ABSTRACT
Hepatitis C virus (HCV) particles associate with lipoproteins and infect cells by using at least four cell entry factors. These factors include scavenger receptor class B type I (SR-BI), CD81, claudin 1 (CLDN1), and occludin (OCLN). Little is known about specific functions of individual host factors during HCV cell entry and viral domains that mediate interactions with these factors. Hypervariable region 1 (HVR1) within viral envelope protein 2 (E2) is involved in the usage of SR-BI and conceals the viral CD81 binding site. Moreover, deletion of this domain alters the density of virions. We compared lipoprotein interaction, surface attachment, receptor usage, and cell entry between wild-type HCV and a viral mutant lacking this domain. Deletion of HVR1 did not affect CD81, CLDN1, and OCLN usage. However, unlike wild-type HCV, HVR1-deleted viruses were not neutralized by antibodies and small molecules targeting SR-BI. Nevertheless, modulation of SR-BI cell surface expression altered the infection efficiencies of both viruses to similar levels. Analysis of affinity-purified virions revealed comparable levels of apolipoprotein E (ApoE) incorporation into viruses with or without HVR1. However, ApoE incorporated into these viruses was differentially recognized by ApoE-specific antibodies. Thus, SR-BI has at least two functions during cell entry. One of them can be neutralized by SR-BI-targeting molecules, and it is critical only for wild-type HCV. The other one is important for both viruses but apparently is not inactivated by the SR-BI binding antibodies and small molecules evaluated here. In addition, HVR1 modulates the conformation and/or epitope exposure of virus particle-associated ApoE.
IMPORTANCE HCV cell entry is SR-BI dependent irrespective of the presence or absence of HVR1. Moreover, this domain modulates the properties of ApoE on the surface of virus particles. These findings have implications for the development of SR-BI-targeting antivirals. Furthermore, these findings highlight separable functions of SR-BI during HCV cell entry and reveal a novel role of HVR1 for the properties of virus-associated lipoproteins.
INTRODUCTION
Hepatitis C virus (HCV) is an enveloped, hepatotropic virus with a single-stranded RNA genome of positive polarity that belongs to the family Flaviviridae (1). Chronic HCV infection is associated with severe liver disease, including hepatitis, liver cirrhosis, and hepatocellular carcinoma, and it is one of the most frequent indications for liver transplantation (2). A hallmark of HCV is its high degree of sequence variability that likely contributes to its ability to establish chronic infections. Patient isolates are grouped into seven genotypes, which differ from each other by ca. 31 to 33% at the nucleotide level (3). The highest degree of sequence variability within the HCV genome can be found in hypervariable region 1 (HVR1), a 27-amino-acid (aa) domain at the N terminus of the viral glycoprotein E2 (3). Notably, HVR1 contains epitopes that are recognized by patients' antibodies (4–7). However, since this domain tolerates substantial variability, it permits continuous evolution of viral escape variants. As a consequence, antibodies targeting this viral domain are rather strain specific and not broadly cross-neutralizing. It has been shown that the HVR1 sequence does not evolve in a gammaglobulin-deficient patient, supporting the notion that sequence diversity within this region is driven primarily by humoral immune pressure (8). Of note, sequence variability of HVR1 is not random, and several basic residues conserved across viral genotypes have been identified (9), suggesting that functional constraints limit the evolution of HVR1. Furthermore, recent studies from others and us suggest that HVR1 is an essential viral domain that shields highly conserved virus-neutralizing epitopes and thus facilitates immune escape (10, 11). Besides the involvement in immune escape, HVR1 has been reported to be important for infectivity of low-density particles and to be involved in viral entry (10, 11).
The formation of virus particles and their release from infected cells require essential components of the cellular very-low-density lipoprotein (VLDL) machinery (12–14). As a consequence, HCV particles circulating in serum are highly enriched with triglycerides and cholesterol and are tightly associated with apolipoprotein E (ApoE) and ApoB (summarized in reference 15). HCV particles released from hepatocytes vary in their degree of lipid and apolipoprotein association as well as in their buoyant densities (16).
Not only virus assembly but also virus entry is linked to lipid metabolism of hepatocytes, since three lipid transfer molecules on the cellular surface have been implicated in viral entry. First, the low-density lipoprotein receptor (LDL-R) mediates cellular uptake of HCV RNA but may be nonessential for productive infection (17). Second, the cholesterol transporter Niemann-Pick-C1-like 1 (NPC1L1) has been shown to support HCV entry, but its exact role remains to be determined (18). Finally, scavenger receptor class B type I (SR-BI), which is best known for cholesteryl ester uptake from high-density lipoproteins (HDLs), is essential for HCV infection in vitro and in vivo (19–22). SR-BI acts at different steps during the HCV entry process. SR-BI might first interact with the lipoprotein component of the lipoviral particle in an E2 binding-independent manner (23, 24). However, later during entry, the interaction between SR-BI and the lipoviroparticle becomes E2 dependent, with HVR1 playing a major role (23). Besides the lipid transport molecules, the tetraspanin molecule CD81 and the tight junction proteins claudin 1 (CLDN1) and occludin (OCLN) are essential for HCV entry (25–27).
To further shed light into the role of HVR1 during the complex HCV entry process, we analyzed the influence of this domain on HCV interactions with SR-BI, CD81, CLDN1, and OCLN. Interestingly, we and others recently showed that HVR1 is important for production and infectivity of low-density HCV particles (10, 11). Since this may indicate that HVR1 modulates HCV interaction with lipoproteins, in this work, we also explored the role of HVR1 for lipoprotein association.
MATERIALS AND METHODS
Plasmids.
Plasmids pFK-Jc1 (28), pFK-Luc-Jc1 (29), pFK-Jc1/ΔHVR1, and pFK-Luc-Jc1/ΔHVR1 (10), encoding the Jc1 chimera with or without HVR1 and with or without a firefly luciferase reporter gene, were described previously. The Jc1 derivative Jc1/FlagE2, encoding E2 N-terminally tagged with a Flag epitope, was described previously (16). Jc1/FlagE2/ΔHVR1, JcR-2a/FlagE2, and JcR-2a/FlagE2/ΔHVR1 were constructed by standard PCR-based techniques and verified by sequencing. Detailed sequence information is available upon request.
Cell culture.
The Huh7 cell-derived cell clone Huh-7.5 is highly permissive for HCV replication (30) and was used for virus production, virus titrations with a limiting-dilution assay, and a luciferase reporter virus infection assay if not stated otherwise.
Huh-7.5/shSRBIkd cells expressing a doxycycline-inducible lentiviral vector with a short hairpin RNA (shRNA) targeting the 3′ untranslated region of SR-BI mRNA were described previously (31). To achieve SR-BI downregulation, Huh-7.5/shSRBIkd cells were incubated with 100 ng/ml doxycycline (Sigma) for 96 h. Huh-7.5-SRBIhigh cells were created by stable transduction with pWPI hSRBI BLR and selected by 5 μg/ml blasticidin.
Huh-7.5/OCLNlow cells were described recently (32). Huh-7.5 and Huh-7.5-derived cell lines were grown in Dulbecco's modified Eagle's medium (DMEM) (Invitrogen, Karlsruhe, Germany) supplemented with 2 mM l-glutamine (Invitrogen), nonessential amino acids (Invitrogen), 100 U/ml penicillin (Invitrogen), 100 μg/ml streptomycin (Invitrogen), and 10% fetal calf serum (PAA, Coelbe, Germany). Chinese hamster ovary (CHO) cells were maintained in RPMI supplemented with HEPES, 2 mM l-glutamine, nonessential amino acids, 100 U/ml penicillin, 100 μg/ml streptomycin, and 10% fetal calf serum.
Compounds and antibodies.
The monoclonal anti-CLDN1 antibody (OM-7D3-B3) was described recently (33). The CD81-specific monoclonal antibody (MAb) (JS81) was purchased from BD Biosciences. Monoclonal anti-SR-BI antibodies NK-8HE-E3 and C16-71 were described previously (20, 34), and C8 and C15 were a kind gift from Alfredo Nicosia. The SR-BI-specific small molecules ITX5061 and ITX7650 as well as the negative-control small molecule ITX7874 were kind gifts from Flossi Wong-Staal and were described previously (35). The goat anti-ApoE polyclonal antibody was obtained from Calbiochem (Cb). The mouse monoclonal anti-ApoE antibody was obtained from Progen (Pg).
In vitro transcription and electroporation.
Methods for in vitro transcription and electroporation were described previously (29, 36).
Quantification of HCV infection.
Authentic viruses were titrated by using a limiting-dilution assay (37). The 50% tissue culture infectious dose (TCID50) was calculated based on methods described previously by Spearman and Kärber (55, 56). Luciferase reporter virus-associated infectivity was determined as described previously (38).
Enzyme-linked immunosorbent assay.
The amount of human ApoE in purified virus particles was determined by an ApoE-specific enzyme-linked immunosorbent assay (ELISA) (Mabtech, Nacka Strand, Sweden) as recommended by the manufacturer. HCV core protein was quantified by using a diagnostic kit (Architect HCV Ag assay; Abbott, Abbott Park, IL).
Immunoblot analysis.
