did not lead to expression of a reporter gene (Fig. 6, PmfsA-6R). OP3 in the mfsA promoter may act as an auxiliary repressor-binding site, which possibly further reduces background transcription from the mfsA promoter by acting as a transcriptional roadblock (38). Indeed, expression from the mfsA promoter was consistently lower in all strain backgrounds than that from the mfsR promoter (Fig. 3). The MfsR system with its two distantly regulated promoters is somewhat unusual for TetR-like systems such as tetR-tetA or ttgR-ttgABC with a single divergent regulatory control, but other systems have been characterized in which the TetR member regulates several promoters at a distance (see, for example, Fig. 8 in reference 15).
Interpretation of our experimental data in comparison to analogous existing systems suggests that the dual regulation by MfsR on ICEclc is a result of recent gene acquisition and rearrangements. The closest neighbors of the ICEclc mfs system cluster within a separate phylogenetic branch (see Fig. S1 in the supplemental material). Their MfsR analogs are Ajs_2925 of Acidovorax sp. strain JS42 (93% amino acid identity; GenBank accession number CP000539.1) and HMPREF1171_02726 in Aeromonas hydrophila SSU (93% amino acid identity; GenBank accession number AGWR01000022.1) (see Fig. S4). In these strains the mfsR gene is located directly upstream and divergently oriented from the corresponding mfsABC cluster (Fig. 7). The mfsR-mfsABC genes in Acidovorax sp. strain JS42 are present on ICE(Tn4371)6039 from the Tn4371 family (39, 40). Whether the mfsR-mfsABC genes in A. hydrophila SSU are also part of an ICE cannot be determined yet, since this genome has not been finished. Putative MfsR binding motifs are present in the JS42 and SSU mfs analogs and are well conserved with the ICEclc mfsR and mfsA promoters (Fig. 7; see also Fig. S4). The conserved relative order of OP1 and OP2/OP3 motifs in both JS42 and SSU mfs systems points to OP1 being closer to mfsR and OP2/OP3 being closer to the divergently oriented target mfsA gene (see Fig. S4). When assuming that an ancestor ICEclc mfs system would have looked like mfs of JS42 or SSU, a direct duplication of the mfsR-mfsA intergenic region plus a gene insertion (e.g., of the amn genes on ICEclc) would have resulted in twice the OP1/(OP2/OP3) relative order, with OP1 being located to the mfsR side and OP2/OP3 to the mfsA side. Indeed, small regions of overall 70% conserved nucleotide identity are still present between mfsR and the mfsA upstream region, suggestive of past duplication and rearrangement events (Fig. 7A, shaded regions; see also Fig. S2). The current state of affairs of mfsR and mfsA regulation on ICEclc is thus likely the result of a duplication and subsequent divergence of an ancestor mfs region with only a single pair of OP1 and OP2/OP3 sequences. Selection maintained the promoter in the direction of the mfsR gene but divergence eroded the promoter in the opposite direction, and vice versa for the mfsA gene (see Fig. S2).
FIG 7.
mfsR/mfsABC synteny comparisons. (A) mfsR and mfsABC on ICEclc, where they are separated by 12.7 kb. Details of the promoter regions are illustrated within boxes. Gray shaded boxes point to regions of 70% nucleotide identity between the mfsR and mfsA promoters. (B) mfsR and mfsABC homologs in Acidovorax sp. strain JS42 (ajs_2925). Note that the annotated mfsR in JS42 is longer than its ICEclc counterpart and overlaps with the predicted OP1 box (see details in Fig. S4 in the supplemental material). A transposase of the IS116/IS10/IS902 family has disrupted the sequence of mfsB, and another transposase (of the IS4 family) is located downstream of mfsR. (C) mfsR and mfsABC in A. hydrophila SSU (HMPREF1171_02726). Sequences highlight the similarity between the OP boxes in the different systems (for further details, see Fig. S4).
Taken together, our results show that MfsR controls expression of two transcription units on ICEclc: mfsR-marR-tciR and a putative mfs-type efflux system gene cassette, both through interactions with conserved DNA binding sites. The function of the putative efflux system in antimicrobial compound resistance remains unknown, as does the nature of the chemical signal that may depress MfsR operator binding. We conclude that MfsR must have originally controlled mfsA gene expression in a cis configuration but has acquired a new and additional role as a global regulator of ICE activation through its transcriptional fusion with marR-tciR (13). The new role seems to have arisen as a consequence of recombination events upon the acquisition of the genes for 2-aminophenol degradation (amn) on ICEclc. As we speculated previously, the new MfsR regulatory control may have been instrumental for ICEclc to reach higher transfer rates than for similar ICE in Proteobacteria (13).
