Abstract
The ubiquitous opportunistic pathogen Pseudomonas aeruginosa has five aerobic terminal oxidases: bo3-type quinol oxidase (Cyo), cyanide-insensitive oxidase (CIO), aa3-type cytochrome c oxidase (aa3), and two cbb3-type cytochrome c oxidases (cbb3-1 and cbb3-2). These terminal oxidases are differentially regulated under various growth conditions and are thought to contribute to the survival of this microorganism in a wide variety of environmental niches. Here, we constructed multiple mutant strains of P. aeruginosa that express only one aerobic terminal oxidase to investigate the enzymatic characteristics and in vivo function of each enzyme. The Km values of Cyo, CIO, and aa3 for oxygen were similar and were 1 order of magnitude higher than those of cbb3-1 and cbb3-2, indicating that Cyo, CIO, and aa3 are low-affinity enzymes and that cbb3-1 and cbb3-2 are high-affinity enzymes. Although cbb3-1 and cbb3-2 exhibited different expression patterns in response to oxygen concentration, they had similar Km values for oxygen. Both cbb3-1 and cbb3-2 utilized cytochrome c4 as the main electron donor under normal growth conditions. The electron transport chains terminated by cbb3-1 and cbb3-2 generate a proton gradient across the cell membrane with similar efficiencies. The electron transport chain of aa3 had the highest proton translocation efficiency, whereas that of CIO had the lowest efficiency. The enzymatic properties of the terminal oxidases reported here are partially in agreement with their regulatory patterns and may explain the environmental adaptability and versatility of P. aeruginosa.
INTRODUCTION
Pseudomonas aeruginosa is an opportunistic pathogen responsible for severe nosocomial infections in immunocompromised individuals and chronic lung infections in patients with the genetic disease cystic fibrosis. The bacterium is ubiquitously distributed in soil and water environments and infects not only humans but also a wide variety of animals, insects, and plants (1). The ubiquity of P. aeruginosa is partly attributed to its ability to utilize various carbon sources and generate energy through multiple pathways (2–5). P. aeruginosa grows by aerobic respiration in the presence of oxygen or by dissimilatory nitrate respiration (denitrification) and arginine fermentation under anaerobic conditions (6, 7).
The genome of P. aeruginosa encodes at least five aerobic terminal oxidases, bo3-type oxidase (Cyo), aa3-type oxidase (aa3), cyanide-insensitive quinol oxidase (CIO), and two isoforms of cbb3-type oxidase (cbb3-1 and cbb3-2), which catalyze the four-electron reduction of molecular oxygen to water as the final reaction of the aerobic respiratory chain (5, 8). Cyo is encoded by the cyoABCDE genes (PA1317 to -1321), which are highly homologous with the corresponding genes of Escherichia coli, while aa3 is encoded by the coxBA-coxC (coIII) genes (PA0105 to -0108). The cioAB genes (PA3930-PA3929) encode CIO, which has homology to bd-type quinol oxidases (9), while the ccoN1O1Q1P1 (cco1 gene cluster; PA1554 to -1552) and ccoN2O2Q2P2 genes (cco2 gene cluster; PA1557 to -1555) encode cbb3-1 and cbb3-2, respectively. The known features of the five terminal oxidases of P. aeruginosa are summarized in Table S1 in the supplemental material.
Two groups of terminal oxidases are associated with aerobic respiration: the cytochrome bd family and the heme-copper oxidoreductase superfamily. CIO belongs to the cytochrome bd family, and although its primary structure is similar to those of canonical bd oxidases, it is phylogenetically distinct (9–11). Recent analyses in Gluconobacter oxydans showed that the CIO contains hemes b558, b595, and d, as in the case of canonical bd oxidases, and has a high turnover rate (12, 13). The cytochrome bd oxidase of E. coli has high affinity for oxygen, whereas CIOs of Campylobacter jejuni and G. oxydans have low affinity for oxygen (13, 14). In contrast, the CIO of P. aeruginosa was predicted to have high affinity for oxygen because a cco1 cco2 double mutant, which lacked cbb3-1 and cbb3-2, was able to grow under microaerobic conditions (2% O2), but a cco1 cco2 cio triple mutant, which lacked cbb3-1, cbb3-2, and CIO, did not grow under such conditions (15). However, the affinity of P. aeruginosa CIO for oxygen has not been determined to date.
The expression of CIO in P. aeruginosa is induced in the stationary phase or under low-oxygen or copper limitation conditions. Inactivation of the other terminal oxidases by respiratory inhibitors or gene disruption also leads to significant upregulation of the cio genes (15–21). The upregulation of CIO in the stationary phase is probably because P. aeruginosa produces cyanide, which inhibits heme-copper oxidases, in the stationary phase. The cio operon is positively regulated by RoxSR, which is a two-component regulatory system that may sense respiratory chain electron flow or redox status of the quinone pool, and is negatively regulated by ANR (anaerobic regulator of arginine deiminase and nitrate reductase), which is a low-oxygen-responsive global regulator (17, 18). The stationary-phase sigma factor RpoS also activates expression of the cio genes (21).
The terminal oxidases Cyo, aa3, cbb3-1, and cbb3-2 belong to the heme-copper oxidoreductase superfamily, whose members have a conserved catalytic core subunit, which contains a low-spin heme and a binuclear catalytic center, consisting of a high-spin heme and copper atom (CuB). Enzymes of this family are classified into three main types (A, B, and C) based on features of the core subunit (22). Cyo and aa3 of P. aeruginosa are type A enzymes, whereas cbb3-1 and cbb3-2 are type C enzymes. Cyo does not carry a CuA center, which receives electrons from cytochrome c, and is therefore considered to be a quinol oxidase. The cyo genes are expressed at low levels under normal growth conditions in LB medium but are upregulated by iron starvation (21). aa3 is closely related to mitochondrial terminal oxidases. aa3-type oxidases typically have low affinity for oxygen and function as main enzymes under high-oxygen conditions in many bacterial species (23–26). However, in P. aeruginosa, aa3 is expressed at very low levels in nutrient-rich medium under high-oxygen conditions and is upregulated only in response to nutrient starvation (21).
cbb3-1 and cbb3-2 are exclusively found in bacteria and are the most phylogenetically distant members of the heme-copper oxidoreductase superfamily (27). cbb3-type oxidases have very high affinity for oxygen. For example, the reported Km values for cbb3 oxidases of Bradyrhizobium japonicum and Campylobacter jejuni are 7 and 40 nM, respectively (14, 28). cbb3-type oxidases are induced under low-oxygen conditions in many bacteria or are the only terminal oxidase in several obligately microaerophilic bacteria, indicating that cbb3 oxidases are the major terminal oxidases in low-oxygen environments (14, 29–32). Similar to cbb3 oxidases from other bacteria, cbb3-2 of P. aeruginosa is highly induced under low-oxygen conditions. cbb3-2 is also induced in the stationary phase under aerobic conditions, probably because of low dissolved oxygen concentration due to high cell density. In contrast, cbb3-1 is constitutively expressed and is the major oxidase that functions in the exponential phase under high-oxygen conditions (15, 17). Thus, P. aeruginosa is unique in that it expresses cbb3-type oxidases as the dominant respiratory enzymes, even under high-oxygen conditions, under normal laboratory growth conditions.
