Abstract
Pseudomonas aeruginosa is a Gram-negative opportunistic human pathogen and a threat for immunocompromised and cystic fibrosis patients. It is responsible for acute and chronic infections and can switch between these lifestyles upon taking an informed decision involving complex regulatory networks. The RetS/LadS/Gac/Rsm network and the cyclic-di-GMP (c-di-GMP) signaling pathways are both central to this phenomenon redirecting the P. aeruginosa population toward a biofilm mode of growth, which is associated with chronic infections. While these two pathways were traditionally studied independently from each other, we recently showed that cellular levels of c-di-GMP are increased in the hyperbiofilm retS mutant. Here, we have formally established the link between the two networks by showing that the SadC diguanylate cyclase is central to the Gac/Rsm-associated phenotypes, notably, biofilm formation. Importantly, SadC is involved in the signaling that converges onto the RsmA translational repressor either via RetS/LadS or via HptB/HsbR. Although the level of expression of the sadC gene does not seem to be impacted by the regulatory cascade, the production of the SadC protein is tightly repressed by RsmA. This adds to the growing complexity of the signaling network associated with c-di-GMP in P. aeruginosa. While this organism possesses more than 40 c-di-GMP-related enzymes, it remains unclear how signaling specificity is maintained within the c-di-GMP network. The finding that SadC but no other diguanylate cyclase is related to the formation of biofilm governed by the Gac/Rsm pathway further contributes to understanding of this insulation mechanism.
INTRODUCTION
Bacteria adopt different lifestyles in response to the fluctuating conditions that they encounter in the environment. They can form a biofilm, which is a sessile community of bacteria, and they can switch between a motile and a sessile lifestyle (1, 2). In a biofilm, bacteria are engulfed in an extracellular matrix composed of exopolysaccharides, extracellular DNA, and proteins (3–6). This helps protect bacteria from various stresses, harsh antimicrobial treatments, or eradication by the immune system (7). In the case of the opportunistic Gram-negative pathogen Pseudomonas aeruginosa, several exopolysaccharides have been described. Whereas mucoid strains isolated from cystic fibrosis patients overproduce alginate (8), nonmucoid strains, such as PAO1, PA14, or PAK, can produce the Pel exopolysaccharide, a glucose-rich polymer (9–11), and/or the Psl exopolysaccharide, a polymer of a repeating pentamer containing d-mannose, l-rhamnose, and d-glucose (12).
The switch in lifestyle and the development of biofilms are based on informed decisions relayed via complex regulatory networks. This results in the control of various cellular processes at the transcriptional, posttranscriptional, or posttranslational level. In the last decade, key networks, such as the Gac/Rsm pathway or the second messenger, cyclic-di-GMP (c-di-GMP) signaling pathway, have been proven to be central in modulating the transition to biofilms (13, 14). In P. aeruginosa, the Gac/Rsm pathway includes the RetS and LadS sensors that antagonistically impact the activity of the GacS histidine kinase (15–18). RetS can form heterodimers with GacS, thus preventing phosphorylation of the cognate response regulator GacA (19), which otherwise promotes the transcription of two small RNAs, RsmY and RsmZ (20). More recently, it was shown that RetS activity could be counteracted by the PA1611 sensor histidine kinase (21). Ultimately, the small RNAs sequester RsmA, a repressor that inhibits the translation of target mRNAs by binding to single-stranded GGA motifs formed near the ribosome binding site (22–24). Among the more than 500 targets of RsmA are the transcripts of the pel and psl genes (22, 25). Hence, a retS or an rsmA mutant has a hyperbiofilm phenotype due to the stimulation of the Gac pathway and the relieved RsmA repression on exopolysaccharide production.
In parallel to the Gac/Rsm pathway, signaling networks mediated by c-di-GMP are also related to the transition to a biofilm (14, 26, 27). This cyclic dinucleotide is ubiquitous among bacteria, with low intracellular levels of c-di-GMP promoting a motile lifestyle and high levels promoting biofilm formation (28). Proteins involved in the synthesis of c-di-GMP harbor a GGDEF domain and are known as diguanylate cyclases (DGCs) (29). Proteins with an EAL or HD-GYP domain are called phosphodiesterases (PDEs) and are involved in c-di-GMP hydrolysis (29).
Recently, it was shown that a retS mutant displays elevated levels of c-di-GMP, suggesting a link between the two networks (30). In the present work, we have attempted to pinpoint the molecular basis of this link. We report that the membrane-associated DGC known as SadC (31) is central to the Gac/Rsm pathway and that additional elements, such as SadB, also play a role in the signaling cascade.
MATERIALS AND METHODS
Strains, plasmids, and growth conditions.
The bacterial strains, plasmids, and primers used in this study are listed in Table S1 in the supplemental material. Bacteria were cultured at 37°C with shaking in LB medium or M9 minimal medium containing 22 mM glucose, 2 mM MgSO4, and 0.1 mM CaCl2. Culture media were supplemented with antibiotics at the following concentrations, when appropriate: for Escherichia coli, 50 μg/ml ampicillin (Ap) and 50 μg/ml kanamycin (Km), and for P. aeruginosa, 500 μg/ml carbenicillin (Cb) for selection or 300 μg/ml Cb for maintenance, 2,000 μg/ml streptomycin (Sm) for selection, 150 μg/ml gentamicin (Gm) for selection and 100 μg/ml Gm for maintenance, and 50 μg/ml tetracycline (Tet) for maintenance. A Congo red staining assay was performed at 30°C on tryptone (10 g/liter) agar (1%) plates supplemented with 40 μg/ml Congo red and 20 μg/ml Coomassie brilliant blue. Plates for swimming motility assays were prepared using 0.3% LB agar and incubated at 30°C as described previously (32). Escherichia coli OmniMAX and TOP10 were used for standard genetic manipulations. PCR products were cloned into pCR2.1-TA and subcloned into pBBR1MCS-4 or pKNG101. Transfer of plasmids into P. aeruginosa strains was carried out by triparental mating using the conjugative properties of plasmid pRK2013. Transconjugants were isolated on Pseudomonas isolation agar (Difco) supplemented with the appropriate antibiotics. Deletion mutants were selected in 5% sucrose after 3 days of incubation at room temperature. Mutator fragments were constructed by PCR amplification of upstream and downstream fragments of approximately 500 bp flanking the chromosomal region to be mutated. Deletions were confirmed by sequencing using external primers.
Biofilm assays.
Visualization of biofilm formation was carried out in 14-ml borosilicate tubes. Briefly, LB (3 ml) or M9 medium supplemented with appropriate antibiotics was inoculated to a final optical density at 600 nm (OD600) of 0.1 and incubated at 37°C. Biofilms were stained with 0.1% crystal violet (CV), and tubes were washed with water to remove unbound dye. Quantification of biofilm formation was performed in 24-well polystyrene microtiter plates. LB (1 ml/well) and antibiotics, when appropriate, were inoculated to a final OD600 of 0.01. The plates were incubated for 6 h or 14 h at 30°C. Biofilms were stained with 100 μl of CV and washed twice with water before being solubilized in 96% ethanol. CV staining was measured by reading the optical density at 600 nm.
RNA extraction and qRT-PCR.
