Abstract
Binding of herpes simplex virus 1 (HSV-1) envelope glycoprotein D (gD) to the receptor 3-O-sulfated heparan sulfate (3-OS HS) mediates viral entry. 3-O-Sulfation of HS is catalyzed by the 3-O-sulfotransferase (3-OST) enzyme. Multiple isoforms of 3-OST are differentially expressed in tissues of zebrafish (ZF) embryos. Here, we performed a comprehensive analysis of the role of ZF 3-OST isoforms (3-OST-1, 3-OST-5, 3-OST-6, and 3-OST-7) in HSV-1 entry. We found that a group of 3-OST gene family isoforms (3-OST-2, -3, -4, and -6) with conserved catalytic and substrate-binding residues of the enzyme mediates HSV-1 entry and spread, while the other group (3-OST-1, -5, and -7) lacks these properties. These results demonstrate that HSV-1 entry can be recapitulated by certain ZF 3-OST enzymes, a significant step toward the establishment of a ZF model of HSV-1 infection and tissue-specific tropism.
TEXT
Herpes simplex virus 1 (HSV-1) entry into the host cells requires interactions between viral envelope glycoprotein D (gD) and host cell receptors (1). An important gD receptor is 3-O-sulfated heparan sulfate (3-OS HS) (2, 3). Synthesis and modifications of HS is a multistep process (4–8), of which the last step involves O-sulfation at different positions of the molecule, catalyzed by various sulfotransferases (4). 3-O-Sulfotransferases (3-OST-1, -2, -3A, -3B, -4, -5, and -6 isoforms; also called Hs3st1, -2, -3A, -3B, -4, -5, and -6, respectively) catalyze sulfation at the C-3 of the glucosamine units of HS, generating specific protein-binding sites for HSV-1 gD (2, 3, 9, 10).
Recent studies of zebrafish (ZF) provided a functional analysis of 3-OST-generated HS (11–15). Out of the characterized eight 3-OST family members in ZF, seven genes show homology to known 3-OST genes in mice and humans (11). These ZF enzyme isoforms are differentially expressed in a tissue-specific manner, suggesting the possibility of creating a tool to study HSV tissue-specific tropism, whose study is hampered by the lack of available convenient tools. Our previous preliminary studies suggested that ZF 3-OST may mediate HSV entry (15). In this study, we investigated the ability of ZF 3-OST-1, -5, -6, and -7 isoforms, comparatively to their human analogue, to facilitate HSV-1 entry and cell-cell fusion. Our previous studies have shown that ZF-encoded 3-OST isoforms (3-OST-2, -3, and -4) allow HSV-1 entry into host cells (14, 16, 17). Here, we provide a comprehensive analysis of the differential abilities of the remaining ZF 3-OST isoforms to mediate HSV-1 entry and propose a unique model for future studies of HSV-1 tropism using ZF as a convenient tool.
Analysis and cloning of zebrafish-encoded 3-OST enzymes.
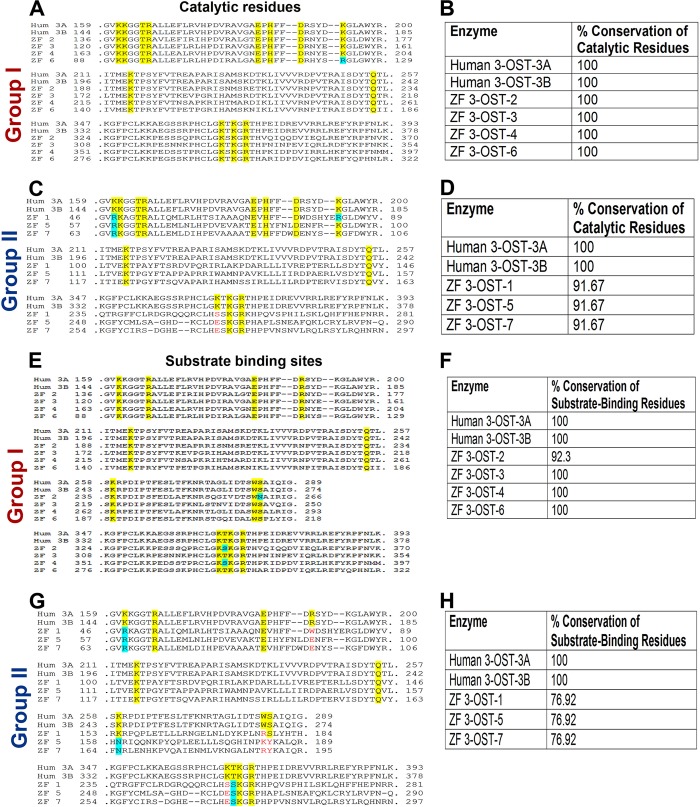
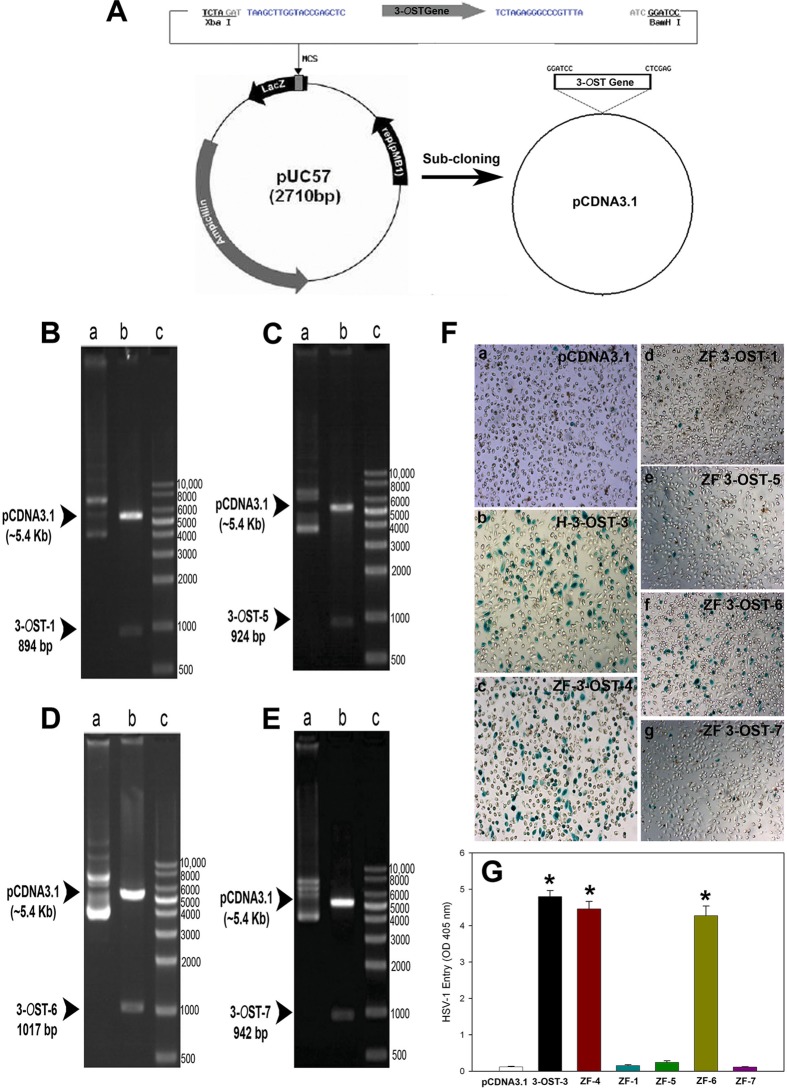
To characterize ZF 3-OST isoforms relative to their human counterparts, we first performed an amino acid sequence alignment of the ZF-encoded 3-OST enzymes to the previously characterized human 3-OST-3A and 3-OST-3B isoforms (2, 11). Interestingly, we found that the amino acid sequences of the ZF 3-OST enzymes show various degrees of homology to the human 3-OST-3 isoforms (Fig. 1). Comparing the conservation of the catalytic residues of the enzymes characterized for human 3-OST-3, we found that ZF enzymes 3-OST-2, -3, -4, and -6 show 100% conservation of the catalytic residues; these were designated group