In P. aeruginosa, the five known terminal oxidases are differentially regulated in response to growth conditions. ANR, RoxSR, the iron-responsive regulator Fur, and the stationary-phase sigma factor RpoS are involved in the regulation of these enzymes in response to oxygen concentration, presence of respiratory inhibitors, iron availability, and nutrient conditions (see Table S1 in the supplemental material) (5, 21). The presence of multiple terminal oxidases with distinct enzymatic features and the ability to regulate the expression of the enzymes that are most suitable for the growth conditions likely contribute to the ubiquity of P. aeruginosa in the environment. To better understand the mechanisms that allow P. aeruginosa to survive in changing environments, detailed analyses of the enzymatic features and in vivo function of each terminal oxidase are necessary. However, because multiple terminal oxidases are simultaneously expressed in P. aeruginosa cells, in vivo functional characterization of individual enzymes is difficult. In this work, we constructed multiple mutant strains of P. aeruginosa that express only one terminal oxidase to characterize the features of each enzyme.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains and plasmids used in this study are described in Table 1. P. aeruginosa PAO1ut, which was used as a wild-type strain in this work, is a PAO1 strain that is maintained in our laboratory (21). Cells were routinely grown in LB medium at 37°C. For aerobic cultivation, Erlenmeyer flasks or test tubes with gas-permeable plugs were used. For microaerobic cultivation, square bottles were used and the medium was continuously bubbled through a sparger with a gas mixture consisting of 2% O2 and 98% N2. For anaerobic cultivation, test tubes sealed with butyl rubber stoppers were used, the medium was supplemented with 100 mM sodium nitrate, and the gas phase was replaced with argon. For aerobic cultivation of the derivatives of E. coli MB43, square bottles were used and LB medium was continuously bubbled with air through a sparger. Bacterial colonies were isolated on LB agar plates or Pseudomonas isolation agar (BD Difco Laboratories, Sparks, MD). The AnaeroPack System (Mitsubishi Gas Chemical, Tokyo, Japan) was used for the anaerobic incubation of plates. The concentrations of the antibiotics were as follows: 100 μg/ml ampicillin and 12.5 μg/ml tetracycline (Tc) (for E. coli) and 200 μg/ml carbenicillin (Cb) and 150 μg/ml Tc (for P. aeruginosa). When necessary, 5% (wt/vol) sucrose or 0.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG) was added to the medium.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s)a | Reference or source |
|---|---|---|
| Strains | ||
| P. aeruginosa | ||
| PAO1ut | Our laboratory strain of PAO1 used as a wild type | 21 |
| QXBo | Deletion of cioAB, coxBA-coxC, ccoN1O1Q1P1, and ccoN2O2Q2P2 | This study |
| QXCi | Deletion of cyoABCDE, coxBA-coxC, ccoN1O1Q1P1, and ccoN2O2Q2P2 | This study |
| QXAa | Deletion of cyoABCDE, cioAB, ccoN1O1Q1P1, and ccoN2O2Q2P2 | This study |
| QXAaS2 | Suppressor mutant of QXAaS2; able to grow aerobically | This study |
| QXCb1 | Deletion of cyoABCDE, cioAB, coxBA-coxC, and ccoN2O2Q2P2 | This study |
| QXCb2 | Deletion of cyoABCDE, cioAB, coxBA-coxC, and ccoN1O1Q1P1 | This study |
| PTO5 | Deletion of cyoABCDE, cioAB, coxBA-coxC, ccoN1O1Q1P1, and ccoN2O2Q2P2 | This study |
| QXCb1Δcc4 | cc4 mutant of QXCb1 | This study |
| QXCb1ΔcycB | cycB mutant of QXCb1 | This study |
| QXCb1ΔPA4571 | PA4571 mutant of QXCb1 | This study |
| QXCb1ΔPA5491 | PA5491 mutant of QXCb1 | This study |
| QXCb2Δcc4 | cc4 mutant of QXCb2 | This study |
| QXCb2ΔcycB | cycB mutant of QXCb2 | This study |
| QXCb2ΔPA4571 | PA4571 mutant of QXCb2 | This study |
| QXCb2ΔPA5491 | PA5491 mutant of QXCb2 | This study |
| E. coli | ||
| JM109 | Host strain for DNA manipulation | 33 |
| S17-1 | C600::RP-4 2-(Tc::Mu)(Kan::Tn7) thi pro hsdR hsdM+ recA | 55 |
| MB43 | cydB cyoB appB nuoB, nonmarker derivative of MB44 | 43 |
| Plasmids | ||
| pUC18 | Cloning vector; Apr | 33 |
| pEX18Ap | Gene replacement vector; Apr Cbr oriT+ sacB+ | 34 |
| pFLP2 | Flp recombinase-expressing plasmid; Apr | 34 |
| pMMB67EH | IncQ, expression vector; Apr Cbr | 56 |
| pPS854tet | tet-FRT cassette, Apr Tcr | 35 |
| pEX-Δcyo | Plasmid for cyoABCDE mutation, a derivative of pEX18Ap, Tcr | This study |
| pEX-Δcio | Plasmid for cioAB mutation, a derivative of pEX18Ap | This study |
| pEX-Δcox | Plasmid for coxBA-coxC mutation, a derivative of pEX18Ap | This study |
| pEX-Δcco1 | Plasmid for ccoN1O1Q1P1 mutation, a derivative of pEX18Ap, Tcr | This study |
| pEX-Δcco2 | Plasmid for ccoN2O2Q2P2 mutation, a derivative of pEX18Ap, Tcr | This study |
| pEX-Δcc4 | Plasmid for cc4 mutation, a derivative of pEX18Ap | This study |
| pEX-ΔcycB | Plasmid for cycB mutation, a derivative of pEX18Ap | This study |
| pEX-ΔPA4571 | Plasmid for PA4571 mutation, a derivative of pEX18Ap | This study |
| pEX-ΔPA5491 | Plasmid for PA5491 mutation, a derivative of pEX18Ap | This study |
| pMMB-cc4 | cc4 on pMMB67EH | This study |
| pMMB-PA5491 | PA5491 on pMMB67EH | This study |
| pMMB-cioAB | cioAB on pMMB67EH | This study |
| pUC-cioAB | cioAB on pUC18 | This study |
| pUC-cydAB | cydAB of E. coli on pUC18 | This study |
Apr, ampicillin resistant; Cbr, carbenicillin resistant; Tcr, tetracycline resistant.
DNA manipulations.
Recombinant DNA experiments were performed using standard methods (33). DNA was introduced into P. aeruginosa strains by transconjugation with E. coli strain S17-1 or by electroporation. Ex Taq (TaKaRa, Kyoto, Japan) and KOD Plus polymerases (Toyobo, Osaka, Japan) were used for PCRs. Synthetic oligonucleotides were obtained from Sigma-Genosys (Hokkaido, Japan) or Greiner Bio One (Tokyo, Japan). The primers used for PCR are listed in Table S2 in the supplemental material.
Construction of mutant strains.
The cyoABCDE, ccoN1O1Q1P1, and ccoN2O2Q2P2 gene clusters were knocked out using the Flp-FRT recombination system (34) with constructs pEX-Δcyo, pEX-Δcco1, and pEX-Δcco2, respectively, according to the method described previously (35). For construction of pEX-Δcyo, 0.9- and 1.4-kb fragments containing the upstream and downstream flanking regions of the cyoABCDE gene cluster, respectively, were amplified by PCR from PAO1ut chromosomal DNA using primer sets BoR1/BoR2 and BoR3/BoR4, respectively, and were then tandemly inserted into the EcoRI-BamHI and BamHI-PstI sites, respectively, of pEX18Ap (34). The resulting construct was digested with BamHI, blunt ended by the Klenow reaction, and then ligated with a blunt-ended SacI-excised fragment containing the tet-FRT cassette from pPS854tet (35), resulting in pEX-Δcyo. pEX-Δcco1 and pEX-Δcco2 were constructed in the same manner using primer sets cbR3/cbR4 and cbR5/cbR6 and primer sets cbR1/cbR2 and cbR3/cbR4, respectively.
The cioAB and coxBA-PA0107-coxC gene clusters were knocked out by in-frame deletion with plasmids pEX-Δcio and pEX-Δcox, respectively, according to the method described previously (21). These plasmids were constructed using the same method as that for pEX-Δcyo, except that the tet-FRT cassette was not inserted. The primer sets used for construction of pEX-Δcio and pEX-Δcox were ciR1/ciR2 and ciR3/ciR4 and sets aaR1/aaR2 and aaR3/aaR4, respectively.
Mutant strains lacking the cc4, cycB, PA4571, or PA5491 gene were constructed by in-frame deletion using the same method as that for the cio and cox mutants with plasmids pEX-Δcc4, pEX-ΔcycB, pEX-ΔPA4571, and pEX-ΔPA5491, respectively. The primer sets used to amplify the upstream and downstream flanking regions of the respective genes for the construction of pEX-Δcc4, pEX-ΔcycB, and pEX-ΔPA4571 were cc4-d/cc4-c and cc4-b/cc4-a, cycB-d/cycB-c and cycB-b/cycB-a, and PA4571-a/PA4571-b and PA4571-c/PA4571-d, respectively. For construction of pEX-ΔPA5491, 1.4-kbp PCR fragments containing the upstream and downstream flanking regions of PA5491 were amplified from PAO1ut chromosomal DNA using primer sets PA5491-d/PA5491-c and PA5491-b/PA5491-a, respectively. The amplified fragments were used as the templates for a second PCR using the primer set PA5491-d/PA5491-a. The amplified fragment, which carried the upstream and downstream flanking regions of PA5491 but lacked the PA5491 coding region, was inserted into the XbaI-HindIII sites of pEX18Ap, resulting in pEX-ΔPA5491.
Strains with multiple mutations were constructed by stepwise mutation of the terminal oxidase genes. Because strains QXAa and PTO5, which lacked the cyo, cio, cco1, and cco2 gene clusters, could not be obtained by aerobic cultivation, they were isolated under anaerobic cultivation conditions.
