ABSTRACT
Capsid-associated tegument proteins have been identified in alpha- and betaherpesviruses to play an essential role in viral DNA packaging. Whether and how such tegument proteins exist in gammaherpesviruses have been mysteries. Here, we report a 6-Å-resolution cryo-electron microscopy (cryo-EM) structure of Kaposi's sarcoma-associated herpesvirus (KSHV) virion, a member of the oncogenic gammaherpesvirus subfamily. The KSHV virion structure reveals, for the first time, how capsid-associated tegument proteins are organized in a gammaherpesvirus, with five tegument densities capping each penton vertex, a pattern highly similar to that in alphaherpesvirus but completely different from that in betaherpesvirus. Each KSHV tegument density can be divided into three prominent regions: a penton-binding globular region, a helix-bundle stalk region, and a β-sheet-rich triplex-binding region. Fitting of the crystal structure of the truncated HSV-1 UL25 protein (the KSHV ORF19 homolog) and secondary structure analysis of the full-length ORF19 established that ORF19 constitutes the globular region with an N-terminal, 60-amino-acid-long helix extending into the stalk region. Matching secondary structural features resolved in the cryo-EM density with secondary structures predicted by sequence analysis identifies the triplex-binding region to be ORF32, a homolog of alphaherpesvirus UL17. Despite the high level of tegument structural similarities between KSHV and alphaherpesvirus, an ORF19 monomer in KSHV, in contrast to a UL25 dimer in alphaherpesviruses, binds each penton subunit, an observation that correlates with conformational differences in their pentons. This newly discovered organization of triplex-ORF32-ORF19 also resolves a long-standing mystery surrounding the virion location and conformation of alphaherpesvirus UL25 protein.
IMPORTANCE Several capsid-associated tegument proteins have been identified in the alpha- and betaherpesvirus subfamilies of the Herpesviridae. These tegument proteins play essential roles in viral propagation and are potential drug targets for curbing herpesvirus infections. However, no such tegument proteins have been identified for gammaherpesviruses, the third herpesvirus subfamily, which contains members causing several human cancers. Here, by high-resolution cryo-EM, we show the three-dimensional structure of the capsid-associated tegument proteins in the prototypical member of gammaherpesviruses, KSHV. The cryo-EM structure reveals that the organization of KSHV capsid-associated tegument proteins is highly similar to that in alphaherpesvirus but completely different from that in betaherpesvirus. Structural analyses further localize ORF19 and ORF32 proteins (the alphaherpesvirus UL25 and UL17 homologs in KSHV, respectively) in the KSHV capsid-associated tegument cryo-EM structure. These findings also resolve a long-standing mystery regarding the location and conformation of alphaherpesvirus UL25 protein inside the virion.
INTRODUCTION
Kaposi's sarcoma-associated herpesvirus (KSHV), also known as human herpesvirus 8 (HHV-8), is the causative agent of Kaposi's sarcoma (1, 2), a cancer commonly occurring in AIDS patients (3). It is also associated with certain B cell lymphoproliferative disorders, like primary effusion lymphoma and multicentric Castleman's disease (4). KSHV is a member of the gammaherpesvirus subfamily of the Herpesviridae. Like all herpesviruses, the KSHV virion is composed of a double-stranded, linear DNA genome encased within an icosahedral capsid, which in turn is wrapped in a proteinaceous layer, called the tegument and, last, a lipid bilayer membrane called the envelope. Although the tegument compartment of herpesvirus is pleomorphic and has no distinguishable substructures, it is roughly divided into outer tegument and inner tegument layers (5, 6). Upon cell entry, the outer tegument proteins are released into the host cell cytoplasm where some of them perform functions needed early in the infection process, while the inner tegument proteins remain associated with the nucleocapsid and participate in its transport to the nucleus via the microtubule/dynein motor system (7–12).
Of particular interest, some inner tegument proteins associate with the nucleocapsid in an ordered manner that enables the visualization of tegument densities in three-dimensional (3D) reconstructions of the virion by cryo-electron microscopy (cryo-EM). In herpes simplex virus 1 (HSV-1) and pseudorabies virus (PRV), two representative members of the alphaherpesvirus subfamily, tegument densities are present at the icosahedral vertices of the capsid; thus, they are named capsid vertex-specific components (CVSC) (13–17). In human cytomegalovirus (HCMV) and simian cytomegalovirus (SCMV), members of the betaherpesvirus subfamily, the capsid-associated tegument protein pp150 forms a net enclosing the entire capsid and presumably stabilizes the capsid to contain the large viral genome (18–21). These capsid-associated tegument components identified in alpha- and betaherpesviruses have been demonstrated to be essential for viral propagation (22–24), but the underlying mechanisms are not clear.
Structural studies of gammaherpesviruses are lagging behind due to more difficulties of sample preparation. In particular, whether and how capsid-associated tegument proteins exist in gammaherpesviruses have not been addressed.
Recently, a new KSHV bacterial artificial chromosome (BAC) plasmid, BAC16, was generated to facilitate production of KSHV infectious virions and genetic modification of the KSHV genome (25). A high titer of BAC16-derived virus stock can be obtained with the use of a cell line, iSLK-puro, engineered to express a doxycycline (DOX)-inducible immediate-early viral protein, RTA, that drives the lytic replication of KSHV (25, 26). Here, by using this iSLK-KSHVBAC16 system for KSHV virion production, we obtained a 6-Å-resolution structure of the KSHV virion, which reveals, for the first time, that KSHV also has capsid-associated tegument densities around the capsid vertices. Two proteins in the KSHV tegument density were identified, ORF19 and ORF32, which are homologs of alphaherpesvirus UL25 and UL17, respectively. ORF32 mediates the assembly of tegument proteins by anchoring to the capsid on penton-proximal triplexes and connecting to ORF19 through its N-terminal helix and possibly to other tegument proteins. This result also clarifies the previous mystery surrounding the locations and structures of alphaherpesvirus UL25 and UL17 proteins.
MATERIALS AND METHODS
Sample preparation of KSHV virion.
