Abstract
The mycobacterial cell wall frequently has been used as a target for drug development, and d-glutamate, synthesized by glutamate racemase (MurI), is an important component of peptidoglycan. While the essentiality of the murI gene has been shown in several bacterial species, including Escherichia coli, Bacillus anthracis, and Streptococcus pneumoniae, studies in mycobacteria have not yet provided definitive results. This study aimed to determine whether murI is indeed essential and can serve as a possible target for structure-aided drug design. We have achieved this goal by creating a ΔmurI strain of Mycobacterium smegmatis, a close relative of Mycobacterium tuberculosis. The deletion of the murI gene in M. smegmatis could be achieved only in minimal medium supplemented with d-glutamate, demonstrating that MurI is essential for growth and that glutamate racemase is the only source of d-glutamate for peptidoglycan synthesis in M. smegmatis.
INTRODUCTION
d-Glutamate is an essential component of the bacterial cell wall, and there are two enzymes in bacteria capable of synthesizing d-glutamate, glutamate racemase (MurI) and d-amino acid aminotransferase (DaaT). In Escherichia coli and Streptococcus pneumoniae, whose genomes contain only murI, genetic deletion studies have shown that MurI is essential for growth in the absence of external d-glutamate supplementation (1–4). Other bacteria, which contain DaaT, such as Staphylococcus haemolyticus and Bacillus sphaericus, are able to produce sufficient d-glutamate in the absence of glutamate racemase (5). There are also bacteria, including some other Bacillus species, that contain two copies of murI, as well as a copy of daaT, yet, at least in B. subtilis, murI is required for these bacteria to grow in both rich and minimal medium (6–8). MurI also has been implicated in several species as having a role in moderating the activity of DNA gyrase, which itself is an important drug target and an essential enzyme in bacteria (9–11). Therefore, the essentiality of murI is a complex question that appears to be species specific.
In mycobacteria, where the identification and validation of new targets for drug design is an ongoing priority, the essentiality of glutamate racemase is unclear. The inspection of the genome of Mycobacterium tuberculosis reveals only one copy of murI and no copy of daaT (12). Therefore, it seems likely that MurI is essential, as it is the only enzyme capable of d-glutamate biosynthesis in this organism. However, in a transposon mutagenesis study carried out in M. tuberculosis, murI was not identified as likely to be essential (13). However, further investigation of that particular strain revealed the transposon had integrated at the penultimate position of the gene sequence and may not have inactivated the gene. In support of this interpretation, a more recent investigation of this issue employing transposon mutagenesis coupled with deep sequencing did conclude that murI was likely to be an essential gene (14). Moreover, a different set of experiments investigating glutamate metabolism in M. tuberculosis found that a murI deletion strain grew in 7H9 media when supplemented with 2% glucose, a known osmoprotectant (15).
In order to study this issue more fully in mycobacteria, we have created a murI deletion in Mycobacterium smegmatis, a nonpathogenic relative of M. tuberculosis that has been used successfully as a drug development surrogate for pathogenic mycobacteria (16). Like M. tuberculosis, M. smegmatis contains only one copy of murI and no daaT gene. We have studied the growth characteristics of this murI deletion strain in minimal medium as well as in media containing osmotically active agents, and we report that in minimal medium the murI gene is indeed essential for growth in the absence of d-glutamate supplementation.
MATERIALS AND METHODS
Bacterial growth and culture conditions.
The plasmids, strains, and oligonucleotides used in this study are listed in Table 1. E. coli was routinely cultured in LB containing 0.5% NaCl; M. smegmatis was cultured in LB medium with 0.05% Tween 80 (LBT). A low-salt version (0.05% NaCl) was used where specified. For solid media, 1.5% agar and 0.05% Tween were added. For the ΔmurI strains, Sauton's minimal medium [0.1% l-asparagine, 0.2% citric acid, 0.05% K2HPO4, 0.05% MgSO4, 0.005% ammonium iron(III) citrate, 0.2% glycerol, 0.05% Tween 80], supplemented with 15 to 60 mM d-glutamate and adjusted to pH 7.4 with NaOH, was used. Sucrose (10%, wt/vol) also was included as needed during the knockout (KO) selection procedure. Unless otherwise stated, bacteria were grown at 37°C. During the murI deletion process, M. smegmatis harboring the gene replacement vector, pKKYL02, was grown at 28°C for temperature-sensitive vector propagation and then placed at 40°C for allelic exchange mutagenesis. All cloning procedures were performed in E. coli DH10B. Kanamycin, hygromycin, and gentamicin were used at final concentrations of 25, 50, and 5 μg/ml, respectively, for M. smegmatis mc2155 and 50, 100, and 7 μg/ml, respectively, for E. coli.
TABLE 1.
