Abstract
The septal association of Mycobacterium tuberculosis MtrB, the kinase partner of the MtrAB two-component signal transduction system, is necessary for the optimal expression of the MtrA regulon targets, including ripA, fbpB, and ftsI, which are involved in cell division and cell wall synthesis. Here, we show that MtrB, irrespective of its phosphorylation status, interacts with Wag31, whereas only phosphorylation-competent MtrB interacts with FtsI. We provide evidence that FtsI depletion compromises the MtrB septal assembly and MtrA regulon expression; likewise, the absence of MtrB compromises FtsI localization and, possibly, FtsI activity. We conclude from these results that FtsI and MtrB are codependent for their activities and that FtsI functions as a positive modulator of MtrB activation and MtrA regulon expression. In contrast to FtsI, Wag31 depletion does not affect MtrB septal assembly and MtrA regulon expression, whereas the loss of MtrB increased Wag31 localization and the levels of PknA/PknB (PknA/B) serine-threonine protein kinase-mediated Wag31 phosphorylation. Interestingly, we found that FtsI decreased levels of phosphorylated Wag31 (Wag31∼P) and that MtrB interacted with PknA/B. Overall, our results indicate that MtrB interactions with FtsI, Wag31, and PknA/B are required for its optimal localization, MtrA regulon expression, and phosphorylation of Wag31. Our results emphasize a new role for MtrB in cell division and cell wall synthesis distinct from that regulating the MtrA phosphorylation activities.
INTRODUCTION
Mycobacterium tuberculosis, the causative agent of tuberculosis (TB), accounts for nearly 1. 3 million deaths per year globally, and one-third of the world's population is latently infected with M. tuberculosis (http://www.who.int/tb). An increase in the numbers of multidrug-resistant M. tuberculosis strains has been seen recently, and this has led to the pursuit of new antimicrobials to control tuberculosis. M. tuberculosis has 11 paired two-component signal transduction systems (two-component regulatory systems [TCRS]), 180 transcriptional regulators, and 11 eukaryotic-like serine/threonine protein kinases for the regulation of various cellular processes (http://tuberculist.epfl.ch). Cell wall synthesis and cell division are critical for the duplication of organisms; hence, a detailed understanding of these processes could provide attractive targets for the development of new antibiotics (1). The components involved in these processes are conserved in all mycobacterial members (http://tuberculist.epfl.ch).
The MtrAB system is one of the two essential TCRS in M. tuberculosis, wherein MtrB is the sensor kinase and MtrA is the cognate response regulator (2, 3). In vitro phosphorylation experiments revealed that MtrB phosphorylates MtrA (4) and that phosphorylated MtrA (MtrA∼P) binds to the origin of replication (oriC) and the promoters of fbpB, ripA, and dnaA (5–7). The mtrB of Mycobacterium smegmatis, a nonpathogen and rapid grower, is not essential, and the loss of MtrB leads to defects in MtrA regulon expression, cell division, and cell shape maintenance processes (8). M. tuberculosis MtrB protein complements the M. smegmatis ΔmtrB mutant defect, indicating that MtrB functions are conserved in the two species (8). MtrB associates with the septa in an FtsZ-dependent manner, and MtrB septal assembly is critical for the optimal expression of the MtrA regulon (8). These studies support the idea that MtrB plays an indirect role in cell division and cell shape maintenance by affecting the expression levels of the MtrA regulon targets involved in these processes.
In addition to FtsZ and the MtrB proteins, previous studies have shown that FtsI, FtsW, FipA, Wag31, CrgA, CwsA, RipA, and penicillin-binding protein A (PBPA) are other necessary septasomal components for the cell division in mycobacteria (6, 8–16). These studies also established pairwise interactions between FtsI and Wag31, FtsI and FtsW, FtsI and CrgA, CwsA and Wag31, and CrgA and PBPA. It is unknown whether MtrB interacts with any of the above septasomal proteins and, if so, what the consequences associated with these interactions are. Additionally, it is unknown whether MtrB exerts a direct role in cell division and cell shape maintenance, besides affecting the modulation of the expression levels of the MtrA targets involved in these processes. The present study was undertaken to address these issues. Given the filamentation and multiseptate phenotypes of the mtrB mutant (8), we considered that MtrB may associate with some septasomal components and that such interactions could modulate the activities of MtrB, its interaction partners, or both. The results presented in this study are consistent with a view that the interactions of MtrB with FtsI and Wag31 are crucial for optimal MtrA regulon expression, phosphorylation of Wag31, and cell division.
MATERIALS AND METHODS
Strains, bacterial growth conditions, and molecular cloning.
The Escherichia coli strain Top10 was used for cloning purposes, E. coli BTH101 was used for bacterial two-hybrid (BACTH) protein-protein interaction assays, and E. coli BL21 (DE3)pLysS was used for the production of recombinant proteins. E. coli strains were propagated in Luria-Bertani (LB) broth or LB agar supplemented with ampicillin (Amp; 50 μg/ml), kanamycin (Km; 50 μg/ml), or hygromycin (Hyg; 200 μg/ml). M. smegmatis strains were grown in Middlebrook 7H9 broth or 7H10 agar supplemented with 0.05% Tween 80 and albumin and dextrose (AD), with the appropriate antibiotics (Km at 25 μg/ml or Hyg at 50 μg/ml). Growth was monitored by measuring the absorbance at 600 nm. The plasmids used in this study and the oligonucleotide primers used for cloning are listed in Tables S1 and S2, respectively, in the supplemental material. The coding regions of various genes were PCR amplified with Phusion DNA polymerase (NE Biolabs, MA), cloned into various plasmids (see Table S1), and confirmed by sequencing.
BACTH assays.
A BACTH system was used to investigate MtrB interactions with other cell division proteins, as described previously (17). The tested proteins were expressed in various combinations as N- or C-terminal fusions to Bordetella pertussis T18 or T25 adenylate cyclase fragments in the E. coli BTH101 strain (see Tables S1 and S2 in the supplemental material). The cotransformants were spotted on minimal medium agar supplemented with 0.004% 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal), 100 μg/ml Amp, and 50 μg/ml Km (18, 19). The interaction strength was determined, as needed, by measuring the extent of β-galactosidase activity in the broth-grown cultures (17). Beta-galactosidase activity of at least 5-fold or higher than that measured for the E. coli BTH101 strain carrying a single gene and empty vector was considered indicative of a positive interaction (18, 19).
Western blotting.
Recombinant protein fractions or cell lysates from exponentially grown cultures were prepared and processed for immunoblotting using appropriate antibodies (anti-MtrB, anti-M. tuberculosis FtsZ [FtsZTB], anti-Wag31TB, anti-SigATB, anti-FtsITB, and anti-phospho-Ser/Thr) as previously described (20). SigA was used as a loading control (20). The immunoblots were processed using an ECF Western blotting kit (GE Life Sciences, Piscataway, NJ) and scanned using a Bio-Rad Molecular Imager (FX). The protein levels were determined using the volume analysis function of QuantityOne software. Phosphorylated Wag31 (Wag∼P) was detected with phospho-Ser/Thr antibodies (Cell Signaling Technology) (21). Various proteins were quantified from three independent experiments. For various tested proteins, protein-to-SigA (protein/SigA) ratios were determined. For calculating the Wag31∼P/Wag31 ratio, the blots were first probed with anti-phospho-Ser/Thr, stripped, and reprobed with anti-Wag31. The FtsI levels in the conditional ftsI strain following growth in the absence of acetamide were determined by immunoblotting with anti-FtsI antibodies and normalized to SigA as described above.
Pulldown and coimmunoprecipitation assays.
