Abstract
Here we describe biosensors that provide readouts for protein stability in the cytosolic compartment of prokaryotes. These biosensors consist of tripartite sandwich fusions that link the in vitro stability or aggregation susceptibility of guest proteins to the in vivo resistance of host cells to the antibiotics kanamycin, spectinomycin, and nourseothricin. These selectable markers confer antibiotic resistance in a wide range of hosts and are easily quantifiable. We show that mutations within guest proteins that affect their stability alter the antibiotic resistances of the cells expressing the biosensors in a manner that is related to the in vitro stabilities of the mutant guest proteins. In addition, we find that polyglutamine tracts of increasing length are associated with an increased tendency to form amyloids in vivo and, in our sandwich fusion system, with decreased resistance to aminoglycoside antibiotics. We demonstrate that our approach allows the in vivo analysis of protein stability in the cytosolic compartment without the need for prior structural and functional knowledge.
INTRODUCTION
Protein folding has been intensely studied in vitro, providing us with a detailed understanding of this process. However, the simplified conditions typically used in vitro (i.e., the use of single purified proteins at very low concentrations) differ substantially from the conditions present in the crowded environment in living cells (1). Although many proteins can fold in vitro without the assistance of other proteins, most proteins in vivo appear to depend on the help of molecular chaperones to fold into their native conformation (2). Protein misfolding in vivo has been linked to various disease states, including Alzheimer's disease (3), Parkinson's disease (4), and cystic fibrosis (5), emphasizing the urgent need to better understand the folding process that occurs inside the cell.
In an attempt to get a better understanding of the in vivo folding process, several in vitro methods have been developed to mimic intracellular conditions. However, replicating the complexity of the cytoplasm in a test tube is a challenging task (6, 7). The difficulty in reproducing the complex environment of living cells has led to the development of methods that allow the measurement of protein stability and folding kinetics inside the cell (8–10). Our lab recently developed biosensors that link protein stability to antimicrobial resistance, allowing the investigation of the periplasmic folding environment (11, 12). Here, we describe the establishment of new protein folding sensors based on proteins involved with resistance to spectinomycin, kanamycin, and nourseothricin (ClonNAT) that expand our approach beyond the periplasm into the more complex folding environment of the cytosol. Using the three guest proteins immunity protein 7 (Im7), human muscle acylphosphatase (AcP), and polyglutamine (polyQ) tracts, we show that the antibiotic resistance conferred by the biosensors is highly correlated with the in vitro stability and aggregation susceptibility of the guest proteins. These new biosensors thus provide additional tools to investigate in vivo protein folding in the cytosol of a variety of hosts.
MATERIALS AND METHODS
Bacterial strains and expression vectors.
Escherichia coli strain NEB10β (New England BioLabs) was used for cloning and protein expression. To develop a sandwich fusion system (Fig. 1) based on kanamycin resistance, we used a vector (pAM18) derived from pCR-Blunt II-TOPO (Invitrogen), which carries the gene (aphA-2) for the kanamycin resistance protein aminoglycoside-3′-phosphotransferase IIa (APH; GenBank accession number WP_004614937). To eliminate expression of the toxic CcdB product present on this plasmid, we introduced a stop codon (TAA) by mutating the tyrosine at amino acid position 5 of the ccdB gene. This was done via site-directed mutagenesis using primers P1 and P2 (see Table S1 in the supplemental material). To increase the proportion of the plasmid taken up by the aphA-2 gene, which encodes kanamycin resistance, the size of the plasmid was decreased by NsiI digestion followed by religation, which removed a 3.4-kb NsiI fragment. The resulting 3.4-kbp plasmid vector was named pAM15.
FIG 1.
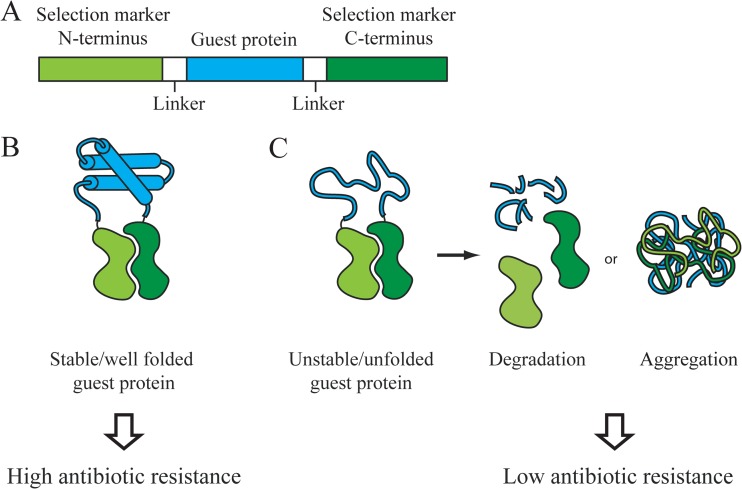
Working principle of the tripartite fusion biosensor for the cytosolic folding environment. (A) The guest protein (blue) is inserted into a permissive site in the antibiotic resistance protein (green), dividing the host protein into an N terminus and a C terminus. The guest protein and the host protein are connected through GS linkers (white), which increase flexibility and allow both proteins to fold into their native conformation. The antibiotic resistance of cells expressing the biosensor is directly correlated to the stability of the guest protein. (B) If the guest protein is stable and well folded, the tripartite system will confer high levels of antibiotic resistance to cells expressing the construct. (C) If the guest protein is unfolded or unstable, the biosensor is rendered nonfunctional by either degradation by cellular proteases or aggregation. Cells expressing this construct have significantly lower antibiotic resistance.
To develop a sandwich fusion system based on streptomycin/spectinomycin resistance, the aadA gene, which encodes resistance to streptomycin and spectinomycin via an adenylyltransferase [ANT(3″)(9); GenBank accession number ABF67771], was cloned from pBAD43 into pBR322 using the restriction enzyme HindIII, creating pAMS1.
To develop a sandwich fusion system based on nourseothricin resistance, the nat1 gene encoding nourseothricin acetyltransferase (NAT; GenBank accession number CAA51674), present on plasmid p4339 (obtained from Amy Chang, University of Michigan), was PCR amplified using primers P134 and P135 to introduce EcoRI sites and then ligated into EcoRI-cleaved pBR322 to generate the plasmid pAM103. The ability of the AM101 strain harboring pAM103 to confer nourseothricin resistance was maximal on Terrific broth plates, so this medium was used in experiments involving nourseothricin.
Linker scanning mutagenesis of resistance genes.
The overall mutagenesis screen strategy is illustrated in Fig. 2. To identify sites within antibiotic resistance genes that would permit the insertion of pentapeptides, we used a GPS-LS linker scanning system with the conditions recommended by the manufacturer (New England BioLabs). The following describes the procedure used for the aphA-2 gene (GenBank accession number WP_004614937), which encodes kanamycin resistance. A similar but not identical protocol was followed to identify sites permissive for the aadA gene (GenBank accession number ABF67771), which encodes spectinomycin resistance, and the nat1 gene, which encodes nourseothricin resistance (see the information in the supplemental material).
FIG 2.
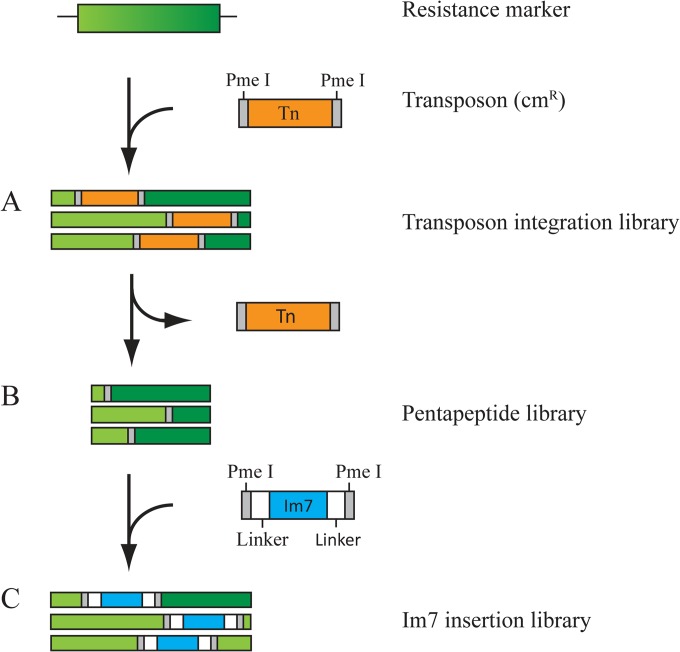
Identification of permissive sites in the resistance marker using the GPS-LS linker scanning system. (A) The transposon (orange) integrates randomly into the coding sequence of the resistance marker (green), creating a transposon integration library. Clones containing the desired transposon insertions in the resistance marker are identified by resistance to chloramphenicol and sensitivity to spectinomycin, kanamycin, or nourseothricin. (B) Digestion of the library with the restriction enzyme PmeI removes the transposon and after religation leaves behind a pentapeptide insertion at the former insertion site. E. coli is transformed with the pentapeptide library and selected for regained spectinomycin, kanamycin, or nourseothricin resistance, allowing the growth of cells that express the resistance marker with pentapeptide insertions in permissive sites. (C) The guest protein Im7 is cloned into the pentapeptide library using PmeI, creating the Im7 insertion library. The Im7 library is transformed into E. coli and selected for spectinomycin, kanamycin, or nourseothricin resistance.
