ABSTRACT
Hepatitis C virus (HCV) assembles its replication complex on cytosolic membrane vesicles often clustered in a membranous web (MW). During infection, HCV NS5A protein activates PI4KIIIα enzyme, causing massive production and redistribution of phosphatidylinositol 4-phosphate (PI4P) lipid to the replication complex. However, the role of PI4P in the HCV life cycle is not well understood. We postulated that PI4P recruits host effectors to modulate HCV genome replication or virus particle production. To test this hypothesis, we generated cell lines for doxycycline-inducible expression of short hairpin RNAs (shRNAs) targeting the PI4P effector, four-phosphate adaptor protein 2 (FAPP2). FAPP2 depletion attenuated HCV infectivity and impeded HCV RNA synthesis. Indeed, FAPP2 has two functional lipid-binding domains specific for PI4P and glycosphingolipids. While expression of the PI4P-binding mutant protein was expected to inhibit HCV replication, a marked drop in replication efficiency was observed unexpectedly with the glycosphingolipid-binding mutant protein. These data suggest that both domains are crucial for the role of FAPP2 in HCV genome replication. We also found that HCV significantly increases the level of some glycosphingolipids, whereas adding these lipids to FAPP2-depleted cells partially rescued replication, further arguing for the importance of glycosphingolipids in HCV RNA synthesis. Interestingly, FAPP2 is redistributed to the replication complex (RC) characterized by HCV NS5A, NS4B, or double-stranded RNA (dsRNA) foci. Additionally, FAPP2 depletion disrupts the RC and alters the colocalization of HCV replicase proteins. Altogether, our study implies that HCV coopts FAPP2 for virus genome replication via PI4P binding and glycosphingolipid transport to the HCV RC.
IMPORTANCE Like most viruses with a positive-sense RNA genome, HCV replicates its RNA on remodeled host membranes composed of lipids hijacked from various internal membrane compartments. During infection, HCV induces massive production and retargeting of the PI4P lipid to its replication complex. However, the role of PI4P in HCV replication is not well understood. In this study, we have shown that FAPP2, a PI4P effector and glycosphingolipid-binding protein, is recruited to the HCV replication complex and is required for HCV genome replication and replication complex formation. More importantly, this study demonstrates, for the first time, the crucial role of glycosphingolipids in the HCV life cycle and suggests a link between PI4P and glycosphingolipids in HCV genome replication.
INTRODUCTION
Hepatitis C virus (HCV) is a positive-strand RNA virus responsible for about 170 million cases of chronic liver disease worldwide and at least 350,000 annual deaths due to cirrhosis and hepatocellular carcinoma (1, 2). HCV belongs to the Flaviviridae family (3, 4), which includes Dengue virus and West Nile virus. The error-prone nature of its polymerase (5) has given rise to at least 7 HCV genotypes and more than 50 subtypes (6, 7). The virus genome, about 9.6 kb long, is flanked by 5′- and 3′-untranslated regions (UTR), both of which are required for HCV genome replication. Additionally, an internal ribosome entry site in the 5′UTR regulates translation of the virus genome, which gives rise to three structural proteins (core, E1, and E2), the p7 viroporin, and six nonstructural (NS) proteins (NS2-3-4A-4B-5A-5B) (8). The NS proteins NS3 to NS5B are sufficient for HCV genome replication in cell culture (9, 10). However, many of these NS proteins (NS3, NS4B, and NS5A) recently were shown to regulate HCV particle production (11–16), consistent with the multifunctional roles of these proteins during HCV infection.
Like most viruses with a positive-strand RNA genome, HCV RNA replication takes place on cytosolic, double-membrane vesicles clustered into a membranous web (MW) (17). Previous studies suggested that HCV NS4B expression was sufficient for MW vesicle formation (17–20). The MW typically is seen as foci in microscopy, and disruption of these foci impedes HCV RNA replication efficiency (19, 21–24). Hence, in cells actively replicating the HCV genome, NS4B foci colocalize with the components of the HCV replication complex, including the replicase proteins (NS3, NS4A, NS4B, NS5A, and NS5B), host factors (19, 25), and viral RNA. NS4B interacts with nonstructural proteins involved in HCV RNA synthesis (17, 19, 26–30), implying that NS4B provides the scaffold for recruiting replicase proteins to the HCV replication complex. Recent reports show an equally crucial role for HCV NS5A in the formation of the MW vesicles. Indeed, NS5A binds to and activates the endoplasmic reticulum-derived phosphatidylinositol-4 kinase III alpha (PI4KIIIα), leading to increased production and redistribution of phosphatidylinositol 4-phosphate (PI4P) lipid to the HCV replication complex (31). Transient depletion of PI4KIIIα or dephosphorylation of PI4P impedes HCV replication efficiency (31–33) and disrupts the MW structure. However, the role of the PI4P lipid in HCV replication is not well understood. We hypothesized that PI4P recruits host adaptor proteins to the HCV replication complex to modulate HCV genome replication or virus particle production.
We found that FAPP2, a PI4P adaptor and glycosphingolipid-binding protein, is recruited to the HCV replication complex. Furthermore, FAPP2 depletion resulted in attenuation of HCV infectivity and impeded HCV RNA synthesis. Further analysis suggests that FAPP2 has a direct role in HCV genome replication via its PI4P-binding domain, glycosphingolipid binding, and transport to the replication complex. The significance of these novel findings will be discussed.
MATERIALS AND METHODS
Cell culture.
Huh7.5 cells were kindly provided by Apath, LLC (St. Louis, MO), and propagated in advanced Dulbecco's modified Eagle's medium (DMEM; Invitrogen, Carlsbad, CA) containing 1.5% fetal bovine serum (FBS; Atlanta Bio, Lawrenceville, GA). Con1 cells, kindly provided by Charles Rice, Rockefeller University, carry the self-replicating HCV 1b subgenomic replicon and were grown in the same medium containing 0.5 mg/ml G418 (Sigma-Aldrich, St. Louis, MO). Human kidney 293T cells (kind gift of Carlos de Noronha, Albany Medical College, Albany, NY) were cultured in DMEM (Invitrogen) supplemented with 10% calf serum. The cells were propagated in media supplemented with l-glutamine, 100 U/ml penicillin, and 100 μg/ml streptomycin (Life Tech Corp., Grand Island, NY) at 37°C in a 5% CO2 incubator.
Antibodies.
Rabbit polyclonal antibody specific for FAPP2 was obtained from Abcam Inc., (Cambridge MA). Mouse monoclonal antibody to SPTLC1 was obtained from Santa Cruz Biotechnology Inc. (Dallas, TX). Mouse monoclonal antibody for HCV NS5A (9E10) was kindly provided by Charles Rice (The Rockefeller University, New York, NY). Rabbit polyclonal antibody to HCV NS4B was produced by Covance (Denver, CO). Rabbit polyclonal antibody to calnexin was purchased from Cell Signaling (Danvers, MA). Mouse J2 (double-stranded RNA [dsRNA]) and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) monoclonal antibodies were obtained from English & Scientific Consulting (Szirák, Hungary) and Fitzgerald (Acton, MA), respectively. Mouse monoclonal antibody to lactosylceramide (LacCer) was obtained from Glycobiotech GmbH (Kuekels, Germany). Horseradish peroxidase-conjugated secondary antibodies, used for chemiluminescence, were obtained from Vector Laboratories (Burlingame, CA). Alexa Fluor 488- and 594-conjugated secondary antibodies (used in immunofluorescence) were from Invitrogen.
Reagents.
Puromycin and doxycycline (Dox) were purchased from Calbiochem (Billerica, MA) and Enzo (Farmingdale, NY), respectively. N-Butyldeoxynojirimycin (NB-DNJ), low-density lipoprotein (LDL), and acetyl-d-sphingosine were obtained from Sigma (St. Louis, MO), whereas d-threo-PDMP, glucosylceramide (GlcCer), lactosylceramide, and globotriaosylceramide (Gb3) were purchased from Matreya (Pleasant Gap, PA).
Plasmids construction.
The pJc1 (34) virus and pLuc-Con1 replicon (35) constructs were kindly provided by Ralf Bartenschlager (University of Heidelberg, Germany). The pJ6/JFH1-mcherry virus (36) construct was kindly provided by Jens Bukh (Copenhagen University Hospital, Hvidovre, Denmark). The pLuc-JFH1-mCherry construct was engineered by replacing the ∼2.2-kb fragment between RsrII and XbaI sites in the pLuc-JFH1 plasmid with the corresponding fragment from pJ6/JFH1-mCherry. To construct the pTRIPZ-GFP plasmid, pEGFP-N2 (Clontech) was digested with BamHI and NotI, and the purified green fluorescent protein (GFP) fragment was blunt ended with T4 DNA polymerase (NEB). The resulting GFP fragment then was ligated into AgeI- and EcoRI-digested and blunt-ended pTRIPZ vector (GE Healthcare, Pittsburgh, PA).
To engineer pTRIPZ-GFP-fused wild-type (WT) or mutant FAPP2 vector, we used plasmids kindly provided by M. Antonietta De Matteis and Kai Simons (37, 38). Vectors containing GFP-FAPP2 and GFP-FAPP2 ΔPH were digested with AgeI and SmaI, and the purified fragments were inserted into AgeI- and HpaI-cut pTRIPZ. To generate the pTRIPZ-GFP FAPP2 W407A vector, the FAPP2 W407A fragment was amplified from pGEX-6P-1-FAPP2 W407A using the following primers: 5′ GACGTGGTACCGAGGGGGTGCTGTACAAGTGGA 3′ and 5′ GCGCCCTCGAGACTAGTTTATCATACCACCTCATCAGATTCCAG 3′ (restriction sites are underlined). The resulting PCR product was digested with KpnI and XhoI and inserted with GFP (AgeI- and KpnI-cut) into AgeI- and XhoI-cleaved pTRIPZ vector.
All of the shRNA clones, in a PTRIPZ vector, were purchased from GE Healthcare (Pittsburgh, PA). The FAPP2 shRNA (TCCATTCCATCTTCCTTCC; targets the FAPP2 open reading frame) and SPTLC1 shRNA (TAAACATCAGTTATACACT; targets the SPTLC1 3′UTR sequence) clones were used to successfully knock down FAPP2 or SPTLC1 in stable Huh7.5 cells.
Generation of stable and doxycycline-inducible cells.
Human kidney 293T cells (7.5 × 106) were grown overnight to obtain 60 to 70% confluence. Media were replaced 1 h before transfection. Four micrograms of pTRIPZ vector (with nontargeting, host-specific shRNA or host-specific gene) were mixed with 4 μg each of HIV pTat, pRev, pGag/pol, and pVSV-G vectors (kindly supplied by Carlos de Noronha, Albany Medical College, NY) and CaCl2 to produce lentivirus and transfected by a standard calcium phosphate protocol to produce lentivirus. After 72 h of transfection, the cell culture supernatant was harvested and used to infect Huh7.5 cells. The resulting cells were grown under selection with puromycin at a concentration of 3 μg/ml for 2 to 3 weeks. Doxycycline-inducible expression of the gene, or knockdown of the host factor, was confirmed via immunoblotting. The doxycycline-inducible promoter (TRE), in the pTRIPZ vector, also drives the expression of a red fluorescent protein (RFP) reporter immediately following the shRNA sequence. The induced RFP fluorescence allows for a quick evaluation of the basal expression of the shRNA or the lentiviral titer.
In vitro transcription and electroporation of viral RNA into Huh7.5 cells or stable cell lines.
Plasmid DNA constructs containing the full-length genome or a subgenomic replicon were linearized with MluI (Jc1), XbaI (Luc-JFH1), or ScaI (Luc-Con1) (New England BioLabs, Ipswich, MA, USA), purified by a Cycle Pure kit (Omega Bio-Tek, Norcross, GA), and used for in vitro transcription of HCV genomic RNA with T7 Express large-scale RiboMAX (Promega, Madison, WI). The RNA transcription reaction mixture (20 μl) contained 2 μl enzyme mix T7 Express (T7 RNA polymerase, recombinant RNasin RNase inhibitor, and recombinant inorganic pyrophosphatase), 10 μl RiboMAX Express T7 2× buffer, 10 μg linearized DNA template, and nuclease-free water. The reaction mixture was incubated at 37°C for 1 h, and the synthesized RNA was DNase treated with RQ1 RNase-free DNase (Promega) for 30 min at 37°C. The RNA then was isolated by an RNeasy minikit (Qiagen GmbH, Hilden, Germany). RNA concentration was measured by NanoDrop, and aliquots were stored at −80°C until use. For RNA transfection, subconfluent cells (Huh7.5 or shRNA-expressing Huh7.5) were washed with phosphate-buffered saline (PBS), trypsinized, and resuspended in complete growth medium. The cells subsequently were washed three times with ice-cold PBS and resuspended at a concentration of 1.25 × 107 cells/ml in ice-cold PBS. Briefly, 5 μg of Jc1 and 10 μg of JFH1-mCherry-Luc RNA were mixed with the desired cells (2.5 × 106) in 0.2 ml ice-cold PBS and electroporated with an Electro Square Porator (BTX) in a 0.2-mm-gap cuvette. The electroporator was set at 820 V and 99 μs, with 1.1-s intervals and 4 pulses. The actual voltage was around 690 V for each sample. The cells were left to recover for 10 min at room temperature before being diluted into 10 ml of complete media. The cells then were seeded into a 10-cm dish, and virus samples were harvested at 24 h and 48 h or, for Luc-expressing replicons, put in 24-well plates and then harvested at 4 h, 48 h, or 72 h for luciferase (Luc) assay.
HCV titration.
Supernatant virus titers were determined by endpoint dilution assays as described previously (16, 20). Huh7.5 cells were seeded into 96-well plates at 7 × 103 cells/well overnight. Viral supernatant was serially diluted 10-fold in complete media and used to infect the cultured cells. After 3 days of incubation, the cells were immunostained with HCV NS5A-specific antibody. Foci positive for HCV NS5A protein were counted, and the infectivity titer was calculated from the average of the number of foci counted in the last and second-to-last wells of the serial dilution that had positive foci. The viral titer was expressed as focus-forming units (FFU)/ml.
CellTiter-Glo luminescent cell viability assay.