Pulldown of Wag31 by MtrB and FtsZ was performed using maltose-binding protein (MBP)-Wag31 and His10-MtrBsol (where MtrBsol is a soluble C-terminal MtrB fragment) (see Table S1 in the supplemental material) or His10-FtsZ, as described elsewhere (14, 17). Briefly, E. coli lysates with MBP-Wag31 were mixed with a Ni-nitrilotriacetic acid (NTA)-bound His-tagged protein (His10-MtrBsol/His10-FtsZ) in 1× binding buffer [50 mM sodium phosphate, pH 7.8, 10% glycerol, 5 mM 2-mercaptoethanol, 150 mM NaCl, 0.1% CHAPS (3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate), and 1% Nonidet P-40] and incubated with rocking at 4°C for 1 h. The supernatant was removed following centrifugation, the resin was washed five times with binding buffer, and the bound proteins were eluted with buffers containing 0.1, 0.3, and 1 M imidazole. The load, last wash, and eluted proteins were resolved by SDS-PAGE, transferred to polyvinylidene difluoride (PVDF) membranes, and immunodetected with anti-MBP/anti-MtrB/anti-Wag31 antibodies. For the immunoprecipitation (IP) experiments, M. smegmatis lysates were prepared by bead-beating cell pellets from a 30-ml culture in 1 ml of lysis buffer (50 mM Tris-HCl, pH 7.5, 1% NP-40, 100 mM NaCl, 5% glycerol, 1 mM EDTA, and 1 mM phenylmethylsulfonyl fluoride) in a mini-bead-beater (Disruptor Genie; Scientific Industries, Inc.) for 30 s 10 times with 5 min of cooling on ice after each beating. Whole-cell lysates were spun at 4°C for 15 min at 13,000 rpm, and supernatants were used for IP experiments. One milliliter of untreated lysate or lysate enriched with ∼60 μg of purified MBP-MtrBsol (86 kDa) (8) was incubated overnight at 4°C with anti-MtrB or anti-Wag31 antibody coupled to magnetic beads (BioMag Plus amine particles). The addition of purified MBP-MtrBsol served as an internal control and also facilitated the IP reaction as sufficient amounts of full-length membrane-bound MtrB could not be solubilized in IP lysis buffer. The beads were washed five times with IP wash buffer (50 mM Tris-HCl, pH 7. 4, 0.05% Triton X-100, 100 mM NaCl, and 0.5 mM EDTA), and the MtrB-Wag31 immunoprecipitates were eluted with 100 mM citrate (pH 3.1) and neutralized with 1 M NaOH, and Wag31 and MtrB were visualized following immunoblotting.
Microscopy.
M. smegmatis strains were grown and visualized by bright-field and fluorescence microscopy as previously described (8). Nascent peptidoglycan (PG) at the septa and poles was visualized by staining with 1.1 μg/ml BODIPY FL (difluoro-4-bora-3a,4a-diaza-s-indacene)-labeled vancomycin (Van-FL) as previously described (22). A Nikon Eclipse 600 microscope equipped with a 100× Nikon Plan Fluor oil immersion objective with a numerical aperture of 1.4 and the appropriate filter sets (Chroma) was used for bright-field and fluorescence microscopy. The images were acquired using a Photometrics Coolsnap ES camera and Metapmorph, version 6.2, imaging software (Universal Imaging Corporation). The images were optimized using Adobe Photoshop CS4. The quantitated microscopy data are averages ± standard errors from two independent experiments. The number of cells counted is indicated with the respective data sets and included at least 100 cells, with the exception of Van-FL staining data for FtsI depletion as the latter presented with serious clumping issues.
Antibiotic sensitivity assays.
Overnight cultures of M. smegmatis wild type (WT) and a ΔmtrB strain were diluted to an A600 of 0.05 and grown for 6 h, and 1 × 105 cells were spread with sterile cotton-tipped swabs. Next, Etest strips (vancomycin and ampicillin; bioMérieux, Inc.) or filter discs containing cycloserine (100 μg) were placed on the culture plates and incubated at 37°C for 4 days, and results were recorded (23). The MICs of ampicillin and vancomycin for the WT and ΔmtrB strains were determined following the Etest strip manufacturer's instructions.
RNA extraction, reverse transcription, and qRT-PCR.
Total RNA was extracted from the 7H9 broth-grown cultures of M. smegmatis, and quantitative real-time PCR (qRT-PCR) was performed as previously described (5) in a Bio-Rad iQ5 Real-Time PCR detection system using 2× iQ SYBR green Supermix (Bio-Rad). The primers for qRT-PCR are listed in Table S3 in the supplemental material. The threshold cycle (CT) value of each gene of interest was normalized to the CT value of sigA, and the fold expression was calculated using the following formula: fold change = 2−Δ(ΔCT) (5). The presented expression data are the averages from experiments with three independent RNA preparations that were reverse transcribed and quantified by real-time PCR in triplicate. Differences in expression levels of 2-fold or more were considered significant (5, 22).
Statistical analysis.
Data presented are means ± standard errors from three independent experiments. Control and test data sets were compared and analyzed using Student's t test.
RESULTS
MtrB interacts with Wag31 and FtsI.
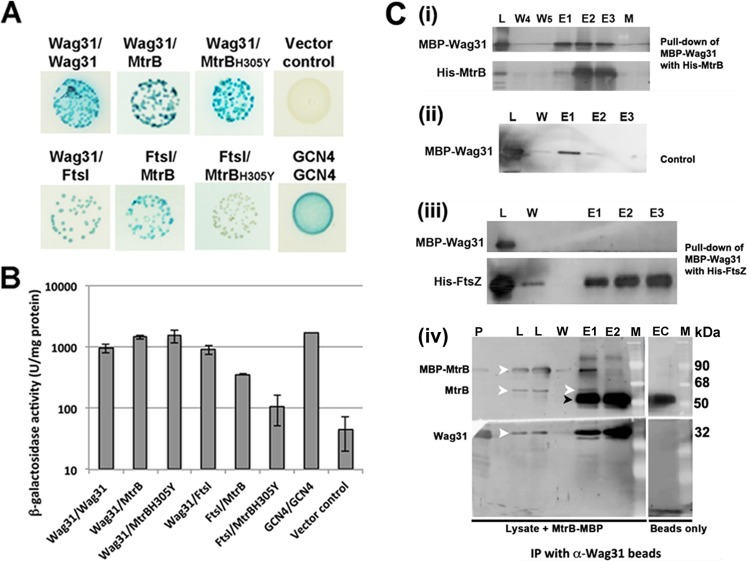
BACTH assays were performed to evaluate MtrB interactions with select septasomal components (19). When E. coli BTH101 cells were cotransformed with plasmids expressing mtrB in combination with the genes involved in cell division and cell wall synthesis and were spread on minimal agar medium containing the chromogenic substrate X-Gal as previously described (8), intense blue-color colonies indicative of interactions were noted with the constructs expressing ftsI and wag31, similar to the results with positive-control plasmids expressing either GCN4/GCN4, Wag31/Wag31, or Wag31/FtsI in two different vectors (Fig. 1A). In these assays, the MtrB fusions to both the T25 and T18 domains of adenylate cyclase (19) interacted with the appropriate FtsI and Wag31 fusions (data not shown). The β-galactosidase activity in the broth-grown cultures of cotransformant colonies validated the interactions and provided a measure for the strength of the interactions (Fig. 1B). For example, cultures expressing the MtrB and Wag31 pair showed ∼32-fold higher β-galactosidase activities than the vector control, similar to the GCN4/GCN4 pair, whereas that expressing the MtrB and FtsI pair showed ∼8-fold higher activity than the control. Thus, the interactions between MtrB and Wag31 are more robust than those between MtrB and FtsI. MtrB did not interact with RipA, ClpX (see Fig. S1 in the supplemental material), CrgA, CwsA, PBPA, RodA, or FtsW protein (data not shown).
FIG 1.
