Abstract
RNA metabolism is a critical but frequently overlooked control element affecting virtually every cellular process in bacteria. RNA processing and degradation is mediated by a suite of ribonucleases having distinct cleavage and substrate specificity. Here, we probe the role of two ribonucleases (RNase III and RNase J) in the emerging model system Streptomyces venezuelae. We show that each enzyme makes a unique contribution to the growth and development of S. venezuelae and further affects the secondary metabolism and antibiotic production of this bacterium. We demonstrate a connection between the action of these ribonucleases and translation, with both enzymes being required for the formation of functional ribosomes. RNase III mutants in particular fail to properly process 23S rRNA, form fewer 70S ribosomes, and show reduced translational processivity. The loss of either RNase III or RNase J additionally led to the appearance of a new ribosomal species (the 100S ribosome dimer) during exponential growth and dramatically sensitized these mutants to a range of antibiotics.
INTRODUCTION
Ribonucleases (RNases) are enzymes that process and degrade RNA molecules; consequently, they are critical for RNA maturation, RNA stability, and posttranscriptional regulation (1). In bacterial cells, the finely tuned balance between RNA synthesis and RNA degradation allows for rapid adaptation to changing environments, proper processing of noncoding RNAs, and efficient recycling of ribonucleotides (2).
Our understanding of RNA metabolism to date is based largely on studies of Escherichia coli and Bacillus subtilis, which employ distinct enzymes for RNA processing and degradation. In E. coli, RNase E is the central component of the RNA degradosome; as such, it is responsible for much of the RNA decay in this organism. B. subtilis lacks this enzyme, and acting in its place are three other nucleases: RNase J1/J2 and RNase Y (3, 4). Virtually all bacteria contain at least one of RNase E (or its paralog, RNase G), RNase J, or RNase Y (5). These RNases are unrelated in primary sequence and mechanism of catalysis but have similar substrate specificity: they all have single-strand-specific endonuclease activity and preferentially cleave AU-rich regions (4, 6, 7). Unlike RNase E and Y, however, RNase J has the capacity to act both as an endonuclease and as a 5′ exonuclease (8, 9). The analysis of available genome sequences suggests that more than half of all bacteria, and over two-thirds of Archaea, possess an RNase J homologue (6, 8). In addition to these diverse single-strand-specific RNases, most bacteria also encode the double-strand-specific RNase III (10). This enzyme is highly conserved in bacteria and eukaryotes and has a critical role in posttranscriptional regulation, where it cleaves double-stranded substrates, such as those resulting from the base pairing of an mRNA and noncoding RNA (11), or those associated with highly structured RNAs, such as ribosomal RNAs (rRNAs) (12, 13). There can be considerable functional interplay between RNases, with a classic example being the generation of mature 23S, 16S, and 5S rRNAs from a single 30S transcript. RNase III typically functions initially to liberate pre-16S, pre-23S, and pre-5S rRNAs from the full-length primary transcript (12, 14). These precursor rRNAs are processed further by a variety of RNases that may include (depending on the organism) RNase E, RNase G, RNase J, and other yet-to-be-identified ribonucleases (15–17). Without the concerted and precisely coordinated activity of these RNases during ribosome assembly, complete rRNA processing cannot be achieved and ribosome function is compromised.
As investigations into RNase activity begin to extend beyond the E. coli and B. subtilis model systems, it is becoming apparent that there is considerable diversity in the arsenal of RNases employed by any given bacterium (18). The actinobacteria, a group of Gram-positive bacteria that include Streptomyces and Mycobacterium, encode a set of RNases that include not only RNase III but also RNase E and RNase J (17). While the mycobacteria are best known for their pathogenic potential, the predominantly soil-dwelling Streptomyces species are renowned for their ability to produce a vast array of useful secondary metabolites, including antibiotics, antifungals, and chemotherapeutic agents. The streptomycetes also are known for their multicellular life cycle that encompasses morphologically and metabolically distinct developmental stages. Their life cycle initiates with spore germination, and subsequent hyphal tip extension and branching leads to the formation of vegetative mycelial networks. Reproductive growth initiates with the emergence of aerial hyphae during solid culture growth (or hyphal fragmentation for those strains that differentiate in liquid culture) and culminates with the subdivision of the aerial cells/hyphal fragments into chains of uniformly sized exospores (19).
Previous investigations into RNase III in Streptomyces coelicolor have revealed it to be essential for normal sporulation and the production of the antibiotics actinorhodin and undecylprodigiosin (20, 21). Its fundamental importance to RNA metabolism in Streptomyces is further illustrated by the fact that up to 10% of all transcripts synthesized during vegetative growth are affected (directly or indirectly) by RNase III activity (22). More recently, studies have begun to explore the biochemical and biological role of RNase J in Streptomyces. The S. coelicolor enzyme, like its B. subtilis counterpart, has dual endo/exonuclease activity (8, 23), and its deletion from the S. coelicolor chromosome results in altered antibiotic production (23).
Here, we probe the roles of RNase III and RNase J in Streptomyces venezuelae. We provide evidence that both RNases are pleiotropic regulators required for normal development and antibiotic production, with each enzyme making distinct contributions to these processes. While defects in mRNA turnover almost certainly contribute to these phenotypic abnormalities, we propose that defects in translation stemming from altered ribosome structure and activity also play a role. We show that both RNase III and RNase J mutants have far greater numbers of translationally inactive ribosome dimers (100S ribosomes) than their wild-type counterpart. RNase III mutants are further defective in rRNA processing, assemble fewer mature 70S ribosomes than the wild type, and exhibit reduced ribosomal processivity (based on polysome profiles).
MATERIALS AND METHODS
Bacterial strains and culture conditions.
Streptomyces strains, E. coli strains, and all plasmids/cosmids used in this study are summarized in Table 1. S. venezuelae ATCC 10712 typically was grown on the surface of maltose-yeast extract-malt extract (MYM) agar medium (24) or in shaken flasks containing liquid MYM at 20°C or 30°C. During conjugation with E. coli, S. venezuelae was grown on soy flour-mannitol agar medium (25), while Difco nutrient agar medium was used in screening for double crossover recombinants when creating RNase mutant strains. Finally, when assessing antibiotic production, specifically jadomycin B, by S. venezuelae, strains were grown in jadomycin B production medium (26), as detailed below. E. coli strains were grown in or on LB (Luria Bertani) medium or in SOB (super optimal broth) medium, with DH5α and ET12567/pUZ8002 strains grown at 37°C and BW25113/pIJ790 grown at 30°C or 37°C.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Genotype, description, or use | Reference(s) or source |
|---|---|---|
| Streptomyces venezuelae | ||
| ATCC 10712 | ||
| E325 | ATCC 10712 ΔSVEN_5265(rnc)::[aac(3)IV-oriT] | This work |
| E326 | ATCC 10712 ΔSVEN_5394(rnj)::[aac(3)IV-oriT] | This work |
| E. coli | ||
| DH5α | Plasmid construction and general subcloning | Invitrogen |
| ET12567/pUZ8002 | Generation of methylation-free plasmid DNA and conjugation into Streptomyces | 95, 96 |
| BW25113/pIJ790 | Construction of cosmid-based knockouts | 27 |
| Plasmids or cosmids | ||
| pIJ790 | Temperature-sensitive plasmid carrying λ-RED genes | 27 |
| pIJ82 | Integrating plasmid vector; complementation of mutant strains | Gift from H. Kieser |
| pIJ773 | Plasmid carrying the apramycin knockout cassette | 27 |
| 3B03 | S. venezuelae cosmid carrying SVEN_5265 (rnc) | Gift from M. Bibb and M. Buttner |
| 3J08 | S. venezuelae cosmid carrying SVEN_5394 (rnj) | Gift from M. Bibb and M. Buttner |
| pMC110 | pIJ82 + rnc; rnc complementation plasmid | This work |
| pMC111 | pIJ82 + rnj; rnj complementation plasmid | This work |
Dilution plating experiments involved the overnight growth of S. venezuelae in MYM liquid medium and the subsequent use of these cultures to inoculate 10 ml fresh liquid MYM to an optical density at 600 nm (OD600) of ∼0.1. Cultures were grown with shaking at 30°C until they reached an OD600 of 0.4, at which point serial dilutions were made in 10-fold increments. Three-microliter aliquots of each dilution were immediately spotted onto MYM agar plates and incubated at either 20°C or 30°C for 3 to 4 days.
Construction of rnc (RNase III) and rnj (RNase J) deletion strains.
In-frame deletions of rnc-SVEN_5265 (RNase III) and rnj-SVEN_5394 (RNase J) were generated using ReDirect technology (27). For each of rnc and rnj, the coding sequence (from start codon to stop codon) was replaced with an oriT-containing apramycin resistance cassette. RNase gene deletions were verified by PCRs performed using combinations of primers located upstream, downstream, and internal to the deleted genes (see Table S1 in the supplemental material).
The rnc mutant phenotype was complemented using a DNA fragment encompassing the wild-type rnc gene and associated upstream (278 bp) and downstream (212 bp) sequences (see Table S1 in the supplemental material for primer information) and cloned into the integrating plasmid vector pIJ82. The same complementation strategy was used for the rnj mutant, only the complementing DNA fragment contained the wild-type rnj gene, along with 280-bp upstream and 153-bp downstream flanking sequences (see Table S1). In each case, the complementing fragment was expected to include the native rnc-rnj promoter region and all associated upstream and downstream regulatory elements. To control for any phenotypic effects stemming from plasmid integration, pIJ82 alone was introduced into wild-type and rnc-rnj mutant strains, and these strains served as the basis for phenotypic comparisons during complementation experiments.
Light microscopy and scanning electron microscopy (SEM).
Samples for light microscopy were obtained by growing strains on MYM agar for 3 days and pressing sterile coverslips against the growing colonies or by applying a 20-μl aliquot of liquid MYM-grown cultures to a slide and overlaying with a coverslip. Images were obtained using a Nikon Eclipse TE2000-S inverted microscope, and, where appropriate, spore lengths were determined using ImageJ software (28). For each strain, a minimum of 400 spores were measured for each of three replicates (≥1,200 spores in total).
SEM was used to examine strains grown for 6 days on MYM agar. Samples were prepared and visualized using a TEMSCAM LSU scanning electron microscope as described previously (29).
Heat shock assay.
Heat shock assays were performed as described previously (29), with minor modifications. Briefly, spore suspensions were diluted in distilled water to a final concentration of ∼250 spores/100 μl. Aliquots (100 μl) were heated at 50°C for 10 to 40 min before being spread on MYM agar plates and incubated at 30°C for 4 days. Survival rates were calculated by dividing the number of colonies on plates after heat treatment by the number of colonies on plates without heat treatment; the resulting values were converted to percentages.
Jadomycin B production assay.
Jadomycin B production was assayed as described previously (30), with minor alterations to the published protocol. S. venezuelae was streaked for single colonies on MYM agar, and plates were grown for a minimum of 3 days. A single colony then was used to inoculate 125 ml of liquid MYM, and the culture was grown with shaking at 30°C for 22 h. Four to six milliliters of culture then was used to inoculate 100 ml glucose/isoleucine-based jadomycin production medium (26) to an OD600 of 0.3. Cultures were shaken at 30°C until an OD600 of 0.6 was reached (∼5 h), at which point 95% ethanol (3%, vol/vol) was added to the flasks to induce jadomycin B production. Samples for jadomycin B analyses were taken immediately after ethanol shock and after 16, 36, and 64 h. Jadomycin B was quantified by measuring the absorbance of culture supernatant at 526 nm and normalizing to culture growth (OD600) at that time point.
RNA isolation.
S. venezuelae was grown at 30°C in shaken flasks containing liquid MYM. Cells were harvested either at time points corresponding to vegetative growth (6 to 8 h) for rRNA analysis and hpf expression analysis or 23 h after ethanol shock for analysis of jadomycin B biosynthetic cluster transcripts. RNA was extracted as described previously (31), using a modified guanidium thiocyanate protocol (32). Cells were lysed by vortexing for 2 min with glass beads in a solution containing 4 M guanidium thiocyanate, 25 mM trisodium citrate dihydrate, 0.5% (wt/vol) sodium N-lauroylsarcosinate, and 0.8% β-mercaptoethanol. The resulting suspension was subjected to two phenol-chloroform extractions, followed by an acid phenol-chloroform extraction. Total nucleic acids were precipitated overnight using 3 M sodium acetate and an equal volume of isopropanol. Samples then were pelleted, washed in 70% ethanol, and resuspended in nuclease-free water. Contaminating DNA was removed using Turbo DNase (Life Technologies), and RNA quantity and purity were determined using a Nanodrop spectrophotometer. RNA quality was analyzed by agarose gel electrophoresis prior to reverse transcription-PCR (RT-PCR) and primer extension analyses.