Cell viability was determined using the CellTiter-Glo luminescent cell viability assay (Promega, Madison, WI) according to the manufacturer's instructions. The assay was performed on Huh7.5 cells treated with various drugs or on shRNA-expressing cells with or without lipid treatment. Typically, 7 × 103 cells were grown in each well of a 96-well plate for 2 days (for shRNA-expressing cells) or 3 days (for drug-treated cells). Prior to the assay, the plates were incubated at room temperature for approximately 30 min. One hundred microliters of CellTiter-Glo reagent then was added to 100 μl of culture medium in each well, and the cells were incubated on an orbital shaker for 2 min to induce lysis. The cells then were incubated at room temperature for 10 min to develop luminescent signal. Luminescence was recorded in a luminometer (Centro LB microplate luminometer; Berthold Technologies, Bad Wildbad, Germany).
Luciferase assay.
Luciferase activity was measured with the Luc assay substrate kit (Promega) and a luminometer (Centro LB microplate luminometer; Berthold Technologies, Bad Wildbad, Germany). Before the Luc assay, the medium was removed from triplicate wells for each transfected construct, and the cells were briefly washed twice in PBS. Fifty microliters of 1× cell culture lysis reagent (CCLR; Promega) buffer (1:5 diluted in PBS) was added to lyse the cells in each well of the 24-well plate, and the plates were shaken gently at room temperature for 15 min. The lysate was removed from the plate and transferred to a 1.5-ml Eppendorf tube. The supernatant was transferred to a fresh tube after a 1-min spin at 12,000 × g in a microcentrifuge. Five microliters of the lysate then was added to 50 μl of Luc assay substrate (Promega) and briefly mixed by vortexing before measuring Luc activity.
Phosphatidylcholine assay.
Huh7.5 cells were mock infected or infected with HCV Jc1. Alternatively, control or FAPP2 shRNA cells were treated with doxycycline. At 48 h postinfection or posttreatment, a phosphatidylcholine assay kit (Sigma-Aldrich, St. Louis, MO) was used to determine the phosphatidylcholine level in 2 × 106 cells according to the manufacturer's instructions.
Immunoblotting of HCV and host proteins.
Transfected or infected cells were lysed in radioimmunoprecipitation assay (RIPA) buffer (150 mM NaCl, 50 mM Tris, pH 8.0, 1 mM EDTA, 1% NP-40, 0.1% SDS, 1 mM phenylmethylsulfonyl fluoride, and complete protease inhibitor cocktail [Roche]). Typically, 100 μg of protein was resuspended in 4× sample buffer (240 mM Tris [pH 6.8], 4% SDS, 40% glycerol, 4% β-mercaptoethanol, 0.01% bromophenol blue) and boiled for 10 min. The proteins were resolved on 10 to 12% SDS-PAGE, followed by transfer onto an Immobilon-P membrane (polyvinylidene difluoride [PVDF]; Millipore). Following incubation with the respective primary antibody and horseradish peroxidase (HRP)-conjugated secondary antibody, proteins were visualized by an enhanced chemiluminescence detection method (clarity Western ECL substrate; Bio-Rad, Hercules, CA).
Indirect immunofluorescence.
Transfected cells were seeded on glass coverslips and placed in 6-well dishes. At 48 h posttransfection or postseeding, cells on coverslips were washed with PBS twice and fixed for 15 min with 4% paraformaldehyde in PBS. After washing twice with PBS, the cells were permeabilized for 5 min at room temperature in 0.05% Triton X-100-PBS. Cells then were incubated in blocking buffer (3% BSA in PBS) for 30 min at room temperature, followed by staining of proteins with NS4B-, NS5A-, or FAPP2-specific antibody and Alexa Fluor 488- or 594-conjugated secondary antibody. When indicated, nuclei were counterstained with 4′,6-diamidino-2-phenylindole (DAPI; Sigma-Aldrich, St. Louis, MO). After three washes with PBS, the cells were mounted on glass slides with Vectashield (Vector Laboratories, Inc., Burlingame, CA) mounting medium and sealed with nail polish. Cell samples were imaged with an Olympus FV1000 confocal microscope.
Membrane floatation and detergent resistance membrane analysis.
Con1 replicon cells were grown for 48 h in 6- to 8- by 100-mm dishes (1 × 106 cells/dish). The isolation of detergent-resistant membrane fractions was performed as described by Aizaki et al. (39), with some modifications. The cells were resuspended in 1 ml hypotonic buffer (10 mM Tris-HCl [pH 7.5], 10 mM KCl, 5 mM MgCl2 1 tablet of Complete Mini; Roche, Nutley, NJ) for 30 min and passed through a 25-gauge needle 20 times (39). Cell lysates were spun at 1,000 × g for 10 min at 4°C to pellet cellular debris and nuclei. A discontinuous OptiPrep gradient (5%, 25%, and 30% in 50 mM Tris-HCl [pH 7.5], 25 mM KCl, and 5 mM MgCl2) was layered on top of the lysate mixed with 2 ml of 60% and 80 μl 5% OptiPrep, and the samples were spun at 41,000 rpm for 4 h, 45 min at 4°C in an SW41Ti rotor. A total of 8 fractions (1,374 μl each) were collected from top to bottom. Each fraction was precipitated with 15% trichloroacetic acid (TCA), and pooled fractions 1 to 4 (membrane-bound proteins) and 5 to 8 (soluble proteins) were separated on 10% SDS-PAGE and processed for Western blotting as described above. For NP-40 treatment, cells lysates were treated with 1% NP-40 for 15 min at 4°C, followed by membrane floatation.
Processing of samples for ultrastructural analysis.
Control and FAPP2 shRNA-expressing cells (7.5 × 106) were electroporated with 10 μg of HCV Jc1 RNA, collected at 48 h postinfection in PBS, and spun at 900 rpm. The cell pellet was fixed with 3% gluteraldehyde in PBS at room temperature (RT) for 2 h, washed with PBS, and incubated with 1% osmium tetroxide (in 0.1 M cacodylate buffer) for 40 min at RT. The cells then were washed once with 0.1 M cacodylate buffer, washed once with 80% acetone, and incubated overnight at 4°C in 2% uranyl acetate–80% acetone. Before visualizing the cells, they were dehydrated in an acetone series and infiltrated with 50%, 75%, and 100% Epon before final embedding and polymerization in fresh 100% Epon at 60°C for 72 h. Thin sections were cut on a Leica UC7 microtome, placed on carbon-coated cooper 100-mesh grids, and stained with 2% uranyl acetate and 1% lead citrate. The cells were imaged at 80 kV in a Technai F30 transmission electron microscope and processed via Adobe Photoshop.
Extraction and analysis of cellular GSLs.
Total glycosphingolipids (GSLs) were extracted from cultured cells as described by Hug et al. (40), with some modifications. Briefly, cells were induced with 3 μg/ml of doxycycline for 48 h or were infected with Jc1 virus (multiplicity of infection [MOI] of 30) for 48 h. After trypsinization, the cells were counted, washed with PBS, and pelleted at 800 × g for 10 min. The cell pellet was resuspended in 0.5 ml of water, and the lysate was added to 2 ml of chloroform-methanol (CHCl3-CH3OH [2:1, vol/vol]). After vortexing, equal volumes (0.5 ml) of CHCl3 and water (H2O) were added to the lysate. The suspension was vortexed and centrifuged at 800 × g for 10 min to separate the three phases: the lower organic phase contains GSLs, the upper aqueous phase contains the remaining GSLs, and the interface contains proteins and other lipids. The GSL extract in the lower phase was removed for storage on ice, and the CHCl3-H2O extraction step was repeated twice with the aqueous phase. Extracted GSLs were pooled, dried, and resuspended in 200 μl of CHCl3-CH3OH (2:1, vol/vol). The extracted GSLs were separated on a thin-layer chromatography (TLC) plate in CHCl3-CH3OH-H2O (60:35:8, vol/vol/vol). At the end of the run, the plate was air dried, and GSLs were visualized by spraying the TLC plate with orcinol-sulfuric acid reagent and heating at 110°C until bands appeared. GSL bands were evaluated with ImageJ software.
Statistical analysis.
A two-tailed unpaired Student's t test was utilized to determine the statistical significance of our data. P < 0.001 was considered extremely statistically significant, P < 0.01 was considered extremely significant, and P < 0.05 was considered statistically significant. Quantitative data are show as means ± standard deviations.
RESULTS
Knockdown of FAPP2 impedes HCV genome replication.
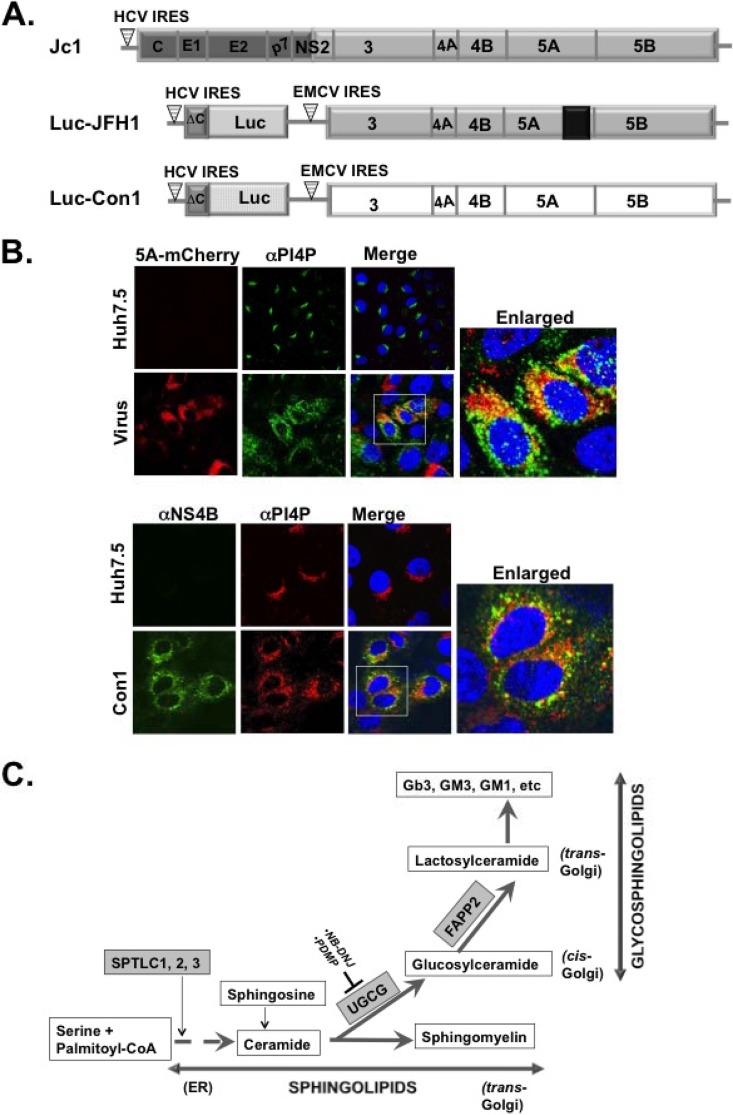
HCV infection is characterized by massive production and retargeting of phosphatidylinositol 4-phosphate (PI4P) lipid to the virus replication complex (Fig. 1A and B) (31, 32, 41–44). However, the role of PI4P in the HCV life cycle is not completely understood. We hypothesized that PI4P recruits host effectors to modulate HCV replication. One of these effectors, four-phosphate adaptor protein 2, or FAPP2, is found in the pathway involved in glycosphingolipid production (Fig. 1C). FAPP2 is crucial for the nonvesicular transport of glucosylceramide (GlcCer) to the trans-Golgi network, where it is converted into more complex glycosphingolipids, such as lactosylceramide (LacCer), globotriaosylceramide (Gb3), and gangliosides (GM3/GM1) (Fig. 1C). To determine the role of FAPP2 protein in HCV production, we generated a stable hepatoma (Huh7.5) cell line with inducible expression of FAPP2 shRNA or nontargeting (control) shRNA. For this study, high-titer HCV Jc1 (genotype 2a or G2a) (34) was the source of the infectious virus, whereas the luciferase-expressing JFH1 (Luc-JFH1; G2a) (36) or Con1 (Luc-Con1; G1b) (35) replicon (Fig. 1A) was used to measure HCV replication efficiency. As seen in Fig. 2A, FAPP2 shRNA induction with doxycycline (Dox) led to a marked decrease (at least 60%) in FAPP2 protein level relative to control shRNA cells. Additionally, FAPP2 shRNA induction had no impact on cell viability as measured by CellTiter-Glo luminescent cell viability assay (Fig. 2B). We also found some decrease (ca. 20%) in FAPP2 protein level in cells without FAPP2 shRNA induction, implying the leakiness of the pTRIPZ shRNA expression vector (Fig. 2A).
FIG 1.
(A) Schematic of the Jc1 (genotype 2a) virus and luciferase reporter replicons (Luc-JFH1 [genotype 2a] and Luc-Con1 [genotype 1b]) used to determine the role of the glycosphingolipid machinery in HCV replication. Note that NS5A has a C-terminal mCherry fusion (in Luc-JFH1 and virus used for panel B) as reported by Gottwein et al. (36). (B) Huh7.5 cells were mock infected or infected with HCV J6/JFH1 (MOI of 0.1) with a C-terminal mCherry fusion to NS5A as described for panel A (36). At 48 h postinfection, the cells were processed for confocal microscopy with mouse monoclonal αPI4P antibody (green). NS5A was detected via mCherry fluorescence. Alternatively, HCV Con1 replicon cells were grown for 48 h and stained with mouse monoclonal α-PI4P antibody (red) and rabbit polyclonal antibody against NS4B (green). The boxed areas are a magnified view for colocalization (yellow) of HCV NS5A or NS4B protein with PI4P. (C) Diagram of the de novo biosynthetic pathway leading to sphingolipids (e.g., ceramide and sphingomyelin) and glycosphingolipids (e.g., glucosylceramide and lactosylceramide) production. The SPTLC1, 2, 3 complex encodes the subunit of SPT (highlighted in gray), the first enzyme in the pathway leading to ceramide production. The SPTLC1 subunit interacts with SPTLC2 or SPTLC3 to form two distinct enzymatic functional complexes. Notice that ceramide is an intermediate product for generating both sphingolipids and glycosphingolipids. UGCG is highlighted in gray and codes for glucosylceramide synthase, a rate-limiting enzyme in glycosphingolipid synthesis. NB-DNJ and PDMP (54–59) are two pharmacological inhibitors of UGCGC. FAPP2 is highlighted in gray and carries glucosylceramide from the cis-Golgi to the trans-Golgi network for conversion into lactosylceramide and other glycosphingolipids. CoA, coenzyme A.