Construction of complementation plasmids.
pMMB-cc4 and pMMB-PA5491 were constructed by insertion of PCR fragments carrying the cc4 and PA5491 genes, which were amplified with primer sets cc4-F/cc4-R and PA5491-F/PA5491-R, respectively, into the BamHI-HindIII and SacI-BamHI sites of pMMB67EH, respectively. pUC-cioAB was constructed by insertion of a PCR fragment carrying the cioAB genes, which was amplified with primers cio1 and cio2 from PAO1ut chromosomal DNA, into the EcoRI-XbaI sites of pUC18. pMMB-cioAB was constructed by insertion of the same fragment into pMMB67EH. pUC-cydAB was constructed by insertion of a PCR fragment carrying the cydAB genes, which was amplified with primers cyd1 and cyd2 from E. coli JM109 chromosomal DNA, into the BamHI-HindIII sites of pUC18.
Measurement of oxygen consumption.
Preparation of membrane fractions and measurements of oxygen consumption were performed according to the methods described by Cunningham and Williams (10). Briefly, P. aeruginosa strains were grown in LB medium until the optical density at 600 nm (OD600) of the culture reached approximately 0.85. Cells were pelleted by centrifugation at 6,000 × g, washed twice with 50 mM potassium phosphate buffer (pH 7.0), and resuspended in 10 mM potassium phosphate buffer (pH 7.0) containing 5 mM MgCl2. DNase at 0.01 mg/ml (final concentration) was added to the cell suspension, and the cells were then disrupted by passing the suspension twice through a French pressure cell at 16,000 lb/in2. Unbroken cells were removed by centrifugation at 6,000 × g. The membrane fraction was collected by centrifugation at 100,000 × g for 1 h at 4°C and resuspended in 10 mM potassium phosphate buffer (pH 7.0) containing 5 mM MgCl2 to give a protein concentration between 10 and 20 mg/ml. The protein concentration was determined by the Bradford method using a protein assay kit (Bio-Rad, Hercules, CA) with bovine serum albumin as the standard.
Oxygen consumption activity was measured amperometrically using an Apollo 4000 free radical analyzer equipped with a 2-mm Iso-Oxy-2 O2 electrode (WPI, Sarasota, FL). The reaction was performed in a multiport chamber (WPI) at 25°C. The reaction mixture (2.5 ml) contained 33 mM potassium phosphate (pH 7.0) and 0.5 mM NADH or 0.1 mM tetramethylene phenylene diamine (TMPD) and 5 mM ascorbate. When necessary, 1 mM KCN or 10 μg/ml antimycin A was added to the reaction mixture. The reaction was started by adding a volume of the membrane fraction equivalent to 0.5 mg protein to the air-saturated reaction mixture (O2 concentration was assumed to be 240 μM at 25°C), and the oxygen consumption activity was calculated by the decline of the O2 concentration.
Determination of oxygen affinity.
For determination of the Km values for oxygen using an oxygen electrode, membrane fractions from each of the terminal oxidase mutants were prepared as described above. The reaction was performed in an anaerobic vial (68-ml total volume) equipped with an Iso-Oxy-2 O2 electrode (WPI), and the reaction mixture (64 ml) contained 33 mM potassium phosphate (pH 7.0) and 30 mM malate. After most of the oxygen in the vial was removed by bubbling argon gas for 10 min, a volume of the membrane fraction equivalent to 0.5 mg protein was added to the reaction buffer. The consumption of dissolved oxygen was monitored with an Apollo 4000 free radical analyzer (WPI) and the Km values for oxygen were determined from Hanes-Woolf plots (see Fig. S1 in the supplemental material).
Determination of the Km values using deoxygenation kinetics of oxymyoglobin or oxyleghemoglobin was performed according to the method described previously (36–38). Membrane fractions were prepared as described above except that 20 mM potassium phosphate buffer (pH 7.0) containing 1 mM EDTA was used for washing and resuspension of the cells. Equine heart myoglobin (Sigma-Aldrich, St. Louis, MO) and partially purified leghemoglobin from the nodules of pea (Pisum sativum), which was kindly provided by N. Suganuma (Aichi University of Education), were reduced with dithionite and desalted by passage through a PD-10 column (GE Healthcare). The concentrations of myoglobin and leghemoglobin were determined by the spectrum of alkaline pyridine hemochrome using an extinction coefficient value for pyridine hemochrome of 33.9 mM−1 cm−1 at 556 nm (39). The reaction was performed in a sealed cuvette containing 5 ml of the degassed reaction mixture, which consisted of 20 mM potassium phosphate (pH 7.0), 1 mM EDTA, 15 mM d,l-malate, and 19.3 mM oxymyoglobin or oxyleghemoglobin. After addition of the membrane fraction sample (20 to 70 μl), the deoxygenation of oxymyoglobin or oxyleghemoglobin was continuously monitored at 581 and 563 nm or 572 and 560 nm, respectively, using a U-3210 or U-2910 spectrophotometer (Hitachi, Tokyo, Japan). The concentration of oxygen in the reaction mixture and the oxygen consumption rate at several time points were calculated according to the method described by Bergersen and Turner (36). The values for the dissociation constant used in the calculations were 0.786 and 0.05 μM for oxymyoglobin and oxyleghemoglobin, respectively. The Km values for oxygen were determined from Eadie-Hofstee plots (see Fig. S2 and S3 in the supplemental material).
Determination of proton translocation efficiency.
P. aeruginosa strains were grown under aerobic conditions in LB medium until the OD600 reached 1.3 to 1.6. E. coli MB43 derivatives used for determination of the H+/O ratio of CIO and the bd oxidase 1 were grown for 24 h. All cells were collected by centrifugation at 10,200 × g for 10 min at 4°C and washed twice with wash buffer, consisting of 300 mM sucrose and 10 mM MgCl2. The cell pellet was resuspended in wash buffer at an OD600 of 200 to 300. The H+/O ratio of the cell suspension was determined by measuring the changes of pH upon oxygen pulse using a pH electrode (6066-10C; Horiba, Kyoto, Japan) connected to a pH meter (F-53; Horiba). The oxygen pulse reaction was performed at 25°C in an anaerobic vessel equipped with the pH electrode. The reaction mixture contained 0.5 mM morpholinepropanesulfonic acid (MOPS) (pH 7.0), 100 mM KCl, 100 mM KSCN, 30 mM sucrose, 1 mM MgCl2, and 800 μl of the cell suspension in a total volume of 8 ml. Electron donors were not added to the reaction mixture because the cells had measurable oxygen consumption activity using endogenous electron donors and the addition of exogenous substrates had no effect on the activity, as was reported for Rhodobacter species (40). After the mixture was flushed with argon and the pH was adjusted to 7.0 with 1 N KOH, the reaction was started by adding a defined amount of oxygen with 60 μl of air-equilibrated water. Calibration of the pH electrode was done by the addition of 50 μl of deoxygenated 3 mM HCl. The H+/O ratio was calculated from the pH change and the amount of added oxygen.
RESULTS
Growth profile of multiple terminal oxidase knockout mutants.
Five terminal oxidase quadruple mutant strains, QXBo, QXCi, QXAa, QXCb1, and QXCb2, which carried only one complete terminal oxidase gene cluster, encoding Cyo, CIO, aa3, cbb3-1, and cbb3-2, respectively, and a quintuple mutant strain, PTO5, which lacked all five terminal oxidase gene clusters, were constructed by repeated homologous recombination (Table 1). Strains QXBo, QXCi, QXCb1, and QXCb2 were able to grow aerobically, whereas strains QXAa and PTO5 did not grow under this condition (Fig. 1A; also see Fig. S4A in the supplemental material). These results indicated that Cyo, CIO, cbb3-1, and cbb3-2 independently supported the growth of P. aeruginosa. The growth rate of QXCb2 under aerobic conditions was lower than that of QXCb1, QXBo, or QXCi (Fig. 1A). When Cyo or CIO was utilized as the terminal oxidase, the final OD600 was lower than that in the strains with only functional cbb3-1 or cbb3-2. Under microaerobic (2% O2) conditions, QXCb1 and QXCb2 had growth rates similar to that of the wild-type strain, but QXBo and QXCi exhibited poor growth (Fig. 1B; see also Fig. S4B).
FIG 1.
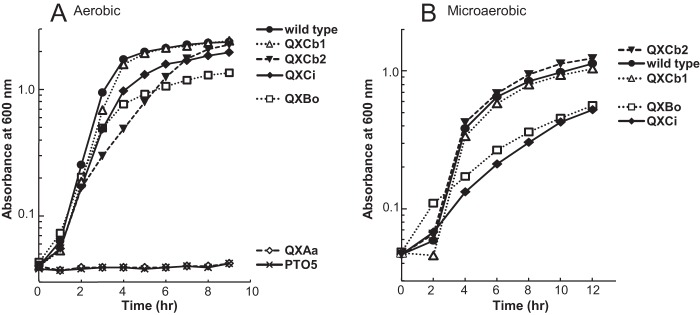
Growth profiles of the quadruple terminal oxidase mutant strains of P. aeruginosa. (A) Growth under aerobic conditions. The strains were cultivated in 40 ml LB medium in 300-ml Erlenmeyer flasks with shaking at 180 rpm. (B) Growth under microaerobic conditions (2% O2). The strains were cultivated in 150 ml LB medium in 300-ml square bottles. The medium was bubbled with a gas mixture consisting of 2% O2 and 98% N2 at a gas flux rate of 150 ml/min. The data shown are representatives from at least three independent experiments. The same plots in linear scale are shown in Fig. S4 in the supplemental material.
