Abstract
The Gram-positive bacterium Cellulomonas fimi produces a large array of carbohydrate-active enzymes. Analysis of the collection of carbohydrate-active enzymes from the recent genome sequence of C. fimi ATCC 484 shows a large number of uncharacterized genes for glycoside hydrolase (GH) enzymes potentially involved in biomass utilization. To investigate the enzymatic activity of potential β-glucosidases in C. fimi, genes encoding several GH3 enzymes and one GH1 enzyme were cloned and recombinant proteins were expressed in Escherichia coli. Biochemical analysis of these proteins revealed that the enzymes exhibited different substrate specificities for para-nitrophenol-linked substrates (pNP), disaccharides, and oligosaccharides. Celf_2726 encoded a bifunctional enzyme with β-d-xylopyranosidase and α-l-arabinofuranosidase activities, based on pNP-linked substrates (CfXyl3A). Celf_0140 encoded a β-d-glucosidase with activity on β-1,3- and β-1,6-linked glucosyl disaccharides as well as pNP-β-Glc (CfBgl3A). Celf_0468 encoded a β-d-glucosidase with hydrolysis of pNP-β-Glc and hydrolysis/transglycosylation activities only on β-1,6-linked glucosyl disaccharide (CfBgl3B). Celf_3372 encoded a GH3 family member with broad aryl-β-d-glycosidase substrate specificity. Celf_2783 encoded the GH1 family member (CfBgl1), which was found to hydrolyze pNP-β-Glc/Fuc/Gal, as well as cellotetraose and cellopentaose. CfBgl1 also had good activity on β-1,2- and β-1,3-linked disaccharides but had only very weak activity on β-1,4/6-linked glucose.
INTRODUCTION
The Gram-positive soil bacterium Cellulomonas fimi degrades a variety of complex polysaccharides in plant cell walls, including cellulose and hemicellulose, using the synergistic catalysis of carbohydrate-active enzymes (1–3). To better understand the organism's cellulolytic strategy, the genome of C. fimi was recently sequenced (4), and analysis of the total carbohydrate-active enzyme (CAZyme) content (5) revealed that C. fimi harbors a large number of putative glycoside hydrolase (GH) genes, many of which are not yet biochemically characterized. It should be stressed that placement of a gene into a glycoside hydrolase family does not predict what the actual substrate of the encoded enzyme will be, so the biochemical analysis is absolutely required to determine the substrates for glycoside hydrolases. Over 30 years ago, there was an observation that C. fimi had both cellobiase and aryl-β-glucosidase activity (6). Since that time, there has been no other published work on these multiple β-glucosidase activities. It is the access to the recently determined genome sequence that permitted us to follow up on this initial observation about β-glucosidase expression in C. fimi, and we focused on multiple GH3 genes and the GH1 gene, as these CAZy families are known to encode β-glucosidases. The goal of the current study was to express these genes from C. fimi and to characterize and compare the biochemical properties of their gene products.
The GH3 family is one of the most abundant in CAZy, comprising over 6,000 enzymes that are widely distributed in bacteria, fungi, and plants. The family contains various activities such as exoacting β-d-glucosidases, α-l-arabinofuranosidases, β-d-xylopyranosidases, and N-acetyl-β-d-glucosaminidases, all of which use a retaining glycosidase mechanism (7–9). In addition to hydrolytic activities, some GH3 enzymes may catalyze glycosidic bond formation via either a thermodynamically controlled reverse hydrolysis or a kinetically controlled transglycosylation (10, 11). Together, these enzymes are responsible for cellulosic biomass degradation, plant and bacterial cell wall remodeling, energy metabolism, and defense against pathogens (7, 12). GH3 β-glucosidases catalyze the hydrolysis of alkyl- and aryl-β-glucosides, as well as di- and oligosaccharides. Interest in β-glucosidases arose from their critical involvement in the conversion of cellulose to glucose for biofuel applications, whereby they catalyze the removal of the end product inhibitor cellobiose (13–15). The α-l-arabinofuranosidases and β-d-xylopyranosidases from GH3 are involved in the degradation of hemicellulosic xylan and arabinoxylan (9), which is the second most abundant polysaccharide in the plant cell wall. GH3 enzymes also play important roles in many other biological processes, such as synthesis of functional glucosides from glycoside precursors (16) and cyanide-based biological defense mechanisms in plants (17).
The GH3 N-acetyl-β-d-glucosaminidases from Gram-positive bacteria are thought to be involved in cell wall remodeling or chitin degradation, and C. fimi has an interesting GH3 of this type with dual β-glucosidase and N-acetyl-β-d-glucosaminidase activity (18). Many of the enzymes of GH3 have dual or broad substrate specificities with respect to monosaccharide residues, linkage position, and chain length of the substrate (8). Additionally, bacteria frequently possess multiple genes for GH3 enzymes; in fact, C. fimi has 10 members of GH3. It is not clear whether these enzymes are overlapping in specificity or have distinct specificities, so biochemical investigations of the GH3 enzymes are required to understand the full spectrum of glycoside hydrolase activity in the C. fimi GH3 family members.
Another candidate gene for a functional β-glucosidase in C. fimi is in the CAZy GH1 family. GH1 enzymes are also characterized with a retaining glycosidase mechanism, and the most common activity for GH1 enzymes are β-glucosidases and β-galactosidases, where typically they work on both substrates but with higher Km values for the galactosides (19). Despite there being a published nucleotide sequence for a C. fimi GH1 gene (GenBank AAA23091.1), there are no published biochemical data of which we are aware. The family of GH1 enzymes is itself quite diverse; however, a detailed characterization of the retaining glycosidase mechanism has been performed for an enzyme from the Agrobacterium sp. GH1 (20, 21).
In this study, we report the biochemical properties of four GH3 enzymes and one GH1 enzyme from C. fimi and show that the examined genes encode nonredundant enzyme specificities, which will aid the biochemical characterization of the many unknown activities in these important enzyme families.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
C. fimi ATCC 484 was grown on low-salt Luria-Bertani medium (only 1 g NaCl per liter instead of 5 g) at 30°C for 48 h. Escherichia coli AD202 (CGSC 7297) and the plasmid pCWori+ were used as the host and vector for subcloning, respectively (22). Recombinant E. coli strains were grown in 2YT broth (16 g yeast extract, 10 g Bacto-tryptone, 5 g NaCl per liter) with 150 mg/liter ampicillin at 37°C.
Basic recombinant DNA methods.
Genomic DNA of C. fimi was isolated using the DNeasy Tissue kit (Qiagen Inc., Mississauga, ON, Canada). PCR was performed using Phusion polymerase and the following program: 98°C for 30 s, 30 successive cycles of 98°C for 10 s, 68°C for 45 s, and 72°C for 1 min, and finally 72°C for 10 min. Primers for the GH3 and GH1genes are shown in Table 1. An N-terminal His6 tag was introduced when designing the primers for amplification.
