Abstract
Staphylococcus aureus responds to changing extracellular environments in part by adjusting its proteome through alterations of transcriptional priorities and selective degradation of the preexisting pool of proteins. In Bacillus subtilis, the proteolytic adaptor protein MecA has been shown to play a role in assisting with the proteolytic degradation of proteins involved in competence and the oxidative stress response. However, the targets of TrfA, the MecA homolog in S. aureus, have not been well characterized. In this work, we investigated how TrfA assists chaperones and proteases to regulate the proteolysis of several classes of proteins in S. aureus. By fusing the last 3 amino acids of the SsrA degradation tag to Venus, a rapidly folding yellow fluorescent protein, we obtained both fluorescence-based and Western blot assay-based evidence that TrfA and ClpCP are the adaptor and protease, respectively, responsible for the degradation of the SsrA-tagged protein in S. aureus. Notably, the impact of TrfA on degradation was most prominent during late log phase and early stationary phase, due in part to a combination of transcriptional regulation and proteolytic degradation of TrfA by ClpCP. We also characterized the temporal transcriptional regulation governing TrfA activity, wherein Spx, a redox-sensitive transcriptional regulator degraded by ClpXP, activates trfA transcription while repressing its own promoter. Finally, the scope of TrfA-mediated proteolysis was expanded by identifying TrfA as the adaptor that works with ClpCP to degrade antitoxins in S. aureus. Together, these results indicate that the adaptor TrfA adds temporal nuance to protein degradation by ClpCP in S. aureus.
INTRODUCTION
The Gram-positive human pathogen Staphylococcus aureus causes invasive infections of the skin and bloodstream that can lead to life-threatening sepsis, endocarditis, and toxin-mediated syndrome (1). Unfortunately, transmission and infection are commonplace in hospitals and are increasing in frequency in community settings (2). This scenario is in part due to the ability of S. aureus to survive and spread from abiotic surfaces, such as gloves (3) and clothing (4), as well as from biotic niches, like the anterior nares of healthy adults (5).
The transcriptional regulation of numerous stress response mechanisms that facilitate its survival is well described (6, 7). However, posttranslational modifications of the proteome in S. aureus are poorly characterized in comparison, even though they play a significant role in the stress response and general housekeeping in many prokaryotes (8). These adjustments include recovery of denatured or aggregated proteins via GroEL- and ClpB-mediated refolding (9), tuning of the temporal and spatial activities of downstream regulatory networks (10), offsetting of starvation by increasing the available pools of amino acids (11), and hedging against unregulated cell death via toxin-antitoxin system activation (12, 13).
In S. aureus, proteolytic degradation is carried out by the intracellular proteases ClpP, FtsH, and ClpQ (also known as HslV). Of the three, ClpP appears to have the greatest impact in shaping the S. aureus proteome (14), and when disrupted, it causes significant defects in growth, survival, virulence gene expression, and cell wall maintenance (15, 16). FtsH has a modest role in regulating growth, stress resistance, starvation survival, and pathogenicity (17), but with a putative localization to the inner membrane, it is thought to mainly degrade membrane-bound targets. Very little is known about ClpQ, in part because a ΔclpQ mutant of S. aureus exhibits few phenotypes (18) and shows few alterations to stress responses.
The manner in which a protein is specifically degraded by one of these proteases is initiated by the identification of a degradation motif during partial ATP-dependent unwinding of the target (19). FtsH contains its own intrinsic unwinding domain to facilitate this process, but ClpP and ClpQ must associate with a member of the AAA+ ATPase family of chaperones (19) to accomplish this. This association with ClpP or ClpQ allows chaperones to unwind and deliver identified targets into the cognate protease's catalytic chamber.
S. aureus has three known chaperones that aid in ClpP- and ClpQ-mediated proteolysis, while a related fourth chaperone, ClpL, lacks the structures necessary to interact with ClpP or ClpQ (20). The chaperone ClpY (also known as HslU) assists ClpQ in recognition of target motifs, and strains lacking clpY have phenotypes similar to those of strains lacking clpQ (18), suggesting that few other partners exist for this chaperone/protease pair. The ClpC and ClpX chaperones associate with ClpP, with each selecting different groups of proteins for ClpP-mediated proteolysis. ClpX is primarily involved in virulence regulation (16), while ClpC tunes how the cell regulates its oxidative stress response (21), tricarboxylic acid cycle (21), toxin-antitoxin system activity (22), and survival in resource-limited medium (23).
Members of a second class of proteins called adaptors assist chaperones in controlling the proteolytic degradation of target substrates (8). These proteins lack the AAA+ ATPase activity found in chaperones and instead facilitate motif identification either through direct interactions with a target (24), through selective motif occlusion (25), or by modification of a protein to alter its degradation rate (26). On the basis of homology to adaptors identified in Bacillus subtilis, there are at least three adaptors in S. aureus, namely, McsB (SA0482), YjbH (SA0860), and TrfA (SA0857). Studies have indicated the involvement of each in various stress responses (27–29), but many gaps remain in our understanding of the various roles played by these adaptors in S. aureus. Details of the protein targets of these S. aureus adaptors are limited, and to date, only two target proteins have been described to be regulated by S. aureus adaptors. McsB targets CtsR for ClpCP degradation (30), and the levels of the redox-sensing transcriptional regulator Spx have been shown to be controlled by the adaptor YjbH (31).
In this work, we investigated how the chaperones and adaptors of S. aureus assist in the proteolysis of several classes of proteins. More specifically, we found that the ClpCP proteolytic system and not the ClpXP system described in Escherichia coli (32) and B. subtilis (33) is essential for SsrA-tagged protein destruction in S. aureus. The adaptor TrfA was discovered to mediate this breakdown, primarily during late-logarithmic- and early-stationary-phase growth. This regulation is likely a result of the transcriptional regulation as well as the growth phase-dependent proteolytic degradation of TrfA by ClpCP. Additionally, a regulatory feedback loop was uncovered for a positive regulator of trfA, whereby Spx represses one of its promoters, which in turns affects trfA transcription. TrfA was also identified as the adaptor involved in the ClpCP-mediated degradation of antitoxins, revealing a role for the modulation of toxin activity in S. aureus (34). Together, these results reveal how the adaptor TrfA temporally tunes the ClpCP-mediated protein degradation of several classes of proteins involved in S. aureus physiology.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
Table 1 contains a list of the bacterial strains used in these studies. E. coli strains were routinely grown in LB, and S. aureus strains were routinely grown in Trypticase soy broth (TSB). Both strains were grown with shaking at 250 rpm at 37°C. Cell density was measured by determination of the absorbance (optical density [OD]) at 650 nm using an 18-mm borosilicate glass tube in a Spectronic 20D+ spectrophotometer. Ampicillin (Amp; 50 μg/ml), chloramphenicol (Cm; 10 μg/ml), or erythromycin (Erm; 2.5 μg/ml) was added to the appropriate medium when required by a particular strain. To disrupt transcription or protein synthesis, rifampin (200 μg/ml) or erythromycin (50 μg/ml) was added to the cultures, respectively. Promoter induction of pEPSA5 was performed using 0.1 to 0.5% xylose, when necessary. Thiol stress was induced by the addition of 5 mM diamide [1,1-azo-bis(N,N-dimethylformamide); Sigma-Aldrich].
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Genotype or characteristics | Source or reference |
|---|---|---|
| Strains | ||
| S. aureus | ||
| RN4220 | Heavily mutagenized derivative of NCTC 8325-4 | 63 |
| SH1000 | NCTC 8325-4 with repaired rsbU gene | 64 |
| JM27 | SH1000 ftsH::tetL | 17 |
| NCTC 8325-4 Δspx | NCTC 8325-4 Δspx | 60 |
| NCTC 8325-4 spx complement | NCTC 8325-4 spx mutant complemented with spx | 60 |
| ALC4976 | SH1000 ΔclpB | 22 |
| ALC4977 | SH1000 ΔclpC | 22 |
| ALC4978 | SH1000 ΔclpL | 22 |
| ALC4979 | SH1000 ΔclpQ | 22 |
| ALC4980 | SH1000 ΔclpX | 22 |
| ALC4981 | SH1000 ΔclpY | 22 |
| ALC5105 | SH1000 ΔclpP | 22 |
| ALC6490 | SH1000 clpC mutant complemented with clpC | 22 |
| ALC6491 | SH1000 clpP mutant complemented with clpP | 22 |
| ALC7010 | SH1000 ΔmcsB | This work |
| ALC7011 | SH1000 ΔtrfA | This work |
| ALC7137 | SH1000 ΔyjbH | This work |
| ALC7272 | SH1000 trfA mutant complemented with trfA | This work |
| E. coli XL1-Blue | General cloning strain | Agilent |
| Plasmids | ||
| pMAD | E. coli/S. aureus shuttle plasmid with the ori pE194ts; bgaB Ampr Ermr | 38 |
| pEPSA5 | Xylose-inducible shuttle vector; Ampr Cmr | 37 |
| pSK236 | S. aureus/E. coli shuttle vector with pUC19 cloned into the HindIII site of pC194 | 65 |
| pALC1484 | pSK236 with promoterless gfp-uvr, a cycle 3 gfp allele | 66 |
| pALC6188 | pEPSA5 with mazE with a 3′ myc tag and sarA ribosome binding site | 22 |
| pALC6486 | pEPSA5 with axe1 with a 5′ myc tag and sarA ribosome binding site | 22 |
| pALC6489 | pEPSA5 with axe2 with a 5′ myc tag and sarA ribosome binding site | 22 |
| pALC6954 | pSK236 with a phage repressor promoter and codon-optimized venus | This work |
| pALC6955 | pSK236 with a phage repressor promoter and codon-optimized venus-VAA | This work |
| pALC7016 | pEPSA5 with codon-optimized venus-VAA and sarA ribosome binding site | This work |
| pALC7113 | pEPSA5 with spx with a 5′ myc tag and sarA ribosome binding site | This work |
| pALC7151 | pEPSA5 with trfA with a 5′ myc tag and sarA ribosome binding site | This work |
| pALC7257 | pMAD::trfA (sa0857) complement | This work |
| pALC7422 | pSK236 with a phage repressor promoter and trfA with a 5′ myc tag | This work |
| pALC7867 | pMAD::ΔtrfA (sa0857) | This work |
| pALC7868 | pMAD::ΔmcsB (sa0482) | This work |
| pALC7869 | pMAD::ΔyjbH (sa0860) | This work |
| pALC7803 | pALC1484 with the spx P1 promoter in EcoRI and XbaI sites | This work |
| pALC7804 | pALC1484 with the spx P2 promoter in EcoRI and XbaI sites | This work |
| pALC7901 | pSK236 with a phage repressor promoter and codon-optimized venus-SsrA | This work |
DNA manipulations.