Although QXAa was unable to grow aerobically, a spontaneous suppressor mutant strain of QXAa, QXAaS2, which displayed slow growth under aerobic conditions in LB medium, was obtained after repeated aerobic subculturing. No mutation was found in the structural genes encoding aa3 by nucleotide sequencing analysis of the cox genes of QXAaS2, indicating that aa3 had the enzymatic ability to support the aerobic growth of P. aeruginosa (data not shown). QXAaS2 was therefore used for further enzymatic characterization of aa3. The detailed isolation procedure and characterization of QXAaS2 will be published elsewhere.
Electron donors of terminal oxidases.
aa3, cbb3-1, and cbb3-2 are thought to function as cytochrome c oxidases, whereas Cyo and CIO have been characterized as quinol oxidases. Here, we examined the physiological electron donor for each terminal oxidase by measuring the oxygen consumption activity in the membrane fractions of each mutant strain grown under aerobic conditions (Fig. 2). When NADH was used as an electron donor, all strains that expressed only one terminal oxidase showed high oxygen consumption activity. The activities of QXBo and QXCi were higher than those of QXAaS2, QXCb1, and QXCb2 (Fig. 2A). Strain PTO5 grown under anaerobic denitrification conditions had no detectable oxygen consumption activity (data not shown). When 1 mM cyanide was added to the reaction mixture, only QXCi displayed high activity, indicating that CIO exclusively mediates cyanide-insensitive respiration under the tested conditions (Fig. 2B). Antimycin A, which is an inhibitor of the cytochrome bc1 complex, significantly inhibited the oxygen consumption activities of QXAaS2, QXCb1, and QXCb2 but had only a minimal effect on QXBo and QXCi (Fig. 2C). In the presence of ascorbate-reduced TMPD, which specifically reduces cytochrome c and was used as an artificial electron donor, QXAaS2, QXCb1, and QXCb2 showed high oxygen-reducing activities, whereas QXBo and QXCi had almost no detectable activity (Fig. 2D). Together, these results clearly indicated that aa3, cbb3-1, and cbb3-2 function as cytochrome c oxidases and that Cyo and CIO function as quinol oxidases in vivo.
FIG 2.
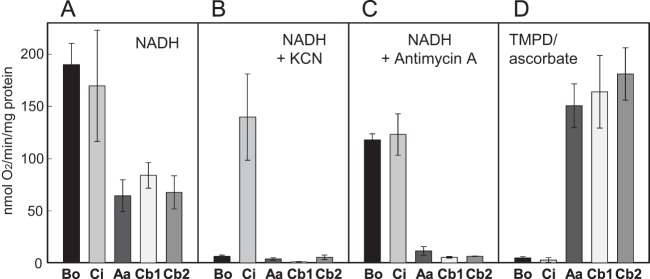
Oxygen consumption activities of the membrane fractions of the quadruple terminal oxidase mutant strains. Bo, Ci, Aa, Cb1, and Cb2 indicate membrane fractions of QXBo, QXCi, QXAaS2, QXCb1, and QXCb2, respectively. A 0.5 mM concentration of NADH (A, B, and C) and a 0.1 mM concentration of TMPD reduced by 5 mM ascorbate (D) were used as electron donors. A 1 mM concentration of KCN (B) or 10 μg/ml antimycin A (C) was added to the reaction mixture to inhibit the heme-copper oxidases or the cytochrome bc1 complex, respectively. The data are means of results from three independent experiments. Error bars indicate standard deviations from the means.
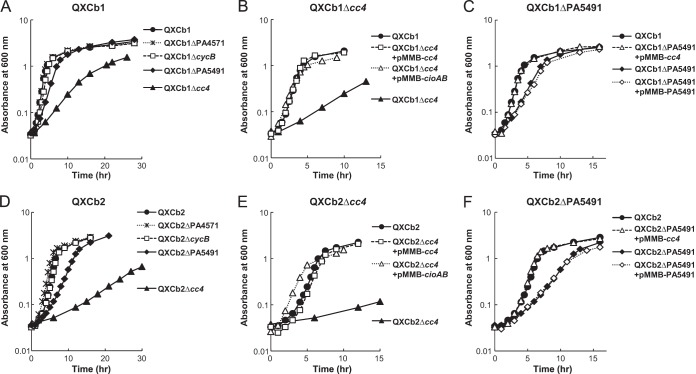
The genome of P. aeruginosa contains several cytochrome c genes. In previous and unpublished microarray experiments, we showed that cc4 (PA5490) and cycB (PA5300), encoding cytochromes c4 and c5, respectively, and PA4571 and PA5491, encoding uncharacterized cytochromes c, are expressed at relatively high levels (21). cc4 and PA5491 were constitutively expressed probably as an operon. cycB was largely expressed in the exponential phase, and its expression level decreased in the stationary phase. PA4571 showed ANR-dependent expression. To identify which cytochromes c are utilized as physiological electron donors for cbb3-1 and cbb3-2, these four cytochrome c genes were disrupted in the mutant strains QXCb1 and QXCb2, and the growth profiles of the resulting mutants were investigated (Fig. 3A and D). Disruption of cycB or PA4571 had no effect on the growth rates of either of QXCb1 and QXCb2; however, disruption of cc4 or PA5491 severely or slightly inhibited, respectively, the growth of these strains. The impaired growth of the cc4 mutants of QXCb1 and QXCb2 was restored by the introduction of a plasmid expressing cc4 (Fig. 3B and E). The poor growth of the PA5491 mutants was also restored by complementation with cc4 but not with PA5491 (Fig. 3C and F). These results indicated that the two cbb3-type oxidases preferentially utilize cytochrome c4 as an electron donor in vivo under the tested conditions. As PA5491 is located upstream of cc4 and is predicted to be transcribed with cc4 as part of an operon, disruption of PA5491 might affect the expression of cc4. The impaired growth of the cc4 mutants was not due to a deficiency in the function of other enzymes that use cytochrome c4 as an electron donor or acceptor, because the growth of these mutants was restored by complementation with the cioAB genes encoding the alternative quinol oxidase CIO (Fig. 3B and E).
FIG 3.
Effect of knockout of the cytochrome c genes on growth rates. Growth profiles of the derivatives of QXCb1 (A) and QXCb2 (D). Complementation of QXCb1Δcc4 (B), QXCb1ΔPA5491 (C), QXCb2Δcc4 (E), and QXCb2ΔPA5491 (F). The strains were grown aerobically in 40 ml LB medium in 100-ml Erlenmeyer flasks with shaking at 200 rpm. The data shown are representatives from three independent experiments. The same plots in linear scale are shown in Fig. S5 in the supplemental material.
Oxygen affinity of terminal oxidases.
The Km values of the terminal oxidases for oxygen were determined by the amperometric method using an oxygen electrode and by the spectrophotometric method with myoglobin or leghemoglobin as oxygen reporters (Table 2). Plots of the representative results for both methods are shown in Fig. S1, S2, and S3 in the supplemental material. The Km value of aa3 could not be determined using myoglobin, because oxymyoglobin was immediately oxidized to metmyoglobin when the membrane fraction of QXAaS2 was added to the reaction mixture. The Km values determined by measurements using an oxygen electrode were significantly higher than those determined using myoglobin, likely because of the relatively low sensitivity and slow response of the electrode to change in oxygen concentration (41). Although the oxygen electrode data underestimated the actual affinity of the terminal oxidases for oxygen, the relative difference of the estimated Km values was similar to that of the spectrophotometric data. Relative comparisons of the data showed that the Km values of Cyo, CIO, and aa3 were similar to each other and higher than those of cbb3-1 and cbb3-2, clearly indicating that Cyo, CIO, and aa3 are low-affinity enzymes.
TABLE 2.