TABLE 1.
Genes for cloning and primers used for amplification
| Gene | Length (bp) | Protein | Molecular mass (Da) | Primera |
|
|---|---|---|---|---|---|
| Orientation | Sequence (5′→3′) | ||||
| Celf_2726 | 2,283 | CfXyl3A | 82,018 | Forward | GGGGGGGAATTCCATATGCACCATCACCATCACCATGTGACGGATCTGAAGCTGTC |
| Reverse | GGGGGGAAGCTTTCATCAGCGCACCACGGACGACGTCCAGCGCGCG | ||||
| Celf_0140 | 2,277 | CfBgl3A | 82,305 | Forward | GGGGGGGAATTCCATATGCACCATCACCATCACCATGTGCCGCACCCGTACCAGGAC |
| Reverse | GGGGGGAAGCTTTCATCAGGCGACGGTGAACCGGCCGCGGAGCAGGACCTC | ||||
| Celf_0468 | 2,529 | CfBgl3B | 88,781 | Forward | GGGGGGGAATTCCATATGCACCATCACCATCACCATGTGTCGACGCAGACACCCCG |
| Reverse | GGGGGGAAGCTTTCATCAGGGGGTGCCGGCCGCGGGGGCCGACGGG | ||||
| Celf_3372 | 2,262 | CfBgl3C | 80,298 | Forward | GGGGGGGAATTCCATATGCACCATCACCATCACCATCCGCACGCGCCGTTCGACG |
| Reverse | GGGGGGAAGCTTTCATCAGGCGCGGGCCTTCTCCACGAGCGCG | ||||
| Celf_2783 | 1,454 | CfBgl1 | 53,518 | Forward | GGGGGGGAATTCCATATGCACCAT CACCATCACCATACCACACGCGCCCC TCGG |
| Reverse | GGGGGGAAGCTTTCATCAGGGCTG GTAGGTCGCGGCGTCCTCGGC | ||||
Restriction enzyme sites are underlined.
Genes digested with NdeI and HindIII were ligated into pCW (22) and used to transform E. coli AD202 cells by electroporation, performed on Bio-Rad MicroPulser. Plasmids were isolated using the High Pure plasmid isolation kit (Roche Diagnostics, Laval, QC, Canada) and verified by restriction analysis, and the gene sequence was confirmed by DNA sequencing performed on the Applied Biosystems model 3100 automated DNA sequencer (Montreal, QC, Canada). All enzymes used were from New England BioLabs (Beverly, MA). The DNA isolations, restriction enzyme digestions, ligations, and transformations were performed according to the suppliers' recommendations.
Expression and purification of enzymes.
E. coli AD202 containing recombinant plasmids were grown in 2YT broth with 150 mg/liter ampicillin at 37°C. When the optical density at 600 nm (OD600) reached 0.5, the culture was induced with 0.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG) and then grown for another 24 h at 20°C. Subsequently, the cells were harvested and disrupted by the Avestin C5 Emulsiflex cell disrupter (Avestin, Ottawa, ON, Canada) in 25 mM sodium phosphate buffer, pH 7.4. Cell debris was removed by centrifugation (14,000 × g, 4°C, 30 min) to obtain the crude extract.
Celf_2726 was purified by a HisTrap HP column (GE Healthcare, Montreal, QC, Canada). The column of 5 ml was preequilibrated by 25 mM sodium phosphate buffer, pH 7.4, and eluted by a 70-ml linear gradient from 0 to 0.4 M imidazole in the same buffer. The flow rate was 1 ml/min, and fractions of 1 ml were collected. All other proteins were purified on 5 ml HiTrap Q HP anion-exchange chromatography. The gradient was 75 ml from 0 to 0.8 M NaCl in 25 mM sodium phosphate buffer, pH 7.4, at a flow rate of 1 ml/min, and 1-ml fractions were collected. The active fractions were pooled, desalted by membrane ultrafiltration with the Amicon YM30 Centriprep device (Amicon, Millipore, Bedford, MA), and used for subsequent characterizations.
Purified enzymes were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) on 10% separating gel. Protein concentrations were determined using the bicinchoninic acid (BCA) assay (Pierce, Rockford, IL).
Hydrolysis of pNP-linked compounds.
The hydrolysis of para-nitrophenol-linked substrates (pNP)-linked compounds (obtained from Sigma-Aldrich Chemicals, Mississauga, ON, Canada) by each enzyme was assayed by a Bio-Tek Power Wave X microplate reader (Winooski, VT, USA). Briefly, the reaction mixture was 200 μl (final volume) and contained 2 mM substrate. One hundred fifty microliters of the substrate in 50 mM sodium phosphate buffer, pH 7.4, and 50 μl of diluted enzyme (5 to 20 μg/ml) were incubated at 30°C for 1 h, and the level of released pNP was determined by monitoring the absorbance at 410 nm (A410). One unit (U) of enzyme activity was defined as the amount of enzyme required to liberate 1 μmol pNP min−1 at 30°C and pH 7.4. The extinction coefficient for pNP at pH 7.4 was assumed to be 13,400 liter−1 M−1.
To determine kinetic parameters against various pNP-glycosides, substrate concentration was varied (0.05 mM to 10 mM) and a reaction time of 15 min was chosen to ensure initial rates. The buffer and enzyme concentrations used are identical to those stated above. The Km and Vmax values were calculated from GraphPad Prism V5.
Hydrolysis of disaccharides and oligosaccharides.
To assess the catalytic activities of C. fimi GH enzymes on glucose disaccharides, neutral xylo- and cello-oligosaccharides (obtained from Sigma-Aldrich Chemicals, Mississauga, ON, Canada), the enzymes were incubated with the sugars and the products were analyzed by thin-layer chromatography (TLC). The 100-μl reaction mixture was assembled with 50 μl of 10 mg/ml substrate in water, 48 μl of 50 mM HEPES buffer (pH 7.4), and 2 μl of enzyme (final concentration, 10 to 20 μg/ml), which was then incubated at 37°C. Portions of 15 μl were removed at 0, 5, 15, 30, and 60 min, the reaction was stopped by heating at 90°C for 5 min, and 4-μl portions were spotted onto a 4-cm by 5-cm aluminum-backed Silica gel 60 TLC plate (Merck, Kirkland, QC, Canada). The plates were developed using the following solvent system: chloroform-methanol-acetic acid-water (50:50:15:5, vol/vol). After developing, the plates were air dried, and the carbohydrates were visualized by dipping the plate in 5% sulfuric acid in ethanol and draining excess liquid and then heating over a hot plate until spots were visible. The hydrolysis products were compared with the sugar standards G1 to G5 for cello-oligosaccharides and X1, X2, X4, and X5 for xylo-oligosaccharides. To assess activity, we used a qualitative estimate of activity as visualized on the TLC plates, which was scored as follows: +++, hydrolysis near complete; ++, hydrolysis near 50% complete; +, hydrolysis less than 50% complete; +/−, detectable but weak hydrolysis.
