Abstract
Thiopeptides are small (12- to 17-amino-acid), heavily modified peptides of bacterial origin. This antibiotic family, with more than 100 known members, is characterized by the presence of sulfur-containing heterocyclic rings and dehydrated residues within a macrocyclic peptide structure. Thiopeptides, including micrococcin P1, have garnered significant attention in recent years for their potent antimicrobial activity against bacteria, fungi, and even protozoa. Micrococcin P1 is known to target the ribosome; however, like those of other thiopeptides, its biosynthesis and mechanisms of self-immunity are poorly characterized. We have discovered an isolate of Staphylococcus epidermidis harboring the genes for thiopeptide production and self-protection on a 24-kb plasmid. Here we report the characterization of this plasmid, identify the antimicrobial peptide that it encodes, and provide evidence of a target replacement-mediated mechanism of self-immunity.
INTRODUCTION
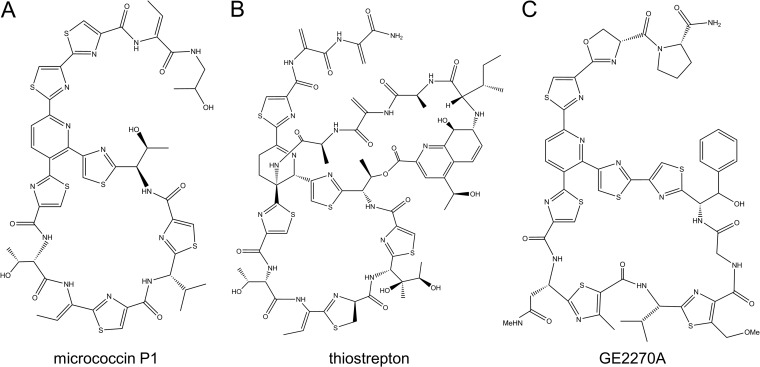
Thiopeptides are an intriguing class of peptide antibiotics that have been investigated for more than 6 decades for their antimicrobial activity. Widely produced by bacteria in nature, thiopeptides possess complex structures that arise from extensive posttranslational tailoring (Fig. 1). A hallmark of thiopeptide structure is the presence of numerous thiazolyl groups arranged around a macrocyclic structure with a pyridine, piperidine, or dehydropiperidine core (1). It is also common for thiopeptides to possess dehydro-amino acids (Dha and Dhb) and other modifications directed by specialized biosynthetic enzymes. Detailed reviews of thiopeptide biosynthesis can be found elsewhere (2–5). These alterations make thiopeptides some of the most densely posttranslationally modified peptides known (6).
FIG 1.
Structures of various thiopeptide antibiotics. Panels: A, MP1; B, thiostrepton; C, GE2270A.
Thiopeptides are potent inhibitors of protein synthesis, particularly among Gram-positive bacteria. In general, thiopeptides have been shown to act by two distinct mechanisms, depending on the size of the macrocycle; 26-membered macrocycles (e.g., micrococcin P1 [MP1] and thiostrepton) (Fig. 1) bind at a cleft between ribosomal protein L11 and the 23S rRNA known as the GTPase-associated center (7–9). This region is a critical binding site for elongation factor G (EF-G), which is necessary for ribosome translocation along template mRNA. When thiopeptides are bound here, EF-G is prevented from binding and protein synthesis is halted (10–13). Thiopeptides with 29-membered rings (e.g., GE2270A) (Fig. 1) bind to elongation factor Tu (EF-Tu), blocking its aminoacyl-tRNA binding site, thus preventing peptide elongation (14–16). Beyond their intrinsic antibacterial properties, thiopeptides (especially thiostrepton and MP1) have been investigated for a variety of other applications, including antifungal (17), antiplasmodial (18–21), and anticancer properties (22–24).
Mechanisms of thiopeptide self-immunity are poorly understood. The majority of thiopeptide-producing bacteria have been shown to antagonize closely related species or even members of the same species, implying that mechanisms of immunity are encoded alongside the genes for biosynthesis. Methylation of 23S rRNA has been shown to confer self-immunity in thiostrepton-producing strains of Streptomyces azureus and nosiheptide-producing strains of Streptomyces actuosus (25). However, for many other thiopeptide producers, the precise self-immunity mechanisms remain unknown or untested.
Here we report the characterization of a novel thiopeptide gene cluster and mechanism of self-immunity in an MP1-producing isolate of Staphylococcus epidermidis, strain 115. First discovered in 1983, this avian isolate was reported to produce an interfering compound that inhibited virulent strains of S. aureus (26). In subsequent studies, researchers devised means to extract the compound in order to evaluate certain physical characteristics (27). However, they were unable to identify the compound or map the genes required for antimicrobial production. Using this strain, we have identified the compound as MP1, defined the MP1 biosynthetic gene cluster, and addressed the mechanism by which strain 115 protects itself from MP1 toxicity.
MATERIALS AND METHODS
Target range assays.
The S. epidermidis 115 target spectrum was assessed by using either the flanking-patch assay or the spot-on-lawn assay. Precise medium types varied according to the strain being tested; for details, see Table S1 in the supplemental material. Flanking-patch assays were performed by growing S. epidermidis 115 (source patch) for 18 h at 37°C before patching indicator strains 5 mm from the source patch (see Fig. 2A). Strains were incubated for a further 12 to 24 h before assessment of resistance or sensitivity. For assays with purified MP1, spot-on-lawn assays were performed. These assays were conducted by spreading 150 μl of a 1:50-diluted overnight culture of the test strain on a compatible growth medium. Plates were allowed to dry before applying 5-μl spots of MP1 dilutions. Spots were allowed to dry before incubation of the plates for 18 to 24 h at 37°C. Strains were deemed sensitive when a clear zone of inhibition was present surrounding MP1 spots but absent around vehicle-only spots.
FIG 2.
