ABSTRACT
Neutralizing antibodies (nAbs) are a high priority for vaccines that aim to prevent the acquisition of HIV-1 infection. Vaccine effectiveness will depend on the extent to which induced antibodies neutralize the global diversity of circulating HIV-1 variants. Using large panels of genetically and geographically diverse HIV-1 Env-pseudotyped viruses and chronic infection plasma samples, we unambiguously show that cross-clade nAb responses are commonly induced in response to infection by any virus clade. Nonetheless, neutralization was significantly greater when the plasma clade matched the clade of the virus being tested. This within-clade advantage was diminished in older, more-diverse epidemics in southern Africa, the United States, and Europe compared to more recent epidemics in Asia. It was most pronounced for circulating recombinant form (CRF) 07_BC, which is common in China and is the least-divergent lineage studied; this was followed by the slightly more diverse Asian CRF01_AE. We found no evidence that transmitted/founder viruses are generally more susceptible to neutralization and are therefore easier targets for vaccination than chronic viruses. Features of the gp120 V1V2 loop, in particular, length, net charge, and number of N-linked glycans, were associated with Env susceptibility and plasma neutralization potency in a manner consistent with neutralization escape being a force that drives viral diversification and plasma neutralization breadth. The overall susceptibility of Envs and potencies of plasma samples were highly predictive of the neutralization outcome of any single virus-plasma combination. These findings highlight important considerations for the design and testing of candidate HIV-1 vaccines that aim to elicit effective nAbs.
IMPORTANCE An effective HIV-1 vaccine will need to overcome the extraordinary variability of the virus, which is most pronounced in the envelope glycoproteins (Env), which are the sole targets for neutralizing antibodies (nAbs). Distinct genetic lineages, or clades, of HIV-1 occur in different locales that may require special consideration when designing and testing vaccines candidates. We show that nAb responses to HIV-1 infection are generally active across clades but are most potent within clades. Because effective vaccine-induced nAbs are likely to share these properties, optimal coverage of a particular clade or combination of clades may require clade-matched immunogens. Optimal within-clade coverage might be easier to achieve in regions such as China and Thailand, where the epidemic is more recent and the virus less diverse than in southern Africa, the United States, and Europe. Finally, features of the first and second hypervariable regions of gp120 (V1V2) may be critical for optimal vaccine design.
INTRODUCTION
HIV-1 exhibits extraordinary sequence variation in its surface gp120 and transmembrane gp41 envelope glycoproteins (Env), which are the sole targets for neutralizing antibodies (nAbs) (1). This variation contributes to the diversity manifested in multiple genetic subtypes, or clades, and a growing number of circulating recombinant forms (CRFs) that differ in prevalence by geographic region (2, 3). Escape from the immune response to the virus during infection fuels the generation of much of this diversity. A vaccine that aims to elicit an effective nAb response will need to overcome this genetic variation at the antigenic level, at least within a given subtype or geographic region, but preferentially across all major subtypes in regions where HIV-1 is most prevalent. Unfortunately, despite an ability of current vaccines to induce moderate to high titers of nAbs against highly neutralization-sensitive strains of the virus (i.e., tier 1 phenotype) (4), these antibodies exhibit little if any neutralizing activity against the less sensitive but more common tier 2 circulating strains (5–10) and have failed to demonstrate a significant level of protection in phase 3 clinical trials of Env-containing vaccines (11–13). In addition, there is no clear evidence that the relatively weak nAb response seen in the RV144 Thai trial (5, 14) was associated with the modest protection observed in this trial (15).
The relatively weak and ineffective vaccine-elicited nAb responses in clinical trials have stimulated a heightened interest in studies of broadly neutralizing antibody (bnAb) responses during chronic HIV-1 infection. This focus provides a way to gain information on what should ultimately be possible to elicit by vaccination, helps refine our knowledge regarding viral epitopes that are particularly susceptible to bnAbs, and enables common characteristics of bnAbs to be defined, thus informing strategies for designing improved vaccine immunogens (16). During chronic infection, most people make antibodies that cross-neutralize multiple tier 2 variants of different clades (17). A small number of individuals make high titers of antibodies that neutralize most tier 2 strains evaluated (17–24). Studies of monoclonal antibodies from the latter set of individuals have shown that at least five regions on the Env trimer spike are vulnerable to bnAbs. These regions include conformational epitopes in the CD4 binding site (CD4bs) of gp120, glycan-dependent conformational epitopes in the V1V2 loop and at the base of V3/C3 of gp120, linear epitopes in the membrane-proximal external region (MPER) of gp41 (reviewed in references 25, 26, 27, and 28), and a more recently described conformational epitope at the gp120-gp41 interface (29). These epitope regions and contacts span both variable and highly conserved amino acids in Env; the breadth exhibited by these bnAbs is enabled by their capacity to recognize conserved aspects of the epitopes across diverse viruses despite some intrinsic local diversity of the virus in epitope regions. Other regions of vulnerability that have yet to be identified with current monoclonal antibodies and epitope mapping strategies might exist.
Increasing knowledge of the biological processes that initiate and govern the production of bnAbs is leading to new ideas for immunogen design. The evolutionary trajectory of clonal lineages of antibodies that eventually develop breadth has successfully been reconstructed for some of the most potent bnAbs isolated to date (30–32), allowing inference of their germ line precursors and design of antigens that trigger B cell precursors known to initiate likely lineages (33, 34). Furthermore, immune selection drives viral escape, resulting in coevolving populations of virus and antibodies in serum and a sustained dynamic of escape and pursuit. The study of subjects followed longitudinally from acute viremia through bnAb development in chronic infection has enabled identification of stages involved in antibody-virus coevolution. In a study by Liao et al. (35), epitope diversification due to escape from autologous neutralization preceded breadth and heterologous neutralization in a bnAb clonal lineage. This observation supports the idea that exposure to epitope variants through vaccination may foster the evolution of antibody breadth (36). It also provides key data to allow a vaccine approach that attempts to reproduce the evolutionary trajectory of bnAb responses by inducing ancestral intermediates of bnAbs (37, 38). This information, combined with other strategies that aim to mimic the native structure of functional Env spikes (39–41) and to overcome self-tolerance mechanisms (42) and the immunosuppressive properties of Env (43–45) that impede B cell maturation and survival, are just a few examples of the potential to discover improved vaccine immunogens in the near future. Vaccines that elicit bnAbs against one or more highly conserved epitopes across multiple clades of the virus are desirable; however, a more feasible and still useful goal may be to elicit multiple responses to less-conserved epitopes that exhibit moderate variability within and between clades (17).
As newer immunogens are developed and tested, it will be important to consider how Env diversity will impact the ability of the vaccine-elicited Abs to neutralize virus variants in different parts of the world. Early studies found little evidence that the different genetic subtypes of HIV-1 corresponded to distinct neutralization serotypes (46–50); the only exception was an ability to distinguish clade B viruses from CRF01 viruses using HIV-1 plasma samples of both clades (47, 48). These early studies were performed with relatively small numbers of samples and were confounded by the use of nonclonal viruses. A subsequent study with a larger number of molecularly cloned Env-pseudotyped viruses and a single pool of HIV-1 plasma samples for each of six subtypes showed that the clade C plasma pool was superior to the others for neutralization of multiple subtypes of the virus (51). This study also found multiple cases of preferential intrasubtype neutralization when the corresponding uncloned virus quasispecies were assayed in human peripheral blood mononuclear cells (PBMC), and it yielded supporting evidence that clade B and CRF01 viruses form two antigenically distinct neutralization serotypes relative to each other. Another study found evidence for regional clustering of shared neutralization determinants on clade C viruses from South Africa (52). Finally, some of the less potent bnAbs exhibit preferential neutralization of clade-matched viruses (31, 53). Two of the most dramatic examples are 2G12 and 2F5, which came from clade B-infected individuals and exhibit substantial breadth against clade B viruses but rarely neutralize clade C viruses (52–56). Differences in neutralization breadth also have been observed with clonally related bnAbs targeting a single epitope (31, 57).
In contrast to the extensive recent work characterizing bnAb breadth, here we inform a broader view of shared characteristics and patterns of serological potency and breadth in natural infection and begin to define determinants of tier 2 virus neutralization sensitivity. This was done by utilizing an exceptionally large neutralization data set comprised of 219 molecularly cloned Env-pseudotyped viruses assayed with 205 plasma samples from HIV-1 infections, where both the Envs and plasma samples were chosen to represent the diversity of predominant M-group clades and circulating recombinant forms. A recent evaluation of this data set helped to clarify the spectrum of neutralization breadth seen during chronic HIV-1 infection (17) and was used to identify a global panel of HIV-1 Env reference strains for standardized assessments of vaccine-elicited nAb responses across multiple vaccine platforms in different parts of the world (58). Here we present an analysis to address the relative contributions of cross-reactive interactions within and between clades for both the plasma samples and viruses, the effects of clade diversity and divergence on neutralization susceptibility, and the contributions of gp120 hypervariable regions to neutralization.
MATERIALS AND METHODS
Plasma samples and viruses.
All plasma samples and Env-pseudotyped viruses used in this study were described previously (17, 58). Briefly, plasma samples were obtained from 205 chronically infected individuals who were antiretroviral drug naive and infected with HIV-1 subtypes A (n = 8), B (n = 59), C (n = 58), and D (n = 2); with circulating recombinant forms CRF01_AE (n = 14), CRF07_BC (n = 16), CFR02_AG (n = 2), and CRF10_CD (n = 1); and with noncirculating recombinants AC (n = 4), AD (n = 3), BD (n = 1), CD (n = 1), and ABCD (n = 2). Sequences could not be obtained for 34 individuals due to low levels of plasma viremia, so the infecting clade of the virus was undetermined in these cases. HIV-1 genetic subtypes of plasma samples were determined by single-genome amplification (SGA) and sequencing of a single gp160 gene. All plasma samples were tested for nonspecific activity in neutralization assays with murine leukemia virus (MLV) Env-pseudotyped virus. Samples that tested positive (50% inhibitory dose [ID50] > 1:40) were not included. For assay results that did not give ID50 values, a small constant placeholder value of 10 was recorded; we refer to these as “censored” values, because the ID50 values were not directly observed, being below the limit of detection.
Functional full-length rev-env cassettes used for Env-pseudotyped viruses were cloned by SGA from plasma viral RNA of 219 subjects infected with HIV-1 clades A (n = 10), B (n = 54), C (n = 67), D (n = 5), G (n = 8), CRF01_AE (n = 21), CRF02_AG (n = 16), CRF06 (n = 1), CRF07_BC (n = 14), AC (n = 6), AD (n = 5), ACD (n = 1), BC (n = 4), BG (n = 1), and CD (n = 6). Because Fiebig stages (59) were frequently unknown or ranged in value, we defined three infection stage categories: early (Fiebig I through IV), intermediate (Fiebig V through early VI), and late (late Fiebig VI, 6 or more months postinfection through chronic infection), where known. As described throughout the text, some analysis considered only viruses with early or late designations, although we also tried some comparisons that grouped intermediate with early and some that grouped intermediate with late. This was done because we did not know a priori whether the intermediate-stage group should be treated as early, late, or independent, and we were mindful of biases due to uneven sampling across clades (Table 1).
TABLE 1.
Summary of HIV-1 Env-pseudotyped viruses and plasma samples by clade, infection stage, and screening status
| Clade | No. of viruses |
No. ofplasma samples |
||||||
|---|---|---|---|---|---|---|---|---|
| Total | At virus infection stage: |
Total | Not screened | Screenedd | ||||
| Ea | Ib | Lc | Unknownf | |||||
| A | 10 | 6 | 2 | 1 | 1 | 8 | 4 | 4 |
| B | 53 | 32 | 7 | 11 | 3 | 59 | 53 | 6 |
| C | 67 | 30 | 26 | 7 | 4 | 58 | 7 | 51 |
| D | 5 | 4 | 0 | 0 | 1 | 2 | 1 | 1 |
| G | 8 | 0 | 1 | 6 | 2 | 0 | 0 | 0 |
| CRF01 | 21 | 8 | 0 | 10 | 3 | 14 | 14 | 0 |
| CRF02 | 16 | 0 | 0 | 15 | 1 | 2 | 2 | 0 |
| CRF07 | 14 | 2 | 0 | 12 | 0 | 16 | 0 | 16 |
| Othere | 25 | 3 | 9 | 6 | 7 | 12 | 6 | 6 |
| Unknownf | 0 | 0 | 0 | 0 | 0 | 34 | 30 | 4 |
| Total | 219 | 85 | 45 | 67 | 22 | 205 | 117 | 88 |
Fiebig I through IV (early).
Fiebig V through early Fiebig VI (intermediate).
Late Fiebig VI through chronic.
Number of plasma samples screened for neutralization potency and breadth.
URFs and uniquely represented CRFs.
Clade unknown because sequence amplification was unsuccessful.
Neutralization assay.
Neutralization of Env-pseudotyped viruses was measured in 96-well culture plates by using Tat-regulated firefly luciferase (Luc) reporter gene expression to quantify reductions in virus infection in TZM-bl cells (60). This assay has been formally optimized and validated (61). Briefly, heat-inactivated (56°C, 1 h) plasma samples were assayed at 3-fold dilutions starting at 1:20. Neutralization titers (ID50) are the plasma dilutions at which relative luminescence units (RLU) were reduced by 50% compared to RLU in virus control wells after subtraction of background RLU in cell control wells. Assay stocks of Env-pseudotyped viruses were produced by transfection in 293T cells and titrated in TZM-bl cells as described previously (60). Because the assays were conducted in three different laboratories (Duke University, Harvard University, and NIH), all plasma samples that were assayed in more than one laboratory were evaluated with a standard set of three viruses (Bal.26, SF162.LS, and Bx08.16) to document the equivalency of sample performance across these laboratories. Any samples that did not meet the predefined criteria for equivalency (ID50 agreement within a 3-fold difference range among the laboratories for all 3 viruses) were not included. All three laboratories also participated in a formal proficiency testing program for the TZM-bl assay (62) during the course of this study. As an additional control for uniform assay performance, HIVIG (polyclonal IgG from a pool of HIV-1-positive plasma samples) was assayed with a single virus (SS1196.1) for each batch of assays performed. The titer obtained was required to be within a 3-fold difference range of a preestablished reference value. Assay batches that did not meet this prespecified standard were repeated.
Sequence analysis.
We aligned the 434 env sequences and HXB2 with GeneCutter (hiv.lanl.gov/content/sequence/GENE_CUTTER/cutter.html), which aims to maintain intact codons for translation and uses HMMer (63) with probability profiles trained specifically for HIV-1. We then manually refined the alignment with Se-Al (64), computed phylogenies using PhyML, version 3.0 (65), with the HIVb+Γ4+I substitution model (66), and rendered them in R, v3.0.2 (67), using the package APE, v3.0-11 (68). We excluded hypervariable loops from phylogenetic analysis and similarity comparisons to emphasize the phylogenetic signal. We computed similarity between each pair of plasma sample and virus Envs as percent identity over the length of the gp160 sequence, with gapped sites excluded. The alignments and neutralization data are available online (http://hiv.lanl.gov/content/sequence/HIV/USER_ALIGNMENTS/Montefiori2014).
For population level alignments, an insertion/deletion mode of mutation predominates in hypervariable loops. Length polymorphisms in alignments of the hypervariable regions within the V loops from unlinked infections lack phylogenetic signal identified by established base substitution models. For this reason, we treated them separately and instead summarized hypervariable region lengths, the number of putative N-linked glycosylation sites (PNGs), and the net charge excised from the hypervariable regions in V1, V2, V4, and V5. To define the boundaries of the hypervariable regions, we identified the borders demarking transitions between locations where the alignment is reasonable and variation is most commonly generated by point mutations and the short hypervariable regions where insertions and deletions dominate sequence variation (69). To characterize hypervariable regions, we excised the regions between the more conserved, easily aligned stretches. A web tool at the Los Alamos HIV-1 database excises and characterizes either complete V loops or the hypervariable regions within them, given an HIV Env alignment (http://hiv.lanl.gov/content/sequence/VAR_REG_CHAR/). The complete details of our strategy are provided there and in Text S1 in the supplemental material. The regions bounding the hypervariable stretches also are noted in the annotation at the HIV database (http://hiv.lanl.gov/content/sequence/HIV/MAP/annotation.html).
To test for dual CRF01 infections in the B-clade plasma samples, 30 sequences were obtained from each plasma sample (T500108_503963 and T500105_286588) to test for the presence of minority CRF01 populations. A sample size of 30 was chosen to obtain power to detect minority variants as follows: for a minority population of 15%, 30 sequences give over 99% chance of sampling it. For a 10% minority population, 30 sequences give 95.8% chance of sampling; a 5% minority population has 78.5% probability of detection with 30 sequences, and a 1% population gives 26% detection probability (70).
