Abstract
Raman spectroscopy was used to study the time course of phenotypic responses of Escherichia coli (DH5α) to 1-butanol exposure (1.2% [vol/vol]). Raman spectroscopy is of interest for bacterial phenotyping because it can be performed (i) in near real time, (ii) with minimal sample preparation (label-free), and (iii) with minimal spectral interference from water. Traditional off-line analytical methodologies were applied to both 1-butanol-treated and control cells to draw correlations with Raman data. Here, distinct sets of Raman bands are presented that characterize phenotypic traits of E. coli with maximized correlation to off-line measurements. In addition, the observed time course phenotypic responses of E. coli to 1.2% (vol/vol) 1-butanol exposure included the following: (i) decreased saturated fatty acids levels, (ii) retention of unsaturated fatty acids and low levels of cyclopropane fatty acids, (iii) increased membrane fluidity following the initial response of increased rigidity, and (iv) no changes in total protein content or protein-derived amino acid composition. For most phenotypic traits, correlation coefficients between Raman spectroscopy and traditional off-line analytical approaches exceeded 0.75, and major trends were captured. The results suggest that near-real-time Raman spectroscopy is suitable for approximating metabolic and physiological phenotyping of bacterial cells subjected to toxic environmental conditions.
INTRODUCTION
The quest for biofuels through fermentation is well established, and numerous studies have focused on the role of product toxicity to the host culture (1–5). In the United States, production of 36 billion gallons of renewable fuel will be required by 2022, with about 44% to be obtained from cellulosic ethanol (6). Currently, approximately 90% of the renewable liquid biofuels market is represented by biodiesel and ethanol (5). However, 1-butanol has long been an alternative biofuel of interest. Unlike ethanol, 1-butanol has a similar energy density as gasoline (27 versus 32 MJ/liter), and its hygroscopicity allows for storage and transport with existing infrastructures (7, 8). In addition, advances in synthetic biology and de novo metabolic pathway engineering are enabling new routes to 1-butanol and other potential liquid biofuels, such as isobutanol, 2-methyl-1-butanol, alkanes, and fatty alcohols (2, 4, 5, 9–11). The broader approach involves the creation of microbial cell factories that are used as biorefineries to produce advanced biofuels and value-added chemicals from renewable substrates (2, 12–14). Major obstacles to this approach, however, include product toxicity to the host microbe, ultimately resulting in low yields (3, 13, 15). For example, growth of Escherichia coli is arrested at concentrations as low as 1% (vol/vol) isobutanol (16). In order to become a viable source for biofuels and chemicals, the host toxicity mechanisms of microbial products must be understood so that rational metabolic engineering strategies can be derived to confer adequate product tolerance and ultimately improve yield.
Microbial toxicity mechanisms of alcohols.
Microbes turn on several genetic programs in response to changing environmental conditions (e.g., alcohol stress) in order to maintain homeostasis and optimize the use of resources (17–20). The cell membrane plays a significant role in the ability of the cell to sense these changes as well as in the adaptation to stress by counteracting alcohol toxicity (15, 21). Both short-chain (<C4) and long-chain (>C4) alcohols are known to cause membrane disruption by mechanisms of desiccation (short-chain alcohols) or intercalation (long-chain alcohols) of lipophilic side chains into the membrane lipid bilayer (15, 16, 22). In general, increased membrane fluidity has been observed as a result of 1-butanol exposure for both E. coli and a natural 1-butanol producer, Clostridium acetobutylicum (23–26). This fluidizing effect has been proposed to result from several host response mechanisms, including the following: (i) an altered ratio of saturated versus unsaturated fatty acids in the cell membrane (27), (ii) denatured protein structure and changed cell surface protein composition (26, 28), (iii) increased use of efflux pumps in several Gram-negative bacteria (17), (iv) disrupted protein-lipid interactions (25), (v) upregulated synthesis of other protective metabolites and macromolecules (26, 28), and (vi) decreased central carbon metabolic activity by inhibition of glucose and nutrient transport (16, 21, 26, 28, 29). The general consensus is that long-chain alcohols have the ability to intercalate further into the membrane lipid bilayer and disrupt hydrogen bonding between hydrophobic tails, causing relatively more toxicity than short-chain alcohols (15, 23, 27, 30). However, this proposed mechanism does not always hold true for 1-butanol, for which toxicity appears to be strain dependent (31).
Major factors found to influence alcohol toxicity include (i) solvent hydrophobicity, (ii) the octanol-water partition coefficient (Kow), (iii) length of alkyl chain, and (iv) degree of saturation (15). Organic solvents with log Kow values between 1.5 and 5.0 have been found to be extremely toxic to microbes, as they preferentially partition the cytoplasmic membrane by disorganizing structure and function (e.g., loss of ions, metabolites, lipids, proteins, etc.), which leads invariably to cell lysis and death (27). The effect of alcohol on microbial cell membrane fluidity has also been studied widely (23, 30, 32, 33). It has been observed that E. coli responds to ethanol exposure with an initial alteration of fatty acid composition as a short-term response, allowing for de novo biosynthesis of membrane components as a permanent and long-term response (23, 27, 34, 35). A net increase in total protein from cells exposed to growth-inhibitory levels of ethanol has also been observed (34). Commonly, biological studies of the phenotypic response to alcohol toxicity have been carried out using genetic and biochemical approaches that utilize standardized procedures, such as thin-layer chromatography (TLC) (36) and gas chromatography-mass spectrometry (GC-MS) (36, 37). By nature, these methods require lipid extraction (cell destruction) and extensive sample preparation. While these methods have proven reliable, the time required for analysis is on the order of hours to days. Real-time methods of analysis are needed to further understand the dynamics of the microbial phenotypic responses to alcohol toxicity. With this information, it is likely that metabolic engineering and process control strategies can be developed to confer product tolerance and optimize cell productivity.
Monitoring dynamic phenotypes with Raman spectroscopy.
Raman spectroscopy is a powerful analytical technique that can be applied to a wide variety of solid, liquid, and gas samples, including biological tissues (38–41). The sample is excited by a monochromatic laser, and the resulting spectrum shows the intensity of Raman scattered radiation (arising from chemical bond rotations, stretching, and bending) as a function of wave number (42). The Raman spectrum of a biological sample is usually complex and contains bands (i.e., peaks) that result from the thousands (or more) of molecules comprising the sample, each having its own specific Raman signature. Biological sample analysis by Raman spectroscopy can be performed on dried or liquid samples. Raman scanning of dried samples currently returns more reliable data than scanning of liquid samples and has the significant advantages of (i) sample preparation that is minimal (involves drying microliters of sample on a metal surface) and label-free, (ii) Raman data are returned in near-real time, and (iii) there is minimal spectral interference from water. Liquid-phase scanning of bacterial cultures remains a topic of research and offers the additional advantages of (iv) scanning directly through glass containers (enabling near-real-time in situ analysis of living cultures) and (v) being nondestructive in nature, it enables continued single-cell monitoring. By virtue of their unique Raman signals, biological molecules, such as nucleic acids, proteins, lipids, and carbohydrates, can also be used to generate fingerprints of whole cells (43, 44).
