Abstract
Peptidoglycan (PG) is an extracytoplasmic glycopeptide matrix essential for the integrity of the envelope of most bacteria. The PG building block is a disaccharide-pentapeptide that is synthesized as a lipid-linked precursor called lipid II. The translocation of the amphipathic lipid II across the cytoplasmic membrane is required for subsequent incorporation of the disaccharide-pentapeptide into PG. In Escherichia coli, the essential inner membrane protein MurJ is the lipid II flippase. Previous studies showed that 8 charged residues in the central cavity region of MurJ are crucial for function. Here, we completed the functional analysis of all 57 charged residues in MurJ and demonstrated that the respective positive or negative charge of the 8 aforementioned residues is required for proper MurJ function. Loss of the negative charge in one of these residues, D39, causes a severe defect in MurJ biogenesis; by engineering an intragenic suppressor mutation that restores MurJ biogenesis, we found that this charge is also essential for MurJ function. Because of the low level of homology between MurJ and putative orthologs from Gram-positive bacteria, we explored the conservation of these 8 charged residues in YtgP, a homolog from Streptococcus pyogenes. We found that only 3 positive charges are similarly positioned and essential in YtgP; YtgP possesses additional charged residues within its predicted cavity that are essential for function and conserved among Gram-positive bacteria. From these data, we hypothesize that some charged residues in the cavity region of MurJ homologs are required for interaction with lipid II and/or energy coupling during transport.
INTRODUCTION
Most bacteria produce a rigid peptidoglycan (PG) layer that is essential for defining cell shape and providing protection against osmotic lysis (1, 2). This mesh-like PG macromolecule consists of glycan strands of repeating N-acetylglucosamine (GlcNAc) and N-acetylmuramic acid (MurNAc) disaccharide units that are bridged by the cross-linking of peptide stems extending from the MurNAc moiety. The PG layer is localized outside the cytoplasmic membrane, and as the cell grows, newly synthesized PG material must be incorporated into the preexisting polymer. Since the disaccharide-pentapeptide building blocks are made in the cytoplasm, the cytoplasmic membrane separates PG precursor synthesis from its utilization. Therefore, transport of the disaccharide-pentapeptide across the membrane is an essential step in PG biogenesis.
In Escherichia coli, PG is present in the periplasm, the aqueous compartment between the inner and outer membranes (3). Like in all PG-producing bacteria, the disaccharide-pentapeptide PG precursor is synthesized onto the lipid carrier undecaprenol at the inner leaflet of the inner membrane (IM) (reviewed in references 4–7). This lipid-linked precursor, known as lipid II (undecaprenyl-pyrophosphoryl-MurNAc-[pentapeptide]-GlcNAc), must be flipped across the IM and delivered to the outer leaflet of the IM. Once lipid II is exposed to the periplasm, the disaccharide-pentapeptide is removed from the undecaprenyl-pyrophosphate carrier and linked to a nascent glycan chain by transglycosylases; stem peptides in adjacent glycan chains then are cross-linked by transpeptidases. The lipid carrier is recycled back to the inner leaflet of the IM for use in another round of lipid II synthesis or in another glycopolymer biosynthetic pathway.
The translocation of lipid II across the cytoplasmic membrane is poorly understood. Because of the amphipathic nature of lipid II, its transport through the membrane must be mediated by a membrane protein called a flippase, which is thought to protect the hydrophilic moiety of lipid II from the hydrophobic core of the IM (8). The IM protein MurJ is essential for PG biogenesis and functions as the lipid II flippase (9–11). In agreement with this function, (i) MurJ depletion leads to the accumulation of PG precursors and cell lysis (10–12); (ii) MurJ belongs to the MVF family of the multidrug/oligosaccharidyl-lipid/polysaccharide (MOP) exporter superfamily, which includes other flippases (13); (iii) MurJ contains a central hydrophilic cavity within the hydrophobic core of the membrane (14); (iv) residues within this hydrophilic cavity are essential for MurJ function (14); and (v) chemical inactivation of MurJ function rapidly stops lipid II flipping and leads to the accumulation of lipid II at the inner leaflet of the IM (9). Although we do not understand how MurJ functions in lipid II translocation, it likely shares functional features with members of the MOP exporter superfamily.
The best understood members of the MOP exporter superfamily are multidrug transporters that export substrates via a substrate-cation antiport mechanism (13, 15). The crystal structures for several MOP exporters have been reported, demonstrating a conserved V-shaped structure comprised of 12 transmembrane domains arranged in 2 six-helical bundles around a central, hydrophilic cavity that is essential for efflux (16–19). Comparison of structures in different states of cation and/or substrate binding suggests that the mechanistic details of antiport are not well conserved. These exporters differ in where their substrate-binding site is located, as well as in their predicted mechanism for coupling substrate export to ion influx. Further, comparison of essential residues identified in MOP exporters reveals little conservation (20). Thus, even if all MOP exporters function by a cation-driven antiport mechanism, each is likely to have specific requirements because of the diversity of substrates translocated by these proteins.
Given that lipid II flippases from different PG-producing bacteria export very similar substrates, we expect some conservation of functional requirements between these proteins. Recent work has demonstrated that a MurJ homolog from the Gram-negative betaproteobacterium Burkholderia cenocepacia (MurJBC) is essential for PG biogenesis and predicted to be a structural homolog of E. coli MurJ (12). Furthermore, the two proteins are functionally interchangeable and share 54% identity (determined by Clustal Omega alignment [21]). The essentiality of 5 charged residues that are homologous to essential residues located within the cavity of E. coli MurJ (14) has been investigated in MurJBC; of these residues, only 3 are essential for MurJBC function in B. cenocepacia (12). Thus, evidence exists for both conserved and organism-specific requirements for MurJ activity.
YtgP, a distant MurJ ortholog from the Gram-positive bacterium Streptococcus pyogenes, also complements depletion of MurJ in E. coli (22) despite having low homology to MurJ at the amino acid sequence level (21.7% identity as determined by Clustal Omega alignment [21]). This experimental system provides an opportunity to test for functional requirements in dissimilar lipid II flippases. Here, we show that the only charged residues required for the function of E. coli MurJ are located within its cavity region. By comparing MurJ and YtgP, we further demonstrate that both proteins require specific charges for function, some of which are unique to each protein. Our findings have implications for how MurJ might function to translocate lipid II across the membrane.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Strains are listed in Table S3 in the supplemental material. With the exception of DH5α, all strains are derived from NR754, an araD+ revertant of MC4100 (23, 24). Liquid cultures were grown under aeration at 37°C in lysogeny broth (LB) or glucose M63 minimal broth, as indicated. Growth was monitored by optical density at 600 nm (OD600). LB and glucose M63 minimal broth and agar were prepared as described previously (25). Yeast tryptone (YT) agar contains tryptone (10 g/liter), yeast extract (5 g/liter), and agar (15 g/liter). When appropriate, chloramphenicol (20 μg/ml), arabinose (0.02%), isopropyl-β-d-thiogalactopyranoside (IPTG; 0.04 mM or 0.06 mM, as indicated), and 5-bromo-4-chloro-indolyl-β-d-galactopyranoside (X-Gal; 20 μg/ml) were added.
Plasmid construction.
All primers used in this study are listed in Table S4 in the supplemental material. Plasmid pFLAGMurJΔCys encodes a functional N-terminally FLAG-tagged MurJ with the two native Cys residues altered to Ser (C314S and C419S), as required for our previous topology study (14). The gene encoding Spy49_0320 (YP_002285353.1) from S. pyogenes NZ131 (GenBank accession no. NC_011375.1) was introduced into pCA24Not as described previously (22) to generate pHIS-YtgP.