MtrB interactions with FtsI and Wag31. (A) BACTH analysis of MtrB interactions with FtsI and Wag31. E. coli BTH101 cotransformants with plasmids expressing Wag31/Wag31, Wag31/MtrB, Wag31/MtrBH305Y, MtrB/FtsI, and MtrBH305Y/FtsI were spotted on indicator agar plates as previously described (18), wherein blue and white spots indicate positive and negative interactions, respectively. Cotransformants with GCN4/GCN4 and empty vector/MtrB, representing positive and negative controls, respectively, were also spotted. (B) Recombinant colonies expressing the indicated combinations of proteins were propagated in LB broth, and β-galactosidase activity was measured as described in the text. Data shown are means ± standard deviations from three independent experiments. (C) Pulldown assays. Wag31 interacts with MtrB and not with FtsZ. E. coli lysates with MBP-Wag31 were incubated with His10-MtrBsol (i) or His10-FtsZ (iii) bound to Ni-NTA resin with rocking at 4°C. The Ni-NTA resin was washed five times, and the bound proteins were eluted with buffer containing imidazole (see below and Materials and Methods) (17). MBP-Wag31 was incubated with Ni-NTA (ii) and processed as described for panels i and iii. Various fractions were separated in SDS-polyacrylamide gels and transferred to PVDF membranes, and the proteins were visualized by immunoblotting using anti-MtrB, anti-Wag31, anti-FtsZ, or anti-MBP antibody (see Fig. S2 in the supplemental material). L, load; W, wash; E, elution fractions; W4 and W5, washes 4 and 5, respectively; E1, E2, and E3 are elutions with buffers containing 100 mM, 300 mM, and 1 M imidazole, respectively. For co-IP assays (iv) 1 ml of M. smegmatis lysate mixed with 60 μg of purified MBP-MtrB was incubated overnight with anti-Wag31 antibodies coupled to magnetic beads (BioMag Plus amine particles). The beads were washed five times with IP wash buffer, and the MtrB immunoprecipitates were eluted with 100 mM citrate (pH 3.1). The eluted proteins were neutralized with 1 N NaOH, separated by SDS-PAGE, and visualized following immunoblotting with anti-Wag31 or anti-MtrB antibody. P, pure protein markers for MBP-MtrB and His-Wag31; L, load; W, wash 5; E1 and E2, elutions 1 and 2; EC, elution from buffer control where beads were incubated with buffer instead of lysate. White arrowhead, MBP-MtrB, MtrB, or Wag31; black arrowhead, IgG band. M, molecular mass marker. α, anti.
The interactions between MtrB and Wag31 were further confirmed by in vitro pulldown assays using the recombinant fusion proteins MBP-Wag31 and His10-MtrBsol (see Table S1 in the supplemental material). Because Wag31 is a soluble protein, we assumed that it would interact with the cytosolic MtrB domain and therefore used MtrBsol protein in these experiments. When the MBP-Wag31-containing lysate was added to the Ni-NTA-bound His10-MtrBsol, followed by elution and immunoblotting analyses using an anti-MBP antibody, MBP-Wag31 was detected in the eluted fractions of His10-MtrB, indicating the pulldown of Wag31 from the reaction mix (Fig. 1C, panel i; see also Fig. S2, panel i, in the supplemental material). Furthermore, in a similar experiment, MBP-Wag31 did not bind to Ni-NTA beads (Fig. 1C, panel ii; see also Fig. S2, panel ii). A small amount of loosely bound and possibly denatured MBP-Wag31 came out in the 100 mM imidazole wash in both the test and control experiment sets (Fig. 1C, panels i and ii, lanes E1); however, no significant amount of MBP-Wag31 was detected in the 300 mM and 1 M imidazole elutions in the control experiment (Fig. 1C, compare panels i and ii, lanes E2 and E3). MBP-Wag31 also did not bind to His10-FtsZ bound to Ni-NTA beads, supporting the specificity of its interaction with MtrB (Fig. 1C, panel iii; see also Fig. S2, panel iii). To demonstrate in vivo interactions between MtrB and Wag31, we carried out co-IP and pulldown experiments with M. smegmatis cell-free lysates (Fig. 1C, panel iv; see also Fig. S2, panels iv and v, in the supplemental material). For IP experiments, M. smegmatis cell-free lysates supplemented with purified MBP-MtrBsol (86 kDa) were incubated with anti-Wag31 antibody-coupled BioMag amine beads. Immunoblotting with anti-MtrB and anti-Wag31 antibodies identified MtrB (both 86-kDa MBP-MtrBsol and full-length 61-kDa MtrB) and Wag31 (28 kDa) protein-specific bands in elution fraction E1 (eluted with 100 mM citrate), confirming in vivo interactions between the two proteins (Fig. 1C, panel iv, white arrowheads). The abundant band (∼50 kDa) below the native MtrB in the E1 fraction is likely IgG as the elution fraction from the buffer control (no-lysate) reaction with anti-Wag31 antibody-coupled beads also contained a similarly sized band (Fig. 1C, panel iv, black arrowhead; also see Fig. S2, panel iv, lane EC). Reciprocal experiments with M. smegmatis lysates and anti-MtrB-coupled beads showed coelution of Wag31 in the MtrB-containing fraction (see Fig. S2, panel iv). Pulldown experiments with M. smegmatis lysates containing Wag31-His (see Table S1 in the supplemental material) further confirmed the in vivo interactions between MtrB and Wag31. In these experiments, MtrB copurified with Wag31-His on an Ni-NTA column (see Fig. S2, panel v). Similar types of studies could not be carried out with FtsI because our attempts to overexpress and purify the full-length recombinant FtsI, an integral membrane protein, were unsuccessful.
MtrBH305Y defective for autokinase activity is not proficient for interactions with FtsI.
We showed previously that MtrB with the histidine residue at position 305 replaced by tyrosine (MtrBH305Y) is defective for phosphorylation and for reversal of the mtrB mutant phenotype but is proficient for localization to the septal sites, indicating that MtrB autokinase activity is essential for functional complementation but not necessary for MtrB septal association (8). BACTH assays revealed strong interactions between MtrBH305Y and Wag31 (∼33-fold higher β-galactosidase activity over the vector control) but not between MtrBH305Y and FtsI (∼2.3-fold higher over the control) (Fig. 1A and B). These results suggest either that MtrBH305Y is not proficient for interaction with FtsI or that the interaction is transient and cannot be detected using the BACTH assay. Such transient interactions between MtrBH305Y and FtsI, if any, could be stabilized by hitherto unidentified factors present in vivo. Nonetheless, the ability of MtrB to undergo phosphorylation is important for its stable association with FtsI.
The interactions of MtrB with FtsI are important for MtrA regulon expression.
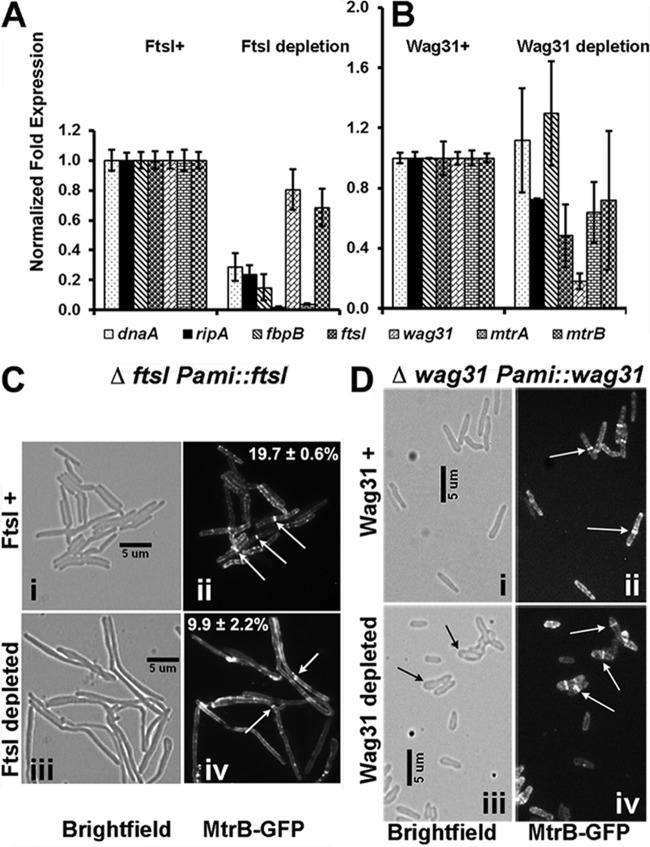
The observed interactions of MtrB with FtsI and Wag31 lead to the prediction that they are required for optimal MtrA regulon expression and, possibly, for balanced cell division and cell wall synthesis. Accordingly, we measured MtrA regulon expression in wag31 (12) and ftsI conditional expression strains with depletion of the respective proteins (see Fig. S3 in the supplemental material and the supplemental data for methodology for the creation of the ftsI strain). The conditional expression strains were used because Wag31 is essential in M. smegmatis (12), and M. smegmatis chromosomal ftsI could be deleted only in the presence of an integrated copy of ftsI (see Fig. S3). Culturing the ftsI conditional strain in acetamide-free medium for 12 to 14 h reduced the expression level of ftsI by >10-fold (Fig. 2A) and led to an ∼50% reduction in the FtsI protein level (see Fig. S3D in the supplemental material). Depletion of FtsI also increased the cell length, indicating cell division defects (see Fig. S3F). In addition, the expression of mtrA along with the MtrA targets dnaA, ripA, and fbpB was decreased (Fig. 2A). As with the ftsI strain, culturing the wag31 conditional strain in acetamide-free medium for 15 h led to a reduction in wag31 expression; however, unlike the ftsI strain, the expression levels of dnaA, ripA, mtrA, and fbpB were not significantly affected (Fig. 2B). Together, these results suggested that MtrB interactions with FtsI are necessary for the optimal expression of the MtrA regulon and that interactions with Wag31 may have unidentified consequences (see below). The expression of mtrB was not significantly altered under both conditions (Fig. 2A and B).