Twenty nanograms of the transposon donor plasmid pGPS4, which contains transprimer-4, which carries the cat gene encoding chloramphenicol resistance (Cmr) (GenBank accession number WP_002361567), was mixed with 80 ng of the recipient plasmid pAM15, which carries the aphA-2 gene encoding kanamycin resistance, and the TnsABC transposase enzyme to initiate in vitro transposition. The transposase acts to integrate the transposon randomly in the recipient plasmid sequence. NEB10β cells were then transformed with the transposition reaction by electroporation. Cells were incubated for 1 h at 37°C and plated onto plates containing chloramphenicol (15 μg/ml) to select for plasmids that were successful recipients in the in vitro transposition reaction. The donor plasmid contains the R6K origin, whose replication requires the π protein to be supplied in cis. This plasmid is thus not able to replicate in NEB10β cells. Therefore, only those events that result in successful transposition into the recipient plasmid result in chloramphenicol-resistant colonies.
To screen for those reactions that resulted in insertion of transprimer-4 into aphA-2 and had thus abolished kanamycin resistance, we simply replica plated the transformants onto Luria-Bertani (LB) plates containing either chloramphenicol alone (15 μg/ml; LB Cm) or both chloramphenicol (15 μg/ml) and kanamycin (100 μg/ml).
Because subsequent steps required the identification of sites that could tolerate the insertion of an open reading frame (ORF) into aphA-2, it was very important to eliminate all kanamycin-resistant bacteria at this stage to prevent contamination in subsequent steps. Thus, the kanamycin-sensitive (Kans) clones were purified to single colonies and subjected to four rounds of replica plating.
The transposons inserted by the GPS-LS linker scanning system were flanked by PmeI restriction enzyme sites. This design allows the facile removal of nearly all of the inserted cat gene, which encodes chloramphenicol resistance, by PmeI digestion followed by religation. The remaining 15-bp transposition scar contains a PmeI restriction site and encodes a tiny 5-amino-acid ORF. This approach allowed us to identify sites in aphA-2 that would tolerate a 5-amino-acid insertion and to easily introduce test protein ORFs via the PmeI site. The Kans clones were grown overnight with shaking at 37°C in 100 μl LB Cm contained in 96-well microtiter plates that had been covered with AirPore tape sheets (Qiagen). Seventy-microliter aliquots from each of these Kans cultures were pooled, and plasmid DNA was prepared and digested with PmeI. The linearized DNA from the Kans library was gel purified and religated. This library contained the linker scanners and was therefore named the KanLS library. It was transformed into NEB10β cells and selected on LB plates containing 200 μg/ml kanamycin, followed by sequencing using primer P3 in order to determine which sites within aphA-2 could sustain 5-amino-acid insertions and still cause kanamycin resistance.
Cloning of Im7 into the KanLS library.
The ceiE7 gene (GenBank accession number Q03708) (11), altered so that it would encode the Im7 F84A mutant flanked by glycine-serine (GS) linkers, was amplified from pAM120 using primers P4 and P5. The GS linkers consisted of 17 amino acids each (upstream sequence, GLNGSGSGSGSGSGSSGSGS; downstream sequence, GSSSGSSGSGSGGGGSGLNG). The primers were designed to introduce PmeI restriction sites at both ends of the gene encoding Im7, ceiE7. The PCR product was ligated into the pCR-Blunt II-TOPO vector (Invitrogen). This clone was then digested with PmeI to isolate the Im7 F84A ORF and subsequently ligated into the KanLS library, which had been PmeI digested and dephosphorylated. Those sites within aphA-2 that allowed phenotypic Kanr even after the insertion of the Im7 ORF were isolated by transformation into electrocompetent NEB10β cells, followed by plating on LB plates that contained 100 μg/ml kanamycin. The sites within aphA-2 that were permissive for the Im7 F84A insertion were identified by sequencing using primer P3. The Im7 F84A mutation inserted into amino acid position 55 of aphA-2 (aphA-2aa55; present on pAM108) was reversed to generate the wild-type (WT) Im7 gene ceiE7 using primers P6 and P7, generating pAM107. To further optimize APH as a biosensor, we evaluated strains containing either Im7 F84A (strain AM236) or wild-type Im7 (strain AM235) for differences in antibiotic resistance after growth on various media (LB, Terrific broth, MacConkey agar, nutrient broth, and M63), at various temperatures (37°C and 42°C), in various strain backgrounds (strains BL21 and MG1655), and with 4 different kanamycin-related antibiotics (kanamycin, neomycin, paramomycin, and gentamicin). The conditions that showed the largest difference in antibiotic resistance between tripartite fusions containing the unstable Im7 F84A mutant and wild-type Im7 were those in which the BL21(DE3)-RIPL strain was grown in LB medium at 37°C, where a very satisfying 5-log-unit difference in cell survival was seen over a nearly 2,000-μg/ml range (see Fig. S2 in the supplemental material). Subsequently, individual point mutants of known thermodynamic stability (F15A, V33E, L34A, L53A, I54V, N26K-S58R, V69A, N26K-T30N-S58R, L3A, D35N-D63N, V27A-D63N, L18F-D35N-D63N, L18A) were introduced into the Im7 WT tripartite fusion by site-directed mutagenesis using primers P8-P9, P10-P11, P12-P13, P14-P15, P16-P17, P46-P47-P48-P49, P52-P53, P44-P45-P48-P49, P30-P31, P25-P26-P36-P37, P25-P26-P40-P41, P38-P39-P25-P26-P36-P37, and P42-P43, respectively. Various variants of Im7 were introduced into the pentapeptide permissive sites in the APH and NAT proteins in a similar manner. The derivation of these plasmids is described in Table S5 in the supplemental material.
Replacing Im7 with another guest protein (AcP) in the spectinomycin resistance marker.
Digestion of the pBR322 plasmid that contains the aadA gene with the ceiE7 gene encoding the guest protein Im7 inserted after amino acid 155 of ANT (pAMS2) with XhoI and SacI removes only ceiE7 from the plasmid and leaves the majority of the GS linker in the plasmid, creating pAMS3 (see Fig. S1 in the supplemental material). This simplifies the swapping of different guest proteins in the biosensor. Note that the digest shortens the original upstream GS linker in pAMS3 from 17 to 11 amino acids, followed by the XhoI restriction site (which encodes Leu and Glu); the downstream linker is similarly shortened from 17 to 13 amino acids and is preceded by a SacI restriction site (which encodes Glu and Leu). The acpY2 gene, encoding the enzyme AcP (GenBank accession number P14621), was thus cloned into pAMS3 using XhoI and SacI, creating pAMS34. This construct was further modified using primers P52 and P53 in order to create a cysteine-less variant of AcP (C21S) called pAMS35. The use of this cysteine-less variant, which was present in all subsequent AcP constructs, facilitates comparison with previously published stability data (13). This pseudo wild-type variant is referred to as the WT throughout the study. pAMS35 therefore has the biosensor with acpY2 inserted after amino acid position 155 of aadA. AcP variants Y11F, V20A, M61A, L65V, and E83D all contain mutation C21S and were created using site-directed mutagenesis with primers P56-P57, P58-P59, P60-P61, P62-P63, and P64-P65, respectively (plasmids pAMS36 to pAMS40; see Table S5 in the supplemental material).
Cloning of polyglutamines at position 55 in the aphA-2 gene, which encodes APH.