Statistical inference.
For each of the clade A, B, C, CRF01, CRF02, and CRF07 plasma samples, we computed the geometric mean within- and between-clade ID50s using assay results from non-URF (unique recombinant forms) Env pseudoviruses. We excluded the URFs to reduce noise from mixed specificities due to recombination breakpoints at varied locations over gp160. Between-clade comparisons excluded those between CRF07 and clade C viruses, due to the predominance of clade C Env in CRF07. We compared within- and between-clade geometric mean titers per plasma with paired, two-sided Wilcoxon tests. We used R, version 3.0.2 (67), to perform all tests. For comparisons between V-loop attributes (length, number of putative N-linked glycosylation sites, and net charge), we used Kendall's τ as implemented in the package Kendall, v 2.2 (71). We conducted 360 tests: 3 response variables (50% inhibitory concentration [IC50], IC80, and pAUC), 5 hypervariable regions (V1 and V2 combined, V1, V2, V4, and V5) × 3 loop attributes (length, net charge, number of glycosylation sites) × 2 Env sources (the pseudoviruses and plasma-derived Envs) × 2 with and without low-titer plasma samples (to adjust for screening) × 2 with and without censored values as placeholders (i.e., all assay results versus only reactive-assay results). We computed false-discovery rates with the q-value package v 1.36.0 (72) after the method of Storey and Tibshirani (73) and required a q of <0.1 for significant support. Curiously, although the direction and trends were consistent between the methods, the pAUC scoring was less sensitive than ID50 in terms of the significance of correlations between loop attributes and neutralization sensitivity. This was true even when we excluded censored data from the ID50 analysis, so it was not due to a statistical artifact introduced by inclusion of censored data that resulted in calculating inappropriate P values. The difference may be due at least in part to noise introduced by assigning ranks to pAUC scores with low values and larger proportional error.
We used mixed-effects linear regression to model neutralization outcomes due to fixed effects of biological interest (matched plasma and virus clades, whether the virus was obtained from an early or late infection, and V1V2 lengths from plasma and virus Envs) while considering random effects of the experimental design (plasma and virus), as described in Text S2 in the supplemental material. Logistic regression uses the logit function ln [P/(1 − P)], where P is the binomial-outcome probability, to model factors as additive effects to the (natural) log of the odds of the binary outcome. Even odds correspond to a parameter estimate of zero (ln 1:1).
Model selection was performed via comparison with simpler, reduced models by comparing Akaike Information Criterion scores computed in analysis of variance (ANOVA) (74). Random-effects grouping factors were virus and plasma. The checkerboard layout of experimental interactions between each virus and plasma gives a completely crossed design. Treating plasma and virus as random effects prevents the risk of pseudoreplication by adjusting the model matrix during parameter estimation (74). That is, the random effects represent the variance-covariance structure of the crossed-factor experimental design rather than treat as independent all 44,758 assays. To estimate model parameters, we used the glmer method of the package lme4 (v 1.1-0) (75).
Row/column effects.
Computing expected values (geometric means) provided individual pseudovirus Env susceptibility scores and individual plasma potency scores such that each combined virus-plasma (row/column) score approximates the log-transformed neutralization titers for each pair of pseudovirus and plasma. Given a table of neutralization ID50s measured for a panel of plasma samples against a panel of viruses, we express the log titers as the sum of two terms and a constant:
| (1) |
which holds for all pairs (virus x, plasma y) in the table.
Minimizing the mean-square error of the above approximation across the table yields the following:
| (2) |
Exponentiation yields
| (3) |
In the above expressions, geometric mean (virus x) is the geometric mean ID50 of any selected virus x with all plasma samples in the table. The geometric mean is used as an unbiased estimator of central tendency because neutralization assay results exhibit log-normal, rather than normal, distributions. Similarly, geometric mean (plasma y) is the geometric mean ID50 of any selected plasma sample y with all viruses in the table. The overall geometric mean xy serves as a normalization constant. We validated correlation findings statistically using a permutation test, to ensure that the correlation coefficient between observed ID50s and ID50s predicted by the row/column approximation was greater than by chance alone. For any particular neutralization data set, we randomized (by permutation without replacement) the log-transformed ID50 data 10,000 times, recomputed expected values from row/column averages of the randomized data, and computed correlation coefficients between permuted data and the approximated values at each iteration (see Table S5 in the supplemental material). This gave a null distribution of correlation coefficients, which we compared with the original r2 to obtain a resampled P value.
Nucleotide sequence accession numbers.
Sequences analyzed in this study have the GenBank accession numbers listed below.
Pseudovirus Envs, U15121, AF259954, AF407152, AF407156, AF407158, AF407160, AY423984, AY424079, AY424138, AY835434, AY835437 to AY835441, AY835443 to AY835452, DQ187010, DQ388514 to DQ388517, DQ411850 to DQ411854, DQ435682, DQ435683, EF042692, EF117255, EF117258 to EF117266, EF117268 to EF117274, EF210726 to EF210730, EF210732, EF210733, EF210735, EU166866, EU289183 to EU289194, EU289198 to EU289202, EU513182 to EU513185, EU513187 to EU513190, EU513193 to EU513196, EU513198, EU575148, EU575170, EU575786, EU575870, EU576296, EU576299, EU577190, EU577213, EU885760 to EU885766, FJ443575, FJ443670, FJ443744, FJ443808, FJ443999, FJ444017, FJ444047, FJ444059, FJ444103, FJ444215, FJ444325, FJ444395, FJ444421, FJ444437, FJ444529, FJ444561, FJ444600, FJ444612, FJ496194, FJ496204, FJ817366, FJ817369, FJ817370, HM215260, HM215264 to HM215266, HM215269, HM215270, HM215272 to HM215276, HM215278 to HM215281, HM215283, Plasma Envs, HQ595767 to HQ595806, HQ615962 to HQ615988, HQ625565 to HQ625585, HQ625587 to HQ625590, HQ625592 to HQ625595, HQ625598, HQ625600, HQ625602 to HQ625605, HQ690973, JF297221 to JF297233, JF346900 to JF346919, JF680905, JF680937, KC748972 to KC749062.
RESULTS
Genetic variation in plasma and pseudovirus Envs.
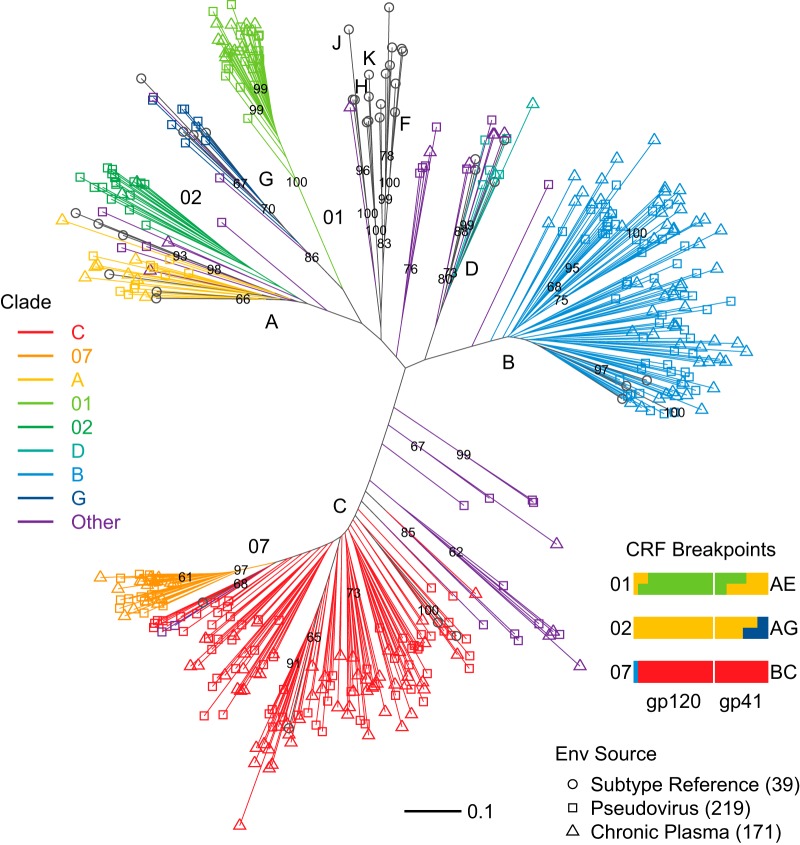
Full-length gp160 sequences were obtained for the 219 Env-pseudotyped viruses and for a single SGA-derived Env from each of 170 plasma samples used for neutralization assays. Table 1 lists the clade distribution of all Envs and the infection stage of the pseudovirus Envs. Figure 1 shows the phylogenetic diversity of plasma and pseudovirus Envs. We could not extract an Env sequence from 34 plasma samples, so in those cases the clade of the infecting strain was unknown. As expected, the nonrecombinant subtype sequences clustered with nonrecombinant Envs of the subtype reference alignment (Fig. 1). With gap-rich hypervariable loops excised, none of the 389 Env sequences studied shared over 93% amino acid identity, and the most divergent sequences shared only 70% amino acid identity.
FIG 1.
Phylogenetic diversity of Env sequences in the pseudoviruses and plasma samples used for neutralization assays. Env-pseudotyped virus (n = 219) and plasma-derived (n = 171) sequence phylogeny followed the HIV-1 M group subtype reference Env distribution (n = 39), with addition of CRFs 01, 02, and 07 reference strains. CRFs 01, 02, and 07 are important epidemic lineages in Thailand and other parts of Asia, West Africa, and China, respectively (36). Unique recombinant forms, not known to be circulating because the particular combination of breakpoints that they exhibit has been identified only in the given sequence, are indicated as “Other” (gray). Bootstrap support is indicated for nodes present in 60% or more of 500 resampled replicates. Note that the evolutionary distances indicated by the branch lengths within the more recent CRF01 and CRF07 clades are substantially shorter than within the older epidemic clades (A, B, C, D, G, and CRF02) (79).
In addition to common CRFs 01 (Asia), 02 (West Africa), and 07 (China), we identified two CRFs (06 and 14) and 23 URFs among virus Envs (6 AC, 5 AD, 4 BC, 6 CD, 1 ACD, and 1 BG) and, among plasma Envs, one CRF10 and 11 URFs (4 AC, 3AD, 2 ABCD, 1 BD, and 1 CD) (76). Because URFs cluster with established nonrecombinant clades of the phylogeny when comprised primarily of one clade, we confirmed the URFs using the Recombinant Identification Program (RIP) (77) and in each case identified Env regions with over 90% similarity to more than one distinct M-group subtype (not shown). CRF07, a recombinant lineage of clades B and C, carries a predominantly C-clade Env (78) and forms a branch within clade C in Env phylogenies (Fig. 1). Similarly, CRF02 is a recombinant of A and G clades, mostly clade A in Env, and associated with the A clade in Env phylogenies. CRF02 is a much older lineage than CRF07 and branches outside clade A, with similar levels of diversity found in our samples of CRF02 and the A clade (Fig. 1) (79). CRF01 has traditionally been designated clade E in Env (76, 79, 80), though no parental, full-genome E-clade virus was ever isolated, making its genetic origins particularly difficult to resolve (79, 81).
Adjustments for prescreened plasma effects.
Prior to inclusion in this study, a subset of plasma samples was selected from larger plasma collections after screening for neutralization activity. Plasma samples from some clades were screened, while others were not. For example, most (68 of 75, or 90%) of the clade C and CRF07 plasma samples were chosen after they had been screened for potency, versus few (6 of 74, or 8%) from clade B and none from CRF01 (Table 1). Screening of plasma was conducted based on the reasoning that plasma samples with little or no neutralization activity would be of little value to identify neutralization serotypes, a major initial goal of this study. Screening was performed only when an excessive number of samples for a desired clade or geographic region was available for downselection. Screening was not performed when sample availability was limited; in these cases, all available samples were included. Samples from all study sites were brought together in this collection to enable a global study. To analyze a data set with reduced impact of screening selection bias, we applied a retroactive postscreening strategy across all plasma samples. The postscreening strategy used a cutoff that approximated the screening criteria to exclude plasma samples with the lowest activity, apply similar criteria across all collection sites and clades, and provide more-uniform sampling for comparative analysis.
To assess the postscreening strategy, we tested whether neutralization potency was greater against clade-matched viruses for screened plasma samples than for nonscreened plasma samples and found a significant difference (two-sided Wilcoxon test, P = 0.013; see Fig. S1A in the supplemental material). Similarly, comparing plasma neutralization potency against clade-mismatched viruses indicated greater potency among screened than nonscreened plasma (see Fig. S1A; P = 0.0077). Excluding the low-titer plasma samples reduced these differences to lose statistical significance (see Fig. S1B; P = 0.73 for clade-matched viruses; P = 0.31 for clade-mismatched viruses). Thus, excluding weakly neutralizing plasma samples helped to mitigate screening bias when testing for clade effects.
Cross-reactive neutralization within and between clades.
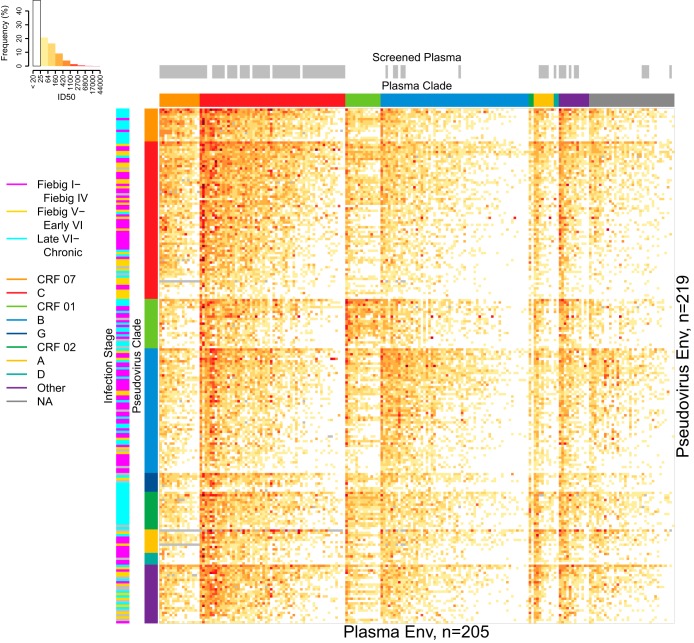
A heatmap representation of ID50 neutralization results for all 44,758 plasma/pseudovirus combinations tested is shown in Fig. 2, sorted first by Env clade and then by plasma potency and pseudovirus sensitivity within clades. This figure provides a comprehensive view of the neutralization data and illustrates the relationships between screened samples and clades, as well as the time of sampling of Env for pseudoviruses, some of which were collected early in infection and some during chronic infection. An alternative view of the complete data, organized by hierarchical clustering of shared reactivity and sensitivity patterns, is presented in Fig. S2 in the supplemental material. In general, plasma potencies were significantly greater against clade-matched than mismatched viruses (paired Wilcoxon test, P < 2.2 × 10−16). This was tested by calculating two geometric mean ID50s for each plasma sample, one for the comparisons to all clade-matched Envs and one for the comparisons to all clade-mismatched Envs. The same outcome was obtained with low-titer postscreened plasma samples excluded (Wilcoxon test, P < 2.2 × 10−16). We also tested whether screened plasma samples alone were more potent against clade-matched than mismatched viruses and again found a significant difference (paired Wilcoxon test, P = 0.00085).
FIG 2.
Plasma neutralization potency and virus sensitivity from 205 chronically infected plasma samples against 219 Tier 2 pseudoviruses. Assay results are presented as a heatmap of 44,758 ID50s. Rows and columns were sorted by geometric mean ID50 per clade, with more-sensitive viruses toward the top and more-potent plasma samples toward the left. The color key histogram summarizes the percentage of observations in each given interval of ID50 values. Annotation bands indicate Env pseudovirus clades (middle-left column), the infection stage from which virus was sampled or NA if unknown (left column), plasma clades or NA if unknown (row above heatmap), and plasma samples screened against smaller panels of 6 to 18 viruses to ensure that neutralization responses were detected to select from a larger plasma sample repository (gray bars, top row). Missing ID50 data are indicated by gray cells in the heatmap.