FIG 2.
MtrB activity and localization are compromised in the absence of FtsI. (A and B) qRT-PCR analysis of MtrA targets. Total RNA was extracted from ΔftsI::Pami-ftsI (A) and Δwag31::Pami-wag31 (B) strains grown without and with 0.2% acetamide (see details in text) and reverse transcribed, and qRT-PCR was performed as described in Materials and Methods. The expression levels of select genes, that is, dnaA, ripA, fbpB, wag31, mtrA, mtrB, and ftsI, relative to the housekeeping gene sigA were compared, and the values are presented as fold difference. In panel A fold expression levels upon FtsI depletion (growth in the absence of acetamide for 12 h) were normalized to those in the presence of acetamide (FtsI+). In panel B fold expression levels upon Wag31 depletion (growth in the absence of acetamide for 15 h) were normalized to those in the presence of acetamide (Wag31+). Wag31 depletion beyond 15 h led to extreme cell distortion and eventually cell lysis; hence, expression studies were not carried out beyond 15 h. (C and D) MtrB-GFP localization. M. smegmatis ΔftsI::Pami-ftsI (C) and Δwag31::Pami-wag31 (D) strains expressing Ptet::mtrB-gfp were grown without and with 0.2% acetamide, as for panels A and B. For the visualization of MtrB-GFP, anhydrotetracycline was added at 10 ng/ml for 1 h. Bright-field (i and iii) and fluorescence (ii and iv) microscopy and imaging were carried out as described in the text. The ΔftsI::Pami-ftsI or Δwag31::Pami-wag31 strain was grown with (i and ii) or without (iii and iv) acetamide. For panel C, percent septal MtrB-GFP localizations from two independent experiments were scored, and averages ± standard errors are given in the respective fluorescent panels. Each experiment included >100 cells from each condition. White arrows, MtrB-GFP septal localization; black arrows, distorted cell shape upon Wag31 depletion.
MtrB septal localization is compromised upon FtsI depletion.
Next, we examined MtrB septal localization under FtsI and Wag31 depletion conditions. To this end, we created an M. smegmatis ΔftsI Pami::ftsI strain expressing Ptet::mtrB-gfp from an inducible tetracycline promoter (see Table S1 in the supplemental material) and evaluated MtrB-green fluorescent protein (GFP) septal assembly following 14 h of growth in acetamide-free medium. Anhydrotetracycline at 10 ng/ml was added 1 h before samples were removed for fluorescence microscopy (Fig. 2C and D). Under FtsI depletion conditions, an ∼50% reduction in MtrB-GFP septal localization was noted (from 19.7% ± 0.6% [n = 231 cells] to 9.9% ± 2.2% [n = 141]) (Fig. 2C, compare panels ii and iv; see also Fig. S3G in the supplemental material), and the MtrB-GFP structures included both sharp (∼6. 3%) (Fig. 2C, panel iv) and diffuse (∼3. 0%) (see Fig. S3G) bands. Wag31 depletion affected cell shape (Fig. 2D, compare panels i and iii) but did not affect MtrB septal assembly (Fig. 2D, panels ii and iv). These data agree with the previous results that indicated the lack of requirement of Wag31 for MtrA regulon expression (Fig. 2B). Together, these data suggest that FtsI promotes MtrB septal assembly.
FtsI and Wag31 localizations are altered in the absence of MtrB.
Next, we investigated FtsI and Wag31 localizations in the mtrB strain background producing GFP-FtsI and Wag31-mCherry, respectively (see Table S1 in the supplemental material). Consistent with the published data (8), the absence of MtrB increased cell length and altered cell shape (Fig. 3A and B, panels iii). Weak GFP-FtsI septal and membrane fluorescence signals that were readily bleached were observed in the absence of MtrB (Fig. 3A, compare panels ii and iv). These data indicate that, although compromised, FtsI can localize to septa independent of MtrB and that FtsI and MtrB interactions are likely necessary for stable/optimal septal FtsI association. It is known that ftsI expression is compromised in the absence of MtrB (8). Thus, the weak and readily bleached GFP-FtsI localization signals could also be due to decreased ftsI expression. Next, we evaluated the consequences of the FtsI interactions with MtrB on FtsI activities. FtsI, also known as PBP3, is a transpeptidase; hence, it is necessary for optimal PG cross-linking, which we evaluated as a measure for FtsI activity in two independent assays. First, the binding of Van-FL to d-Ala-d-Ala residues in un-cross-linked PG was measured. As shown previously, ∼ 30% of the WT cells showed distinct polar staining, and ∼3% showed septal staining with Van-FL (6) (Fig. 3B, panel ii). In contrast, nearly all of the chain-linked filamentous ΔmtrB cells (99%; n = 100) showed distinct staining at the poles and at the presumptive septa, indicating increased Van-FL staining (Fig. 3B, panel iv) and, hence, reduced PG cross-linking. Second, the sensitivity of the ΔmtrB strain to cell wall-targeting antibiotics was examined. MIC determinations indicated that the ΔmtrB strain was ∼16-fold more sensitive to vancomycin than the wild type (WT) (Fig. 3C) but resembled the WT with respect to the sensitivity to ampicillin (Fig. 3C) and cycloserine (data not shown). We also found that an ∼50% reduction in FtsI levels in the ftsI conditional strain (see Fig. S3 in the supplemental material) led to an ∼13-fold increase in septal (with FtsI depletion, ∼42.6%; with FtsI expression, ∼ 3.1%) and ∼ 2-fold increase in polar (with FtsI depletion, ∼50%; with FtsI expression, ∼ 25%) Van-FL staining (see Fig. S3E), indicating that the reduced FtsI levels in the ΔmtrB strain could partially account for the defective PG cross-linking. In sum, these data support the idea that FtsI function is compromised in the absence of MtrB and that interactions between FtsI and MtrB are important for their stable localizations and optimal activities.
FIG 3.
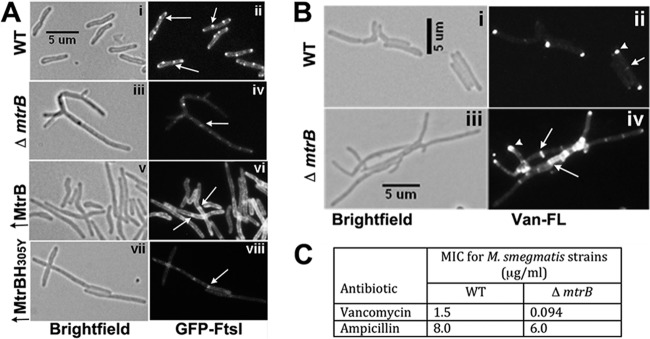
FtsI localization and activity are altered in the absence of MtrB. (A) GFP-FtsI localization was examined in M. smegmatis WT (i and ii), the ΔmtrB strain (iii and iv), and merodiploid strains overproducing (↑) MtrB (v and vi) or MtrBH305Y (vii and viii). In all of the strains, the GFP-FtsI fusion protein was produced from Pami::gfp-ftsI following induction with 0.2% acetamide for 3 h, visualized by bright-field (left panels) and fluorescent (right panels) microscopy, and imaged as described in the text. (B) The exponential cultures of the M. smegmatis WT strain (i and ii) and the ΔmtrB strain (iii and iv) were grown in the presence of Van-FL for 2 h and were imaged by bright-field (i and iii) and fluorescence (ii and iv) microscopy. (C) Loss of MtrB increases sensitivity to vancomycin. M. smegmatis WT and ΔmtrB strains were grown for 6 h, and 1 × 105 cells were spread on 7H10 agar plates. Etest antibiotic strips (ampicillin or vancomycin) were placed on the agar plates, plates were incubated for 4 days at 37°C, and MICs were measured as per the supplier's protocol.
