Abstract
Legionella pneumophila is an intracellular human pathogen that utilizes the Icm/Dot type IVB secretion system to translocate a large repertoire of effectors into host cells. For most of these effectors, there is no information regarding their regulation. Therefore, the aim of this study was to examine the involvement of the three L. pneumophila Fis homologs in the regulation of effector-encoding genes. Deletion mutants constructed in the genes encoding the three Fis regulators revealed that Fis1 (lpg0542 gene) and Fis3 (lpg1743) but not Fis2 (lpg1370) are partially required for intracellular growth of L. pneumophila in Acanthamoeba castellanii. To identify pathogenesis-related genes directly regulated by Fis, we established a novel in vivo system which resulted in the discovery of numerous effector-encoding genes directly regulated by Fis. Further examination of these genes revealed that Fis1 and Fis3 repress the level of expression of effector-encoding genes during exponential phase. Three groups of effector-encoding genes were identified: (i) effectors regulated mainly by Fis1, (ii) effectors regulated mainly by Fis3, and (iii) effectors regulated by both Fis1 and Fis3. Examination of the upstream regulatory region of all of these effector-encoding genes revealed multiple putative Fis regulatory elements, and site-directed mutagenesis confirmed that a few of these sites constitute part of a repressor binding element. Furthermore, gel mobility shift assays demonstrated the direct relation between the Fis1 and Fis3 regulators and these regulatory elements. Collectively, our results demonstrate for the first time that two of the three L. pneumophila Fis regulators directly repress the expression of Icm/Dot effector-encoding genes.
INTRODUCTION
Legionella pneumophila is an opportunistic human pathogen that multiplies within alveolar macrophages and causes a severe pneumonia known as Legionnaires' disease (1–3). In the environment L. pneumophila thrives in many different protozoan cells, and these cells probably serve as their training ground for pathogenesis (4–6). Inside its hosts L. pneumophila avoids degradation by the endocytic pathway, and instead the bacterium remodels its phagosome into an endoplasmic reticulum (ER)-like compartment (7, 8). In order to establish this replicative niche inside eukaryotic cells, L. pneumophila utilizes the Icm/Dot type IV secretion system to deliver a cohort of about 300 effector proteins which modulate host-cell functions during infection (reviewed in references 9 to 12). The numerous effectors that take part in the establishment of the L. pneumophila-containing vacuole (LCV), the stepwise process that occurs during the establishment of the LCV inside host cells (7), and the many host cell pathways manipulated by L. pneumophila effectors (13, 14) suggest that Icm/Dot effectors will most likely be subjected to many levels of regulation, one of which occurs at the level of gene expression.
To date, three regulatory systems have been shown to directly regulate the expression of effector-encoding genes: (i) the PmrAB two-component system (TCS), which includes the PmrA response regulator and the PmrB sensor-histidine kinase, was shown to directly activate the expression of 43 effector-encoding genes (15, 16); (ii) the CpxRA TCS, which includes the CpxR response regulator and the CpxA sensor-histidine kinase, was shown to directly activate or repress the expression of 11 effector-encoding genes, as well as four genes encoding Icm/Dot components (17, 18); and (iii) the LetAS-RsmYZ-CsrA regulatory cascade, which includes the LetA response regulator, the LetS sensor-histidine kinase, the two small RNAs RsmY and RsmZ, and the posttranscriptional repressor CsrA, was shown to posttranscriptionally repress the translation of 26 effector-encoding genes (19–23). These three regulatory systems control the expression of about a quarter of the known L. pneumophila effectors (24), suggesting that additional regulators of gene expression which directly control the expression of effector-encoding genes in L. pneumophila remain to be found.
The three regulatory systems described above were also shown to participate in the regulation of pathogenesis-related genes in other bacteria. The PmrAB TCS was studied extensively in Salmonella enterica, where it functions as the major regulator of lipopolysaccharide modification genes (25). A similar function was found to be mediated by the TCS homologous to PmrAB in other bacteria such as Escherichia coli (the BasRS TCS) (26) and Pseudomonas aeruginosa (27). In addition, the S. enterica PmrAB TCS was found to be active when the bacteria are inside macrophages and during infection of mice (28). The CpxRA TCS was shown to be required for host cell invasion in several bacterial species. In S. enterica a constitutively active CpxA mutation inhibits adherence to cultured cells and reduces virulence in mice (29). In pathogenic E. coli, a cpxR deletion mutant exhibited decreased formation of bundle-forming pili and decreased adherence to cultured cells (30). In Shigella spp., the CpxRA TCS directly controls the expression of virF, which encodes a positive regulator of type III secretion genes required for virulence (31). A homologous regulatory cascade functionally similar to the L. pneumophila LetAS-RsmYZ-CsrA was found in several bacteria. The S. enterica BarA-SirA-CsrBC-CsrA system was found to posttranscriptionally regulate hilD expression by CsrA directly binding to the hilD mRNA. HilD regulates the expression of hilA, located in Salmonella pathogenicity island (SPI-1), and of the ssrAB operon, located in SPI-2. These genes encode HilA and the SsrAB TCS, which are the positive regulators of the SPI-1 and SPI-2 regulons, respectively, that play key roles in the pathogenesis of S. enterica (32). In addition, in enteropathogenic E. coli (EPEC) CsrA acts as an activator or repressor of the locus of enterocyte effacement (LEE) pathogenicity island (33). However, unlike the case of the enteric bacteria, components of the L. pneumophila LetAS-RsmYZ-CsrA cascade were shown to be involved in cellular differentiation of this bacterium from a replicating form to a virulent form (34).
The observation that homologous regulatory systems control the expression of pathogenesis-related genes in many gammaproteobacteria raises the possibility that additional systems which were shown to control the expression of pathogenesis-related genes in these bacteria regulate virulence gene expression in L. pneumophila as well. One such regulator is Fis (factor for inversion stimulation). In S. enterica, Fis was found to affect the expression of many SPI-1 and SPI-2 genes by binding SPI genes directly, by binding to upstream regions of SPI regulators, or by binding to the gene encoding OmpR which affects SPI gene expression by controlling SPI regulators SsrA and HilD (described above) (35). In EPEC, the transcription of the LEE4 operon consisting of espADB and the virulence activator, Ler, were found to be Fis dependent (36). In Shigella, Fis was shown to bind to four specific sites in the promoter region of the virF regulator (described above) (37).
The involvement of Fis in the regulation of pathogenesis-related genes in different pathogenic bacteria and the observation that L. pneumophila contains three Fis homologs led us to explore the involvement of these three Fis regulators in the regulation of effector-encoding genes. Our results clearly indicate that two of the L. pneumophila Fis regulators are required for maximal intracellular multiplication in amoebae, and these two Fis regulators were found to directly repress the expression of numerous L. pneumophila effector-encoding genes during exponential growth.
MATERIALS AND METHODS
Bacterial strains and media.
The L. pneumophila wild-type strain used in this work was JR32, a streptomycin-resistant, restriction-negative mutant of L. pneumophila Philadelphia-1, which is a wild-type strain in terms of intracellular growth (38). In addition, mutant strains derived from JR32 which contain a kanamycin (Km) cassette instead of the icmT gene (GS3011) (39), the fis1 (lpg0542) gene (ZT-Fis1) (this study), the fis2 (lpg1370) gene (ZT-Fis2) (this study), and the fis3 (lpg1743) gene (ZT-Fis3) (this study) were used. The E. coli strains used in this work were MC1022 and MC1061 (40). Bacterial media, plates, and antibiotic concentrations used were as described previously (41).
Plasmid construction.
To construct lacZ translational fusions, the 300-bp regulatory regions of 100 effector-encoding genes and five regulator-encoding genes (see Data set S1 in the supplemental material) were amplified by PCR using the primers listed in Data set S2. The PCR products were then digested with BamHI and EcoRI (or only with BamHI if an EcoRI site was present in the regulatory region amplified), cloned into pGS-lac-02, and sequenced. The 105 new lacZ fusions generated as well as the 77 lacZ fusions that were constructed before and used in this study are listed in Data set S1.
To construct substitutions in the putative Fis binding sites in the regulatory regions of the legA9, legA12, mavT, ravI, ravN, ceg20, mavU, legC4, cegC4, lem21, sidM, sdbB, sdeD, sidC, legK3, lem28, legU2, legC8, lpg0634, and lpg1967 genes, site-directed mutagenesis was performed by the PCR overlap extension approach (42), in a similar way as described before (16). In all the mutations constructed in the putative Fis binding site, the G and C nucleotides of the consensus were changed to C and G, respectively. The primers used for the mutagenesis are listed in Data set S2 in the supplemental material, and the plasmids resulting from the site-directed mutagenesis are listed in Data set S1.
To construct isopropyl β-d-1-thiogalactopyranoside (IPTG)-inducible Fis regulators, the L. pneumophila fis1, fis2, and fis3 genes were amplified by PCR using the primers listed in Data set S2 in the supplemental material. The PCR products were then digested with EcoRI and BamHI for fis1 and fis2 and with PstI and BamHI for fis3 and cloned into pUC-18 to generate the plasmids listed in Data set S1 and sequenced. The resulting plasmids were digested with the same enzymes, and the fragments containing the three fis genes were cloned into pMMB207 downstream from the Ptac promoter to generate the plasmids listed in Data set S1; these plasmids contain the Fis regulators under Ptac control, and they were used for intracellular growth complementation. In addition, the resulting plasmids were then digested with EheI and BamHI for fis1 and fis2 and with EheI and PstI for fis3, and the fragments containing Ptac-fis together with the lacI gene were cloned into the pHG-165 vector digested with SmaI and BamHI for fis1 and fis2 and with SmaI and PstI for fis3, resulting in the plasmids listed in Data set S1. These plasmids were used for the library screen performed in E. coli.