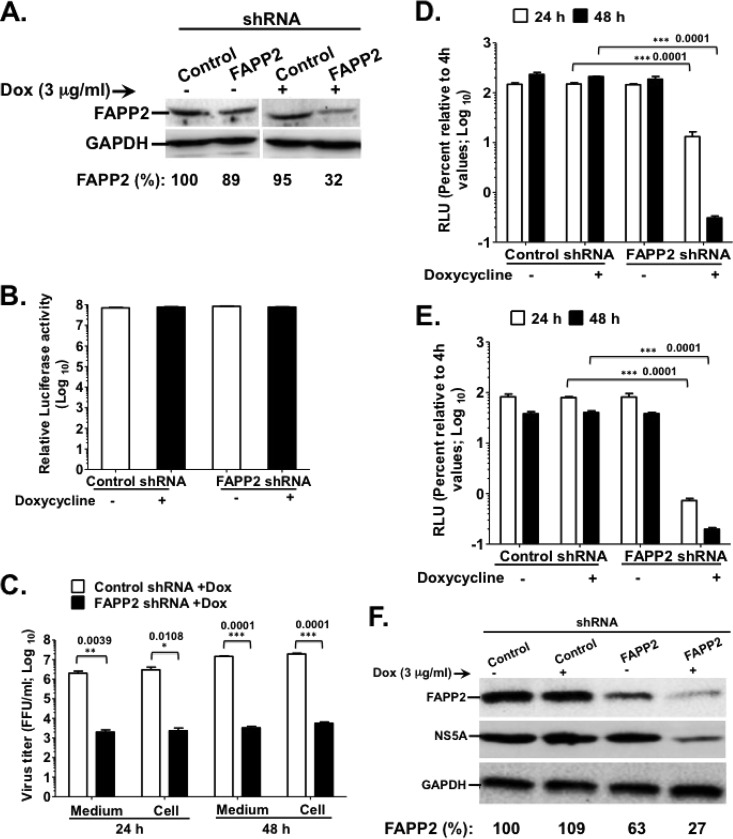
FIG 2.
FAPP2 function is required for HCV genome replication. (A) Control and FAPP2 shRNA-expressing cells were treated with 3 μg/ml doxycycline or left untreated. Forty-eight h posttreatment, cell lysates were separated by SDS-PAGE, followed by immunoblotting with αFAPP2 (1:1,000) and αGAPDH (1:8,000) antibodies. (B) Control and FAPP2 shRNA cells were treated as described for panel A. At 48 h posttreatment, cell viability was determined using the CellTiter-Glo luminescent cell viability assay. (C) Control and FAPP2 shRNA cells were induced with 3 μg/ml doxycycline. At 48 h postinduction, the cells were electroporated with 10 μg of HCV Jc1 RNA. At 24 h and 48 h posttransfection, cell-associated (Cell) and extracellular (Medium) viruses were collected. Virus titers were measured using a limiting-dilution assay (16, 20), and the results are expressed as focus-forming units (FFU)/ml. (D and E) Control and FAPP2 shRNA cells were grown for 48 h with or without doxycycline, followed by transfection with 10 μg of Luc-JFH1 (D) or Luc-Con1 (E) replicon RNA. At 4 h, 24 h, and 48 h posttransfection, cell lysates were collected and HCV replication efficiency was measured by luciferase reporter activity as reported previously (16, 20). RLU, relative light units. The values represent percent luciferase activity relative to 4-h values. (F) The cell extracts also were collected at 48 h posttransfection (D) for immunoblotting with FAPP2 (1:1,000)-, NS5A (1:8,000)-, or GAPDH (1:8,000)-specific antibody. The data are representative of at least two independent experiments with triplicate samples for panels B to E. *, P < 0.05 (statistically significant); **, P < 0.01 (very significant); ***, P < 0.001 (extremely significant).
To determine the impact of FAPP2 knockdown on HCV production, Dox-treated control and FAPP2 shRNA cells were electroporated with HCV Jc1 RNA. Cell-associated and supernatant viruses were collected at 24 h and 48 h posttransfection, followed by Jc1 virus titration. As seen in Fig. 2C, FAPP2 knockdown (FAPP2 KD) led to a more than 100-fold drop in intracellular and cell-associated Jc1 virus titers relative to those of control shRNA cells. These data suggest that FAPP2 protein function is required for infectious HCV production. To test whether the attenuation in Jc1 virus titers could be due to a decrease in virus genome replication, Luc-JFH1 (G2a) or Luc-Con1 (G1b) replicon RNA was transfected into the shRNA stable cells, and HCV replication efficiency was measured at 24 h and 48 h posttransfection. Relative luciferase activity was similar in control and FAPP2 shRNA cells at 4 h posttransfection (data not shown). These data imply that FAPP2 KD has no significant impact on HCV RNA translation. Interestingly, by 48 h posttransfection, JFH1 or Con1 replication efficiency was at least 100-fold lower in Dox-induced FAPP2 than in control shRNA cells (Fig. 2D and E). Consistent with the replication results, FAPP2 KD led to a marked decrease in HCV NS5A expression at 48 h posttransfection (Fig. 2F). Together, these data imply that FAPP2 plays a role in HCV genome replication. However, there is no stringent requirement for FAPP2 protein level, as an approximately 35% decrease in FAPP2 expression (uninduced FAPP2 shRNA) has no significant impact on HCV replication or NS5A expression (Fig. 2F).
Knockdown of SPT impedes HCV genome replication in a manner similar to that of FAPP2 silencing.
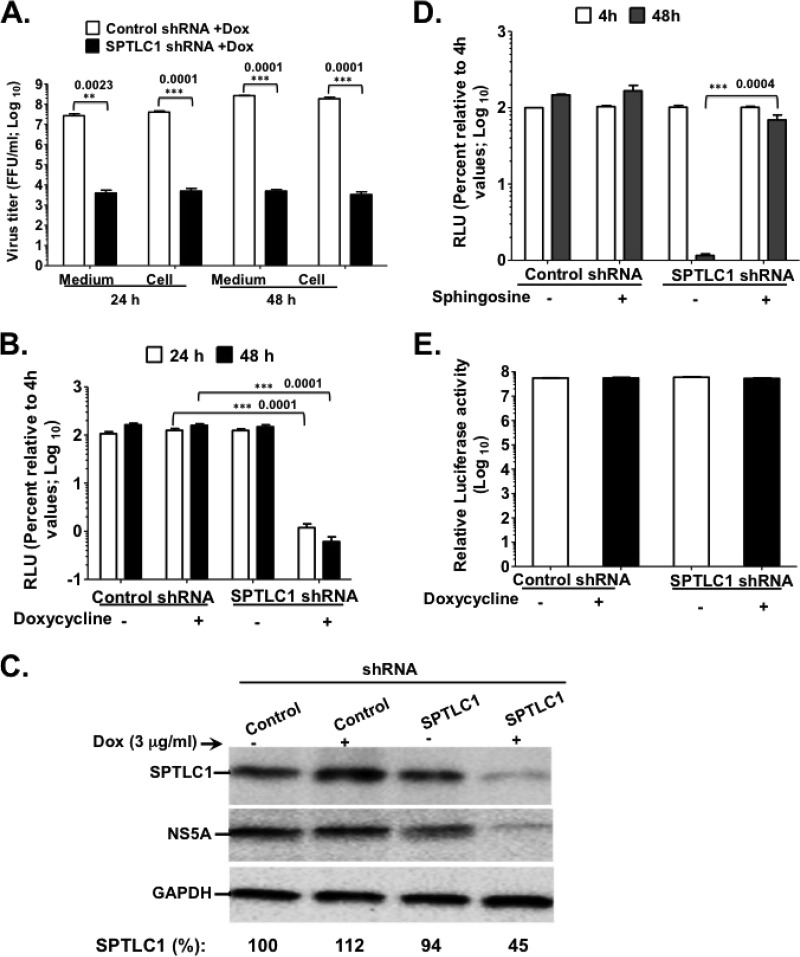
Serine palmitoyltransferase (SPT) is the rate-limiting enzyme in the de novo biosynthesis of ceramide, which can be converted into sphingomyelin (a sphingolipid) or glucosylceramide (a glycosphingolipid) (Fig. 1C) (45, 46). Hence, we predicted that depletion of SPT would negatively impact the cellular synthesis of sphingomyelin and glucosylceramide. To determine the role of SPT in HCV genome replication, we engineered Huh7.5 cells with Dox-inducible shRNA knockdown of SPTLC1, a subunit crucial for SPTLC1-2 or SPTPLC1-3 complex enzymatic activity (47, 48). Indeed, SPTLC1 knockdown led to an at least 100-fold decrease in cell-associated and extracellular Jc1 virus titers compared to those of Dox-induced control shRNA (Fig. 3A). Additionally, SPTLC1 depletion resulted in a more than 100-fold decrease in JFH1 replication efficiency (Fig. 3B) and a marked drop in HCV NS5A protein levels (Fig. 3C). To determine whether SPTLC1 knockdown was responsible mainly for the reduced HCV replication efficiency shown in Fig. 3B, a precursor to ceramide, called sphingosine (Fig. 1C), was added to the Dox-induced culture media of both control and SPTLC1 shRNA cells. Three hundred micrograms/ml of sphingosine was chosen, because this concentration leads to a significant rescue of HCV replication efficiency in cells treated with the SPT inhibitor myriocin (49). As seen in Fig. 3D, supplementing SPTLC1 knockdown cells with 300 μg/ml of sphingosine led to a ca. 60-fold increase in HCV genome replication, suggesting the specificity of SPTLC1 knockdown. Additionally, SPTLC1 knockdown has no marked impact on cell viability, as shown in Fig. 3E.
FIG 3.
SPTLC1 knockdown impedes HCV genome replication. (A) Control and SPTLC1 shRNA-expressing cells were electroporated with 10 μg of HCV Jc1 RNA as described in the legend to Fig. 2B. At 24 h and 48 h posttransfection, virus titers were determined as described for Fig. 2C. (B and C) Control and SPTLC1 shRNA cells were grown for 48 h with or without doxycycline, followed by transfection with 10 μg of Luc-JFH1 replicon RNA. At 4 h, 24 h, 48 h, and 72 h posttransfection, HCV replication efficiency (B) was measured as described for Fig. 2C. (C) The cell extracts also were collected at 48 h posttransfection for immunoblotting with αSPTLC1 (1:500)-, αNS5A (1:8,000)-, or αGAPDH (1:8,000)-specific antibody. (D) Control and SPTLC1 shRNA cells were transfected with Luc-JFH1 replicon RNA as described for panel B. At 4 h posttransfection, 300 μg/ml sphingosine (49) was added to transfected cells, and HCV replication efficiency was measured at 48 h posttransfection. (E) Control and SPTLC1 shRNA cells were left untreated or were treated with doxycycline. At 48 h posttreatment, cell viability was determined as described for Fig. 2B. The data are representative of at least two independent experiments with triplicate samples for panels B to D and F. *, P < 0.05 (statistically significant); **, P < 0.01 (very significant); ***, P < 0.001 (extremely significant).
Functional FAPP2 domains are required for HCV genome replication.
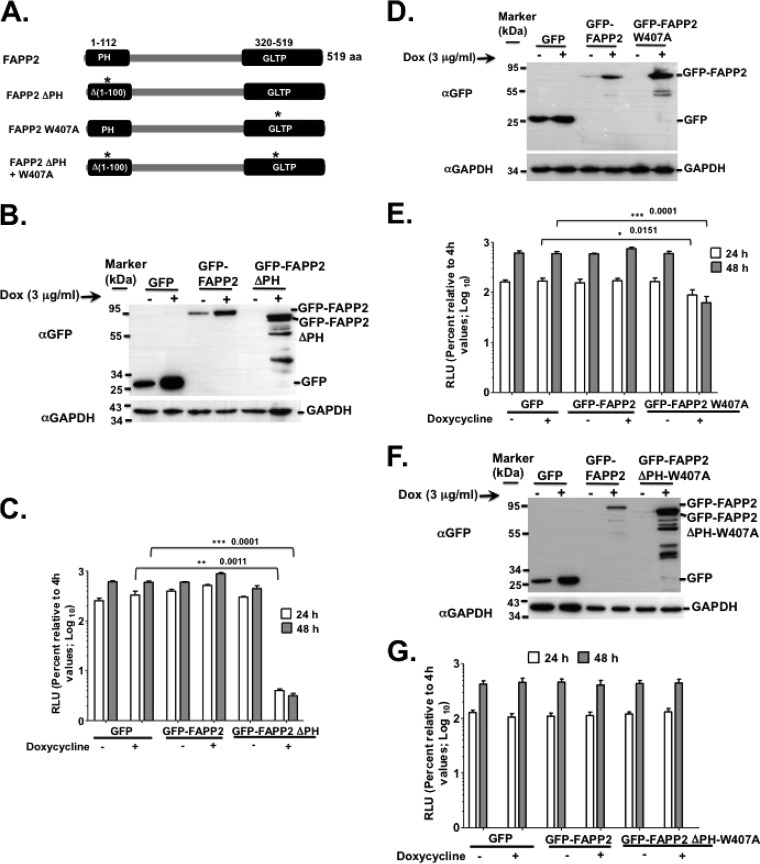
FAPP2 has at least two domains (37, 38, 50, 51). The pleckstrin homology (PH) domain binds to PI4P, Arf1 GTPase, and tubulates membranes (37, 38) (Fig. 4A), while the glycolipid transfer protein (GLTP) domain binds to glucosylceramide, a glycosphingolipid, for transport to the trans-Golgi network, where glucosylceramide is converted into more complex glycosphingolipids (Fig. 1C and 4A). Since PI4P and Arf1 are required for HCV genome replication (33, 52, 53), we reasoned that disrupting the interaction of FAPP2 with PI4P and Arf1 would inhibit HCV RNA synthesis. To test this hypothesis, we generated a stable Huh7.5 cell line expressing a previously reported FAPP2 PH domain deletion mutant (FAPP2 ΔPH) (Fig. 4A) (37, 38, 50) with an N-terminal GFP fusion. For controls, we made Huh7.5 cell lines expressing control GFP vector or WT FAPP2 with the N-terminal GFP fusion. Note that these stable cells do not express FAPP2 shRNA. The cells were grown in the presence or absence of Dox, followed by electroporation with the luciferase-expressing JFH1 replicon RNA. As shown in Fig. 4B, GFP, WT GFP-FAPP2, and GFP-FAPP2 ΔPH proteins were expressed following Dox induction. Some breakdown products also were observed in the GFP-FAPP2 ΔPH (Fig. 4B) cells, suggesting the instability of FAPP2 ΔPH relative to full-length FAPP2 protein. In addition, Luc-JFH1 RNA replication efficiency was measured at 24 h and 48 h postelectroporation. As shown in Fig. 4C, by 48 h postelectroporation, WT FAPP2 protein slightly enhanced HCV genome replication (by ca. 2-fold) compared to that of the GFP vector control. As expected, the expression of the FAPP2 ΔPH mutant led to a significant decrease (ca. 100-fold) in HCV replication (Fig. 4C). We also engineered stable Huh7.5 cells expressing an FAPP2 GLTP domain mutation, W407A, which is defective in glucosylceramide binding (Fig. 4A) (38, 50). Dox-induced expression of FAPP2 W407A mutant protein (Fig. 4D) led to a ca. 10-fold decrease in HCV RNA replication efficiency (Fig. 4E) but had no impact on cell viability (data not shown). Finally, we engineered stable Huh7.5 cells expressing FAPP2 with a mutation in each domain. As seen in Fig. 4F and G, the resulting FAPP2 mutant protein no longer impeded HCV genome replication. Altogether, these data imply that both FAPP2 PH and GLTP domains play a crucial role in HCV replication.