Primer extension.
Gene-specific primers (see Table S1 in the supplemental material) internal to the mature sequence of rRNA genes were radiolabeled with [γ-32P]ATP using T4 polynucleotide kinase (New England BioLabs). Two micrograms of total RNA was heated at 80°C for 3 min before being added to an annealing mix containing 1 μl of a 10 mM deoxynucleoside triphosphate mixture and 2 pmol radiolabeled primer. The mixture then was incubated at 65°C for 8 min before being chilled on ice for 5 min. RT was performed using SuperScript III reverse transcriptase (Invitrogen) according to the manufacturer's instructions, with minor modifications. Briefly, 4 μl 5× first-strand buffer, 1 μl 0.1 M dithiothreitol (DTT), 1 μl RNaseOUT, and 1 μl (200 U) SuperScript III were mixed and incubated at 55°C for 1 h, followed by incubation at 70°C for 15 min to inactivate the enzyme. Reaction products were mixed with 2× loading dye and heated to 95°C for 5 min, and 2 to 12 μl was loaded and separated on a 6% denaturing polyacrylamide gel. The primer ladder was generated by radiolabeling 2 pmol of each of five differently sized primers (see Table S1) as described above.
RT-PCR.
RT-PCR was conducted as described by Hindra and Elliot (33). Specifically, RT was performed as outlined above for the primer extension experiment, with the only change being the use of 0.9 μg total RNA as the template. The resulting cDNA in turn served as the template for PCR amplification using Taq DNA polymerase and gene-specific primers (see Table S1 in the supplemental material). The number of cycles was optimized to ensure products were detected in the linear range of amplification. Negative controls containing nuclease-free water instead of reverse transcriptase were included to ensure RNA samples, and reverse transcription/amplification reagents, did not contain residual or contaminating genomic DNA. cDNA corresponding to the vegetative sigma factor-encoding gene hrdB was amplified as a positive control for RNA levels and RNA integrity. Ten microliters of each PCR mixture was separated on a 2% agarose gel and visualized by staining with ethidium bromide. All reactions were conducted in triplicate, using at least two independently isolated RNA samples.
Ribosome profiles.
Overnight cultures of S. venezuelae were used to inoculate 300 ml MYM in 2-liter flasks to an OD600 of 0.05, and these cultures were grown until they reached an OD600 of ∼0.4. All subsequent steps were carried out at 4°C. Cultures were harvested by centrifugation at 7,798 × g for 10 min. Cell pellets were washed and resuspended in 20 ml of 10 mM Tris-HCl, pH 7.5, 10 mM MgCl2, and 60 mM KCl. The cell resuspension then was centrifuged again at 3,400 × g for 15 min. The resulting cell pellet was resuspended in 7 ml lysis buffer containing 10 mM Tris-HCl, pH 7.5, 10 mM MgCl2, 60 mM KCl, 1 mM DTT, together with 35 μl of Tween 20, a Roche cOmplete mini protease inhibitor cocktail tablet, and 20 μl DNase I (1 mg/ml). Resuspended cells were lysed by three passages through a French press at 20,000 lbs/in2. The cell lysate was clarified by centrifugation at 16,000 × g for 10 min, and the resulting supernatant was further spun at 135,000 × g for 145 min. Resuspension of the pelleted ribosomes was allowed to proceed for 30 min on ice in buffer containing 20 mM Tris-HCl, pH 7.5, 6 mM MgCl2, 30 mM NH4Cl, and 1 mM DTT. An equal amount of 20 mM Tris-HCl, pH 7.5, 6 mM MgCl2, 800 mM NH4Cl, and 1 mM DTT then was added, and incubation on ice was continued for another hour. The mixture was clarified by centrifugation at 22,000 × g for 20 min, and the crude ribosomes were pelleted by centrifuging the supernatant at 135,000 × g, again for 145 min. The resulting ribosome pellet was resuspended on ice for 1 h in 20 mM Tris-HCl, pH 7.5, 10 mM MgCl2, 30 mM NH4Cl, and 1 mM DTT. After another clarifying spin, 10 to 15 A260 units of resuspended crude ribosomes were applied to 10.5 ml 10% to 30% (wt/vol) sucrose gradients prepared with buffer B (20 mM Tris-HCl, pH 7.5, 10 mM MgCl2, 50 mM NH4Cl, 1 mM DTT). The gradients were spun for 16 h at 40,000 × g in a Beckman SW41 Ti rotor. Gradients were fractionated using a Brandel fractionator apparatus and an AKTA Prime fast protein liquid chromatography system (GE Healthcare). Elution profiles were monitored by UV absorbance at 260 nm, and fractions corresponding to 70S and 100S subunit peaks were individually pooled and collected for negative-staining electron microscopy. The subunits were removed from the sucrose buffer by centrifugation at 135,000 × g for 145 min, after which the pellet was rinsed and resuspended in buffer B prior to storage at −80°C.
The proportion of free 30S to bound 30S (30S subunits contained within 70S and 100S ribosomes) and free 50S to bound 50S (50S subunits contained within 70S and 100S ribosomes) in the wild-type and mutant strains was calculated using peak areas obtained from the sucrose gradient profiles. The area of the 30S peak was divided by the area corresponding to the total number of 30S subunits (30S peak plus one-third the area of the 70S peak and 100S peak when observed) to obtain the percentage of free 30S subunits. To obtain the percentage of free 50S subunits, the area of the 50S peak was divided by the area corresponding to the total number of 50S subunits (50S peak plus two-thirds the area of the 70S peak and 100S peak when observed). Prior to obtaining these percentages, we normalized all ribosome profiles by making the entire area under the 30S, 50S, 70S, and 100S (when present) peaks constant; this allowed for direct comparison of the peaks representing the 70S and 100S particles between strains. These experiments were conducted using two (mutant) or three (wild-type) independently grown cultures, with a minimum of three technical replicates conducted for each biological sample.
Polysome profiles.
Polysome profiles were generated as described in Leong et al. (34), with minor adjustments. Overnight cultures of S. venezuelae were used to inoculate 300 ml MYM in 2-liter flasks to an OD600 of 0.05, and cultures were grown by shaking at 30°C until reaching an OD600 of ∼0.4. Polysomes typically are stabilized by adding chloramphenicol (35), but S. venezuelae can produce chloramphenicol and consequently is resistant to the effects of this antibiotic. Therefore, we attempted to use clindamycin to stabilize polysomes (this targets the same site/process as chloramphenicol and is not reported to cause ribosome distortion); however, we found that S. venezuelae polysomes were most stable without antibiotic addition. Cells were harvested by centrifugation at 7,798 × g for 10 min, and each pellet was resuspended in 0.5 ml cold resuspension buffer (20 mM Tris-HCl, pH 7.5, 15 mM MgCl2), with lysozyme added to a final concentration of 1 mg/ml. Tubes were incubated on ice for 15 min, and the suspensions were subjected to three cycles of freezing in liquid nitrogen and thawing at 30°C. Cell lysis was completed by adding 15 μl 10% sodium deoxycholate and 10 μl DNase I (1 mg/ml). The mixtures were centrifuged at 5,000 × g for 15 min, after which the supernatants were transferred to new tubes. Sample concentrations were measured by absorbance at 260 nm, and 10 A260 units of resuspended crude ribosome particles were added to buffer containing 10 mM Tris-HCl, pH 7.5, 50 mM KCl, 10 mM MgCl2, and 6 mM β-mercaptoethanol before being loaded onto a 10-ml 10 to 40% (wt/vol) sucrose gradient prepared in the same buffer. The gradients were spun for 16 h at 40,000 × g in a Beckman SW41 Ti rotor at 4°C. Gradients were fractionated using a Brandel fractionator apparatus and a syringe pump, and the elution profiles were monitored by UV absorbance at A260. All polysome profiles were conducted using at least two independent cell cultures and three technical replicates per strain. The obtained profiles were normalized before plotting them by making the area under each profile constant.
Electron microscopy.
Fractions corresponding to 70S and 100S subunit peaks were diluted to 18 μg/ml in buffer B. Electron microscopy grids freshly coated with a continuous layer of carbon were floated in 5-μl drops of sample immediately following glow discharge application to the grid (5 mA for 15 s). Grids then were blotted and floated for 1 min in a 5-μl drop of 1% uranyl acetate for staining. Excess stain was removed by blotting, and the grids were air dried. Specimens were visualized in a JEOL 1200EX electron microscope operated at 80 kV at a magnification of ×200,000. Images were acquired with an AMT 4-megapixel digital camera (Advanced Microscopy Techniques, Woburn, MA). Between 800 and 1,000 particles per sample were counted, and each particle was assigned to one of the following three categories: 70S (monomer), 100S (dimer), or polysome (3 or more 70S ribosomes in a row).
Antibiotic sensitivity assays.
Overnight cultures of S. venezuelae were used to inoculate 10 ml liquid MYM in 25-ml flasks to an OD600 of 0.04. Antibiotics were added to each flask to allow for between 15% and 50% growth relative to that of the wild-type strain in the absence of antibiotic. The final antibiotic concentrations used were 2.5 μg/ml hygromycin B, 2.5 μg/ml spectinomycin, 0.5 μg/ml viomycin, 0.5 μg/ml tetracycline, 0.1 μg/ml clindamycin, 0.5 μg/ml erythromycin, 0.025 μg/ml rifampin, and 0.5 μg/ml vancomycin. Following antibiotic addition, cultures were grown with shaking at 30°C for 6 h, and relative survival rates were calculating by dividing OD600 values after growth in the presence of antibiotics by OD600 values of cultures without antibiotics, with results being presented as percentages.
RESULTS
Bioinformatic and expression analyses of RNase J and RNase III in S. venezuelae.
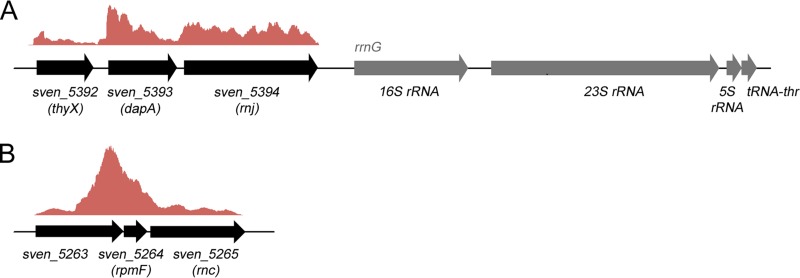
To begin exploring the function of RNase J (SVEN_5394) and RNase III (SVEN_5265) in S. venezuelae, their sequences and genomic context were compared to those of other well-studied bacteria. They each shared >90% amino acid sequence identity with their S. coelicolor counterparts. RNase J also shared 41% full-length identity (59% similarity) with RNase J1 and 38% identity (60% similarity) with RNase J2 from B. subtilis. S. venezuelae appears representative of the streptomycetes in encoding a single RNase J protein, and in all species examined, its corresponding gene (rnj) is found immediately upstream of an rRNA operon and directly downstream of genes encoding a dihydropicolinate synthetase (dapA) and thymidylate synthase (thyX) (Fig. 1A). Analysis of data from previously published RNA-sequencing (RNA-seq) experiments (32) suggested that each of rnj, dapA, and thyX was expressed from a distinct promoter but also shared low-level readthrough transcription, particularly between dapA and rnj (Fig. 1A). The genetic connection between rnj and dapA is conserved in many bacteria, including Mycobacterium species and B. subtilis (in the case of rnjB encoding RNase J2), while the rnj association with rRNA operons seems limited to the streptomycetes.
FIG 1.
Genetic organization and transcript profiles of rnc and rnj. (A) Organization of the genes flanking the RNase J-encoding rnj. Above the three protein-encoding genes (thyX, dapA, and rnj) is shown a graph of relative transcript levels throughout this region, as determined using RNA-seq data from RNA samples representing all stages of S. venezuelae growth (RNA was harvested at three distinct growth stages, vegetative, fragmentation, and sporulation, and was combined prior to sequencing). (B) Genetic organization of the RNase III-encoding rnc region. As described for panel A, above these genes is depicted a graph of relative transcript levels for this region.