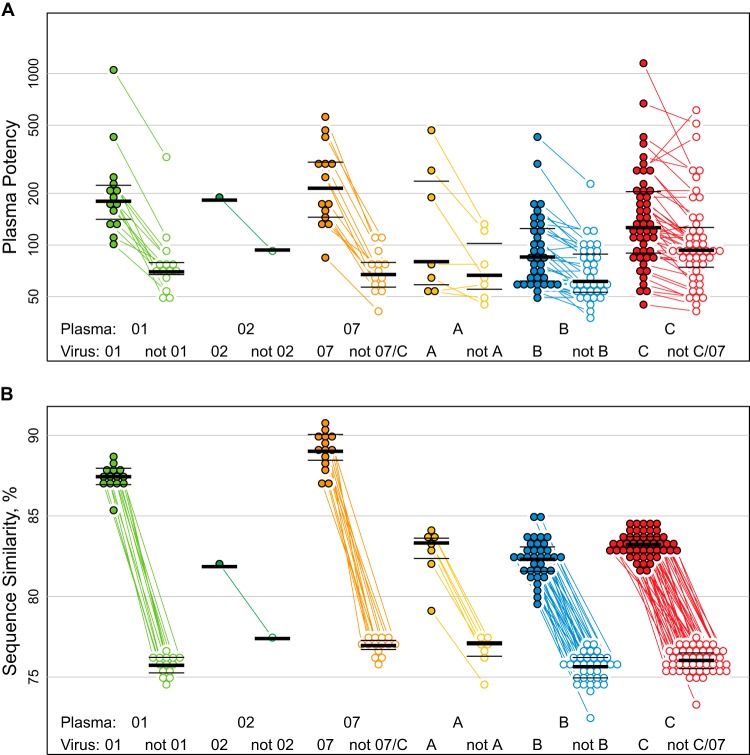
For a more detailed analysis of cross-reactive neutralization effects, we identified the clades with the largest sample sizes for both plasma and virus Envs and used these (A, B, C, CRF07, and CRF01) for statistical comparisons. Consistently for each clade, plasma neutralization potencies were significantly greater against clade-matched than mismatched viruses (Fig. 3A). Paired, two-sided comparisons indicated significant differences in geometric mean ID50s for 4 of the 5 clades, i.e., B, C, CRF01, and CRF07, even with Bonferroni's correction applied to accommodate five tests (all had P values of <0.01; specific Wilcoxon P values appear in Fig. 3A). Clade A had fewer samples and shared the trend for greater potency within than between clades (Fig. 3A).
FIG 3.
Greater plasma potency within than between clades. (A) Plasma potency, computed as geometric mean ID50, was significantly greater against clade-matched viruses (filled symbols, left group per plasma clade) than against clade-mismatched viruses (open symbols, right group per clade). Test results from URFs were omitted, as were weakly neutralizing, screened plasma samples (see the text). Lines connect paired means from the same plasma sample. Paired, two-sided Wilcoxon tests gave P values of 0.00012 for CRF07 (n = 14) and also for CRF01 (n = 14), 0.078 for A (n = 7), 3.3 × 10−7 for B (n = 35), and 8.9 × 10−7 for C (n = 48). CRF02 is also shown (n = 1). Clade C viruses were excluded from analysis of CRF07 plasma potency, and vice versa, as CRF07 is an outgrowth of C. (B) Average similarity between plasma and virus Envs was greater within (filled symbols, left group per clade) than between (open symbols, right) clades. Thick lines show the median, and thin lines show interquartile ranges (25th and 75th percentiles) per distribution.
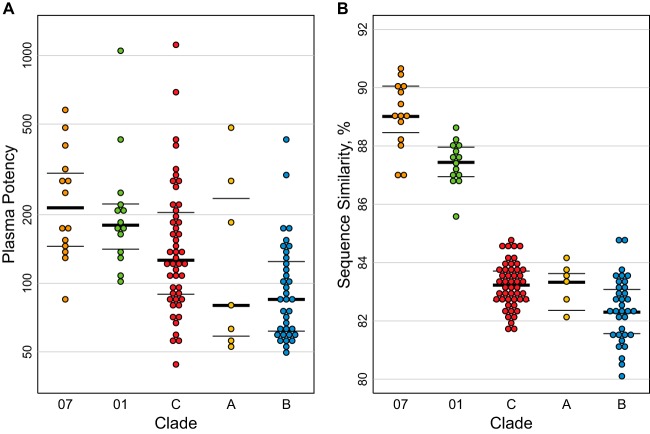
The average sequence similarity between pseudovirus Envs and plasma Envs, essentially by definition, was greater within than between clades and particularly high for within-clade comparisons of CRF07 and CRF01 (Fig. 1 and 3B). CRF07 originated in the 1990s in China (79). It is essentially clade C in Env (Fig. 1), and it is the least diversified lineage that we sampled. The CRF01 lineage in Asia originated in Thailand in the late 1980s (80) and was more diverse than CRF07. The A, B, and C clades are geographically widespread and much older and more diversified clades (79, 111). Rank ordering of clades by within-clade plasma potency corresponded to the ranking by mean per-plasma Env similarity with clade-matched Env pseudotyped viruses, except for clade A, which had fewer samples than the others (Fig. 4). Essentially, less diverse clades had greater within-clade potency, and the level of diversity within clade reflected the age of the lineage. We discuss the implications of this more fully in Discussion below and provide more detail regarding what is known about the timing of the origins of these clades. We think these observations are relevant to potential vaccine trial populations, as our CRF07 and CRF01 samples were representative of the known diversity of these important lineages in the Asian HIV epidemic. Both CRF07 and CRF01 were collected from multiple geographically distant locations in China, (Beijing, Yunnan, and Henan), and CRF01 samples were also obtained in Thailand. To further verify that we did not have a biased sampling of CRF01 and CRF07 strains, we compared them using maximum likelihood trees to all CRF01 and CRF07 sequences available in the database at Los Alamos (representing viruses obtained from 322 CRF07 and 421 CRF01 distinctly infected individuals). The sequences of the CRF01 and CRF07 pseudovirus and plasma Envs studied here were widely dispersed throughout their respective genetic clades, with no evidence of clustering in our sample (data not shown).
FIG 4.
Within-clade plasma potency distributions followed rank order of similarity distributions between plasma and virus Envs. Within-clade data from Fig. 3 are ordered by decreasing values. (A) Plasma potency, computed as geometric mean ID50, was greatest against clade-matched pseudoviruses for CRF07, followed by CRF01 and then clades C, A, and B. (B) Mean similarity between plasma and clade-matched pseudovirus Envs roughly followed the rank ordering of plasma potencies per clade, with greater similarities within more recently emergent CRF07 and CRF01 than nonrecombinant clades. Thick lines show the median, and thin lines show interquartile ranges (25th and 75th percentiles) per distribution.
A simple, overall test for associations between virus infection stage (i.e., early, intermediate, and late HIV-1 infection; the criteria used for these categories are defined in Materials and Methods) and nAb sensitivity was vulnerable to bias from differences in sample size and cross-reactivity (Table 1). Rather than risk confounding cross-reactivity effects with infection stage, we compared plasma potency against viruses stratified by infection stage (early versus late) and with several alternative representations of the data: (i) with and without low-titer plasma samples, i.e., using the full data set or the postscreened subset; (ii) with and without censored ID50s; and (iii) with intermediate-stage viruses excluded or grouped with either early or late stages, for a total of eight different representations. Because we did 16 Wilcoxon tests, we required an uncorrected P value of <0.003 for statistical significance to allow for Bonferroni's correction.
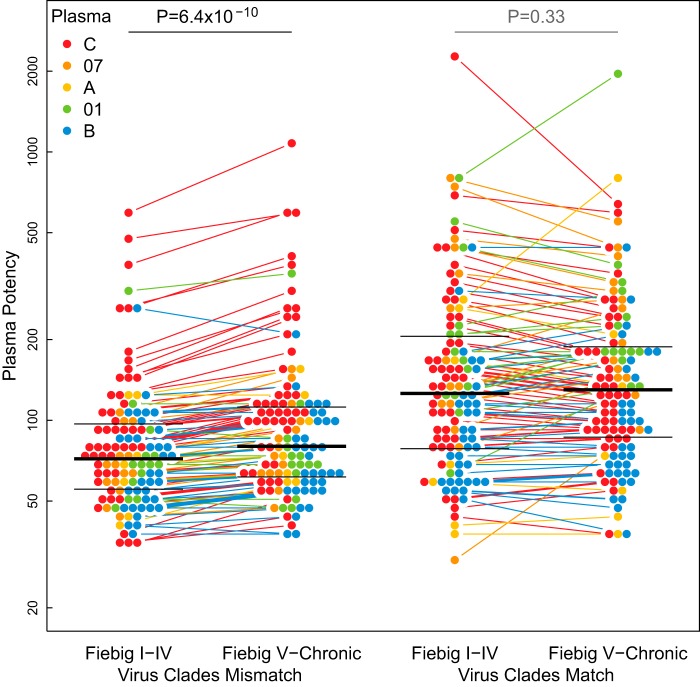
Surprisingly, there was a clear distinction in the sensitivity of early and late viruses when virus and plasma Env clades did not match, but no distinction was seen when they did match (a representative example appears in Fig. 5). When plasma potencies against clade-mismatched viruses from early and late infections were compared, late viruses were more sensitive than early viruses (P = 6.4 × 10−10 by paired, two-sided Wilcoxon test). This effect was consistently highly significant regardless of which data representation was involved (every paired, two-sided Wilcoxon test yielded P values of ≤7.1 × 10−7, among the 16 comparisons described in the paragraph above, though only a representative example is shown in Fig. 5). Moreover, it remained significant even when we excluded the 11 most potently neutralizing clade C plasma samples (P = 5.2 × 10−8), the highest paired values shown in Fig. 5, which could be considered outliers. In contrast, we were unable to detect a difference in sensitivity between early and late viruses assayed against clade-matched plasma samples. This effect was not confirmed in the logistic regression analysis described below, though a tendency for late viruses to be more sensitive to neutralization was again evident. Notably, no comparison provided statistically significant support that early viruses were more sensitive to neutralization than late viruses.
FIG 5.
Virus clade interacted with infection stage effects on plasma potency. Among clade-mismatched viruses, plasma potency (geometric mean ID50) was significantly greater against viruses from late (Fiebig V through chronic) than early (Fiebig I-IV) infections (paired, two-sided Wilcoxon test, P = 6.4 × 10−10, left). However, among clade-matched viruses, no evidence was found for differences in neutralization between early and late infections (paired, two-sided Wilcoxon test, P = 0.33, right). Low-titer plasma and censored values were excluded. Colored lines connect pairs of plasma samples. Thick black lines indicate median ID50s. Thin black lines indicate 25th and 75th percentiles.
Influence of hypervariable regions on virus susceptibility and plasma potency.
We examined how neutralization titers vary with length, net charge, and the number of putative N-linked glycosylation sites in plasma and virus Env V1, V2, V4, and V5 hypervariable regions. The hypervariable regions are embedded within the variable loops of gp120. The hypervariable regions mutate very rapidly, evolve in a manner dominated by insertions and deletions, and carry different numbers and locations of potential N-linked glycosylation sites and charged residues. The boundaries between hypervariable regions and regions that are more readily aligned in Env are indicated in the Los Alamos HIV database available at http://hiv.lanl.gov/content/sequence/HIV/MAP/annotation.html. We also combined V1 and V2 (V1V2) hypervariable regions for analyses, because they are spatially juxtaposed and surround regions known to contain critical epitopes for PG9-like bnAbs (82, 83).
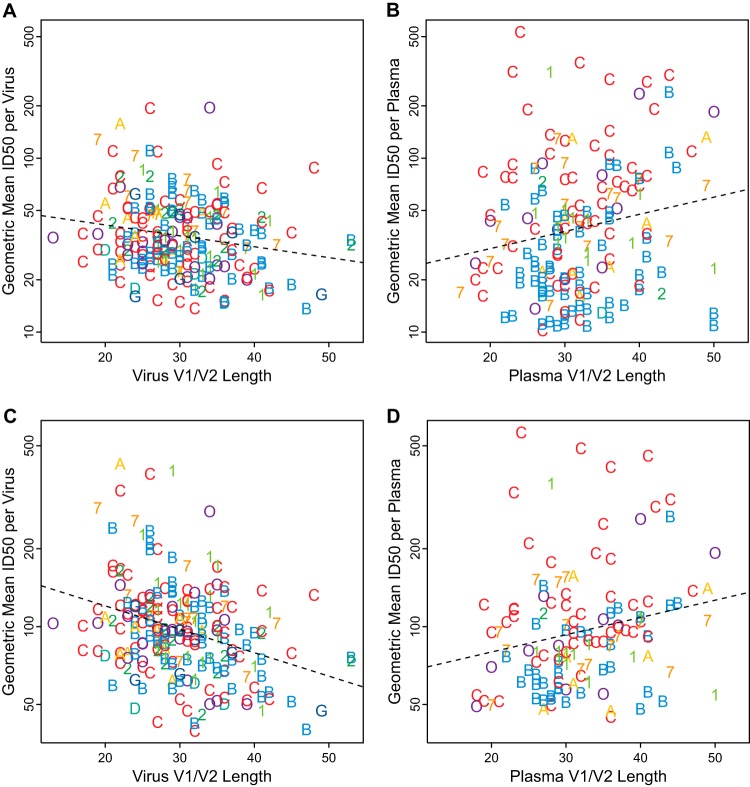
Virus V1V2 lengths were negatively correlated with geometric mean neutralization ID50 per virus (Fig. 6A; Table 2) such that viruses with longer V1V2 hypervariable regions were generally less sensitive to neutralization than viruses with shorter loops. This outcome remained significant whether or not the low-titer plasma samples were excluded and whether or not the subset of only reactive results was used (Fig. 6C). Lower levels of sensitivity were also associated with lower net negative V1V2 charge and greater numbers of potential N-linked glycosylation sites in the hypervariable regions of V1V2 (Table 2). Statistically significant associations were also found when V1 and V2 were considered independently, though these were not as profound as when combined. We were unable to detect associations between viral sensitivity determined by ID50s and V4 or V5 hypervariable region characteristics across the full data set (Table 2).
FIG 6.
Short V1V2 loops were associated with increased virus sensitivity and reduced plasma potency. (A and B) Negative association between virus V1V2 lengths and geometric mean ID50 per virus (Kendall's τ = −0.127, P = 0.006, n = 219) (A) contrasts with a positive association between plasma V1V2 lengths and geometric mean ID50 per plasma (τ = 0.128, P = 0.0156, n = 170) (B). (C and D) Similarly, with low-titer plasma and nonreactive (censored) ID50 assay results excluded, the negative association for virus Envs (τ = −0.213, P = 4.44 × 10−6, n = 219) (C) and positive association for plasma Envs (τ = 0.172, P = 0.00462, n = 129) (D) remain. Symbols indicate the clade of each virus (left) or plasma sample (right), using the same colors for clades as in previous figures (“O” indicates other CRF/URF). Dashed lines were fit by least-squares regression.
TABLE 2.
Correlation of virus Env hypervariable loop properties (length, net charge, and number of potential N-linked glycosylation sites) with geometric mean plasma ID50 per virus (n = 219 viruses)
| Property | All dataa |
Positive valuesb |
No-low-titer plasma samplesc |
Positive and no-low-titer plasma samplesd |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| τe | pf | qg | τ | p | q | τ | p | q | τ | p | q | |
| V1 lengthh | −0.08 | 0.101 | 0.213 | −0.14 | 0.003 | 0.026** | −0.09 | 0.049 | 0.129 | −0.13 | 0.004 | 0.031** |
| V1 netchgi | 0.19 | 2e-04 | 0.003*** | 0.13 | 0.011 | 0.048** | 0.20 | 1e-04 | 0.002*** | 0.13 | 0.007 | 0.036** |
| V1 glycosj | −0.01 | 0.800 | 0.739 | −0.06 | 0.274 | 0.404 | −0.02 | 0.704 | 0.699 | −0.05 | 0.351 | 0.468 |
| V2 length | −0.06 | 0.182 | 0.311 | −0.13 | 0.006 | 0.034** | −0.06 | 0.174 | 0.305 | −0.13 | 0.007 | 0.036** |
| V2 netchg | 0.03 | 0.555 | 0.619 | 0.10 | 0.042 | 0.119 | 0.04 | 0.434 | 0.545 | 0.10 | 0.049 | 0.129 |
| V2 glycos | −0.05 | 0.344 | 0.464 | −0.15 | 0.004 | 0.031** | −0.05 | 0.314 | 0.440 | −0.14 | 0.007 | 0.036** |
| V1V2 length | −0.13 | 0.006 | 0.034** | −0.22 | 3e-06 | 8e-05*** | −0.14 | 0.002 | 0.025** | −0.21 | 4e-06 | 1e-04*** |
| V1V2 netchg | 0.13 | 0.006 | 0.034** | 0.13 | 0.006 | 0.034** | 0.14 | 0.004 | 0.031** | 0.14 | 0.004 | 0.031** |
| V1V2 glycos | −0.05 | 0.308 | 0.438 | −0.15 | 0.003 | 0.029** | −0.06 | 0.229 | 0.363 | −0.14 | 0.007 | 0.036** |
| V4 length | 0.00 | 0.999 | 0.782 | −0.07 | 0.160 | 0.291 | −0.01 | 0.909 | 0.763 | −0.05 | 0.261 | 0.394 |
| V4 netchg | 0.00 | 0.962 | 0.770 | 0.04 | 0.461 | 0.555 | 0.01 | 0.840 | 0.743 | 0.04 | 0.452 | 0.552 |
| V4 glycos | 0.01 | 0.785 | 0.736 | −0.05 | 0.322 | 0.444 | 0.01 | 0.820 | 0.741 | −0.04 | 0.412 | 0.528 |
| V5 length | −0.04 | 0.463 | 0.555 | −0.10 | 0.047 | 0.129 | −0.04 | 0.393 | 0.508 | −0.09 | 0.070 | 0.166 |
| V5 netchg | −0.04 | 0.458 | 0.555 | −0.03 | 0.537 | 0.606 | −0.04 | 0.426 | 0.539 | −0.04 | 0.482 | 0.566 |
Censored values taken as given, i.e., placeholder constants of 10 for ID50s below 20.