I members (Fig. 1A and B). The other isoforms (3-OST-1, -5, and -7), which do not show conservation of an essential catalytic residue, lysine 352 (K352) of human 3-OST-3B, were classified as group II (Fig. 1C and D). In addition, in studying the substrate-binding sites, we observed that ZF 3-OST-2, -3, -4, and -6 isoforms showed conservation of all or most amino acid residues responsible for substrate binding, whereas the other isoforms showed significantly less conservation of these residues (Fig. 1E to H). Both the catalytic residues and the substrate-binding sites have been shown to be important for HSV-1 entry-mediating activity of 3-OSTs. The residue K162 in human 3-OST-3A is considered key to the catalytic activity of the enzyme, and it is highly conserved among entry-mediating human and ZF isoforms (2). The significant homology of ZF to human 3-OST isoforms provided a strong rationale to investigate these multiple ZF 3-OST isoforms for HSV entry. The open reading frame (ORF) of ZF, encoding sequences of the 3-OST-1, -5, -6, and -7 genes, was blunt-end cloned in the pUC57 vector via an EcoRV strategy. The pUC57 vector with an insert was then subcloned into the pcDNA3.1 vector for mammalian expression by BamHI and Xhol (Fig. 2A). Successful cloning of the ZF enzymes was verified by enzymatic digestion with restriction endonucleases (Fig. 2B to E) as previously described (14, 16, 17).
FIG 1.
(A to D) Comparison of catalytic residues between human- and zebrafish (ZF)-encoded multiple isoforms of 3-O sulfotransferase (3-OST) via amino acid sequence alignment. Human (Hum) 3-OST-3A was used as a template for alignment of ZF 3-OST enzymes. Previous structural analysis of human 3-OST-3A demonstrated the catalytic residues of human 3-OST-3A (41). In panels A and C, the numbers to the left of the sequence represent the amino acid numbering (N-terminal Met = 1) in the protein. Yellow boxes represent the conserved catalytic residues; a cyan box indicates a nonidentical but similar residue (e.g., positively charged K-to-R) replacement. Red residues represent replacement of a catalytic residue with a nonsimilar residue. In panels B and D, the table represents percent conservation of catalytic residues of human and group I (B) or group II (D) ZF 3-OSTs. Percent conservation of catalytic residues was calculated as the percent conserved catalytic residues of the total number of catalytic residues. (E to H) Comparison of substrate-binding residues between human- and zebrafish (ZF)-encoded multiple isoforms of 3-O sulfotransferase (3-OST) via amino acid sequence alignment. Human 3-OST-3A was used as a template for alignment of ZF 3-OST enzymes. Substrate-binding residues of human 3-OST-3A were previously reported (41). Numbers to the left of a sequence represent the amino acid numbering (N-terminal Met = 1) in the protein. Yellow boxes indicate the conserved catalytic residues; a cyan box indicates a nonidentical but similar residue (e.g., positively charged K-to-R) replacement. Red residues represent replacement of a substrate-binding residue with a nonsimilar residue. In panels F and H, the table represents conservation of substrate-binding residues of human and group I (F) or group II (H) ZF 3-OSTs. Percent conservation of substrate-binding residues was calculated as the percent conserved substrate-binding residues of the total number of substrate-binding residues.
FIG 2.