Bacterial strains, plasmids, and primers used in this study
| Strain, plasmid, or primer | Relevant information or sequence | Restriction site | Reference and/or source |
|---|---|---|---|
| Escherichia coli | |||
| DH10B | F− araD139 Δ(ara, leu)7697 ΔlacX74 galU galK rpsL deoR ϕ80dlacZΔM15 endA1 nupG recA1 mcrA Δ(mrr hsdRMS mcrBC) | 30 | |
| MB3360 | pET28-TEV; M. tuberculosis murI in NdeI and BamHI in DH10B | This study | |
| Mycobacterium smegmatis | |||
| mc2155 wild type | ept-1 | 31 | |
| KK104-10 | mc2155 ΔmurI::aphA-3 Kanr; selected at 15 mM d-Glu | This study | |
| KK104-11 | mc2155 ΔmurI::aphA-3 Kanr; selected at 30 mM d-Glu | This study | |
| KK104-12 | mc2155 ΔmurI::aphA-3 Kanr; selected at 30 mM d-Glu | This study | |
| KK104-13 | mc2155 ΔmurI::aphA-3 Kanr; selected at 60 mM d-Glu | This study | |
| KK104-16 | mc2155 ΔmurI::aphA-3 Kanr; selected at 125 mM d-Glu | This study | |
| KK104-19 | mc2155 ΔmurI::aphA-3 murI+ Kanr; selected at 125 mM d-Glu | This study | |
| KK104-09 | KK104-10 harboring pKKYL03; Hygr | This study | |
| Plasmids | |||
| pBluescript II SK(+) | E. coli cloning vector; f1 Ori; Ampr | Stratagene | |
| pDM3 | Source of the mycobacterial alr promoter | 18 | |
| pKKYL02 | pPR23 harboring ΔmurI::aphA-3; Kanr Gmr | This study | |
| pKKYL03 | pUHA267 expressing murI; Hygr | This study | |
| pPR23 | E. coli-mycobacterial shuttle vector; oriM(Ts) sacB Gmr | 32 | |
| pUHA267 | E. coli-mycobacterial shuttle vector; Hygr | AgResearch, Wallaceville, New Zealand; 19 | |
| Primers | |||
| MurISM KO-3.1 | 5′-TTTAAAAAGCTTCCGAATTGGATTTGCTGCGA-3′ | HindIII | This study |
| MurISM KO-4.2 | 5′-AGACTAGTGTCTTCTCGTCTGGACACGT-3′ | SpeI | This study |
| MurISM KO-1.3 | 5′-CTCGTTCGACGCGGCCGCCCGCACCCGTGAACTCGTCA-3′ | NotI | This study |
| MurISM KO-2.2 | 5′-AAATTTGAATTCGCGCGGATCTGCGGGATGGT-3′ | EcoRI | This study |
| 5alr2 | 5′-AAATTTCCATGGTGGGGCAGTACTACAACTTC-3′ | KpnI | 18 |
| 3alrpro | 5′-GTGGTCTGCATATGCATAATCTCCGGCGCCCATT-3′ | NdeI | This study |
| MurISM EX-1.0 | 5′-TTTAAACATATGAGCGATCGACTCGCGCC-3′ | NdeI | This study |
| MurISM EX-2.1 | 5′-TTTAAAGGATCCGGTGCCAAGACATGCCCGGT-3′ | BamHI | This study |
Mutagenesis vector construction.
Following a method derived for use in this strain of Mycobacterium (17), a shuttle vector was created to delete murI by homologous recombination. Briefly, primers MurISM KO-1.3, -2.2, -3.1, and -4.2 were used to amplify the left and right flanks of murI from strain MB3360, generating two 1,080- and 1,043-bp amplicons. These flanking regions were ligated to an EcoRI- and HindIII-treated kanamycin resistance cassette (aphA-3) and then inserted into the shuttle vector pPR23 using NotI and SpeI, creating plasmid pKKYL02. This plasmid carries a temperature-sensitive origin of replication, gentamicin resistance, and the sucrose sensitivity-conveying sacB gene for counterselection to be utilized in the screening of the double crossover event. Following the allelic exchange, ΔmurI strains have 60% of the murI gene removed and are kanamycin resistant.
Selection of knockout strains and Southern blotting.
The pKKYL02 gene replacement vector was transformed into M. smegmatis mc2155 via electroporation (0.2-cm-gap cuvette at 25 kV, 1 kΩ, and 25 μF). The transformants were grown in Sauton's medium supplemented with 15 mM d-glutamate at 28°C to allow propagation of the temperature-sensitive vector and allow for the gene crossover to take place. The culture then was plated on Sauton's medium supplemented with 15 mM d-glutamate, 10% sucrose, and kanamycin at 40°C to select for the murI deletion. After 5 days of growth, the colonies were streaked out on Sauton's medium containing gentamicin and 15 mM d-glutamate; the gentamicin sensitivity of these colonies verified the loss of the pKKYL02 vector backbone. The genotype of these putative ΔmurI mutant strains was verified by Southern blotting in which genomic DNA was digested by SmaI, separated by agarose gel electrophoresis, and then blotted onto a nylon membrane. The radioactively labeled right flank then was used as a probe to detect homologous restriction fragments on the membrane by autoradiography (Fig. 1).
FIG 1.