Censored values treated as missing, i.e., only positive assay results were used.
Plasmas with geometric mean ID50 below 20 were excluded from these comparisons.
Plasmas with low geometric mean ID50s and censored values were excluded from these tests.
Kendall's τ as computed by the eponymous R package.
Two-sided P value on the null hypothesis of no correlation.
False-discovery rates computed from 360 P values by the q-value package. Significance levels: *,* q < 0.05; ***, q < 0.01.
Hypervariable loop boundaries are defined in Text S2 in the supplemental material and are not simply cysteine-cysteine.
netchg, net charge, i.e., number of K, H, and R sites minus the number of D and E sites.
glycos, number of potential N-linked glycosylation sites following the Nx[ST] motif, with x not P.
Significant associations were found also for geometric mean ID80 per virus with V1 and V2 properties, both separately and combined (see Table S1 in the supplemental material). Some support for negative associations between virus ID80s with V5 loop length and number of glycosylation sites was found when nonreactive plasma samples were excluded, though this outcome was not supported by ID50s with virus sensitivity computed in aggregate across plasma samples (see Table S1 in the supplemental material).
Interestingly, the direction of neutralization-response associations was reversed when Env sequences in the plasma samples were considered. Positive associations were found between the geometric mean neutralization ID50 per plasma and the V1V2 length of plasma Envs (Fig. 6B and D), regardless of whether or not the low-titer plasma samples and nonreactive assay results were included (Table 3). That is, plasma samples from individuals with Envs that had longer V1V2 loops tended to be more potent than plasma samples from which the Envs contained shorter V1V2 loops. Because long V1V2 loops were associated with resistance in the pseudotyped Envs (Fig. 6A and C), this raises the hypothesis that viruses from individuals with more-potent nAbs carried Envs with greater overall resistance as a consequence of immune escape from effective antibodies and that this resistance was at least in part manifested by longer V1V2 loops. This agrees with our finding that Envs with longer loops tend to be more resistant to plasma neutralization. We found no robust, statistically significant support for associations between average neutralization response and plasma Env V4 or V5 properties across all data (Table 3; see also Table S1 in the supplemental material).
TABLE 3.
Correlation of plasma Env hypervariable loop properties (length, net charge, and number of potential N-linked glycosylation sites) with geometric mean ID50 per plasma sample
| Property | All dataa |
Positive valuesb |
No-low-titer plasma samplesc |
Positive and no-low-titer plasma samplesd |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| τe | pf | qg | τ | p | q | τ | p | q | τ | p | q | |
| V1 lengthh | 0.08 | 0.144 | 0.273 | 0.06 | 0.258 | 0.394 | 0.15 | 0.017 | 0.062* | 0.13 | 0.027 | 0.088* |
| V1 netchgi | −0.02 | 0.696 | 0.696 | 0.01 | 0.804 | 0.739 | 0.00 | 0.961 | 0.770 | 0.01 | 0.827 | 0.742 |
| V1 glycosj | 0.09 | 0.118 | 0.240 | 0.11 | 0.064 | 0.155 | 0.18 | 0.006 | 0.034** | 0.18 | 0.005 | 0.031** |
| V2 length | 0.10 | 0.068 | 0.163 | 0.10 | 0.068 | 0.163 | 0.04 | 0.561 | 0.623 | 0.03 | 0.609 | 0.646 |
| V2 netchg | 0.15 | 0.011 | 0.048** | 0.12 | 0.038 | 0.111 | 0.16 | 0.012 | 0.051* | 0.13 | 0.053 | 0.136 |
| V2 glycos | 0.01 | 0.931 | 0.763 | −0.01 | 0.898 | 0.763 | −0.03 | 0.691 | 0.694 | −0.04 | 0.569 | 0.627 |
| V1V2 length | 0.13 | 0.016 | 0.061* | 0.13 | 0.017 | 0.062* | 0.17 | 0.004 | 0.031** | 0.17 | 0.005 | 0.031** |
| V1V2 netchg | 0.06 | 0.259 | 0.394 | 0.07 | 0.180 | 0.310 | 0.10 | 0.107 | 0.223 | 0.10 | 0.135 | 0.261 |
| V1V2 glycos | 0.09 | 0.110 | 0.225 | 0.09 | 0.108 | 0.223 | 0.16 | 0.016 | 0.061* | 0.14 | 0.030 | 0.093* |
| V4 length | −0.03 | 0.591 | 0.633 | −0.07 | 0.205 | 0.336 | −0.04 | 0.486 | 0.568 | −0.07 | 0.228 | 0.363 |
| V4 netchg | −0.01 | 0.835 | 0.742 | 0.02 | 0.747 | 0.729 | 0.00 | 0.987 | 0.775 | 0.03 | 0.658 | 0.682 |
| V4 glycos | −0.07 | 0.265 | 0.395 | −0.08 | 0.165 | 0.294 | −0.01 | 0.925 | 0.763 | −0.01 | 0.852 | 0.746 |
| V5 length | 0.01 | 0.876 | 0.758 | −0.01 | 0.906 | 0.763 | 0.08 | 0.227 | 0.363 | 0.05 | 0.475 | 0.566 |
| V5 netchg | −0.01 | 0.814 | 0.739 | −0.01 | 0.927 | 0.763 | 0.03 | 0.650 | 0.679 | 0.01 | 0.920 | 0.763 |
Censored values taken as given, i.e., placeholder constants of 10 for ID50s below 20 (n = 170).
Censored values treated as missing, i.e., only positive assay results were used (n = 170).
Plasma samples with geometric mean ID50 below 20 were excluded from these comparisons (n = 129).
Both plasma samples with low geometric mean ID50s and censored values were excluded (n = 129).
Kendall's τ as computed by the eponymous R package.
Two-sided P value for the null hypothesis of no correlation.
False-discovery rates computed from 360 P values by the q-value package. Significance levels: *, q < 0.10; **, q < 0.05.
Hypervariable loop boundaries are defined in Text S2 in the supplemental material and are not simply cysteine-cysteine.
Net charge, i.e., number of K, H, and R sites minus the number of D and E sites.
Number of potential N-linked glycosylation sites following the Nx[ST] motif, with x not P.
Anomalous profile among Thai plasma samples.
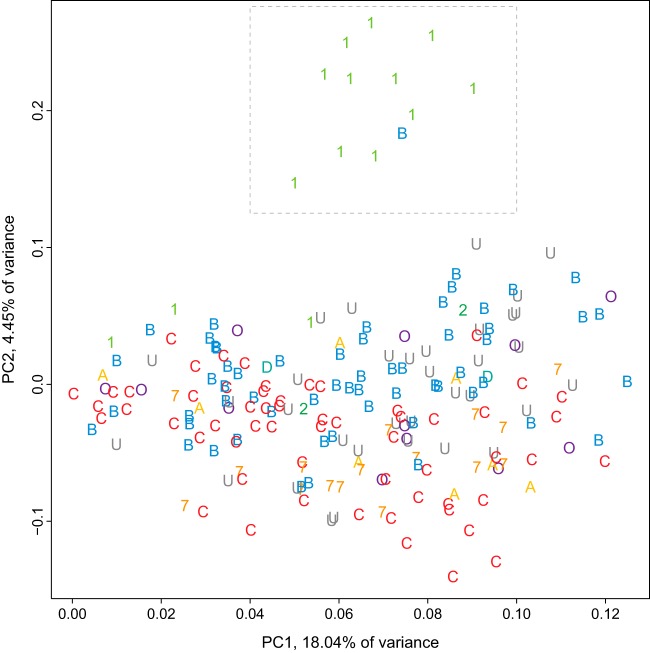
Plasma samples from Thailand, primarily from CRF01 infections but also from two B-clade infections, shared a distinctive neutralization profile relative to the other plasma samples. Principal components analysis (PCA) clustered a B-clade plasma with 11 CRF01 plasma samples, all from Thailand (Fig. 7). Similarly, hierarchical clustering of the ID50 values formed two distinct groupings of plasma samples from Thailand, one group comprised of a B-clade plasma together with 10 CRF01 plasma samples and another bootstrap-supported group of a B-clade plasma with two from CRF01 (see Fig. S2 in the supplemental material). Hierarchical clustering of neutralization values joined 14 of 15 plasma samples from Thailand together into one cluster with less-robust bootstrap support (not shown, but see Fig. 1 in reference 58). While other serological clustering patterns are evident in the heatmaps shown in Fig. 2 and in Fig. S2 in the supplemental material, the Thai B and CRF01 plasma samples had a particularly distinctive pattern. Of note, none of the plasma samples from Thailand had been screened for enhanced neutralization activity.
FIG 7.
Clustering plasma samples by neutralization outcomes with principal components analysis (PCA) shows Thai plasma grouping. The boxed region identifies 12 Thai plasma samples with distinct, shared profiles, including T500108_503963 from a B-clade infection clustered with CRF01 plasma samples (T500204_502281, T500207_503006, T500104_276248, T500105_293735, T500206_614109, T500208_504258, T500105_500617, T500207_509989, T500108_503963, T500107_535902, T500207_502102, and T500106_501602). Binary-transformed ID50s (1 for above and 0 for below the TZM-bl assay limit of detection) per plasma are projected onto orthogonal axes that together capture 22.49% of the total variance. This rendering gives a simplified summary of plasma neutralization profiles on uncorrelated axes, which include as much of the overall variation as possible, so plasma samples with similar profiles appear close together. Symbols indicate plasma clade (“O” for other CRFs and URFs, “U” for unknown due to inability to amplify from the plasma). PCA of neutralization data with log rank transformations yielded similar prominent groupings of the Thai plasma samples (not shown).
While diverse Env-pseudotyped viruses were susceptible to neutralization by these plasma samples, k-means clustering (k = 2) of 219 virus ID50s for the 10 CRF01 and 1 B-clade plasma from Thailand yielded virus groups with high (n = 86) or low (n = 107) susceptibility supported by over 70% of bootstrap replicates (26 viruses were not stably assigned to either group). CRF01 viruses were enriched in the susceptible group (15 CRF01s versus 71 non-CRF01s) relative to the group resistant (3 CRF01s versus 104 non-CRF01s) to these plasma samples from Thailand (Fisher's exact test, P = 0.00007, odds ratio [OR] = 0.14, 95% confidence interval [CI] = 0.025 to 0.51). Given this, we hypothesized that the CRF01 viruses may be exposing a distinctive CRF01 clade-specific epitope resulting in this shared serological response and that the two Thai donors with this serotype that had a B-clade Env-infecting sequence might have been dually infected with undetected CRF01 that could be contributing to this serotype.
To seek evidence of mixed CRF01/B clade coinfections in these two cases, approximately 30 (range, 29 to 32) additional sequences were obtained from each plasma sample. Only B clade sequences were found. If dual-clade infections were present, they were rare in both plasma samples. Additional analysis with the recombination identification program (RIP) did not show significant support for between-clade recombination among any of the B-clade sequences (not shown). Phylogenetic analysis of the full-length env sequences (see Fig. S3 in the supplemental material) showed no evidence for cross-clade coinfection among sequences from the CRF01 infection or either of the B-clade infections. Aside from the possibility of transient or rare CRF01 superinfecting subpopulations in both B-clade plasma samples, which we could not eliminate, this suggests that the shared serological pattern may not be clade related, as it is found in both CRF01 and B-clade infections among plasma samples only from Thailand. Rather, this finding might be explained by a host-driven serological response, possibly due to host genetics causing shared patterns of cross-reactive responses to similar immunogens, or by preexisting immunity that has a shared regional aspect (84).
Corroboration of major findings by mixed-effects logistic regression.
Analyzing average neutralization responses across plasma samples or viruses with different characteristics identified the correlations with neutralization sensitivity described above. Here we used mixed-effects logistic regression to evaluate whether the significant outcomes could be corroborated by an alternative analysis method that can represent variation due to random effects; in these models, we treated the plasma and the virus as random variables. Because 19,169 of 44,758 (42.8%) ID50 measurements were below the level of detection, we used a logistic model that modeled neutralization outcome as a binary variable, based on whether or not the neutralization score was above the median ID50 for the full data set. These analyses also allowed us to explore the relative impacts of different variables on neutralization and to test for possible interactions between them. A complete description appears in Text S2 in the supplemental material; the main findings are summarized here.
We considered several models to accommodate nuances of the experimental design. In the first model, we defined four match levels based on degrees of Env similarity between viruses and plasma, in accordance with what is shown in Fig. 4B. Level 1 referred to CRF07 plasma and CRF07 virus, which were the most similar, level 2 to CRF01 plasma and CRF01 virus, level 3 to other within-clade neutralization scores (including C plasma against CRF07 virus and vice versa), and level 4 to between-clade neutralization. We also included in the model the V1V2 lengths of plasma and virus Envs, the stage of the infection from which the virus was sampled (early or late), and whether or not the plasma sample was screened (see Table S2 in the supplemental material). The most highly significant effect was for matching CRF01 plasma with virus (P = 1.2 × 10−7), followed by the impact of screening (P = 0.002). Matches between CRF07 plasma and virus were still significant (P = 0.013), and the most negative association was the log-odds ratio seen for between-clade responses. In this model, we found no statistical support for an interaction between the stage of the virus and the match level, despite the result shown in Fig. 5, in which geometric mean ID50s per plasma differed significantly only against clade-mismatched (level 4) viruses when computed separately for early and late viruses. However, we did find that late viruses were significantly more sensitive to neutralization than isolates from early infections (P = 0.015).
Consistent with the results shown in Fig. 6, longer total V1V2 loops were associated with more-resistant viruses (P = 0.02), suggesting that insertions in these regions facilitated acquisition of resistance. Also, Envs with longer V1V2 loops were associated with more potent plasma neutralization activity (P = 0.03), supporting the idea that potent plasma samples had selected for more resistant contemporaneous viruses with longer V1V2 hypervariable regions. Of note, the estimated standard deviations of the random effects for plasma and virus were 1.8 and 0.99, respectively. Thus, the random effect for plasma is about twice that of viruses. Further, the magnitude of the estimated fixed effects was typically smaller (compared with the standard error) than the random effects (see Table S2 in the supplemental material).
We confirmed that these outcomes did not result from the screening effect by implementing a second model that used a simplified representation of clade matching and virus infection stages, together with exclusion of censored values and application of the postscreening criterion described earlier. Again, effects of clade matching, virus stage, plasma Env V1V2 length, and virus V1V2 length were supported by statistical significance (see Table S3 in the supplemental material). The estimated standard deviations for random effects of plasma and virus were 1.2 and 1.0, respectively, suggesting that the greater contribution of plasma random effects in the previous model was influenced by the low-titer ID50s, particularly for low-titer plasma samples that satisfied the postscreening criterion.