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to J. M. Enteza and M. Giddey for their help with the MIC assays. S. Beggah is thanked for her help with testing the possible sensitivities of wild-type and mutant strains to chromium chloride. H. P. Schweizer and L. Randall from Colorado State University are thanked for supplying P. aeruginosa PAO509.
N.P. was supported by a fellowship of the Faculty of Biology and Medicine from the University of Lausanne, Lausanne, Switzerland. This work was supported by grants 3100A0-108199 and 31003A_124711 from the Swiss National Science Foundation.
Footnotes
Published ahead of print 2 September 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.02129-14.
REFERENCES
- 1.Martinez JL, Sanchez MB, Martinez-Solano L, Hernandez A, Garmendia L, Fajardo A, Alvarez-Ortega C. 2009. Functional role of bacterial multidrug efflux pumps in microbial natural ecosystems. FEMS Microbiol. Rev. 33:430–449. 10.1111/j.1574-6976.2008.00157.x. [DOI] [PubMed] [Google Scholar]
- 2.Misra R, Bavro VN. 2009. Assembly and transport mechanism of tripartite drug efflux systems. Biochim. Biophys. Acta 1794:817–825. 10.1016/j.bbapap.2009.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rodríguez-Rojas A, Rodriguez-Beltran J, Couce A, Blazquez J. 2013. Antibiotics and antibiotic resistance: a bitter fight against evolution. Int. J. Med. Microbiol. 303:293–297. 10.1016/j.ijmm.2013.02.004. [DOI] [PubMed] [Google Scholar]
- 4.Van Houdt R, Monchy S, Leys N, Mergeay M. 2009. New mobile genetic elements in Cupriavidus metallidurans CH34, their possible roles and occurrence in other bacteria. Antonie van Leeuwenhoek 96:205–226. 10.1007/s10482-009-9345-4. [DOI] [PubMed] [Google Scholar]
- 5.Norberg P, Bergström M, Jethava V, Dubhashi D, Hermansson M. 2011. The IncP-1 plasmid backbone adapts to different host bacterial species and evolves through homologous recombination. Nat. Commun. 2:268. 10.1038/ncomms1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bouzat JL, Hoostal MJ. 2013. Evolutionary analysis and lateral gene transfer of two-component regulatory systems associated with heavy-metal tolerance in bacteria. J. Mol. Evol. 76:267–279. 10.1007/s00239-013-9558-z. [DOI] [PubMed] [Google Scholar]
- 7.Gaillard M, Vallaeys T, Vorhölter FJ, Minoia M, Werlen C, Sentchilo V, Pühler A, van der Meer JR. 2006. The clc element of Pseudomonas sp. strain B13, a genomic island with various catabolic properties. J. Bacteriol. 188:1999–2013. 10.1128/JB.188.5.1999-2013.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miyazaki R, Minoia M, Pradervand N, Sulser S, Reinhard F, van der Meer JR. 2012. Cellular variability of RpoS expression underlies subpopulation activation of an integrative and conjugative element. PLoS Genet. 8:e1002818. 10.1371/journal.pgen.1002818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miyazaki R, Bertelli C, Benaglio P, Canton J, De Coi N, Gharib WH, Gjoksi B, Goesmann A, Greub G, Harshman K, Linke B, Mikulic J, Mueller L, Nicolas D, Robinson-Rechavi M, Rivolta C, Roggo C, Roy S, Sentchilo V, Siebenthal AV, Falquet L, van der Meer JR. 2014. Comparative genome analysis of Pseudomonas knackmussii B13, the first bacterium known to degrade chloroaromatic compounds. Environ. Microbiol. 10.1111/1462-2920.12498. [DOI] [PubMed] [Google Scholar]
- 10.Ravatn R, Studer S, Springael D, Zehnder AJB, van der Meer JR. 1998. Chromosomal integration, tandem amplification, and deamplification in Pseudomonas putida F1 of a 105-kilobase genetic element containing the chlorocatechol degradative genes from Pseudomonas sp. strain B13. J. Bacteriol. 180:4360–4369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Müller TA, Werlen C, Spain J, van der Meer JR. 2003. Evolution of a chlorobenzene degradative pathway among bacteria in a contaminated groundwater mediated by a genomic island in Ralstonia. Environ. Microbiol. 5:163–173. 10.1046/j.1462-2920.2003.00400.x. [DOI] [PubMed] [Google Scholar]