One of the challenges of working with Raman data is the assignment of chemical species to spectral bands. Due to the complex nature of biological samples, most Raman bands result from the overlapping bands of several individual molecules. Researchers have often relied on literature sources for Raman band identification; however, (i) the interpretation of spectral data differs significantly among the various sources, and (ii) band assignment remains ambiguous, as a single Raman band can have several assignments (42, 45, 46). Recently, a Raman database was published that provides the band assignments for several individual biological molecules (42). Though useful, expert user input is still required for accurate band assignments. For example, different band assignments (980, 1,443, 1,447, 1,449, 1,655 to 1,680, 1,656, 2,883, 2,900, 2,915, and 2,940 cm−1) have been reported for both proteins and lipids. Deconvolution of a set of Raman spectra to obtain chemical composition information is not trivial and represents a unique challenge. With advancement in this area, Raman spectroscopy will ultimately be useful as a means of obtaining chemical composition data for biological samples in near-real time without disrupting the system.
Studying the response of E. coli to 1-butanol exposure via Raman spectroscopy.
The practical use of Raman spectroscopy is demonstrated in this research as an analytical method for studying the phenotypic responses of E. coli cells to growth-inhibitory 1-butanol exposure. Traditional methods of analysis, including (i) GC coupled with flame ionization detection (FID) and MS, (ii) ultraperformance liquid chromatography (UPLC), and (iii) fluorescence anisotropy were used to elucidate the 1-butanol toxicity responses of E. coli. By correlating these measurements with specific Raman bands, a methodology was developed that monitored changing phenotypes and cell chemical compositions in near-real time. Also, Raman bands were identified that characterized the fluidizing effect of 1-butanol on the E. coli cell membrane, thus connecting molecular changes to measurable physiological phenotypes. The use of Raman spectroscopy to monitor culture phenotypes is advantageous as it is relatively fast and noninvasive, unlike traditional analysis methods, which are both resource- and time-intensive. The methods described in this research have potentially wide-reaching applications for industrial and clinical microbiology; however, the Raman band assignments reported in this research should be considered valid for E. coli only until larger-scale studies have been completed.
MATERIALS AND METHODS
Bacteria strain.
E. coli DH5α cells were obtained from Invitrogen Life Technologies (Grand Island, NY) and used in all experiments. Frozen cells were stored as glycerol stocks at −80°C and were thawed and plated onto solid agar plates to select a single colony for experiments in liquid culture medium.
Chemicals and reagents.
All chemicals and reagents (1-butanol, methanol, chloroform, ethanol, 1 N methanolic-HCl, borate buffer, yeast extract, tryptone, sodium chloride, and cis-9,cis-12-octadecadienoic acid [C18:2]) were purchased from Sigma-Aldrich (St. Louis, MO) or included as part of a kit. cis-9,cis-12-octadecadienoic acid (C18:2) was used as an internal standard to account for losses during GC analysis of fatty acids as fatty acid methyl esters (FAMEs). It was chosen because E. coli does not produce it natively. All solvents used for extractions were LC or GC grade.
Culture media, growth conditions, and harvesting.
Overnight cultures were prepared by inoculating 15 ml liquid Luria-Bertani (LB) growth medium with E. coli cells from a solid agar plate. Cultures were placed in an incubator with a rotary shaker set to 210 rpm and 37°C. For solid LB medium, agar was used at 15 g/liter. Cell growth in liquid medium was monitored based on the optical density measured at 600 nm (OD600). An aliquot sample was then used to prepare a subculture by diluting 10 ml of cell culture with 1 liter of fresh LB medium in a sterilized culture flask. The culture was then grown to the start of the exponential growth phase (OD600 of 0.4 to 0.5). At this point, the culture was split into two equal portions (∼500 ml each), with one serving as the negative control. 1-Butanol was added to the experimental culture to a concentration of 1.2% (vol/vol), and both cultures were incubated at 210 rpm and 37°C for 1 h prior to sampling. Sample volumes of 50 ml (for FAME analysis), 5 ml (for membrane leakage analysis), 1 ml (for cell viability analysis), and 1 ml (for analysis by Raman spectroscopy) were taken every half hour from both cultures until a constant OD600 reading was observed. Cells were harvested following centrifugation at 10,000 rpm at 4°C for 5 min. Cells were washed with ice-cold purified water (except cells to be used for membrane leakage analysis), and this procedure was repeated. Cells for Raman analysis were resuspended in 1 ml of type I purified water for analysis. Cells for FAME and metabolite analyses were frozen in liquid nitrogen and lyophilized.
Raman spectroscopy.
To prepare samples for Raman analysis, 2-μl aliquots of washed cells were dried on an aluminum surface at room temperature. Dried cells were analyzed using a Bruker Senterra dispersive Raman spectrometer equipped with a confocal microscope and objective lens with ×100 magnification (Bruker Optics, Billerica, MA). Measurements were carried out using laser excitation of 532 nm (20 mW) for 25 s with spectral resolution of 9 to 15 cm−1. A similar method has been described elsewhere (47). A minimum of 50 individual spectra was acquired per sample prior to data analysis. Software programs enabling data collection as well as data normalization and processing are discussed in the next section.
Raman data processing.
Raman data were processed and analyzed using two different software platforms: (i) OPUS (Bruker Optics, Billerica, MA) and (ii) MATLAB (R2012A; MathWorks, Natick, MA) with a custom Raman data analysis (RDA) toolbox (A. I. M. Athamneh and R. S. Senger, submitted for publication) containing functions for comparative peak analysis and multivariate statistics. The OPUS platform allowed for interaction with the Raman instrument and provided initial baseline correction of spectra. Raman spectra of biological samples are commonly corrupted by the influence of (i) background fluorescence, (ii) charge-coupled-device background noise, (iii) Gaussian noise, and (iv) cosmic spikes (40). During acquisition, spectra with cosmic spikes were identified through manual inspection and discarded. The following spectra analysis was performed in MATLAB. The RDA toolbox provided a more convenient graphical user interface, but it was not essential for this analysis. For consistency, data from all spectra (i.e., intensities at all wave numbers) were normalized by using vector normalization over the entire spectral range (300 cm−1 to 3,600 cm−1). In vector normalization, the signal intensity at each wave number (i.e., band) was divided by the norm of the spectrum intensities. This allowed intensities from several spectra to be compared directly. Thus, only the vector-normalized intensities of specified bands were used for comparisons throughout.
Fatty acid analysis by GC-MS/FID.
Lipid extractions were performed on 1 mg of lyophilized cells by using biphasic chloroform-methanol-water extractions and 10 μg of C18:2 as an internal standard, according to a standard protocol (48). Fatty acid methyl esters were prepared by transesterification in the presence of methanolic HCl and analyzed on an Agilent 7890A series GC equipped with an FID (Agilent Technologies, Santa Clara, CA). The FAME separation was achieved on an Agilent 30-m DB-23 column (0.25 mm by 0.25 μm), and the identities of the individual fatty acids were confirmed by analyzing spectral information of FAME in selected samples on the same GC instrument coupled to an Agilent 5975C series MS apparatus (Agilent Technologies) (48).