Wag31 localizes in the WT cells primarily to the cell poles, with ∼7% of cells showing a midcell localization (6, 12) (Fig. 4A, panel ii). Similar to the WT, Wag31-mCherry was found at the poles of a majority of ΔmtrB cells (90% ± 2.6%; n = 111). However, unlike in the WT strain, Wag31-mCherry protein was at septa in more than two-thirds of ΔmtrB cells (70.5% ± 2.6%; n = 111) (Fig. 4, panels iii and iv; presumptive septa and the corresponding Wag31-mCherry localizations are marked with black and white arrows, respectively). These data indicate aberrant Wag31-mCherry localization at the unsplit septa in the absence of MtrB. Wag31-mCherry septal localization was restored to WT levels upon the production of MtrB in the ΔmtrB strain background (polar localization, 87.4% ± 2.6%; septal localization, 4.0% ± 1.1%; n = 672). Together, these data indicate that MtrB activity is required for septum resolution at steps subsequent to the Wag31 localization. It was recently shown that Wag31 localizes at septa after cytokinesis but prior to daughter cell separation (24). Accordingly, we envisage that MtrB and Wag31 together contribute to cell wall synthesis and septum resolution. The Wag31-mCherry localization pattern in the ΔmtrB strain producing MtrBH305Y was, however, similar to that in the ΔmtrB strain (see Fig. S4 in the supplemental material). These results suggest that MtrB interactions with Wag31 have possible consequences on Wag31 activities (see below). Finally, FtsZ-GFP localization was unaffected in the ΔmtrB strain (Fig. 4B, compare panels ii and iv), indicating that the absence of MtrB has no effect on the FtsZ ring assembly.
FIG 4.
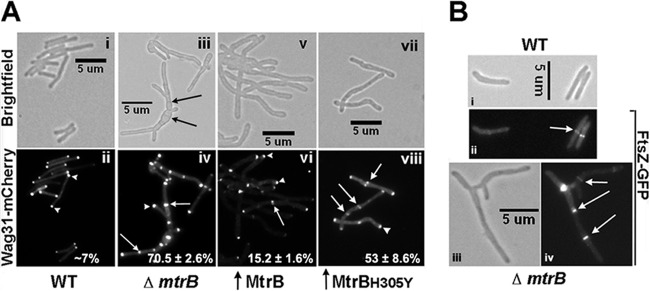
Wag31 localization is altered in the absence of MtrB. (A) Pami::wag31-mCherry expressing M. smegmatis WT (i and ii), the ΔmtrB strain (iii and iv), and M. smegmatis overproducing MtrB (v and vi) or MtrBH305Y (vii and viii) were grown with 0.2% acetamide for 3 h and visualized as described in the legend to Fig. 3A. Top panels are bright-field images; bottom panels are fluorescence images. Arrow, septal localization; arrowhead, polar localization. Percent septal Wag31-mCherry localizations from two independent experiments were scored, and averages ± standard errors are given in the respective fluorescent panels. Each experiment included >132 cells from each strain. Black arrows, septa in multiseptate ΔmtrB strain; white arrows and arrowheads, septal and polar localizations, respectively. (B) FtsZ-GFP localization in WT (i and ii) and the ΔmtrB strain (iii and iv). Arrow, FtsZ-GFP rings.
Overexpression of phosphorylation-proficient MtrB promotes FtsI localization.
Next, we evaluated the FtsI and Wag31 localizations under MtrB overproduction conditions. Overexpression of mtrB for 6 h increased the cellular MtrB levels by ∼3-fold (see Fig. S5A in the supplemental material), increased cell length, and produced bud-like structures, indicating cell shape defects (Fig. 3A, panel v; see also Fig. S5B). Additionally, an overall increase in GFP-FtsI localization at membranes and distinct midcell bands instead of foci at the septa was noted (Fig. 3A, compare panels ii and vi). When quantitated, 21.3% ± 3.3% of cells (n = 218) showed septal bands compared to ∼2% in the WT strain (25). MtrBH305Y overproduction led to increased cell length and cell shape defects (see Fig. S5B) and compromised GFP-FtsI localization (Fig. 3A, panel viii), similar to the situation observed with the ΔmtrB strain (Fig. 3A, panel iv).
MtrBH305Y overproduction led to an ∼7-fold increase in Wag31-mCherry septal association (Fig. 4A, compare panels viii and ii). In contrast, the septal Wag31-mCherry localization in the cells overproducing MtrB was only ∼2-fold higher than that of the WT strain (Fig. 4A, compare panels ii and vi). Together, these data suggest that while FtsI activity and localization are responsive to MtrB levels and phosphorylation, Wag31 localization was markedly increased in the absence of MtrB activity and/or in cells enriched for the production of the phosphorylation-defective MtrB.
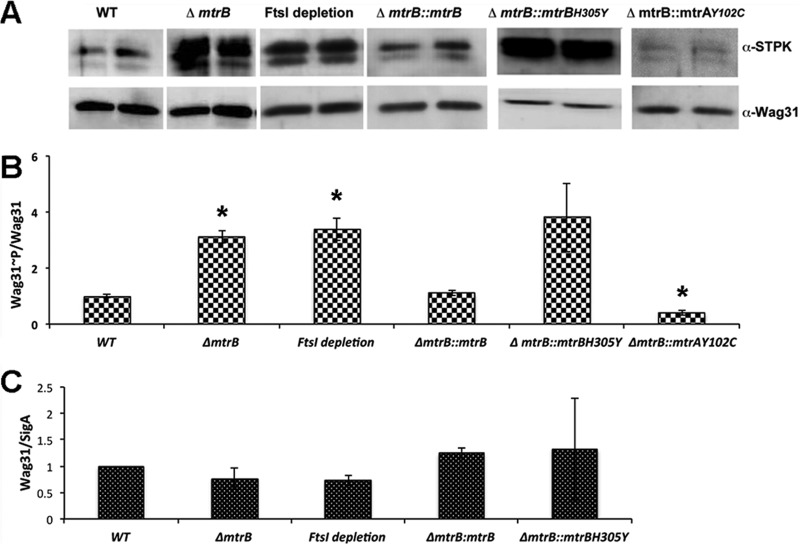
MtrB modulates Wag31 phosphorylation.
Wag31 is phosphorylated directly by serine-threonine protein kinase (STPK) PknA or indirectly by PknB through PknA (21). Alterations in the Wag31 levels and its phosphorylation status are associated with defects in Wag31 localization, cell wall synthesis, and cell shape maintenance (21). The filamentous and bulgy phenotype of the mtrB mutant combined with sustained Wag31-mCherry localization at the unsplit septa in the ΔmtrB strain background indicates that the Wag31∼P/Wag31 level (Wag31∼P-to-Wag31 ratio) may be altered. Accordingly, the Wag31∼P/Wag31 levels were determined in whole-cell lysates of ΔmtrB cells by immunoblotting with anti-phospho-Ser/Thr and anti-Wag31 antibodies. A 3-fold increase in the Wag31∼P/Wag31 level in the ΔmtrB strain compared to that in the WT strain was noted (Fig. 5A and B). Wag31∼P was restored to WT levels upon production of the phosphorylation-competent MtrB in the ΔmtrB strain background (Fig. 5A and B; see also Table S1 in the supplemental material). However, overproduction of the phosphorylation-defective MtrBH305Y did not restore WT Wag31∼P levels, indicating that MtrB∼P activity is required for maintaining optimal Wag31∼P levels (Fig. 5A and B).
FIG 5.
Loss of MtrB or depletion of FtsI increases phosphorylation of Wag31. (A) M. smegmatis WT, the ΔmtrB strain, FtsI-depleted cultures (FtsI depletion for 12 h), and the ΔmtrB Pami::mtrB, ΔmtrB Pami::mtrAY102C (where mtrAY102C is the Y-to-C change at position 102 encoded by mtrA) and ΔmtrB Pami::mtrBH305Y strains were grown as described in the legends to Fig. 3 and 4. The complemented ΔmtrB strains were grown with 0.2% acetamide. Wag31∼P/Wag31 ratios were determined by immunoblotting with anti-phospho-Ser/Thr and anti-Wag31 antibodies. Whole-cell lysates (5 μg protein) from the above strains were resolved by SDS-PAGE in 12% gels, and immunoblotting was performed with anti-phospho-Ser/Thr antibodies. The blots were then stripped and reprobed with anti-Wag31 antibodies. Wag31∼P/Wag31 (anti-phospho-Ser/Thr and anti-Wag31) ratios for various strains were normalized to those in the WT and plotted (B). The data shown are represented as the averages ± standard errors from three independent experiments. *, P < 0.05. (C) The Wag31 levels in the indicated strains were measured by immunoblotting, as previously described (36), and normalized to SigA levels. The data are represented as the means ± standard errors from three independent experiments.