The iSLK-KSHVBAC16 cells (kind gift from Jae U. Jung) were cultured in Dulbecco's modified Eagle medium (DMEM) with 10% fetal bovine serum (FBS), 1 μg/ml puromycin, 250 μg/ml G418, and 1,200 μg/ml hygromycin. When cells reached ∼80% confluence, the media were replaced with DMEM plus 10% FBS, 1 mM sodium butyrate, and 1 μg/ml doxycycline. After 3 to 5 days, when 80% of the cells were round, the supernatant was collected to purify KSHV virion by following a procedure described previously (19).
Cryo-EM imaging and data processing.
An aliquot of 2.5 μl purified KSHV virion suspension in phosphate-buffered saline (pH 7.4) was applied to a 300-mesh copper grid coated with holey carbon film, blotted with filter paper, and plunge-frozen in liquid ethane. The sample was imaged at liquid nitrogen temperature in an FEI Titan Krios cryo-electron microscope operated at 300 kV with the Leginon software (27). The images were recorded on a Gatan K2 summit direct detection camera on super-resolution mode with an effective magnification of ×24,300 (nominal magnification, ×14,000), corresponding to a pixel size of 1.03 Å/pixel at the specimen level. Each selected sample area was exposed at a dose rate of ∼8e−/pixel/s on camera for 13 s, resulting in a total dose of ∼25e−/Å2 on specimen. Each image was fractioned into 26 frames, with a 500-ms accumulation time for each frame. All of the frames were aligned and averaged with the drift-correction software described in reference 28. The averaged images were used for monitoring data processing and 3D reconstruction.
Defocus values of the data set were determined with CTFFIND3 (29) and were in the range of 1 to 2 μm underfocus. A total of ∼40,000 particles were picked manually using EMAN (30). Orientation and center parameters of each particle were determined and refined iteratively with the common-line method implemented in the IMIRS software package (31, 32), and 3D reconstructions were carried out with the GPU program eLite3D (33). The final reconstruction was obtained by averaging the 20,395 best particles, which were selected on the basis of phase residues between raw images and model projections. The resolution of the density map was measured to be 6 Å based on the 0.143 Fourier shell correlation (FSC) criterion (34). The visualization of the density map was carried out with UCSF Chimera (35).
Fitting of crystal structures into the cryo-EM density map was done with the “Fit in Map” tool of UCSF Chimera (35). During the fitting, a simulated map was generated from the crystal structure at the same resolution as that of the experimental map. A cross-correlation coefficient between the simulated map and the experimental map was calculated and reported as a quantitative estimate of the quality of fitting.
Secondary structure predictions were performed with PSIPRED using the Protein Structure Prediction Server (36).
Cryo-EM accession number.
The cryo-EM density map has been deposited in EMDB under the accession number EMD-6038.
RESULTS
Structure of KSHV virion at 6-Å resolution.
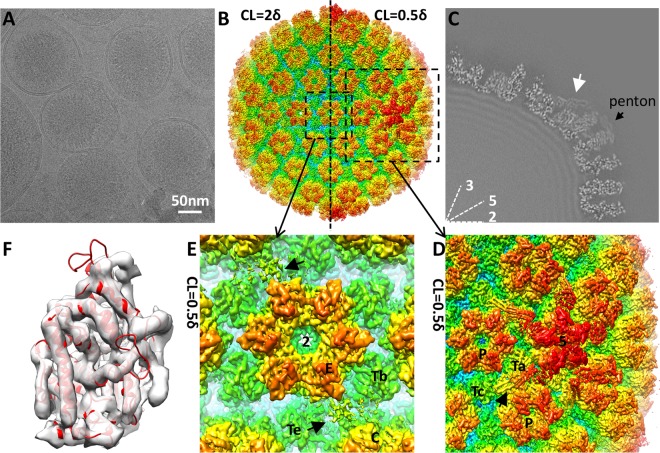
With the aim of resolving the tegument, we chose to purify intact virion particles from viral culture media for this study. Protected by a viral envelope, a virion encounters less structural disturbance and should have better preservation of its tegument structure compared to nucleocapsid purified from infected cell nuclei used in previous structural studies. We imaged the purified KSHV virions embedded in vitreous ice (Fig. 1A) with a 300-kV Titan Krios electron microscope equipped with an electron-counting direct detection camera (Gatan K2 Summit) operated at super-resolution mode. By averaging over 20,000 images of virion particles, we reconstructed the KSHV virion to 6-Å resolution (Fig. 1B).
FIG 1.
Cryo-EM and 3D reconstruction of KSHV virions. (A) Cryo-EM image of purified KSHV virion. (B) 3D reconstruction of the KSHV virion at 6-Å resolution. The structure is rainbow-colored radially. The left half of the capsid is rendered at a contour level (CL) of 2δ (δ is the standard deviation), while the right half is rendered at 0.5δ to show the presence of tegument densities. (C) One-quarter of a central slice of the KSHV virion reconstruction. The white arrow points to a capsid-associated tegument density surrounding the penton. The intensity of the tegument density is measured to be ∼30% of the peak intensity of the capsid shell. Lines 2, 3, and 5, icosahedral symmetry axes. (D and E) Zoomed-in views of capsid surface areas as denoted with dashed squares in panel B. The black arrow in panel D points to a tegument density around the capsid vertex. Black arrows in panel E point to tegument density close to the 2-fold axis. 5, 5-fold axis; P, periphery-hexon; Ta and Tc, triplexes; 2, 2-fold axis; E, edge hexon; C, central hexon; Tb and Te, triplexes. (F) KSHV MCP upper domain (MCPud) density map (transparent gray) fitted with the HSV-1 MCPud atomic model (red ribbon; PDB entry 1NO7) (37).
The capsid structure of KSHV is similar to that of other published herpesvirus capsid reconstructions. It is composed of major capsid protein (MCP; ORF25) pentamers and hexamers joined by triplexes (heterotrimers of one ORF62 and two ORF26 proteins) and decorated by the smallest capsid proteins (SCP; ORF65). A density of the MCP upper domain (MCPud) can be fitted well with the atomic model of HSV-1 MCPud (PDB entry 1NO7) (37) (cross-correlation coefficient, 0.47; see Materials and Methods), demonstrating good quality of the reconstruction and also indicating structural conservation between MCP molecules of the two viruses (Fig. 1F).