Total protein content, protein hydrolysis, and amino acid analysis by UPLC.
The total protein contents of 1-butanol-exposed and control cultures were determined by the Coomassie Plus (Bradford) assay (Thermo Fisher Scientific, Rockford, IL) according to the manufacturer's protocol. Proteins and free metabolites are contained in the aqueous phase and the insoluble pellet remaining after the removal and reextraction of lipids used in FAME analysis. It was found that free amino acids are present at very low levels in E. coli cells and do not interfere with the analysis of protein-derived amino acids, so there was no need to separate proteins by acidic precipitation from the rest of the polar metabolites present in the aqueous phase. Because some proteins are not soluble in water and are present in the insoluble pellet, total protein hydrolysis was performed in the same tube as the extraction. The solvents (water and methanol) were dried, and proteins were hydrolyzed under vacuum at 110°C for at least 16 h in a custom-made Teflon hydrolysis chamber containing 4 ml of 6 N HCl. The resulting protein-derived amino acids were dissolved in 1 ml of water, and 1 μl of sample was derivatized in a 50-μl total volume by using the AccQ•Tag Ultra amino acid kit according to the manufacturer's recommendations (Waters Corporation, Milford, MA). The derivatized amino acid samples (0.5 μl) were injected on an H-class Acquity UPLC and detected by fluorescence as described for analysis of protein hydrolysates (Waters Corporation, Milford, MA).
Membrane fluidity.
Changes in membrane fluidity induced by 1-butanol were monitored by analyzing fluorescence anisotropy from E. coli cells labeled with the membrane probe 1,3-diphenyl-1,3,5-hexatriene (DPH), as described previously (49, 50). Stock 8 mM DPH solution in tetrahydrofuran was diluted 1,000-fold in vigorously stirred 50 mM NaCl (50). The dispersion was mixed 1:1 with E. coli cells, washed three times with chilled phosphate-buffered saline (pH 7.4), and resuspended to an OD600 of 4.0. Fluorescence anisotropy was measured at room temperature by using a SpectraMax M5 plate reader (Molecular Devices, Sunnyvale, CA) with excitation and emission wavelengths of 360 and 428 nm, respectively. Background fluorescence from DPH-free E. coli cells was measured and subtracted from the DPH-labeled samples. The fluorescence anisotropy (r) of a fluorescent probe is inversely proportional to cell membrane fluidity, as defined in the following equation: r = (Ivv − Ivh)/(Ivv + 2Ivh). Here, Ivv and Ivh represent fluorescence intensities measured through a polarizer oriented parallel and perpendicular, respectively, to the plane of polarization of the excitation beam (50). Fluorescence anisotropy results for 1-butanol-treated cultures are presented as the percent change from control (i.e., untreated) cultures (49).
RESULTS
Raman spectroscopy of growing and growth-inhibited cells.
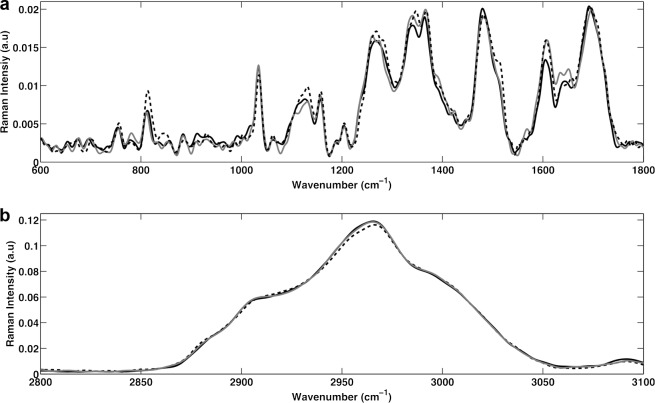
The baseline-corrected and vector-normalized Raman spectra for E. coli cells at the start of 1.2% (vol/vol) 1-butanol treatment (time zero) and at the end of the experiment (time = 180 min) for both the 1-butanol-treated and control (untreated) cells are shown in Fig. 1. The three spectra were superimposed to show potential differences in the biologically relevant spectral region (600 to 1,800 cm−1) (Fig. 1a) and the CH region (2,800 to 3,100 cm−1) (Fig. 1b) (40). No significant changes were noted in the CH region (Fig. 1b). However, noticeable signal intensities were observed (without the use of statistical analyses) in Fig. 1a for bands assigned to (i) phosphodiester bonds in DNA (∼788 cm−1) and in RNA (∼813 cm−1) (indicative of nucleic acids) (42, 46); (ii) symmetric PO2− stretching of DNA (∼1,070 to 1,090 cm−1) (indicative of nucleic acids); (iii) C—C chain stretch (∼1,060 to 1,075 cm−1) (indicative of fatty acids) (45, 46); (iv) amide III bands, =CH bend, and nucleic acid bases (1,220 to 1,284 cm−1) (indicative of proteins, lipids, and nucleic acids) (42, 51); (v) C—H deformation and guanine (∼1,320 cm−1) (indicative of lipids and nucleic acids) (42, 45, 46); (vi) C—H vibrations (∼1,449 cm−1) (indicative of proteins, lipids, and nucleic acids) (42, 46); and (vii) C=C bands (∼1,607 cm−1) (indicative of aromatics and unsaturated lipids) (42, 46). Significant changes in Raman spectra were observed between the 1-butanol-treated and control cells with both time and treatment. Thus, the Raman spectroscopy results suggested that cellular chemical composition and physiology changed significantly in response to both factors. The standard deviation of the averaged spectra (not shown in Fig. 1) was found to be very close to zero, confirming high reproducibility of scans.
FIG 1.
Raman spectra of the biological region (600 to 1,800 cm−1) (a) and the CH region (2,800 to 3,100 cm−1) (b) at time zero (before the application of 1-butanol; solid black line), time equal to 180 min for the control cells (gray line), and time equal to 180 min after treatment for the 1-butanol-treated cells (dashed black line).