Amino acid substitutions in FLAG-MurJΔCys and His-YtgP were generated by site-directed mutagenesis (SDM) PCR (95°C for 2 min, followed by 18 cycles of 96°C for 1 min, 56°C or 60°C for 1 min, and 72°C for 12 min, and then a final extension of 12 min at 72°C) using Pfu Turbo polymerase (Agilent Technologies) per the manufacturer's instructions, with pFLAGMurJΔCys (14) and pHIS-YtgP as templates. Variants generated in pFLAGMurJΔCys were introduced into strain NR2117; variants generated in pHIS-YtgP were introduced into DH5α. All transformants were selected on media containing chloramphenicol.
Testing for function of FLAG-murJΔcys and His-ytgP alleles.
NR2117 strains carrying pFLAGMurJΔCys derivatives were evaluated for complementation as described previously (14). Briefly, NR2117 carries the plasmid pRC7MurJ (14), which encodes wild-type murJ and has a partitioning defect that causes its rapid loss from the population in the absence of selection. Since murJ is essential for viability, daughter cells of a ΔmurJ (pRC7MurJ) strain, such as NR2117, that do not inherit a copy of pRC7MurJ do not survive; however, this selection can be alleviated in the presence of a compatible plasmid encoding a functional murJ allele. Therefore, mutant alleles carried by pFLAGMurJΔCys were assessed for complementation after they were introduced into NR2117 by monitoring the loss of pRC7MurJ. Because pRC7MurJ also encodes β-galactosidase, its loss was monitored by the appearance of white colonies in the presence of X-Gal. Derivatives of pFLAGMurJΔCys encoding a functional murJ allele yield white colonies, while those that encode a nonfunctional murJ allele yield blue colonies. Strains expressing functional FLAG-murJΔcys alleles first were evaluated for growth defects in low-osmolarity medium (YT agar) by examining relative growth and colony morphology compared to those of the parent strain after overnight growth. Strains demonstrating growth defects were evaluated further by efficiency-of-plating assay as follows. Overnight cultures were grown in LB or glucose minimal media as indicated in the text. Approximately 2 μl of 10-fold serial dilutions of these cultures was spotted onto LB and YT agar plates using a 48-pin manifold. Following overnight growth, efficiency-of-plating values were calculated by dividing the last dilution yielding growth for each strain under each condition by the number of the last dilution of the parent culture that yielded growth on LB agar. Data represent the averages ± standard errors of the means from three independent experiments.
To test for functionality of His-ytgP alleles, pHIS-YtgP-derived plasmids were introduced into the MurJ depletion strain NR1152, which requires the inducer arabinose to grow (11). Viability was compared to that of NR1152 carrying the parent plasmid pHIS-YtgP and the vector control pCA24Not (22), which is a derivative of pCA24N (26) lacking gfp. Cells grown overnight in LB containing arabinose were washed once with LB and used for efficiency of plating as described above under conditions permissive for murJ expression (LB containing arabinose) and under MurJ depletion conditions with the induction of expression of the plasmid-carried His-ytgP alleles (LB with 0.04 or 0.06 mM IPTG).
Immunoblotting.
For detection of FLAG-MurJ variants, cells were grown overnight in LB. Culture volumes (in μl) normalized by dividing 400 by OD600 values were collected by centrifugation, and cells were resuspended in 100 μl of 1× AB buffer (3.43 mM Na2HPO4, 1.58 mM NaH2PO4, 25 mM Tris-HCl, pH 6.8, 3 M urea, 0.5% β-mercaptoethanol, 1.5% SDS, 5% glycerol, 0.05% bromophenol blue) (14). Samples were incubated for 30 min at 45°C prior to loading onto a 12% SDS-polyacrylamide gel for electrophoresis. Proteins were transferred to nitrocellulose membranes in a semidry transfer apparatus (Bio-Rad) and probed with mouse anti-FLAG M2 (1:10,000; Sigma-Aldrich) and anti-mouse horseradish peroxidase (HRP; 1:10,000; GE Amersham)-conjugated antibodies. Signal was developed using the Clarity Western ECL substrate according to the manufacturer's instructions (Bio-Rad) and detected using a ChemiDoc XRS+ system (Bio-Rad). Without further treatment, membranes then were reprobed with rabbit anti-LptB (1:50,000; our laboratory collection) and HRP-conjugated anti-rabbit (1:10,000; GE Amersham) antibodies to control for sample loading, and signal was developed as described above.
For detection of His-YtgP variants, cells were collected from logarithmically growing cultures which had been induced for His-ytgP expression for 1 h by addition of 0.04 mM IPTG. Cells were normalized by OD600, resuspended in 50 μl of 1× AB buffer, and incubated for 30 min at 45°C. SDS-polyacrylamide gel electrophoresis and immunoblotting were performed as described above using a mouse anti-His monoclonal (1:1,000; GE Healthcare Life Sciences) and HRP-conjugated anti-mouse (1:10,000; GE Amersham) antibodies.
In silico modeling and structural alignments.
The MurJ model structure was previously described (14). The amino acid sequence of S. pyogenes YtgP (YP_002285353.1) was submitted to the I-TASSER web server (http://zhanglab.ccmb.med.umich.edu/I-TASSER/) on 13 February 2014 (27). PDB files from I-TASSER were used to generate figures in the PyMOL molecular graphics system (Schrödinger, LLC, Portland, Oregon). CEalign was used to perform structure alignments (28).
RESULTS
Charged residues that are essential in MurJ are restricted to the cavity region.
We previously conducted a structure-function analysis of 110 variants of FLAG-MurJΔCys (which, for simplicity, we refer to as FLAG-MurJ) bearing single Cys substitutions (14). We found only 8 substitutions that resulted in either a total or a severe loss of MurJ function. These 8 residues are charged and located in the central cavity region of MurJ. To determine whether a role for charged amino acids in MurJ function is restricted to those localized in the cavity region, we analyzed the remaining 31 (out of 57 total) R, K, E, or D residues within MurJ (see Fig. S1A in the supplemental material).
Derivatives of pFLAGMurJΔCys carrying alleles that encode single Cys substitutions in FLAG-MurJ were tested for their ability to complement a ΔmurJ chromosomal allele in an established complementation system described in Materials and Methods (14). Through this complementation analysis, we previously reported that 5 variants carrying substitutions at R18C, R24C, D39C, R52C, and R270C were nonfunctional (14). Here, we did not find any additional total-loss-of-function alleles among the new 31 alleles tested.
We further screened haploid strains producing each of the 31 complementing Cys substitution variants for a partial loss of function by monitoring their growth under PG stress. By comparing growth and colony morphology on YT agar, a low-osmolarity media, we previously found that the K46C, D269C, and E273C substitutions resulted in a partial loss of function (14). Here, we identified R312C in transmembrane domain (TMD) 9 as an additional substitution that partially reduces FLAG-MurJ function (see Fig. S1A and B in the supplemental material). The observed defect in FLAG-MurJ/R312C biogenesis likely underlies the partial loss-of-function phenotype (see Fig. S1C); therefore, we did not pursue further characterization of R312 in this work.