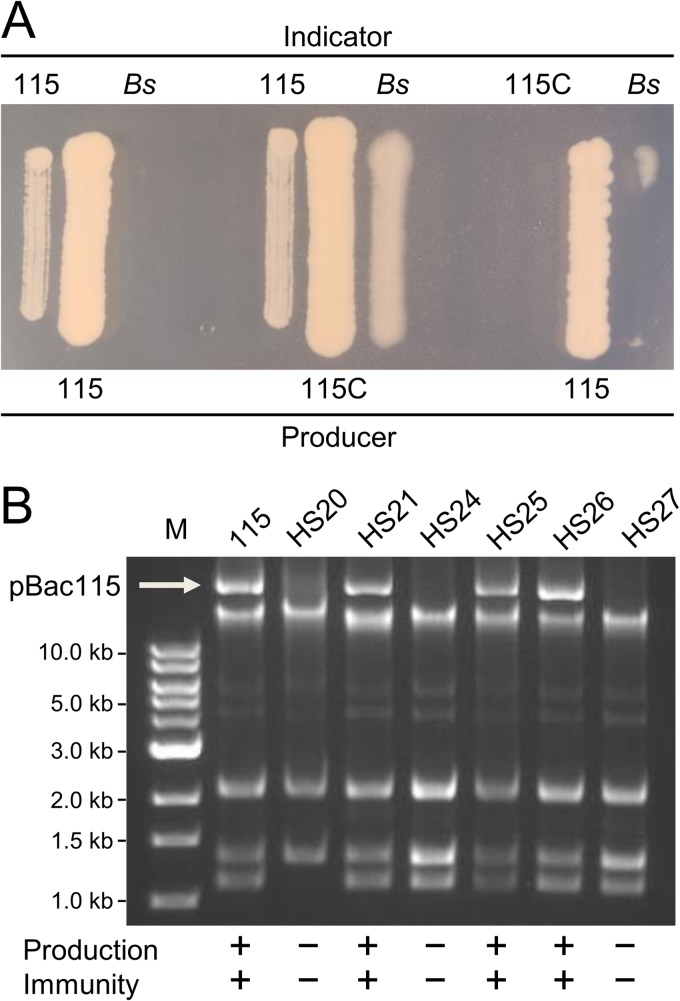
The antimicrobial activity of S. epidermidis strain 115 is plasmid encoded. (A) Examples of flanking-patch assays for production and immunity. In each triplet, the central patch was applied to the agar 18 h prior to the flanking patches. 115C is a pBac115-cured derivative. Bs corresponds to B. subtilis 168, a susceptible indicator strain. (B) Plasmid preparations from S. epidermidis 115 and HS derivatives. Lane M, molecular size markers.
MP1 purification.
S. epidermidis strain 115 was cultured on 2×YT agar and then harvested by scraping plates into 0.9% NaCl. Harvested cells were pelleted in 50-ml Oak Ridge centrifuge tubes (Nalgene), weighed, and then resuspended in 7 M urea (4 ml/1-g cell pellet). The suspension was incubated at 25°C for 5 min with intermittent vortexing and then centrifuged at 12,000 × g for 10 min. MP1-containing supernatant was transferred to a new tube and recentrifuged to remove remaining particulate material. The final supernatant was heat treated to kill residual cells (65°C, 20 min) and then tested for activity by spot-on-lawn assay.
Urea-extracted MP1 was further purified with a Sep-Pak C18 Plus Long cartridge (Waters). The cartridge was equilibrated with 50% and then 20% acetonitrile (ACN). Approximately 30 ml of extract was applied to the cartridge before it was washed with 30% ACN. Elution was accomplished with 50% ACN. All solvents were acidified with 0.1% formic acid (FA).
Final purification was performed by reversed-phase high-performance liquid chromatography (RP-HPLC) on an Agilent 300SB-C8 column. MP1 was eluted isocratically with 35% (vol/vol) ACN–0.1% (vol/vol) FA over 16 min at a flow rate of 0.8 ml/min. The active fraction (retention time, 10 to 12.5 min) was collected for further analysis. All purification steps were performed at room temperature. In parallel, the same steps were followed for pBac115-cured S. epidermidis strain 115C. This isogenic control strain is incapable of antimicrobial compound production (see below).
Plasmid curing.
To generate plasmid-cured derivatives, overnight cultures of S. epidermidis 115 were diluted 4 × 104-fold into fresh tryptic soy broth containing 0.004% SDS and cultured with shaking (225 rpm) at 45°C overnight. Cultures were plated on tryptic soy agar and incubated for 18 to 24 h at 37°C. Heat/SDS-surviving (HS) strains were tested by flanking-patch assay for MP1 production and sensitivity. The plasmid profiles of HS strains were evaluated by standard alkaline lysis miniprep after treatment of cells with lysostaphin (Sigma) at a concentration of 120 μg/ml for 30 min at 37°C. The resulting DNA was analyzed on a 1% agarose gel.
Plasmid sequencing and annotation.
Plasmid minipreps of strain 115 were generated as described above and submitted for 454 shotgun sequencing (Roche) at the Brigham Young University DNA Sequencing Center. Sequence assembly was accomplished with GS De Novo Assembler (Roche) with localized PCR and Sanger sequencing implemented to resolve ambiguous areas. Contigs representing portions of pBac115 were identified on the basis of successful PCR amplification from strain 115 and no amplification from pBac115-cured strain 115C. These tests are summarized in Table S2 in the supplemental material. Annotation of pBac115 was accomplished by using GeneMark.hmm prokaryotic gene prediction software (28) to find open reading frames (ORFs) and BLAST analysis (http://www.ncbi.nlm.nih.gov/blast) to assign predicted functions. We confirmed the sequence of the MP1 biosynthetic gene cluster by resequencing the entire region by Sanger sequencing.
MS and NMR analysis.
The identity of purified MP1 was determined by a combination of high-resolution electrospray ionization mass spectrometry (ESI-MS) and high-resolution nuclear magnetic resonance (NMR) spectroscopy. ESI-MS was performed at the MS Core Facility (Brigham Young University) with an Agilent MSD time-of-flight instrument with ESI. Theoretical MP1 masses were calculated by mMass (29). One- and two-dimensional NMR spectra were collected on a Varian NMR System 500-MHz spectrometer equipped with a OneNMR probe and ProTune (NMR Facility, Brigham Young University). All NMR experiments are summarized in Table S3 in the supplemental material. The sample used for NMR analysis was composed of approximately 14 mg of MP1 dissolved in 0.8 ml of hexadeuterodimethyl sulfoxide in a 5-mm NMR tube (∼10 mM). Both the 13C and 15N spectra were acquired at natural abundance. The spectra were analyzed with VNMRJ 3.2 software.