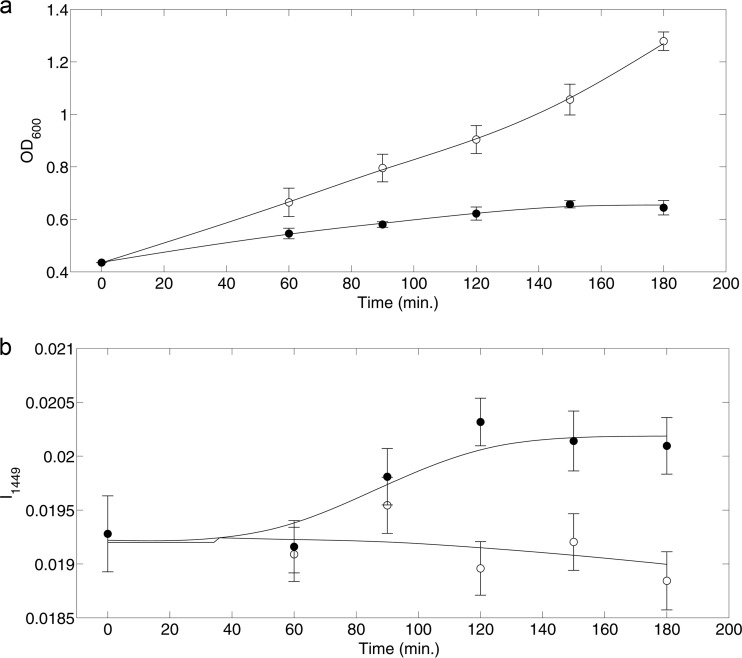
The time course OD600 measurements for both 1-butanol-treated and control cultures are shown in Fig. 2a. Upon exposure to 1.2% (vol/vol) 1-butanol, E. coli cells showed arrested cell growth over time, consistent with expected toxicity responses. The Raman signal intensity at 1,449 cm−1 (I1449) over the time course is shown in Fig. 2b. The Raman band at 1,449 cm−1 corresponds to C—H vibrations (42), which are abundant in all biomass components (e.g., lipids, proteins, nucleic acids, carbohydrates, etc.) of bacterial cells. The Raman signal intensity at 1,449 cm−1 can provide insight into the overall metabolic activity of the cells. As seen in Fig. 2b, the Raman signal intensity at 1,449 cm−1 increased with 1-butanol exposure. This suggests the cells may have upregulated several metabolic programs (i.e., a toxicity response) with 1-butanol treatment. Because Raman spectra were normalized, an increase in the I1449 suggests that the number of molecules exhibiting C—H vibration signals (e.g., lipids, proteins, etc.) was increased per cell. This is also consistent with the increased nucleotide abundances (per cell) noted at 788 cm−1 and 813 cm−1. Thus, with 1-butanol exposure, it is likely that metabolic activity shifts from growth-related functions to toxicity response programs for survival. The following analyses are aimed at identifying the changes in cell composition and phenotype brought on by these metabolic programs.
FIG 2.
Culture growth (OD600) (a) and Raman (I1449) measure of broader metabolic activity (b) as functions of time for 1-butanol-treated cells (black circles) and control cells (open circles). 1-Butanol (1.2% [vol/vol]) was added to the treated cells at 0 min. Error bars represent 1 standard deviation for data from at least 3 biological replicates.
The Raman signal intensities for both 1-butanol-treated and control cells were investigated for potential candidate bands that could explain the observed physiological changes. The current methodology for determining chemical composition of cells includes several off-line analytical measurements. These often involve long sample preparation times, and analyses cannot be performed in real time. In order to establish a new near-real-time analysis protocol using Raman spectroscopy, it was necessary to correlate Raman results with the existing well-established methodologies. In this approach, correlations were determined by calculating the correlation coefficient (R) between time course Raman signal intensities and off-line experimental measurements. Often, multiple Raman band assignments exist in the literature for a single phenotypic trait (e.g., unsaturated fatty acid content). The goal of this research was to determine the Raman band assignment(s) that best correlates with experimental measurements.
Correlation of Raman spectroscopy and GC-FID results for fatty acids analysis.
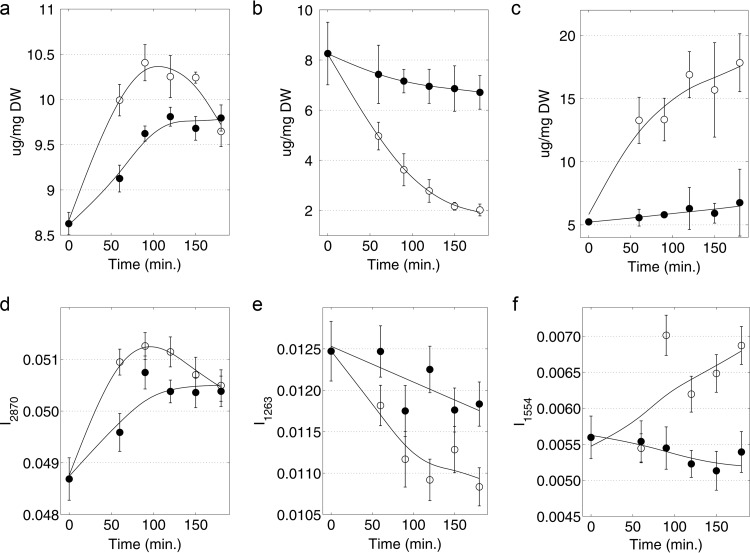
Time course Raman signal intensities (at assigned bands taken from the literature [42, 45, 46, 52–54]) were correlated with the corresponding changes in membrane-derived fatty acid levels and composition, as determined by GC-FID. Correlation coefficients (R values) between these two methods are presented in Table S1 in the supplemental material. Several, but not all, of the previously assigned Raman bands from the literature showed correlation with the actual changes in the composition of different fatty acid types. While GC-FID provides quantification of absolute levels of the individual fatty acids, Raman enabled the identification of three different classes of fatty acids that have great relevance to changes in membrane physical properties: (i) saturated, (ii) unsaturated, and (iii) cyclopropane fatty acids. From the results (see Table S1), Raman bands for saturated fatty acids (I2870; i.e., the Raman signal intensity at 2,870 cm−1), unsaturated fatty acids (I1263), and cyclopropane fatty acids (I1554) showed high correlations with results obtained by GC-FID. The correlation coefficients between Raman spectroscopy and GC-FID data were 0.78, 0.76, and 0.79, respectively. The time course (over 180 min) Raman and FAME analyses data were plotted for both 1-butanol-treated and control cells, and results are shown in Fig. 3. In general, the trends were conserved in all three cases; however, it was obvious that GC-FID analyses produced data with less experimental error and better-defined trends over the time course. Relative to the control, the 1-butanol-treated cells (i) produced fewer saturated fatty acids (Fig. 3a), (ii) conserved the presence of unsaturated fatty acids (Fig. 3b), and (iii) largely resisted the formation of cyclopropane fatty acids over the time course (Fig. 3c). On the other hand, the untreated control cells reduced the amount of unsaturated fatty acids by about 75% over the time course, in favor of saturated fatty acids (∼20% increase) and cyclopropane fatty acids (∼250% increase). In summary, the dynamics of fatty acid composition were resolved well by GC-FID and were approximated with correlation coefficient levels greater than 0.75 by using Raman spectroscopy. While some accuracy is sacrificed with Raman measurements, results can be obtained in near-real time in a nondisruptive manner.
FIG 3.