We attempted kinetic assays on glucose disaccharides performed as described above, except that glucose production was used as a measure of activity instead of visual inspection of the TLC plates. Glucose release was measured with a glucose assay kit (Sigma Chemical Company) as recommended by the manufacturer.
Effects of pH and temperature.
To examine the effect of pH on the enzyme activity, the pH was varied from 4.0 to 8.0 using 25 mM citrate phosphate buffer. The 50-μl reaction mixture containing 2 mM substrate, 10 μl of diluted enzyme (10 to 20 μg/ml), and 35 μl of buffer at various pHs was incubated at 30°C for 15 min. Then, the reaction was stopped by adding 150 μl of 50 mM NaOH, and the absorbance was read at 410 nm. The molar extinction coefficient for pNP at this pH was 18,000 liter−1 M−1. The effect of temperature on the enzyme activity was investigated at temperatures ranging from 25 to 80°C. The reactions were performed in 25 mM citrate phosphate buffer (pH 7) containing 2 mM substrate and diluted enzyme (10 to 20 μg/ml).
RESULTS
Sequence alignment and analysis of the GH3 and the GH1 enzymes.
The amino acid sequences encoded by the four genes Celf_2726, Celf_0140, Celf_0468, and Celf_3372 were found in CAZy GH3. None of these genes encoded signal sequences or other accessory modules like carbohydrate binding domains; however, BLAST searches suggested that all of them contain a C-terminal fibronectin type III-like domain of unknown function (5). Signal sequences were analyzed by the Signal 4.1P server (23), and the lack of signal sequence suggests their intracellular location in C. fimi ATCC 484. The multiple amino acid sequence alignment of these four GH3 family enzymes from C. fimi ATCC 484 is shown in Fig. S1 in the supplemental material. A BLAST search with all 4 proteins shows that related genes from other bacteria are very common, but none of the top BLAST hits had any biochemical data to suggest a function (data not shown), and all were in fact annotated as simply β-d-glucoside glucohydrolase. The C-terminal fibronectin type III-like domain was also seen in these BLAST hits and seems to be part of the GH3 family protein structure, albeit of unknown function. Celf_0468, Celf_2726, and Celf_3372 show a much lower sequence homology with other GH3s than does Celf_0140 in the NCBI protein database.
The Celf_2783 gene product shows some homology to many members of GH1, but we have limited the sequence alignment to three other GH1 sequences from cellulomonads (see Fig. S2 in the supplemental material). The C. fimi and Cellulomonas flavigena proteins are 83% identical, but they both share only 55% identity to the Cellulomonas biazotea GH1 (GenBank AEM45802.1) and 66% and 52%, respectively, to the entry named as a C. fimi GH1 gene (GenBank AAA23091.1).
Expression and purification of the enzymes.
To systematically study the functional diversity of the four GH3 enzymes and the GH1 enzyme, the genes were cloned into the expression vector pCW, which allowed overexpression of the C. fimi enzymes in E. coli. The four GH3 enzymes possess similar molecular masses (from 80,300 Da to 89,000 Da) and theoretical pIs of 4.95 to 5.09, as calculated by the online server at http://web.expasy.org/protparam. The GH1 protein has a molecular mass of 53,000 Da and a theoretical pI of 4.98. Even though the proteins contained a polyhistidine (6×His) tag for immobilized-metal affinity chromatography (IMAC), we found that purification on ion exchange yielded purer proteins, except for Celf_2726 (data not shown). Typically, about 50 to 150 mg of pure proteins were obtainable from 1 liter of 2YT culture. The detailed purification results from a representative 200-ml culture are summarized for each enzyme in Table 2.
TABLE 2.
Summary of purification of β-glycoside hydrolases
| Enzyme | Purification step | Total activitya (U) | Total protein (mg) | Sp act (U/mg) | Purification (fold) | Recovery (%) |
|---|---|---|---|---|---|---|
| CfBgl1 | Crude enzyme extract | 189.5 | 141 | 1.34 | 1 | 100 |
| HiTrap Q HP column | 49.9 | 9.64 | 5.18 | 3.9 | 26.3 | |
| CfXyl3 | Crude enzyme extract | 94.6 | 124.8 | 0.76 | 1.0 | 100 |
| HisTrap HP column | 84.2 | 15.5 | 5.43 | 7.14 | 89 | |
| CfBgl3A | Crude enzyme extract | 286.1 | 179.1 | 1.60 | 1.0 | 100 |
| HiTrap HP Q column | 100.0 | 7.1 | 14.08 | 8.8 | 35.0 | |
| CfBgl3B | Crude enzyme extract | 8.0 | 73.5 | 0.11 | 1.0 | 100 |
| HiTrap HP Q column | 2.7 | 5.1 | 0.53 | 4.8 | 33.8 | |
| CfBgl3C | Crude enzyme extract | 103.2 | 97.0 | 1.06 | 1.0 | 100 |
| HiTrap HP Q column | 42.0 | 12.8 | 3.28 | 3.1 | 40.7 |
All the activities are reported as activity on pNP-β-Glc, except for CfXyl3A, which was assayed with pNP-β-Xyl.
GH family 3 enzymes from C. fimi exhibited different specificities on pNP substrates.
To determine the substrate specificities for the GH3 enzymes, the recombinant proteins were screened for activity with a library of pNP-β-glycosides (Table 3). Of all four GH3 enzymes, Celf_2726 is the only protein that did not hydrolyze pNP-β-Glc; it is clearly a β-xylosidase with some activity on pNP-α-l-Ara. Using the CAZy enzyme naming scheme, we assigned to the Celf_2726 protein the designation CfXyl3. Celf_0140 (CfBgl3A) was also fairly specific, with robust hydrolysis of pNP-β-Glc and little activity on pNP-β-cellobioside and pNP-β-Xyl. Celf_0468 (CfBgl3B) had the least activity, as the only substrate hydrolyzed was pNP-β-Glc and its activity was 6 and 28 times lower than that of CfBgl3C and CfBgl3A, respectively. In contrast to the other GH3 enzymes, Celf_3372 (CfBgl3C) was quite promiscuous with pNP substrates, as it exhibited multiple activities against pNP-β-Glc, pNP-β-Xyl, pNP-α-l-Ara, and pNP-β-Fuc (Table 3). These results indicated that each of the four GH3 genes encodes a functional glycoside hydrolase enzyme, three of which possess some overlapping enzyme activity for pNP-β-Glc but have different properties for hydrolysis of various substrates.