- 12.Chain PS, Denef VJ, Konstantinidis KT, Vergez LM, Agullo L, Reyes VL, Hauser L, Cordova M, Gomez L, Gonzalez M, Land M, Lao V, Larimer F, LiPuma JJ, Mahenthiralingam E, Malfatti SA, Marx CJ, Parnell JJ, Ramette A, Richardson P, Seeger M, Smith D, Spilker T, Sul WJ, Tsoi TV, Ulrich LE, Zhulin IB, Tiedje JM. 2006. Burkholderia xenovorans LB400 harbors a multi-replicon, 9.73-Mbp genome shaped for versatility. Proc. Natl. Acad. Sci. U. S. A. 103:15280–15287. 10.1073/pnas.0606924103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pradervand N, Sulser S, Delavat F, Miyazaki R, Lamas I, van der Meer JR. 2014. An operon of three transcriptional regulators controls horizontal gene transfer of the integrative and conjugative element ICEclc in Pseudomonas knackmussii B13. PLoS Genet. 10:e1004441. 10.1371/journal.pgen.1004441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grkovic S, Brown MH, Skurray RA. 2002. Regulation of bacterial drug export systems. Microbiol. Mol. Biol. Rev. 66:671–701. 10.1128/MMBR.66.4.671-701.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ramos JL, Martinez-Bueno M, Molina-Henares AJ, Teran W, Watanabe K, Zhang X, Gallegos MT, Brennan R, Tobes R. 2005. The TetR family of transcriptional repressors. Microbiol. Mol. Biol. Rev. 69:326–356. 10.1128/MMBR.69.2.326-356.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shimada T, Hirao K, Kori A, Yamamoto K, Ishihama A. 2007. RutR is the uracil/thymine-sensing master regulator of a set of genes for synthesis and degradation of pyrimidines. Mol. Microbiol. 66:744–757. 10.1111/j.1365-2958.2007.05954.x. [DOI] [PubMed] [Google Scholar]
- 17.Quiñones B, Pujol CJ, Lindow SE. 2004. Regulation of AHL production and its contribution to epiphytic fitness in Pseudomonas syringae. Mol. Plant Microbe Interact. 17:521–531. 10.1094/MPMI.2004.17.5.521. [DOI] [PubMed] [Google Scholar]
- 18.Duque E, Segura A, Mosqueda G, Ramos JL. 2001. Global and cognate regulators control the expression of the organic solvent efflux pumps TtgABC and TtgDEF of Pseudomonas putida. Mol. Microbiol. 39:1100–1106. 10.1046/j.1365-2958.2001.02310.x. [DOI] [PubMed] [Google Scholar]
- 19.Terán W, Krell T, Ramos JL, Gallegos MT. 2006. Effector-repressor interactions, binding of a single effector molecule to the operator-bound TtgR homodimer mediates derepression. J. Biol. Chem. 281:7102–7109. 10.1074/jbc.M511095200. [DOI] [PubMed] [Google Scholar]
- 20.Mima T, Joshi S, Gomez-Escalada M, Schweizer HP. 2007. Identification and characterization of TriABC-OpmH, a triclosan efflux pump of Pseudomonas aeruginosa requiring two membrane fusion proteins. J. Bacteriol. 189:7600–7609. 10.1128/JB.00850-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gerhardt P, Murray RGE, Costilow RN, Nester EW, Wood WA, Krieg NR, Briggs Phillips G. (ed). 1981. Manual of methods for general bacteriology. American Society for Microbiology, Washington, DC. [Google Scholar]
- 22.Sambrook J, Russell DW. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 23.Miyazaki R, van der Meer JR. 2011. A dual functional origin of transfer in the ICEclc genomic island of Pseudomonas knackmussii B13. Mol. Microbiol. 79:743–758. 10.1111/j.1365-2958.2010.07484.x. [DOI] [PubMed] [Google Scholar]
- 24.Choi KH, Gaynor JB, White KG, Lopez C, Bosio CM, Karkhoff-Schweizer RR, Schweizer HP. 2005. A Tn7-based broad-range bacterial cloning and expression system. Nat. Methods 2:443–448. 10.1038/nmeth765. [DOI] [PubMed] [Google Scholar]