E. coli plasmid purification was performed using Qiagen miniprep kits (Qiagen) per the manufacturer's instructions, while plasmid isolation from S. aureus was performed as described previously (35). Plasmid transformations in S. aureus were achieved via electroporation with a MicroPulser apparatus (Bio-Rad), using RN4220 as an intermediate between E. coli and relevant S. aureus strains.
Codon optimization of venus.
To create a construct that yields a rapidly maturing fluorescent protein, the Venus gene was codon optimized for S. aureus using the codon frequencies calculated from the S. aureus COL genome, while avoiding the generation of additional restriction enzyme sites. Subsequently, 42-bp primers staggered to each neighboring sequence by 21 bp were combined in assembly reactions using HiFi PCR Supermix (Invitrogen) to create ∼300-bp fragments of venus, followed by the addition of flanking primers at the 5′ and 3′ ends of each 300-bp fragment using a new PCR. Following gel purification, equimolar amounts of each fragment were combined for a second assembly reaction with primers at the 5′ and 3′ ends of venus, and the PCR product was purified and sequenced. Both the primers and S. aureus codon-optimized venus sequences are available upon request.
Plasmid construction.
To ensure the robust expression of our codon-optimized venus gene, we first cloned a strong phage repressor promoter of saOUHSC_02234 from a lysogenized phage (36) into the EcoRI-XbaI restriction sites of shuttle plasmid pSK236. To place venus under the control of the phage repressor promoter, we cloned venus together with an upstream sarA ribosomal binding site into the SalI-PstI sites of pSK236. To add the S. aureus SsrA tag GKSNNNFAVAA (25) to the C terminus of Venus (in which the construct was designated venus-SsrA), the following primers were used: 5′-AACTGCAGTTATGCTGCAACCTTGTAAAGTTCATCCATTCC-3′ and 5′-AACTGCAGTTAGGCAGCTACTGCGAAATTATTGTTTGATTTGCCAGTCTTGTAAAGTTCATCCATTCCT-3′. This sequence diverges from that of Flynn et al. (25) by its lack of an alanine at its N terminus, as this residue was not found in genomic sequences of clinical and laboratory isolates (data not shown). To add the VAA tag to the C terminus of Venus (in which the construct was designated venus-VAA), the following primer was used in place of the second primer listed above: 5′-ACGCGTCGACGGAGGTTTTAAACATGGTTTCTAAAGGTGAA-3′. To construct a separate recombinant plasmid for the induction of Venus expression, we cloned venus and the venus-VAA variant into the EcoRI and BamHI sites of pEPSA5 containing a xylose-inducible promoter (37).
For plasmid constructs capable of expressing trfA or spx under induction, we used a cloning strategy in pEPSA5 similar to the one used for venus. Briefly, trfA or spx was amplified by PCR with a 5′ codon-optimized myc tag, and the amplified product was cloned into the EcoRI and BamHI sites of pEPSA5. In addition, S. aureus SH1000 strains capable of overexpressing myc-tagged antitoxin genes from S. aureus under the control of the xylose promoter of pEPSA5 were constructed as previously described (22).
To generate an spx promoter reporter construct, we cloned the distal P1 promoter or proximal P2 promoter region of spx (starting 245 bp or 102 bp before the spx start codon, respectively; amplification primers are available on request) into the EcoRI and XbaI sites of pALC1484, a shuttle plasmid containing a green fluorescent protein (GFP) reporter gene. Following sequencing verification, the plasmids were used to transform RN4220 and subsequent strains.
Construction of deletion strains.
All sequenced S. aureus genomes contain the following three adaptor genes: mcsB (sa0482), trfA (sa0857), and yjbH (sa0860). As our relevant clp deletion strains are constructed in the S. aureus SH1000 background, we proceeded to construct deletion mutants of these adaptor genes in strain SH1000. Deletion mutants were constructed using the temperature-sensitive allelic replacement plasmid pMAD (38). In brief, chromosomal regions 1,000 bp up- and downstream of the gene of interest were amplified by PCR with primers proximal to the deletion site, joined by gene sewing, ligated into pMAD, and transformed into RN4220 and then into SH1000. The mutant was then generated by a temperature shift strategy to yield the mutant, as described previously (38). To generate a pMAD construct to complement a trfA deletion mutant, we inserted a DNA fragment with the intact gene flanked by 1,000 bp on each side into pMAD, followed by allelic replacement in the mutant. All constructs were verified by colony PCR and chromosomal sequencing.
Isolation of RNA and Northern blot hybridization.
Overnight cultures of S. aureus were each diluted 1:1,000 and grown at 37°C with shaking at 250 rpm in 100 ml of TSB inside 500-ml sidearm flasks (flask/medium ratio, 5:1). Cell density was monitored by determination of the absorbance at 650 nm, and at the appropriate OD, aliquots of cells were pelleted and flash frozen at −80°C. RNA extraction was accomplished by lysing the cells with 1 ml of cold TRIzol reagent (Invitrogen) and 0.25 ml of 0.1-mm-diameter silica/glass beads in a Mini-BeadBeater 8 apparatus (Biospec Products) at its maximum setting for two 1-min pulses. RNA purification and subsequent Northern analysis were performed as described previously (39).
Cell lysate preparation and Western blot analyses.
Protein lysates were prepared as described before (22) from cells grown in a fashion similar to that for RNA extraction described above. At an OD of 650 nm (OD650) of 1.1, growth was stalled with antibiotics (either 200 μg/ml rifampin or 50 μg/ml erythromycin, depending on the experiment), and a series of cells (10 ml each at a 15- or 30-min interval) were pelleted at 1°C, flash frozen in liquid nitrogen, and stored at −80°C. For lysis, the pellets were thawed on wet ice, washed twice with ice-cold sample buffer (50 mM Tris-HCl, pH 7.6, 5 mM EDTA), and resuspended in 200 μl of sample buffer to which 100 μl of 0.1-mm-diameter silica/glass beads was added. The cells were then disrupted in a Mini-BeadBeater 8 apparatus at its maximum setting for two 1-min pulses with a 1-min rest, followed by centrifugation at 20,000 × g for 15 min at 1°C. Halt protease inhibitors (1×; Thermo Fisher) were then added to the lysates, and their protein concentrations were determined by Bradford assay using an FL600 microplate reader (BioTek Instruments).
For Western blot assays, SDS-PAGE was first performed with 30 μg of lysate proteins, and then the proteins were transferred to polyvinylidene difluoride (PVDF) membranes using an iBlot Western blot transfer system (Invitrogen). Following blocking in 5% milk in Tris-buffered saline with Tween (TBS-T; 150 mM NaCl and 10 mM Tris, pH 8.0, with 0.1% [vol/vol] Tween 20) for at least 30 min, the membranes were then incubated with either mouse anti-GFP (Abcam) or mouse anti-Myc (Cell Signaling) antibodies in TBS-T for 2 h. The membranes were then washed in TBS-T and incubated with horseradish peroxidase-conjugated goat anti-mouse secondary antibody (Jackson ImmunoResearch). After additional washing, enhanced chemiluminescence (ECL) reagent (GE Healthcare) was added and the membrane was exposed to film. Scanned images from nonsaturated exposed film were then analyzed densitometrically using ImageJ software, and the half-lives of the proteins of interest were calculated using Prism (version 5) software (GraphPad).
Fluorescence analysis.
To analyze the fluorescence of S. aureus cells expressing Venus or its variants, cells were monitored using one of two machines. For single time points, results for 200 μl of cells at various time points were read in a 96-well plate (Costar plate 3632) in an FL600 plate reader (BioTek) with 515-nm excitation and 540-nm emission filters (Omega Optical). In experiments with multiple time points, 150 μl of cells was shaken in a 96-well plate (Costar plate 3632) with a heated top plate to prevent condensation at 250 rpm and 37°C in a Tecan Infinite M1000 Pro apparatus (Tecan). Fluorescence was recorded using 515-nm excitation and 528-nm emission filters and 650-nm absorbance filters. In both cases, data are reported as the level of fluorescence per absorbance unit to normalize the cell density.
RESULTS
Establishing an in vivo assay for SsrA-tagged proteins in S. aureus using Venus fluorescence.