For Western blotting, cells were scraped into sample buffer containing 2% sodium dodecyl sulfate (SDS), boiled for 5 min at 98°C, and loaded onto a 12% SDS gel. After electrophoresis, proteins were transferred onto a polyvinylidene difluoride membrane with a semidry blotter. The membrane was blocked with 5% milk in phosphate-buffered saline (PBS) containing 0.5% Tween. Proteins were detected with SR-BI-specific (NB104; Novus Biology) (1:1,000 dilution), OCLN-specific (clone OC-3F10; Zymed/Invitrogen, Darmstadt, Germany) (1:1,000), or beta-actin-specific (Sigma-Aldrich, Munich, Germany) (1:1,000) antibody and a secondary antibody coupled to horseradish peroxidase (Sigma-Aldrich) (1:20,000) or coupled to fluorescent molecules (IRDye-800RD; Li-Cor) (1:15,000). The antibody signal was detected with either the ECL Prime detection reagent (GE Healthcare Europe, Freiburg, Germany) or an Odyssey imager (Li-Cor).
Neutralization of HCV infection.
For inhibition of HCV infection, 200 μl of an Huh-7.5 cell suspension (3 × 105 cells per ml) was seeded into each well of a 96-well plate 24 h prior to inoculation. Luciferase reporter viruses were mixed with serial dilutions of antibodies indicated in the text and were used to inoculate cells for 72 h.
Binding of HCVcc to CHO cells.
CHO cells, mock transduced or transduced with SR-BI, were incubated with cell culture-derived HCV (HCVcc) particles for 2 h at 37°C. After binding, the cells were washed extensively with PBS, and total RNA was isolated.
Quantitative measurement of viral RNA.
RNA was isolated from cells by using a Nucleo Spin RNAII kit (Macherey-Nagel) as recommended by the manufacturer. Two microliters of the RNA sample was used for quantitative reverse transcription-PCR (qRT-PCR) analysis using a LightCycler 480 device (Roche, Mannheim, Germany). HCV-specific RT-PCRs were conducted in duplicate by utilizing a one-step RT-PCR LightCycler 480 RNA master hydrolysis probes kit (Roche, Mannheim, Germany) and the following HCV-specific probe (TIB Molbiol, Berlin, Germany) and primers (MWG-Biotech, Martinsried, Germany): A-195 (5′-6-FAM [6-carboxyfluorescein]-AAA GGA CCC AGT CTT CCC GGC AAT T-TAMRA (tetra-chloro-6-carboxyfluorescein]-3′), S-146 (5′-TCT GCG GAA CCG GTG AGT A-3′), and A-219 (5′-GGG CAT AGA GTG GGT TTA TCC A-3′). Reactions were performed in three stages under the following conditions: stage 1 for 3 min at 63°C (reverse transcription), stage 2 for 30 s at 95°C (initial denaturation), and stage 3 for 35 cycles of 15 s at 95°C and 30 s at 60°C (amplification). The amount of HCV RNA was calculated in comparison to serially diluted in vitro transcripts.
Production of soluble E2 glycoproteins.
Expression plasmids encoding HCV E2 (aa 384 to 661) protein genotype 1a (H77) or 2a (J6) and containing a His6 tag were transfected into 293T cells by using polyethyleneimine (PEI). At 48 h posttransfection, cell supernatants were harvested and passed through 0.45-μm-pore-size filters.
Measurement of E2 binding to SR-BI.
Soluble recombinant E2 glycoprotein was added to CHO cells expressing SR-BI (CHO-SR-BI cells) or an empty vector (CHO-BLR cells) for 2 h at room temperature, and binding was quantified by flow cytometry after incubation with anti-His MAb (Qiagen).
Statistical analysis.
Statistical analyses were performed by using Welch's 2-sample t test, the Kolmogorov-Smirnoff test, and, alternatively, the Wilcoxon test (for >5 biological replicates). P values of <0.1 were considered marginally significant (indicated by single asterisks in the figure legends), P values of <0.05 were considered statistically significant (indicated by double asterisks), and P values of <0.01 were considered highly significant (indicated by triple asterisks).
RESULTS
Deletion of HVR1 does not affect usage of CD81, CLDN1, and OCLN during HCV cell entry.
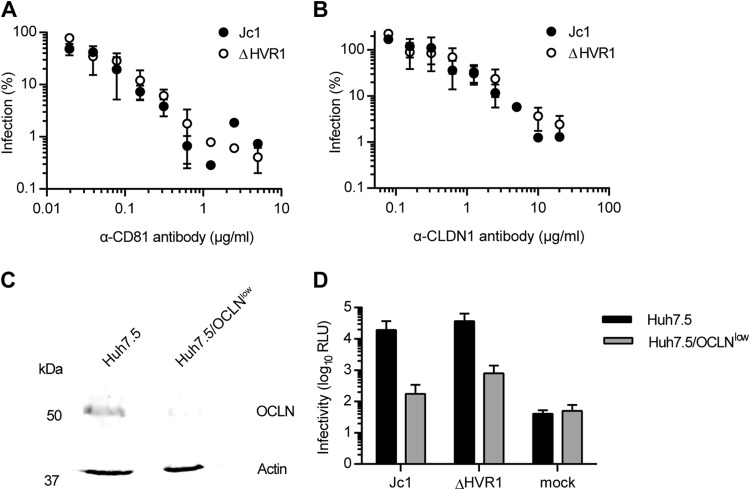
To analyze the role of HVR1 for receptor usage and lipoprotein association, we used the Jc1 chimera, which grows to high virus titers in tissue culture (32), and compared Jc1 and a Jc1 mutant lacking HVR1, i.e., the 27 amino-terminal residues of E2 (10, 11). Removal of HVR1 did not affect utilization of CD81 or CLDN1 since antibodies specific for these receptors competed with HCV infection to a similar extent irrespective of whether cells were challenged with wild-type (wt) Jc1 or Jc1/ΔHVR1 (Fig. 1A and B). Usage of OCLN was also not altered by deletion of HVR1, since a knockdown of OCLN impaired the infectivity of Jc1 with and without HVR1 ∼10-fold (Fig. 1C and D).
FIG 1.
HCV particles lacking HVR1 are not affected in their usage of CD81, CLDN1, or OCLN. (A and B) Huh-7.5 cells were inoculated with Jc1 or Jc1/ΔHVR1 (ΔHVR1) luciferase reporter viruses mixed with increasing doses of CD81-specific antibodies (A) or CLDN1-specific antibodies (B) for 72 h at 37°C. Luciferase activity was determined and is expressed relative to infections performed in the absence of antibodies. (C) Expression of OCLN and actin in cell extracts of Huh-7.5 and Huh-7.5/OCLNlow cells determined by Western blotting using specific antibodies. (D) Luciferase activity determined in given cell lines at 72 h postinoculation with given viruses. Mean values of triplicate measurements, including standard deviations, are given. RLU, relative light units.
Deletion of HVR1 confers viral resistance to SR-BI-targeting antibodies and compounds.
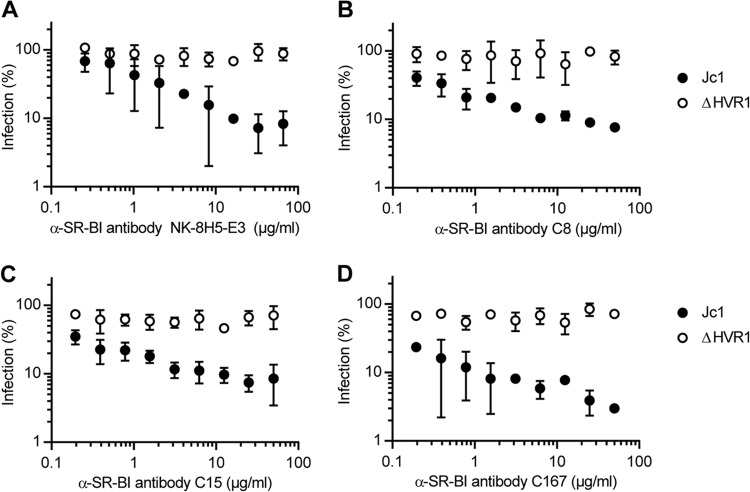
Recently, we reported that HCV particles lacking HVR1 were no longer neutralized by an SR-BI-specific antiserum that efficiently competed with infection of the parental virus (10). To explore if ΔHVR1 particles are generally resistant to SR-BI-targeting antibodies and/or compounds, we took advantage of a set of monoclonal antibodies (C8, C15, C167, and NK-8H5-E3) that potently repress infection by HCV in vitro as well as in humanized mice (20, 34, 39, 40). Strikingly, all these antibodies inhibited infection only with wild-type HCV but not with the mutant lacking HVR1 (Fig. 2). In parallel, we investigated the effect of the SR-BI-specific small-molecule inhibitors ITX5061 and ITX1650 on Jc1 and Jc1/ΔHVR1 infectivity (Fig. 3). Administration of ITX5061 to mice or humans increased plasma levels of HDL, probably through inhibiting SR-BI-mediated HDL lipid transfer (41). Moreover, it was shown previously that both ITX compounds inhibit HCVcc infection of human hepatoma cell lines and HCV pseudoparticle (HCVpp) infection of primary human hepatocytes (35). Similar to what we found for all SR-BI-specific antibodies tested, only Jc1 was inhibited by ITX5061 and ITX1650 in a dose-dependent manner, whereas removal of HVR1 rendered the virus resistant to both inhibitors (Fig. 3A and B). Together, these results indicate that deletion of HVR1 ablated or strongly reduced HCV neutralization by molecules targeting SR-BI.
FIG 2.