Affinity of terminal oxidases for oxygena
| Terminal oxidaseb | Oxygen electrode |
Myoglobin |
Leghemoglobin |
|||
|---|---|---|---|---|---|---|
| Km (μM) | Vmax (nmol/min/mg) | Km (μM) | Vmax (nmol/min/mg) | Km (nM) | Vmax (nmol/min/mg) | |
| bo3 | 3.2 ± 1.3 | 117 ± 14 | 0.25 ± 0.04 | 92 ± 13 | NDc | ND |
| CIO | 4.0 ± 2.1 | 213 ± 34 | 0.41 ± 0.10 | 130 ± 34 | ND | ND |
| aa3 | 4.3 ± 1.0 | 91 ± 38 | ND | ND | ND | ND |
| cbb3-1 | 0.25 ± 0.02 | 77 ± 30 | 0.044 ± 0.022 | 51 ± 15 | 6.6 | 7.7 |
| cbb3-2 | 0.23 ± 0.08 | 82 ± 35 | 0.032 ± 0.021 | 53 ± 25 | 6.5 | 8.5 |
The Km values for oxygen were determined using an oxygen electrode or deoxygenation kinetics of oxymyoglobin or oxyleghemoglobin. Values for the oxygen electrode and myoglobin methods are presented as means ± standard deviations of results from at least three independent experiments. Values for the leghemogbobin method were determined from only one experiment.
Membrane fractions of the strains QXBo, QXCi, QXAaS2, QXCb1, and QXCb2 were used for determination of the oxygen affinity of bo3, CIO, aa3, cbb3-1, and ccb3-2 oxidases, respectively.
ND, not determined.
The Km values of cbb3-1 and cbb3-2 for oxygen did not significantly differ and were approximately 1 order of magnitude lower than those of the other three enzymes when evaluated by either method (Table 2). The Km values of cbb3-1 and cbb3-2 determined by the myoglobin method were comparable to the reported values of the cbb3-type oxidases from B. japonicum (7 nM) and C. jejuni (40 nM) (14, 28), indicating that both cbb3-1 and cbb3-2 are high-affinity enzymes. However, because these values were lower than the working range (0.1 to 10 μM) for myoglobin (37), we also determined the Km values of cbb3-1 and cbb3-2 using partially purified pea nodule leghemoglobin (working range, 3 to 100 nM). The determined Km values for cbb3-1 and cbb3-2 using leghemoglobin were 6.6 and 6.5 nM, respectively. However, the experiments using leghemoglobin were performed only once due to the limited amount of leghemoglobin that was available.
CIO of P. aeruginosa was previously predicted to have high affinity for oxygen (15). However, the Km value of CIO for oxygen measured here (Table 2) and the observed growth profile of QXCi under microaerobic conditions (Fig. 1B) clearly showed that P. aeruginosa CIO is a low-affinity enzyme, similar to the CIOs of C. jejuni and G. oxydans (13, 14). The low affinity of these CIOs is in contrast to the high affinity of the canonical bd oxidase of E. coli (42). To identify the functional difference between P. aeruginosa CIO and the canonical bd oxidase of E. coli, the P. aeruginosa cioAB genes or E. coli cydAB genes, which encode cytochrome bd oxidase 1, were transformed into E. coli MB43 (43), which lacks all three terminal oxidases and proton-pumping NADH dehydrogenase (NDH), and the growth profiles of the recombinant strains, MB43(pUC-cioAB) and MB43(pUC-cydAB), were compared under different oxygen conditions. MB43 does not grow by aerobic respiration because it has no proton-pumping activity, but aerobic growth was restored by the introduction of cioAB, indicating that CIO was functional in E. coli (Fig. 4; also see Fig. S6 in the supplemental material). Although the lag period of MB43(pUC-cioAB) under aerobic conditions was longer than that of MB43(pUC-cydAB), the maximum growth rates of the two strains were nearly identical. In contrast, the growth rate of MB43(pUC-cioAB) under microaerobic conditions was lower than that of MB43(pUC-cydAB). These results clearly showed that CIO was not optimally functional under low-oxygen conditions.
FIG 4.
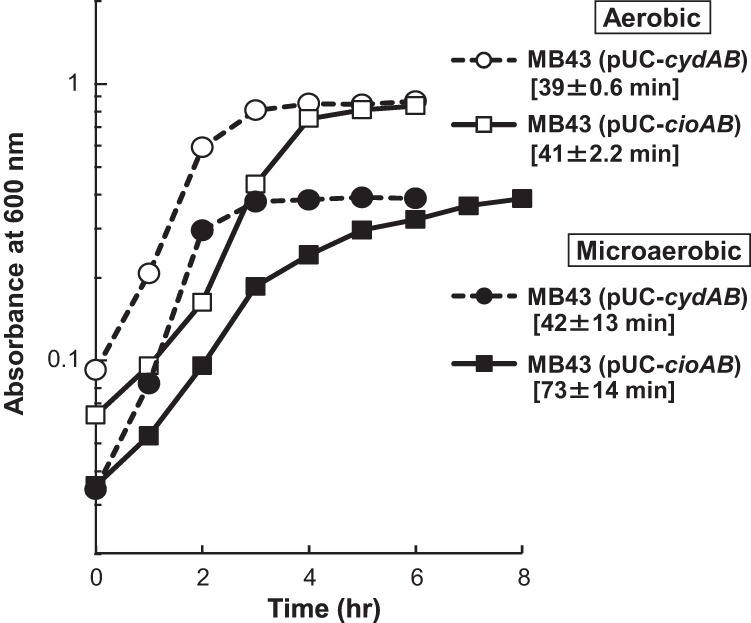
Functional heterologous expression of CIO in E. coli MB43, which lacks type 1 NADH dehydrogenase and three quinol oxidases. MB43 was transformed with pUC-cioAB or pUC-cydAB, and the transformants were cultivated in 100 ml LB medium in 300-ml square bottles. The medium was bubbled with air at a gas flux rate of 500 ml/min for aerobic conditions or with a gas mixture consisting of 2% O2 and 98% N2 at a gas flux rate of 150 ml/min for microaerobic conditions. The data shown are representatives from two independent experiments. The doubling times ± standard deviations are indicated in brackets. The same plots in linear scale are shown in Fig. S6 in the supplemental material.
Proton translocation efficiency of the ETC of terminal oxidases.
Aerobic respiration utilizes the proton motive force for ATP synthesis. Therefore, the ATP generation efficiency of aerobic respiration closely correlates with the efficiency of the electron transport chain (ETC) to create a proton gradient across the cell membrane. Protons are translocated across the cell membrane during the transfer of electrons through NDH, cytochrome bc1 complex (complex III), and terminal oxidases. As P. aeruginosa has proton-pumping type I NDH (NDH-1) and non-proton-pumping type II NDH (NDH-2), the net number of protons translocated across the cell membrane per oxygen atom consumed (H+/O ratio) depends on the proportion of NDH-1 and NDH-2 and the composition of terminal oxidases. Here, the H+/O ratio was determined by the oxygen pulse method using a cell suspension of each mutant to evaluate the contribution of each terminal oxidase for generation of ATP. The H+/O ratios of the ETCs terminated by Cyo, CIO, aa3, cbb3-1, and cbb3-2 were approximately 4, 3, 6, 4, and 4, respectively (Table 3).
TABLE 3.
Proton translocation efficiency of whole cells expressing only one terminal oxidase
| Strain | Terminal oxidase | H+/O ratioa |
|---|---|---|
| QXBo | bo3 | 4.04 ± 0.190 |
| QXCi | CIO | 2.83 ± 0.211 |
| QXAaS2 | aa3 | 6.28 ± 0.281 |
| QXCb1 | cbb3-1 | 4.07 ± 0.134 |
| QXCb2 | cbb3-2 | 4.16 ± 0.235 |
| MB43(pUC-cioAB) | CIO | 1.85 ± 0.111 |
| MB43(pUC-cydAB) | E. coli bd oxidase 1 | 2.04 ± 0.176 |
The H+/O ratio was determined by the oxygen pulse method. The aerobically grown stationary-phase cells were used. Values are means ± standard deviations of results from at least three independent experiments.
To compare the proton translocation stoichiometry of CIO with that of canonical cytochrome bd oxidase, the H+/O ratios of E. coli MB43(pUC-cioAB) and MB43(pUC-cydAB) were also determined. Because MB43 does not have NDH-1, the values represent the H+/O ratios of the quinol oxidase enzymes. The H+/O ratio of CIO in E. coli MB43 was approximately 2, a ratio that was nearly identical with that of the bd oxidase 1 of E. coli (Table 3). This result indicated that the proton translocation stoichiometry of CIO was identical with that of the canonical cytochrome bd oxidase, irrespective of their differing affinities for oxygen.