Overnight PAK and PAK ΔretS cultures were subcultured in LB medium with a starting OD600 of 0.1 and incubated at 37°C with shaking for 6 h. Cells were then harvested into RNAlater stabilization solution (Ambion), and RNA was extracted using an RNeasy extraction kit (Qiagen). To remove DNA, a Turbo DNA-free kit (Applied Biosystems) was used, and the RNA was repurified using an RNeasy kit, following the supplier's indications. cDNA was synthesized from 200 ng of RNA template by adding 20 U of Protector RNase inhibitor from Roche, 10 pmol of Pd(N)6 random hexamer oligonucleotides from Amersham, and 10 pmol deoxynucleoside triphosphates from Bioline to the reaction mix. Quantitative real-time reverse transcription-PCR (qRT-PCR) was performed on an ABI 7300 real-time PCR system using ABI SYBR green PCR master mix.
Quantification of c-di-GMP.
The reporter PcdrA-gfp plasmid, obtained from Matthew Parsek (33), was introduced into the P. aeruginosa strains of interest by electroporation, and overnight cultures were subcultured in LB medium supplemented with the appropriate antibiotic to a starting OD600 of 0.1. After shaking incubation at 37°C for 6 h, 1 ml of culture was harvested and cells were resuspended in 1× phosphate-buffered saline (PBS) before the optical density at 600 nm and fluorescence (excitation, 485 nm; emission, 520 nm) were measured in a black 96-well plate with a see-through bottom (Falcon) using a FLUOstar Optima plate reader (BMG Labtech). Quantifications were performed in triplicate, and data are presented as relative fluorescent units (RFU), which are arbitrary fluorescent units corrected for cell density.
Construction of chromosomal Flag fusions.
The PAO1 strains encoding DGC proteins with a C-terminal Flag tag were engineered using a strategy previously described (34). Briefly, the Flag fusion constructs were produced by splicing by overhang extension PCR using the primers listed in Table S1 in the supplemental material and contained 500- to 700-bp homologous flanking regions, with the Flag tag positioned in the middle. This construct was ligated into pME3087 between BamHI and HindIII/EcoRI restrictions sites. The resulting vector was then used to introduce the Flag tag fusion by a two-step allelic exchange. Following biparental conjugation (donor S17-1) into the target strain, single crossovers were selected on Tet and restreaked. Cultures from single crossovers were grown overnight in LB medium and diluted 1:100 into fresh medium. After 2 h, 20 μg/ml Tet was added to inhibit the growth of cells that had lost the Tet resistance cassette. After a further hour of growth, 2,000 μg/ml Cb was added to select against growing bacteria. Cultures were grown for a further 4 to 6 h, before cells were harvested by centrifugation, washed once in LB, and used to inoculate an overnight culture. This counterselection was done twice, before plating a dilution series of the final samples onto LB agar. Individual colonies were patched onto LB plates with or without Tet, and Tet-sensitive colonies were tested for Flag insertion by colony PCR.
Western blots.
Strains with chromosomal Flag-tagged DGCs containing a vector with either rsmY or rsmA under the control of an inducible promoter were grown overnight, diluted 1:100 in the morning, and grown to an OD600 of 1.5. This culture was used to inoculate 20 ml prewarmed LB supplemented with 0.05% Triton X-100. At an OD600 of 0.4, 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) was added for induction of rsmY or rsmA. After induction, samples were taken every hour. Samples were separated on 12% Tris-HCl gels and blotted onto 0.45-μm-pore-size polyvinylidene difluoride (PVDF) membranes (Millipore). The membranes were incubated overnight in blocking solution (1× PBS, pH 7.4, 0.01% Tween 20, 5% milk powder), after which proteins were detected with 1:7,000-diluted and 1:10,000-diluted rabbit anti-mouse M2-specific antiserum (DakoCytomation). Bound antibodies were visualized using ECL chemiluminescent detection reagent (PerkinElmer).
RESULTS
The RsmA-dependent diguanylate cyclase PA0338 is not involved in the retS mutant hyperbiofilm phenotype.
It has previously been shown that in a P. aeruginosa PAK retS mutant, the levels of c-di-GMP are increased (30). We hypothesized that this could be a consequence of either (i) an upregulation or activation of proteins with DGC activity or (ii) a downregulation or inactivation of proteins with PDE activity. Since a retS mutation results in the relief of RsmA repression, the RsmA regulon (22) was screened for putative upregulated DGCs or downregulated PDEs. Analysis of the data published by Brencic and Lory in 2009 (22) revealed that only one putative DGC, PA0338, was upregulated (2.8-fold) in the rsmA mutant compared to its level of regulation in the PAK wild-type strain, and no putative PDE was downregulated. Using qRT-PCR, we were able to show that PA0338 mRNA levels are 3.8-fold higher in a retS background than in the parental PAK strain (Fig. 1A). RetS is also known to inversely regulate the type III secretion system (T3SS) and type VI secretion system (T6SS) via RsmA (30), and our qRT-PCR confirmed this observation (Fig. 1A). In light of the PA0338 upregulation, we tested whether a mutation in PA0338 resulted in the suppression of the retS hyperbiofilm phenotype. A retS PA0338 mutant was engineered and spotted on Congo red agar plates. Both the retS and retS PA0338 mutants displayed a wrinkly red phenotype that contrasts with the smooth white appearance of the PAK wild-type strain, suggesting that the double mutant is still a hyperbiofilm former (Fig. 1B). The level of c-di-GMP was also monitored in these strains using the PcdrA-gfp reporter fusion (33). As shown in Fig. 1C, the retS and retS PA0338 mutants displayed elevated levels of c-di-GMP. Therefore, these data suggest that PA0338 is not involved in the synthesis of c-di-GMP that leads to the hyperbiofilm phenotype of the retS mutant.
FIG 1.
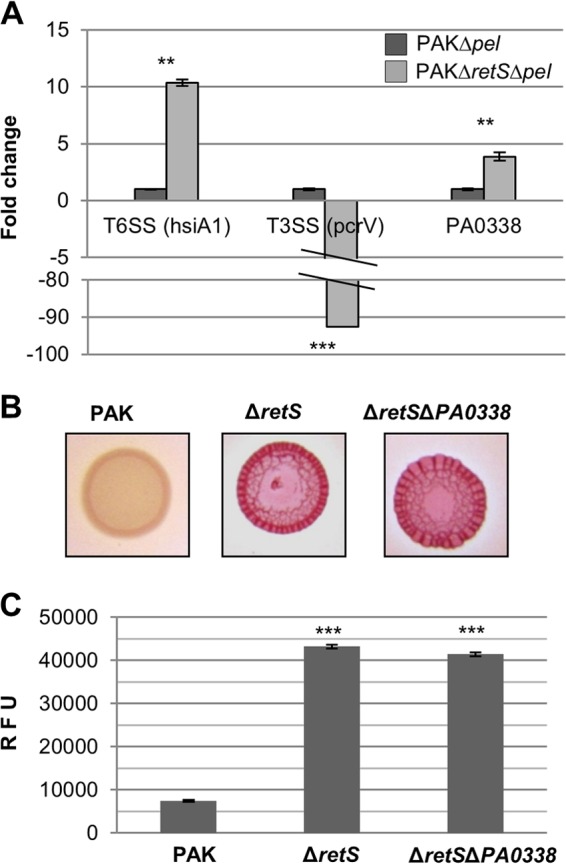
PA0338 is upregulated in a retS mutant background. (A) Transcript levels of hsiA1 (positive control), pcrV (negative control), and PA0338 measured by qRT-PCR and normalized to those of gyrA. Statistical Student's t-test analysis was based on three replicates, and significant changes are indicated (**, P < 0.001; ***, P < 0.0001). (B) Congo red binding of the indicated strains after 2 days of incubation. (C) Intracellular levels of c-di-GMP measured with the transcriptional PcdrA-gfp reporter. Relative fluorescence units (RFU) are arbitrary fluorescence intensity units corrected for cell density. At least three independent experiments were performed. Statistically significant changes were calculated using the Student t test and are indicated (***, P < 0.0001).