HCV particles devoid of HVR1 are more resistant to neutralization with SR-BI-specific antibodies. Huh-7.5 cells were inoculated with the indicated luciferase reporter viruses in the presence of increasing doses of SR-BI-specific monoclonal antibodies. The efficiency of infection was determined 72 h later by a luciferase reporter assay and is expressed relative to infections performed in the absence of antibodies. Means ± standard deviations of data from three independent experiments are shown.
FIG 3.
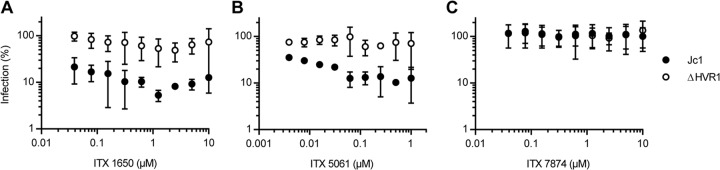
HCV particles devoid of HVR1 are more resistant to inhibition with SR-BI-specific inhibitors. Huh-7.5 cells were inoculated with the indicated luciferase reporter viruses in the presence of increasing doses of SR-BI-specific inhibitors (A and B) or a control (C). The efficiency of infection was determined 72 h later by a luciferase reporter assay and is expressed relative to efficiencies of infections performed in the absence of antibodies. Means ± standard deviations of data from three independent experiments are shown.
Modulation of the SR-BI expression level affects infectivity of HCV with and without HVR1.
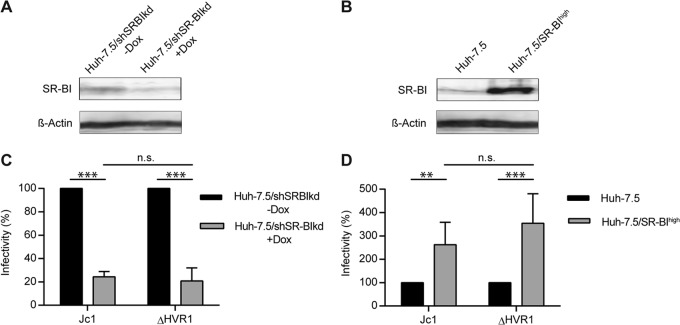
The findings described above in turn suggested that viruses lacking HVR1 may be independent of SR-BI usage for cell entry. To test this hypothesis and to evaluate possible direct and indirect roles of SR-BI for infection by HCV, we modulated SR-BI expression on the surface of Huh-7.5 cells by overexpression or RNA interference-mediated silencing. SR-BI overexpression was shown to enhance HCV infectivity 4-fold (21). In agreement with this, Jc1 infection of cells overexpressing SR-BI was increased ca. 2- to 3-fold compared to infection of parental cells expressing endogenous levels of SR-BI (P value of 0.023) (Fig. 4B and D). Unexpectedly, infection of ΔHVR viruses was increased to a similar extent by overexpression of SR-BI (P value of 0.007). This finding suggested that SR-BI surface density influences HCV infectivity in both the presence and absence of HVR1 (Fig. 4B and D). In agreement with this notion, downregulation of SR-BI expression using a doxycycline-regulated SR-BI-targeting shRNA (31) reduced permissiveness of these cells to infection by both parental HCV Jc1 as well as the Jc1/ΔHVR1 mutant (P values of 0.001 and 0.005 for Jc1 and ΔHVR1, respectively) (Fig. 4A and C). Thus, modulation of SR-BI expression clearly modifies the permissiveness of cells to HCV lacking HVR1. Importantly, silencing or overexpression of SR-BI did not influence replication of HCV replicons, indicating that SR-BI abundance does not affect RNA replication of HCV (data not shown). Therefore, we concluded that these viruses still rely on SR-BI functions during their cell entry. Of note, however, these critical functions do not seem to be ablated by SR-BI-targeting antibodies or small molecules. Therefore, these results suggest that SR-BI has at least two functions during cell entry. One of these functions can be neutralized by SR-BI-targeting molecules, and it is critical for wild-type HCV only. The other one is important for both viruses but is not inactivated by the SR-BI-specific antibodies and small molecules evaluated here.
FIG 4.
HCV infection of hepatoma cells with down- or upregulated SR-BI. (A and B) SR-BI expression levels in Huh-7.5/shSRBIkd cells pretreated with or without doxycycline (Dox) for 96 h (A) and Huh-7.5 and Huh-7.5/SR-BIhigh cells (B) were determined by Western blotting. (C) Jc1 or Jc1/ΔHVR1 (ΔHVR1) was added to Huh-7.5/shSRBIkd cells pretreated or not with doxycycline, and HCV infection was determined at 72 h postinfection (hpi) by luciferase reporter activity. Infectivity in the absence of doxycycline is set to 100%. (D) Jc1 or Jc1/ΔHVR1 (ΔHVR1) was added to Huh-7.5 or Huh-7.5/SR-BIhigh cells, and HCV infection was determined at 72 h postinfection by luciferase reporter activity. Infection of Huh-7.5 cells is set to 100%. Means ± standard deviations of data from three independent experiments are shown. n.s., not significant.
Expression of human SR-BI enhances binding of HCVcc with and without HVR1 to CHO cells.
Previous studies indicated that recombinant soluble E2 devoid of HVR1 exhibits increased CD81 binding yet decreased interactions with SR-BI compared to wild-type E2 comprising this domain (42). Combined with the finding that HCVpp as well as HCVcc lacking HVR1 were not neutralized by SR-BI-targeting antibodies (10, 43), we and others speculated that HCV particles lacking HVR1 may no longer bind and use SR-BI. The results described above, however, indicate that viruses lacking HVR1 are still dependent on SR-BI.
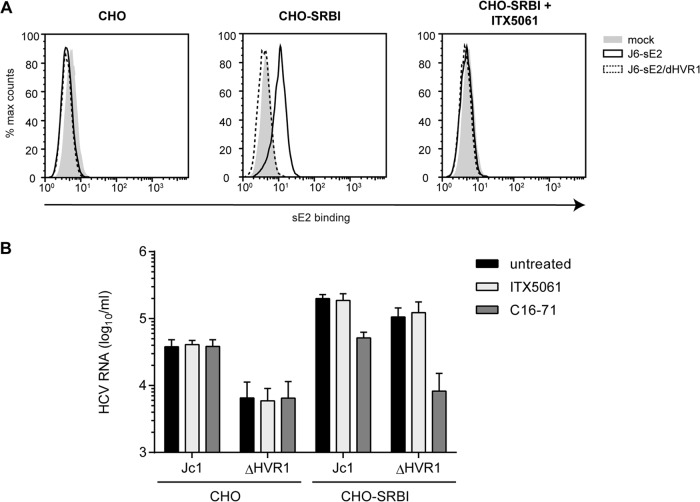
Therefore, we wished to determine if this function of SR-BI was independent of an interaction of the ΔHVR1 virus with SR-BI. To test this, we employed a cell binding assay similar to the one described previously by Evans et al. (25). Specifically, binding of soluble E2 or HCVcc particles with or without HVR1 to parental Chinese hamster ovary (CHO) cells or to CHO cells ectopically overexpressing SR-BI was determined by a fluorescence-activated cell sorter (FACS) assay or quantitative RT-PCR, respectively. In order to reduce the background from E2 or virus adherence to glycosaminoglycans (GAGs), we used a CHO cell line with defective xylosyltransferase activity for glycan processing, resulting in strongly reduced cell surface-expressed GAGs (44). Ectopic expression of SR-BI at the cell surface of these cells was confirmed by FACS analysis (data not shown). As expected, soluble J6CF-derived E2 (note that in Jc1, the structure genes, including E1-E2, are encoded by genotype 2a isolate J6CF [28, 45]) bound to CHO cells in an SR-BI-dependent fashion, and this binding was inhibited by ITX5061 (Fig. 5A). In contrast, E2-ΔHVR1 did not bind these cells irrespective of whether SR-BI was expressed (Fig. 5A). Notably, both recombinant proteins bound to CHO cells overexpressing CD81, and this binding was neutralized by the addition of CD81-specific antibodies (data not shown). Thus, we can rule out that E2-ΔHVR1 was globally misfolded. In fact, as reported previously, E2-ΔHVR1 bound to CHO-CD81 cells much more efficiently than did parental soluble E2, confirming the previously reported observations that deletion of HVR1 exposes the CD81 binding site and increases the binding of soluble E2 to cell surface-expressed CD81 (data not shown) (10, 42).
FIG 5.
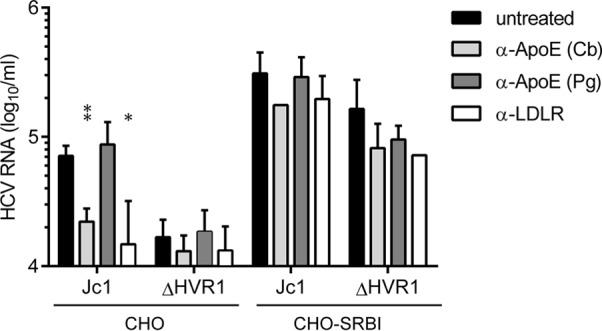
Attachment of HCVcc particles and soluble E2 to CHO cells expressing SR-BI or not. (A) CHO cells expressing SR-BI (CHO-SRBI) or not (CHO) were incubated with soluble E2 (sE2)-J6 or soluble E2-J6/ΔHVR1 for 2 h at room temperature. Binding was performed in the presence or absence of SR-BI-inhibiting compound ITX5061 (1 μM). Bound soluble E2 was detected by using an anti-His antibody and FACS measurements. (B) Equal amounts of Jc1 and Jc1/ΔHVR1 (ΔHVR1) were incubated with CHO cells or CHO-SRBI cells for 2 h at 37°C. Incubation was performed in either the presence or absence of SR-BI-inhibiting compound ITX5061 (1 μM) or monoclonal anti-SRBI antibody C16-71 (5 μg/ml). The number of bound HCVcc particles was measured by quantifying HCV RNA genomes in lysates of virus-incubated cells. Means ± standard deviations of data from three independent experiments are shown.