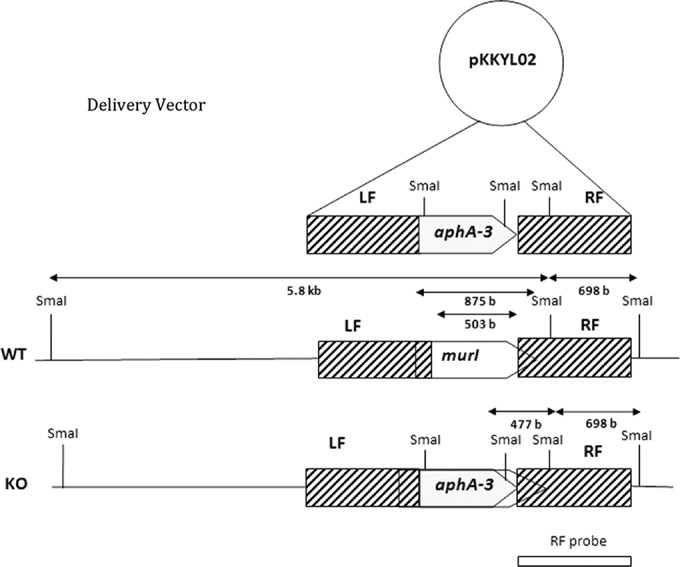
Vector map of the ΔmurI mutant of Mycobacterium smegmatis mc2155. The construction of the delivery vector pKKYL02 was described in Materials and Methods. Homologous crossover at the left flank (LF) and right flank (RF) replaces 60% of murI with a nonpolar kanamycin resistance cassette (aphA-3). SmaI digestion sites utilized in the Southern blot are displayed. The right flank was used as a probe for hybridization.
Construction of a complementation vector for ΔmurI mutants.
A vector expressing a copy of murI in trans was designed to complement the ΔmurI mutant. The murI gene lies in an operon and its promoter is unknown; therefore, the promoter of the alr gene from M. smegmatis was ligated to the murI gene (18). Primers MurISM EX-1.0 and MurISM EX-2.1 (Table 1) were used to amplify murI. The alr promoter from pDM3 was excised with NcoI and NdeI, ligated with murI, and then ligated into the E. coli-mycobacterial shuttle vector pUHA267 (19), forming plasmid vector pKKYL03.
RESULTS AND DISCUSSION
Selection of murI deletion mutants in M. smegmatis at 5 mM d,l-glutamate.
The murI gene lies in an operon and overlaps MSMEG_4904 at its 5′ end by 4 bp. It is closely followed by MSMEG_4902 at its 3′ end. To ensure that the disruption of the murI gene did not affect these two flanking genes, only 60% of the murI coding sequence was replaced with a nonpolar kanamycin cassette (20). However, both cysteines known to be essential for MurI activity were removed in this process (21, 22). The flanking DNA regions (1,080 bp and 1,043 bp, respectively) first were ligated with the kanamycin resistance-encoding aphA-3 cassette and cloned in pBluescript II SK(+). The complete murI::aphA-3 insert then was excised and ligated into the gene delivery vector pPR23 (Fig. 1). pPR23 contains a temperature-sensitive origin of replication that prohibits plasmid replication past 40°C, a gentamicin resistance cassette, and a sucrose sensitivity gene (sacB) for counterselection purposes. Thus, the initial selection attempt was carried out at 40°C in the presence of kanamycin and sucrose and 5 mM d,l-glutamate or 1 mM d-glutamate. Six colonies from this attempt were tested by pick and patch. However, these six colonies grew normally on LBT medium without d,l-glutamate supplementation. The selection was repeated, and all additional colonies tested by pick and patch remained independent of d-glutamate.
This result was consistent with the formation of a strain containing the expected murI disruption by aphA-3 but that had also retained a wild-type (WT) copy of murI, either through gene duplication or because the plasmid had not properly integrated into the chromosome. Duplications in the M. smegmatis genome have previously been reported both in wild-type strains (23) and strains under genetic selection (17). If duplication of murI did occur during this experiment, it suggests that it was disadvantageous for the organism to lose its murI gene under the selection conditions containing 5 mM d,l-glutamate. Since d-glutamate is known to be poorly transported across bacterial membranes (24, 25), we concluded that larger amounts of d-glutamate supplementation likely were required for true ΔmurI mutants to survive. This conclusion is consistent with studies in E. coli showing that a ΔmurI strain could not be obtained until a mutant strain (WM335) was isolated that contained a mutation allowing for increased transport of d-glutamate across the plasma membrane (24, 26).
Disruption of the murI gene in M. smegmatis at high d-glutamate concentrations.
To address the above-described hypothesis, the selection experiment was repeated in Sauton's minimal medium supplemented with 15 to 125 mM d-glutamate. Eight clones obtained under these conditions were chosen for screening by Southern blotting. Genomic DNA was extracted from these eight mutant colonies and from wild-type colonies and then digested by SmaI. For the wild type, hybridization with a probe for the right flank should have yielded bands at 5.8 kbp and 698 bp, respectively. For the ΔmurI mutant, on the other hand, the 5.8-kbp band should disappear, leaving bands only at 698 bp and 477 bp. An incomplete removal of the gene replacement vector, pKKYL02, from the cell should yield bands at 477 bp and 2.5 kbp (Fig. 1).
As shown in Fig. 2, strains KK104-10 (selected at 15 mM d-glutamate), KK104-11 and KK104-12 (both selected at 30 mM d-glutamate), KK104-13 (selected at 60 mM d-glutamate), and KK104-16 (selected at 125 mM d-glutamate) all had undergone the double crossover event; therefore, they were genuine murI deletion mutants. Strain KK104-19 (selected at 125 mM D-glutamate) showed the typical wild-type band of 5.8 kbp and a typical vector band of 2.5 kbp, indicating incomplete gene disruption. When retested after the original selection, strain KK104-19 did not require any exogenous d-glutamate for growth on minimal medium. In addition, all six clones originally selected on 1 and 5 mM d-glutamate had maintained the two characteristic genomic DNA and vector DNA bands upon Southern hybridization (data not shown).