Additionally, we found that an approximately 50% reduction in FtsI levels in the ftsI conditional strain led to a ∼3.5 -fold increase in Wag31∼P, but Wag31 protein levels were not affected significantly (Fig. 5A to C). These results indicate that FtsI activity is important for Wag31∼P levels. Because mtrB transcript levels were not significantly affected upon FtsI depletion (Fig. 2C) whereas those of ftsI were compromised in the mtrB mutant background (8), it is likely that the reduced pools of FtsI in the mtrB strain background contribute to the unregulated PknA/PknB (PknA/B)-mediated phosphorylation of Wag31 (Fig. 5A). To further explore this idea, we measured Wag31∼P/Wag31 in the ΔmtrB strain producing MtrAY102C; expression of MtrAY102C results in the restoration of ftsI expression (8). Under these conditions, a nearly 60% reduction in Wag31∼P/Wag31 was noted compared to that in the WT (Fig. 5A and B). These data suggest that FtsI, in the absence of MtrB, exerts a negative regulatory effect and that MtrB and FtsI together play a critical role in maintaining balanced levels of Wag31∼P. Wag31/SigA showed only modest changes in levels under all tested conditions (Fig. 5C).
MtrB also interacts with PknA and PknB proteins.
Because PknA/B proteins phosphorylate Wag31, we next examined whether these proteins interact with either or both MtrB and FtsI through BACTH assays. MtrB was found to interact with PknA and PknB, whereas FtsI did not interact with either protein (Fig. 6). PknB also interacted with MtrBH305Y (data not shown). These results suggest that the ability of MtrB to interact with PknA/B along with Wag31 and FtsI is a necessary feature for promoting optimal Wag31 phosphorylation.
FIG 6.
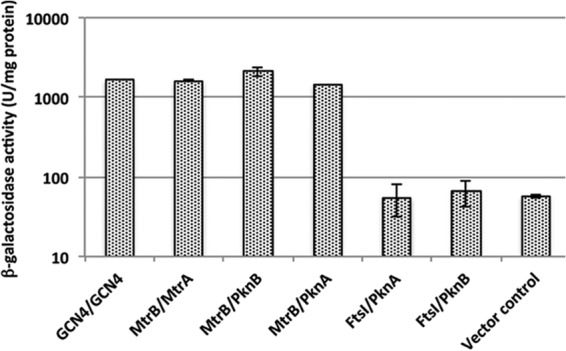
MtrB interacts with PknA and PknB. Interactions of MtrB and MtrA with PknA or PknB were examined using BACTH assays, as previously described (17). MtrA, MtrB, FtsI, PknA, and PknB fusions to the T18 and T25 fragments of adenylate cyclase in BACTH vectors (see Table S1 in the supplemental material) were used to transform E. coli BTH101, and the recombinants were plated on LB agar supplemented with X-Gal and isopropyl-β-d-thiogalactopyranoside. Green-blue colonies, indicative of positive interactions, were subsequently propagated in LB broth, and β-galactosidase activity was measured as described in the text. GCN4/GCN4 and MtrB/MtrA are shown as positive controls, and MtrB/empty vector (vector control) is shown as the negative control. The data shown are the means ± standard deviations from three independent experiments.
DISCUSSION
Cell division and cell wall synthesis are well-orchestrated events that involve an ordered assembly of multiple proteins at the septa (26). Published data indicate that in mycobacteria, as in other Gram-positive bacteria, not only the known cell division proteins, such as FtsZ, FtsI, and Wag31, but also those of the sensor kinases belonging to the TCRS and STPK system, e.g., MtrB and PknA/B, and novel small membrane proteins, e.g., CwsA and CrgA, localize at the septa (6, 8, 10, 12, 14, 27). Our data take these observations to the next level, define interactions among select septasomal components, namely, MtrB with FtsI and Wag31, and evaluate the consequences of these interactions. These studies led to two important conclusions. First, FtsI promotes MtrB activation and MtrA regulon expression and, hence, acts as a positive modulator of the MtrAB TCRS. This statement is based on the fact that the interactions of FtsI with MtrB are necessary for optimal MtrB activities, i.e., MtrB septal assembly and MtrA regulon expression. Second, MtrB interactions with FtsI, Wag31, and PknA/B are necessary for optimal Wag31∼P levels, and FtsI likely exerts a negative regulatory effect on PknA/B-mediated phosphorylation of Wag31 (see below). The widely accepted role of a histidine sensor kinase is to affect the response regulator phosphorylation and, hence, the associated target gene expression levels (28). The finding that MtrB activity is necessary for optimal PknA/B-mediated phosphorylation of Wag31 signals an additional role for the MtrB sensor kinase in addition to affecting MtrA regulon expression. A further implication of the above results is that the signal transduction processes mediated by the divergent kinases of the STPK system and TCRS converge to affect cell division and cell wall synthesis in mycobacteria (see below).
FtsI is a positive modulator of the MtrAB system.
Our results provided important clues to the order and nature of the septal assembly of FtsI, Wag31, and MtrB. For example, while FtsI and Wag31 can localize to the septa independent of MtrB, the association with MtrB influences the extent and, possibly, the stability of the localized FtsI and Wag31 proteins. In support of this finding, we showed that FtsI septal assembly and FtsI activities were compromised in the absence of MtrB and vice versa (Fig. 2A and C and 3). These results indicate that FtsI and MtrB are codependent on each other for their optimal activities. For example, the depletion of FtsI, which is a PBP with transpeptidase activity, is associated with compromised septal PG cross-linking and increased sensitivity to vancomycin (25, 29) (see also Fig. S3 in the supplemental material). FtsI depletion also compromised MtrA regulon expression (Fig. 2A), implying compromised MtrB activity under these conditions. Additionally, the absence of MtrB was found to increase both vancomycin sensitivity and Van-FL staining ability, again implying compromised FtsI activity.
On the other hand, Wag31 localization was aberrant and unregulated both in the absence of MtrB and in the mtrB mutant producing MtrBH305Y; however, MtrB septal assembly and MtrA regulon expression were unaffected under Wag31 depletion (Fig. 2 and 4; see also Fig. S4 in the supplemental material). These results imply that although Wag31 does not influence MtrB localization and activity, MtrB activity is necessary for optimal Wag31 localization. The distinct multiseptate phenotype of the ΔmtrB strain combined with the aberrant septal localization of Wag31 in the mutant cells (Fig. 4A, panels iii and iv) (8) suggests that MtrB exerts a role(s) in cell division at steps subsequent to septation and Wag31 localization. Whether this role(s) requires the interactions of MtrB with other septal proteins and/or the MtrB-mediated modulation of MtrA regulon expression remains to be determined. With regard to the latter, it is significant that the MtrA regulon includes ripA (8), rpfB, and other cell wall hydrolases (P. Gorla et al., unpublished data) needed for septum resolution. It should be noted that the expression levels of FtsI, which is an MtrA target, as per the TB database (http://www.tbdb.org/) and our recent chromatin immunoprecipitation with high-throughput sequencing (ChIP-Seq) data (30; also Gorla et al., unpublished), are shown to be compromised in the absence of MtrB (8). Thus, the possibility that compromised FtsI levels in the MtrB background contribute to altered Wag31 localization remains open. An evaluation of Wag31 localization under FtsI depletion conditions would help resolve this issue; however, our attempts to create a recombinant ftsI strain producing Wag31-mCherry to address this hypothesis have not been successful.