We identified extra densities other than capsid proteins in the KSHV virion reconstruction that are attributable to capsid-associated tegument components (CATC). The tegument densities are distributed mainly around the capsid vertices, with five sets binding each penton and its surrounding triplexes, Ta and Tc (as in the nomenclature of reference 38) (Fig. 1D). These densities are weaker than capsid proteins. They can be visualized only when the map is displayed at a low contour level (∼0.5δ above the mean; δ is the standard deviation) but are hardly discernible when other capsid components are displayed at a contour level of 2δ (Fig. 1B; also see Fig. S1 in the supplemental material). A central slice of the 3D reconstruction shows that the intensity of these tegument densities is only about 30% of the highest intensity of capsid proteins (Fig. 1C). The weak intensity of these tegument densities explains why we failed to detect them previously in a low-resolution cryo-EM reconstruction of murine herpesvirus 68 (MHV-68) virion with only a few hundred particles (5).
Other minor tegument densities also are observed around the 2-fold symmetric axis, roughly at the midpoint between triplexes Tb and Te, edge hexon, and central hexon (Fig. 1E). These minor densities are very weak and broken, resulting from either very low occupancy or high flexibility. Given their similarity in bridging over two triplexes, we propose that these densities are the same kind of tegument proteins as those surrounding the capsid vertices but occupying a secondary, lower-affinity binding site.
Capsid-associated tegument components in KSHV are similar to those in alphaherpesviruses but different from those in betaherpesviruses.
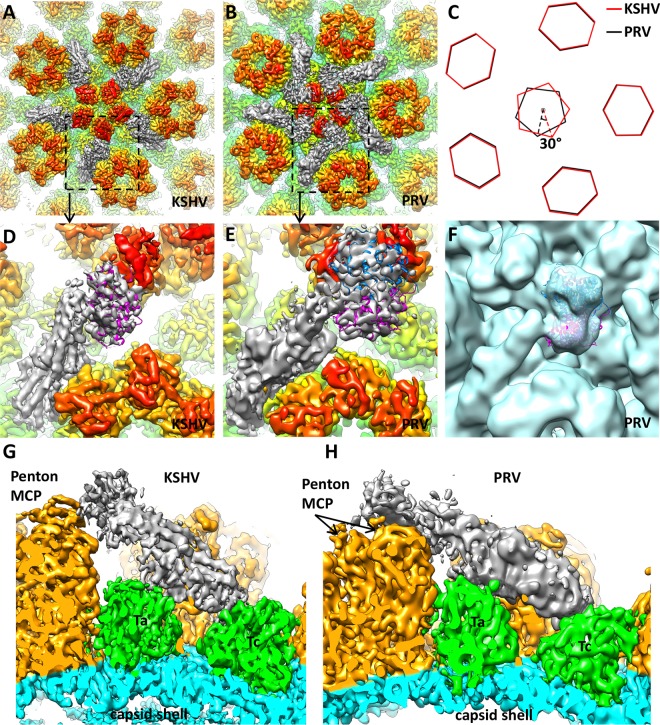
Comparison of KSHV CATC with tegument components in alpha- and betaherpesvirus reconstructions revealed its similarity to those in alphaherpesviruses and difference from those in betaherpesviruses. The distribution of visible tegument densities in published HSV-1 and PRV reconstructions is limited to particle vertices; thus, these densities were named capsid vertex-specific component, or CVSC (16). The binding pattern and overall shape of KSHV CATC densities are similar to those of HSV-1 and PRV CVSC. To illustrate these similarities, we compare our KSHV structure side by side with that of a recently published 9-Å-resolution reconstruction of PRV C capsid (EMDB-5650) (17) (Fig. 2A, B, D, E, G, and H). The tegument density in both KSHV and PRV can be divided into three regions in the same way: a globular region in the upper end binds the penton MCP; a bridge-shaped region in the lower end binds two triplexes closest to the penton, triplexes Ta and Tc; and a stalk region in the middle connects the penton-binding region to the triplex-binding region (Fig. 2G and H and 3A).
FIG 2.
Comparison of capsid-associated tegument densities in KSHV and pseudorabies virus (PRV). (A and B) Densities around the capsid vertex in the 6-Å KSHV virion reconstruction (A) and 9-Å PRV C-capsid reconstruction (B) (rendered from EMDB-5650; published by Homa et al. [17]). In both structures, capsid proteins are rainbow colored radially and tegument densities are gray, and they are displayed at a lower contour level than that of the capsid because they are weaker than capsid densities. (C) Schematic representation of pentons and periphery hexons in KSHV and PRV. The PRV penton has a 30° clockwise rotation when its periphery hexons are aligned to those of KSHV. (D and E) Zoomed-in views of a single capsid-associated tegument density in KSHV and PRV. Their positions are denoted with dashed squares in panels A and B. Note the slightly different orientations of the tegument density relative to the penton MCP. In KSHV (D), a single copy of the HSV-1 UL25 atomic model (magenta ribbon; PDB entry 2F5U) (39) can be fitted into the tegument density, while in PRV (E) two copies (magenta and blue ribbons) can be accommodated. (F) Two copies of the HSV-1 UL25 atomic model (magenta and blue ribbons) fitted in the tegument density of the PRV virion reconstruction (light blue) (EMDB-5655; Homa et al. [17]). Note the good match between the overall shape of the density map and that of the model. (G and H) Side views of tegument density interacting with penton and triplexes Ta and Tc in KSHV (G) and PRV (H), respectively.
FIG 3.