TABLE 3.
Screening for enzyme activity on p-nitrophenyl-glycosides
| Substrate | Relative activitya (%) |
||||
|---|---|---|---|---|---|
| Celf_2783 (CfBgl1) | Celf_2726 (CfXyl3A) | Celf_0140 (CfBgl3A) | Celf_0468 (CfBgl3B) | Celf_3372 (CfBgl3C) | |
| p-Nitrophenyl-β-d-glucopyranoside | 100 | —b | 100 | 100 | 100 |
| p-Nitrophenyl-β-d-cellobioside | 21 | — | 7 | — | — |
| p-Nitrophenyl-α-l-arabinofuranoside | — | 21 | — | — | 11 |
| p-Nitrophenyl-β-d-xylopyranoside | 0.5 | 100 | 3 | — | 50 |
| p-Nitrophenyl-β-d-glucosaminide | — | — | — | — | — |
| p-Nitrophenyl-β-d-galactopyranoside | 5 | — | — | — | — |
| p-Nitrophenyl-β-d-fucopyranoside | 99 | — | — | — | 23 |
| p-Nitrophenyl-β-d-mannopyranoside | — | — | — | — | — |
| p-Nitrophenyl-β-d-lactoside | 3 | — | — | — | — |
| p-Nitrophenyl-β-d-glucuronide | — | — | — | — | — |
Reactions were performed with 2 mM substrate, pH 7.4, at 30°C for 60 min. The highest level of activity was taken as 100%, and the other activity was made relative to that value.
—, no activity was observed.
GH3 enzymes from C. fimi release monosaccharides from plant cell wall polysaccharides.
β-1,4-Linked xylosides and glucosides are common moieties found in plant cell walls and are of particular importance for biomass conversion for biofuel production (24). The activity with pNP-linked sugars suggested that the four GH3 enzymes may have activity against β-1,4-linked xylosides or glucosides. To evaluate whether the four GH3 proteins can contribute to the hydrolysis of plant cell wall polysaccharides, the activity of each protein was measured against a panel of cello- and xylo-oligosaccharides.
As expected from the pNP hydrolysis data, CfXyl3A produced xylose from xylobiose (Table 4), confirming that it is indeed a β-xylopyranosidase, with α-arabinofuranosidase activity. Somewhat surprisingly, it was not active on xylotetraose or xylopentaose, suggesting a rather specific activity.
TABLE 4.
Catalysis of oligosaccharides by four GH3 enzymes and one GH1 enzyme
| Saccharide | Linkage | Activitya |
||||
|---|---|---|---|---|---|---|
| CfXyl3A | CfBgl3A | CfBgl3B | CfBgl3C | CfBgl1 | ||
| Sophorose | β-1,2- | − | + | − | − | +++ |
| Laminaribiose | β-1,3- | − | +++ | − | − | +++ |
| Gentiobiose | β-1,6- | − | ++ | +++b | − | +/− |
| Cellobiose (G2) | β-1,4- | − | + | − | − | + |
| Cellotriose (G3) | β-1,4- | − | ++ | − | − | +++ |
| Cellotetraose (G4) | β-1,4- | − | + | − | − | +++ |
| Cellopentaose (G5) | β-1,4- | − | + | − | − | ++ |
| Xylobiose (X2) | β-1,4- | +++ | − | − | − | − |
| Xylotetraose (X4) | β-1,4- | − | − | − | − | − |
| Xylopentaose (X5) | β-1,4- | − | − | − | − | − |
| Lactose | β-1,4- | − | − | − | − | +/− |
Activity scoring: +++, near-complete hydrolysis; ++, hydrolysis near 50% complete; +, hydrolysis less than 50%; +/−, detectable but weak hydrolysis.
Reaction showed transglycosylation product.
CfBgl3A, which had the highest specific activity on pNP-β-Glc of all the enzymes (Table 2), hydrolyzed all glucose-containing plant cell wall polysaccharides, regardless of linkage (Table 4). The enzyme displayed a clear preference for laminaribiose and gentiobiose, giving it a linkage preference as follows: β-1,3 = β-1,6 > β-1,2 = β-1,4. Analysis of the reaction products from cellopentaose and cellotetraose revealed predominant reaction products of glucose and a cello-oligosaccharide that is one residue shorter than the starting material (Fig. 1). Few shorter cello-oligosaccharides were observed, suggesting that CfBgl3A cleaves the nonreducing terminal residue of cellodextrins, making it an exo-β-glucosidase. Although CfBgl3A exhibited activity on pNP-β-Xyl, activity against xylo-oligosaccharides was not observed.
FIG 1.
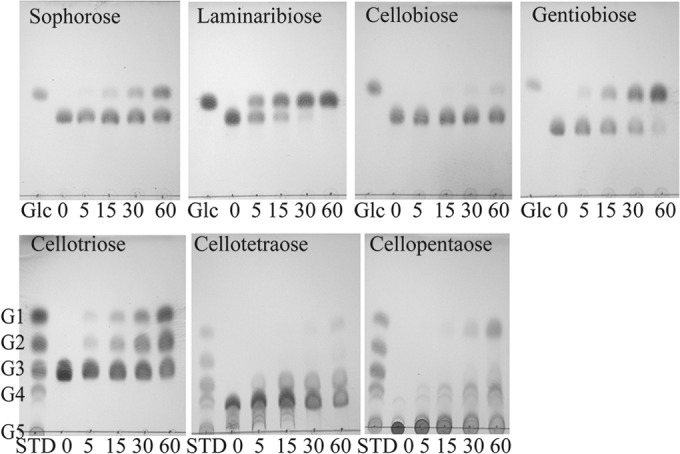
CfBgl3A-catalyzed hydrolysis of β-linked disaccharides (top) and cello-oligosaccharides (bottom) examined by TLC. Standard sugars (STD) include glucose (G1), cellobiose (G2), cellotriose (G3), cellotetraose (G4), and cellopentaose (G5).
Purified CfBgl3B, which had the lowest specific activity and no promiscuity on the pNP substrates, hydrolyzed only gentiobiose (Glc-β-1,6-Glc), without touching the other disaccharides and oligosaccharides tested. However, substrate transglycosylation also occurs simultaneously with hydrolysis (Fig. 2). With increasing concentrations of gentiobiose, the rate of substrate transglycosylation increased while the rate of hydrolysis decreased. No transglycosylation activity was observed with cellobiose as the substrate (data not shown). The transglycosylation activity can be inhibited with increasing concentrations of glucose in the starting reaction mixture, from 5 to 25 mg/ml, without affecting hydrolysis (see Fig. S3 in the supplemental material).