SadC is responsible for the retS mutant hyperbiofilm phenotype.
In the initial characterization of the retS mutant (15), a transposon mutagenesis screen for suppressors of the retS hyperbiofilm phenotype was performed, and insertions were mapped to the gacS, gacA, rsmZ, and PA4332 (sadC) genes. SadC is a membrane protein and has a cytoplasmic C-terminal GGDEF domain with known DGC activity (31, 35). Herein, we engineered a retS sadC mutant and examined the biofilm phenotype using crystal violet staining. As a control, a sadC single mutant was also included. Interestingly, deletion of sadC in the retS background readily resulted in the loss of the retS mutant hyperbiofilm phenotype (Fig. 2A). This loss of phenotype was more pronounced than the loss caused by the deletion of sadC alone (Fig. 2A) and could be complemented by introducing the sadC gene in trans (Fig. 2B). Moreover, the reduced ability of the retS sadC mutant to form biofilms was accompanied by an increase in the ability of the strain to swim in soft agar plates (Fig. 2C). Finally, the retS sadC mutant displayed levels of c-di-GMP similar to those displayed by the wild-type strain (Fig. 2D), suggesting that the SadC diguanylate cyclase is active in the retS mutant and is responsible for the hyperbiofilm phenotype.
FIG 2.
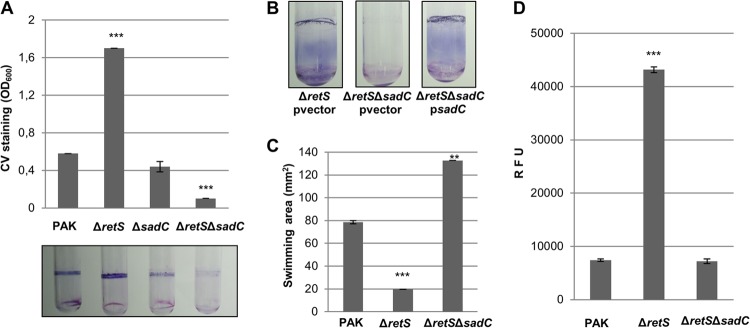
SadC is required for the retS mutant hyperbiofilm phenotype. (A) Quantification of the crystal violet staining of biofilms grown in microtiter plates for 14 h. A photo of the test tubes was taken prior to the addition of ethanol for quantification purposes. (B) Crystal violet staining of biofilms grown in test tubes for 6 h. The empty vector pBBR1MCS-4 (pvector) and pBBR1MCS-4-sadC (psadC) were conjugated into PAK ΔretS or PAK ΔretS ΔsadC. (C) Results of a swimming motility assay performed in 0.3% LB agar plates. (D) Intracellular levels of c-di-GMP measured with the transcriptional PcdrA-gfp reporter. Relative fluorescence units (RFU) are arbitrary fluorescence intensity units corrected for cell density. At least three independent experiments were performed. (A, C, and D) Statistically significant changes were calculated using the Student t test and are indicated (**, P < 0.001; ***, P < 0.0001).
SadC is responsible for the hptB mutant hyperbiofilm phenotype.
We next investigated the role of SadC in a known regulatory pathway that converges onto the Gac/Rsm pathway and involves the histidine phosphotransfer module HptB (36–38). It was shown previously that an hptB mutant displays a hyperbiofilm phenotype and this phenotype is milder than the one observed for the retS mutant (36). Analysis of the c-di-GMP levels and Congo red binding showed that the intermediate level of biofilm formation of the hptB mutant corresponds to an intermediate level of intracellular c-di-GMP, and both phenotypes were abrogated in an hptB sadC mutant (Fig. 3A and B). To further establish the pivotal role of SadC in these RsmA-dependent pathways, an inverse approach was used whereby the RetS and HptB antagonists, namely, LadS (17) and HsbR (36, 39), respectively, were overexpressed in a sadC mutant. For this purpose, plasmids encoding either the sensor LadS or the response regulator HsbR were introduced by conjugation into the relevant strains. While overexpression of ladS and hsbR in the wild-type strain induced Congo red binding, a similar phenotype could not be obtained in the sadC mutant (Fig. 3C). Overall, these data suggest that the increase of c-di-GMP levels and the induction of biofilm-related phenotypes observed upon activation of the Gac/Rsm cascade rely entirely on the diguanylate cyclase SadC.
FIG 3.
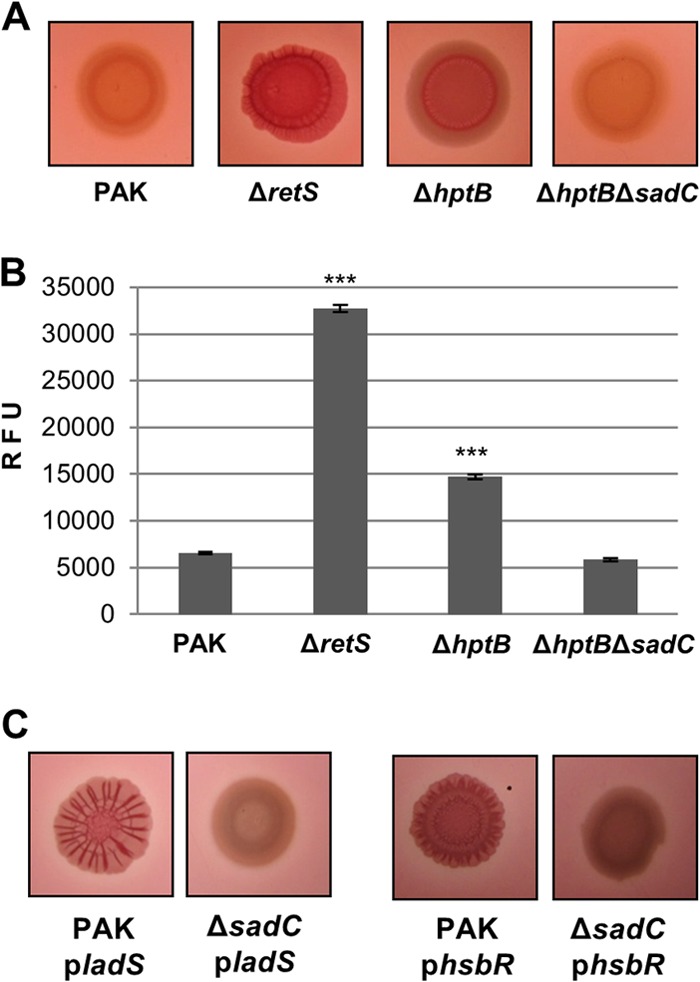
SadC is required for the hptB mutant hyperbiofilm phenotype. (A) Congo red binding of the indicated strains after 2 days of incubation. (B) Intracellular levels of c-di-GMP measured with the transcriptional PcdrA-gfp reporter. Relative fluorescence units (RFU) are arbitrary fluorescence intensity units corrected for cell density. At least three independent experiments were performed. Statistically significant changes are indicated (***, P < 0.0001, Student's t test). (C) PAK and PAK ΔsadC were conjugated with pBBR1-MCS4-ladS (pladS) or pBBR1-MCS4-hsbR (phsbR). Congo red binding is from day 2 of incubation.