FIG 2.
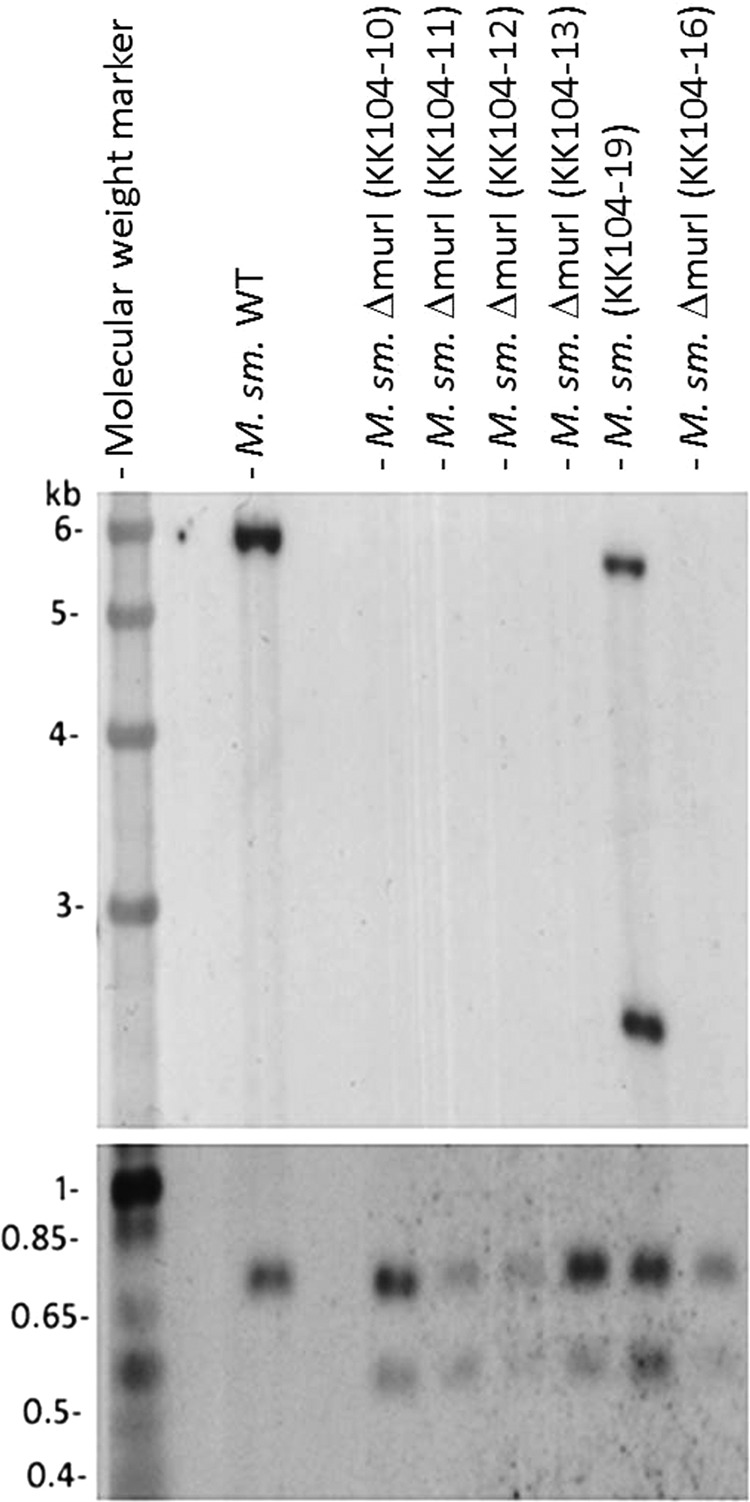
Southern blot of potential ΔmurI mutants. Genomic DNA was isolated and digested with SmaI, and identical sample aliquots were loaded on low-percentage (top) and high-percentage (bottom) agarose gels. M. sm., M. smegmatis.
Phenotype of the ΔmurI mutant strains on solid minimal medium.
The phenotype of a representative selection of ΔmurI mutants was characterized on solid Sauton's medium (Fig. 3). It was found that the ΔmurI mutants KK104-10, KK104-11, KK104-12, KK104-13, and KK104-16 did not grow on Sauton's medium in the absence of external d-glutamate supplementation. Below 5 mM d-glutamate, no appreciable growth was observed. With increasing d-glutamate supplementation, increased growth occurred. At 60 mM d-glutamate or higher, the ΔmurI mutants grew indistinguishably from the wild-type strain. Strains that had undergone incomplete gene replacement events, such as KK104-19, did not require external d-glutamate supplementation to grow on solid Sauton's medium.
FIG 3.