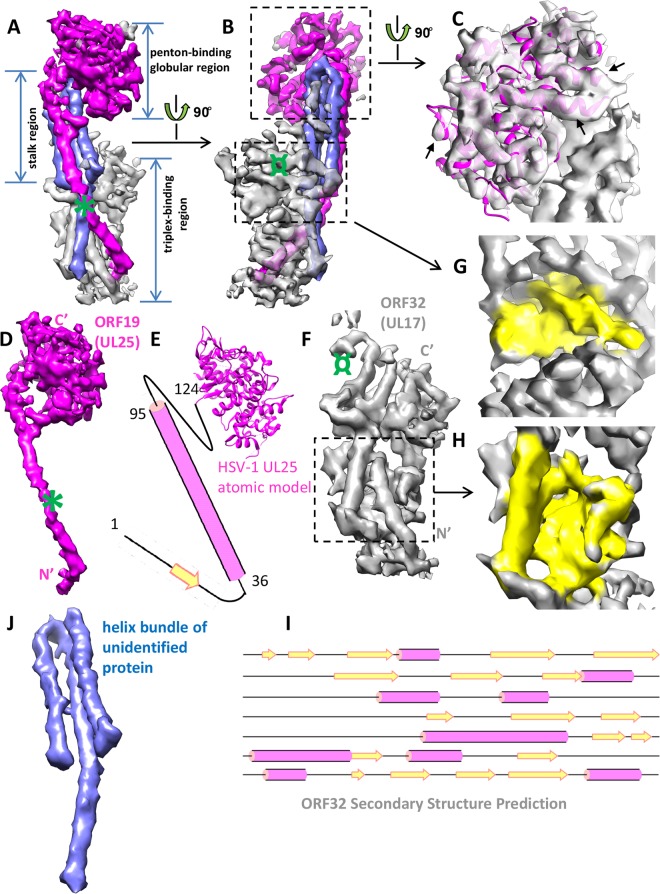
Identity of constituent proteins in KSHV capsid-associated tegument component. (A and B) Different views of the KSHV tegument density. For convenience of description, the density is divided into a penton-binding globular region, a stalk region, and a triplex-binding region. The density is colored by its putative identity (magenta, ORF19; gray, ORF32; blue, unidentified tegument protein). The green symbol in panel A roughly corresponds to the position of extra GFP density in the HSV-1 UL25 GFP-labeling study as described by Conway et al. (15). The green symbol in panel B roughly corresponds to the position of GFP label inserted at the HSV-1 UL17 C terminus as described by Toropova et al. (16). (C) Zoomed-in view of the penton-binding globular region. The density map is presented as semitransparent and from an orientation different from that shown in panels A and B to appreciate its satisfactory fitting with the HSV-1 UL25 atomic model (magenta ribbon). This evidence strongly supports our assignment of ORF19 (UL25 homolog) to this region. The black arrows point to several well-fitted alpha-helices. (D) In situ, full-length structure of ORF19. N′ and C′ denote protein amino and carboxyl termini, respectively. The green mark is the same as that in panel A. (E) Schematic model of ORF19. Secondary structure prediction suggests that the N-terminal segment (aa 1 to 124 in KSHV ORF19, corresponding to aa 1 to 133 in HSV-1 UL25) missing from the HSV-1 UL25 crystal structure (aa 134 to 577) has a long helix and a short β-strand. (F) Structure of ORF32 (UL17 homolog). The green mark is the same as that described for panel B. N′ and C′ denote protein amino and carboxyl termini, respectively. (G and H) Zoomed-in views of the two domains of ORF32 to show its β-sheet-rich structural feature (colored yellow). (I) Secondary structure prediction of the ORF32 sequence. Note the abundance of β-strands, correlating with structural features shown in panels G and H. (J) Structure of the three-helix bundle in the stalk region.
In contrast, the KSHV and PRV tegument densities bear no similarity to the tegument densities of cytomegalovirus (betaherpesvirus subfamily). In cytomegaloviruses, three filamentous, pp150-containing densities bind to each triplex and its surrounding capsomers (compare Fig. 2A and B to Fig. 1H in our previously published paper [19]).
These observations suggest that the KSHV inner tegument is very similar to that of alphaherpesvirus but has nothing in common with that of betaherpesvirus.
In situ structure of full-length ORF19 revealed in the KSHV virion reconstruction.
In alphaherpesviruses, one of the capsid-associated tegument components was shown to be UL25, a homolog of KSHV ORF19 (15, 17). The structure of the N-terminally truncated (missing 133 amino acids [aa]) UL25 of HSV-1 has been solved by crystallography (PDB entry 2F5U) (39). We find that this HSV-1 UL25 atomic model fits well with the penton-binding globular region of the KSHV tegument density (cross-correlation coefficient, 0.49; see Materials and Methods) (Fig. 3C; also see Movie S1 in the supplemental material). This observation indicates that the penton-binding globular region is made up of a monomer of the ORF19 protein. The structural agreement between the HSV-1 UL25 model and KSHV tegument density further demonstrates structural conservation between gamma- and alphaherpesvirus tegument proteins up to the level of secondary structures.
The N-terminal segment of aa 1 to 133 is missing from the UL25 crystal structure. Secondary structure predictions of the full-length KSHV ORF19 and HSV-1 UL25 sequences reveal similar secondary structures for their N-terminal segments. Both of them have a short β strand connected to a long helix of about 60 residues (Fig. 3E). We suggest that this predicted long helix corresponds to the magenta helix observed at the surface of the stalk region (Fig. 3A). This helix spans ∼100 Å from the penton-binding globular region to the bottom-right portion of the triplex-binding region, with a kink in the middle (Fig. 3A, B, and D; also see Movie S2 in the supplemental material). The length of this helix agrees with that expected for the predicted N-terminal helix containing 60 amino acids (∼90 Å in length, given an axial rise of 1.5 Å/residue). At 6-Å resolution, the β-strand and loop predicted at the very end of the N-terminal segment are not resolved in our structure but could be part of the large β-sheet in the triplex-binding region (the one in Fig. 3H).
Localization of ORF32 in KSHV virion.
A second constituent of alphaherpesvirus tegument density was determined to be UL17, a homolog of KSHV ORF32 (16). It was assigned to occupy mainly the stalk region and a small portion of the triplex-binding region closest to the penton (16).
Based on the correlation of the secondary structural feature of the remaining unassigned density in the KSHV tegument to the secondary structure prediction of ORF32, we assign it to be the entire triplex-binding region (Fig. 3A, B, and F; see Movie S2 in the supplemental material). The density of the triplex-binding region in KSHV tegument is well-separated into two domains, with each domain having a large β-sheet in the core (Fig. 3F, G, and H). This β-sheet-rich structural feature correlates well with secondary structure prediction of ORF32, which is also rich in β-strands (Fig. 3I).