DISCUSSION
P. aeruginosa has two tandemly located cco gene clusters encoding the cbb3-type cytochrome c oxidases cbb3-1 and cbb3-2. The former is constitutively expressed, whereas the latter is induced under low-oxygen conditions and during the stationary growth phase (15, 17, 21). In the present study, we demonstrated that the growth rates of the quadruple terminal oxidase mutant strains QXCb1 and QXCb2, which expressed only cbb3-1 or cbb3-2, respectively, were comparable to or slightly higher than that of the wild-type strain under aerobic and microaerobic conditions, respectively (Fig. 1; see also Fig. S4 in the supplemental material). These results indicate that cbb3-1 and cbb3-2 are the main functioning terminal oxidases in P. aeruginosa under aerobic and microaerobic conditions, respectively, although the two enzymes have similar high affinities for oxygen (Table 2). High-affinity terminal oxidases are typically induced under low-oxygen conditions in many bacterial species and are often repressed under high-oxygen conditions, in which low-affinity terminal oxidases are the dominant enzymes (5, 44). Thus, the finding that high-affinity terminal oxidases predominantly function in P. aeruginosa even under high-oxygen conditions indicates that this is a characteristic feature of this ubiquitous organism.
Many pseudomonads, including Pseudomonas putida, Pseudomonas fluorescens, and Pseudomonas stutzeri, have two sets of gene clusters encoding cbb3 oxidases corresponding to cbb3-1 and cbb3-2, suggesting that carrying these two isoforms is advantageous for survival. Recently, Xie et al. (45) purified and characterized the two cbb3 isoforms from P. stutzeri and found that they differed in thermal stability but did not have significant difference in the UV-visible spectrum or oxygen reductase and catalase activities. Here, we also found no significant difference in oxygen affinity or proton translocation efficiency between the two cbb3 isoforms of P. aeruginosa. In addition, both cbb3-1 and cbb3-2 utilized cytochrome c4 as the main electron donor under the tested aerobic conditions in rich medium. However, a difference was found between the aerobic growth profiles of strains QXCb1 and QXCb2, which express only cbb3-1 and cbb3-2, respectively, as the growth rate of QXCb2 was lower than that of QXCb1 (Fig. 1A; see also Fig. S4A in the supplemental material). The main reason for the difference was likely due to the low expression level of cbb3-2 during the exponential phase (21), but it also is possible that cbb3-2 is sensitive to high intracellular oxygen tension or oxidative stress.
The ETCs of P. aeruginosa terminated by cbb3-1 and cbb3-2 had nearly identical proton translocation stoichiometry (Table 3). Compared to type A family enzymes, cbb3 oxidases have only one of two possible proton-conducting channels (46), and their proton-pumping efficiency remains a point of debate (40, 47). The H+/O ratios determined in the present study were reflective of all active ETCs using endogenous electron donors. When respiratory chain inhibitors and artificial electron donors were added in our experimental system, clear results could not be obtained, likely due to the low permeability of the outer membrane of P. aeruginosa cells or the presence of endogenous electron donors. Therefore, the enzymatic proton-pumping efficiency of each terminal oxidase could not be determined. Although the H+/O ratio of the ETC might be affected by the proportion of electron flow through NDH-1 and NDH-2 and the constitution of endogenous electron donors, the present results clearly indicate that P. aeruginosa cells produce less energy when cbb3 oxidases are utilized than when aa3 is utilized (Table 3).
aa3 is a close phylogenetic relative of the mitochondrial terminal oxidase. The ETC of aa3 generated a proton gradient across the cell membrane with the highest efficiency of all five examined terminal oxidases, indicating that aa3 most efficiently generates ATP per molecule of nutrient (Table 3). This result is in good agreement with the fact that aa3 is induced under nutrient starvation conditions (21). However, it was unexpected that the quadruple mutant strain QXAa was unable to grow aerobically (Fig. 1A), because aa3-type oxidases play major roles in aerobic growth in many bacterial species (23–26). Transcription of the cox genes encoding aa3 is repressed by the two-component regulator RoxSR and is dependent on the stationary-phase sigma factor RpoS (21). RoxSR is homologous to RegBA/PrrBA-type regulators, which regulate the transcription of photosynthesis genes of purple nonsulfur photosynthetic bacteria and are speculated to sense the electron flow of ETC or the redox status of the quinone pool (48, 49). One reason for the inability of strain QXAa to grow under aerobic conditions is that the cox genes may be tightly repressed in nutrient-rich LB medium.
The quinol oxidase Cyo was found to have low affinity for oxygen (Table 2). The gene arrangement and sequence of Cyo are similar to those of the E. coli bo3 quinol oxidase, which is the main terminal oxidase in E. coli under aerobic conditions. The bo3 oxidase also plays a major role under aerobic conditions in P. putida, which is a nonpathogenic relative of P. aeruginosa that also has five terminal oxidases corresponding to those of P. aeruginosa (50). In P. putida, knockout of the cyo genes significantly changes the transcriptome profile and relieves catabolite repression of the phenol and alkane degradation genes (51–53). In contrast to P. putida, Cyo of P. aeruginosa seems to play a minor role under normal growth conditions in LB medium because the expression level of the cyo genes is low (21) and aerobic growth is not affected in QXCb1, which expresses cbb3-1 but lacks Cyo (Fig. 1). This difference in the major terminal oxidases of P. aeruginosa and P. putida might reflect the unique characteristics of each species. The cyo genes are upregulated in P. aeruginosa by the nitric oxide (NO)-generating reagent S-nitrosoglutathione (GSNO) or by iron starvation, indicating that Cyo is utilized under these stress conditions.
The copper-free quinol oxidase CIO belongs to the cytochrome bd family but is phylogenetically and characteristically distinct from the canonical bd oxidases (9–14). The amino acid sequences of the two subunits of CIO, CioA and CioB, are highly similar to those of the canonical bd oxidases CydA and CydB, respectively, but the conserved sequence of the periplasmic loop (Q loop) that contains the putative quinol-oxidizing site is significantly shorter in CioA than in CydA (9). CIO of P. aeruginosa was predicted to have high affinity for oxygen, but its Km value for oxygen determined in this study was comparable to those of aa3 and Cyo, indicating that it is a low-affinity enzyme, similar to the CIOs of C. jejuni and G. oxydans (Table 2) (13, 14). The low affinity of CIOs for oxygen is in contrast to the high affinity of the canonical bd oxidase of E. coli (42, 43). The growth rate of strain QXCi under the microaerobic conditions was lower than those of the strains that expressed the cbb3 oxidases (Fig. 1B). It was also reported that CIO did not support the growth of P. aeruginosa under 0.4% O2 conditions (15), indicating that CIO does not function under very low O2 conditions. A cco1 cco2 double mutant was able to grow under 2% O2 conditions, but a cco1 cco2 cio triple mutant did not grow under the conditions in synthetic medium (15). The inability of the triple mutant to grow under the conditions was probably because of the sensitivity of Cyo and aa3 to cyanide, which would be produced under low-O2 conditions. However, Cyo was able to support the growth under 2% O2 conditions in LB medium in the present study (Fig. 1B; see also Fig. S4B in the supplemental material). One of the reasons for the contradictory results might be the different stress resistances of the cells grown in different media.
Despite differences in oxygen affinities, the proton translocation stoichiometry of CIO was nearly identical with that of the E. coli bd oxidase (Table 3). The bd oxidases do not pump protons, but the oxidation of the quinol at the periplasmic side of the membrane and subsequent uptake of protons from the cytoplasm for formation of water result in the H+/O stoichiometry of 2 (43). The H+/O ratio of the entire ETC terminated by CIO was lowest among the ETCs involving the other terminal oxidases, indicating that the utilization of CIO is not efficient for the generation of ATP. CIO is induced by inhibitors of the respiratory chain, cyanide and sodium nitroprusside (SNP), or by copper starvation (16, 18, 19, 21). Thus, CIO acts as a complementary enzyme when the heme-copper oxidases are not functioning or are inhibited. The low proton translocation efficiency of CIO might also be advantageous to P. aeruginosa for maintaining the redox balance of the ETC when the respiratory electron flow is not consistent.
Conclusions and perspectives.
Modulating the expression of multiple terminal oxidases with unique properties likely contributes to survival and proliferation of P. aeruginosa in diverse environmental niches. In this study, we characterized the enzymatic properties of the five known terminal oxidases. The type A heme-copper superfamily enzymes, aa3 and Cyo, had low affinity for oxygen. The aa3 and bo3 oxidases typically play a major role in respiration under aerobic conditions in many bacterial species (23–26, 50, 54). However, aa3 and Cyo are specifically utilized in P. aeruginosa under starvation and specific stress conditions, respectively. Expression of aa3 under starvation conditions is consistent with the fact that the ETC terminated by aa3 has the highest energy generation efficiency among the five terminal oxidases examined here. The non-heme-copper oxidase CIO was also found to have low affinity for oxygen, and its energy generation efficiency was identical to that of the canonical bd oxidase. P. aeruginosa has two cbb3 oxidases, cbb3-1 and cbb3-2, which belong to the type C family of heme-copper oxidases. Although both oxidases had high affinity for oxygen and the two were functionally similar, cbb3-1 and cbb3-2 played major roles in respiration under aerobic and microaerobic conditions, respectively. Utilization of cbb3-1 as the main terminal oxidase under aerobic conditions may contribute to the resistance of P. aeruginosa to reactive oxygen species. Identification of the enzymatic differences between the two cbb3 oxidase isoforms should be a future target for investigation.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to M. Bekker (University of Amsterdam) and N. Suganuma (Aichi University of Education) for kindly providing E. coli MB43 and pea nodule leghemoglobin, respectively. We thank T. Matsuno and I. Yumoto (AIST) for their technical advice on the measurement of proton translocation efficiency and T. Uchiumi (Kagoshima University) for his help with the preparation of leghemoglobin.