Genetic manipulation of Bacillus subtilis.
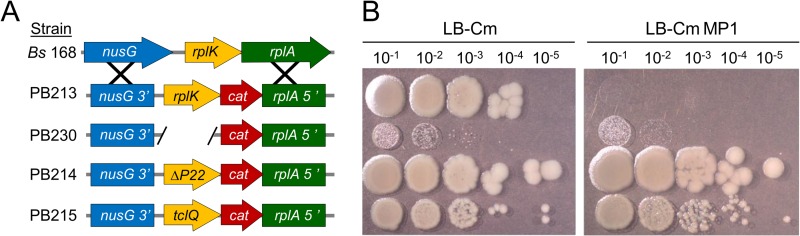
Plasmid pRB014 (see Fig. S1A in the supplemental material) was used to generate the B. subtilis strain expressing tclQ in trans at the amyE locus. A similarly constructed plasmid (pRB015) (see Fig. S1B) was used to introduce the tclWX cassette into the amyE locus. These preliminary experiments were performed with B. subtilis strain SCK6, which is modified for xylose-inducible competence. The more definitive allelic exchange strains depicted in Fig. 6 were created with B. subtilis strain 168. Strains PB213, PB230, PB214, and PB215 resulted from rplK allelic replacement with plasmids pRB029, pRB031, pRB030, and pRB024, respectively. For the full plasmid sequences, see Fig. S1. The allelic replacements were designed to maintain the native overall transcriptional unit so that the expression of introduced alleles and downstream genes would be left unperturbed.
FIG 6.
Allelic-exchange experiments at the L11-encoding rplK locus. (A) Map of rplK alterations of the B. subtilis 168 (Bs) chromosome, with strain names indicated. ΔP22 is an abbreviation for the B. subtilis rplKΔP22 allele, which is known to give resistance to MP1. The tclQ allele was amplified from S. epidermidis 115. (B) Spot dilution assays showing growth at 24 h on LB agar with or without MP1 (18 μg/ml). The rows correspond to the genotypes indicated in panel A. Cm, chloramphenicol.
Plasmids were constructed by standard techniques with enzymes purchased from New England BioLabs. The high-fidelity polymerases Q5 (New England BioLabs) and Pfx50 (Invitrogen) were used for insert amplification. All constructs and genetic insertions or replacements were confirmed by Sanger sequencing. Custom oligonucleotides were purchased from Invitrogen.
Spot dilution assays.
Overnight liquid cultures of B. subtilis mutants were normalized to an optical density at 600 nm of 0.1 before 10-fold serial dilutions were performed. Five-microliter spots were made on LB agar supplemented with chloramphenicol (5 μg/ml), with or without MP1 (18 μg/ml). The spots were allowed to dry before the plates were incubated at 37°C for 24 h.
Nucleotide sequence accession number.
The complete sequence of pBac115 has been deposited in the GenBank database (http://www.ncbi.nlm.nih.gov/GenBank/) under accession number KM613043.
RESULTS
S. epidermidis 115 produces a potent, broad-spectrum antimicrobial compound.
In order to test the target range of strain 115, we devised a flanking-patch assay (Fig. 2A), which we used on a broad panel of bacterial isolates in our possession. We observed that strain 115 secretes a diffusible compound that inhibits the growth of diverse Gram-positive bacteria (Table 1). Of particular interest was the observation that many clinically relevant pathogens were inhibited, including vancomycin-resistant Enterococcus faecalis (VRE), methicillin-resistant S. aureus (MRSA), Clostridium difficile, Streptococcus agalactiae, Bacillus anthracis, Listeria monocytogenes, and Streptococcus pyogenes. Interestingly, none of the Gram-negative bacteria tested were sensitive. This activity profile is consistent with that of many known thiopeptides (4).
TABLE 1.
Inhibitory spectrum of strain 115a
| Species | No. of strains |
|
|---|---|---|
| Sensitive | Resistant | |
| Gram-positive bacteria | ||
| Staphylococcus spp. | ||
| S. aureus | 2 | 0 |
| S. aureus (MRSA) | 6 | 0 |
| S. epidermidis | 2 | 0 |
| Enterococcus spp. | ||
| E. faecalis | 1 | 0 |
| E. faecalis (VRE) | 1 | 0 |
| Streptococcus spp. | ||
| S. agalactiae | 1 | 0 |
| S. pyogenes | 2 | 0 |
| S. pneumoniae | 1 | 0 |
| Bacillus spp. | ||
| B. anthracis | 1 | 0 |
| B. brevis | 1 | 0 |
| B. cereus | 0 | 1 |
| B. licheniformis | 1 | 0 |
| B. megaterium | 1 | 0 |
| B. mycoides | 1 | 0 |
| B. sphaericus | 1 | 0 |
| B. subtilis | 4 | 1 |
| B. thuringiensis | 2 | 0 |
| Clostridium spp. | ||
| C. difficile | 1 | 0 |
| C. perfringens | 1 | 0 |
| Listeria monocytogenes | 2 | 0 |
| Paenibacillus polymyxa | 1 | 0 |
| Gram-negative bacteria | ||
| Escherichia coli | 0 | 2 |
| Yersinia pseudotuberculosis | 0 | 1 |
| Burkholderia thailandensis | 0 | 1 |
| Acid-fast bacterium Mycobacterium smegmatis | 1 | 0 |
For detailed strain information, see Table S1 in the supplemental material.
The antimicrobial activity of S. epidermidis 115 is plasmid encoded.
Earlier speculation suggested that antimicrobial production by strain 115 may be plasmid mediated (27). We investigated this hypothesis by screening for derivatives of S. epidermidis 115 that lose antimicrobial production under conditions that promote plasmid curing. Following growth at elevated temperatures and in the presence of low concentrations of SDS, we successfully screened for HS derivative strains that had lost the ability to inhibit the growth of B. subtilis 168. The loss of antimicrobial production always coincided with loss of resistance to the compound (strain 115C in Fig. 2). Plasmid preparations from HS strains revealed that derivatives deficient in both the production and immunity phenotypes were devoid of a high-molecular-weight plasmid that we have named pBac115 (Fig. 2B). We were able to replicate this phenomenon on repeated occasions; in every instance, the loss of antimicrobial production and immunity was perfectly correlated with the absence of pBac115.
pBac115 contains thiopeptide biosynthetic genes.