To facilitate the insertion of additional peptides at permissive position 55 within the APH protein, a unique BamHI site was introduced at the corresponding position of the aphA-2 gene in pAM15 by site-directed mutagenesis using primers P18 and P19. Different lengths of polyQ-encoding DNA flanked by BamHI ends was amplified from p416/PQ103 (14) using primers P20 and P21. PCR products ranging in size from 50 to 500 bp were isolated from a 1% preparative agarose gel. Subclones of polyQ-encoding DNA of various lengths were made in the pCR-Blunt II-TOPO vector. Three different lengths of polyQ inserts (20, 45, and 87 Q residues), which were isolated by BamHI digestion and ligated into a BamHI-linearized dephosphorylated vector followed by transformation into NEB10β cells, were identified by sequencing. The corresponding plasmids were called pAM79, pAM80, and pAM81. We noticed that the polyQ87 tract in pAM81 was somewhat unstable, readily generating N-terminal 40-nucleotide deletions in APH. To prevent these specific recombinations, several silent mutations were generated at positions 41 to 43 of APH in pAM81 using primers P22 and P23 to generate pAM180. This plasmid was stable, so the changes appear to have reduced homologous recombinations between the ORF and its promoter.
MIC determination in E. coli.
Antibiotic resistance was measured in terms of the MIC, i.e., the lowest concentration of an antibiotic that inhibits the growth of a microorganism. MIC experiments were performed as described by Foit et al. (11) with the following modifications. Prior to spot titer experiments for kanamycin or nourseothricin resistance, cultures were made by inoculating a single colony into 5 ml LB containing either 25 μg/ml zeocin or 200 μg/ml ampicillin, followed by incubation overnight at 37°C without shaking to obtain an optical density (OD) at 600 nm (OD600) of ∼0.1 to 0.2. Note that this overnight selection was not for the partially crippled antibiotic resistance marker but, rather, was for the alternative antibiotic resistance marker present on the plasmid. Following this standing overnight growth, tubes were transferred to a shaking incubator at 37°C, where cells rapidly entered log-phase growth and after 2 to 3 h had reached late log phase (OD600 = 1 to 1.5). Spot titer experiments were done on prewarmed LB agar plates containing increasing concentrations of kanamycin or nourseothricin. The growth of cultures for determining MICs for spectinomycin resistance was done in a slightly different fashion. Single colonies were inoculated from plates and grown overnight in culture tubes containing 5 ml LB and ampicillin (100 μg/ml) in a rotary drum incubator at 37°C. Overnight cultures were diluted 1:100 and grown to log phase under the same conditions. Cells were pelleted and adjusted to an OD600 of 1 with phosphate-buffered saline (PBS). Cells were serially diluted, and dilutions of 100 to 10−5 were spotted on LB plates containing increasing concentrations of spectinomycin.
After incubation overnight at 37°C, MICs for every variant were determined by evaluating the growth of each dilution on antibiotic at each concentration. The average of the MICs of dilutions of 10−1 to 10−5 was calculated. Experiments were repeated at least three times, and the results are shown as means ± standard errors of the means.
Fractionation of E. coli extracts.
Cultures expressing aphA-2aa55::ceiE7 variants were grown overnight in LB containing 25 μg/ml zeocin. The culture density was then adjusted to an OD600 of 3.5 by adding 10 mM Tris, pH 8.0. For soluble protein extraction, 0.5 ml of these cultures was pelleted at 15,700 × g for 10 min, and the pellets were resuspended in 500 μl lysis buffer (1 mg/ml lysozyme, 5 mM EDTA in Tris-buffered saline). One hundred microliters of 0.1-mm-diameter glass beads (BioSpec) was added to each tube, and the tubes were incubated on ice for 30 min, during which time they were vortexed four times at top speed for 30 s each time to lyse the cells and break up big aggregates. Aggregates were separated by centrifugation at 15,700 × g for 10 min. Aggregated proteins in the pellet were extracted by boiling the pellet for 5 min in 600 μl of 1× SDS loading buffer. Two hundred microliters of the supernatant was mixed with 40 μl of 5× SDS loading buffer and denatured by boiling for 3 min at 95°C. This fraction was designated the soluble fraction. Samples from the soluble and insoluble fractions (15 μl each) were loaded on precast 4 to 12% bis-Tris polyacrylamide gels. Proteins were blotted on an iBlot system (Invitrogen) according to the manufacturer's instructions. Protein fusions containing the kanamycin resistance protein were detected with Western blots using anti-neomycin phosphotransferase II (Sigma) antibody at a 1:10,000 dilution. Horseradish peroxidase-conjugated goat anti-rabbit IgG at a 1:10,000 dilution was used as the secondary antibody. SuperSignal West Pico chemiluminescent substrate (Pierce) was used to visualize antibody binding.
For fractionation of the polyQ sandwich fusion proteins, 0.2-ml samples from the overnight cultures (OD600 = 5) were incubated with lysis buffer on ice. To break the cells, freezing and thawing were performed for 3 rounds by cycling between a dry ice ethanol bath and a water bath held at 37°C. The supernatant obtained after centrifugation at 15,700 × g for 10 min was considered the soluble fraction. The insoluble pellet was dissolved in 240 μl 1× SDS loading buffer, and 40 μl 5× SDS loading buffer was added to the 200-μl supernatant. Samples were denatured by boiling at 95°C for 3 min, and 20 μl from both fractions was loaded onto precast 4 to 12% bis-Tris gels. Western blotting and development were done as described above.
Cloning of aphA-2aa55::ceiE7 variants and polyQ sandwich fusions in Saccharomyces cerevisiae.
Primers P24 and P25 were designed to amplify sandwich fusions from the pTOPO vector, and these fusions were cloned into the pYES2.1 TOPO TA expression shuttle vector. Clones were sequenced with primers P26 and P27.
MIC determination in S. cerevisiae.
Sandwich fusions were transformed into the INVSc1 strain of S. cerevisiae expressed in synthetic complete (SC) minimal broth medium minus uracil (SC−U) supplemented with 2% galactose under the control of the GAL promoter for 48 h. All cultures were adjusted to an OD600 of 0.2. Fully induced cultures (3.2 μl) were inoculated in 100 μl yeast extract-peptone dextrose (YPD) medium containing various concentrations of G418 in 96-well plate formats. Microtiter plates were sealed with tape pads (Qiagen) and incubated at 30°C for 20 h. The OD600 was measured with a Synergy HT plate reader (BioTek Instruments, Inc.).
Analysis of sandwich fusions in S. cerevisiae.
Sandwich fusions with Im7 variants or polyQ were expressed in SC−U supplemented with 2% galactose for 48 h. Duplicate 0.2-ml samples from the cultures were adjusted to an OD600 of 5. Cells were pelleted by centrifuging at 6,500 × g for 5 min. For soluble protein extraction, pellets were treated with 200 μl yeast lysis solution (Amersham) according to the manufacturer's instructions. For total protein analysis, the pellet was dissolved in 240 μl 1× SDS loading buffer. Glass beads (∼100 μl; diameter, 0.5 mm) were added to each of the tubes. The tubes were incubated on ice and vortexed at high speed six times for 30 s each time. For soluble protein extraction, centrifugation was done at 15,700 × g for 10 min. One hundred microliters supernatant was aspirated, and 20 μl 5× SDS loading buffer was added. Samples were boiled at 95°C for 3 min. For the loading control, 15-μl samples were loaded on 7% Tris-acetate gels, and for Western blot analysis, 20-μl samples were loaded on 4 to 12% bis-Tris gels. Blotting and development were done as described above.
RESULTS
Development of a protein folding biosensor.
The principle of a sandwich folding biosensor is illustrated in Fig. 1. It involves the insertion of a guest protein (shown in blue) into the middle of a selectable marker. The concept is that if the guest protein is folded properly, it should bring the N- and C-terminal halves of the selectable marker close together, enabling the marker to fold properly and thus function. However, if the guest protein is unstable, it will be cleaved by the plethora of proteases present in vivo or it will be prone to aggregation. Both proteolysis and aggregation should, in theory, decrease the activity of the biosensor. In principle, if one is able to couple in vivo proteolysis or aggregation to an appropriate selectable marker, one should be able to study protein stability and aggregation propensity in vivo. If the selectable marker is efficient enough, one should also be able to select for more stable or less aggregation-prone variants and to identify host mutants that show improved folding environments. We previously developed biosensors based on two periplasmic proteins: the β-lactamase protein, which confers ampicillin resistance, and the DsbA protein, which catalyzes disulfide bond formation and confers cadmium resistance. The selective power of this approach allowed the facile isolation of stabilized protein variants (11) and the discovery of a new periplasmic chaperone called Spy (11, 15). Although powerful, these systems cannot be utilized to investigate the folding environment of the cytoplasm because export to the periplasm is essential for β-lactamase function and for the cadmium resistance encoded by the DsbA gene. We therefore decided to search for selectable markers that could expand our approach into the cytosolic compartment and potentially into other organisms as well.