Cloning and characterization of the zebrafish 3-OST gene. (A to E) The open reading frame (ORF) of ZF, encoding sequences of the 3-OST-1, -5, -6, and -7 gene, was blunt-end cloned in the pUC57 vector via an EcoRV strategy, and the pUC57 vector with an insert was then subcloned into the pcDNA3.1 vector for mammalian expression by BamHI and Xhol (A). Successful cloning of the ZF enzymes was verified by enzymatic digestion with restriction endonucleases (B to E). ZF-encoded 3-OST-1 (894 bp) (B), 3-OST-5 (924 bp) (C), 3-OST-6 (1,017 bp) (D), and 3-OST-7 (942 bp) (E) isoforms cloned into pcDNA3.1 (5.4 Kb) are shown. ZF encoding 3-OST plasmids were constructed by inserting the open reading frame of 3-OSTs into pcDNA3.1, and the construct was designated the pcDNA3.1-ZF-3-OST specific isoform. The inserted sequence of 3-OST-1, -5, -6, and -7 was verified after digestion using BamHI and XhoI. (F) Wild-type Chinese hamster ovary (CHO-K1) cells expressing ZF 3-OST-6 (subpanel f), but not 3-OST-1 (subpanel d), 3-OST-5 (subpanel e), and 3-OST-7 (subpanel g), are susceptible to HSV-1 entry. CHO-K1 cells expressing various 3-OST isoforms grown (4 × 106 cells) in a separate six-well dishes were challenged with β-galactosidase-expressing recombinant HSV-1 (gL86) at 100 PFU/cell. Wild-type CHO-K1 cells transfected with the empty vector (pcDNA3.1; subpanel a) and the human isoform of 3-OST-3 (subpanel b) and ZF 3-OST-4 (subpanel c) were also infected in parallel as negative and positive controls, respectively. After 6 h of infection at 37°C, cells were washed with PBS, fixed and permeabilized, and incubated with X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactosidase) at 1.0 mg/ml, which yields an insoluble blue-stained product upon hydrolysis by β-galactosidase. Blue cells (representing viral entry) were seen as shown. Microscopy was performed using a 20× objective with a Zeiss Axiovert 100. (G) HSV-1 entry into ZF expressing individual 3-OSTs in CHO-K1 cells was further confirmed by using a soluble-substrate o-nitrophenyl-β-d-galactopyranoside (ONPG) assay. Resistant wild-type CHO-K1 cells were transfected with various ZF 3-OST isoforms (3-OST-1, -4, -5, -6, and -7) at 2.5 μg DNA. Cells transfected with empty vector pcDNA3.1 at 2.5 μg DNA and human 3-OST-3 were used as a negative and positive control, respectively. Cultured cells were plated in 96-well plates and inoculated with β-galactosidase-expressing recombinant virus HSV-1 (KOS) gL86 at 50 PFU/cell. After 6 h, the cells were washed, permeabilized, and incubated with ONPG substrate (3.0 mg/ml) for quantitation of β-galactosidase activity expressed from the input viral genome. The enzymatic activity was measured at an optical density of 405 nm (OD405).
Zebrafish 3-OST-6, but not 3-OST-1, -5, and -7, supports HSV-1 entry into resistant CHO-K1 cells.
We investigated whether the ZF 3-OST enzymes can support HSV-1 entry into resistant Chinese hamster ovary (CHO-K1) cells. The ability of ZF-encoded multiple isoforms (3-OST-1, -5, -6, and -7) was determined by transiently transfecting CHO-K1 cells with an expression plasmid or a control pcDNA3.1 plasmid as a negative control. An HSV-1 entry assay was performed using reporter HSV-1 virus (HSV-1 [KOS] gL86). As shown in Fig. 2F, subpanels b to g, X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactosidase) staining was found to be positive for ZF 3-OST-6 cells (subpanel f), similar to the positive control for human 3-OST-3B (subpanel b) and ZF 3-OST-4 (subpanel c). In contrast, CHO-K1 cells expressing ZF 3-OST-1 (subpanel d), -5 (subpanel e), and -7 (subpanel g) were resistant to HSV entry and hence colorless, similar to the wild-type CHO-K1 cells expressing the pcDNA3.1 empty vector (Fig. 2F, subpanel a). The results from the X-Gal assay were further confirmed by a quantitative ONPG (o-nitrophenyl-β-d-galactopyranoside) assay (Fig. 2G). As shown in Fig. 2G, HSV-1 entry into CHO-K1 cells expressing ZF 3-OST-6 was similar to that for human 3-OST-3 cells and also for the previously reported ZF 3-OST isoforms (3-OST-2, -3, and -4; data shown only for 3-OST-4) (14, 16, 17), while no HSV-1 entry was observed for CHO-K1 cells expressing the empty vector plasmid pcDNA3.1. Interestingly, ZF-encoded 3-OST isoforms (3-OST-1, -5, and -7) were similar to the pcDNA3.1 control. It is possible that loss of HSV-1 entry-mediating activity results from the lack of certain functionally important residues in 3-OST-1, -5, and -7 isoforms since these enzymes are less homologous to entry-supporting enzymes, such as ZF 3-OST-2, -3, and -4 and human 3-OST-3 (Fig. 1). Future mutagenesis studies using various 3-OSTs may help address this interesting possibility.
Expression of HSV-1 glycoprotein D (gD) in zebrafish 3-OST-6-expressing cells inhibits HSV entry.