SadC production is controlled by RsmA.
Both the RetS- and HptB-dependent pathways ultimately relay onto the translational repressor RsmA, and therefore, the sadC deletion was also engineered in an rsmA mutant background. Note that the rsmA mutant displayed a wrinkly red phenotype and this corresponded to an increased level of c-di-GMP (Fig. 4A and B). As was observed for the retS sadC and hptB sadC double mutants, hyperbiofilm formation and the increase in c-di-GMP levels were also lost in the rsmA sadC mutant (Fig. 4A and B). In addition, a crystal violet assay showed that the overexpression of sadC resulted in hyperbiofilm formation in the wild-type or sadC mutant strains but failed to do so in the rsmYZ mutant (Fig. 5A). This suggests that sadC overexpression cannot relieve the repression by RsmA and is in favor of a link between these two signaling pathways, yet it is a possibility that the lack of biofilm is due to the absence of rsmYZ, a reduced level of c-di-GMP, or both. In order to address this further, we then used an alternative strategy. As discussed above, previous studies identified PA0338 as a direct target for RsmA (22), but no RsmA-dependent control on SadC production was reported. Here, we used an original approach to look at the production of SadC in a genetic background where either rsmA or one of its regulatory antagonists, the small RNA rsmY, is overexpressed. To do this, the chromosomally encoded sadC was genetically modified to encode a FLAG-tagged version of the protein, and the levels of SadC were monitored by Western blotting using antibodies against the tag. Strikingly, SadC was readily detected when the RsmA repression was released by RsmY overproduction, whereas SadC production was drastically reduced upon RsmA induction (Fig. 5B). The same was observed for PA0338 but not for two additional DGCs that were taken as negative controls, namely, PA0847 and PA5487 (Fig. 5B).
FIG 4.
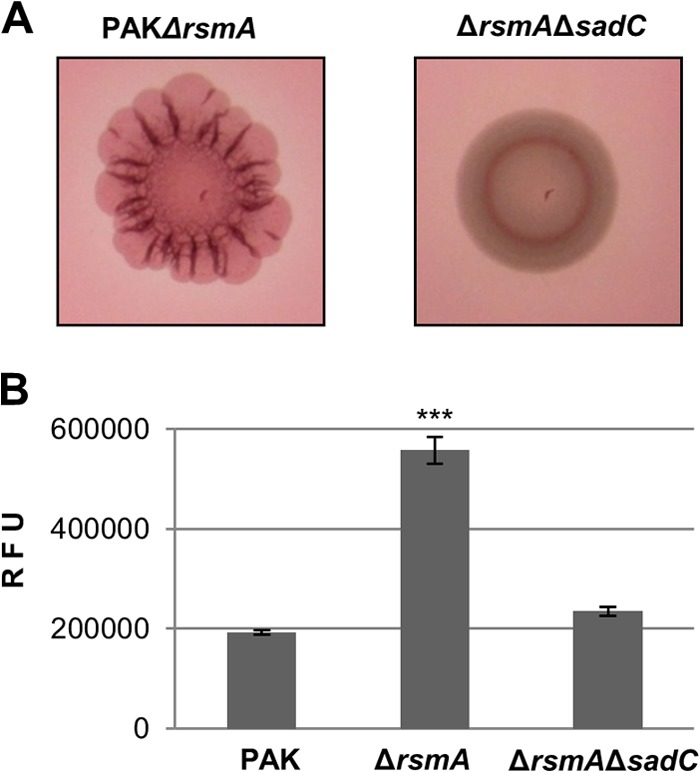
Deletion of sadC abrogates the hyperbiofilm and high c-di-GMP phenotypes of an rsmA mutant. (A) Congo red binding phenotypes visualized on day 2 of incubation. (B) Intracellular levels of c-di-GMP measured with the transcriptional PcdrA-gfp reporter. Relative fluorescence units (RFU) are arbitrary fluorescence intensity units corrected for cell density. Statistically significant changes are indicated (***, P < 0.0001, Student's t test).
FIG 5.
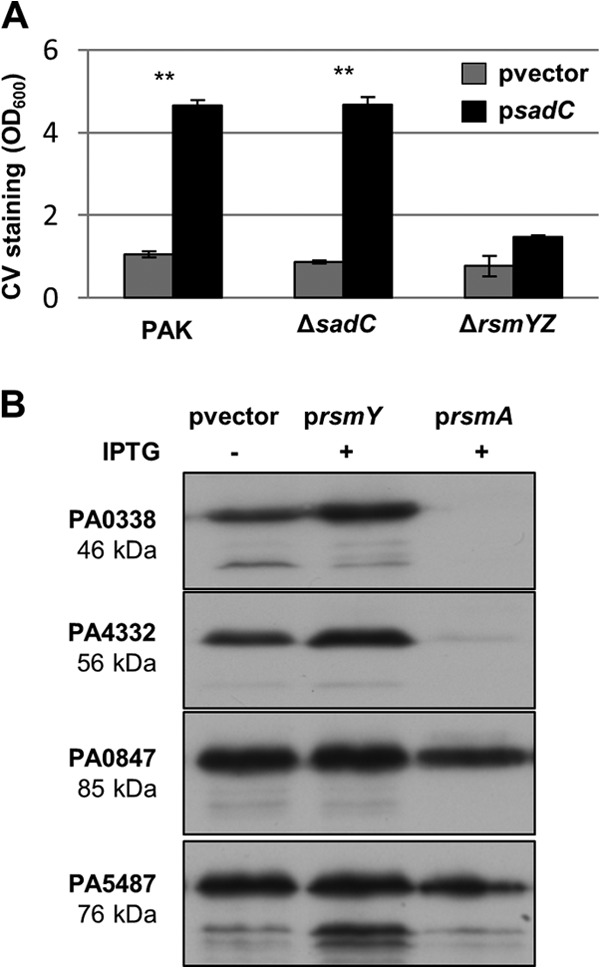
SadC production is controlled by RsmA. (A) Quantification of the crystal violet staining of biofilms grown in microtiter plates for 6 h. Statistically significant changes are indicated (**, P < 0.001, Student's t test). (B) Immunoblots with M2 antiserum showing the levels of DGC-Flag variants in whole-cell lysates. The PAO1 DGC-Flag strains indicated on the left contain different plasmids: pvector (empty vector), prsmY (IPTG-inducible rsmY), and prsmA (IPTG-inducible rsmA). Cells were harvested after 1 h induction with 1 mM IPTG. Shown here is a representative immunoblot from at least three independent experiments.
SadB is also central to the Gac/Rsm pathway.
Previous work (31) has placed SadB and SadC in the same genetic pathway regulating biofilm formation. Although SadB is a protein of unknown function (40), it was shown that it is required for SadC signaling. We thus tested whether a sadB mutation was able to suppress the phenotype associated with retS or hptB mutations, as was observed for a sadC mutation. In both cases, the hyperbiofilm phenotype was significantly reduced upon introduction of the sadB deletion (Fig. 6). Altogether, our results indicate that the Gac/Rsm pathway is tightly linked to c-di-GMP signaling via the SadC/SadB pathway.