Antibiotic resistance markers are ideal for our purposes because antibiotics very effectively kill cells or inhibit their growth, their resistance markers often provide very high levels of protection, and they often work on a wide variety of species. Antimicrobial resistance can easily be determined in spot titer experiments, and the effects of different conditions or mutations on protein stability or aggregation propensity can be compared. We decided to investigate markers for kanamycin, spectinomycin, and nourseothricin resistance. The aminoglycoside-3′-phosphotransferase IIa (APH) gene, which encodes kanamycin resistance in bacteria and G418 resistance in yeast, was chosen because its structure is known and it is commonly used as a selection marker (16, 17). The streptomycin/spectinomycin adenylyltransferase ANT(3″)(9) protein (18) was similarly chosen because spectinomycin and streptomycin are also commonly used, inexpensive, and readily available (19). Resistance to the antibiotic nourseothricin (20) is conferred by nourseothricin acetyltransferase (NAT). This antibiotic, though much more expensive and less available than kanamycin or spectinomycin, was chosen because it is effective not just against Gram-negative bacteria but also against Gram-positive bacteria, mycobacteria, mycoplasmas, protista, yeasts, and plants (21–24).
For the sandwich fusion approach to be successful, we needed to find sites within these antibiotic resistance markers that would tolerate the insertion of folded proteins and be responsive to the stability or aggregation susceptibility of the inserted protein, allowing us to link antibiotic resistance to protein stability or aggregation susceptibility. These types of sites are presumably quite rare, as most insertions are disruptive to protein function (25). We decided to develop a general protocol to find sites within selectable markers that would tolerate insertions and then to find within this set those in which the stability of the inserted protein determined the selectable marker's activity. To accomplish this, we first constructed a library of insertions within the antibiotic resistance genes, screened them for sites that would tolerate short amino acid insertions, and then screened these for sites that would tolerate the insertion of a complete protein. Finally, we screened these sites for those in which the stability of the inserted protein determined the level of antibiotic resistance. This procedure is diagramed in Fig. 2 and described in detail in the Materials and Methods.
To construct libraries of insertion sites within the antibiotic resistance genes, the GPS-LS linker scanning system, a transposon-based mutagenesis system commercially available from New England BioLabs, was used. This system enables the rapid construction of an extensive set of random insertions of a minitransposon consisting of the chloramphenicol resistance marker flanked by PmeI sites into plasmid DNA.
The development of a kanamycin resistance-based stability biosensor is described here in detail. Stability biosensors based on other antibiotic resistance markers were generated in a similar fashion and are described in the Materials and Methods and in the information in the supplemental material.
We first transposed a chloramphenicol resistance (Cmr) marker from the transposon donor plasmid pGPS4 into a recipient plasmid, pAM15, which contains the kanamycin resistance marker. We screened the ∼30,000 chloramphenicol-resistant insertions that we obtained for those that were kanamycin sensitive in order to obtain a library of clones containing insertion sites within the aphA-2 gene, which normally encodes kanamycin resistance. After four rounds of purification, 1,800 insertions verified to be kanamycin sensitive were obtained. The insertion sites were determined for 50 of these insertions. All were within aphA-2 and its promoter region, and all were in different sites, consistent with previous observations that the Tn9 transposase, on which the GPS-LS linker scanning system is based, inserts with very little sequence specificity. Because the number of insertions that we obtained (n = 1,800) exceeded the number of codons (n = 264) in the aphA-2 gene by a factor of ∼6, we reasoned that our insertion library was likely reasonably complete.
The transposons were excised from the pooled library, leaving behind a pentapeptide insertion carrying a PmeI site. Those sites within APH that could tolerate insertions of 5 amino acids and still confer kanamycin resistance were readily obtained simply by plating the religated library pool on medium containing 200 μg/ml kanamycin (26). Determination of the insertion site within 46 randomly picked Kanr clones (see Fig. S2 in the supplemental material) indicates that the majority of the pentapeptide permissible insertion sites are present in surface loops in the N-terminal domain of the APH protein. We then discovered which of these sites could tolerate larger insertions by simply recleaving the Cmr Kans library of all 1,800 clones with PmeI, followed by the kanamycin-resistant insertional cloning of a larger open reading frame. For a guest protein, we initially picked immunity protein 7 (Im7), a 10-kDa, helical E. coli protein. We chose Im7 as a model protein because of the ready availability of Im7 variants with a wide spectrum of known thermodynamic stabilities (11, 27). This facilitated the following step in our procedure: namely, to test different permissive sites for their correlation between antibiotic resistance and protein stability. For initial experiments, we chose to investigate a very destabilized variant, Im7 F84A. Im7 F84A was chosen as a guest protein because it is unstable in vivo and in vitro but is still capable of expressing significant resistance to penicillin when tested in the β-lactamase stability biosensors (11, 27). We ligated the plasmids with pAM108, a construct consisting of Im7 F84A and flanking glycine-serine (GS) linkers at its C and N termini (see Fig. S1 in the supplemental material). The linkers were designed to provide flexibility and reduce the steric interference of the fusion proteins, allowing them to fold independently into their native conformations.
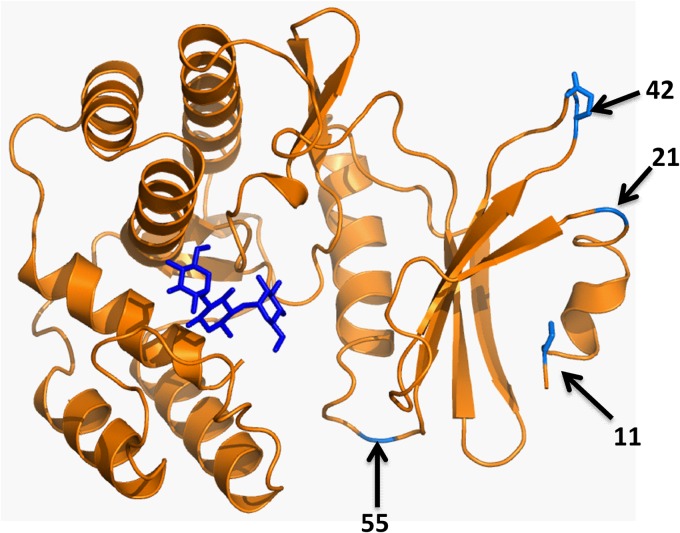
Four sites at positions 11, 21, 42, and 55 within the APH protein that tolerate the insertion of Im7 F84A and still exhibit resistance to at least 100 μg/ml kanamycin were found (Fig. 3; see also Fig. S2 in the supplemental material). The APH protein, present in plasmid pAM108, with insertions of Im7 at position 55 was selected for further analysis.
FIG 3.
(A) Sites in the structure of aminoglycoside-3′-phosphotransferase IIa (PDB accession number 1ND4), which confers resistance to kanamycin, that we found to be suitable for the insertion of proteins.
Optimization of insertion sites.
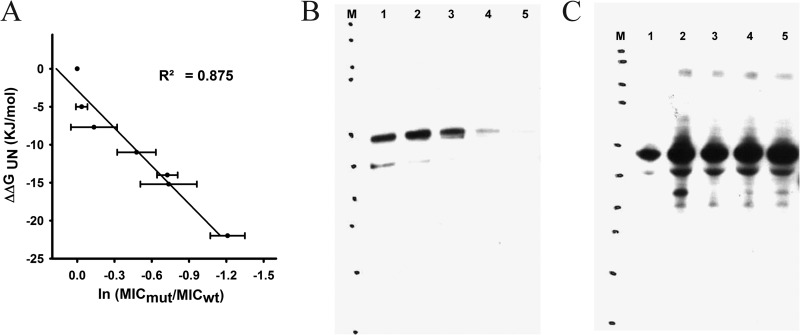
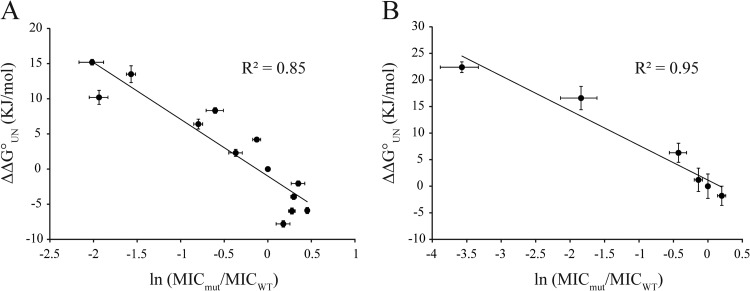
For a stability biosensor to be effective, it should give a linear readout over a wide range of stabilities of the guest protein. For biosensors derived from the APH protein, this means that there should be a large difference in kanamycin resistance for sandwich fusions containing inserted guest proteins of various stabilities, and their kanamycin resistance should be directly related to the stabilities of the guest proteins. Thus, we restored the wild-type Im7 sequence in our Im7 F84A construct and then introduced into Im7 a wide range of mutations that are known to affect Im7 protein stability (see Tables S5 and S2 in supplemental material). The relative MICs conferred by different sandwich fusions showed a very nice relationship with the stabilities of Im7 variants for all the destabilized variants and the wild-type protein (Fig. 4A). Variants of Im7 with stabilities greater than the stability of the wild type, however, showed MIC values that scattered around the wild-type MIC, indicating that the biosensors' capacity to measure stabilities may saturate for proteins with stabilities greater than that of wild-type Im7 (−24.9 kJ/mol) (27).