RNase III has been reasonably well studied in S. coelicolor (20–22, 36), and given the extensive sequence similarity shared with RNase III in S. venezuelae, these proteins are expected to function in a similar manner. For all Streptomyces species in the StrepDB database (http://strepdb.streptomyces.org.uk/), rnc appears to be the third gene in an operon that includes genes encoding a conserved, hypothetical protein and the large ribosomal subunit protein L32 (rpmF) (Fig. 1B). The investigation of available RNA-seq data (32) suggested that while these three genes do indeed appear to be cotranscribed in S. venezuelae, there exists an additional strong promoter upstream of rnc (and rpmF) within the first gene in the operon (Fig. 1B). This observation is further supported by our complementation experiments described below.
RNase J and RNase III deletions affect growth and morphology in S. venezuelae.
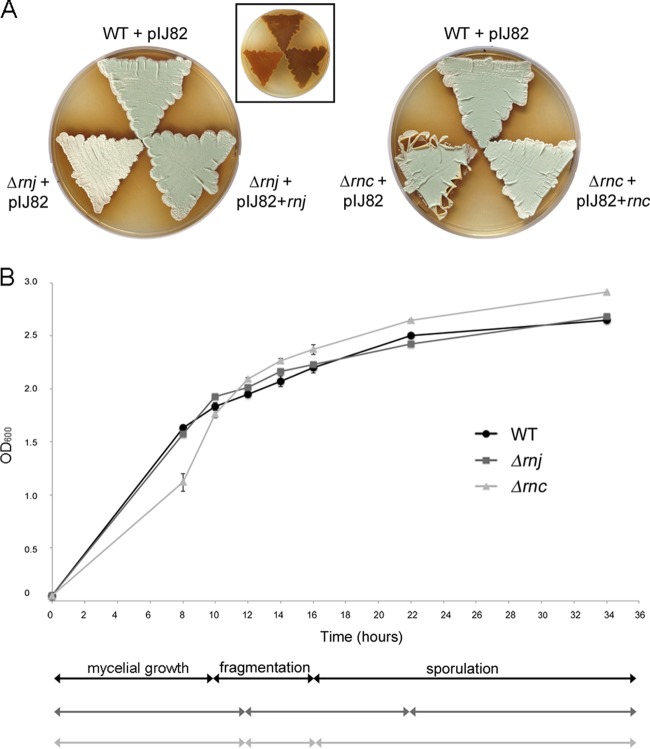
As a first step in investigating the biological role of RNase III and RNase J in S. venezuelae, we replaced their respective genes with an antibiotic resistance cassette. Growth for 4 days on MYM agar revealed both mutant strains to be phenotypically distinct from their wild-type parent (Fig. 2A). The Δrnj strain had reduced melanin production and produced little of the green pigment associated with wild-type S. venezuelae spores, suggesting a potential sporulation defect. In contrast, the Δrnc mutant appeared to sporulate normally, although it did display unusual plate growth properties in that the edges of growth curled away from the growth medium, suggesting that the vegetative cells are unusually hydrophobic (Fig. 2A). The morphological defects of both mutant strains could be restored upon complementation with wild-type copies of their respective genes on the integrating plasmid vector pIJ82 compared with wild-type S. venezuelae carrying pIJ82 alone (Fig. 2A). We observed effective complementation of the S. venezuelae rnc mutant phenotype with the rnc coding sequence and ∼300 nucleotides (nt) of upstream sequence; this sequence encompassed the upstream rpmF gene and the C-terminal half of SVEN_5263 and included the promoter region indicated in our RNA-seq data (Fig. 1B).
FIG 2.
Phenotypic comparison of wild-type S. venezuelae and the Δrnc and Δrnj mutants. (A) Colony morphologies of wild-type (WT) S. venezuelae and RNase mutants (carrying the integrating plasmid vector pIJ82) and the corresponding complemented strains grown on MYM agar medium for 4 days. The inset image shows the underside of the plate to the left, bearing the rnj mutant and wild-type-complemented mutant strains in the same order and orientation as that for the larger rnj panel (mutant, bottom left; wild type, top; complemented, bottom right), showing relative levels of the brown melanin pigment. (B) Comparison of growth rates and transitions between life cycle stages of wild-type, Δrnj, and Δrnc strains. Cultures were inoculated to an OD600 of 0.05 and incubated with shaking at 30°C for 34 h. The optical density (OD600) was measured at the indicated times, and life cycle stages (indicated below the graph) were assessed using light microscopy. Each OD600 value represents an average from three to four replicates; the standard error for growth density was calculated at each time point.
We next took advantage of the fact that S. venezuelae fully differentiates in liquid culture in a relatively synchronous and dispersed manner and examined the effects of these two RNase deletions on S. venezuelae growth and development in liquid culture (Fig. 2B). We found that while the Δrnj strain had growth kinetics similar to those of the wild type, microscopic analyses revealed that it took longer to transition from mycelial growth to fragmentation, and that the onset of sporulation was significantly delayed (∼6 h), consistent with the putative sporulation defects seen during growth on solid medium (lack of mature spore pigment) (Fig. 2A). The Δrnc strain initially grew more slowly than either the wild type or the rnj mutant, but by early stationary phase its growth kinetics matched that of the wild type. Despite this initial lag in growth, the Δrnc strain exhibited only a modest delay in fragmentation and initiated sporulation at the same time as the wild type (Fig. 2B).
RNase III and RNase J mutants form defective spores.
The developmental defects of the Δrnj strain and the unusual growth of the Δrnc strain on solid medium prompted us to investigate the morphology of these strains using SEM (see Fig. S1 in the supplemental material). After 6 days of growth, we observed few spore chains for the Δrnj strain, suggesting that sporulation in this strain was an infrequent occurrence. For the Δrnc mutant, spore chains were abundant, but the spores themselves were irregularly shaped compared with the more uniformly sized spores of wild-type S. venezuelae.
A rigorous assessment of spore size for mutant and wild-type strains then was undertaken (n ≥ 1,200 spores for each strain) using light microscopy images of spore chains adhering to glass coverslips. We found the Δrnj strain had slightly shorter spores than the wild type (P < 0.001), with the average spore length for the Δrnj strain being 0.62 μm, compared with 0.7 μm for the wild type (see Fig. S1 in the supplemental material). While the average spore length for both Δrnc and wild-type strains was comparable (Δrnc mutant, 0.67 μm; wild type, 0.7 μm), the size range was far greater for the Δrnc strain, with the standard deviation being 28% greater than that of the wild type (P < 0.001) (see Fig. S1), further validating our SEM observations that this mutant formed irregular spores.
Given that both RNase mutant strains appeared to form defective spores with shorter or variable spore lengths, we wanted to assess how these strains responded to heat stress, as heat sensitivity is suggestive of defects in spore maturation (29, 37, 38). We conducted heat shock assays using wild-type and RNase mutant spores and found the Δrnj strain to be the most sensitive, displaying an ∼5-fold increase in heat sensitivity after a 20-min heat shock relative to the wild type (see Fig. S1 in the supplemental material). The Δrnc strain was less impacted, appearing more heat resistant after 10 min but showing a slight increase (∼1.3-fold) in heat sensitivity after 20 min (see Fig. S1).
RNase deletion abolishes/reduces jadomycin B production.
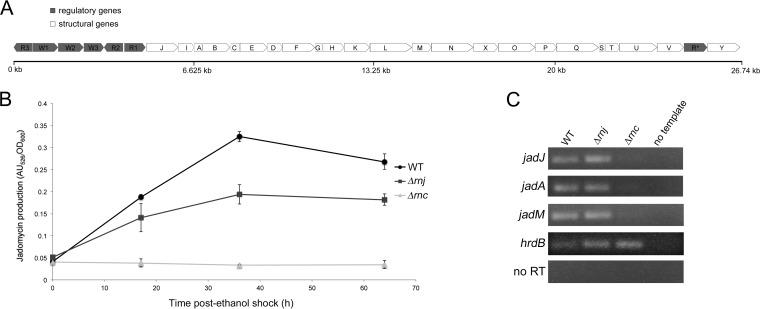
RNase III is required for antibiotic production in several Streptomyces species (20, 21, 39, 40), while RNase J was recently shown to have varied effects on antibiotic production in S. coelicolor (delayed actinorhodin, increased undecylprodigiosin, and reduced calcium-dependent antibiotic) (23). The melanin defect associated with the rnj mutant in S. venezuelae suggested that RNase J impacts secondary metabolite production in this species as well. To quantitatively probe the effect of losing either RNase III or RNase J on antibiotic production in S. venezuelae, we assayed production of the polyketide-derived anguacycline glycoside antibiotic jadomycin B, a secondary metabolite with antimicrobial, antifungal, and antitumor activity (41). While S. venezuelae is predicted to encode upwards of 30 secondary metabolic clusters (as determined using antiSMASH [42]), jadomycin B (whose biosynthetic cluster is shown in Fig. 3A) is the only antibiotic produced by this species for which a specific production assay has been developed. We grew wild-type and mutant cultures in jadomycin B production medium and found the jadomycin B levels of the Δrnj mutant typically were 20 to 30% lower than that of the wild-type strain after controlling for growth. For the Δrnc mutant, jadomycin B was never detected (Fig. 3B), confirming an essential role for this RNase in promoting antibiotic (or at least jadomycin B) production in S. venezuelae.
FIG 3.
RNase J and RNase III are required for normal antibiotic production in S. venezuelae. (A) Organization of the jadomycin B biosynthetic cluster. Regulatory genes are shown in gray, and structural genes are shown in white. (B) Wild-type and RNase mutant strains were grown under jadomycin B-inducing conditions, and jadomycin B production at each time point was calculated by measuring the units of absorbance of the culture supernatant at 526 nm (AU526) and normalizing to growth density (OD600). These results reflect the averages from three independent cultures, and error bars denote standard errors for each time point. (C) Semiquantitative RT-PCR using RNA isolated from the wild-type strain and the RNase mutants during jadomycin B production (24 h post-ethanol shock). The vegetative sigma factor hrdB served as a positive control for RNA loading and RNA integrity, while no-RT reactions (using RNA as template) and no-template reactions were included as negative controls to ensure a lack of DNA contamination in both RNA preparations and all PCR reagents.
Transcriptional analysis of the jadomycin cluster.
The underlying basis for the jadomycin B production defects observed for RNase III and RNase J mutants, and the antibiotic production defects observed for equivalent mutants in other Streptomyces species, has yet to be clearly established. There is some evidence supporting a transcriptional mechanism in the case of RNase III, as reduced expression of several pathway-specific antibiotic regulators has been shown previously for rnc mutants (21, 33, 36). To determine whether the jadomycin B production defects seen here were due to transcriptional abnormalities, we isolated RNA from wild-type and mutant strains induced to produce jadomycin B and examined the expression of the jadomycin B pathway-specific regulator-encoding gene jadJ (43, 44), along with two biosynthetic genes (jadA and jadM), using semiquantitative RT-PCR. These three genes appear to be part of a long operon and likely are expressed from the same promoter (Fig. 3A) (note that the jadomycin B cluster organization in the sequenced S. venezuelae ATCC 10712 strain differs slightly from that reported in other studies [43, 45, 46]). The Δrnj strain had approximately wild-type transcript levels for all three genes tested, while the Δrnc strain had virtually undetectable expression of these same genes (Fig. 3C). This suggested that the failure of an RNase III mutant to produce jadomycin B is due to the lack of transcription of its biosynthetic genes (or dramatically reduced transcript stability); however, such an explanation does not hold for the RNase J mutant, which reproducibly yielded at least wild-type levels of expression for the three genes tested yet produced significantly less jadomycin B (Fig. 3B and C).
RNase deletion strains are cold sensitive.
Given that a transcriptional mechanism could not explain the reduced levels of jadomycin B observed for the RNase J mutant in particular, we wondered whether translation plays a role, given that RNase J and RNase III have been implicated in rRNA processing in different bacterial species (8, 16, 47), and their respective genes are genetically linked to ribosomal components, where rnc is cotranscribed with a ribosomal protein and rnj is located upstream of an rRNA operon (Fig. 1) (21, 48, 49). mRNA stability also can be influenced by ribosome activity and the associated protection that translating ribosomes provide.