When Jc1 and Jc1/ΔHVR1 HCVcc preparations normalized for equal HCV RNA titers were incubated with parental CHO cells, the parental virus bound more efficiently than Jc1/ΔHVR1, as evidenced by ca. 5-fold-higher levels of viral RNA associated with CHO cells (Fig. 5B). Interestingly, when CHO-SR-BI cells were incubated with these particle types, in both cases, substantially higher levels of viral RNA were associated with the cells, indicating that binding of both viruses was facilitated by expression of SR-BI. Notably, under these circumstances, binding of Jc1/ΔHVR1 particles was essentially as effective as the binding of parental Jc1 particles. Strikingly, the addition of SR-BI-specific monoclonal antibody C16-71 repressed both Jc1 as well as Jc1/ΔHVR1 virus binding to the basal level observed in the absence of SR-BI (Fig. 5B). This finding indicates that the gain of virus binding observed upon ectopic expression of SR-BI in these CHO cells was due to binding of these particles to SR-BI. Notably, addition of ITX5061 at a dose that neutralizes ca. 80 to 90% of Jc1 infection (1 μM) (Fig. 3B) reduced neither Jc1 nor Jc1/ΔHVR1 binding to CHO-SR-BI cells (Fig. 5B). Collectively, these observations confirm that HVR1 is critical for binding of soluble E2 to cell surface-expressed SR-BI. However, this domain is dispensable for binding of HCVcc particles to CHO-SR-BI cells. Therefore, features other than HVR1 likely mediate SR-BI-dependent cell surface attachment of HCV particles in this cellular model. Moreover, virus particle binding, unlike binding of soluble E2, is not inhibited by ITX5061, further confirming a different mode of SR-BI interaction between soluble E2 and virus particles. Finally, since viruses lacking HVR1 displayed lower levels of cell surface binding to parental CHO cells, this viral domain modulates interactions with the cell surface in the absence of SR-BI.
HCV particles lacking HVR1 display reduced ApoE and LDL-R dependence in cell surface attachment to CHO cells.
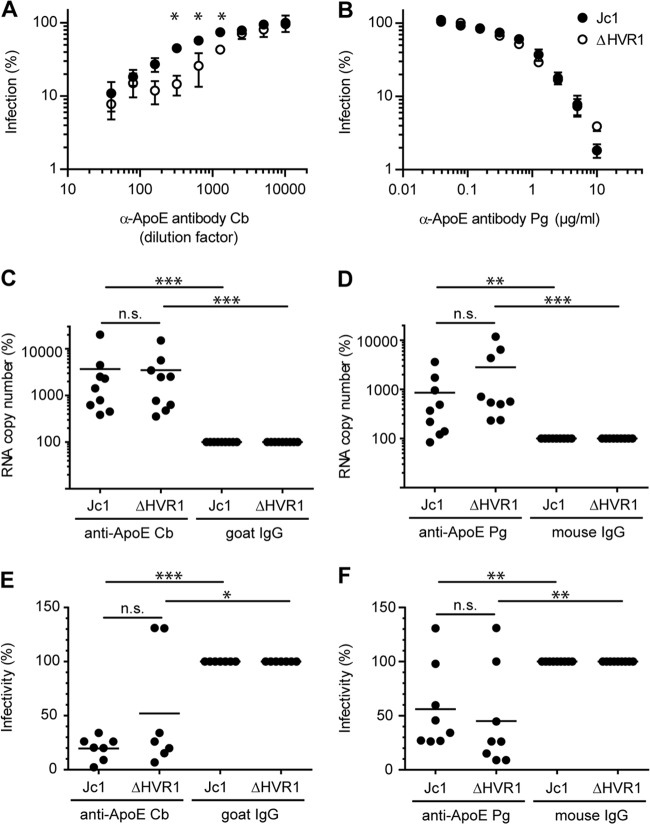
Since HCV particles associate with lipoproteins, and as ΔHVR1 particles have an altered buoyant density (10, 11), we speculated that different particle compositions with regard to lipoproteins and/or lipids may be responsible for the lower level of binding of ΔHVR1 particles to parental CHO cells. Moreover, recent evidence indicates that virus-associated ApoE facilitates cell surface attachment through binding to the low-density lipoprotein receptor (LDL-R) (46) and cell surface-resident GAGs (47). Thus, we repeated the binding assay in the presence of two different ApoE-specific antibodies and an anti-LDL-R antibody to assess the role of ApoE and LDL-R in cell surface binding of parental Jc1 and Jc1/ΔHVR1 particles (Fig. 6). Jc1 binding to these cells was indeed reduced with comparable efficiencies by polyclonal anti-ApoE antibody Cb (ca. 3-fold; P value of 0.015) and the anti-LDL-R antibody (ca. 3-fold; P value of 0.072), suggesting that ApoE and LDL-R were involved in the binding of these viruses to naive CHO cells, possibly through a direct interaction between virus-associated ApoE and cell surface-resident LDL-R. Of note, monoclonal anti-ApoE antibody Pg did not significantly block binding of the virus variants to CHO or CHO-SR-BI cells (P values of 0.591 and 0.863, respectively), probably because the antibody recognizes a different epitope than the Cb antibody. Interestingly, binding of Jc1/ΔHVR1 particles to parental CHO cells was not significantly repressed by the ApoE- or the LDL-R-specific antibodies (P values of 0.439 and 0.547, respectively), suggesting that these viruses lacking HVR1 display reduced levels of ApoE- and LDL-R-dependent cell surface binding. As described above, both Jc1 and Jc1/ΔHVR1 particles showed increased binding to SR-BI-expressing CHO cells (Fig. 5 and 6). Interestingly, cell surface binding of HCV particles with or without HVR1 to CHO-SR-BI cells was poorly neutralized by ApoE- and LDL-R-specific antibodies, and there was no statistically significant difference in binding in the presence or absence of ApoE- or LDL-R-specific antibodies. Therefore, we conclude that SR-BI binding is dominant over attachment via ApoE and LDL-R in this cell system.
FIG 6.
ApoE- and LDL-R-mediated attachment of HCVcc particles to CHO cells in the presence or absence of SR-BI. Equal amounts of Jc1 and ΔHVR1 were incubated with CHO cells expressing SR-BI (CHO-SR-BI) or a vector control (CHO) for 2 h at 37°C. Infection was performed in either the presence or absence of anti-ApoE antibodies from Calbiochem (Cb) (1/40 dilution) or Progen (Pg) (2 μg/ml) or an anti-LDL-R antibody (5 μg/ml). After washing, the number of bound HCVcc particles was assessed by determining the number copies of viral RNA in lysates of virus-incubated cells. Means ± standard deviations of data from three independent experiments are shown. Statistical significance was tested relative to untreated cells.
Combined with the previously reported observation that ΔHVR1 particles display a different buoyant density than wild-type HCV particles (10, 11), these results suggest that ΔHVR1 particles may incorporate different levels (or types) of lipids or lipoproteins. This in turn may alter usage of cellular lipoprotein receptors (LDL-R and SR-BI) during cell entry.
Deletion of HVR1 does not change the abundance of ApoE in HCV lipoviroparticles.
To test the two hypotheses mentioned above and to analyze incorporation of ApoE into Jc1 and Jc1/ΔHVR1 particles, we took advantage of a recently described virus variant carrying a Flag tag at the N terminus of E2 (Jc1/FlagE2) (16, 48). In parallel, we created a novel Jc1/ΔHVR1 variant comprising a Flag epitope at the N terminus of the truncated E2 protein (Jc1/FlagE2/ΔHVR1). Importantly, insertion of the tag into both viruses was well tolerated (16, 48) and did not decrease infectivity of the particles (data not shown). Moreover, similar to the untagged viruses, Flag-tagged ΔHVR1 particles also displayed an altered distribution in density gradients (data not shown).
Using Flag-specific antibodies, we affinity purified Jc1/FlagE2 and Jc1/FlagE2/ΔHVR1 particles and determined the amounts of ApoE and core in the purified viruses by ELISA. Although Jc1/FlagE2 and Jc1/ΔHVR1/FlagE2 virus preparations were normalized to equal core protein levels before affinity purification, Jc1/ΔHVR1/FlagE2 particles were consistently more efficiently precipitated, probably because they bind more efficiently to anti-Flag antibodies and therefore are captured more readily (Fig. 7). Consequently, about two times more core and two times more ApoE were detected in the precipitation of Jc1/ΔHVR1/FlagE2 particles than in the precipitation of parental Jc1/FlagE2. Importantly, the amount of coprecipitated ApoE was commensurately increased for the Jc1/FlagE2/ΔHVR1 particles compared to the parental viruses. Thus, in conclusion, these experiments indicate that deletion of HVR1 does not grossly affect the amount of ApoE incorporated into HCV particles.
FIG 7.