FIG 4.
FAPP2 domains are required for HCV genome replication. (A) Diagram of FAPP2 protein (519 amino acids [aa]) and its functional domains. The PH domain (aa 1 to 112) and the GLTP domain (aa 320 to 519) bind to PI4P and glucosylceramide, respectively. The mutation in the PH domain (FAPP2 ΔPH; aa 1 to 100) or GLTP domain (FAPP2 W407A) is indicated by an asterisk. (B) Stable Huh7.5 cells expressing control GFP, WT GFP-FAPP2, or GFP-FAPP2 ΔPH were grown for 48 h with or without doxycycline, followed by immunoblotting with αGFP (1:2,000) or αGAPDH (1:8,000) antibody. Note that these cells express no shRNA. (C) The stable cells shown in panel B were transfected with 10 μg of Luc-JFH1 replicon RNA, and HCV replication efficiency was determined as described for Fig. 2C. (D and E) Stable cells, with FAPP2 W407A mutation or controls, were treated and processed for immunoblotting (D) and HCV replication efficiency assay (E), respectively. (F and G) Stable cells, with FAPP2 ΔPH-W407A mutations or controls, were treated and processed for immunoblotting (F) and HCV replication efficiency assay (G), respectively. The data are representative of at least two independent experiments with triplicate samples for panels C and D. *, P < 0.05 (statistically significant); **, P < 0.01 (very significant); ***, P < 0.001 (extremely significant).
Pharmacological inhibition of glucosylceramide synthase impedes HCV genome replication.
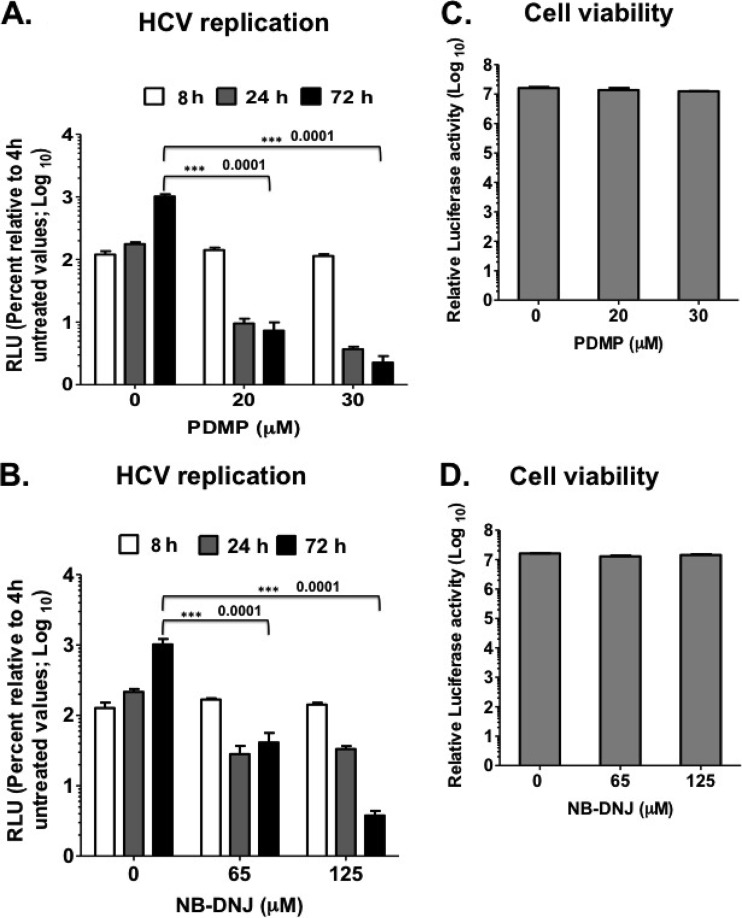
In Fig. 4, the data from genetic analysis suggest that the FAPP2 GLTP domain, or glucosylceramide, is required for HCV genome replication. Here, we took advantage of pharmacological inhibitors of glucosylceramide synthase (UGCG), N-butyldeoxynojirimycin (NB-DNJ) (54–57) and d,l-threo-1-phenyl-2-decanoylamino-3-morpholino-1-propanol (PDMP) (58, 59), to determine the role of glucosylceramide in HCV RNA synthesis. Since UGCG activity is required to generate the glucosylceramide (Fig. 1C) bound to the FAPP2 GLTP domain, we reasoned that UGCG inhibition would have an impact on HCV genome replication similar to that of the FAPP2 W407A mutant protein (38, 50). Thus, Luc-JFH1 replicon RNA was electroporated into Huh7.5 cells. At 4 h postelectroporation, the cells were treated with previously reported concentrations of the drugs (54–59), and treatment was kept until the end of the experiment. As shown in Fig. 5A and B, PDMP or NB-DNJ inhibited HCV genome replication in a dose-dependent manner at 24 h and 72 h posttransfection. Since UGCG activity is crucial for glucosylceramide synthesis, these data are consistent with the finding that the FAPP2 GLTP domain plays a role in HCV genome replication. Note that treating the Huh7.5 cells for 72 h with PDMP or NB-DNJ had a negligible impact on cell viability (less than 10% drop at the highest concentration) (Fig. 5C and D).
FIG 5.
Glucosylceramide synthase (UGCG) inhibitors impede HCV replication efficiency. (A and B) Huh7.5 cells were transfected with 10 μg of JFH1-Luc replicon RNA and were left untreated or treated with various concentrations of PDMP or NB-DNJ. HCV replication efficiency was determined at 8 h, 24 h, and 72 h posttransfection. Note that the cells were treated at 4 h postelectroporation. (C and D) Impact of PDMP and NB-DNJ on cell viability. The cells were treated with 20 and 30 μM PDMP or 63 and 125 μM NB-DNJ for 72 h, followed by cell viability measurement as described in Materials and Methods. The results are representative of three independent experiments with triplicate samples for data from panels A to D. *, P < 0.05 (statistically significant); **, P < 0.01 (very significant); ***, P < 0.001 (extremely significant).
HCV genome replication in FAPP2 knockdown cells is rescued to near completion by some glycosphingolipids.
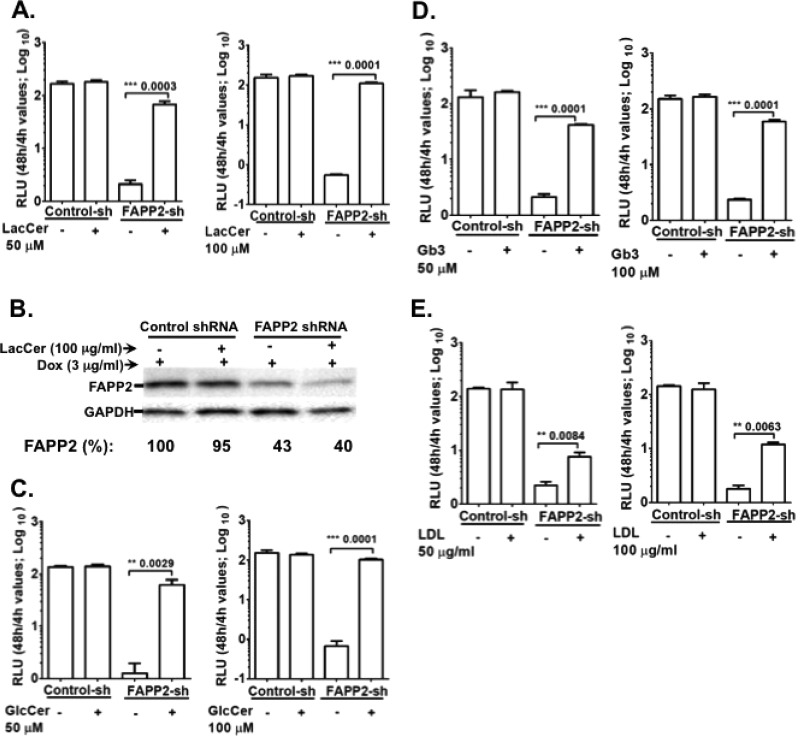
Based on the findings shown in Fig. 4 and 5, we postulated that FAPP2 brings GlcCer into the HCV replication complex to enhance virus RNA synthesis. Alternatively, GlcCer is converted into complex glycosphingolipids, such as LacCer or Gb3, to facilitate HCV genome replication. To test this hypothesis, Dox-induced control and FAPP2 shRNA cells were electroporated with Luc-JFH1 replicon RNA. At 4 h posttransfection, the cells were supplemented with 0, 50, or 100 μM GlcCer, LacCer, or Gb3. We also treated the cells with cholesterol in the form of LDL, as cholesterol is required for HCV genome replication (39, 60). As seen in Fig. 6A, C, and D, GlcCer, LacCer, or Gb3 could increase HCV replication efficiency in FAPP2 knockdown cells. The biggest rescue of replication was observed with 100 μM LacCer (ca. 200-fold), followed by 100 μM GlcCer (ca. 100-fold) and 100 μM Gb3 (ca. 40-fold). Additionally, LacCer (Fig. 6B) or GlcCer (data not shown) did not increase the FAPP2 protein level in the FAPP2 knockdown cells, implying a direct role for these glycosphingolipids in HCV genome replication. Thus, we did not determine the FAPP2 level in Gb3- and LDL-supplemented cells. LDL treatment increased HCV genome replication in FAPP2 shRNA cells (Fig. 6E) by only ca. 3-fold, suggesting the specificity of glycosphingolipid rescue in this study.
FIG 6.
HCV replication is restored to near completion by some glycosphingolipids in FAPP2 knockdown cells. (A and B) Dox-induced control and FAPP2 shRNA cells were transfected with 10 μg of Luc-JFH1 replicon RNA as described in Materials and Methods. (A) At 4 h posttransfection, 0, 50, or 100 μM LacCer was added to transfected cells, and HCV replication efficiency was determined at 4 h and 48 h posttransfection. (B) Additionally, cell lysates were subjected to immunoblotting with αFAPP2 (1:1,000) or αGAPDH (1:8000) antibody. (C) Control and FAPP2 shRNA cells were transfected with replicon RNA as described for panel A but treated with 0, 50, or 100 μM GlcCer. HCV replication efficiency was determined as described for panel A. (D and E) Control and FAPP2 shRNA cells were transfected with replicon RNA as described for panel A but treated with various concentrations of Gb3 (D) or cholesterol as a low-density lipoprotein (LDL) (E). HCV replication efficiency was determined as described for panel A. The data are representative of three independent experiments with triplicate samples for panels A, C, D, and E. *, P < 0.05 (statistically significant); **, P < 0.01 (very significant); ***, P < 0.001 (extremely significant).
FAPP2 expression regulates glycosphingolipid levels in virus-infected cells.