This work was supported in part by a Grant-in-Aid for Scientific Research from the Japan Society for the Promotion of Science (JSPS) and by a research grant from the Sapporo Bioscience Foundation.
Footnotes
Published ahead of print 2 September 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.02176-14.
REFERENCES
- 1.Lyczak JB, Cannon CL, Pier GB. 2002. Lung infections associated with cystic fibrosis. Clin. Microbiol. Rev. 15:194–222. 10.1128/CMR.15.2.194-222.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Williams HD, Zlosnik JEA, Ryall B. 2007. Oxygen, cyanide and energy generation in the cystic fibrosis pathogen Pseudomonas aeruginosa. Adv. Microb. Physiol. 52:1–71. 10.1016/S0065-2911(06)52001-6. [DOI] [PubMed] [Google Scholar]
- 3.Schobert M, Jahn D. 2010. Anaerobic physiology of Pseudomonas aeruginosa in the cystic fibrosis lung. Int. J. Med. Microbiol. 300:549–556. 10.1016/j.ijmm.2010.08.007. [DOI] [PubMed] [Google Scholar]
- 4.Schobert M, Tielen P. 2010. Contribution of oxygen-limiting conditions to persistent infection of Pseudomonas aeruginosa. Future Microbiol. 5:603–621. 10.2217/fmb.10.16. [DOI] [PubMed] [Google Scholar]
- 5.Arai H. 2011. Regulation and function of versatile aerobic and anaerobic respiratory metabolism in Pseudomonas aeruginosa. Front. Microbiol. 2:103. 10.3389/fmicb.2011.00103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vander Wauven C, Piérard A, Kley-Raymann M, Haas D. 1984. Pseudomonas aeruginosa mutants affected in anaerobic growth on arginine: evidence for a four-gene cluster encoding the arginine deiminase pathway. J. Bacteriol. 160:928–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zumft WG. 1997. Cell biology and molecular basis of denitrification. Microbiol. Mol. Biol. Rev. 61:533–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stover CK, Pham XQ, Erwin AL, Mizoguchi SD, Warrener P, Hickey MJ, Brinkman FSL, Hufnagle WO, Kowalik DJ, Lagrou M, Garber RL, Goltry L, Tolentino E, Westbrock-Wadman S, Yuan Y, Brody LL, Coulter SN, Folger KR, Kas A, Larbig K, Lim R, Smith K, Spencer D, Wong GK-S, Wu Z, Paulsen IT, Reizer J, Saier MH, Hancock REW, Lory S, Olson MV. 2000. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 406:959–964. 10.1038/35023079. [DOI] [PubMed] [Google Scholar]
- 9.Cunningham L, Pitt M, Williams HD. 1997. The cioAB genes from Pseudomonas aeruginosa code for a novel cyanide-insensitive terminal oxidase related to the cytochrome bd quinol oxidases. Mol. Microbiol. 24:579–591. 10.1046/j.1365-2958.1997.3561728.x. [DOI] [PubMed] [Google Scholar]
- 10.Cunningham L, Williams HD. 1995. Isolation and characterization of mutants defective in the cyanide-insensitive respiratory pathway of Pseudomonas aeruginosa. J. Bacteriol. 177:432–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Matsushita K, Yamada M, Shinagawa E, Adachi O, Ameyama M. 1983. Membrane-bound respiratory chain of Pseudomonas aeruginosa grown aerobically. A KCN-insensitive alternate oxidase chain and its energetics. J. Biochem. 93:1137–1144. [DOI] [PubMed] [Google Scholar]
- 12.Mogi T, Ano Y, Nakatsuka T, Toyama H, Muroi A, Miyoshi H, Migita CT, Ui H, Shiomi K, Omura S, Kita K, Matsushita K. 2009. Biochemical and spectroscopic properties of cyanide-insensitive quinol oxidase from Gluconobacter oxydans. J. Biochem. 146:263–271. 10.1093/jb/mvp067. [DOI] [PubMed] [Google Scholar]
- 13.Miura H, Mogi T, Ano Y, Migita CT, Matsutani M, Yakushi T, Kita K, Matsushita K. 2013. Cyanide-insensitive quinol oxidase (CIO) from Gluconobacter oxydans is a unique terminal oxidase subfamily of cytochrome bd. J. Biochem. 153:535–545. 10.1093/jb/mvt019. [DOI] [PubMed] [Google Scholar]
- 14.Jackson RJ, Elvers KT, Lee LJ, Gidley MD, Wainwright LM, Lightfoot J, Park SF, Poole RK. 2007. Oxygen reactivity of both respiratory oxidases in Campylobacter jejuni: the cydAB genes encode a cyanide-resistant, low-affinity oxidase that is not of the cytochrome bd type. J. Bacteriol. 189:1604–1615. 10.1128/JB.00897-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alvarez-Ortega C, Harwood CS. 2007. Responses of Pseudomonas aeruginosa to low oxygen indicate that growth in the cystic fibrosis lung is by aerobic respiration. Mol. Microbiol. 65:153–165. 10.1111/j.1365-2958.2007.05772.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Comolli JC, Donohue TJ. 2002. Pseudomonas aeruginosa RoxR, a response regulator related to Rhodobacter sphaeroides PrrA, activates expression of the cyanide-insensitive terminal oxidase. Mol. Microbiol. 45:755–768. 10.1046/j.1365-2958.2002.03046.x. [DOI] [PubMed] [Google Scholar]
- 17.Comolli JC, Donohue TJ. 2004. Differences in two Pseudomonas aeruginosa cbb3 cytochrome oxidases. Mol. Microbiol. 51:1193–1203. 10.1046/j.1365-2958.2003.03904.x. [DOI] [PubMed] [Google Scholar]
- 18.Cooper M, Tavankar GR, Williams HD. 2003. Regulation of expression of the cyanide-insensitive terminal oxidase in Pseudomonas aeruginosa. Microbiology 149:1275–1284. 10.1099/mic.0.26017-0. [DOI] [PubMed] [Google Scholar]
- 19.Frangipani E, Slaveykova VI, Reimmann C, Haas D. 2008. Adaptation of aerobically growing Pseudomonas aeruginosa to copper starvation. J. Bacteriol. 190:6706–6717. 10.1128/JB.00450-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Frangipani E, Haas D. 2009. Copper acquisition by the SenC protein regulates aerobic respiration in Pseudomonas aeruginosa PAO1. FEMS Microbiol. Lett. 298:234–240. 10.1111/j.1574-6968.2009.01726.x. [DOI] [PubMed] [Google Scholar]
- 21.Kawakami T, Kuroki M, Ishii M, Igarashi Y, Arai H. 2010. Differential expression of multiple terminal oxidases for aerobic respiration in Pseudomonas aeruginosa. Environ. Microbiol. 12:1399–1412. 10.1111/j.1462-2920.2009.02109.x. [DOI] [PubMed] [Google Scholar]
- 22.Pereira MM, Santana M, Teixeira M. 2001. A novel scenario for the evolution of haem-copper oxygen reductases. Biochim. Biophys. Acta 1505:185–208. 10.1016/S0005-2728(01)00169-4. [DOI] [PubMed] [Google Scholar]
- 23.Bosma G, Braster M, Stouthamer AH, van Verseveld HW. 1987. Isolation and characterization of ubiquinol oxidase complexes from Paracoccus denitrificans cells cultured under various limiting growth conditions in the chemostat. Eur. J. Biochem. 165:657–663. 10.1111/j.1432-1033.1987.tb11491.x. [DOI] [PubMed] [Google Scholar]
- 24.Gabel C, Maier RJ. 1993. Oxygen-dependent transcriptional regulation of cytochrome aa3 in Bradyrhizobium japonicum. J. Bacteriol. 175:128–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Winstedt L, von Wachenfeldt C. 2000. Terminal oxidases of Bacillus subtilis strain 168: one quinol oxidase, cytochrome aa3 or cytochrome bd, is required for aerobic growth. J. Bacteriol. 182:6557–6564. 10.1128/JB.182.23.6557-6564.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Arai H, Roh JH, Kaplan S. 2008. Transcriptome dynamics during the transition from anaerobic photosynthesis to aerobic respiration in Rhodobacter sphaeroides 2.4.1. J. Bacteriol. 190:286–299. 10.1128/JB.01375-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pitcher RS, Watmough NJ. 2004. The bacterial cytochrome cbb3 oxidases. Biochim. Biophys. Acta 1655:388–399. 10.1016/j.bbabio.2003.09.017. [DOI] [PubMed] [Google Scholar]