In a third model, we considered each of the clades independently, instead of grouping them into sequence similarity-matched levels (see Table S4 in the supplemental material). Each of the clades had statistically significant positive interaction estimates for within-clade responses (Table 4). The relative impact of the within-clade advantage ranked highest for CRF07 and CRF01 and lowest for clade C (Table 4). Close examination of Fig. 3A suggests that the minimal impact of the within-clade advantage for clade C may be explained by the observation that a small subset of plasma samples from individuals with C-clade infections retained potent cross-clade reactivity. Most of the between-clade mismatched plasma and virus combinations had negative interactions, with a few interesting exceptions. Clade C and CRF07 plasma samples and viruses, not surprisingly, had a positive interaction, consistent with CRF07 essentially being clade C in Env, a recent outgrowth of the clade C lineage (Fig. 1). More unexpectedly, CRF02 viruses had a positive interaction with CRF01 plasma samples, and the capacity to neutralize CRF02 viruses relatively potently may be an aspect of the distinctive serological profile associated with Thai viruses. Plasma samples from clade B had a slightly enhanced ability to recognize clade A viruses relative to other clades (see Table S4 in the supplemental material).
TABLE 4.
Estimated log-odds ratios for interactions between virus and plasma cladea
| Virus clade | Plasma sample-infecting strain clade |
||||||
|---|---|---|---|---|---|---|---|
| CRF07 | CRF01 | CRF02 | Clade A | Clade B | Clade C | Clade D | |
| CRF07 | 1.288a | −0.532 | NSb | NS | −0.494 | 0.700 | 0.389c |
| CRF01 | −0.546 | 1.295 | −0.739 | NS | NS | NS | −0.137 |
| CRF02 | −0.473 | 0.877 | 1.184 | NS | −0.304 | −0.363 | 0.680 |
| Clade A | NS | −1.184 | NS | 0.895 | 0.316 | NS | 0.245 |
| Clade B | −0.224 | −0.750 | NS | NS | 0.802 | −0.230 | −0.795 |
| Clade C | 0.742 | −0.680 | NS | NS | −0.219 | 0.430 | 0.087 |
| Clade D | −1.116 | NS | NS | NS | NS | NS | −1.346 |
| Clade G | −0.501 | −0.460 | −0.265 | 0.440 | −0.085 | −0.007 | −0.878 |
Bold values indicate where the clade of the infecting strain of the plasma samples matches that of the test viruses. Only statistically significant interactions are shown. Positive values among within-clade interactions indicate that plasma samples and viruses from the same clade typically increase the proportion of ID50 values above the median value of 28; the value indicates the magnitude of the effect. Most off-diagonal estimates are negative, which indicates that a mismatch between plasma sample and virus clades decreases the proportion of ID50 values above the median ID50. Because the estimated effects for each factor total zero, some estimates are determined by the others; italicized values are implied by the zero-total contrast and do not have an estimated standard deviation or P value.
NS, not significant.
Modeling neutralization outcomes as row/column effects.
It is well known that some viruses (e.g., tier 1 viruses) are highly susceptible to neutralization by HIV-1-positive plasma samples, while others (e.g., tier 2 viruses) are generally more resistant (4, 85). It is also well known that some plasma samples are generally more potent across a range of HIV-1 viruses, while other plasma samples are generally less potent (16, 17). What is not well known is the degree to which the variation (technically, the variance) of log-transformed titers in a table (i.e., a tabular data matrix, with rows and columns representing viruses and plasma samples, respectively) of checkerboard-style HIV-1 neutralization assay results can be represented simply as the sum of a susceptibility score for each Env and a potency score for each plasma sample. The susceptibility score of an Env is the log-transformed geometric mean titer (GMT) across plasma samples for that Env. Similarly, the plasma potency score is the log-transformed GMT across Envs for that plasma. Residual ID50s, obtained by subtracting approximated from observed ID50s, represent neutralization signal specific to each interaction not captured fully by the approximation based on the expected behavior of the plasma and the virus (see Fig. S4 in the supplemental material).
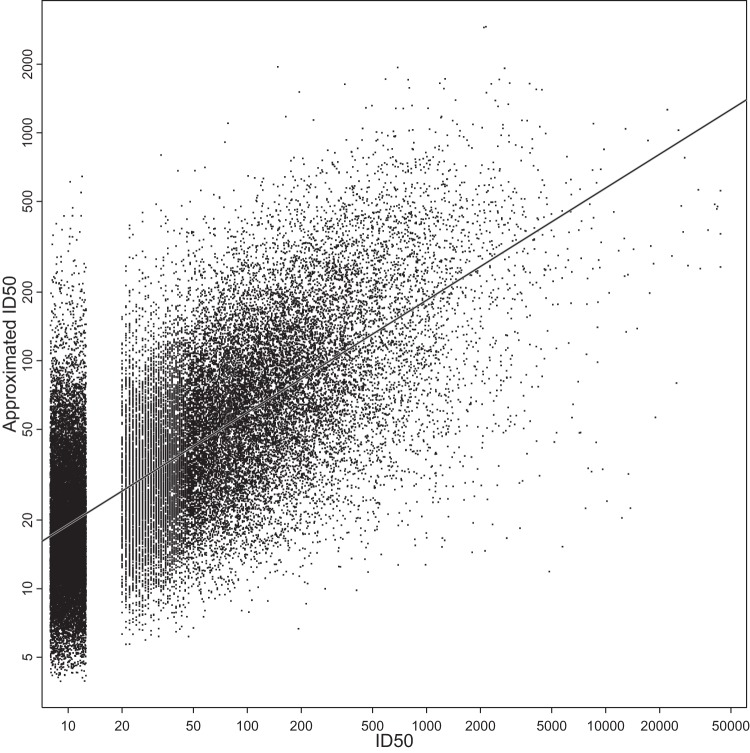
A simple approximation for log-transformed titers based on the sum of the Env susceptibility and plasma potency scores (equation 2) accounted for roughly one-half of the variance in the neutralization data presented herein, as the r2 (Pearson correlation coefficient, squared) between approximated and measured log-ID50 values was 0.49. This was significantly greater than would be expected if the same values were redistributed randomly over the neutralization data table (P < 0.0001 by permutation testing, based on 10,000 randomizations of the data; see Materials and Methods). The randomized data, as expected, gave very low r2 values, between 0.006 and 0.014 (see Table S5 in the supplemental material). In other words, roughly one-half of the variance in the table of log-transformed titers was accounted for by simply adding the overall scores from Env susceptibility and plasma potency (Fig. 8).
FIG 8.
Approximated neutralization ID50s were correlated with observed ID50 values and explained one-half of the observed variation. ID50s were approximated as geometric mean ID50 per plasma sample times geometric mean ID50 per virus, divided by overall geometric mean ID50. The solid line indicates least-squares fit (P < 2.2 × 10−12, Pearson's r = 0.702). The r2 corresponds to 49.3% of all variance. To reduce overplotting, jitter was applied when rendering the censored ID50s (<20, 42.8% of all ID50s).
We confirmed this unexpected observation with other, previously published checkerboard-style HIV-1 neutralization data sets. In each case, the “row/column approximation” accounted for roughly one-half to two-thirds of the variance, depending on the neutralization table and the extent of phylogenetic diversity represented among assays performed. Analysis of neutralization data based on plasma samples from clade C infections in India compared to Env-pseudotyped viruses of diverse clades, including the C isolates from India used in the study by Kulkarni et al. (54), produced an r2 of 0.57. For the diverse multiclade neutralization table used by Seaman et al. (4), r2 was 0.46. For B-clade plasma samples tested against a multiclade panel in the neutralization table used by Doria-Rose et al. (86), r2 was 0.66, i.e., two-thirds of the variance of log-transformed titers is accounted for by simply adding susceptibility and potency scores. All of these r2 scores were significantly greater than expected from random data by permutation testing (P < 0.0001; see Table S5 in the supplemental material).
Thus, the results for the row/column effect based on the current neutralization data were confirmed with independent neutralization data and demonstrate that specific interactions between Envs and constituent antibodies of plasma are not the exclusive dominating factor determining neutralization titers. Individual properties of an Env, as summarized by its susceptibility score, and of antibodies in plasma, as summarized by the potency score, are at least equally, if not more important.
DISCUSSION
This large study permitted an unambiguous resolution of whether there is a within-clade enhancement of neutralization sensitivity and enabled us to detect a dependency of the magnitude of this effect on the age of the HIV-1 regional epidemic. Neutralization potency was generally greater when plasma from a person infected with a given clade was tested against Envs from that same clade, and the within-clade advantage was more pronounced for the more recent, less diverse CRF07 and CRF01 Asian clades than for the older A, B, and C clades (Fig. 1). Thus, the potential benefit of a within-clade regional vaccine is diminishing substantially on the time scale of decades. CRF07 is thought to have originated in Yunnan in 1993 (79, 87, 88) and subsequently spread throughout China to become an epidemiologically significant lineage. The Thai CRF01 epidemic dates to the late 1980s, going from no detected cases in 1986 to over 5,000 cases by 1988 (89, 90). The A, B, and C clades originated much earlier (91), with estimates for the most recent ancestor of the B and C clades in the 1960s and the A clade even earlier (89, 92). The potential impact of the within-clade advantage seen here may be relevant for antibodies in general, not only nAbs. A possible consequence of the diminishing impact with age of the epidemic is that the modest protective effect observed in the RV144 Thai vaccine trial (15), which correlated with antibodies to the V2 and V3 regions of gp120 (14, 93–95), may be difficult to emulate in other populations, where the virus is more diverse, even if the HIV-1 epidemic in the targeted population is dominated by a single clade. Despite the within-clade advantage, most plasma samples had extensive cross-clade neutralizing activity.
A cluster of 12 plasma samples from Thailand had a particularly distinctive serologic pattern, shared by plasma from 11 CRF01 infections and a single B-clade infection (Fig. 7; see also Fig. S3 in the supplemental material). Another small Thai serologic cluster (see Fig. S3) included both one B-clade and two CRF01 plasma samples. Plasma samples from B-clade infections outside Thailand did not share these patterns. One hypothesis to explain the clustering of B and CRF01 plasma samples from Thailand is that subjects with B-clade Env sequences may have been coinfected with CRF01 (96). We sequenced 30 viruses from each of the Thai individuals having B-clade infections and found no traces of CRF01 coinfection; however, CRF01 may have been present at levels too low to detect by this method. Also, transiently detected superinfections have been reported (97, 98). Alternatively, host factors, whether genetic or environmental (e.g., exposure to an HIV-1 cross-reactive epitope from an organism in the Thai communities where the samples were obtained), might explain the shared serological profiles (84).
Our study also permitted an examination of whether transmission selects viruses that are generally more sensitive to heterologous neutralization than viruses isolated in chronic infection. Presumably, transmitted/founder (T/F) viruses undergo selection for transmission and replication fitness prior to the onset of adaptive immunity. A manifestation of this may be the shorter hypervariable loop lengths that tend to be selected at transmission (99–101), which in turn may better expose key neutralization epitopes. Also, selection of mutations associated with increased Env expression levels might enhance both infectivity and neutralization sensitivity (102–104). Chronic viruses, on the other hand, might acquire general features associated with neutralization resistance under continuous selection by autologous immune responses. Previous studies have yielded conflicting results as to whether T/F viruses are slightly more sensitive to neutralization than chronic viruses (99, 105, 106). Our results do not support the notion that T/F viruses are more sensitive to neutralization. Indeed, we found early viruses to be less sensitive than late viruses when assayed with clade-mismatched plasma samples; however, no difference was found when the viruses were assayed with clade-matched plasma samples. We do not have a clear biological explanation for these statistically supported observations, which were contrary to our expectations. The collective genetic features required for transmission fitness and initial replication in a new host may have a negative impact on neutralization in ways not yet understood, despite the fact that early variants have features that one might expect a priori to favor neutralization sensitivity.
We also explored the relationship between neutralization and gp120 hypervariable regions. While the characteristics of these regions have not been considered in the immunogen selection process of earlier vaccine efficacy trials, they may be important for both vaccine design and the interpretation of vaccine-elicited antibody responses. The evolution of hypervariable regions is dominated by insertion and deletion events (69), and they are under intense selective pressure in HIV-infected individuals (35, 69, 107). Consequently, they are subject to extreme length variation (e.g., the V1 hypervariable region varies between 5 and 42 amino acids among the Envs in this study). Given this, they are not readily aligned, and they violate model assumptions inherent in most phylogenetic methods. The V1, V2, V4, and V5 loops contain hypervariable regions. The V3 loop, while variable, does not contain a hypervariable region.
Among hypervariable regions, the V1V2 loops, taken together, were most influential for both neutralization susceptibility of Env-pseudotyped viruses and overall neutralization activity in plasma samples. Env-pseudotyped viruses with greater resistance to plasma neutralization had V1V2 loops that were longer, with more N-linked glycosylation sites and more negatively charged residues, suggesting that these features facilitate immune evasion. These same features, when present in plasma Envs, were associated with broader, more potent plasma neutralization activity, suggesting that V1V2-mediated escape from autologous nAbs helped drive the response to greater neutralization breadth (108). No associations were found with V4 characteristics, and only a moderately significant association was seen between greater neutralization sensitivity and viruses with shorter and less-glycosylated V5 regions. As part of the contact region for CD4 binding site neutralizing antibodies (30), the V5 hypervariable region is of particular interest. These observations suggest that V1V2 plays a more dominant role than V4 and V5 in shaping overall neutralization susceptibility of viruses and nAb responses in plasma. Specific bnAbs may, however, have distinctive patterns of variable loop characteristic sensitivity that were not evident based on this study of polyvalent plasma samples.
Interestingly, a general neutralization susceptibility score assigned to each Env based on its average sensitivity, combined with the average potency of the plasma, approximated with surprisingly good accuracy the individual neutralization titers in our data. We had not anticipated such a finding, given that a priori the most important factor that influences the level of neutralization against specific sets of Envs would be differential exposure and conservation of the particular epitopes recognized by the dominant antibody responses in the plasma samples. Our findings may be related to recent independent work linking the general neutralization susceptibility of an Env to its physical properties (109). The intrinsic Env reactivity (ER), defined as the degree of neutralization and binding seen with soluble CD4, when combined with an antibody perturbation factor (PF), which characterizes the general neutralization potency of a nAb in terms of certain physical properties, was shown to approximate the actual neutralization potency of a particular HIV-1-nAb combination (109, 110). How our computationally defined susceptibility and potency scores for Envs and plasma samples relate to the physically defined ER and PF characteristics of Envs and nAbs, respectively, is under investigation.
The results presented here may help to inform vaccine design in several ways. In particular, the impact of V1V2 attributes should be carefully considered. Future studies could explore whether multivalent vaccine candidates that comprise a range of V1V2 characteristics, including viruses with resistance-associated loop characteristics, might be advantageous for driving neutralization breadth. Alternatively, it may be best to optimize V1V2 for sensitivity, as this might improve access to key epitopes. The proximity of these hypervariable regions to PG9-like broadly neutralizing epitopes (82, 83) and to an epitope of a nonneutralizing antibody response associated with a reduced risk of infection in RV144 (14, 93–95) emphasizes their importance. The fact that transmission events tend to select for short hypervariable regions (99–101) provides an additional reason to emphasize short loops in vaccine strains, so that they better match their primary target. We note that the V1V2 features identified here did not impart a highly neutralization-sensitive tier 1 phenotype on any of the viruses studied. Rather, these features influenced a spectrum of neutralization sensitivities within the boundaries of the tier 2 phenotype that is common for most circulating strains. A short V5 domain with few glycosylation sites was also associated with Env neutralization, but only when using ID80s as the measure of responses. Because CD4 and CD4 binding site bnAbs make direct contact in the region around the hypervariable part of V5 (30), it may also be prudent to choose vaccine immunogens with minimal V5 hypervariable stretches.
These data also indicate that including Env immunogens that match the genetic subtype of the major circulating variants in the region where the vaccine will be given may afford additional advantages for optimal within-clade neutralization. However, the high prevalence of cross-clade neutralization seen here suggests that clade-mismatched Envs are also useful for generating neutralization responses, though the average responses are reduced. Thus, the idea of a more global vaccine need not be dismissed, particularly if vaccine antigens are designed specifically to enhance cross-reactive neutralization responses through inclusion of polyvalent mixtures or by focusing the immune response on the most highly conserved epitopes.
The results presented here also have implications for assessments and interpretations of vaccine-elicited nAb responses. Appropriate clade representatives and realistic distributions of the intrinsic reactivity of Env reagents both might be important for selection of assay reference strains that best represent the target population. In this regard, a new global panel of reference strains was shown to be comparable to clade-specific panels for predicting clade-specific nAb responses in HIV-1 infection (58). However, it is not yet known how well this panel will predict vaccine-elicited clade responses. The small but statistically significant advantage for clade-matched neutralization seen here suggests that clade-matched reference strains may be an important adjunct to the global panel for optimal detection of neutralization potency and breadth. The age of the epidemic might be another important consideration for reference strain selection, especially for the more recent epidemics in Thailand and China, where inclusion of contemporary regional viruses may best reflect the potential value of vaccine-elicited nAbs. Finally, the interesting and potentially host-driven serotype found among Thai samples suggests that host factors and examination of preexisting immune cross-reactive memory cells may be an important determinant, which might sometimes impact patterns of immune response.