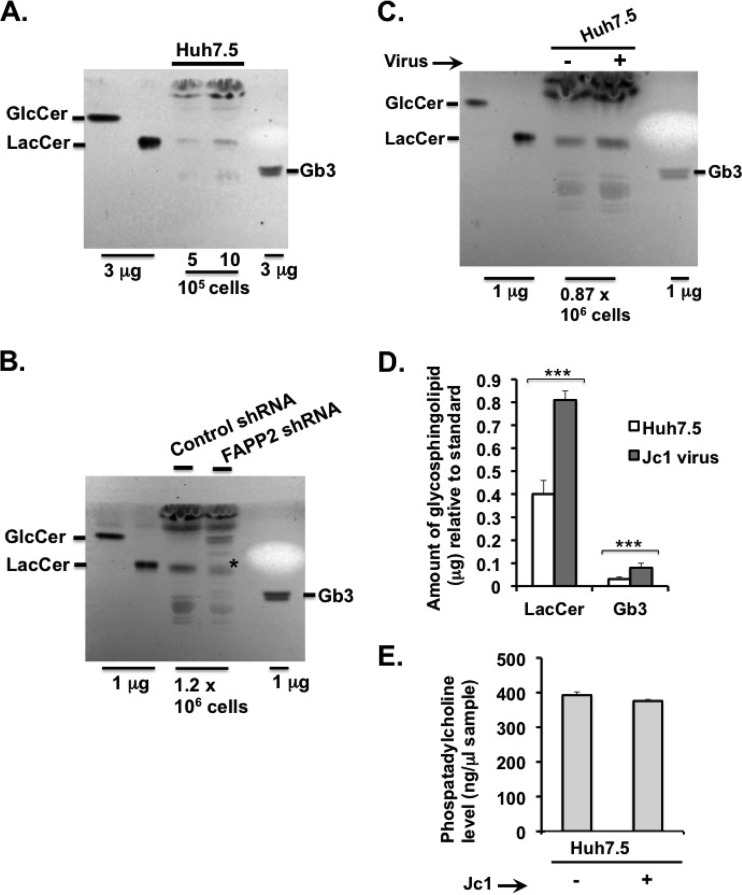
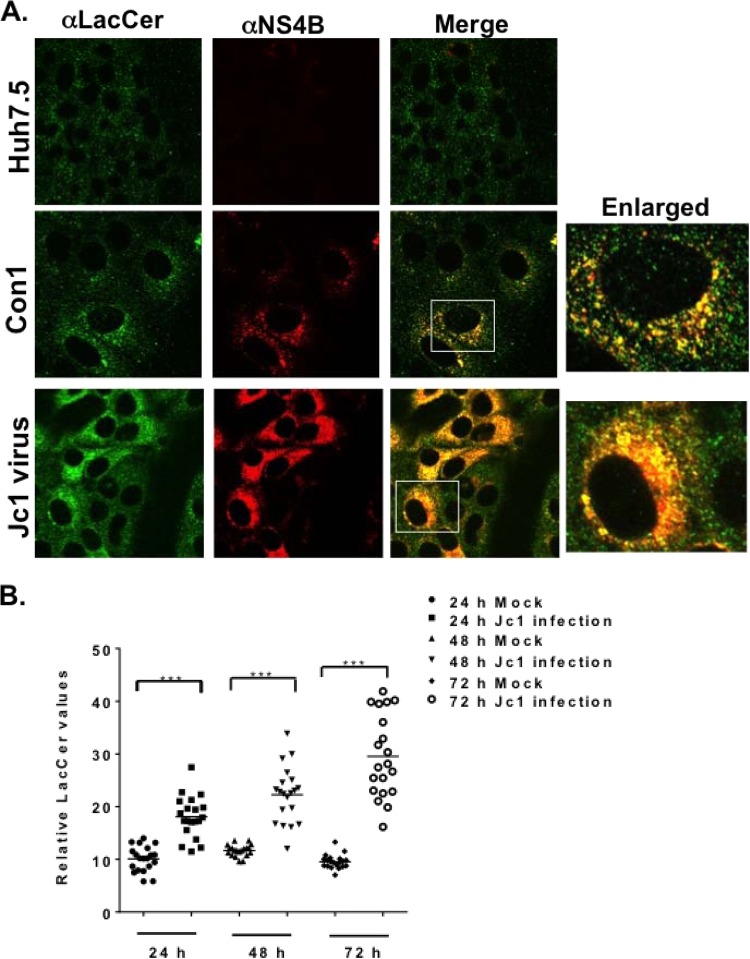
Since GlcCer and LacCer restore HCV genome replication to near completion in FAPP2 knockdown cells (Fig. 6A to C), we sought to detect these lipids in the various cells used in this study. Thus, lipid extract from equal numbers of Huh7.5, control, or FAPP2 shRNA cells was spotted onto TLC plates. The resolved glycolipids then were stained by the orcinol method. As shown in Fig. 7A, LacCer and Gb3 could be detected in extracts from 5 × 105 to 10 × 105 Huh7.5 cells, whereas the GlcCer level was too low to detect. We also detected LacCer and Gb3 in extracts from Dox-induced control shRNA cells. Interestingly, while LacCer and Gb3 levels were reduced by ca. 2-fold (data not shown) in Dox-induced FAPP2 shRNA cells (Fig. 7B; the asterisk indicates LacCer), GlcCer accumulated significantly in FAPP2 shRNA cells relative to control shRNA cells (Fig. 7B). These data are consistent with the role of FAPP2 in the transport of GlcCer and its conversion into more complex glycosphingolipids, such as LacCer and Gb3. We also sought to determine glycosphingolipid levels during HCV infection. Hence, Huh7.5 cells were mock infected or infected with HCV Jc1 at a multiplicity of infection of 30, and glycosphingolipids were extracted at 48 h postinfection. As shown in Fig. 7C and D, LacCer and Gb3 levels increased by 2- to 3-fold in Jc1 virus-infected cells compared to control Huh7.5 cells. No change in GlcCer (Fig. 7C) or phosphatidylcholine (Fig. 7E) level was detected in Jc1 virus-infected cells. Altogether, these data suggest that changes in glycosphingolipid levels regulate HCV genome replication. We also determined the subcellular distribution of LacCer in control, HCV Con1 replicon, or Jc1 virus-infected cells. As seen in Fig. 8A, LacCer colocalizes with HCV NS4B replicase protein in both replicon and virus-infected cells. Additionally, we confirm the finding that the LacCer level increases during the course of HCV infection (Fig. 8B).
FIG 7.
Changes in glycosphingolipid levels in various HCV-expressing cells. (A) Huh7.5 cells were grown for 48 h, followed by glycosphingolipid extraction from 107 cells, as discussed in Materials and Methods. Extracted lipids, from 5 × 105 to 10 × 105 cells, were separated on TLC plates in a chloroform-methanol-water (60:35:8) solvent system and visualized by spraying with orcinol-sulfuric acid reagent. (B) Control and FAPP2 shRNA cells were treated with 3 μg/ml Dox for 48 h. Glycosphingolipids were extracted from 107 cells, and aliquots that correspond to 1.2 × 106 cells were separated on TLC plates and processed as described for panel A. (C) Huh7.5 cells (9 × 106) were mock infected or infected with HCV Jc1 at an MOI of 30 to ensure at least 95% infection efficiency. At 48 h postinfection, glycosphingolipids were extracted from 7 × 106 cells, and aliquots that correspond to 0.87 × 106 cells were separated on TLC plates as described for panel A. (D) The amount of glycosphingolipids relative to the standard (1 μg of LacCer or Gb3), in mock-infected and Jc1 virus-infected Huh7.5 cells shown in panel C, was determined with ImageJ software. ***, P < 0.001 (extremely significant). (E) Two million mock-infected and Jc1 virus-infected cells from panel C were processed for phosphatidylcholine assay as described in Materials and Methods. The results shown in panels D and E are representative of at least two independent experiments, each containing data from 3 TLC runs.
FIG 8.
Lactosylceramide is associated with HCV NS4B protein. (A) Parental Huh7.5 and HCV Con1 (genotype 1b) replicon cells were grown for 48 h and processed for confocal microscopy with mouse monoclonal αLacCer antibody (1:500; green) and rabbit polyclonal αNS4B antibody (1:150; red). Huh7.5 cells also were infected with HCV Jc1 (MOI of 1) and processed for confocal microscopy as described above. The boxed areas represent a magnified view for colocalization (yellow) of lactosylceramide and HCV NS4B protein. (B) Huh7.5 cells were infected with HCV Jc1 (MOI of 1) as described for panel A and processed for confocal microscopy 24 h, 48 h, and 72 h postinfection. For each infection time point, confocal images were taken of 20 representative cells. The intensity of lactosylceramide pixels was calculated with the JACoP plugin in ImageJ software. Each filled circle or square (24 h; mock or Jc1 infected), upper or lower triangle (48 h; mock or Jc1 infected), diamond (72 h; mock), or open circle (72 h; Jc1 infected) represents one cell.
FAPP2 is associated with HCV NS5A and viral dsRNA.
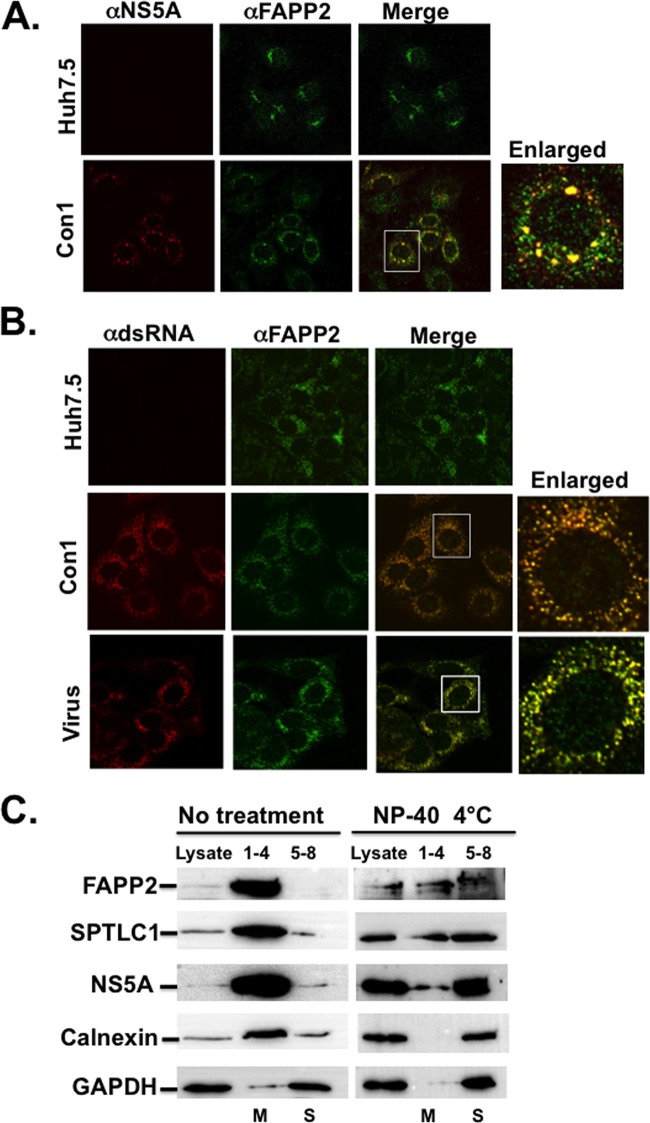
FAPP2 is responsible for the nonvesicular transport of GlcCer from the cis-Golgi to the trans-Golgi network, where FAPP2 binds to PI4P (Fig. 1C) (37, 50, 51, 61). Thus, in naive Huh7.5 cells, FAPP2 is expected to have a Golgi network-like distribution. However, given the massive production and redistribution of PI4P in HCV Con1 replicon- and Jc1-infected cells (Fig. 1B), we predicted an alteration of FAPP2 subcellular distribution in these cells. Indeed, while FAPP2 displays a perinuclear and Golgi network-like distribution in Huh7.5 cells, there was a redistribution and colocalization of FAPP2 with HCV NS5A replicase protein in Con1 replicon cells (Fig. 9A). Additionally, FAPP2 colocalized with HCV dsRNA, the site of virus genome replication, in both Con1 replicon and virus-infected cells (Fig. 9B). We also performed membrane floatation assay of Con1 replicon cell lysates with or without 1% NP-40 treatment. This approach has been used in several studies to demonstrate the association of the HCV replication complex with detergent-resistant membrane fractions (32, 39, 62, 63). As seen in Fig. 9C, NS5A, FAPP2, SPTLC1, and calnexin are enriched in membrane (M) fractions (1–4) in untreated Con1 lysate. As expected, GAPDH was found mostly in soluble fractions (5–8). Following NP-40 treatment, these proteins became soluble, but a fraction of NS5A, FAPP2, or SPTLC1 protein cofractionated with the detergent-resistant membranes. Altogether, these data suggest that FAPP2 is recruited to the HCV replication complex.
FIG 9.
FAPP2 colocalizes with HCV NS5A and viral dsRNA. (A) Parental Huh7.5 and HCV Con1 (genotype 1b) replicon cells were grown for 48 h and processed for confocal microscopy with mouse monoclonal αNS5A antibody (1:1,000; red) and rabbit polyclonal αFAPP2 antibody (1:100; green). (B) Huh7.5, Con1 replicon, or Jc1 (34) virus-infected cells were grown as described for panel A and processed for confocal microscopy with mouse monoclonal antibody against dsRNA (1: 200; red) and rabbit polyclonal αFAPP2 antibody (1:100; green). The boxed areas indicate the magnified view for colocalization (yellow color) of HCV NS5A (A) or dsRNA (B) with FAPP2 protein. (C) FAPP2 cofractionates with HCV replicase NS5A protein in the detergent-resistant membrane fraction. Con1 replicon cell lysates were left untreated or were treated with 1% NP-40 on ice and subjected to membrane floatation. Proteins from pooled fractions (1 to 4 and 5 to 9) were separated by SDS-PAGE, followed by immunoblotting with antibodies against FAPP2, SPTLC1, NS5A, calnexin, or GAPDH. Numbers 1 to 4 refer to membrane (M) fractions, and numbers 5 to 8 refer to soluble (S) fractions.
FAPP2 is required for HCV replication complex formation.
HCV genome replication takes place on membranous web (MW) vesicles (17–19, 27) typically seen as NS4B or NS5A foci in confocal microscopy (19, 20, 27, 64). The disruption of these foci impedes HCV RNA replication efficiency (19, 21–24). Since FAPP2 colocalizes with HCV NS5A and dsRNA, we postulated that FAPP2 supports HCV replication complex formation. To test this hypothesis, control and FAPP2 shRNA-expressing cells were infected with Jc1 virus. At 48 h postinfection, the virus titer in the supernatants was measured to confirm the impact of FAPP2 shRNA knockdown on HCV production (data not shown). Additionally, cells were collected and processed for electron microscopy.
As seen in Fig. 10A and B, virus-infected control shRNA cells accumulate vesicles, ranging from 150 nm to 200 nm in size, often close to each other and the nucleus. Such vesicles were observed in more than 50% of the processed samples. Some of the vesicles have darker membrane staining (arrowhead), suggestive of the double membrane vesicles associated with the MW. In contrast, most of the virus-infected FAPP2 shRNA cells did not accumulate such vesicles, and the darker membrane staining was not apparent (Fig. 10C and D). Additionally, a few larger vesicles, ranging from 250 to 300 nm in size (Fig. 10D, arrows), were found in approximately 50% of the infected FAPP2 shRNA cells. These data imply that FAPP2 knockdown interferes with proper MW vesicle formation. However, to rule out the impact of genome replication on MW vesicles or HCV replication complex, we transfected Dox-induced control and FAPP2 shRNA cells with a construct expressing nonreplicating HCV polyprotein NS3-4A-4B-5A-5B, followed by confocal microscopy at 48 h posttransfection. As shown in, Fig. 11A control shRNA cells display NS5A foci typically associated with functional HCV replication complex (19, 20, 65). However, NS5A foci appear to be clustered or diffuse in at least 50% of the FAPP2 shRNA cells expressing the HCV polyprotein (Fig. 11A). Additionally, while NS4B and NS5A foci colocalize in control shRNA cells (Fig. 11B, i to x; Pearson coefficient, 0.79), the diffuse subcellular distribution of NS4B and NS5A made it difficult to accurately determine their colocalization status in FAPP2 shRNA cells (Fig. 11B, xi to xx). Together, these data suggest that FAPP2 plays a role in the formation of a functional HCV replication complex.