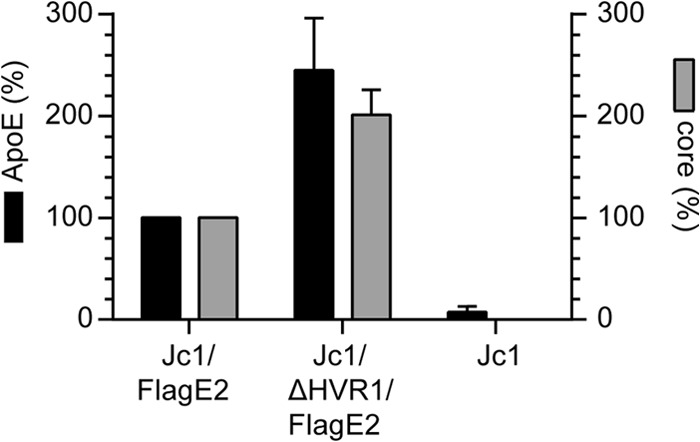
ApoE incorporation into HCV particles is not altered by deletion of HVR1. The amounts of ApoE and core in affinity-purified Jc1/FlagE2 or Jc1/ΔHVR1/FlagE2 particles were assessed by ELISA. The amounts of ApoE and core in affinity purified Jc1/FlagE2 particles are set to 100%. Means ± standard deviations of data from three independent experiments are shown.
HCV particles with or without HVR1 are differentially recognized and neutralized by ApoE-specific antibodies.
Next, we assessed whether the differences between Jc1 and Jc1/ΔHVR1 binding to CHO cells correlated with divergent properties of virus-associated ApoE. To this end, we utilized the anti-ApoE-specific antibodies described above to neutralize infection of parental Jc1 and Jc1/ΔHVR1 particles. Interestingly, although polyclonal anti-ApoE antibody Cb prevented attachment of Jc1 particles to parental CHO cells more efficiently than binding of Jc1/ΔHVR1 viruses, in the infection assay, Jc1/ΔHVR1 was more efficiently neutralized. Indeed, smaller amounts of the antibodies were required for a greater reduction of infection (Fig. 8A). Also, monoclonal anti-ApoE antibody Pg, which had not efficiently inhibited cell surface binding of these viruses to parental CHO cells (Fig. 6), interfered with infection of both viruses in a dose-dependent fashion (Fig. 8B). Therefore, these data indicate that the inhibition of cell surface binding of HCV particles to CHO cells by these antibodies does not directly correlate with their ability to neutralize infection of Huh-7.5 cells by HCV particles. Moreover, these results provide additional evidence that Jc1 and Jc1/ΔHVR1 particles may be differentially targeted by ApoE-specific antibodies (Fig. 6 and 8A).
FIG 8.
ApoE exposure on HCV particles with or without HVR1. (A and B) Huh-7.5 cells were inoculated with the indicated luciferase reporter viruses in the presence of increasing doses of a polyclonal anti-ApoE antibody (Cb) (A) and a monoclonal ApoE-specific antibody (Pg) (B). The efficiency of infection was determined 72 h later by a luciferase reporter assay and is expressed relative to infections performed in the absence of antibodies. Results of one representative experiment out of five independent repetitions are shown. Statistical analysis was performed on the five independent repetitions, and the statistical significances of the differences between Jc1 and ΔHVR1 were tested with a paired t test. (C and D) Equal amounts of Jc1 and Jc1/ΔHVR1 particles were subjected to an immunocapture assay by using Cb (C) or Pg (D) anti-ApoE antibodies. HCV RNA contained in each captured sample was quantified by qRT-PCR. RNA copy numbers captured by control IgGs are set to 100%. (E and F) The infectivity of the postcapture supernatant of the samples shown in panels C and D was determined by a TCID50 assay. Data points and means of data from seven to nine independent experiments are shown.
To further corroborate this notion, we investigated the binding of these ApoE-specific antibodies to Jc1 and Jc1/ΔHVR1 particles. To this end, equal amounts of these particles (normalized for equal viral RNA levels) were incubated with different ApoE-specific antibodies or isotype-matched control antibodies. Subsequently, antibody complexes were precipitated by using protein A beads, and the amount of coprecipitating viral RNA was determined by quantitative RT-PCR (Fig. 8C and D). Moreover, the amount of residual HCV infectivity remaining in the supernatant was determined (Fig. 8E and F). Using this approach, we observed comparable coprecipitations of viral RNA between Jc1 and Jc1/ΔHVR1 in the case of polyclonal anti-ApoE antibody Cb particles (Fig. 8C). Likewise, these polyclonal antibodies depleted viral infectivity to similar extents when incubated with virus-containing supernatants (Fig. 8E). These results therefore suggest that despite the differential neutralization of Jc1 and Jc1/ΔHVR1 particles, there is no evidence for differential binding of virus particles by these antibodies. In contrast, the monoclonal anti-ApoE Pg antibody neutralized these virus particle types with a similar efficacy but precipitated slightly larger quantities of viral RNA and infectious virus in the case of Jc1/ΔHVR1 than in the case of Jc1 (Fig. 8D and F). Collectively, these results further support the notion that ApoE associated with Jc1/ΔHVR1 particles is differentially targeted by ApoE-specific antibodies compared to ApoE on Jc1 viruses. Thus, it is conceivable that ApoE, although incorporated into both particles types to similar levels, may display different epitopes and/or adopt a slightly different conformation between Jc1 and Jc1/ΔHVR1 particles, thus resulting in differential recognition and neutralization by distinct ApoE-targeting antibodies.
DISCUSSION
In this study, we explored the role of HVR1 during HCV cell entry, thereby focusing on the interaction of HCV with entry receptors and lipoproteins. When analyzing inhibition of infection by HCV with or without HVR1 by antibodies specific for CD81 or CLDN1 or by silencing of OCLN expression, we observed similar reductions of virus infection between these virus types (Fig. 1). Since these treatments decrease the cell surface availability of targeted entry factors, these results indicate that HCV particles with or without HVR1 similarly depend on the availability of accessible CD81, CLDN1, and OCLN molecules for productive infection of Huh-7.5 cells. Therefore, deletion of HVR1 apparently does not modulate the requirement for utilization of these three critical HCV entry factors. In contrast, our results indicate that HVR1 is important for certain but not all functions that SR-BI plays during the entry process. Moreover, we provide evidence suggesting that deletion of HVR1 changes the properties of ApoE on the virus particle without affecting the amount of ApoE incorporated into the virion (Fig. 6 to 8).
We previously reported that HCV particles lacking HVR1, unlike wild-type particles, were resistant to neutralization by a polyclonal SR-B1-specific antiserum, suggesting that this viral surface domain modulated SR-B1 dependency (10). In this study, we compared the neutralization of these viruses by a panel of different SR-BI-specific monoclonal antibodies as well as two small molecules (ITX5061 and ITX7650) targeting this HCV entry factor. Of note, the monoclonal anti-SR-BI antibodies have different modes of action. While the mechanism by which the C8 and C15 antibodies preclude infection by wild-type HCV has to our knowledge not been explored, the NK-8H5-E3 antibody was reported to interfere with a postbinding step during HCV entry without interfering with soluble envelope glycoprotein E2 or HCVcc particle binding to the target cell surface (34). In contrast, the C167 antibody, recognizing a conformational epitope on the receptor, was shown to inhibit binding of E2 and HDL to SR-BI and to interfere with SR-BI-mediated lipid transfer (20). In addition, the two SR-BI specific inhibitors ITX7650 and ITX5061 both interfere with HDL-mediated lipid transfer of SR-BI (35) and block the interaction of soluble E2 with SR-BI. Despite their different modes of action, these antibodies and small molecules targeting SR-BI consistently inhibited Jc1 infection in a dose-dependent fashion, whereas they did not decrease infection by the Jc1/ΔHVR1 variant (Fig. 2 and 3). These results therefore extend and refine observations previously reported by us and others (10, 49) and indicate that HVR1 modulates HCV utilization of SR-BI and that Jc1 particles lacking this domain display modified SR-BI dependence during cell entry, as they are no longer neutralized by these SR-BI-targeting molecules.
These experiments involving SR-BI binding compounds suggested that Jc1/ΔHVR1 particles may be completely independent of SR-BI usage for infection. To test this, we next modulated SR-BI cell surface levels by overexpression or shRNA-mediated silencing. Using these approaches, we noted that the infection efficiencies of both wild-type Jc1 and the variant lacking HVR1 were significantly increased by SR-BI overexpression and decreased by SR-BI downregulation (Fig. 4). Therefore, we concluded that infection by both viruses was similarly dependent on SR-BI surface expression. This indicates that deletion of HVR1 does not ablate SR-BI dependence of HCV cell entry. Nevertheless, these results support a model where wild-type Jc1 particles, ΔHVR1 virions, require an additional SR-BI function(s) that is inhibited by the SR-BI binding molecules tested by us.