During translation, ribosomes may stall for a variety of reasons, including stress, starvation, or premature degradation of the template mRNA. To reset the translational machinery, a hybrid RNA called transfer-messenger RNA (tmRNA) enters the open A site of the ribosome, shifting translation from the degraded or truncated mRNA to a small open reading frame with a stop codon within the tmRNA (40). The resulting 11-amino-acid tag, called the SsrA tag, is appended to the C terminus of the nascent but truncated protein and is then recognized as a signal for rapid degradation by ClpAP and ClpXP in E. coli (32) or ClpXP in B. subtilis (33). As the half-lives of SsrA-tagged proteins are short, reportedly <10 min (41), the tracking of such proteins is difficult in vivo. To investigate this system in S. aureus and to obtain an improved sensitivity compared to that obtained with the slowly folding non-codon-optimized cycle 3 gfp (42), we utilized a fast-folding yellow fluorescent protein (YFP)-tagged Venus variant (SEYFP-F46L) (43) which exhibits an accelerated oxidation step as a result of the F46L mutation, leading to rapid and enhanced fluorescence (43). In S. aureus, the SsrA tag has been predicted to be AGKSNNNFAVAA (25), but it has never been tested. Given that the N-terminal alanine of the predicted S. aureus sequence is not conserved in sequenced genomes (data not shown), we chose instead to append the sequence GKSNNNFAVAA to the C terminus of Venus. Furthermore, identifying the minimal unit of SsrA necessary for degradation was also of interest. Based on prior work in other organisms (44) suggesting that the final 3 amino acids are sufficient as a degradation signal, we created an additional construct appending the amino acids VAA to the C terminus of Venus.
Following introduction of the recombinant plasmids into S. aureus strain SH1000, initial work revealed significant fluorescence, while the cells exhibited no retardation of growth in the presence of any of these plasmids (data not shown). To verify that the SsrA tag and the abbreviated VAA tag on Venus were produced and functional, cells with the venus construct (pALC6954), the venus-VAA construct (pALC6955), or the venus-SsrA construct (pALC7901) were grown in TSB as described in Materials and Methods until the OD650 reached 1.1, whereupon 200 μg/ml of rifampin was added to the culture and samples were withdrawn at predetermined time points for 2 h. Western blot analysis of whole-cell lysates probed with a mouse anti-GFP antibody capable of detecting Venus showed that the Venus protein levels rapidly decreased in both the strain containing Venus-VAA (pALC6955) and the strain containing Venus-SsrA (pALC7901), but no equivalent decrease was observed in cells producing untagged Venus (pALC6954) (Fig. 1A). This finding implied that Venus is stable in S. aureus and that addition of SsrA, notably, just the last 3 amino acids of such a tag (VAA), was sufficient to destabilize it. Densitometric analysis indicated that the expressed Venus-VAA construct had a half-life of 47 min, while the Venus-SsrA construct had a half-life of 27 min, suggesting that while VAA is sufficient for degradation, other residues of the SsrA tag may play a role in proteolytic degradation.
FIG 1.
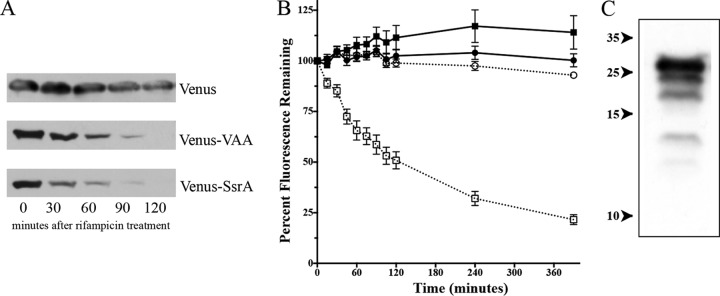
Venus and Venus-VAA protein levels in S. aureus SH1000 wild-type cells following rifampin treatment. (A) Western blot analysis of lysates of wild-type SH1000 with either pALC6954 (producing Venus), pALC6955 (producing Venus-VAA), or pALC7901 (producing Venus-SsrA) grown to an OD650 of 1.1 after rifampin treatment (200 μg/ml). The blot was probed with an anti-GFP primary antibody and then a secondary horseradish peroxidase-conjugated goat anti-mouse antibody. The experiments were repeated three times, and a representative blot of each set of lysates is displayed. (B) Venus fluorescence emitted from strains ALC6954 (producing Venus; circles) and ALC6955 (producing Venus-VAA; squares) either stalled by 50 μg/ml erythromycin after reaching an OD650 of 1.1 (open symbols) or not treated with drug (closed symbols). The fluorescence at 540 nm was observed following excitation at 515 nm and normalized to the cell density at the OD650. Values were then compared to the fluorescence at time zero. The result for each time point is the mean and standard deviation of three biological replicates. (C) Degradative laddering of Venus-VAA. The lysate from wild-type SH1000 expressing Venus-VAA from pALC6955 was resolved and immunoblotted with an anti-GFP primary antibody and a secondary horseradish peroxidase-conjugated goat anti-mouse antibody. Exposure to film was extended to reveal lower-molecular-mass bands. Numbers on the left are molecular masses (in kilodaltons).
To pursue our observation that the VAA residues of SsrA were sufficient for proteolytic degradation, we examined whether there was a functional correlation of protein degradation to fluorescence loss. To accomplish this, we used erythromycin (50 μg/ml) at an OD650 of 1.1 to stop protein synthesis (rifampin interferes with the fluorescence spectrum), followed by monitoring of cell density and fluorescence over 6 h. As seen in Fig. 1B, Venus and Venus-VAA levels remained largely unchanged over time in the absence of erythromycin. However, when erythromycin was added, the fluorescence levels of Venus-VAA (ALC6955) began to decrease with a half-life of 75 min, whereas the levels in ALC6954 (Venus) remained unchanged. This result suggested that the fluorescence associated with the degradation of Venus with the SsrA tag was reduced in the cell, albeit more slowly than the reduction of the protein (half-life, 47 min), as determined by Western blotting (Fig. 1A). The reason for this discrepancy is unknown but may be due to either Venus-VAA molecules retaining the capacity for emitting fluorescence even when partially degraded or the formation of in vivo foci that affect overall fluorescence (45). In support of the former possibility, we observed a laddering of lower-molecular-weight bands in Western blots of the Venus-VAA lanes following addition of rifampin (Fig. 1C), suggesting that proteolysis occurred in a stepwise fashion.
clpCP instead of clpXP is involved in SsrA degradation in S. aureus.
With the availability of a reporter construct capable of displaying the relative amount of an SsrA- or VAA-tagged protein in a cell, we examined the role of ATP-dependent proteases and their cognate chaperones in the breakdown of SsrA-tagged protein in various clp mutants in our collection using the Venus-VAA construct. As shown in Fig. 2A, Venus-VAA levels were higher in overnight cultures of ΔclpC and ΔclpP strains than in those of wild-type and other protease and chaperone mutant strains. Similar results were also observed in clpC and clpP mutants when grown for 6 h (data not shown). Interestingly, the amount of Venus-VAA that accumulated in the ΔclpP mutant was not as high as that which accumulated in the ΔclpC mutant. Given that ΔclpP strains exhibit lower growth rates, smaller colony sizes, and altered metabolic intracellular environments compared to those of the wild type (16), it is conceivable that these altered phenotypes may have accounted for the lower fluorescence in the clpP mutant than the clpC mutant. We next examined whether this difference in fluorescence between the ΔclpC and ΔclpP mutants was actually reflected in degradation of the Venus-VAA protein. Accordingly, we cloned the venus-VAA gene into pEPSA5, which allows Venus-VAA expression upon xylose induction, and then introduced this plasmid, pALC7016, into SH1000, ΔclpC, and ΔclpP strains. Western blots of cell lysates taken from these xylose-induced cells following rifampin treatment were probed with anti-GFP antibody, revealing increased half-lives of Venus-VAA in the clpC and clpP mutants during 2 h of transcriptional stalling, while degradation was restored in the complemented clpC and clpP mutants (Fig. 2B). In contrast, a similar effect was not seen in the clpX mutant. Taken together, these data indicate that ClpC is the chaperone involved in SsrA-mediated degradation in S. aureus, contrary to the finding in E. coli (32) and B. subtilis, where ClpX is the primary chaperone that facilitates SsrA-mediation degradation (33).
FIG 2.
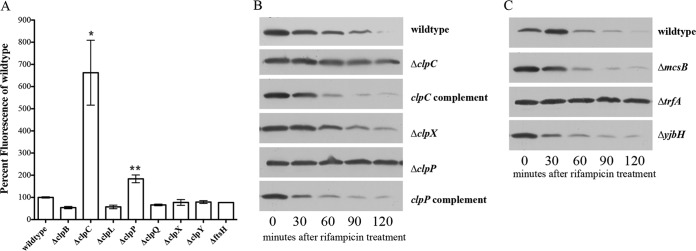
Venus levels in clp and adaptor mutant strains of SH1000. (A) Venus fluorescence analysis of overnight cultures of S. aureus strains containing pALC6955 (producing Venus-VAA). Venus fluorescence values (excitation and emission, 515 nm and 540 nm, respectively) were normalized to the OD650 values and then reported as a percentage of the wild-type levels. Each value is the mean and standard deviation of three biological replicates. *, statistically significant differences (P < 0.005) compared to the wild type measured by Student's unpaired t test. (B) Western blot of Venus-VAA protein levels in S. aureus SH1000 wild-type and Δclp cells containing pALC7016 with a xylose-inducible promoter (producing Venus-VAA) after transcriptional arrest by rifampin (200 μg/ml). Strains were grown to an OD650 of 1.1 and then treated with rifampin (200 μg/ml), and samples were taken every 30 min. Whole-cell lysates of these samples were then separated by SDS-PAGE on a 20% gel and transferred to PVDF for immunoblotting with an anti-GFP primary antibody and subsequently with a horseradish peroxidase-conjugated goat anti-mouse antibody. The experiments were repeated three times, and representative blots are displayed. (C) Western blot of Venus-VAA protein levels in S. aureus SH1000 wild-type and adaptor mutant strains containing pALC7016 (producing Venus-VAA). Whole-cell lysates of 0.5% xylose-induced samples obtained at different time points after rifampin treatment were resolved, blotted, and probed as described above for panel B. The experiments were repeated three times, and representative blots are displayed.
TrfA is necessary to break down SsrA in S. aureus.