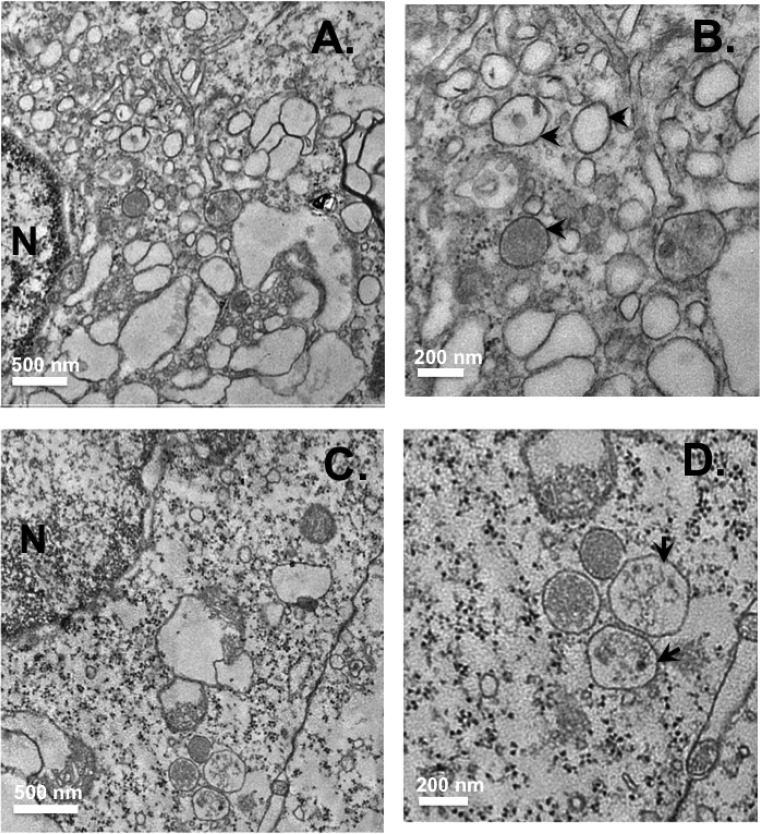
FIG 10.
Ultrastructural analysis of shRNA cells infected with Jc1 virus. Control (A and B) and FAPP2 (C and D) shRNA cells were treated with Dox as described in the legend to Fig. 7, followed by Jc1 virus infection as described in Materials and Methods. The cells were processed for electron microscopy at 48 h postinfection. (B and D) Magnified images of panels A and C, respectively. (B) Arrowheads indicate vesicles of 150 to 200 nm in size. (D) Arrows show vesicles larger than 250 nm in size. Scale bars indicate the magnification for each image.
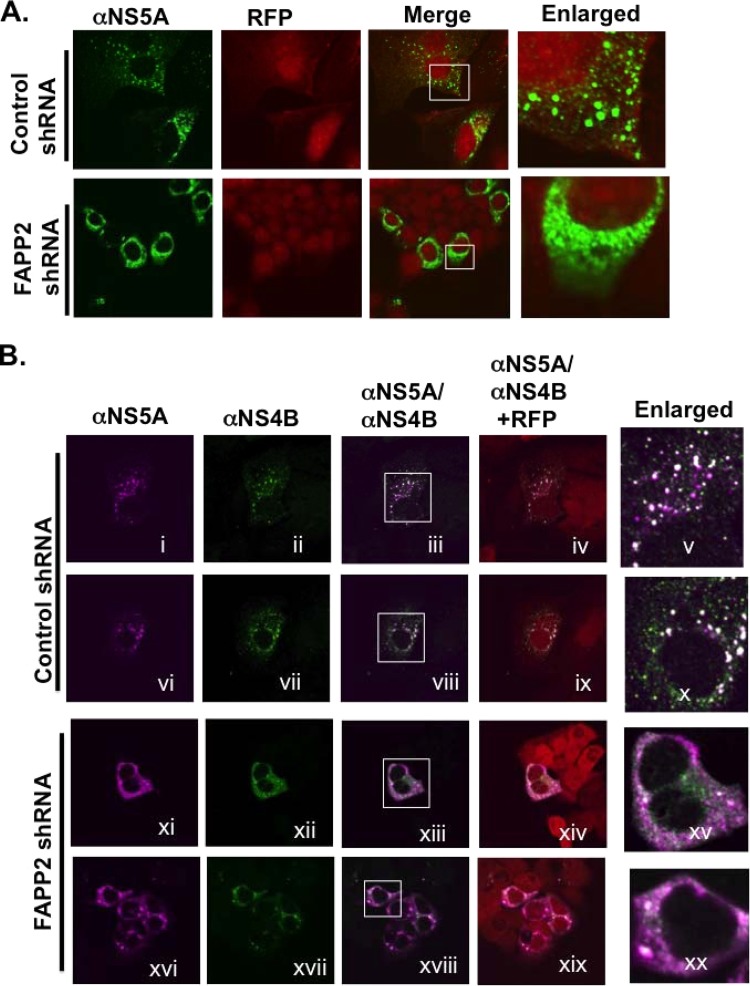
FIG 11.
FAPP2 knockdown disrupts HCV NS4B and NS5A focus formation. (A) Control and FAPP2 shRNA cells were treated with 3 μg/ml doxycycline as described in the legend to Fig. 7, followed by transfection with pIRES vector expressing HCV NS3-4A-4B-5A-5B polyprotein in the presence of doxycycline. At 48 h posttransfection, the cells were fixed and processed for confocal microscopy with mouse monoclonal αNS5A antibody (1:1,000; green). Red fluorescent protein (RFP) indicates control and FAPP2 shRNA cells. Magnified areas, with NS5A subcellular distribution, are shown by rectangles. (B) Control (i to x) and FAPP2 (xi to xx) shRNA cells were treated as described for panel A and processed for confocal microscopy with mouse monoclonal αNS5A antibody (1:1,000; magenta) and rabbit polyclonal antibody against NS4B (1:25; green). RFP indicates shRNA-expressing cells as described for panel A. Magnified areas, with putative NS4B and NS5A colocalization (white), are indicated by rectangles.
DISCUSSION
Like poliovirus (33), HCV infection is characterized by phosphatidylinositol 4-kinase activation, increased production, and redistribution of PI4P lipid to HCV replication complex (31, 32, 41–44). However, the role of PI4P in HCV production is not well defined. As an integral component of host membranes, PI4P may contribute to the curvature or integrity of the MW vesicles, the site of HCV genome replication (17–19, 27). PI4P also may interact with HCV replicase proteins to stimulate their function or keep them anchored in the MW. Alternatively, PI4P may bind to host factors which modulate the transition from HCV genome replication to virus particle production. Indeed, several host factors, also called effectors, have been reported to bind to PI4P. They include oxysterol binding protein (OSBP), ceramide transfer protein (CERT), four-phosphate adaptor proteins 1 and 2 (FAPP1 and FAPP2), and Golgi phosphoprotein 3 (GOLPH3) (66). Recent reports indicate that knockdown of OSBP or GOLPH3 leads to a marked reduction in HCV secretion (67, 68), implying that some PI4P effector proteins are crucial for infectious HCV particle production.
In the current study, we demonstrate that FAPP2, a glycosphingolipids transport protein, is required for HCV genome replication. We postulated that the FAPP2 pleckstrin homology (PH) domain, which binds to PI4P, is required for HCV RNA synthesis. Consistent with this hypothesis, the FAPP2 PH domain mutant (FAPP2 ΔPH) protein impedes HCV replication efficiency. Notably, several lines of evidence suggest that the FAPP2 glycolipid-binding domain (GLTP) (50) also is required for HCV replication: (i) a mutation in the GLTP domain inhibits HCV RNA synthesis; (ii) inhibition of glucosylceramide synthesis hampers HCV replication; and (iii) some glycosphingolipids (e.g., lactosylceramide) are markedly induced during HCV infection and restore to near completion virus replication in FAPP2 knockdown cells. More importantly, we observed a redistribution and colocalization of FAPP2 and lactosylceramide with HCV replicase protein (or viral dsRNA), implying a direct role of FAPP2 and glycosphingolipids in HCV RNA synthesis. In addition, FAPP2 knockdown interferes with MW vesicle formation in virus-infected cells and alters the subcellular distribution of HCV replicase proteins. These findings imply that FAPP2 modulates the organization of functional HCV replication complex. Our data also indicate that FAPP2 does not regulate HCV RNA translation. Nevertheless, we do not completely rule out a role for FAPP2 in HCV entry, virus particle assembly, or release. Note that FAPP2 knockdown has no impact on cell viability in vitro or in vivo (61). Finally, poliovirus induces PI4P production (33) but does not require FAPP2 for genome replication (data not shown), implying the specificity of the FAPP2 requirement in the HCV life cycle.
FAPP2 is a rate-limiting protein in the de novo biosynthesis of glycosphingolipids (Fig. 1C), which are tightly linked to ceramide (a sphingolipid) production and SPT enzyme complex (SPTLC1 and SPTLC2 or SPTLC1 and SPTLC3) activity (45, 47, 48, 69) (Fig. 1C). A recent report indicates the upregulation of ceramide levels during HCV infection, and the pharmacological inhibition of SPT enzyme impedes HCV genome replication (70). Additionally, another sphingolipid, sphingomyelin (Fig. 1C), was reported to bind to and stimulate HCV RNA-dependent RNA polymerase activity (71). These findings have led to the proposal that sphingomyelin is the crucial sphingolipid regulator of HCV genome replication. Hence, since ceramide transfer protein (CERT) is involved in sphingomyelin production (72–76), we predicted that CERT knockdown would inhibit HCV genome replication in a manner similar to that of SPTLC1 knockdown. Surprisingly, CERT knockdown led to ca. 3-fold drop in HCV replication (data not shown), whereas SPTLC1 depletion resulted in a decrease in HCV replication efficiency similar to that of FAPP2 knockdown (ca.100-fold). Thus, we conclude that the sphingolipid pathway, leading to glycosphingolipid production, plays a major role in HCV genome replication.
To demonstrate how FAPP2 modulates HCV RNA synthesis, we have shown that FAPP2 is associated with components of the HCV replication complex (RC) (Fig. 9). FAPP2 appears to colocalize completely with the dsRNA but only partially with HCV NS5A (Fig. 9). While this needs to be further investigated, we have identified putative RNA-binding motifs in FAPP2 protein, implying potential FAPP2 interaction with HCV RNA. Nevertheless, we propose that the FAPP2 PH domain binds to PI4P and plays a role in the colocalization of FAPP2 with the HCV replication complex. The redistribution of FAPP2 during HCV infection also may require Arf1 GTPase, which interacts with FAPP2 and facilitates HCV genome replication (37, 52, 53). Finally, another PI4P effector, oxysterol binding protein, binds to HCV NS5A and recently was shown to play a role in HCV replication via cholesterol transport to the HCV replication complex (67, 77). Hence, studies are under way to determine putative interactions between FAPP2 and HCV proteins and their biological significance.
Our study indicates that the FAPP2 ΔPH mutant protein has a more dominant-negative impact on HCV RNA synthesis than the expression of the FAPP2 W407A protein (100-fold versus 10-fold decrease) (Fig. 4). A likely explanation is that the FAPP2 ΔPH mutant protein cannot be recruited to the HCV RC but would compete against endogenous FAPP2 for glycosphingolipid binding, whereas FAPP2 W407A mutant protein is recruited to the HCV RC, taking up either too much PI4P or too much space on the RC but unable to bring the glycosphingolipids. If correct, the transported glycosphingolipids are major determinants in the role of FAPP2 in HCV replication. Hence, we propose the model shown in Fig. 12. Following infection and translation of the virus genome, HCV activates PI4KIIIα to produce PI4P (Fig. 12A). PI4P binds to FAPP2, recruiting glycosphingolipids to the initiating RC. Localized membrane accumulation causes the glycosphingolipids to stimulate membrane curvature (Fig. 12B), the first step in MW vesicle and HCV RC formation. In this scenario, the induction of the double membrane vesicles (Fig. 12C) may involve the concerted action of HCV NS4B, Rab5, and autophagic proteins as previously reported (23, 78, 79). Alternatively, glycosphingolipids may regulate the size of the nascent MW vesicles or retention of the replicase proteins in these vesicles. While we favor a role for glycosphingolipids in the formation of the MW vesicles, it is conceivable that glycosphingolipids stimulate the activity of some replicase proteins, as recently reported for sphingomyelin (71).
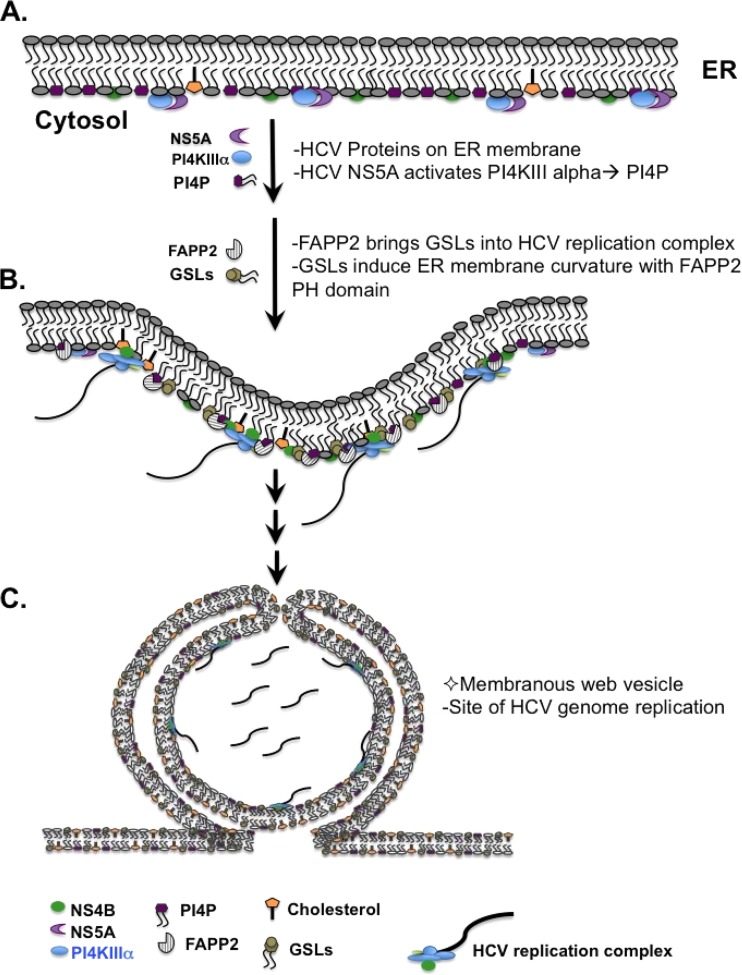
FIG 12.