In previous green fluorescent protein (GFP)-labeling studies of HSV-1 UL17, the GFP tag was attached to the C terminus and produced extra density at the penton-proximal end of the triplex-binding region in the cryo-EM reconstruction (Fig. 3B and F, green symbols) (16). Because the cryo-EM densities for the triplex-binding regions of capsid-associated tegument are similar in KSHV and HSV-1, by analogy, we assign this penton-proximal domain in KSHV to be the C-terminal half of ORF32 and the penton-distal domain to be the N-terminal half (Fig. 3F).
Other constituents in KSHV capsid-associated tegument structure.
The remaining three helices in the stalk region have no obvious density connectivity to either the penton-binding region ORF19 or triplex-binding region ORF32 (Fig. 3A, B, and J; also see Movie S2 in the supplemental material). Given the still-limited (6-Å) resolution of the current structure and the lack of biochemical data, we cannot make a definitive protein assignment to these three helices in the helix bundle.
Previously, it was proposed for alphaherpesviruses that VP1/2 (encoded by UL36) is present in the capsid-associated tegument densities and constitutes the penton-binding region (17, 40). Biochemical studies of HSV-1 and PRV also showed that VP1/2 was recruited into nuclear assemblons and specifically bound capsids via its C terminus, and this binding required UL25 (41–43). However, as shown above, the proposed location of VP1/2, the penton-binding region, has been determined unambiguously to be occupied by UL25. Whether the three-helix bundle in the stalk region could be part of ORF64, the KSHV homolog of VP1/2, awaits further investigations.
Stoichiometric difference between capsid-associated tegument proteins of alpha- and gammaherpesviruses.
When applying the above-determined organization of tegument proteins in KSHV to PRV, we further identified a stoichiometric difference in the capsid-associated tegument proteins between the two viruses. The penton-binding globular region of PRV tegument is roughly twice the size of that in KSHV (Fig. 2D and E). By fitting with the HSV-1 UL25 atomic model, we confirmed that this region in PRV indeed can accommodate two UL25 molecules (Fig. 2E). We double-checked the fitting in the PRV virion reconstruction (EMDB-5655) (17), which has poorer resolution but better preservation of tegument densities than the capsid reconstruction. As expected, the overall shape of the density in the penton-binding globular region matches that of the two UL25 models (Fig. 2F). We further examined tegument densities in HSV-1 reconstructions published to date. Two bulbs of density similar to that in PRV virion reconstruction can be easily identified (compare Fig. 2F to Fig. 3 from reference 13 and Fig. 4A in reference 40). We propose that it is a conserved feature among all alphaherpesviruses that two UL25 molecules, in contrast to only one ORF19 in KSHV, are present in each capsid-associated tegument component. In support of our proposal, Conway et al. had found in their HSV-1 UL25 GFP-labeling study that the GFP density could accommodate two GFP molecules (15). Moreover, approximately 2:1 ratios of UL25 to UL17 were reported for HSV-1 and PRV C capsids, respectively (see Table S2 in reference 14 and Fig. 4b in reference 17). The UL25 homolog in HCMV, UL77, also was demonstrated to form a dimer (44).
FIG 4.
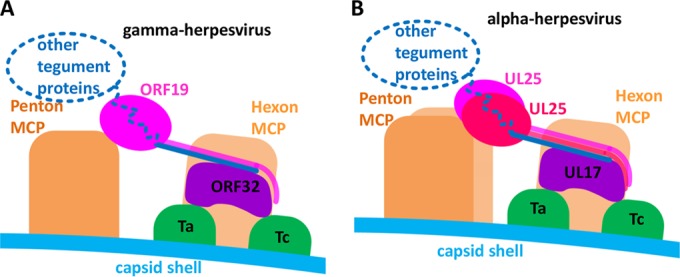
Schematic presentation of capsid-associated tegument component organizations in gammaherpesvirus (A) and alphaherpesvirus (B). The main differences are the following: (i) only one ORF19 molecule is present in each gammaherpesvirus tegument density, compared to two copies of UL25 in each alphaherpesvirus tegument density; (ii) ORF19 binds only one penton MCP in gammaherpesvirus, while each UL25 in alphaherpesvirus binds two penton MCPs due to a 30° clockwise rotation of the penton, denoted as described in the legend to Fig. 2C.
In contrast, the stoichiometries of alphaherpesvirus UL17 and KSHV ORF32 appear to be the same. In KSHV, an ORF32 monomer occupies the triplex-binding region of the capsid-associated tegument component. The similar size of this region in KSHV and PRV (Fig. 2D, E, G, and H) indicates that only one copy of UL17 exists in each alphaherpesvirus CVSC as well.
The stoichiometric differences between the UL25/ORF19 proteins in alpha- and gammaherpesviruses may correlate with structural differences in their pentons. ORF19 in KSHV and UL25 in PRV bind pentons in different ways. More specifically, KSHV ORF19 binds only one penton MCP, while each of the two PRV UL25 molecules binds two MCPs at the same time, and the binding area on penton MCP is different from that in KSHV (Fig. 2D, E, G, and H). By superimposing the 5-fold axis of the KSHV and PRV reconstructions, we find that the pentons in the two structures do not fit with each other, although their periphery hexons and triplexes are aligned well. By fitting the HSV-1 MCPud atomic model (PDB entry 1NO7) (37) into the penton MCPs and marking positions of the same structural element in KSHV and PRV, we measured that there is roughly a 30° clockwise rotation of the PRV penton compared to that in KSHV (Fig. 2C). Conceivably, rotation of the PRV penton exposes alternative binding areas on the MCP to its UL25 protein and also results in more space between the penton and its adjacent hexon to accommodate a UL25 dimer than that required for an ORF19 monomer in KSHV.
DISCUSSION
Alphaherpesvirus tegument has been studied extensively, and cryo-EM of HSV-1/PRV capsids with GFP-tagged UL25 confirmed its identity as one contributor to the CVSC. However, both the exact location and in situ conformation of the UL25 protein inside alphaherpesvirus virions remained hitherto puzzling. It was mysterious why the crystal structure of the HSV-1 N-terminally truncated UL25 did not fit the density designated UL25 in a 9-Å-resolution cryo-EM reconstruction of PRV C capsids (17). Two possibilities were offered to explain this discrepancy: first, PRV and HSV-1 UL25 proteins have different structures, and second, UL25 has a conformation inside the virion that is different from that in the crystal (17). One can safely rule out the first possibility, given the rule of thumb of structural biology that sequence identity of over 30% usually leads to nearly identical folds and the high sequence identity between PRV and HSV-1 UL25 proteins (50% identity and 60% similarity). The structure presented here has allowed us to rule out the second possibility too, resolving the long-standing mystery surrounding the exact location and the in situ structure of UL25 and its homolog in gammaherpesvirus.