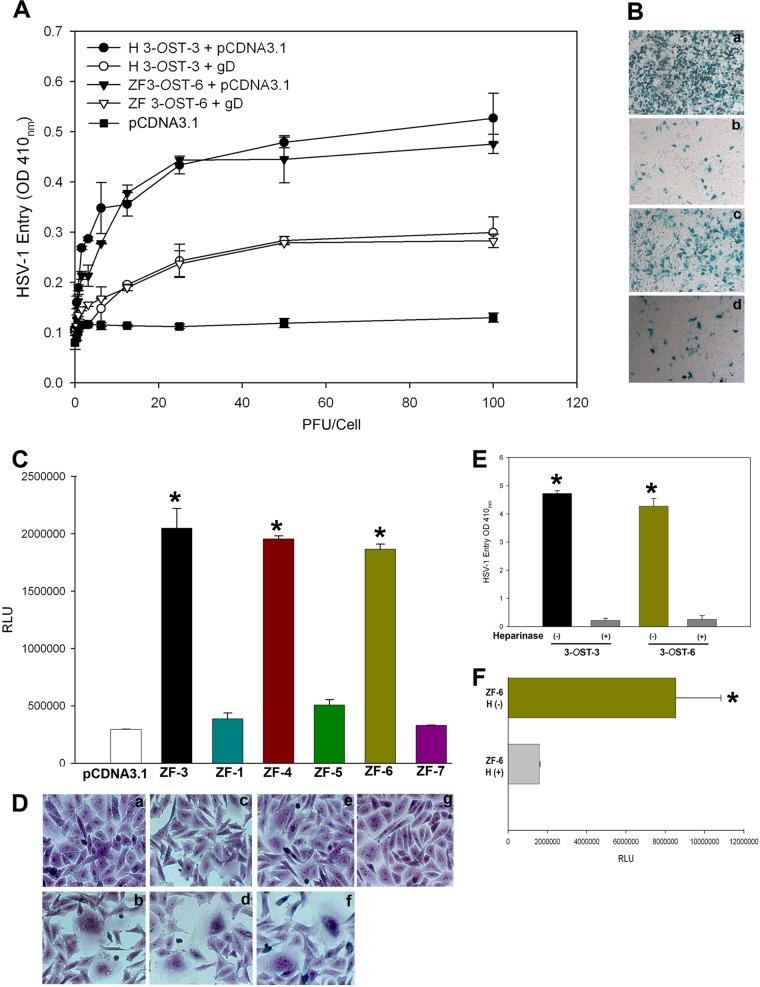
To validate the idea that 3-OST-mediated HSV-1 entry is gD dependent, we utilized the gD-mediated interference assay previously described (18). This assay is based on the principle that cells that are normally susceptible to viral entry become resistant upon expression of viral gD because of sequestration of gD receptors by cell-expressed gD. To carry out the assay, CHO-K1 cells were transiently cotransfected with an HSV-1 gD-expressing plasmid with ZF 3-OST-6 (or an equal amount of the empty vector, pcDNA3.1, as a control), followed by infection with serial dilutions of β-galactosidase-expressing HSV-1 (KOS) gL86. In this experiment, we used CHO-K1 cells coexpressing the human 3-OST-3 isoform with either pcDNA3.1 or HSV-1 gD as a control as previously described (18). As shown in Fig. 3A, HSV-1 entry into gD-expressing ZF 3-OST-6 was suppressed compared to results with the 3-OST-6 control. The ONPG experiment was repeated using an X-Gal assay which clearly showed less entry for cells coexpressing gD and human 3-OST-3 or ZF 3-OST-6, respectively (i.e., fewer blue cells) (Fig. 3B, subpanels b and d) compared to that for cells expressing human 3-OST-3 or ZF 3-OST-6 with pcDNA3.1 (Fig. 3B, subpanels a and c).
FIG 3.
Zebrafish 3-OST-mediated HSV-1 entry is glycoprotein D (gD) dependent. (A) Wild-type CHO-K1 cells were cotransfected with an HSV-1 gD expression plasmid and ZF 3-OST-6 (∇) infected with HSV-1. In parallel, CHO-K1 cells were cotransfected with ZF 3-OST-6 with pcDNA3.1 (▼). In parallel, CHO-K1 cells were infected with HSV-1 expressing empty vector pcDNA3.1 with human 3-OST-3 (●) or cells coexpressing gD with human 3-OST-3 (○). Reporter enzyme activity, as a measurement of viral entry, was measured with a spectrophotometer at 410 nm. (B) Inhibition of HSV-1 entry during gD interference assay confirmed by X-Gal assay. CHO-K1 cells expressing human 3-OST-3 with pcDNA3.1 (subpanel a) showed a greater number of blue cells than did CHO-K1 cells coexpressing gD with human 3-OST-3 (subpanel b). Similarly, a greater number of stained cells were observed for CHO-K1 cells expressing ZF 3-OST-6 (subpanel c) than that for cells coexpressing gD with ZF 3-OST-6 (subpanel d). Microscopy was performed using a 20× objective with a Zeiss Axiovert 100. (C) ZF-encoded 3-OST-6, but not ZF 3-OST-1, -5, and -7, mediated cell-to-cell fusion with HSV-1 glycoprotein-expressing cells. The effector CHO-K1 cells were transfected with HSV-1 glycoproteins gB, gD, gH, and gL and T7 RNA polymerase. The target CHO-K1 cells were transfected with plasmids expressing ZF 3-OST-1, -5, -6, and -7 and the luciferase reporter gene. The target cells expressing ZF 3-OST-4 and human 3-OST-3 and control plasmid pcDNA3.1 were used as a positive and negative control, respectively. A luciferase reporter assay was performed 24 h after the two cell populations were mixed together. Cell fusion was measured in relative light units (RLUs) using a Sirius luminometer (Berthold detection system). (D) Similarly, visual observation resulted in multinucleated giant cells, with CHO-K1 cells expressing ZF 3-OST-6 mixed with effector cells expressing HSV-1 glycoproteins (subpanel f), similar to the positive human 3-OST-3 control and ZF 3-OST-4 (subpanels b and d). No multinucleated giant cells were observed with ZF 3-OST-1 (subpanel c), -5 (subpanel e), and -7 (subpanel g). (E and F) Enzymatic removal of cell surface heparan sulfate (HS) by heparinase treatment of ZF 3-OST-6-expressing CHO-K1 cells reduces HSV-1 infection and cell fusion. CHO-K1 cells expressing ZF 3-OST-6 or human 3-OST-3 were treated with heparinase II/III (1.5 U ml−1) or mock treated (for 3-OST-3 and ZF 3-OST-6) followed by exposure of cells to HSV-1 (KOS) gL86 at 20 PFU/cell or target cells expressing 3-OSTs. Viral entry was quantitated 6 h later by an ONPG assay using a spectrophotometer at 410 nm while cell fusion was measured in relative luciferase units (RLUs) using a Sirius luminometer (Berthold detection system).