FIG 2.
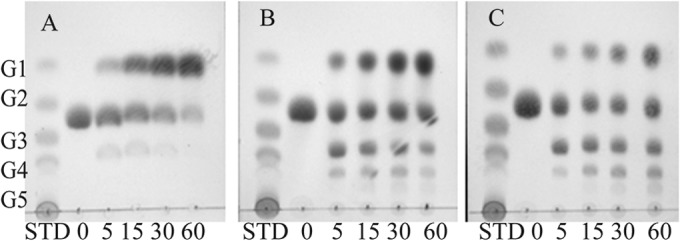
CfBgl3B-catalyzed hydrolysis and transglycosylation of gentiobiose examined by TLC under different substrate concentrations: 5 mg/ml (A), 25 mg/ml (B), and 50 mg/ml (C). Standard sugars (STD) include glucose (G1), cellobiose (G2), cellotriose (G3), cellotetraose (G4), and cellopentaose (G5).
For purified CfBgl3C, no hydrolysis or transglycosylation activity was observed on the disaccharides and oligosaccharides tested, which indicated that CfBgl3C had a strict aryl-β-glycosidase activity but with a broad specificity for the sugar moiety.
Kinetic parameters of four GH3 enzymes.
pNP-glycosides were used to determine the kinetic parameters for each of the GH3 enzymes (Table 5). CfXyl3A was highly active on pNP-β-Xyl and pNP-α-l-Ara: the kcat/Km of 175 s−1 mM−1 for pNP-β-Xyl suggests that this is indeed a relevant substrate; the much higher Km for pNP-α-l-Ara produces a very reduced specificity constant. The hydrolysis of xylobiose by CfXyl3A did not appear to show saturation kinetics when using the reducing sugar assay to measure xylose release, so we were unable to calculate kinetic parameters for xylobiose. For the other three GH3 enzymes, the kcat/Km values for pNP substrates showed that CfBgl3A had the highest specificity for pNP-β-Glc and that CfBgl3B is a relatively poor enzyme with a very slow hydrolysis rate for pNP-β-Glc, whereas CfBgl3C had its highest specificity for pNP-β-Xyl. The hydrolysis rate for CfBgl3A with pNP-β-Xyl was too low to calculate kinetic parameters even when increasing the enzyme concentration to 2 mg/ml in the reaction mixture. For CfBgl3C, the kinetic parameters for pNPα-l-Ara could not be obtained since the hydrolysis rate was too low to give accurate values. Using the detection of glucose release from disaccharides did not yield interpretable kinetic parameters for any of the enzymes that we analyzed, as these reactions also did not show saturation kinetics with substrate concentrations as high as 15 mM.
TABLE 5.
Kinetic values of GH enzymes on synthetic substratesa
| GH enzyme and substrate | Km (mM) | kcat (s−1) | kcat/Km (s−1 mM−1) |
|---|---|---|---|
| CfXyl3A | |||
| pNP-β-d-xylopyranoside | 0.13 ± 0.03 | 21.05 ± 2.80 | 175.01 ± 48.83 |
| pNP-α-l-arabinofuranoside | 7.12 ± 2.58 | 29.45 ± 8.32 | 4.27 ± 0.42 |
| CfBgl3A | |||
| pNP-β-d-glucopyranoside | 5.14 ± 0.85 | 172.29 ± 12.80 | 34.32 ± 5.61 |
| pNP-β-d-xylopyranoside | —b | — | — |
| CfBgl3B | |||
| pNP-β-d-glucopyranoside | 1.25 ± 0.02 | 0.71 ± 0.01 | 0.57 ± 0.01 |
| CfBgl3C | |||
| pNP-β-d-glucopyranoside | 3.52 ± 0.91 | 32.03 ± 3.66 | 9.59 ± 2.43 |
| pNP-β-d-xylopyranoside | 0.27 ± 0.07 | 7.98 ± 0.34 | 31.65 ± 6.84 |
| pNP-β-d-fucopyranoside | 2.29 ± 0.33 | 10.18 ± 0.42 | 4.51 ± 0.52 |
| pNP-α-l-arabinofuranoside | — | — | — |
| CfBgl1 | |||
| pNP-β-d-galactopyranoside | No saturation | — | — |
| pNP-β-d-fucopyranoside | 1.26 ± 0.30 | 84.14 ± 17.81 | 67.52 ± 6.83 |
| pNP-β-d-glucopyranoside | 0.44 ± 0.09c | 32.27 ± 3.27 | 74.65 ± 7.80 |
Data are reported as means ± standard errors of the means from three independent experiments.
—, kinetic data were not determined because the rates were not high enough to accurately measure kinetic parameters.
Some substrate inhibition was seen at substrate concentrations above 2 mM.
Substrate specificity of CfBgl1.
We assayed the GH1 enzyme on a variety of substrates and found it to have β-glucosidase, β-galactosidase, and β-fucosidase activity when assayed with pNP-glycosides (Table 3). The GH1 enzyme had good activity on sophorose and laminaribiose but weak activity on cellobiose and very weak activity on gentiobiose. The linkage preference for disaccharides was as follows: β-1,2 > β-1,3 ≫ β-1,4 >≫ β-1,6. Cellodextrins G3, G4, and G5 were also hydrolyzed very well with the release of glucose and cellobiose (Fig. 3). Kinetic parameters showed the best kcat/Km for pNP-β-Fuc, followed by pNP-β-Glc, and no saturation for pNP-β-Gal. For the glucose disaccharides, saturation was not seen with substrate concentrations as high as 15 mM.
FIG 3.
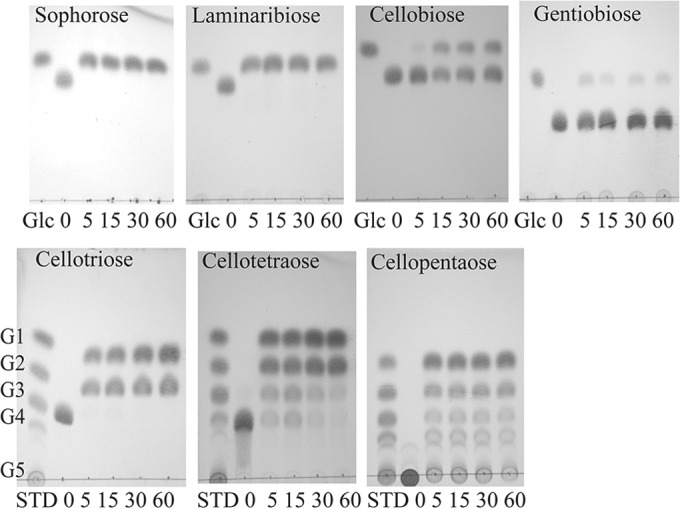
CfBgl1A-catalyzed hydrolysis of β-linked disaccharides (top) and cello-oligosaccharides (bottom) examined by TLC. Standard sugars (STD) include glucose (G1), cellobiose (G2), cellotriose (G3), cellotetraose (G4), and cellopentaose (G5).