(a) Saturated fatty acids measured by GC-FID; (b) unsaturated fatty acids measured by GC-FID, (c) cyclopropane fatty acids measured by GC-FID, (d) saturated fatty acids measured by Raman spectroscopy (I2870), (e) unsaturated fatty acids measured by Raman spectroscopy (I1263), and (f) cyclopropane fatty acids measured by Raman spectroscopy (I1554) as functions of time for 1-butanol-treated cells (black circles) and control cells (open circles). 1-Butanol (1.2% [vol/vol]) was added to the treated cells at 0 min. Error bars represent 1 standard deviation for data from at least 3 biological replicates.
Correlation of Raman spectroscopy and UPLC results for amino acid analysis.
Total amino acid content and composition did not change in the control and 1-butanol-treated cultures over the time course when measured by UPLC. The steady-state levels of total amino acids are shown in Table 1. However, different amounts of each amino acid were observed, and these relative abundances remained constant in all samples. The same was observed when total protein was measured (Fig. 4). Raman band intensities (using previously published bands for individual amino acids) were correlated with experimental UPLC measurements. In the literature (55, 56), between 9 and 29 distinct Raman bands (with medium to strong intensities) were cited for each amino acid (given in Table S2 in the supplemental material). A unique set of Raman bands (one band for each amino acid) was identified computationally to fully characterize the amino acid composition of E. coli to be consistent with UPLC experimental results and published literature values (Table 1). Given the large number of possible Raman bands per amino acid, 1.4 × 1021 possible band combinations exist. A set leading to good correlation with UPLC-derived data (R = 0.93) was identified after examining 5 × 109 possibilities in a stochastic simulation. Table 1 presents (i) the optimum set of Raman bands, (ii) the amino acid composition determined by Raman, (iii) the amino acid composition determined by UPLC, and (iv) published amino acid compositions (55). Good correlation was observed between Raman spectroscopy, UPLC, and values in the published data, suggesting that Raman spectroscopy may be used as a near-real-time analytical method for determining total amino acid compositions of E. coli cells. Among the Raman band assignments listed in Table 1, the assignments for Ala (1,308 cm−1) and Ile (1,309 cm−1) overlap and result in the same amino acid composition values when determined by Raman spectroscopy. In addition, Trp (759 cm−1) and Met (765 cm−1) are also close to having overlapping bands. Thus, additional sets of amino acid band assignment solutions have been included in Tables S3 to S5 in the supplemental material. Each of these band assignment solutions represents a different alternative that can be employed if specific amino acids are of critical interest. However, the band assignment set in Table 1 returned the overall optimum solution.
TABLE 1.
Amino acid compositions determined by Raman spectroscopy and UPLC and as reported in the literature
| Amino acid | Optimum Raman band (cm−1) | Amino acid composition (%) based on: |
||
|---|---|---|---|---|
| Raman spectroscopy | UPLC | Published valueb | ||
| Ala | 1,308 | 11.1 | 11.1 | 9.60 |
| Arg | 1,199 | 2.46 | 3.39 | 5.53 |
| Asp/Asn | 1,695 | 6.41 | 8.98 | 9.01 |
| Cys | 678 | 1.24 | 1.64a | 1.64 |
| Glu/Gln | 1,319 | 10.8 | 9.16 | 9.84 |
| Gly | 1,332 | 11.3 | 12.0 | 11.5 |
| His | 731 | 2.16 | 0.890 | 1.77 |
| Ile | 1,309 | 11.1 | 8.76 | 5.43 |
| Leu | 1,243 | 9.67 | 11.4 | 8.42 |
| Lys | 1,072 | 3.44 | 5.24 | 6.41 |
| Met | 765 | 1.14 | 0.467 | 2.87 |
| Phe | 1,214 | 5.78 | 4.59 | 3.46 |
| Pro | 843 | 0.727 | 1.02 | 4.13 |
| Ser | 1,010 | 4.37 | 4.42 | 4.03 |
| Thr | 1,116 | 3.38 | 6.26 | 4.74 |
| Trp | 759 | 1.41 | 1.06a | 1.06 |
| Tyr | 798 | 1.70 | 0.390 | 2.58 |
| Val | 1,454 | 11.7 | 9.22 | 7.91 |
| Sum | 100 | 100 | 100 | |
Values for Cys and Trp could not be resolved by the UPLC method. Cys is converted to several oxidation products, while Trp is destroyed during the acidic protein hydrolysis method used in this research. The published literature values were used for Cys and Trp.
Amino acid composition is defined as the fraction of each amino acid among E. coli total protein. The published values were obtained from reference 55.
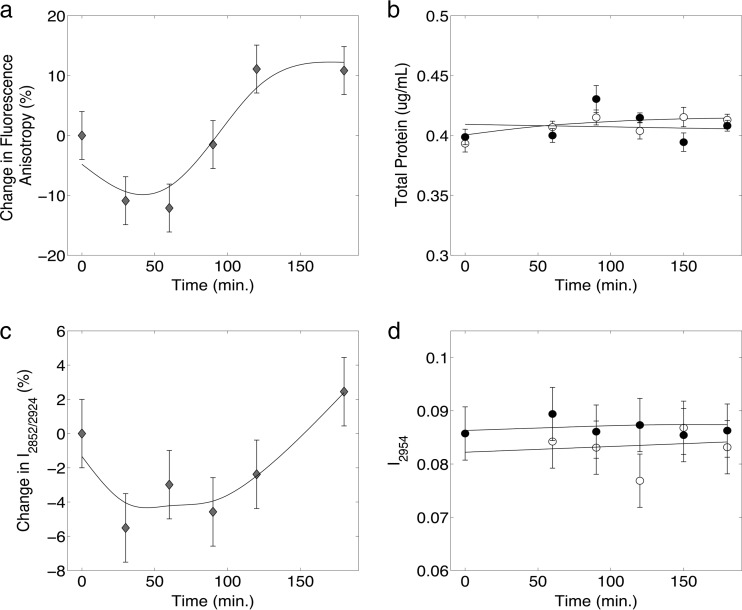
FIG 4.
(a) Change in fluorescence anisotropy (as a percentage) for 1-butanol-treated cells relative to the control cells, (b) experimentally measured total protein content, (c) membrane fluidity (I2852/I2924) measured by Raman spectroscopy, and (d) total protein content measured by Raman spectroscopy (I2954) as functions of time for 1-butanol-treated cells (black circles) and control cells (open circles) (b and d); diamonds indicate the percent changes between 1-butanol-treated and control cells (a and c). 1-Butanol (1.2% [vol/vol]) was added to the treated cells at 0 min. Error bars represent 1 standard deviation for data from at least 3 biological replicates.
Correlation of Raman spectroscopy and fluorescence anisotropy results for cell membrane fluidity analysis.