Zebrafish-encoded 3-OST enzymes facilitate HSV-1 glycoprotein-mediated cell-to-cell spread.
We next examined the role of ZF 3-OST isoforms (3-OST-1, -5, -6, and -7) in HSV-1 spread using the previously described HSV-1 glycoprotein-mediated cell-to-cell fusion assay (19, 20). We selected wild-type CHO-K1 cells because they lack the endogenous glycoprotein D (gD) receptor required for HSV-1 entry (2). Wild-type CHO-K1 cells were transiently transfected with each of four glycoprotein plasmids—pPEP98 (gB), pPEP99 (gD), pPEP100 (gH), and pPEP101 (gL)—as well as the plasmid pT7EMCLuc that expresses a luciferase reporter gene and that was considered an “effector” cell. In parallel, “target” cells were transfected with individual and separate pools of 3-OST plasmid expressing ZF-encoded 3-OST-1, -5, -6, and -7 along with the plasmid pCAGT7, which expresses T7 RNA polymerase, to induce expression of the luciferase gene. For a negative control, cells were transfected with T7 RNA polymerase and control plasmid pcDNA3.1. The cells expressing ZF 3-OST-3 and T7 RNA polymerase served as a positive control (20). As shown in Fig. 3C, a high level of fusion occurred in ZF-encoded isoforms, such as 3-OST-3-, 3-OST-4-, and 3-OST-6-expressing cells, compared to levels seen with the negative pcDNA3.1 control. Similarly, ZF 3-OST isoforms (3-OST-1, -5, and -7) were unable to induce cell fusion. We further confirmed our results by visualizing polykaryocyte formation as a means of viral spread (20). As indicated in Fig. 3D, subpanel b, no multinucleated giant cells or syncytial formation was observed in ZF 3-OST-1 (subpanel c), -5 (subpanel e), and -7 (subpanel g), while large number of giant cells were formed in ZF 3-OST-3 (subpanel b), -4 (subpanel d), and -6 (subpanel f). These results reinforce our findings that CHO-K1 cells expressing ZF 3-OST-6 allow cell fusion to occur and thus potentially could facilitate the spread of HSV-1 in a ZF model.
Enzymatic heparinase treatment of zebrafish 3-OST-6 cells significantly reduces HSV-1 entry and cell-to-cell fusion.
We also evaluated if enzymatic removal of ZF 3-OST-6 cells affects both HSV-1 entry and viral glycoprotein-mediated cell fusion. In the viral entry experiment, CHO-K1 cells expressing ZF 3-OST-6 were treated separately with heparinase I and heparinase II (1.5 U ml−1) or mock treated with 1× phosphate-buffered saline (PBS) for 45 min before infection with reporter β-galactosidase expressing HSV-1 gL86 (2). These enzymes selectively degrade HS chains by cleaving them (21). Mock-treated CHO-K1 cells expressing human 3-OST-3 and heparinase-treated cells were used as positive and negative controls, respectively, as previously described (18). As indicated in Fig. 3E, heparinase-treated cells showed a significant reduction in HSV-1 entry (Fig. 3D) compared to that for mock-treated 3-OST-3 (Fig. 3E) or ZF 3-OST-6 cells (Fig. 3E). Similar results were obtained with cells expressing ZF 3-OST-2, -3, and -4 isoforms (data not shown) as previously reported (14, 16, 17). We next determined the effect of enzymatic removal of 3-O-sulfated heparan sulfate (3-OS HS) generated by ZF-encoded 3-OST-6 on HSV-1 glycoprotein-mediated cell fusion by treating effector cells with a mixture of heparinase II and III (1.5 U · ml/liter). As shown in Fig. 3F, ZF 3-OST-6 cells treated with heparinase showed a significant reduction in fusion compared to that for mock-treated cells (Fig. 3F). Similarly, heparinase treatment reduced HSV glycoprotein-mediated cell fusion with cells expressing ZF 3-OST-2, -3, and -4 isoforms (data not shown) as previously reported (14, 16, 17).
Potential of zebrafish 3-OST-3 cells to spread HSV-1.
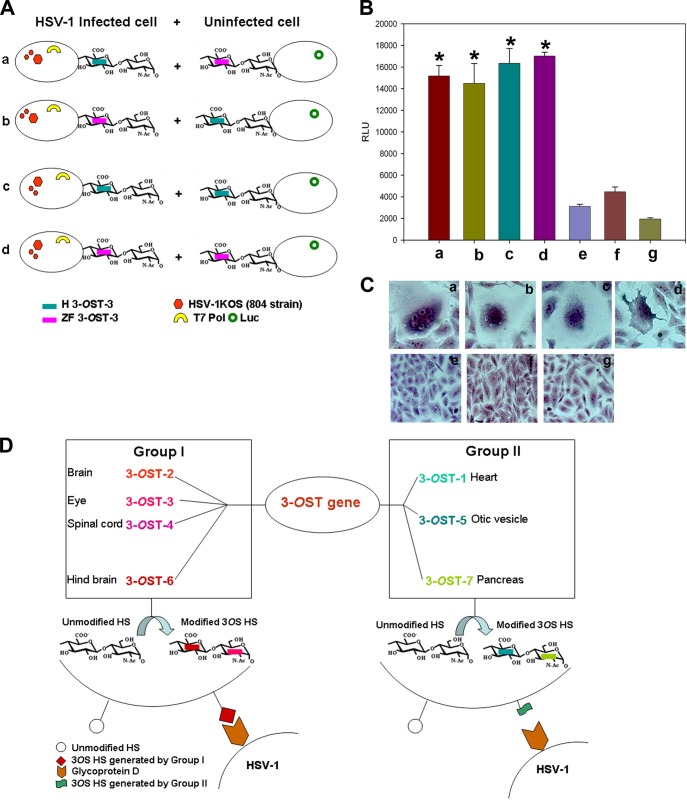
Finally, we tested the ability of ZF and human 3-OST-3 to spread HSV-1 via coculturing HSV-1 (KOS-804 strain)-infected cells coexpressing either the human or ZF 3-OST-3 isoform with T7 polymerase with uninfected cells coexpressing either the human or ZF 3-OST-3 isoform with the luciferase gene (Fig. 4A). As shown in Fig. 4B, the potential of ZF 3-OST-3-generated HS was similar to that of human 3-OST-3 in spreading the virus, which was confirmed by a luciferase-based reporter assay. Three negative controls (HSV-1-infected ZF 3-OST-3 cells cocultured with empty vector CHO cells, HSV-1-infected human 3-OST-3 cells cocultured with empty vector CHO cells, and HSV-1-infected empty vector cells cocultured with empty vector CHO cells) used in the experiments did not provide signal for fusion. Similar results were recorded in coculture experiments when visual documentation of syncytium formation was carried out (Fig. 4C).