To determine if other additional components were involved in degrading SsrA-tagged proteins in S. aureus, we examined the role(s) played by various adaptor proteins. The S. aureus genome contains genes for three known adaptors: mcsB (sa0482), trfA (sa0857), and yjbH (sa0860). We first transformed the pEPSA5-Venus-VAA construct pALC7016 into mcsB, trfA, and yjbH mutants. Western blot analysis of Venus-VAA breakdown in these adaptor mutants with pALC7016, grown to an OD650 of 1.1, after which rifampin was added, showed that only trfA was involved in degrading Venus-VAA (Fig. 2C). These data indicate that TrfA, together with ClpCP, controls SsrA-mediated degradation in S. aureus through the 3 terminal amino acids of the SsrA tag.
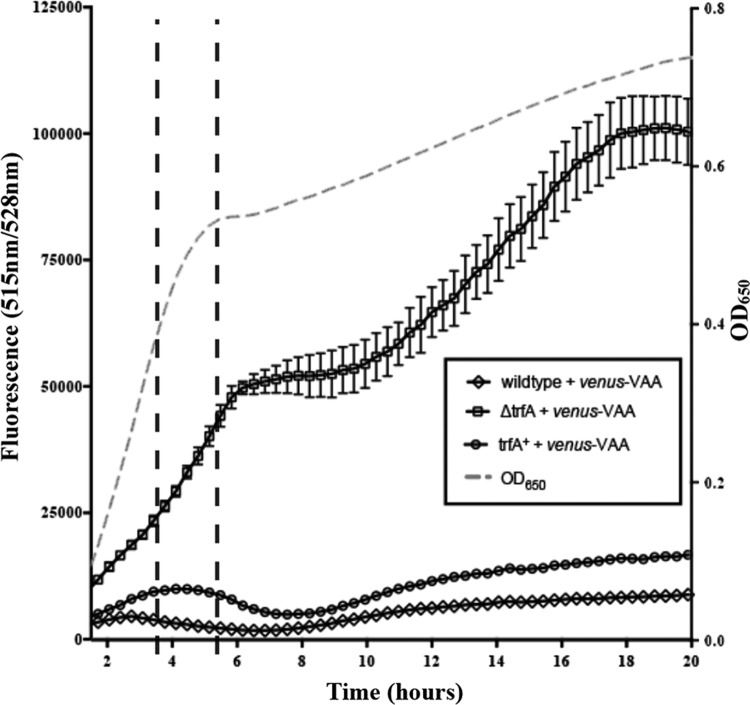
To examine how deletion of trfA impacted SsrA degradation at various phases of growth, we sampled fluorescence levels during 20 h of growth in TSB medium for wild-type, ΔtrfA, and trfA-complemented strains expressing Venus-VAA (Fig. 3). Starting from overnight cultures, strains were back-diluted 1/100 and grown to an OD650 of 0.6, whereupon they were normalized to an OD650 of 0.1, and 150 μl of this culture was aliquoted into a well of a 96-well plate. Cell growth was comparable among the three strains when incubated at 37°C with shaking for 20 h (data not shown). Analysis of the strains at these time points revealed that the ΔtrfA strain (Fig. 3, squares) produced much higher levels of both raw and normalized fluorescence at all time points than the wild-type (Fig. 3, diamonds) and trfA-complemented (Fig. 3, circles) strains. This suggests that Venus-VAA levels are generally elevated in a ΔtrfA strain relative to those in the wild-type and complemented strains.
FIG 3.
Venus-VAA levels in the wild type and the ΔtrfA mutant over 20 h of growth. The raw Venus fluorescence of the S. aureus SH1000 wild-type, ΔtrfA, and trfA-complemented (trfA+) strains containing pALC6955 (producing Venus-VAA) over 20 h is shown. Cell densities at an OD650 were measured, while Venus fluorescence was detected at 540 nm (black lines), with each value representing the mean of three biological replicates.
The fluorescence intensity of Venus-VAA and its 47-min half-life make it is possible to uncover conditional changes of fluorescence between strains. Specifically, the raw fluorescence in all strains (Fig. 3) increased at various rates in early logarithmic phase up through 3.5 h, but at late logarithmic phase (3.5 to 5.5 h), Venus-VAA levels began to decrease in the wild-type and trfA-complemented strains but not a ΔtrfA strain (Fig. 3). This decrease or flattening of both the raw and normalized fluorescence of strains containing trfA was maintained well into stationary phase, while the fluorescence of Venus-VAA continued to climb in the ΔtrfA strain. This suggests that trfA has a lesser role in SsrA regulation in early and mid-logarithmic phase (when there was no decrease in fluorescence in the wild type) but gains in importance as cells begin to exit logarithmic phase and enter stationary phase.
Transcriptional regulation of trfA.
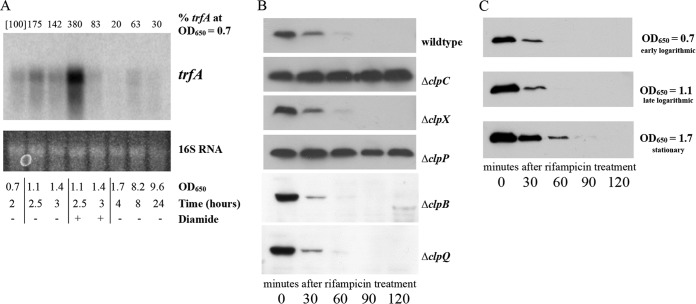
trfA is transcribed from three promoters, generating transcripts of closely related sizes of 751, 818, and 898 bases, which can be seen as a single transcript in a 0.7% agarose gel (28). The changes to trfA transcription during various growth phases remain unclear, however. To investigate trfA transcript levels over time, we conducted Northern blot analyses using RNA taken from SH1000 cells grown to various time points. Our data demonstrate (Fig. 4A, unstimulated time points) that the trfA transcripts in SH1000 increased from early log phase and peaked at late log phase (OD650, 1.1), followed by a significant decrease in stationary phase to a point where transcription was all but halted during deep-stationary-phase culture (at 24 h).
FIG 4.
Transcriptional and posttranslational regulation of trfA. (A) Northern blot of trfA during growth and thiol stress. S. aureus SH1000 was grown to various ODs under conditions with and without 5 mM diamide for thiol stress. The RNAs obtained from these cells (20 μg each) were resolved on a denaturing agarose gel, blotted to a Hybond XL membrane, and hybridized with a 300-bp 32P-radiolabeled trfA DNA fragment. The 16S rRNA of the ethidium bromide-stained gel was used as the loading control for the blot. The percentages of the trfA transcript at a given time point relative to the amount of trfA transcript at an OD650 of 0.7 (set equal to 100%, as indicated in brackets), determined by densitometric analysis, are reported at the top of the image. (B) Western blots of whole-cell lysates of various SH1000 strains transformed with pALC7151 (producing TrfA with an N-terminal myc tag). Cells were induced with 0.1% xylose at an OD650 of 1.1 and then treated with rifampin (200 μg/ml). Samples taken every 30 min were immunoblotted with a mouse anti-Myc primary antibody, followed by a horseradish peroxidase-conjugated goat anti-mouse antibody. The experiments were repeated three times, and representative blots are displayed. (C) Western blot analyses of TrfA in wild-type strain SH1000 at various time points following rifampin treatment. Wild-type strain SH1000 containing pEPSA5 expressing N-terminally Myc-tagged TrfA was induced with 0.1% xylose at early logarithmic phase (OD650 = 0.7), late logarithmic phase (OD650 = 1.1), or stationary phase (OD650 = 1.7) and then treated with rifampin (200 μg/ml). Samples were then immunoblotted with an anti-Myc primary antibody, followed by a horseradish peroxidase-conjugated goat anti-mouse antibody. The experiments were repeated three times, and representative blots are displayed.
trfA has also been shown to be responsive to certain stresses (28). Reverse transcription-PCR (RT-PCR) analysis showed that one or more of the trfA transcripts were elevated following antibiotic or thiol stress. To evaluate the resiliency and duration of this trfA induction, we examined trfA levels following the addition of 5 mM diamide. S. aureus cells were grown to an OD650 of 0.7 and then split for continued growth or challenge with 5 mM diamide. Samples were withdrawn 30 and 60 min later (corresponding to OD650s of 1.1 and 1.4, respectively). As seen in Fig. 4A, comparing the results at time points with and without diamide, the level of transcription of trfA more than doubled at 30 min after diamide addition relative to that of untreated cells at an analogous OD. Remarkably, this induction of trfA transcripts nearly vanished at 60 min after diamide induction (i.e., at an OD650 of 1.4), presumably due in part to the rapid inactivation of diamide within 20 to 30 min after addition (Ana Posada, personal communication).
TrfA is degraded by ClpCP.
The adaptor MecA of B. subtilis shares 36% amino acid sequence identity to TrfA, primarily in the first 80 amino acids. MecA controls competence by targeting the transcriptional regulator ComK for degradation through ClpCP (46), and MecA is itself proteolytically regulated by this system (46). Although S. aureus does not contain the competence machinery of B. subtilis, it is of interest to ascertain if S. aureus utilizes a similar method of posttranslational regulation. Accordingly, we induced the expression of TrfA with a codon-optimized N-terminal Myc tag using the xylose-inducible plasmid pEPSA5. The Myc tag was chosen because it has been demonstrated not to alter the pattern of proteolysis in other proteins (22). At an OD650 of 0.6, S. aureus cells were induced with 0.2% xylose and grown to an OD650 of 1.1. At this point, rifampin was added to stall transcription and samples were withdrawn at predetermined time points for 2 h. Whole-cell lysates of these aliquots were then examined by Western blotting with anti-Myc antibodies. As observed in Fig. 4B, TrfA levels decreased rapidly in the wild-type strain SH1000 with an ∼22-min half-life, as determined by densitometric analysis. Similar rates were seen for the clpB, clpQ, and clpX deletion mutants. However, TrfA levels remained relatively steady in the ΔclpC and ΔclpP strains, indicating that the ClpCP protease system controls the degradation of TrfA, as it does in B. subtilis. These data, combined with the transcriptional transiency of trfA induction after thiol stress, suggest that TrfA levels can be rapidly induced under specific conditions and just as rapidly recede to preinduced levels following the cessation of a specific stress.