Previous reports have established that HVR1 is critical for binding of soluble E2 to SR-BI (42), suggesting that viruses lacking HVR1 may no longer interact with this entry factor. It was conceivable that viruses lacking HVR1 could indirectly rely on SR-BI for infection without direct binding of E2 being involved. Therefore, we explored whether deletion of HVR1 abrogated binding of HCVcc particles to this entry factor. First, we confirmed that deletion of HVR1 in the context of the J6CF (GT2a) isolate, as for H77 (GT1a)-derived E2 (42), prevented the interaction of soluble E2 with SR-BI (Fig. 5A). Next, we utilized an established HCVcc cell binding assay that involves CHO cells that lack endogenous expression of human HCV entry factors. Using these cells, Evans et al. recently reported that HCVcc particles attach to the cell surface in an SR-BI-dependent fashion (25). Accordingly, we observed ca. 5-fold-increased binding of Jc1 particles to CHO-SR-BI cells compared to parental cells lacking SR-BI expression (Fig. 5B). Moreover, this additional binding was inhibited by SR-BI-specific antibody C16-71 (Fig. 5B). In contrast, ITX5061 was not able to inhibit this SR-BI-dependent binding, suggesting that the mechanism by which this molecule interferes with HCV infection is not a block of virus attachment to SR-BI. Strikingly, cell surface binding of Jc1/ΔHVR1 particles to these CHO cells was increased >10-fold by overexpression of SR-BI, and this additional viral cell binding in the presence of SR-BI was fully ablated by the addition of the SR-BI-specific monoclonal antibody (Fig. 5B). These data indicate that HCVcc particles lacking HVR1, unlike soluble E2 lacking this domain, attach to the cell surface in an SR-BI-dependent manner. Therefore, viral domains other than HVR1 and/or host constituents of HCV particles are important for the interaction with SR-BI. Finally, we observed that wild-type Jc1 particles bound ca. 5-fold more efficiently to parental CHO cells than did Jc1/ΔHVR1 particles (Fig. 5B). Since these cells lack human HCV cell entry factors and are unable to produce GAGs, we speculate that this may be due to increased interactions of wild-type particles with hamster cell-derived cell surface proteins. At present, we do not know which cellular factor is responsible for the differential binding of Jc1 and Jc1/ΔHVR1 particles to the surface of CHO cells. However, recent reports indicate that HCV particles lacking HVR1 have a higher buoyant density than wild-type particles (10, 11) and that virus-associated ApoE facilitates attachment by way of interaction with LDL-R (46). Moreover, these CHO cells do express LDL-R (50). Therefore, we tested the influence of different ApoE-specific antibodies as well as of an antibody targeting LDL-R on the binding of HCVs to CHO cells. One of the two ApoE-specific antibodies as well as the LDL-R-targeting antibody significantly decreased cell surface attachment of wild-type Jc1 particles to parental CHO cells, suggesting that an interplay between virus-resident ApoE and cell surface-expressed LDL-R measurably contributes to attachment of these viruses to parental CHO cells. We do not know why the monoclonal anti-ApoE Pg antibody did not interfere with HCV binding, but this may be because the epitope targeted by this antibody is not involved in ApoE binding to LDL-R. Interestingly, binding of Jc1 wild-type particles to parental CHO cells was repressed by the polyclonal anti-ApoE Cb antibodies and the LDL-R-specific antibody approximately to the level of binding achieved by Jc1/ΔHVR1 particles in the absence of antibodies. Moreover, addition of these antibodies to Jc1/ΔHVR1 particle binding assay mixtures only weakly reduced attachment of these viruses (<2-fold). Finally, once SR-BI was overexpressed, inhibition of virus binding by the ApoE- and LDL-R-specific antibodies was less pronounced. Taken together, these observations are consistent with a model where virus binding to SR-BI occurs for viruses with and without HVR1 and may be the dominant mode of cell surface attachment. Moreover, the ApoE-specific antibodies used here do not efficiently block this interaction. Finally, wild-type Jc1 particles attach to parental CHO cells in an ApoE- and LDL-R-dependent fashion. Of note, our binding assays were conducted with CHO cells that are unable to produce GAGs. Therefore, this system does not reflect the contribution of protein interactions with GAGs, which have been shown to additionally contribute to virus attachment to human hepatocytes (29, 47, 51, 52).
It is currently unclear why Jc1/ΔHVR1 particles show reduced ApoE–LDL-R-dependent binding to parental CHO cells compared to parental Jc1 particles. However, it is possible that either HVR1 may contribute to LDL-R binding and/or the abundances or the properties of ApoE between wild-type and HVR1-deleted viruses may differ, thus causing differential attachment through LDL-R. Clearly, more work will be necessary to prove, or disprove, a role of HVR1 in binding to LDL-R. However, given that HVR1-deleted particles display an aberrant distribution in density gradients that is characterized by reduced numbers of particles with low density (11, 53), we speculated that aberrant loading of these particles with lipids and/or lipoproteins may be responsible.
To obtain evidence supporting this hypothesis, we compared the abundances and the properties of virus particle-associated ApoE of parental Jc1 and Jc1/ΔHVR1 particles. Using Flag-E2-tagged versions of both viral variants, we affinity purified equal numbers of these particles using Flag-specific antibodies. Importantly, insertion of the Flag epitope at the N terminus of E2 is well tolerated (16, 48) and did not alter the differential distribution of these particle types in density gradients (data not shown). While we consistently observed greater affinity purification of the Jc1/FlagE2/ΔHVR1 virus than of the parental virus with HVR1, we coprecipitated almost identical relative amounts of ApoE protein (Fig. 7). Therefore, these results exclude gross differences in the abundances of virus-incorporated ApoE between these particle types. Finally, we compared binding and neutralization of Jc1 viruses in the presence and in the absence of HVR1 by the above-mentioned mono- or polyclonal ApoE-specific antibodies. Using this approach, we observed striking differences between these particle types with regard to their interactions with these ApoE-specific antibodies. First, the polyclonal anti-ApoE Cb antibody neutralized Jc1/ΔHVR1 particles more efficiently than parental Jc1 particles, although it bound both particle types with comparable efficiencies. Second, the monoclonal anti-ApoE Pg antibody neutralized both particle types to similar levels; however, it bound viruses lacking HVR1 slightly more effectively (Fig. 8). Based on these findings, we conclude that the ability of these ApoE-specific antibodies to bind HCV particles does not directly correlate with their HCV neutralization properties. Moreover, the ability of these antibodies to prevent HCV particle binding to CHO cells does not predict their capability to neutralize infection. The latter finding may suggest that, in the CHO cell binding assay, HCV associates primarily via ApoE-independent routes and that ApoE may exert postattachment functions during cell entry. Finally, these results provide evidence supporting the conclusion that Jc1/ΔHVR1 particles incorporate ApoE with differential conformation and/or epitope exposure compared to parental Jc1 particles. Moreover, different amounts of lipids present in wild-type compared with HVR1-deleted viruses may result in differential epitope exposures of ApoE, thus explaining the different interplays of these particle types with these antibodies. Interestingly, the conformation of ApoE is known to be influenced by the lipoprotein and lipid composition on a given lipoprotein particle, and a large body of literature supports the assumption that conformational opening of the N-terminal domain of ApoE modulates its receptor binding activity (summarized in reference 54). Finally, it is possible that an altered conformation of virus-associated ApoE in HVR1-deleted particles is in part responsible for the reduced infectiousness of these viruses (10). Careful lipid and protein profiling of Jc1/ΔHVR1 particles may help to validate this assumption.
Collectively, our data indicate that HVR1 contributes to efficient cell surface attachment of HCV Jc1 particles. This function of HVR1 may be linked to its involvement in the incorporation of ApoE, a natural ligand of LDL-R. Our data support a model where HVR1 modulates the conformation/epitope exposure of virus-resident ApoE but not its total abundance on the virus particle. These altered properties of ApoE, which we documented by the differential interplays between these viruses and ApoE-specific antibodies, could influence virus attachment through LDL-R and possibly postbinding entry steps facilitated by ApoE. As a consequence, they may be responsible for the observed lower level of infectiousness of Jc1/ΔHVR1 particles. Finally, we show that viruses lacking HVR1 remain SR-BI dependent in infection, highlighting the important role of SR-BI for HCV cell entry. Notably, SR-BI-targeting antibodies and small molecules inhibit wild-type but not HVR1-deleted Jc1 in vitro. This finding confirms that SR-BI fulfills additional “druggable” functions that are specifically required by wild-type HCV and that these functions are targeted by the antibodies and small molecules described above. Whether viral variants independent of these SR-BI functions and, thus, possibly resistant to these SR-BI-targeting modalities can arise remains to be shown.
ACKNOWLEDGMENTS
We are grateful to Takaji Wakita and Jens Bukh for JFH1 and J6CF isolates, respectively; Charles Rice for Huh-7.5 cells and the E9E10 antibodies; Sandra Ciesek and Thomas von Hahn for Huh-7.5/OCLNlow cells; and Flossie Wong-Staal for providing the ITX compounds. We also thank all members of the Institute of Experimental Virology for helpful suggestions and discussions.
This work was supported by a grant from the Deutsche Forschungsgemeinschaft (SFB 900, project A6) and by a grant from the Helmholtz Association, SO-024, to T.P. T.F.B. acknowledges funding through the ANRS and Laboratoire d'Excellence LabEx HEPSYS (Investissement d'Avenir; ANR-10-LAB-28).