FIG 4.
Comparison between human and zebrafish 3-OST-3 for potential to spread HSV-1. (A) Subpanels a to d show combinations of 3-OST-3- and T7 polymerase-coexpressing HSV-1-infected cells used to mix with 3-OST and luciferase gene-coexpressing uninfected cells. (B) The results were confirmed using the previously descried luciferase-based assay. The bars a to d mimic the combinations described for panel A. The bars e to g were negative controls which included either ZF (bar e) or human (bar f) 3-OST, and T7 polymerase coexpressing HSV-1 (KOS-804 strain)-infected cells was mixed with a receptor-negative cell line or receptor-negative cells were challenged with HSV-1 and then mixed with receptor-negative cells (bar g). (C) Subpanels a to d, visualization of syncytial formation from the combinations from panels A and B are presented. Subpanels e to g, negative controls had no syncytial formation. (D) Illustration of the differential role of the various ZF 3-OST enzymes in HSV-1 entry and their tissue-specific distribution pattern in the zebrafish embryo model. Cell- and tissue-specific ZF 3-OST genes fall into two groups based on their ability to generate HSV-1 entry receptor. Note that group I allows HSV-1 entry and spread.
A deep understanding 3-OST-generated HS and its significance to HSV entry is challenged by the lack of tools (22). The ZF embryo model has been recognized to express a wide range of heparan sulfate-modifying 3-OST enzymes during physiological development (11). In order to develop an intact ZF model to study 3-OS HS-dependent HSV interaction, characterization of all known 3-OST isoforms in ZF in a cell culture model is a first critical step. In this study, we characterized ZF3-OST-1, -5, -6, and -7 isoforms for HSV-1 entry and spread.
We first cloned multiple ZF 3-OST isoforms encoding regions into empty vector pcDNA3.1 (Fig. 1A to D) (14, 16, 17). The resultant construct allowed us to successfully express 3-OST-1, -5, -6, and -7 in resistant CHO-K1 cells. Only the ZF-encoded 3-OST-6 isoform, and not other isoforms (3-OST-1, -5, and -7), allowed HSV-1 entry (Fig. 2F and G). Similarly, CHO-K1 cells expressing 3-OST-6 but not other isoforms allowed cell-to-cell fusion as an indicator of HSV-1 spread (Fig. 3C). Both HSV-1 entry and spread were gD and HS dependent, as was evident from a gD interference (Fig. 3A and B) assay and from an enzymatic treatment of cells which resulted in a significant decrease in HSV-1 infection (Fig. 3E and F). Taken together, our current findings suggest that the ZF 3-OST-6 isoform, but not 3-OST-1, -5, and -7, mediates HSV-1 entry and cell fusion in a manner similar to that of the previously reported ZF 3-OST isoforms (3-OST-2, -3, and -4) (14–17). Therefore, based on the ZF 3-OST gene characterizations for HSV-1 entry, we categorize them into two groups, in which group I (3-OST-2, -3, -4, and -6) allows HSV entry, while group II (3-OST-1, -5, and -7) does not generate a gD receptor; hence, cells expressing the latter 3-OST isoforms are not susceptible to HSV-1 entry (Fig. 4D).
Currently, the ZF model is an emerging model in infectious diseases, including those caused by HSV and other medically important viruses (23–32). The usage of the ZF model to understand in vivo HSV infection in the brain and eye is of particular interest because of the expression of multiple 3-OST enzymes during development and tissue-specific expression of 3-OST (11). A similar 3-OST expression profile for human and mouse models has been suggested (3). Therefore, the tight regulation of GAG modification might show variability in susceptibility to HSV-1 infection, which in turn could shed new light on the role of the modifications within HS in viral infectivity. Similarly, HSV-1 tropism in the ZF embryo may very well be guided by 3-OST expression, including that of 3-OST-6, especially in the brain or in eye tissues (11).
Overall, the characterization of all of the ZF 3-OST isoform-generated modified HSs has provided the structural and functional aspects of ZF 3-OS HS during HSV entry. Additionally, since HS and regulating enzymes are now emerging as markers of inflammation and facilitators of certain types of infection (33–37), it is worth dissecting the sulfated forms of HS involved during HSV-associated infection in the ZF model (11, 37) and also identifying crucial proinflammatory markers of susceptible locations (21, 38). Our initiative urges an investigation of the in vivo significance of 3-OST-generated HS and associated distinct syndecans during HSV infection in the ZF model by using 3-OST-specific knockouts (KO) (12, 13, 39, 40). Further study of how HSV infection affects 3-OS HS expression, including that of cell- and tissue-specific pathologies associated with clinically relevant HSV strains in a 3-OST-specific ZF model, is warranted.
ACKNOWLEDGMENTS
This work was supported by NIH grants to V.T. (AI088429-01A1) and D.S. (AI081869). J.B. and N.R. (10-2014-8172) were supported by Midwestern University (Downers Grove, IL)-sponsored Kenneth A. Suarez Summer Research Fellowships.