To construct deletion substitutions in the L. pneumophila fis1, fis2, and fis3 genes, a 1-kb DNA fragment located on each side of the planned deletion was amplified by PCR using the primers listed in Data set S2 in the supplemental material. The primers were designed to contain an SalI site at the place of the deletion. The two fragments that were amplified for each gene were cloned into pUC-18 digested with suitable enzymes, and the inserts were sequenced to generate the plasmids listed in Data set S1. The resulting plasmids were digested with suitable enzymes, and the inserts were used for a four-way ligation containing the Km resistance cassette (Pharmacia) digested with SalI and the pUC-18 vector digested with suitable enzymes. The correct plasmids were identified by plating the bacteria after transformation on plates containing ampicillin (Amp) and Km, and after plasmid preparation the correct clones were identified by restriction digests. The three plasmids generated (see Data set S1 in the supplemental material) were digested with PvuII (this enzyme cuts on both sides of the pUC-18 polylinker), and the resulting fragments were cloned into the pLAW344 allelic exchange vector digested with EcoRV to generate the plasmids listed in Data set S1 that were used for allelic exchange, as described previously (41).
To overexpress the Fis1 and Fis3 proteins for gel mobility shift assays, a fragment containing the L. pneumophila fis1 or fis3 gene was amplified by PCR using the primers listed in Data set S2 in the supplemental material. The resulting fragments were digested with NdeI and BamHI and cloned into the pET-21a vector digested with the same enzymes to generate the plasmids listed in Data set S1. The resulting plasmids express a full-length Fis1 or Fis3 protein fused to a His6 tag on the C terminus.
Library screen to identify genes regulated by the three Fis regulators.
A screen allowing identification of genes that are directly regulated by the Fis regulators was performed, using a plasmid containing the Fis regulators under the IPTG-inducible Ptac promoter (described above) and a pooled library of 182 lacZ translational fusions. The library included the following lacZ fusions: 160 effector-encoding genes, 10 regulator-encoding genes, and 12 icm/dot genes, which are all listed in Data set S1 in the supplemental material.
A plasmid containing an inducible Fis regulator and the pooled library plasmids were coelectroporated into E. coli MC1061, and the bacteria were plated on LB plates containing Amp and chloramphenicol (Cm). Single colonies were suspended in each well of a 96-well microtiter plate containing LB medium supplemented with Amp and Cm. From each bacterial suspension, 10 μl was transferred to a plate containing LB medium supplemented with Amp and Cm, and another 10 μl was transferred into a plate containing LB medium supplemented with Amp, Cm, and IPTG (0.05 mM for fis1, 0.1 mM for fis2, and 0.5 mM for fis3). For each of the fis genes, the highest IPTG concentration which did not inhibit E. coli growth was found and used in the analysis. The three plates were incubated overnight at 37°C with agitation. Plates with and without IPTG were subjected to a β-galactosidase assay by transferring an aliquot from each well into 100 μl of Z buffer (60 mM Na2HPO4, 40 mM NaH2PO4, 10 mM KCl, 1 mM MgSO4 and 50 mM β-mercaptoethanol at pH 7.0) containing 1.25% toluene and mixed well. The toluene was evaporated, and the plates were placed at room temperature for 5 min. To start the reaction, 22 μl of P buffer (60 mM Na2HPO4, 40 mM NaH2PO4 at pH 7.0) containing 4 mg/ml o-nitrophenyl-β-d-galactopyranoside (ONPG) was added. The reaction was stopped by the addition of 55 μl of 1 M Na2CO3. The optical density (OD) of the cultures was determined at 600 nm (OD600), and the β-galactosidase activities of the fusions were determined at 420 nm. The β-galactosidase specific activity was calculated in arbitrary units (OD420/OD600), and candidates that showed considerably different values (more than a 2.5-fold change in the levels of expression) between the plates with and without IPTG were plated on selective LB medium. The positive genes were identified by sequencing of the regulatory region located upstream from the lacZ gene. The levels of expression of the individual lacZ fusions were examined in the L. pneumophila wild-type and mutant strains at exponential and stationary phases as described before (16).
Protein purification and gel mobility shift assay.
Fis1-His6 and Fis3-His6 were purified from E. coli BL21(DE3) using nickel bead columns (Qiagen) according to the manufacturer's instructions. After purification, the fractions containing the protein were dialyzed against a buffer containing 20 mM Tris-HCl (pH 7.5), 0.5 M NaCl, 1 mM EDTA, 0.5 mM dithiothreitol, and 10% glycerol overnight. The glycerol concentration was increased to 50%, and the purified protein was then stored at −20°C. A gel mobility shift assay was performed as previously described (16), with few modifications. The regulatory regions of sidC, legA12, and ceg20 (∼150 bp) were amplified by PCR using the primers listed in Data set S2 in the supplemental material and 3′ end labeled with digoxigenin (DIG) by using DIG-11-ddUTP (Roche). Increasing amounts of the purified proteins (Fis1 between 0.25 and 2 μM and Fis3 between 0.05 and 0.4 μM) were mixed with a 1.6 nM concentration of the DIG-labeled probe in buffer containing 20 mM Tris-HCl (pH 7.5), 80 mM NaCl, 10 mM KCl, 5 mM MgCl2, 1 mM EDTA, 0.5 mM dithiothreitol, 60 μg/ml poly-l-lysine, 60 μg/ml poly(dI-dC), and 30% glycerol. For the competition experiments, the unlabeled probe or mutated unlabeled probe was allowed to bind the Fis1 and Fis3 proteins for 15 min before addition of the DIG-labeled probe. A binding reaction was carried out for 20 min at room temperature, and samples were then loaded onto a 6% polyacrylamide–0.5× Tris-boric acid-EDTA gel in 0.5× Tris-boric acid-EDTA running buffer. Following electrophoresis, the gel was transferred to a nylon membrane and fixed by UV cross-linking. The DIG-labeled DNA fragments were detected by following the manufacturer's instructions (Roche).
Intracellular growth in Acanthamoeba castellanii.
Intracellular growth assays of L. pneumophila strains in Acanthamoeba castellanii were performed as described before (43). Briefly, A. castellanii (ATCC 30234) (1.5 × 105 organisms) in proteose peptone-yeast extract-glucose (PYG) medium was added to wells of a 24-well microtiter plate, and the amoebae were incubated for 1 h at 37°C to let the amoebae adhere. Then, the PYG medium was aspirated, and the wells were washed once with 0.5 ml of warm (37°C) Acanthamoeba buffer, and 0.5 ml of warm Acanthamoeba buffer was added to the wells. Then, L. pneumophila in Acanthamoeba buffer was added to the wells at a multiplicity of infection (MOI) of ∼0.1. The plate was incubated for 30 min at 37°C, the Acanthamoeba buffer was aspirated, the wells were washed three times with 0.5 ml of warm Acanthamoeba buffer, and 0.6 ml of warm Acanthamoeba buffer was added to the wells. The supernatant of each well was sampled at intervals of 24 h, and the numbers of CFU were determined by plating samples on N-(2-acetamido)-2-aminoethanesulfonic acid (ACES)-buffered charcoal yeast extract (CYE). For complementation analysis, the plates on which the bacteria were grown were supplemented with 1 mM IPTG (no addition of IPTG or 0.1 mM IPTG did not result with complementation), and the Acanthamoeba buffer in which the infection was performed was supplemented with the same concentration of IPTG.
Intracellular growth in HL-60-derived human macrophages.
Intracellular growth assays of L. pneumophila strains in HL-60-derived human macrophages were performed as described before (43). Briefly, wells of a 24-well microtiter dish containing 4 × 105 differentiated HL-60-derived macrophages were used for infection. L. pneumophila was added to the wells at an MOI of approximately 0.01 and incubated for 1 h, and cells were washed three times. The infected HL-60 cells were incubated at 37°C under CO2 (5%), and bacterial CFU counts were determined at 0, 24, 48, 72, and 96 h postinfection. The number of CFU was determined by plating samples on CYE plates.
RESULTS
L. pneumophila contains three Fis regulators.
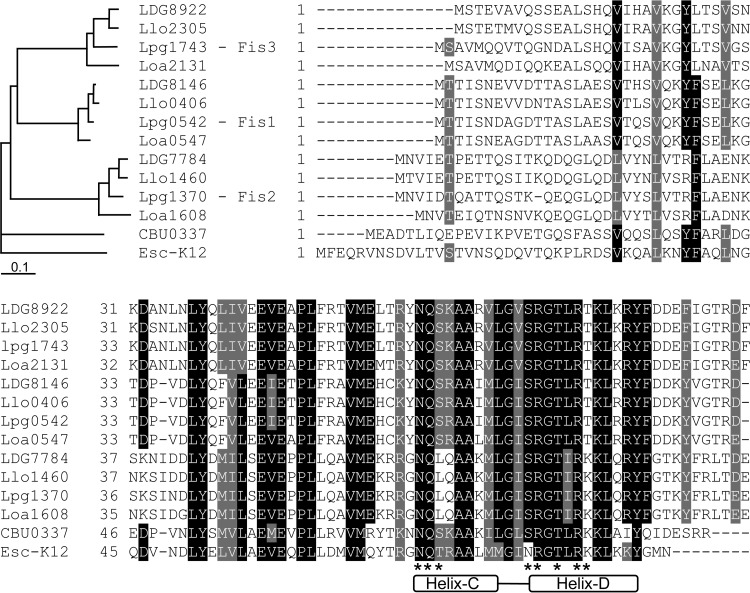
Fis orthologs were found to be present in almost all the members of the gammaproteobacteria, which usually contain a single Fis protein (44). Examination of the available Legionella genomic sequences revealed that all Legionella species contain three Fis paralogs: Fis1 (Lpg0542), Fis2 (Lpg1370), and Fis3 (Lpg1743). Sequence alignment and reconstruction of the Fis protein evolutionary tree from four Legionella species strongly suggest that two duplication events took place before the divergence of the Legionella genus, implying that the three Fis paralogs were already present in the last common ancestor of the Legionella species (Fig. 1). The three Fis homologs in each genome were clustered together with their homologs from the other species. Fis1 and Fis3 were found to be evolutionarily more closely related to each other than to Fis2 as well as to the single Fis orthologs present in bacteria such as E. coli and Coxiella burnetii. This result indicates that Fis1 and Fis3 probably arise from a more recent gene duplication of the Fis regulator present in the gammaproteobacteria.