Membrane fluidity has long been used to assess the effects of solvents on microbial cell membranes (15, 21, 23, 27), and membrane fluidity has been observed to increase in E. coli cells upon 1-butanol exposure (23). Previous research has also closely tied cell membrane fluidity with changes in fluorescence anisotropy (21, 23, 54). Experimental fluorescence anisotropy results obtained in this research are in agreement with this observation. However, an initial decrease in fluorescence anisotropy was observed before a short “lag” phase followed by an increased long-term response. These results are shown in Fig. 4a. The molecular mechanisms behind this initial decrease and lag are unknown and remain a topic for further investigation. To assess membrane fluidity using Raman spectroscopy, three signal intensity ratios (I2870/I2954, I2850/I2880, and I2852/I2924) were analyzed based on previously published results (53, 54, 57, 58). The identification of Raman peaks corresponding to symmetric and asymmetric stretching of cell membranes was used to derive these ratios. In all three cases examined, the signature decrease, lag, and the eventual increase in signal were observed. In addition, the I2852/I2924 Raman signal intensity ratio was found to best correlate with fluorescence anisotropy results. The percent changes in fluorescence anisotropy between 1-butanol-treated and control cells over the time course are shown in Fig. 4a, and the percent change for the I2852/I2924 ratio between 1-butanol-treated and control cells is shown in Fig. 4c. While a perfect correlation was not observed, a strong resemblance in general trend was observed between changes in Raman peak intensities obtained in near-real-time and off-line fluorescence anisotropy measurements. To determine if total cell protein was involved in the observed changes in membrane fluidity, the total cell protein content measured; no change was detected (Fig. 4b). These results corresponded with the Raman I2954 value cited in the literature (54).
DISCUSSION
Use of Raman spectroscopy to study cell physiology in near real time.
In this research, the applicability of Raman spectroscopy for studying E. coli cell physiology and chemical composition in near real time was demonstrated. It has been documented previously that upon exposure to solvents, bacteria initiate response mechanisms to revert induced physiological changes (17, 26). In bacteria, the physiological responses upon alcohol exposure are often measured by (i) quantifying the fluidizing effects on the cell membrane and (ii) measuring the degree of saturation of membrane lipids. 1-Butanol exposure has been found to result in increased membrane fluidity of E. coli (23, 26). This observed increase has been shown to be a result of disruptions in fatty acid and protein structure of the cell membrane, which also affects protein-lipid interactions (26, 59). In the analysis presented here, exposure of E. coli cells to 1-butanol resulted in an increased level of saturated fatty acids, while maintaining a relatively constant level of unsaturated fatty acids. Low levels of cyclopropane fatty acids were also observed with 1-butanol exposure. These trends were first measured by an established GC-FID methodology, and Raman bands were identified that showed good correlation with these results. In addition to fatty acid content, Raman bands were justified that showed good correlation with (i) cellular amino acid composition, (ii) fluorescence anisotropy (a measure of membrane fluidity), and (iii) total protein content of the cell. While “perfect” correlations were not identified in this research, several strong correlations were found, indicating that Raman spectroscopy can be successfully and reliably used as a diagnostic tool that has the distinct advantage of offering near-real-time analyses. However, superior accuracies were obtained from elaborate standard methods of analysis.
Peak assignment observations and discrepancies.
The main objective of this research was to further develop Raman spectroscopy as a near-real-time measurement tool for a quick, reliable diagnosis of changes in metabolic and physiological phenotypes. Currently, researchers commonly rely on published Raman band assignments. However, band selection can be a daunting task, as assignments are not always consistent within the published literature. Several published studies have compiled useful databases of band assignments for biological samples; however, questions regarding band selection remain, since assignments of the same molecule/group are often made to several Raman bands. In addition, current reported assignments do not differentiate between cyclopropane fatty acids and saturated fatty acids. To provide additional experimental evidence to select among the several Raman band assignments related to fatty acids, correlations were found among saturated, unsaturated, and cyclopropane fatty acids. Raman spectroscopy was found to be able to capture the induced variances due to 1-butanol treatment and bacterial cell development over time. Currently, the Raman band assignments identified in this research are limited to E. coli, and similar studies can be performed to establish useful sets of bands for other organisms. Ultimately, the goal of establishing a set of universally applied Raman bands will require a large-scale study involving several different organisms and treatments.
Potential applications.
The applications of real-time analysis of biological samples with Raman spectroscopy are numerous, and the method is capable of delivering near-real-time phenotyping. The results of this research demonstrate the power of Raman for phenotypic profiling of E. coli cells. As with the application of 1-butanol exposure in this research, Raman spectroscopy can be used to monitor cell culture responses to metabolically engineered products and be used to signal culture toxicity or even identify optimally productive cell states. In addition, real-time phenotypic monitoring has tremendous application to the field of biosensing, as cell composition changes may be observed in response to small quantities of environmental toxins as well as chemical or biological warfare agents. Future research will determine whether phenotypic profiling can rival the sensitivity of other analytical methods.
Supplementary Material
ACKNOWLEDGMENTS
Fluorescence anisotropy data were obtained with the help of Pablo Sobrado and Tijana Grove at Virginia Tech.
We acknowledge financial support from the Institute of Critical Technologies and Applied Science (ICTAS) at Virginia Tech.
Footnotes
Published ahead of print 25 August 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.01590-14.
REFERENCES