Specifically, the newly resolved structure of the N-terminal segment not present in the crystal structure of HSV-1 UL25 explains why the previous labeling studies had led to the mystery and the eventual incorrect assignment of UL25 to the triplex-binding region of CVSC (15, 17, 45). The crystal structure of UL25 contains the globular domain encompassing aa 134 to 577 but not the extended N-terminal segment (Fig. 3D and E). In the GFP-labeling studies of HSV-1 and PRV UL25, the GFP tag was inserted between residues 50 and 51 and produced an extra density on top of the CVSC densities (15, 17), roughly at the position marked by an asterisk in our KSHV structure (Fig. 3A and D). Similarly, the position of the N terminus of HSV-1 UL25 was labeled by a tandem affinity purification tag and visualized by cryo-EM (45). A small extra density was identified at one side of the triplex-binding region (see Fig. 8 of reference 45), which agrees with our assignment of the UL25 N terminus extending to the triplex-binding region. In all of these cases, the N terminus of UL25 was correctly identified by labeling; however, the poor resolutions of the cryo-EM structures unfortunately have led to the misassignment of the globular domain of UL25 to the density largely belonging to UL17.
The direct structural comparison presented here shows that a similar, but not identical, organization of capsid-associated tegument proteins exists in alpha- and gammaherpesviruses, as depicted in Fig. 4. This organization suggests an important structural role for ORF32/UL17. This tegument protein mediates assembly of tegument proteins by anchoring to the capsid on penton-proximal triplexes and connecting to ORF19/UL25 through its N-terminal helix and possibly to other tegument proteins as well. This notion is supported by the previous finding in HSV-1 that UL17 is required for efficient binding of UL25 to capsid (46).
UL17 and UL25 genes are conserved among all three herpesvirus subfamilies. We show here that the capsid association pattern of UL17 and UL25 homologs is similar in gammaherpesviruses and alphaherpesviruses. In addition to binding to pentons as visualized here, HSV-1 UL25 also has been shown to bind the portal protein UL6 (47), a component of the DNA-packaging machinery. The portal occupies only 1 of the 12 capsid vertices and is not visible in the reconstruction shown here due to imposition of icosahedral symmetry. The capability of UL25 homologs to bind DNA-packaging machinery suggests that they are directly involved in the processes of genome packaging, retention, and/or releasing (23, 47–50). The presence of UL25 improved the efficiency of packaging the full-length genome (48, 49, 51). In the betaherpesvirus HCMV, the UL17 and UL25 homologs (UL93 and UL77, respectively) also are required for efficient virus growth in cultured fibroblasts (52, 53). We have shown previously that the most abundant capsid-associated tegument protein in HCMV is pp150 (18, 19), a betaherpesvirus-specific tegument protein not found in alpha- and gammaherpesviruses. The binding of pp150 might have prevented the association of the UL17/UL25 homologs around the penton locations to those in alpha- and gammaherpesviruses. However, the UL25 homolog in HCMV was demonstrated to be a structural protein associated with the capsid and to interact with portal protein UL104 (the alphaherpesvirus UL6 homolog) (44). It is conceivable that in HCMV, the HSV-1 UL17 and UL25 homologs also could bind to the portal-specific vertex, which, as in the case of alpha- and gammaherpesvirus virions, were not visible in the icosahedral reconstructions due to the imposition of symmetry during cryo-EM reconstruction.
Supplementary Material
ACKNOWLEDGMENTS
We thank Jae U. Jung of the University of Southern California for providing us with the iSLK-KSHVBAC16 cell line used for viral culture.
This research was supported in part by the NIH (AI046420/AI094386 and GM071940).
We acknowledge the use of instruments at the Electron Imaging Center for Nanomachines, supported by UCLA and by instrumentation grants from the NIH (1S10RR23057 and 1S10OD018111) and NSF (DBI-1338135).
Footnotes
Published ahead of print 20 August 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.01509-14.
REFERENCES
- 1.Chang Y, Cesarman E, Pessin MS, Lee F, Culpepper J, Knowles DM, Moore PS. 1994. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science 266:1865–1869. 10.1126/science.7997879. [DOI] [PubMed] [Google Scholar]
- 2.Moore PS, Chang Y. 1995. Detection of herpesvirus-like DNA sequences in Kaposi's sarcoma in patients with and without HIV infection. N. Engl. J. Med. 332:1181–1185. 10.1056/NEJM199505043321801. [DOI] [PubMed] [Google Scholar]
- 3.Boshoff C, Weiss R. 2002. AIDS-related malignancies. Nat. Rev. Cancer 2:373–382. 10.1038/nrc797. [DOI] [PubMed] [Google Scholar]
- 4.Moore PS, Chang Y. 2001. Kaposi's sarcoma-associated herpesvirus, p 2803–2833 In Knipe D, Howley P, Griffin D, Lamb R, Martin M, Straus S. (ed), Fields virology, 4 ed, vol 2 Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 5.Dai W, Jia Q, Bortz E, Shah S, Liu J, Atanasov I, Li X, Taylor KA, Sun R, Zhou ZH. 2008. Unique structures in a tumor herpesvirus revealed by cryo-electron tomography and microscopy. J. Struct. Biol. 161:428–438. 10.1016/j.jsb.2007.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grunewald K, Desai P, Winkler DC, Heymann JB, Belnap DM, Baumeister W, Steven AC. 2003. Three-dimensional structure of herpes simplex virus from cryo-electron tomography. Science 302:1396–1398. 10.1126/science.1090284. [DOI] [PubMed] [Google Scholar]
- 7.Smith GA, Enquist LW. 2002. Break ins and break outs: viral interactions with the cytoskeleton of mammalian cells. Annu. Rev. Cell Dev. Biol. 18:135–161. 10.1146/annurev.cellbio.18.012502.105920. [DOI] [PubMed] [Google Scholar]