Temperature and pH profiles of GH3 and GH1 enzymes.
All of the enzymes were assayed with their best pNP substrate at pH intervals between 4 and 8 and at temperature intervals between 25 and 80°C (data not shown). These data are summarized in Table S1 in the supplemental material. The pH optima were all between pH 6.5 and 7.5, usually with a clear peak of activity. None of the enzymes had appreciable activity at pH 5. CfXyl3A and CfBgl3B were broadly active between pH 6 and 8, whereas the other proteins had more-dramatic changes in activity, as a 30 to 50% increase in activity was seen with a 0.5 pH unit increase on the acidic side of the optimum. The CfBgl3B/C proteins showed the most temperature sensitivity, as anything over 30°C resulted in dramatic loss of activity. We did not attempt to measure the melting temperature (Tm) for any of these proteins.
DISCUSSION
In the postgenomic era, our identification of enzymes from genome sequence projects still depends heavily on biochemical characterization of the proteins. The genus Cellulomonas has been studied for over 50 years as a model bacterial cellulase system, and much fundamental knowledge about the secreted enzymes involved in biomass degradation has been gathered, especially once molecular cloning was a routine technique (1, 3, 25, 26). The problem that we are faced with now is that despite having cloned, expressed, and studied a long list of extracellular glycoside hydrolase enzymes, there remains much about how this bacterium degrades biomass that we do not know. The focus of this work was to revisit the question of the β-glucosidase enzymes that have been shown to be critical for removing cellobiose, which is an end product inhibitor of many cellulases (14). In the early work on Cellulomonas, we identified multiple β-glucosidases using a native gel analysis (6) but were unable to generate a recombinant clone for this activity using an activity-screening procedure. Access to the genome sequence for C. fimi ATCC 484 prompted us to undertake the characterization of putative β-glucosidase genes.
β-Glucosidase genes are found primarily in two families in the CAZy database, GH1 and GH3. C. fimi ATCC 484 contains a single GH1 gene, Celf_2783, and 10 GH3 genes. Celf_2983 is the CfNag3 β-N-acetylglucosaminidase/β-glucosidase previously characterized by Mayer et al. (18). We designed PCR primers for all the remaining GH3 genes but were unable to amplify many of them from the genomic DNA (data not shown). The genes that yielded distinct PCR products were Celf_0140, Celf_0468, Celf_2726, Celf_2783, and Celf_3372. Clones with these 5 genes yielded good protein expression and were straightforward to purify using anion-exchange chromatography for 4 of the 5 genes and IMAC for 1 of them. Our screening assay was based on a panel of pNP-β-glycosides based on the known specificities of the GH1 and GH3 families.
Celf_2726 was a β-xylosidase/α-l-arabinosidase with a similar substrate specificity as that of a GH3 enzyme purified from barley (9), Prevotella ruminicola (PrXyl3A [GenBank ACN78955.1]) (27) and Thermotoga thermarum (TtXyl3, Theth_0138) (28). Recently, a trifunctional GH3 from a rumen metagenomic screen Clostridium saccharoperbutylacetonicum N1–4(HMT); GenBank YP_007457327.1; CsBgl3] showed activity against pNP-β-Glc/Xyl and pNP-α-Ara as well as activity on xylan (29). BLAST searches of these other characterized GH3 enzymes against the C. fimi predicated proteome showed that they had no significant sequence similarities to any C. fimi proteins (data not shown). A major difference between CfXyl3A, TtXyl3, and PrXyl3A was the lack of hydrolysis of longer xylo-oligosaccharides by the CfXyl3A, which suggests that the active-site architectures are quite different among these enzymes, despite having some common activity on synthetic pNP-linked substrates.
In Prevotella bryantii B14, four other Xyl3 proteins have been characterized, but all of these had dual specificity on xylo- and gluco-oligosaccharide (30). None of the C. fimi GH3 enzymes that we examined had this dual specificity for the xylo- and gluco-oligosaccharides. It is likely that CfXyl3A is involved in the degradation of xylan, together with endoxylanases for cleavage of end product xylobiose groups. The α-l-arabinofuranosidase activity from CfXyl3A is likely to be of minor importance, as it was measured only on a pNP substrate and because there are also seven GH43 and four GH51 enzymes, some of which are likely more potent α-l-arabinofuranosidases.
The enzyme CfBgl3A has a broad activity for different linkage types in disaccharides and polymeric β-d-glucan substrates. The β-linked glucosyl disaccharides were all substrates, but we were unable to measure kinetic constants for their hydrolysis, as we could not achieve saturation up to 15 mM substrate. CfBgl3A also catalyzes the release of glucose from cello-oligosaccharides G3/G4/G5 but is not very efficient under the conditions we used. Therefore, CfBgl3A is primarily an exo-β-d-1,3/1,6-glucosidase.
CfBgl3B had the narrowest substrate specificity of the proteins we examined, having only very weak activity on pNP-β-Glc, where the kcat was 242-fold lower than CfBgl3A and 45-fold lower than CfBgl3C. The only activity on a disaccharide was with gentiobiose, and it was accompanied by a strong transglycosylation activity, which was not seen with the other proteins in our study. It is unlikely that this activity plays a role in vivo, as the substrate concentrations in the cell would never reach the ∼70 mM that was used in the transglycosylation assay.
CfBgl3C had a very broad spectrum of activities on pNP-linked glycosides. It hydrolyzed pNP-β-Glc/Xyl/Fuc, and pNP-α-l-Ara; however, no hydrolysis of any natural disaccharide or xylo-/gluco-oligosaccharide was observed. The kcat/Km values suggest the preferred substrate to be pNP-β-Xyl, but it had only 25% of the turnover number seen with pNP-β-Glc. The question of the natural substrate for this enzyme remains open, as there are many possible aryl-glycosides from plants such as flavonoids (31) and cyanogenic glycosides (32) and many other potential aryl-glycosides in the soil environment, so the role of this enzyme in biomass degradation remains uncertain.