- 1.Lennen RM, Braden DJ, West RA, Dumesic JA, Pfleger BF. 2010. A process for microbial hydrocarbon synthesis: overproduction of fatty acids in Escherichia coli and catalytic conversion to alkanes. Biotechnol. Bioeng. 106:193–202. 10.1002/bit.22660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Atsumi S, Cann AF, Connor MR, Shen CR, Smith KM, Brynildsen MP, Chou KJ, Hanai T, Liao JC. 2008. Metabolic engineering of Escherichia coli for 1-butanol production. Metab. Eng. 10:305–311. 10.1016/j.ymben.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 3.Atsumi S, Hanai T, Liao JC. 2008. Non-fermentative pathways for synthesis of branched-chain higher alcohols as biofuels. Nature 451:86–89. 10.1038/nature06450. [DOI] [PubMed] [Google Scholar]
- 4.Peralta-Yahya PP, Keasling JD. 2010. Advanced biofuel production in microbes. Biotechnol. J. 5:147–162. 10.1002/biot.200900220. [DOI] [PubMed] [Google Scholar]
- 5.Colin VL, Rodriguez A, Cristobal HA. 2011. The role of synthetic biology in the design of microbial cell factories for biofuel production. J. Biomed. Biotechnol. 2011:601834–601843. 10.1155/2011/601834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sissine F. 2008. Energy Independence and Security Act of 2007: a summary of major provisions. CRS Report for Congress, RL34294. National Agricultural Law Center, Fayetteville, AR: http://nationalaglawcenter.org/wp-content/uploads/assets/crs/RL34294.pdf. [Google Scholar]
- 7.Jin C, Yao M, Liu H, Lee C-F, Ji J. 2011. Progress in the production and application of n-butanol as a biofuel. Renew. Sust. Energ. Rev. 15:4080–4106. 10.1016/j.rser.2011.06.001. [DOI] [Google Scholar]
- 8.Rakopoulos D, Rakopoulos C, Hountalas D, Kakaras E, Giakoumis E, Papagiannakis R. 2010. Investigation of the performance and emissions of bus engine operating on butanol/diesel fuel blends. Fuel 89:2781–2790. 10.1016/j.fuel.2010.03.047. [DOI] [Google Scholar]
- 9.Connor MR, Atsumi S. 2010. Synthetic biology guides biofuel production. J. Biomed. Biotechnol. 2010:pii=541698. 10.1155/2010/541698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dellomonaco C, Fava F, Gonzalez R. 2010. The path to next generation biofuels: successes and challenges in the era of synthetic biology. Microb. Cell Fact. 9:3. 10.1186/1475-2859-9-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee SK, Chou H, Ham TS, Lee TS, Keasling JD. 2008. Metabolic engineering of microorganisms for biofuels production: from bugs to synthetic biology to fuels. Curr. Opin. Biotechnol. 19:556–563. 10.1016/j.copbio.2008.10.014. [DOI] [PubMed] [Google Scholar]
- 12.Moon TS, Yoon SH, Lanza AM, Roy-Mayhew JD, Prather KL. 2009. Production of glucaric acid from a synthetic pathway in recombinant Escherichia coli. Appl. Environ. Microbiol. 75:589–595. 10.1128/AEM.00973-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dunlop MJ, Dossani ZY, Szmidt HL, Chu HC, Lee TS, Keasling JD, Hadi MZ, Mukhopadhyay A. 2011. Engineering microbial biofuel tolerance and export using efflux pumps. Mol. Syst. Biol. 7:487. 10.1038/msb.2011.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leonard E, Nielsen D, Solomon K, Prather KJ. 2008. Engineering microbes with synthetic biology frameworks. Trends Biotechnol. 26:674–681. 10.1016/j.tibtech.2008.08.003. [DOI] [PubMed] [Google Scholar]
- 15.Rutherford BJ, Dahl RH, Price RE, Szmidt HL, Benke PI, Mukhopadhyay A, Keasling JD. 2010. Functional genomic study of exogenous n-butanol stress in Escherichia coli. Appl. Environ. Microbiol. 76:1935–1945. 10.1128/AEM.02323-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brynildsen MP, Liao JC. 2009. An integrated network approach identifies the isobutanol response network of Escherichia coli. Mol. Syst. Biol. 5:277. 10.1038/msb.2009.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dunlop MJ, Keasling JD, Mukhopadhyay A. 2010. A model for improving microbial biofuel production using a synthetic feedback loop. Syst. Synth. Biol. 4:95–104. 10.1007/s11693-010-9052-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Freddolino PL, Tavazoie S. 2012. Beyond homeostasis: a predictive-dynamic framework for understanding cellular behavior. Annu. Rev. Cell Dev. Biol. 28:363–384. 10.1146/annurev-cellbio-092910-154129. [DOI] [PubMed] [Google Scholar]
- 19.Nicolaou SA, Gaida SM, Papoutsakis ET. 2010. A comparative view of metabolite and substrate stress and tolerance in microbial bioprocessing: from biofuels and chemicals, to biocatalysis and bioremediation. Metab. Eng. 12:307–331. 10.1016/j.ymben.2010.03.004. [DOI] [PubMed] [Google Scholar]
- 20.Sinensky M. 1974. Homeoviscous adaptation: a homeostatic process that regulates the viscosity of membrane lipids in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 71:522–525. 10.1073/pnas.71.2.522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ingram LO. 1976. Adaptation of membrane lipids to alcohols. J. Bacteriol. 125:670–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Papoutsakis ET. 2008. Engineering solventogenic clostridia. Curr. Opin. Biotechnol. 19:420–429. 10.1016/j.copbio.2008.08.003. [DOI] [PubMed] [Google Scholar]
- 23.Huffer S, Clark ME, Ning JC, Blanch HW, Clark DS. 2011. Role of alcohols in growth, lipid composition, and membrane fluidity of yeasts, bacteria, and archaea. Appl. Environ. Microbiol. 77:6400–6408. 10.1128/AEM.00694-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baer SH, Blaschek HP, Smith TL. 1987. Effect of butanol challenge and temperature on lipid composition and membrane fluidity of butanol-tolerant Clostridium acetobutylicum. Appl. Environ. Microbiol. 53:2854–2861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Borden JR, Papoutsakis ET. 2007. Dynamics of genomic-library enrichment and identification of solvent tolerance genes for Clostridium acetobutylicum. Appl. Environ. Microbiol. 73:3061–3068. 10.1128/AEM.02296-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reyes LH, Almario MP, Kao KC. 2011. Genomic library screens for genes involved in n-butanol tolerance in Escherichia coli. PLoS One 6:e17678. 10.1371/journal.pone.0017678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ramos JL, Duque E, Gallegos MT, Godoy P, Ramos-Gonzalez MI, Rojas A, Teran W, Segura A. 2002. Mechanisms of solvent tolerance in gram-negative bacteria. Annu. Rev. Microbiol. 56:743–768. 10.1146/annurev.micro.56.012302.161038. [DOI] [PubMed] [Google Scholar]
- 28.Minty JJ, Lesnefsky AA, Lin F, Chen Y, Zaroff TA, Veloso AB, Xie B, McConnell CA, Ward RJ, Schwartz DR, Rouillard JM, Gao Y, Gulari E, Lin XN. 2011. Evolution combined with genomic study elucidates genetic bases of isobutanol tolerance in Escherichia coli. Microb. Cell Fact. 10:18. 10.1186/1475-2859-10-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Heipieper HJ, Neumann G, Cornelissen S, Meinhardt F. 2007. Solvent-tolerant bacteria for biotransformations in two-phase fermentation systems. Appl. Microbiol. Biotechnol. 74:961–973. 10.1007/s00253-006-0833-4. [DOI] [PubMed] [Google Scholar]