In concert with the finding that nearly all infected subjects have the potential to develop antibodies that neutralize a substantial proportion of heterologous viruses rather than an exclusive few (17), the prospect for vaccine immunogens to stimulate bnAbs against diverse HIV-1 Envs remains a promising and reasonable objective.
Supplementary Material
ACKNOWLEDGMENTS
We thank George Shaw, Julie Overbaugh, Dennis Ellenberger, Salvatore Butera, Kunxue Hong, Yiming Shao, Eric Sanders-Buell, Jesus Salazar, Denise Kothe, Xiping Wei, Cynthia Derdeyn, Gama Bandawe, Ana Revilla, Elena Delgado, Yolanda Vega, Amit Kumar, Li-Hua Ping, Jeffrey Anderson, Elizabeth Russell, and Ruwayhida Thebus for additional env clones and sequences. For plasma samples, we thank Lynn Morris, Guy de Bruyn, Salim Abdool Karim, and Koleka Mlisana (South Africa), Leonard Maboko and Michael Hoelscher (Mbeya Medical Research Programme, Tanzania), Ramesh Paranjape and Pachamuthu Balakrishnan (India), Yiming Shao, Kunxue Hong, Hao Wu, Ning Li, Linqi Zhang, and Hong Shang (China), Aine McKnight (Europe), Ruengpung Sutthent and Parichart Permpikul (Thailand), Lindsey Baden and Ray Dolin (United States), CHAVI (South Africa, Malawi, and Senegal), CAPRISA (South Africa), IAVI (South Africa, Kenya, Uganda, and Rwanda), HVTN (South Africa), HPTN (South Africa), Southern African National Blood Services (South Africa), HISIS (Tanzania), Partners in AIDS (USA), US MHRP (Thailand, Uganda, and Tanzania), Malawi and HIV-1 in Pregnancy Program (Malawi), and the Zambia-Emory HIV Research Project (Zambia). We thank Nicole Doria-Rose and Mike Seaman for access to and permission to present analysis of results of their published neutralization data, Hillel Haim and Joe Sodroski for insightful discussions, Thomas Donn, Mark Bollenbeck, and Sarah Ramsay for assay data management, and Marcella Sarzotti-Kelsoe for quality assurance oversight.
This work was funded by grants from the Bill & Melinda Gates Foundation, which established the Comprehensive Antibody Vaccine Immune Monitoring Consortium as part of the Collaboration for AIDS Vaccine Discovery (grants 38619 and 1032144).
F.M. is recently retired from the Bill & Melinda Gates Foundation, Seattle, WA, USA.
The opinions expressed herein are those of the authors and do not purport to reflect the official views of the U.S. Department of the Army or Department of Defense.
Footnotes
Published ahead of print 20 August 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.01705-14
REFERENCES
- 1.Wyatt R, Sodroski J. 1998. The HIV-1 envelope glycoproteins: fusogens, antigens, and immunogens. Science 280:1884–1888. 10.1126/science.280.5371.1884. [DOI] [PubMed] [Google Scholar]
- 2.Hemelaar J. 2013. Implications of HIV diversity for the HIV-1 pandemic. J. Infect. 66:391–400. 10.1016/j.jinf.2012.10.026. [DOI] [PubMed] [Google Scholar]
- 3.Hemelaar J, Gouws E, Ghys PD, Osmanov S. 2011. Global trends in molecular epidemiology of HIV-1 during 2000-2007. AIDS 25:679–689. 10.1097/QAD.0b013e328342ff93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Seaman MS, Janes H, Hawkins N, Grandpre LE, Devoy C, Giri A, Coffey RT, Harris L, Wood B, Daniels MG, Bhattacharya T, Lapedes A, Polonis VR, McCutchan FE, Gilbert PB, Self SG, Korber BT, Montefiori DC, Mascola JR. 2010. Tiered categorization of a diverse panel of HIV-1 Env pseudoviruses for assessments of neutralizing antibodies. J. Virol. 84:1439–1452. 10.1128/JVI.02108-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Montefiori DC, Karnasuta C, Huang Y, Ahmed H, Gilbert P, de Souza MS, McLinden R, Tovanabutra S, Laurence-Chenine A, Sanders-Buell E, Moody MA, Bonsignori M, Ochsenbauer C, Kappes J, Tang H, Greene K, Gao H, LaBranche CC, Andrews C, Polonis VR, Rerks-Ngarm S, Pitisuttithum P, Nitayaphan S, Kaewkungwal J, Self SG, Berman PW, Francis D, Sinangil F, Lee C, Tartaglia J, Robb ML, Haynes BF, Michael NL, Kim JH. 2012. Magnitude and breadth of the neutralizing antibody response in the RV144 and Vax003 HIV-1 vaccine efficacy trials. J. Infect. Dis. 206:431–441. 10.1093/infdis/jis367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gilbert P, Wang M, Wrin T, Petropoulos C, Gurwith M, Sinangil F, D'Souza P, Rodriguez-Chavez IR, DeCamp A, Giganti M, Berman PW, Self SG, Montefiori DC. 2010. Magnitude and breadth of a nonprotective neutralizing antibody response in an efficacy trial of a candidate HIV-1 gp120 vaccine. J. Infect. Dis. 202:595–605. 10.1086/654816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Spearman P, Lally MA, Elizaga M, Montefiori D, Tomaras GD, McElrath MJ, Hural J, De Rosa SC, Sato A, Huang Y, Frey SE, Sato P, Donnelly J, Barnett S, Corey LJ, HIV Vaccine Trials Network of NIAID 2011. A trimeric, V2-deleted HIV-1 envelope glycoprotein vaccine elicits potent neutralizing antibodies but limited breadth of neutralization in human volunteers. J. Infect. Dis. 203:1165–1173. 10.1093/infdis/jiq175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bures R, Gaitan A, Zhu T, Graziosi C, McGrath KM, Tartaglia J, Caudrelier El Habib R, Klein M, Lazzarin A, Stablein DM, Deers M, Corey L, Greenberg ML, Schwartz DH, Montefiori DC. 2000. Immunization with recombinant canarypox vectors expressing membrane-anchored gp120 followed by gp160 protein boosting fails to generate antibodies that neutralize R5 primary isolates of human immunodeficiency virus type 1. AIDS Res. Hum. Retroviruses 16:2019–2035. 10.1089/088922200750054756. [DOI] [PubMed] [Google Scholar]
- 9.Mascola JR, Snyder SW, Weislow OS, Belay SM, Belshe RB, Schwartz DH, Clements ML, Dolin R, Graham BS, Gorse GJ, Keefer MC, McElrath MJ, Walker MC, Wagner KF, McNeil JG, McCutchan FE, Burke DS. 1996. Immunization with envelope subunit vaccine products elicits neutralizing antibodies against laboratory-adapted but not primary isolates of human immunodeficiency virus type 1. J. Infect. Dis. 173:340–348. 10.1093/infdis/173.2.340. [DOI] [PubMed] [Google Scholar]
- 10.Belshe RB, Graham BS, Keefer MC, Gorse GJ, Wright P, Dolin R, Matthews T, Weinhold K, Bolognesi DP, Sposto R, Stablein DM, Twaddell T, Berman PW, Gregory T, Izu AE, Walker MC, Fast P. 1994. Neutralizing antibodies to HIV-1 in seronegative volunteers immunized with recombinant gp120 from the MN strain of HIV-1. JAMA 272:475–480. 10.1001/jama.272.6.475. [DOI] [PubMed] [Google Scholar]
- 11.Pitisuttithum P, Gilbert P, Gurwith M, Heyward W, Martin M, van Griensven F, Hu D, Tappero JW, Choopanya K, Bangkok Vaccine Evaluation Group 2006. Randomized, double-blind, placebo-controlled efficacy trial of a bivalent recombinant glycoprotein 120 HIV-1 vaccine among injecting drug users in Bangkok, Thailand. J. Infect. Dis. 194:1661–1671. 10.1086/508748. [DOI] [PubMed] [Google Scholar]
- 12.Flynn NM, Forthal DN, Harro CD, Judson FN, Mayer KH, Para MF, rgp120 HIV Vaccine Study Group 2005. Placebo-controlled phase 3 trial of recombinant glycoprotein 120 vaccine to prevent HIV-1 infection. J. Infect. Dis. 191:654–665. 10.1086/428404. [DOI] [PubMed] [Google Scholar]
- 13.Hammer SM, Sobieszczyk ME, Janes H, Karuna ST, Mulligan MJ, Grove D, Koblin BA, Buchbinder SP, Keefer MC, Tomaras GD, Frahm N, Hural J, Anude C, Graham BS, Enama ME, Adams E, DeJesus E, Novak RM, Frank I, Bentley C, Ramirez S, Fu R, Koup RA, Mascola JR, Nabel GJ, Montefiori DC, Kublin J, McElrath MJ, Corey L, Gilbert PB. 2013. Efficacy trial of a DNA/rAd5 HIV-1 preventive vaccine. N. Engl. J. Med. 369:2083–2092. 10.1056/NEJMoa1310566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haynes BF, Gilbert PB, McElrath MJ, Zolla-Pazner S, Tomaras GD, Alam SM, Evans DT, Montefiori DC, Karnasuta C, Sutthent R, Liao H-X, DeVico AL, Lewis GK, Williams C, Pinter A, Fong Y, Janes H, DeCamp A, Huang Y, Rao M, Billings E, Karasavvas N, Robb ML, Ngauy V, de Souza MS, Paris R, Ferrari G, Bailer RT, Soderberg KA, Andrews C, Berman PW, Frahm N, De Rosa SC, Alpert MD, Yates NL, Shen X, Koup RA, Pitisuttithum P, Kaewkungwal J, Nitayaphan S, Rerks-Ngarm S, Michael NL, Kim JH. 2012. Immune correlates analysis of an HIV-1 vaccine efficacy trial. N. Engl. J. Med. 366:1275–1286. 10.1056/NEJMoa1113425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rerks-Ngarm S, Pitisuttithum P, Nitayaphan S, Kaewkungwal J, Chiu J, Paris R, Premsri N, Namwat C, de Souza M, Adams E, Benenson M, Gurunathan S, Tartaglia J, McNeil JG, Francis DP, Stablein D, Birx DL, Chunsuttiwat S, Khamboonruang C, Thongcharoen P, Robb ML, Michael NL, Kunasol P, Kim JH. 2009. Vaccination with ALVAC and AIDSVAX to prevent HIV-1 infection in Thailand. N. Engl. J. Med. 361:2209–2220. 10.1056/NEJMoa0908492. [DOI] [PubMed] [Google Scholar]
- 16.Stamatatos L, Morris L, Burton DR, Mascola JR. 2009. Neutralizing antibodies generated during natural HIV-1 infection: good news for an HIV-1 vaccine? Nat. Med. 15:866–870. 10.1038/nm.1949. [DOI] [PubMed] [Google Scholar]
- 17.Hraber P, Seaman MS, Bailer RT, Mascola JR, Montefiori DC, Korber BT. 2014. Prevalence of broadly neutralizing antibody responses during chronic HIV-1 infection. AIDS 28:163–169. 10.1097/QAD.0000000000000106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gray ES, Madiga MC, Hermanus T, Moore PL, Wibmer CK, Tumba NL, Werner L, Mlisana K, Sibeko S, Williamson C, Abdool Karim SS, Morris L, the CAPRISA 002 Study Team 2011. The neutralization breadth of HIV-1 develops incrementally over four years and is associated with CD4 T cell decline and high viral load during acute infection. J. Virol. 85:4828–4840. 10.1128/JVI.00198-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tomaras GD, Binley JM, Gray ES, Crooks ET, Osawa K, Moore PL, Tumba N, Tong T, Shen X, Yates NL, Decker J, Wibmer CK, Gao F, Alam SM, Easterbrook P, Abdool-Karim S, Kamanga G, Crump JA, Cohen M, Shaw GM, Mascola JR, Haynes BF, Montefiori DC, Morris L. 2011. Polyclonal B cell responses to conserved neutralization epitopes in a subset of HIV-1-infected individuals. J. Virol. 85:11502–11519. 10.1128/JVI.05363-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Simek MD, Rida W, Priddy FH, Pung P, Carrow E, Laufer DS, Lehrman JK, Boaz M, Tarragona-Fiol T, Miiro G, Birungi J, Pozniak A, McPhee DA, Manigart O, Karita E, Inwoley A, Jaoko W, Dehovitz J, Bekker LG, Pitisuttithum P, Paris R, Walker LM, Poignard P, Wrin T, Fast PE, Burton DR, Koff WC. 2009. Human immunodeficiency virus type 1 elite neutralizers: individuals with broad and potent neutralizing activity identified by using a high-throughput neutralization assay together with an analytical selection algorithm. J. Virol. 83:7337–7348. 10.1128/JVI.00110-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sather DN, Armann J, Ching LK, Mavrantoni A, Sellhorn G, Caldwell Z, Yu X, Wood B, Self S, Kalams S, Stamatatos L. 2009. Factors associated with the development of cross-reactive neutralizing antibodies during human immunodeficiency virus type 1 infection. J. Virol. 83:757–769. 10.1128/JVI.02036-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li Y, Svehla K, Louder MK, Wycuff D, Phogat S, Tang M, Migueles SA, Wu X, Phogat A, Shaw GM, Connors M, Hoxie J, Mascola JR, Wyatt R. 2009. Analysis of neutralization specificities in polyclonal sera derived from human immunodeficiency virus type 1-infected individuals. J. Virol. 83:1045–1059. 10.1128/JVI.01992-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Doria-Rose NA, Klein RM, Manion MM, O'Dell S, Phogat A, Chakrabarti B, Hallahan CW, Migueles SA, Wrammert J, Ahmed R, Nason M, Wyatt RT, Mascola JR, Connors M. 2009. Frequency and phenotype of human immunodeficiency virus envelope-specific B cells from patients with broadly cross-neutralizing antibodies. J. Virol. 83:188–199. 10.1128/JVI.01583-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Binley JM, Lybarger EA, Crooks ET, Seaman MS, Gray E, Davis KL, Decker JM, Wycuff D, Harris L, Hawkins N, Wood B, Nathe C, Richman D, Tomaras GD, Bibollet-Ruche F, Robinson JE, Morris L, Shaw GM, Montefiori DC, Mascola JR. 2008. Profiling the specificity of neutralizing antibodies in a large panel of plasmas from patients chronically infected with human immunodeficiency virus type 1 subtypes B and C. J. Virol. 82:11651–11668. 10.1128/JVI.01762-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Corti D, Lanzavecchia A. 2013. Broadly neutralizing antiviral antibodies. Annu. Rev. Immunol. 31:705–742. 10.1146/annurev-immunol-032712-095916. [DOI] [PubMed] [Google Scholar]