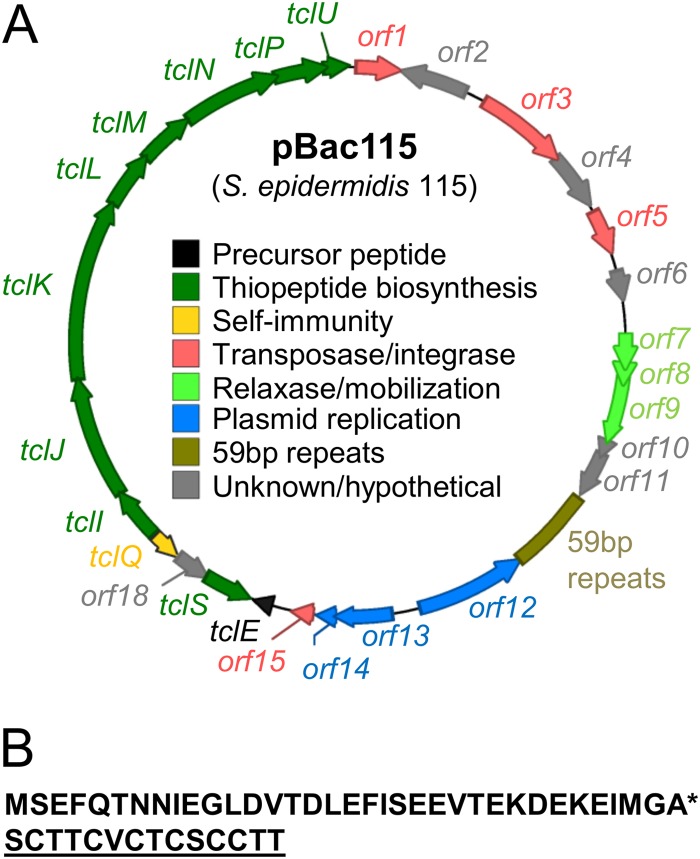
In order to identify the plasmid-encoded biosynthetic genes, we sequenced DNA from an alkaline-lysis plasmid preparation derived from S. epidermidis 115. This preparation appeared to contain at least three low-molecular-weight plasmids, as well as pBac115. A second high-molecular-weight band was attributed to chromosomal DNA fragments (Fig. 2B). The resulting 454 pyrosequencing data allowed us to discern a circular 24-kb plasmid consistent with the properties of pBac115. First, in our panel of HS strains (Fig. 2B), those harboring the pBac115 band also tested positive for the 24-kb molecule by PCR, whereas those lacking the pBac115 band tested negative for the 24-kb molecule. Second, annotation of the 24-kb plasmid revealed 27 putative ORFs, including an 11-gene cluster predicted to encode thiopeptide biosynthetic functions (tcl genes) (Fig. 3A).The tcl designations are according to reference 6. For a detailed report of predicted pBac115 gene functions, see Table S4 in the supplemental material.
FIG 3.
pBac115 contains thiopeptide biosynthetic (tcl) genes. (A) Plasmid map of pBac115 with annotation based upon results from BLASTx and direct alignments with previously identified tcl genes. (B) Amino acid sequence of the TclE precursor peptide. The asterisk indicates the predicted cleavage site; the core peptide, which undergoes modification to produce the mature thiopeptide, is underlined.
Thiopeptide biosynthesis consists of extensive posttranslational tailoring of a structural peptide by multiple biosynthetic enzymes. Although the biosynthesis of thiopeptides is incompletely understood, thiopeptide maturation is thought to occur on a precursor peptide scaffold from which the mature modified peptide is cleaved by a final macrocyclization reaction (30). Peptide modification steps include dehydration of Ser and Thr residues, Cys cyclization to give thiazolyl groups, and the joining of Dha pairs to form the central macrocycle, consequently cleaving the leader peptide (1, 31). On pBac115, the short ORF tclE drew our attention as potentially encoding a precursor peptide (Fig. 3B) because the C-terminal 14 amino acids are identical to the core peptide from which a certain family of thiopeptides (thiocillins) are derived in other organisms (6).
Purification of the antimicrobial activity of S. epidermidis 115.
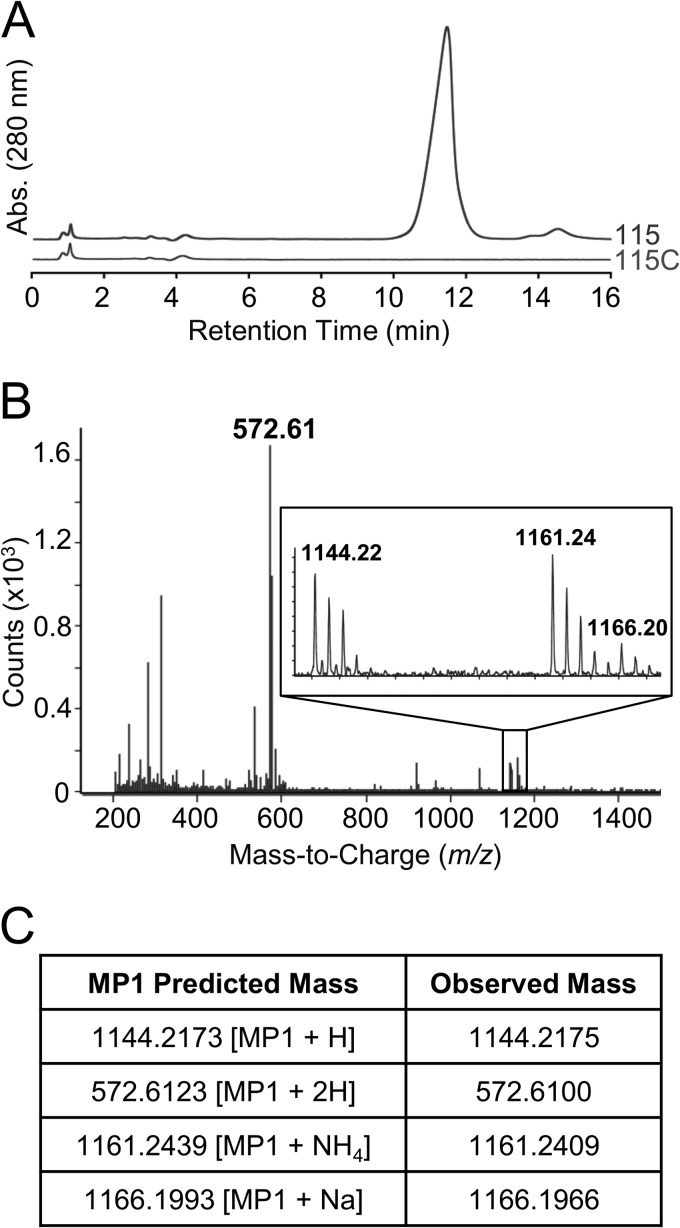
According to previous reports, the antimicrobial compound produced by strain 115 can be extracted by suspending cell pellets in 7 M urea (27). We used this approach to obtain high yields of a urea-extracted compound with high biological activity. We further enriched this extract by reversed-phase cartridge chromatography. We hypothesized that this purified activity would be found in strain 115 but not in pBac115-cured strain 115C. To test this, we performed parallel purification procedures with both strains. As expected, the strain 115 extract retained antimicrobial activity in a spot-on-lawn assay even when diluted 1,000-fold, whereas the strain 115C sample exhibited no detectable activity. We analyzed the strain 115 and 115C samples by HPLC and observed a very prominent peak present in the strain 115 sample that was absent from the strain 115C sample (Fig. 4A). In spot-on-lawn assays, fractions corresponding to this peak exhibited antimicrobial activity while all other fractions were devoid of activity.