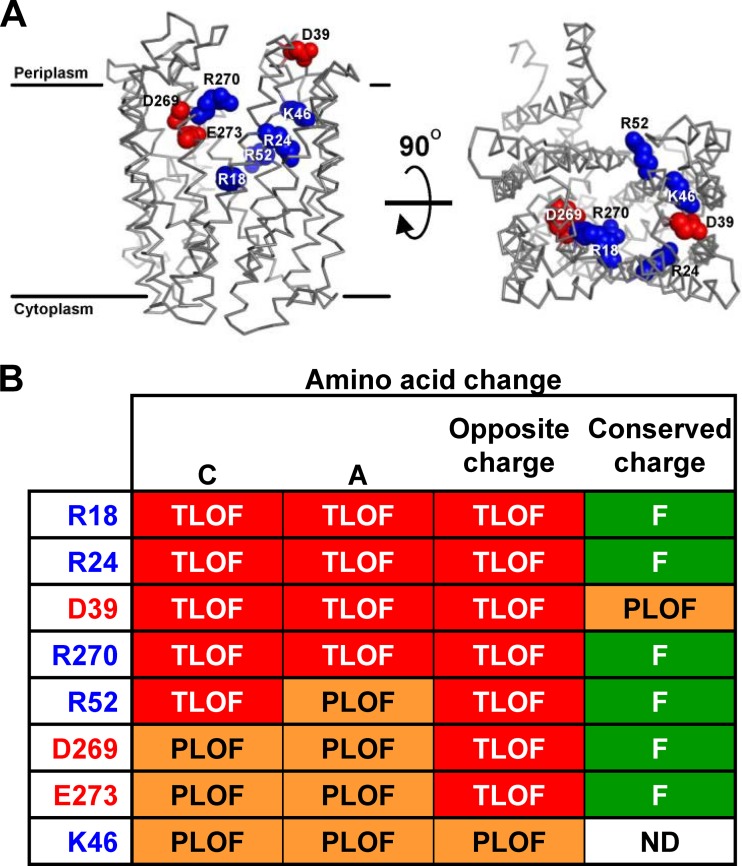
The first 12 TMDs of MurJ form a structure homologous to the canonical V-shaped structure of MOP exporters, containing a solvent-exposed, central cavity primarily defined by TMDs 1, 2, 7, and 8 (Fig. 1A) (14). Seven of the 13 charged residues within those cavity-lining TMDs are important for MurJ function (R18, R24, K46, R52, D269, R270, and E273), while another essential residue, D39, lies in periplasmic loop 1 (Fig. 1). Thus, charged residues that are critical for MurJ function are located strictly in the cavity region. Because the hydrophilic cavity of flippases is likely to serve as the conduit for substrate transport (8, 29), we investigated further the requirement for these charged residues.
FIG 1.
Specific charges in the cavity region of MurJ are required for function. (A) Structural model of MurJ (14) showing charged residues in the cavity region that are critical for function. Shown are the front view from the membrane plane with approximate membrane boundaries marked (left) and the top view from the periplasm (right). Relevant positively (blue) and negatively (red) charged side chains are rendered as spheres. (B) Summary of the effect on MurJ function conferred by Ala and Cys substitutions (14) and substitutions to the opposite (R/K to E or D/E to R/K) or conserved (R to K or D/E to E/D) charge at each residue (left column) shown in panel A. Phenotypes are given as total loss of function (TLOF) if the allele does not complement a ΔmurJ chromosomal deletion, partial loss of function (PLOF) if the allele complements a ΔmurJ chromosomal deletion but confers sensitivity to low-osmolarity conditions, and functional (F) if the allele does not confer any mutant phenotype. Phenotypes were assessed as described in Materials and Methods. Data for Cys substitutions are described by Butler et al. (14); data for changes to Ala and the opposite and conserved charge are described in Results and Tables S1 and S2 in the supplemental material. ND, not done.
The native charges of specific residues in MurJ are required for function.
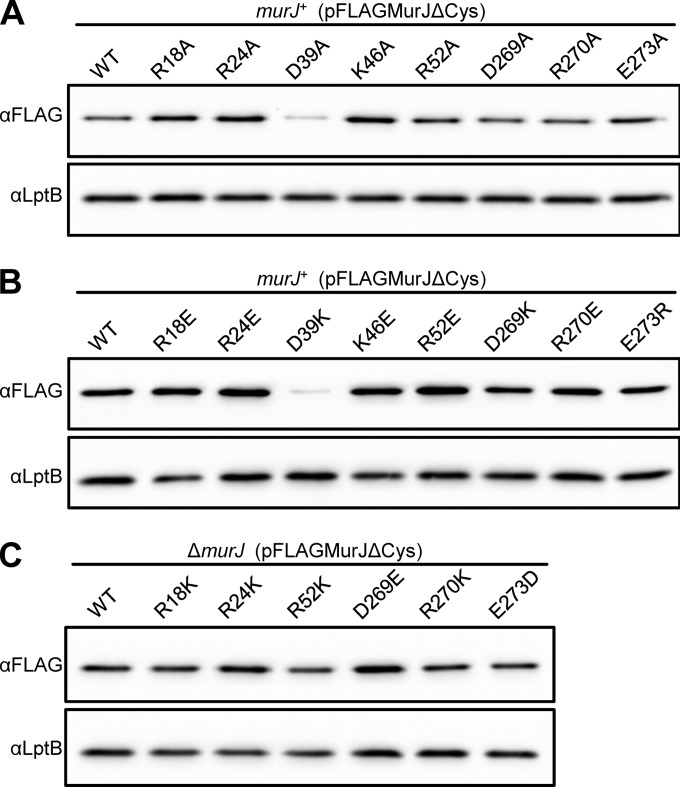
The replacement of the aforementioned 8 residues within the cavity region of MurJ with either Cys or Ala severely compromises MurJ function (Fig. 1B) (14). Of these, only alterations of D39 to Cys (14) or Ala (Fig. 2A) caused a reduction in FLAG-MurJ protein levels. Thus, R18, R24, K46, R52, D269, R270, and E273 play a crucial role in MurJ function but not biogenesis. However, the total loss of function caused by the D39A change (Fig. 1B) cannot be attributed solely to the observed decrease in protein levels because, as seen with the R312C variant (see Fig. S1C in the supplemental material) and other examples to follow, FLAG-MurJ variants that are present at levels lower than those of the D39A variant still can be partially functional. Therefore, although D39 is important for MurJ biogenesis, it also seems to be involved in MurJ activity (see below).
FIG 2.
Detection of FLAG-MurJ variants with substitutions at charged residues crucial for MurJ function. Samples prepared from overnight cultures of merodiploid (murJ+) or haploid (ΔmurJ) strains producing FLAG-MurJ variants bearing substitutions to Ala (A), an opposite charge (B), or a conserved charge (C) were subjected to anti-FLAG immunoblotting and compared to the wild-type (WT) parent. Data are representative of at least three independent experiments. Anti-LptB immunoblotting was performed to control for sample loading.
In order to test whether the positive or negative character of the native charge of R18, R24, D39, K46, R52, D269, R270, and E273 is important for MurJ function, we made changes to residues bearing the opposite and the same charge. Changes to the opposite charge at positions R18, R24, D39, R52, D269, R270, and E273 each resulted in total loss of function, while changes to the same charge were functional (Fig. 1B). The only variant with an alteration to the opposite charge that could support viability in haploid cells encoded the K46E change (Fig. 1B). The failure of most variants to complement is not caused by defects in biogenesis, since the assessment of the effect of these changes on protein levels revealed that only the D39K substitution caused a decrease in protein levels (Fig. 2B and C).
To further evaluate the effect of functional substitutions on MurJ function, we determined the sensitivity of haploid strains to low osmolarity during growth on YT medium (14). We note that haploid strains producing K46A and D39E variants grow as small, flat colonies and accrue fast-growing suppressors on LB agar but do not exhibit growth defects in glucose minimal media; therefore, we maintained these two haploid strains and, for reference, the one producing the K46E variant on minimal media until testing on LB and YT was performed. These functionality studies revealed that haploid strains producing variants with a conserved charge at positions R18, R24, R52, D269, and R270 grew similarly to the parent strain, although a marginal defect was observed for the mutant producing the R270K variant (see Table S1 in the supplemental material). In contrast, all complementing variants with Ala substitutions and K46E (the only haploid strain that was viable with a change to the opposite charge) caused profound defects on low-osmolarity medium (see Tables S1 and S2 in the supplemental material). As expected from the growth defects described above, haploid strains producing the K46A and D39E variants also exhibited sensitivity to low osmolarity (see Table S2). Together, these data indicate that the specific native charge of cavity residues R18, R24, D39, K46, R52, D269, R270, and E273 is required for proper MurJ function (Fig. 1B).
The dual role of D39.