FIG 1.
Legionella species contain three Fis regulators. Sequence alignment of Fis regulatory proteins from different bacteria is shown. Abbreviations: Lpg, L. pneumophila; Llo, L. longbeachae; LDG, L. drancourtii; Loa, L. oakridgensis; CBU, C. burnetii; and Esc-K12, E. coli K-12. The location of the helix-turn-helix DNA binding domain of these proteins is indicated at the bottom of the alignment (helix C and helix D). Amino acids that were shown before to form direct contact with the Fis regulatory element are marked by asterisks at the bottom of the alignment. A rectangular cladogram generated by the sequences of the Fis proteins is also presented.
Two of the L. pneumophila Fis regulators are required for optimal intracellular growth.
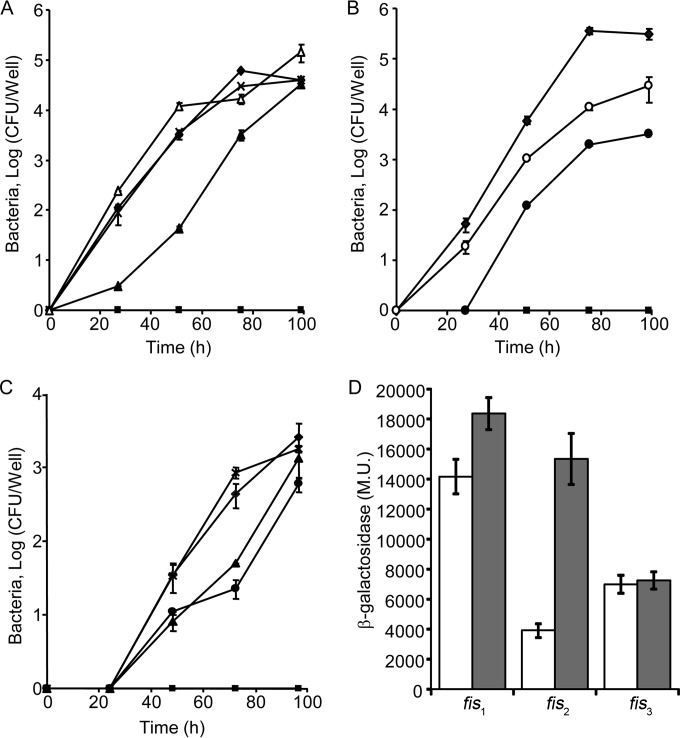
Previously, examination of L. pneumophila mutants with deletions of genes encoding regulators of effector-encoding genes such as PmrA and LetA resulted in an intracellular growth phenotype (16, 45). Therefore, we constructed deletion mutants in each of the three Fis regulators and examined them for intracellular growth in the amoeba host Acanthamoeba castellanii. Examination of these mutants revealed that fis2 (lpg1370) had no intracellular growth defect in A. castellanii (Fig. 2A). In contrast, the fis1 and fis3 deletion mutants were found to be partially defective for intracellular growth in A. castellanii (Fig. 2A and B). The intracellular growth phenotype of the fis1 and fis3 deletion mutants was complemented by introducing a plasmid containing the fis1 and fis3 genes, respectively, cloned under control of the Ptac promoter (induced by IPTG) (Fig. 2A and B). These three deletion mutants were also examined for intracellular growth in HL-60-derived human macrophages (Fig. 2C). In these cells, both the fis1 and fis3 deletion mutants showed mild intracellular growth phenotypes, and the fis2 deletion mutant had no intracellular phenotype (Fig. 2C).
FIG 2.
Fis1 and Fis3 are partially required for L. pneumophila intracellular growth in A. castellanii. (A) Intracellular growth assays of fis1 and fis2 deletion mutants in A. castellanii. (B) Intracellular growth assays of the fis3 deletion mutant in A. castellanii. (C) Intracellular growth assays of fis1, fis2, and fis3 deletion mutants in HL-60-derived human macrophages. Symbols: diamond, L. pneumophila wild type (JR32) containing the vector pMMB207; square, icmT deletion mutant containing the vector pMMB207; filled triangle, fis1 deletion mutant containing the vector pMMB207; open triangle, fis1 deletion mutant containing the complementing plasmid pZT-207-Ptac-0542; X, fis2 deletion mutant containing the vector pMMB207; filled circle, fis3 deletion mutant containing the vector pMMB207; open circle, fis3 deletion mutant containing the complementing plasmid pZT-207-Ptac-fis3. The experiment was performed as described in Materials and Methods. The experiments were performed three times, and similar results were obtained. (D) The expression levels of fis1, fis2, and fis3 translational lacZ fusions were examined in the wild-type strain (JR32) at the exponential phase (white bars) and at the stationary phase (gray bars). β-Galactosidase activity was measured as described in Materials and Methods. Data (expressed in Miller units [M.U.]) are the averages ± standard deviations (error bars) of the results of at least three different experiments.
To obtain additional information about the three fis genes, we determined their levels of expression at the exponential and stationary phases (Fig. 2D). The three fis genes were found to have high levels of expression. Fis1 had the highest level of expression, which was slightly (1.3-fold) higher at stationary phase, Fis2 had a higher (4-fold) level of expression at stationary phase, and Fis3 had the same levels of expression at both exponential and stationary phases (Fig. 2D).
Collectively, these results indicate that the Fis1 and Fis3 regulators, which are evolutionarily closely related to one another, are expressed similarly at the exponential and stationary phases and probably participate in the regulation of pathogenesis-related genes in L. pneumophila.
Identification of effector-encoding genes regulated by Fis.
To further explore the involvement of the three Fis regulators in L. pneumophila effector gene expression, we aimed to identify effector-encoding genes directly regulated by the Fis regulators. Fis was previously shown in other bacteria to function as a nucleoid-associated protein (NAP) and as a direct regulator of gene expression (46, 47). Since we wanted to identify effector-encoding genes directly regulated by Fis, we developed a new procedure in order to identify such genes. We generated a library of 182 upstream regulatory regions fused to the lacZ reporter, including the following: 160 effector-encoding genes (100 of which were constructed for this study; the others were described previously [16, 17, 19, 20, 48, 49]), 12 icm/dot genes, and 10 regulator-encoding genes (see Data set S1 in the supplemental material). In addition, three plasmids in which each of the Fis regulators was cloned under the control of the Ptac promoter (induced by IPTG) were constructed (see Data set S1 in the supplemental material). For each screen, the pool of 182 lacZ fusions and one of the Fis regulators were cotransformed into E. coli, and individual colonies were examined for differences in their β-galactosidase levels under inducing and noninducing conditions (with and without IPTG, respectively). Clones showing marked changes in their levels of expression due to the expression of a certain Fis regulator were collected, the plasmids were prepared, and the genes regulated were indentified by sequencing of the regulatory region found upstream from the lacZ gene (for additional details, see the Materials and Methods section).
This analysis uncovered numerous effector-encoding genes, icm/dot genes, and regulator-encoding genes as potentially regulated by Fis. After genes that came up only once in the three screens were eliminated, 14 effector-encoding genes that came up multiple times with one, two, or all three of the Fis regulators were identified (Table 1). Three of the effector-encoding genes identified were previously shown to be regulated by the PmrAB TCS, and two others were regulated by the CpxRA TCS (Table 1); the other nine effector-encoding genes have no known regulators. The regulation of the effector-encoding genes by the three Fis regulators was further characterized as described below.
TABLE 1.
Genes identified in the screen using the Fis regulators
| Locus | Gene | Known regulator | No. of hits in screen with:a |
||
|---|---|---|---|---|---|
| Fis1 | Fis2 | Fis3 | |||
| lpg0402 | legA9 | PmrA | 2 | ||
| lpg0483 | legA12 | 2 | |||
| lpg0634 | 1 | 2 | 2 | ||
| lpg0926 | ravI | 1 | 1 | 1 | |
| lpg1111 | ravN | 2 | |||
| lpg1137 | ceg20 | PmrA | 1 | 4 | |
| lpg2200 | cegC4 | CpxR | 2 | 1 | |
| lpg2464 | sidM | CpxR | 1 | 1 | |
| lpg2482 | sdbB | PmrA | 3 | ||
| lpg2509 | sdeD | 2 | |||
| lpg2511 | sidC | 2 | |||
| lpg2556 | legK3 | 2 | |||
| lpg2603 | lem28 | 1 | 2 | ||
| lpg2862 | legC8 | 3 | |||
Number of times each gene was identified in each screen.
The effector-encoding genes identified are expressed at higher levels at stationary phase.