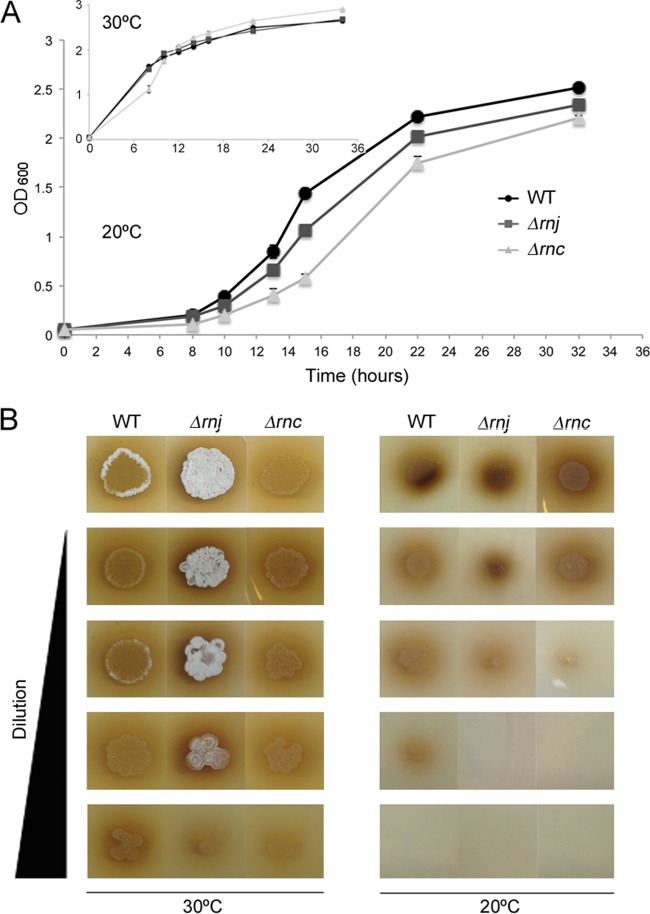
While heat sensitivity is indicative of sporulation defects, cold sensitivity is a hallmark of ribosome defects (50, 51); thus, we tested the cold sensitivity of our mutant strains, comparing the growth of these strains to the wild type at 30°C and 20°C. Both RNase deletion strains displayed greater cold sensitivity than the wild type during growth at 20°C in liquid medium. The ratio between log-phase growth rates of wild-type and Δrnj mutant strains was 1.0 at 30°C, and this diverged to 1.6 at 20°C (i.e., the Δrnj mutant was ∼30% more cold sensitive than the wild type), while the equivalent ratios for the wild-type and Δrnc strains were 1.1 at 30°C and 1.4 at 20°C (i.e., the Δrnc mutant was ∼20% more cold sensitive than the wild type) (Table 2 and Fig. 4A). The cold sensitivity of wild-type and mutant strains also was analyzed during growth on solid medium, where dilution plating assays showed that wild-type and mutant strains grew equally robustly at 30°C but that at 20°C growth of the two RNase mutant strains was compromised (Fig. 4B).
TABLE 2.
Doubling time and growth rates of S. venezuelae strains in MYM liquid mediaa
| Strain | DT (h) at: |
Growth rate (h−1) atb: |
||
|---|---|---|---|---|
| 30°C | 20°C | 30°C | 20°C | |
| Wild type | 1.59 ± 0.01 | 2.43 ± 0.09 | 0.436 ± 0.002 | 0.286 ± 0.010 |
| rnj mutant | 1.61 ± 0.01 | 3.93 ± 0.1 | 0.431 ± 0.002 | 0.177 ± 0.005 |
| rnc mutant | 1.79 ± 0.04 | 3.42 ± 0.08 | 0.387 ± 0.009 | 0.203 ± 0.005 |
Standard deviations were calculated from 3 to 4 replicates per strain. DT, doubling time.
Growth rate was determined by the equation k = ln2/DT.
FIG 4.
Cold-sensitive phenotype of the RNase mutant strains. (A) Growth profiles of the wild-type and RNase mutant strains in MYM liquid medium at 20°C and 30°C (inset). Cultures were inoculated to an OD600 of 0.05 and incubated with shaking at 30°C for 34 h. Each value is an average from three to four replicates, and the standard error for growth density was calculated at each time point. (B) Dilution plating of wild-type and mutant strains. Cultures were grown with shaking at 30°C to an OD600 of 0.4 before they were diluted in 10-fold increments. Three microliters of each dilution was spotted onto MYM agar plates, and plates were incubated at either 20°C or 30°C for 4 days. The Δrnj strain occasionally raised aerial hyphae more rapidly than either of the other strains when inoculated from liquid-grown culture (as seen here for the culture grown at 30°C), but this phenotype was not reproducibly observed.
RNase deletion strains are defective in ribosome assembly and form 100S ribosomes.
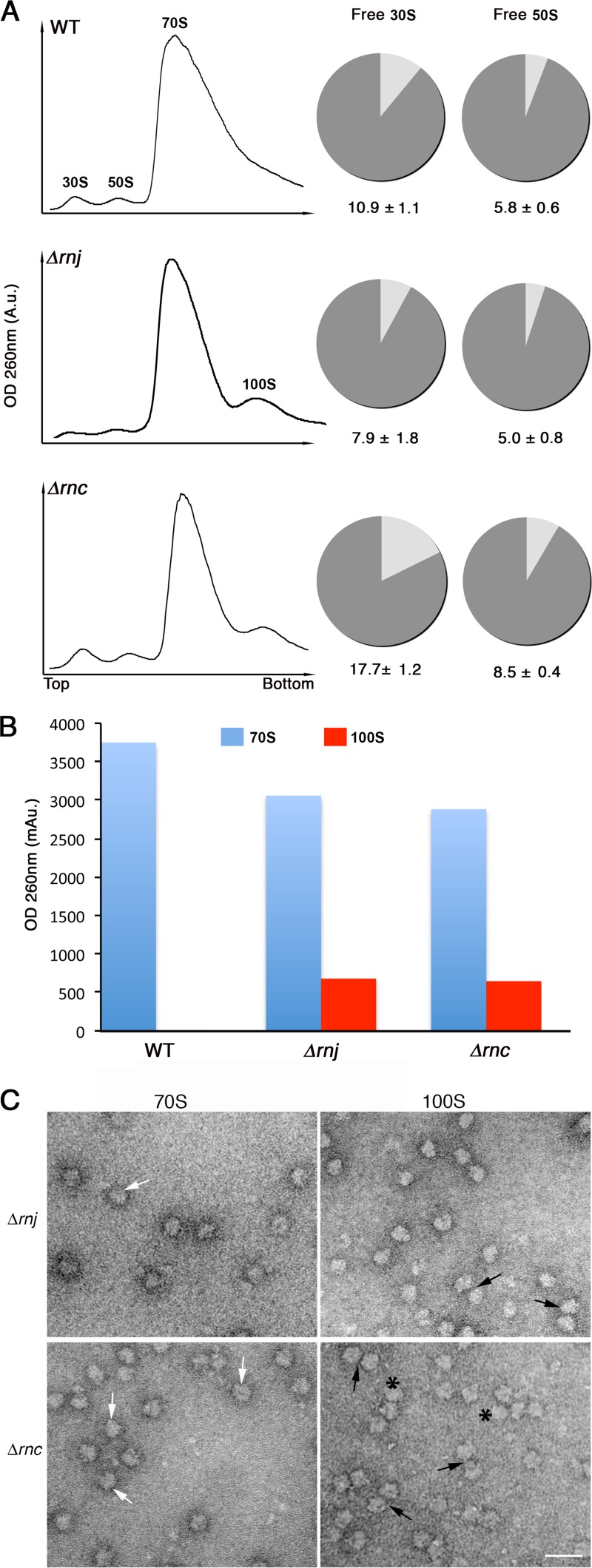
The cold-sensitive phenotypes of both RNase mutant strains suggested they have defective ribosomes. To determine whether ribosome assembly was affected by mutations in rnc and rnj, we obtained ribosome profiles of wild-type and mutant strains during early-exponential growth using sucrose density gradient centrifugation, a technique that separates free 30S subunits, free 50S subunits, and fully assembled 70S ribosomes. We found wild-type and Δrnj strains had similar profiles, with roughly equivalent proportions of free 30S and 50S subunits. In contrast, the Δrnc strain had increased levels of free 30S (approximately 2-fold) and 50S subunits (Fig. 5A).
FIG 5.
Ribosome profiling of wild-type and RNase mutant strains. (A) Ribosomes from wild-type (top), Δrnj (middle), and Δrnc (bottom) strains were fractionated on 10 to 30% sucrose gradients, and sedimentation profiles were plotted (left). Peaks corresponding to 30S and 50S ribosomal subunits, 70S ribosomes, and 100S ribosome dimers are indicated. The pie graphs to the right depict the proportion of free and complexed subunits in wild-type and RNase mutant strains in the same order as that on the left. The proportion of free 30S to bound 30S (i.e., 30S subunits in 70S and 100S ribosomes) and free 50S to bound 50S (i.e., 50S subunits in 70S and 100S ribosomes) in the wild-type and mutant strains was calculated using peak areas obtained from the sucrose gradient profiles (left). A.u., arbitrary units. (B) Plot representing the amount of 70S and 100S particles obtained from the profiles shown in panel A. (C) Electron micrographs of images corresponding to 70S peaks and the additional peaks from Δrnj and Δrnc strains. Peak fractions were pooled separately (i.e., 70S fraction and 100S fraction), purified, and visualized using transmission electron microscopy. 70S and 100S ribosomal particles are indicated by white and black arrows, respectively. Polysomes are indicated by asterisks. Scale bars, 50 nm.
Intriguingly, the sedimentation profiles of Δrnc and Δrnj mutants but not the wild type revealed an extra peak of ∼100S beyond the expected 30S, 50S, and 70S fractions (Fig. 5A), and this represented a significant proportion of the assembled ribosomes (Fig. 5B). Previous studies in E. coli showed that 70S ribosomes could dimerize to form translationally inactive 100S ribosomes during stationary phase (52). To determine whether the larger peak detected in the RNase mutants corresponded to 100S ribosomes, we examined the content of the 100S fraction using transmission electron microscopy and compared these with the particles obtained from the 70S fraction. Even in the absence of cross-linking (100S particles are not stable complexes [53, 54]), we observed abundant ribosome dimers in the 100S fraction (∼10 to 15% of all ribosome particles) for both RNase mutants relative to the 70S fraction (3 to 4% dimers) (n > 800 for each 100S and 70S fraction) (Fig. 5C).
Given the sequestration of some of the 70S ribosomes into 100S particles in the Δrnj and Δrnc strains, there was a proportional decrease in the level of 70S ribosomes in these strains (Fig. 5B), with the number of 70S ribosomes further reduced in the Δrnc strain, consistent with these cells having a larger proportion of free 30S and 50S particles (Fig. 5A and B).
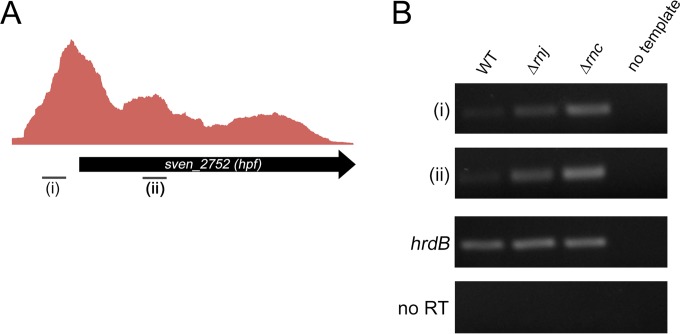
In E. coli, ribosome dimer formation occurs in stationary phase and is promoted by the ribosome modulation factor Rmf, in conjunction with the so-called hibernation promoting factor Hpf (52, 55). Rmf is confined to the gammaproteobacteria, and in most other bacteria it is a longer form of Hpf that directs ribosome dimerization (56–58). The streptomycetes possess a gene encoding this longer Hpf form, and as such, we sought to determine whether the rnc and rnj mutant strains showed higher levels of expression of this gene (SVEN_2752) during exponential growth than the wild type. We have RNA-seq data suggesting that hpf was expressed from a promoter located considerably (∼150 to 200 nt) upstream of the start codon (Fig. 6A), so we investigated the expression of both its 5′ untranslated region (UTR) and its coding sequence (to account for any transcriptional attenuation that may occur between the transcription and translation start sites), using RNA isolated from the wild type and the two RNase mutant strains during exponential-phase growth. We observed similar profiles for both coding sequence- and UTR-associated transcripts in all strains and found that hpf transcripts were present at much higher levels in both rnc and rnj mutants than in the wild-type (Fig. 6B), suggesting that increased expression of hpf in these strains contributes to 100S dimer formation at a time when these are not present (or at least detected) in the wild type (Fig. 5A and B).