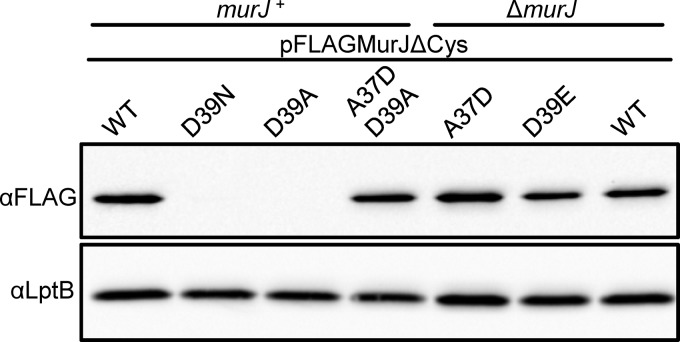
As described above, alterations of D39 to Ala or Cys cause total loss of function and a significant decrease in cellular levels of FLAG-MurJ (Fig. 1B and 2A) (14). The D39E change does not affect protein levels, but it causes a partial loss of FLAG-MurJ function (Fig. 1B and 3) (see Table S2 in the supplemental material). These results imply that D39 is important for both the biogenesis and the function of MurJ. To investigate whether the negative charge at position 39 is essential for MurJ function, we made a conserved substitution to the polar, uncharged Asn, thinking that it might not affect protein levels. The D39N variant did not complement and still had a severe defect in biogenesis (Fig. 3).
FIG 3.
Biogenesis of MurJ requires the presence of a negatively charged residue in loop 1. Protein levels of FLAG-MurJ variants in overnight cultures of merodiploid (murJ+) or haploid (ΔmurJ) strains were compared via anti-FLAG immunoblotting. The presence of a negative charge at position 39 in loop 1 is required for proper biogenesis of MurJ (compare WT and D39E to D39N and D39A). The reduction in MurJ levels caused by the D39A substitution is suppressed by the introduction of a negative charge at a different position in loop 1 (compare D39A to A37D/D39A). Data are representative of at least three independent experiments. Anti-LptB immunoblotting was performed to control for sample loading.
Negatively charged residues in periplasmic loops have been shown to be required for proper membrane insertion of some integral membrane proteins (30–33). Because D39 is located in periplasmic loop 1, we considered that this could be the reason for the decreased levels of FLAG-MurJ derivatives bearing uncharged substitutions at position 39. We reasoned that if this was the case, the addition of a negatively charged residue in loop 1 might suppress the biogenesis defect conferred by the D39A substitution. We tested an A37D substitution because, on its own, it does not affect either MurJ function or levels (Fig. 3). Remarkably, the A37D change rescued the biogenesis defect caused by D39A, as the levels of FLAG-MurJ/A37D/D39A were observed at nearly wild-type levels (Fig. 3). Importantly, the FLAG-MurJ/A37D/D39A variant did not complement the loss of murJ. As such, the biogenesis defect observed for the D39A variant can be unlinked from its defect in MurJ function. Therefore, we conclude that the negative charge in D39 has two crucial roles: to provide a negative charge in loop 1 that is necessary for proper MurJ biogenesis and to perform a yet-to-be-defined role that is essential for MurJ function.
Conservation of charge requirements across distant MurJ homologs.
The lipid-pyrophosphate-disaccharide portion of lipid II is conserved among PG-producing bacteria. However, some key differences in the composition of the stem peptide exist between lipid II from Gram-positive and Gram-negative bacteria (5, 34). In addition, there is a low level of conservation between MurJ and YtgP, the putative MurJ ortholog in Gram-positive bacteria (22, 35). Given that we have shown that 8 residues within the MurJ cavity region are critical for MurJ function in E. coli, we tested whether these requirements are conserved in YtgP by taking advantage of the fact that ytgP from S. pyogenes complements the depletion of MurJ in E. coli (22).
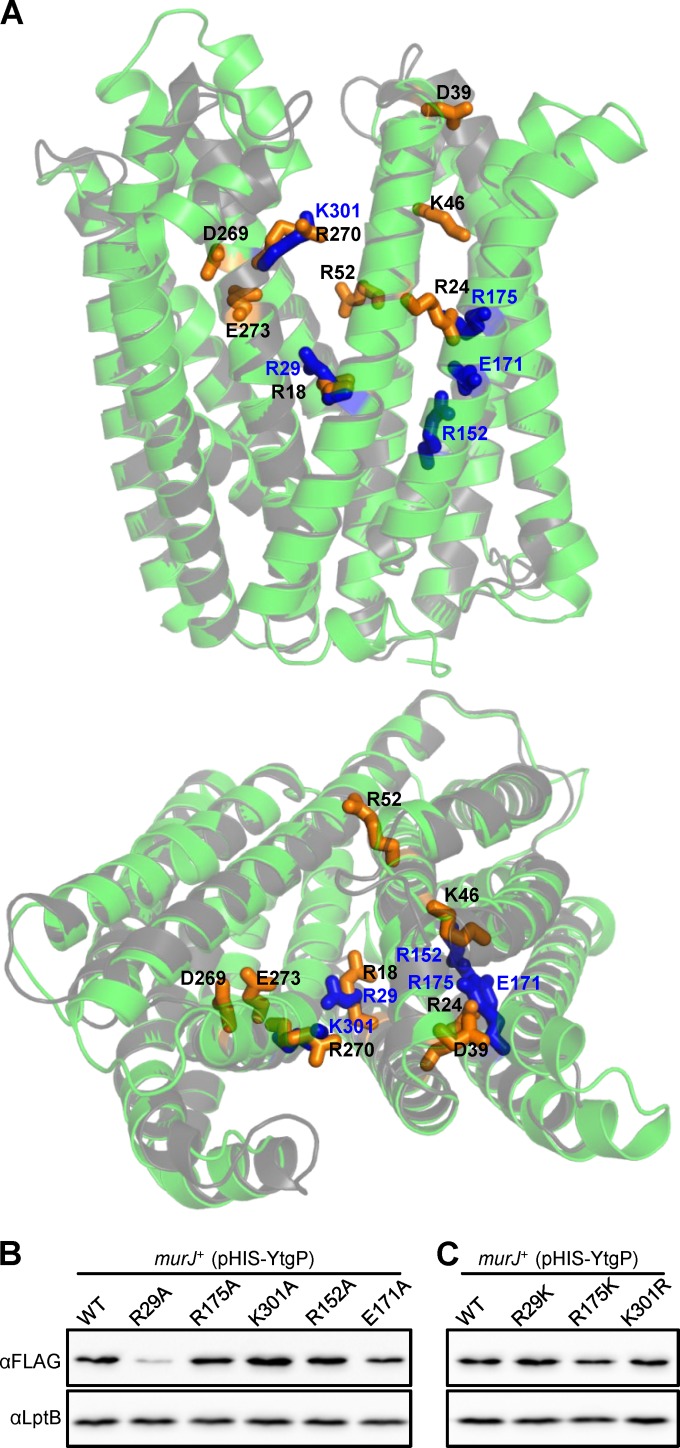
A Clustal Omega (21) amino acid sequence alignment predicts that R29 in YtgP is equivalent to R18 in MurJ; however, conservation of other charged essential residues is not apparent (see Fig. S2A in the supplemental material). Because some MOP exporters that share a low level of conservation at the amino acid sequence level have remarkably similar three-dimensional structures (16–19), we obtained a structural model of YtgP using I-TASSER (27) and compared it to that of MurJ (14). Despite the fact that MurJ and YtgP share only 21.7% sequence identity, these models reveal a high degree of structural homology in their first 12 TMDs (see Fig. S2B in the supplemental material). When aligning their structural models, we found that R29 of YtgP indeed is positioned similarly to R18 in MurJ (Fig. 4A). Further, K301 of YtgP is located in TMD 8 in the same position as R270 of MurJ. However, MurJ residues R24, D39, K46, R52, D269, and E273 are not conserved in YtgP. Interestingly, we noted that R175 in TMD 5 of YtgP is oriented such that its side chain occupies a space between TMDs 1 and 5 comparable to that of the side chain of R24 in TMD 1 of MurJ (Fig. 4A), suggesting that these two residues are functionally equivalent.
FIG 4.