FIG 4.
Relationship between stabilities of Im7 guest proteins and kanamycin resistance in E. coli BL21(DE3)-RIPL. (A) Position 55 in APH (which causes kanamycin resistance) was evaluated by inserting different thermodynamically destabilized mutants of Im7. The kanamycin resistances of cells containing the fusions with the destabilized Im7 mutants relative to those of cells containing WT Im7 correlated well with the stabilities of the guest proteins. From left to right, the strains used were AM235, AM240, AM258, AM243, AM242, AM247, and AM236, respectively. UN, unfolding; MICmut, MIC for the mutant; MICwt, MIC for the WT. (B and C) Western blots showing the amount and solubility of the sandwich fusions in the soluble fraction (B) and in whole-cell lysates (C). Lanes: M, molecular size marker; 1, WT Im7 fused at position 55 (AM235); 2, Im7 V33E fused at position 55 (AM240); 3, Im7 L34A fused at position 55 (AM258); 4, Im7 F15A fused at position 55 (AM247); and 5, Im7 F84A fused at position 55 (AM236).
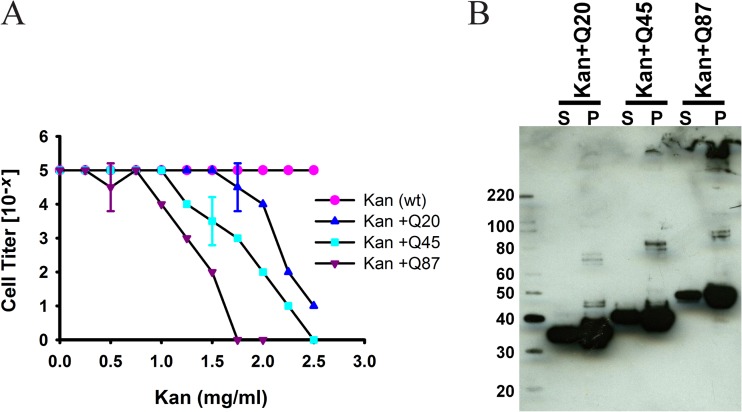
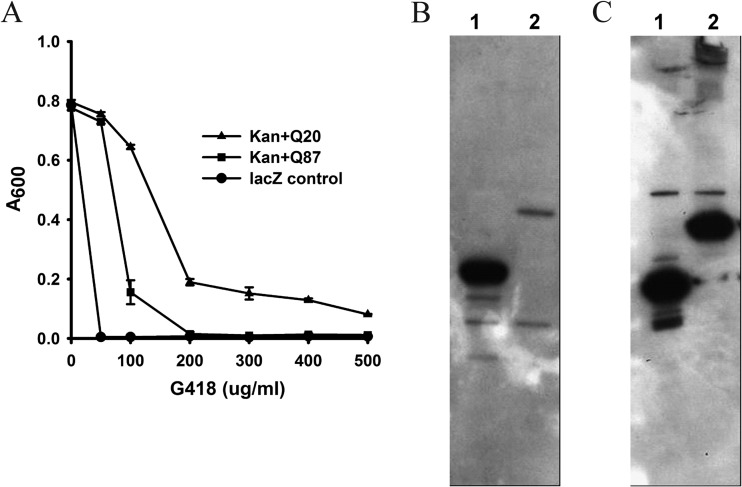
The amount and solubility of the sandwich fusions were evaluated via Western blotting using both whole-cell lysates and soluble fractions (Fig. 4B and C; see also Fig. S3A and B in supplemental material). We observed increased aggregation of the sandwich fusion as the stability of the guest protein inserted into the tripartite fusion decreased. It has previously been observed that when polyglutamine tracts longer than ∼35 residues are present in proteins, they have a strong tendency to form amyloids in vitro and in vivo, with an increasing length of polyglutamine being associated with more severe amyloidogenesis (28). We inserted polyglutamine tracts of three different lengths (20, 45, and 87 Q residues) at position 55 in APH and evaluated their effect on the solubility of the resulting sandwich fusions and on their ability to cause kanamycin resistance. As shown in Fig. 5, as the length of the polyglutamine tracts within sandwich fusions increased, the kanamycin resistance of cells expressing these fusions decreased (Fig. 5A); the amount of soluble material also decreased (Fig. 5B). Conversely, the proportion of SDS-insoluble material expressed increased with increasing polyglutamine tract length.
FIG 5.
Correlation between amyloidogenicity and kanamycin resistance in E. coli. (A) Polyglutamine tracts of three different lengths (20, 45, and 87 Q residues, designated by +Q20, +Q45, and +Q87) were directly fused at position 55 in APH, and the resulting sandwich fusions were expressed in NEB10β. Cell survival at maximum dilutions was scored for different concentrations of kanamycin. The Kan WT is strain AM144, Q20 is strain AM200, Q45 is strain AM201, and Q87 is strain AM308. (B) Western blots showing the distribution of polyQ sandwich fusion proteins in the soluble fraction (lanes S) and in the corresponding cell pellets of the strains (lanes P). Numbers on the left are molecular size markers (in kilodaltons).
This APH biosensor thus appears to function as a readout for the aggregation propensity of polyglutamine tracts in the range of 20 to 87 amino acids and as a readout for Im7 stability in the range of 0 to −24.9 kJ/mol. The periplasmic β-lactamase-based stability biosensor that we previously developed was sensitive over a broader range. We wondered if biosensors based on resistance to another antibiotic might be responsive to a broader range of protein stabilities, so we repeated the biosensor development process using the streptomycin/spectinomycin adenylyltransferase ANT(3″) (9) protein, which confers spectinomycin/streptomycin resistance.
Identification of pentapeptide permissive sites in ANT.
Pentapeptide permissive sites could be identified in a fashion similar to that described for APH by selecting colonies for regained spectinomycin resistance and identifying permissive sites by sequencing. While the transposon insertions appeared to be randomly distributed throughout aadA, pentapeptide permissive sites were found in only three distinct hot spots in the protein (see Fig. S4 in the supplemental material).
Im7 permissive sites in ANT show a distribution pattern similar to that of pentapeptide permissive sites.
We again reasoned that only sites that tolerate a pentapeptide insertion would be likely to accept the insertion of a model protein. Im7 with linker sequences was thus cloned in the PmeI site carried by the pentapeptide insertion. The library containing Im7 in ANT pentapeptide permissive sites was transformed and selected on plates containing 100 μg/ml spectinomycin. Plasmids from resistant colonies were sequenced in order to identify Im7 permissive sites. Sequencing of resistant colonies showed that sites tolerating Im7 insertions also accumulated in three hot spots in ANT in a pattern very similar to that found for the pentapeptide library (see Fig. S4C and D in the supplemental material), allowing us to conclude that many sites that allow the insertion of a pentapeptide also allow the insertion of a full-length protein. The three hot spots in ANT that permitted Im7 insertions are positions 75 to 81, 152 to 157, and 289 to 317 (see Fig. S4C and D in the supplemental material). The last two groups of insertions were, as expected, in frame. Surprisingly, many of the insertions located in the N terminus (positions 75 to 81) caused frameshifts.
To further investigate this result, we screened 500 clones for spectinomycin resistance by letting them grow overnight in increasing concentrations of spectinomycin. By measuring the final ODs of the overnight cultures, we separated the insertions into two subgroups: (i) those with high spectinomycin resistance (for which MICs were indistinguishable from the MIC obtained for wild-type ANT) and (ii) those with moderate spectinomycin resistance. All the clones with high spectinomycin resistance had in-frame insertions and were located in the N terminus of ANT (see Fig. S4C in the supplemental material). The subgroup with significantly lower resistance had insertions in the N terminus, the middle, or the C terminus of the protein (see Fig. S4D in the supplemental material). We reasoned that clones with frameshifts in the N terminus might express truncated but still fully functional versions of ANT that use an alternate initiation codon that lies downstream of the Im7 insertion site. If this were the case, it seemed unlikely that the insertion of proteins with various stabilities at these positions would alter spectinomycin sensitivity. Therefore, we focused on insertion sites in the middle and the C terminus of the protein.