Proposed model for the role of FAPP2 in HCV genome replication. (A) After translation and processing on endoplasmic reticulum (ER) membranes, HCV NS5A activates PI4KIIIα to produce PI4P. (B) FAPP2 brings glycosphingolipids (GSLs) into the HCV RC in part via interaction with PI4P. GSLs and the FAPP2 PH domain regulate ER membrane curvature to form the MW vesicles. Alternatively, GSLs control the size of the MW vesicles or the local concentration of the replicase proteins. (C) Schematic of the double-membrane vesicles, as depicted by Paul et al. (79); these vesicles are clustered into an MW structure.
The order of glycosphingolipid preference for HCV replication rescue in FAPP2 KD cells is LacCer (ca. 200-fold), followed by GlcCer (ca. 100-fold) and Gb3 (ca. 40-fold). In addition, LacCer is upregulated and retargeted to the HCV RC (Fig. 7 and 8), further implying a direct role for LacCer in HCV replication. Hence, we propose that the supplemented LacCer contributes to HCV replication via nonvesicular and vesicular trafficking, as a recent report indicates that FAPP2 also binds to LacCer (80, 81). However, we were surprised that GlcCer also would rescue HCV replication, as FAPP2 KD slightly increased the GlcCer level (Fig. 7B). Since FAPP2 KD leads to a decrease in LacCer production (Fig. 7B), we propose that the excess GlcCer is transported via vesicular trafficking to the trans-Golgi network and utilized for more LacCer production. Alternatively, the added GlcCer and, to a lesser extent, Gb3, may substitute for LacCer utilization during HCV replication. Studies are under way to further define the relative contribution of these glycosphingolipids, and their cognate enzymes, to HCV replication efficiency.
Our model does not rule out a direct role for the FAPP2 PH domain in HCV replication. Indeed, the FAPP2 PH domain has been reported to induce membrane tubulation or wedging, and mutations in the PH domain impede this activity (38, 82). Although speculative, the FAPP2 PH domain also may contribute to membrane curvature to facilitate MW vesicle formation. Hence, in addition to competing for glycosphingolipid binding, we propose that FAPP2 ΔPH mutant protein engages in an unproductive complex with endogenous FAPP2 protein. Consistent with this hypothesis, studies with size-exclusion chromatography and analytical ultracentrifugation suggest that FAPP2 exists as a dimer (38). Additionally, our preliminary study with FAPP2 coimmunoprecipitation also implies that FAPP2 dimerizes (data not shown).
In conclusion, this study has revealed, for the first time, the crucial role of glycosphingolipids and FAPP2 protein in HCV RNA synthesis. Future investigation will focus on combining genetic, biochemical, and ultrastructural approaches to further define the roles of glycosphingolipids, and FAPP2 protein, in the HCV life cycle.
ACKNOWLEDGMENTS
We are grateful to Jens Bukh, Ralf Bartenschlager, Charles Rice, Carlos de Noronha, Jim Drake, Cara Pager, John Wills, Toshiyuki Yamaji, and Raul Andino for reagents, suggestions, and critical readings of the manuscript.
This work was supported by 1R56AI087769 (K.V.K.) and 1R21AI097858-01 (K.V.K.), from the National Institutes of Health, and by the Takeda Science Foundation (to K.H.).
Footnotes
Published ahead of print 13 August 2014
REFERENCES
- 1.Averhoff FM, Glass N, Holtzman D. 2012. Global burden of hepatitis C: considerations for healthcare providers in the United States. Clin. Infect. Dis. 55(Suppl 1):S10–S15. 10.1093/cid/cis361. [DOI] [PubMed] [Google Scholar]
- 2.Lavanchy D. 2011. Evolving epidemiology of hepatitis C virus. Clin. Microbiol. Infect. 17:107–115. 10.1111/j.1469-0691.2010.03432.x. [DOI] [PubMed] [Google Scholar]
- 3.Miller RH, Purcell RH. 1990. Hepatitis C virus shares amino acid sequence similarity with pestiviruses and flaviviruses as well as members of two plant virus supergroups. Proc. Natl. Acad. Sci. U. S. A. 87:2057–2061. 10.1073/pnas.87.6.2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Simmonds P, Bukh J, Combet C, Deleage G, Enomoto N, Feinstone S, Halfon P, Inchauspe G, Kuiken C, Maertens G, Mizokami M, Murphy DG, Okamoto H, Pawlotsky JM, Penin F, Sablon E, Shin IT, Stuyver LJ, Thiel HJ, Viazov S, Weiner AJ, Widell A. 2005. Consensus proposals for a unified system of nomenclature of hepatitis C virus genotypes. Hepatology 42:962–973. 10.1002/hep.20819. [DOI] [PubMed] [Google Scholar]
- 5.Bartenschlager R, Lohmann V. 2000. Replication of hepatitis C virus. J. Gen. Virol. 81(Part 7):1631–1648 http://vir.sgmjournals.org/content/81/7/1631.long. [DOI] [PubMed] [Google Scholar]
- 6.Kuiken C, Simmonds P. 2009. Nomenclature and numbering of the hepatitis C virus. Methods Mol. Biol. 510:33–53. 10.1007/978-1-59745-394-3_4. [DOI] [PubMed] [Google Scholar]
- 7.Hajarizadeh B, Grebely J, Dore GJ. 2013. Epidemiology and natural history of HCV infection. Nat. Rev. Gastroenterol. Hepatol. 10:553–562. 10.1038/nrgastro.2013.107. [DOI] [PubMed] [Google Scholar]
- 8.Bartenschlager R, Sparacio S. 2007. Hepatitis C virus molecular clones and their replication capacity in vivo and in cell culture. Virus Res. 127:195–207. 10.1016/j.virusres.2007.02.022. [DOI] [PubMed] [Google Scholar]
- 9.Blight KJ, Kolykhalov AA, Rice CM. 2000. Efficient initiation of HCV RNA replication in cell culture. Science 290:1972–1974. 10.1126/science.290.5498.1972. [DOI] [PubMed] [Google Scholar]
- 10.Lohmann V, Korner F, Koch J, Herian U, Theilmann L, Bartenschlager R. 1999. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science 285:110–113. 10.1126/science.285.5424.110. [DOI] [PubMed] [Google Scholar]
- 11.Appel N, Zayas M, Miller S, Krijnse-Locker J, Schaller T, Friebe P, Kallis S, Engel U, Bartenschlager R. 2008. Essential role of domain III of nonstructural protein 5A for hepatitis C virus infectious particle assembly. PLoS Pathog. 4:e1000035. 10.1371/journal.ppat.1000035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Masaki T, Suzuki R, Murakami K, Aizaki H, Ishii K, Murayama A, Date T, Matsuura Y, Miyamura T, Wakita T, Suzuki T. 2008. Interaction of hepatitis C virus nonstructural protein 5A with core protein is critical for the production of infectious virus particles. J. Virol. 82:7964–7976. 10.1128/JVI.00826-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tellinghuisen TL, Foss KL, Treadaway J. 2008. Regulation of hepatitis C virion production via phosphorylation of the NS5A protein. PLoS Pathog. 4:e1000032. 10.1371/journal.ppat.1000032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ma Y, Yates J, Liang Y, Lemon SM, Yi M. 2008. NS3 helicase domains involved in infectious intracellular hepatitis C virus particle assembly. J. Virol. 82:7624–7639. 10.1128/JVI.00724-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yi M, Ma Y, Yates J, Lemon SM. 2007. Compensatory mutations in E1, p7, NS2, and NS3 enhance yields of cell culture-infectious intergenotypic chimeric hepatitis C virus. J. Virol. 81:629–638. 10.1128/JVI.01890-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Han Q, Manna D, Belton K, Cole R, Konan KV. 2013. Modulation of hepatitis C virus genome encapsidation by nonstructural protein 4B. J. Virol. 87:7409–7422. 10.1128/JVI.03523-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Egger D, Wolk B, Gosert R, Bianchi L, Blum HE, Moradpour D, Bienz K. 2002. Expression of hepatitis C virus proteins induces distinct membrane alterations including a candidate viral replication complex. J. Virol. 76:5974–5984. 10.1128/JVI.76.12.5974-5984.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Konan KV, Giddings TH, Jr, Ikeda M, Li K, Lemon SM, Kirkegaard K. 2003. Nonstructural protein precursor NS4A/B from hepatitis C virus alters function and ultrastructure of host secretory apparatus. J. Virol. 77:7843–7855. 10.1128/JVI.77.14.7843-7855.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aligo J, Jia S, Manna D, Konan KV. 2009. Formation and function of hepatitis C virus replication complexes require residues in the carboxy-terminal domain of NS4B protein. Virology 393:68–83. 10.1016/j.virol.2009.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Han Q, Aligo J, Manna D, Belton K, Chintapalli SV, Hong Y, Patterson RL, van Rossum DB, Konan KV. 2011. Conserved GXXXG- and S/T-like motifs in the transmembrane domains of NS4B protein are required for hepatitis C virus replication. J. Virol. 85:6464–6479. 10.1128/JVI.02298-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Elazar M, Liu P, Rice CM, Glenn JS. 2004. An N-terminal amphipathic helix in hepatitis C virus (HCV) NS4B mediates membrane association, correct localization of replication complex proteins, and HCV RNA replication. J. Virol. 78:11393–11400. 10.1128/JVI.78.20.11393-11400.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lundin M, Monne M, Widell A, Von Heijne G, Persson MA. 2003. Topology of the membrane-associated hepatitis C virus protein NS4B. J. Virol. 77:5428–5438. 10.1128/JVI.77.9.5428-5438.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stone M, Jia S, Heo WD, Meyer T, Konan KV. 2007. Participation of rab5, an early endosome protein, in hepatitis C virus RNA replication machinery. J. Virol. 81:4551–4563. 10.1128/JVI.01366-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Targett-Adams P, Boulant S, McLauchlan J. 2008. Visualization of double-stranded RNA in cells supporting hepatitis C virus RNA replication. J. Virol. 82:2182–2195. 10.1128/JVI.01565-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Manna D, Aligo J, Xu C, Park WS, Koc H, Do Heo W, Konan KV. 2010. Endocytic Rab proteins are required for hepatitis C virus replication complex formation. Virology 398:21–37. 10.1016/j.virol.2009.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.El-Hage N, Luo G. 2003. Replication of hepatitis C virus RNA occurs in a membrane-bound replication complex containing nonstructural viral proteins and RNA. J. Gen. Virol. 84:2761–2769. 10.1099/vir.0.19305-0. [DOI] [PubMed] [Google Scholar]
- 27.Gosert R, Egger D, Lohmann V, Bartenschlager R, Blum HE, Bienz K, Moradpour D. 2003. Identification of the hepatitis C virus RNA replication complex in Huh-7 cells harboring subgenomic replicons. J. Virol. 77:5487–5492. 10.1128/JVI.77.9.5487-5492.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ishido S, Fujita T, Hotta H. 1998. Complex formation of NS5B with NS3 and NS4A proteins of hepatitis C virus. Biochem. Biophys. Res. Commun. 244:35–40. 10.1006/bbrc.1998.8202. [DOI] [PubMed] [Google Scholar]
- 29.Ali N, Tardif KD, Siddiqui A. 2002. Cell-free replication of the hepatitis C virus subgenomic replicon. J. Virol. 76:12001–12007. 10.1128/JVI.76.23.12001-12007.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Alter MJ. 1997. Epidemiology of hepatitis C. Hepatology 26:62S–65S. 10.1002/hep.510260711. [DOI] [PubMed] [Google Scholar]
- 31.Reiss S, Rebhan I, Backes P, Romero-Brey I, Erfle H, Matula P, Kaderali L, Poenisch M, Blankenburg H, Hiet MS, Longerich T, Diehl S, Ramirez F, Balla T, Rohr K, Kaul A, Buhler S, Pepperkok R, Lengauer T, Albrecht M, Eils R, Schirmacher P, Lohmann V, Bartenschlager R. 2011. Recruitment and activation of a lipid kinase by hepatitis C virus NS5A is essential for integrity of the membranous replication compartment. Cell Host Microbe 9:32–45. 10.1016/j.chom.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Berger KL, Cooper JD, Heaton NS, Yoon R, Oakland TE, Jordan TX, Mateu G, Grakoui A, Randall G. 2009. Roles for endocytic trafficking and phosphatidylinositol 4-kinase III alpha in hepatitis C virus replication. Proc. Natl. Acad. Sci. U. S. A. 106:7577–7582. 10.1073/pnas.0902693106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hsu NY, Ilnytska O, Belov G, Santiana M, Chen YH, Takvorian PM, Pau C, van der Schaar H, Kaushik-Basu N, Balla T, Cameron CE, Ehrenfeld E, van Kuppeveld FJ, Altan-Bonnet N. 2010. Viral reorganization of the secretory pathway generates distinct organelles for RNA replication. Cell 141:799–811. 10.1016/j.cell.2010.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pietschmann T, Kaul A, Koutsoudakis G, Shavinskaya A, Kallis S, Steinmann E, Abid K, Negro F, Dreux M, Cosset FL, Bartenschlager R. 2006. Construction and characterization of infectious intragenotypic and intergenotypic hepatitis C virus chimeras. Proc. Natl. Acad. Sci. U. S. A. 103:7408–7413. 10.1073/pnas.0504877103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Friebe P, Lohmann V, Krieger N, Bartenschlager R. 2001. Sequences in the 5′ nontranslated region of hepatitis C virus required for RNA replication. J. Virol. 75:12047–12057. 10.1128/JVI.75.24.12047-12057.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gottwein JM, Jensen TB, Mathiesen CK, Meuleman P, Serre SB, Lademann JB, Ghanem L, Scheel TK, Leroux-Roels G, Bukh J. 2011. Development and application of hepatitis C reporter viruses with genotype 1 to 7 core-nonstructural protein 2 (NS2) expressing fluorescent proteins or luciferase in modified JFH1 NS5A. J. Virol. 85:8913–8928. 10.1128/JVI.00049-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Godi A, Di Campli A, Konstantakopoulos A, Di Tullio G, Alessi DR, Kular GS, Daniele T, Marra P, Lucocq JM, De Matteis MA. 2004. FAPPs control Golgi-to-cell-surface membrane traffic by binding to ARF and PtdIns(4)P. Nat. Cell Biol. 6:393–404. 10.1038/ncb1119. [DOI] [PubMed] [Google Scholar]