Charged residues within the cavity region are required for YtgP function. (A) Alignment of the first 12 transmembrane domains of YtgP (green) and MurJ (gray) structure models showing the side chains (stick representation) of functionally important residues in MurJ (orange sticks, black labels) and YtgP (blue sticks, blue labels). The aligned structures are shown as viewed from the membrane plane (top) and from the periplasm (bottom). Refer to Fig. S2 in the supplemental material and Materials and Methods for alignment details. (B and C) Anti-His immunoblots of nonfunctional (B) and functional (C) His-YtgP variants show relative levels that are similar to those of the wild-type (WT) parent in all cases except for the R29A variant, which is present at lower levels. Data are representative of at least three independent experiments. Anti-LptB immunoblotting was performed to control for sample loading.
We addressed whether R29, R175, and K301 play a role in YtgP by constructing respective Ala substitutions in pHIS-YtgP. Variants were introduced into a MurJ depletion strain (11) and assessed for viability under conditions where MurJ was depleted and the parental His-ytgP allele complemented. We found that alleles encoding YtgP variants with the R29A, R175A, or K301A substitution did not complement (Table 1). All proteins were detected, although the levels of the R29A variant were reduced by approximately 80% (Fig. 4B). As in MurJ, we tested whether substitutions that conserve the positive charge in R29, R175, and K301 in YtgP would allow complementation. We found that the R29K, R175K, and K301R changes did not affect protein levels (Fig. 4C) and that the R175K and K301R variants complemented comparably to wild-type YtgP; the R29K variant also complemented, but it required higher levels of expression for full complementation (Table 1). Thus, these data support that R29 and K301 in YtgP are equivalent to R18 and R270, respectively, in MurJ. Moreover, although residue R175 is localized in TMD 5 of YtgP, its charge requirement for function and the predicted position of its side chain suggest that it is equivalent to R24 in TMD 1 of MurJ.
TABLE 1.
Complementation analysis of MurJ depletion by S. pyogenes ytgP alleles
| Strain | Relevant genotypeb | Efficiency of platinga |
||
|---|---|---|---|---|
| MurJ+ YtgP− | MurJ− YtgP+ (40 μM IPTG) | MurJ− YtgP+ (60 μM IPTG) | ||
| NR1305 | NR1152(pCA24Not) | 1.0 ± 0.2 | <1.2 × 10−4 | <1.2 × 10−4 |
| NR3064 | NR1152(pHIS-YtgP) | 1.0 | 0.4 ± 0.1 | 2.1 ± 1.6 |
| NR3067 | NR1152(pHIS-YtgP/R29A) | 0.9 ± 0.3 | <10−4 | <10−4 |
| NR3068 | NR1152(pHIS-YtgP/R29K) | 0.4 ± 0. 3 | 2.0 (± 1.8) × 10−3 | 0.1 ± 0.0 |
| NR3073 | NR1152(pHIS-YtgP/R152A) | 5.7 ± 4.3 | <10−4 | <10−4 |
| NR3074 | NR1152(pHIS-YtgP/E171A) | 0.9 ± 0.4 | <10−4 | <10−4 |
| NR3069 | NR1152(pHIS-YtgP/R175A) | 2.3 ± 1.5 | <10−4 | <10−4 |
| NR3070 | NR1152(pHIS-YtgP/R175K) | 1.1 ± 0.1 | 2.2 ± 1.8 | 2.0 ± 1.8 |
| NR3071 | NR1152(pHIS-YtgP/K301A) | 4.2 ± 2.9 | <10−4 | <10−4 |
| NR3072 | NR1152(pHIS-YtgP/K301R) | 2.0 ± 1.8 | 0.4 ± 0.3 | 2.0 ± 1.6 |
Efficiency of plating values for growth of the arabinose-dependent MurJ depletion strain (NR1152) expressing His-ytgP alleles carrying the indicated substitutions were calculated as described in Materials and Methods. A value of 1 (set by the wild-type allele) indicates full complementation by His-ytgP alleles; a value of <1.2 × 10−4 indicates no complementation, as this is the highest value obtained from the strain carrying the control plasmid pCA24Not; a value of <1 to >1.2 × 10−4 indicates partial complementation. For MurJ+ YtgP− conditions, cells were grown on LB plates with arabinose to induce the expression of wild-type E. coli murJ. For MurJ− YtgP+ conditions, cells were grown on LB plates with either 40 or 60 μM IPTG to induce the expression of the S. pyogenes His-ytgP alleles.
NR1152 is the arabinose-dependent MurJ depletion strain. Plasmid pHIS-YtgP expresses His-ytgP upon induction with IPTG; pCA24Not is the vector control lacking His-ytgP.
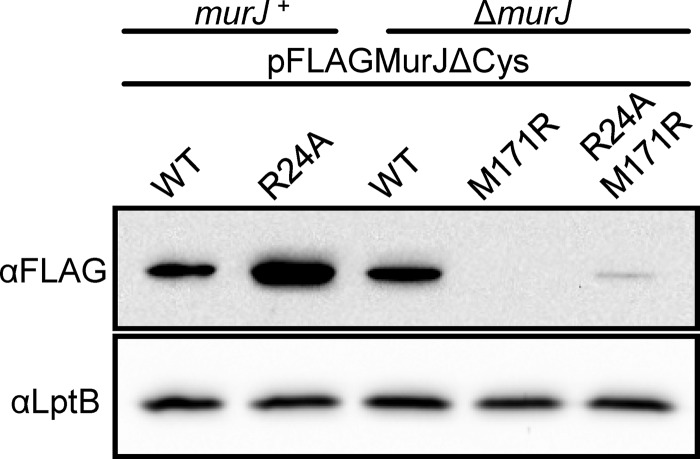
M171R suppresses the loss of a positive charge at position 24 in MurJ, confirming that R24 in MurJ is functionally equivalent to R175 in YtgP.
We reasoned that if, as suggested above, R175 in TMD 5 of YtgP is functionally equivalent to R24 in TMD 1 of MurJ, an Arg positioned similarly within TMD 5 of E. coli MurJ might suppress the total loss of function conferred by the R24A substitution. We tested this, as it could shed light onto how these two proteins have diverged while maintaining the same activity.
The structural alignment of YtgP and MurJ reveals that M171 in MurJ is positioned similarly to R175 in YtgP, such that the side chain in an M171R substitution in MurJ might overlap that of R24 (see Fig. S3A in the supplemental material). Therefore, we introduced the M171R substitution into wild-type FLAG-MurJ and its R24A variant and tested for complementation. The allele carrying the single M171R substitution complemented, although it was partially defective (Table 2), likely because of a defect in biogenesis (Fig. 5; also see Fig. S3B). Remarkably, the M171R substitution completely rescued function of the R24A variant (Table 2) despite being present at low levels (Fig. 5). Possibly, the difference in protein levels of these two M171R variants explains why haploid cells producing the single M171R variant are more sensitive to low osmolarity than those producing the R24A/M171R mutant protein (Fig. 5). Moreover, we demonstrated that the suppression of R24A by M171R is specific, because the introduction of the M171R substitution in the respective R18A, D39A, R52A, or R270A loss-of-function variant did not rescue MurJ function. Thus, these genetic data are consistent with the positive charge in TMD 1 provided by R24 in MurJ performing the same role as that provided by R175 in TMD 5 of YtgP.
TABLE 2.
Effect of the M171R substitution on FLAG-MurJ function
| Strain | Relevant genotype | Efficiency of plating ina: |
|
|---|---|---|---|
| LB | YT | ||
| NR2131 | ΔmurJ(pFLAGMurJΔCys) | 1.0 | 1.0 ± 0.3 |
| NR3075 | ΔmurJ(pFLAGMurJΔCys/M171R) | 0.0 ± 0.0 | <10−5 |
| NR3058 | ΔmurJ(pFLAGMurJΔCys/R24A/M171R) | 0.8 ± 0.5 | 0.4 ± 0.3 |
Efficiency of plating values for growth of haploid strains expressing FLAG-murJ alleles carrying the indicated substitutions were calculated as described in Materials and Methods. A value of 1 (set by the wild-type allele) indicates full complementation by FLAG-murJ alleles; a value of <1 indicates partial complementation. The haploid strain producing FLAG-MurJ/R24A is not viable, because the R24A substitution renders MurJ nonfunctional.