A spectinomycin-based biosensor links antibiotic resistance and in vitro stability for different guest proteins.
Initially, we chose an insertion site in the middle of ANT (at position 155) present on plasmid pAMS2, to test the relationship between the in vitro stability of the Im7 guest protein and the antibiotic resistance of cells expressing the fusion protein. Using site-directed mutagenesis and pAMS2 as a template, we generated Im7 variants (pAMS4 to pAM15) covering a wide stability range (see Table S2 in the supplemental material). We then measured the MICs of cells expressing those fusion proteins (AMS2 and AMS4 to AMS15). Figure 6A shows that there is a very good correlation between the in vitro stability of the tested Im7 variants and the antibiotic resistance of cells expressing the corresponding fusion proteins. Variants that are destabilized compared to the stability of the WT in vitro show low antibiotic resistance, and in contrast to the kanamycin resistance-based protein stability biosensor, stabilized variants have resistances higher than the resistance of the wild type. Thus, it appears that the spectinomycin-based stability biosensors have a significantly broader range than the kanamycin-based ones.
FIG 6.
Correlation between antibiotic resistance and protein stabilities of Im7 and AcP variants in ANT. The in vitro stabilities of Im7 variants (A) and AcP variants (B) were plotted against the MICs for bacteria expressing the respective variants in the tripartite fusion Im7, which were (from left to right) strains AMS6, AMS15, AMS11, AMS5, AMS4, AMS8, AMS10, AMS2, AMS14, AMS12, AMS13, AMS7, and AMS9, and AcP, which were (from left to right) strains AMS37, AMS40, AMS38, AMS39, AMS35, and AMS36. The in vitro protein stabilities were taken from the literature (13, 27). MICs for cells expressing the constructs were measured in spot titer experiments. For both guest proteins, we see a significant correlation between stability and spectinomycin resistance.
The spectinomycin-based system contains XhoI and SacI restriction sites, placed close to Im7 within the GS linker sequence, to facilitate the easy replacement of Im7 with different guest proteins (Fig. S1 in the supplemental material). To determine if this spectinomycin-based system functions as a stability biosensor with a different guest protein (in addition to Im7), we tested its functionality with human muscle acylphosphate (AcP). This 98-amino-acid enzyme was chosen because the in vitro stabilities of many of its variants are known (13, 29). We switched the Im7 protein in our ANT tripartite fusion (pAMS2) with wild-type AcP (creating pAMS34) (see Fig. S1 in the supplemental material) and then used site-directed mutagenesis to generate a series of AcP variants (pAMS35 to pAMS40; see Table S5 in the supplemental material) with a wide range of in vitro stabilities (see Table S4 in the supplemental material). The spectinomycin MICs of these constructs measured in spot titer experiments were very well correlated to the in vitro stabilities of the inserted AcP protein variants (13) (Fig. 6B), showing that this system is not limited to Im7.
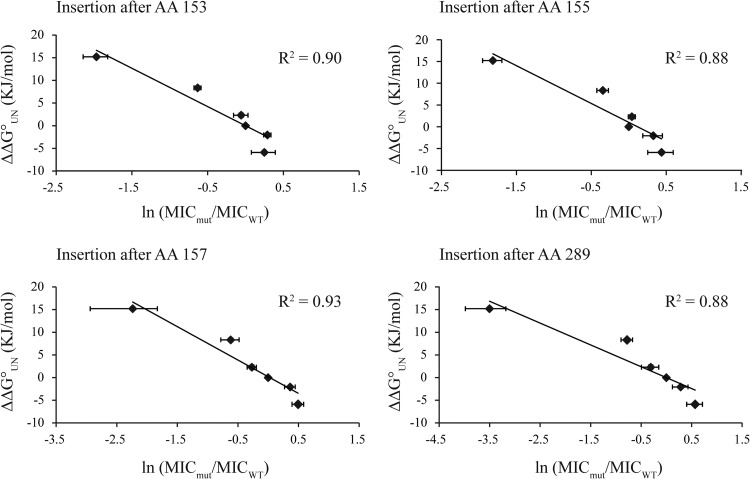
To test whether our spectinomycin-based biosensor results depend on the position of the insertion site, we repeated the Im7 experiments for three additional permissive sites: two in the middle (positions 153 and 157) and one at the C terminus (position 289) of ANT (Fig. 7). MICs were measured in strains AMS2, AMS4, and AMS6 to AMS9 for insertion site 155, AMS16 to AMS21 for insertion site 153, AMS22 to AMS27 for insertion site 157, and AMS28 to AMS33 for insertion site 289 (see Table S5 in the supplemental material). For all tested sites, a very clear correlation between the in vitro stability of the insert and the in vivo antibiotic resistance of the corresponding construct was shown. For all four sites tested, there was a drop-off in apparent sensitivity for the most stable Im7 variant (Y11F; ΔΔG0, 1.8 kJ/mol) (13). This suggests that the spectinomycin-based stability biosensors may, like the kanamycin-based sensors, also work over a broader but still limited range of corresponding in vitro stabilities.
FIG 7.
Four different permissive sites in ANT were evaluated with six Im7 variants spanning a wide range of in vitro stabilities. The MIC values were determined for the following strains, from left to right: for the insertion site at amino acid (AA) 153, strains AMS18, AMS19, AMS17, AMS16, AMS20, and AMS21; for the insertion site at amino acid 155, AMS6, AMS4, AMS8, AMS2, AMS7, and AMS9; for the insertion site at amino acid 157, AMS24, AMS25, AMS23, AMS22, AMS26, and AMS27; and for the insertion site at amino acid 289, AMS30, AMS31, AMS29, AMS28, AMS32, and AMS33. Independently of the exact location of the permissive site in ANT, we found that all biosensors showed a clear correlation between the in vitro stabilities of the Im7 inserts and the antibiotic resistance of the corresponding fusions.
Toward the development of a stability biosensor that functions in the eukaryote S. cerevisiae.
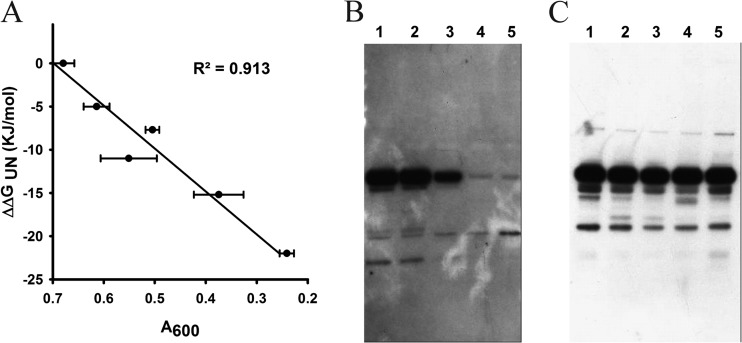
Finally, we attempted to develop a stability biosensor that functions in yeast. The APH protein confers resistance to the aminoglycoside antibiotic G418 in S. cerevisiae (30). Six different APH sandwich fusions containing Im7 guest proteins with various stabilities were cloned behind the GAL promoter and grown with induction using 2% galactose for 48 h. Following induction, ∼2 ×104 yeast cells were inoculated into 100 μl YPD medium containing increasing concentrations of G418 (0 to 53 mg/ml) and grown at 30°C for 20 h; growth was monitored by measuring the OD600 (see Fig. S5 in the supplemental material). Growth inhibition by G418 of S. cerevisiae cells expressing these different sandwich fusions was linearly correlated with the stabilities of the Im7 guest proteins in the fusion; i.e., fusions containing proteins with greater stability showed greater antibiotic resistance (Fig. 8A).
FIG 8.
Relationship between stabilities of guest proteins and growth inhibition of S. cerevisiae. (A) APH sandwich fusions containing Im7 guest proteins with different stabilities were cloned behind the GAL promoter, and cells were grown using 2% galactose. Twenty thousand fully induced yeast cells were inoculated into 100 μl YPD containing increasing concentrations of the antibiotic G418 and incubated at 30°C for 20 h. The growth of S. cerevisiae at 400 μg/ml G418 was plotted against the in vitro stabilities of the Im7 variants. From top to bottom, the strains were AYC29, AYC34, AYC35, AYC39, AYC30, and AYC42, carrying the WT and Im7 variants V33E, L34A, I54V, F15A, and F84A, respectively. (B and C) Western blots quantifying sandwich fusion material obtained from soluble fractions (B) and whole-cell lysates (C). Lanes 1 to 5, strains AYC29 (lanes 1), AYC34 (lanes 2), AYC35 (lanes 3), AYC30 (lanes 4), and AYC42 (lanes 5), with WT Im7 and Im7 variants V33E, L34A, F15A, and F84A at position 55 in APH.