- 38.Cao X, Coskun U, Rossle M, Buschhorn SB, Grzybek M, Dafforn TR, Lenoir M, Overduin M, Simons K. 2009. Golgi protein FAPP2 tubulates membranes. Proc. Natl. Acad. Sci. U. S. A. 106:21121–21125. 10.1073/pnas.0911789106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Aizaki H, Lee KJ, Sung VM, Ishiko H, Lai MM. 2004. Characterization of the hepatitis C virus RNA replication complex associated with lipid rafts. Virology 324:450–461. 10.1016/j.virol.2004.03.034. [DOI] [PubMed] [Google Scholar]
- 40.Hug P, Lin HM, Korte T, Xiao X, Dimitrov DS, Wang JM, Puri A, Blumenthal R. 2000. Glycosphingolipids promote entry of a broad range of human immunodeficiency virus type 1 isolates into cell lines expressing CD4, CXCR4, and/or CCR5. J. Virol. 74:6377–6385. 10.1128/JVI.74.14.6377-6385.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Berger KL, Kelly SM, Jordan TX, Tartell MA, Randall G. 2011. Hepatitis C virus stimulates the phosphatidylinositol 4-kinase III alpha-dependent phosphatidylinositol 4-phosphate production that is essential for its replication. J. Virol. 85:8870–8883. 10.1128/JVI.00059-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reiss S, Harak C, Romero-Brey I, Radujkovic D, Klein R, Ruggieri A, Rebhan I, Bartenschlager R, Lohmann V. 2013. The lipid kinase phosphatidylinositol-4 kinase III alpha regulates the phosphorylation status of hepatitis C virus NS5A. PLoS Pathog. 9:e1003359. 10.1371/journal.ppat.1003359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tai AW, Benita Y, Peng LF, Kim SS, Sakamoto N, Xavier RJ, Chung RT. 2009. A functional genomic screen identifies cellular cofactors of hepatitis C virus replication. Cell Host Microbe 5:298–307. 10.1016/j.chom.2009.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tai AW, Salloum S. 2011. The role of the phosphatidylinositol 4-kinase PI4KA in hepatitis C virus-induced host membrane rearrangement. PLoS One 6:e26300. 10.1371/journal.pone.0026300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hanada K. 2003. Serine palmitoyltransferase, a key enzyme of sphingolipid metabolism. Biochim. Biophys. Acta 1632:16–30. 10.1016/S1388-1981(03)00059-3. [DOI] [PubMed] [Google Scholar]
- 46.Hanada K, Kumagai K, Tomishige N, Yamaji T. 2009. CERT-mediated trafficking of ceramide. Biochim. Biophys. Acta 1791:684–691. 10.1016/j.bbalip.2009.01.006. [DOI] [PubMed] [Google Scholar]
- 47.Hornemann T, Wei Y, von Eckardstein A. 2007. Is the mammalian serine palmitoyltransferase a high-molecular-mass complex? Biochem. J. 405:157–164. 10.1042/BJ20070025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yasuda S, Nishijima M, Hanada K. 2003. Localization, topology, and function of the LCB1 subunit of serine palmitoyltransferase in mammalian cells. J. Biol. Chem. 278:4176–4183. 10.1074/jbc.M209602200. [DOI] [PubMed] [Google Scholar]
- 49.Ciesek S, Steinmann E, Manns MP, Wedemeyer H, Pietschmann T. 2008. The suppressive effect that myriocin has on hepatitis C virus RNA replication is independent of inhibition of serine palmitoyl transferase. J. Infect. Dis. 198:1091–1093. 10.1086/591463. [DOI] [PubMed] [Google Scholar]
- 50.D'Angelo G, Polishchuk E, Di Tullio G, Santoro M, Di Campli A, Godi A, West G, Bielawski J, Chuang CC, van der Spoel AC, Platt FM, Hannun YA, Polishchuk R, Mattjus P, De Matteis MA. 2007. Glycosphingolipid synthesis requires FAPP2 transfer of glucosylceramide. Nature 449:62–67. 10.1038/nature06097. [DOI] [PubMed] [Google Scholar]
- 51.D'Angelo G, Rega LR, De Matteis MA. 2012. Connecting vesicular transport with lipid synthesis: FAPP2. Biochim. Biophys. Acta 1821:1089–1095. 10.1016/j.bbalip.2012.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Goueslain L, Alsaleh K, Horellou P, Roingeard P, Descamps V, Duverlie G, Ciczora Y, Wychowski C, Dubuisson J, Rouille Y. 2010. Identification of GBF1 as a cellular factor required for hepatitis C virus RNA replication. J. Virol. 84:773–787. 10.1128/JVI.01190-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang L, Hong Z, Lin W, Shao RX, Goto K, Hsu VW, Chung RT. 2012. ARF1 and GBF1 generate a PI4P-enriched environment supportive of hepatitis C virus replication. PLoS One 7:e32135. 10.1371/journal.pone.0032135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bieberich E, Freischutz B, Suzuki M, Yu RK. 1999. Differential effects of glycolipid biosynthesis inhibitors on ceramide-induced cell death in neuroblastoma cells. J. Neurochem. 72:1040–1049. [DOI] [PubMed] [Google Scholar]
- 55.Platt FM, Neises GR, Reinkensmeier G, Townsend MJ, Perry VH, Proia RL, Winchester B, Dwek RA, Butters TD. 1997. Prevention of lysosomal storage in Tay-Sachs mice treated with N-butyldeoxynojirimycin. Science 276:428–431. 10.1126/science.276.5311.428. [DOI] [PubMed] [Google Scholar]
- 56.Platt FM, Reinkensmeier G, Dwek RA, Butters TD. 1997. Extensive glycosphingolipid depletion in the liver and lymphoid organs of mice treated with N-butyldeoxynojirimycin. J. Biol. Chem. 272:19365–19372. 10.1074/jbc.272.31.19365. [DOI] [PubMed] [Google Scholar]
- 57.Rawat SS, Johnson BT, Puri A. 2005. Sphingolipids: modulators of HIV-1 infection and pathogenesis. Biosci. Rep. 25:329–343. 10.1007/s10540-005-2894-5. [DOI] [PubMed] [Google Scholar]
- 58.Casson L, Howell L, Mathews LA, Ferrer M, Southall N, Guha R, Keller JM, Thomas C, Siskind LJ, Beverly LJ. 2013. Inhibition of ceramide metabolism sensitizes human leukemia cells to inhibition of BCL2-like proteins. PLoS One 8:e54525. 10.1371/journal.pone.0054525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Komori H, Ichikawa S, Hirabayashi Y, Ito M. 1999. Regulation of intracellular ceramide content in B16 melanoma cells. Biological implications of ceramide glycosylation. J. Biol. Chem. 274:8981–8987. [DOI] [PubMed] [Google Scholar]
- 60.Ye J, Wang C, Sumpter R, Jr, Brown MS, Goldstein JL, Gale M., Jr 2003. Disruption of hepatitis C virus RNA replication through inhibition of host protein geranylgeranylation. Proc. Natl. Acad. Sci. U. S. A. 100:15865–15870. 10.1073/pnas.2237238100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.D'Angelo G, Uemura T, Chuang CC, Polishchuk E, Santoro M, Ohvo-Rekila H, Sato T, Di Tullio G, Varriale A, D'Auria S, Daniele T, Capuani F, Johannes L, Mattjus P, Monti M, Pucci P, Williams RL, Burke JE, Platt FM, Harada A, De Matteis MA. 2013. Vesicular and non-vesicular transport feed distinct glycosylation pathways in the Golgi. Nature 501:116–120. 10.1038/nature12423. [DOI] [PubMed] [Google Scholar]
- 62.Shi ST, Lee KJ, Aizaki H, Hwang SB, Lai MM. 2003. Hepatitis C virus RNA replication occurs on a detergent-resistant membrane that cofractionates with caveolin-2. J. Virol. 77:4160–4168. 10.1128/JVI.77.7.4160-4168.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang H, Perry JW, Lauring AS, Neddermann P, De Francesco R, Tai AW. 2014. Oxysterol-binding protein is a phosphatidylinositol 4-kinase effector required for HCV replication membrane integrity and cholesterol trafficking. Gastroenterology 146:1373–1385. 10.1053/j.gastro.2014.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Manna D, Aligo J, Xu C, Park WS, Koc H, Heo WD, Konan KV. 2010. Endocytic Rab proteins are required for hepatitis C virus replication complex formation. Virology 398:21–37. 10.1016/j.virol.2009.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wolk B, Buchele B, Moradpour D, Rice CM. 2008. A dynamic view of hepatitis C virus replication complexes. J. Virol. 82:10519–10531. 10.1128/JVI.00640-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Santiago-Tirado FH, Bretscher A. 2011. Membrane-trafficking sorting hubs: cooperation between PI4P and small GTPases at the trans-Golgi network. Trends Cell Biol. 21:515–525. 10.1016/j.tcb.2011.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Amako Y, Sarkeshik A, Hotta H, Yates J, III, Siddiqui A. 2009. Role of oxysterol binding protein in hepatitis C virus infection. J. Virol. 83:9237–9246. 10.1128/JVI.00958-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bishe B, Syed GH, Field SJ, Siddiqui A. 2012. Role of phosphatidylinositol 4-phosphate (PI4P) and its binding protein GOLPH3 in hepatitis C virus secretion. J. Biol. Chem. 287:27637–27647. 10.1074/jbc.M112.346569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hanada K, Nishijima M. 2003. Purification of mammalian serine palmitoyltransferase, a hetero-subunit enzyme for sphingolipid biosynthesis, by affinity-peptide chromatography. Methods Mol. Biol. 228:163–174. 10.1385/1-59259-400-X:163. [DOI] [PubMed] [Google Scholar]
- 70.Hirata Y, Ikeda K, Sudoh M, Tokunaga Y, Suzuki A, Weng L, Ohta M, Tobita Y, Okano K, Ozeki K, Kawasaki K, Tsukuda T, Katsume A, Aoki Y, Umehara T, Sekiguchi S, Toyoda T, Shimotohno K, Soga T, Nishijima M, Taguchi R, Kohara M. 2012. Self-enhancement of hepatitis C virus replication by promotion of specific sphingolipid biosynthesis. PLoS Pathog. 8:e1002860. 10.1371/journal.ppat.1002860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Weng L, Hirata Y, Arai M, Kohara M, Wakita T, Watashi K, Shimotohno K, He Y, Zhong J, Toyoda T. 2010. Sphingomyelin activates hepatitis C virus RNA polymerase in a genotype-specific manner. J. Virol. 84:11761–11770. 10.1128/JVI.00638-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kawano M, Kumagai K, Nishijima M, Hanada K. 2006. Efficient trafficking of ceramide from the endoplasmic reticulum to the Golgi apparatus requires a VAMP-associated protein-interacting FFAT motif of CERT. J. Biol. Chem. 281:30279–30288. 10.1074/jbc.M605032200. [DOI] [PubMed] [Google Scholar]
- 73.Kumagai K, Yasuda S, Okemoto K, Nishijima M, Kobayashi S, Hanada K. 2005. CERT mediates intermembrane transfer of various molecular species of ceramides. J. Biol. Chem. 280:6488–6495. 10.1074/jbc.M409290200. [DOI] [PubMed] [Google Scholar]
- 74.Hanada K, Kumagai K, Tomishige N, Kawano M. 2007. CERT and intracellular trafficking of ceramide. Biochim. Biophys. Acta 1771:644–653. 10.1016/j.bbalip.2007.01.009. [DOI] [PubMed] [Google Scholar]
- 75.Yamaji T, Kumagai K, Tomishige N, Hanada K. 2008. Two sphingolipid transfer proteins, CERT and FAPP2: their roles in sphingolipid metabolism. IUBMB Life 60:511–518. 10.1002/iub.83. [DOI] [PubMed] [Google Scholar]
- 76.Hanada K, Kumagai K, Yasuda S, Miura Y, Kawano M, Fukasawa M, Nishijima M. 2003. Molecular machinery for non-vesicular trafficking of ceramide. Nature 426:803–809. 10.1038/nature02188. [DOI] [PubMed] [Google Scholar]
- 77.Wang H, Perry JW, Lauring AS, Neddermann P, De Francesco R, Tai AW. 2014. Oxysterol-binding protein is a phosphatidylinositol 4-kinase effector required for HCV replication membrane integrity and cholesterol trafficking. Gastroenterology 146:1373–1385. 10.1053/j.gastro.2014.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Su WC, Chao TC, Huang YL, Weng SC, Jeng KS, Lai MM. 2011. Rab5 and class III phosphoinositide 3-kinase Vps34 are involved in hepatitis C virus NS4B-induced autophagy. J. Virol. 85:10561–10571. 10.1128/JVI.00173-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Paul D, Hoppe S, Saher G, Krijnse-Locker J, Bartenschlager R. 2013. Morphological and biochemical characterization of the membranous hepatitis C virus replication compartment. J. Virol. 87:10612–10627. 10.1128/JVI.01370-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kamlekar RK, Simanshu DK, Gao YG, Kenoth R, Pike HM, Prendergast FG, Malinina L, Molotkovsky JG, Venyaminov SY, Patel DJ, Brown RE. 2013. The glycolipid transfer protein (GLTP) domain of phosphoinositol 4-phosphate adaptor protein-2 (FAPP2): structure drives preference for simple neutral glycosphingolipids. Biochim. Biophys. Acta 1831:417–427. 10.1016/j.bbalip.2012.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tuuf J, Mattjus P. 2014. Membranes and mammalian glycolipid transferring proteins. Chem. Phys. Lipids 178:27–37. 10.1016/j.chemphyslip.2013.10.013. [DOI] [PubMed] [Google Scholar]
- 82.Lenoir M, Coskun U, Grzybek M, Cao X, Buschhorn SB, James J, Simons K, Overduin M. 2010. Structural basis of wedging the Golgi membrane by FAPP pleckstrin homology domains. EMBO Rep. 11:279–284. 10.1038/embor.2010.28. [DOI] [PMC free article] [PubMed] [Google Scholar]