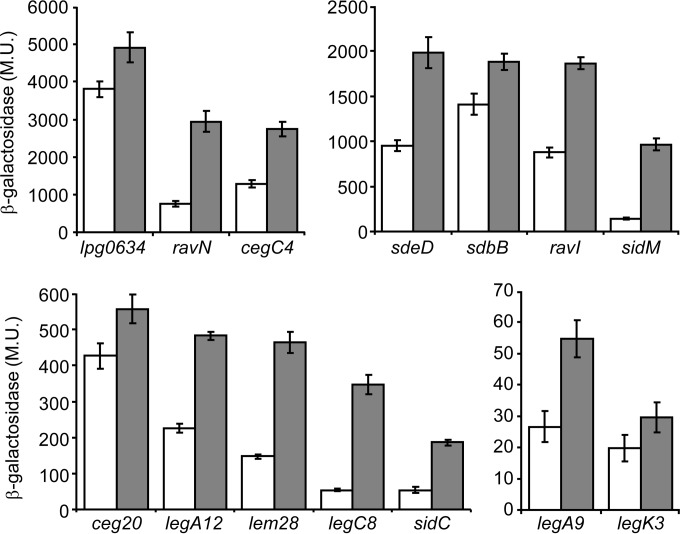
To learn about the expression patterns of the 14 effector-encoding genes identified in the screen, the levels of expression of their lacZ fusions were examined in the L. pneumophila wild-type strain JR32 at exponential and stationary growth phases. These lacZ fusions were found to have different levels of expression, and all of them were expressed at higher levels at stationary phase than at exponential phase (Fig. 3). The expression level at stationary phase was found to be between 1.3- and 7-fold higher than that at exponential phase. This increase was not very strong, and it was similar to the increase that was observed with effector-encoding genes regulated by PmrA (between no effect and up to 5-fold) and much lower than the increase that was observed at stationary phase with genes regulated by the LetAS-RsmYZ-CsrA regulatory cascade (between 5- and 20-fold) (19, 20).
FIG 3.
Effector-encoding genes regulated by Fis are expressed at higher levels at stationary phase. The expression levels of effector translational lacZ fusions (the effectors examined are indicated below the bars) were examined in the wild-type strain (JR32) at the exponential phase (white bars) and at the stationary phase (gray bars). β-Galactosidase activity was measured as described in Materials and Methods. Data (expressed in Miller units [M.U.]) are the averages ± standard deviations (error bars) of the results of at least three different experiments. The effector-encoding genes were divided according to their levels of expression.
Fis1 and Fis3 repress the expression of effector-encoding genes.
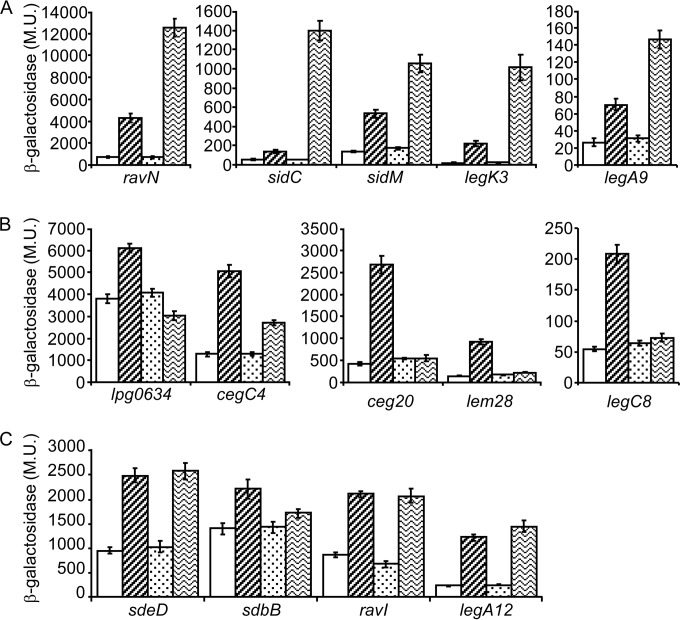
In order to determine if one or more of the three Fis regulators control the expression of effector-encoding genes in L. pneumophila, the levels of expression of the 14 lacZ fusions were examined in the strains containing a deletion in each of the three fis genes. Since Fis1 and Fis3 are expressed similarly at both exponential and stationary phases (Fig. 2D), we wanted to determine the growth phase in which these regulators affect effector gene expression. To this end, we determined the effect of the three fis deletion mutants on several of these genes at both exponential and stationary phases. This examination indicated that the deletion of the Fis regulators had a more pronounced effect at exponential phase (the effect at stationary phase was less than 2-fold with most of the genes examined [data not shown]). These results led us to examine the effect of the three fis deletion mutants on the expression of these 14 effector-encoding genes at exponential phase (Fig. 4). The results obtained made it possible to divide the 14 genes into three groups. The first group contained five genes (ravN, sidC, sidM, legK3, and legA9) whose levels of expression increased (between 5- and 50-fold) in the fis3 deletion mutant, and they were all moderately affected (between 2.6- and 11.3-fold) by the fis1 deletion mutant (Fig. 4A). The second group also contained five genes (lpg0634, cegC4, ceg20, lem28, and legC8), and the expression pattern of this group was the opposite of that of the first group. With these genes a stronger effect was observed in the fis1 deletion mutant (between 1.6- and 6.3-fold), and much weaker effect (between no effect and 2.1-fold) was present in the fis3 deletion mutant (Fig. 4B). The third group contained four genes (sdeD, sdbB, ravI, and legA12), and they were affected similarly by the fis1 and the fis3 deletion mutants (between 1.3- and 6.4-fold) (Fig. 4C). It is important to note that the fis2 deletion mutant had no effect on the levels of expression of the effector-encoding genes examined. This result fits our previous observations showing that Fis2 is evolutionarily distantly related to Fis1 and Fis3 and that it had no effect on L. pneumophila intracellular growth.
FIG 4.
Numerous L. pneumophila effector-encoding genes are repressed by the Fis1 and Fis3 regulators. The expression levels of effector translational lacZ fusions (the effectors examined are indicated below the bars) were examined at exponential phase in the wild-type strain (JR32) (white bars), in the fis1 deletion mutant (ZT-Fis1) (bars with diagonal stripes), in the fis2 deletion mutant (ZT-Fis2) (dotted bars), and in the fis3 deletion mutant (ZT-Fis3) (bars with wavy stripes). (A) effector-encoding genes affected strongly by the fis3 deletion mutant and weakly by the fis1 deletion mutant. (B) Effector-encoding genes affected strongly by the fis1 deletion mutant and weakly by the fis3 deletion mutant. (C) Effector-encoding genes affected similarly by the fis1 and fis3 deletion mutants. β-Galactosidase activity was measured as described in Materials and Methods. Data (expressed in Miller units [M.U.]) are the averages ± standard deviations (error bars) of the results of at least three different experiments. In each panel the effector-encoding genes were divided according to their levels of expression.
Collectively, the results presented clearly indicate that Fis1 and Fis3 function as repressors of effector-encoding genes at exponential phase and that they affect the expression of these genes differently.
Identification of Fis regulatory elements of effector-encoding genes.
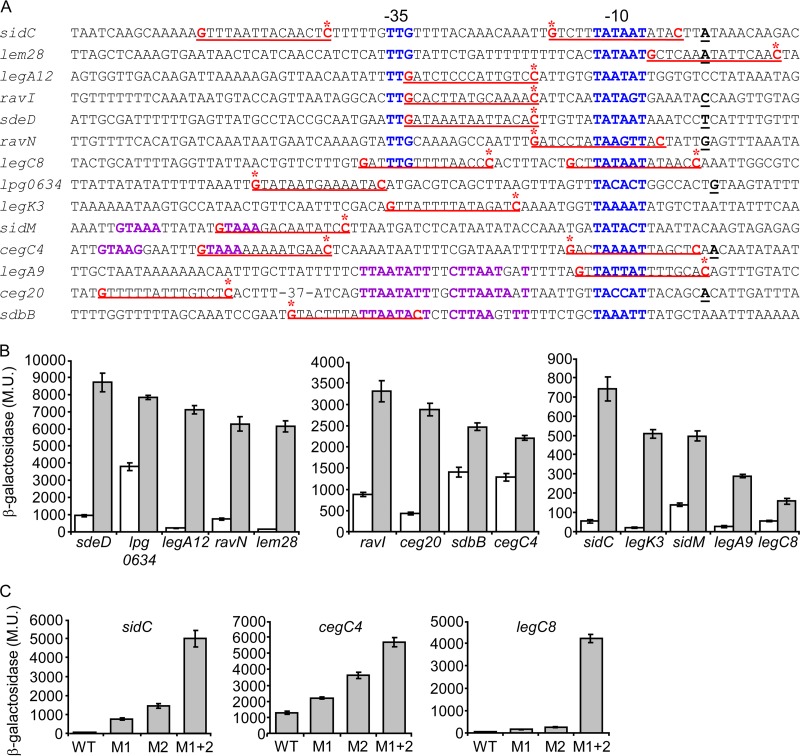
The effect of the Fis regulators on the levels of expression of these 14 effector-encoding genes in E. coli and the results obtained with the L. pneumophila fis1 and fis3 deletion mutants suggest that Fis1 and Fis3 directly regulate the expression of these effector-encoding genes. Therefore, we looked for potential Fis regulatory elements in their upstream regulatory regions. Previous work performed with the E. coli Fis regulator, as well as with Fis regulators from other bacteria, indicated that the Fis regulators usually bind a 17-bp regulatory element which is mostly AT rich, except at positions 2 and 15, where G and C nucleotides are commonly found (50, 51). When we looked for such regulatory elements in the regulatory regions of the 14 effector-encoding genes described, each of them was found to contain at least one and up to four such elements. In addition, it was previously shown that when Fis functions as a repressor, its regulatory elements are usually located very close to the promoter element of the genes repressed (52). Therefore, we utilized the information regarding the transcription start sites available for most of the L. pneumophila genes (53) and focused on putative Fis regulatory elements which overlap or are located very close to the −10 or −35 promoter elements or other known activator binding sites (PmrA and CpxR) of the genes investigated. In order to examine the functionality of these Fis sites, we preformed site-directed mutagenesis of the G and C nucleotides of one putative Fis regulatory element in each of these genes (marked by asterisks in Fig. 5A). We expected to obtain higher levels of expression than in the wild-type fusions due to at least partial relief of the Fis repression from these mutated fusions. As expected, the mutagenesis performed in the regulatory regions of 12 of these genes resulted in an increase in the levels of expression of the mutated lacZ fusions in comparison to the wild-type lacZ fusions (Fig. 5B). For two genes (sidM and legK3) the mutations constructed had no effect on their levels of expression (data not shown). Therefore, we examined a second putative Fis regulatory element present in their regulatory regions, and the mutagenesis of these sites resulted in an increase in the levels of expression of the mutated lacZ fusions (Fig. 5B). As can be seen in Fig. 5B, with some of the effector-encoding genes (such as legA12, lem28, and legK3), very strong increases in the levels of expression (more than 10-fold) were observed after a single nucleotide change in their regulatory regions, indicating that these genes are subjected to very strong repression by Fis. With other effector-encoding genes the increases in the levels of expression observed with the mutated regulatory regions were much lower (around 2-fold) than in the wild-type regulatory region. Therefore, a mutation in a second putative Fis site was constructed in three genes (sidC, legC8, and cegC4), and the second sites were also found to be functional Fis sites (Fig. 5C). Furthermore, mutagenesis of both Fis sites together indicated that these genes are also subjected to very strong repression by Fis since the levels of expression observed with the double mutants were very high in comparison to those of the wild-type lacZ fusions (up to 90-fold) (Fig. 5C).