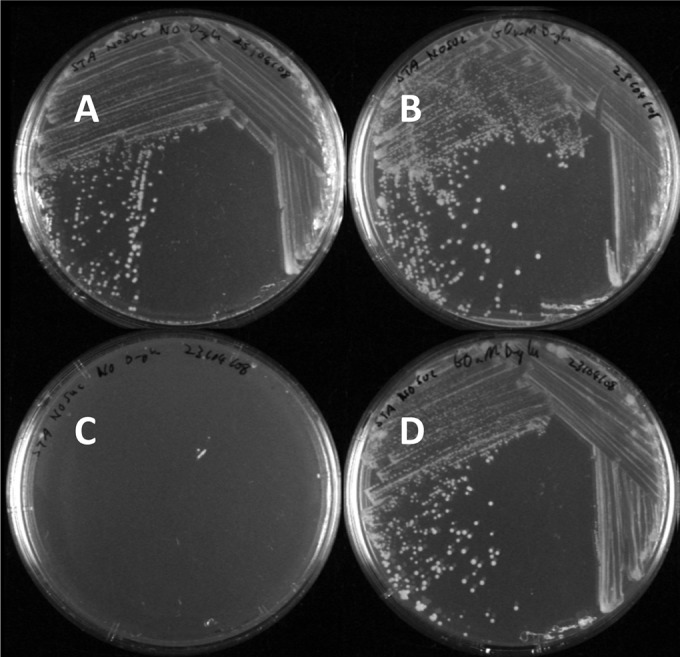
Phenotype of wild-type M. smegmatis cells grown on Sauton's medium in the absence (A) and presence (B) of 60 mM d-glutamate and ΔmurI (KK104-10) mutant cells grown on Sauton's medium in the absence (C) and presence (D) of 60 mM d-glutamate.
Growth of ΔmurI mutant strains in other growth media.
As described above, ΔmurI mutant strains require d-glutamate supplementation for growth on Sauton's (agar) solid medium. These ΔmurI deletion mutants also are unable to grow in low-salt LB broth without d-glutamate supplementation. However, supplementation of low-salt LB broth with either KCl, sucrose, or additional NaCl was found, at some concentrations, to complement the ΔmurI phenotype. Using sucrose supplementation, minimal growth could be detected at 1 to 2% (wt/vol) sucrose supplementation, but 10% sucrose was required for wild-type-like growth. With NaCl supplementation, growth of the ΔmurI mutant occurred at concentrations greater than 200 mM, but wild-type growth was never observed. Notably, LB solid medium containing 5% NaCl (855 mM) was able to support the growth of the ΔmurI mutant.
These phenotypes have been observed in cell wall deletion mutants of other microorganisms and often are ascribed to osmotic stabilization effects (27, 28, and M. Benedik, personal communication). Strikingly, the ΔmurI mutant does not require d-glutamate supplementation to grow in liquid Sauton's medium, although the growth rate is slower than that of the wild type. The difference in osmotic conditions between solid Sauton's medium and liquid medium is thought to contribute to these observations, with the salt and 0.2% glycerol present in liquid Sauton's medium allowing the ΔmurI strain to grow in the absence of d-glutamate supplementation.
In order to demonstrate that the phenotype of the ΔmurI strain was due to the lack of d-glutamate racemase (MurI), a copy of murI was provided in trans via the integrative shuttle vector pUHA267, forming strain KK-09 MurI+. This complementation allowed KKYL09 to grow normally in low-salt LB broth (Fig. 4), suggesting that the ΔmurI strain phenotype in low-salt LB is due to insufficient d-glutamate.
FIG 4.
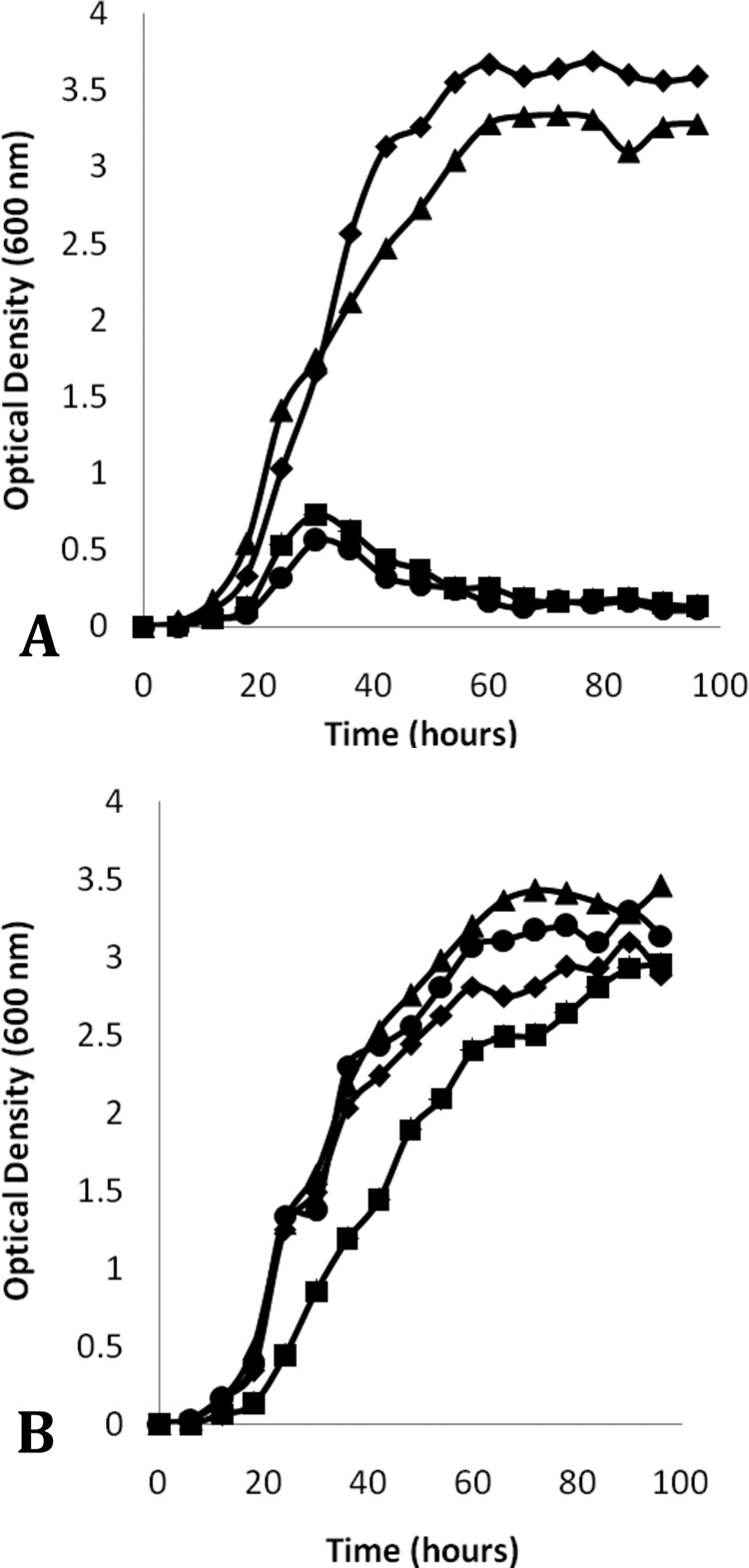
Growth curves of the wild-type strain (◆) and the KK104-10 ΔmurI strain (●), KK104-13 (ΔmurI) strain (■), and K-09 (murI complemented in trans) strain (▲). The growth curve was performed in low-salt LB liquid broth without d-glutamate supplementation (A) and with 60 mM d-glutamate supplementation (B), and growth was recorded over 96 h.