Footnotes
Published ahead of print 20 August 2014
REFERENCES
- 1.Lindenbach BD, Thiel HJ, Rice CM. 2007. Flaviviridae: the viruses and their replication, p 1101–1152 In Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE. (ed), Fields virology, 5th ed. Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 2.Brown RS. 2005. Hepatitis C and liver transplantation. Nature 436:973–978. 10.1038/nature04083. [DOI] [PubMed] [Google Scholar]
- 3.Simmonds P, Bukh J, Combet C, Deleage G, Enomoto N, Feinstone S, Halfon P, Inchauspe G, Kuiken C, Maertens G, Mizokami M, Murphy DG, Okamoto H, Pawlotsky JM, Penin F, Sablon E, Shin IT, Stuyver LJ, Thiel HJ, Viazov S, Weiner AJ, Widell A. 2005. Consensus proposals for a unified system of nomenclature of hepatitis C virus genotypes. Hepatology 42:962–973. 10.1002/hep.20819. [DOI] [PubMed] [Google Scholar]
- 4.Kato N, Sekiya H, Ootsuyama Y, Nakazawa T, Hijikata M, Ohkoshi S, Shimotohno K. 1993. Humoral immune response to hypervariable region 1 of the putative envelope glycoprotein (gp70) of hepatitis C virus. J. Virol. 67:3923–3930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kato N, Ootsuyama Y, Sekiya H, Ohkoshi S, Nakazawa T, Hijikata M, Shimotohno K. 1994. Genetic drift in hypervariable region 1 of the viral genome in persistent hepatitis C virus infection. J. Virol. 68:4776–4784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Taniguchi S, Okamoto H, Sakamoto M, Kojima M, Tsuda F, Tanaka T, Munekata E, Muchmore EE, Peterson DA, Mishiro S. 1993. A structurally flexible and antigenically variable N-terminal domain of the hepatitis C virus E2/NS1 protein: implication for an escape from antibody. Virology 195:297–301. 10.1006/viro.1993.1378. [DOI] [PubMed] [Google Scholar]
- 7.Weiner AJ, Geysen HM, Christopherson C, Hall JE, Mason TJ, Saracco G, Bonino F, Crawford K, Marion CD, Crawford KA. 1992. Evidence for immune selection of hepatitis C virus (HCV) putative envelope glycoprotein variants: potential role in chronic HCV infections. Proc. Natl. Acad. Sci. U. S. A. 89:3468–3472. 10.1073/pnas.89.8.3468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kumar U, Monjardino J, Thomas HC. 1994. Hypervariable region of hepatitis C virus envelope glycoprotein (E2/NS1) in an agammaglobulinemic patient. Gastroenterology 106:1072–1075. [DOI] [PubMed] [Google Scholar]
- 9.Penin F, Combet C, Germanidis G, Frainais PO, Deleage G, Pawlotsky JM. 2001. Conservation of the conformation and positive charges of hepatitis C virus E2 envelope glycoprotein hypervariable region 1 points to a role in cell attachment. J. Virol. 75:5703–5710. 10.1128/JVI.75.12.5703-5710.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bankwitz D, Steinmann E, Bitzegeio J, Ciesek S, Friesland M, Herrmann E, Zeisel MB, Baumert TF, Keck ZY, Foung SK, Pecheur EI, Pietschmann T. 2010. Hepatitis C virus hypervariable region 1 modulates receptor interactions, conceals the CD81 binding site, and protects conserved neutralizing epitopes. J. Virol. 84:5751–5763. 10.1128/JVI.02200-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Prentoe J, Jensen TB, Meuleman P, Serre SB, Scheel TK, Leroux-Roels G, Gottwein JM, Bukh J. 2011. Hypervariable region 1 differentially impacts viability of hepatitis C virus strains of genotypes 1 to 6 and impairs virus neutralization. J. Virol. 85:2224–2234. 10.1128/JVI.01594-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chang KS, Jiang J, Cai Z, Luo G. 2007. Human apolipoprotein E is required for infectivity and production of hepatitis C virus in cell culture. J. Virol. 81:13783–13793. 10.1128/JVI.01091-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang H, Sun F, Owen DM, Li W, Chen Y, Gale M, Jr, Ye J. 2007. Hepatitis C virus production by human hepatocytes dependent on assembly and secretion of very low-density lipoproteins. Proc. Natl. Acad. Sci. U. S. A. 104:5848–5853. 10.1073/pnas.0700760104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gastaminza P, Cheng G, Wieland S, Zhong J, Liao W, Chisari FV. 2008. Cellular determinants of hepatitis C virus assembly, maturation, degradation, and secretion. J. Virol. 82:2120–2129. 10.1128/JVI.02053-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Felmlee DJ, Hafirassou ML, Lefevre M, Baumert TF, Schuster C. 2013. Hepatitis C virus, cholesterol and lipoproteins—impact for the viral life cycle and pathogenesis of liver disease. Viruses 5:1292–1324. 10.3390/v5051292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Merz A, Long G, Hiet MS, Brugger B, Chlanda P, Andre P, Wieland F, Krijnse-Locker J, Bartenschlager R. 2011. Biochemical and morphological properties of hepatitis C virus particles and determination of their lipidome. J. Biol. Chem. 286:3018–3032. 10.1074/jbc.M110.175018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Albecka A, Belouzard S, Op de Beeck A, Descamps V, Goueslain L, Bertrand-Michel J, Terce F, Duverlie G, Rouille Y, Dubuisson J. 2012. Role of low-density lipoprotein receptor in the hepatitis C virus life cycle. Hepatology 55:998–1007. 10.1002/hep.25501. [DOI] [PubMed] [Google Scholar]
- 18.Sainz B, Jr, Barretto N, Martin DN, Hiraga N, Imamura M, Hussain S, Marsh KA, Yu X, Chayama K, Alrefai WA, Uprichard SL. 2012. Identification of the Niemann-Pick C1-like 1 cholesterol absorption receptor as a new hepatitis C virus entry factor. Nat. Med. 18:281–285. 10.1038/nm.2581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zeisel MB, Koutsoudakis G, Schnober EK, Haberstroh A, Blum HE, Cosset FL, Wakita T, Jaeck D, Doffoel M, Royer C, Soulier E, Schvoerer E, Schuster C, Stoll-Keller F, Bartenschlager R, Pietschmann T, Barth H, Baumert TF. 2007. Scavenger receptor class B type I is a key host factor for hepatitis C virus infection required for an entry step closely linked to CD81. Hepatology 46:1722–1731. 10.1002/hep.21994. [DOI] [PubMed] [Google Scholar]
- 20.Catanese MT, Graziani R, von Hahn T, Moreau M, Huby T, Paonessa G, Santini C, Luzzago A, Rice CM, Cortese R, Vitelli A, Nicosia A. 2007. High-avidity monoclonal antibodies against the human scavenger class B type I receptor efficiently block hepatitis C virus infection in the presence of high-density lipoprotein. J. Virol. 81:8063–8071. 10.1128/JVI.00193-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grove J, Huby T, Stamataki Z, Vanwolleghem T, Meuleman P, Farquhar M, Schwarz A, Moreau M, Owen JS, Leroux-Roels G, Balfe P, McKeating JA. 2007. Scavenger receptor BI and BII expression levels modulate hepatitis C virus infectivity. J. Virol. 81:3162–3169. 10.1128/JVI.02356-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dorner M, Rice CM, Ploss A. 2013. Study of hepatitis C virus entry in genetically humanized mice. Methods 59:249–257. 10.1016/j.ymeth.2012.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dao Thi VL, Granier C, Zeisel MB, Guerin M, Mancip J, Granio O, Penin F, Lavillette D, Bartenschlager R, Baumert TF, Cosset FL, Dreux M. 2012. Characterization of hepatitis C virus particle subpopulations reveals multiple usage of the scavenger receptor BI for entry steps. J. Biol. Chem. 287:31242–31257. 10.1074/jbc.M112.365924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maillard P, Huby T, Andreo U, Moreau M, Chapman J, Budkowska A. 2006. The interaction of natural hepatitis C virus with human scavenger receptor SR-BI/ClaI is mediated by ApoB-containing lipoproteins. FASEB J. 20:735–737. 10.1096/fj.05-4728fje. [DOI] [PubMed] [Google Scholar]
- 25.Evans MJ, von Hahn T, Tscherne DM, Syder AJ, Panis M, Wolk B, Hatziioannou T, McKeating JA, Bieniasz PD, Rice CM. 2007. Claudin-1 is a hepatitis C virus co-receptor required for a late step in entry. Nature 446:801–805. 10.1038/nature05654. [DOI] [PubMed] [Google Scholar]
- 26.Ploss A, Evans MJ, Gaysinskaya VA, Panis M, You H, de Jong YP, Rice CM. 2009. Human occludin is a hepatitis C virus entry factor required for infection of mouse cells. Nature 457:882–886. 10.1038/nature07684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pileri P, Uematsu Y, Campagnoli S, Galli G, Falugi F, Petracca R, Weiner AJ, Houghton M, Rosa D, Grandi G, Abrignani S. 1998. Binding of hepatitis C virus to CD81. Science 282:938–941. 10.1126/science.282.5390.938. [DOI] [PubMed] [Google Scholar]