Our data enable us to envisage a scenario in which FtsI plays a critical role in affecting MtrB activation and MtrA regulon expression. For example, in response to unrecognized signals, MtrB is activated at the septa (8), and this could lead to de novo ftsI transcription. The FtsI produced could in turn associate with the septa and interact with and increase the half-life of MtrB∼P to trigger optimal MtrA regulon expression. When the signals promoting MtrB kinase activation are diminished, resulting in reduced levels of MtrB∼P, FtsI may disassociate from MtrB, and this could, in turn, lead to compromised MtrA activation and MtrA regulon expression. We believe that this type of braking system ensures the optimal activation of the MtrAB TCRS. Noteworthy in this regard is the recent finding that the lipoprotein LpqB interacts with MtrB and modulates MtrA phosphorylation activity (31). Together, these data indicate that FtsI and LpqB are two MtrB-interacting proteins that promote MtrB activities. While our results emphasize the importance of the MtrB and FtsI interactions, we do not yet know whether other proteins affect these interactions. Further studies are required to identify the potential mediators that stabilize MtrB and FtsI interactions at septa.
Wag31 and MtrAB system.
Our data showing that Wag31∼P levels are elevated in the backgrounds of either the ΔmtrB mutant or the ΔmtrB mutant producing MtrBH305Y or under FtsI depletion conditions but are reduced in the ΔmtrB strain overproducing MtrAY102C (Fig. 5) suggest that FtsI functions as a negative regulator of PknA/B-mediated Wag31 phosphorylation, whereas MtrB (MtrB∼P) functions to negate FtsI interference effects by interacting with it. Thus, another consequence of the MtrB interactions with FtsI and Wag31 is the modulation of the Wag31 phosphorylation potential. Because MtrB also interacts with PknA/B proteins (Fig. 6), we conclude that MtrB mediates optimal Wag31 phosphorylation by virtue of its interactions with FtsI, PknA/B, and Wag31.
Alterations in Wag31 levels and activities are associated with defects in cell shape and cell division in mycobacteria (12, 13), and Wag31∼P promotes protein-protein interactions between the Wag31 molecules and therefore promotes Wag31 localization, regulated cell wall synthesis, and growth rate (21, 32). Accordingly, we envisage that the observed cell division and cell shape maintenance defects in the mtrB strain background are, in part, due to aberrant Wag31 localization and Wag31∼P. Thus, modulation of Wag31∼P by MtrB signals that MtrB kinase has a direct role in cell division and cell wall synthesis that is distinct from its role in MtrA regulon expression.
Published data indicate that the PknA/B STPK and the MtrAB TCRS signal transduction pathways individually affect cell division and cell wall synthesis in mycobacteria (8, 21, 31). The results presented in this study go further and show how these two divergent signal transduction pathways converge at the Wag31 phosphorylation step and affect cell division and cell wall synthesis. PknB has many targets, including PBPA (27), and it remains to be tested whether MtrB's effects are specific to Wag31 or also extend to other proteins as well. Unraveling the regulation and interactions of these pathways will provide valuable insights into M. tuberculosis proliferation and pathogenesis.
The YycFG (also called WalKR) TCRS is similar to the MtrAB TCRS and is well conserved in low-G+C rich Gram-positive bacteria, including Bacillus subtilis, Staphylococcus aureus, Streptococcus pneumoniae, Streptococcus pyogenes, Listeria monocytogenes, and Enterococcus faecalis (reviewed in reference 33). YycG kinase, like MtrB, associates with the septa, and its septal association is essential for its activation and for controlling the expression of several genes involved in cell wall metabolism and turnover (34, 35). Pertinent to this study is the observation that the YycG kinase interacts with PbpB (FtsI) and with other proteins, such as FtsL and DivIB (34). These studies also showed that the septal localization of YycG is not affected by the depletion of DivIB and FtsL and that studies with PbpB depletion are not conclusive. Nonetheless, our data show distinct differences between the MtrB and YycG kinases. For example, unlike YycG, MtrB interacts with and modulates Wag31 (DivIVA is the Wag31 counterpart in low-G+C Gram-positive bacteria) phosphorylation, and, importantly, both FtsI and MtrB are codependent on each other for their activities and the septal localizations. Thus, the MtrB kinase and the MtrAB system have evolved in mycobacteria to fit their complex life style.
Supplementary Material
ACKNOWLEDGMENTS
We thank Joyoti Basu for antibodies to FtsI and C. Kang for the wag31 conditional mutant strain.
This work was supported, in whole or in part, by National Institutes of Health grants AI48417 (to M.R.) and AI084734 (to M.V.M.) and by Polish Ministry of Science and Higher Education grant IP2011 042571 (to R.P).
Footnotes
Published ahead of print 15 September 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.01795-14.
REFERENCES
- 1.Lock RL, Harry EJ. 2008. Cell-division inhibitors: new insights for future antibiotics. Nat. Rev. Drug Discov. 7:324–338. 10.1038/nrd2510. [DOI] [PubMed] [Google Scholar]
- 2.Haydel SE, Malhotra V, Cornelison GL, Clark-Curtiss JE. 2012. The prrAB two-component system is essential for Mycobacterium tuberculosis viability and is induced under nitrogen-limiting conditions. J. Bacteriol. 194:354–361. 10.1128/JB.06258-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zahrt TC, Deretic V. 2000. An essential two-component signal transduction system in Mycobacterium tuberculosis. J. Bacteriol. 182:3832–3838. 10.1128/JB.182.13.3832-3838.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Al Zayer M, Stankowska D, Dziedzic R, Sarva K, Madiraju MV, Rajagopalan M. 2011. Mycobacterium tuberculosis mtrA merodiploid strains with point mutations in the signal-receiving domain of MtrA exhibit growth defects in nutrient broth. Plasmid 65:210–218. 10.1016/j.plasmid.2011.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fol M, Chauhan A, Nair NK, Maloney E, Moomey M, Jagannath C, Madiraju MV, Rajagopalan M. 2006. Modulation of Mycobacterium tuberculosis proliferation by MtrA, an essential two-component response regulator. Mol. Microbiol. 60:643–657. 10.1111/j.1365-2958.2006.05137.x. [DOI] [PubMed] [Google Scholar]
- 6.Plocinski P, Arora N, Sarva K, Blaszczyk E, Qin H, Das N, Plocinska R, Ziolkiewicz M, Dziadek J, Kiran M, Gorla P, Cross TA, Madiraju M, Rajagopalan M. 2012. Mycobacterium tuberculosis CwsA interacts with CrgA and Wag31 and the CrgA-CwsA complex is involved in peptidoglycan synthesis and cell shape determination. J. Bacteriol. 194:6398–6409. 10.1128/JB.01005-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rajagopalan M, Dziedzic R, Al Zayer M, Stankowska D, Ouimet MC, Bastedo DP, Marczynski GT, Madiraju MV. 2010. Mycobacterium tuberculosis origin of replication and the promoter for immunodominant secreted antigen 85B are the targets of MtrA, the essential response regulator. J. Biol. Chem. 285:15816–15827. 10.1074/jbc.M109.040097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Plocinska R, Purushotham G, Sarva K, Vadrevu IS, Pandeeti EV, Arora N, Plocinski P, Madiraju MV, Rajagopalan M. 2012. Septal localization of the Mycobacterium tuberculosis MtrB sensor kinase promotes MtrA regulon expression. J. Biol. Chem. 287:23887–23899. 10.1074/jbc.M112.346544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Datta P, Dasgupta A, Bhakta S, Basu J. 2002. Interaction between FtsZ and FtsW of Mycobacterium tuberculosis. J. Biol. Chem. 277:24983–24987. 10.1074/jbc.M203847200. [DOI] [PubMed] [Google Scholar]