The GH1 family of enzymes is as large as the GH3 family (GH1 currently has 6,154 members) but contains far more substrate diversity than GH3, with 24 known specificities according to the CAZy database, compared to 10 for GH3 (www.cazy.org). Enzymes from GH1 are good candidates for true cellobiase activity from a variety of organisms (33–35), so we reasoned that the Celf_2783 protein was a candidate cellobiase. A GH1 cellobiase from C. biazotea has recently been examined, but the complete substrate specificity profile and enzymatic characterization have not been published (36). There is also a GenBank entry for a GH1 from C. fimi (AAA23091.1), which differs in amino acid sequence from the protein encoded by Celf_2783 (the genome-derived GH1), and we were unable to find published biochemical data about this protein. Our examination of CfBgl1 shows that it is not a very efficient cellobiase, as it shows much better activity on β-1,2/3-linked glucose than β-1,4-linked glucose. It does have good activity on the longer cello-oligosaccharide but generates cellobiose as a major end product, unlike other members of this family, which generate glucose. So, it appears that there must be other proteins that are responsible for the efficient hydrolysis of cellobiose, perhaps in the group of GH3 genes that we were unable to clone. An alternative possibility is that Cellulomonas uses cellobiose phosphorylase (Celf_0317 [GH94]) as its main strategy for removing cellobiose from the cellulose hydrolysis mixture, and that the other enzymes contribute, like CfBgl3A and CfBgl1, in that they work on cellodextrins generating some glucose or cellobiose. The gene for cellobiose phosphorylase is found in C. fimi, Cellulomonas flavigena, and Cellulomonas uda, and the induction of GH94 during growth on carboxymethylcellulose in related cellulomonads has been demonstrated (37).
In conclusion, we have demonstrated that the five new genes that we expressed all encode active β-glycoside hydrolase enzymes, with a variety of substrate specificities that are consistent with their activity in some aspect of biomass metabolism. Our data also clearly demonstrate that the natural substrates utilized by these multigene families need to be identified in order to assign a biological role to family members. This will require the availability of more structurally defined oligosaccharide fragments. Of course, having more protein structures of the more diverse family members (preferably with bound substrates) would provide a better comprehension of the determinants of substrate specificity. When more of these GH3/GH1 enzymes have been characterized and the corresponding structures have been solved, there may be some signature sequences that might provide a better means of subdividing these large families.
Supplementary Material
ACKNOWLEDGMENTS
W.W. acknowledges the financial support of a discovery grant from the Natural Sciences and Engineering Research Council of Canada. J.G. acknowledges a visiting student stipend to perform this work from the Government of the People's Republic of China.
We thank Melissa J. Schur of the National Research Council for help with primer design for the PCRs. We thank R. A. J. Warren, Lyann Sim, and Lisa Willis for critical reading of the manuscript.
Footnotes
Published ahead of print 15 September 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.02194-14.
REFERENCES
- 1.Béguin P, Eisen Hroupas A. 1977. Free and cellulose-bound cellulases in a Cellulomonas species. J. Gen. Microbiol. 101:191–196. 10.1099/00221287-101-2-191. [DOI] [Google Scholar]
- 2.Han YW, Srinivasan VR. 1968. Isolation and characterization of a cellulose-utilizing bacterium. Appl. Microbiol. 16:1140–1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Langsford ML, Gilkes NR, Wakarchuk WW, Kilburn DG, Miller RC, Warren RAJ. 1984. The cellulase system of Cellulomonas fimi. J. Gen. Microbiol. 130:1367–1376. [Google Scholar]
- 4.Christopherson MR, Suen G, Bramhacharya S, Jewell KA, Aylward FO, Mead D, Brumm PJ. 2013. The genome sequences of Cellulomonas fimi and “Cellvibrio gilvus” reveal the cellulolytic strategies of two facultative anaerobes, transfer of “Cellvibrio gilvus” to the genus Cellulomonas, and proposal of Cellulomonas gilvus sp. nov. PLoS One 8:e53954. 10.1371/journal.pone.0053954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cantarel BL, Coutinho PM, Rancurel C, Bernard T, Lombard V, Henrissat B. 2009. The Carbohydrate-Active EnZymes database (CAZy): an expert resource for glycogenomics. Nucleic Acids Res. 37:D233–D238. 10.1093/nar/gkn663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wakarchuk WW, Kilburn DG, Miller RC, Warren RAJ. 1984. The preliminary characterization of the β-glucosidases of Cellulomonas fimi. J. Gen. Microbiol. 130:1385–1389. [Google Scholar]
- 7.Fincher G, Mark B, Brumer H. 2014. Glycoside hydrolase family 3. http://www.cazypedia.org/ Accessed 4 July 2014.
- 8.Harvey AJ, Hrmova M, De Gori R, Varghese JN, Fincher GB. 2000. Comparative modeling of the three-dimensional structures of family 3 glycoside hydrolases. Proteins 41:257–269. . [DOI] [PubMed] [Google Scholar]
- 9.Lee RC, Hrmova M, Burton RA, Lahnstein J, Fincher GB. 2003. Bifunctional family 3 glycoside hydrolases from barley with α-l-arabinofuranosidase and β-d-xylosidase activity: characterization, primary structures, and COOH-terminal processing. J. Biol. Chem. 278:5377–5387. 10.1074/jbc.M210627200. [DOI] [PubMed] [Google Scholar]
- 10.Hancock SM, Corbett K, Fordham-Skelton AP, Gatehouse JA, Davis BG. 2005. Developing promiscuous glycosidases for glycoside synthesis: residues W433 and E432 in Sulfolobus solfataricus beta-glycosidase are important glucoside- and galactoside-specificity determinants. Chembiochem 6:866–875. 10.1002/cbic.200400341. [DOI] [PubMed] [Google Scholar]
- 11.Kawai R, Igarashi K, Kitaoka M, Ishii T, Samejima M. 2004. Kinetics of substrate transglycosylation by glycoside hydrolase family 3 glucan (1–>3)-beta-glucosidase from the white-rot fungus Phanerochaete chrysosporium. Carbohydr. Res. 339:2851–2857. 10.1016/j.carres.2004.09.019. [DOI] [PubMed] [Google Scholar]
- 12.Faure D. 2002. The family-3 glycoside hydrolases: from housekeeping functions to host-microbe interactions. Appl. Environ. Microbiol. 68:1485–1490. 10.1128/AEM.68.4.1485-1490.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lynd LR, Laser MS, Bransby D, Dale BE, Davison B, Hamilton R, Himmel M, Keller M, McMillan JD, Sheehan J, Wyman CE. 2008. How biotech can transform biofuels. Nat. Biotechnol. 26:169–172. 10.1038/nbt0208-169. [DOI] [PubMed] [Google Scholar]
- 14.Bhatia Y, Mishra S, Bisaria VS. 2002. Microbial beta-glucosidases: cloning, properties, and applications. Crit. Rev. Biotechnol. 22:375–407. 10.1080/07388550290789568. [DOI] [PubMed] [Google Scholar]
- 15.Uchiyama T, Miyazaki K, Yaoi K. 2013. Characterization of a novel beta-glucosidase from a compost microbial metagenome with strong transglycosylation activity. J. Biol. Chem. 288:18325–18334. 10.1074/jbc.M113.471342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nomura T, Quesada AL, Kutchan TM. 2008. The new beta-D-glucosidase in terpenoid-isoquinoline alkaloid biosynthesis in Psychotria ipecacuanha. J. Biol. Chem. 283:34650–34659. 10.1074/jbc.M806953200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou J, Hartmann S, Shepherd BK, Poulton JE. 2002. Investigation of the microheterogeneity and aglycone specificity-conferring residues of black cherry prunasin hydrolases. Plant Physiol. 129:1252–1264. 10.1104/pp.010863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mayer C, Vocadlo DJ, Mah M, Rupitz K, Stoll D, Warren RAJ, Withers SG. 2006. Characterization of a β-N-acetylhexosaminidase and a β-N-acetylglucosaminidase/β-glucosidase from Cellulomonas fimi. FEBS J. 273:2929–2941. 10.1111/j.1742-4658.2006.05308.x. [DOI] [PubMed] [Google Scholar]
- 19.Withers S. 2014. Glycoside hydrolase family 1. http://www.cazypedia.org/ Accessed 4 July 2014.