FIG 4.
Biochemical analysis of S. epidermidis 115 antimicrobial activity. (A) RP-HPLC analysis of purified material from strain 115 and material similarly prepared from pBac115-cured strain 115C. Abs, absorbance. (B) ESI-MS fragmentation of the major HPLC peak from strain 115. The inset shows a closeup of the region from m/z 1,142 to m/z 1,168. (C) Comparison of observed m/z values with those predicted for MP1.
Structural analysis identifies the unknown compound as MP1.
The antimicrobial compound was analyzed by MS and NMR. By high-resolution ESI-MS, we observed dominant peaks at m/z ratios of 572.61, 1,144.22, 1,161.24, and 1,166.20 (Fig. 4B). These peaks are remarkably consistent with the thiopeptide MP1 in various ionization states: the dually protonated form (572.61), the singly protonated form (1,144.22), the ammonium adduct (1,161.24), and the sodium adduct (1,166.20) (Fig. 4C). MP1, also produced by Bacillus cereus strain ATCC 14579 (6), is known to be synthesized on the peptide scaffold SCTTCVCTCSCCTT, which we noted above is encoded by tclE on pBac115. The pBac115-dependent compound was finally analyzed by NMR, which confirmed its identity as MP1 (see Tables S3 and S5 and Fig. S2 in the supplemental material). The structure of MP1 is shown in Fig. 1A.
The plasmid pBac115 harbors only a single immunity gene.
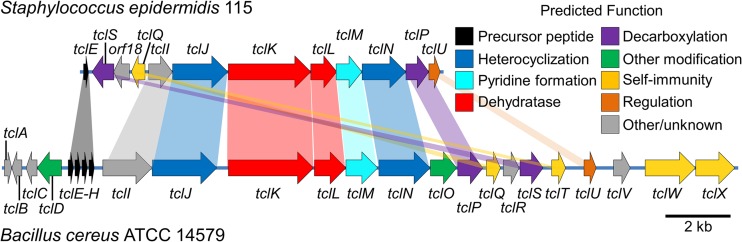
Having identified the antimicrobial compound known as MP1, we compared the putative MP1-encoding gene cluster on pBac115 with the only other empirically tested MP1 biosynthetic gene cluster, the tcl cluster from B. cereus ATCC 14579 (6). Composed of 24 tcl genes within a 22-kb region of the B. cereus chromosome, the tcl cluster reportedly gives rise to eight different thiopeptides (including MP1) from the SCTTCVCTCSCCTT scaffold. Genetic comparisons of pBac115 and the B. cereus gene cluster identified 11 genes clustered within an 11-kb region of pBac115 with homology to tcl genes (Fig. 5; see Table S4 in the supplemental material). Of these 11, only one copy of the precursor peptide was identified (tclE), in contrast to the four tandem copies (tclE to -H) in B. cereus. Additionally, of the four genes presumed to encode immunity functions in B. cereus (tclQ, tclT, tclW, and tclX), the pBac115 cluster contains only a single gene; a homolog of tclQ and tclT (tclQ and tclT are identical copies of the same gene in B. cereus). This observation led us to hypothesize that a single gene (here referred to as tclQ) may be sufficient to confer self-immunity in S. epidermidis 115.
FIG 5.
Comparisons of the tcl gene clusters from S. epidermidis 115 and B. cereus ATCC 14579. Connectors indicate homologous genes based on BLASTp comparisons of their predicted protein products. Predicted functions are shown in the color-coded key.
TclQ confers resistance to MP1 in B. subtilis.
TclQ is a homolog of 50S ribosomal protein L11, which is located near the ribosomal GTPase center (32, 33) and is known to be involved in the mechanism of action of MP1 (9). We hypothesized that TclQ confers immunity by replacing native L11 to protect ribosomes from MP1. To test our “target replacement” hypothesis, we evaluated whether TclQ could confer MP1 resistance in the model B. subtilis strain 168. This strain is sensitive to MP1, and we have determined that ribosomal protein L11 is a major target, since spontaneous resistance is invariably associated with mutations in the L11-encoding gene rplK (data not shown). In multiple independent MP1-resistant isolates, we see the same rplK mutation that results in deletion of Pro-22 (referred to here as the ΔP22 allele).