- 8.Luxton GW, Haverlock S, Coller KE, Antinone SE, Pincetic A, Smith GA. 2005. Targeting of herpesvirus capsid transport in axons is coupled to association with specific sets of tegument proteins. Proc. Natl. Acad. Sci. U. S. A. 102:5832–5837. 10.1073/pnas.0500803102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Luxton GW, Lee JI, Haverlock-Moyns S, Schober JM, Smith GA. 2006. The pseudorabies virus VP1/2 tegument protein is required for intracellular capsid transport. J. Virol. 80:201–209. 10.1128/JVI.80.1.201-209.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Antinone SE, Smith GA. 2010. Retrograde axon transport of herpes simplex virus and pseudorabies virus: a live-cell comparative analysis. J. Virol. 84:1504–1512. 10.1128/JVI.02029-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zaichick SV, Bohannon KP, Smith GA. 2011. Alphaherpesviruses and the cytoskeleton in neuronal infections. Viruses 3:941–981. 10.3390/v3070941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zaichick SV, Bohannon KP, Hughes A, Sollars PJ, Pickard GE, Smith GA. 2013. The herpesvirus VP1/2 protein is an effector of dynein-mediated capsid transport and neuroinvasion. Cell Host Microbe 13:193–203. 10.1016/j.chom.2013.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou ZH, Chen DH, Jakana J, Rixon FJ, Chiu W. 1999. Visualization of tegument-capsid interactions and DNA in intact herpes simplex virus type 1 virions. J. Virol. 73:3210–3218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Trus BL, Newcomb WW, Cheng N, Cardone G, Marekov L, Homa FL, Brown JC, Steven AC. 2007. Allosteric signaling and a nuclear exit strategy: binding of UL25/UL17 heterodimers to DNA-filled HSV-1 capsids. Mol. Cell 26:479–489. 10.1016/j.molcel.2007.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Conway JF, Cockrell SK, Copeland AM, Newcomb WW, Brown JC, Homa FL. 2010. Labeling and localization of the herpes simplex virus capsid protein UL25 and its interaction with the two triplexes closest to the penton. J. Mol. Biol. 397:575–586. 10.1016/j.jmb.2010.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Toropova K, Huffman JB, Homa FL, Conway JF. 2011. The herpes simplex virus 1 UL17 protein is the second constituent of the capsid vertex-specific component required for DNA packaging and retention. J. Virol. 85:7513–7522. 10.1128/JVI.00837-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Homa FL, Huffman JB, Toropova K, Lopez HR, Makhov AM, Conway JF. 2013. Structure of the pseudorabies virus capsid: comparison with herpes simplex virus type 1 and differential binding of essential minor proteins. J. Mol. Biol. 425:3415–3428. 10.1016/j.jmb.2013.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yu X, Shah S, Lee M, Dai W, Lo P, Britt W, Zhu H, Liu F, Zhou ZH. 2011. Biochemical and structural characterization of the capsid-bound tegument proteins of human cytomegalovirus. J. Struct. Biol. 174:451–460. 10.1016/j.jsb.2011.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dai X, Yu X, Gong H, Jiang X, Abenes G, Liu H, Shivakoti S, Britt WJ, Zhu H, Liu F, Zhou ZH. 2013. The smallest capsid protein mediates binding of the essential tegument protein pp150 to stabilize DNA-containing capsids in human cytomegalovirus. PLoS Pathog. 9:e1003525. 10.1371/journal.ppat.1003525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen DH, Jiang H, Lee M, Liu F, Zhou ZH. 1999. Three-dimensional visualization of tegument/capsid interactions in the intact human cytomegalovirus. Virology 260:10–16. 10.1006/viro.1999.9791. [DOI] [PubMed] [Google Scholar]
- 21.Trus BL, Gibson W, Cheng N, Steven AC. 1999. Capsid structure of simian cytomegalovirus from cryoelectron microscopy: evidence for tegument attachment sites. J. Virol. 73:2181–2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Salmon B, Cunningham C, Davison AJ, Harris WJ, Baines JD. 1998. The herpes simplex virus type 1 U(L)17 gene encodes virion tegument proteins that are required for cleavage and packaging of viral DNA. J. Virol. 72:3779–3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McNab AR, Desai P, Person S, Roof LL, Thomsen DR, Newcomb WW, Brown JC, Homa FL. 1998. The product of the herpes simplex virus type 1 UL25 gene is required for encapsidation but not for cleavage of replicated viral DNA. J. Virol. 72:1060–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meyer HH, Ripalti A, Landini MP, Radsak K, Kern HF, Hensel GM. 1997. Human cytomegalovirus late-phase maturation is blocked by stably expressed UL32 antisense mRNA in astrocytoma cells. J. Gen. Virol. 78(Part 10):2621–2631. [DOI] [PubMed] [Google Scholar]
- 25.Brulois KF, Chang H, Lee AS, Ensser A, Wong LY, Toth Z, Lee SH, Lee HR, Myoung J, Ganem D, Oh TK, Kim JF, Gao SJ, Jung JU. 2012. Construction and manipulation of a new Kaposi's sarcoma-associated herpesvirus bacterial artificial chromosome clone. J. Virol. 86:9708–9720. 10.1128/JVI.01019-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Myoung J, Ganem D. 2011. Generation of a doxycycline-inducible KSHV producer cell line of endothelial origin: maintenance of tight latency with efficient reactivation upon induction. J. Virol. Methods 174:12–21. 10.1016/j.jviromet.2011.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Suloway C, Pulokas J, Fellmann D, Cheng A, Guerra F, Quispe J, Stagg S, Potter CS, Carragher B. 2005. Automated molecular microscopy: the new Leginon system. J. Struct. Biol. 151:41–60. 10.1016/j.jsb.2005.03.010. [DOI] [PubMed] [Google Scholar]
- 28.Li X, Mooney P, Zheng S, Booth CR, Braunfeld MB, Gubbens S, Agard DA, Cheng Y. 2013. Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM. Nat. Methods 10:584–590. 10.1038/nmeth.2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mindell JA, Grigorieff N. 2003. Accurate determination of local defocus and specimen tilt in electron microscopy. J. Struct. Biol. 142:334–347. 10.1016/S1047-8477(03)00069-8. [DOI] [PubMed] [Google Scholar]