ΔmurI mutant cells display a morphology different from the wild-type cells.
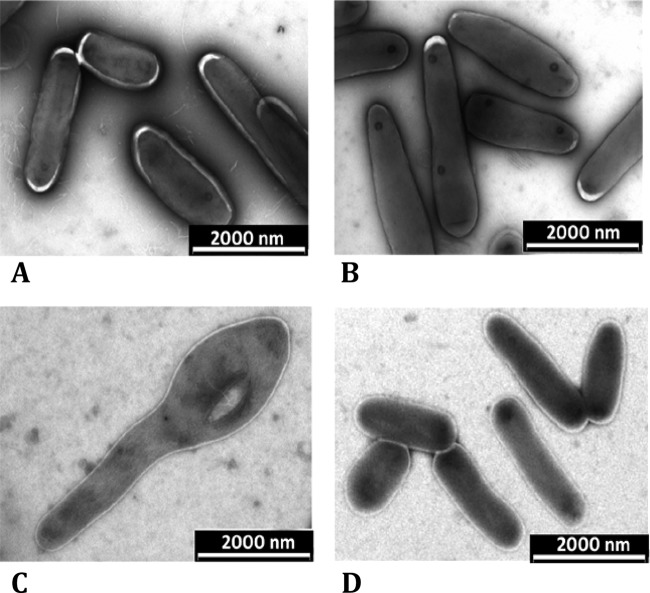
At the end of a growth curve in low-salt LB broth, a representative sample of stationary-phase cells of the ΔmurI mutant strain was examined using electron microscopy and compared to the wild type. It was found that the ΔmurI cells grown without d-glutamate supplementation grew to 2 to 3 times the size of wild-type cells (Fig. 5) and often displayed pear-shaped swellings at their polar ends, while in the presence of d-glutamate the phenotype was unchanged from that of the wild type (Fig. 5). A similar phenotype also was reported for M. smegmatis Δalr mutants, which grew to triple the length of wild-type strains (18).
FIG 5.
Electron microscopy of representative stationary-phase wild-type (WT) and mutant (KK104-10) M. smegmatis cells at the end of the growth curve in low-salt LB broth. Shown are the WT without d-glutamate (A) and with 60 mM d-glutamate supplementation (B) and KK104-10 mutants without d-glutamate (C) and with 60 mM d-glutamate supplementation (D).
Summary and conclusions.
In this study, we demonstrate that the murI gene encoding glutamate racemase in M. smegmatis is essential for growth in Sauton's minimal liquid medium. This essentiality is overcome by adding >15 mM d-glutamate to this liquid growth medium. In other media containing osmoprotectants (salt, sugars, etc.), murI is nonessential, a result that is consistent with other studies on cell wall deletion mutants (27, 28). In fact, microorganisms that are completely lacking a cell well, such as L-forms of eubacteria, can be grown under iso-osmolar conditions (29). Our results also may help explain a recent report that murI is nonessential in M. tuberculosis (15). In this study, the deletion strain was studied only under one condition, 7H9 medium containing 2% glucose. It is tempting to propose that glucose was able to serve as an osmoprotectant, as we found in our studies on M. smegmatis. Certainly, our results show that murI deletion mutants do not grow in minimal medium. The relevance of these findings to human disease is difficult to ascertain, because it is unknown if conditions in the macrophage or a pulmonary granuloma would allow an organism with this cell wall defect to persist and to multiply. Studies with murI deletion mutants of M. tuberculosis are needed to investigate this possibility.