- 28.Pietschmann T, Kaul A, Koutsoudakis G, Shavinskaya A, Kallis S, Steinmann E, Abid K, Negro F, Dreux M, Cosset FL, Bartenschlager R. 2006. Construction and characterization of infectious intragenotypic and intergenotypic hepatitis C virus chimeras. Proc. Natl. Acad. Sci. U. S. A. 103:7408–7413. 10.1073/pnas.0504877103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Koutsoudakis G, Kaul A, Steinmann E, Kallis S, Lohmann V, Pietschmann T, Bartenschlager R. 2006. Characterization of the early steps of hepatitis C virus infection by using luciferase reporter viruses. J. Virol. 80:5308–5320. 10.1128/JVI.02460-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Blight KJ, McKeating JA, Rice CM. 2002. Highly permissive cell lines for subgenomic and genomic hepatitis C virus RNA replication. J. Virol. 76:13001–13014. 10.1128/JVI.76.24.13001-13014.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Catanese MT, Ansuini H, Graziani R, Huby T, Moreau M, Ball JK, Paonessa G, Rice CM, Cortese R, Vitelli A, Nicosia A. 2010. Role of scavenger receptor class B type I in hepatitis C virus entry: kinetics and molecular determinants. J. Virol. 84:34–43. 10.1128/JVI.02199-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ciesek S, Westhaus S, Wicht M, Wappler I, Henschen S, Sarrazin C, Hamdi N, Abdelaziz AI, Strassburg CP, Wedemeyer H, Manns MP, Pietschmann T, von Hahn T. 2011. Impact of intra- and interspecies variation of occludin on its function as coreceptor for authentic hepatitis C virus particles. J. Virol. 85:7613–7621. 10.1128/JVI.00212-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fofana I, Krieger SE, Grunert F, Glauben S, Xiao F, Fafi-Kremer S, Soulier E, Royer C, Thumann C, Mee CJ, McKeating JA, Dragic T, Pessaux P, Stoll-Keller F, Schuster C, Thompson J, Baumert TF. 2010. Monoclonal anti-claudin 1 antibodies prevent hepatitis C virus infection of primary human hepatocytes. Gastroenterology 139:953–964. 10.1053/j.gastro.2010.05.073. [DOI] [PubMed] [Google Scholar]
- 34.Zahid MN, Turek M, Xiao F, Thi VL, Guerin M, Fofana I, Bachellier P, Thompson J, Delang L, Neyts J, Bankwitz D, Pietschmann T, Dreux M, Cosset FL, Grunert F, Baumert TF, Zeisel MB. 2013. The postbinding activity of scavenger receptor class B type I mediates initiation of hepatitis C virus infection and viral dissemination. Hepatology 57:492–504. 10.1002/hep.26097. [DOI] [PubMed] [Google Scholar]
- 35.Syder AJ, Lee H, Zeisel MB, Grove J, Soulier E, Macdonald J, Chow S, Chang J, Baumert TF, McKeating JA, McKelvy J, Wong-Staal F. 2011. Small molecule scavenger receptor BI antagonists are potent HCV entry inhibitors. J. Hepatol. 54:48–55. 10.1016/j.jhep.2010.06.024. [DOI] [PubMed] [Google Scholar]
- 36.Wakita T, Pietschmann T, Kato T, Date T, Miyamoto M, Zhao Z, Murthy K, Habermann A, Krausslich HG, Mizokami M, Bartenschlager R, Liang TJ. 2005. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat. Med. 11:791–796. 10.1038/nm1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vieyres G, Pietschmann T. 2013. Entry and replication of recombinant hepatitis C viruses in cell culture. Methods 59:233–248. 10.1016/j.ymeth.2012.09.005. [DOI] [PubMed] [Google Scholar]
- 38.Owsianka A, Clayton RF, Loomis-Price LD, McKeating JA, Patel AH. 2001. Functional analysis of hepatitis C virus E2 glycoproteins and virus-like particles reveals structural dissimilarities between different forms of E2. J. Gen. Virol. 82:1877–1883. [DOI] [PubMed] [Google Scholar]
- 39.Meuleman P, Catanese MT, Verhoye L, Desombere I, Farhoudi A, Jones CT, Sheahan T, Grzyb K, Cortese R, Rice CM, Leroux-Roels G, Nicosia A. 2012. A human monoclonal antibody targeting scavenger receptor class B type I precludes hepatitis C virus infection and viral spread in vitro and in vivo. Hepatology 55:364–372. 10.1002/hep.24692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lacek K, Vercauteren K, Grzyb K, Naddeo M, Verhoye L, Slowikowski MP, Fafi-Kremer S, Patel AH, Baumert TF, Folgori A, Leroux-Roels G, Cortese R, Meuleman P, Nicosia A. 2012. Novel human SR-BI antibodies prevent infection and dissemination of HCV in vitro and in humanized mice. J. Hepatol. 57:17–23. 10.1016/j.jhep.2012.02.018. [DOI] [PubMed] [Google Scholar]
- 41.Masson D, Koseki M, Ishibashi M, Larson CJ, Miller SG, King BD, Tall AR. 2009. Increased HDL cholesterol and apoA-I in humans and mice treated with a novel SR-BI inhibitor. Arterioscler. Thromb. Vasc. Biol. 29:2054–2060. 10.1161/ATVBAHA.109.191320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Roccasecca R, Ansuini H, Vitelli A, Meola A, Scarselli E, Acali S, Pezzanera M, Ercole BB, McKeating J, Yagnik A, Lahm A, Tramontano A, Cortese R, Nicosia A. 2003. Binding of the hepatitis C virus E2 glycoprotein to CD81 is strain specific and is modulated by a complex interplay between hypervariable regions 1 and 2. J. Virol. 77:1856–1867. 10.1128/JVI.77.3.1856-1867.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bartosch B, Vitelli A, Granier C, Goujon C, Dubuisson J, Pascale S, Scarselli E, Cortese R, Nicosia A, Cosset FL. 2003. Cell entry of hepatitis C virus requires a set of co-receptors that include the CD81 tetraspanin and the SR-B1 scavenger receptor. J. Biol. Chem. 278:41624–41630. 10.1074/jbc.M305289200. [DOI] [PubMed] [Google Scholar]
- 44.Esko JD, Stewart TE, Taylor WH. 1985. Animal cell mutants defective in glycosaminoglycan biosynthesis. Proc. Natl. Acad. Sci. U. S. A. 82:3197–3201. 10.1073/pnas.82.10.3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yanagi M, Purcell RH, Emerson SU, Bukh J. 1999. Hepatitis C virus: an infectious molecular clone of a second major genotype (2a) and lack of viability of intertypic 1a and 2a chimeras. Virology 262:250–263. 10.1006/viro.1999.9889. [DOI] [PubMed] [Google Scholar]
- 46.Owen DM, Huang H, Ye J, Gale M., Jr 2009. Apolipoprotein E on hepatitis C virion facilitates infection through interaction with low-density lipoprotein receptor. Virology 394:99–108. 10.1016/j.virol.2009.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jiang J, Cun W, Wu X, Shi Q, Tang H, Luo G. 2012. Hepatitis C virus attachment mediated by apolipoprotein E binding to cell surface heparan sulfate. J. Virol. 86:7256–7267. 10.1128/JVI.07222-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Menzel N, Fischl W, Hueging K, Bankwitz D, Frentzen A, Haid S, Gentzsch J, Kaderali L, Bartenschlager R, Pietschmann T. 2012. MAP-kinase regulated cytosolic phospholipase A2 activity is essential for production of infectious hepatitis C virus particles. PLoS Pathog. 8:e1002829. 10.1371/journal.ppat.1002829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Prentoe J, Serre SB, Ramirez S, Nicosia A, Gottwein JM, Bukh J. 2014. Hypervariable region 1 deletion and required adaptive envelope mutations confer decreased dependency on scavenger receptor class B type I and low-density lipoprotein receptor for hepatitis C virus. J. Virol. 88:1725–1739. 10.1128/JVI.02017-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ji ZS, Brecht WJ, Miranda RD, Hussain MM, Innerarity TL, Mahley RW. 1993. Role of heparan sulfate proteoglycans in the binding and uptake of apolipoprotein E-enriched remnant lipoproteins by cultured cells. J. Biol. Chem. 268:10160–10167. [PubMed] [Google Scholar]
- 51.Barth H, Schafer C, Adah MI, Zhang F, Linhardt RJ, Toyoda H, Kinoshita-Toyoda A, Toida T, Van Kuppevelt TH, Depla E, Von Weizsacker F, Blum HE, Baumert TF. 2003. Cellular binding of hepatitis C virus envelope glycoprotein E2 requires cell surface heparan sulfate. J. Biol. Chem. 278:41003–41012. 10.1074/jbc.M302267200. [DOI] [PubMed] [Google Scholar]
- 52.Jiang J, Wu X, Tang H, Luo G. 2013. Apolipoprotein E mediates attachment of clinical hepatitis C virus to hepatocytes by binding to cell surface heparan sulfate proteoglycan receptors. PLoS One 8:e67982. 10.1371/journal.pone.0067982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Anggakusuma , Colpitts CC, Schang LM, Rachmawati H, Frentzen A, Pfaender S, Behrendt P, Brown RJ, Bankwitz D, Steinmann J, Ott M, Meuleman P, Rice CM, Ploss A, Pietschmann T, Steinmann E. 2014. Turmeric curcumin inhibits entry of all hepatitis C virus genotypes into human liver cells. Gut 63:1137–1149. 10.1136/gutjnl-2012-304299. [DOI] [PubMed] [Google Scholar]
- 54.Narayanaswami V, Ryan RO. 2000. Molecular basis of exchangeable apolipoprotein function. Biochim. Biophys. Acta 1483:15–36. 10.1016/S1388-1981(99)00176-6. [DOI] [PubMed] [Google Scholar]
- 55.Spearman C. 1908. The method of “right and wrong cases” (“constant stimuli”) without Gauss's formulae. Br. J. Psychol. 2:227–242. [Google Scholar]
- 56.Kärber G. 1931. Beitrag zur kollektiven Behandlung pharmakologischer Reihenversuche. Arch. Exp. Pathol. Pharmakol. 162:480–487. [Google Scholar]