- 26.Klein F, Mouquet H, Dosenovic P, Scheid JF, Scharf L, Nussenzweig MC. 2013. Antibodies in HIV-1 vaccine development and therapy. Science 341:1199–1204. 10.1126/science.1241144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kwong PD, Mascola JR. 2012. Human antibodies that neutralize HIV: identification, structures, and B cell ontogenies. Immunity 37:412–425. 10.1016/j.immuni.2012.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Burton DR, Ahmed R, Barouch DH, Butera ST, Crotty S, Godzik A, Kaufmann DE, McElrath MJ, Nussenzweig MC, Pulendran B, Scanlan CN, Schief WR, Silvestri G, Streeck H, Walker BD, Walker LM, Ward AB, Wilson IA, Wyatt R. 2012. A blueprint for HIV vaccine discovery. Cell Host Microbe 12:396–407. 10.1016/j.chom.2012.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Blattner C, Lee JH, Sliepen K, Derking R, Falkowska E, de la Peña AT, Cupo A, Julien JP, van Gils M, Lee PS, Peng W, Paulson JC, Poignard P, Burton DR, Moore JP, Sanders RW, Wilson IA, Ward AB. 2014. Structural delineation of a quaternary, cleavage-dependent epitope at the gp41-gp120 interface on intact HIV-1 Env trimers. Immunity 40:669–680. 10.1016/j.immuni.2014.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou T, Georgiev I, Wu X, Yang ZY, Dai K, Finzi A, Kwon YD, Scheid JF, Shi W, Xu L, Yang Y, Zhu J, Nussenzweig MC, Sodroski J, Shapiro L, Nabel GJ, Mascola JR, Kwong PD. 2010. Structural basis for broad and potent neutralization of HIV-1 by antibody VRC01. Science 329:811–817. 10.1126/science.1192819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bonsignori M, Hwang KK, Chen X, Tsao CY, Morris L, Gray E, Marshall DJ, Crump JA, Kapiga SH, Sam NE, Sinangil F, Pancera M, Yongping Y, Zhang B, Zhu J, Kwong PD, O'Dell S, Mascola JR, Wu L, Nabel GJ, Phogat S, Seaman MS, Whitesides JF, Moody MA, Kelsoe G, Yang X, Sodroski J, Shaw GM, Montefiori DC, Kepler TB, Tomaras GD, Alam SM, Liao HX, Haynes BF. 2011. Analysis of a clonal lineage of HIV-1 envelope V2/V3 conformational epitope-specific broadly neutralizing antibodies and their inferred unmutated common ancestors. J. Virol. 85:9998–10009. 10.1128/JVI.05045-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou T, Zhu J, Wu X, Moquin S, Zhang B, Acharya P, Georgiev IS, Altae-Tran HR, Chuang GY, Joyce MG, Do Kwon Y, Longo NS, Louder MK, Luongo T, McKee K, Schramm CA, Skinner J, Yang Y, Yang Z, Zhang Z, Zheng A, Bonsignori M, Haynes BF, Scheid JF, Nussenzweig MC, Simek M, Burton DR, Koff WC, NISC Comparative Sequencing Program. Mullikin JC, Connors M, Shapiro L, Nabel GJ, Mascola JR, Kwong PD. 2013. Multidonor analysis reveals structural elements, genetic determinants, and maturation pathway for HIV-1 neutralization by VRC01-class antibodies. Immunity 39:245–258. 10.1016/j.immuni.2013.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jardine J, Julien JP, Menis S, Ota T, Kalyuzhniy O, McGuire A, Sok D, Huang PS, MacPherson S, Jones M, Nieusma T, Mathison J, Baker D, Ward AB, Burton DR, Stamatatos L, Nemazee D, Wilson IA, Schief WR. 2013. Rational HIV immunogen design to target specific germline B cell receptors. Science 340:711–716. 10.1126/science.1234150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McGuire AT, Hoot S, Dreyer AM, Lippy A, Stuart A, Cohen KW, Jardine J, Menis S, Scheid JF, West AP, Schief WR, Stamatatos L. 2013. Engineering HIV envelope protein to activate germline B cell receptors of broadly neutralizing anti-CD4 binding site antibodies. J. Exp. Med. 210:655–633. 10.1084/jem.20122824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liao H-X, Lynch R, Zhou T, Gao F, Alam SM, Boyd SD, Fire AZ, Roskin KM, Schramm CA, Zhang Z, Zhu J, Shapiro L, NISC Comparative Sequencing Program. Mullikin JC, Gnanakaran S, Hraber P, Wiehe K, Kelsoe G, Yang G, Xia SM, Montefiori DC, Parks R, Lloyd KE, Scearce RM, Soderberg KA, Cohen M, Kamanga G, Louder MK, Tran LM, Chen Y, Cai F, Chen S, Moquin S, Du X, Joyce MG, Srivatsan S, Zhang B, Zheng A, Shaw GM, Hahn BH, Kepler TB, Korber BT, Kwong PD, Mascola JR, Haynes BF. 2013. Co-evolution of a broadly neutralizing HIV-1 antibody and founder virus. Nature 496:469–476. 10.1038/nature12053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Korber B, Gnanakaran S. 2009. The implications of patterns in HIV diversity for neutralizing antibody induction and susceptibility. Curr. Opin. HIV AIDS 4:408–417. 10.1097/COH.0b013e32832f129e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Haynes BF, Kelsoe G, Harrison SC, Kepler TB. 2012. B-cell–lineage immunogen design in vaccine development with HIV-1 as a case study. Nat. Biotechnol. 30:423–433. 10.1038/nbt.2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kwong PD, Mascola JR, Nabel GJ. 2013. Broadly neutralizing antibodies and the search for an HIV-1 vaccine: the end of the beginning. Nat. Rev. Immunol. 13:693–701. 10.1038/nri3516. [DOI] [PubMed] [Google Scholar]
- 39.Burton DR, Desrosiers RC, Doms RW, Koff WC, Kwong PD, Moore JP, Nabel GJ, Sodroski J, Wilson IA, Wyatt RT. 2004. HIV vaccine design and the neutralizing antibody problem. Nat. Immunol. 5:233–236. 10.1038/ni0304-233. [DOI] [PubMed] [Google Scholar]
- 40.Douek DC, Kwong PD, Nabel GJ. 2006. The rational design of an AIDS vaccine. Cell 124:677–681. 10.1016/j.cell.2006.02.005. [DOI] [PubMed] [Google Scholar]
- 41.Sanders RW, Derking R, Cupo A, Julien J-P, Yasmeen A, de Val N, Kim HJ, Blattner C, de la Peña AT, Korzun J, Golabek M, de los Reyes K, Ketas TJ, van Gils MJ, King CR, Wilson IA, Ward AB, Klasse PJ, Moore JP. 2013. A next-generation cleaved, soluble HIV-1 Env trimer, BG505 SOSIP.664 gp140, expresses multiple epitopes for broadly neutralizing but not non-neutralizing antibodies. PLoS Pathog. 9(9):e1003618. 10.1371/journal.ppat.1003618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Haynes BF, Moody MA, Verkoczy L, Kelsoe G, Alam SM. 2005. Antibody polyspecificity and neutralization of HIV-1: a hypothesis. Hum. Antibodies 14:59–67. [PMC free article] [PubMed] [Google Scholar]
- 43.Fernando K, Hu H, Ni H, Hoxie JA, Weissman D. 2007. Vaccine-delivered HIV envelope inhibits CD4+ T-cell activation, a mechanism for poor HIV vaccine responses. Blood 109:2538–2544. 10.1182/blood-2006-08-038661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shan M, Klasse PJ, Banerjee K, Dey AK, Iyer SPN, Dionisio R, Charles D, Campbell-Gardener L, Olson WC, Sanders RW, Moore JP. 2007. HIV-1 gp120 mannoses induce immunosuppressive responses from dendritic cells. PLoS Pathog. 3(11):e169. 10.1371/journal.ppat.0030169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.He B, Qiao X, Klasse PJ, Chiu A, Chadburn A, Knowles DM, Moore JP, Cerutti A. 2006. HIV-1 envelope triggers polyclonal IgG class switch recombination through a CD40-independent mechanism involving BAFF and C-type lectin receptors. J. Immunol. 176:3931–3941. 10.4049/jimmunol.176.7.3931. [DOI] [PubMed] [Google Scholar]
- 46.Moore JP, Cao Y, Leu J, Qin L, Korber B, Ho DD. 1996. Inter- and intraclade neutralization of human immunodeficiency virus type 1: genetic clades do not correspond to neutralization serotypes but partially correspond to gp120 antigenic serotypes. J. Virol. 70:427–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mascola JR, Louder MK, Surman SR, VanCott TC, Yu XF, Bradac J, Porter KR, Nelson KE, Girard M, McNeil JG, McCutchan FE, Birx DL, Burke DS. 1996. Human immunodeficiency virus type 1 neutralizing antibody serotyping using serum pools and an infectivity reduction assay. AIDS Res. Hum. Retroviruses 12:1319–1328. 10.1089/aid.1996.12.1319. [DOI] [PubMed] [Google Scholar]
- 48.Mascola JR, Louwagie J, McCutchan FE, Fischer CL, Hegerich PA, Wagner KF, Fowler AK, McNeil JG, Burke DS. 1994. Two antigenically distinct subtypes of human immunodeficiency virus type 1: viral genotype predicts neutralization serotype. J. Infect. Dis. 169:48–54. 10.1093/infdis/169.1.48. [DOI] [PubMed] [Google Scholar]
- 49.Nyambi PN, Nkengasong J, Lewi P, Andries K, Janssens W, Fransen K, Heyndrickx L, Piot P, van der Groen G. 1996. Multivariate analysis of human immunodeficiency virus type 1 neutralization data. J. Virol. 70:6235–6243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Weber J, Fenyö EM, Beddows S, Kaleebu P, Björndal Å. 1996. Neutralization serotypes of human immunodeficiency virus type 1 field isolates are not predicted by genetic subtype. J. Virol. 70:7827–7832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Brown BK, Wiecaorek L, Sanders-Buell E, Borges AR, Robb ML, Birx DL, Michael NL, McCutchan FE, Polonis VR. 2008. Cross-clade neutralization patterns among HIV-1 strains from the six major clades of the pandemic evaluated and compared in two different models. Virology 375:529–538. 10.1016/j.virol.2008.02.022. [DOI] [PubMed] [Google Scholar]
- 52.Bures R, Morris L, Williamson C, Ramjee G, Deers M, Fiscus SA, Karim SA, Montefiori DC. 2002. Regional clustering of shared neutralization determinants on primary isolates of clade C human immunodeficiency virus type 1 from South Africa. J. Virol. 76:2233–2244. 10.1128/jvi.76.5.2233-2244.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Binley J, Wrin T, Korber B, Zwick M, Wang M, Chappey C, Stiegler G, Kunert R, Zolla-Pazner S, Katinger H, Petropoulos C, Burton D. 2004. Comprehensive cross-subtype neutralization analysis of a panel of anti-human immunodeficiency virus type 1 monoclonal antibodies. J. Virol. 78:13232–13252. 10.1128/JVI.78.23.13232-13252.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kulkarni SS, Lapedes A, Tang H, Gnanakaran S, Daniels MG, Zhang M, Bhattacharya T, Li M, Polonis VR, McCutchan FE, Morris L, Ellenberger D, Butera ST, Bollinger RC, Korber BT, Paranjape RS, Montefiori DC. 2009. Highly complex neutralization determinants on a monophyletic lineage of newly transmitted subtype C human immunodeficiency virus type 1 env clones from India. Virology 385:505–520. 10.1016/j.virol.2008.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li M, Salazar-Gonzalez JF, Derdeyn CA, Morris L, Williamson C, Robinson JE, Decker JM, Li Y, Salazar MG, Polonis VR, Mlisana K, Karim SA, Hong K, Greene KM, Bilska M, Zhou JT, Allen S, Chomba E, Mulenga J, Vwalika C, Gao F, Zhang M, Korber BTM, Hunter E, Hahn BH, Montefiori DC. 2006. Genetic and neutralization properties of subtype C human immunodeficiency virus type 1 molecular env clones from acute and early heterosexually acquired infections in southern Africa. J. Virol. 80:11776–11790. 10.1128/JVI.01730-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Walker LM, Phogat SK, Chan-Hui PY, Wagner D, Phung P, Goss JL, Wrin T, Simek MD, Fling S, Mitcham JL, Lehrman JK, Priddy FH, Olsen OA, Frey SM, Hammond PW, Kaminsky S, Zamb T, Moyle M, Koff WC, Poignard P, Burton DR. 2009. Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science 326:285–289. 10.1126/science.1178746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Walker LM, Huber M, Doores KJ, Falkowska E, Pejchal R, Julien J-P, Wang S-K, Ramos A, Chan-Hui P-Y, Moyle M, Mitcham JL, Hammond PW, Olsen OA, Phung P, Fling S, Wong C-H, Phogat S, Wrin T, Simek MD, Protocol G Principal Investigators. Koff WC, Wilson IA, Burton DR, Poignard P. 2011. Broad neutralization coverage of HIV by multiple highly potent antibodies. Nature 477:466–478. 10.1038/nature10373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.deCamp A, Hraber P, Bailer RT, Seaman MS, Ochsenbauer C, Kappes J, Gottardo R, Edlefsen P, Self S, Tang H, Greene K, Gao H, Daniell X, Sarzotti-Kelsoe M, Gorny MK, Zolla-Pazner S, LaBranche CC, Mascola JR, Korber BT, Montefiori DC. 2014. Global panel of HIV-1 Env reference strains for standardized assessments of vaccine-elicited neutralizing antibodies. J. Virol. 88:2489–2507. 10.1128/JVI.02853-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fiebig EW, Wright DJ, Rawal BD, Garrett PE, Schumacher RT, Peddada L, Heldebrant C, Smith R, Conrad A, Kleinman SH, Busch MP. 2003. Dynamics of HIV viremia and antibody seroconversion in plasma donors: Implications for diagnosis and staging of primary HIV infection. AIDS 17:1871–1879. 10.1097/00002030-200309050-00005. [DOI] [PubMed] [Google Scholar]
- 60.Montefiori DC. 2009. Measuring HIV neutralization in a luciferase reporter gene assay. Methods Mol. Biol. 485:395–405. 10.1007/978-1-59745-170-3_26. [DOI] [PubMed] [Google Scholar]
- 61.Sarzotti-Kelsoe M, Bailer RT, Turk E, Lin CL, Bilska M, Greene KM, Gao H, Todd CA, Ozaki DA, Seaman MS, Mascola JR, Montefiori DC. 2014. Optimization and validation of the TZM-bl assay for standardized assessments of neutralizing antibodies against HIV-1. J. Immunol. Methods 409C:131–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Todd CA, Greene KM, Yu X, Ozaki DA, Gao H, Huang Y, Wang M, Li G, Brown R, Wood B, D'Souza MP, Gilbert P, Montefiori DC, Sarzotti-Kelsoe M. 2012. Development and implementation of an international proficiency testing program for a neutralizing antibody assay for HIV-1 in TZM-bl cells. J. Immunol. Methods 375:57–67. 10.1016/j.jim.2011.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Eddy SR. 1998. Profile hidden Markov models. Bioinformatics 14:755–763. 10.1093/bioinformatics/14.9.755. [DOI] [PubMed] [Google Scholar]
- 64.Rambaut A. 2002. Se-Al. Sequence Alignment Editor v2.0a11. http://tree.bio.ed.ac.uk/software/seal/.
- 65.Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59:307–321. 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 66.Nickle DC, Heath L, Jensen MA, Gilbert PB, Mullins JI, Kosakovsky-Pond SL. 2007. HIV-specific probabilistic models of protein evolution. PLoS One 2:e503. 10.1371/journal.pone.0000503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.R Core Team. 2013. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria: http://www.r-project.org. [Google Scholar]
- 68.Paradis E, Claude J, Strimmer K. 2004. APE: analyses of phylogenetics and evolution in R language. Bioinformatics 20:289–290. 10.1093/bioinformatics/btg412. [DOI] [PubMed] [Google Scholar]
- 69.Wood N, Bhattacharya T, Keele BF, Giorgi E, Liu M, Gaschen B, Daniels M, Ferrari G, Haynes BF, McMichael A, Shaw GM, Hahn BH, Korber B, Seoighe C. 2009. HIV evolution in early infection: selection pressures, patterns of insertion and deletion, and the impact of APOBEC. PLoS Pathog. 5:e1000414. 10.1371/journal.ppat.1000414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Keele BF, Giorgi EE, Salazar-Gonzalez JF, Decker JM, Pham KT, Salazar MG, Sun C, Grayson T, Wang S, Li H, Wei X, Jiang C, Kirchherr JL, Gao F, Anderson JA, Ping LH, Swanstrom R, Tomaras GD, Blattner WA, Goepfert PA, Kilby JM, Saag MS, Delwart EL, Busch MP, Cohen MS, Montefiori DC, Haynes BF, Gaschen B, Athreya GS, Lee HY, Wood N, Seoighe C, Perelson AS, Bhattacharya T, Korber BT, Hahn BH, Shaw GM. 2008. Identification and characterization of transmitted and early founder virus envelopes in primary HIV-1 infection. Proc. Natl. Acad. Sci. U. S. A. 105:7552–7557. 10.1073/pnas.0802203105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.McLeod AI. 2011. Kendall: Kendall rank correlation and Mann-Kendall trend test. R package version 2.2. http://cran.r-project.org/package=Kendall.
- 72.Dabney A, Storey JD. 2013. qvalue: q-value estimation for false discovery rate control. http://bioconductor.org/packages/release/bioc/html/qvalue.html.