- 28.Preisig O, Zufferey R, Thöny-Meyer L, Appleby CA, Hennecke H. 1996. A high-affinity cbb3-type cytochrome oxidase terminates the symbiosis-specific respiratory chain of Bradyrhizobium japonicum. J. Bacteriol. 178:1532–1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nagata K, Tsukita S, Tamura T, Sone N. 1996. A cb-type cytochrome-c oxidase terminates the respiratory chain in Helicobacter pylori. Microbiology 142:1757–1763. 10.1099/13500872-142-7-1757. [DOI] [PubMed] [Google Scholar]
- 30.Mouncey NJ, Kaplan S. 1998. Oxygen regulation of the ccoN gene encoding a component of the cbb3 oxidase in Rhodobacter sphaeroides 2.4.1T: involvement of the FnrL protein. J. Bacteriol. 180:2228–2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Otten MF, Stork DM, Reijnders WN, Westerhoff HV, Van Spanning RJM. 2001. Regulation of expression of terminal oxidases in Paracoccus denitrificans. Eur. J. Biochem. 268:2486–2497. 10.1046/j.1432-1327.2001.02131.x. [DOI] [PubMed] [Google Scholar]
- 32.Swem DL, Bauer CE. 2002. Coordination of ubiquinol oxidase and cytochrome cbb3 oxidase expression by multiple regulators in Rhodobacter capsulatus. J. Bacteriol. 184:2815–2820. 10.1128/JB.184.10.2815-2820.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 34.Hoang TT, Karkhoff-Schweizer RR, Kutchma AJ, Schweizer HP. 1998. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212:77–86. 10.1016/S0378-1119(98)00130-9. [DOI] [PubMed] [Google Scholar]
- 35.Arai H, Hayashi M, Kuroi A, Ishii M, Igarashi Y. 2005. Transcriptional regulation of the flavohemoglobin gene for aerobic nitric oxide detoxification by the second nitric oxide-responsive regulator of Pseudomonas aeruginosa. J. Bacteriol. 187:3960–3968. 10.1128/JB.187.12.3960-3968.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bergersen FJ, Turner GL. 1979. Systems utilizing oxygenated leghemoglobin and myoglobin as sources of free dissolved O2 at low concentrations for experiments with bacteria. Anal. Biochem. 96:165–174. 10.1016/0003-2697(79)90569-4. [DOI] [PubMed] [Google Scholar]
- 37.Bergersen FJ, Turner GL. 1980. Properties of terminal oxidase systems of bacteroids from root-nodules of soybean and cowpea and of N2-fixing bacteria grown in continuous culture. J. Gen. Microbiol. 118:235–252. [Google Scholar]
- 38.D'Mello R, Hill S, Poole RK. 1994. Determination of the oxygen affinities of terminal oxidases in Azotobacter vinelandii using the deoxygenation of oxyleghemoglobin and oxymyoglobin: cytochrome bd is a low-affinity oxidase. Microbiology 140:1395–1402. 10.1099/00221287-140-6-1395. [DOI] [Google Scholar]
- 39.Appleby CA, Bergersen FJ. 1980. Preparation and experimental use of leghaemoglobin, p 315–335 In Bergersen FJ. (ed), Methods of evaluating biological nitrogen fixation. John Wiley and Sons, Chichester, United Kingdom. [Google Scholar]
- 40.Han H, Hemp J, Pace LA, Ouyang H, Ganesan K, Roh JH, Daldal F, Blanke SR, Gennis RB. 2011. Adaptation of aerobic respiration to low O2 environments. Proc. Natl. Acad. Sci. U. S. A. 108:14109–14114. 10.1073/pnas.1018958108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.D'Mello R, Hill S, Poole RK. 1995. The oxygen affinity of cytochrome bo′ in Escherichia coli determined by the deoxygenation of oxyleghemoglobin and oxymyoglobin: Km values for oxygen are in the submicromolar range. J. Bacteriol. 177:867–870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kita K, Konishi K, Anraku Y. 1984. Terminal oxidases of Escherichia coli aerobic respiratory chain. II. Purification and properties of cytochrome b558-d complex from cells grown with limited oxygen and evidence of branched electron-carrying systems. J. Biol. Chem. 259:3375–3381. [PubMed] [Google Scholar]
- 43.Bekker M, de Vries S, Ter Beek A, Hellingwerf KJ, Teixeira de Mattos MJ. 2009. Respiration of Escherichia coli can be fully uncoupled via the nonelectrogenic terminal cytochrome bd-II oxidase. J. Bacteriol. 191:5510–5517. 10.1128/JB.00562-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Morris RL, Schmidt TM. 2013. Shallow breathing: bacterial life at low O2. Nat. Rev. Microbiol. 11:205–212. 10.1038/nrmicro2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xie H, Buschmann S, Langer JD, Ludwig B, Michel H. 2014. Biochemical and biophysical characterization of the two isoforms of cbb3-type cytochrome c oxidase from Pseudomonas stutzeri. J. Bacteriol. 196:472–482. 10.1128/JB.01072-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Buschmann S, Warkentin E, Xie H, Langer JD, Ermler U, Michel H. 2010. The structure of cbb3 cytochrome oxidase provides insights into proton pumping. Science 329:327–330. 10.1126/science.1187303. [DOI] [PubMed] [Google Scholar]
- 47.Rauhamäki V, Bloch DA, Wikström M. 2012. Mechanistic stoichiometry of proton translocation by cytochrome cbb3. Proc. Natl. Acad. Sci. U. S. A. 109:7286–7291. 10.1073/pnas.1202151109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim YJ, Ko IJ, Lee JM, Kang HY, Kim YM, Kaplan S, Oh JI. 2007. Dominant role of the cbb3 oxidase in regulation of photosynthesis gene expression through the PrrBA system in Rhodobacter sphaeroides 2.4.1. J. Bacteriol. 189:5617–5625. 10.1128/JB.00443-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wu J, Bauer CE. 2010. RegB kinase activity is controlled in part by monitoring the ratio of oxidized to reduced ubiquinones in the ubiquinone pool. mBio 1(5):e00272–10. 10.1128/mBio.00272-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ugidos A, Morales G, Rial E, Williams HD, Rojo F. 2008. The coordinate regulation of multiple terminal oxidases by the Pseudomonas putida ANR global regulator. Environ. Microbiol. 10:1690–1702. 10.1111/j.1462-2920.2008.01586.x. [DOI] [PubMed] [Google Scholar]
- 51.Petruschka L, Burchhardt G, Müller C, Weihe C, Herrmann H. 2001. The cyo operon of Pseudomonas putida is involved in carbon catabolite repression of phenol degradation. Mol. Genet. Genomics 266:199–206. 10.1007/s004380100539. [DOI] [PubMed] [Google Scholar]
- 52.Dinamarca MA, Ruiz-Manzano A, Rojo F. 2002. Inactivation of cytochrome o ubiquinol oxidase relieves catabolic repression of the Pseudomonas putida GPo1 alkane degradation pathway. J. Bacteriol. 184:3785–3793. 10.1128/JB.184.14.3785-3793.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Morales G, Ugidos A, Rojo F. 2006. Inactivation of the Pseudomonas putida cytochrome o ubiquinol oxidase leads to a significant change in the transcriptome and to increased expression of the CIO and cbb3-1 terminal oxidases. Environ. Microbiol. 8:1764–1674. 10.1111/j.1462-2920.2006.01061.x. [DOI] [PubMed] [Google Scholar]
- 54.Cotter PA, Chepuri V, Gennis RB, Gunsalus RP. 1990. Cytochrome o (cyoABCDE) and d (cydAB) oxidase gene expression in Escherichia coli is regulated by oxygen, pH, and the fnr gene product. J. Bacteriol. 172:6333–6338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Simon R, Priefer U, Puhler A. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram-negative bacteria. Biotechnology 1:784–791. 10.1038/nbt1183-784. [DOI] [Google Scholar]
- 56.Fürste JP, Pansegrau W, Frank R, Blöcker H, Scholz P, Bagdasarian M, Lanka E. 1986. Molecular cloning of the plasmid RP4 primase region in a multi-host-range tacP expression vector. Gene 48:119–131. 10.1016/0378-1119(86)90358-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.