FIG 5.
M171R substitution causes defects in MurJ biogenesis. Anti-FLAG immunoblot of samples prepared from overnight cultures of merodiploid (murJ+) or haploid (ΔmurJ) strains reveals that the M171R substitution is responsible for the reduced levels of functional M171R and R24A/M171R FLAG-MurJ variants. Data are representative of at least three independent experiments. Anti-LptB immunoblotting was performed to control for sample loading.
The central cavities of MurJ and YtgP have unique charge requirements.
The 8 critical residues identified in MurJ are located in TMDs 1, 2, and 8 and periplasmic loop 1. Surprisingly, as described above, only 2 of them are conserved in TMDs 1 and 8 of YtgP. In addition, R175 in YtgP is analogous to R24 in MurJ even though it is located in TMD 5 (Fig. 4A). Interestingly, TMDs 4 and 5 in YtgP possess additional charges (R152 and E171) that are absent from MurJ. We thought these charges could be relevant for YtgP function because of the essentiality of R175 (TMD 5). In addition, solved structures of multidrug MOP exporters have revealed that although TMDs 1, 2, 7, and 8 line the central solvent-exposed cavity, TMDs 4 to 6 also can interact with substrates and cations during transport (17, 19). Therefore, we tested whether R152 and E171 are important for YtgP function and found that although their change to Ala did not affect the levels of their respective His-YtgP mutant proteins (Fig. 4B), both substitutions rendered His-YtgP nonfunctional (Table 1).
From these studies, we conclude that both YtgP and MurJ require specific charges for function. Although both proteins share 3 analogous charged positions (R29, R175, and K301 in YtgP; R18, R24, and R270 in MurJ), each also has unique requirements to function in E. coli.
DISCUSSION
The translocation of lipid II across the cytoplasmic membrane is an essential step in PG biogenesis. In E. coli, this step is mediated by the lipid II flippase MurJ (9). In MurJ, the first 12 (of 14) TMDs are predicted to form a V-shaped structure around a central, solvent-exposed cavity (14). Charged residues within this cavity are required for MurJ function, supporting a model where this cavity serves to protect the hydrophilic portion of lipid II during flipping (8, 14). Here, we focused on defining the requirement for all charged residues in MurJ from E. coli. We found that specific charges important for MurJ activity are limited to residues within the cavity-lining TMDs 1, 2, and 8 and the periplasmic loop between TMDs 1 and 2. Our studies also showed that D39 (periplasmic loop 1) and R312 (TMD 9) are required for proper MurJ biogenesis. We further asked whether those 8 charges are found and relevant in YtgP, the distant MurJ homolog from S. pyogenes. We found that YtgP also requires charged residues within its cavity; however, positional conservation with MurJ is limited to 3 positively charged residues.
MurJ is a member of the MOP exporter superfamily (13), and its predicted structure resembles that of multidrug MOP exporters (14, 16–19). In these exporters, 12 TMDs adopt a V-shaped structure that is thought to reflect an alternating access mode of transport where the central cavity undergoes conformational changes that switch the side of the membrane which is open in order to translocate substrates across the membrane. In addition, these conformational changes are driven by the coupling of substrate efflux to the influx of a counter ion. Interestingly, transporters of the MOP exporter superfamily accomplish cation/substrate interchange across the membrane through divergent mechanisms. For example, in the drug efflux antiporter NorM, Na+ binds within the C-terminal lobe (C-lobe) of the V-shaped molecule, coordinating movement of TMDs 7 and 8 which, in turn, mediate substrate binding within the central cavity (18). On the other hand, substrate extrusion by PfMATE is driven by interconversion of TMD 1 between bent and straight conformations, which affect the distinct drug and cation binding sites within the N-terminal lobe (N-lobe) cavity region between TMDs 1 and 2 (17). In contrast, the exporter DinF was found to lack pseudosymmetry, having TMDs 7 and 8 bent inward to occlude the central cavity and expose an outward-open crevice in the C-lobe. A mechanism was proposed in which TMDs 9 to 12 of the C-lobe of DinF rotate from outward- to inward-facing conformations depending on the competitive binding of substrate or H+ to an acidic residue (17). How MurJ flips lipid II is not understood. We predict that at least the hydrophilic portion of lipid II interacts with the hydrophilic cavity of MurJ during transport (14). However, details of these interactions are unknown. We also do not know if lipid II flipping is coupled to the transport of a counter ion. Given the variety of mechanisms for MOP exporter antiport, we do not have a model to predict the specific functional requirements of MurJ. However, we can extrapolate that if MurJ functions by a substrate-cation antiport mechanism, critical residues could play a role in substrate binding, cation binding, and translocation.
MurJ is predicted to have a V-shaped structure comprised of an N-lobe (TMDs 1 to 6) and C-lobe (TMDs 7 to 12) arranged pseudosymmetrically around a central hydrophilic cavity (lined mainly by TMDs 1, 2, 7, and 8) that extends into both lobes. Here and in a previous study (14), we have reported the functional analysis to define the requirement for all charged residues in MurJ from E. coli. Alterations of Cys resulted in detectable defects in only 9 of its 57 charged residues. These changes altered MurJ biogenesis and/or function, and all but one of the residues localize to the periplasmic-facing half of the cavity region. Among those residues important for MurJ biogenesis, we found R312. The side chain of R312 is predicted to point into the membrane in the MurJ model structure. This structural prediction is supported by the observation that Lys residues are positioned similarly in the crystal structures of MOP exporters PfMATE (PBD entry 3VVN, residue K325) and DinF (PBD entry 4LZ6, residue K324). Indeed, in TMDs, arginine and lysine residues can “snorkel” toward the membrane interface (36). Furthermore, all of the residues discussed in this study, including R312, are present within allowed energy fields in the structural model (see Fig. S4 in the supplemental material). It is also possible that R312 contacts a yet-to-be-identified partner required for stability or a biogenesis factor important for membrane insertion and/or folding; alternatively, R312 could mediate intramolecular interactions with TMDs 13 to 14, since the structural model cannot predict the position of these TMDs with confidence (Fig. 1A) (14). Regardless, the phenotypes caused by the R312C substitution likely are related to the observed biogenesis defect. We must note, however, that it is unclear how many functional MurJ proteins a cell must have to properly build PG and whether this requirement varies with growth conditions. In our system, we observe phenotypes when MurJ variants are present at low levels (e.g., FLAG-MurJ/R312C and MurJ/M171R) in rich medium (LB); however, we cannot detect wild-type, fully functional FLAG-MurJ in minimal medium through immunoblotting.
The loss of a negative charge in periplasmic loop 1 at position 39 (D39) also compromises MurJ biogenesis. The fact that this defect can be suppressed by introducing a nearby negative charge into periplasmic loop 1 suggests that D39 is crucial for the insertion of MurJ into the membrane, a role that might be related to the unusually high content of hydrophilic residues in TMDs 1 and 2 (30, 33). Importantly, this suppressor of the biogenesis defect demonstrated that D39 also is essential for MurJ activity, as previously suggested (14). Therefore, these studies indicate that in E. coli, the contribution of charged residues to MurJ activity is limited to 8 residues (R18, R24, D39, K46, R52, D269, R270, and E273) located within the cavity-lining TMDs 1, 2, and 8 and periplasmic loop 1. The fact that the nature of their native charge is required for proper MurJ function demonstrates that a hydrophilic environment in the central cavity of MurJ is not sufficient for lipid II translocation. Instead, MurJ activity requires specific charges in the cavity region. Interestingly, no single charged residue in the cytoplasmic-facing half of the protein has been identified as being essential for MurJ function. It remains unknown whether this is because of redundancy or because charged residues in the cytoplasmic-facing half of the protein do not play a role in the flipping process.