We appreciate the University of Illinois (UIC) ophthalmology core facility (core grant number EY001792) for confocal imaging.
Footnotes
Published ahead of print 20 August 2014
REFERENCES
- 1.Spear PG, Longnecker R. 2003. Herpesvirus entry: an update. J. Virol. 77:10179–10185. 10.1128/JVI.77.19.10179-10185.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shukla D, Liu J, Blaiklock P, Shworak NW, Bai X, Esko JD, Cohen GH, Eisenberg RJ, Rosenberg RD, Spear PG. 1999. A novel role for 3-O-sulfated heparan sulfate in herpes simplex virus 1 entry. Cell 99:13–22. 10.1016/S0092-8674(00)80058-6. [DOI] [PubMed] [Google Scholar]
- 3.Shukla D, Spear PG. 2001. Herpesviruses and heparan sulfate: an intimate relationship in aid of viral entry. J. Clin. Invest. 108:503–510. 10.1172/JCI13799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lindahl U, Kusche-Gullberg M, Kjellén L. 1998. Regulated diversity of heparan sulfate. J. Biol. Chem. 273:24979–24982. 10.1074/jbc.273.39.24979. [DOI] [PubMed] [Google Scholar]
- 5.Esko JD, Selleck SB. 2002. Order out of chaos: assembly of ligand binding sites in heparan sulfate. Annu. Rev. Biochem. 71:435–471. 10.1146/annurev.biochem.71.110601.135458. [DOI] [PubMed] [Google Scholar]
- 6.Esko JD, Lindahl U. 2001. Molecular diversity of heparan sulfate. J. Clin. Invest. 108:169–173. 10.1172/JCI200113530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grobe K, Ledin J, Ringvall M, Holmborn K, Forsberg E, Esko JD, Kjellén L. 2002. Heparan sulfate and development: differential roles of the N-acetylglucosamine N-deacetylase/N-sulfotransferase isozymes. Biochim. Biophys. Acta 1573:209–215. 10.1016/S0304-4165(02)00386-0. [DOI] [PubMed] [Google Scholar]
- 8.Kreuger J, Kjellén L. 2012. Heparan sulfate biosynthesis: regulation and variability. J. Histochem. Cytochem. 60:898–907. 10.1369/0022155412464972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shworak NW, Liu J, Fritze LM, Schwartz JJ, Zhang L, Logeart D, Rosenberg RD. 1997. Molecular cloning and expression of mouse and human cDNAs encoding heparan sulfate D-glucosaminyl 3-O-sulfotransferase. J. Biol. Chem. 272:28008–28019. 10.1074/jbc.272.44.28008. [DOI] [PubMed] [Google Scholar]
- 10.Liu J, Shworak NW, Sinaÿ P, Schwartz JJ, Zhang L, Fritze LM, Rosenberg RD. 1999. Expression of heparan sulfate D-glucosaminyl 3-O-sulfotransferase isoforms reveals novel substrate specificities. J. Biol. Chem. 274:5185–5192. 10.1074/jbc.274.8.5185. [DOI] [PubMed] [Google Scholar]
- 11.Cadwallander AB, Yost HJ. 2006. Combinatorial expression patterns of heparan sulfate sulfotransferases in zebrafish. I. The 3-O-sulfotransferase family. Dev. Dyn. 235:3423–3431. 10.1002/dvdy.20991. [DOI] [PubMed] [Google Scholar]
- 12.Samson SC, Ferrer T, Jou CJ, Sachse FB, Shankaran SS, Shaw RM, Chi NC, Tristani-Firouzi M, Yost HJ. 2013. 3-OST-7 regulates BMP-dependent cardiac contraction. PLoS Biol. 11:e1001727. 10.1371/journal.pbio.1001727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Neugebauer JM, Cadwallader AB, Amack JD, Bisgrove BW, Yost HJ. 2013. Differential roles for 3-OSTs in the regulation of cilia length and motility. Development 140:3892–3902. 10.1242/dev.096388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Antoine TE, Yakoub A, Maus E, Shukla D, Tiwari V. 2014. Zebrafish 3-O-sulfotransferase-4 generated heparan sulfate mediates HSV entry and spread. PLoS One 9:e87302. 10.1371/journal.pone.0087302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Antoine TE, Jones KS, Dale RM, Shukla D, Tiwari V. 2014. Zebrafish: modeling for herpes simplex virus infection. Zebrafish 11:17–25. 10.1089/zeb.2013.0920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hubbard S, Darmani NA, Thrush GR, Dey D, Burnham L, Thompson JM, Jones K, Tiwari V. 2010. Zebrafish-encoded 3-O-sulfotransferase-3 isoform mediates herpes simplex virus type 1 entry and spread. Zebrafish 7:181–187. 10.1089/zeb.2009.0621. [DOI] [PubMed] [Google Scholar]
- 17.Baldwin J, Antoine TE, Shukla D, Tiwari V. 2013. Zebrafish encoded 3-O-sulfotransferase-2 generated heparan sulfate serves as a receptor during HSV-1 entry and spread. Biochem. Biophys. Res. Commun. 432:672–676. 10.1016/j.bbrc.2013.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tiwari V, Clement C, Xu D, Valyi-Nagy T, Yue BY, Liu J, Shukla D. 2006. Role for 3-O-sulfated heparan sulfate as the receptor for herpes simplex virus type 1 entry into primary human corneal fibroblasts. J. Virol. 80:8970–8980. 10.1128/JVI.00296-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pertel P, Fridberg A, Parish M, Spear PG. 2001. Cell fusion induced by herpes simplex virus glycoproteins gB, gD, and gH-gL requires a gD receptor but not necessarily heparan sulfate. Virology 279:313–324. 10.1006/viro.2000.0713. [DOI] [PubMed] [Google Scholar]
- 20.Tiwari V, Clement C, Duncan MB, Chen J, Liu J, Shukla D. 2004. A role for 3-O-sulfated heparan in cell fusion induced by herpes simplex virus type 1. J. Gen. Virol. 85:805–809. 10.1099/vir.0.19641-0. [DOI] [PubMed] [Google Scholar]