FIG 6.
Transcription of hpf, a ribosome dimer-promoting factor. (A) Graph depicting the relative transcript levels of hpf, including what appears to be a long 5′ untranslated region (5′UTR), as determined by analysis of RNA isolated from all growth stages, combined and subjected to RNA-seq. (i) and (ii) indicate the location of products analyzed by RT-PCR. (B) Semiquantitative RT-PCR analysis of the 5′UTR of hpf (i) and the hpf coding sequence (ii) in wild-type and RNase mutant strains using RNA isolated from early-exponential-phase cultures (OD600 of ∼0.4). hrdB was used as a positive control for RNA abundance and RNA integrity, while no-RT reactions (using RNA as template) and no-template reactions served as negative controls, ensuring there was no DNA contamination of either RNA preparations or any PCR reagents.
RNase deletion strains have reduced translation efficiency.
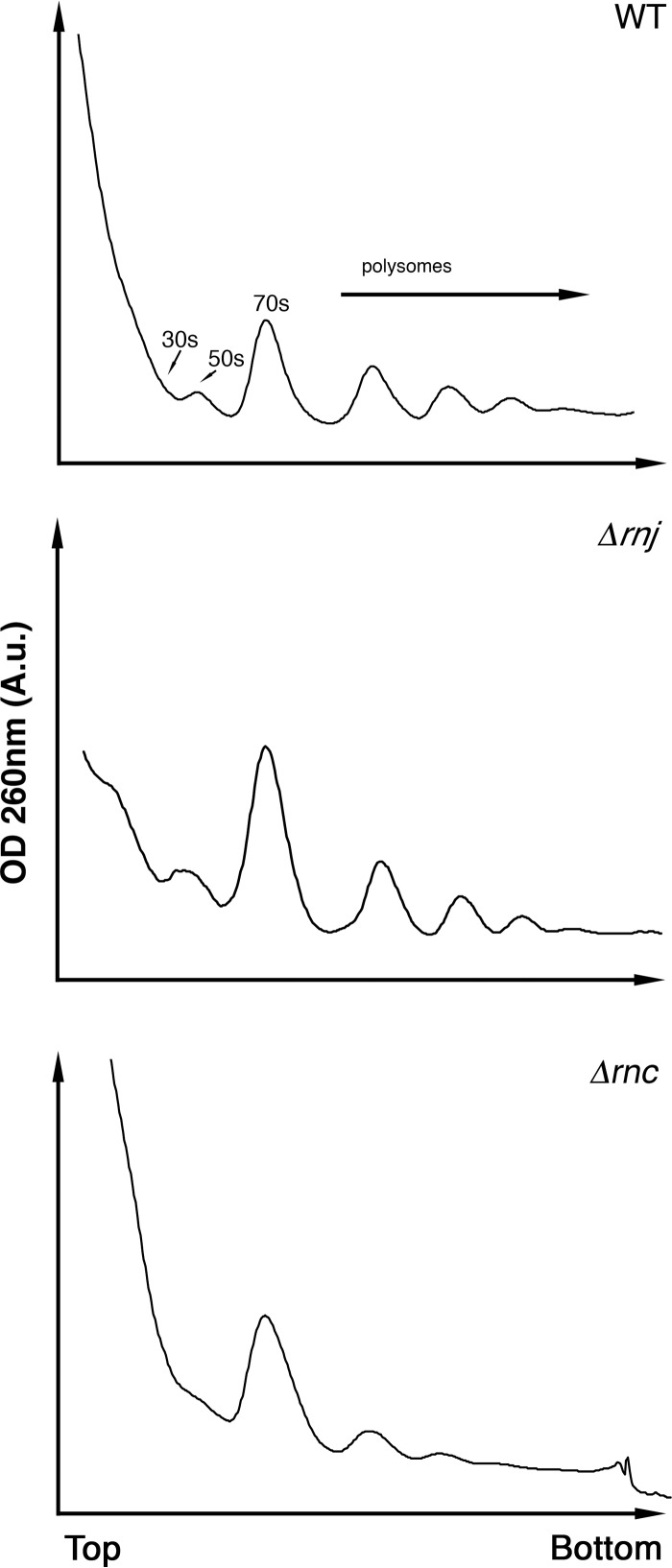
The propensity of the two RNase mutants to form translationally inactive 100S ribosomes and the cold sensitivity phenotype of the Δrnc strain collectively suggested that RNase III and RNase J are required to produce ribosomes capable of efficient translation. To probe how these RNases affected global translation in S. venezuelae, we conducted polysome profiling for wild-type and mutant strains using sucrose density gradient ultracentrifugation. Polysome profiles provide a snapshot of translation activity and reveal the density of actively translating ribosomes on mRNA transcripts (59). Wild-type profiles revealed three clear polysome peaks, similar to that of the Δrnj strain. However, the Δrnc strain profile revealed only two small polysome peaks, indicating that translation activity and ribosome density on mRNA transcripts were severely reduced in this mutant strain (Fig. 7).
FIG 7.
Polysome profiles of wild-type and RNase mutant strains. Polysome profiles for each of the three strains were generated following fractionation on a 10 to 40% sucrose gradient. Peaks corresponding to 30S subunits, 50S subunits, and 70S ribosomes are labeled, and the locations of polysomes are indicated.
RNase III but not RNase J is involved in rRNA processing at the 5′ end.
Having established that rnc and rnj mutants formed abnormal ribosomes with reduced processivity, we considered the possibility that these defects stemmed from altered rRNA processing. RNase J1 is involved in 16S rRNA processing in B. subtilis (16) and in 16S and 23S rRNA maturation in Mycobacterium smegmatis, while RNase III has a conserved role in rRNA processing in nearly all bacteria, cleaving the stem-loop structures flanking mature 16S and 23S species (reviewed in reference 60). rRNA processing in Streptomyces has not been studied in any detail, although there is some data suggesting that 30S rRNA (full-length) transcripts accumulate in rnc null mutants in S. coelicolor (21); the role of RNase J in rRNA processing has not been examined.
S. venezuelae has seven rRNA operons dispersed throughout its linear genome, and of these, only two (rrnC and rrnF) have been fully sequenced. The two sequenced operons differ only in the spacer regions between 16S and 23S (the rrnC spacer is 10 nt longer than that of rrnF) and between 23S and 5S (the rrnF spacer is 1 nt longer than that of rrnC) (see Fig. S2 in the supplemental material).
We set out to map rRNA processing sites using primer extension analyses on total RNA isolated from wild-type and RNase mutant strains during exponential growth and radiolabeled oligonucleotides complementary to regions near the 5′ end of the mature 16S and 23S transcripts (p16 and p23, respectively) (see Fig. S2 in the supplemental material). The extension of primer p16 revealed the primary processing site to be +105/106 in all three strains relative to the transcription start site of the rRNA operon (see Fig. S2). This suggested that neither RNase III nor RNase J was involved in processing the 5′ end of pre-16S rRNA. The extension of primer p23 revealed the primary 5′ pre-23S processing site to be located at +1896 (relative to the start site for transcription of the rRNA operon). The Δrnc strain lacked this processing site, while processing appeared wild type in the Δrnj strain, indicating that RNase III, but not RNase J, was needed for cleavage of 5′ pre-23S rRNA (see Fig. S2).
RNase deletion strains display differential sensitivities to antibiotics that target the ribosome.
Collectively, our results showed that both RNase mutant strains had proportionally greater numbers of translationally inactive 100S ribosome dimers and exhibited various defects in ribosome assembly and translational efficiency. For the Δrnc mutant, this may be explained in part by the abnormal processing of the 23S rRNA, but no such processing defect was observed for the Δrnj mutant. In an attempt to gain further insight into the ribosome defects of the two RNase mutants, we took advantage of recent structural studies that have revealed the molecular basis by which diverse antibiotics target the bacterial ribosome (reviewed in references 61 and 62).
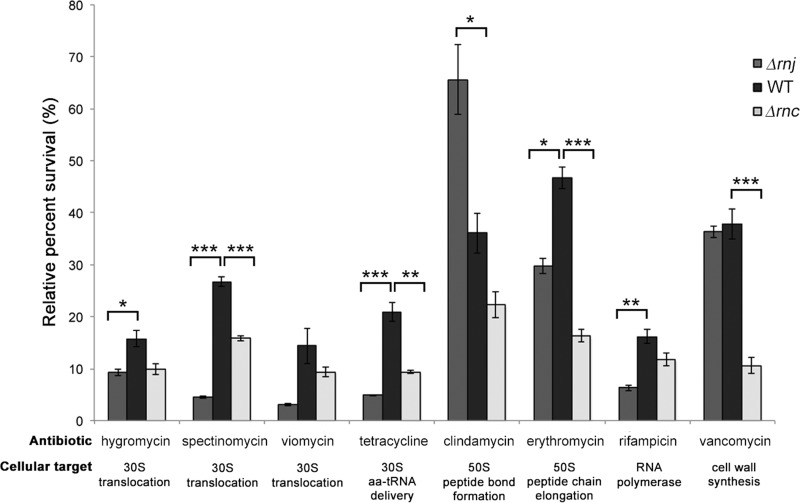
We analyzed the sensitivities of wild-type and RNase mutant strains to antibiotics that specifically target different sites in the ribosome: tetracycline, which binds the 30S subunit and inhibits delivery of aminoacylated tRNAs to the A site; the aminoglycosides viomycin (63), spectinomycin (64), and hygromycin B (65), which bind the 30S subunit and inhibit translocation of tRNAs from the A site to the P site; the macrolide erythromycin, which binds the 50S subunit and inhibits elongation of the peptide chain (66); and the lincosamide clindamycin, which binds the 50S subunit and inhibits peptide bond formation between the tRNA-bound amino acid at the P site of the ribosome and the growing tRNA-anchored polypeptide chain in the A site (66). For all antibiotics, we used antibiotic concentrations that inhibited the growth of the wild type by ∼50 to 85%.
We determined that both RNase mutants were hypersensitive to the majority of ribosome-targeting antibiotics tested (Fig. 8), but that the rnj mutant generally was more sensitive than the rnc mutant to antibiotics targeting the 30S subunit (tetracycline and the aminoglycosides hygromycin, spectinomycin, and viomycin). Unexpectedly, the Δrnj strain exhibited increased resistance to clindamycin: ∼35% of wild-type cells survived clindamycin treatment, while 65% of Δrnj mutant cells survived exposure to this antibiotic (Fig. 8).
FIG 8.
RNase mutant sensitivity to ribosome-targeting antibiotics. Wild-type, rnj, and rnc strains were tested for their resistance to a variety of different antibiotics, including 2.5 μg/ml hygromycin B, 2.5 μg/ml spectinomycin, 0.5 μg/ml viomycin, 0.5 μg/ml tetracycline, 0.1 μg/ml clindamycin, 0.5 μg/ml erythromycin, 0.025 μg/ml rifampin, and 0.5 μg/ml vancomycin. Relative survival rates were calculating by dividing the optical density (OD600) after growth in the presence of antibiotics by the optical density (OD600) of cultures without antibiotics and are shown as percentages. Each value is an average from three replicates, and the standard error was calculated for the percent survival at each time point. Asterisks indicate statistically significant differences (*, P value of 0.05 to 0.01; **, P value of 0.01 to 0.005; ***, P value below 0.005).
To probe the effect of antibiotics targeting other cellular systems, we assessed the sensitivities of all strains to rifampin (inhibits RNA polymerase activity) (67) and vancomycin (inhibits cell wall synthesis) (68). Interestingly, both strains exhibited increased sensitivity to rifampin, with the Δrnj strain in particular being 2.7-fold more sensitive than the wild type (Fig. 8). Wild-type and Δrnj strains were equally sensitive to vancomycin, while the Δrnc strain had heightened sensitivity.
DISCUSSION
Developmental defects associated with RNase deletion.