FIG 6.
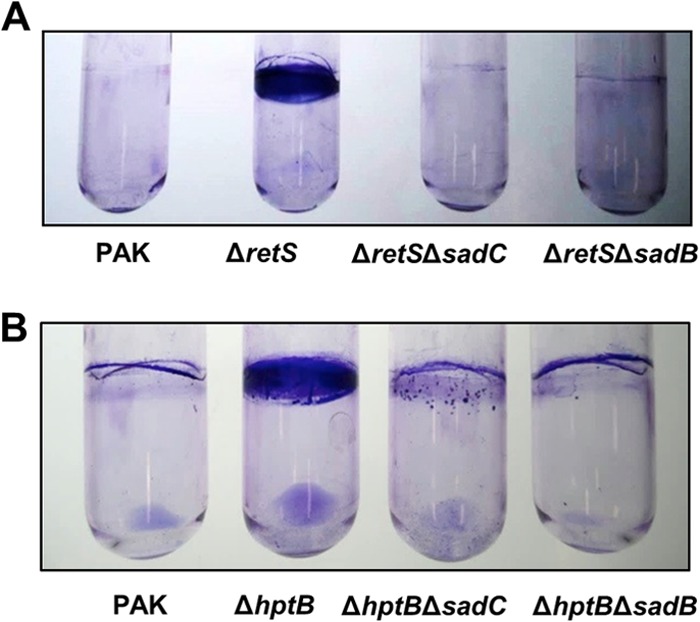
SadB is central to the RetS- and HptB-dependent signaling pathways. Crystal violet staining of biofilms grown under shaking conditions for 8 h (A) and 18 h (B) is shown.
DISCUSSION
In P. aeruginosa there are more than 40 putative proteins involved in the metabolism of c-di-GMP, and this is thought to form an intricate intracellular network that specifically regulates biological processes crucial for bacterial adaptation and virulence (41). However, many of the studies about this second messenger have used genetic approaches whereby a particular DGC or PDE is overexpressed, and this seems to have a general impact in dictating the lifestyle fate of a bacterial population, failing to establish a more specific function. Here, we sought to identify the protein or proteins specifically responsible for the elevated levels of c-di-GMP in a retS mutant, thus physically connecting the two networks (Fig. 7). RetS is a sensor protein that represses Gac/Rsm signaling by forming heterodimers with the GacS sensor (15). Although the complexity of the Gac/Rsm system varies between different species, the GacA/GacS two-component system, the cognate small RNA targets and the translational repressor, are conserved in Gammaproteobacteria (24). In some species, a specific link between the Gac/Rsm pathway and c-di-GMP signaling has been reported in the last few years. In E. coli, the GacA/GacS two-component system is known as BarA/UvrY and controls the expression of the small RNAs CsrB and CsrC. These, in turn, modulate the activity of the CsrA translational repressor, which targets, among many other genes, two genes encoding proteins with a GGDEF domain, YdeH and YcdT (42). In Salmonella enterica, the GacA/GacS two-component system is known as BarA/SirA and modulates the CsrA translational repressor via CsrB and CsrC. In this case, CsrA is known to regulate eight genes encoding GGDEF, GGDEF/EAL, or EAL domain proteins, and five of these are regulated by direct binding of CsrA to the mRNA (43). In Xanthomonas campestris, RsmA has been shown to control posttranscriptionally at least three GGDEF domain proteins, and the three contribute additively to the elevated levels of c-di-GMP in the rsmA mutant (44). In P. aeruginosa, direct evidence that the Gac system and c-di-GMP signaling were interlinked came from the observation that the retS mutant displays high levels of c-di-GMP and that the c-di-GMP-induced T3SS/T6SS switch is dependent on the two sRNAs RsmY and RsmZ (30).
FIG 7.
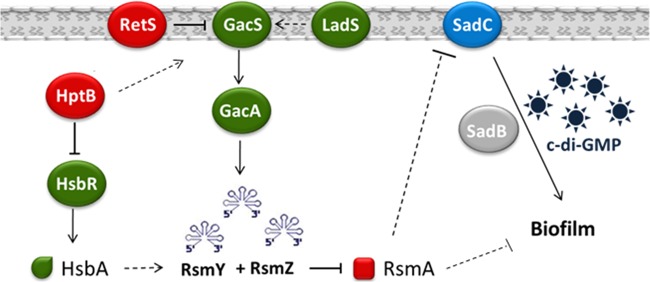
The SadC and the Gac/Rsm pathways are interlinked. The Gac/Rsm system is a complex signaling cascade that regulates several biological functions, including biofilm formation. The proteins belonging to this cascade that negatively impact the formation of biofilms are indicated in red. All the proteins of this cascade indicated in green have a positive impact on biofilm formation. The hyperbiofilm phenotype induced by the deletion of the retS, hptB, or rsmA gene is dependent on an intact sadC gene product to produce c-di-GMP (represented by stars). SadC levels are under the control of RsmA, but the exact mechanism by which it occurs is still unknown. Downstream of SadC is SadB, whose function remains obscure.
On the basis of the published literature, two proteins with a GGDEF domain were considered of interest in this study. On the one hand, PA0338 was reported to be upregulated in an rsmA mutant (22), and on the other hand, a PA4332 (sadC) transposon mutant had been identified as a suppressor of the retS mutant hyperbiofilm phenotype (15). By engineering mutants with deletions of these genes, we were able to conclude that the activity of SadC is directly responsible for the hyperbiofilm phenotype and the elevated levels of c-di-GMP observed in a retS mutant. To further understand the importance of SadC in the Gac/Rsm pathway, deletion of sadC was also introduced into hptB and rsmA mutant backgrounds. The HptB pathway is known to converge onto RsmA, possibly via the transcriptional repression of RsmY (36). In addition, HptB can be phosphorylated by the sensor PA1611 (37), which is able to interact with RetS to counteract its repressing role upon GacS (21). Once it is phosphorylated, HptB phosphorylates HsbR, whose output domain has both a phosphatase activity and a kinase activity (36, 45). HsbR controls the phosphorylation state of HsbA, an anti-anti-sigma factor that is likely to control the availability of a sigma factor required for rsmY expression. For the flagellum biogenesis, it has recently been shown that this anti-anti-sigma factor regulates the anti-sigma factor FlgM, releasing σ28 (45). In both the htpB sadC and rsmA sadC mutants, we observed the abrogation of the hyperbiofilm phenotype and a restoration of the c-di-GMP levels.
Even though PA0338 does not seem to account for the production of c-di-GMP in the retS mutant, both qRT-PCR and Western blot analysis indicate that the expression and production of PA0338 are under the control of RsmA. This may suggest that the catalytic activity of PA0338 is too low to be a major contributor to biofilm formation or that it might be connected to other RsmA-dependent phenotypes, yet it was previously shown that overexpression of PA0338 is able to increase the biofilm levels in PA14 (46), a genuine ladS mutant (47). In the case of SadC, RsmA control was detected only at the level of protein production, while no regulation of transcript abundance was observed by qRT-PCR (data not shown).