As an initial test of our target replacement hypothesis, the tclQ gene from pBac115 was expressed in trans from its native promoter at the B. subtilis amyE locus. This strain was modestly (10-fold) more resistant to MP1 than the wild type was. In a similar experiment, tclWX, the putative efflux system from B. cereus, did not confer detectable resistance (data not shown). We reasoned that TclQ expression in trans may not confer very high resistance in B. subtilis because of competition for ribosomal incorporation with the native L11 protein. According to this logic, replacement of rplK (the L11-encoding gene) with tclQ would lead to much higher levels of resistance, as competition for ribosomal incorporation is eliminated. As shown in Fig. 6, replacement of rplK with tclQ allows nearly normal growth and high-level resistance to MP1. MP1 dilution experiments show that the replacement strain was ≥200-fold more resistant than the wild type. This increased resistance suggests that TclQ substitutes for L11 on the ribosome, but we could not rule out the possibility that removal of L11 would lead to high resistance regardless of what replaces it. To address this, we constructed an isogenic strain in which the rplK ORF was simply removed from the chromosome and not replaced with tclQ. This strain was viable but very slow growing. It was, however, noticeably resistant to MP1 (Fig. 6B). The robust growth observed in the tclQ replacement strain shows that tclQ functionally complements the rplK mutant for normal protein synthesis and growth. This strongly suggests that the TclQ protein is incorporated into the ribosome in place of L11 and also confers resistance to MP1.
DISCUSSION
We have shown that S. epidermidis 115 produces the broad-spectrum antibiotic MP1. This is the first reported instance of MP1 being produced by a member of this species and the second reported example of thiopeptide production in Staphylococcus spp. (34). S. epidermidis is an animal-derived bacterial isolate, whereas the majority of thiopeptide-producing bacteria isolated to date were derived from environmental samples (31). In their early work, Jensen and colleagues (26, 27, 35) demonstrated that strain 115 confers protective benefits on host animals, suggesting that thiopeptide-producing microbiota may play an important role in shaping microbial communities in animal hosts.
We have also provided evidence for a remarkably simple yet efficient mechanism of self-immunity in S. epidermidis 115: TclQ replaces L11 at the ribosome and prevents the action of MP1. In the annotation of the thiopeptide gene cluster from B. cereus, the authors predicted that tclQ, as well as tclWX, might contribute to self-immunity (6). In our B. subtilis bioassay, we did not observe tclWX-based immunity. With this observation and the fact that tclWX is not found in the S. epidermidis MP1 gene cluster, it appears that tclQ may provide the dominant mechanism of self-immunity in both organisms.
The plasmid-encoded nature of the S. epidermidis MP1 biosynthetic gene cluster is also of interest. Beyond pBac115, only one other plasmid-encoded thiopeptide gene cluster has been reported to date (36). pBac115 shows strong sequence similarity to plasmid pMCCL1 from Macrococcus caseolyticus JCSC5402 (37). These plasmids show more than 94% sequence identity through an 8.7-kb stretch of DNA that includes replication- and mobilization-associated genes. However, all of the tcl genes on pBac115 are absent from pMCCL1, suggesting that they are gained and lost as a single functional unit.
With few exceptions, the biosynthetic functions of many thiopeptide-modifying enzymes remain largely undemonstrated. The B. cereus TclM enzyme has been implicated in what is thought to be the culminating macrocyclization step of thiopeptide maturation (38). In a few other cases, researchers have modified the gene encoding the precursor peptide and detected resultant biochemical changes in the mature thiopeptide, including expansion of macrocyclic ring size (39–41). The tcl gene cluster on pBac115 provides a unique platform for continuing these kinds of studies because it is half the size of the homologous gene cluster from B. cereus and it controls the biosynthesis of a single thiopeptide product.
Access to a simpler thiopeptide gene cluster may also facilitate pharmaceutical development of these potent antibiotics. At present, poor pharmacokinetics and poor aqueous solubility have limited the translation of thiopeptide technology from laboratory to clinic. Thiostrepton and nosiheptide remain the only successfully commercialized thiopeptides to date; both are used in veterinary applications (31). Some attempts have been made to improve thiopeptide solubility and generate synthetic derivatives with improved pharmacokinetics (42–44), but this has been difficult. The discovery of this streamlined MP1 gene cluster opens the door to effectively addressing these issues and improving synthetic strategies.
MP1 is considered the optimal platform to explore the medicinal chemistry of thiopeptides for a number of reasons: its mechanism of action has been well addressed, it is produced by a variety of bacterial species, and its structure is relatively simple compared with those of other thiopeptides, yet it retains all of the attractive biochemical properties (potency, stability, etc.) of this chemical family (4). Furthermore, the availability of a simple system of self-immunity, which we have demonstrated here, enables further development with model bacterial hosts such as B. subtilis.
Supplementary Material
ACKNOWLEDGMENTS
We thank Marcus Jensen for providing S. epidermidis strain 115 and for helpful insights into earlier work. We also acknowledge technical assistance provided by Jared Balaich, Joseph Thiriot, Cody Ashcroft, Joann Diray-Arce, Brendan Coutu, Andrew Mathis, Jordon March, and Kaylee Bennallack. We sincerely appreciate Petra Levin for her generous assistance with Bacillus genetics, as well as Jiping Zou for his assistance with HPLC.
This work was supported by a grant from the BYU College of Life Sciences Vaccine Royalties Fund.
Footnotes
Published ahead of print 13 October 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.02243-14.