The combination of TrfA's proteolytic degradation and its transcriptional peak during late logarithmic phase gives a sense of how TrfA operates in the cell. However, to fully understand how TrfA changes during growth in medium, we were interested to determine whether the TrfA degradation rate was maintained at other growth phases. Similar to our previous time point experiments, we grew wild-type SH1000 expressing N-terminally Myc-tagged TrfA from pEPSA5 to various ODs under conditions of xylose induction and, following the addition of rifampin, took samples at various time points over 2 h. Western blots of lysates of these cells were probed with anti-Myc antibody and revealed that while cells in early logarithmic phase degraded TrfA similarly to those in late logarithmic phase (half-life, ∼22 min), cells in stationary phase took longer to degrade TrfA (half-life, ∼37 min) (Fig. 4C). This suggests that even with the decreased transcription of trfA in stationary phase (Fig. 4A), TrfA may still be present in stationary-phase cells.
spx activates trfA but represses its own promoter.
Previous studies of spx transcription by RT-PCR have shown that the gene product of spx likely activates one or more of these transcripts of trfA (28). Spx is a thiol-sensitive transcriptional regulator that is itself proteolytically regulated and is responsible for controlling the adaptation and survival of S. aureus in environments with oxidative stress.
To examine whether the transcriptional regulation of trfA by spx was subject to any autoregulation, we constructed transcriptional fusions of the spx P1 (distal) and P2 (proximal) promoters linked to GFP, as described in Materials and Methods, to yield pALC7803 and pALC7804, respectively. These plasmid constructs were used to transform the Δspx mutant and its complemented strain of S. aureus. The results revealed that for the P2 promoter construct pALC7804, GFP levels were equivalent between the Δspx and complemented strains (data not shown). However, GFP fluorescence levels were consistently higher in the Δspx strain with the P1 promoter (Fig. 5A) at multiple time points than in its complemented strain. These data indicate that spx represses its own distal (P1) promoter but does not regulate its proximal (P2) promoter.
FIG 5.
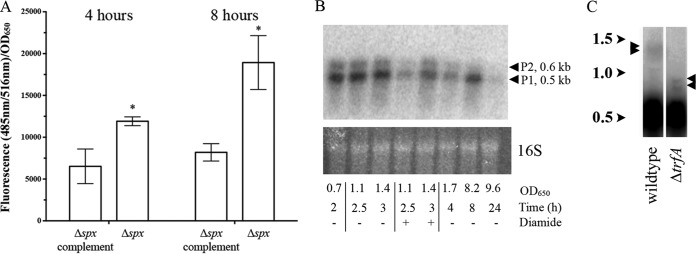
spx regulation of trfA and spx transcription. (A) GFP fluorescence driven by the spx P1 promoter on a shuttle plasmid containing a GFP reporter (pALC7803) in the Δspx and spx-complemented strains at the 4- and 8-h time points (late log and early stationary phases, respectively). To minimize variations in fluorescence attributable to cell density, the data are presented as the average number of fluorescence units for triplicate samples per unit of absorbance at 650 nm. *, a statistically significant difference (P <0.05) compared to the results for spx-complemented strain, as measured by Student's unpaired t test. (B) Northern blot of spx transcription from S. aureus SH1000 at various growth phases with and without 5 mM diamide. RNA (20 μg each) was resolved on a denaturing agarose gel, blotted as described above, and probed with a 300-bp 32P-radiolabeled spx DNA probe. As a loading control, ethidium bromide-stained 16S rRNA was included, and the results are shown at the bottom. Arrowheads, sizes of the transcripts generated from the P1 and P2 spx promoters. (C) Northern blots of RNA extracted from the S. aureus SH1000 and ΔtrfA strains at the late exponential phase of growth (OD650 = 1.1) and probed with a 300-bp 32P-radiolabeled spx probe. Faint bands are identified by arrowheads to the side of each lane. The numbers to the left indicate kilobases.
To better assess the growth conditions under which Spx represses its own promoter, we examined spx transcription at the same times of growth used for measuring trfA transcription (Fig. 4A). Surprisingly, spx transcription was relatively constant through log and early stationary phases under unstimulated conditions (Fig. 5B). However, following the addition of 5 mM diamide, spx transcription was greatly reduced after 30 min (OD650 = 1.1 with diamide) and recovered after 60 min as the diamide effect tapered (OD650 = 1.4 with diamide). Interestingly, protein levels of Spx actually increase following diamide treatment in both B. subtilis and S. aureus (31, 47). This increase in Spx levels alongside a corresponding decrease in spx transcript levels following diamide treatment (Fig. 5B), as well as the increase in spx promoter fusion activity in a Δspx strain in general (Fig. 5A), supports our notion that the Spx protein represses its own transcription in S. aureus.
We also examined the amount of transcriptional read-through that occurs from the spx locus into the adjacent gene, trfA (Fig. 5C). A rho-independent terminator was predicted between spx and the adjacent trfA gene (−ΔG = 14.5 kcal/mol) (48), so to examine its effect on trfA transcription during cultured growth, we isolated RNA from wild-type strains grown to mid-log phase (OD650 = 1.1). In addition to the two previously noted spx transcripts (28), several putative read-through bands at ∼1.4 kb were also observed with extended exposure (Fig. 5C, arrowheads on the left). Relative to the smaller spx transcripts, these were estimated by densitometric analysis to be 3 to 4% of total spx transcripts. Importantly, these larger bands decreased in size in the ΔtrfA strain, corresponding to the reduction of trfA (720 bp) in the mutant. Therefore, the terminator between spx and trfA is strong and able to terminate transcription in ∼95% of cases.
TrfA does not regulate Spx protein levels in S. aureus.
Spx has been shown to be proteolytically regulated by YjbH in S. aureus (31) and B. subtilis (49). However, YpbH, a MecA paralog with which TrfA shares 26% identity, also recognizes and degrades Spx in B. subtilis (50). To determine whether YjbH and TrfA share Spx as a substrate or whether another adaptor, such as McsB, was involved, we exogenously expressed Spx with an N-terminal myc tag from pALC7113 in various clp and adaptor deletion strains and tracked the degradation rates in transcriptionally stalled cells. As shown in Fig. 6, only in the yjbH mutant did the level of Spx persist through 120 min after rifampin treatment, whereas Spx was barely detectable at 30 min after rifampin treatment in the wild-type, ΔmcsB, and ΔtrfA strains. Furthermore, Spx levels remained high in the ΔclpX and ΔclpP strains but not the ΔclpC strains. Together, these data indicate that only yjbH acts as the facilitator of Spx breakdown in S. aureus through the ClpXP system.
FIG 6.
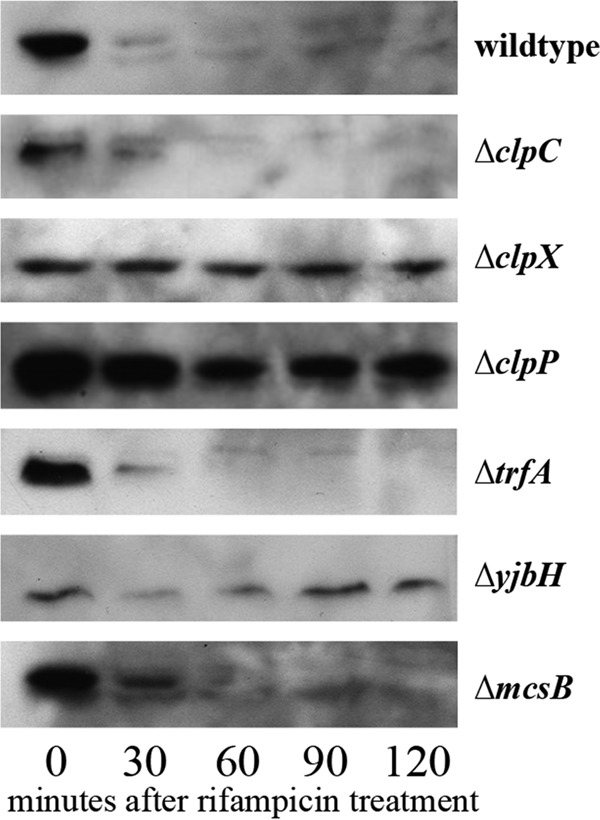
Role of S. aureus adaptors in Spx degradation. Western blots of whole-cell lysates of the SH1000 wild-type, ΔtrfA, ΔyjbH, ΔclpC, ΔclpX, and ΔclpP strains with pALC7113 (producing Spx with an N-terminal myc tag). Cells were grown to an OD650 of 1.1 with 0.1% xylose and then treated with rifampin (200 μg/ml). Cell lysates taken every 30 min were immunoblotted with a mouse anti-Myc primary antibody, followed by a horseradish peroxidase-conjugated goat anti-mouse antibody. The experiments were repeated three times, and representative blots are displayed.
TrfA is the adaptor regulating S. aureus antitoxin degradation.