- 20.Trimbur DE, Warren RA, Withers SG. 1992. Region-directed mutagenesis of residues surrounding the active site nucleophile in beta-glucosidase from Agrobacterium faecalis. J. Biol. Chem. 267:10248–10251. [PubMed] [Google Scholar]
- 21.Wang Q, Trimbur D, Graham R, Warren RAJ, Withers SG. 1995. Identification of the acid/base catalyst in Agrobacterium faecalis β-glucosidase by kinetic analysis of mutants. Biochemistry 34:14554–14562. 10.1021/bi00044a034. [DOI] [PubMed] [Google Scholar]
- 22.Wakarchuk WW, Campbell RL, Sung WL, Davoodi J, Yaguchi M. 1994. Mutational and crystallographic analyses of the active site residues of the Bacillus circulans xylanase. Protein Sci. 3:467–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Petersen TN, Brunak S, von Heijne G, Nielsen H. 2011. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods 8:785–786. 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- 24.Gowen CM, Fong SS. 2010. Exploring biodiversity for cellulosic biofuel production. Chem. Biodivers. 7:1086–1097. 10.1002/cbdv.200900314. [DOI] [PubMed] [Google Scholar]
- 25.Gilkes NR, Kilburn DG, Langsford ML, Miller RC, Wakarchuk WW, Warren RAJ, Whittle DJ, Wong WKR. 1984. Isolation and characterization of Escherichia coli clones expressing cellulase genes from Cellulomonas fimi. J. Gen. Microbiol. 130:1377–1384. 10.1099/00221287-130-6-1377. [DOI] [Google Scholar]
- 26.Whittle DJ, Kilburn DG, Warren RA, Miller RC., Jr 1982. Molecular cloning of a Cellulomonas fimi cellulase gene in Escherichia coli. Gene 17:139–145. 10.1016/0378-1119(82)90066-X. [DOI] [PubMed] [Google Scholar]
- 27.Dodd D, Kocherginskaya SA, Spies MA, Beery KE, Abbas CA, Mackie RI, Cann IKO. 2009. Biochemical analysis of a β-D-xylosidase and a bifunctional xylanase-ferulic acid esterase from a xylanolytic gene cluster in Prevotella ruminicola 23. J. Bacteriol. 191:3328–3338. 10.1128/JB.01628-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shi H, Li X, Gu H, Zhang Y, Huang Y, Wang L, Wang F. 2013. Biochemical properties of a novel thermostable and highly xylose-tolerant beta-xylosidase/alpha-arabinosidase from Thermotoga thermarum. Biotechnol. Biofuels 6:27. 10.1186/1754-6834-6-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gruninger RJ, Gong X, Forster RJ, McAllister TA. 2014. Biochemical and kinetic characterization of the multifunctional beta-glucosidase/beta-xylosidase/alpha-arabinosidase, Bgxa1. Appl. Microbiol. Biotechnol. 98:3003–3012. 10.1007/s00253-013-5191-4. [DOI] [PubMed] [Google Scholar]
- 30.Dodd D, Kiyonari S, Mackie RI, Cann IKO. 2010. Functional diversity of four glycoside hydrolase family 3 enzymes from the rumen bacterium Prevotella bryantii B14. J. Bacteriol. 192:2335–2345. 10.1128/JB.01654-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xiao J, Chen T, Cao H. 2014. Flavonoid glycosylation and biological benefits. Biotechnol. Adv. 2014:pii:S0734-9750(14)00092-5. 10.1016/j.biotechadv.2014.05.004. [DOI] [PubMed] [Google Scholar]
- 32.Gleadow RM, Moller BL. 2014. Cyanogenic glycosides: synthesis, physiology, and phenotypic plasticity. Annu. Rev. Plant Biol. 65:155–185. 10.1146/annurev-arplant-050213-040027. [DOI] [PubMed] [Google Scholar]
- 33.Wakarchuk W, Kilburn D, Miller R, Jr, Warren RA. 1986. The molecular cloning and expression of a cellobiase gene from an Agrobacterium in Escherichia coli. Mol. Gen. Genet. 205:146–152. 10.1007/BF02428044. [DOI] [Google Scholar]
- 34.Love DR, Fisher R, Bergquist PL. 1988. Sequence structure and expression of a cloned beta-glucosidase gene from an extreme thermophile. Mol. Gen. Genet. 213:84–92. 10.1007/BF00333402. [DOI] [PubMed] [Google Scholar]
- 35.Grabnitz F, Seiss M, Rucknagel KP, Staudenbauer WL. 1991. Structure of the beta-glucosidase gene bglA of Clostridium thermocellum. Sequence analysis reveals a superfamily of cellulases and beta-glycosidases including human lactase/phlorizin hydrolase. Eur. J. Biochem. 200:301–309. [DOI] [PubMed] [Google Scholar]
- 36.Chan AK, Wang YY, Ng KL, Fu Z, Wong WK. 2012. Cloning and characterization of a novel cellobiase gene, cba3, encoding the first known beta-glucosidase of glycoside hydrolase family 1 of Cellulomonas biazotea. Gene 493:52–61. 10.1016/j.gene.2011.11.027. [DOI] [PubMed] [Google Scholar]
- 37.Schimz KL, Broll B, John B. 1983. Cellobiose phosphorylase (EC 2.4.1.20) of Cellulomonas: occurrence, induction, and its role in cellobiose metabolism. Arch. Microbiol. 135:241–249. 10.1007/BF00413475. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