REFERENCES
- 1.Arnison PG, Bibb MJ, Bierbaum G, Bowers AA, Bugni TS, Bulaj G, Camarero JA, Campopiano DJ, Challis GL, Clardy J, Cotter PD, Craik DJ, Dawson M, Dittmann E, Donadio S, Dorrestein PC, Entian K-D, Fischbach MA, Garavelli JS, Goransson U, Gruber CW, Haft DH, Hemscheidt TK, Hertweck C, Hill C, Horswill AR, Jaspars M, Kelly WL, Klinman JP, Kuipers OP, Link AJ, Liu W, Marahiel MA, Mitchell DA, Moll GN, Moore BS, Muller R, Nair SK, Nes IF, Norris GE, Olivera BM, Onaka H, Patchett ML, Piel J, Reaney MJT, Rebuffat S, Ross RP, Sahl H-G, Schmidt EW, Selsted ME, Severinov K, Shen B, Sivonen K, Smith L, Stein T, Sussmuth RD, Tagg JR, Tang G-L, Truman AW, Vederas JC, Walsh CT, Walton JD, Wenzel SC, Willey JM, van der Donk WA. 2013. Ribosomally synthesized and post-translationally modified peptide natural products: overview and recommendations for a universal nomenclature. Nat. Prod. Rep. 30:108–160. 10.1039/c2np20085f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liao R, Duan L, Lei C, Pan H, Ding Y, Zhang Q, Chen D, Shen B, Yu Y, Liu W. 2009. Thiopeptide biosynthesis featuring ribosomally synthesized precursor peptides and conserved posttranslational modifications. Chem. Biol. 16:141–147. 10.1016/j.chembiol.2009.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kelly WL, Pan L, Li C. 2009. Thiostrepton biosynthesis: prototype for a new family of bacteriocins. J. Am. Chem. Soc. 131:4327–4334. 10.1021/ja807890a. [DOI] [PubMed] [Google Scholar]
- 4.Bagley MC, Dale JW, Merritt EA, Xiong X. 2005. Thiopeptide antibiotics. Chem. Rev. 105:685–714. 10.1021/cr0300441. [DOI] [PubMed] [Google Scholar]
- 5.Ciufolini MA, Lefranc D. 2010. Micrococcin P1: structure, biology and synthesis. Nat. Prod. Rep. 27:330–342. 10.1039/b919071f. [DOI] [PubMed] [Google Scholar]
- 6.Wieland Brown LC, Acker MG, Clardy J, Walsh CT, Fischbach MA. 2009. Thirteen posttranslational modifications convert a 14-residue peptide into the antibiotic thiocillin. Proc. Natl. Acad. Sci. U. S. A. 106:2549–2553. 10.1073/pnas.0900008106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rosendahl G, Douthwaite S. 1994. The antibiotics micrococcin and thiostrepton interact directly with 23S rRNA nucleotides 1067A and 1095A. Nucleic Acids Res. 22:357–363. 10.1093/nar/22.3.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Porse BT, Leviev I, Mankin AS, Garrett RA. 1998. The antibiotic thiostrepton inhibits a functional transition within protein L11 at the ribosomal GTPase centre. J. Mol. Biol. 276:391–404. 10.1006/jmbi.1997.1541. [DOI] [PubMed] [Google Scholar]
- 9.Porse BT, Cundliffe E, Garrett RA. 1999. The antibiotic micrococcin acts on protein L11 at the ribosomal GTPase centre. J. Mol. Biol. 287:33–45. 10.1006/jmbi.1999.2600. [DOI] [PubMed] [Google Scholar]
- 10.Harms JM, Wilson DN, Schluenzen F, Connell SR, Stachelhaus T, Zaborowska Z, Spahn CMT, Fucini P. 2008. Translational regulation via L11: molecular switches on the ribosome turned on and off by thiostrepton and micrococcin. Mol. Cell 30:26–38. 10.1016/j.molcel.2008.01.009. [DOI] [PubMed] [Google Scholar]
- 11.Baumann S, Schoof S, Harkal SD, Arndt HD. 2008. Mapping the binding site of thiopeptide antibiotics by proximity-induced covalent capture. J. Am. Chem. Soc. 130:5664–5666. 10.1021/ja710608w. [DOI] [PubMed] [Google Scholar]
- 12.Walter JD, Hunter M, Cobb M, Traeger G, Spiegel PC. 2012. Thiostrepton inhibits stable 70S ribosome binding and ribosome-dependent GTPase activation of elongation factor G and elongation factor 4. Nucleic Acids Res. 40:360–370. 10.1093/nar/gkr623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cundliffe E, Thompson J. 1981. Concerning the mode of action of micrococcin upon bacterial protein synthesis. Eur. J. Biochem. 118:47–52. 10.1111/j.1432-1033.1981.tb05484.x. [DOI] [PubMed] [Google Scholar]
- 14.Selva E, Montanini N, Stella S, Soffientini A, Gastaldo L, Denaro M. 1997. Targeted screening for elongation factor Tu binding antibiotics. J. Antibiot. (Tokyo) 50:22–26. 10.7164/antibiotics.50.22. [DOI] [PubMed] [Google Scholar]
- 15.Heffron SE, Jurnak F. 2000. Structure of an EF-Tu complex with a thiazolyl peptide antibiotic determined at 2.35 A resolution: atomic basis for GE2270A inhibition of EF-Tu. Biochemistry 39:37–45. 10.1021/bi9913597. [DOI] [PubMed] [Google Scholar]
- 16.Parmeggiani A, Nissen P. 2006. Elongation factor Tu-targeted antibiotics: four different structures, two mechanisms of action. FEBS Lett. 580:4576–4581. 10.1016/j.febslet.2006.07.039. [DOI] [PubMed] [Google Scholar]
- 17.Mizuhara N, Kuroda M, Ogita A, Tanaka T, Usuki Y, Fujita K. 2011. Antifungal thiopeptide cyclothiazomycin B1 exhibits growth inhibition accompanying morphological changes via binding to fungal cell wall chitin. Bioorg. Med. Chem. 19:5300–5310. 10.1016/j.bmc.2011.08.010. [DOI] [PubMed] [Google Scholar]
- 18.Rogers MJ, Cundliffe E, McCutchan TF. 1998. The antibiotic micrococcin is a potent inhibitor of growth and protein synthesis in the malaria parasite. Antimicrob. Agents Chemother. 42:715–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dahl EL, Rosenthal PJ. 2007. Multiple antibiotics exert delayed effects against the Plasmodium falciparum apicoplast. Antimicrob. Agents Chemother. 51:3485–3490. 10.1128/AAC.00527-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Clough B, Strath M, Preiser P, Denny P, Wilson IR. 1997. Thiostrepton binds to malarial plastid rRNA. FEBS Lett. 406:123–125. 10.1016/S0014-5793(97)00241-X. [DOI] [PubMed] [Google Scholar]
- 21.Clough B, Rangachari K, Strath M, Preiser PR, Wilson RJ. 1999. Antibiotic inhibitors of organellar protein synthesis in Plasmodium falciparum. Protist 150:189–195. 10.1016/S1434-4610(99)70021-0. [DOI] [PubMed] [Google Scholar]