- 73.Storey JD, Tibshirani R. 2003. Statistical significance for genome-wide experiments. Proc. Natl. Acad. Sci. U. S. A. 100:9440–9445. 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Crawley MJ. 2002. Statistical computing: an introduction to data analysis using S-Plus. John Wiley and Sons, Inc., New York, NY. [Google Scholar]
- 75.Bates D, Maechler M, Bolker B, Walker S. 2013. lme4: linear mixed-effects models using Eigen and S4. https://github.com/lme4/lme4/. [Google Scholar]
- 76.Robertson DL, Anderson JP, Bradac JA, Carr JK, Foley B, Funkhouser RK, Gao F, Hahn BH, Kalish ML, Kuiken C, Learn GH, Leitner T, McCutchan F, Osmanov S, Peeters M, Pieniazek D, Salminen M, Sharp PM, Wolinsky S, Korber B. 2000. HIV-1 nomenclature proposal. Science 288:55–56. 10.1126/science.288.5463.55d. [DOI] [PubMed] [Google Scholar]
- 77.Siepel AC, Halpern AL, Macken C, Korber BT. 1995. A computer program designed to screen rapidly for HIV type 1 intersubtype recombinant sequences. AIDS Res. Hum. Retroviruses 11:1413–1416. 10.1089/aid.1995.11.1413. [DOI] [PubMed] [Google Scholar]
- 78.Su L, Graf M, Zhang Y, von Briesen H, Xing H, Köstler J, Melzl H, Wolf H, Shao Y, Wagner R. 2000. Characterization of a virtually full-length human immunodeficiency virus type 1 genome of a prevalent intersubtype (C/B') recombinant strain in China. J. Virol. 74:11367–11376. 10.1128/JVI.74.23.11367-11376.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhang M, Foley B, Schultz AK, Macke JP, Bulla I, Stanke M, Morgenstern B, Korber B, Leitner T. 2010. The role of recombination in the emergence of a complex and dynamic HIV epidemic. Retrovirology 7:25. 10.1186/1742-4690-7-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Carr JK, Salminen MO, Koch C, Gotte D, Artenstein AW, Hegerich PA, St Louis D, Burke DS, McCutchan FE. 1996. Full-length sequence and mosaic structure of a human immunodeficiency virus type 1 isolate from Thailand. J. Virol. 70:5935–5943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Anderson JP, Rodrigo AG, Learn GH, Madan A, Delahunty C, Coon M, Girard M, Osmanov S, Hood L, Mullins JI. 2000. Testing the hypothesis of a recombinant origin of human immunodeficiency virus type 1 subtype E. J. Virol. 74:10752–10765. 10.1128/JVI.74.22.10752-10765.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Pancera M, Shahzad-Ul-Hussan S, Doria-Rose NA, McLellan JS, Bailer RT, Dai K, Loesgen S, Louder MK, Staupe RP, Yang Y, Zhang B, Parks R, Eudailey J, Lloyd KE, Blinn J, Alam SM, Haynes BF, Amin MN, Wang LX, Burton DR, Koff WC, Nabel GJ, Mascola JR, Bewley CA, Kwong PD. 2013. Structural basis for diverse N-glycan recognition by HIV-1-neutralizing V1–V2-directed antibody PG16. Nat. Struct. Mol. Biol. 20:804–813. 10.1038/nsmb.2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.McLellan JS, Pancera M, Carrico C, Gorman J, Julien JP, Khayat R, Louder R, Pejchal R, Sastry M, Dai K, O'Dell S, Patel N, Shahzad-ul Hussan S, Yang Y, Zhang B, Zhou T, Zhu J, Boyington JC, Chuang GY, Diwanji D, Georgiev I, Kwon YD, Lee D, Louder MK, Moquin S, Schmidt SD, Yang ZY, Bonsignori M, Crump JA, Kapiga SH, Sam NE, Haynes BF, Burton DR, Koff WC, Walker LM, Phogat S, Wyatt R, Orwenyo J, Wang LX, Arthos J, Bewley CA, Mascola JR, Nabel GJ, Schief WR, Ward AB, Wilson IA, Kwong PD. 2011. Structure of HIV-1 gp120 V1/V2 domain with broadly neutralizing antibody PG9. Nature 480:336–343. 10.1038/nature10696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Liao H-X, Chen X, Munshaw S, Zhang R, Marshall DJ, Vandergrift N, Whitesides JF, Lu X, Yu JS, Hwang KK, Gao F, Markowitz M, Heath SL, Bar KJ, Goepfert PA, Montefiori DC, Shaw GC, Alam SM, Margolis DM, Denny TN, Boyd SD, Marshal E, Egholm M, Simen BB, Hanczaruk B, Fire AZ, Voss G, Kelsoe G, Tomaras GD, Moody MA, Kepler TB, Haynes BF. 2011. Initial antibodies binding to HIV-1 gp41 in acutely infected subjects are polyreactive and highly mutated. J. Exp. Med. 208:2237–2249. 10.1084/jem.20110363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Mascola JR, D'Souza P, Gilbert P, Hahn BH, Haigwood NL, Morris L, Petropoulos CJ, Polonis VR, Sarzotti M, Montefiori DC. 2005. Recommendations for the design and use of standard virus panels to assess neutralizing antibody responses elicited by candidate human immunodeficiency virus type 1 vaccines. J. Virol. 79:10103–10107. 10.1128/JVI.79.16.10103-10107.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Doria-Rose NA, Klein RM, Daniels MG, O'Dell S, Nason M, Lapedes A, Bhattacharya T, Migueles SA, Wyatt RT, Korber BT, Mascola JR, Connors M. 2010. Breadth of human immunodeficiency virus-specific neutralizing activity in sera: clustering analysis and association with clinical variables. J. Virol. 84:1631–1636. 10.1128/JVI.01482-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Meng Z, Xin R, Zhong P, Zhang C, Abubakar YF, Li J, Liu W, Zhang X, Xu J. 2012. A new migration map of HIV-1 CRF07_BC in China: analysis of sequences from 12 provinces over a decade. PLoS One 7:e52373. 10.1371/journal.pone.0052373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Tee KK, Pybus OG, Li XJ, Han X, Shang H, Kamarulzaman A, Takebe Y. 2008. Temporal and spatial dynamics of human immunodeficiency virus type 1 circulating recombinant forms 08_BC and 07_BC in Asia. J. Virol. 82:9206–9215. 10.1128/JVI.00399-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Korber B, Muldoon M, Theiler J, Gao F, Gupta R, Lapedes A, Hahn BH, Wolinsky S, Bhattacharya T. 2000. Timing the ancestor of the HIV-1 pandemic strains. Science 288:1789–1796. 10.1126/science.288.5472.1789. [DOI] [PubMed] [Google Scholar]
- 90.Kalish ML, Korber BT, Pillai S, Robbins KE, Leo YS, Saekhou A, Verghese I, Gerrish P, Goh CL, Lupo D, Tan BH, Brown TM, Chan R. 2002. The sequential introduction of HIV-1 subtype B and CRF01AE in Singapore by sexual transmission: accelerated V3 region evolution in a subpopulation of Asian CRF01 viruses. Virology 304:311–329. 10.1006/viro.2002.1691. [DOI] [PubMed] [Google Scholar]
- 91.Kalish ML, Robbins KE, Pieniazek D, Schaefer A, Nzilambi N, Quinn TC, St Louis ME, Youngpairoj AS, Phillips J, Jaffe HW, Folks TM. 2004. Recombinant viruses and early global HIV-1 epidemic. Emerg. Infect. Dis. 10:1227–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Worobey M, Gemmel M, Teuwen DE, Haselkorn T, Kunstman K, Bunce M, Muyembe JJ, Kabongo JM, Kalengayi RM, Van Marck E, Gilbert MT, Wolinsky SM. 2008. Direct evidence of extensive diversity of HIV-1 in Kinshasa by 1960. Nature 455:661–664. 10.1038/nature07390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zolla-Pazner S, deCamp A, Gilbert PB, Williams C, Yates NL, Williams WT, Howington R, Fong Y, Morris DE, Soderberg KA, Irene C, Reichman C, Pinter A, Parks R, Pitisuttithum P, Kaewkungwal J, Rerks-Ngarm S, Nitayaphan S, Andrews C, O'Connell RJ, Yang ZY, Nabel GJ, Kim JH, Michael NL, Montefiori DC, Liao H-X, Haynes BF, Tomaras GD. 2014. Vaccine-induced IgG antibodies to V1V2 regions of multiple 1 HIV-1 subtypes correlate with decreased risk of HIV-1 infection. PLoS One 9:e87572. 10.1371/journal.pone.0087572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Gottardo R, Bailer RT, Korber BT, Gnanakaran S, Phillips J, Shen X, Tomaras GD, Turk E, Imholte G, Eckler L, Wenschuh H, Zerweck J, Greene K, Gao H, Berman PW, Francis D, Sinangil F, Lee C, Nitayaphan S, Rerks-Ngarm S, Kaewkungwal J, Pitisuttithum P, Tartaglia J, Robb ML, Michael NL, Kim JH, Zolla-Pazner S, Haynes BF, Mascola JR, Self S, Gilbert P, Montefiori DC. 2013. Plasma IgG to linear epitopes in the V2 and V3 regions of HIV-1 gp120 correlate with a reduced risk of infection in the RV144 vaccine efficacy trial. PLoS One 8:e75665. 10.1371/journal.pone.0075665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Yates NL, Liao H-X, Fong Y, deCamp A, Vandergrift NA, Williams WT, Alam SM, Ferrari G, Yang ZY, Seaton KE, Berman PW, Alpert MD, Evans DT, O'Connell RJ, Francis D, Sinangil F, Lee C, Nitayaphan S, Rerks-Ngarm S, Kaewkungwal J, Pitisuttithum P, Tartaglia J, Pinter A, Zolla-Pazner S, Gilbert PB, Nabel GJ, Michael NL, Kim JH, Montefiori DC, Haynes BF, Tomaras GD. 2014. Vaccine-induced Env V1–V2 IgG3 correlates with lower HIV-1 infection risk and declines soon after vaccination. Sci. Transl. Med. 6:228ra39. 10.1126/scitranslmed.3007730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hu DJ, Subbarao S, Vanichseni S, Mock PA, Ramos A, Nguyen L, Chaowanachan T, Griensven Fv Choopanya K, Mastro TD, Tappero JW. 2005. Frequency of HIV-1 dual subtype infections, including intersubtype superinfections, among injection drug users in Bangkok, Thailand. AIDS 19:303–308. [PubMed] [Google Scholar]
- 97.Yerly S, Jost S, Monnat M, Telenti A, Cavassini M, Chave JP, Kaiser L, Burgisser P, Perrin L, Swiss HIV Cohort Study 2004. HIV-1 co/superinfection in intravenous drug users. AIDS 18:1413–1421. 10.1097/01.aids.0000131330.28762.0c. [DOI] [PubMed] [Google Scholar]
- 98.Templeton AR, Kramer MG, Jarvis J, Kowalski J, Gange S, Schneider MF, Shao Q, Zhang GW, Yeh MF, Tsai HL, Zhang H, Markham RB. 2009. Multiple-infection and recombination in HIV-1 within a longitudinal cohort of women. Retrovirology 6:54. 10.1186/1742-4690-6-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Derdeyn CA, Decker JM, Bibollet-Ruche F, Mokili JL, Muldoon M, Denham SA, Heil ML, Kasolo F, Musonda R, Hahn BH, Shaw GM, Korber BT, Allen S, Hunter E. 2004. Envelope-constrained neutralization-sensitive HIV-1 after heterosexual transmission. Science 303:2019–2022. 10.1126/science.1093137. [DOI] [PubMed] [Google Scholar]
- 100.Chohan B, Lang D, Sagar M, Korber B, Lavreys L, Richardson B, Overbaugh J. 2005. Selection for human immunodeficiency virus type 1 envelope glycosylation variants with shorter V1–V2 loop sequences occurs during transmission of certain genetic subtypes and may impact viral RNA levels. J. Virol. 79:6528–6531. 10.1128/JVI.79.10.6528-6531.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Liu Y, Curlin ME, Diem K, Zhao H, Ghosh AK, Zhu H, Woodward AS, Maenza J, Stevens CE, Stekler J, Collier AC, Genowati I, Deng W, Zioni R, Corey L, Zhu T, Mullins JI. 2008. Env length and N-linked glycosylation following transmission of human immunodeficiency virus type 1 subtype B viruses. Virology 374:229–233. 10.1016/j.virol.2008.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Parrish NF, Gao F, Li H, Giorgi EE, Barbian HJ, Parrish EH, Zajic L, Iyer SS, Decker JM, Kumar A, Hora B, Berg A, Cai F, Hopper J, Denny TN, Ding H, Ochsenbauer C, Kappes JC, Galimidi RP, West AP, Jr, Bjorkman PJ, Wilen CB, Doms RW, O'Brien M, Bhardwaj N, Borrow P, Haynes BF, Muldoon M, Theiler JP, Korber B, Shaw GM, Hahn BH. 2013. Phenotypic properties of transmitted founder HIV-1. Proc. Natl. Acad. Sci. U. S. A. 110:6626–6633. 10.1073/pnas.1304288110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Gnanakaran S, Bhattacharya T, Daniels M, Keele BF, Hraber PT, Lapedes AS, Shen T, Gaschen B, Krishnamoorthy M, Li H, Decker JM, Salazar-Gonzalez JF, Wang S, Jiang C, Gao F, Swanstrom R, Anderson JA, Ping LH, Cohen MS, Markowitz M, Goepfert PA, Saag MS, Eron JJ, Hicks CB, Blattner WA, Tomaras GD, Asmal M, Letvin NL, Gilbert PB, Decamp AC, Magaret CA, Schief WR, Ban YE, Zhang M, Soderberg KA, Sodroski JG, Haynes BF, Shaw GM, Hahn BH, Korber B. 2011. Recurrent signature patterns in HIV-1 B clade envelope glycoproteins associated with either early or chronic infections. PLoS Pathog. 7:e1002209. 10.1371/journal.ppat.1002209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Asmal M, Hellmann I, Liu W, Keele BF, Perelson AS, Bhattacharya T, Gnanakaran S, Daniels M, Haynes BF, Korber BT, Hahn BH, Shaw GM, Letvin NL. 2011. A signature in HIV-1 envelope leader peptide associated with transition from acute to chronic infection impacts envelope processing and infectivity. PLoS One 6:e23673. 10.1371/journal.pone.0023673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Zhang H, Rola M, West JT, Tully DC, Kubis P, He J, Kankasa C, Wood C. 2010. Functional properties of the HIV-1 subtype C envelope glycoprotein associated with mother-to-child transmission. Virology 400:164–174. 10.1016/j.virol.2009.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Wilen CB, Parrish NF, Pfaff JM, Decker JM, Henning EA, Haim H, Petersen JE, Wojcechowskyj JA, Sodroski J, Haynes BF, Montefiori DC, Tilton JC, Shaw GM, Hahn BH, Doms RW. 2011. Phenotypic and immunologic comparison of clade B transmitted/founder and chronic HIV-1 envelope glycoproteins. J. Virol. 85:8514–8527. 10.1128/JVI.00736-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Bar KJ, Tsao CY, Iyer SS, Decker JM, Yang Y, Bonsignori M, Chen X, Hwang KK, Montefiori DC, Liao HX, Hraber P, Fischer W, Li H, Wang S, Sterrett S, Keele BF, Ganusov VV, Perelson AS, Korber BT, Georgiev I, McLellan JS, Pavlicek JW, Gao F, Haynes BF, Hahn BH, Kwong PD, Shaw GM. 2012. Early low-titer neutralizing antibodies impede HIV-1 replication and select for virus escape. PLoS Pathog. 8:e1002721. 10.1371/journal.ppat.1002721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Wibmer CK, Bhiman JN, Gray ES, Tumba N, Abdool Karim SS, Williamson C, Morris L, Moore PL. 2013. Viral escape from HIV-1 neutralizing antibodies drives increased plasma neutralization breadth through sequential recognition of multiple epitopes and immunotypes. PLoS Pathog. 9:e1003738. 10.1371/journal.ppat.1003738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Haim H, Strack B, Kassa A, Madani N, Wang L, Courter JR, Princiotto A, McGee K, Pacheco B, Seaman MS, Smith AB, III, Sodroski J. 2011. Contribution of intrinsic reactivity of the HIV-1 envelope glycoproteins to CD4-independent infection and global inhibitor sensitivity. PLoS Pathog. 7:e1002101. 10.1371/journal.ppat.1002101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Haim H, Salas I, McGee M, Eichelberger N, Winter E, Pacheco B, Sodroski J. 2013. Modeling virus- and antibody-specific factors to predict human immunodeficiency virus neutralization efficiency. Cell Host Microbe 14:547–558. 10.1016/j.chom.2013.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Robbins KE, Lemey P, Pybus OG, Jaffe HW, Youngpairoj AS, Brown TM, Salemi M, Vandamme AM, Kalish ML. 2003. U.S. human immunodeficiency virus type 1 epidemic: date of origin, population history, and characterization of early strains. J. Virol. 77:6359–6366. 10.1128/JVI.77.11.6359-6366.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.