- 30.Ludtke SJ, Baldwin PR, Chiu W. 1999. EMAN: semiautomated software for high-resolution single-particle reconstructions. J. Struct. Biol. 128:82–97. 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- 31.Liang Y, Ke EY, Zhou ZH. 2002. IMIRS: a high-resolution 3D reconstruction package integrated with a relational image database. J. Struct. Biol. 137:292–304. 10.1016/S1047-8477(02)00014-X. [DOI] [PubMed] [Google Scholar]
- 32.Liu H, Cheng L, Zeng S, Cai C, Zhou ZH, Yang Q. 2008. Symmetry-adapted spherical harmonics method for high-resolution 3D single-particle reconstructions. J. Struct. Biol. 161:64–73. 10.1016/j.jsb.2007.09.016. [DOI] [PubMed] [Google Scholar]
- 33.Zhang X, Zhang X, Zhou ZH. 2010. Low cost, high performance GPU computing solution for atomic resolution cryoEM single-particle reconstruction. J. Struct. Biol. 172:400–406. 10.1016/j.jsb.2010.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rosenthal PB, Henderson R. 2003. Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. J. Mol. Biol. 333:721–745. 10.1016/j.jmb.2003.07.013. [DOI] [PubMed] [Google Scholar]
- 35.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. 2004. UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 25:1605–1612. 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 36.Jones DT. 1999. Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol. 292:195–202. 10.1006/jmbi.1999.3091. [DOI] [PubMed] [Google Scholar]
- 37.Bowman BR, Baker ML, Rixon FJ, Chiu W, Quiocho FA. 2003. Structure of the herpesvirus major capsid protein. EMBO J. 22:757–765. 10.1093/emboj/cdg086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhou ZH, Prasad BV, Jakana J, Rixon FJ, Chiu W. 1994. Protein subunit structures in the herpes simplex virus A-capsid determined from 400 kV spot-scan electron cryomicroscopy. J. Mol. Biol. 242:456–469. 10.1006/jmbi.1994.1594. [DOI] [PubMed] [Google Scholar]
- 39.Bowman BR, Welschhans RL, Jayaram H, Stow ND, Preston VG, Quiocho FA. 2006. Structural characterization of the UL25 DNA-packaging protein from herpes simplex virus type 1. J. Virol. 80:2309–2317. 10.1128/JVI.80.5.2309-2317.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cardone G, Newcomb WW, Cheng N, Wingfield PT, Trus BL, Brown JC, Steven AC. 2012. The UL36 tegument protein of herpes simplex virus 1 has a composite binding site at the capsid vertices. J. Virol. 86:4058–4064. 10.1128/JVI.00012-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee JI, Luxton GW, Smith GA. 2006. Identification of an essential domain in the herpesvirus VP1/2 tegument protein: the carboxy terminus directs incorporation into capsid assemblons. J. Virol. 80:12086–12094. 10.1128/JVI.01184-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Coller KE, Lee JI, Ueda A, Smith GA. 2007. The capsid and tegument of the alphaherpesviruses are linked by an interaction between the UL25 and VP1/2 proteins. J. Virol. 81:11790–11797. 10.1128/JVI.01113-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Leelawong M, Lee JI, Smith GA. 2012. Nuclear egress of pseudorabies virus capsids is enhanced by a subspecies of the large tegument protein that is lost upon cytoplasmic maturation. J. Virol. 86:6303–6314. 10.1128/JVI.07051-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Meissner CS, Koppen-Rung P, Dittmer A, Lapp S, Bogner E. 2011. A “coiled-coil” motif is important for oligomerization and DNA binding properties of human cytomegalovirus protein UL77. PLoS One 6:e25115. 10.1371/journal.pone.0025115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cockrell SK, Huffman JB, Toropova K, Conway JF, Homa FL. 2011. Residues of the UL25 protein of herpes simplex virus that are required for its stable interaction with capsids. J. Virol. 85:4875–4887. 10.1128/JVI.00242-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thurlow JK, Murphy M, Stow ND, Preston VG. 2006. Herpes simplex virus type 1 DNA-packaging protein UL17 is required for efficient binding of UL25 to capsids. J. Virol. 80:2118–2126. 10.1128/JVI.80.5.2118-2126.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pasdeloup D, Blondel D, Isidro AL, Rixon FJ. 2009. Herpesvirus capsid association with the nuclear pore complex and viral DNA release involve the nucleoporin CAN/Nup214 and the capsid protein pUL25. J. Virol. 83:6610–6623. 10.1128/JVI.02655-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stow ND. 2001. Packaging of genomic and amplicon DNA by the herpes simplex virus type 1 UL25-null mutant KUL25NS. J. Virol. 75:10755–10765. 10.1128/JVI.75.22.10755-10765.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Klupp BG, Granzow H, Keil GM, Mettenleiter TC. 2006. The capsid-associated UL25 protein of the alphaherpesvirus pseudorabies virus is nonessential for cleavage and encapsidation of genomic DNA but is required for nuclear egress of capsids. J. Virol. 80:6235–6246. 10.1128/JVI.02662-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cockrell SK, Sanchez ME, Erazo A, Homa FL. 2009. Role of the UL25 protein in herpes simplex virus DNA encapsidation. J. Virol. 83:47–57. 10.1128/JVI.01889-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Preston VG, Murray J, Preston CM, McDougall IM, Stow ND. 2008. The UL25 gene product of herpes simplex virus type 1 is involved in uncoating of the viral genome. J. Virol. 82:6654–6666. 10.1128/JVI.00257-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yu D, Silva MC, Shenk T. 2003. Functional map of human cytomegalovirus AD169 defined by global mutational analysis. Proc. Natl. Acad. Sci. U. S. A. 100:12396–12401. 10.1073/pnas.1635160100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dunn W, Chou C, Li H, Hai R, Patterson D, Stolc V, Zhu H, Liu F. 2003. Functional profiling of a human cytomegalovirus genome. Proc. Natl. Acad. Sci. U. S. A. 100:14223–14228. 10.1073/pnas.2334032100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.