- 22.Kwok JM, Myatt SS, Marson CM, Coombes RC, Constantinidou D, Lam EW. 2008. Thiostrepton selectively targets breast cancer cells through inhibition of forkhead box M1 expression. Mol. Cancer Ther. 7:2022–2032. 10.1158/1535-7163.MCT-08-0188. [DOI] [PubMed] [Google Scholar]
- 23.Hegde NS, Sanders DA, Rodriguez R, Balasubramanian S. 2011. The transcription factor FOXM1 is a cellular target of the natural product thiostrepton. Nat. Chem. 3:725–731. 10.1038/nchem.1114. [DOI] [PubMed] [Google Scholar]
- 24.Bhat UG, Halasi M, Gartel AL. 2009. Thiazole antibiotics target FoxM1 and induce apoptosis in human cancer cells. PLoS One 4(5):e5592. 10.1371/journal.pone.0005592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li C, Kelly WL. 2010. Recent advances in thiopeptide antibiotic biosynthesis. Nat. Prod. Rep. 27:153–164. 10.1039/b922434c. [DOI] [PubMed] [Google Scholar]
- 26.Meyers CM, Jensen MM. 1987. Staphylococcosis of turkeys. 3. Bacterial interference as a possible means of control. Avian Dis. 31:74–79. [PubMed] [Google Scholar]
- 27.Wilkinson DM, Jensen MM. 1987. Staphylococcosis of turkeys. 4. Characterization of a bacteriocin produced by an interfering Staphylococcus. Avian Dis. 31:80–84. [PubMed] [Google Scholar]
- 28.Lukashin AV, Borodovsky M. 1998. GeneMark.hmm: new solutions for gene finding. Nucleic Acids Res. 26:1107–1115. 10.1093/nar/26.4.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Niedermeyer TH, Strohalm M. 2012. mMass as a software tool for the annotation of cyclic peptide tandem mass spectra. PLoS One 7(9):e44913. 10.1371/journal.pone.0044913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Walsh CT, Malcolmson SJ, Young TS. 2012. Three ring posttranslational circuses: insertion of oxazoles, thiazoles, and pyridines into protein-derived frameworks. ACS Chem. Biol. 7:429–442. 10.1021/cb200518n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Just-Baringo X, Albericio F, Alvarez M. 2014. Thiopeptide antibiotics: retrospective and recent advances. Mar. Drugs 12:317–351. 10.3390/md12010317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Agrawal RK, Linde J, Sengupta J, Nierhaus KH, Frank J. 2001. Localization of L11 protein on the ribosome and elucidation of its involvement in EF-G-dependent translocation. J. Mol. Biol. 311:777–787. 10.1006/jmbi.2001.4907. [DOI] [PubMed] [Google Scholar]
- 33.Bausch SL, Poliakova E, Draper DE. 2005. Interactions of the N-terminal domain of ribosomal protein L11 with thiostrepton and rRNA. J. Biol. Chem. 280:29956–29963. 10.1074/jbc.M504182200. [DOI] [PubMed] [Google Scholar]
- 34.Carnio MC, Holtzel A, Rudolf M, Henle T, Jung G, Scherer S. 2000. The macrocyclic peptide antibiotic micrococcin P(1) is secreted by the food-borne bacterium Staphylococcus equorum WS 2733 and inhibits Listeria monocytogenes on soft cheese. Appl. Environ. Microbiol. 66:2378–2384. 10.1128/AEM.66.6.2378-2384.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nicoll TR, Jensen MM. 1987. Staphylococcosis of turkeys. 5. Large-scale control programs using bacterial interference. Avian Dis. 31:85–88. [PubMed] [Google Scholar]
- 36.Donia MS, Cimermancic P, Schulze CJ, Wieland Brown LC, Martin J, Mitreva M, Clardy J, Linington RG, Fischbach MA. 2014. A systematic analysis of biosynthetic gene clusters in the human microbiome reveals a common family of antibiotics. Cell 158:1402–1414. 10.1016/j.cell.2014.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Baba T, Kuwahara-Arai K, Uchiyama I, Takeuchi F, Ito T, Hiramatsu K. 2009. Complete genome sequence of Macrococcus caseolyticus strain JCSCS5402, [corrected] reflecting the ancestral genome of the human-pathogenic staphylococci. J. Bacteriol. 191:1180–1190. 10.1128/JB.01058-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bowers AA, Walsh CT, Acker MG. 2010. Genetic interception and structural characterization of thiopeptide cyclization precursors from Bacillus cereus. J. Am. Chem. Soc. 132:12182–12184. 10.1021/ja104524q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bowers AA, Acker MG, Young TS, Walsh CT. 2012. Generation of thiocillin ring size variants by prepeptide gene replacement and in vivo processing by Bacillus cereus. J. Am. Chem. Soc. 134:10313–10316. 10.1021/ja302820x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bowers AA, Acker MG, Koglin A, Walsh CT. 2010. Manipulation of thiocillin variants by prepeptide gene replacement: structure, conformation, and activity of heterocycle substitution mutants. J. Am. Chem. Soc. 132:7519–7527. 10.1021/ja102339q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Acker MG, Bowers AA, Walsh CT. 2009. Generation of thiocillin variants by prepeptide gene replacement and in vivo processing by Bacillus cereus. J. Am. Chem. Soc. 131:17563–17565. 10.1021/ja908777t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Trzasko A, Leeds JA, Praestgaard J, Lamarche MJ, McKenney D. 2012. Efficacy of LFF571 in a hamster model of Clostridium difficile infection. Antimicrob. Agents Chemother. 56:4459–4462. 10.1128/AAC.06355-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Leeds JA, Sachdeva M, Mullin S, Dzink-Fox J, Lamarche MJ. 2012. Mechanism of action of and mechanism of reduced susceptibility to the novel anti-Clostridium difficile compound LFF571. Antimicrob. Agents Chemother. 56:4463–4465. 10.1128/AAC.06354-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Naidu BN, Sorenson ME, Zhang Y, Kim OK, Matiskella JD, Wichtowski JA, Connolly TP, Li W, Lam KS, Bronson JJ, Pucci MJ, Clark JM, Ueda Y. 2004. Nocathiacin I analogues: synthesis, in vitro and in vivo biological activity of novel semi-synthetic thiazolyl peptide antibiotics. Bioorg. Med. Chem. Lett. 14:5573–5577. 10.1016/j.bmcl.2004.08.058. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.