- 30.Dombek KM, Ingram LO. 1984. Effects of ethanol on the Escherichia coli plasma membrane. J. Bacteriol. 157:233–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ingram LO, Buttke TM. 1984. Effects of alcohols on micro-organisms. Adv. Microb. Physiol. 25:253–300. [DOI] [PubMed] [Google Scholar]
- 32.Ingram LO, Eaton LC, Erdos GW, Tedder TF, Vreeland NL. 1982. Unsaturated fatty acid requirement in Escherichia coli: mechanism of palmitate-induced inhibition of growth of strain WN1. J. Membr. Biol. 65:31–40. 10.1007/BF01870466. [DOI] [PubMed] [Google Scholar]
- 33.Ingram LO. 1982. Regulation of fatty acid composition in Escherichia coli: a proposed common mechanism for changes induced by ethanol, chaotropic agents, and a reduction of growth temperature. J. Bacteriol. 149:166–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chiou RY, Phillips RD, Zhao P, Doyle MP, Beuchat LR. 2004. Ethanol-mediated variations in cellular fatty acid composition and protein profiles of two genotypically different strains of Escherichia coli O157:H7. Appl. Environ. Microbiol. 70:2204–2210. 10.1128/AEM.70.4.2204-2210.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Segura A, Duque E, Mosqueda G, Ramos JL, Junker F. 1999. Multiple responses of gram-negative bacteria to organic solvents. Environ. Microbiol. 1:191–198. 10.1046/j.1462-2920.1999.00033.x. [DOI] [PubMed] [Google Scholar]
- 36.Kalscheuer R, Stolting T, Steinbuchel A. 2006. Microdiesel: Escherichia coli engineered for fuel production. Microbiology 152:2529–2536. 10.1099/mic.0.29028-0. [DOI] [PubMed] [Google Scholar]
- 37.Lu X, Vora H, Khosla C. 2008. Overproduction of free fatty acids in E. coli: implications for biodiesel production. Metab. Eng. 10:333–339. 10.1016/j.ymben.2008.08.006. [DOI] [PubMed] [Google Scholar]
- 38.Crow P, Stone N, Kendall CA, Uff JS, Farmer JA, Barr H, Wright MP. 2003. The use of Raman spectroscopy to identify and grade prostatic adenocarcinoma in vitro. Br. J. Cancer 89:106–108. 10.1038/sj.bjc.6601059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mahadevan-Jansen A, Mitchell MF, Ramanujam N, Malpica A, Thomsen S, Utzinger U, Richards-Kortum R. 1998. Near-infrared Raman spectroscopy for in vitro detection of cervical precancers. Photochem. Photobiol. 68:123–132. 10.1111/j.1751-1097.1998.tb03262.x. [DOI] [PubMed] [Google Scholar]
- 40.Bocklitz T, Walter A, Hartmann K, Rosch P, Popp J. 2011. How to pre-process Raman spectra for reliable and stable models? Anal. Chim Acta 704:47–56. 10.1016/j.aca.2011.06.043. [DOI] [PubMed] [Google Scholar]
- 41.Pilaniya K, Chandrawanshi HK, Pilaniya U, Manchandani P, Jain P, Singh N. 2010. Recent trends in the impurity profile of pharmaceuticals. J. Adv. Pharm. Technol. Res. 1:302–310. 10.4103/0110-5558.72422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Movasaghi Z, Rehman S, Rehman IU. 2007. Raman spectroscopy of biological tissues. Appl. Spectrosc. Rev. 42:493–541. 10.1080/05704920701551530. [DOI] [Google Scholar]
- 43.Dutta RK, Sharma PK, Pandey AC. 2009. Surface enhanced Raman spectra of Escherichia coli cell using ZnO nanoparticles. Dig. J. Nanomater. Biostruct. 4:83–87. [Google Scholar]
- 44.She C, Dinh N, Tu AT. 1974. Laser raman scattering of glucosamine N-acetylglucosamine, and glucuronic acid. Biochim. Biophys. Acta 372:345–357. 10.1016/0304-4165(74)90196-2. [DOI] [Google Scholar]
- 45.Wu H, Volponi JV, Oliver AE, Parikh AN, Simmons BA, Singh S. 2011. In vivo lipidomics using single-cell Raman spectroscopy. Proc. Natl. Acad. Sci. U. S. A. 108:3809–3814. 10.1073/pnas.1009043108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Notingher I. 2007. Raman spectroscopy cell-based biosensors. Sensors 7:1343–1358. 10.3390/s7081343. [DOI] [Google Scholar]
- 47.Athamneh AI, Alajlouni RA, Wallace RS, Seleem MN, Senger RS. 2014. Phenotypic profiling of antibiotic response signatures in Escherichia coli using Raman spectroscopy. Antimicrob. Agents Chemother. 58:1302–1314. 10.1128/AAC.02098-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Collakova E, Aghamirzaie D, Fang Y, Klumas C, Tabataba F, Kakumanu A, Myers E, Heath LS, Grene R. 2013. Metabolic and transcriptional reprogramming in developing soybean (Glycine max) embryos. Metabolites 3:347–372. 10.3390/metabo3020347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Binenbaum Z, Klyman E, Fishov I. 1999. Division-associated changes in membrane viscosity of Escherichia coli. Biochimie 81:921–929. 10.1016/S0300-9084(99)00202-3. [DOI] [PubMed] [Google Scholar]
- 50.Shinitzky M, Barenholz Y. 1974. Dynamics of the hydrocarbon layer in liposomes of lecithin and sphingomyelin containing dicetylphosphate. J. Biol. Chem. 249:2652–2657. [PubMed] [Google Scholar]
- 51.Kong K, Rowlands CJ, Elsheikha H, Notingher I. 2012. Label-free molecular analysis of live Neospora caninum tachyzoites in host cells by selective scanning Raman micro-spectroscopy. Analyst 137:4119–4122. 10.1039/c2an35640f. [DOI] [PubMed] [Google Scholar]
- 52.Lee TH, Chang JS, Wang HY. 2013. Rapid and in vivo quantification of cellular lipids in Chlorella vulgaris using near-infrared Raman spectrometry. Anal. Chem. 85:2155–2160. 10.1021/ac3028118. [DOI] [PubMed] [Google Scholar]
- 53.Cherney DP, Conboy JC, Harris JM. 2003. Optical-trapping Raman microscopy detection of single unilamellar lipid vesicles. Anal. Chem. 75:6621–6628. 10.1021/ac034838r. [DOI] [PubMed] [Google Scholar]
- 54.Schultz ZD, Levin IW. 2011. Vibrational spectroscopy of biomembranes. Annu. Rev. Anal. Chem. 4:343–366. 10.1146/annurev-anchem-061010-114048. [DOI] [PubMed] [Google Scholar]
- 55.Neidhardt FC, Ingraham JL, Schaechter M. 1990. Physiology of the bacterial cell: a molecular approach. Sinauer Associates, Sunderland, MA. [Google Scholar]
- 56.Zhu G, Zhu X, Fan Q, Wan X. 2011. Raman spectra of amino acids and their aqueous solutions. Spectrochim Acta A Mol. Biomol. Spectrosc. 78:1187–1195. 10.1016/j.saa.2010.12.079. [DOI] [PubMed] [Google Scholar]
- 57.Gardikis K, Hatziantoniou S, Viras K, Wagner M, Demetzos C. 2006. A DSC and Raman spectroscopy study on the effect of PAMAM dendrimer on DPPC model lipid membranes. Int. J. Pharm. 318:118–123. 10.1016/j.ijpharm.2006.03.023. [DOI] [PubMed] [Google Scholar]
- 58.Mendelsohn R, Koch CC. 1980. Deuterated phospholipids as Raman spectroscopic probes of membrane structure: phase diagrams for the dipalmitoyl phosphatidylcholine (and its d62 derivative)-dipalmitoyl phosphatidylethanolamine system. Biochem. Biophys. Acta 598:260–271. 10.1016/0005-2736(80)90004-8. [DOI] [PubMed] [Google Scholar]
- 59.Weber FJ, de Bont JA. 1996. Adaptation mechanisms of microorganisms to the toxic effects of organic solvents on membranes. Biochim. Biophys. Acta 1286:225–245. 10.1016/S0304-4157(96)00010-X. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.