SadC was previously characterized for its role in swarming motility and put in the same genetic pathway as BifA (48), a PDE, and SadB, a protein of unknown function (40). In addition, RoeA, a DGC, has been shown to have an additive effect with SadC in P. aeruginosa strain PA14 (35, 49). Since SadB has been demonstrated to act downstream of SadC, the impact of a sadB deletion in the retS and hptB mutants was also investigated, and the results confirmed the position of SadB downstream of SadC in the signaling cascade, although the function of SadB is not clear. It has previously been shown that both the N-terminal YbaK domain of SadB and the C-terminal HD domain of SadB are required for function (40, 50). Hypothetically, it was considered that SadB could act upstream of c-di-GMP synthesis, somehow assisting SadC to achieve its DGC activity, or that SadB could function as a c-di-GMP receptor, somehow transmitting the c-di-GMP signal to downstream targets. In addition to this, SadB is known to act upstream of the Pil-Chp chemotactic cluster (50), and this cluster is required for type IV pilus (T4P) biogenesis (51). Interestingly, in Pseudomonas fluorescens, a link between SadB and the Gac system in which it exerts a negative regulation on flagellum-driven motility during exponential phase has recently been established (52). In this bacterium, the Gac system comprises three translational repressors, RsmA, RsmE, and RsmI (53), and the two signaling cascades were shown to intersect in the cooperative regulation of the σ22 sigma factor, also known as AlgT or AlgU. On the one hand, SadB is required for the transcription of σ22, and on the other hand, RsmA and RsmE act as translational repressors of σ22. Once it is produced, σ22 is necessary for the expression of the transcriptional regulator amrZ (also referred to as algZ) (54, 55), which functions to downregulate fleQ, a c-di-GMP binding protein (56) and the master regulator of the flagellar components. In P. aeruginosa, it is not known if σ22 is a member of the RsmA regulon, but AmrZ belongs to the σ22 regulon and has been shown to be involved in both the downregulation of flagella and Psl exopolysaccharide and the upregulation of twitching motility and alginate production (57–60).
In conclusion, the regulatory pathways involved in the control of P. aeruginosa lifestyles are increasingly complex. The connection between Gac/Rsm signaling and c-di-GMP is unlikely to be restricted to biofilm control or the T3SS/T6SS switch, and recent studies have also highlighted that iron uptake is coordinately regulated by these two networks (61). It is thus important to study in further depth the direct connections existing between each component and preferably avoid an overexpression context. Our future work will aim at establishing what the downstream targets of c-di-GMP signaling via SadC are. More particularly, we will investigate what the function of SadB is, although it is likely that other important players in the cascade are still to be identified.
Supplementary Material
ACKNOWLEDGMENTS
We thank Qi Pan for helping with the characterization of the different mutants and Matt Parsek for providing the PcdrA-gfp reporter.
Joana A. Moscoso was supported by a grant from the Fundação para a Ciência e a Tecnologia (FCT), and Martina Valentini is supported by a Swiss National Science Foundation early postdoc mobility grant. Work in Alain Filloux's laboratory is supported by BBSRC grants BB/F019645/1 and BB/I019871/1. Work in the laboratory of Urs Jenal was supported by a grant from the Gebert Rüf Foundation on Rare Diseases (GRS-035/12).
Footnotes
Published ahead of print 15 September 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.01850-14.
REFERENCES
- 1.Coggan KA, Wolfgang MC. 2012. Global regulatory pathways and cross-talk control Pseudomonas aeruginosa environmental lifestyle and virulence phenotype. Curr. Issues Mol. Biol. 14:47–70. [PMC free article] [PubMed] [Google Scholar]
- 2.Jimenez PN, Koch G, Thompson JA, Xavier KB, Cool RH, Quax WJ. 2012. The multiple signaling systems regulating virulence in Pseudomonas aeruginosa. Microbiol. Mol. Biol. Rev. 76:46–65. 10.1128/MMBR.05007-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hall-Stoodley L, Costerton JW, Stoodley P. 2004. Bacterial biofilms: from the natural environment to infectious diseases. Nat. Rev. Microbiol. 2:95–108. 10.1038/nrmicro821. [DOI] [PubMed] [Google Scholar]
- 4.Mann EE, Wozniak DJ. 2012. Pseudomonas biofilm matrix composition and niche biology. FEMS Microbiol. Rev. 36:893–916. 10.1111/j.1574-6976.2011.00322.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Okshevsky M, Meyer RL. 4 December 2013. The role of extracellular DNA in the establishment, maintenance and perpetuation of bacterial biofilms. Crit. Rev. Microbiol. 10.3109/1040841X.2013.841639. [DOI] [PubMed] [Google Scholar]
- 6.Wei Q, Ma LZ. 2013. Biofilm matrix and its regulation in Pseudomonas aeruginosa. Int. J. Mol. Sci. 14:20983–21005. 10.3390/ijms141020983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hoiby N, Ciofu O, Johansen HK, Song ZJ, Moser C, Jensen PO, Molin S, Givskov M, Tolker-Nielsen T, Bjarnsholt T. 2011. The clinical impact of bacterial biofilms. Int. J. Oral Sci. 3:55–65. 10.4248/IJOS11026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hoiby N, Ciofu O, Bjarnsholt T. 2010. Pseudomonas aeruginosa biofilms in cystic fibrosis. Future Microbiol. 5:1663–1674. 10.2217/fmb.10.125. [DOI] [PubMed] [Google Scholar]
- 9.Coulon C, Vinogradov E, Filloux A, Sadovskaya I. 2010. Chemical analysis of cellular and extracellular carbohydrates of a biofilm-forming strain Pseudomonas aeruginosa PA14. PLoS One 5:e14220. 10.1371/journal.pone.0014220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Friedman L, Kolter R. 2004. Two genetic loci produce distinct carbohydrate-rich structural components of the Pseudomonas aeruginosa biofilm matrix. J. Bacteriol. 186:4457–4465. 10.1128/JB.186.14.4457-4465.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vasseur P, Vallet-Gely I, Soscia C, Genin S, Filloux A. 2005. The pel genes of the Pseudomonas aeruginosa PAK strain are involved at early and late stages of biofilm formation. Microbiology 151:985–997. 10.1099/mic.0.27410-0. [DOI] [PubMed] [Google Scholar]
- 12.Byrd MS, Sadovskaya I, Vinogradov E, Lu H, Sprinkle AB, Richardson SH, Ma L, Ralston B, Parsek MR, Anderson EM, Lam JS, Wozniak DJ. 2009. Genetic and biochemical analyses of the Pseudomonas aeruginosa Psl exopolysaccharide reveal overlapping roles for polysaccharide synthesis enzymes in Psl and LPS production. Mol. Microbiol. 73:622–638. 10.1111/j.1365-2958.2009.06795.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mikkelsen H, Sivaneson M, Filloux A. 2011. Key two-component regulatory systems that control biofilm formation in Pseudomonas aeruginosa. Environ. Microbiol. 13:1666–1681. 10.1111/j.1462-2920.2011.02495.x. [DOI] [PubMed] [Google Scholar]