The amount and solubility of the sandwich fusions were quantified via Western blotting using both whole-cell lysates and soluble fractions (Fig. 8B and C; see also Fig. S6 in the supplemental material). The amount of soluble material varied directly with the stability of the inserted Im7 protein, whereas the total amount of sandwich fusion remained constant. We next tested the solubility and activity of the APH sandwich fusions containing polyglutamine tracts. S. cerevisiae cells expressing polyQ inserted at amino acid 20 (polyQ20) or polyQ87 under galactose control were inoculated into 100 μl YPD containing 0 to 53 mg/ml of G418. After 20 h of incubation at 30°C, the growth of S. cerevisiae cells expressing fusions containing polyQ87 was significantly lower in the presence of G418 than that of yeast cells expressing fusions containing polyQ20 (Fig. 9A). Western blots (Fig. 9B and C) revealed that the total concentration of material was similar for both sandwich fusions, but their solubility differed, with the fusion containing polyQ87 being considerably less soluble (Fig. 9B). We do not have any specific structural information about how the polyQ tracts behave in the context of the fusion; we do know, however, that as the polyQ tracts get longer, the antibiotic resistances caused by the corresponding fusions go down and the solubility of the corresponding fusions also goes down.
FIG 9.
Relationship between the length of polyQ tracts in the fusion and growth inhibition of S. cerevisiae. (A) APH sandwich fusions containing polyQ20 or polyQ87 inserts were grown in yeast cells under galactose control. Twenty thousand fully induced cells were inoculated into 100 μl YPD medium containing increasing concentrations of G418. Growth was monitored after 20 h of incubation at 30°C. The strains used were AYC44 for Kan::Q20 and AYC46 for Kan::Q87. (B and C) Western blots quantifying sandwich fusion material obtained from soluble fractions (B) and whole-cell lysates (C). Lanes 1, APH with the polyQ20 insert at position 55 (AYC44); lanes 2, APH with the polyQ87 insert at position 55 (AYC46).
These results suggested that APH/NAT-based biosensors could be used to monitor protein stability and the aggregation tendency in yeast. The differences in OD exhibited in Fig. 9A, however, are substantially less than the >5-log-unit difference in the numbers of CFU seen with the kanamycin selection in E. coli (Fig. 3B). We therefore attempted to develop a stability biosensor based on nourseothricin acetyltransferase, which causes resistance to the antibiotic nourseothricin in a wide range of prokaryotic as well as eukaryotic hosts. Using a procedure similar to that used for the kanamycin and spectinomycin resistance markers, we identified two permissive sites in the NAT protein that resulted in a very significant drop in resistance to nourseothricin in E. coli compared to that obtained with the protein with wild-type Im7 when they contained the unstable Im7 F15A insert (see Fig. S7 in the supplemental material). In both cases, cells containing wild-type and hyperstable variants of Im7 had similar nourseothricin resistances, suggesting that, similar to kanamycin resistance in E. coli, the in vitro stability exhibited by wild-type Im7 (15.2 kJ/mol) may represent the limit of the assay. Unfortunately, yeast cells containing our various tripartite fusions were equally and extremely nourseothricin resistant. Concentrations of nourseothricin as high as 5,000 μg/ml on plates showed no effect on yeast growth, independently of the NAT Im7 fusion that it contained. This was true when these fusions were cloned on both high-copy-number (pYES-derived) and low-copy-number (pYC-derived) plasmids. In liquid culture, growth could not be completely inhibited even at 33,000 μg/ml nourseothricin, and tripartite fusions showed very similar growth curves under nourseothricin selection pressure, irrespective of the in vitro stability of the Im7 that they contained.
DISCUSSION
Levels of protein expression are influenced by the folding capacity of the host and the intrinsic stability or aggregation propensity of the protein of interest (11, 31, 32). To study protein stability in vivo, we previously developed a selection system based on sandwich fusions to the TEM-1 β-lactamase protein, which causes resistance to β-lactam antibiotics (11). This system proved to be effective in analyzing the in vivo stability of periplasmic proteins. This paper describes the development of a similar system based on several cytosolically expressed genes that encode resistance to aminoglycoside antibiotics. These sandwich fusion-based systems directly link the stability and solubility of model proteins to a phenotype that is easily selectable in E. coli and, in the case of G418, are screenable in the yeast S. cerevisiae. Insertions are usually deleterious, frequently disrupting the protein structure at the insertion site in catastrophic ways (25, 33). It is difficult to predict those rare sites within a protein that can accommodate insertions without severe functional disruption (34).
We therefore developed an approach to experimentally determine insertion-permissive sites within selectable markers. This protocol is very straightforward. It functioned well with all three antibiotic resistance markers that we tested and is potentially generalizable to any selectable marker carried by a plasmid. Sites within a host protein's structure that permit the insertion of a guest protein have historically been most commonly found in linker regions on the surface of the host protein. This is presumably because at these locations, the insertion of a guest protein does not interfere with the overall fold of the host protein (35, 36). This localization also exposes the guest protein to cellular proteases that recognize unstable and unfolded protein variants and degrade the fusion protein in a rate dependent on the guest protein's stability (37). The structure of the kanamycin resistance protein is known, and our permissive insertion sites do generally occur on surface regions in some (but not all) loops, but not within linker regions. Thus, even for proteins whose structure is known, like the APH protein, it would have been difficult to predict a priori where permissive insertion sites might lie, emphasizing the need to take an experimental approach. This is even more important for selectable markers like the ANT and NAT proteins, whose structures have not yet been solved.
The sandwich biosensor assays described here were developed to monitor in vivo protein stability and aggregation propensity in the cytosolic folding environment. Using these methods, we demonstrate a striking correlation between in vivo antibiotic resistances (the selectable traits) and in vitro stability for two different proteins, immunity protein 7 from E. coli and human muscle phosphatase. We also show that polyQ tracts of increasing length are associated with an increased tendency to form amyloids in vivo and are associated with a decreased resistance to aminoglycoside antibiotics in our sandwich fusion system. We demonstrate that our approach allows the quantitative analysis of protein stability in the cytosolic compartment without the need for prior structural and functional knowledge. These various selection systems will likely be useful in identifying folding modulators that enhance the stability of very unstable proteins (15).
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the Howard Hughes Medical Institute, for which J.C.A.B. is an investigator.
We thank Linda Foit and Ursula Jakob for useful discussions.
Footnotes
Published ahead of print 29 September 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.02215-14.