We observed striking but distinct developmental defects for each of the rnc and rnj mutant strains in S. venezuelae. The heterogenous spore sizes observed for the RNase III mutant are consistent with those seen for an equivalent mutant in S. coelicolor (20); however, the unusual peeling phenotype observed here has not been noted before. These morphological defects may stem from misregulation of the important developmental regulator AdpA. Previous work in S. coelicolor has shown that loss of RNase III function (due to an rnc point mutation) results in increased expression of AdpA (69). AdpA controls the expression of the RamR-encoding gene, and RamR in turn promotes the synthesis of SapB, an aerial hypha-promoting, amphipathic peptide (69, 70). Enhanced SapB production may contribute to the peeling phenotype of the rnc mutant. This phenotype may be further exacerbated by the lack of agarase produced by S. venezuelae; in S. coelicolor, agarase production leads to vegetative hyphal growth into the agar substrate, and this would likely prevent the colony peeling seen here for S. venezuelae. AdpA was first identified in Streptomyces griseus, and in this system it also controls the expression of ssgA (71, 72), which encodes a key determinant of spore septum placement (73). It will be interesting to see whether ssgA levels are altered in an rnc mutant strain and if this contributes to the irregular placement of sporulation septa seen for this mutant in S. venezuelae.
Unlike the rnc mutant, rnj mutants failed to achieve robust sporulation, and those spores that did form generally were smaller and less heat resistant than the wild type, suggesting defects in spore formation and spore maturation. In Bacillus, where RNase J has been best studied, mutation and/or depletion of one or both rnj genes results in profound sporulation defects (74, 75). Transcriptional and proteomic analyses in Bacillus have revealed dramatic alterations in the expression of at least two sporulation genes in an rnj mutant: higher spo0E transcript levels were observed by Durand and colleagues (76), where Spo0E is a phosphatase responsible for dephosphorylating the sporulation checkpoint protein Spo0A (77), alongside increased levels of spoVG transcripts and its corresponding protein (75, 76), where SpoVG inhibits asymmetric septation (78). While homologues of these gene products are not found in Streptomyces species, these results support the idea that RNase J is important for promoting sporulation and adoption of a dormant state in evolutionarily divergent bacteria.
Metabolic defects associated with RNase deletion.
RNase III has long been associated with antibiotic production defects in S. coelicolor (20, 21, 79), and more recent work has revealed a role for RNase III in promoting actinomycin production in Streptomyces antibioticus (40). Here, we show that S. venezuelae also requires RNase III for jadomycin B production. In S. coelicolor, and now S. venezuelae, these antibiotic production defects are due at least in part to aberrant transcription (or dramatically reduced transcript stability) of the corresponding biosynthetic genes and cognate pathway-specific regulators (36, 80). While this cannot be a direct effect, how it is mediated remains to be determined.
RNase J has not been as well studied in the streptomycetes as RNase III, but its effects on antibiotic production extend to both S. coelicolor (23) and S. venezuelae. In S. venezuelae, the loss of RNase J resulted in reduced jadomycin B production, but unlike the situation for RNase III, the biosynthetic cluster (or at least the three genes tested here) was transcribed at nearly wild-type levels. This suggested the jadomycin B production defect was not due to transcriptional misregulation, although we cannot exclude the possibility that biosynthetic genes outside this operon were not expressed at wild-type levels. Our observation that there were increased numbers of presumably translationally inactive 100S ribosomes in this strain during exponential growth, coupled with phenotypic characteristics consistent with ribosome defects (cold sensitivity and a notable response to ribosome-targeting antibiotics), instead suggested a role for the translational regulation of jadomycin B production. Given that ribosomal defects also were seen for the RNase III mutant, it is not unreasonable to presume that translational defects (in conjunction with RNA metabolism defects) also contribute to reduced antibiotic production in this strain. Antibiotic biosynthetic genes typically are organized in large operons, with many genes being translationally coupled. Studies in Saccharomyces cerevisiae (81) and E. coli (82) have revealed strong negative correlations between translation ratios (the amount of protein produced per mRNA) and mRNA lengths, with longer mRNAs having lower rates of translation initiation and correspondingly decreased ribosome densities. As translational efficiency would be adversely affected by the long mRNAs associated with many antibiotic biosynthetic clusters, and RNase mutation could further compromise ribosome activity, these transcripts may be translated less effectively than in a wild-type strain.
Interestingly, ribosome alterations have been broadly linked with changes in antibiotic levels in a multitude of streptomycetes, although in different ways than shown here. Specifically, mutations in rpsL (encoding ribosomal protein S12) and rsmG (encoding a 16S rRNA methyltransferase) significantly enhance antibiotic production (83–85); mutations in these genes also are associated with increased antibiotic (specifically streptomycin) resistance. A consistent effect stemming from the rsmG mutation is enhanced S-adenosyl methionine (SAM) production due to increased transcription of the SAM synthetase-encoding gene metK. In the streptomycetes, overexpression or exogenous application of SAM has found broad-spectrum utility in stimulating antibiotic production (86). In B. subtilis, RNase J mutations have been associated with reduced levels of metK (75). Whether an analogous expression defect for metK exists in the streptomycetes remains to be seen, but reduced levels of metK in either RNase mutant could further contribute to the antibiotic defects observed for these strains.
Role of RNase III and RNase J in the assembly of functional ribosomes in the streptomycetes.
The ribosomal defects associated with the loss of RNase III include abnormal 5′ end processing of 23S rRNA, a reduced ability to assemble 70S ribosomes, and a propensity to form 100S ribosome dimers. Collectively, these characteristics indicate this strain has serious translational defects, a prediction borne out by the dramatic reduction in polysome peaks observed for the rnc mutant relative to the wild type. The final 23S rRNA processing step occurs in polysomes (87), suggesting that only when the ribosomes are associated with mRNAs do they adopt a conformation conducive to the final stages of maturation. Fewer polysomes were detected in the rnc mutant than in the wild type (and rnj mutant), which could contribute to a further reduction in the number of fully mature ribosomes present in this strain, excluding those sequestered as 100S dimers. The situation is less clear-cut for RNase J, where its loss led to cold sensitivity and accumulated 100S ribosome dimers but no enrichment in ribosome subunits or any obvious rRNA processing defects, at least at the 5′ ends of the 16S and 23S rRNAs. In M. smegmatis, RNase E acts alongside RNase J to process all three rRNA molecules (17), and it is conceivable that RNase E has an equivalent role in S. venezuelae.
The accumulation of 100S ribosome dimers for both rnc and rnj mutants was unexpected, as such a phenomenon has not been observed for equivalent mutants in other bacteria. All evidence to date suggests that 100S ribosomes are translationally inactive: their peptidyl transferase centers and peptide exit tunnels are inaccessible, and they are not associated with tRNAs or mRNAs (53, 88, 89). These inactive dimers typically form during stationary phase, and their formation has been tied to the stringent response (56, 90), although it is worth noting that not all bacteria adopt such ribosome configurations. In Mycobacterium, for example, the essential dormancy regulator DosR promotes the formation of translationally inactive stabilized 70S ribosomes during conditions of stress, and in M. smegmatis, this is mediated by a protein termed RafH (MSMEG_3935) (91). The Hpf homologue (MSMEG_1878) was found associated with ribosomes in stationary phase, but its presence was not sufficient to promote dimerization. Our observations here suggest that the streptomycetes differ from their mycobacterial relatives in forming 100S ribosome dimers, and this may be mediated at least in part by Hpf proteins. It is conceivable that losing either RNase J or RNase III in S. venezuelae results in a cellular stress response analogous to that occurring during stationary phase, leading to the downregulation of translation.
RNase activity and antibiotic sensitivity.
One of the most profound phenotypic changes observed for the rnc and rnj mutants was significantly increased sensitivity to a range of antibiotics targeting not only the ribosome but also RNA polymerase (particularly for the Δrnj mutant) and the cell wall (for the Δrnc mutant). The response to ribosome-specific antibiotics may stem from the fact that ribosomes in these strains are already defective; consequently, they are hypersensitive to antibiotic effects. It is also possible that the inability of these strains to effectively turn over aberrant ribosome precursors formed during antibiotic treatment contributes to the observed increase in antibiotic sensitivity (92). An exception to this sensitization comes with clindamycin, where we observed increased resistance for an rnj mutant. This antibiotic binds to the base of the peptide tunnel, in part through extensive hydrogen bonding with the 23S rRNA in this region (93), a region not expected to be modified or processed by RNase J given its central position within the 23S rRNA. Hence, it is currently unclear why this strain exhibits increased clindamycin resistance.
Our work suggests that RNase III and RNase J contribute to the formation of fully functional ribosomes. In addition to directing translation, ribosomes also contribute directly to RNA polymerase processivity through the close coupling of transcription and translation in bacteria (94). rnc and rnj appear to have reduced ribosomal processivity, and this may lead to less effective transcription elongation and an exacerbated effect of any antibiotic targeting RNA polymerase, such as was observed following rifampin treatment here.
While an rnj mutant had an essentially wild-type response to vancomycin, the rnc mutant again was more sensitive. The mechanism underlying this observation is not immediately obvious, but previous RNA-seq and chromatin immunoprecipitation analyses in S. coelicolor revealed a number of membrane proteins, lipoproteins, and other secreted protein-encoding genes to be affected by the loss of RNase III (22), and it is conceivable that one or more of these contribute to increased vancomycin sensitivity or, indeed, general antibiotic sensitivity through enhanced uptake/reduced efflux.
It is clear from this work, and from that of others, that RNase III and RNase J play important but distinct roles in morphological development, secondary metabolism, and ribosome assembly and function in the streptomycetes. Further investigations into the cellular targets of these RNases and the accompanying downstream effects of their activities will allow us to better understand the role that these enzymes play in the regulatory and metabolic networks governing these processes and others in the cell.
Supplementary Material
ACKNOWLEDGMENTS
We thank members of the Elliot and Ortega laboratories for helpful suggestions and discussions and Mark Buttner, Maureen Bibb, and the Buttner group for their very helpful introduction to S. venezuelae.
This work was supported by funding from the Natural Sciences and Engineering Research Council of Canada (NSERC Discovery Grant no. 312495 to M.A.E and NSERC Discovery Grant no. 288327 to J.O.), the Canada Research Chairs program (to M.A.E.), and an NSERC Canada Graduate Scholarship (to S.E.J.).
Footnotes
Published ahead of print 29 September 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.02205-14.