Toxin-antitoxin systems are small genetic loci that serve a variety of functions, from plasmid maintenance, chromosomal stabilization, and persister formation to a bacterial form of programmed cell death (51). Several classes of toxin-antitoxin systems exist, based in part on whether the antitoxin is RNA (type I and type III) or protein (type II). In all cases, the antitoxin is short-lived due to its rapid degradation but is stabilized by its binding to its cognate toxin. The antitoxin also inactivates the toxin, protecting the bacteria until levels of antitoxin drop in response to a variety of environmental stresses (52). There are three known type II toxin-antitoxin systems in S. aureus, Axe1/Txe1, Axe2/Txe2 and MazEF, all of which are known to be proteolytically regulated by ClpCP (22). Given that TrfA works with ClpCP to degrade SsrA-tagged proteins in S. aureus, we wanted to determine whether TrfA also contributed to antitoxin degradation. For this purpose, the wild-type strain SH1000 and its isogenic trfA mutant were individually transformed with recombinant pEPSA5 plasmids carrying the antitoxin gene mazE, axe1, or axe2. For detection, we attached either a 5′ or 3′ myc tag (depending on the success of expression) to the antitoxin genes (Table 1) (22). At an OD650 of 0.6, cells were induced with 0.2% xylose and grown to an OD650 of 1.1, at which point the cells were transcriptionally stalled by addition of rifampin. Western blots of whole-cell lysates from these time points were probed with an anti-Myc antibody, disclosing that the levels of S. aureus MazE (MazEsa), Axe1, and Axe2 all remained constant in the ΔtrfA strain for 120 min after rifampin treatment, while those of the parental strain decreased rapidly after 30 to 60 min (Fig. 7). Degradation rates comparable to the rate for the wild type were also observed in the mcsB and yjbH mutants of S. aureus (data not shown). Together, these results demonstrate that the adaptor TrfA facilitates antitoxin degradation via ClpCP in S. aureus (22), hence linking a new set of proteins to adaptor-mediated proteolysis in bacteria.
FIG 7.
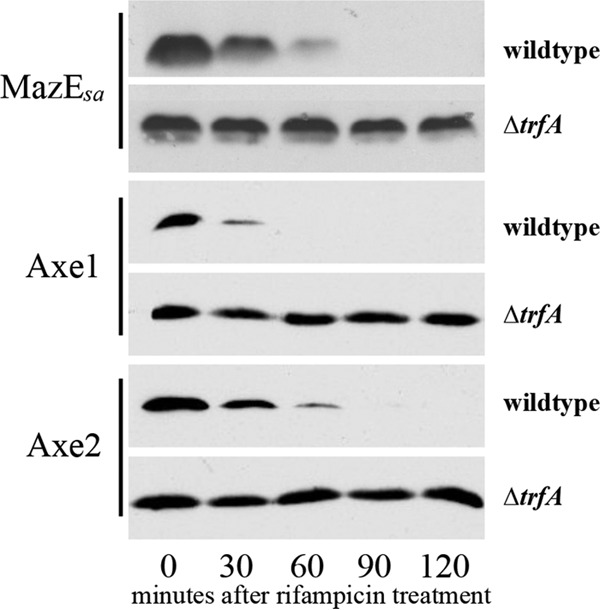
Degradation of antitoxins in trfA mutants. The SH1000 wild-type and ΔtrfA strains with either pALC6188 (mazEsa), pALC6486 (axe1), or pALC6489 (axe2) were grown to an OD650 of 1.1 with 0.1% xylose and then treated with rifampin (200 μg/ml). Cell lysates taken every 30 min were immunoblotted with an anti-GFP primary antibody, followed by a horseradish peroxidase-conjugated goat anti-mouse antibody. The experiments were repeated three times, and representative blots are displayed.
DISCUSSION
S. aureus cells adjust the levels of their intracellular proteins through alterations in either transcriptional priorities or degradation rates. Chaperones and adaptors assist with the latter by tuning the substrate specificity of various proteases, allowing proteins to be selectively removed. One way that regulated proteolysis is utilized by many bacteria is to recover from errors in translation that may result in stalled ribosomes. In this process, tmRNA, a tRNA/mRNA hybrid that contains a small open reading frame for an 11-amino-acid tag called SsrA, enters the A site of the stalled ribosome, and as translation restarts, it is appended to the C terminus of the nascent protein. When newly tagged protein is freed from the ribosome, SsrA is then recognized as a signal for degradation. This is performed by both ClpXP and ClpAP (32, 53) in E. coli, while in B. subtilis only ClpXP is used (33).
Interestingly, we discovered that SsrA-mediated proteolysis in S. aureus is performed not by ClpXP but by ClpCP (Fig. 2A and B). Previous work has shown that ClpX is involved in regulating toxin production and virulence in S. aureus (16, 54), while ClpC plays a modulatory role in responding to stress, metabolic changes, and starvation (23). Therefore, it is not surprising that the degradation of SsrA-tagged proteins in S. aureus falls under the regulation of ClpCP (Fig. 8). Furthermore, SsrA degradation by proteases other than ClpXP and ClpAP is not without precedent, as Lon is used in such a capacity in the small genome of Mycoplasma (55), and Tsp (56), Lon (57), and FtsH (58) are all used in E. coli degradation of the SsrA tag, albeit at a lower frequency than ClpXP and ClpAP. While the reason that S. aureus swapped ClpXP for ClpCP in such a role is not known, ClpC in S. aureus shares some homology to ClpA in E. coli (36% identity, 56% similarity). It is possible that S. aureus (or a progenitor of it) utilized both ClpC/ClpA and ClpX for SsrA-mediated breakdown in a manner similar to that in which they are utilized in E. coli. However, as this bacterium evolved and ClpC became dominant in controlling the stress response and ClpX became dominant in controlling virulence, the ability of ClpX to degrade SsrA-tagged proteins may have become less of a necessity and ultimately lost entirely.
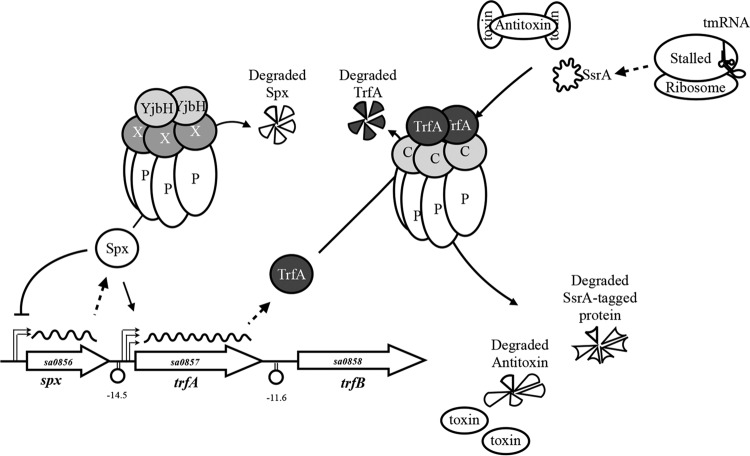
FIG 8.
Proposed model for TrfA regulation and activity in S. aureus. The adaptor TrfA associates with ClpCP to selectively degrade SsrA-tagged proteins and class II antitoxins, such as MazEsa. TrfA is itself degraded by ClpCP, which gives TrfA temporal nuance. The transcription of trfA is initiated from three upstream promoters, at least one of which is positively regulated by Spx. Spx activity is controlled by the degradation of the Spx protein via ClpXP and YjbH under normal conditions and with decreased proteolysis during oxidative, thiol, or antibiotic stress. Spx represses its own distal P1 promoter, creating a negative-feedback loop where less spx is transcribed during such stresses. X, ClpX; P, ClpP; C, ClpC.
In E. coli, adaptors have long been known to complement chaperones in controlling SsrA-mediated degradation. For example, the adaptor SspB normally competes for the same SsrA motif as ClpA, blocking its recognition of SsrA-tagged proteins (25). SspB levels increase during heat shock (25), effectively redirecting ClpAP to degrade other targets, including misfolded proteins. No homologs of SspB or any other E. coli adaptors exist in S. aureus, so the mechanism by which TrfA mediates SsrA-mediated degradation in S. aureus may be novel (Fig. 8). Although the 3 C-terminal amino acids of the SsrA tag are sufficient for TrfA-mediated degradation of SsrA-tagged proteins in S. aureus, it is plausible that an additional factor(s) may mediate recognition of the entire native SsrA tag by ClpCP since Venus-SsrA appears to be degraded faster than Venus-VAA (Fig. 1A). Furthermore, as TrfA seems to have the greatest influence on SsrA degradation during late logarithmic and early stationary phases, the yet unidentified adaptor(s) may assist with degradation during other growth states.
The regulation of TrfA has also begun to be unraveled by our studies. Similar to MecA in B. subtilis (46), TrfA is degraded by the very protease that it assists in proteolysis. The 22-min half-life of TrfA likely ties its activity to its transcription rate, suggesting that TrfA functions largely during logarithmic and early stationary growth phases, as well as to the oxidative stress response. In the last case, the transcription of trfA is upregulated in an Spx-dependent manner (28, 59), but it is quickly restored to baseline levels following abatement of thiol-related stresses (Fig. 4A). Such a pulse of transcription during these oxidative stresses could temporarily redirect ClpCP to process more TrfA-specific targets.
The thiol stress-sensitive transcriptional regulator Spx is degraded by ClpXP under normal growth conditions (60), but it is stabilized during oxidative stress in S. aureus (28, 31), allowing levels to rise. YjbH has been shown to contribute to this regulation in S. aureus (31), but YpbH, an adaptor in B. subtilis that has 26% identity to TrfA, has been shown to also control Spx degradation (50). We clarified this uncertainty in S. aureus by determining that only YjbH regulates Spx degradation (Fig. 6 and 8).
In B. subtilis, one of the three promoters of spx is upregulated following thiol stress (61), which further contributes to an increase in Spx levels. On the basis of our data, an alternative system appears to exist in S. aureus, where Spx (directly or indirectly) represses its own promoter. Accordingly, following the increase in Spx levels after exposure to 5 mM diamide, the transcription of spx is repressed. Considering the spx-dependent activation of trfA, this suggests that Spx can act as both an activator and a repressor in S. aureus. This brings the Spx function in line with that in B. subtilis, wherein it functions both to enhance RNA polymerase activity (47) and to act as an anti-alpha factor repressing transcription (62).