ACKNOWLEDGMENTS
We are grateful to Eric Rubin and Jyothi Rengarajan, Harvard Medical School, for information on the murI strain containing the transposon insertion and for helpful discussions regarding this project.
We are thankful for financial support from the University of Otago and the Thrash Foundation, Houston, Texas.
Footnotes
Published ahead of print 22 September 2014
REFERENCES
- 1.Baltz RH, Hoskins JA, Solenberg PJ, Treadway PJ. November 1999. Method for knockout mutagenesis in Streptococcus pneumoniae. US patent 5,981,281.
- 2.Hoffmann B, Messer W, Schwarz U. 1972. Regulation of polar cap formation in the life cycle of Escherichia coli. J. Supramol. Struct. 1:29–37. 10.1002/jss.400010105. [DOI] [PubMed] [Google Scholar]
- 3.Lugtenberg EJ, Wijsman HJ, van Zaane D. 1973. Properties of a D-glutamic acid-requiring mutant of Escherichia coli. J. Bacteriol. 114:499–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Song JH, Ko KS, Lee JY, Baek JY, Oh WS, Yoon HS, Jeong JY, Chun J. 2005. Identification of essential genes in Streptococcus pneumoniae by allelic replacement mutagenesis. Mol. Cells 19:365–374. [PubMed] [Google Scholar]
- 5.Kimura K, Tran LS, Itoh Y. 2004. Roles and regulation of the glutamate racemase isogenes, racE and yrpC, in Bacillus subtilis. Microbiology 150:2911–2920. 10.1099/mic.0.27045-0. [DOI] [PubMed] [Google Scholar]
- 6.Ashiuchi M, Soda K, Misono H. 1999. Characterization of yrpC gene product of Bacillus subtilis IFO 3336 as glutamate racemase isozyme. Biosci. Biotechnol. Biochem. 63:792–798. 10.1271/bbb.63.792. [DOI] [PubMed] [Google Scholar]
- 7.Kunst F, Ogasawara N, Moszer I, Albertini AM, Alloni G, Azevedo V, Bertero MG, Bessières P, Bolotin A, Borchert S, Borriss R, Boursier L, Brans A, Braun M, Brignell SC, Bron S, Brouillet S, Bruschi CV, Caldwell B, Capuano V, Carter NM, Choi SK, Cordani JJ, Connerton IF, Cummings NJ, Daniel RA, Denziot F, Devine KM, Düsterhöft A, Ehrlich SD, Emmerson PT, Entian KD, Errington J, Fabret C, Ferrari E, Foulger D, Fritz C, et al. 1997. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature 390:249–256. 10.1038/36786. [DOI] [PubMed] [Google Scholar]
- 8.Shatalin KY, Neyfakh AA. 2005. Efficient gene inactivation in Bacillus anthracis. FEMS Microbiol. Lett. 245:315–319. 10.1016/j.femsle.2005.03.029. [DOI] [PubMed] [Google Scholar]
- 9.Ashiuchi M, Kuwana E, Komatsu K, Soda K, Misono H. 2003. Differences in effects on DNA gyrase activity between two glutamate racemases of Bacillus subtilis, the poly-gamma-glutamate synthesis-linking Glr enzyme and the YrpC (MurI) isozyme. FEMS Microbiol. Lett. 223:221–225. 10.1016/S0378-1097(03)00381-1. [DOI] [PubMed] [Google Scholar]
- 10.Ashiuchi M, Kuwana E, Yamamoto T, Komatsu K, Soda K, Misono H. 2002. Glutamate racemase is an endogenous DNA gyrase inhibitor. J. Biol. Chem. 277:39070–39073. 10.1074/jbc.C200253200. [DOI] [PubMed] [Google Scholar]
- 11.Sengupta S, Ghosh S, Nagaraja V. 2008. Moonlighting function of glutamate racemase from Mycobacterium tuberculosis: racemization and DNA gyrase inhibition are two independent activities of the enzyme. Microbiology 154:2796–2803. 10.1099/mic.0.2008/020933-0. [DOI] [PubMed] [Google Scholar]
- 12.Cole ST, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon SV, Eiglmeier K, Gas S, Barry CE, III, Tekaia F, Badcock K, Basham D, Brown D, Chillingworth T, Connor R, Davies R, Devlin K, Feltwell T, Gentles S, Hamlin N, Holroyd S, Hornsby T, Jagels K, Krogh A, McLean J, Moule S, Murphy L, Oliver K, Osborne J, Quail MA, Rajandream MA, Rogers J, Rutter S, Seeger K, Skelton J, Squares R, Squares S, Sulston JE, Taylor K, Whitehead S, Barrell BG. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393:537–544. [DOI] [PubMed] [Google Scholar]
- 13.Sassetti CM, Boyd DH, Rubin EJ. 2003. Genes required for mycobacterial growth defined by high density mutagenesis. Mol. Microbiol. 48:77–84. 10.1046/j.1365-2958.2003.03425.x. [DOI] [PubMed] [Google Scholar]
- 14.Griffin JE, Gawronski JD, Dejesus MA, Ioerger TR, Akerley BJ, Sassetti CM. 2011. High-resolution phenotypic profiling defines genes essential for mycobacterial growth and cholesterol catabolism. PLoS Pathog. 7:e1002251. 10.1371/journal.ppat.1002251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harth G, Maslesa-Galic S, Tullius MV, Horwitz MA. 2005. All four Mycobacterium tuberculosis glnA genes encode glutamine synthetase activities but only GlnA1 is abundantly expressed and essential for bacterial homeostasis. Mol. Microbiol. 58:1157–1172. 10.1111/j.1365-2958.2005.04899.x. [DOI] [PubMed] [Google Scholar]