FIG 5.
Fis regulatory elements identified in the regulatory regions of effector-encoding genes. (A) The regulatory regions of the effectors found to be repressed by the Fis1 and Fis3 regulators are presented. The nucleotides representing the putative Fis consensus are in red and underlined, the transcription start sites are in bold and underlined, the −10 and −35 promoter elements are in blue, the CpxR (sidM and cegC4) and PmrA (legA9, ceg20 and sdbB) consensus sequences are in purple, and the nucleotides that were mutated are marked by asterisks. The effector designations are indicated on the left. (B and C) Mutations constructed in the putative Fis regulatory elements resulted in elevated levels of expression at exponential phase. (B) The expression levels of effector (indicated below the bars) wild-type lacZ fusions (white bars) and lacZ fusions of the same effector containing a mutation in a putative Fis binding site (gray bars) were examined at the exponential phase in the L. pneumophila wild-type strain. The mutations constructed are marked by asterisks in panel A. (C) In three genes (sidC, cegC4, and legC8) two individual mutations in two Fis sites were generated (M1 and M2) as well as a double mutation in both sites together (M1 + 2). For sidC and cegC4 the upstream Fis site was named M1, and the downstream Fis site was named M2. For legC8 the downstream Fis site was named M1, and the upstream Fis site was named M2. β-Galactosidase activity was measured as described in Materials and Methods. Data (expressed in Miller units [M.U.]) are the averages ± standard deviations (error bars) of the results of at least three different experiments. The effector-encoding genes were divided according to their levels of expression.
The results presented demonstrate that the regulatory regions of effector-encoding genes harbor multiple Fis regulatory elements which are used to repress the expression of these effector-encoding genes during exponential phase.
Fis1-His6 and Fis3-His6 proteins directly bind to the regulatory regions of effector-encoding genes.
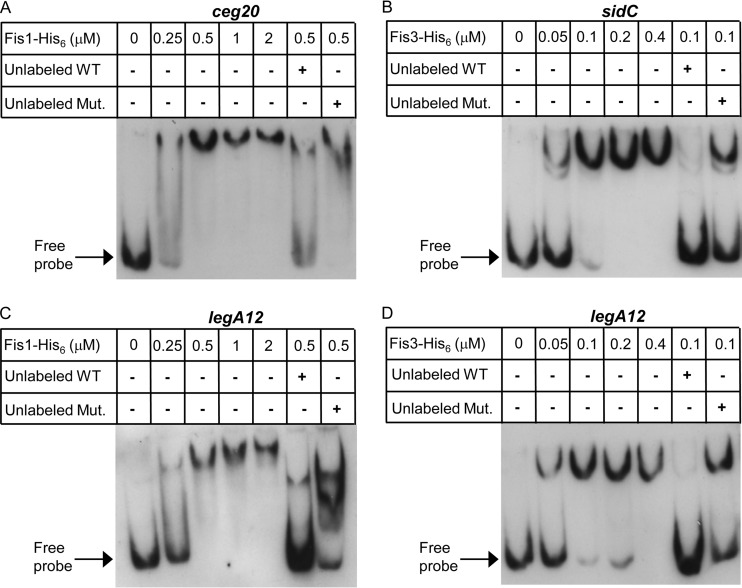
To further support the results presented, the L. pneumophila Fis1 and Fis3 proteins were His tagged, overexpressed, purified, and used for gel mobility shift assays with 150-bp fragments that covered the legA12, ceg20, and sidC regulatory regions. These three genes were chosen for the analysis as representatives of the three groups of genes described above (Fig. 4). sidC represents the group of effectors that were affected strongly by Fis3 (Fig. 4A), ceg20 represents the group of effectors that were affected strongly by Fis1 (Fig. 4B), and legA12 represents the group of effectors that were affected similarly by Fis1 and Fis3 (Fig. 4C). The L. pneumophila Fis1-His6 and Fis3-His6 proteins were found to bind to the regulatory regions of these genes, as evidenced by a shift in the migration of the DNA probe (Fig. 6). The degree of the band shift as well as the amount of the shifted probe correlated with the increasing amounts of the Fis1-His6 and Fis3-His6 proteins (Fig. 6). In addition, competition with unlabeled probe reduced the band shift (Fig. 6, compare the 3rd and 6th lanes in each panel). To further validate the specificity of the binding, we performed competition assays also with unlabeled probes containing mutations in the Fis sites (the mutations examined in the experiment shown in Fig. 5). When the unlabeled mutated probes were used, a dramatic decrease in the competition was observed in comparison to the unlabeled wild-type probes (Fig. 6, compare the 6th and 7th lanes in each panel).
FIG 6.
The L. pneumophila Fis1 and Fis3 proteins bind the regulatory regions of the ceg20, sidC, and legA12 genes. Mobility shift assays were performed with L. pneumophila purified Fis1-His6 (A and C) and Fis3-His6 (B and D) proteins and the DIG-labeled probe (1.6 nM) of ceg20 (A), sidC (B), and legA12 (C and D) regulatory regions. The first lane in each panel did not contain any protein. The rest of the lanes contained increasing amounts of the relevant proteins in 2-fold increments, starting from 0.25 μM for Fis1-His6 and 0.05 μM for Fis3-His6. Competition was performed by incubating the protein amount indicated with a 100-fold excess of unlabeled probe as a specific competitor (Unlabeled WT) or a probe containing mutations in the Fis sites (Unlabeled Mut.) for 15 min prior to the addition of the DIG-labeled probe. For each of the genes examined, the corresponding mutated probe was used.
The mobility shift assays, together with the examination of gene expression in the fis1 and fis3 deletion mutants, and the analysis of mutations in the Fis consensus sequence establish Fis1 and Fis3 as direct regulators of effector-encoding genes in L. pneumophila.
Identification of additional effectors regulated by Fis.
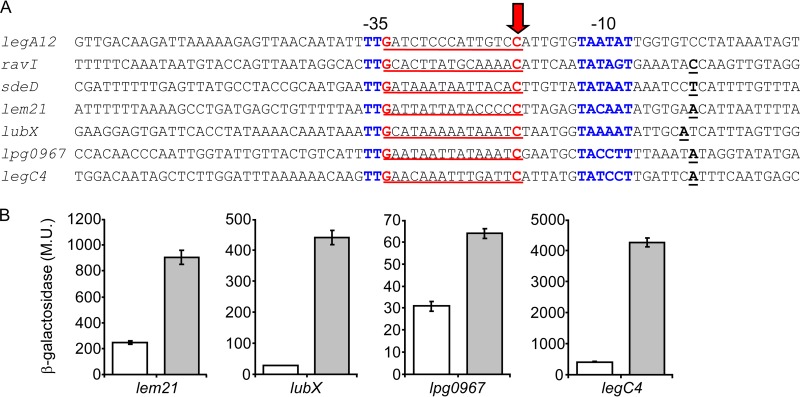
Examination of the regulatory regions of the 14 effector-encoding genes that were found to be regulated by the Fis1 and Fis3 regulators and to harbor a validated Fis regulatory element revealed that in three of these genes (legA12, ravI, and sdeD), the Fis regulatory element overlaps the −35 promoter element in a way that the G residue of the −35 promoter constitutes also the G residue of the Fis regulatory element (Fig. 5A). This observation made it possible to perform a bioinformatics search and to identify additional effector-encoding genes for which the transcription start site was determined (53) and for which the positioning of the −35 promoter, the −10 promoter, and a putative Fis site is similar. This bioinformatics search resulted in the identification of six additional effector-encoding genes (lem21, lubX, lpg0967, mavU, legC4, and mavT) (Fig. 7A and data not shown) which feature a similar organization. To determine if these genes also harbor functional Fis regulatory elements, we mutagenized the C residues of the putative Fis sites in all of these genes and compared the levels of expression of the lacZ fusions containing the wild-type regulatory regions to those of lacZ fusions containing the mutated putative Fis sites. The mutated fusions of four of these genes (lem21, lubX, lpg0967, and legC4) were found to have higher levels of expression than the wild-type lacZ fusions (Fig. 7B). The mutated and wild-type fusions of the two other genes (mavT and mavU) had similar levels of expression (data not shown). This result might be due to additional putative Fis sites present in the upstream regulatory regions of these genes (two additional putative Fis sites were found in the vicinity of the promoter elements of both mavT and mavU), which might compensate for the Fis site mutated, as was shown in the case of legC8 (Fig. 5C).
FIG 7.
Fis regulatory elements identified using a bioinformatics search. (A) The regulatory regions that were found to contain a putative Fis site which overlaps the −35 promoter element of the effector-encoding genes are presented. The nucleotides representing the putative Fis consensus are in red and underlined, the transcription start sites are in bold and underlined, the −10 and −35 promoter elements are in blue, and the C nucleotides that were mutated are marked with an arrow. The effector designations are indicated on the left. (B) Mutations constructed in the putative Fis regulatory elements resulted in elevated levels of expression at exponential phase. The expression levels of effector (indicated below the bars) wild-type lacZ fusions (white bars) and lacZ fusions of the same genes containing a mutation in a putative Fis binding site (gray bars) were examined at the exponential phase in the L. pneumophila wild-type strain. The mutations constructed are marked with a red arrow in panel A. β-Galactosidase activity was measured as described in Materials and Methods. Data (expressed in Miller units [M.U.]) are the averages ± standard deviations (error bars) of the results of at least three different experiments.