REFERENCES
- 1.Laalami S, Zig L, Putzer H. 2014. Initiation of mRNA decay in bacteria. Cell. Mol. Life Sci. 71:1799–1828. 10.1007/s00018-013-1472-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arraiano CM, Andrade JM, Domingues S, Guinote IB, Malecki M, Matos RG, Moreira RN, Pobre V, Reis FP, Saramago M, Silva IJ, Viegas SC. 2010. The critical role of RNA processing and degradation in the control of gene expression. FEMS Microbiol. Rev. 34:883–923. 10.1111/j.1574-6976.2010.00242.x. [DOI] [PubMed] [Google Scholar]
- 3.Lehnik-Habrink M, Lewis RJ, Mäder U, Stülke J. 2012. RNA degradation in Bacillus subtilis: an interplay of essential endo- and exoribonucleases. Mol. Microbiol. 84:1005–1017. 10.1111/j.1365-2958.2012.08072.x. [DOI] [PubMed] [Google Scholar]
- 4.Shahbabian K, Jamalli A, Zig L, Putzer H. 2009. RNase Y, a novel endoribonuclease, initiates riboswitch turnover in Bacillus subtilis. EMBO J. 28:3523–3533. 10.1038/emboj.2009.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kaberdin VR, Singh D, Lin-Chao S. 2011. Composition and conservation of the mRNA-degrading machinery in bacteria. J. Biomed. Sci. 18:23. 10.1186/1423-0127-18-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Even S, Pellegrini O, Zig L, Labas V, Vinh J, Bréchemmier-Baey D, Putzer H. 2005. Ribonucleases J1 and J2: two novel endoribonucleases in B. subtilis with functional homology to E. coli RNase E. Nucleic Acids Res. 33:2141–2152. 10.1093/nar/gki505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Callaghan AJ, Marcaida MJ, Stead JA, McDowall KJ, Scott WG, Luisi BF. 2005. Structure of Escherichia coli RNase E catalytic domain and implications for RNA turnover. Nature 437:1187–1191. 10.1038/nature04084. [DOI] [PubMed] [Google Scholar]
- 8.Mathy N, Bénard L, Pellegrini O, Daou R, Wen T, Condon C. 2007. 5′-To-3′ exoribonuclease activity in bacteria: role of RNase J1 in rRNA maturation and 5′ stability of mRNA. Cell 129:681–692. 10.1016/j.cell.2007.02.051. [DOI] [PubMed] [Google Scholar]
- 9.Li de la Sierra-Gallay I, Zig L, Jamalli A, Putzer H. 2008. Structural insights into the dual activity of RNase J. Nat. Struct. Mol. Biol. 15:206–212. 10.1038/nsmb.1376. [DOI] [PubMed] [Google Scholar]
- 10.Nicholson AW. 2013. Ribonuclease III mechanisms of double-stranded RNA cleavage. Wiley Interdiscip. Rev. RNA 5:31–48. 10.1002/wrna.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Viegas SC, Silva IJ, Saramago M, Domingues S, Arraiano CM. 2011. Regulation of the small regulatory RNA MicA by ribonuclease III: a target-dependent pathway. Nucleic Acids Res. 39:2918–2930. 10.1093/nar/gkq1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nikolaev N, Silengo L. 1973. A role for ribonuclease III in processing of ribosomal ribonucleic acid and messenger ribonucleic acid precursors in Escherichia coli. J. Biol. Chem. 248:7967–7969. [PubMed] [Google Scholar]
- 13.Westphal H, Crouch RJ. 1975. Cleavage of adenovirus messenger RNA and of 28S and 18S ribosomal RNA by RNase III. Proc. Natl. Acad. Sci. U. S. A. 72:3077–3081. 10.1073/pnas.72.8.3077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ginzburg D, Steitz A. 1975. The 30 S ribosomal precursor RNA from Escherichia coli. A primary transcript containing 23 S, 16 S, and 5 S sequences. J. Biol. Chem. 250:5647–5654. [PubMed] [Google Scholar]
- 15.Li Z, Pandit S, Deutscher MP. 1999. RNase G (CafA protein) and RNase E are both required for the 5′ maturation of 16S ribosomal RNA. EMBO J. 18:2878–2885. 10.1093/emboj/18.10.2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Britton RA, Wen T, Schaefer L, Pellegrini O, Uicker WC, Mathy N, Tobin C, Daou R, Szyk J, Condon C. 2007. Maturation of the 5′ end of Bacillus subtilis 16S rRNA by the essential ribonuclease YkqC/RNase J1. Mol. Microbiol. 63:127–138. 10.1111/j.1365-2958.2006.05499.x. [DOI] [PubMed] [Google Scholar]
- 17.Taverniti V, Forti F, Ghisotti D, Putzer H. 2011. Mycobacterium smegmatis RNase J. is a 5′-3′ exo-/endoribonuclease and both RNase J. and RNase E are involved in ribosomal RNA maturation. Mol. Microbiol. 82:1260–1276. 10.1111/j.1365-2958.2011.07888.x. [DOI] [PubMed] [Google Scholar]
- 18.Condon C, Putzer H. 2002. The phylogenetic distribution of bacterial ribonucleases. Nucleic Acids Res. 30:5339–5346. 10.1093/nar/gkf691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Flärdh K, Buttner MJ. 2009. Streptomyces morphogenetics: dissecting differentiation in a filamentous bacterium. Nat. Rev. Microbiol. 7:36–49. 10.1038/nrmicro1968. [DOI] [PubMed] [Google Scholar]
- 20.Sello JK, Buttner MJ. 2008. The gene encoding RNase III in Streptomyces coelicolor is transcribed during exponential phase and is required for antibiotic production and for proper sporulation. J. Bacteriol. 190:4079–4083. 10.1128/JB.01889-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Price B, Adamidis T, Kong R, Champness W. 1999. A Streptomyces coelicolor antibiotic regulatory gene, absB, encodes an RNase III homolog. J. Bacteriol. 181:6142–6151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gatewood ML, Bralley P, Weil MR, Jones GH. 2012. RNA-Seq and RNA immunoprecipitation analyses of the transcriptome of Streptomyces coelicolor identify substrates for RNase III. J. Bacteriol. 194:2228–2237. 10.1128/JB.06541-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bralley P, Aseem M, Jones GH. 2014. SCO5745, a bifunctional RNase J. ortholog, affects antibiotic production in Streptomyces coelicolor. J. Bacteriol. 196:1197–1205. 10.1128/JB.01422-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang K, Han L, He J, Wang L, Vining LC. 2001. A repressor-response regulator gene pair controlling jadomycin B production in Streptomyces venezuelae ISP5230. Gene 279:165–173. 10.1016/S0378-1119(01)00723-5. [DOI] [PubMed] [Google Scholar]
- 25.Kieser T, Bibb MJ, Buttner MJ, Chater KF, Hopwood DA. 2000. Practical Streptomyces genetics. The John Innes Foundation, Norwich, England. [Google Scholar]
- 26.Brooks M, Burdock SJT, Ghaly EA. 2012. Changes in cell structure, morphology and activity of Streptomyces venezuelae during the growth, shocking and jadomycin production stages. J. Microb. Biochem. Technol. 4:63–75. 10.4172/1948-5948.1000073. [DOI] [Google Scholar]
- 27.Gust B, Challis GL, Fowler K, Kieser T, Chater KF. 2003. PCR-targeted Streptomyces gene replacement identifies a protein domain needed for biosynthesis of the sesquiterpene soil odor geosmin. Proc. Natl. Acad. Sci. U. S. A. 100:1541–1546. 10.1073/pnas.0337542100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Abràmoff MD, Magalhães PJ, Ram SJ. 2004. Image processing with ImageJ. Biophotonics Int. 11:33–42. [Google Scholar]
- 29.Haiser HJ, Yousef MR, Elliot MA. 2009. Cell wall hydrolases affect germination, vegetative growth, and sporulation in Streptomyces coelicolor. J. Bacteriol. 191:6501–6512. 10.1128/JB.00767-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jakeman DL, Graham CL, Young W, Vining LC. 2006. Culture conditions improving the production of jadomycin B. J. Ind. Microbiol. Biotechnol. 33:767–772. 10.1007/s10295-006-0113-4. [DOI] [PubMed] [Google Scholar]
- 31.Chomczynski P. 1987. Single-step method of RNA isolation by acid guanidinium extraction. Anal. Biochem. 159:156–159. [DOI] [PubMed] [Google Scholar]
- 32.Moody MJ, Young RA, Jones SE, Elliot MA. 2013. Comparative analysis of non-coding RNAs in the antibiotic-producing Streptomyces bacteria. BMC Genomics 14:558. 10.1186/1471-2164-14-558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hindra, Pak P, Elliot MA. 2010. Regulation of a novel gene cluster involved in secondary metabolite production in Streptomyces coelicolor. J. Bacteriol. 192:4973–4982. 10.1128/JB.00681-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Leong V, Kent M, Jomaa A, Ortega J. 2013. Escherichia coli rimM and yjeQ null strains accumulate immature 30S subunits of similar structure and protein complement. RNA 19:789–802. 10.1261/rna.037523.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brow DA, Noller HF. 1983. Protection of ribosomal RNA from kethoxal in polyribosomes. Implication of specific sites in ribosome function. J. Mol. Biol. 163:27–46. [DOI] [PubMed] [Google Scholar]
- 36.Huang J, Shi J, Molle V, Sohlberg B, Weaver D, Bibb MJ, Karoonuthaisiri N, Lih C-J, Kao CM, Buttner MJ, Cohen SN. 2005. Cross-regulation among disparate antibiotic biosynthetic pathways of Streptomyces coelicolor. Mol. Microbiol. 58:1276–1287. 10.1111/j.1365-2958.2005.04879.x. [DOI] [PubMed] [Google Scholar]
- 37.Mazza P, Noens EE, Schirner K, Grantcharova N, Mommaas A, Koerten MHK, Muth G, Flärdh K, van Wezel GP, Wohlleben W. 2006. MreB of Streptomyces coelicolor is not essential for vegetative growth but is required for the integrity of aerial hyphae and spores. Mol. Microbiol. 60:838–852. 10.1111/j.1365-2958.2006.05134.x. [DOI] [PubMed] [Google Scholar]
- 38.Molle V, Palframan WJ, Findlay KC, Buttner MJ. 2000. WhiD and WhiB, homologous proteins required for different stages of sporulation in Streptomyces coelicolor A3(2). J. Bacteriol. 182:1286–1295. 10.1128/JB.182.5.1286-1295.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Adamidis T, Champness W. 1992. Genetic analysis of absB, a Streptomyces coelicolor locus involved in global antibiotic regulation. J. Bacteriol. 174:4622–4628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee J-H, Gatewood ML, Jones GH. 2013. RNase III is required for actinomycin production in Streptomyces antibioticus. Appl. Environ. Microbiol. 79:6447–6451. 10.1128/AEM.02272-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zheng J-T, Rix U, Zhao L, Mattingly C, Adams V, Chen Q, Rohr J, Yang K-Q. 2005. Cytotoxic activities of new jadomycin derivatives. J. Antibiot. 58:405–408. 10.1038/ja.2005.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Blin K, Medema MH, Kazempour D, Fischbach MA, Breitling R, Takano E, Weber T. 2013. antiSMASH 2.0–a versatile platform for genome mining of secondary metabolite producers. Nucleic Acids Res. 41:W204–W212. 10.1093/nar/gkt449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang Y, Pan G, Zou Z, Fan K, Yang K, Tan H. 2013. JadR*-mediated feed-forward regulation of cofactor supply in jadomycin biosynthesis. Mol. Microbiol. 90:884–897. 10.1111/mmi.12406. [DOI] [PubMed] [Google Scholar]
- 44.Wang L, Tian X, Wang J, Yang H, Fan K, Xu G, Yang K, Tan H. 2009. Autoregulation of antibiotic biosynthesis by binding of the end product to an atypical response regulator. Proc. Natl. Acad. Sci. U. S. A. 106:8617–8622. 10.1073/pnas.0900592106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang L. 2003. Control of growth, secondary metabolism and sporulation in Streptomyces venezuelae ISP5230 by jadW1, a member of the afsA family of gamma-butyrolactone regulatory genes. Microbiology 149:1991–2004. 10.1099/mic.0.26209-0. [DOI] [PubMed] [Google Scholar]
- 46.Zheng J-T, Wang S-L, Yang K-Q. 2007. Engineering a regulatory region of jadomycin gene cluster to improve jadomycin B production in Streptomyces venezuelae. Appl. Microbiol. Biotechnol. 76:883–888. 10.1007/s00253-007-1064-z. [DOI] [PubMed] [Google Scholar]
- 47.Madhugiri R, Evguenieva-Hackenberg E. 2009. RNase J is involved in the 5′-end maturation of 16S rRNA and 23S rRNA in Sinorhizobium meliloti. FEBS Lett. 583:2339–2342. 10.1016/j.febslet.2009.06.026. [DOI] [PubMed] [Google Scholar]
- 48.Chang SA, Bralley P, Jones GH. 2005. The absB gene encodes a double strand-specific endoribonuclease that cleaves the read-through transcript of the rpsO-pnp operon in Streptomyces coelicolor. J. Biol. Chem. 280:33213–33219. 10.1074/jbc.M503440200. [DOI] [PubMed] [Google Scholar]