- 21.Ernst S, Langer R, Cooney CL, Sasisekharan R. 1995. Enzymatic degradation of glycosaminoglycans. Crit. Rev. Biochem. Mol. Biol. 30:387–444. 10.3109/10409239509083490. [DOI] [PubMed] [Google Scholar]
- 22.Thacker BE, Xu D, Lawrence R, Esko JD. 2014. Heparan sulfate 3-O-sulfation: a rare modification in search of a function. Matrix Biol. 35:60–72. 10.1016/j.matbio.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Burgos JS, Ripoll-Gomez J, Alfaro JM, Sastre I, Valdivieso F. 2008. Zebrafish as a new model for herpes simplex virus type 1 infection. Zebrafish 5:323–333. 10.1089/zeb.2008.0552. [DOI] [PubMed] [Google Scholar]
- 24.van der Sar AM, Appelmelk BJ, Vandenbroucke-Grauls CM, Bitter W. 2004. A star with stripes: zebrafish as an infection model. Trends Microbiol. 12:451–457. 10.1016/j.tim.2004.08.001. [DOI] [PubMed] [Google Scholar]
- 25.Allen JP, Neely MN. 2010. Trolling for the ideal model host: zebrafish take the bait. Future Microbiol. 5:563–569. 10.2217/fmb.10.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sullivan C, Kim CH. 2008. Zebrafish as a model for infectious disease and immune function. Fish Shellfish Immunol. 25:341–350. 10.1016/j.fsi.2008.05.005. [DOI] [PubMed] [Google Scholar]
- 27.Meijer AH, Spaink HP. 2011. Host-pathogen interactions made transparent with the zebrafish model. Curr. Drug Targets 12:1000–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Crim MJ, Riley LK. 2012. Viral diseases in zebrafish: what is known and unknown. ILAR J. 53:135–143. 10.1093/ilar.53.2.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Goody MF, Sullivan C, Kim CH. 2014. Studying the immune response to human viral infections using zebrafish. Dev. Comp. Immunol. 46:84–95. 10.1016/j.dci.2014.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Levraud JP, Palha N, Langevin C, Boudinot P. 2014. Through the looking glass: witnessing host-virus interplay in zebrafish. Trends Microbiol. pii:S0966-842X(14)00096-1. 10.1016/j.tim.2014.04.014. [DOI] [PubMed] [Google Scholar]
- 31.Briolat V, Jouneau L, Carvalho R, Palha N, Langevin C, Herbomel P, Schwartz O, Spaink HP, Levraud JP, Boudinot P. 2014. Contrasted innate responses to two viruses in zebrafish: insights into the ancestral repertoire of vertebrate IFN-stimulated genes. J. Immunol. 192:4328–4341. 10.4049/jimmunol.1302611. [DOI] [PubMed] [Google Scholar]
- 32.Palha N, Guivel-Benhassine F, Briolat V, Lutfalla G, Sourisseau M, Ellett F, Wang CH, Lieschke GJ, Herbomel P, Schwartz O, Levraud JP. 2013. Real-time whole-body visualization of Chikungunya virus infection and host interferon response in zebrafish. PLoS Pathog. 9:e1003619. 10.1371/journal.ppat.1003619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang X, Wang B, Li JP. 2014. Implications of heparan sulfate and heparanase in neuroinflammation. Matrix Biol. 35:174–181. 10.1016/j.matbio.2013.12.009. [DOI] [PubMed] [Google Scholar]
- 34.Ferro V. 2013. Heparan sulfate inhibitors and their therapeutic implications in inflammatory illnesses. Expert Opin. Ther. Targets 17:965–975. 10.1517/14728222.2013.811491. [DOI] [PubMed] [Google Scholar]
- 35.Lindahl U, Kjellén L. 2013. Pathophysiology of heparan sulphate: many diseases, few drugs. J. Intern. Med. 273:555–571. 10.1111/joim.12061. [DOI] [PubMed] [Google Scholar]
- 36.Tiwari V, Maus E, Sigar IM, Ramsey KH, Shukla D. 2012. Role of heparan sulfate in sexually transmitted infections. Glycobiology 22:1402–1412. 10.1093/glycob/cws106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang F, Zhang Z, Thistle R, McKeen L, Hosoyama S, Toida T, Linhardt RJ, Page-McCaw P. 2009. Structural characterization of glycosaminoglycans from zebrafish in different ages. Glycoconj. J. 26:211–218. 10.1007/s10719-008-9177-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zou M, De Koninck P, Neve RL, Friedrich RW. 2014. Fast gene transfer into the adult zebrafish brain by herpes simplex virus 1 (HSV-1) and electroporation: methods and optogenetic applications. Front. Neural Circuits 8:41. 10.3389/fncir.2014.00041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hofmeister W, Devine CA, Key B. 2013. Distinct expression patterns of syndecans in the embryonic zebrafish brain. Gene Expr. Patterns 13:126–132. 10.1016/j.gep.2013.02.002. [DOI] [PubMed] [Google Scholar]
- 40.Poulain FE, Chien CB. 2013. Proteoglycan-mediated axon degeneration corrects pretarget topographic sorting errors. Neuron 78:49–56. 10.1016/j.neuron.2013.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moon AF, Edavettal SC, Krahn JM, Munoz EM, Negishi M, Linhardt RJ, Liu J, Pedersen LC. 2004. Structural analysis of the sulfotransferase (3-O-sulfotransferase isoform 3) involved in the biosynthesis of an entry receptor for herpes simplex virus 1. J. Biol. Chem. 279:45185–45193. 10.1074/jbc.M405013200. [DOI] [PMC free article] [PubMed] [Google Scholar]