Collectively, these results demonstrate that at least 18 effector-encoding genes are repressed by Fis in L. pneumophila.
DISCUSSION
Fis regulators were found to be present in many bacterial species, and they belong to a group of bacterial regulators called nucleoid-associated proteins (NAPs). This group of regulators plays a key role in genome organization, replication, and gene expression (54). Several NAPs have been identified in bacteria, and the most abundant and studied ones are histone-like nucleoid structuring protein (H-NS), integration host factor (IHF), HU, and Fis (54). The involvement of these NAPs in expression of pathogenesis genes was studied in several bacteria, and three of them were studied also in L. pneumophila.
The first of these NAPs, H-NS, is probably the most studied NAP, and it was shown to recognize AT-rich sequences and to function as a homodimer. In Salmonella H-NS was shown to bind to genomic regions that were probably acquired by horizontal gene transfer (55). Many L. pneumophila effectors were shown to be homologous to eukaryotic proteins or to contain eukaryotic domains, and they were probably acquired by horizontal gene transfer from amoebae (11, 49). However, no H-NS-homologous protein was found in L. pneumophila, and in this bacterium silencing of horizontally acquired genes is probably mediated by another regulator. The second NAP, IHF (integration host factor), was shown before in E. coli to recognize a specific sequence [(A/T)ATCAANNNNTT(A/G)] and to function as a heterodimer of two subunits, IHFα and IHFβ. IHF was shown to be involved in regulation of the expression of pathogenesis genes in several bacteria, including Salmonella (56) and EPEC (57). In L. pneumophila, deletion mutants of both IHF subunits (Lpg2709 and Lpg2955) were found to be required for intracellular growth in amoebae but dispensable for intracellular growth in HeLa cells (58). In addition, the L. pneumophila IHF was shown to control the expression of the two small sRNAs, rsmY and rsmZ, which are part of the LetAS-RsmYZ-CsrA regulatory cascade that controls the expression of effector-encoding genes (59). The third NAP, HU, seems to interact with DNA in a nonspecific manner, but it has a preference for binding to distorted regions of the DNA (60). In E. coli HU functions as a heterodimer (HUα and HUβ) or a homodimer (61), and it was shown to control the expression of genes involved in Salmonella virulence (62). L. pneumophila harbors only one HU subunit, HupB (Lpg1858), and it probably functions as a homodimer. The L. pneumophila gene encoding HU was found to be essential and to be expressed during exponential phase (58), but its involvement in L. pneumophila intracellular growth and virulence gene expression was not determined. The fourth NAP, which was the subject of this study, is Fis (factor for inversion stimulation). Fis was shown in E. coli to bind as a homodimer to a consensus sequence that is usually 17 bp in length and AT rich, except at positions 2 and 16, where G and C residues are commonly found (50). Fis was shown to regulate transcription initiation at specific promoters and also to function together with other transcription factors (52). Fis was shown to control the expression of pathogenesis-related genes in several bacteria such as Salmonella and EPEC (35, 36). Most bacteria contain only a single fis gene, but different Legionella species were found to encode three Fis homologs (Fig. 1). The Fis1 and Fis3 regulators are the first L. pneumophila NAPs that were found to directly control the expression of effector-encoding genes (Fig. 4 to 7). Similar to what has been previously shown with several mutants of L. pneumophila regulators, such as rpoS, letA, and pmrA, the fis1 and fis3 regulators were found to have more severe intracellular growth phenotypes in A. castellanii than in HL-60-derived human macrophages (16, 45, 63) (Fig. 2). In addition, Fis1 and Fis3 were found to affect effector gene expression mainly at exponential phase even though they were found to have similar levels of expression at both exponential and stationary phases. This result might indicate that during stationary phase the Fis1 and Fis3 regulators are mainly involved in maintaining the structure of the chromosome as NAPs and less as regulators of gene expression.
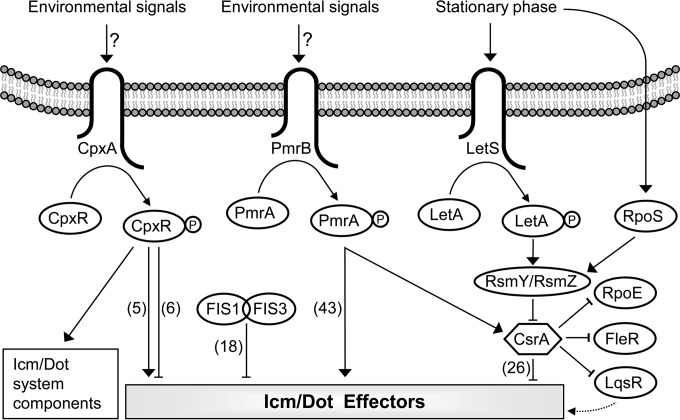
L. pneumophila was found to encode about 300 effectors that are expected to be regulated at the level of gene expression (as well as other levels) in order to result in a successful infection and intracellular growth in host cells. Until now L. pneumophila effector-encoding genes were found to be regulated by four regulatory systems (Fig. 8): (i) the PmrAB TCS, which regulates 43 effector-encoding genes (15, 16); (ii) the CpxRA TCS, which regulates 11 effector-encoding genes (17, 18); (iii) the LetAS-RsmYZ-CsrA regulatory cascade, which regulates 26 effector-encoding genes (19–23); and (iv) the Fis1 and Fis3 repressors, which regulate 18 effector-encoding genes (this study). In addition, several effector-encoding genes were found to be regulated by two of these regulatory systems. PmrA and CpxR were found to regulate together the expression of three effector-encoding genes (ceg7, ceg18, and ceg33); PmrA was found to function as an activator of all these genes, and CpxR was found to function as an activator of two of them and as a repressor of the third (17). CpxR and CsrA were found to regulate together two effector-encoding genes (cegC1 and lpg2461), and with both genes they were found to function as repressors (17, 19; unpublished results). Fis and PmrA were found to control together the expression of three effector-encoding genes (legA9, ceg20, and sdbB); with all of them PmrA was found to function as an activator, and Fis functioned as a repressor (16; this study). Fis and CpxR were also found to control together the expression of two effector-encoding genes (cegC4 and sidM); with both of them CpxR was found to function as an activator, and Fis functioned as a repressor (17; this study). In the two cases where Fis was found to regulate effector-encoding genes together with CpxR and PmrA, Fis might fine-tune the gene expression regulation mediated by these two regulators. Two combinations of coregulation have not yet been found: Fis together with CsrA and PmrA together with CsrA. The first combination might be present since both Fis and CsrA repress the expression of their target genes during exponential phase, and therefore simultaneous regulation at both the transcriptional (Fis) and the translational (CsrA) levels is possible. However, in both cases where Fis was found to function together with another regulator in L. pneumophila, it was found to repress the expression of genes subjected to activation at the level of transcription (by CpxR or PmrA) (see above); functioning together with CsrA would result in regulation in the same direction (repression) at both the transcriptional (Fis) and the translational (CsrA) levels. The absence of genes regulated by both PmrA and CsrA is expected since PmrA was shown to activate the transcription of CsrA (16) that represses the translation of its target genes. Thus, a gene that will be regulated by these two regulators will be activated at the level of transcription (PmrA) and repressed at the level of translation (CsrA) simultaneously, which is not a likely scenario.
FIG 8.
Model of the regulatory systems that control the expression of the L. pneumophila icm/dot genes and effector-encoding genes. The three TCSs (CpxRA, PmrAB, and LetAS), the components of the LetAS-RsmYZ-CsrA regulatory cascade, and the two Fis regulators (Fis1 and Fis3) are schematically illustrated. The environmental signals sensed by CpxA and PmrB are currently not known, and the phosphorylation of these components is expected to be activated by transfer of the phosphate group to their cognate response regulators CpxR and PmrA, respectively, which then directly activate or repress the transcription of their target effector-encoding genes. During stationary phase, the LetAS TCS activates the expression of the small RNAs RsmY and RsmZ that thus sequester CsrA from its target mRNAs and relieve the CsrA posttranscriptional repression. The csrA gene was shown to be under the regulation of the PmrA transcriptional regulator. The numbers of effector-encoding genes which were shown to be regulated by each of the regulatory systems are indicated in parentheses. Solid lines and dashed lines indicate direct and indirect regulation, respectively. Arrows and T-shaped symbols indicate activation and repression, respectively.
Besides coregulation with CpxR and PmrA (Table 1), Fis was also found to function as a sole regulator of effector gene expression (according to our current knowledge). Examination of the effect of Fis deletion mutants on the levels of expression of effector-encoding genes revealed that genes strongly repressed by Fis, such as sidC, legK3, and ravN, were all found to be affected mainly by Fis3 (and to a lesser extent also by Fis1), and no additional regulators are currently known to control their expression. These results suggest that the L. pneumophila Fis regulators probably regulate the expression of effector-encoding genes by themselves or together with other regulators. These two types of regulation by Fis were also shown in other bacteria where Fis was shown to repress the expression of genes by binding to promoter elements or by preventing an activator from binding to its regulatory element (52).