- 16.Andries K, Verhasselt P, Guillemont J, Gohlmann HW, Neefs JM, Winkler H, Van Gestel J, Timmerman P, Zhu M, Lee E, Williams P, de Chaffoy D, Huitric E, Hoffner S, Cambau E, Truffot-Pernot C, Lounis N, Jarlier V. 2005. A diarylquinoline drug active on the ATP synthase of Mycobacterium tuberculosis. Science 307:223–227. 10.1126/science.1106753. [DOI] [PubMed] [Google Scholar]
- 17.Tran SL, Cook GM. 2005. The F1Fo-ATP synthase of Mycobacterium smegmatis is essential for growth. J. Bacteriol. 187:5023–5028. 10.1128/JB.187.14.5023-5028.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Milligan DL, Tran SL, Strych U, Cook GM, Krause KL. 2007. The alanine racemase of Mycobacterium smegmatis is essential for growth in the absence of D-alanine. J. Bacteriol. 189:8381–8386. 10.1128/JB.01201-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee MH, Pascopella L, Jacobs WR, Jr, Hatfull GF. 1991. Site-specific integration of mycobacteriophage L5: integration-proficient vectors for Mycobacterium smegmatis, Mycobacterium tuberculosis, and bacille Calmette-Guerin. Proc. Natl. Acad. Sci. U. S. A. 88:3111–3115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Menard R, Sansonetti PJ, Parsot C. 1993. Nonpolar mutagenesis of the ipa genes defines IpaB, IpaC, and IpaD as effectors of Shigella flexneri entry into epithelial cells. J. Bacteriol. 175:5899–5906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gallo KA, Knowles JR. 1993. Purification, cloning, and cofactor independence of glutamate racemase from Lactobacillus. Biochemistry 32:3981–3990. [DOI] [PubMed] [Google Scholar]
- 22.Gallo KA, Tanner ME, Knowles JR. 1993. Mechanism of the reaction catalyzed by glutamate racemase. Biochemistry 32:3991–3997. [DOI] [PubMed] [Google Scholar]
- 23.Springer B, Sander P, Sedlacek L, Hardt WD, Mizrahi V, Schar P, Bottger EC. 2004. Lack of mismatch correction facilitates genome evolution in mycobacteria. Mol. Microbiol. 53:1601–1609. 10.1111/j.1365-2958.2004.04231.x. [DOI] [PubMed] [Google Scholar]
- 24.Doublet P, van Heijenoort J, Bohin JP, Mengin-Lecreulx D. 1993. The murI gene of Escherichia coli is an essential gene that encodes a glutamate racemase activity. J. Bacteriol. 175:2970–2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yabu K. 1970. Amino acid transport in Mycobacterium smegmatis. J. Bacteriol. 102:6–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dougherty TJ, Thanassi JA, Pucci MJ. 1993. The Escherichia coli mutant requiring D-glutamic acid is the result of mutations in two distinct genetic loci. J. Bacteriol. 175:111–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kriangkripipat T, Momany M. 2009. Aspergillus nidulans protein O-mannosyltransferases play roles in cell wall integrity and developmental patterning. Eukaryot. Cell 8:1475–1485. 10.1128/EC.00040-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shen B, Hohmann S, Jensen RG, Bohnert H. 1999. Roles of sugar alcohols in osmotic stress adaptation. Replacement of glycerol by mannitol and sorbitol in yeast. Plant Physiol. 121:45–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dienes L, Sharp JT. 1956. The role of high electrolyte concentration in the production and growth of L forms of bacteria. J. Bacteriol. 71:208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Grant SG, Jessee J, Bloom FR, Hanahan D. 1990. Differential plasmid rescue from transgenic mouse DNAs into Escherichia coli methylation-restriction mutants. Proc. Natl. Acad. Sci. U. S. A. 87:4645–4649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Snapper SB, Melton RE, Mustafa S, Kieser T, Jacobs WR., Jr 1990. Isolation and characterization of efficient plasmid transformation mutants of Mycobacterium smegmatis. Mol. Microbiol. 4:1911–1919. [DOI] [PubMed] [Google Scholar]
- 32.Pelicic V, Jackson M, Reyrat JM, Jacobs WR, Jr, Gicquel B, Guilhot C. 1997. Efficient allelic exchange and transposon mutagenesis in Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U. S. A. 94:10955–10960. [DOI] [PMC free article] [PubMed] [Google Scholar]