- 14.Romling U, Galperin MY, Gomelsky M. 2013. Cyclic di-GMP: the first 25 years of a universal bacterial second messenger. Microbiol. Mol. Biol. Rev. 77:1–52. 10.1128/MMBR.00043-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goodman AL, Kulasekara B, Rietsch A, Boyd D, Smith RS, Lory S. 2004. A signaling network reciprocally regulates genes associated with acute infection and chronic persistence in Pseudomonas aeruginosa. Dev. Cell 7:745–754. 10.1016/j.devcel.2004.08.020. [DOI] [PubMed] [Google Scholar]
- 16.Jing X, Jaw J, Robinson HH, Schubot FD. 2010. Crystal structure and oligomeric state of the RetS signaling kinase sensory domain. Proteins 78:1631–1640. 10.1002/prot.22679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ventre I, Goodman AL, Vallet-Gely I, Vasseur P, Soscia C, Molin S, Bleves S, Lazdunski A, Lory S, Filloux A. 2006. Multiple sensors control reciprocal expression of Pseudomonas aeruginosa regulatory RNA and virulence genes. Proc. Natl. Acad. Sci. U. S. A. 103:171–176. 10.1073/pnas.0507407103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vincent F, Round A, Reynaud A, Bordi C, Filloux A, Bourne Y. 2010. Distinct oligomeric forms of the Pseudomonas aeruginosa RetS sensor domain modulate accessibility to the ligand binding site. Environ. Microbiol. 12:1775–1786. 10.1111/j.1462-2920.2010.02264.x. [DOI] [PubMed] [Google Scholar]
- 19.Goodman AL, Merighi M, Hyodo M, Ventre I, Filloux A, Lory S. 2009. Direct interaction between sensor kinase proteins mediates acute and chronic disease phenotypes in a bacterial pathogen. Genes Dev. 23:249–259. 10.1101/gad.1739009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brencic A, McFarland KA, McManus HR, Castang S, Mogno I, Dove SL, Lory S. 2009. The GacS/GacA signal transduction system of Pseudomonas aeruginosa acts exclusively through its control over the transcription of the RsmY and RsmZ regulatory small RNAs. Mol. Microbiol. 73:434–445. 10.1111/j.1365-2958.2009.06782.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kong W, Chen L, Zhao J, Shen T, Surette MG, Shen L, Duan K. 2013. Hybrid sensor kinase PA1611 in Pseudomonas aeruginosa regulates transitions between acute and chronic infection through direct interaction with RetS. Mol. Microbiol. 88:784–797. 10.1111/mmi.12223. [DOI] [PubMed] [Google Scholar]
- 22.Brencic A, Lory S. 2009. Determination of the regulon and identification of novel mRNA targets of Pseudomonas aeruginosa RsmA. Mol. Microbiol. 72:612–632. 10.1111/j.1365-2958.2009.06670.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heurlier K, Williams F, Heeb S, Dormond C, Pessi G, Singer D, Camara M, Williams P, Haas D. 2004. Positive control of swarming, rhamnolipid synthesis, and lipase production by the posttranscriptional RsmA/RsmZ system in Pseudomonas aeruginosa PAO1. J. Bacteriol. 186:2936–2945. 10.1128/JB.186.10.2936-2945.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lapouge K, Schubert M, Allain FH, Haas D. 2008. Gac/Rsm signal transduction pathway of gamma-proteobacteria: from RNA recognition to regulation of social behaviour. Mol. Microbiol. 67:241–253. 10.1111/j.1365-2958.2007.06042.x. [DOI] [PubMed] [Google Scholar]
- 25.Irie Y, Starkey M, Edwards AN, Wozniak DJ, Romeo T, Parsek MR. 2010. Pseudomonas aeruginosa biofilm matrix polysaccharide Psl is regulated transcriptionally by RpoS and post-transcriptionally by RsmA. Mol. Microbiol. 78:158–172. 10.1111/j.1365-2958.2010.07320.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hengge R. 2009. Principles of c-di-GMP signalling in bacteria. Nat. Rev. Microbiol. 7:263–273. 10.1038/nrmicro2109. [DOI] [PubMed] [Google Scholar]
- 27.Schirmer T, Jenal U. 2009. Structural and mechanistic determinants of c-di-GMP signalling. Nat. Rev. Microbiol. 7:724–735. 10.1038/nrmicro2203. [DOI] [PubMed] [Google Scholar]
- 28.Simm R, Morr M, Kader A, Nimtz M, Romling U. 2004. GGDEF and EAL domains inversely regulate cyclic di-GMP levels and transition from sessility to motility. Mol. Microbiol. 53:1123–1134. 10.1111/j.1365-2958.2004.04206.x. [DOI] [PubMed] [Google Scholar]
- 29.Jenal U, Malone J. 2006. Mechanisms of cyclic-di-GMP signaling in bacteria. Annu. Rev. Genet. 40:385–407. 10.1146/annurev.genet.40.110405.090423. [DOI] [PubMed] [Google Scholar]
- 30.Moscoso JA, Mikkelsen H, Heeb S, Williams P, Filloux A. 2011. The Pseudomonas aeruginosa sensor RetS switches type III and type VI secretion via c-di-GMP signalling. Environ. Microbiol. 13:3128–3138. 10.1111/j.1462-2920.2011.02595.x. [DOI] [PubMed] [Google Scholar]
- 31.Merritt JH, Brothers KM, Kuchma SL, O'Toole GA. 2007. SadC reciprocally influences biofilm formation and swarming motility via modulation of exopolysaccharide production and flagellar function. J. Bacteriol. 189:8154–8164. 10.1128/JB.00585-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mikkelsen H, Ball G, Giraud C, Filloux A. 2009. Expression of Pseudomonas aeruginosa CupD fimbrial genes is antagonistically controlled by RcsB and the EAL-containing PvrR response regulators. PLoS One 4:e6018. 10.1371/journal.pone.0006018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rybtke MT, Borlee BR, Murakami K, Irie Y, Hentzer M, Nielsen TE, Givskov M, Parsek MR, Tolker-Nielsen T. 2012. Fluorescence-based reporter for gauging cyclic di-GMP levels in Pseudomonas aeruginosa. Appl. Environ. Microbiol. 78:5060–5069. 10.1128/AEM.00414-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Voisard C, Bull CT, Keel C, Laville J, Maurhofer M, Schnider U, Defago G, Haas D. 1994. Biocontrol of root diseases by Pseudomonas fluorescens CHA0: current concepts and experimental approaches, p 67–89 In O'Gara F, Dowling DN, Boesten B. (ed), Molecular ecology of rhizosphere microorganisms: biotechnology and the release of GMOs. VCH, Weinheim, Germany. [Google Scholar]
- 35.Merritt JH, Ha DG, Cowles KN, Lu W, Morales DK, Rabinowitz J, Gitai Z, O'Toole GA. 2010. Specific control of Pseudomonas aeruginosa surface-associated behaviors by two c-di-GMP diguanylate cyclases. mBio 1(4):e00183-10. 10.1128/mBio.00183-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bordi C, Lamy MC, Ventre I, Termine E, Hachani A, Fillet S, Roche B, Bleves S, Mejean V, Lazdunski A, Filloux A. 2010. Regulatory RNAs and the HptB/RetS signalling pathways fine-tune Pseudomonas aeruginosa pathogenesis. Mol. Microbiol. 76:1427–1443. 10.1111/j.1365-2958.2010.07146.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hsu JL, Chen HC, Peng HL, Chang HY. 2008. Characterization of the histidine-containing phosphotransfer protein B-mediated multistep phosphorelay system in Pseudomonas aeruginosa PAO1. J. Biol. Chem. 283:9933–9944. 10.1074/jbc.M708836200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lin CT, Huang YJ, Chu PH, Hsu JL, Huang CH, Peng HL. 2006. Identification of an HptB-mediated multi-step phosphorelay in Pseudomonas aeruginosa PAO1. Res. Microbiol. 157:169–175. 10.1016/j.resmic.2005.06.012. [DOI] [PubMed] [Google Scholar]