Besides SsrA-tagged proteins, our studies here also expand the set of proteins regulated by TrfA to include the labile antitoxins of the type II toxin-antitoxin systems (Fig. 7 and 8). This finding is of interest to us because we have shown previously that ClpCP is the major proteolytic system for the degradation of class II antitoxins in S. aureus (22). While these protein-based antitoxins have been known to be degraded by a variety of ATP-dependent proteases and chaperones (52), to our knowledge, this is the first time that an adaptor has been linked to antitoxin degradation. As toxin-antitoxin systems are intimately linked to environmental and antibiotic-induced stresses, our results indicate that an adaptor such as TrfA may also participate in this stress response by modulating the degradation of antitoxins in S. aureus.
ACKNOWLEDGMENT
This work was supported by NIH grant RO1 AI91801 (to A.L.C.).
Footnotes
Published ahead of print 15 September 2014
REFERENCES
- 1.Lowy FD. 1998. Staphylococcus aureus infections. N. Engl. J. Med. 339:520–532. 10.1056/NEJM199808203390806. [DOI] [PubMed] [Google Scholar]
- 2.Kluytmans JAJW, van Belkum A, Verbrugh HA. 1997. Nasal carriage of Staphylococcus aureus: epidemiology, underlying mechanisms, and associated risks. Clin. Microbiol. Rev. 10:505–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moore G, Dunnill CW, Wilson APR. 2013. The effect of glove material upon the transfer of methicillin-resistant Staphylococcus aureus to and from a gloved hand. Am. J. Infect. Control 41:19–23. 10.1016/j.ajic.2012.03.017. [DOI] [PubMed] [Google Scholar]
- 4.Wong D, Nye K, Hollis P. 1991. Microbial flora on doctors' white coats. BMJ 303:1602–1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ellis MW, Hospenthal DR, Dooley DP, Gray PJ, Murray CK. 2004. Natural history of community-acquired methicillin-resistant Staphylococcus aureus colonization and infection in soldiers. Clin. Infect. Dis. 39:971–979. 10.1086/423965. [DOI] [PubMed] [Google Scholar]
- 6.Chastanet A, Derré I, Nair S, Msadek T. 2004. clpB, a novel member of the Listeria monocytogenes CtsR regulon, is involved in virulence but not in general stress tolerance. J. Bacteriol. 186:1165–1174. 10.1128/JB.186.4.1165-1174.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Anderson KL, Roberts C, Disz T, Vonstein V, Hwang K, Overbeek R, Olson PD, Projan SJ, Dunman PM. 2006. Characterization of the Staphylococcus aureus heat shock, cold shock, stringent, and SOS responses and their effects on log-phase mRNA turnover. J. Bacteriol. 188:6739–6756. 10.1128/JB.00609-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kirstein J, Molière N, Dougan DA, Turgay K. 2009. Adapting the machine: adaptor proteins for Hsp100/Clp and AAA+ proteases. Nat. Rev. Microbiol. 7:589–599. 10.1038/nrmicro2185. [DOI] [PubMed] [Google Scholar]
- 9.Laport M, de Castro A, Villardo A, Lemos JAC, Bastos M, Giambiagi-deMarval M. 2001. Expression of the major heat shock proteins DnaK and GroEL in Streptococcus pyogenes: a comparison to Enterococcus faecalis and Staphylococcus aureus. Curr. Microbiol. 42:264–268. 10.1007/s002840110215. [DOI] [PubMed] [Google Scholar]
- 10.Gottesman S. 2003. Proteolysis in bacterial regulatory circuits. Annu. Rev. Cell Dev. Biol. 19:565–587. 10.1146/annurev.cellbio.19.110701.153228. [DOI] [PubMed] [Google Scholar]
- 11.Michalik S, Liebeke M, Zühlke D, Lalk M, Bernhardt J, Gerth U, Hecker M. 2009. Proteolysis during long-term glucose starvation in Staphylococcus aureus COL. Proteomics 9:4468–4477. 10.1002/pmic.200900168. [DOI] [PubMed] [Google Scholar]
- 12.Aizenman E, Engelberg-Kulka H, Glaser G. 1996. An Escherichia coli chromosomal “addiction module” regulated by guanosine 3′,5′-bispyrophosphate: a model for programmed bacterial cell death. Proc. Natl. Acad. Sci. U. S. A. 93:6059–6063. 10.1073/pnas.93.12.6059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fu Z, Tamber S, Memmi G, Donegan NP, Cheung AL. 2009. Overexpression of MazFsa in Staphylococcus aureus induces bacteriostasis by selectively targeting mRNAs for cleavage. J. Bacteriol. 191:2051–2059. 10.1128/JB.00907-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Feng J, Michalik S, Varming AN, Andersen JH, Albrecht D, Jelsbak L, Krieger S, Ohlsen K, Hecker M, Gerth U, Ingmer H, Frees D. 2013. Trapping and proteomic identification of cellular substrates of the ClpP protease in Staphylococcus aureus. J. Proteome Res. 12:547–558. 10.1021/pr300394r. [DOI] [PubMed] [Google Scholar]
- 15.Michel A, Agerer F, Hauck C, Herrmann M, Ullrich J, Hacker J, Ohlsen K. 2006. Global regulatory impact of ClpP protease of Staphylococcus aureus on regulons involved in virulence, oxidative stress response, autolysis, and DNA repair. J. Bacteriol. 188:5783–5796. 10.1128/JB.00074-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Frees D, Qazi SN, Hill PJ, Ingmer H. 2003. Alternative roles of ClpX and ClpP in Staphylococcus aureus stress tolerance and virulence. Mol. Microbiol. 48:1565–1578. 10.1046/j.1365-2958.2003.03524.x. [DOI] [PubMed] [Google Scholar]
- 17.Lithgow JK, Ingham E, Foster SJ. 2004. Role of the hprT-ftsH locus in Staphylococcus aureus. Microbiology 150:373–381. 10.1099/mic.0.26674-0. [DOI] [PubMed] [Google Scholar]
- 18.Frees D, Thomsen LE, Ingmer H. 2005. Staphylococcus aureus ClpYQ plays a minor role in stress survival. Arch. Microbiol. 183:286–291. 10.1007/s00203-005-0773-x. [DOI] [PubMed] [Google Scholar]
- 19.Neuwald AF, Aravind L, Spouge JL, Koonin EV. 1999. AAA+: a class of chaperone-like ATPases associated with the assembly, operation, and disassembly of protein complexes. Genome Res. 9:27–43. [PubMed] [Google Scholar]
- 20.Frees D, Chastanet A, Qazi SN, Sørensen K, Hill PJ, Msadek T, Ingmer H. 2004. Clp ATPases are required for stress tolerance, intracellular replication and biofilm formation in Staphylococcus aureus. Mol. Microbiol. 54:1445–1462. 10.1111/j.1365-2958.2004.04368.x. [DOI] [PubMed] [Google Scholar]
- 21.Chatterjee I, Becker P, Grundmeier M, Bischoff M, Somerville GA, Peters G, Sinha B, Harraghy N, Proctor RA, Herrmann M. 2005. Staphylococcus aureus ClpC is required for stress resistance, aconitase activity, growth recovery, and death. J. Bacteriol. 187:4488–4496. 10.1128/JB.187.13.4488-4496.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Donegan NP, Thompson ET, Fu Z, Cheung AL. 2010. Proteolytic regulation of toxin-antitoxin systems by ClpPC in Staphylococcus aureus. J. Bacteriol. 192:1416–1422. 10.1128/JB.00233-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chatterjee I, Schmitt S, Batzilla CF, Engelmann S, Keller A, Ring MW, Kautenburger R, Ziebuhr W, Hecker M, Preissner KT, Bischoff M, Proctor RA, Beck HP, Lenhof H-P, Somerville GA, Herrmann M. 2009. Staphylococcus aureus ClpC ATPase is a late growth phase effector of metabolism and persistence. Proteomics 9:1152–1176. 10.1002/pmic.200800586. [DOI] [PubMed] [Google Scholar]
- 24.Chan CM, Garg SK, Lin AA, Zuber P. 2012. Geobacillus thermodenitrificans YjbH recognizes the C-terminal end of Bacillus subtilis Spx to accelerate Spx proteolysis by ClpXP. Microbiology 158:1268–1278. 10.1099/mic.0.057661-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Flynn JM, Levchenko I, Seidel M, Wickner SH, Sauer RT, Baker TA. 2001. Overlapping recognition determinants within the ssrA degradation tag allow modulation of proteolysis. Proc. Natl. Acad. Sci. U. S. A. 98:10584–10589. 10.1073/pnas.191375298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Elsholz AKW, Michalik S, Zühlke D, Hecker M, Gerth U. 2010. CtsR, the Gram-positive master regulator of protein quality control, feels the heat. EMBO J. 29:3621–3629. 10.1038/emboj.2010.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sitthisak S, Kitti T, Boonyonying K, Mongkolsuk S, Jayaswal RK. 2012. McsA and the roles of metal-binding motif in Staphylococcus aureus. FEMS Microbiol. Lett. 327:126–133. 10.1111/j.1574-6968.2011.02468.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jousselin A, Kelley WL, Barras C, Lew DP, Renzoni A. 2013. The Staphylococcus aureus thiol/oxidative stress global regulator Spx controls trfA, a gene implicated in cell wall antibiotic resistance. Antimicrob. Agents Chemother. 57:3283–3292. 10.1128/AAC.00220-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Göhring N, Fedtke I, Xia G, Jorge AM, Pinho MG, Bertsche U, Peschel A. 2011. New role of the disulfide stress effector YjbH in β-lactam susceptibility of Staphylococcus aureus. Antimicrob. Agents Chemother. 55:5452–5458. 10.1128/AAC.00286-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Elsholz AKW, Hempel K, Michalik S, Gronau K, Becher D, Hecker M, Gerth U. 2011. Activity control of the ClpC adaptor McsB in Bacillus subtilis. J. Bacteriol. 193:3887–3893. 10.1128/JB.00079-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Engman J, Rogstam A, Frees D, Ingmer H, von Wachenfeldt C. 2012. The YjbH adaptor protein enhances proteolysis of the transcriptional regulator Spx in Staphylococcus aureus. J. Bacteriol. 194:1186–1194. 10.1128/JB.06414-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gottesman S, Roche E, Zhou Y, Sauer RT. 1998. The ClpXP and ClpAP proteases degrade proteins with carboxy-terminal peptide tails added by the SsrA-tagging system. Genes Dev. 12:1338–1347. 10.1101/gad.12.9.1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wiegert T, Schumann W. 2001. SsrA-mediated tagging in Bacillus subtilis. J. Bacteriol. 183:3885–3889. 10.1128/JB.183.13.3885-3889.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fu Z, Donegan NP, Memmi G, Cheung AL. 2007. Characterization of MazFSa, an endoribonuclease from Staphylococcus aureus. J. Bacteriol. 189:8871–8879. 10.1128/JB.01272-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schenk S, Laddaga RA. 1992. Improved method for electroporation of Staphylococcus aureus. FEMS Microbiol. Lett. 73:133–138. [DOI] [PubMed] [Google Scholar]