Although structure-function analysis of MurJ orthologs has been limited to MurJ from E. coli and the related Gram-negative betaproteobacterium B. cenocepacia and to YtgP from the distant Gram-positive S. pyogenes, these data reveal that the specific charges required for MurJ activity appear to fall into two types: those that are widely conserved and those that are specific to different MurJ orthologs. Explicitly, the function of these three proteins requires positively charged residues that structural models predict to be equivalent to R18, R24, and R270 in E. coli MurJ. We propose that these conserved cationic residues interact with lipid II, given its anionic character. We note that in the transglycosylase domain of PBP1A and PBP1B, positively charged residues also have been proposed to interact with lipid II (37–40).
In addition, we have shown that the cavity region in MurJ from E. coli has specific requirements for charged residues that are absent from YtgP and vice versa. Notably, those residues specific to MurJ are highly conserved among MurJ homologs in Gram-negative bacteria, while those specific to YtgP are highly conserved among YtgP homologs in Gram-positive bacteria (data not shown). The observed differing requirements of these proteins may reflect adaptations of Gram-negative and Gram-positive bacteria to suit specific PG synthesis systems, such as modifications to the peptide stem or different levels of demand of flippase activity. Nevertheless, despite their different specific locations, we note that in both MurJ and YtgP, these residues are located either in the central cavity or its extension into the N-lobe, suggesting this lobe is of critical importance to the flipping mechanism in these proteins. These requirements resemble those found in Wzx from Pseudomonas aeruginosa, a MOP exporter superfamily member that flips a trisaccharide linked to undecaprenol-pyrophosphate across the IM. Movement of the anionic trisaccharide moiety through a hydrophilic channel in this flippase is thought to be coordinated by cationic residues in its N-lobe and cavity region (29, 41).
Based on the overall mechanism of transport that has been proposed for Wzx flippases and for multidrug exporters and flippases of polyisoprenoid polar lipids that are members of the MOP exporter superfamily (8, 16–20), it is possible that MurJ couples lipid II flipping to the import of a cation and that the undecaprenyl moiety of lipid II stays embedded in the membrane during flipping while the hydrophilic moiety travels through the lumen. In some MOP exporters, acidic residues have been shown to coordinate cation binding, and these residues are found at diverse positions within their structures (16–19). Although we have identified some critical acidic residues at different positions in MurJ and YtgP, we do not know if they mediate cation binding. Lastly, if the undecaprenyl moiety of lipid II stays in the hydrophobic core of the membrane during flipping, the flippase would have to have an open slot between two TMDs. The structural models of MurJ from E. coli (14) and B. cenocepacia (12) and of YtgP predict two of these features that are conserved in all three proteins. These open slots are present between TMDs 1 and 8 and TMDs 1 and 5 (see Fig. S5 in the supplemental material). Interestingly, the three positively charged residues that are conserved and essential in these proteins are located in TMDs 1 (R18 in E. coli MurJ, R29 in YtgP) and 8 (R270 in E. coli MurJ, K301 in YtgP) with side chains facing the main cavity and in either TMD 1 (R24 in E. coli MurJ) or TMD 5 (R175 in YtgP) with side chains pointing into TMD 5 or 1, respectively.
As we find both conserved and organism-specific requirements for the function of MurJ and YtgP, the mechanism for lipid II flipping is not wholly conserved, as in Wzx flippases and multidrug MOP exporters (16–20, 41). However, taken together, our data support that charged residues within the central cavity and its extension into the N-lobes of MurJ and YtgP play critical roles that allow functional interchangeability in E. coli. We propose that the pyrophosphate-disaccharide-pentapeptide moiety of lipid II likely is engaged into the flippase channel, where positively charged residues in the N-lobe coordinate its movement to the periplasmic face of the IM.
Supplementary Material
ACKNOWLEDGMENTS
We thank Rebecca Davis for technical assistance.
This research was supported by funds from the National Institute of General Medical Sciences of the National Institutes of Health under award number R01GM100951 (to N.R.).
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Published ahead of print 15 September 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.02172-14.
REFERENCES
- 1.Typas A, Banzhaf M, Gross CA, Vollmer W. 2012. From the regulation of peptidoglycan synthesis to bacterial growth and morphology. Nat. Rev. Microbiol. 10:123–136. 10.1038/nrmicro2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vollmer W, Bertsche U. 2008. Murein (peptidoglycan) structure, architecture and biosynthesis in Escherichia coli. Biochim. Biophys. Acta 1778:1714–1734. 10.1016/j.bbamem.2007.06.007. [DOI] [PubMed] [Google Scholar]
- 3.Silhavy TJ, Kahne D, Walker S. 2010. The bacterial cell envelope. Cold Spring Harb. Perspect. Biol. 2:a000414. 10.1101/cshperspect.a000414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barreteau H, Kovač A, Boniface A, Sova M, Gobec S, Blanot D. 2008. Cytoplasmic steps of peptidoglycan biosynthesis. FEMS Microbiol. Rev. 32:168–207. 10.1111/j.1574-6976.2008.00104.x. [DOI] [PubMed] [Google Scholar]
- 5.Bouhss A, Trunkfield AE, Bugg TDH, Mengin-Lecreulx D. 2008. The biosynthesis of peptidoglycan lipid-linked intermediates. FEMS Microbiol. Rev. 32:208–233. 10.1111/j.1574-6976.2007.00089.x. [DOI] [PubMed] [Google Scholar]
- 6.Den Blaauwen T, De Pedro MA, Nguyen-Distèche M, Ayala JA. 2008. Morphogenesis of rod-shaped sacculi. FEMS Microbiol. Rev. 32:321–344. 10.1111/j.1574-6976.2007.00090.x. [DOI] [PubMed] [Google Scholar]
- 7.Lovering AL, Safadi SS, Strynadka NCJ. 2012. Structural perspective of peptidoglycan biosynthesis and assembly. Annu. Rev. Biochem. 81:451–478. 10.1146/annurev-biochem-061809-112742. [DOI] [PubMed] [Google Scholar]
- 8.Sanyal S, Menon AK. 2009. Flipping lipids: why an' what's the reason for? ACS Chem. Biol. 4:895–909. 10.1021/cb900163d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sham L-T, Butler EK, Lebar MD, Kahne D, Bernhardt TG, Ruiz N. 2014. MurJ is the flippase of lipid-linked precursors for peptidoglycan biogenesis. Science 345:220–222. 10.1126/science.1254522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Inoue A, Murata Y, Takahashi H, Tsuji N, Fujisaki S, Kato J-I. 2008. Involvement of an essential gene, mviN, in murein synthesis in Escherichia coli. J. Bacteriol. 190:7298–7301. 10.1128/JB.00551-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ruiz N. 2008. Bioinformatics identification of MurJ (MviN) as the peptidoglycan lipid II flippase in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 105:15553–15557. 10.1073/pnas.0808352105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mohamed YF, Valvano MA. 2014. A Burkholderia cenocepacia MurJ (MviN) homolog is essential for cell wall peptidoglycan synthesis and bacterial viability. Glycobiology 24:564–576. 10.1093/glycob/cwu025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hvorup RN, Winnen B, Chang AB, Jiang Y, Zhou X-F, Saier MH. 2003. The multidrug/oligosaccharidyl-lipid/polysaccharide (MOP) exporter superfamily. Eur. J. Biochem. 270:799–813. 10.1046/j.1432-1033.2003.03418.x. [DOI] [PubMed] [Google Scholar]