- 25.Clinical and Laboratory Standards Institute. 2006. Performance standards for antimicrobial susceptibility testing; approved standard. Sixteenth informational supplement. Document M100-S16. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 26.Christensen BB, Sternberg C, Andersen JB, Eberl L, Moller S, Givskov M, Molin S. 1998. Establishment of new genetic traits in a microbial biofilm community. Appl. Environ. Microbiol. 64:2247–2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gaillard M, Pradervand N, Minoia M, Sentchilo V, Johnson DR, van der Meer JR. 2010. Transcriptome analysis of the mobile genome ICEclc in Pseudomonas knackmussii B13. BMC Microbiol. 10:153. 10.1186/1471-2180-10-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dubey AK, Baker CS, Suzuki K, Jones AD, Pandit P, Romeo T, Babitzke P. 2003. CsrA regulates translation of the Escherichia coli carbon starvation gene, cstA, by blocking ribosome access to the cstA transcript. J. Bacteriol. 185:4450–4460. 10.1128/JB.185.15.4450-4460.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bi D, Xu Z, Harrison EM, Tai C, Wei Y, He X, Jia S, Deng Z, Rajakumar K, Ou HY. 2012. ICEberg: a web-based resource for integrative and conjugative elements found in bacteria. Nucleic Acids Res. 40:D621–D626. 10.1093/nar/gkr846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Abbott JC, Aanensen DM, Bentley SD. 2007. WebACT: an online genome comparison suite. Methods Mol. Biol. 395:57–74. 10.1007/978-1-59745-514-5_4. [DOI] [PubMed] [Google Scholar]
- 31.Krell T, Teran W, Mayorga OL, Rivas G, Jiménez M, Daniels C, Molina-Henares AJ, Martinez-Bueno M, Gallegos MT, Ramos JL. 2007. Optimization of the palindromic order of the TtgR operator enhances binding cooperativity. J. Mol. Biol. 369:1188–1199. 10.1016/j.jmb.2007.04.025. [DOI] [PubMed] [Google Scholar]
- 32.Wenzel M, Lang K, Günther T, Bhandari A, Weiss A, Luichev P, Szentgyörgyi E, Kranzunsch B, Göttfert M. 2012. Characterization of the flavonoid-responsive regulator FrrA and its binding sites. J. Bacteriol. 194:2363–2370. 10.1128/JB.06567-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Czechowska K, Reimmann C, van der Meer JR. 2013. Characterization of a MexAB-OprM efflux system necessary for productive metabolism of Pseudomonas azelaica HBP1 on 2-hydroxybiphenyl. Front. Microbiol. 4:203. 10.3389/fmicb.2013.00203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reinhard F, Miyazaki R, Pradervand N, van der Meer JR. 2013. Cell differentiation to “mating bodies” induced by an integrating and conjugative element in free-living bacteria. Curr. Biol. 23:255–259. 10.1016/j.cub.2012.12.025. [DOI] [PubMed] [Google Scholar]
- 35.Hillen W, Berens C. 1994. Mechanisms underlying expression of Tn10 encoded tetracycline resistance. Annu. Rev. Microbiol. 48:345–369. 10.1146/annurev.mi.48.100194.002021. [DOI] [PubMed] [Google Scholar]
- 36.Terán W, Felipe A, Segura A, Rojas A, Ramos JL, Gallegos MT. 2003. Antibiotic-dependent induction of Pseudomonas putida DOT-T1E TtgABC efflux pump is mediated by the drug binding repressor TtgR. Antimicrob. Agents Chemother. 47:3067–3072. 10.1128/AAC.47.10.3067-3072.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hammar P, Leroy P, Mahmutovic A, Marklund EG, Berg OG, Elf J. 2012. The lac repressor displays facilitated diffusion in living cells. Science 336:1595–1598. 10.1126/science.1221648. [DOI] [PubMed] [Google Scholar]
- 38.He B, Zalkin H. 1992. Repression of Escherichia coli purB is by a transcriptional roadblock mechanism. J. Bacteriol. 174:7121–7127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Haigler BE, Wallace WH, Spain JC. 1994. Biodegradation of 2-nitrotoluene by Pseudomonas sp. strain JS42. Appl. Environ. Microbiol. 60:3466–3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ryan MP, Pembroke JT, Adley CC. 2009. Novel Tn4371-ICE like element in Ralstonia pickettii and genome mining for comparative elements. BMC Microbiol. 9:242. 10.1186/1471-2180-9-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.