One of the most intriguing observations about the Fis regulators was that L pneumophila harbors three Fis homologs. A similar phenomenon was also found in other Legionella species (Fig. 1). Examination of the other direct regulators of L. pneumophila effector-encoding genes (PmrA, CpxR, and CsrA) (Fig. 8) revealed that Fis is not the only L. pneumophila regulator for which more than a single copy is present in the Legionella genomes. The CsrA posttranscriptional repressor, which is part of the LetAS-RsmYZ-CsrA regulatory cascade, was also found to have several homologs in different Legionella species: L. pneumophila contains five CsrA homologs (Lpg0781, Lpg1593, Lpg1003, Lpg1257, and Lpg2094), L. longbeachae contains four homologs (Llo2071, Llo2874, Llo1850, and Llo1813), L. drancourtii contains seven homologs (LDG5259, LDG8306, LDG7476, LDG7118, LDG6018, LDG5119, and LDG7862), and L. oakridgensis contains three homologs (Loa01097, Loa00186, and Loa01513). Unlike the Fis regulator, where three Fis homologs were found to be present in all the Legionella species examined, the number of CsrA homologs varies between the different Legionella species, but at least three homologs were found in all of them. Reconstruction of the CsrA evolutionary tree revealed that, as in the case of Fis, the three CsrA homologs which are present in all the Legionella genomes examined probably resulted from two duplication events that occurred before the divergence of the Legionella genus (data not shown), implying that, similar to Fis, the three CsrA paralogs were already present in the last common ancestor of the Legionella species. The CpxRA TCS was found to be present in the L. pneumophila genome in a single copy, and it was previously shown to control the expression of icm/dot genes as well as effector-encoding genes (17, 18, 64). However, examination of the available genomic data of other Legionella species revealed that three homologs of the CpxRA TCS are found in L. longbeachae (Llo1781, Llo2778, and Llo1157), one of which (Llo2278) is located inside the icm/dot region II (this copy of the cpxRA operon is not the paralog that was shown to regulate effector-encoding genes in L. pneumophila, which is Llo1781). A similar situation was also found in Legionella dumoffii. These multiple copies of regulators from which at least one was shown to participate in the regulation of effector-encoding genes are intriguing, and it is tempting to speculate that these duplication events occurred in order to fit these regulatory systems to the large number of effectors present in the different Legionella species and to allow fine-tuning of the expression of their target genes or to make it possible to respond to multiple stimuli. Further study is required in order to decipher the involvement of all of these homologous regulatory systems in the regulation of effector-encoding genes in different Legionella species.
Supplementary Material
ACKNOWLEDGMENT
This research was supported by Israeli Science Foundation grant 479/11 (to G.S.).
Footnotes
Published ahead of print 15 September 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.02017-14.
REFERENCES
- 1.Fields BS, Benson RF, Besser RE. 2002. Legionella and Legionnaires' disease: 25 years of investigation. Clin. Microbiol. Rev. 15:506–526. 10.1128/CMR.15.3.506-526.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Steinert M, Hentschel U, Hacker J. 2002. Legionella pneumophila: an aquatic microbe goes astray. FEMS Microbiol. Rev. 26:149–162. 10.1111/j.1574-6976.2002.tb00607.x. [DOI] [PubMed] [Google Scholar]
- 3.Cianciotto NP. 2001. Pathogenicity of Legionella pneumophila. Int. J. Med. Microbiol. 291:331–343. 10.1078/1438-4221-00139. [DOI] [PubMed] [Google Scholar]
- 4.Gimenez G, Bertelli C, Moliner C, Robert C, Raoult D, Fournier PE, Greub G. 2011. Insight into cross-talk between intra-amoebal pathogens. BMC Genomics 12:542. 10.1186/1471-2164-12-542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Franco IS, Shuman HA, Charpentier X. 2009. The perplexing functions and surprising origins of Legionella pneumophila type IV secretion effectors. Cell Microbiol. 11:1435–1443. 10.1111/j.1462-5822.2009.01351.x. [DOI] [PubMed] [Google Scholar]
- 6.Fields BS. 1996. The molecular ecology of legionellae. Trends Microbiol. 4:286–290. 10.1016/0966-842X(96)10041-X. [DOI] [PubMed] [Google Scholar]
- 7.Horwitz MA. 1983. Formation of a novel phagosome by the Legionnaires' disease bacterium (Legionella pneumophila) in human monocytes. J. Exp. Med. 158:1319–1331. 10.1084/jem.158.4.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tilney LG, Harb OS, Connelly PS, Robinson CG, Roy CR. 2001. How the parasitic bacterium Legionella pneumophila modifies its phagosome and transforms it into rough ER: implications for conversion of plasma membrane to the ER membrane. J. Cell Sci. 114:4637–4650. [DOI] [PubMed] [Google Scholar]
- 9.Ensminger AW, Isberg RR. 2009. Legionella pneumophila Dot/Icm translocated substrates: a sum of parts. Curr. Opin. Microbiol. 12:67–73. 10.1016/j.mib.2008.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shin S, Roy CR. 2008. Host cell processes that influence the intracellular survival of Legionella pneumophila. Cell Microbiol. 10:1209–1220. 10.1111/j.1462-5822.2008.01145.x. [DOI] [PubMed] [Google Scholar]
- 11.Gomez-Valero L, Rusniok C, Cazalet C, Buchrieser C. 2011. Comparative and functional genomics of Legionella identified eukaryotic like proteins as key players in host-pathogen interactions. Front. Microbiol. 2:208. 10.3389/fmicb.2011.00208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lifshitz Z, Burstein D, Peeri M, Zusman T, Schwartz K, Shuman HA, Pupko T, Segal G. 2013. Computational modeling and experimental validation of the Legionella and Coxiella virulence-related type-IVB secretion signal. Proc. Natl. Acad. Sci. U. S. A. 110:E707–E715. 10.1073/pnas.1215278110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dorer MS, Kirton D, Bader JS, Isberg RR. 2006. RNA interference analysis of Legionella in Drosophila cells: exploitation of early secretory apparatus dynamics. PLoS Pathog. 2:e34. 10.1371/journal.ppat.0020034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.O'Connor TJ, Boyd D, Dorer MS, Isberg RR. 2012. Aggravating genetic interactions allow a solution to redundancy in a bacterial pathogen. Science 338:1440–1444. 10.1126/science.1229556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Al-Khodor S, Kalachikov S, Morozova I, Price CT, Abu Kwaik Y. 2009. The PmrA/PmrB two-component system of Legionella pneumophila is a global regulator required for intracellular replication within macrophages and protozoa. Infect. Immun. 77:374–786. 10.1128/IAI.01081-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zusman T, Aloni G, Halperin E, Kotzer H, Degtyar E, Feldman M, Segal G. 2007. The response regulator PmrA is a major regulator of the icm/dot type IV secretion system in Legionella pneumophila and Coxiella burnetii. Mol. Microbiol. 63:1508–1523. 10.1111/j.1365-2958.2007.05604.x. [DOI] [PubMed] [Google Scholar]
- 17.Altman E, Segal G. 2008. The response regulator CpxR directly regulates the expression of several Legionella pneumophila icm/dot components as well as new translocated substrates. J. Bacteriol. 190:1985–1996. 10.1128/JB.01493-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gal-Mor O, Segal G. 2003. Identification of CpxR as a positive regulator of icm and dot virulence genes of Legionella pneumophila. J. Bacteriol. 185:4908–4919. 10.1128/JB.185.16.4908-4919.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nevo O, Zusman T, Rasis M, Lifshitz Z, Segal G. 2014. Identification of Legionella pneumophila effectors regulated by the LetAS-RsmYZ-CsrA regulatory cascade, many of which modulate vesicular trafficking. J. Bacteriol. 196:681–692. 10.1128/JB.01175-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rasis M, Segal G. 2009. The LetA-RsmYZ-CsrA regulatory cascade, together with RpoS and PmrA, post-transcriptionally regulates stationary phase activation of Legionella pneumophila Icm/Dot effectors. Mol. Microbiol. 72:995–1010. 10.1111/j.1365-2958.2009.06705.x. [DOI] [PubMed] [Google Scholar]
- 21.Hammer BK, Tateda ES, Swanson MS. 2002. A two-component regulator induces the transmission phenotype of stationary-phase Legionella pneumophila. Mol. Microbiol. 44:107–118. 10.1046/j.1365-2958.2002.02884.x. [DOI] [PubMed] [Google Scholar]
- 22.Sahr T, Bruggemann H, Jules M, Lomma M, Albert-Weissenberger C, Cazalet C, Buchrieser C. 2009. Two small ncRNAs jointly govern virulence and transmission in Legionella pneumophila. Mol. Microbiol. 72:741–762. 10.1111/j.1365-2958.2009.06677.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Molofsky AB, Swanson MS. 2003. Legionella pneumophila CsrA is a pivotal repressor of transmission traits and activator of replication. Mol. Microbiol. 50:445–461. 10.1046/j.1365-2958.2003.03706.x. [DOI] [PubMed] [Google Scholar]
- 24.Segal G. 2014. The Legionella pneumophila two-component regulatory systems that participate in the regulation of Icm/Dot effectors. Curr. Top. Microbiol. Immunol. 376:35–52. 10.1007/82_2013_346. [DOI] [PubMed] [Google Scholar]
- 25.Gunn JS. 2008. The Salmonella PmrAB regulon: lipopolysaccharide modifications, antimicrobial peptide resistance and more. Trends Microbiol. 16:284–290. 10.1016/j.tim.2008.03.007. [DOI] [PubMed] [Google Scholar]
- 26.Hagiwara D, Yamashino T, Mizuno T. 2004. A Genome-wide view of the Escherichia coli BasS-BasR two-component system implicated in iron-responses. Biosci. Biotechnol. Biochem. 68:1758–1567. 10.1271/bbb.68.1758. [DOI] [PubMed] [Google Scholar]