- 14.Butler EK, Davis RM, Bari V, Nicholson PA, Ruiz N. 2013. Structure-function analysis of MurJ reveals a solvent-exposed cavity containing residues essential for peptidoglycan biogenesis in Escherichia coli. J. Bacteriol. 195:4639–4649. 10.1128/JB.00731-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kuroda T, Tsuchiya T. 2009. Multidrug efflux transporters in the MATE family. Biochim. Biophys. Acta 1794:763–768. 10.1016/j.bbapap.2008.11.012. [DOI] [PubMed] [Google Scholar]
- 16.He X, Szewczyk P, Karyakin A, Evin M, Hong W-X, Zhang Q, Chang G. 2010. Structure of a cation-bound multidrug and toxic compound extrusion transporter. Nature 467:991–994. 10.1038/nature09408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lu M, Radchenko M, Symersky J, Nie R, Guo Y. 2013. Structural insights into H+-coupled multidrug extrusion by a MATE transporter. Nat. Struct. Mol. Biol. 20:1310–1317. 10.1038/nsmb.2687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lu M, Symersky J, Radchenko M, Koide A, Guo Y, Nie R, Koide S. 2013. Structures of a Na+-coupled, substrate-bound MATE multidrug transporter. Proc. Natl. Acad. Sci. U. S. A. 110:2099–2104. 10.1073/pnas.1219901110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tanaka Y, Hipolito CJ, Maturana AD, Ito K, Kuroda T, Higuchi T, Katoh T, Kato HE, Hattori M, Kumazaki K, Tsukazaki T, Ishitani R, Suga H, Nureki O. 2013. Structural basis for the drug extrusion mechanism by a MATE multidrug transporter. Nature 496:247–251. 10.1038/nature12014. [DOI] [PubMed] [Google Scholar]
- 20.Islam ST, Lam JS. 2013. Wzx flippase-mediated membrane translocation of sugar polymer precursors in bacteria. Environ. Microbiol. 15:1001–1015. 10.1111/j.1462-2920.2012.02890.x. [DOI] [PubMed] [Google Scholar]
- 21.Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Soding J, Thompson JD, Higgins DG. 2011. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7:539. 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ruiz N. 2009. Streptococcus pyogenes YtgP (Spy_0390) complements Escherichia coli strains depleted of the putative peptidoglycan flippase, MurJ. Antimicrob. Agents Chemother. 53:3604–3605. 10.1128/AAC.00578-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ruiz N, Gronenberg LS, Kahne D, Silhavy TJ. 2008. Identification of two inner-membrane proteins required for the transport of lipopolysaccharide to the outer membrane of Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 105:5537–5542. 10.1073/pnas.0801196105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Casadaban MJ. 1976. Transposition and fusion of the lac genes to selected promoters in Escherichia coli using bacteriophage lambda and Mu. J. Mol. Biol. 104:541–555. 10.1016/0022-2836(76)90119-4. [DOI] [PubMed] [Google Scholar]
- 25.Silhavy TJ, Berman ML, Enquist LW. 1984. Experiments with gene fusions. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. [Google Scholar]
- 26.Kitagawa M, Ara T, Arifuzzaman M, Ioka-Nakamichi T, Inamoto E, Toyonaga H, Mori H. 2005. Complete set of ORF clones of Escherichia coli ASKA library (a complete set of E. coli K-12 ORF archive): unique resources for biological research. DNA Res. 12:291–299. 10.1093/dnares/dsi012. [DOI] [PubMed] [Google Scholar]
- 27.Roy A, Kucukural A, Zhang Y. 2010. I-TASSER: a unified platform for automated protein structure and function prediction. Nat. Protoc. 5:725–738. 10.1038/nprot.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shindyalov IN, Bourne PE. 1998. Protein structure alignment by incremental combinatorial extension (CE) of the optimal path. Protein Eng. 11:739–747. 10.1093/protein/11.9.739. [DOI] [PubMed] [Google Scholar]
- 29.Islam ST, Fieldhouse RJ, Anderson EM, Taylor VL, Keates RAB, Ford RC, Lam JS. 2012. A cationic lumen in the Wzx flippase mediates anionic O-antigen subunit translocation in Pseudomonas aeruginosa PAO1. Mol. Microbiol. 84:1165–1176. 10.1111/j.1365-2958.2012.08084.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Delgado-Partin VM, Dalbey RE. 1998. The proton motive force, acting on acidic residues, promotes translocation of amino-terminal domains of membrane proteins when the hydrophobicity of the translocation signal is low. J. Biol. Chem. 273:9927–9934. 10.1074/jbc.273.16.9927. [DOI] [PubMed] [Google Scholar]
- 31.Kiefer D, Hu X, Dalbey R, Kuhn A. 1997. Negatively charged amino acid residues play an active role in orienting the Sec-independent Pf3 coat protein in the Escherichia coli inner membrane. EMBO J. 16:2197–2204. 10.1093/emboj/16.9.2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rutz C, Rosenthal W, Schülein R. 1999. A single negatively charged residue affects the orientation of a membrane protein in the inner membrane of Escherichia coli only when it is located adjacent to a transmembrane domain. J. Biol. Chem. 274:33757–33763. 10.1074/jbc.274.47.33757. [DOI] [PubMed] [Google Scholar]
- 33.Zhu L, Wasey A, White SH, Dalbey RE. 2013. Charge composition features of model single-span membrane proteins that determine selection of YidC and SecYEG translocase pathways in Escherichia coli. J. Biol. Chem. 288:7704–7716. 10.1074/jbc.M112.429431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vollmer W, Blanot D, De Pedro MA. 2008. Peptidoglycan structure and architecture. FEMS Microbiol. Rev. 32:149–167. 10.1111/j.1574-6976.2007.00094.x. [DOI] [PubMed] [Google Scholar]
- 35.Huber J, Donald RGK, Lee SH, Jarantow LW, Salvatore MJ, Meng X, Painter R, Onishi RH, Occi J, Dorso K, Young K, Park YW, Skwish S, Szymonifka MJ, Waddell TS, Miesel L, Phillips JW, Roemer T. 2009. Chemical genetic identification of peptidoglycan inhibitors potentiating carbapenem activity against methicillin-resistant Staphylococcus aureus. Chem. Biol. 16:837–848. 10.1016/j.chembiol.2009.05.012. [DOI] [PubMed] [Google Scholar]
- 36.Schow E, Freites JA, Cheng P, Bernsel A, von Heijne G, White S, Tobias D. 2011. Arginine in membranes: the connection between molecular dynamics simulations and translocon-mediated insertion experiments. J. Membrane Biol. 239:35–48. 10.1007/s00232-010-9330-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huang C-Y, Shih H-W, Lin L-Y, Tien Y-W, Cheng T-JR, Cheng W-C, Wong C-H, Ma C. 2012. Crystal structure of Staphylococcus aureus transglycosylase in complex with a lipid II analog and elucidation of peptidoglycan synthesis mechanism. Proc. Natl. Acad. Sci. U. S. A. 109:6496–6501. 10.1073/pnas.1203900109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sung M-T, Lai Y-T, Huang C-Y, Chou L-Y, Shih H-W, Cheng W-C, Wong C-H, Ma C. 2009. Crystal structure of the membrane-bound bifunctional transglycosylase PBP1b from Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 106:8824–8829. 10.1073/pnas.0904030106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Terrak M, Sauvage E, Derouaux A, Dehareng D, Bouhss A, Breukink E, Jeanjean S, Nguyen-Distèche M. 2008. Importance of the conserved residues in the peptidoglycan glycosyltransferase module of the class A penicillin-binding protein 1b of Escherichia coli. J. Biol. Chem. 283:28464–28470. 10.1074/jbc.M803223200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yuan Y, Barrett D, Zhang Y, Kahne D, Sliz P, Walker S. 2007. Crystal structure of a peptidoglycan glycosyltransferase suggests a model for processive glycan chain synthesis. Proc. Natl. Acad. Sci. U. S. A. 104:5348–5353. 10.1073/pnas.0701160104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Islam ST, Eckford PDW, Jones ML, Nugent T, Bear CE, Vogel C, Lam JS. 2013. Proton-dependent gating and proton uptake by Wzx support O-antigen-subunit antiport across the bacterial inner membrane. mBio 4:e00678–13. 10.1128/mBio.00678-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.