- 39.Houot L, Fanni A, de Bentzmann S, Bordi C. 2012. A bacterial two-hybrid genome fragment library for deciphering regulatory networks of the opportunistic pathogen Pseudomonas aeruginosa. Microbiology 158:1964–1971. 10.1099/mic.0.057059-0. [DOI] [PubMed] [Google Scholar]
- 40.Caiazza NC, O'Toole GA. 2004. SadB is required for the transition from reversible to irreversible attachment during biofilm formation by Pseudomonas aeruginosa PA14. J. Bacteriol. 186:4476–4485. 10.1128/JB.186.14.4476-4485.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lory S, Merighi M, Hyodo M. 2009. Multiple activities of c-di-GMP in Pseudomonas aeruginosa. Nucleic Acids Symp. Ser. (Oxf.) 2009:51–52. 10.1093/nass/nrp026. [DOI] [PubMed] [Google Scholar]
- 42.Jonas K, Edwards AN, Simm R, Romeo T, Romling U, Melefors O. 2008. The RNA binding protein CsrA controls cyclic di-GMP metabolism by directly regulating the expression of GGDEF proteins. Mol. Microbiol. 70:236–257. 10.1111/j.1365-2958.2008.06411.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jonas K, Edwards AN, Ahmad I, Romeo T, Romling U, Melefors O. 2010. Complex regulatory network encompassing the Csr, c-di-GMP and motility systems of Salmonella Typhimurium. Environ. Microbiol. 12:524–540. 10.1111/j.1462-2920.2009.02097.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lu XH, An SQ, Tang DJ, McCarthy Y, Tang JL, Dow JM, Ryan RP. 2012. RsmA regulates biofilm formation in Xanthomonas campestris through a regulatory network involving cyclic di-GMP and the Clp transcription factor. PLoS One 7:e52646. 10.1371/journal.pone.0052646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bhuwan M, Lee HJ, Peng HL, Chang HY. 2012. Histidine-containing phosphotransfer protein-B (HptB) regulates swarming motility through partner-switching system in Pseudomonas aeruginosa PAO1 strain. J. Biol. Chem. 287:1903–1914. 10.1074/jbc.M111.256586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kulasakara H, Lee V, Brencic A, Liberati N, Urbach J, Miyata S, Lee DG, Neely AN, Hyodo M, Hayakawa Y, Ausubel FM, Lory S. 2006. Analysis of Pseudomonas aeruginosa diguanylate cyclases and phosphodiesterases reveals a role for bis-(3′-5′)-cyclic-GMP in virulence. Proc. Natl. Acad. Sci. U. S. A. 103:2839–2844. 10.1073/pnas.0511090103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mikkelsen H, McMullan R, Filloux A. 2011. The Pseudomonas aeruginosa reference strain PA14 displays increased virulence due to a mutation in ladS. PLoS One 6:e29113. 10.1371/journal.pone.0029113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kuchma SL, Brothers KM, Merritt JH, Liberati NT, Ausubel FM, O'Toole GA. 2007. BifA, a cyclic-di-GMP phosphodiesterase, inversely regulates biofilm formation and swarming motility by Pseudomonas aeruginosa PA14. J. Bacteriol. 189:8165–8178. 10.1128/JB.00586-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bernier SP, Ha DG, Khan W, Merritt JH, O'Toole GA. 2011. Modulation of Pseudomonas aeruginosa surface-associated group behaviors by individual amino acids through c-di-GMP signaling. Res. Microbiol. 162:680–688. 10.1016/j.resmic.2011.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Caiazza NC, Merritt JH, Brothers KM, O'Toole GA. 2007. Inverse regulation of biofilm formation and swarming motility by Pseudomonas aeruginosa PA14. J. Bacteriol. 189:3603–3612. 10.1128/JB.01685-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bertrand JJ, West JT, Engel JN. 2010. Genetic analysis of the regulation of type IV pilus function by the Chp chemosensory system of Pseudomonas aeruginosa. J. Bacteriol. 192:994–1010. 10.1128/JB.01390-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Martinez-Granero F, Navazo A, Barahona E, Redondo-Nieto M, Rivilla R, Martin M. 2012. The Gac-Rsm and SadB signal transduction pathways converge on AlgU to downregulate motility in Pseudomonas fluorescens. PLoS One 7:e31765. 10.1371/journal.pone.0031765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Humair B, Wackwitz B, Haas D. 2010. GacA-controlled activation of promoters for small RNA genes in Pseudomonas fluorescens. Appl. Environ. Microbiol. 76:1497–1506. 10.1128/AEM.02014-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jones CJ, Newsom D, Kelly B, Irie Y, Jennings LK, Xu B, Limoli DH, Harrison JJ, Parsek MR, White P, Wozniak DJ. 2014. ChIP-Seq and RNA-Seq reveal an AmrZ-mediated mechanism for cyclic di-GMP synthesis and biofilm development by Pseudomonas aeruginosa. PLoS Pathog. 10:e1003984. 10.1371/journal.ppat.1003984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Martinez-Granero F, Redondo-Nieto M, Vesga P, Martin M, Rivilla R. 2014. AmrZ is a global transcriptional regulator implicated in iron uptake and environmental adaption in P. fluorescens F113. BMC Genomics 15:237. 10.1186/1471-2164-15-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hickman JW, Harwood CS. 2008. Identification of FleQ from Pseudomonas aeruginosa as a c-di-GMP-responsive transcription factor. Mol. Microbiol. 69:376–389. 10.1111/j.1365-2958.2008.06281.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Baynham PJ, Ramsey DM, Gvozdyev BV, Cordonnier EM, Wozniak DJ. 2006. The Pseudomonas aeruginosa ribbon-helix-helix DNA-binding protein AlgZ (AmrZ) controls twitching motility and biogenesis of type IV pili. J. Bacteriol. 188:132–140. 10.1128/JB.188.1.132-140.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jones CJ, Ryder CR, Mann EE, Wozniak DJ. 2013. AmrZ modulates Pseudomonas aeruginosa biofilm architecture by directly repressing transcription of the psl operon. J. Bacteriol. 195:1637–1644. 10.1128/JB.02190-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tart AH, Wolfgang MC, Wozniak DJ. 2005. The alternative sigma factor AlgT represses Pseudomonas aeruginosa flagellum biosynthesis by inhibiting expression of fleQ. J. Bacteriol. 187:7955–7962. 10.1128/JB.187.23.7955-7962.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wozniak DJ, Sprinkle AB, Baynham PJ. 2003. Control of Pseudomonas aeruginosa algZ expression by the alternative sigma factor AlgT. J. Bacteriol. 185:7297–7300. 10.1128/JB.185.24.7297-7300.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Frangipani E, Visaggio D, Heeb S, Kaever V, Camara M, Visca P, Imperi F. 2014. The Gac/Rsm and cyclic-di-GMP signalling networks coordinately regulate iron uptake in Pseudomonas aeruginosa. Environ. Microbiol. 16:676–688. 10.1111/1462-2920.12164. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