- 49.Xu W, Huang J, Cohen SN. 2008. Autoregulation of AbsB (RNase III) expression in Streptomyces coelicolor by endoribonucleolytic cleavage of absB operon transcripts. J. Bacteriol. 190:5526–5530. 10.1128/JB.00558-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bylund GO, Wipemo LC, Lundberg LA, Wikström PM. 1998. RimM and RbfA are essential for efficient processing of 16S rRNA in Escherichia coli. J. Bacteriol. 180:73–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Connolly K, Culver G. 2009. Deconstructing ribosome construction. Trends Biochem. Sci. 34:256–263. 10.1016/j.tibs.2009.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wada A, Yamazaki Y, Fujita N, Ishihama A. 1990. Structure and probable genetic location of a “ribosome modulation factor” associated with 100S ribosomes in stationary-phase Escherichia coli cells. Proc. Natl. Acad. Sci. U. S. A. 87:2657–2661. 10.1073/pnas.87.7.2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kato T, Yoshida H, Miyata T, Maki Y, Wada A, Namba K. 2010. Structure of the 100S ribosome in the hibernation stage revealed by electron cryomicroscopy. Structure 18:719–724. 10.1016/j.str.2010.02.017. [DOI] [PubMed] [Google Scholar]
- 54.Krokowski D, Gaccioli F, Majumder M, Mullins MR, Yuan CL, Papadopoulou B, Merrick WC, Komar AA, Taylor DJ, Hatzoglou M. 2011. Characterization of hibernating ribosomes in mammalian cells. Cell Cycle 10:2691–2702. 10.4161/cc.10.16.16844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ueta M, Yoshida H, Wada C, Baba T, Mori H, Wada A. 2005. Ribosome binding proteins YhbH and YfiA have opposite functions during 100S formation in the stationary phase of Escherichia coli. Genes Cells 10:1103–1112. 10.1111/j.1365-2443.2005.00903.x. [DOI] [PubMed] [Google Scholar]
- 56.Tagami K, Nanamiya H, Kazo Y, Maehashi M, Suzuki S, Namba E, Hoshiya M, Hanai R, Tozawa Y, Morimoto T, Ogasawara N, Kageyama Y, Ara K, Ozaki K, Yoshida M, Kuroiwa H, Kuroiwa T, Ohashi Y, Kawamura F. 2012. Expression of a small (p)ppGpp synthetase, YwaC, in the (p)ppGpp(0) mutant of Bacillus subtilis triggers YvyD-dependent dimerization of ribosome. Microbiologyopen 1:115–134. 10.1002/mbo3.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ueta M, Wada C, Wada A. 2010. Formation of 100S ribosomes in Staphylococcus aureus by the hibernation promoting factor homolog SaHPF. Genes Cells 15:43–58. 10.1111/j.1365-2443.2009.01364.x. [DOI] [PubMed] [Google Scholar]
- 58.Ueta M, Wada C, Daifuku T, Sako Y, Bessho Y, Kitamura A, Ohniwa RL, Morikawa K, Yoshida H, Kato T, Miyata T, Namba K, Wada A. 2013. Conservation of two distinct types of 100S ribosome in bacteria. Genes Cells 18:554–574. 10.1111/gtc.12057. [DOI] [PubMed] [Google Scholar]
- 59.Qin D, Fredrick K. 2013. Analysis of polysomes from bacteria. Methods Enzymol. 530:159–172. 10.1016/B978-0-12-420037-1.00008-7. [DOI] [PubMed] [Google Scholar]
- 60.Deutscher MP. 2009. Maturation and degradation of ribosomal RNA in bacteria. Prog. Mol. Biol. Transl. Sci. 85:369–391. 10.1016/S0079-6603(08)00809-X. [DOI] [PubMed] [Google Scholar]
- 61.Wilson DN. 2014. Ribosome-targeting antibiotics and mechanisms of bacterial resistance. Nat. Rev. Microbiol. 12:35–48. 10.1038/nrmicro3155. [DOI] [PubMed] [Google Scholar]
- 62.Lambert T. 2012. Antibiotics that affect the ribosome. Rev. Sci. Tech. 31:57–64. [DOI] [PubMed] [Google Scholar]
- 63.Modolell J, Vázquez D. 1977. The inhibition of ribosomal translocation by viomycin. Eur. J. Biochem. 81:491–497. 10.1111/j.1432-1033.1977.tb11974.x. [DOI] [PubMed] [Google Scholar]
- 64.Thom G, Prescott CD. 1997. The selection in vivo and characterization of an RNA recognition motif for spectinomycin. Bioorg. Med. Chem. 5:1081–1086. 10.1016/S0968-0896(97)00060-6. [DOI] [PubMed] [Google Scholar]
- 65.Brodersen DE, Clemons WM, Carter AP, Morgan-Warren RJ, Wimberly BT, Ramakrishnan V. 2000. The structural basis for the action of the antibiotics tetracycline, pactamycin, and hygromycin B on the 30S ribosomal subunit. Cell 103:1143–1154. 10.1016/S0092-8674(00)00216-6. [DOI] [PubMed] [Google Scholar]
- 66.Tenson T, Lovmar M, Ehrenberg M. 2003. The mechanism of action of macrolides, lincosamides and streptogramin B reveals the nascent peptide exit path in the ribosome. J. Mol. Biol. 330:1005–1014. 10.1016/S0022-2836(03)00662-4. [DOI] [PubMed] [Google Scholar]
- 67.Hartmann G, Honikel KO, Knusel F, Nuesch J. 1967. The specific inhibition of the DNA-directed RNA synthesis by rifamycin. Biochim. Biophys. Acta 145:843–844. 10.1016/0005-2787(67)90147-5. [DOI] [PubMed] [Google Scholar]
- 68.Hammes WP, Neuhaus FC. 1974. On the mechanism of action of vancomycin: inhibition of peptidoglycan synthesis in Gaffkya homari. Antimicrob. Agents Chemother. 6:722–728. 10.1128/AAC.6.6.722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Xu W, Huang J, Lin R, Shi J, Cohen SN. 2010. Regulation of morphological differentiation in S. coelicolor by RNase III (AbsB) cleavage of mRNA encoding the AdpA transcription factor. Mol. Microbiol. 75:781–791. 10.1111/j.1365-2958.2009.07023.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wolanski M, Donczew R, Kois-Ostrowska A, Masiewicz P, Jakimowicz D, Zakrzewska-Czerwińska J. 2011. The level of AdpA directly affects expression of developmental genes in Streptomyces coelicolor. J. Bacteriol. 193:6358–6365. 10.1128/JB.05734-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yamazaki H, Ohnishi Y, Horinouchi S. 2003. Transcriptional switch on of ssgA by A-factor, which Is essential for spore septum formation in Streptomyces griseus. J. Bacteriol. 185:1273–1283. 10.1128/JB.185.4.1273-1283.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Higo A, Hara H, Horinouchi S, Ohnishi Y. 2012. Genome-wide distribution of AdpA, a global regulator for secondary metabolism and morphological differentiation in Streptomyces, revealed the extent and complexity of the AdpA regulatory network. DNA Res. 19:259–273. 10.1093/dnares/dss010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Willemse J, Mommaas AM, van Wezel GP. 2012. Constitutive expression of ftsZ overrides the whi developmental genes to initiate sporulation of Streptomyces coelicolor. Antonie Van Leeuwenhoek 101:619–632. 10.1007/s10482-011-9678-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Figaro S, Durand S, Gilet L, Cayet N, Sachse M, Condon C. 2013. Bacillus subtilis mutants with knockouts of the genes encoding ribonucleases RNase Y and RNase J1 are viable, with major defects in cell morphology, sporulation, and competence. J. Bacteriol. 195:2340–2348. 10.1128/JB.00164-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mäder U, Zig L, Kretschmer J, Homuth G, Putzer H. 2008. mRNA processing by RNases J1 and J2 affects Bacillus subtilis gene expression on a global scale. Mol. Microbiol. 70:183–196. 10.1111/j.1365-2958.2008.06400.x. [DOI] [PubMed] [Google Scholar]
- 76.Durand S, Gilet L, Bessières P, Nicolas P, Condon C. 2012. Three essential ribonucleases-RNase Y, J1, and III-control the abundance of a majority of Bacillus subtilis mRNAs. PLoS Genet. 8:e1002520. 10.1371/journal.pgen.1002520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ohlsen KL, Grimsley JK, Hoch JA. 1994. Deactivation of the sporulation transcription factor Spo0A by the Spo0E protein phosphatase. Proc. Natl. Acad. Sci. U. S. A. 91:1756–1760. 10.1073/pnas.91.5.1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Matsuno K, Sonenshein AL. 1999. Role of SpoVG in asymmetric septation in Bacillus subtilis. J. Bacteriol. 181:3392–3401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gravenbeek ML, Jones GH. 2008. The endonuclease activity of RNase III is required for the regulation of antibiotic production by Streptomyces coelicolor. Microbiology 154:3547–3555. 10.1099/mic.0.2008/022095-0. [DOI] [PubMed] [Google Scholar]
- 80.Aceti DJ, Champness WC. 1998. Transcriptional regulation of Streptomyces coelicolor pathway-specific antibiotic regulators by the absA and absB loci. J. Bacteriol. 180:3100–3106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ciandrini L, Stansfield I, Romano MC. 2013. Ribosome traffic on mRNAs maps to gene ontology: genome-wide quantification of translation initiation rates and polysome size regulation. PLoS Comput. Biol. 9:e1002866. 10.1371/journal.pcbi.1002866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Valleriani A, Zhang G, Nagar A, Ignatova Z, Lipowsky R. 2011. Length-dependent translation of messenger RNA by ribosomes. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 83:042903. 10.1103/PhysRevE.83.042903. [DOI] [PubMed] [Google Scholar]
- 83.Hosaka T, Ohnishi-Kameyama M, Muramatsu H, Murakami K, Tsurumi Y, Kodani S, Yoshida M, Fujie A, Ochi K. 2009. Antibacterial discovery in actinomycetes strains with mutations in RNA polymerase or ribosomal protein S12. Nat. Biotechnol. 27:462–464. 10.1038/nbt.1538. [DOI] [PubMed] [Google Scholar]
- 84.Tanaka Y, Komatsu M, Okamoto S, Tokuyama S, Kaji A, Ikeda H, Ochi K. 2009. Antibiotic overproduction by rpsL and rsmG mutants of various actinomycetes. Appl. Environ. Microbiol. 75:4919–4922. 10.1128/AEM.00681-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Nishimura K, Hosaka T, Tokuyama S, Okamoto S, Ochi K. 2007. Mutations in rsmG, encoding a 16S rRNA methyltransferase, result in low-level streptomycin resistance and antibiotic overproduction in Streptomyces coelicolor A3(2). J. Bacteriol. 189:3876–3883. 10.1128/JB.01776-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Okamoto S, Lezhava A, Hosaka T, Okamoto-Hosoya Y, Ochi K. 2003. Enhanced expression of S-adenosylmethionine synthetase causes overproduction of actinorhodin in Streptomyces coelicolor A3(2). J. Bacteriol. 185:601–609. 10.1128/JB.185.2.601-609.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Srivastava AK, Schlessinger D. 1988. Coregulation of processing and translation: mature 5′ termini of Escherichia coli 23S ribosomal RNA form in polysomes. Proc. Natl. Acad. Sci. U. S. A. 85:7144–7148. 10.1073/pnas.85.19.7144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wada A, Igarashi K, Yoshimura S, Aimoto S, Ishihama A. 1995. Ribosome modulation factor: stationary growth phase-specific inhibitor of ribosome functions from Escherichia coli. Biochem. Biophys. Res. Commun. 214:410–417. 10.1006/bbrc.1995.2302. [DOI] [PubMed] [Google Scholar]
- 89.Yoshida H, Maki Y, Kato H, Hisao F, Izutsu K, Wada C, Wada A. 2002. The ribosome modulation factor (RMF) binding site on the 100S ribosome of Escherichia coli. J. Biochem. 132:983–989. 10.1093/oxfordjournals.jbchem.a003313. [DOI] [PubMed] [Google Scholar]
- 90.Izutsu K, Wada C, Komine Y, Sako T, Ueguchi C, Nakura S, Wada A. 2001. Escherichia coli ribosome-associated protein SRA, whose copy number increases during stationary phase. J. Bacteriol. 183:2765–2773. 10.1128/JB.183.9.2765-2773.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Trauner A, Lougheed KE, Bennett MH, Hingley-Wilson SM, Williams HD. 2012. The dormancy regulator DosR controls ribosome stability in hypoxic mycobacteria. J. Biol. Chem. 287:24053–24063. 10.1074/jbc.M112.364851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Maguire BA. 2009. Inhibition of bacterial ribosome assembly: a suitable drug target? Microbiol. Mol. Biol. Rev. 73:22–35. 10.1128/MMBR.00030-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Tu D, Blaha G, Moore PB, Steitz TA. 2005. Structures of MLSBK antibiotics bound to mutated large ribosomal subunits provide a structural explanation for resistance. Cell 121:257–270. 10.1016/j.cell.2005.02.005. [DOI] [PubMed] [Google Scholar]
- 94.Proshkin S, Rahmouni AR, Mironov A, Nudler E. 2010. Cooperation between translating ribosomes and RNA polymerase in transcription elongation. Science 328:504–508. 10.1126/science.1184939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.MacNeil DJ, Gewain KM, Ruby CL, Dezeny G, Gibbons PH, MacNeil T. 1992. Analysis of Streptomyces avermitilis genes required for avermectin biosynthesis utilizing a novel integration vector. Gene 111:61–68. 10.1016/0378-1119(92)90603-M. [DOI] [PubMed] [Google Scholar]
- 96.Paget MS, Chamberlin L, Atrih A, Foster SJ, Buttner MJ. 1999. Evidence that the extracytoplasmic function sigma factor sigmaE is required for normal cell wall structure in Streptomyces coelicolor A3(2). J. Bacteriol. 181:204–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.