- 10.Datta P, Dasgupta A, Singh AK, Mukherjee P, Kundu M, Basu J. 2006. Interaction between FtsW and penicillin-binding protein 3 (PBP3) directs PBP3 to mid-cell, controls cell septation and mediates the formation of a trimeric complex involving FtsZ, FtsW and PBP3 in mycobacteria. Mol. Microbiol. 62:1655–1673. 10.1111/j.1365-2958.2006.05491.x. [DOI] [PubMed] [Google Scholar]
- 11.Hett EC, Chao MC, Steyn AJ, Fortune SM, Deng LL, Rubin EJ. 2007. A partner for the resuscitation-promoting factors of Mycobacterium tuberculosis. Mol. Microbiol. 66:658–668. 10.1111/j.1365-2958.2007.05945.x. [DOI] [PubMed] [Google Scholar]
- 12.Kang CM, Nyayapathy S, Lee JY, Suh JW, Husson RN. 2008. Wag31, a homologue of the cell division protein DivIVA, regulates growth, morphology and polar cell wall synthesis in mycobacteria. Microbiology 154:725–735. 10.1099/mic.0.2007/014076-0. [DOI] [PubMed] [Google Scholar]
- 13.Nguyen L, Scherr N, Gatfield J, Walburger A, Pieters J, Thompson CJ. 2007. Antigen 84, an effector of pleiomorphism in Mycobacterium smegmatis. J. Bacteriol. 189:7896–7910. 10.1128/JB.00726-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Plocinski P, Ziolkiewicz M, Kiran M, Vadrevu SI, Nguyen HB, Hugonnet J, Veckerle C, Arthur M, Dziadek J, Cross TA, Madiraju M, Rajagopalan M. 2011. Characterization of CrgA, a new partner of the Mycobacterium tuberculosis peptidoglycan polymerization complexes. J. Bacteriol. 193:3246–3256. 10.1128/JB.00188-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rajagopalan M, Atkinson MA, Lofton H, Chauhan A, Madiraju MV. 2005. Mutations in the GTP-binding and synergy loop domains of Mycobacterium tuberculosis ftsZ compromise its function in vitro and in vivo. Biochem. Biophys. Res. Commun. 331:1171–1177. 10.1016/j.bbrc.2005.03.239. [DOI] [PubMed] [Google Scholar]
- 16.Sureka K, Hossain T, Mukherjee P, Chatterjee P, Datta P, Kundu M, Basu J. 2010. Novel role of phosphorylation-dependent interaction between FtsZ and FipA in mycobacterial cell division. PLoS One 5:e8590. 10.1371/journal.pone.0008590. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 17.Dziedzic R, Kiran M, Plocinski P, Ziolkiewicz M, Brzostek A, Moomey M, Vadrevu IS, Dziadek J, Madiraju M, Rajagopalan M. 2010. Mycobacterium tuberculosis ClpX interacts with FtsZ and interferes with FtsZ assembly. PLoS One 5:e11058. 10.1371/journal.pone.0011058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Daniel RA, Noirot-Gros MF, Noirot P, Errington J. 2006. Multiple interactions between the transmembrane division proteins of Bacillus subtilis and the role of FtsL instability in divisome assembly. J. Bacteriol. 188:7396–7404. 10.1128/JB.01031-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Karimova G, Dautin N, Ladant D. 2005. Interaction network among Escherichia coli membrane proteins involved in cell division as revealed by bacterial two-hybrid analysis. J. Bacteriol. 187:2233–2243. 10.1128/JB.187.7.2233-2243.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chauhan A, Madiraju MV, Fol M, Lofton H, Maloney E, Reynolds R, Rajagopalan M. 2006. Mycobacterium tuberculosis cells growing in macrophages are filamentous and deficient in FtsZ rings. J. Bacteriol. 188:1856–1865. 10.1128/JB.188.5.1856-1865.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kang CM, Abbott DW, Park ST, Dascher CC, Cantley LC, Husson RN. 2005. The Mycobacterium tuberculosis serine/threonine kinases PknA and PknB: substrate identification and regulation of cell shape. Genes Dev. 19:1692–1704. 10.1101/gad.1311105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chauhan A, Lofton H, Maloney E, Moore J, Fol M, Madiraju MV, Rajagopalan M. 2006. Interference of Mycobacterium tuberculosis cell division by Rv2719c, a cell wall hydrolase. Mol. Microbiol. 62:132–147. 10.1111/j.1365-2958.2006.05333.x. [DOI] [PubMed] [Google Scholar]
- 23.Joloba ML, Bajaksouzian S, Jacobs MR. 2000. Evaluation of Etest for susceptibility testing of Mycobacterium tuberculosis. J. Clin. Microbiol. 38:3834–3836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Santi I, Dhar N, Bousbaine D, Wakamoto Y, McKinney JD. 2013. Single-cell dynamics of the chromosome replication and cell division cycles in mycobacteria. Nat. Commun. 4:2470. 10.1038/ncomms3470. [DOI] [PubMed] [Google Scholar]
- 25.Sauvage E, Kerff F, Terrak M, Ayala JA, Charlier P. 2008. The penicillin-binding proteins: structure and role in peptidoglycan biosynthesis. FEMS Microbiol. Rev. 32:234–258. 10.1111/j.1574-6976.2008.00105.x. [DOI] [PubMed] [Google Scholar]
- 26.Adams DW, Errington J. 2009. Bacterial cell division: assembly, maintenance and disassembly of the Z ring. Nat. Rev. Microbiol. 7:642–653. 10.1038/nrmicro2198. [DOI] [PubMed] [Google Scholar]
- 27.Dasgupta A, Datta P, Kundu M, Basu J. 2006. The serine/threonine kinase PknB of Mycobacterium tuberculosis phosphorylates PBPA, a penicillin-binding protein required for cell division. Microbiology 152:493–504. 10.1099/mic.0.28630-0. [DOI] [PubMed] [Google Scholar]
- 28.Stock AM, Robinson VL, Goudreau PN. 2000. Two-component signal transduction. Annu. Rev. Biochem. 69:183–215. 10.1146/annurev.biochem.69.1.183. [DOI] [PubMed] [Google Scholar]
- 29.Vollmer W, Blanot D, de Pedro MA. 2008. Peptidoglycan structure and architecture. FEMS Microbiol. Rev. 32:149–167. 10.1111/j.1574-6976.2007.00094.x. [DOI] [PubMed] [Google Scholar]
- 30.Galagan JE, Minch K, Peterson M, Lyubetskaya A, Azizi E, Sweet L, Gomes A, Rustad T, Dolganov G, Glotova I, Abeel T, Mahwinney C, Kennedy AD, Allard R, Brabant W, Krueger A, Jaini S, Honda B, Yu WH, Hickey MJ, Zucker J, Garay C, Weiner B, Sisk P, Stolte C, Winkler JK, Van de Peer Y, Iazzetti P, Camacho D, Dreyfuss J, Liu Y, Dorhoi A, Mollenkopf HJ, Drogaris P, Lamontagne J, Zhou Y, Piquenot J, Park ST, Raman S, Kaufmann SH, Mohney RP, Chelsky D, Moody DB, Sherman DR, Schoolnik GK. 2013. The Mycobacterium tuberculosis regulatory network and hypoxia. Nature 499:178–183. 10.1038/nature12337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nguyen HT, Wolff KA, Cartabuke RH, Ogwang S, Nguyen L. 2010. A lipoprotein modulates activity of the MtrAB two-component system to provide intrinsic multidrug resistance, cytokinetic control and cell wall homeostasis in Mycobacterium. Mol. Microbiol. 76:348–364. 10.1111/j.1365-2958.2010.07110.x. [DOI] [PubMed] [Google Scholar]
- 32.Jani C, Eoh H, Lee JJ, Hamasha K, Sahana MB, Han JS, Nyayapathy S, Lee JY, Suh JW, Lee SH, Rehse SJ, Crick DC, Kang CM. 2010. Regulation of polar peptidoglycan biosynthesis by Wag31 phosphorylation in mycobacteria. BMC Microbiol. 10:327. 10.1186/1471-2180-10-327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dubrac S, Bisicchia P, Devine KM, Msadek T. 2008. A matter of life and death: cell wall homeostasis and the WalKR (YycGF) essential signal transduction pathway. Mol. Microbiol. 70:1307–1322. 10.1111/j.1365-2958.2008.06483.x. [DOI] [PubMed] [Google Scholar]
- 34.Fukushima T, Furihata I, Emmins R, Daniel RA, Hoch JA, Szurmant H. 2011. A role for the essential YycG sensor histidine kinase in sensing cell division. Mol. Microbiol. 79:503–522. 10.1111/j.1365-2958.2010.07464.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fukushima T, Szurmant H, Kim EJ, Perego M, Hoch JA. 2008. A sensor histidine kinase co-ordinates cell wall architecture with cell division in Bacillus subtilis. Mol. Microbiol. 69:621–632. 10.1111/j.1365-2958.2008.06308.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Plocinski P, Martinez L, Sarva K, Plocinska R, Madiraju M, Rajagopalan M. 2013. Mycobacterium tuberculosis CwsA overproduction modulates cell division and cell wall synthesis. Tuberculosis 93(Suppl):S21–S27. 10.1016/S1472-9792(13)70006-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.