REFERENCES
- 1.Hartl FU, Hayer-Hartl M. 2009. Converging concepts of protein folding in vitro and in vivo. Nat. Struct. Mol. Biol. 16:574–581. 10.1038/nsmb.1591. [DOI] [PubMed] [Google Scholar]
- 2.Ellis RJ, Hartl FU. 1999. Principles of protein folding in the cellular environment. Curr. Opin. Struct. Biol. 9:102–110. 10.1016/S0959-440X(99)80013-X. [DOI] [PubMed] [Google Scholar]
- 3.Selkoe DJ. 2002. Alzheimer's disease is a synaptic failure. Science 298:789–791. 10.1126/science.1074069. [DOI] [PubMed] [Google Scholar]
- 4.Narhi L, Wood SJ, Steavenson S, Jiang Y, Wu GM, Anafi D, Kaufman SA, Martin F, Sitney K, Denis P, Louis J-C, Wypych J, Biere AL, Citron M. 1999. Both familial Parkinson's disease mutations accelerate α-synuclein aggregation. J. Biol. Chem. 274:9843–9846. 10.1074/jbc.274.14.9843. [DOI] [PubMed] [Google Scholar]
- 5.Powell K, Zeitlin PL. 2002. Therapeutic approaches to repair defects in ΔF508 CFTR folding and cellular targeting. Adv. Drug Deliv. Rev. 54:1395–1408. 10.1016/S0169-409X(02)00148-5. [DOI] [PubMed] [Google Scholar]
- 6.Cheung MS, Klimov D, Thirumalai D. 2005. Molecular crowding enhances native state stability and refolding rates of globular proteins. Proc. Natl. Acad. Sci. U. S. A. 102:4753–4758. 10.1073/pnas.0409630102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stagg L, Christiansen A, Wittung-Stafshede P. 2011. Macromolecular crowding tunes folding landscape of parallel α/β protein, apoflavodoxin. J. Am. Chem. Soc. 133:646–648. 10.1021/ja107638e. [DOI] [PubMed] [Google Scholar]
- 8.Ebbinghaus S, Dhar A, McDonald JD, Gruebele M. 2010. Protein folding stability and dynamics imaged in a living cell. Nat. Methods 7:319–323. 10.1038/nmeth.1435. [DOI] [PubMed] [Google Scholar]
- 9.Ignatova Z, Krishnan B, Bombardier JP, Marcelino AMC, Hong J, Gierasch LM. 2007. From the test tube to the cell: exploring the folding and aggregation of a β-clam protein. Pept. Sci. 88:157–163. 10.1002/bip.20665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ghaemmaghami S, Oas TG. 2001. Quantitative protein stability measurement in vivo. Nat. Struct. Mol. Biol. 8:879–882. 10.1038/nsb1001-879. [DOI] [PubMed] [Google Scholar]
- 11.Foit L, Morgan GJ, Kern MJ, Steimer LR, von Hacht AA, Titchmarsh J, Warriner SL, Radford SE, Bardwell JCA. 2009. Optimizing protein stability in vivo. Mol. Cell 36:861–871. 10.1016/j.molcel.2009.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hailu TT, Foit L, Bardwell JCA. 2013. In vivo detection and quantification of chemicals that enhance protein stability. Anal. Biochem. 434:181–186. 10.1016/j.ab.2012.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chiti F, Taddei N, White PM, Bucciantini M, Magherini F, Stefani M, Dobson CM. 1999. Mutational analysis of acylphosphatase suggests the importance of topology and contact order in protein folding. Nat. Struct. Mol. Biol. 6:1005–1009. 10.1038/14890. [DOI] [PubMed] [Google Scholar]
- 14.Krobitsch S, Lindquist S. 2000. Aggregation of huntingtin in yeast varies with the length of the polyglutamine expansion and the expression of chaperone proteins. Proc. Natl. Acad. Sci. U. S. A. 97:1589–1594. 10.1073/pnas.97.4.1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Quan S, Koldewey P, Tapley T, Kirsch N, Ruane KM, Pfizenmaier J, Shi R, Hofmann S, Foit L, Ren G, Jakob U, Xu Z, Cygler M, Bardwell JCA. 2011. Genetic selection designed to stabilize proteins uncovers a chaperone called Spy. Nat. Struct. Mol. Biol. 18:262–269. 10.1038/nsmb.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Karimi R, Ehrenberg M. 1996. Dissociation rates of peptidyl-tRNA from the P-site of E. coli ribosomes. EMBO J. 15:1149. [PMC free article] [PubMed] [Google Scholar]
- 17.Nurizzo D, Shewry SC, Perlin MH, Brown SA, Dholakia JN, Fuchs RL, Deva T, Baker EN, Smith CA. 2003. The crystal structure of aminoglycoside-3′-phosphotransferase-IIa, an enzyme responsible for antibiotic resistance. J. Mol. Biol. 327:491–506. 10.1016/S0022-2836(03)00121-9. [DOI] [PubMed] [Google Scholar]
- 18.Clark NC, Olsvik Ø, Swenson JM, Spiegel CA, Tenover FC. 1999. Detection of a streptomycin/spectinomycin adenylyltransferase gene (aadA) in Enterococcus faecalis. Antimicrob. Agents Chemother. 43:157–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hollingshead S, Vapnek D. 1985. Nucleotide sequence analysis of a gene encoding a streptomycin/spectinomycin adenyltransferase. Plasmid 13:17–30. 10.1016/0147-619X(85)90052-6. [DOI] [PubMed] [Google Scholar]
- 20.Krügel H, Fiedler G, Smith C, Baumberg S. 1993. Sequence and transcriptional analysis of the nourseothricin acetyltransferase-encoding gene nat1 from Streptomyces noursei. Gene 127:127–131. 10.1016/0378-1119(93)90627-F. [DOI] [PubMed] [Google Scholar]
- 21.Alshahni MM, Makimura K, Yamada T, Takatori K, Sawada T. 2010. Nourseothricin acetyltransferase: a new dominant selectable marker for the dermatophyte Trichophyton mentagrophytes. Med. Mycol. 48:665–668. 10.3109/13693780903330555. [DOI] [PubMed] [Google Scholar]
- 22.Kochupurakkal BS, Iglehart JD. 2013. Nourseothricin N-acetyl transferase: a positive selection marker for mammalian cells. PLoS One 8:e68509. 10.1371/journal.pone.0068509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Van Driessche B, Tafforeau L, Hentges P, Carr AM, Vandenhaute J. 2005. Additional vectors for PCR-based gene tagging in Saccharomyces cerevisiae and Schizosaccharomyces pombe using nourseothricin resistance. Yeast 22:1061–1068. 10.1002/yea.1293. [DOI] [PubMed] [Google Scholar]
- 24.Van TT, Rooney PJ, Knoll LJ. 2006. Nourseothricin acetyltransferase: a positive selectable marker for Toxoplasma gondii. J. Parasitol. 92:668–670. 10.1645/GE-706R.1. [DOI] [PubMed] [Google Scholar]
- 25.Cutler TA, Mills BM, Lubin DJ, Chong LT, Loh SN. 2009. Effect of interdomain linker length on an antagonistic folding-unfolding equilibrium between two protein domains. J. Mol. Biol. 386:854–868. 10.1016/j.jmb.2008.10.090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Betton J-M, Jacob JP, Hofnung M, Broome-Smith JK. 1997. Creating a bifunctional protein by insertion of β-lactamase into the maltodextrin-binding protein. Nat. Biotechnol. 15:1276–1279. 10.1038/nbt1197-1276. [DOI] [PubMed] [Google Scholar]
- 27.Capaldi AP, Kleanthous C, Radford SE. 2002. Im7 folding mechanism: misfolding on a path to the native state. Nat. Struct. Biol. 9:209–216. 10.1038/nsb757. [DOI] [PubMed] [Google Scholar]
- 28.Reddy PH, Williams M, Tagle DA. 1999. Recent advances in understanding the pathogenesis of Huntington's disease. Trends Neurosci. 22:248–255. 10.1016/S0166-2236(99)01415-0. [DOI] [PubMed] [Google Scholar]
- 29.Van Nuland NA, Chiti F, Taddei N, Raugei G, Ramponi G, Dobson CM. 1998. Slow folding of muscle acylphosphatase in the absence of intermediates. J. Mol. Biol. 283:883–891. 10.1006/jmbi.1998.2009. [DOI] [PubMed] [Google Scholar]
- 30.Lang-Hinrichs C, Berndorff D, Seefeldt C, Stahl U. 1989. G418 resistance in the yeast Saccharomyces cerevisiae: comparison of the neomycin resistance genes from Tn5 and Tn903. Appl. Microbiol. Biotechnol. 30:388–394. 10.1007/BF00296629. [DOI] [Google Scholar]
- 31.Espargaró A, Castillo V, de Groot NS, Ventura S. 2008. The in vivo and in vitro aggregation properties of globular proteins correlate with their conformational stability: the SH3 case. J. Mol. Biol. 378:1116–1131. 10.1016/j.jmb.2008.03.020. [DOI] [PubMed] [Google Scholar]
- 32.Mayer S, Rüdiger S, Ang HC, Joerger AC, Fersht AR. 2007. Correlation of levels of folded recombinant p53 in Escherichia coli with thermodynamic stability in vitro. J. Mol. Biol. 372:268–276. 10.1016/j.jmb.2007.06.044. [DOI] [PubMed] [Google Scholar]
- 33.Collinet B, Hervé M, Pecorari F, Minard P, Eder O, Desmadril M. 2000. Functionally accepted insertions of proteins within protein domains. J. Biol. Chem. 275:17428–17433. 10.1074/jbc.M000666200. [DOI] [PubMed] [Google Scholar]
- 34.Ruth N, Quinting B, Mainil J, Hallet B, Frère J-M, Huygen K, Galleni M. 2008. Creating hybrid proteins by insertion of exogenous peptides into permissive sites of a class A β-lactamase. FEBS J. 275:5150–5160. 10.1111/j.1742-4658.2008.06646.x. [DOI] [PubMed] [Google Scholar]
- 35.Charbit A, Ronco J, Michel V, Werts C, Hofnung M. 1991. Permissive sites and topology of an outer membrane protein with a reporter epitope. J. Bacteriol. 173:262–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Manoil C, Bailey J. 1997. A simple screen for permissive sites in proteins: analysis of Escherichia coli lac permease. J. Mol. Biol. 267:250–263. 10.1006/jmbi.1996.0881. [DOI] [PubMed] [Google Scholar]
- 37.Parsell DA, Sauer RT. 1989. The structural stability of a protein is an important determinant of its proteolytic susceptibility in Escherichia coli. J. Biol. Chem. 264:7590–7595. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.