- 36.Rooijakkers SHM, van Wamel W, Ruyken M, van Kessel KPM, van Strijp JAG. 2005. Anti-opsonic properties of staphylokinase. Microbes Infect. 7:476–484. 10.1016/j.micinf.2004.12.014. [DOI] [PubMed] [Google Scholar]
- 37.Forsyth RA, Haselbeck RJ, Ohlsen KL, Yamamoto RT, Xu H, Trawick JD, Wall D, Wang L, Brown-Driver V, Froelich JM, Kedar GC, King P, McCarthy M, Malone CL, Misiner B, Robbins D, Tan Z, Zhu Zy Z-Y, Carr G, Mosca DA, Zamudio C, Foulkes JG, Zyskind JW. 2002. A genome-wide strategy for the identification of essential genes in Staphylococcus aureus. Mol. Microbiol. 43:1387–1400. 10.1046/j.1365-2958.2002.02832.x. [DOI] [PubMed] [Google Scholar]
- 38.Arnaud M, Chastanet A, Débarbouillé M. 2004. New vector for efficient allelic replacement in naturally nontransformable, low-GC-content, gram-positive bacteria. Appl. Environ. Microbiol. 70:6887–6891. 10.1128/AEM.70.11.6887-6891.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cheung AL, Eberhardt K, Fischetti VA. 1994. A method to isolate RNA from gram-positive bacteria and mycobacteria. Anal. Biochem. 222:511–514. 10.1006/abio.1994.1528. [DOI] [PubMed] [Google Scholar]
- 40.Moore SD, Sauer RT. 2007. The tmRNA system for translational surveillance and ribosome rescue. Annu. Rev. Biochem. 76:101–124. 10.1146/annurev.biochem.75.103004.142733. [DOI] [PubMed] [Google Scholar]
- 41.Lies M, Maurizi MR. 2008. Turnover of endogenous SsrA-tagged proteins mediated by ATP-dependent proteases in Escherichia coli. J. Biochem. 283:22918–22929. 10.1074/jbc.M801692200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Crameri A, Whitehorn EA, Tate E, Stemmer WP. 1996. Improved green fluorescent protein by molecular evolution using DNA shuffling. Nat. Biotechnol. 14:315–319. 10.1038/nbt0396-315. [DOI] [PubMed] [Google Scholar]
- 43.Nagai T, Ibata K, Park ES, Kubota M, Mikoshiba K, Miyawaki A. 2002. A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat. Biotechnol. 20:87–90. 10.1038/nbt0102-87. [DOI] [PubMed] [Google Scholar]
- 44.McGinness KE, Baker TA, Sauer RT. 2006. Engineering controllable protein degradation. Mol. Cell 22:701–707. 10.1016/j.molcel.2006.04.027. [DOI] [PubMed] [Google Scholar]
- 45.Landgraf D, Okumus B, Chien P, Baker TA, Paulsson J. 2012. Segregation of molecules at cell division reveals native protein localization. Nat. Methods 9:480–482. 10.1038/nmeth.1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Turgay K, Hahn J, Burghoorn J, Dubnau D. 1998. Competence in Bacillus subtilis is controlled by regulated proteolysis of a transcription factor. EMBO J. 17:6730–6738. 10.1093/emboj/17.22.6730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nakano S, Küster-Schöck E, Grossman AD, Zuber P. 2003. Spx-dependent global transcriptional control is induced by thiol-specific oxidative stress in Bacillus subtilis. Proc. Natl. Acad. Sci. U. S. A. 100:13603–13608. 10.1073/pnas.2235180100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.de Hoon M, Makita Y, Nakai K, Miyano S. 2005. Prediction of transcriptional terminators in Bacillus subtilis and related species. PLoS Comput. Biol. 1:e25. 10.1371/journal.pcbi.0010025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Garg SK, Kommineni S, Henslee L, Zhang Y, Zuber P. 2009. The YjbH protein of Bacillus subtilis enhances ClpXP-catalyzed proteolysis of Spx. J. Bacteriol. 191:1268–1277. 10.1128/JB.01289-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nakano S, Zheng G, Nakano MM, Zuber P. 2002. Multiple pathways of Spx (YjbD) proteolysis in Bacillus subtilis. J. Bacteriol. 184:3664–3670. 10.1128/JB.184.13.3664-3670.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Magnuson RD. 2007. Hypothetical functions of toxin-antitoxin systems. J. Bacteriol. 189:6089–6092. 10.1128/JB.00958-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gerdes K, Christensen SK, Løbner-Olesen A. 2005. Prokaryotic toxin-antitoxin stress response loci. Nat. Rev. Microbiol. 3:371–382. 10.1038/nrmicro1147. [DOI] [PubMed] [Google Scholar]
- 53.Farrell CM, Grossman AD, Sauer RT. 2005. Cytoplasmic degradation of ssrA-tagged proteins. Mol. Microbiol. 57:1750–1761. 10.1111/j.1365-2958.2005.04798.x. [DOI] [PubMed] [Google Scholar]
- 54.Jelsbak L, Ingmer H, Valihrach L, Cohn MT, Christiansen MHG, Kallipolitis BH, Frees D. 2010. The chaperone ClpX stimulates expression of Staphylococcus aureus protein A by Rot dependent and independent pathways. PLoS One 5:e12752. 10.1371/journal.pone.0012752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gur E, Sauer RT. 2008. Evolution of the ssrA degradation tag in Mycoplasma: specificity switch to a different protease. Proc. Natl. Acad. Sci. U. S. A. 105:16113–16118. 10.1073/pnas.0808802105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Keiler KC, Waller PR, Sauer RT. 1996. Role of a peptide tagging system in degradation of proteins synthesized from damaged messenger RNA. Science 271:990–993. 10.1126/science.271.5251.990. [DOI] [PubMed] [Google Scholar]
- 57.Choy JS, Aung LL, Karzai AW. 2007. Lon protease degrades transfer-messenger RNA-tagged proteins. J. Bacteriol. 189:6564–6571. 10.1128/JB.00860-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Herman C, Thévenet D, Bouloc P, Walker GC, D'Ari R. 1998. Degradation of carboxy-terminal-tagged cytoplasmic proteins by the Escherichia coli protease HflB (FtsH). Genes Dev. 12:1348–1355. 10.1101/gad.12.9.1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Renzoni A, Andrey DO, Jousselin A, Barras C, Monod A, Vaudaux P, Lew DP, Kelley WL. 2011. Whole genome sequencing and complete genetic analysis reveals novel pathways to glycopeptide resistance in Staphylococcus aureus. PLoS One 6:e21577. 10.1371/journal.pone.0021577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pamp SJ, Frees D, Engelmann S, Hecker M, Ingmer H. 2006. Spx is a global effector impacting stress tolerance and biofilm formation in Staphylococcus aureus. J. Bacteriol. 188:4861–4870. 10.1128/JB.00194-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Leelakriangsak M, Zuber P. 2007. Transcription from the P3 promoter of the Bacillus subtilis spx gene is induced in response to disulfide stress. J. Bacteriol. 189:1727–1735. 10.1128/JB.01519-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nakano S, Nakano MM, Zhang Y, Leelakriangsak M, Zuber P. 2003. A regulatory protein that interferes with activator-stimulated transcription in bacteria. Proc. Natl. Acad. Sci. U. S. A. 100:4233–4238. 10.1073/pnas.0637648100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kreiswirth BN, Lofdahl S, Betley M, O'Reilly M, Schlievert PM, Bergdoll M, Novick RP. 1983. The toxic shock syndrome exotoxin structural gene is not detectably transmitted by a prophage. Nature 305:709–712. 10.1038/305709a0. [DOI] [PubMed] [Google Scholar]
- 64.Horsburgh M, Aish J, White I, Shaw LN, Lithgow JK, Foster SJ. 2002. sigmaB modulates virulence determinant expression and stress resistance: characterization of a functional rsbU strain derived from Staphylococcus aureus 8325-4. J. Bacteriol. 184:5457–5467. 10.1128/JB.184.19.5457-5467.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gaskill ME, Khan SA. 1988. Regulation of the enterotoxin B gene in Staphylococcus aureus. J. Biochem. 263:6276–6280. [PubMed] [Google Scholar]
- 66.Kahl BC, Goulian M, van Wamel W, Herrmann M, Simon SM, Kaplan G, Peters G, Cheung AL. 2000. Staphylococcus aureus RN6390 replicates and induces apoptosis in a pulmonary epithelial cell line. Infect. Immun. 68:5385–5392. 10.1128/IAI.68.9.5385-5392.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]