- 27.McPhee JB, Bains M, Winsor G, Lewenza S, Kwasnicka A, Brazas MD, Brinkman FS, Hancock RE. 2006. Contribution of the PhoP-PhoQ and PmrA-PmrB two-component regulatory systems to Mg2+-induced gene regulation in Pseudomonas aeruginosa. J. Bacteriol. 188:3995–4006. 10.1128/JB.00053-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Merighi M, Ellermeier CD, Slauch JM, Gunn JS. 2005. Resolvase-in vivo expression technology analysis of the Salmonella enterica serovar Typhimurium PhoP and PmrA regulons in BALB/c mice. J. Bacteriol. 187:7407–7416. 10.1128/JB.187.21.7407-7416.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Humphreys S, Rowley G, Stevenson A, Anjum MF, Woodward MJ, Gilbert S, Kormanec J, Roberts M. 2004. Role of the two-component regulator CpxAR in the virulence of Salmonella enterica serotype Typhimurium. Infect. Immun. 72:4654–4661. 10.1128/IAI.72.8.4654-4661.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nevesinjac AZ, Raivio TL. 2005. The Cpx envelope stress response affects expression of the type IV bundle-forming pili of enteropathogenic Escherichia coli. J. Bacteriol. 187:672–686. 10.1128/JB.187.2.672-686.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nakayama S, Watanabe H. 1998. Identification of cpxR as a positive regulator essential for expression of the Shigella sonnei virF gene. J. Bacteriol. 180:3522–3528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Martinez LC, Yakhnin H, Camacho MI, Georgellis D, Babitzke P, Puente JL, Bustamante VH. 2011. Integration of a complex regulatory cascade involving the SirA/BarA and Csr global regulatory systems that controls expression of the Salmonella SPI-1 and SPI-2 virulence regulons through HilD. Mol. Microbiol. 80:1637–1656. 10.1111/j.1365-2958.2011.07674.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bhatt S, Romeo T, Kalman D. 2011. Honing the message: post-transcriptional and post-translational control in attaching and effacing pathogens. Trends Microbiol. 19:217–224. 10.1016/j.tim.2011.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Molofsky AB, Swanson MS. 2004. Differentiate to thrive: lessons from the Legionella pneumophila life cycle. Mol. Microbiol. 53:29–40. 10.1111/j.1365-2958.2004.04129.x. [DOI] [PubMed] [Google Scholar]
- 35.Wang H, Liu B, Wang Q, Wang L. 2013. Genome-wide analysis of the salmonella Fis regulon and its regulatory mechanism on pathogenicity islands. PLoS One 8:e64688. 10.1371/journal.pone.0064688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Goldberg MD, Johnson M, Hinton JC, Williams PH. 2001. Role of the nucleoid-associated protein Fis in the regulation of virulence properties of enteropathogenic Escherichia coli. Mol. Microbiol. 41:549–559. 10.1046/j.1365-2958.2001.02526.x. [DOI] [PubMed] [Google Scholar]
- 37.Falconi M, Prosseda G, Giangrossi M, Beghetto E, Colonna B. 2001. Involvement of FIS in the H-NS-mediated regulation of virF gene of Shigella and enteroinvasive Escherichia coli. Mol. Microbiol. 42:439–452. 10.1046/j.1365-2958.2001.02646.x. [DOI] [PubMed] [Google Scholar]
- 38.Sadosky AB, Wiater LA, Shuman HA. 1993. Identification of Legionella pneumophila genes required for growth within and killing of human macrophages. Infect. Immun. 61:5361–5373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zusman T, Yerushalmi G, Segal G. 2003. Functional similarities between the icm/dot pathogenesis systems of Coxiella burnetii and Legionella pneumophila. Infect. Immun. 71:3714–3723. 10.1128/IAI.71.7.3714-3723.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Casadaban MJ, Cohen SN. 1980. Analysis of gene control signals by DNA fusion and cloning in Escherichia coli. J. Mol. Biol. 138:179–207. 10.1016/0022-2836(80)90283-1. [DOI] [PubMed] [Google Scholar]
- 41.Segal G, Shuman HA. 1997. Characterization of a new region required for macrophage killing by Legionella pneumophila. Infect. Immun. 65:5057–5066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ho SN, Hunt HD, Horton RM, Pullen JK, Pease LR. 1989. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77:51–59. 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 43.Segal G, Shuman HA. 1999. Legionella pneumophila utilize the same genes to multiply within Acanthamoeba castellanii and human macrophages. Infect. Immun. 67:2117–2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brickman E, Soll L, Beckwith J. 1973. Genetic characterization of mutations which affect catabolite-sensitive operons in Escherichia coli, including deletions of the gene for adenyl cyclase. J. Bacteriol. 116:582–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gal-Mor O, Segal G. 2003. The Legionella pneumophila GacA homolog (LetA) is involved in the regulation of icm virulence genes and is required for intracellular multiplication in Acanthamoeba castellanii. Microb. Pathog. 34:187–194. 10.1016/S0882-4010(03)00027-5. [DOI] [PubMed] [Google Scholar]
- 46.Duprey A, Reverchon S, Nasser W. 2014. Bacterial virulence and Fis: adapting regulatory networks to the host environment. Trends Microbiol. 22:92–99. 10.1016/j.tim.2013.11.008. [DOI] [PubMed] [Google Scholar]
- 47.Dorman CJ. 2013. Co-operative roles for DNA supercoiling and nucleoid-associated proteins in the regulation of bacterial transcription. Biochem. Soc. Trans. 41:542–547. 10.1042/BST20120222. [DOI] [PubMed] [Google Scholar]
- 48.Zusman T, Degtyar E, Segal G. 2008. Identification of a hypervariable region containing new Legionella pneumophila Icm/Dot translocated substrates by using the conserved icmQ regulatory signature. Infect. Immun. 76:4581–4591. 10.1128/IAI.00337-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Degtyar E, Zusman T, Ehrlich M, Segal G. 2009. A Legionella effector acquired from protozoa is involved in sphingolipids metabolism and is targeted to the host cell mitochondria. Cell Microbiol. 11:1219–1135. 10.1111/j.1462-5822.2009.01328.x. [DOI] [PubMed] [Google Scholar]
- 50.Cho BK, Knight EM, Barrett CL, Palsson BO. 2008. Genome-wide analysis of Fis binding in Escherichia coli indicates a causative role for A-/AT-tracts. Genome Res. 18:900–910. 10.1101/gr.070276.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hengen PN, Bartram SL, Stewart LE, Schneider TD. 1997. Information analysis of Fis binding sites. Nucleic Acids Res. 25:4994–5002. 10.1093/nar/25.24.4994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Browning DF, Grainger DC, Busby SJ. 2010. Effects of nucleoid-associated proteins on bacterial chromosome structure and gene expression. Curr. Opin. Microbiol. 13:773–780. 10.1016/j.mib.2010.09.013. [DOI] [PubMed] [Google Scholar]
- 53.Sahr T, Rusniok C, Dervins-Ravault D, Sismeiro O, Coppee JY, Buchrieser C. 2012. Deep sequencing defines the transcriptional map of L. pneumophila and identifies growth phase-dependent regulated ncRNAs implicated in virulence. RNA Biol. 9:503–519. 10.4161/rna.20270. [DOI] [PubMed] [Google Scholar]
- 54.Dillon SC, Dorman CJ. 2010. Bacterial nucleoid-associated proteins, nucleoid structure and gene expression. Nat. Rev. Microbiol. 8:185–195. 10.1038/nrmicro2261. [DOI] [PubMed] [Google Scholar]
- 55.Navarre WW, Porwollik S, Wang Y, McClelland M, Rosen H, Libby SJ, Fang FC. 2006. Selective silencing of foreign DNA with low GC content by the H-NS protein in Salmonella. Science 313:236–238. 10.1126/science.1128794. [DOI] [PubMed] [Google Scholar]
- 56.Fass E, Groisman EA. 2009. Control of Salmonella pathogenicity island-2 gene expression. Curr. Opin. Microbiol. 12:199–204. 10.1016/j.mib.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jeong JH, Kim HJ, Kim KH, Shin M, Hong Y, Rhee JH, Schneider TD, Choy HE. 2012. An unusual feature associated with LEE1 P1 promoters in enteropathogenic Escherichia coli (EPEC). Mol. Microbiol. 83:612–622. 10.1111/j.1365-2958.2011.07956.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Morash MG, Brassinga AK, Warthan M, Gourabathini P, Garduno RA, Goodman SD, Hoffman PS. 2009. Reciprocal expression of integration host factor and HU in the developmental cycle and infectivity of Legionella pneumophila. Appl. Environ. Microbiol. 75:1826–1837. 10.1128/AEM.02756-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pitre CA, Tanner JR, Patel P, Brassinga AK. 2013. Regulatory control of temporally expressed integration host factor (IHF) in Legionella pneumophila. Microbiology 159:475–492. 10.1099/mic.0.062117-0. [DOI] [PubMed] [Google Scholar]
- 60.Castaing B, Zelwer C, Laval J, Boiteux S. 1995. HU protein of Escherichia coli binds specifically to DNA that contains single-strand breaks or gaps. J. Biol. Chem. 270:10291–10296. 10.1074/jbc.270.17.10291. [DOI] [PubMed] [Google Scholar]
- 61.Tanaka H, Goshima N, Kohno K, Kano Y, Imamoto F. 1993. Properties of DNA-binding of HU heterotypic and homotypic dimers from Escherichia coli. J. Biochem. 113:568–572. [DOI] [PubMed] [Google Scholar]
- 62.Mangan MW, Lucchini S, Fitzgerald T OCS, Hinton JC, Dorman CJ. 2011. Nucleoid-associated protein HU controls three regulons that coordinate virulence, response to stress and general physiology in Salmonella enterica serovar Typhimurium. Microbiology 157:1075–1087. 10.1099/mic.0.046359-0. [DOI] [PubMed] [Google Scholar]
- 63.Hales LM, Shuman HA. 1999. The Legionella pneumophila rpoS gene is required for growth within Acanthamoeba castellanii. J. Bacteriol. 181:4879–4889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Feldman M, Segal G. 2007. A pair of highly conserved two-component systems participates in the regulation of the hypervariable FIR proteins in different Legionella species. J. Bacteriol. 189:3382–3391. 10.1128/JB.01742-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.